Abstract
The Chinese hamster cell lines E36 and CHOK1 dramatically differ in susceptibility to amphotropic murine leukemia virus (A-MuLV) and gibbon ape leukemia virus (GALV); E36 cells are highly susceptible to both viruses, CHOK1 cells are not. We have previously shown that GALV can infect E36 cells by using both its own receptor, HaPit1, and the A-MuLV receptor, HaPit2. Given that the two cell lines are from the same species, the loss of function of both of these receptors in CHOK1 cells is surprising. Other studies have shown that CHOK1 cells secrete proteins that block A-MuLV entry into CHOK1 as well as E36, suggesting the two A-MuLV receptors are functionally identical. However, CHOK1 conditioned medium does not block GALV entry into E36, indicating the secreted inhibitors do not block HaPit1. HaPit1 and ChoPit1 therefore differ as receptors for GALV; ChoPit1 is either inactivated by secreted factors or intrinsically nonfunctional. To determine why GALV cannot infect CHOK1, we cloned and sequenced ChoPit1 and ChoPit2. ChoPit2 is almost identical to HaPit2, which explains why CHOK1 conditioned medium blocks A-MuLV entry via both receptors. Although ChoPit1 and HaPit1 are 91% identical, a notable difference is at position 550 in the fourth extracellular region, shown by several studies to be crucial for GALV infection. Pit1 and HaPit1 have aspartate at 550, whereas ChoPit1 has threonine at this position. We assessed the significance of this difference for GALV infection by replacing the aspartate 550 in Pit1 with threonine. This single substitution rendered Pit1 nonfunctional for GALV and suggests that threonine at 550 inactivates ChoPit1 as a GALV receptor. Whether native ChoPit1 functions for GALV was determined by interference assays using Lec8, a glycosylation-deficient derivative of CHOK1 that is susceptible to both viruses and that has the same receptors as CHOK1. Unlike with E36, GALV and A-MuLV exhibited reciprocal interference when infecting Lec8, suggesting that they use the same receptor. We conclude both viruses can use ChoPit2 in the absence of the inhibitors secreted by CHOK1 and ChoPit1 is nonfunctional.
Gibbon ape leukemia virus (GALV) and amphotropic murine leukemia virus (A-MuLV) use related cell surface proteins as receptors to infect mammalian cells. Both proteins are sodium-dependent phosphate symporters predicted to have 10 transmembrane helices, five extracellular regions, and a large intracellular portion (8, 14, 17–19, 27, 28, 31). The receptors from human, mouse, rat, and hamster cells have been cloned and characterized, and the human receptors for GALV and A-MuLV have been designated Pit1 and Pit2, respectively (7, 14, 18, 25, 27–29). The human and rat receptors for the two viruses are functionally distinct; each virus uses only its own receptor to infect these cells. A hallmark property of A-MuLV is that it strictly retains Pit2 receptor specificity for infecting cells from various species; no naturally occurring GALV receptor that is also permissive for A-MuLV is known. However, receptor usage by GALV is not restricted to Pit1; the virus can infect the murine cell line MMMol and the Chinese hamster lung fibroblast line E36 via both Pit1 and Pit2 (28, 29). All feline leukemia virus subgroup B isolates use Pit1 as the receptor (26), and some can also use Pit2 (1). 10A1 murine leukemia virus can infect cells via both Pit1 and Pit2 (17, 28).
Previously we showed that GALV and A-MuLV exhibit nonreciprocal interference when infecting E36 cells; GALV-infected cells become resistant to A-MuLV, but A-MuLV-infected cells remain susceptible to GALV. This finding suggested that GALV infects these cells via its own receptor, as well as the A-MuLV receptor. We cloned and characterized the receptors from E36 and found that GALV indeed uses both its own receptor (HaPit1) and the A-MuLV receptor (HaPit2) (5). Thus, dual-receptor usage by GALV sets E36 cells phenotypically apart from human, rat, and most other cells.
