Abstract
Tralokinumab, a fully human mAb specifically targeting the IL-13 cytokine, has demonstrated clinical efficacy and safety in patients with moderate-to-severe atopic dermatitis. Tralokinumab binds IL-13 with high affinity, which prevents the interaction of IL-13 with IL-13Rα1 and subsequent signaling. Similarly, tralokinumab-bound IL-13 cannot bind to IL-13Rα2, a proposed decoy receptor that is reported to bind IL-13 with extraordinarily high affinity. It has however not been fully elucidated to what extent tralokinumab interferes with the endogenous regulation of IL-13 through IL-13Rα2. In this mechanistic study, we used biophysical, biochemical, and cellular assays to investigate the effect of tralokinumab on the interaction between IL-13 and IL-13Rα1 and IL-13Rα2, respectively, as well as the effects on IL-13Rα2–mediated IL-13 internalization. We demonstrate that IL-13Rα2 binds IL-13 with exceptionally high affinity and that tralokinumab is unable to displace IL-13 from IL-13Rα2. In contrast to this, tralokinumab is able to disrupt the IL-13/IL-13Rα1 and IL-13Rα1/IL-13/IL-4Rα complex. Furthermore, we demonstrate that whereas the IL-13/tralokinumab complex is unable to bind IL-13Rα2, any IL-13 that is not bound by tralokinumab (i.e., free IL-13) can be bound by IL-13Rα2 and subsequently internalized, regardless of the presence of tralokinumab. In summary, our study indicates that tralokinumab does not interfere with endogenous IL-13Rα2–mediated regulation of free IL-13.
Introduction
IL-13 is a key type 2 cytokine that is found overexpressed in the skin of patients with atopic dermatitis (AD) and drives many disease hallmarks, including recruitment of immune cells, impairment of the epidermal barrier, and changes in the lipid composition of the stratum corneum (Bieber, 2020). IL-13 signals through the so-called type II receptor complex, which consists of IL-13Rα1 and IL-4Rα (Aman et al., 1996; Hilton et al., 1996). Assembly of the type II signaling complex occurs in a stepwise fashion, where IL-13 first binds to the IL-13Rα1 subunit after which the IL-4Rα subunit is recruited, leading to subsequent signal transducer and activator of transcription 6 signaling.
IL-13 can also bind to IL-13Rα2 (Donaldson et al., 1998). In contrast to IL-13Rα1, IL-13Rα2 is not able to form a signaling complex with IL-4Rα and has a truncated intracellular domain that lacks the C-terminal box I motif that is found in IL-13Rα1 and required for signal transducer and activator of transcription 6 signaling (Donaldson et al., 1998). The affinity for IL-13 to IL-13Rα2 is reported to be much higher than the affinity for IL-13 to IL-13Rα1 (Andrews et al., 2002; Donaldson et al., 1998; Lupardus et al., 2010; Miloux et al., 1997). Owing to the combination of a high-affinity interaction and the inability to induce canonical Jak/signal transducer and activator of transcription signaling, IL-13Rα2 has been proposed to act as a decoy receptor that attenuates IL-13 signaling (Hershey, 2003; Lupardus et al., 2010). To this end, IL-13Rα2 is able to reduce extracellular IL-13 levels through receptor-mediated internalization in vitro (Kasaian et al., 2011). In line with a role as a decoy receptor, high expression of IL-13Rα2 correlates with reduced IL-13 signaling in a plethora of in vitro human cellular model systems as well as in vivo mouse models, whereas absence or inhibition of IL-13Rα2 results in increased IL-13–mediated signaling (Andrews et al., 2006; Badalyan et al., 2014; Chandriani et al., 2014; Daines and Hershey, 2002; Donaldson et al., 1998; McKenzie and Fallon, 2003; Rahaman et al., 2002; Sivaprasad et al., 2010; Wood et al., 2003). In contrast, some reports have described noncanonical and signal transducer and activator of transcription 6–independent signaling roles of IL-13Rα2 (Chandriani et al., 2014; Chen et al., 2013; Fichtner-Feigl et al., 2006; Yang et al., 2019). Moreover, chitinase-3-like 1 (YKL-40), an inflammation-associated protein that has been correlated with the severity and with late onset of AD, was found to interact with the extracellular domain of IL-13Rα2 in a complex with IL-13 and mediate cellular responses through IL-13Rα2 (Dežman et al., 2017; He et al., 2013; Kwak et al., 2019; Salomon et al., 2017). IL-13Rα2 expression and localization are tightly regulated by various proinflammatory cytokines, including IL-13 itself (Chiaramonte et al., 2003; Daines and Hershey, 2002; Fichtner-Feigl et al., 2006). IL-13Rα2 is found significantly overexpressed in skin biopsies from patients with AD and correlates with IL-13 expression (Tollenaere et al., 2021; Tsoi et al., 2019). Because increased IL-13Rα2 expression desensitizes cells for IL-13 signaling, these data suggest that cells possess intrinsic negative feedback loops to limit IL-13 signaling. In line with this, IL-13Rα2 expression is normalized upon pharmacological inhibition of IL-4 and/or IL-13 signaling in patients with AD (Hamilton et al., 2014).
Several biologics targeting the IL-13 and/or IL-4 signaling pathway, with distinct mechanisms of action, are approved or are in clinical development for the treatment of AD. Tralokinumab is a fully human IgG4 mAb that specifically binds the IL-13 cytokine with high affinity and has shown efficacy and safety in patients with moderate-to-severe AD (Popovic et al., 2017; Wollenberg et al., 2021). The tralokinumab-binding epitope on IL-13 overlaps with the binding sites for IL-13Rα1 and IL-13Rα2. Thus, by binding to IL-13, tralokinumab prevents IL-13 from interacting with either of these receptors (Popovic et al., 2017). IL-13 has been shown to bind to IL-13Rα1 with moderate affinity (5–30 nM) (LaPorte et al., 2008; Lupardus et al., 2010; Miloux et al., 1997), whereas it binds to tralokinumab with high affinity (58 pM) (Popovic et al., 2017). Tralokinumab is a potent inhibitor of IL-13 signaling through IL-13Rα1 in vitro and is efficacious in patients with moderate-to-severe AD, indicating that the high-affinity interaction between tralokinumab and IL-13 is sufficient to inhibit IL-13 binding to IL-13Rα1 and effectively prevent IL-13–mediated signaling (Popovic et al., 2017; Tollenaere et al., 2021; Wollenberg et al., 2021). However, considering the higher affinity of IL-13 for binding IL-13Rα2 compared with that of IL-13Rα1 and even tralokinumab, it is currently not completely understood what the effect of tralokinumab is on the association of IL-13 with IL-13Rα2 or its subsequent internalization by cells.
In this mechanistic study, we combined a range of biophysical, biochemical, and cellular assays to confirm the binding affinities of IL-13 with its receptors and assessed to what extent tralokinumab impacts the binding of IL-13 to IL-13Rα1 and IL-13Rα2 and its subsequent internalization through IL-13Rα2.
