Highlights
-
•
Non-encapsulated pneumococcus increased in the post-vaccine era in Japan.
-
•
Antimicrobial susceptibility was not worsened in the post-vaccine era.
-
•
A non-vaccine type 35B showed less susceptibility to β-lactams and fluoroquinolones.
Keywords: Streptococcus pneumoniae, Capsular serotype, Serotype replacement, Null capsule clade, Antimicrobial susceptibility, Resistant genes
Abstract
Objectives
It is feared that the serotype replacement of Streptococcus pneumoniae occurred by the introduction of pneumococcal vaccines as periodical inoculation leads to reduced efficacy of the approved vaccines and altered antimicrobial susceptibility.
Methods
We determined serotypes of 351 S. pneumoniae isolates collected at a commercial clinical laboratory in Hokkaido prefecture, Japan, from December 2018 to February 2019 by using the polymerase chain reaction procedure of the US Centers for Disease Control and Prevention. Antimicrobial susceptibility and resistance gene profiles were also examined.
Results
Vaccine coverage rates were 7.9% for 13-valent conjugate vaccine, and 32.5% for 23-valent polysaccharide vaccine, respectively. Non-typable strains were 19.7%. cpsA-positive isolates (group I), and null capsule clade (NCC)1, NCC2 and NCC3 (group II) comprised 31.3%, 28.4%, 32.8%, and 7.5% of the 69 non-typable strains, respectively. No penicillin-resistant/intermediate isolates were found; however, serotypes 35B and 15A/F showed low susceptibility to β-lactams. Only five strains (1.4%) were levofloxacin-resistant, and all were from the older persons, and three strains were serotype 35B.
Conclusion
The progression of serotype replacement in non-invasive pneumococcal infections has occurred during the post-vaccine era in Japan, and non-encapsulated isolates, such as NCC, have increased. Antimicrobial susceptibility is not worsened.
Introduction
Streptococcus pneumoniae is a major causative agent of community pneumonia in the older persons and of upper respiratory infectious diseases, such as acute otitis media and sinusitis, in children. Invasive infections, such as meningitis and sepsis, often occurs. Pneumococcus colonizes in the nasopharyngeal cavity of healthy children, and the colonized bacteria are the source of infections [1].
Pneumococcus is classified into more than 100 serotypes based on the diverse structures of the capsular polysaccharides [2]. The capsular polysaccharides are a major pathogenic factor that mediate resistance to host complement, whereas they also act as a protective antigen by eliciting production of antibodies that trigger opsonophagocytosis by host immune cells. Two types of pneumococcal vaccines, the 13-valent conjugate vaccine (PCV13) and the 23-valent polysaccharide vaccine (PPSV23) are approved for routine vaccination of infants and the older persons, respectively, in Japan. The introduction of PCV13 and its prototype 7-valent conjugate vaccine (PCV7) for children led to a reduction in invasive pneumococcal infections not only in children but also in adults and the older persons via herd immunity [3]. However, pneumococcal infections occurred by strains of non-vaccine serotypes have been increasing, and the phenomenon is known as the serotype replacement [4], [5], [6].
In addition, non-typable (NT), namely, non-capsulated strains, have been identified [7,8]. Group I NT isolates carry csp locus, and the locus is similar to that of conventional encapsulated isolates; however, the locus is unfunctional to generate capsular polysaccharides [7]. Group II NT isolates, which lack a cps locus, are classified into three null capsule clades (NCC): NCC1, NCC2, and NCC3. NCC1 carries the pspK gene instead of the cps locus. pspK encodes pneumococcal surface protein K, which allows bacteria to adhere to host epithelial cells before nasopharyngeal colonization [9]. NCC2 shares both aliC and aliD, and NCC3 shares aliD only [7,8]. aliC and aliD encode oligopeptide transporters, which regulate expression of several genes, including the gene encoding choline-binding protein AC (CbpAC). Cell surface expression of CbpAC mediated by AliC and AliD protects cells against deposition of C-reactive protein (CRP), C1q, and C3b, thereby escaping opsonophagocytosis [10]. These activities are considered to compensate for capsular functions.
