Abstract
The matrix (M) protein of rhabdoviruses has been shown to play a key role in virus assembly and budding; however, the precise mechanism by which M mediates these processes remains unclear. We have associated a highly conserved, proline-rich motif (PPxY or PY motif, where P denotes proline, Y represents tyrosine, and x denotes any amino acid) of rhabdoviral M proteins with a possible role in budding mediated by the M protein. Point mutations that disrupt the PY motif of the M protein of vesicular stomatitis virus (VSV) have no obvious effect on membrane localization of M but instead lead to a decrease in the amount of M protein released from cells in a functional budding assay. Interestingly, the PPxY sequence within rhabdoviral M proteins is identical to that of the ligand which interacts with WW domains of cellular proteins. Indeed, results from two in vitro binding assays demonstrate that amino acids 17 through 33 and 29 through 44, which contain the PY motifs of VSV and rabies virus M proteins, respectively, mediate interactions with WW domains of specific cellular proteins. Point mutations that disrupt the consensus PY motif of VSV or rabies virus M protein result in a significant decrease in their ability to interact with the WW domains. These properties of the PY motif of rhabdovirus M proteins are strikingly analogous to those of the late (L) budding domain identified in the gag-specific protein p2b of Rous sarcoma virus. Thus, it is possible that rhabdoviruses may usurp host proteins to facilitate the budding process and that late stages in the budding process of rhabdoviruses and retroviruses may have features in common.
Viruses belonging to the Rhabdoviridae family cause disease in a wide variety of species including vertebrates, invertebrates, and plants. Two prototypic members of the Rhabdoviridae family include vesicular stomatitis virus (VSV) and rabies virus. Rhabdoviruses possess a negative-sense RNA genome, replicate exclusively in the cytoplasm of infected cells, and derive their lipid envelope via budding through the cytoplasmic membrane (for a review, see reference 51). While many aspects of the budding and assembly process of rhabdoviruses remain unclear, the major structural protein of rhabdoviruses (matrix [M]) is thought to play a key role in viral assembly and release (10, 11, 22, 27, 31, 33–36, 38, 61). Indeed, when the M protein of VSV was expressed in mammalian cells or in a baculovirus system in the absence of any other viral protein, M protein was released from the cells in the form of lipid vesicles by budding through the cytoplasmic membrane (20, 29). The N-terminal portion of the VSV M protein has been shown to be important for membrane localization and release from cells (10, 11, 20, 28, 60, 62). However, the precise mechanism of how M protein is released from cells and the potential function(s) of host proteins in the budding process remain unclear.
Recently, the role of the M protein in rhabdovirus assembly was compared to that of the Gag protein in retroviral assembly (27). The Gag protein of Rous sarcoma virus (RSV) and the M protein of VSV both have the ability to associate with the cellular membrane and to bud from cells independently of other viral proteins (20, 29, 55). In addition to the membrane association (MA) domain of RSV Gag, a late (L) budding domain was identified in the p2b protein of RSV Gag and shown to play an essential role in the late stage of budding (55, 58). Interestingly, the sequence of the RSV L domain (PPPY) matches the sequence of the consensus motif required for interacting with WW domains of cellular proteins (8, 9, 32, 39, 50). While L domains have been identified in the Gag proteins of other retroviruses, only the Gag proteins of the oncoviruses appear to have the PPxY motif conserved (17, 19, 37, 41, 55, 59). The recently described WW domain is (i) a highly structured, modular domain that mediates protein-protein interactions, (ii) present in a wide range of cellular proteins with unrelated functions, and (iii) functionally similar to, but structurally distinct from, Src homology-3 (SH3) domains (for a review see reference 48). The biology of the WW domain and its interacting ligand have been implicated as playing a role in a number of disease states including Liddle’s syndrome (a genetic form of hypertension), muscular dystrophy, and Alzheimer’s disease (5, 14, 46, 48). In addition, the WW domain has also been implicated in the biology of retroviral budding and assembly (15, 48). Indeed, the L domain of RSV Gag mentioned above has been shown recently to interact with the WW domain of the cellular Yes-kinase associated protein (YAP) (15, 47, 58).
