Abstract
To uncover the role of satellite cells (SCs) in paravertebral muscle development and aging, we constructed a single-nucleus transcriptomic atlas of mouse paravertebral muscle across seven timepoints spanning the embryo (day 16.5) to old (month 24) stages. Eight cell types, including SCs, fast muscle cells, and slow muscle cells, were identified. An energy metabolism-related gene set, TCA CYCLE IN SENESCENCE, was enriched in SCs. Forty-two skeletal muscle disease-related genes were highly expressed in SCs and exhibited similar expression patterns. Among them, Pdha1 was the core gene in the TCA CYCLE IN SENESCENCE; Pgam2, Sod1, and Suclg1 are transcription factors closely associated with skeletal muscle energy metabolism. Transcription factor enrichment analysis of the 42 genes revealed that Myod1 and Mef2a were also highly expressed in SCs, which regulated Pdha1 expression and were associated with skeletal muscle development. These findings hint that energy metabolism may be pivotal in SCs development and aging. Three ligand-receptor pairs of extracellular matrix (ECM)-receptor interactions, Lamc1-Dag1, Lama2-Dag1, and Hspg2-Dag1, may play a vital role in SCs interactions with slow/fast muscle cells and SCs self-renewal. Finally, we built the first database of a skeletal muscle single-cell transcriptome, the Musculoskeletal Cell Atlas (http://www.mskca.tech), which lists 630,040 skeletal muscle cells and provides interactive visualization, a useful resource for revealing skeletal muscle cellular heterogeneity during development and aging. Our study could provide new targets and ideas for developing drugs to inhibit skeletal muscle aging and treat skeletal muscle diseases.
Keywords: Niche, Paraspinal muscle, Satellite cells, Senescence, Single-nucleus RNA sequencing
Introduction
Skeletal muscle homeostasis disequilibrium is a major mechanism in skeletal muscle disease pathogenesis. Adult stem cells in skeletal muscle are a core component in maintaining skeletal muscle homeostasis. These cells, also known as muscle satellite cells (SCs), are located between the muscle fiber basal lamina and sarcolemma. They often remain quiescent and play an essential part in skeletal muscle regeneration and homeostasis. When muscle tissues are damaged, SCs can be activated and undergo self-renewal and differentiation. Activated SCs can differentiate into progenitor cells capable of repairing muscle fibers.1
SCs numbers gradually decrease with age and the progression of the disease course, leading to a progressive decline in skeletal muscle regeneration and repair capacity. This promotes the development of skeletal muscle diseases, e.g., sarcopenia,2 Duchenne muscular dystrophies,3 and degenerative lumbar scoliosis,4 seriously affecting the quality of life. Hence, SCs may be potential targets for developing drugs that inhibit skeletal muscle aging and treat skeletal muscle diseases. However, the dynamics of SCs gene transcription during skeletal muscle development and aging are not fully understood. Furthermore, skeletal muscle regeneration and repair capacity depends on the interactions between SCs and other cells [e.g., immune cells, endothelial cells (ECs), fibroblasts] in skeletal muscle.5,6 Yet, such cell–cell interactions in skeletal muscle during development and aging remain unclear, which impedes further SCs research and utilization.
Paravertebral muscle constitutes an important part of skeletal muscle and forms a vital structure for maintaining spine stability. Paravertebral muscle degeneration is mainly manifested by decreased muscle volume, diminished muscle cross-sectional area, and reduced muscle mass with age.7,8 Consequently, analyzing the transcriptional characteristics of SCs in paravertebral muscle development and aging contributes to the understanding of the prominent role of SCs in skeletal muscle development and aging.
Therefore, we constructed a single-cell atlas of mouse paravertebral muscle across seven timepoints from the embryo (day 16.5) to old (month 24) stages based on single-nucleus RNA sequencing (snRNA-seq). We focused on the potential SCs biological functions at different developmental stages and their interactions with other skeletal muscle cells. We organized the snRNA-seq data of paravertebral muscle in this study and that related to published skeletal muscle research. Subsequently, we built a website by integrating the available data. Our research not only revealed the prominent role of SCs in paravertebral muscle development and aging but also provided new targets and ideas for regenerative cell therapies, such as reversing skeletal muscle aging and facilitating skeletal muscle damage repair.
Materials and methods
Ethical statement
The Laboratory Animal Welfare and Ethics Committee of Jiangxi Zhonghong Boyuan Biotechnology Co., Ltd. (Institutional Review Board approval number: 2021102601) and the Ethics Committee of BGI (approval letter reference number: BGI-IRB A22014) approved this study. All operations were performed in accordance with the guidance of the Laboratory Animal Welfare and Ethics Committee of Jiangxi Zhonghong Boyuan Biotechnology Co., Ltd. All procedures followed all institutional and national guidelines applicable to the care and use of animals.
Sample collection and nucleus extraction
C57BL/6 mice were obtained from GemPharmatech (Jiangsu, China) and housed in a temperature/light-controlled environment with access to standard mouse chow and water ad libitum. Embryonic day 16.5 (E16.5), E17.5, and postnatal day 0 (P0) embryos were microdissected to obtain the dorsal paraspinal muscles. The female mice were dissected at month 1 (M1), M6, M12, and M24 to obtain the paraspinal muscles.9 The collected tissues were washed with 1× phosphate-buffered saline (PBS), and then snap-frozen and stored in liquid nitrogen prior to nuclear extraction. The nuclei were separated by mechanical extraction.10
snRNA-seq library construction
We performed cDNA amplification and library construction according to a DNBelab C series single-cell library preparation kit (MGI, 1000021082). After library construction, sequencing was performed based on the DNBSEQ-G400 (BGISEQ-500) sequencing platform. Table S1 lists the sample information.
snRNA-seq data pre-processing and quality control
The raw sequencing data of each sample were processed with DNBelab C Series 3.2 single-cell RNA analysis software10 and the gene expression matrix for each sample was obtained (the gene expression matrix consisted of three files, namely, matrix.mtx.gz, features.tsv.gz, and barcodes.tsv.gz). The read number was matched to the mouse genome GRCm38 (Genome Reference Consortium Mouse Build 38). Table S1 lists the quality control results for each sample.
Cell quality control and cluster analysis of snRNA-seq data were performed using the R package Seurat version 4.0.3.11 Next, the matrix.mtx.gz file for each sample was loaded using the Seurat readMM function, and a sparse matrix of UMI (unique molecular identifier) counts was obtained. Then, the barcodes.tsv.gz and features.tsv.gz files for each sample were read with read.table, yielding barcodes and features, respectively, which were assigned to the column and row names of the sparse matrix, respectively. Simultaneously, the CreateSeuratObject function of the Seurat package was used to read the modified sparse matrix and create a Seurat object. For this Seurat object, low-quality cells and low-expression genes were filtered using the following criteria: i) Each gene was expressed in at least three cells; ii) Each cell expressed was no less than 200 genes; iii) The proportion of mitochondrial genes was <20%. Then, the cell depth was corrected with the NormalizeData function of the R package Seurat (normalization.method = “LogNormalize”), and the top 2000 highly variable genes (HVGs) were identified with the FindVariableFeatures function (selection.method = “vst”).
Next, the anchors of all samples were determined through the FindIntegrationAnchors function (reduction = “canonical correlation analysis, cca”). All samples were integrated with the IntegrateData function to obtain the integrated Seurat object. The ScaleData function was used to remove the effect of mitochondrial genes on the gene expression matrix in the integrated Seurat object and to tend the data expression values toward normal distribution. Then, principal component analysis (PCA) was performed on the top 2000 HVGs and the contribution of the top 50 principal components (PCs) to the overall variance was assessed using the JackStraw and ElbowPlot functions of the R package Seurat. The top 30 PCs with the most significant P-values were used for t-distributed stochastic neighbor embedding (t-SNE) dimensionality reduction. Finally, the cells were clustered using the top 30 PCs, for which the cluster analysis resolution was 1.0.
Differentially expressed gene analysis, gene ontology enrichment, cell type annotation
The FindAllMarks function of the R package Seurat (test.use = “wilcox”, logfc.threshold = 1) was used to compare the gene expression of different clusters. Differentially expressed genes (DEGs) in the clusters were filtered out with a corrected P ≤ 0.05. Next, the differential genes in each cluster were used to perform functional enrichment analysis through the R package clusterprofiler (version 4.0.5).12 Statistically significant functional gene sets across clusters were filtered out using an adjusted P ≤ 0.05. Then, the cell types of the clusters were annotated based on the differential genes of each cluster, the enriched functional gene sets, and the marker genes of cell types in published articles. Based on the cell annotation results, the same method was used to calculate the differential genes of different cell types and perform functional enrichment analysis of the differential genes. The statistically significant differential genes and functional gene sets were obtained with adjusted P ≤ 0.05.
Gene set enrichment analysis (GSEA)
Cell death plays a central role in skeletal muscle development and aging. To assess the potential biological functions of different cell types related to cell death, we collected programmed cell death, apoptosis, ferroptosis, pyroptosis, senescence, autophagy, and inflammasome-related gene sets from the Molecular Signatures Database,13 FerrDb,14 and the related literature15, 16, 17, 18 (Table S2) and performed GSEA. To eliminate sample background information bias, we selected single-cell GSEA methods based on expression ranking, namely, AUCell version 1.14.0,19 UCell version 1.1.0,20 singscore version 1.12.0,21 and single-sample GSEA (ssGSEA) version 1.40.1.22 ssGSEA removes the final normalization step, making it closer to single-cell GSEA. To assess whether the gene set was enriched in a certain cell subset, the differential gene set in the enrichment score matrix was calculated by the Wilcox test (corrected P ≤ 0.05, P-value correction method: Bonferroni). The differential analysis results were comprehensively evaluated through robust rank aggregation (RRA) of the RobustRankAggreg package version 1.1.0,23 and genes that were significantly enriched in most GSEA methods were filtered out. The gene sets that were not enriched or significantly not enriched (corrected P ≤ 0.05, P-value correction method: Bonferroni) were displayed in a heat map.