In the present study, we examined the serotype distribution and antimicrobial susceptibility of S. pneumoniae isolates obtained from patients with non-invasive infections in 2018-2019 in Hokkaido prefecture, Japan. A gold standard procedure for serotype determination of S. pneumoniae is the capsular quellung test; however, this method is laborious and expensive. In this study, we used a multiplex polymerase chain reaction (PCR) procedure distributed by the Centers for Disease Control and Prevention (CDC; Atlanta, GA) of the US.
Methods
Bacteria
In total, 351 clinical isolates of S. pneumoniae isolated between December 2018 to February 2019 and stocked in Hokkaido Laboratory, SRL, Inc. (Sapporo, Japan) were analyzed. The clinical samples were from the patients who visited the community clinics and hospitals of otolaryngology, pediatrics, and internal medicine. Analysis of multiple isolates derived from the same patients was avoided. The laboratory serves almost all areas of Hokkaido prefecture, but the strains used were collected mainly from the Sapporo district. Identification of S. pneumoniae was carried out using MicroScan WalkAway (Beckman Coulter, Tokyo, Japan).
Determination of serogroup/serotype and resistance genes
Serotypes/serogroups were determined using a multiplex PCR procedure distributed by the CDC of the US. The protocol for the multiplex PCR—S. pneumoniae SEROTYPING—clinical specimens and pneumococcal isolates (USA set) was obtained from the CDC website [11]. The protocol comprises a conventional multiplex PCR assay using 41 primer pairs for detection of 70 pneumococcal serotypes [12].
The PCR of respective genes [and their primer names] were performed as described previously; lytA [Aly3 and Aly4] [13], ermB [ERM721S and ERM922R], mefE [MEF180S and MEF562R] [14], cpsA [5202 and 3202], aliC [5433 and 3433], aliD [5434 and 3434], and pspK [5743 and 3743] [8]. lytA is a marker of S. pneumoniae [13]. The entire coding region of pspK was amplified by PCR using primer pair: pspK-fullF: ATGAATAATAAGAATATCATCCCGAT, pspK-fullR: CTAATTTTTATGTTTAACAAATGGAAGA. Mutations in the quinolone resistance determining region (QRDR) of topoisomerase IV and DNA gyrase were examined by PCR, followed by direct sequencing, as previously described [15]. DNA sequencing was carried out at Hokkaido System Science Co., Ltd. (Sapporo, Japan).
PCR was performed using the KAPATaq Extra HotStart Ready Mix with dye (Nippon Genetics, Tokyo, Japan). The PCR products were analyzed by agarose electrophoresis, followed by ethidium bromide staining. pHY markers (Takara Bio, Kusatsu, Japan) and 100 bp DNA Ladder PLUS (Nippon Genetics) were used as size markers.
Antibiotic susceptibility testing
The minimum inhibitory concentrations (MICs) of antibiotics were determined by a broth microdilution method using Dryplate 96 (Eiken, Tokyo, Japan) and the MicroScan WalkAway. Susceptibility was determined according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [16]. Breakpoints were according to CLSI [16] and The European Committee on Antimicrobial Susceptibility Testing (EUCAST) [17].
Inhibition of efflux pumps
To evaluate the effect of efflux pumps on the fluoroquinolone resistance, MICs were determined in the presence (10 μg/ml) or absence of reserpine (Daiichi Sankyo, Tokyo, Japan) [18].
Results
Details of isolates
In total, 351 isolates were examined. These were originated from nasal discharge (n = 314), sputum (n = 18), pharyngeal mucus (n = 17), nasal cavity (n = 1), and otorrhea (n = 1). Regarding gender, 182 patients were male, 151 were female, and 18 were unknown. Age ranged 0-4 (n = 218), 5-9 (n = 76), 10-29 (n = 9), 30-49 (n = 12), 50-69 (n = 5), >70 (n = 17), and unknown (n = 14).