In this report, we demonstrate for the first time that a highly conserved PPxY motif present within the M proteins of VSV and rabies virus can interact with WW domains of specific cellular proteins, including YAP. In addition, our results suggest that similar to the role of the retroviral PPxY motif in budding, the rhabdoviral PPxY motif is also likely important to play a role in a late step(s) of rhabdovirus maturation. Mutations that disrupt the rhabdoviral PPxY motifs not only abolish binding to WW domains but also interfere significantly with the release of VSV M protein from cells in a functional budding assay. Thus, in addition to possessing membrane association domains, the M protein of rhabdoviruses and the Gag polyprotein of retroviruses also have a similar, proline-rich motif (L domain) which appears to play a role in the budding process (55, 59). Last, our results suggest that late stages of the budding process of rhabdoviruses may be mediated through interactions with selected host proteins.
MATERIALS AND METHODS
Cells and viruses.
Stocks of CV-1 and BHK-21 cells were maintained in Dulbecco’s minimal essential medium (DMEM) (Life Technologies) supplemented with 10% fetal calf serum (Hyclone). VSV (Indiana serotype) was generously provided by M. Schubert (National Institutes of Health) and propagated in BHK-21 cells.
Plasmids.
The M gene of VSV (Indiana serotype) was cloned by reverse transcriptase (RT)-PCR by using primers flanking the open reading frame and containing EcoRV (5′) and XbaI (3′) restriction endonuclease sites. The PCR product was inserted into the EcoRV/XbaI-digested pSP72 vector containing the bacteriophage T7 promoter (Promega) by using standard protocols (1). Briefly, total RNA was isolated from BHK-21 cells infected with VSV by using the TRIzol reagent and protocol of the manufacturer (Life Technologies). Reverse transcription was performed by using avian myeloblastosis virus (AMV) RT (Life Technologies), and PCR was performed by using standard protocols with Vent DNA polymerase (New England Biolabs). The full-length M gene of rabies virus was generously provided by W. Wunner (Wistar Institute, Philadelphia, Pa.). PCR fragments encoding amino acids 1 through 74 of VSV M and 1 through 202, 1 through 69, and 1 through 52 of rabies virus M protein were inserted into the BamHI and EcoRI restriction sites of the vector pGEX-2TK (Pharmacia) for expression of glutathione S-transferase (GST) fusion proteins. Oligonucleotide primers and standard PCR protocols were utilized to introduce point mutations within the PPxY motifs of VSV and rabies virus GST-M fusion proteins. All plasmids and introduced mutations were confirmed by restriction endonuclease digestion and DNA sequencing by the Sanger method (45). Plasmid DNAs were maintained in either Escherichia coli DH5alpha (Life Technologies) or E. coli SURE2 (Stratagene), and DNA was purified by using the Qiagen purification system.
Purification of M protein from VSV virions.
Briefly, the supernatant was harvested from VSV-infected BHK-21 cells at 36 h postinfection and clarified, first at 2,500 rpm for 10 min and then at 3,200 rpm for 10 min. The supernatant was then centrifuged at 36,000 rpm for 30 min in an SW41 rotor. The virion pellet was then suspended in 400 μl of buffer containing 10 mM Tris (pH 8.0), 0.25 M NaCl, 1.0% Triton X-100, and 0.2-mg/ml dithiothreitol and incubated at room temperature (RT) for 30 min. The sample was then centrifuged at 75,000 rpm for 2 h in a TL-100 ultracentrifuge (Beckman). The supernatant fraction mostly containing the M and G proteins was removed and stored at −70°C.
Purification and radiolabeling of GST fusion proteins.