Results
IL-13 binds to IL-13Rα2 with extraordinarily high affinity
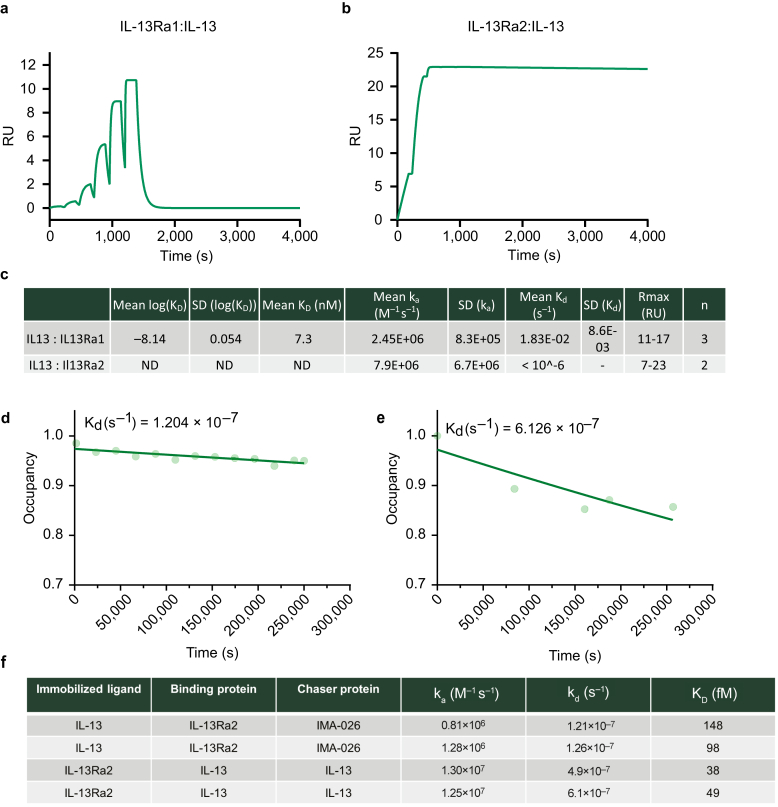
To be able to understand the effect of tralokinumab on IL-13 binding to IL-13Rα1 and IL-13Rα2, we first set out to determine the affinity of IL-13 to IL-13Rα1 and IL-13Rα2 using state-of-the-art techniques. We initially attempted to measure the affinity between IL-13 and both of its receptors (IL-13Rα1 and IL-13Rα2) in a classical surface plasmon resonance (SPR) experiment. This approach confirmed a moderately strong interaction between IL-13 and IL-13Rα1 (∼7 nM) and a very strong interaction between IL-13 and IL-13Rα2 (Figure 1a–c). We were however unable to quantify the affinity between IL-13 and IL-13Rα2 owing to its slow off-rate using classical SPR (Figure 1b and c). We therefore used chaser SPR (Quinn et al., 2018) as an alternative strategy, where the off-rate is calculated by measuring the binding occupancy of IL-13 to the receptor at different time points (Figure 1d and e). This allowed a much more accurate determination of the off-rate, which together with the on-rate from the classical SPR experiment allowed the calculation of the KD (Figure 1f). Our chaser SPR approach yielded KD values between 38 and 148 fM, depending on the orientation. Faster kinetics were observed when IL-13Rα2 was captured on the chip surface (Figure 1e and f), which were possibly caused by steric or matrix effects related to a larger analyte when capturing IL-13 but did not have a major impact on the calculated KD. In conclusion, our measurements confirm a moderately strong interaction between IL-13 and IL-13Rα1 (7 nM) and an exceptionally strong interaction between IL-13 and IL-13Rα2 with a KD in the femtomolar range, which is roughly 1,000-fold stronger than the reported affinity of tralokinumab for IL-13 (58 pM) (Popovic et al., 2017).
Figure 1.
SPR-based affinity measurements of IL-13 for IL-13Rα1 and IL-13Rα2. (a) IL-13Rα1 was captured on the chip surface followed by single-cycle kinetics injections of IL-13 at 30 μl/min. The line displays a fitted SPR sensogram using a 1:1 binding model from a representative experiment from one of the three experiments performed. (b) As in a but using IL-13Rα2 instead of IL-13Rα1 bound on the chip surface. (c) Measured on-rates (ka), off-rates (kd), and affinities (KD) for IL-13 binding to IL-13Rα1 (n = 3) and IL-13Rα2 (n = 2) using classical SPR. ND denotes that affinity could not be determined because the dissociation rate was slower than the limit of detection of the SPR equipment. (d) Chaser-SPR receptor occupancy plot. IL-13 was immobilized on the chip surface and blocked with IL-13Rα2; occupancy was calculated by adding a chaser molecule (IMA-026 Fab) at different time points. Representative occupancy plot from two experiments is shown. (e) As in d where IL-13Rα2 was immobilized on the chip surface and blocked with IL-13. In addition, IL-13 was used as a chase molecule. Representative occupancy plot from two experiments. (f) Measured on-rates (ka), off-rates (kd), and affinities (KD) for IL-13 binding to IL-13Rα2 using a chaser-SPR setup. Each row represents an individual experiment. ND, not determined; Rmax, maximum binding; RU, resonance units; s, second; SPR, surface plasmon resonance.
Tralokinumab is unable to disrupt the IL-13/IL-13Rα2 complex
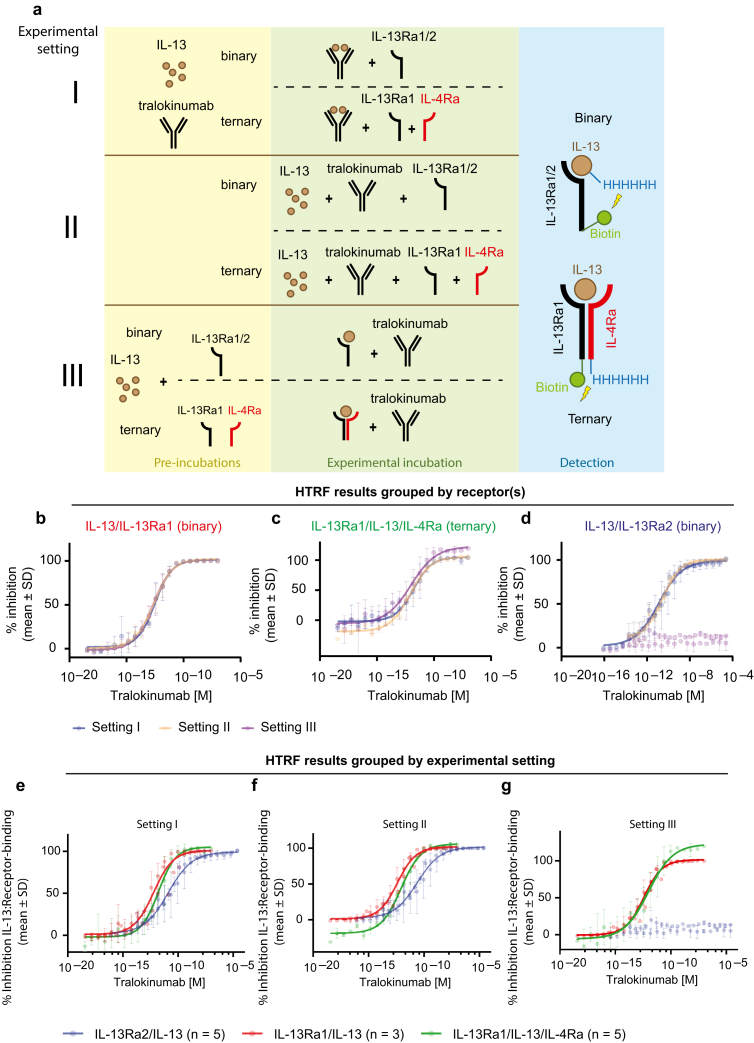
To assess the effect of tralokinumab on the ability of IL-13 to associate with IL-13Rα1 or IL-13Rα2, we developed an in vitro Homogeneous Time-Resolved Fluorescence (HTRF) assay that measures the interaction between IL-13 and each receptor and performed this assay in three different settings (Figure 2a). First, we assessed the effect of tralokinumab on IL-13/IL-13Rα1 and IL-13/IL-13Rα2 association when tralokinumab was allowed to form the IL-13/tralokinumab complex prior to addition of each receptor (setting I). In the second experimental setup, IL-13, tralokinumab, and IL-13Rα1 or IL-13Rα2 were added simultaneously (setting II). In the third experimental setup, IL-13 was allowed to bind IL-13Rα1 or IL-13Rα2 prior to the addition of tralokinumab (setting III). In all three experimental conditions, tralokinumab was able to fully block the interaction between IL-13 and the signaling receptor IL-13Rα1 in a dose-dependent manner with comparable potency (Figure 2b). The finding that tralokinumab had a comparable effect when it was incubated with the preformed IL-13/IL-13Rα1 complex (setting III) suggests that this interaction is reversible in the presence of tralokinumab. To investigate whether tralokinumab is also able to disrupt the IL-13 signaling complex consisting of IL-13, IL-13Rα1, and IL-4Rα, which is reported to be more stable than the binary interaction between IL-13 and IL-13Rα1 (Hilton et al., 1996; Miloux et al., 1997), we developed a third HTRF assay that allowed us to measure ternary complex formation (Figure 2a). Comparable with the results of the binary HTRF assay looking only at IL-13 association with IL-13Rα1, tralokinumab was also able to fully and dose-dependently inhibit the formation of the ternary signaling complex and showed similar potencies between all three individual settings (Figure 2c). These data indicate that tralokinumab is also able to disrupt the IL-13Rα1/IL-13/IL-4Rα signaling complex in addition to binding free IL-13 and preventing it from interacting with IL-13Rα1.
Figure 2.
The effect of tralokinumab on IL-13/receptor interactions. (a) Schematic of the binary and ternary HTRF assays to assess the effect of tralokinumab on IL-13 association with IL-13Rα2 and IL-13Rα1 in the presence or absence of IL-4Rα. Three different experimental settings were utilized: setting I, where IL-13 and tralokinumab were preincubated before addition of the receptor(s); setting II, where IL-13, tralokinumab, and receptor(s) were added simultaneously; and setting III, where IL-13 was preincubated with the receptor(s) prior to addition of tralokinumab. (b) Binary HTRF assays on IL-13 with IL-13Rα1 according to the three experimental settings shown in a. (c) Ternary HTRF assays performed on IL-13 with the IL-13Rα1/IL-4Rα complex according to the three experimental settings shown in a. (d) Binary HTRF assays on IL-13 with IL-13Rα2 according to the three experimental settings shown in a. (e) HTRF assays grouped by experimental setting I as described in a. (f) HTRF assays grouped by experimental setting II as described in a. (g) HTRF assays grouped by experimental setting III as described in a. All data points indicate mean percentage inhibition ± SD. Data were normalized to control samples where 0% inhibition corresponds to samples incubated in the absence of tralokinumab, and 100% inhibition corresponds to samples incubated with the highest concentration of tralokinumab. For all data from binary HTRF using IL-13Rα1, n = 3; for all data from binary HTRF using IL-13Rα2, n = 5; and for all data from ternary HTRF (IL-13Rα1/IL-4Rα), n = 5. HTRF, Homogeneous Time-Resolved Fluorescence.
