Abstract
Hepatitis G virus (HGV) nonstructural protein 3 (NS3) contains amino acid sequence motifs typical of ATPase and RNA helicase proteins. In order to examine the RNA helicase activity of the HGV NS3 protein, the NS3 region (amino acids 904 to 1580) was fused with maltose-binding protein (MBP), and the fusion protein was expressed in Escherichia coli and purified with amylose resin and anion-exchange chromatography. The purified MBP-HGV/NS3 protein possessed RNA-stimulated ATPase and RNA helicase activities. Characterization of the ATPase and RNA helicase activities of MBP-HGV/NS3 showed that the optimal reaction conditions were similar to those of other Flaviviridae viral NS3 proteins. However, the kinetic analysis of NTPase activity showed that the MBP-HGV/NS3 protein had several unique properties compared to the other Flaviviridae NS3 proteins. The HGV NS3 helicase unwinds RNA-RNA duplexes in a 3′-to-5′ direction and can unwind RNA-DNA heteroduplexes and DNA-DNA duplexes as well. In a gel retardation assay, the MBP-HGV/NS3 helicase bound to RNA, RNA/DNA, and DNA duplexes with 5′ and 3′ overhangs but not to blunt-ended RNA duplexes. We also found that the conserved motif VI was important for RNA binding. Further deletion mapping showed that the RNA binding domain was located between residues 1383 and 1395, QRRGRTGRGRSGR. Our data showed that the MBP-HCV/NS3 protein also contains the RNA binding domain in the similar domain.
Hepatitis G virus (HGV) was first cloned from the plasma of a patient with chronic hepatitis in 1996 (14). Three more related viruses, GBV-A, GBV-B, and GBV-C, were also reported elsewhere (18, 21). Amino acid sequence comparison of viral polyproteins revealed that HGV and GBV-C are nearly identical and that HGV and GBV-C share sequence similarity with hepatitis C virus (HCV), GBV-A, and GBV-B (14). All four viruses are closely related and belong to the Flaviviridae family (14, 18, 21). HGV has a positive-stranded RNA genome about 9.4 kb in size (14). The putative genomic structure of HGV is organized as follows: 5′ UTR (untranslated region)-E1-E2-NS2 (nonstructural protein 2)-NS3-NS4A-NS4B-NS5A-NS5B-3′ UTR. The core region of HGV has not yet been identified. Amino acid sequence alignments show that the Flaviviridae family (flaviviruses, pestiviruses, and HCV) has the conserved sequence motifs of a serine-type proteinase and a nucleoside triphosphatase (NTPase)-RNA helicase in each respective NS3 protein (14). Two HGV proteases that are similar to HCV NS2 and NS3 were recently identified (2). The mode of HGV polyprotein cleavage in its NS region appears to be very similar to that of HCV, except for the substrate specificities of the proteases.
The HGV NS3 protein has GKS/T and DEXH motifs which are typical of the DEXH protein subfamily of the DEAD box family. A number of DEAD box family proteins, such as RNA helicase A, mouse eIF-4A, and the vaccinia virus NPHII protein, contain NTPase and RNA helicase activities (13, 16, 19, 20). NTPase and RNA helicase activities have also been reported for the Flaviviridate NS3 proteins. Wengler and Wengler (27) showed that the NS3 protein of the West Nile virus contains an RNA-stimulated NTPase activity. Suzich et al. (22) and Warrener et al. (26) reported that the NS3 proteins of HCV and the yellow fever virus exhibit an NTPase activity. The p80 protein of bovine viral diarrhea virus (BVDV) expressed in insect cells has an RNA-stimulated NTPase activity (24) and an RNA helicase activity (25). The RNA helicase activity of the HCV NS3 protein was demonstrated by our group and others (6, 8, 9, 23).
We sought to determine whether the HGV NS3 protein possessed NTPase and RNA helicase activities. To this end, we expressed the NS3 region of HGV in E. coli and performed biochemical analyses on the purified, recombinant protein. We show that HGV NS3 does indeed display RNA-stimulated NTPase and RNA helicase activities. The protein had certain characteristics that were similar to those of other Flaviviridae RNA helicases (for example, direction of translocation, salt sensitivity, and requirements for ATP and divalent cations). Unique features are found in the kinetic analysis of NTPase activity and substrate specificity of the helicase activity. In addition, we found the RNA binding domain of the MBP-HGV/NS3 protein, and this domain was also important for the RNA binding activity of the HCV NS3 protein.
MATERIALS AND METHODS
Expression and purification of the HGV NS3 protein.
The putative HGV NS3 open reading frame, encompassing amino acids 904 to 1580, was amplified from HGV cDNA PNF2161 by PCR. EcoRI and HindIII sites were created at the 5′ and 3′ ends, respectively, during the PCR process. The PCR product was digested with EcoRI and HindIII and cloned into the EcoRI and HindIII sites of pMAL-c2 vector (New England Biolabs, Beverly, Mass.). The recombinant plasmid was designated pMAL-c2/HGVNS3, and the protein product from this plasmid was designated MBP-HGV/NS3.