All GST fusion proteins were expressed from the plasmid pGEX-2TK in E. coli SURE2 cells by using the GST Gene Fusion System and the protocols of the manufacturer (Pharmacia). The labeling of the fusion proteins with [γ-32P]ATP (6,000 Ci/mmol; NEN Dupont) and far-Western blotting were as described previously (7, 21). Briefly, GST fusion proteins containing a phosphorylation site were radiolabeled by using an in vitro kinase reaction. The probe was purified by using GST-Sepharose beads, and nitrocellulose filters were incubated with the probe overnight at 4°C. Filters were then washed extensively, dried, and exposed to film.
BAP binding assays.
DNA fragments of the VSV and rabies virus M genes encoding amino acids 17 through 33 and 29 through 44, respectively, were amplified by PCR and inserted in-frame with bacterial alkaline phosphatase (BAP) into vector pMY101 (generously provided by B. Kay, University of Wisconsin) digested with SalI/XbaI. All inserts and mutations were confirmed by DNA sequencing. BAP fusion proteins were expressed in E. coli DH5alpha following induction with 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Boehringer Mannheim Biochemicals) for 4 h at 37°C. Briefly, following induction, the cells were pelleted at 5,000 rpm, resuspended in 1× phosphate-buffered saline (PBS), and sonicated briefly to disrupt the cells. Triton X-100 (1.0%) was added, and the lysates were rocked at RT for 30 min. Cell debris was pelleted at 10,000 rpm for 10 min, and the supernatant fraction containing the fusion protein was recovered. Enzymatic activity was assayed by serially diluting the fusion protein in AP buffer (0.1 M Tris-HCl [pH 9.4], 0.1 M NaCl, 50 mM MgCl2) containing 50 mg of nitroblue tetrazolium chloride (NBT) per ml and 50 mg of 5-bromo-4-chloro-3-indolylphosphate toluidinium (BCIP; Gibco BRL) per ml. Nitrocellulose filters containing WW domain fusion proteins were blocked with Superblock (Pierce Biochemicals) for 2 h at RT and then incubated with the BAP fusion protein overnight at 4°C. Filters were washed extensively in 1× PBS with 0.1% Triton X-100 and then incubated in AP buffer containing NBT and BCIP for 15 to 30 min at RT.
Budding assay.
The budding assay was essentially performed as described by Justice et al. (20). Briefly, 35-mm-diameter dishes of CV-1 cells were infected with VVT7 (generously provided by B. Moss, National Institutes of Health) and then transfected with the appropriate plasmid by using the DOTAP reagent (Boehringer Mannheim Corporation). At 2 h posttransfection the cells were metabolically labeled with 150 μCi of [35S]Met-Cys (NEN Dupont), and the cells and media were harvested at various times posttransfection. Cells were lysed in RIPA buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1.0% Nonidet P-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]), while 900 μl of medium was added to 100 μl of 10× NTE buffer (0.5 M Tris-HCl [pH 7.5], 1.5 M NaCl, 1.0% Nonidet P-40, 10 mM EDTA, 2.5% gelatin, and 0.2 M sodium azide). Immunoprecipitation of both cells and clarified media was performed by using either polyclonal antiserum directed against VSV M (kindly provided by Z. Ye, National Institutes of Health) or VSV virions (American Type Culture Collection [ATCC]). Protein samples were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE) and visualized by autoradiography.
Indirect immunofluorescence.
Subcellular localization of the VSV M protein was accomplished by indirect immunofluorescence. CV-1 cells expressing the M protein of VSV were fixed and permeabilized for 15 min in 2.5% formaldehyde–0.5% Triton X-100–PBS. The primary antibody was polyclonal anti-VSV M, while the secondary antibody was affinity-purified goat anti-rabbit antibody conjugated to fluorescein isothiocyanate (Boehringer Mannheim Corporation). Positive cells were visualized with the use of a Leica CLSM confocal microscope.
RESULTS
Conservation of the PY motif in the M protein of rhabdoviruses.