Serotype and genotype of NT isolates
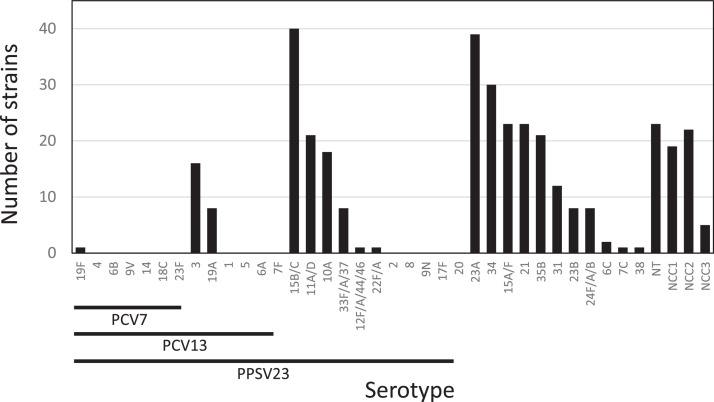
Among the 351 isolates examined, 282 isolates were typed serotypes by PCR using the CDC protocol (Figure 1). Vaccine coverage rates were 0.28% for PCV7, 7.9% for PCV13, and 32.5% for PPSV23, respectively. Only one isolate was serotyped as 19F, which is included in PCV7. With respect to serotypes included in PCV13 other than PCV7, 16 and 8 isolates were serotyped as 3 and 19A, respectively. Strains of serotypes included in PPSV23 (other than those including PCV13) were 89 strains (25.4%), and major serotypes were 15B/C, 11A/D, 10A, and 33F/A/37. Strains of serotypes not included in the approved vaccines were 169 strains (47.9%), and major serotypes were 23A, 34, 15A/F, 21, 35B, and 31.
Figure 1.
Serotype distribution of Streptococcus pneumoniae isolated from patients with non-invasive infections in 2018-2019.
Underlining denotes the serotypes included in each vaccine. PPSV23 does not include serotype 6A.
NCC, Null capsule clade; PCV7, 7-valent conjugate vaccine; PCV13, 13-valent conjugate vaccine; PPSV23, 23-valent polysaccharide vaccine.
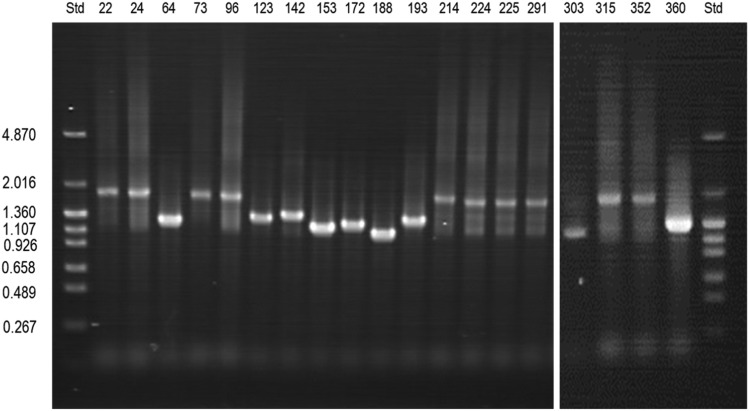
Among the remaining 69 isolates, 23 were positive for cpsA, but any serotypes were not detected by the multiplex PCR (Figure 1). Among the other 46 cps-negative isolates, namely NCC,19 isolates were pspK-positive (NCC1), 22 were positive for both aliC and aliD (NCC2), and five were positive for only aliD (NCC3) (Table 1). The sizes of pspK gene varied among isolates. Agarose electrophoresis revealed that the full-length pspK PCR products were 1-2k base pairs in size (Figure 2).
Table 1.
Genotypes of cpsA-negative non-typable (group II non-typable) strains.
| Genes present |
Null capsule clade (NCC) | Number of strains | |||
|---|---|---|---|---|---|
| pspK | aliC | aliD | lytA | ||
| + | - | - | + | NCC1 | 19 |
| - | + | + | + | NCC2 | 22 |
| - | - | + | + | NCC3 | 3 |
| - | - | + | - | NCC3 | 2 |
Figure 2.
Full sizes of pspK coding region determined by polymerase chain reaction with agarose gel electrophoresis
Full length of pspK polymerase chain reaction products were analyzed by 1%(w/v) agarose gel electrophoresis. The pHY marker was used as a size marker; the numbers on the left refer to x 103 base pairs. The numbers above the gel denote strain names.
Std, size marker.
All strains (14/14) of serotype 3, and 75.0% strains (6/8) of serotype 33F/A/37 appeared colonies with mucoid phenotype on the agar plates.