Skeletal muscle disease-related DEGs
To explore the differential genes related to SCs development and aging, we collected 390 skeletal muscle disease-related genes from the literature.24 These skeletal muscle diseases are associated with abnormal skeletal muscle cell development and aging. The 390 genes intersected with the up-regulated DEGs in SCs in our data and yielded 42 skeletal muscle disease-related DEGs. The expression of these 42 DEGs was displayed through bubble plots, and time series expression pattern cluster analysis of the genes was performed through the R package Mfuzz version 1.14.025 to observe their expression trend at different SCs developmental timepoints. Then, the scoring of the gene set of interest at different SCs timepoints was depicted through violin plots. Next, the genes in the TCA CYCLE IN SENESCENCE gene set were intersected with the skeletal muscle disease-related SCs DEGs, and the expression levels of the intersected genes at the SC developmental timepoints were displayed with violin plots.
Trajectory analysis
The SCs trajectories were constructed via the Monocle version 2.22.026 package. The SCs expression matrix and metadata information were extracted from the integrated Seurat object with the subset function, and a Monocle object was created through the newCellDataSet function (lowerDetectionLimit = 0.5). The size factor in the Monocle object was evaluated with the estimateSizeFactors function, and data dispersion was evaluated by the estimateDispersions function. Low-quality genes (each gene expressed in at least three cells) were filtered using the detectGenes function. Based on the top 2000 SC HVGs, trajectories were constructed using the setOrderingFilter function, reduceDimension function (reduction_method = “DDRTree”, max_components = 2, norm_method = “log”), and orderCells function. E16.5, E17.5, and P0 were combined into the embryo stage; M1 and M6 into the young stage; and M12 and M24 into the old stage according to the mouse life stages.27
The distribution of expression levels of the different timepoints, different stages, the TCA CYCLE IN SENESCENCE gene set enrichment score, and Pdha1 expression were depicted with plot_cell_trajectory function. The expression trends of the 42 skeletal muscle disease-related DEGs at the SC developmental timepoints were displayed through the plot pseudotime heatmap function. To explore whether the 42 genes demonstrated co-expression trends and coordination effects at the SCs developmental timepoints, the expression values of the genes at the timepoints were extracted from the Monocle object and skeletal muscle disease-related DEG pairwise correlation analysis was performed using the ggcorrplot package (ggcorrplot function in version 0.1.3).
Transcription factor (TF) enrichment analysis
To obtain the genes belonging to the TF family among the SCs skeletal muscle disease-related DEGs, the mouse TF list19 was downloaded and the intersection with the 42 skeletal muscle disease-related DEGs was obtained. The expression trends of these intersecting genes at the SCs development timepoints were depicted by violin plots. To search for TFs that may regulate gene expression, TF enrichment analysis of the 42 genes was performed through the R package RcisTarget version 1.12.0,19 and the high-confidence TFs with normalized enrichment score (NES) ≥ 4 were filtered out (Table S9). TFs that might regulate the target genes were filtered out, and the correlation between the expression levels of the target genes and TFs were explored through Pearson correlation analysis. The expression trends of the TFs at the SCs timepoints were depicted through violin plots.
Cellular communication network construction
SCs can differentiate into fast muscle cells (FMCs) or slow muscle cells (SMCs). SCs, FMCs, and SMCs are closely related and play an important role in skeletal muscle development. Here, the intercellular communication network between the three cell types was inferred and visualized through the R package CellChat version 1.0.0.28 The general analysis process was as follows: First, CellChat objects of the embryo (E16.5, E17.5, P0), young (M1 and M6), and old (M12 and M24) periods were created through the createCellChat function of the R package CellChat and the database was set as the human ligand-receptor interaction database. Overexpressed ligands or receptors were identified and the gene expression data were projected onto the protein–protein interaction network. A ligand-receptor interaction was assigned a probability value so that a statistically significant interaction could be inferred by performing a permutation test. Each ligand-receptor interaction was assigned to a signaling pathway, and the communication probability for each signaling pathway was calculated by combining the probabilities of all interactions in each signaling pathway. The number, distribution, and type proportion of ligand-receptor pairs with statistical significance in the timepoints were displayed on bar charts, chord diagrams, and pie charts. Then, we intersected the skeletal muscle developmental disease-related genes, SC differential genes, and statistically significant ligand-receptor pair genes. The intersection genes of the three sets are displayed in a Venn diagram. The intersection genes of these three sets may play an important role in the interaction of SCs, FMCs, and SMCs. All statistically significant ligand-receptor pairs were shown in bubble plots of expression in the timepoints. The structural similarity of all statistically significant signaling pathways was quantified and different signaling pathway groups were identified, which aided the exploration of the synergistic signaling pathways.
Collation of public data
Collection and annotation
PubMed and Google Scholar were searched using the keywords “skeletal muscle” and “single cell RNA-seq”. Articles were filtered based on whether they contained the downloaded data. The downloaded data should contain the standard 10X Cell Ranger processing file filtered_feature_bc_matrix, original count matrix file, and expression matrix file with metadata information. For data containing cell type metadata, the cell type corresponding to the barcode was directly matched when creating the Seurat object for the data. Data that only provided the Cell Ranger result file filtered_feature_bc_matrix and the expression matrix were re-annotated with the marker genes used in the article. The paraspinal muscle single-cell nuclear transcriptome sequencing data in this study were also included in the database. A total of 630,040 cells were included, for which 69 datasets from 19 articles were collected in addition to the nuclear transcriptome sequencing data in the present study.
Processing
For website construction, the metadata information was extracted from the Seurat object and all the data were integrated. For individual data, the DEGs between the different cell types were obtained with the FindAllMarkers function. The individual data should include at least two cell types. Then, the SC expression matrix in these data was extracted and the transcriptional regulatory network was constructed based on the expression matrix of these SCs by pySCENIC software. Finally, the TFs and their target genes and corresponding motif IDs in the SCs were predicted.
Website construction
The Musculoskeletal Cell Atlas (MSKCA, version1.0 Skeletal Muscle Regeneration Cell Atlas, http://www.mskca.tech) uses a Linux-Apache-MySQL-PHP system. Data are stored in a MySQL database and include tissue information on single-cell transcriptome sequencing, library construction method, publication time, cell number, expression matrix, and data visualization.29 The website is built on the powerful PHP framework CodeIgniter, which provides an application programming interface to connect the web to a MySQL database. Dynamic web services were performed using JavaScript libraries including jQuery 2.2.0, jQuery labelauty, and additional plugins. The multi-timepoint data were also processed using the ShinyCell package.30,31
Data availability
The expression matrices of the paraspinal muscle single-cell nuclear transcriptome sequencing data (seven timepoints) in this study are all stored on the MSKCA website. The data reported in this study are also available in the CNGB Nucleotide Sequence Archive (CNSA: https://db.cngb.org/cnsa; accession number CNP0003561). The data of the multi-timepoints research are available from the Zenodo repository (10.5281/zenodo.7294918).
Results
Cell type annotation and differential gene expression
The paravertebral muscle is a vital structure for spine stability and motor function. To elucidate the transcriptomic changes of different cell types in paravertebral muscle development and aging, we analyzed the paravertebral muscle samples collected at seven timepoints during mouse development and aging based on snRNA-seq (Fig. 1A). Quality control of all paravertebral muscle samples yielded 34,589 high-quality cells.
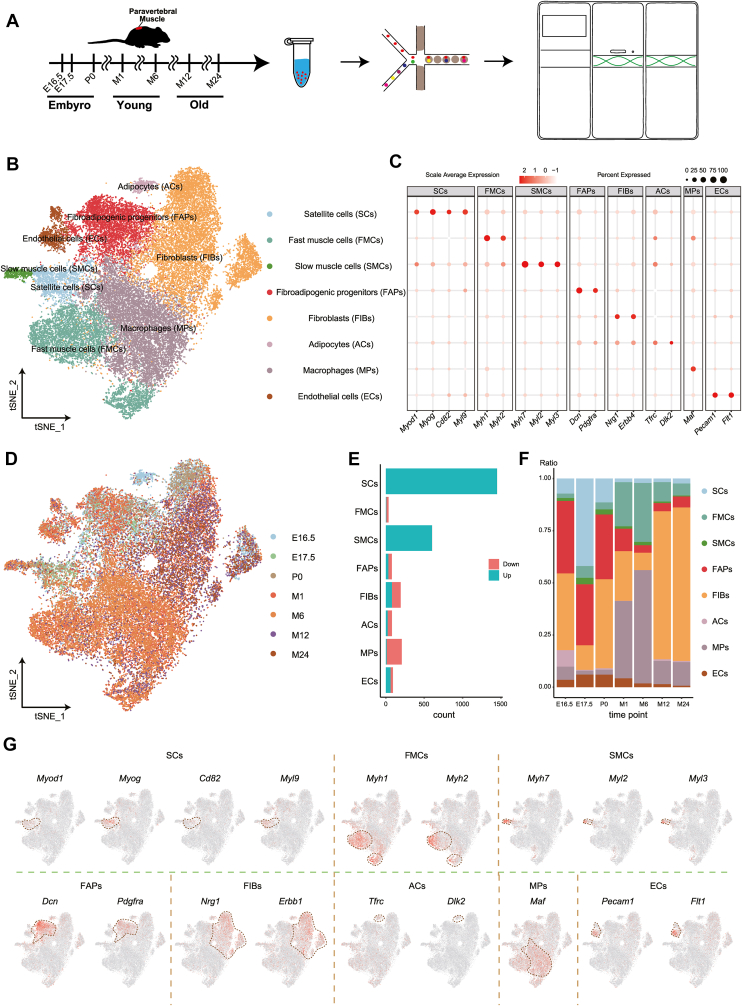
Figure 1.