Unlike E36, the Chinese hamster ovary cell line CHOK1 is resistant to both GALV and A-MuLV. The block to infection is glycosylation dependent and is at the entry level (15, 16). Moreover, the block to GALV and A-MuLV results from the presence of proteinaceous factors that CHOK1 secrete (15, 16). These factors presumably block viral interaction with CHOK1 receptors, rendering the cells resistant. In contrast, Lec2 and Lec8 cells, both glycosylation-deficient derivatives of CHOK1 (23, 24), are susceptible to both viruses (15, 16). Lec2 and Lec8 have the same A-MuLV and GALV receptors as CHOK1, and conditioned medium from CHOK1 inhibits virus entry into both cell lines. The inhibitory effect of the factors that CHOK1 secrete is specific for GALV and A-MuLV entry into CHOK1 and some other hamster cells; the conditioned medium has no effect on virus entry into human, mouse, and rat cells or on virus entry into CHOK1 expressing the receptors from other species (5, 16, 30).
It is not known whether CHOK1 receptors for GALV and A-MuLV, like their isotypes from E36, are both functional for GALV. Because both E36 and CHOK1 were derived from the Chinese hamster, it appeared likely that the two sets of receptors are functionally identical and that GALV can use both CHOK1 receptors in the absence of secreted inhibitors. However, conditioned medium from CHOK1 has little or no effect on GALV entry into E36, which shows that the secreted inhibitors do not inactivate HaPit1. GALV receptor isotypes from CHOK1 and E36 are therefore functionally different. We report here cloning and sequencing of the A-MuLV and GALV receptors from CHOK1. Our results show that while the A-MuLV receptor from CHOK1, like its isotype from E36, functions for both viruses in the absence of secreted inhibitors, the GALV receptor from CHOK1 has an intrinsic lesion that renders it nonfunctional.
MATERIALS AND METHODS
Cells.
The Chinese hamster cell lines CHOK1 and the glycosylation-deficient CHOK1 derivative lines Lec2 and Lec8 were obtained from American Type Culture Collection (CCL 61, CRL 1736, CRL 1737, respectively). E36, a derivative of male Chinese hamster lung fibroblast cell line V79, was kindly provided by Christine Kozak (National Institute of Allergy and Infectious Diseases, Bethesda, Md.). The Mus dunni tail fibroblasts (MDTF; ATCC CRL 2017) were a gift from Olivier Danos (Institut Pasteur, Paris, France). PA317/BSN and PG13/BSN produce vectors with A-MuLV and GALV envelopes, respectively, and their construction has been described elsewhere (10, 11, 13). MDTF expressing Pit1 or Pit1-D550T (MDTF-Pit1 or MDTF-Pit1-D550T) were derived as described before (29). CHOK1, Lec2, and Lec8 were grown in alpha minimal essential medium; all other cell lines were grown in Dulbecco’s modified Eagle’s medium (BioWhittaker, Walkersville, Md.). The growth media contained 10% fetal bovine serum, 4 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml.
ChoPit1 and ChoPit2 cloning.
A cDNA library was constructed by using CHOK1 poly(A)+ RNA. Known receptor sequences were obtained from GenBank, and PCR primers were designed on the basis of conserved regions. Primers were used in a reverse transcription-PCR with CHOK1 mRNA as template to synthesize small cDNA fragments that were then used to screen the CHOK1 λgt 22A (Gibco-BRL, Gaithersburg, Md.) cDNA library. A partial A-MuLV receptor cDNA homolog lacking the 5′ sequences was subcloned into pBluescript II KS(+) (Stratagene, La Jolla, Calif.) and sequenced. Reverse primers based on this partial cDNA were used to amplify a full-length amphotropic cDNA from CHOK1 mRNA. A similar approach was required to obtain a full-length CHOK1 GALV receptor homolog cDNA from a partial cDNA isolated from the same λgt 22A CHOK1 cDNA library. Contiguous sequence files (GenBank accession no. AF063024 for ChoPit1 and AF063025 for ChoPit2) were assembled and aligned to the A-MuLV and GALV receptor cDNA sequences obtained from human and E36 hamster receptor cDNA homologs.