The MBP-HGV/NS3 protein contains a portion of maltose-binding protein (MBP) at the NH2-terminal end for solubility and easier purification. In order to purify MBP-HGV/NS3 protein, JM109 cells harboring pMAL-c2/HGVNS3 were grown in Luria-Bertani medium with 10 μg of ampicillin per ml, and expression of the recombinant protein was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM. After being cultured for 3 additional h at 37°C, the cells were harvested by centrifugation at 6,000 × g for 10 min. The cell pellet was resuspended in 40 ml of lysis buffer (10 mM Tris-HCl [pH 7.4], 0.2 M NaCl, 10 mM β-mercaptoethanol, and 1 mM EDTA) containing 0.25% Tween 20. The resuspended cells were frozen at −70°C for at least 30 min and then thawed. The cells were further disrupted by brief sonication. The resulting mixture was centrifuged at 27,000 × g for 30 min, and the supernatant was loaded onto a 2-ml-bed-volume amylose resin (New England Biolabs) column equilibrated with lysis buffer. Proteins not specifically retained by the resin were removed with 10 column volumes of lysis buffer, whereas the proteins retained by the column were eluted with lysis buffer containing 10 mM maltose. This protein pool was dialyzed against 20 mM Tris-HCl (pH 8.0)–10 mM NaCl and applied to a 2-ml Q-Sepharose column (Pharmacia Biotech, Uppsala, Sweden). The column was eluted stepwise with 20 mM Tris-HCl (pH 8.0) containing 50, 100, 150, 200, 250, 300, 400, 500, 600, 700, 800, and 1,000 mM NaCl. The eluted fractions were dialyzed against 20 mM Tris-HCl (pH 7.2) plus 30% glycerol.
ATPase assay.
Standard ATPase assays were conducted in a total volume of 10 μl containing 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS)–KOH (pH 7.0), 2 mM MgCl2, 0.05 mg of bovine serum albumin (BSA) per ml, 2 mM dithiothreitol (DTT), 5 mM ATP, 1 μCi of [α-32P]ATP, (3,000 Ci/mmol; Amersham Corp., Buckinghamshire, United Kingdom), 10 pmol of the MBP-HGV/NS3 protein and, if needed, 1 μg of poly(U). The buffer systems used for the various pH levels were as follows: pH 5.5 and 6.0, 2-(N-morpholino)ethanesulfonic acid (MES)–HCl; pH 6.5 and 7.0, MOPS-KOH; pH 7.5 and 8.0, HEPES-HCl; and pH 8.5 and 9.0, Tris-HCl. Reactions were incubated at 37°C for 40 min and then terminated by the addition of EDTA to a final concentration of 20 mM. In order to analyze the products of ATP hydrolysis, 1 μl of each reaction mixture was spotted onto a polyethyleneimine-cellulose plate, and the plate was developed with 0.375 M potassium phosphate (pH 3.5). Radioactivity was measured with a FUJIX BAS-1500 (Fuji Photo Film Co., Tokyo, Japan). Values for Km (millimolar) and kcat (per minute) were determined from Lineweaver-Burk plots of hydrolysis activity. Reactions were conducted with NTP or dNTP concentrations of between 0.25 and 2.0 mM.
RNA helicase assays.
The standard RNA helicase assay was conducted in a total volume of 20 μl containing 50 mM HEPES-HCl (pH 7.5), 2 mM MgCl2, 0.05 mg of BSA per ml, 2 mM DTT, 10 mM ATP, 20 U of RNasin, 15 pmol of the MBP-HGV/NS3 protein, and 1 pmol of substrate (see below). The buffer systems used for the various pH levels were as described for the ATPase assay. Reactions were incubated at 37°C for 1.5 h and then terminated by the addition of EDTA and sodium dodecyl sulfate (SDS) to final concentrations of 20 mM and 0.2%, respectively. After the addition of 10 μg of proteinase K, the reactions were incubated for 10 min more at 37°C and then supplemented with 5× RNA loading dye (0.1 M Tris-HCl [pH 7.4], 20 mM EDTA, 0.5% SDS, 0.1% bromophenol blue, 0.1% xylene cyanol, and 50% glycerol). The reaction products were subjected to electrophoresis on a 10% native polyacrylamide gel. The gel was dried and autoradiographed. The ratio of single-stranded products to double-stranded products was quantified by PhosphorImager analysis with a FUJIX BAS-1500.
Preparation of helicase substrates.
For the double-stranded RNA (dsRNA; R/R) substrates, the long unlabeled strands and short 32P-labeled short strands were prepared by in vitro transcription of PvuII-digested pGEM1 and BamHI-digested pSP65 by using SP6 RNA polymerase as described previously (10, 23). The transcripts were treated with DNase; extracted with phenol-chloroform; precipitated with ethanol; combined in a solution containing 20 mM HEPES-KOH (pH 7.6), 0.5 M NaCl, 1 mM EDTA, and 0.1% SDS; boiled for 5 min; and incubated at room temperature for 1 h. The hybridized products were mixed with 5× RNA loading dye and subjected to electrophoresis on 10% native polyacrylamide gel. The duplex band was localized by autoradiography, excised from the gel, and extracted by soaking elution buffer (0.5 M ammonium acetate, 0.1% SDS, 10 mM EDTA) for 2 h at room temperature. The supernatant was extracted with chloroform, and the dsRNA was precipitated with ethanol, recovered by centrifugation, and dissolved in water. For the RNA-DNA heteroduplex substrate (R/D), DNA-RNA substrate (D/R), and DNA-DNA substrate (D/D), all procedures were the same as that described above except that synthesized DNA oligonucleotides were used and the short DNA oligonucleotide was labeled with [γ-32P]ATP and T4 kinase. The RNA substrate containing only a 3′ single-stranded region (3′/3′) consisted of an SP6 transcript from PvuII-digested pGEM1 and a labeled T7 RNA polymerase transcript from RsaI-digested pGEM1. The substrate that contained only a 5′ single-stranded region (5′/5′) consisted of a SP6 labeled transcript from SacI-digested pGEM1 and an SP6 transcript from AccI-digested pSP65. Substrate without a single-stranded region was prepared from the 3′/3′ RNA substrate as follows. A portion (20 pmol) of the 3′/3′ RNA substrate was incubated at 30°C with 40 U of mung bean nuclease (New England Biolabs) in an appropriate buffer. After 30 min, the reaction was terminated by the addition of RNA loading dye. The resulting duplex substrate was gel purified as described above.