The cell type annotation of the single-cell transcriptome atlas of mouse paravertebral muscles' developmental and aging stages. (A) Flow chart showing the experimental design for constructing a single-cell transcriptome atlas of mouse paravertebral muscle during development and aging. Cells were collected at seven timepoints: E16.5, E17.5, P0, M1, M6, M12, and M24. (B)t-SNE plot displaying the cell atlas of mouse paravertebral muscle integrating the seven timepoints. The colors represent different cell types. (C) Bubble chart indicating the expression of marker genes used for cell type annotation. The circle color represents the scale average expression of marker genes and the circle size represents the percentage of marker genes expressed. (D)t-SNE plot demonstrating integrated annotations of cells at the mouse developmental timepoints. The colors represent the timepoints. (E) Bar plot illustrating the count of up-regulated and down-regulated genes in the different cell types. Red indicates down-regulation and blue indicates up-regulation. (F) Bar plot depicting the ratios of cell types at the timepoints. The colors represent different cell types. (G)t-SNE plot presenting the expression of marker genes in the mouse paravertebral muscle cell atlas. Red indicates positive expression in a given cell.
Then, we carried out biological annotations according to the literature and annotated eight cell types (Fig. 1B), SCs (expressing Myod1,32 Myog,33 Cd82,34 Myl935), FMCs (expressing Myh136 and Myh237), SMCs (expressing Myh7,37 Myl2,38 Myl339), fibro/adipogenic progenitors (FAPs, expressing Dcn40 and Pdgfra32), fibroblasts (expressing Nrg141 and Erbb442), adipocytes (expressing Tfrc43 and Dlk244), macrophages (expressing Maf45), and ECs (expressing Pecam146 and Flt147) (Fig. 1B, C, G).
We compared gene expression among the cell types and identified the DEGs in eight cell types with corrected P ≤ 0.05 (Fig. 1E and Tables S5, 6). The cell distribution in a low-dimensional space at the timepoints was illustrated using a t-SNE plot (Fig. 1D; Fig. S1). Moreover, the ratio of each cell type across the timepoints was depicted using a bar plot (Fig. 1F and Table S7).
Gene set functional enrichment
Cell death plays a central role in skeletal muscle development and aging. To assess the potential death-related biological functions of the annotated cell types, we collected gene sets associated with programmed cell death, apoptosis, ferroptosis, pyroptosis, senescence, autophagy, and inflammasome (Table S2). To eliminate sample background information bias, we selected the AUCell, UCell, singscore, and ssGSEA methods for GSEA based on the ranking of single-cell gene expression. In ssGSEA, the final normalization step was removed to make it closer to single-cell GSEA. To assess whether the gene sets were enriched in a certain cell subset, we calculated the DEG sets in the enrichment score matrix using the Wilcox test (adjusted P < 0.05 based on Bonferroni correction) and comprehensively evaluated the differential expression analysis results. The gene sets significantly enriched or depleted in most GSEAs were selected (P < 0.05) and presented in a heatmap (Fig. 2A). Several senescence-related gene sets were enriched in the SCs, FMCs, and SMCs, namely, POSITIVE REGULATION OF AUTOPHAGY OF MITOCHONDRION, TCA CYCLE IN SENESCENCE, GLYCOLYSIS IN SENESCENCE, and POSITIVE REGULATION OF NAD METABOLISM IN ONCOGENE-INDUCED SENESCENCE AND MITOCHONDRIAL DYSFUNCTION-ASSOCIATED SENESCENCE. These gene sets are closely related to energy metabolism, suggesting that energy metabolism may be greatly important in maintaining SCs, FMCs, and SMCs biological functions.
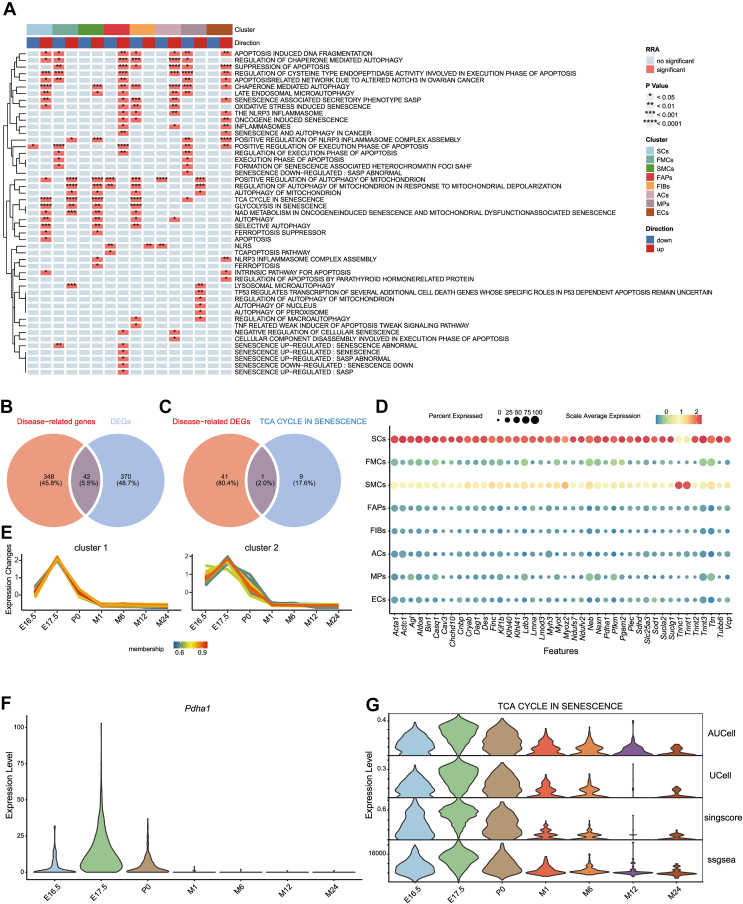
Figure 2.
Gene set functional enrichment of paravertebral muscle development and aging. (A) Heatmap demonstrating the statistical significance of gene set enrichment or depletion among cell types in the comprehensive evaluation. The heatmap mainly comprises red and blue grids, which indicate significant and no significant gene set enrichment or depletion in a given cell type, respectively (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001). The dendrogram on the left of the heatmap demonstrates similar enrichment patterns of gene sets in different cell subsets. In the top color bar above the heatmap, blue and red represent gene set depletion and enrichment, respectively. In the bottom color bar, the colors represent different cell subsets. (B) Venn diagram depicting 42 intersected genes between 390 skeletal muscle disease-related genes and 412 DEGs (log|FC| ≥ 2) in SCs. (C) Venn diagram revealing one intersected gene (Pdha1) between the 42 disease-related DEGs in SCs and the TCA CYCLE IN SENESCENCE gene set. (D) Bubble plot depicting the expression of 42 intersected genes in the cell types. The circle color represents the scale average expression of genes and the circle size represents the percentage of genes expressed. (E) Line chart demonstrating the expression trends of 42 intersected genes in SCs across the timepoints, which can be divided into two patterns that exhibited similar changes, with an initial increase before the decrease in the embryo stage, then a gradual decrease in the postnatal stage. (F) Violin plot depicting Pdha1 expression levels at the SC development and aging timepoints. (G) Violin plot demonstrating the expression trends of the TCA CYCLE IN SENESCENCE gene set in different SC stages based on enrichment scores calculated using AUCell, UCell, singscore, and ssGSEA.
SCs play a considerable role in skeletal muscle disease development and progression. To explore the SCs genes that may be involved in skeletal muscle development and aging, we collected 390 genes related to skeletal muscle diseases from the literature (Table S3). Most skeletal muscle diseases are associated with abnormal skeletal muscle cell development and aging. Therefore, we intersected the 390 disease-related genes with the significantly up-regulated DEGs in SCs (logFC [fold change] ≥ 2) and obtained 42 DEGs that may affect SC development and aging (Fig. 2B). The expression of the 42 disease-related DEGs in eight cell types was displayed using a bubble chart (Fig. 2D) and clustering analysis of time series expression patterns on the 42 genes was performed. The expression changes of the 42 genes with the developmental timepoints could be divided into two patterns (Fig. 2E), which exhibited similar trends with time. Specifically, the gene expression levels initially increased in the embryo development stage, then gradually decreased and remained at stable and low levels in the postnatal stage.