Mutagenesis.
Aspartate at position 550 in human Pit1 was replaced with threonine by PCR mutagenesis. Plasmid DNA from three independent clones was sequenced to confirm the mutation and to detect any unscheduled mutations. The mutant fragment was subcloned into pLNS-Pit1 to derive pLNS-Pit1-D550T. pCRII (Invitrogen Corporation, Carlsbad, Calif.) was used for sequencing to confirm the D550T mutation in Pit1. Primers used for PCR mutagenesis were synthesized by Gibco-BRL. The retroviral expression vector pLNSX, which carries the neo gene as the selectable marker, has been described elsewhere (12).
Expression of receptor cDNAs.
Stable expression of mutant receptor cDNAs was accomplished as described before (5). Plasmids containing mutant receptor cDNAs were transfected into PA317 packaging cells by calcium phosphate-mediated DNA precipitation using a Profection kit (Promega, Madison, Wis.), and the cells were selected with 450 μg of G418 (active; Gibco-BRL) per ml. MDTF cells were then exposed to supernatant from transfected PA317 as previously described (5). The infected MDTF were selected with 600 μg of G418 (active) per ml. For transient expression of receptors, CHOK1 were directly transfected with equal amounts of DNA, and the cells were tested for A-MuLV infection without selection with G418.
Infection assays.
A day before the assay, 20,000 cells per well were seeded in 24-well plates. Supernatant from PA317/BSN or PG13/BSN were added in various dilutions and removed after additional 20 h. Incubation at 37°C was continued for another 48 h, at which time the cells were processed for β-galactosidase activity. Blue cells were counted to determine the apparent titer, reported here as blue-forming units (BFU) per milliliter. Each value is the mean ± standard error of the mean.
Interference assays.
Lec8 and MDTF-Pit1 were productively infected with wild-type GALV(SEATO) or A-MuLV(4070A), and vectors produced by PA317-BSN and PG13-BSN were used as the superinfecting pseudotypes. Uninfected Lec8 and MDTF-Pit1 were used as controls to determine the apparent vector titers. Cells were seeded at a density of 25,000 per well in 24-well plates, and the infections with viral pseudotypes were carried out as described above.
RESULTS
The A-MuLV receptors from CHOK1 and its E36 isotype are similar.
Previously we have reported the cloning and characterization of HaPit2, the A-MuLV receptor from E36 cells (28). The A-MuLV receptor from CHOK1 (ChoPit2) was cloned and sequenced as described in Materials and Methods. The amino acid sequence comparison shows that ChoPit2 and HaPit2 are 97% identical (Fig. 1). The coding sequences for the two receptors are 99% identical. HaPit2 was the first receptor identified that has the unusual property of being functional not only for A-MuLV but also for GALV. Thus, GALV enters E36 cells via its own receptor, HaPit1, as well as HaPit2. This accounts for the nonreciprocal interference the two viruses exhibit when infecting E36; A-MuLV-infected cells remain susceptible to GALV, while GALV-infected cells become resistant to A-MuLV (3, 28, 30).
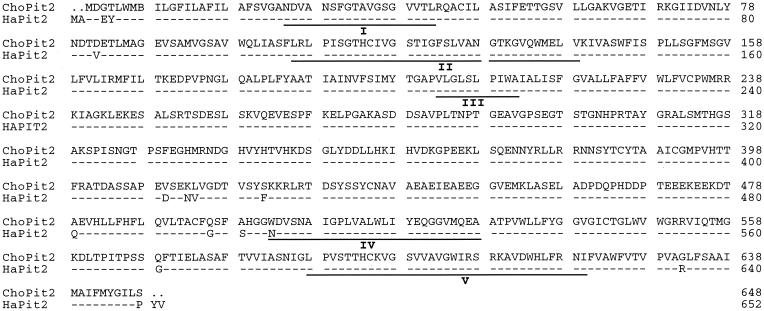
FIG. 1.