RNA binding assay.
In a gel retardation assay, the MBP-HGV/NS3 protein (10 pmol) and substrate (1 pmol of each) were incubated in the helicase reaction buffer (20 μl) without ATP at 37°C for 30 min. The binding reaction was terminated by adding 5× RNA loading dye containing 0.5% Nonidet P-40 instead of SDS, and the reaction products were subjected to electrophoresis on a 3.5% polyacrylamide gel (79:1 acrylamide–bisacrylamide in 1/3× Tris-borate-EDTA) containing 5% glycerol. The protein-nucleic acid complexes were visualized by autoradiography. In Northwestern assays, protein samples (ca. 3 μg of each) were separated by SDS–10% polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose by using a Trans-Blot apparatus (Bio-Rad). The membrane was incubated at room temperature for 1 h in 10 ml of renaturation buffer (HEPES-HCl [pH 7.5], 1 mM EDTA, 100 mM NaCl, 0.1% Triton X-100, 1× Denhardt’s reagent). This step was repeated four times. The membrane was incubated in renaturation buffer containing 32P-labeled single-stranded RNA (ssRNA; 98 nucleotides). ssRNA was prepared by in vitro transcription of PvuII-digested pGEM1 by using SP6 RNA polymerase and was gel purified as described above. After three washes, the membrane was dried in air and exposed to X-ray film.
RESULTS
RNA-stimulated ATPase and RNA helicase activities of the HGV NS3 protein.
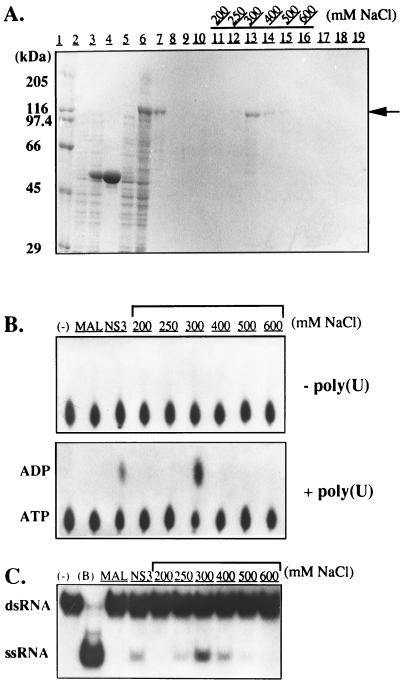
Our first attempt to purify an active form of HGV NS3 protein from E. coli with a pET vector system (Novagen, Madison, Wis.) was unsuccessful because of the extreme insolubility of the recombinant protein. To increase the solubility of the HGV NS3 protein, we expressed it as an MBP fusion protein (see Materials and Methods). The predicted HGV NS3 protein fragment (amino acids 904 to 1580) that we expressed in E. coli by using the pMAL-c2 system contained the protease domain and the putative RNA helicase domain. The recombinant HGV NS3 protein fused with MBP at the NH2 terminus was expressed efficiently as a soluble form and was purified with an amylose resin column (Fig. 1A). This protein was designated MBP-HGV/NS3. The protein fractions from the amylose column were then loaded onto a Q-Sepharose column, and bound protein was eluted with an NaCl gradient. The fractions eluted with 250 to 400 mM NaCl contained an 117-kDa protein that reacted with anti-MBP antiserum. This protein was not detected in IPTG-induced E. coli cells harboring the pMAL-c2 parental vector (see Materials and Methods). We concluded that the 117-kDa protein was MBP-HGV/NS3. Analysis of the final protein preparation by SDS-PAGE revealed that the purity of the 117-kDa protein was >90%. Several other minor protein bands were detectable. It is likely that these contaminating proteins derived from the full-length MBP-HGV/NS3 protein, since they reacted with anti-MBP antiserum and were absent in fractions purified from E. coli cells harboring pMAL-c2 (data not shown). The ATPase assay with or without poly(U) was performed with 200, 250, 300, 400, 500, and 600 mM NaCl-eluted fractions (Fig. 1B). No ATPase activity was detected with affinity-purified MBP. Only the reactions containing MBP-HGV/NS3 protein had ATPase activity, which was stimulated 7- to 10-fold under this condition. A truncated form of HGV NS3 protein (amino acids 1088 to 1580) that was fused with MBP and purified by the same procedures showed neither an RNA-stimulated ATPase activity nor an RNA helicase activity (data not shown). This result indicates that the RNA-stimulated ATPase and RNA helicase activities of HGV NS3 were not derived from contaminating E. coli proteins. RNA helicase activity was detected only in the presence of MBP-HGV/NS3 and was proportional to the amount of MBP-HGV/NS3 protein added to the reaction (Fig. 1C).
FIG. 1.