Tralokinumab was also able to dose-dependently block the interaction between IL-13 and IL-13Rα2 when tralokinumab was either preincubated with IL-13 or when all the three proteins were added simultaneously (Figure 2d) (settings I and II). However, of note, we consistently observed that tralokinumab was less potent at inhibiting IL-13 binding to IL-13Rα2 than IL-13 binding to IL-13Rα1 and the ternary IL-13Rα1/IL-13/IL-4Rα complex (Figure 2e–g). Interestingly, in contrast to the reversible nature of the interaction between IL-13 and IL-13Rα1 and the ternary signaling complex in the presence of tralokinumab, tralokinumab was unable to displace IL-13 from IL-13Rα2. Specifically, we did not measure a significant difference (P = 0.30, two-tailed t-test) in percentage inhibition between the highest (10.2% inhibition, 95% confidence interval = 2.4–18.0) and lowest concentration (5.2% inhibition, 95% confidence interval = –4.5 to 15.0) of tralokinumab, when IL-13 was prebound to IL-13Rα2 (Figure 2d, setting III, and Figure 2g, blue dots). This observation indicates that IL-13 and IL-13Rα2 form a tight and stable complex in vitro, which is unperturbed by tralokinumab. In addition, the lower potency of tralokinumab at inhibiting IL-13 binding to IL-13Rα2 than to IL-13Rα1 correlates well with our finding that the affinity of IL-13 for binding IL-13Rα2 is much higher than for IL-13Rα1.
Free IL-13 but not tralokinumab-bound IL-13 is able to bind IL-13Rα2 on cells
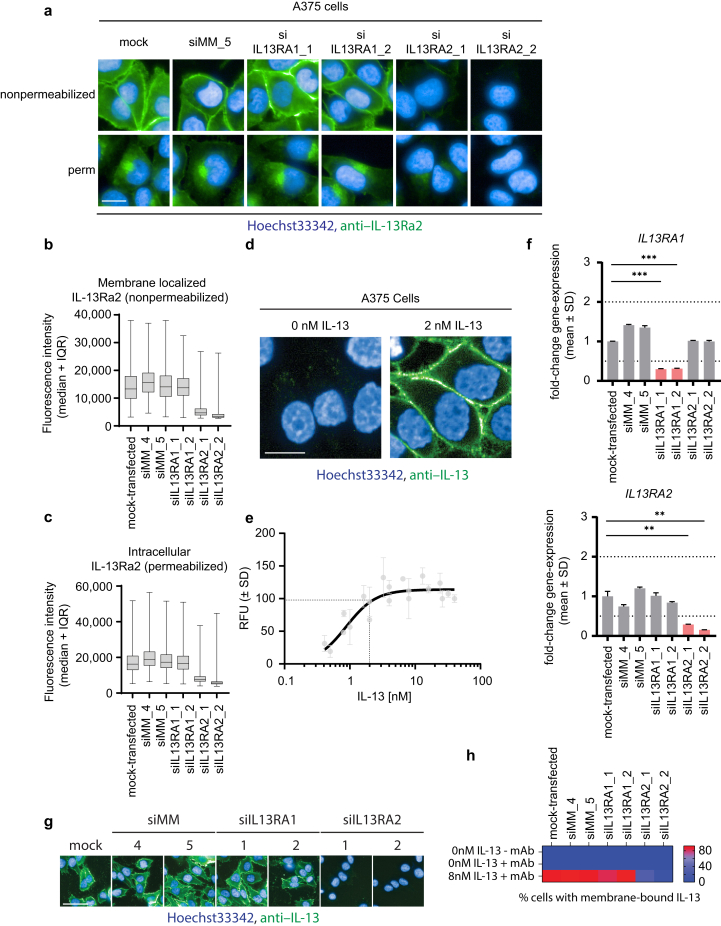
To set the findings from our biophysical and biochemical assays in a more relevant biological context, we next investigated the effects of tralokinumab on IL-13 binding to IL-13Rα2 in a cellular system. Previous studies on IL-13Rα2–mediated IL-13 internalization utilized A375 cells, which have been reported to express high endogenous levels of IL-13Rα2 and no IL-13Rα1 (Kasaian et al., 2011). In line with this report, we detected IL-13Rα2 on the plasma membrane and in the cytoplasm of A375 cells (Figure 3a–c), depending on whether the cells were permeabilized or not, and this subcellular distribution has been described in other cell types as well (Daines and Hershey, 2002). In addition, upon knocking down IL13RA2 with two independent small interfering RNAs (siRNAs), no such immunofluorescent staining was observed, confirming the specificity of the antibody (Figure 3a–c). Upon incubation of chilled A375 cells with recombinant IL-13, we were able to detect association of IL-13 on the plasma membrane (Figure 3d and e). However, upon siRNA-mediated depletion of IL-13Ra2, A375 cells were no longer able to bind IL-13 on their membrane (Figure 3f–h). Combined, these data confirm that A375 cells represent a suitable model system to investigate the effects of tralokinumab on IL-13 binding to IL-13Rα2 in a cellular context, without interference from IL-13Rα1 (Figure 3f–h). Next, we applied the same three experimental approaches as in the previously performed HTRF experiments to assess the effects of tralokinumab in the cellular IL-13–binding assay (Figure 4a). When IL-13 was incubated with tralokinumab before addition to A375 cells (setting I), tralokinumab was able to fully and dose-dependently inhibit IL-13 binding to IL-13Rα2, with a geometric mean half-maximal inhibitory concentration of 0.5 nM (95% CI: 0.3, 0.8) (Figure 4b and c) (setting I). When measuring IL-13 association with IL-13Rα2 upon simultaneous addition of tralokinumab and IL-13 to A375 cells (setting II), we detected a comparable dose-dependent effect (Figure 4b and c) (setting II). However, interestingly, detailed analysis of the images revealed a clear and persistent signal from membrane-bound IL-13 regardless of the presence of tralokinumab, indicating that tralokinumab was unable to fully block IL-13 association with IL-13Rα2 in this setting, even when in high molar excess compared with IL-13 (100 nM tralokinumab vs. 2 nM IL-13) (Figure 4d). This contrasts with setting I, where IL-13 was allowed to prebind to tralokinumab, and no membrane-bound IL-13 could be observed at saturating concentrations of tralokinumab. In the last experimental setting, where IL-13 was allowed to bind IL-13Rα2 on the membrane of A375 cells prior to addition of tralokinumab, we did not observe any effect of tralokinumab on IL-13Rα2–bound IL-13 (Figure 4b and c) (setting III). These observations are in line with our biochemical assays and confirm that the high-affinity interaction between IL-13 and IL-13Rα2 observed in biophysical assays translates to a cellular context.
Figure 3.
Characterization of A375 cells. (a) IL-13Rα1 or IL-13Rα2 was depleted from A375 cells through siRNA-mediated knockdown. Subsequently, cells were fixed and permeabilized as indicated in the figure panel. Cells were subsequently stained using an anti–IL-13Rα2 antibody (green) and Hoechst33342 (blue). Bar = 10 μM. (b) Quantitation of fluorescence intensity from anti–IL-13Rα2 on the plasma membrane of nonpermeabilized, A375 cells as shown in a. The median is represented by the horizontal bar, boxes represent the IQR, and whiskers extend to the lowest and highest values measured. More than 900 individual cells per condition were quantified. (c) Quantitation of intracellular fluorescence intensity from anti–IL-13Rα2 of permeabilized A375 cells as shown in a. The median is represented by the horizontal bar, boxes represent the IQR, and whiskers extend to the lowest and highest values measured (min to max). More than 1,500 individual cells per condition were quantified. (d) Chilled A375 cells were incubated with 2 nM unlabeled recombinant human IL-13 for 4 hours. After cell fixation, fluorescently labeled anti–IL-13 antibody (green) was used to stain for IL-13, and Hoechst33342 (blue) was used to visualize the nucleus. Bar = 10 μM. (e) Chilled A375 cells were incubated with various concentrations of recombinant human IL-13 for 4 hours, fixed, and stained as described in d. Membrane-localized fluorescence intensity was measured and plotted against IL-13 concentration. Data points represent mean RFUs normalized to the RFU at 40 nM IL-13 (100%) and no IL-13 (0%). n = 2. The dotted lines indicate an Emax of nearly 100% at 2 nM IL-13. (f) IL-13Rα1 or IL-13Rα2 was depleted from A375 cells through siRNA knockdown and analyzed by qPCR to confirm the successful knockdown of IL13RA1 and IL13RA2. Bars represent mean fold change versus mock-transfected controls. ∗∗P ≤ 0.01 and ∗∗∗P ≤ 0.001 using a two-tailed t-test versus nontransfected cells. Only samples that reached the twofold upregulated or downregulated threshold (dotted lines) were included for statistical analysis. (g) IL-13Rα1 or IL-13Rα2 was depleted from A375 cells through siRNA knockdown as shown in f. siRNA-transfected cells were subsequently chilled and incubated with 8 nM recombinant human IL-13 for 4 hours and fixed and stained as described in d. Fluorescence intensity on the membrane was quantified, and cells were scored positive or negative with regard to membrane-bound IL-13 (heatmap). Bar = 30 μM. (h) Quantitation of g; heatmap displays the percentage of A375 cells with membrane-associated IL-13. More than 1,500 individual cells per condition were scored. si denotes siRNA, and siMM denotes siRNA MisMatch. IQR, interquartile range; max, maximum; min, minimum; RFU, relative fluorescence unit; siRNA, small interfering RNA.