Sequence comparison of ChoPit2 and HaPit2, the A-MuLV receptors from CHOK1 and E36. For HaPit2, dashes represent residues common with ChoPit2, dots represent spaces introduced for alignment, and the amino acid differences are shown by single-letter codes. The putative extracellular regions are underlined and labeled I to IV.
The putative extracellular regions of ChoPit2 are identical to those of HaPit2 (Fig. 1). Both receptors have glutamate instead of lysine at position 522 in the fourth extracellular region. This position is critical for GALV entry; a lysine at 522 in the Pit2 group receptors and at the corresponding position in the Pit1 group receptors renders them nonfunctional for GALV (28). We have shown before that Pit2, which is nonfunctional for GALV, becomes a highly efficient receptor for this virus when the lysine at 522 is replaced with glutamate (5). While the absence of lysine at position 522 confers GALV receptor function, its presence or absence in the Pit2 group receptors does not affect A-MuLV entry. Thus, ChoPit2, like HaPit2, should function as a receptor not only for A-MuLV but also for GALV.
Threonine at position 550 in the fourth extracellular region of Pit1 is detrimental for GALV entry.
ChoPit1 and HaPit1 are 91% identical in sequence, and the two receptors differ only at five positions in the putative extracellular regions (Fig. 2). One of these differences is in domain II and the other four are in region A, the last nine residues of domain IV (Fig. 3). The position in domain II varies in many functional receptors and is therefore likely not important for GALV entry.
FIG. 2.
Sequence comparison of ChoPit1 and HaPit1, the GALV receptors from CHOK1 and E36. For HaPit1, dashes represent residues common with ChoPit1, dots represent spaces introduced for alignment, and the amino acid differences are shown by single-letter codes. The putative extracellular regions are underlined and labeled I to IV.
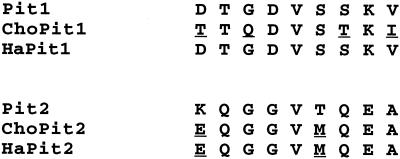
FIG. 3.
Region A sequences of GALV and A-MuLV receptors. The sequence represents residues 550 to 558 in the Pit1 group receptors and 522 to 530 in the Pit2 group. In comparison with Pit1 and Pit2, the residues that differ in each group are underlined.
Region A (residues 550 to 558) of Pit1 group receptors is the only portion of the receptors implicated in GALV infection (6 7, 25). A number of studies have suggested that only positions 550 and 551 are crucial for infection (2, 25). ChoPit1, Pit1, and HaPit1 have threonine at 551 but differ at position 550; ChoPit1 has threonine at this position, whereas Pit1 and HaPit1, both efficient receptors for GALV, have aspartate. Lysine at position 550 in the Pit1 group receptors and at the corresponding position in the Pit2 group receptors blocks GALV infection (6, 7) and is the only known residue at this position that renders the receptors nonfunctional. We thought it likely that threonine at 550 may also be detrimental for GALV entry. To assess the role of threonine at position 550 in GALV infection, we replaced the aspartate at this position in Pit1 with threonine and tested the resulting receptor, Pit1-D550T, in MDTF. This single substitution rendered Pit1 nonfunctional for GALV, strongly suggesting that threonine at 550 also renders ChoPit1 nonfunctional (Table 1). We therefore determined the GALV receptor function of native ChoPit1 by interference assays.
TABLE 1.
Replacement of aspartate at position 550 in the human receptor for gibbon ape leukemia virus renders the receptor nonfunctional
| Receptor | GALV vector titer (BFU/ml) |
|---|---|
| Pit1 | (1.5 ± 0.3) × 105 |
| Pit1-D550T | 0 |
GALV and A-MuLV exhibit reciprocal interference in Lec8.