Skeletal muscle is a vital energy metabolism organ, and the tricarboxylic acid (TCA) cycle is a central hub of energy metabolism. Therefore, we quantified the expression levels of the TCA CYCLE IN SENESCENCE gene set using AUCell, UCell, singscore, and ssGSEA, and observed their expression changes in SCs across time (Fig. 2G). The expression levels of the TCA CYCLE IN SENESCENCE gene set were increased in the embryo stage, then gradually decreased to stable levels in the postnatal stage. We intersected the 42 disease-related DEGs with the TCA CYCLE IN SENESCENCE gene set and obtained one gene, Pdha1 (Fig. 2C). The Pdha1 expression trends in SCs across time were displayed using a violin plot (Fig. 2F). Pdha1 expression levels initially increased in the embryo stage, followed by a gradual decrease to stable levels in the postnatal stage. This suggested that Pdha1 played a vital role in SCs energy metabolism.
Satellite cell development
We constructed the pseudotime trajectory of SCs, then merged E16.5, E17.5, and P0 cells into the embryo stage; M1 and M6 cells into the young stage; and M12 and M24 cells into the old stage. The SCs pseudotime trajectory exhibited consistency with their actual developmental stages (Fig. 3A, B). We also observed the distributions of the Pdha1 gene and the TCA CYCLE IN SENESCENCE gene set quantified using AUCell across the pseudotime trajectory, where they followed consistent trends along the pseudotime trajectory in accordance with the actual SCs developmental stage (Fig. 3C, D).
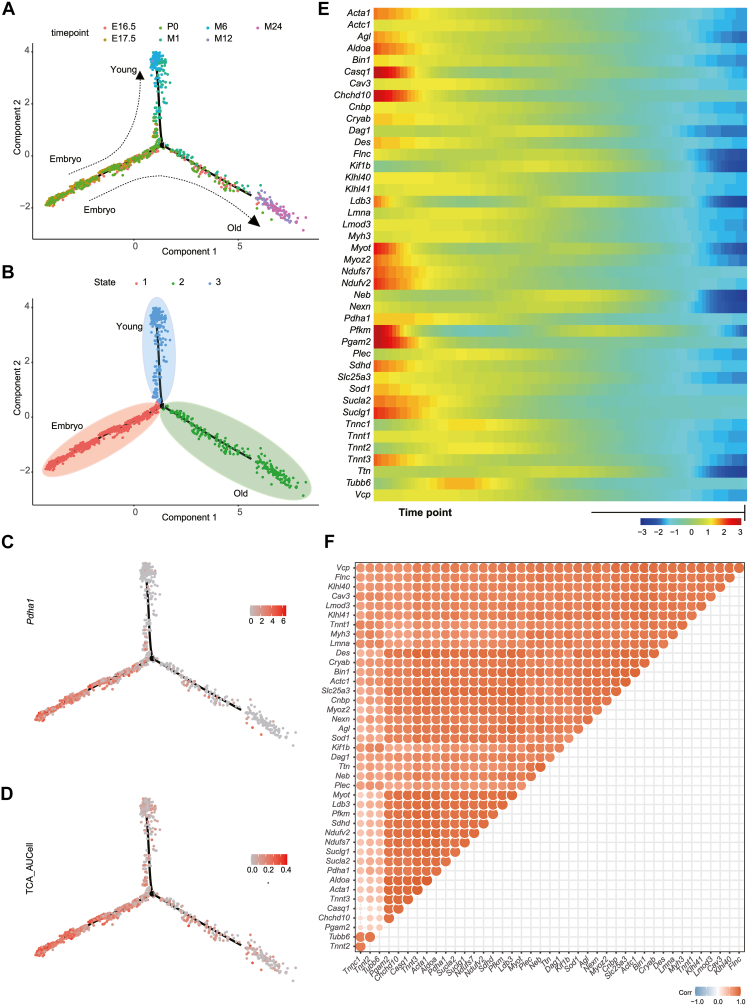
Figure 3.
Single-cell trajectory analysis of the developmental trajectory of satellite cells (SCs). (A) Developmental trajectory of SCs at the seven timepoints. The colors represent the timepoints. (B) Developmental trajectory of SCs in the three stages. The colors represent the different stages. (C) Developmental trajectory of Pdha1 expression in SCs in different stages. Red indicates positive expression in the cells. (D) Developmental trajectory of TCA CYCLE IN SENESCENCE gene set expression calculated using AUCell in SCs in different stages. Red indicates positive expression in the cells. (E) Heatmap inferred from the expression trajectory of 42 disease-related DEGs in SCs in different stages. A redder color indicates a higher gene expression level. (F) Bubble chart depicting the correlations between disease-related DEGS in SCs. A redder color indicates a higher correlation.
Subsequently, we visualized the expression trends of the 42 DEGs across the developmental timepoints (Fig. 3E). Overall, the gene expression levels decreased gradually with the increase in pseudotime values and were maintained at low levels. The expression levels of some genes first increased with the increase in pseudotime values. However, after reaching a maximum level, the gene expression gradually decreased with the increase in pseudotime values. This suggests that the 42 DEGs may play a notable role in SCs development and differentiation. Finally, to explore whether the 42 genes had a co-expression tendency and synergistic effect at the SCs developmental timepoints, we performed pairwise correlation analysis (Fig. 3F and Table S8). In most cases, the correlation coefficients between the quantity of the 42 genes were ≥0.5, suggesting the potential co-expression and synergistic relationships of these genes in SCs development and aging.
Regulatory relationship of TFs in SCs
To annotate the genes of TF families among the DEGs related to SCs development and aging, we downloaded a list of mouse TFs and intersected them with the 42 disease-related DEGs. We obtained three genes annotated as TFs, namely, Pgam2, Sod1, and Suclg1 (Fig. 4A). Then, to search the TFs that may regulate the expression of the 42 DEGs, we performed TF enrichment analysis and selected TFs with high confidence (NES ≥4), identifying a total of 150 TFs (Table S9). Furthermore, we selected the TFs that may regulate the target genes based on a target gene list of high-confidence TFs. We found that two TFs, i.e., Mef2a and Myod1, may regulate Pdha1 expression.
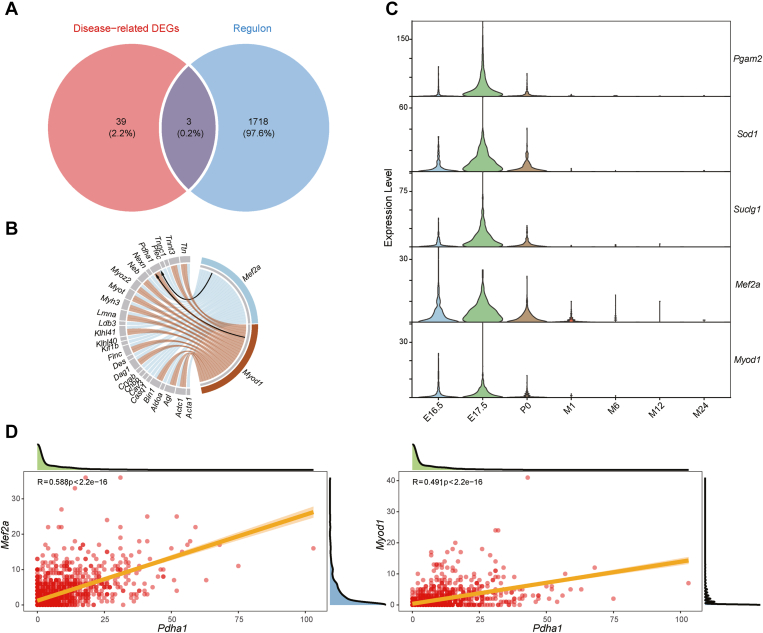
Figure 4.
Transcription factor (TF) enrichment analysis of the skeletal muscle disease-related DEGs. (A) Venn diagram depicting the intersection of 42 disease-related DEGs in SCs and 1721 mouse TFs. (B) Chord diagram revealing possible target genes regulated by Myod1 and Mef2a. The arrows indicate that Myod1 and Mef2a may regulate Pdha1 expression. (C) Violin plot illustrating Pgam2, Sod1, Suclg1, Myod1, and Mef2a expression levels in SCs at the seven timepoints. (D) Scatter plot displaying the correlation between Pdha1 and Myod1 or Mef2a expression levels.
We depicted the trends of Pgam2, Sod1, Suclg1, Mef2a, and Myod1 expression at different SCs developmental timepoints using violin plots. Pgam2, Sod1, Suclg1, Mef2a, and Myod1 exhibited similar expression trends to Pdha1, i.e., increased in the embryo stage before decreasing to stable levels in the postnatal stage (Figure 2, Figure 4). Finally, we explored the correlation between Pdha1 and Mef2a or Myod1 expression by Pearson correlation analysis (Fig. 4D). Mef2a and Pdha1 expression (r = 0.588, P-value <2.2e-16) were highly correlated with each other, as were Myod1 and Pdha1 expression levels (r = 0.491, P-value <2.2e-16). These results suggested that Pdha1 expression may be regulated by Mef2a and Myod1.