Antimicrobial susceptibility
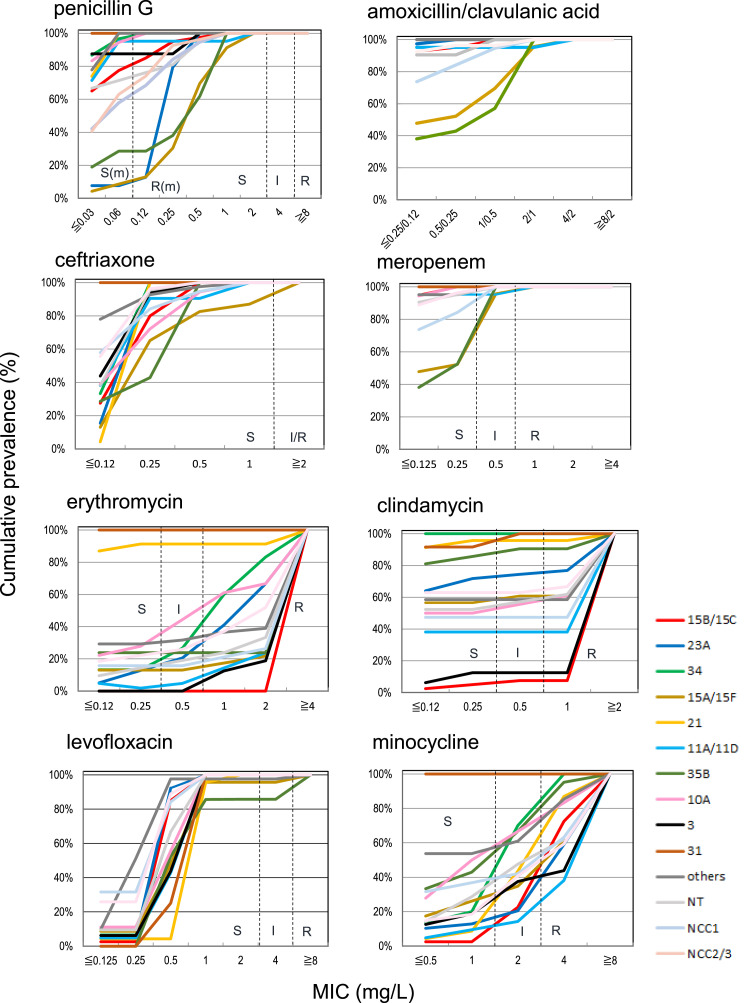
The antimicrobial susceptibility of all isolates and specific serotypes is shown in Table 2 and Figure 3, respectively. No penicillin-G-resistant/intermediate according to CLSI breakpoint strain was found. However, 32.2% of strains showed resistance to penicillin G according to the CLSI and EUCAST breakpoint for meningitis. Only a few strains of resistant/intermediate to clavulanic acid/amoxicillin and cephem antibiotics tested other than cefotiam were observed. Strains of two (0.6%) resistant according to CLSI and EUCAST breakpoint and 27 (7.7%) intermediate according to CLSI breakpoint to meropenem were identified. With respect to β-lactam antibiotics, isolates of serotypes 35B and 15A/F contained frequently less susceptible isolates to penicillin G, meropenem, and amoxicillin/clavulanic acid. In addition, isolates of 23A also contained less susceptible strains to penicillin G (Figure 3).
Table 2.
Antimicrobial susceptibility of Streptococcus pneumoniae clinical isolates.
| Drug | Number of isolates (percentages of total isolates) |
||
|---|---|---|---|
| Resistant | Intermediate | Susceptible | |
| Penicillin G (non-meningitis) | 0 (0%) | 0 (0%) | 351 (100%) |
| Penicillin G (meningitis) | 113 (32.2%) | - | 238 (67.8%) |
| Cefotiam | 33 (9.4%) | 19 (5.4%) | 299 (85.2%) |
| Cefotaxime | 1 (0.3%) | 1 (0.3%) | 349 (99.4%) |
| Cefepime | 0 (0%) | 1 (0.3%) | 350 (99.7%) |
| Ceftriaxone | 0 (0%) | 3 (0.9%) | 348 (99.1%) |
| Cefozopran | 0 (0%) | 4 (1.1%) | 347 (98.9%) |
| Clavulanic acid/amoxicillin | 0 (0%) | 2 (0.6%) | 349 (99.4%) |
| Meropenem | 2 (0.6%) | 27 (7.7%) | 322 (91.7%) |
| Erythromycin | 258 (73.5%) | 13 (3.7%) | 80 (22.8%) |
| Azithromycin | 243 (69.2%) | 23 (6.9%) | 85 (24.2%) |
| Clindamycin | 140 (39.9%) | 7 (2.0%) | 204 (58.1%) |
| Minocycline | 97 (27.6%) | 98 (27.9%) | 156 (44.4%) |
| Chloramphenicol | 11 (3.1%) | - | 340 (96.9%) |
| Levofloxacin | 5 (1.4%) | 0 (0%) | 346 98.6%) |
| Sulfamethoxazole/trimethoprim | 0 (0%) | 38 (10.8%) | 313 (89.2%)) |
| Vancomycin | - | - | 531 (100%) |
Breakpoints are determined according to the Clinical and Laboratory Standards Institute guideline [16]. -, Not defined.