RNA-stimulated ATPase and RNA helicase activities of the MBP-HGV/NS3 protein. (A) Column fractions (2 μl) were subjected to SDS-PAGE (10% acrylamide gel), and the gel was stained with Coomassie blue. Lanes: 2 and 3, cells harboring pMAL-c2 vector at 0 and 3 h, respectively, after induction with IPTG; 5 and 6, cells harboring pMAL-c2 containing the full HGV NS3 region at 0 and 3 h, respectively after induction with IPTG; 4 and 7, amylose column fractions of MBP and HGV NS3 protein fused to MBP. The amylose-purified HGV NS3 MBP fusion protein was loaded onto a Q-Sepharose column and eluted with buffers containing 50, 100, 150, 200, 250, 300, 400, 500, 600, 700, 800, and 1,000 mM NaCl (lanes 8 to 19). Only the fractions containing the NS3 proteins are shown. The arrow indicates the migration position of MBP-HGV/NS3. Molecular size markers (in kilodaltons) are shown at the left. (B) Standard ATPase assays were performed with the column fractions (2 μl) in the presence [+poly(U)] or absence [−poly(U)] of poly(U). Lanes: −, ATPase reaction mixture without added protein; MAL, reaction containing affinity-purified MBP; NS3, reaction containing affinity-purified MBP-HGV/NS3 protein; 200 to 600 mM NaCl, reactions with Q-Sepharose-eluted fractions containing the MBP-HGV/NS3 protein. The migration positions of ADP and ATP are indicated at the left. (C) Standard helicase assay with an RNA hybrid was performed with the same fractions (2 μl) as those shown in panel B. −, No added protein; B, boiled RNA substrate. The migration positions of dsRNA and ssRNA are indicated at the left.
In the presence of poly(U), the ATPase activity was highest at pH 7.0 and approached basal activity levels with increasing or decreasing pH (data not shown). The MBP-HGV/NS3 ATPase activity was dependent on the presence of divalent ions. In addition, in the presence of poly(U), ATPase activity was inhibited by increasing concentrations of NaCl. At 200 mM NaCl, ATPase activity decreased to basal levels. The optimal pH for the RNA helicase activity of MBP-HGV/NS3 was 7.5, which was slightly different from the optimal condition (pH 7.0) for the RNA-stimulated ATPase activity. At pH 7.5 and with 10 mM ATP in the reaction mixture, the RNA helicase activity of MBP-HGV/NS3 was stimulated at low divalent ion concentrations (up to 2 mM); however, the RNA helicase activity decreased in the presence of divalent ion concentrations higher than 2 mM. Helicase activity was inhibited by increasing concentrations of NaCl, and at 100 mM NaCl, the activity fell to basal levels. Thus, the RNA helicase activity was more sensitive to NaCl than was the RNA-stimulated ATPase activity.
Kinetic analysis of NTPase and dNTPase activity of MBP-HGV/NS3 protein.
All NTPs and dNTPs except dTTP were hydrolyzed efficiently by the MBP-HGV/NS3 protein (Table 1). The kinetic parameters kcat (turnover number) and Km (Michaelis constant) were determined in the absence or presence of poly(U). In the presence of poly(U), the kcat values increased 13- to 100-fold. The MBP-HGV/NS3 protein exhibited either no change or a two- to threefold increase in Km in the presence of poly(U). The catalytic efficiency, i.e., the kcat/Km ratios, increased 15- to 40-fold in the presence of poly(U).
TABLE 1.
Kinetics of NTP and dNTP hydrolysis by the HGV NS3 protein
| Substrate | NTP and dNTP hydrolysis kinetics:
|
|||||
|---|---|---|---|---|---|---|
| Without poly(U)
|
With poly(U)
|
|||||
| Km (mM) | kcat (min−1) | kcat/Km | Km (mM) | kcat (min−1) | kcat/Km | |
| ATP | 0.39 | 6.2 | 15.9 | 1.03 | 250.0 | 242.7 |
| GTP | 0.50 | 4.3 | 8.6 | 0.89 | 142.9 | 160.6 |
| UTP | 0.38 | 3.5 | 9.2 | 1.20 | 200.0 | 166.7 |
| CTP | 1.11 | 13.3 | 12.0 | 1.21 | 250.0 | 206.6 |
| dATP | 0.77 | 12.8 | 16.6 | 0.57 | 167.0 | 293.0 |
| dGTP | 0.33 | 1.1 | 3.3 | 1.00 | 111.1 | 111.1 |
| dTTP | –a | – | – | – | – | – |
| dCTP | 0.48 | 2.5 | 5.2 | 1.28 | 250.0 | 195.3 |
–, Extent of hydrolysis of dTTP by the HGV NS3 protein was too low to determine the Km and kcat values.
Substrate specificity and directionality of the HGV helicase.
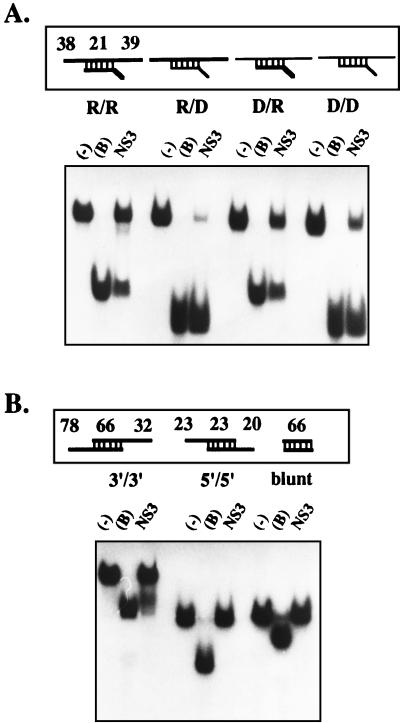
In order to determine the substrate specificity of the MBP-HGV/NS3 helicase, three additional substrates were synthesized. These substrates included an RNA template strand annealed to a DNA release strand (R/D) and a long DNA template strand annealed to a short RNA release strand (D/R) and a short DNA release strand (D/D). All of these substrates were displaced by the MBP-HGV/NS3 protein (Fig. 2A). Therefore, the HGV RNA helicase can unwind RNA-DNA duplexes and DNA-DNA duplexes, as well as RNA-RNA duplexes.