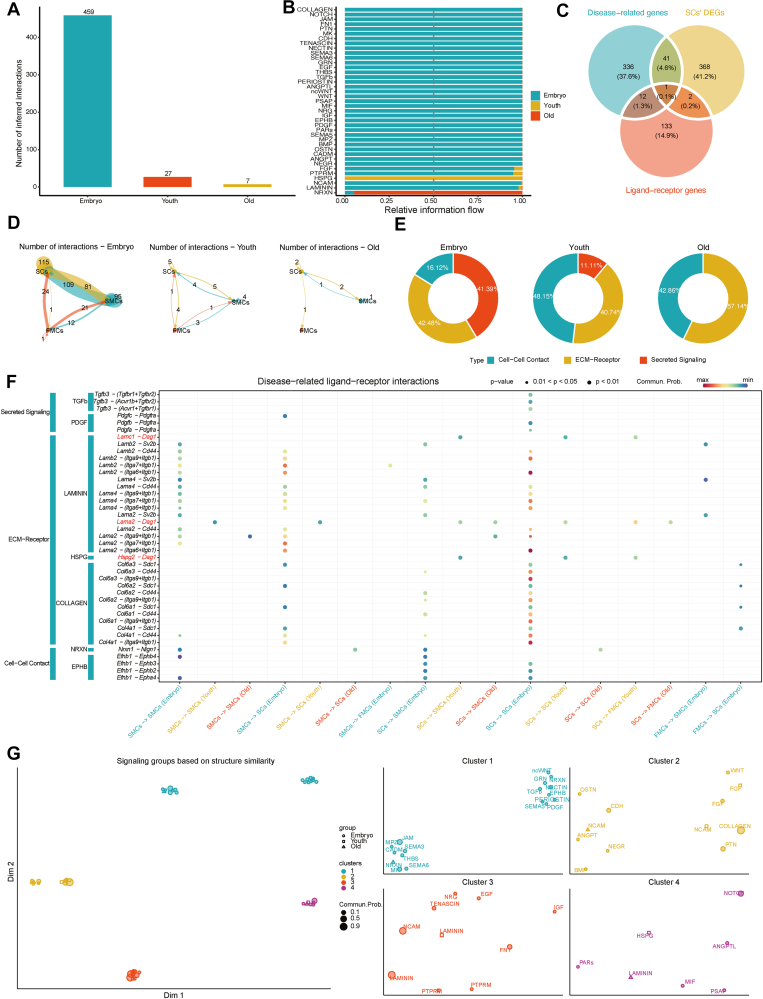
Interactions of SCs with FMCs and SMCs
SCs can differentiate into FMCs or SMCs. These three cell types are tightly linked and play a major role in skeletal muscle development. Therefore, we inferred and visualized the intercellular communication network between the three cell types. We illustrated the number of significant ligand-receptor pairs in the three stages using a bar plot (Fig. 5A) and displayed the proportion of each stage in different signaling pathways (Fig. 5B). We also presented the distribution of the number of significant ligand-receptor pairs in different stages among SCs, FMCs, and SMCs using chord diagrams (Fig. 5D). The number of interactions between SCs and FMCs or SMCs gradually decreased with the progress of natural aging. This prompted the idea that reducing or silencing such cell–cell interactions may play a major role in preventing SC differentiation and skeletal muscle aging progression.
Figure 5.
The cellular communication network between satellite cells (SCs), fast muscle cells (FMCs), and slow muscle cells (SMCs). (A) Bar plot displaying statistically significant ligand-receptor pairs in the three stages. (B) Bar plot demonstrating the proportions of the three stages in distinct signaling pathways. (C) Venn diagram demonstrating the intersection of skeletal muscle disease-related genes, DEGs in SCs, and genes related to ligand-receptor pairs. (D) Chord diagram presenting the distribution of the number of disease-related ligand-receptor pairs among SCs, FMCs, and SMCs in different stages. (E) Doughnut charts showing the proportions of three types of ligand-receptor pairs in the different stages. (F) Bubble plot depicting the specific distribution of statistically significant ligand-receptor pairs among SCs, FMCs, and SMCs. We focused on Dag1-related ligand-receptor interaction pairs. (G) UMAP (uniform manifold approximation and projection) plots indicating signaling pathway groups. The colors represent signaling pathway clusters; the shapes represent stages. The shape sizes are related to the communication probability of the signaling pathways.
We also demonstrated the proportions of three types of ligand-receptor pairs in the different stages using doughnut charts (Fig. 5E). In the embryo, young, and old stages, cell–cell contact interactions accounted for 16.12%, 48.15%, and 42.86% of total interactions, respectively; ECM-receptor interactions accounted for 42.48%, 40.74%, and 57.14%, respectively. It is worth noting that secreted signaling interactions accounted for 41.39%, 11.11%, and 0% of total interactions in the embryo, young, and old stages, respectively. We found that with the progress of natural aging, the proportions of ECM-receptor interactions increased overall, while the proportions of secreted signaling interactions demonstrated a decreasing trend. To identify ligand-receptor pairs that may have an important role in SC development and aging, we demonstrated the intersection of skeletal muscle disease-related genes, SCs DEGs, and ligand-receptor genes in a Venn diagram, which yielded the dystroglycan (Dag1) gene (Fig. 5C). Dag1 plays a major part in skeletal muscle development and aging-related diseases.
We extracted all significant ligand-receptor pairs. If a ligand or receptor gene was assigned to the 390 skeletal muscle disease-related genes, the corresponding ligand-receptor pair was defined as a disease-related ligand-receptor pair. The specific distribution of the disease-related ligand-receptor pairs among the SCs, FMCs, and SMCs was depicted using a bubble chart (Fig. 5F). We focused on displaying Dag1-related ligand-receptor interaction pairs, namely, laminin subunit gamma 1 (Lamc1)-Dag1, laminin subunit alpha 2 (Lama2)-Dag1, and heparan sulfate proteoglycan 2 (Hspg2)-Dag1. Both Lamc1-Dag1 and Lama2-Dag1 belong to the LAMNN pathway, while Hspg2-Dag1 belongs to the HSPG pathway. These ligand-receptor pairs were classified into ECM-receptor interactions, suggesting that ECM-receptor interactions play a vital role in SC development and aging. We also observed the largest number of ligand-receptor interactions between SCs and SCs (embryo stage), with Lamb2-(Itga6+Itgb1), Lama2-(Itga6+Itgb1), and Col4a1-(Itga9+Itgb1) being the most prominent. The number of ligand-receptor interactions between SMCs and SCs (embryo stage) was the second largest, with Lamb2-(Itga7+Itgb1) and Lama4-(Itga7+Itgb1) being the most prominent.
Finally, to explore other signaling pathways that act synergistically with the signaling pathway of interest, we quantified the structural similarity of all significant signaling pathways and clustered them into distinct groups (Fig. 5G), and obtained four signaling pathway clusters. The LAMNN pathway was present in Cluster 3 and 4; the HSPG pathway was present in Cluster 4. Among these, the projections of all pathways in Cluster 4 were closer in a low-dimensional space. This suggested that other Cluster 4 signaling pathways (NOTCH, ANGPTL, PARs, MIF, PSAP) may act synergistically with the LAMNN and HSPG pathways to play a prominent role in SC development and aging.
Public database construction
To facilitate skeletal muscle single-cell data exploration and downloading, we built an online database for skeletal muscle, MSKCA. We obtained a total of 630,040 cells from 70 data sets, with 69 public data sets and one data set obtained in this study. The human data comprised 29 datasets containing 212,920 cells and the mouse data comprised 41 datasets containing 417,120 cells. The mouse data were obtained at seven timepoints and exhibited distinct characteristics such as damage and no damage. In the published data, single-cell library construction mainly involved 10X Genomics and Drop-seq (Table S4).
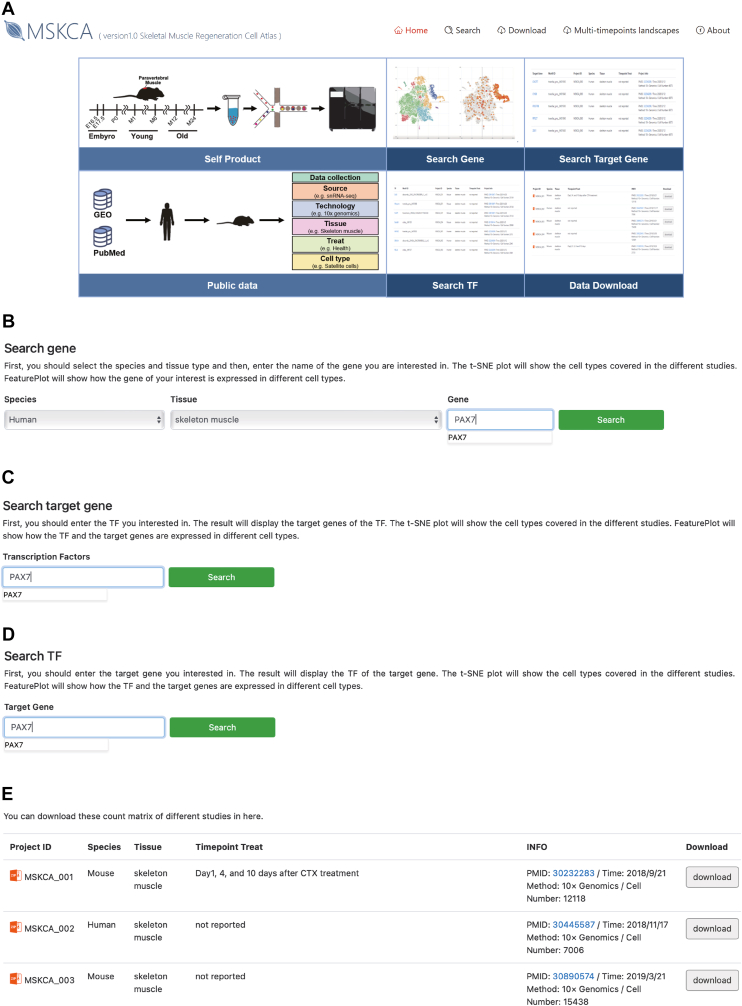
The MSKCA website has five main functions, namely, search gene, search target gene, search TF, multi-timepoints landscapes, and download (Fig. 6A). In the search gene module, the user can select a gene in different human or mouse skeletal muscle, e.g., select Species = “Human”, Tissue = “skeleton muscle”, Gene = “PAX7” (Fig. 6B). First, the results will display the data containing the selected gene in addition to the data sampling timepoint information, PMID, publication time, library construction method used, and a number of cells contained and the abstract (Fig. S3A). Second, the user can select the data of interest and display the cell type clustering results, gene expression feature plot, and marker gene list in the data with t-SNE (Fig. S3B, C). In the search target gene module, the user can enter a TF to obtain the target gene predicted in different data by this TF (Fig. 6C). For example, if selecting TF = “PAX7″, the results will display the target gene predicted by this TF, the motif ID to which the interaction pair belongs, data source, timepoint, and publication-related information. The search TF module has a similar function to the search target gene (Fig. 6D). However, the target gene is input to obtain the predicted TF in addition to the motif ID of the interaction pair, data source, timepoint, and publication-related information. In addition, in “search target gene” and “search TF” functions, users can view the expression of the TF-target genes interaction pairs in the results in the current data (Fig. S3D).