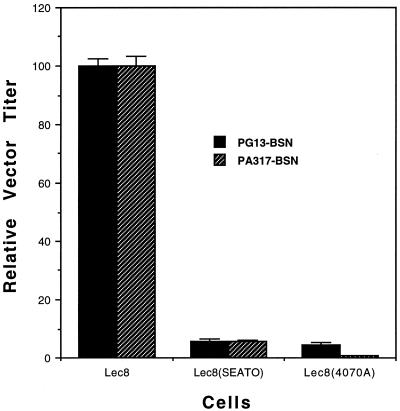
CHOK1 cells resist infection in a glycosylation-dependent manner, evidently because the inhibitory proteins they secrete require glycosylation for activity (15, 16). We therefore used the glycosylation-deficient CHOK1 derivative Lec8 for interference assays (23, 24). Lec8 cells have the same viral receptors as parental CHOK1, and they either secrete no inhibitory proteins or secrete defective ones, presumably due to lack of glycosylation. Lec8 cells are therefore susceptible to both GALV and A-MuLV (16) (Table 2). For interference assays, Lec8 cells were productively infected with A-MuLV (4070A) or GALV (SEATO), and each cell line was then tested for susceptibility to superinfecting vectors. The two viruses exhibited reciprocal interference when infecting Lec8 (Fig. 4), unlike the nonreciprocal interference that they exhibit when infecting E36 (3). GALV and A-MuLV therefore enter Lec8 via the same receptor. Because no naturally occurring GALV receptor that also functions for A-MuLV is known and because ChoPit2 is nearly identical to its isotype HaPit2, a functional receptor for GALV, we conclude that the two viruses infect Lec8 via ChoPit2. Moreover, the results suggest that the native GALV receptor, ChoPit1, is nonfunctional. GALV entry into CHOK1 is therefore blocked by two separate mechanisms; the secreted inhibitory factors block virus entry via ChoPit2, while ChoPit1 is intrinsically nonfunctional.
TABLE 2.
Cell line CHOK1 is resistant to A-MuLV, and this block is glycosylation dependenta
FIG. 4.
GALV and A-MuLV exhibit reciprocal interference when infecting the glycosylation-deficient CHOK1 derivative cell line Lec8. The titers with Lec8 for each vector were considered 100, and the titers afforded by cells productively infected with GALV(SEATO) or A-MuLV(4070A) were normalized to this number. PG13-BSN supernatant gave a titer of (7.1 ± 0.2) × 104 BFU/ml, and PA317-BSN supernatant gave a titer of (9.1 ± 0.3) × 104 BFU/ml. The assays were done as described in Materials and Methods.
As expected, the two viruses exhibited no interference in MDTF-Pit1 (Table 3), consistent with the fact that A-MuLV enters these cells via the native receptor and GALV via Pit1.
TABLE 3.
GALV and A-MuLV exhibit nonreciprocal interference when infecting MDTF-Pit1 cells
| Cells | Relative titer of superinfecting vectora (mean ± SEM)
|
|
|---|---|---|
| PG13-BSN | PA317-BSN | |
| MDTF-Pit1 | 100 ± 3 | 100 ± 6 |
| MDTF-Pit1(SEATO) | 0.034 ± 0.004 | 101 ± 2 |
| MDTF-Pit1(4070A) | 33 ± 4 | 0.0002 ± 0.0001 |
The titers afforded by MDTF-Pit1 for each vector were considered 100, and the titers afforded by cells productively infected with GALV(SEATO) or A-MuLV(4070A) were normalized to this number. PG13-BSN supernatant gave a titer of (3.0 ± 0.1) × 105 BFU/ml, and PA317-BSN supernatant gave a titer of (1.8 ± 0.1) × 106 BFU/ml. The assays were done as described in Materials and Methods.
Presence of threonine at 550 has no effect on receptor transport to cell surface.