FIG. 2.
Substrate specificity and directionality of the MBP-HGV/NS3. (A) Substrate specificity of the MBP-HGV/NS3 helicase activity was determined by using various substrates consisting of RNA or DNA strands. Lanes: −, no protein added to the helicase reaction; B, boiled substrate control; NS3, helicase reaction containing MBP-HGV/NS3. The thick lines indicate RNA strands, and the thin lines indicate DNA strands. (B) The direction of the helicase activity was determined by using various RNA substrates under the condition of the standard helicase assay. The direction of the upper strands of all the substrates shown in this figure is 5′ to 3′. The length (in nucleotides) of each single-stranded and double-stranded region is indicated by the numbers at the top of each substrate schematic.
To determine the directionality of RNA unwinding by the MBP-HGV/NS3 protein, we constructed three different RNA substrates, one with a 3′ single-stranded tail (3′/3′), one with a 5′ single-stranded tail (5′/5′), and one with no single-stranded tail (blunt). As shown in Fig. 2B, only the RNA substrate containing the 3′ single-stranded region (3′/3′) was displaced by MBP-HGV/NS3. Thus, we conclude that the MBP-HGV/NS3 protein unwinds RNA duplexes in a 3′-to-5′ direction.
Binding of the HGV NS3 protein to helicase substrates.
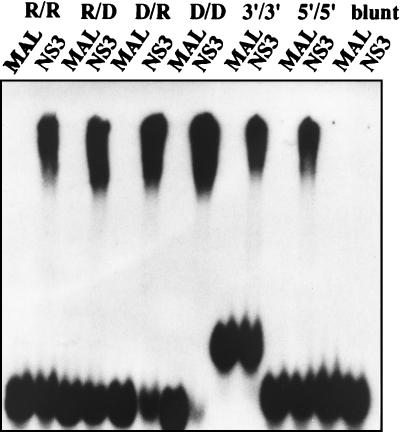
The inability of the MBP-HGV/NS3 protein to unwind the 5′/5′ and blunt RNA substrates may indicate that the NS3 protein does not interact with them. Thus, we measured the ability of each substrate. Substrates were incubated with the MBP-HGV/NS3 protein in the helicase reaction mixture lacking ATP to bind to the MBP-HGV/NS3 protein in vitro, and the reaction mixtures were subjected to electrophoresis through a polyacrylamide gel (Fig. 3). This gel mobility shift assay revealed that the MBP-HGV/NS3 protein was able to bind to all but the blunt RNA substrate. The reactions containing only the MBP as a negative control did not show complex formation.
FIG. 3.
RNA and DNA binding activities of the MBP-HGV/NS3 protein. Gel retardation assays were performed with all of the substrates (R/R, R/D, D/R, D/D, 3′/3′, 5′/5′, and blunt) tested in the helicase reaction. Lanes: MAL, reaction containing affinity-purified MBP; NS3, reaction containing HGV NS3 protein.
Mapping of an RNA binding domain of the MBP-HGV/NS3 protein.
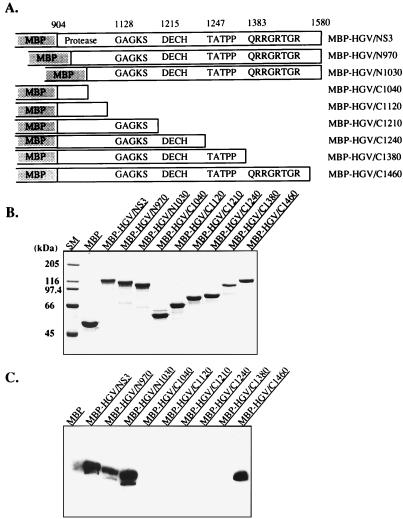
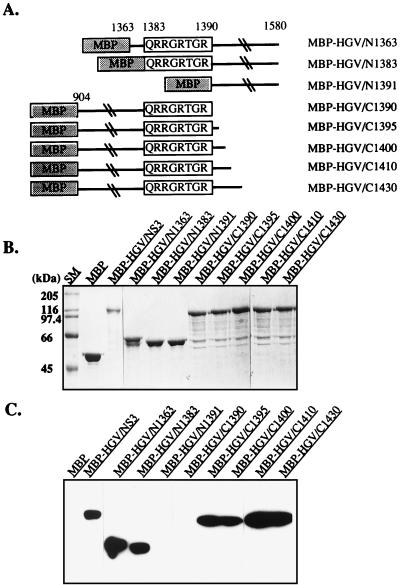
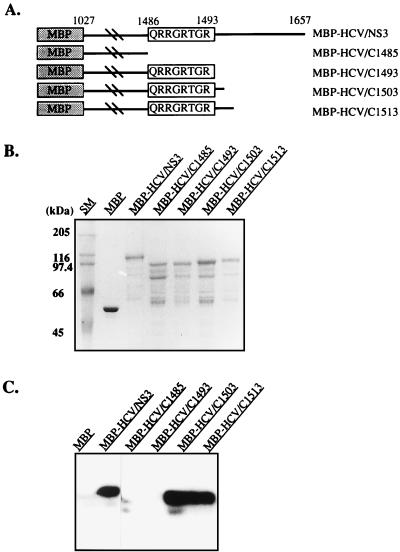
To determine the RNA binding domain of the MBP-HGV/NS3 protein, we constructed various deletion mutants from the amino-terminal and carboxy-terminal ends (Fig. 4A and B). The amino-terminal amino-acid-deleted proteins, MBP-HGV/N970 and N1030, contained the same RNA helicase and RNA-stimulated ATPase activities as the MBP-HGV/NS3 protein, but all of the carboxy-terminal amino-acid-deleted proteins did not have any RNA-stimulated ATPase and helicase activities (data not shown). The RNA binding activities of these proteins were determined by Northwestern assay (Fig. 4C). MBP-HGV/NS3, N970, N1030, and only the MBP-HGV/C1460 among the carboxy-terminal amino-acid-deleted proteins bound to 32P-labeled ssRNA (98 nucleotides). These RNA binding activities were not sequence specific because other ssRNA (e.g., 