Figure 4.
The effect of tralokinumab on IL-13 binding to IL-13Rα2 on A375 cells. (a) Schematic of cellular IL-13/IL-13Rα2 binding assay workflow using three different experimental settings (I, II, and III). Setting I: IL-13 and tralokinumab were preincubated prior to addition to chilled A375 cells. Setting II: IL-13 and tralokinumab were added simultaneously to chilled A375 cells. Setting III: IL-13 was preincubated with chilled A375 cells prior to addition of tralokinumab. (b) Representative images of A375 cells incubated according to the three individual experimental settings (I, II, and III) in the presence of 0–100 nM tralokinumab. IL-13 was visualized by confocal microscopy on fixed A375 cells that were stained using an anti–IL-13 mAb (green) and Hoechst33342 (blue). Bar = 10 μM. (c) Dose–response curves of the inhibitory effect (in percentage, mean ± SD) of tralokinumab on IL-13 binding to IL-13Rα2 on the membrane of A375 cells from the representative experiment as shown in b. Data were normalized to samples incubated in the absence of IL-13 (100% inhibition) and in the absence of tralokinumab (0% inhibition). IC50 values were calculated from a nonlinear four-parameter variable slope from three independent experiments and used to calculate the geometric mean IC50 and 95% CIs. More than 600 individual cells per condition per well were quantified in all experiments. (d) Confocal microscopy images of the representative experiment shown in b, with increased intensity and contrast, demonstrating IL-13Rα2–bound IL-13 on A375 cells in the presence of 100 nM tralokinumab in experimental setting II but not in setting I. Bar = 10 μM. CI, confidence interval; IC50, half-maximal inhibitory concentration; N/A, not applicable.
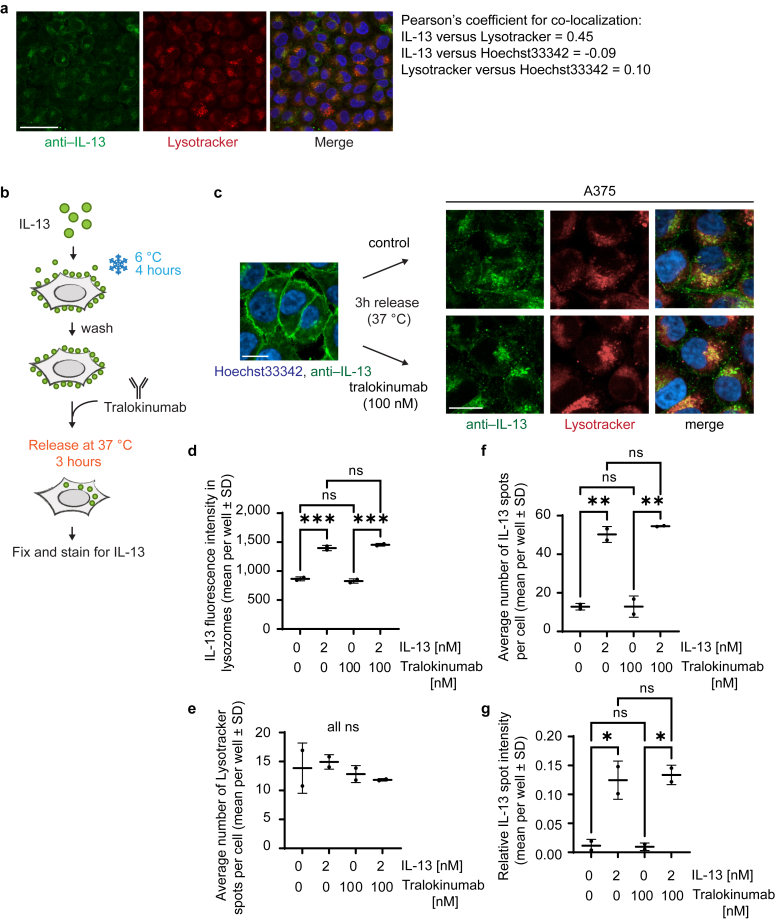
Tralokinumab does not block internalization of IL-13Rα2–associated IL-13
Because IL-13Rα2 has been proposed to regulate extracellular IL-13 levels through receptor-mediated internalization (Kawakami et al., 2001), we next wanted to assess to what extent tralokinumab potentially interferes with IL-13Rα2–mediated internalization. We were able to detect internalization of IL-13 by A375 cells, where it clustered in granules in the vicinity of the nucleus, which partially colocalized with lysosomes (Figure 5a). Owing to the overlapping binding epitope of tralokinumab and IL-13Rα2 on IL-13, we did not detect IL-13Rα2 association or internalization or IL-13–dependent cointernalization of tralokinumab under any experimental condition (data not shown). Because we have shown that anyunbound IL-13 is able to bind IL-13Rα2 and that the interaction between IL-13 and IL-13Rα2 remains stable despite the presence of tralokinumab, we hypothesized that tralokinumab would have no effect on internalization of IL-13 already captured by IL-13Rα2. To test this, IL-13 was bound to IL-13Rα2 on the membrane of A375 cells for 4 hours at 6 °C, after which the cells were released at 37 °C in the presence or absence of 100 nM tralokinumab after which IL-13 internalization was assessed using immunofluorescent staining (Figure 5b). We were able to detect IL-13–positive intracellular granules in cells that were exposed to tralokinumab, indicating that IL-13 internalization still occurred (Figure 5c). We found no significant difference in the intensity of IL-13 granules that colocalized with lysosomes in the presence or absence of tralokinumab (Figure 5d), and tralokinumab had no effect on the number of lysosomal spots detected (Figure 5e). Because not all IL-13 granules showed clear colocalization with the lysosomal marker, we also quantified the total amount of IL-13 spots we could detect per cell as well as their intensity. In line with our findings on the IL-13 granules colocalizing with lysosomes, we did not detect a significant difference in the total amount of IL-13 granules or their intensity in the presence or absence of tralokinumab (Figure 5f and g). Combined, our data indicate that the presence of tralokinumab does not interfere with receptor-mediated internalization of IL-13Rα2–associated IL-13.
Figure 5.
Tralokinumab does not inhibit IL-13Rα2–mediated internalization of receptor-bound IL-13. (a) Chilled A375 cells were incubated with 2 nM recombinant human IL-13 for 4 hours and released at 37 °C for 3 hours. After fixation, cells were stained with anti–IL-13 antibody (green) and Hoechst33342 (blue), and colocalization with lysotracker dye (red) was quantified using Pearson’s coefficient for colocalization. Bar = 30 μM. (b) Schematic of IL-13/IL-13Rα2 binding and internalization assay. (c) A375 cells were treated as depicted in b; fixed; and stained using an anti–IL-13 antibody (green), lysotracker (red), and Hoechst33342 (blue). Images were taken using a confocal microscope. Bar = 10 μM. (d) Quantification of b, showing the mean (±SD) anti–IL-13 fluorescence intensity colocalizing with lysotracker dye per well (n = 2). (e) Quantification of b, showing the mean (±SD) number of lysotracker granules per cell across four individual wells (n = 2). (f) Quantification of b, showing the mean (±SD) number of anti–IL-13–positive granules per cell across four individual wells (n = 2). (g) Quantification of b, showing the relative spot intensity of all anti–IL-13–positive granules across four individual wells (n = 2). ns and nondrawn comparison lines indicate nonsignificant. P > 0.05, ∗∗∗P ≤ 0.001, ∗∗P ≤ 0.01, and ∗P ≤ 0.05 using a one-way ANOVA. ns, not significant.