Although threonine at position 550 renders Pit1 nonfunctional for GALV, it is unclear whether the loss of function is specific for GALV. It is possible, for example, that the mutation has a generically disruptive effect on receptor structure, blocking receptor transport to the cell surface. We therefore determined whether the presence of this mutation reduced receptor expression. One way to assess the expression of a receptor that is nonfunctional for GALV is to express the receptor in CHOK1 and test for A-MuLV infection. This requires that the receptor be functional for the amphotropic virus. We therefore incorporated the D550T mutation in a Pit1/Pit2 chimera, Chi-WT. This chimera contains the extracellular domains I, II, III, and V from Pit2 and IV from Pit1, and it is highly efficient receptor for both GALV and A-MuLV (2). The derivative chimera, Chi-D550T, proved nonfunctional for GALV. Thus, for GALV entry Chi-D550T functionally mimics Pit1-D550T. When expressed in CHOK1, Chi-D550T proved as efficient a receptor for A-MuLV as Chi-WT. This finding shows that the effect of threonine at 550 in Chi-D550T, though disruptive for GALV receptor function, does not hinder receptor transport to the cell surface. We conclude the detrimental effect of threonine at 550 is specific for GALV.
DISCUSSION
Previous studies had shown that CHOK1 cells are resistant to both GALV and A-MuLV because these cells secrete proteins that block receptor usage by these viruses (15, 16). We have shown here that ChoPit2 is functionally similar to HaPit2; it permits both A-MuLV and GALV entry. Unlike with E36 cells, however, GALV cannot use its own receptor from CHOK1 (ChoPit1). Thus, CHOK1 cells are resistant to GALV and A-MuLV not only because the secreted factors block ChoPit2 but also because ChoPit1 is intrinsically nonfunctional for GALV. It is known that conditioned medium from CHOK1 blocks A-MuLV entry into E36 as well as into Lec8. However, while the conditioned medium also blocks GALV entry into Lec8, it fails to block GALV entry into E36. Our current findings account for these observations. The inhibitory effect of the secreted proteins, while definitive for the A-MuLV receptors from both E36 and CHOK1, is absent for GALV entry via HaPit1. Thus, GALV enters E36 cells via its own receptor even in the presence of CHOK1 conditioned medium. On the other hand, GALV fails to infect CHOK1 because the secreted factors block its entry via ChoPit2 and because ChoPit1 is nonfunctional.
The precise nature of the secreted factors that block GALV and A-MuLV is unknown. Previous studies have suggested the inhibitors may be endogenous envelope gene products (15, 16). It is known that CHOK1 cells harbor multiple type C retrovirus sequences, and the full-length genome of one such virus has been isolated (9). The env of this endogenous virus has high homology to env of A-MuLV and very little to env of GALV (9), which may explain why the inhibitory effect of CHOK1 conditioned medium is specific for the A-MuLV receptor isotypes.
Our results demonstrate that the presence of threonine at position 550 in the fourth extracellular region is detrimental for GALV entry; when aspartate at this position in Pit1 is replaced with threonine, the receptor becomes nonpermissive for GALV. Lysine is the only other known residue whose presence at this position blocks GALV infection. We have shown before that GALV does not require an acidic residue at position 550; Pit1 with glycine, isoleucine, or glutamine at this position remains fully permissive for virus entry (2). The exact mechanism by which lysine or threonine at position 550 disrupts receptor function is unclear. ChoPit1 is the only known native receptor that has threonine at 550. The reciprocal interference exhibited by GALV and A-MuLV when infecting Lec8 shows that the two viruses use ChoPit2 and that ChoPit1 is nonfunctional. These cells are therefore unique in that GALV enters them via the A-MuLV receptor but not its own.
The viral interference with Lec8 was not as great as with MDTF-Pit1. This was expected because Lec8 cells are defective in UDP-galactose transport into the Golgi apparatus (3, 23) and therefore fail to properly glycosylate the viral envelope proteins. Consequently, receptor occupancy by such envelope proteins is considerably less, which permits enhanced entry of the superinfecting virus. That improper glycosylation of envelope proteins markedly reduces the severity of interference is well known. Rein et al. (21) have shown that in the presence of the glycosylation inhibitor 2-deoxy-d-glucose, retroviral interference is reduced 2 to 3 orders of magnitude, while tunicamycin treatment completely reverses interference. They further showed that in the presence of glycosylation inhibitors, the envelope proteins are inefficiently processed into SU and TM subunits and fail to reach the cell surface. Consequently, the receptors present on the cell surface remain available for the superinfecting virus. Thus, phenotypically Lec8 would be similar to 2-deoxy-d-glucose-treated cells, not tunicamycin-treated cells, which explains why GALV and A-MuLV exhibited reduced interference instead of no interference.