32P-labeled 28-nucleotide RNA prepared from in vitro transcription of BamHI-digested pSP65 with SP6 RNA polymerase) showed similar results. From these data, we speculated that the conserved helicase motif VI was the RNA binding domain of MBP-HGV/NS3. In order to determine the RNA binding domain more precisely, further deletions were constructed around the motif VI (Fig. 5A and B). The amino-terminal amino-acid-deleted proteins, MBP-HGV/N1363 and N1383, still possessed RNA binding activity, however, MBP-HGV/N1391, which did not contain the conserved motif QRRGRTGR, had no RNA binding activity (Fig. 5C). The carboxy-terminal amino-acid-deleted protein, MBP-HGV/C1390, which ended just after QRRGRTGR motif, could not bind to the RNA. MBP-HGV/C1395 protein, which has five additional carboxy-terminal amino acids from the QRRGRTGR motif, regained the RNA binding activity, and all of the carboxy-terminal amino-acid-deleted proteins with more additional carboxy-terminal amino acids than MBP-HGV/C1395 had the RNA binding activities. In order to determine whether this motif is sufficient for the RNA binding activity, we synthesized three peptides, SNTPHVNHHMPHC, QRRGRTGRGRSGR, and TRGRGSRGGRQRR. SNTPHVNHHMPHC is a 13-amino-acid peptide and has a random sequence. TRGRGSRGGRQRR has the same amino acid composition as QRRGRTGRGRSGR, but it has a randomized sequence. All three peptides did not bind to 98-nucleotide RNA when we test them by using Northwestern assay (data not shown). We conclude that the helicase motif VI is necessary for RNA binding; however, it is not sufficient for the RNA binding activity. To determine whether this fact could also be applied to the HCV NS3 protein, we made several carboxy-terminal amino-acid-deleted MBP-HCV/NS3 proteins (Fig. 6A and B). MBP-HCV/C1485, which had no helicase motif VI, and MBP-HCV/C1493, which had QRRGRTGR motif carboxy-terminal ends, had no RNA binding activity (Fig. 6C). MBP-HCV/C1503 and C1513, which each had 10 or 20 more carboxy-terminal amino acids, respectively, than MBP-HCV/C1493, showed the RNA binding activity.
FIG. 4.
RNA binding activity of MBP-HGV/NS3 protein. (A) Schematic representation of the full and deleted MBP-HGV/NS3 proteins. The HGV cDNA, encompassing the indicated amino acids, was amplified by PCR and cloned into the mMAL-c2 vector as described in Materials and Methods. MBP-HGV/N970 and N1030 were the N-terminal amino-acid-deleted NS3 proteins. MBP-HGV/C1040-C1460 were the C-terminal amino-acid-deleted proteins. The amino acid numbers of the conserved helicase motifs are indicated at the top. (B and C) MBP-HGV/NS3 proteins purified by amylose affinity chromatography, analyzed by SDS-PAGE (B) and then stained with Coomassie blue. (C) RNA binding of MBP-HGV/NS3 proteins (Northwestern assay).
FIG. 5.
Mapping of an RNA binding domains of MBP-HGV/NS3 protein. (A) Schematic representation of the deleted forms of MBP-HGV/NS3 proteins. (B) MBP-HGV/NS3 proteins analyzed by SDS-PAGE. (C) RNA binding of the MBP-HGV/NS3 proteins (Northwestern assay).
FIG. 6.
Mapping of an RNA binding domain of the MBP-HCV/NS3 protein. (A) Schematic representation of the full and C-terminal amino-acid-deleted MBP-HCV/NS3 proteins. (B) MBP-HCV/NS3 proteins analyzed by SDS-PAGE. (C) RNA binding of the MBP-HCV/NS3 proteins (Northwestern assay).