Discussion
In this mechanistic study, we utilized a combination of biophysical, biochemical, and cellular assays to investigate the effect of tralokinumab on the binding of IL-13 to IL-13Rα1 and IL-13Rα2 as well as the internalization of IL-13 through IL-13Rα2 by cells. Using SPR-based experiments, we were able to measure the binding affinity of IL-13 to IL-13Rα1 and IL-13Rα2. Because we observed that IL-13Rα2 binds to IL-13 with an extraordinarily high affinity (in fM range), whereas IL-13 binds IL-13Rα1 with a moderate affinity (in nM range), our results are largely confirmatory of earlier reports (Lupardus et al., 2010; Miloux et al., 1997). Tralokinumab has a high affinity for IL-13 (in pM range), which is around 100-fold higher than for IL-13Rα1 but approximately 1,000-fold lower than for IL-13Rα2 (Popovic et al., 2017). By using HTRF assays, we showed that tralokinumab is able to fully and dose-dependently block the interaction between IL-13 and IL-13Rα1 as well as the formation of the ternary IL-13Rα1/IL-13/IL-4Rα complex. In addition, our experiments demonstrated that the IL-13/IL-13Rα1 and IL-13Rα1/IL-13/IL-4Rα complexes are reversible in the presence of tralokinumab, indicating that tralokinumab is able to disrupt active signaling complexes. In contrast to the reversible nature of the IL-13Rα1/IL-13/IL-4Rα complex, tralokinumab was unable to disrupt the IL-13/IL-13Rα2 complex in our biochemical and cellular assays, which reflects the extraordinarily high-affinity interaction and, more specifically, the slow dissociation rate between these two proteins.
Even though IL-13Rα2 binds IL-13 with a higher affinity than tralokinumab, an earlier report indicated that tralokinumab fully inhibits IL-13 binding to IL-13Rα1 as well as IL-13Rα2 (Popovic et al., 2017). Although the previously reported half-maximal inhibitory concentration values for the effect of tralokinumab on IL-13 binding to IL-13Rα1 and IL-13Rα2 are comparable (660 pM and 716 pM, respectively), it should be noted that the experiments involving IL-13Rα2 contained considerably less receptor and ligand than the assays investigating the effect on IL-13 binding to IL-13Rα1, demonstrating that tralokinumab is less potent in inhibiting the association of IL-13 with IL-13Rα2. In line with this, we also noted that tralokinumab was less potent at inhibiting the interaction between IL-13 and IL-13Rα2 than IL-13 binding to IL-13Rα1. Inhibition of IL-13 binding to IL-13Rα2 was only observed in experimental conditions where tralokinumab was allowed to complex with IL-13 prior to addition of the receptor as well as when tralokinumab, IL-13, and receptor were added simultaneously. In this context, it is likely that IL-13Rα2 is unable to bind IL-13 because its primary binding epitope on IL-13 is already masked by tralokinumab. In contrast, when using a cellular and more biologically relevant system to measure IL-13–IL-13Rα2 interaction, we detected low levels of membrane-localized IL-13 staining even in the presence of 100 nM tralokinumab, which shows that tralokinumab was unable to fully block the association of IL-13 with IL-13Rα2 at this concentration. The minor discrepancy from the full inhibitory effect of tralokinumab in the HTRF assay could arise from various factors such as altered binding dynamics, relative concentrations of the ligand and receptor, the use of recombinant versus endogenously expressed IL-13Rα2, or assay sensitivity. Although the cellular IL-13Rα2 assay is more physiologically relevant, we were unable to assess IL-13 binding to IL-13Rα2 on cells for extended periods of time owing to technical limitations. Importantly, however, the cellular binding experiments were performed at physiologically relevant concentrations of tralokinumab on the basis of estimated skin concentrations (Tollenaere et al., 2021). Although the A375 melanoma cell line is a suitable model system for this mechanistic study, these cells may not represent the exact dynamics and levels of IL-13Rα2 as found in epidermal keratinocytes and dermal fibroblasts influenced by the presence of IL-13Rα1.
Increased total IL-13 serum concentrations have been detected in humans treated with IMA-026, a humanized anti–IL-13 IgG1 (Tiwari et al., 2016). Comparable with tralokinumab, IMA-026 prevents IL-13 from binding to IL-13Rα2 and IL-13Rα1, and IL-13 accumulation in response to IMA-026 was suggested to occur owing to reduced IL-13Rα2–mediated cellular clearance rates (Kasaian et al., 2011). Interestingly, however, our data show that any IL-13 molecules that are not bound or neutralized by tralokinumab are still able to associate with IL-13Rα2 and that tralokinumab does not impact subsequent IL-13Rα2–mediated internalization. Furthermore, because tralokinumab-bound IL-13 is unable to bind to IL-13Rα2, the receptor is never occupied by IL-13 molecules that are already unable to signal through their association with tralokinumab. In addition, because free tralokinumab and IL-13–bound tralokinumab cannot be internalized through IL-13Rα2, tralokinumab is not sequestered by IL-13Rα2–expressing cells and remains available in the extracellular milieu to neutralize any remaining free IL-13 molecules. Thus, two independent pathways may function in parallel to attenuate IL-13 signaling in tralokinumab-treated patients with AD: one primarily through tralokinumab-mediated neutralization of IL-13 and a secondary contribution from IL-13Rα2–mediated internalization of free IL-13. It must however be noted that although IL-13Rα2 expression is upregulated in the skin of patients with AD, high IL-13Rα2 expression by itself appears insufficient in inhibiting pathogenic IL-13 signaling in these patients. In addition, because IL-13Rα2 expression is directly regulated by IL-13, its expression is normalized upon inhibition of this pathway, which potentially reduces the impact of IL-13Rα2 on attenuating IL-13 signaling further (Guttman-Yassky et al., 2019; Tollenaere et al., 2021). Thus, inhibition of IL-13–mediated signaling in tralokinumab-treated patients with AD is therefore predominantly driven by neutralization of IL-13 by tralokinumab. The clinical relevance of the mechanistic studies presented in this paper could be further elaborated by studying patient samples, for example, by measuring and comparing concentrations of free and antibody-bound IL-13 at baseline and during treatment with tralokinumab in patients with AD. Besides tralokinumab, two other antibodies targeting the IL-13 signaling pathway have been developed for treatment of AD: lebrikizumab and dupilumab. The mode of action of lebrikizumab differs from that of tralokinumab because it binds a different epitope on IL-13, which allows IL-13 to associate with IL-13Rα1 and IL-13Rα2, regardless of whether it has already been bound and neutralized by lebrikizumab (Ultsch et al., 2013). In addition, lebrikizumab-bound IL-13 is therefore likely to be internalized by IL-13Rα2–expressing cells. Dupilumab inhibits IL-13 signaling by binding to and inhibiting IL-4Rα–mediated signaling (Le Floc'h et al., 2020). Because dupilumab does not bind IL-13, IL-13Rα1, or IL-13Rα2, dupilumab’s mode of action is unlikely to impact IL-13 binding to either receptor or its subsequent internalization.
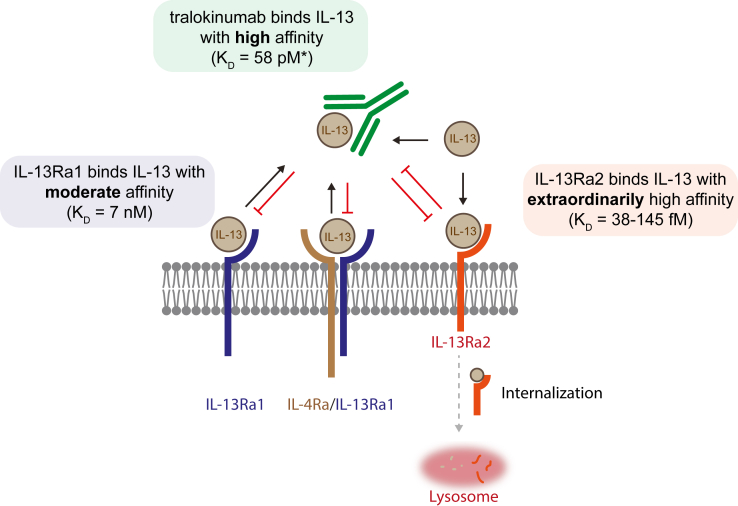
In summary, our data demonstrate that tralokinumab binds to IL-13 with high affinity and prevents IL-13 binding to IL-13Rα1 and IL-13Rα2, whereas any unbound IL-13 (i.e., free IL-13) can still bind to IL-13Rα2. Importantly, the high affinity of tralokinumab to IL-13 is able to prevent IL-13 from binding the signaling receptor IL-13Rα1 as well as disrupt the IL-13Rα1/IL-13/IL-4Rα signaling complex. In contrast, tralokinumab is unable to reverse the exceptionally strong interaction between IL-13 and IL-13Rα2 and does not impact the internalization of IL-13Rα2–bound IL-13 molecules. Our data provide detailed insights into the mechanism of action of tralokinumab, especially with regard to its impact on the association of IL-13 with IL-13Rα1 and IL-13Rα2 and endogenous regulation through IL-13Rα2 (Figure 6).