Figure 6.
Introduction to the functions of the Musculoskeletal Cell Atlas. (A) The plot showing the home page of the online database for skeletal muscle, MSKCA. (B–E) The plot displaying the “Search gene”, “Search target gene”, “Search TF”, and “download” functions of MSKCA.
There are seven functions in the multi-timepoints landscapes module, including CellInfo vs. GeneExpr, CellInfo vs. CellInfo, GeneExpr vs. GeneExpr, Gene coexpression, Violinplot/Boxplot, Proportion plot, and Bubbleplot/Heatmap. The data for this module were generated by integrating single-cell data that consisted of different timepoints. This function is convenient for users who want to explore gene expression in the developing stages. In CellInfo vs. GeneExpr function, the user can choose one or more timepoints or cell types to check the gene expression in UMAP or t-SNE plots (Fig. S4A). In CellInfo vs. CellInfo function, the user can select different timepoints to view the distribution of data at different timepoints or different cell types at the chosen timepoint in UMAP and t-SNE plots (Fig. S4B). In the GeneExpr vs. GeneExpr function, the user can view the different gene expressions in one or more cell types or timepoints in UMAP and t-SNE plots simultaneously (Fig. S4C). In the Gene coexpression function, the user can compare two gene expressions at different timepoints or cell types. The cell numbers and percent of coexpression in different timepoints or cell types were also displayed on the right (Fig. S5A). In the Violinplot/Boxplot functions, the user can test the dynamic changes of gene expression in developing timepoints or different cell types by violin plot or boxplot (Fig. S5B). In the Proportion plot function, the cell proportion of data for each timepoint or cell type can be viewed in the proportion plot (Fig. S5C). In Bubbleplot/Heatmap function, the user can choose some interesting genes. These genes' expressions in different timepoints or cell types will be shown in a bubble plot of a heatmap (Fig. S5D). Users-generated graphs and tables are available for downloading in this module. The last function module is “download” (Fig. 6E). For each data set collected, the expression matrices of different data were organized for users to download and use.
Discussion
The paravertebral muscle is a major component of skeletal muscle and a vital structure that maintains spine stability. Paravertebral muscle degeneration is manifested by decreased muscle volume, muscle cross-sectional area, and muscle mass with increased age.7,8 However, the gene transcription dynamics in SCs during paravertebral muscle development and aging are not completely clarified. To uncover the prominent role of SCs in paravertebral muscle development and aging, we used snRNA-seq to construct a single-cell atlas of mouse paravertebral muscle across seven developmental stages and identified eight cell types (SCs, FMCs, SMCs, FAPs, fibroblasts, adipocytes, macrophages, ECs).
Quiescent SCs are located between the basal lamina and plasma membrane of muscle fibers, maintaining self-renewal and high regenerative potential,48 and can be activated after muscle tissue is stimulated or damaged. Activated SCs enter the cell cycle, differentiate into new myotubes, and participate in muscle tissue repair. Quiescent SCs numbers tend to decrease with age and their regenerative repair ability declines gradually.49, 50, 51 Here, we compared the ratios of SCs across developmental stages and found that the SCs ratios in paravertebral muscle showed a continuous increasing trend in the embryo stage, followed by an inverse trend in the postnatal stage. This indicated that the SCs regeneration and repair capacity in the embryonic stage is relatively active. However, the SCs regeneration and repair capacity gradually diminished with the progress of postnatal development. This finding suggested that SCs play a notable role in paravertebral muscle regeneration and repair.
Skeletal muscle regeneration depends on SCs, but SC numbers and their regeneration capacity both decrease with aging and cellular senescence.52 Senescence and age-related diseases share the same basic biological mechanisms, such as metabolic dysfunction and stem cell deficiency.53,54 Skeletal muscle cell death, clearance, and regeneration are precisely regulated in physiological conditions. However, their dysregulation promotes the development of skeletal muscle diseases. The expression of TCA cycle-related genes is increased in activated SCs,55,56 while oxidative phosphorylation in senescent SCs is inhibited.57 The depletion of mitochondrial energy metabolism proteins (TCA cycle and oxidative phosphorylation) and neuromuscular junction proteins is most prominently associated with sarcopenia.58 This suggests that energy metabolism plays an important part in SC activation and senescence and skeletal muscle-related diseases. Here, we performed quantitative gene set analysis and determined that the TCA CYCLE IN SENESCENCE and energy metabolism-related gene sets were highly enriched in SCs. The expression levels of the TCA CYCLE IN SENESCENCE gene set were elevated in the embryo stage but gradually decreased in the postnatal stage. The expression trends of the TCA CYCLE IN SENESCENCE gene set were consistent with the trends of the SC ratios, suggesting that TCA cycle-related genes may be tightly connected to SC regeneration capacity.
DEG analysis revealed that 42 skeletal muscle disease-related DEGs were highly expressed in SCs and exhibited similar expression patterns. Among the 42 DEGs, Pdha1 was the core gene involved in the TCA CYCLE IN SENESCENCE process, and its close relationship with skeletal muscle energy metabolism has been demonstrated previously. The pyruvate dehydrogenase complex (PDC) is a nuclear-encoded mitochondrial matrix multienzyme complex that forms the primary link between glycolysis and the TCA cycle by catalyzing the irreversible conversion of pyruvate into acetyl-CoA.59 Pdha1 is a critical subunit of the PDC.59 PDC deficiency is a disorder of energy metabolism in the mitochondrial respiratory chain and is mostly caused by mutations in the X-linked Pdha1 gene. Pdha1 gene deficiency leads to decreased PDC activity, disordered pyruvate metabolism in the TCA cycle, and insufficient energy synthesis, leading to complex clinical manifestations such as lactic acidosis and neuromuscular damage.60 Here, the Pdha1 expression pattern in the SCs initially increased before decreasing across the timepoints, and its distribution along the pseudotime trajectory was consistent with the actual SCs developmental stages. These results suggest that Pdha1 may play an essential part in SCs energy metabolism (including glycolysis and the TCA cycle).
We also found that among the 42 disease-related DEGs, Pgam2, Sod1, and Suclg1 were previously identified as TF genes closely associated with energy metabolism. Pgam2 is specifically highly expressed in muscle, playing a major role in muscle growth and development, and performing vital biological functions in skeletal muscle glucose metabolism. The protein encoded by Pgam2 participates in glycolysis and is related to striated muscle contraction.61 Sod1 is a key factor in skeletal muscle antioxidation and participates in energy metabolism, redox regulation, cholesterol metabolism, and regulation of mitochondrial respiration.62 In normal conditions, the muscle antioxidant protection system is maintained in dynamic equilibrium. Sod1 mutations can produce excessive free radicals that mediate toxicity to directly act on skeletal muscle, causing internal environment disturbance and muscle cell dysfunction, leading to myogenic differentiation disorders.63 Suclg1 encodes the succinyl-CoA alpha subunit and participates in the TCA cycle. Suclg1 gene mutations can cause mitochondrial DNA depletion syndromes.64 Taken together, these findings suggest that Pgam2, Sod1, and Suclg1 may affect SC energy metabolism and subsequently influence SCs development and aging, ultimately inducing skeletal muscle diseases.
To find the TFs that may regulate the 42 disease-related DEGs, we carried out TF enrichment analysis and selected 150 high-confidence TFs with NES ≥4. Among them, Mef2a and Myod1 regulated Pdha1 expression. The correlation analysis revealed a high correlation between the Pdha1 and Mef2a or Myod1 expression levels. Skeletal muscle development and regeneration are influenced by multiple extracellular factors (including cytokines and hormones) and regulated by myogenic regulatory factors (MYOD1, MYF5, MYOG, MYF6) in addition to myocyte enhancer factor-2 (MEF2A, MEF2C, MEF2D).65,66 Additionally, MyoD myogenic activity can be enhanced by interaction with MEF2 family members.67,68 Accordingly, we consider that energy metabolism may be a pivotal process in SCs development.