Figure 3.
Cumulative antibiotic MIC curves for each serotype
The serotypes and genotypes of NT strains detected in more than ten isolates are shown.
I, intermediate; MIC, minimum inhibitory concentration; NT, non-typable; R, resistant; R(m), resistant for meningitis; S, susceptible; S(m): susceptible for meningitis. These breakpoints are defined according to the Clinical and Laboratory Standards Institute guideline [16].
More than 70% and approximately 40% of all isolates were resistant to erythromycin and clindamycin, respectively. Approximately 80% of strains shared either one or both ermA or mefE genes, and almost all strains carrying either gene were resistant to erythromycin (Supplementary material S1). All clindamycin-resistant isolates shared ermA, but only 37% of ermA-positive isolates were susceptible to clindamycin.
Approximately 27% and 28% of strains were resistant and intermediate according to CLSI breakpoint to minocycline, respectively. According to EUCAST breakpoint, 78.3% were resistant to minocycline.
Only five isolates (1.4%) were resistant to levofloxacin; all were obtained from older patients (i.e., aged >62 years). Three isolates identified as serotype 35B, and commonly shared S79F in ParC and S81F in GyrA as resistant mutations (Supplementary material S2). The occurrence rate of levofloxacin-resistance in serotype 35B was 14.3% (3/21 strains). One levofloxacin-resistant serotype 15A isolate shared S81F in GyrA as a resistant mutation, and S114G in GyrA, and I460V and G475K in ParE, which amino acid residues are observed in oral streptococci [19]. One levofloxacin-resistant serotype 19A isolate did not detect known resistant mutation(s) in QRDRs, whereas the levofloxacin MIC was reduced (8 to 2 mg/ml) in the presence of the efflux pump inhibitor, reserpine.
No isolates resistant to vancomycin and sulfamethoxazole/trimethoprim (ST) were observed; however, 3.4% of isolates showed intermediate according to CLSI breakpoint to ST. Of these ST-intermediate isolates, 82% (31/38) were serotypes 35B, 31 and 24F/A/B, and NCC1.
Discussion
The present study examined the serotype distribution and antibiotic susceptibility of S. pneumoniae strains isolated from patients with non-invasive infections between 2018 to 2019 in Japan. The introduction of pneumococcal vaccines caused serotype replacement similar to previous observations [4], [5], [6]. With respect to the vaccine serotypes in PCV13, serotype 3, which is mucoid phenotype and associated with invasive infections, especially in adults [20], was the most dominant serotype of PCV13, and this phenomenon was also observed in other studies [5,21,22]. Among the non-vaccine serotypes, 15A, 23A, 6C, and 35B are the top four detected in the post-vaccine era in Japan [5,21,22]; of these, 15A, 23A, and 35B show low susceptibility to penicillin G [21,23]. Consistent with the previous studies, we also found that serotypes 15A/F and 35B were less susceptible to β-lactams, including carbapenem. In addition, serotype 23A showed reduced susceptibility to penicillin G. Although fluoroquinolone-resistant strains were rare (found in only five isolates), three and one of these were serotype 35B and 15A/F, respectively. In addition, 14.3% of 35B isolates were resistant to levofloxacin. All levofloxacin-resistant isolates were isolated from older persons and not from children, and the trends did not alter before the introduction of periodical vaccination [19], even after tosufloxacin was approved for children in 2018 in Japan.
Serotype 35B has emerged and expanded in various regions of the world, including Japan [5,6,[22], [23], [24]]. The increase is indicated to be due to clonal expansion of clonal complex (CC) 558, which is less susceptible to β-lactam antibiotics, and carries transposons containing macrolide- and tetracycline-resistant genes [25] and rrgC gene encoding an adhesin Pilus-1 [23].