Figure 6.
Tralokinumab effectively disrupts the IL-13/IL-13Rα1/IL-4Rα signaling complex but not the IL-13/IL-13Rα2 complex. Model of the effects of tralokinumab on the association of IL-13 with its receptors. IL-13 is able to bind IL-13Rα1 with moderate affinity, tralokinumab with high affinity∗, and IL-13Rα2 with extraordinarily high affinity. When tralokinumab is bound to IL-13, IL-13 is unable to interact with either receptor. The interaction between IL-13 and IL-13Rα1 as well as the ternary IL-13Rα1/IL-13/IL-4Rα signaling complex is reversible in the presence of tralokinumab, but tralokinumab is unable to disrupt the interaction between IL-13 and IL-13Rα2. IL-13 molecules that are not bound to tralokinumab (i.e., free IL-13) are able to bind IL-13Rα2, and tralokinumab does not inhibit internalization of IL-13Rα2–bound IL-13. Because tralokinumab-bound IL-13 is unable to bind to either receptor and cannot be internalized through IL-13Rα2, tralokinumab stays available in the extracellular matrix to bind free IL-13 molecules. The asterisk (∗) denotes reference citation Popovic et al. (2017).
Materials and Methods
Recombinant proteins
Recombinant human IL-13 (213-ILB, R&D Systems, Minneapolis, MN) and IL-4 (204-IL, R&D Systems), expressed in E. coli, were used for cellular studies. For SPR and HTRF experiments, tagged proteins were needed, which were accomplished by custom protein production performed in house or by contract research organizations to ensure suitable tagging strategy and protein quality. IL-13 (aa 35–146) was expressed in insect cells and purified through affinity chromatography (His-tag) and size-exclusion chromatography (SEC). The used IL-13 was fully glycosylated and was produced in the following formats: C-terminally Avi-His-tagged, nonbiotinylated (binary HTRF and analyte for SPR), C-terminally Avi-His-tagged, biotinylated (ligand for SPR), and untagged (ternary HTRF). The extracellular domain (aa 22–343) of human IL-13Rα1 was expressed in Expi293 cells and purified through affinity chromatography (His-tag cleaved off during purification) and SEC. The final protein had a C-terminal biotinylated Avi-tag for use in SPR (ligand) and HTRF experiments. The extracellular domain (aa 29–331) of IL-13Rα2 was expressed in insect cells and purified through affinity chromatography (His-tag cleaved off during purification) and SEC. Two formats of the protein were produced: a C-terminal biotinylated Avi-tag (binary HTRF and ligand for SPR) and a C-terminal Avi-His-tag, nonbiotinylated (analyte in SPR). Protein biotinylation was done using the BirA enzyme followed by purification by SEC. The competitive anti–IL-13 antibody (IMA026) (Tiwari et al., 2016) Fab was expressed without a tag in ExpiCHO cells (Thermo Fisher Scientific, Waltham, MA) and purified using Protein L followed by SEC. His-tagged IL-4Rα was commercially available and procured from ACRO Biosystems (ILR-H5221) (Newark, DE). Protein quality control was generally done using SDS-PAGE, analytical SEC, and SPR for test of functionality.
SPR
SPR experiments were conducted on Biacore T200 and 8K instruments (Cytiva, Marlborough, MA). The general setup was based on capturing biotinylated IL-13, IL-13Rα1, or IL-13Rα2 on CM5 chips (Cytiva) with immobilized neutravidin (amine-coupling using default settings in the T200 and 8K software). Rmax values were kept below 25 RU to avoid impact from mass transport limitations. HBS-P+ was used as a running buffer. No regeneration conditions were needed for the IL-13–IL-13Rα1 interaction as a long enough dissociation period ensured complete surface regeneration. No regeneration conditions were identified for the IL-13–IL-13Rα2 interaction, and a new surface was prepared for each experiment.
The affinity of IL-13 for IL-13Rα1 and IL-13Rα2 was attempted measured by classical (direct) SPR by capturing the receptors on the surface of the chip and performing single-cycle kinetics injections of IL-13 at 30 μl/min. Flow cell 1 functioned as a reference and was subtracted from the active flow cell with the captured receptor protein. In addition to this, running buffer was used for blank injections, which was used to correct for any drift during the measurements. A total of 120–180 seconds was used for association, and up to 3,600 seconds were used for dissociation (for IL-13Rα2). The reference-subtracted, blank-corrected data were fitted and analyzed according to the 1:1 binding model available with the Biacore evaluation software.
The chaser experiments were done with both orientations testing IL-13 and IL-13Rα2 as both ligand and analyte. In both setups, the surface was initially saturated by the analyte by running a single-cycle injection, and chaser injections were made at strategically chosen time points over a time course of up to 72 hours. A flow rate of 30 μl/min was used. To correct for the decrease in surface activity during the course of the experiment, a matched control surface without prior analyte binding was used for parallel evaluation of chaser binding capabilities. For captured IL-13Rα2, 5 nM IL-13 was used as chaser in lack of a validated monoclonal IL-13–competitive anti–IL-13Rα2 antibody or tool compound (time-course was achieved by running multichannel measurements), and for captured IL-13, 50 nM Fab from a faster-dissociating IL-13R–competitive anti–IL-13 antibody was used as chaser (time-course was achievable by repeated injections within the same channel).
Data were analyzed in the Biacore Insight Evaluation software. Theoretical Rmax was calculated according to the following equation:
Occupancy is calculated as 1 – B/C, where B is the chaser response on the saturated surface, and C is the chaser response on the control surface (the experiment was run over 2–3 days to reach quantifiable levels of dissociation, during which time the surface activity was around 30% compared with that at the beginning; the reason why correction hereof was critical). The results are plotted against time in Prism and fitted to a single exponential decay with the plateau constrained to zero. The fitted rate constant K is the off-rate constant kd. The single-cycle kinetics data are fitted to the 1:1 affinity model with kd fixed at the value determined earlier to give the on-rate constant ka. The affinity, KD, is calculated as kd/ka.
HTRF
The binary HTRF experiments were done with His-tagged IL-13 (0.5 nM) combined with either Avi-biotinylated IL-13Rα1 (5 nM) or IL-13Rα2 (5 nM) in PBS buffer with 0.05% Tween-20 and 0.1% BSA. The ternary HTRF experiments were performed with untagged IL-13 (0.5 nM), Avi-biotinylated IL-13Rα1 (5 nM), and His-tagged IL-4Rα (2 nM) in PBS buffer with 0.05% Tween-20 and 0.1% BSA. Preincubations of one or more proteins were performed according to the settings specified in the result section for 4 hours at 21 °C (room temperature), after which the remaining proteins were added and incubated for a further 1 hour at 21 °C (room temperature). Finally, the detection reagents (MAb anti-6HIS-Tb cryptate Gold and Streptavidin-XL665 from Cisbio, Bedford, MA) were added (final concentration of 0.05 ng/μl and 2.5 ng/μl, prediluted in the PPI-Terbium detection buffer from Cisbio), and the mixture was incubated overnight at 4 °C before reading the plate on a Spark Cyto (Tecan, Männedorf, Switzerland). All experiments included quadruple replicates for all assay settings (I, II, and II). Three independent experiments were performed for the binary IL-13–IL-13Rα1 assay, and five independent experiments were performed for the binary IL-13–IL-13Rα2 and ternary IL-13Rα1–IL-13–IL-4Rα experimental setup.
Cell culture and transfection reagents
The human melanoma cell line A375 (CRL-1619, ATCC, Manassas, VA) was cultured in high-glucose (25.0 mM) DMEM with 1.0 mM pyruvate (Gibco, Waltham, MA), supplemented with 10% fetal bovine serum, penicillin, and streptomycin. Cells were cultured at 37 °C in a humidified incubator containing 5% carbon dioxide. siRNA transfections were carried out with Lipofectamine RNAiMAX (Life Technologies, Carlsbad, CA) following the manufacturer's instructions using 50 nM siRNA. siRNA targets (Silencer Select, Thermo Fisher Scientific) used in this study were S7374 (siIL13RA2_1), S7376 (siIL13RA2_2), S7373 (siIL13RA1_1), S7371 (siIL13RA1_2), AM4641 (siMM4), and AM4642 (siMM5).
Cellular IL-13/IL-13Rα2 binding assay
A375 cells (156.000 per cm2) were seeded 1 day prior to the experiment. Appropriate IL-13 and tralokinumab dilutions were prepared for the three different settings (setting I, II, and III) and added to prechilled cells (6 °C) to inhibit receptor trafficking.