A novel globular domain which mediates protein-protein interactions was identified recently and shown to be present in a wide range of cellular proteins involved in signal transduction, gene regulation, and cytoskeletal formation (48). This domain, termed WW domain, is 38 to 40 amino acids long and contains a number of conserved amino acids, including two highly conserved tryptophans spaced 20 to 22 amino acids apart (48). The WW domain was shown to interact with a polyproline ligand having the core consensus sequence PPxY (9, 14). Of interest to us is the fact that this PPxY motif is highly conserved in the M proteins of various rhabdoviruses (Table 1 and references 16, 23, 42, and 43). Not only is the primary sequence conserved but also the relative location within the N termini of these M proteins is maintained (Table 1). In addition to the rhabdoviruses, the putative matrix proteins (VP40) of both Ebola and Marburg viruses (filoviruses that were initially classified as rhabdoviruses) also contain the PPxY motif at their amino termini (Table 1 and references 6 and 44). The highly conserved nature of the sequence and location of the PPxY motif within these viral structural proteins implies that the PY motif is important in the structure and/or function of these major structural proteins.
TABLE 1.
Conservation of the PY motif within the matrix proteins of various rhabdoviruses and filoviruses
| Virus | Protein sequencea | Positionb | GenBank accession no. |
|---|---|---|---|
| VSV (Indiana) | KLGIA PPPY EEDTS | 24–27 | X04452 |
| VSV (New Jersey) | KKMGL PPPY DESCP | 24–27 | M14553 |
| Rabies virusc | DLWLP PPEY VPLKE | 35–38 | M31046 |
| Piry virus | MEWES PPSY NEIKS | 33–36 | D26175 |
| Spring viremia of carp virus | KSKGT PPTY EETLA | 17–20 | K02123 |
| Ebola virus (Zaire strain) | ILPTA PPEY MEAIY | 10–13 | L11365 |
| Marburg virus (Popp strain) | MQYLN PPPY ADHGA | 16–19 | Z29337 |
The sequences are derived from the matrix (M) proteins of VSV, rabies virus, Piry virus, and spring viremia of carp virus and from the VP40 proteins of Ebola and Marburg viruses.
The numbers correspond to amino acid positions of the PY motif within the protein beginning from the N terminus.
This sequence (14 amino acids) of the rabies virus M protein is perfectly conserved in the strains SAD B19, CVS, Nishigahara, ERA, and PV.
VSV M protein interacts with cellular WW domains in vitro.
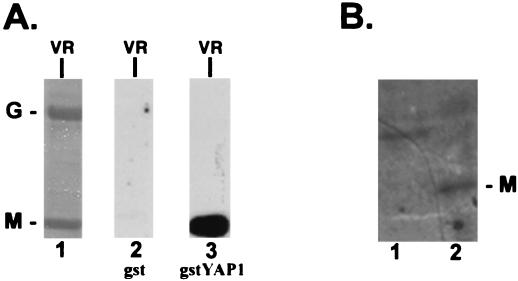
Since the highly conserved PY motif present within rhabdoviral M proteins is identical to the sequence of the ligand which interacts with WW domains, we wanted to determine whether the M protein of VSV could interact with WW domains of cellular proteins. To obtain the M protein from VSV virions, BHK-21 cells were infected with VSV and progeny virions were purified from the supernatant as described (see Materials and Methods). The virion preparation was detergent treated, and the viral proteins were analyzed by SDS-PAGE. As expected, the detergent-soluble sample contained predominantly the two viral envelope-associated proteins M and G (Fig. 1, lane 1). The M and G virion proteins were probed with either GST alone or GST fused to the WW domain of YAP (gstYAPWW1) in a far-Western assay (Fig. 1A, lanes 2 and 3). A strong interaction was observed for gstYAPWW1 with the VSV M protein but not with the