Skeletal muscle myoblasts exhibit plasticity in the postnatal stage, switching between SMCs and FMCs to adapt to specific conditions.67 As myoblastic stem cells, SCs can also differentiate into FMCs or SMCs. The intercellular communication network analysis revealed that the interactions between SCs and FMCs or SMCs gradually decreased during the process of development and aging. This suggests that reducing or silencing interactions between SCs and FMCs or SMCs may be the main reason for the decreased proliferation and differentiation capacity of senescent SCs in vivo, thereby preventing muscle regeneration.69
The ECM plays a crucial role during skeletal muscle growth and repair. SCs exist between the sarcolemma and basal lamina of muscle fibers, which form the niche environment of SCs.48 In healthy tissues, SCs are maintained in a quiescent state via the microenvironment formed by the sarcolemma and intact basal lamina.70 Muscle basal lamina is a supramolecular ECM structure that connects two laminin and collagen polymer networks through the bridging function of glycoproteins and heparan sulfate proteoglycans (nidogen and perlecan).71 The Hspg2 gene encodes perlecan protein.72 Laminins, a family of ECM glycoproteins, are the main non-collagenous components of the basal lamina and are essential for its assembly and function. Laminins comprise α, β, and γ subunits, which bind each other to form a cross-shaped molecule through disulfide bonding. The Lamc1 gene encodes the γ 1 chain; the Lama2 gene encodes the alpha 2 chain.73 Upon activation, SCs can differentiate into myogenic progenitor cells through self-renewal or migration into the ECM. These studies demonstrate that skeletal muscle ECM can regulate SCs activity and renewal, influencing skeletal muscle tissue regeneration and repair. We also found that three ligand-receptor interaction pairs of the ECM-receptor group (Lamc1-Dag1, Lama2-Dag1, Hspg2-Dag1) may play a major part in SCs and SMCs/FMCs interactions. The receptor Dag1 was highly expressed in SCs. Cohn et al74 reported that striated muscle-specific disruption of the Dag1 gene caused muscular dystrophy in mice. This allowed us to conclude that ECM also plays a considerable role in SCs differentiation and biological function.
Finally, we built MSKCA, the first single-cell transcriptome database for skeletal muscle. The MSKCA database contains the snRNA-seq data of paravertebral muscle in the present study and that related to skeletal muscle research in public databases, involving a total of 630,040 cells. The database uses an interactive network that directly displays the coordinate points of each cell and the expression levels of genes in each cell. It also presents superb visualization, which facilitates quick exploration of target gene expression in different skeletal muscle cell subsets for skeletal muscle-related research and provides a useful resource for uncovering cellular heterogeneity in skeletal muscle during development and aging. The database also contains a TF prediction table of SCs, which advances skeletal muscle-related research at the single-cell level.
Nonetheless, this study has limitations. Although we obtained many interesting conclusions at the single-cell level, they were based on preliminary bioinformatics analysis and have not been validated. Second, we only used mouse paravertebral muscle as the research object of skeletal muscle, so the extendibility of our results should be considered carefully. Despite these limitations, our study presents new insights and ideas for uncovering the prominent role of SCs in paravertebral muscle development and aging.
Author contributions
Feng Zhu and Xin Qiu conceived and designed the project. Xin Qiu, Hao-Yu Wang, Zhen-Yu Yang, Li-Ming Sun, and Chui-Qin Fan wrote and revised the manuscript. Hao-Yu Wang and Chui-Qin Fan were responsible for data quality control, bioinformatics analysis, and data visualization. Xin Qiu, Zhen-Yu Yang, and Shu-Nan Liu participated in tissue dissection, sample processing, library construction, and experimental validation. Li-Ming Sun and Feng Zhu contributed to the project discussion and technical support. We also thank Dr. Xiaosen Jiang for his contribution to the design of MSKCA.
Conflict of interests
All the authors declare no conflict of interests with the content of this article. The authors declare no affiliation with or financial involvement in organizations or entities with a direct financial interest in the subject matter or materials discussed in the manuscript.
Funding
The research was supported by the Shenzhen Fundamental Research Program, China (No. JCYJ20180306173518936), Sanming Project of Medicine in Shenzhen, China (No. SZSM201612055), HKU-SZH Fund for Shenzhen Key Medical Discipline (China) (No. SZXK2020084), and China National GeneBank (CNGB).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2023.01.005.
Contributor Information
Chui-Qin Fan, Email: fcq065837@163.com.
Feng Zhu, Email: aaronzhu@hku.hk.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Brack A.S., Rando T.A. Tissue-specific stem cells: lessons from the skeletal muscle satellite cell. Cell Stem Cell. 2012;10(5):504–514. doi: 10.1016/j.stem.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsson L., Degens H., Li M., et al. Sarcopenia: aging-related loss of muscle mass and function. Physiol Rev. 2019;99:427–511. doi: 10.1152/physrev.00061.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dumont N.A., Wang Y.X., von Maltzahn J., et al. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat Med. 2015;21(12):1455–1463. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang Y., Yang S., Chen C., et al. Assessment of the association between paraspinal muscle degeneration and quality of life in patients with degenerative lumbar scoliosis. Exp Ther Med. 2020;20(1):505–511. doi: 10.3892/etm.2020.8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wosczyna M.N., Rando T.A. A muscle stem cell support group: coordinated cellular responses in muscle regeneration. Dev Cell. 2018;46(2):135–143. doi: 10.1016/j.devcel.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentzinger C.F., Wang Y.X., Dumont N.A., et al. Cellular dynamics in the muscle satellite cell niche. EMBO Rep. 2013;14(12):1062–1072. doi: 10.1038/embor.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takayama K., Kita T., Nakamura H., et al. New predictive index for lumbar paraspinal muscle degeneration associated with aging. Spine. 2016;41(2):E84–E90. doi: 10.1097/BRS.0000000000001154. [DOI] [PubMed] [Google Scholar]
- 8.Urrutia J., Besa P., Lobos D., et al. Lumbar paraspinal muscle fat infiltration is independently associated with sex, age, and inter-vertebral disc degeneration in symptomatic patients. Skeletal Radiol. 2018;47(7):955–961. doi: 10.1007/s00256-018-2880-1. [DOI] [PubMed] [Google Scholar]
- 9.Veras M.A., Lim Y.J., Kuljanin M., et al. Protocol for parallel proteomic and metabolomic analysis of mouse intervertebral disc tissues. JOR Spine. 2020;3(3):e1099. doi: 10.1002/jsp2.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C., Wu T., Fan F., et al. A portable and cost-effective microfluidic system for massively parallel single-cell transcriptome profiling. bioRxiv. 2019:818450. [Google Scholar]
- 11.Hao Y., Hao S., Andersen-Nissen E., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu G., Wang L.G., Han Y., et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liberzon A., Birger C., Thorvaldsdóttir H., et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou N., FerrDb Bao J. A manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database (Oxford) 2020;2020 doi: 10.1093/database/baaa021. baaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green D.R. The coming decade of cell death research: five riddles. Cell. 2019;177(5):1094–1107. doi: 10.1016/j.cell.2019.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schroder K., Tschopp J. The inflammasomes. Cell. 2010;140(6):821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 17.Ye Y., Dai Q., Qi H. A novel defined pyroptosis-related gene signature for predicting the prognosis of ovarian cancer. Cell Death Dis. 2021;7:71. doi: 10.1038/s41420-021-00451-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faget D.V., Ren Q., Stewart S.A. Unmasking senescence: context-dependent effects of SASP in cancer. Nat Rev Cancer. 2019;19(8):439–453. doi: 10.1038/s41568-019-0156-2. [DOI] [PubMed] [Google Scholar]
- 19.Aibar S., González-Blas C.B., Moerman T., et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreatta M., Carmona S.J. UCell: robust and scalable single-cell gene signature scoring. Comput Struct Biotechnol J. 2021;19:3796–3798. doi: 10.1016/j.csbj.2021.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foroutan M., Bhuva D.D., Lyu R., et al. Single sample scoring of molecular phenotypes. BMC Bioinf. 2018;19:404. doi: 10.1186/s12859-018-2435-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolde R., Laur S., Adler P., et al. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28(4):573–580. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H., Huang A.Y., Wang L.K., et al. Diagnostic utility of transcriptome sequencing for rare Mendelian diseases. Genet Med. 2020;22(3):490–499. doi: 10.1038/s41436-019-0672-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar L., E Futschik M. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2(1):5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu X., Hill A., Packer J., et al. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14(3):309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ackert-Bicknell C.L., Anderson L.C., Sheehan S., et al. Aging research using mouse models. Curr Protoc Mol Biol. 2015;5(2):95–133. doi: 10.1002/9780470942390.mo140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin S., Guerrero-Juarez C.F., Zhang L., et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen D., Tan C., Ding P., et al. VThunter: a database for single-cell screening of virus target cells in the animal kingdom. Nucleic Acids Res. 2022;50(D1):D934–D942. doi: 10.1093/nar/gkab894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouyang J.F., Kamaraj U.S., Cao E.Y., et al. ShinyCell: simple and sharable visualisation of single-cell gene expression data. Bioinformatics. 2021 doi: 10.1093/bioinformatics/btab209. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y., Zhang X., Peng X., et al. SPEED: single-cell pan-species atlas in the light of ecology and evolution for development and diseases. Nucleic Acids Res. 2023;51(D1):D1150–D1159. doi: 10.1093/nar/gkac930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Micheli A.J., Laurilliard E.J., Heinke C.L., et al. Single-cell analysis of the muscle stem cell hierarchy identifies heterotypic communication signals involved in skeletal muscle regeneration. Cell Rep. 2020;30(10):3583–3595. doi: 10.1016/j.celrep.2020.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganassi M., Badodi S., Wanders K., et al. Myogenin is an essential regulator of adult myofibre growth and muscle stem cell homeostasis. Elife. 2020;9 doi: 10.7554/eLife.60445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander M.S., Rozkalne A., Colletta A., et al. CD82 is a marker for prospective isolation of human muscle satellite cells and is linked to muscular dystrophies. Cell Stem Cell. 2016;19(6):800–807. doi: 10.1016/j.stem.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giordani L., He G.J., Negroni E., et al. High-dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Mol Cell. 2019;74(3):609–621. doi: 10.1016/j.molcel.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 36.Dos Santos M., Backer S., Auradé F., et al. A fast Myosin super enhancer dictates muscle fiber phenotype through competitive interactions with Myosin genes. Nat Commun. 2022;13:1039. doi: 10.1038/s41467-022-28666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han L., Wei X., Liu C., et al. Cell transcriptomic atlas of the non-human primate Macaca fascicularis. Nature. 2022;604(7907):723–731. doi: 10.1038/s41586-022-04587-3. [DOI] [PubMed] [Google Scholar]