Resistant rates of erythromycin and clindamycin were very high, whereas those were low only in serotypes 21 and 31. Although resistant rates of clindamycin were low in serotypes 34 and 35B, those of erythromycin were high. High prevalence of macrolide resistance is observed pre- and post-vaccine era in Eastern Asian countries, such as China, Japan, and Korea, and this is indicated to be closely associated with antibiotic use [26].
A significant proportion of stains (69 isolates; 19.7% of total ones) were not able to identify serotypes. Approximately one-third (33.3%) of these shared the cpsA gene. This suggests that these are either capsule-bearing bacteria whose serotypes were not detectable using the PCR system used in this study, or non-encapsulated bacteria with an unfunctional cps gene (i.e., group I NT). Another one-third (27.5%) of these were NCC1, which carries the pspK gene. The sizes of pspK coding region varied (Figure 2), and this should be due to differences in the number of repeating motifs [(E)EEAK(R/Q)KA] [27]. This suggests the spread of plural NCC1 clones during the post-vaccine era. The remaining one-third (31.9%) were NCC2 (aliC+/aliD+), with the remainder comprising several isolates (7.2%) of NCC3 (aliC-/aliD+). Two strains of NCC3 were negative for lytA, a marker of S. pneumoniae. Phylogeny suggests that NCC3 is linked closely with Streptococcus pseudopneumoniae and Streptococcus mitis [8]. The antimicrobial susceptibility of NCC 1, 2, and 3 was similar to that of other serotypes, but the NCC clades tended to be less susceptible to penicillin G, similar to the findings of a previous report [28]; however, the prevalence of low susceptibility was less noticeable when compared with serotypes 35B, 15A/F, and 23A.
This study has several limitations. First, we examined a relatively limited number of isolates, and most were from children. Because the clinical specimens were from the symptomatic patients visiting clinics of otolaryngology, pediatrics, and internal medicine, the isolates should represent infections rather than colonization. Also, the samples were from a single commercial clinical laboratory; however, the laboratory covers the entire area of Hokkaido prefecture. Serotypes were determined using a conventional PCR established by CDC in the US [11,12]. The method provides limited data regarding subtypes. We did not perform multilocus sequence typing. Although these issues need to be addressed in the future, the data in the present study point to certain trends of pneumococcal serotype distribution and antimicrobial susceptibility in this region.
Serotype replacement, including the increase in non-capsular type isolates, might progress in the future. New pneumococcal vaccines that include other serotype capsules have been approved or are under development [29]. Recent observations, including those reported herein, suggest that the number of non-capsule type isolates has increased significantly in the post-vaccine era. New types of vaccines containing protein antigens common to pneumococci, such as pneumolysin (Ply), pneumococcal surface protein A (PspA), choline-binding protein A (CbpA), pneumococcal choline-binding protein A (PcpA), and polyhistidine triad proteins D (PhtD) are undergoing research and development [30]. These approaches may result in promising prophylaxis against pneumococcal infections caused by NT and non-encapsulated strains.
Declarations of competing interest
The authors have no competing interests to declare.
Acknowledgments
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethics approval
This study was approved by the review board of SRL, Inc.
Acknowledgments
The authors thank Ms A. Shiga for clerical and technical support.
Author contributions
NT and SYokota performed the experiments. SYokota wrote the manuscript. YO, TS, SYamamoto, and NO made technical suggestions and undertook a critical review of the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijregi.2023.07.002.