DISCUSSION
On the basis of sequence motif analysis, the HGV NS3 protein is predictive of a multifunctional protein containing serine protease, ATPase, and RNA helicase activities (2, 14). Recently, it was found that the HGV NS3 protein has the ATPase and DNA helicase activities (12). In this study we confirm that the HGV NS3 protein has an RNA-stimulated ATPase activity and an RNA helicase activity, and we characterize the biochemical properties of the protein in detail. The kinetic analysis of the NTPase activity showed that the MBP-HGV/NS3 protein had several unique properties compared with the other Flaviviridae viral NS3 proteins. In the presence of poly(U), the HGV NTPase, like the HCV and BVDV NTPases, showed a little increase in Km values, while the YFV NTPase showed either no change or a two- to threefold reduction (22, 24, 26). The kcat values of HGV NTPase increased more than the kcat values of any other Flaviviridae NTPase in the presence of poly(U). For ATP hydrolysis, the increase in the kcat values in the presence of cofactors was 40-, 11-, 15-, and 5-fold for the HGV, HCV, BVDV, and yellow fever virus (YFV) NTPases, respectively. Thus, the increases in the catalytic efficiency (kcat/Km) of HGV NTPase were greater than with the HCV, BVDV, and YFV NTPases.
We found that the MBP-HGV/NS3 protein is able to unwind both RNA and DNA duplexes and is thus the fifth such enzyme to be identified. The other four enzymes are simian virus 40 large T antigen, nuclear DNA helicase II, the vaccinia virus protein 18R, and the HCV NS3 protein (1, 17, 7, 29). All of these helicases displace RNA or DNA duplexes in a 3′-to-5′ direction. In addition, they all possess oligonucleotide-stimulated ATPase and helicase activities that are sensitive to monovalent ions. Despite these common characteristics, these enzymes do not share a conserved primary amino acid sequence or an identical mechanism for displacement of the duplexes.
Since it had not yet been proved experimentally that the Flaviviridae viral helicases contained a RNA binding motif, we tried to determine the presence of a RNA binding motif of the HGV helicase. By using deletion mutants and Northwestern assay, we showed that the RNA binding domain of the HGV NS3 protein was located at the conserved helicase domain VI and five additional amino acids after this domain, i.e., QRRGRTGRGRSGR. The amino acid motif QRRGRTGR in the RNA binding domain was well conserved among the Flaviviridae viral helicases. The five-additional-amino-acid motif GRSGR was less conserved, but most Flaviviridae viral helicases contained at least one positively charged amino acid in this domain. We also determined that the MBP-HCV/NS3 protein has the RNA binding domain at the same position, i.e., the helicase motif VI and several additional amino acids at the carboxy-terminal end. The RNA binding domain of the plum pox potyvirus CI protein was previously determined by Northwestern assay to be around the helicase motif VI, although the exact RNA binding motif was not defined in detail (5). The HRIGRXXR region, the conserved motif VI of the eIF-4A, was proved to be required for RNA binding and ATP hydrolysis (15). So it is not surprising that the helicase motif VI of the HGV helicase was necessary for RNA binding activity. However, the RNA binding domain of the HCV NS3 protein is controversial. Yao et al. (28) and Cho et al. (3) proposed from their X-ray crystallography data that the RNA helicase motif VI of the HCV NS3 protein is required for RNA binding and that several additional positively charged amino acids may be involved. This proposal seems well matched with the biochemical data for the other RNA helicases, the plum pox potyvirus CI protein and eIF-4A, which belong to the helicase superfamily II (SF-II), and with our own data (5, 15). However, Kim et al. (11) proposed, based on the X-crystallography data of the HCV NS3 protein and oligonucleotide complex, that the RNA binding domain of the HCV NS3 helicase did not contain the helicase motif VI. We do not know the reason for this discrepancy, but one possibility is that several RNA binding motifs may be dispersed in the HCV NS3 protein. We made 40 different kinds of single-amino-acid-substituted mutation, including the previously published (10) mutation in the conserved amino acid motifs and the putative RNA binding motifs (unpublished data), but we could not find any change in the RNA binding activity. This means that the RNA binding motifs of the HCV helicase are dispersed throughout the protein rather than being limited to a few amino acids that were important for RNA binding. In fact, another RNA binding motif of the plum pox potyvirus CI protein was found with a subtle change of the assay condition assay (e.g., pH) (4). In conclusion, the helicase motif VI of the HCV and the HGV NS3 proteins seems necessary for the RNA binding activity of these proteins, but it could not be excluded that another RNA binding motif exists.
ACKNOWLEDGMENTS
We thank J. S. Kim of Genelabs for supplying the HGV cDNA clone (PNF2161).
This work was supported in parts by grants from the Korea Science and Engineering Foundation (KOSEF) through the Research Center for Cell Differentiation at Seoul National University and from Pohang University of Science and Technology.