VSV G protein. No signal was observed when GST alone was used as the probe (Fig. 1A, lane 2). In addition to virion proteins (VR), cell extracts from mock-infected (Fig. 1B, lane 1) or VSV-infected (Fig. 1B, lane 2) BHK-21 cells were probed with 32P-labeled gstYAPWW1 (Fig. 2). A protein of 30 kDa (VSV M) was detected in VSV-infected cell extracts (Fig. 1B, lane 2) and not in mock-infected cell extracts (Fig. 1B, lane 1). The amount of M protein (approximately 0.1 μg) present in the lane for VSV-infected cells (Fig. 1B, lane 2) is approximately 20 times lower than that shown for purified viral proteins (Fig. 1A, lanes 1, 2, and 3) based on comparison with a known standard of purified virus from Coomassie blue-stained gels (data not shown). A protein of approximately 38 kDa was observed in the mock-infected cell extract probed with gstYAPWW1 (Fig. 1B, lane 1). The presence of this cellular protein was not unexpected since it has been described previously as WBP-2, a cellular protein of unknown function that interacts with the WW domain of YAP (7). Interestingly, WBP-2 was not observed in the VSV-infected cell extract (Fig. 1B, lane 2). This experiment has been repeated three times, and the same result was obtained each time. The potential role of VSV M protein in mediating the apparent absence of WBP-2 in VSV-infected cells is currently under investigation. Neither WBP-2 nor VSV M was detected when an identical blot was probed with 32P-labeled GST protein (data not shown). These data indicate that full-length M protein from VSV virions as well as from VSV-infected cell extracts interacts with WW domain 1 of YAP.
FIG. 1.
Far-Western analysis of VSV virion proteins. (A) Lane 1, Coomassie brilliant blue stain of detergent-soluble VSV virion proteins separated on an SDS–10% polyacrylamide gel (5.0 μg of total protein). G, glycoprotein; M, matrix protein. Lane 2, nitrocellulose filter containing an identical amount of G and M as shown in the Coomassie blue-stained gel in lane 1. This filter was probed with 32P-labeled GST alone. Lane 3, a second nitrocellulose filter containing an identical amount of G and M as shown in the Coomassie blue-stained gel in lane 1. This filter was probed with 32P-labeled gstYAPWW1. (B) Far-Western analysis of mock-infected (lane 1) and VSV-infected (lane 2) cell extracts. Cells were lysed in standard radioimmunoprecipitation assay buffer. Approximately 105 cell equivalents of mock- and VSV-infected cell extracts were loaded per lane. The amount of M protein present in lane 2 is approximately one-twentieth of the amounts loaded in panel A, lanes 1, 2, and 3.
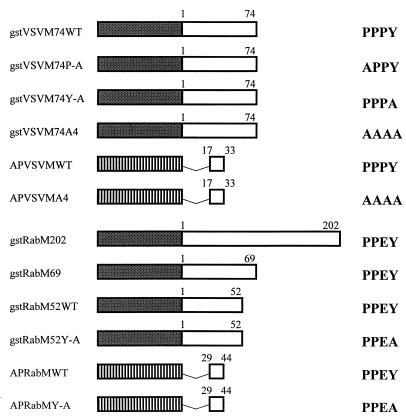
FIG. 2.
Diagram of VSV and rabies virus M fusion proteins. The shaded box represents the GST moiety which was joined in-frame to the indicated number of amino acids from VSV or rabies virus M proteins. The striped box represents the BAP protein joined in-frame to the indicated amino acids from VSV or rabies virus M proteins. The sequence of the PY motif within each of the fusion proteins is shown at the right. The designation WT refers to the PY motif and not the full-length protein. The name of each fusion protein is listed on the left. The construct gstRabM202 contains the full-length rabies virus M protein.
The N terminus of VSV M is sufficient to interact with cellular WW domains.