Appendix. Supplementary materials
References
- 1.Weiser JN, Ferreira DM, Paton JC. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol. 2018;16:355–367. doi: 10.1038/s41579-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganaie F, Saad JS, McGee L, van Tonder AJ, Bentley SD, Lo SW, et al. A new pneumococcal capsule type, 10D, is the 100th serotype and has a large cps fragment from an oral Streptococcus. mBio. 2020;11 doi: 10.1128/mBio.00937-20. e00937-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsaban G, Ben-Shimol S. Indirect (herd) protection, following pneumococcal conjugated vaccines introduction: a systematic review of the literature. Vaccine. 2017;35:2882–2891. doi: 10.1016/j.vaccine.2017.04.032. [DOI] [PubMed] [Google Scholar]
- 4.Ladhani SN, Collins S, Djennad A, Sheppard CL, Borrow R, Fry NK, et al. Rapid increase in non-vaccine serotypes causing invasive pneumococcal disease in England and Wales, 2000–17: a prospective national observational cohort study. Lancet Infect Dis. 2018;18:441–451. doi: 10.1016/S1473-3099(18)30052-5. [DOI] [PubMed] [Google Scholar]
- 5.Ubukata K, Takata M, Morozumi M, Chiba N, Wajima T, Hanada S, et al. Effects of Pneumococcal Conjugate Vaccine on genotypic penicillin resistance and serotype changes, Japan, 2010–2017. Emerg Infect Dis. 2018;24:2010–2020. doi: 10.3201/eid2411.180326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo SW, Gladstone RA, van Tonder AJ, Lees JA, du Plessis M, Benisty R, et al. Pneumococcal lineages associated with serotype replacement and antibiotic resistance in childhood invasive pneumococcal disease in the post-PCV13 era: an international whole-genome sequencing study. Lancet Infect Dis. 2019;19:759–769. doi: 10.1016/S1473-3099(19)30297-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hathaway LJ, Stutzmann Meier P, Bättig P, Aebi S, Mühlemann K. A homologue of aliB is found in the capsule region of nonencapsulated Streptococcus pneumoniae. J Bacteriol. 2004;186:3721–3729. doi: 10.1128/JB.186.12.3721-3729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park IH, Kim KH, Andrade AL, Briles DE, McDaniel LS, Nahm MH. Nontypeable pneumococci can be divided into multiple cps types, including one type expressing the novel gene pspK. mBio. 2012;3 doi: 10.1128/mBio.00035-12. e00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keller LE, Jones CV, Thornton JA, Sanders ME, Swiatlo E, Nahm MH, et al. PspK of Streptococcus pneumoniae increases adherence to epithelial cells and enhances nasopharyngeal colonization. Infect Immun. 2013;81:173–181. doi: 10.1128/IAI.00755-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson CD, Bradshaw JL, Miller WS, Vidal AGJ, Vidal JE, Rosch JW, et al. Oligopeptide transporters of nonencapsulated Streptococcus pneumoniae regulate CbpAC and PspA expression and reduce complement-mediated clearance. mBio. 2023;14 doi: 10.1128/mbio.03325-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention . 2021. Multiplex conventional PCR schemes for pneumococcal serotype deduction. U.S. scheme 2021.https://www.cdc.gov/streplab/downloads/pcr-oligonucleotide-primers.pdf [accessed 16 June 2023] [Google Scholar]
- 12.Centers for Disease Control and Prevention. Conventional PCR serotype deduction protocols. Oligonucleotide primers used for deducing 41 pneumococcal capsular serotypes, https://www.cdc.gov/streplab/downloads/pcr-us-clinical-specimens.pdf; 2021 [accessed 16 June 2023].
- 13.Ubukata K, Muraki T, Igarashi A, Asahi Y, Konno M. Identification of penicillin and other beta-lactam resistance in Streptococcus pneumoniae by polymerase chain reaction. J Infect Chemother. 1997;3:190–197. doi: 10.1007/BF02490033. [DOI] [PubMed] [Google Scholar]
- 14.Ubukata K, Iwata S, Sunakawa K. In vitro activities of new ketolide, telithromycin, and eight other macrolide antibiotics against Streptococcus pneumoniae having mefA and ermB genes that mediate macrolide resistance. J Infect Chemother. 2003;9:221–226. doi: 10.1007/s10156-003-0258-2. [DOI] [PubMed] [Google Scholar]