IL-13/tralokinumab prebinding
Two nM IL-13 was mixed with a range of tralokinumab concentrations (0.01–100 nM) and incubated for 30 minutes at 37 °C to allow IL-13/tralokinumab complex formation. After incubation, the IL-13/tralokinumab mixtures were cooled down to 6 °C and added to prechilled A375 cells and incubated for 4 hours at 6 °C.
Simultaneous addition of IL-13 and tralokinumab
IL-13 and tralokinumab were added to prechilled (6 °C) A375 cells simultaneously with a final concentration of 2 nM IL-13 in combination with a range of tralokinumab concentrations (0.01–100 nM) and incubated for 4 hours at 6 °C.
IL-13/IL-13Rα2 prebinding
Two nM IL-13 was added to prechilled (6 °C) A375 cells and incubated for 4 hours at 6 °C. Subsequently, cells were washed three times with PBS, and fresh DMEM containing a wide concentration range of tralokinumab (0.01–100 nM) was added to the cells, followed by an additional 4–6 hours of incubation at 6 °C.
After the final incubation step at 6 °C, cells were processed for detection of membrane-bound IL-13 by immunofluorescent staining and microscopy. Each data point was generated by calculating the mean membrane localized IL-13 signal from replicate samples (wells), and a minimum of 600 individual cells per well were included in the analysis. To calculate percentage inhibition values, the data were normalized to samples incubated in the absence of IL-13 (100% inhibition) and in the absence of tralokinumab (0% inhibition). Half-maximal inhibitory concentration values were calculated as the geometric mean of three independent experiments.
Immunofluorescent staining and microscopy
For immunofluorescent staining of membrane-bound IL-13, cells were fixed in 3% methanol-free formaldehyde (Thermo Fisher Scientific) for 15 minutes at room temperature and subsequently incubated with fluorescently tagged 4.5 μg/ml monoclonal anti-human IL-13 (WO2005062967A2), 0.6 U rhodamine phalloidin (R415, Thermo Fisher Scientific), and 2 μM Hoechst33342 overnight at 4 °C. Cells were washed three times with PBS prior to imaging. Immunofluorescent staining of internalized IL-13 was performed similarly; however, cells were permeabilized by incubating the cells in 0.2% Triton X-100 in PBS for 5 minutes at room temperature before the addition of fluorescent antibodies and dyes. For immunofluorescent staining of IL-13Rα2, cells were fixed in 3% methanol-free formaldehyde for 15 minutes at room temperature and permeabilized by incubating the cells in 0.2% Triton X-100 in PBS for 5 minutes at room temperature. Samples were stained with 5 μg/ml goat anti–IL13-Rα2 (AF146, Abcam, Cambridge, United Kingdom) diluted in DMEM overnight at 4 °C, followed by staining with 4 μg/ml fluorescent donkey anti-goat secondary antibody (Thermo Fisher Scientific) for 1 hour at room temperature.
All images were acquired using an Operetta CLS High-Content microscope (Perkin Elmer, Waltham, MA) with a ×40 1.1 numerical aperture water-immersed objective (HH14000422, Perkin Elmer) in confocal mode or ×20 1.0 numerical aperture water-immersed objective (HH14000421, Perkin Elmer) in wide-field mode. Image segmentation and analysis were carried out using Perkin Elmer Harmony high-content imaging and analysis software (version 4.8).
qPCR analysis
RNA extraction from A375 cell cultures was followed by cDNA synthesis and amplification of cDNA by quantitative real-time PCR using TaqMan Gene Expression Assays. The following assays were used in this study: RPLP0 (Hs99999902_m1), ACTB (Hs01060665_g1), PPIA (Hs99999904_m1), IL13RA1 (Hs00609815_m1), and IL13RA2 (Hs00152924_m1). Raw Ct values were normalized by subtracting the normalization constant from three independent reference genes from the Ct value of the target of interest (). Subsequently, the ΔCt of the experimental sample was subtracted from the ΔCt of the control sample (ΔΔCt). Fold-change expression values were calculated by raising 2 to the power of the ΔΔCt (). Only samples that reached the twofold upregulated or downregulated threshold in comparison with the control sample were included for statistical analysis (two-tailed t-test vs. control sample). All statistics were performed on ΔΔCt values.
Ethics statement
No animal or human test samples were used in this study.
Data availability statement
No large datasets were generated or analyzed during this study.
ORCIDs
Maxim A.X. Tollenaere: http://orcid.org/0000-0002-2653-0777
Christina Mølck: http://orcid.org/0000-0002-2885-7861
Ian Henderson: http://orcid.org/0000-0001-9431-4745
Scott Pollack: http://orcid.org/0000-0002-8176-0997
Philip Addis: http://orcid.org/0000-0003-1185-651X
Helle Heibroch Petersen: http://orcid.org/0000-0001-9286-6044
Hanne Norsgaard: http://orcid.org/0000-0002-5249-6701
Conflict of Interest
MAXT, CM, HHP, and HN are shareholders and/or employees of LEO Pharma A/S. The remaining authors state no conflict of interest.
Acknowledgments
The authors would like to thank Peter Hansen for his technical expertise in developing the Homogeneous Time-Resolved Fluorescence assays as well as the technicians in the Department of Protein Technology at LEO Pharma A/S for their technical assistance on surface plasmon resonance experiments. This study was fully funded by LEO Pharma A/S.
Author Contributions
Conceptualization: HHP, HN; Data Curation: MAXT, CM; Formal Analysis: MAXT, CM, IH, SP, PA; Investigation: MAXT, CM, IH, SP, PA; Methodology: MAXT, CM, IH, SP, PA, HHP; Project Administration: MAXT; Resources: MAXT, CM, IH, SP, PA; Supervision: HN; Validation: MAXT, CM; Visualization: MAXT; Writing - Original Draft Preparation: MAXT; Writing – Review and Editing: MAXT, CM, IH, SP, PA, HHP, HN
accepted manuscript published online XXX; corrected proof published online XXX
Footnotes
Cite this article as: JID Innovations 2023.100214
References
- Aman M.J., Tayebi N., Obiri N.I., Puri R.K., Modi W.S., Leonard W.J. cDNA cloning and characterization of the human interleukin 13 receptor alpha chain. J Biol Chem. 1996;271:29265–29270. doi: 10.1074/jbc.271.46.29265. [DOI] [PubMed] [Google Scholar]
- Andrews A.L., Holloway J.W., Puddicombe S.M., Holgate S.T., Davies D.E. Kinetic analysis of the interleukin-13 receptor complex. J Biol Chem. 2002;277:46073–46078. doi: 10.1074/jbc.M209560200. [DOI] [PubMed] [Google Scholar]
- Andrews A.L., Nasir T., Bucchieri F., Holloway J.W., Holgate S.T., Davies D.E. IL-13 receptor alpha 2: a regulator of IL-13 and IL-4 signal transduction in primary human fibroblasts. J Allergy Clin Immunol. 2006;118:858–865. doi: 10.1016/j.jaci.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Badalyan V., Thompson R., Addo K., Borthwick L.A., Fisher A.J., Ort T., et al. TNF-α/IL-17 synergy inhibits IL-13 bioactivity via IL-13Rα2 induction. J Allergy Clin Immunol. 2014;134:975–978.e5. doi: 10.1016/j.jaci.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieber T. Interleukin-13: targeting an underestimated cytokine in atopic dermatitis. Allergy. 2020;75:54–62. doi: 10.1111/all.13954. [DOI] [PubMed] [Google Scholar]
- Chandriani S., DePianto D.J., N'Diaye E.N., Abbas A.R., Jackman J., Bevers J., 3rd, et al. Endogenously expressed IL-13Ralpha2 attenuates IL-13-mediated responses but does not activate signaling in human lung fibroblasts. J Immunol. 2014;193:111–119. doi: 10.4049/jimmunol.1301761. [DOI] [PubMed] [Google Scholar]
- Chen W., Sivaprasad U., Gibson A.M., Ericksen M.B., Cunningham C.M., Bass S.A., et al. IL-13 receptor alpha2 contributes to development of experimental allergic asthma. J Allergy Clin Immunol. 2013;132:951–958.e1-6. doi: 10.1016/j.jaci.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaramonte M.G., Mentink-Kane M., Jacobson B.A., Cheever A.W., Whitters M.J., Goad M.E., et al. Regulation and function of the interleukin 13 receptor alpha 2 during a T helper cell type 2-dominant immune response. J Exp Med. 2003;197:687–701. doi: 10.1084/jem.20020903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daines M.O., Hershey G.K. A novel mechanism by which interferon-gamma can regulate interleukin (IL)-13 responses. Evidence for intracellular stores of IL-13 receptor alpha -2 and their rapid mobilization by interferon-gamma. J Biol Chem. 2002;277:10387–10393. doi: 10.1074/jbc.M108109200. [DOI] [PubMed] [Google Scholar]