REFERENCES
- 1.Bayliss C D, Smith G L. Vaccinia virion protein 18R has both DNA and RNA helicase activities: implications for vaccinia virus transcription. J Virol. 1996;70:794–800. doi: 10.1128/jvi.70.2.794-800.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belyaev A S, Chong S, Novikov A, Kongpachith A, Masiarz F R, Lim M, Kim J P. Hepatitis G virus protease activities which can effect processing of the virus putative nonstructural proteins. J Virol. 1998;72:868–872. doi: 10.1128/jvi.72.1.868-872.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho H S, Ha N C, Kang L W, Chung K M, Back S H, Jang S K, Oh B H. Crystal structure of RNA helicase from genotype 1b hepatitis C virus. J Biol Chem. 1998;273:15045–15052. doi: 10.1074/jbc.273.24.15045. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez A, Garcia J A. The RNA helicase CI from plum pox potyvirus has two regions involved in binding to RNA. FEBS Lett. 1996;388:206–210. doi: 10.1016/0014-5793(96)00571-6. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez A, Lain S, Garcia J A. RNA helicase activity of the plum pox potyvirus CI protein expressed in Escherichia coli. Mapping of an RNA binding domain. Nucleic Acids Res. 1995;23:1327–1332. doi: 10.1093/nar/23.8.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gwack Y, Kim D W, Han J H, Choe J. Characterization of RNA binding activity and RNA helicase activity of the hepatitis C virus NS3 protein. Biochem Biophys Res Commun. 1996;225:654–659. doi: 10.1006/bbrc.1996.1225. [DOI] [PubMed] [Google Scholar]
- 7.Gwack Y, Kim D W, Han J H, Choe J. DNA helicase activity of the hepatitis C virus nonstructural protein 3. Eur J Biochem. 1997;250:47–54. doi: 10.1111/j.1432-1033.1997.00047.x. [DOI] [PubMed] [Google Scholar]
- 8.Jin L, Peterson D L. Expression, isolation, and characterization of the hepatitis C virus ATPase/RNA helicase. Arch Biochem Biophys. 1995;323:47–53. doi: 10.1006/abbi.1995.0008. [DOI] [PubMed] [Google Scholar]
- 9.Kim D W, Gwack Y, Han J H, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 10.Kim D W, Kim J, Gwack Y, Han J H, Choe J. Mutational analysis of the hepatitis C virus RNA helicase. J Virol. 1997;71:9400–9409. doi: 10.1128/jvi.71.12.9400-9409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J L, Morgenstern K A, Griffith J P, Dwyer M D, Thomson J A, Murcko M A, Lin C, Caron P R. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 12.Laxton C D, McMillan D, Sullivan V, Ackrill A M. Expression and characterization of the hepatitis G virus helicase. J Viral Hepatol. 1998;5:21–26. doi: 10.1046/j.1365-2893.1998.00084.x. [DOI] [PubMed] [Google Scholar]
- 13.Lee C-G, Hurwitz J. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J Biol Chem. 1992;267:4398–4407. [PubMed] [Google Scholar]
- 14.Linnen J, Wages J, Zhang-Keck Z-Y, Fry K, Krawczynski K Z, Alter H, Koonin E, Gallacher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih J, Young L, Piatak M, Hoover C, Fernandez J, Chen S, Zou J C, Morris T, Hyams K C, Ismay S, Lifson J D, Hess G, Foung S K H, Thomas H, Bradley D, Margolis H, Kim J P. Molecular cloning and disease association of hepatitis G virus: a new transfusion transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 15.Pause A, Methot N, Sonenberg N. The HRIGRXXGR region of the DEAD box RNA helicase eucaryotic translational initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol Cell Biol. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rozen F, Edery I, Meerovich K, Dever T E, Merrick W C, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheffner M, Knippers R, Stahl H. RNA unwinding activity of SV40 large T antigen. Cell. 1989;57:955–963. doi: 10.1016/0092-8674(89)90334-6. [DOI] [PubMed] [Google Scholar]
- 18.Schlauder G G, Dawson G, Simons J N, Pilot-Matias T J, Gutierrez R A, Heynen C A, Knigge M F, Kurpiewski G S, Buijk S L, Leary T P, Muerhoff A S, Desai S M, Mushahwar I K. Molecular and serologic analysis in the transmission of the GB hepatitis agents. J Med Virol. 1995;46:81–90. doi: 10.1002/jmv.1890460117. [DOI] [PubMed] [Google Scholar]
- 19.Shuman S. Vaccinia virus RNA helicase: an essential enzyme related to the DE-H family of RNA-dependent NTPases. Proc Natl Acad Sci USA. 1992;89:10935–10939. doi: 10.1073/pnas.89.22.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuman S. Vaccinia virus RNA helicase. J Biol Chem. 1993;268:11798–11802. [PubMed] [Google Scholar]
- 21.Simons J N, Leary T P, Dawson G J, Pilot-Martias T J, Muerhoff A S, Schlauder G, Desai S M, Mushahwar I K. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 22.Suzich J A, Tamura J K, Palmer-Hill F, Warrener P, Grouki A, Rice C M, Feinstone S M, Collett M S. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tai C-L, Chi W-K, Chen D-S, Hwang L-H. The helicase activity associated with hepatitis C virus nonstructural protein 3. J Virol. 1996;70:8477–8484. doi: 10.1128/jvi.70.12.8477-8484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura J K, Warrener P, Collett M S. RNA stimulated NTPase activity associated with the p80 protein of the pestivirus bovine viral diarrhea virus. Virology. 1993;193:1–10. doi: 10.1006/viro.1993.1097. [DOI] [PubMed] [Google Scholar]
- 25.Warrener P, Collett M S. Pestivirus (p80) protein possesses RNA helicase activity. J Virol. 1995;69:1720–1726. doi: 10.1128/jvi.69.3.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warrener P, Tamura J K, Collett M S. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J Virol. 1993;67:989–996. doi: 10.1128/jvi.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wengler G, Wengler G. The carboxy-terminal part of the NS3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology. 1991;184:707–715. doi: 10.1016/0042-6822(91)90440-m. [DOI] [PubMed] [Google Scholar]
- 28.Yao N, Hesson T, Cable M, Hong Z, Kwong A D, Le H V, Weber P C. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol. 1997;4:463–477. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S, Grosse F. Nuclear DNA helicase II unwinds both DNA and RNA. Biochemistry. 1994;33:3906–3912. doi: 10.1021/bi00179a016. [DOI] [PubMed] [Google Scholar]