- 15.Pan XS, Ambler J, Mehtar S, Fisher LM. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:2321–2326. doi: 10.1128/AAC.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute . 32nd ed. Clinical and Laboratory Standards Institute; Malvern: 2022. M100 Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 17.The European committee on antimicrobial susceptibility testing . Streptococcus pneumoniae. 2023. Clinical breakpoints - breakpoints and guidance.https://view.officeapps.live.com/op/view.aspx?src=https%3A%2F%2Fwww.eucast.org%2Ffileadmin%2Fsrc%2Fmedia%2FPDFs%2FEUCAST_files%2FBreakpoint_tables%2Fv_13.1_Breakpoint_Tables.xlsx&wdOrigin=BROWSELINK [accessed 04 July 2023] [Google Scholar]
- 18.Markham PN. Inhibition of the emergence of ciprofloxacin resistance in Streptococcus pneumoniae by the multidrug efflux inhibitor reserpine. Antimicrob Agents Chemother. 1999;43:988–989. doi: 10.1128/AAC.43.4.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokota S, Sato K, Kuwahara O, Habadera S, Tsukamoto N, Ohuchi H, et al. Fluoroquinolone-resistant Streptococcus pneumoniae strains occur frequently in elderly patients in Japan. Antimicrob Agents Chemother. 2002;46:3311–3315. doi: 10.1128/AAC.46.10.3311-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugimoto N, Yamagishi Y, Hirai J, Sakanashi D, Suematsu H, Nishiyama N, et al. Invasive pneumococcal disease caused by mucoid serotype 3 Streptococcus pneumoniae: a case report and literature review. BMC Res Notes. 2017;10:21. doi: 10.1186/s13104-016-2353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyazaki H, Shibuya R, Midorikawa N, Chang B, Ohnishi M, Matsumoto T. Serotype distribution and antimicrobial susceptibility of Streptococcus pneumoniae strains isolated in Japan after introduction of the routine immunization program. J Infect Chemother. 2017;23:234–240. doi: 10.1016/j.jiac.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Kawaguchiya M, Urushibara N, Aung MS, Ito M, Takahashi A, Habadera S, et al. High prevalence of antimicrobial resistance in non-vaccine serotypes of non-invasive/colonization isolates of Streptococcus pneumoniae: a cross-sectional study eight years after the licensure of conjugate vaccine in Japan. J Infect Public Health. 2020;13:1094–1100. doi: 10.1016/j.jiph.2020.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Miyazaki H, Shibuya R, Chang B, Inukai T, Miyazaki Y, Ubukata K, et al. Genetic characteristics of piliated Streptococcus pneumoniae serotype 35B, increased after introduction of pneumococcal vaccines in Japan. J Infect Chemother. 2020;26:1198–1204. doi: 10.1016/j.jiac.2020.06.016. [DOI] [PubMed] [Google Scholar]
- 24.Cohen R, Varon E, Doit C, Schlemmer C, Romain O, Thollot F, et al. A 13-year survey of pneumococcal nasopharyngeal carriage in children with acute otitis media following PCV7 and PCV13 implementation. Vaccine. 2015;33:5118–5126. doi: 10.1016/j.vaccine.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Shinohara K, Fujisawa T, Chang B, Ito Y, Suga S, Matsumura Y, et al. Frequent transmission of Streptococcus pneumoniae serotype 35B and 35D, clonal complex 558 lineage, across continents and the formation of multiple clades in Japan. Antimicrob Agents Chemother. 2023;67 doi: 10.1128/aac.01083-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berbel D, González-Díaz A, López de Egea G, Càmara J, Ardanuy C. An overview of macrolide resistance in Streptococci: prevalence, mobile elements and dynamics. Microorganisms. 2022;10:2316. doi: 10.3390/microorganisms10122316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Na IY, Baek JY, Park IH, Kim DH, Song JH, Ko KS. pspK gene prevalence and characterization of non-typable Streptococcus pneumonia isolates from Asian countries. Microbiology (Reading) 2015;161:973–979. doi: 10.1099/mic.0.000073. [DOI] [PubMed] [Google Scholar]
- 28.Kawaguchiya M, Urushibara N, Aung MS, Kudo K, Ito M, Sumi A, et al. Clonal lineages and antimicrobial resistance of nonencapsulated Streptococcus pneumoniae in the post-pneumococcal conjugate vaccine era in Japan. Int J Infect Dis. 2021;105:695–701. doi: 10.1016/j.ijid.2021.02.109. [DOI] [PubMed] [Google Scholar]
- 29.Micoli F, Romano MR, Carboni F, Adamo R, Berti F. Strengths and weaknesses of Pneumococcal Conjugate Vaccines. Glycoconj J. 2023;40:135–148. doi: 10.1007/s10719-023-10100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aceil J, Avci FY. Pneumococcal surface proteins as virulence factors, immunogens, and conserved vaccine targets. Front Cell Infect Microbiol. 2022;12 doi: 10.3389/fcimb.2022.832254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.