- Dežman K., Korošec P., Rupnik H., Rijavec M. SPINK5 is associated with early-onset and CHI3L1 with late-onset atopic dermatitis. Int J Immunogenet. 2017;44:212–218. doi: 10.1111/iji.12327. [DOI] [PubMed] [Google Scholar]
- Donaldson D.D., Whitters M.J., Fitz L.J., Neben T.Y., Finnerty H., Henderson S.L., et al. The murine IL-13 receptor alpha 2: molecular cloning, characterization, and comparison with murine IL-13 receptor alpha 1. J Immunol. 1998;161:2317–2324. [PubMed] [Google Scholar]
- Fichtner-Feigl S., Strober W., Kawakami K., Puri R.K., Kitani A. IL-13 signaling through the IL-13alpha2 receptor is involved in induction of TGF-beta1 production and fibrosis. Nat Med. 2006;12:99–106. doi: 10.1038/nm1332. [DOI] [PubMed] [Google Scholar]
- Guttman-Yassky E., Bissonnette R., Ungar B., Suárez-Fariñas M., Ardeleanu M., Esaki H., et al. Dupilumab progressively improves systemic and cutaneous abnormalities in patients with atopic dermatitis. J Allergy Clin Immunol. 2019;143:155–172. doi: 10.1016/j.jaci.2018.08.022. [DOI] [PubMed] [Google Scholar]
- Hamilton J.D., Suárez-Fariñas M., Dhingra N., Cardinale I., Li X., Kostic A., et al. Dupilumab improves the molecular signature in skin of patients with moderate-to-severe atopic dermatitis. J Allergy Clin Immunol. 2014;134:1293–1300. doi: 10.1016/j.jaci.2014.10.013. [DOI] [PubMed] [Google Scholar]
- He C.H., Lee C.G., Dela Cruz C.S., Lee C.M., Zhou Y., Ahangari F., et al. Chitinase 3-like 1 regulates cellular and tissue responses via IL-13 receptor α2 [published correction appears in Cell Rep 2013;5:1156] Cell Rep. 2013;4:830–841. doi: 10.1016/j.celrep.2013.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey G.K. IL-13 receptors and signaling pathways: an evolving web. J Allergy Clin Immunol. 2003;111:677–690. doi: 10.1067/mai.2003.1333. quiz 91. [DOI] [PubMed] [Google Scholar]
- Hilton D.J., Zhang J.G., Metcalf D., Alexander W.S., Nicola N.A., Willson T.A. Cloning and characterization of a binding subunit of the interleukin 13 receptor that is also a component of the interleukin 4 receptor. Proc Natl Acad Sci USA. 1996;93:497–501. doi: 10.1073/pnas.93.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasaian M.T., Raible D., Marquette K., Cook T.A., Zhou S., Tan X.Y., et al. IL-13 antibodies influence IL-13 clearance in humans by modulating scavenger activity of IL-13Rα2. J Immunol. 2011;187:561–569. doi: 10.4049/jimmunol.1100467. [DOI] [PubMed] [Google Scholar]
- Kawakami K., Takeshita F., Puri R.K. Identification of distinct roles for a dileucine and a tyrosine internalization motif in the interleukin (IL)-13 binding component IL-13 receptor alpha 2 chain. J Biol Chem. 2001;276:25114–25120. doi: 10.1074/jbc.M100936200. [DOI] [PubMed] [Google Scholar]
- Kwak E.J., Hong J.Y., Kim M.N., Kim S.Y., Kim S.H., Park C.O., et al. Chitinase 3-like 1 drives allergic skin inflammation via Th2 immunity and M2 macrophage activation. Clin Exp Allergy. 2019;49:1464–1474. doi: 10.1111/cea.13478. [DOI] [PubMed] [Google Scholar]
- LaPorte S.L., Juo Z.S., Vaclavikova J., Colf L.A., Qi X., Heller N.M., et al. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132:259–272. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Floc'h A., Allinne J., Nagashima K., Scott G., Birchard D., Asrat S., et al. Dual blockade of IL-4 and IL-13 with dupilumab, an IL-4Ralpha antibody, is required to broadly inhibit type 2 inflammation. Allergy. 2020;75:1188–1204. doi: 10.1111/all.14151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupardus P.J., Birnbaum M.E., Garcia K.C. Molecular basis for shared cytokine recognition revealed in the structure of an unusually high affinity complex between IL-13 and IL-13Ralpha2. Structure. 2010;18:332–342. doi: 10.1016/j.str.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie A.N., Fallon P.G. Decoy receptors in the regulation of T helper cell type 2 responses. J Exp Med. 2003;197:675–679. doi: 10.1084/jem.20030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miloux B., Laurent P., Bonnin O., Lupker J., Caput D., Vita N., et al. Cloning of the human IL-13R alpha1 chain and reconstitution with the IL4R alpha of a functional IL-4/IL-13 receptor complex. FEBS Lett. 1997;401:163–166. doi: 10.1016/s0014-5793(96)01462-7. [DOI] [PubMed] [Google Scholar]
- Popovic B., Breed J., Rees D.G., Gardener M.J., Vinall L.M., Kemp B., et al. Structural characterisation reveals mechanism of IL-13-neutralising monoclonal antibody tralokinumab as inhibition of binding to IL-13Rα1 and IL-13Rα2. J Mol Biol. 2017;429:208–219. doi: 10.1016/j.jmb.2016.12.005. [DOI] [PubMed] [Google Scholar]
- Quinn J.G., Pitts K.E., Steffek M., Mulvihill M.M. Determination of affinity and residence time of potent drug-target complexes by label-free biosensing. J Med Chem. 2018;61:5154–5161. doi: 10.1021/acs.jmedchem.7b01829. [DOI] [PubMed] [Google Scholar]
- Rahaman S.O., Sharma P., Harbor P.C., Aman M.J., Vogelbaum M.A., Haque S.J. IL-13R(alpha)2, a decoy receptor for IL-13 acts as an inhibitor of IL-4-dependent signal transduction in glioblastoma cells. Cancer Res. 2002;62:1103–1109. [PubMed] [Google Scholar]
- Salomon J., Matusiak Ł., Nowicka-Suszko D., Szepietowski J.C. Chitinase-3-Like Protein 1 (YKL-40) reflects the severity of symptoms in atopic dermatitis. J Immunol Res. 2017;2017 doi: 10.1155/2017/5746031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaprasad U., Warrier M.R., Gibson A.M., Chen W., Tabata Y., Bass S.A., et al. IL-13Ralpha2 has a protective role in a mouse model of cutaneous inflammation. J Immunol. 2010;185:6802–6808. doi: 10.4049/jimmunol.1002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari A., Kasaian M., Heatherington A.C., Jones H.M., Hua F. A mechanistic PK/PD model for two anti-IL13 antibodies explains the difference in total IL-13 accumulation observed in clinical studies. mAbs. 2016;8:983–990. doi: 10.1080/19420862.2016.1172151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollenaere M.A.X., Litman T., Moebus L., Rodriguez E., Stölzl D., Drerup K., et al. Skin barrier and inflammation genes associated with atopic dermatitis are regulated by interleukin-13 and modulated by tralokinumab in vitro. Acta Derm Venereol. 2021;101 doi: 10.2340/00015555-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi L.C., Rodriguez E., Degenhardt F., Baurecht H., Wehkamp U., Volks N., et al. Atopic dermatitis is an IL-13-dominant disease with greater molecular heterogeneity compared to psoriasis. J Invest Dermatol. 2019;139:1480–1489. doi: 10.1016/j.jid.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultsch M., Bevers J., Nakamura G., Vandlen R., Kelley R.F., Wu L.C., et al. Structural basis of signaling blockade by anti-IL-13 antibody Lebrikizumab. J Mol Biol. 2013;425:1330–1339. doi: 10.1016/j.jmb.2013.01.024. [DOI] [PubMed] [Google Scholar]
- Wollenberg A., Blauvelt A., Guttman-Yassky E., Worm M., Lynde C., Lacour J.P., et al. Tralokinumab for moderate-to-severe atopic dermatitis: results from two 52-week, randomized, double-blind, multicentre, placebo-controlled phase III trials (ECZTRA 1 and ECZTRA 2) Br J Dermatol. 2021;184:437–449. doi: 10.1111/bjd.19574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood N., Whitters M.J., Jacobson B.A., Witek J., Sypek J.P., Kasaian M., et al. Enhanced interleukin (IL)-13 responses in mice lacking IL-13 receptor alpha 2. J Exp Med. 2003;197:703–709. doi: 10.1084/jem.20020906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.J., Allahverdian S., Saunders A.D.R., Liu E., Dorscheid D.R. IL-13 signaling through IL-13 receptor alpha2 mediates airway epithelial wound repair. FASEB J. 2019;33:3746–3757. doi: 10.1096/fj.201801285R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No large datasets were generated or analyzed during this study.