CHOK1 cells also resist ecotropic MuLV in a glycosylation-dependent manner. The block to this virus is due to direct inactivation of the viral receptor, not due to secreted factors. The ecotropic virus receptor from murine and hamster cells has a conserved N-linked glycosylation site. When this site in the murine receptor is mutated and the resulting variant is expressed in CHOK1, it renders the cells susceptible to ecotropic virus (4). Both ChoPit2 and HaPit2 have a potential N-linked glycosylation site in the second extracellular domain. Pit2, which renders CHOK1 highly susceptible to A-MuLV, lacks this site. But glycosylation of the two hamster receptors is not a prerequisite for inhibition of GALV and A-MuLV infection by the secreted factors; virus entry into tunicamycin-treated CHOK1 can be blocked with conditioned medium (15, 16). Moreover, E36 cells, despite normal glycosylation, are highly susceptible to both viruses. Thus, the presence of the glycosylation site in ChoPit2 and HaPit2 is irrelevant to inactivation of these receptors by CHOK1-secreted factors. Further, the differences that set the two hamster receptors phenotypically apart from Pit2, a receptor not inactivated by the secreted factors, are elsewhere in their primary sequence.
ACKNOWLEDGMENT
We thank Linda Tang for technical assistance.
REFERENCES
- 1.Boomer S, Eiden M V, Burns C C, Overbaugh J. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J Virol. 1997;71:8116–8123. doi: 10.1128/jvi.71.11.8116-8123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaudry G J, Eiden M V. Mutational analysis of the proposed gibbon ape leukemia virus binding site in Pit1 suggests that other regions are important for infection. J Virol. 1997;71:8078–8081. doi: 10.1128/jvi.71.10.8078-8081.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eglitis M A, Eiden M V, Wilson C A. Gibbon ape leukemia virus and the amphotropic murine leukemia virus 4070A exhibit an unusual interference pattern on E36 Chinese hamster cells. J Virol. 1993;67:5472–5477. doi: 10.1128/jvi.67.9.5472-5477.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eiden M V, Farrell K B, Wilson C A. Glycosylation-dependent inactivation of the ecotropic murine leukemia virus receptor. J Virol. 1994;68:626–631. doi: 10.1128/jvi.68.2.626-631.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eiden M V, Farrell K B, Wilson C A. Substitution of a single amino acid residue is sufficient to allow the human amphotropic murine leukemia virus receptor to function as a gibbon ape leukemia virus receptor. J Virol. 1996;70:1080–1085. doi: 10.1128/jvi.70.2.1080-1085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johann S V, Gibbons J J, O’Hara B. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J Virol. 1992;66:1635–1640. doi: 10.1128/jvi.66.3.1635-1640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johann S V, van Zeijl M, Cekleniak J, O’Hara B. Definition of a domain of GLVR1 which is necessary for infection by gibbon ape leukemia virus and which is highly polymorphic between species. J Virol. 1993;67:6733–6736. doi: 10.1128/jvi.67.11.6733-6736.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kavanaugh M P, Miller D G, Zhang W, Law W, Kozak S L, Kabat D, Miller A D. Cell surface receptors for gibbon ape leukemia virus and amphotropic murine leukemia virus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lie Y S, Penuel E M, Low M-A L, Nguyen T P, Mangahas J O, Anderson K P, Petropoulos C J. Chinese hamster ovary cells contain transcriptionally active full-length type C proviruses. J Virol. 1994;68:7840–7849. doi: 10.1128/jvi.68.12.7840-7849.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLachlin J R, Mittereder N, Daucher M B, Kadan M, Eglitis M A. Factors affecting retroviral function and structural integrity. Virology. 1993;195:1–5. doi: 10.1006/viro.1993.1340. [DOI] [PubMed] [Google Scholar]