- 38.Kazmierczak K., Liang J., Yuan C.C., et al. Slow-twitch skeletal muscle defects accompany cardiac dysfunction in transgenic mice with a mutation in the myosin regulatory light chain. Faseb J. 2019;33(3):3152–3166. doi: 10.1096/fj.201801402R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho I.C., Park H.B., Ahn J.S., et al. A functional regulatory variant of MYH3 influences muscle fiber-type composition and intramuscular fat content in pigs. PLoS Genet. 2019;15(10) doi: 10.1371/journal.pgen.1008279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubenstein A.B., Smith G.R., Raue U., et al. Single-cell transcriptional profiles in human skeletal muscle. Sci Rep. 2020;10:229. doi: 10.1038/s41598-019-57110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogier C., Colombo P.E., Bousquet C., et al. Targeting the NRG1/HER3 pathway in tumor cells and cancer-associated fibroblasts with an anti-neuregulin 1 antibody inhibits tumor growth in pre-clinical models of pancreatic cancer. Cancer Lett. 2018;432:227–236. doi: 10.1016/j.canlet.2018.06.023. [DOI] [PubMed] [Google Scholar]
- 42.Liu W., Zscheppang K., Murray S., et al. The ErbB4 receptor in fetal rat lung fibroblasts and epithelial type II cells. Biochim Biophys Acta. 2007;1772(7):737–747. doi: 10.1016/j.bbadis.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z.Z., Funcke J.B., Zi Z.Z., et al. Adipocyte iron levels impinge on a fat-gut crosstalk to regulate intestinal lipid absorption and mediate protection from obesity. Cell Metabol. 2021;33(8):1624–1639. doi: 10.1016/j.cmet.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivero S., Ruiz-García A., Díaz-Guerra M.J., et al. Characterization of a proximal Sp1 response element in the mouse Dlk2 gene promoter. BMC Mol Biol. 2011;12:52. doi: 10.1186/1471-2199-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li S.Y., Gu X., Heinrich A., et al. Loss of Mafb and Maf distorts myeloid cell ratios and disrupts fetal mouse testis vascularization and organogenesis. Biol Reprod. 2021;105(4):958–975. doi: 10.1093/biolre/ioab098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Müller A.M., Hermanns M.I., Skrzynski C., et al. Expression of the endothelial markers PECAM-1, vWf, and CD34 in vivo and in vitro. Exp Mol Pathol. 2002;72(3):221–229. doi: 10.1006/exmp.2002.2424. [DOI] [PubMed] [Google Scholar]
- 47.Kiss T., Nyúl-Tóth Á., Balasubramanian P., et al. Single-cell RNA sequencing identifies senescent cerebromicrovascular endothelial cells in the aged mouse brain. GeroScience. 2020;42(2):429–444. doi: 10.1007/s11357-020-00177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9(2):493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakkalakal J.V., Jones K.M., Basson M.A., et al. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490(7420):355–360. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cerletti M., Jang Y.C., Finley L.W.S., et al. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell. 2012;10(5):515–519. doi: 10.1016/j.stem.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shefer G., Rauner G., Yablonka-Reuveni Z., et al. Reduced satellite cell numbers and myogenic capacity in aging can be alleviated by endurance exercise. PLoS One. 2010;5(10) doi: 10.1371/journal.pone.0013307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blau H.M., Cosgrove B.D., Ho A.T.V. The central role of muscle stem cells in regenerative failure with aging. Nat Med. 2015;21(8):854–862. doi: 10.1038/nm.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kennedy B.K., Berger S.L., Brunet A., et al. Geroscience: linking aging to chronic disease. Cell. 2014;159(4):709–713. doi: 10.1016/j.cell.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oh J., Lee Y.D., Wagers A.J. Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nat Med. 2014;20(8):870–880. doi: 10.1038/nm.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pallafacchina G., François S., Regnault B., et al. An adult tissue-specific stem cell in its niche: a gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res. 2010;4(2):77–91. doi: 10.1016/j.scr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 56.Purohit G., Dhawan J. Adult muscle stem cells: exploring the links between systemic and cellular metabolism. Front Cell Dev Biol. 2019;7:312. doi: 10.3389/fcell.2019.00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pala F., di Girolamo D., Mella S., et al. Distinct metabolic states govern skeletal muscle stem cell fates during prenatal and postnatal myogenesis. J Cell Sci. 2018;131(14) doi: 10.1242/jcs.212977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J.C., Dong S.S., Shen H., et al. Multi-omics research in sarcopenia: current progress and future prospects. Ageing Res Rev. 2022;76 doi: 10.1016/j.arr.2022.101576. [DOI] [PubMed] [Google Scholar]
- 59.Svensson K., Dent J.R., Tahvilian S., et al. Defining the contribution of skeletal muscle pyruvate dehydrogenase α1 to exercise performance and insulin action. Am J Physiol Endocrinol Metab. 2018;315(5):E1034–E1045. doi: 10.1152/ajpendo.00241.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gupta N., Rutledge C. Pyruvate dehydrogenase complex deficiency: an unusual cause of recurrent lactic acidosis in a paediatric critical care unit. J Crit Care Med (Targu Mures) 2019;5(2):71–75. doi: 10.2478/jccm-2019-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qiu H., Zhao S., Xu X., et al. Assignment and expression patterns of porcine muscle-specific isoform of phosphoglycerate mutase gene. J Genet Genomics. 2008;35(5):257–260. doi: 10.1016/S1673-8527(08)60036-3. [DOI] [PubMed] [Google Scholar]
- 62.Damiano S., Sozio C., La Rosa G., et al. Metabolism regulation and redox state: insight into the role of superoxide dismutase 1. Int J Mol Sci. 2020;21(18):6606. doi: 10.3390/ijms21186606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peggion C., Scalcon V., Massimino M.L., et al. SOD1 in ALS: taking stock in pathogenic mechanisms and the role of glial and muscle cells. Antioxidants. 2022;11(4):614. doi: 10.3390/antiox11040614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nogueira C., Almeida L.S., Nesti C., et al. Syndromes associated with mitochondrial DNA depletion. Ital J Pediatr. 2014;40:34. doi: 10.1186/1824-7288-40-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu N., Nelson B.R., Bezprozvannaya S., et al. Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc Natl Acad Sci USA. 2014;111(11):4109–4114. doi: 10.1073/pnas.1401732111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Snyder C.M., Rice A.L., Estrella N.L., et al. MEF2A regulates the Gtl2-Dio3 microRNA mega-cluster to modulate WNT signaling in skeletal muscle regeneration. Development. 2013;140(1):31–42. doi: 10.1242/dev.081851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Braun T., Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol. 2011;12(6):349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- 68.Tapscott S.J., Davis R.L., Thayer M.J., et al. MyoD1:a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242(4877):405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- 69.Almada A.E., Wagers A.J. Molecular circuitry of stem cell fate in skeletal muscle regeneration, ageing and disease. Nat Rev Mol Cell Biol. 2016;17(5):267–279. doi: 10.1038/nrm.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ishii K., Sakurai H., Suzuki N., et al. Recapitulation of extracellular LAMININ environment maintains stemness of satellite cells in vitro. Stem Cell Rep. 2018;10(2):568–582. doi: 10.1016/j.stemcr.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rayagiri S.S., Ranaldi D., Raven A., et al. Basal lamina remodeling at the skeletal muscle stem cell niche mediates stem cell self-renewal. Nat Commun. 2018;9:1075. doi: 10.1038/s41467-018-03425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guilak F., Hayes A.J., Melrose J. Perlecan in pericellular mechanosensory cell-matrix communication, extracellular matrix stabilisation and mechanoregulation of load-bearing connective tissues. Int J Mol Sci. 2021;22(5):2716. doi: 10.3390/ijms22052716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leivo I., Engvall E. Merosin, a protein specific for basement membranes of Schwann cells, striated muscle, and trophoblast, is expressed late in nerve and muscle development. Proc Natl Acad Sci USA. 1988;85(5):1544–1548. doi: 10.1073/pnas.85.5.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cohn R.D., Henry M.D., Michele D.E., et al. Disruption of Dag1 in differentiated skeletal muscle reveals a role for dystroglycan in muscle regeneration. Cell. 2002;110(5):639–648. doi: 10.1016/s0092-8674(02)00907-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The expression matrices of the paraspinal muscle single-cell nuclear transcriptome sequencing data (seven timepoints) in this study are all stored on the MSKCA website. The data reported in this study are also available in the CNGB Nucleotide Sequence Archive (CNSA: https://db.cngb.org/cnsa; accession number CNP0003561). The data of the multi-timepoints research are available from the Zenodo repository (10.5281/zenodo.7294918).