- 11.Miller A D, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 13.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C W, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine leukemia virus reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller D G, Miller A D. Tunicamycin treatment of CHO cells abrogates multiple blocks to retrovirus infection, one of which is due to a secreted inhibitor. J Virol. 1992;66:78–84. doi: 10.1128/jvi.66.1.78-84.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller D G, Miller A D. Inhibitors of retroviral infection are secreted by several hamster cell lines and are also present in hamster sera. J Virol. 1993;67:5346–5352. doi: 10.1128/jvi.67.9.5346-5352.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller D G, Miller A D. A family of retroviruses that utilize related phosphate transporters for cell entry. J Virol. 1994;68:8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Hara B, Johann S V, Klinger H P, Blair D G, Rubinson H, Dunn K J, Sass P, Vitek S M, Robins T. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 19.Olah Z, Lehel C, Anderson W B, Eiden M V, Wilson C A. The cellular receptor for gibbon ape leukemia virus is a novel high affinity phosphate transporter. J Biol Chem. 1994;269:25426–25431. [PubMed] [Google Scholar]
- 20.Pedersen L, Johann S V, van Zeijl M, Pedersen F S, O’Hara B. Chimeras of receptors for gibbon ape leukemia virus/feline leukemia virus B and amphotropic murine leukemia virus reveal different modes of receptor recognition by retroviruses. J Virol. 1995;69:2401–2405. doi: 10.1128/jvi.69.4.2401-2405.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rein A, Schultz A M, Bader J P, Bassin R H. Inhibitors of glycosylation reverse retroviral interference. Virology. 1982;119:185–192. doi: 10.1016/0042-6822(82)90075-7. [DOI] [PubMed] [Google Scholar]
- 22.Schneiderman R D, Farrell K B, Wilson C A, Eiden M V. The Japanese feral mouse Pit1 and Pit2 homologs lack an acidic residue at position 550 but still function as gibbon ape leukemia virus receptors: implications for virus binding motif. J Virol. 1996;70:6982–6986. doi: 10.1128/jvi.70.10.6982-6986.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanley P. Selection of specific wheat germ agglutinin-resistant (WgaR) phenotypes from Chinese hamster ovary cell populations containing numerous LecR genotypes. Mol Cell Biol. 1981;1:687–696. doi: 10.1128/mcb.1.8.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanley P, Siminovitch L. Complementation between mutants of CHO cells resistant to a variety of plant lectins. Somatic Cell Genet. 1977;3:391–405. doi: 10.1007/BF01542968. [DOI] [PubMed] [Google Scholar]
- 25.Tailor C S, Takeuchi Y, O’Hara B, Johann S V, Weiss R A, Collins M K L. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infection. J Virol. 1993;67:6737–6741. doi: 10.1128/jvi.67.11.6737-6741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeuchi Y, Vile R G, Simpson G, O’Hara B, Collins M K L, Weiss R A. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol. 1992;66:1219–1223. doi: 10.1128/jvi.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Zeijl M, Johann S V, Closs E, Cunningham J, Eddy R, Shows T B, O’Hara B. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson C A, Farrell K B, Eiden M V. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J Virol. 1994;68:7697–7703. doi: 10.1128/jvi.68.12.7697-7703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson C A, Farrell K B, Eiden M V. Comparison of cDNAs encoding the gibbon ape leukemia virus receptor from susceptible and nonsusceptible murine cells. J Gen Virol. 1994;75:1901–1908. doi: 10.1099/0022-1317-75-8-1901. [DOI] [PubMed] [Google Scholar]
- 30.Wilson C A, Eiden M V. Viral and cellular factors governing hamster cell infection by murine and gibbon ape leukemia viruses. J Virol. 1991;65:5975–5982. doi: 10.1128/jvi.65.11.5975-5982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson C A, Eiden M V, Anderson W B, Lehel C, Olah Z. The dual-function hamster receptor for amphotropic murine leukemia virus (MuLV), 10A1 MuLV, and gibbon ape leukemia virus is a phosphate symporter. J Virol. 1995;69:534–537. doi: 10.1128/jvi.69.1.534-537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]