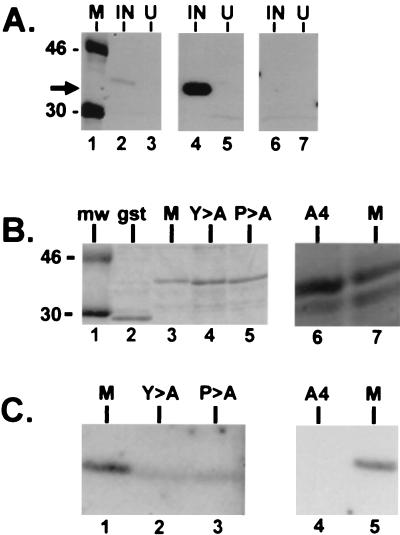
To determine whether the N terminus of VSV M protein, which contains the PPxY motif, was sufficient to mediate the interaction with the WW domains of YAP, the full-length M gene of VSV was first cloned by RT-PCR. Several plasmids were then constructed to express various lengths of the N terminus of VSV M fused to either GST or BAP (Fig. 2). The PPxY motif, which begins at amino acid position 24, was unmodified in plasmid gstVSVM74WT (Fig. 2). Plasmids gstVSVM74P-A, gstVSVM74Y-A, and gstVSVM74A4 were engineered to express proteins with mutations in the PPxY motif (Fig. 2). All of the GST fusion proteins were expressed in E. coli grown under inducing conditions. Equivalent amounts of induced or uninduced bacterial extracts were separated by SDS-PAGE, immobilized onto nitrocellulose filters, and probed with gstYAPWW2 (Fig. 3A, lanes 2 and 3), gstYAPWW1 (lanes 4 and 5), or GST alone (lanes 6 and 7). The gstVSVM74WT fusion protein interacted with both WW domains 1 and 2 from the mouse YAP. A reproducibly stronger interaction was observed with WW domain 1 (Fig. 3A, compare lanes 2 and 4). A similar preference for WW domain 1 was also observed when full-length M protein from purified virions and VSV-infected cell extracts was assayed by far-Western analysis (data not shown). The gstVSVM74WT fusion protein did not interact with GST alone (Fig. 3A, lane 6).
FIG. 3.
Far-Western analysis of gstVSVM74WT and mutants. (A) Three nitrocellulose filters containing equivalent amounts of protein extract (determined by Coomassie blue staining) from E. coli grown under inducing (IN) or noninducing (U) conditions. The filters were probed with radiolabeled gstYAPWW2 (lanes 1, 2, and 3), gstYAPWW1 (lanes 4 and 5), and GST alone (lanes 6 and 7). The arrow indicates the position of gstVSVM74WT. M, 14C-labeled protein standards. (B) Coomassie blue-stained gels containing approximately 1.0 μg of GST alone (lane 2), gstVSVM74WT (lane 3), gstVSVM74Y-A (lane 4), and gstVSVM74P-A (lane 5). In a separate experiment approximately 1.0 μg of gstVSVM74A4 (lane 6) and gstVSVM74WT (lane 7) were also stained with Coomassie blue. (C) Far-Western analysis of the same amount of protein (approximately 1.0 μg/lane) as shown in panel B. The radiolabeled probe was gstYAPWW2. The signals obtained with mutants carrying Y→A (Y>A) and P→A (P>A) in lanes 2 and 3 of panel C were reduced by 90% as compared to that of the wild type (lane 1). No signal was obtained with mutant A4 (lane 4).
To demonstrate further that the PY motif present within the N terminus of VSV M was responsible for this interaction, the fusion proteins containing point mutations within the PY motif were used in a similar far-Western blotting assay. Mutations of the first P or Y in PPxY (substitutions of alanine) have been shown to result in a decrease in the efficiency of binding to WW domains (7, 9). The wild-type and mutant GST fusion proteins were expressed to equivalent levels in E. coli (Fig. 3B) and probed with gstYAPWW2 (Fig. 3C). As expected, WW domain 2 of YAP interacted with gstVSVM74WT (Fig. 3C, lanes 1 and 5). The ability of WW domain 2 of YAP to interact with either of the point mutants was reduced substantially by 90% (Fig. 3C, lanes 2 and 3). This interaction was abolished completely when gstVSVM74A4 was probed with gstYAPWW2 (Fig. 3C, lane 4). The gstYAPWW2 probe did not interact with GST alone, and the GST probe did not interact with the wild-type or mutant gstVSVM74 fusion proteins (data not shown).
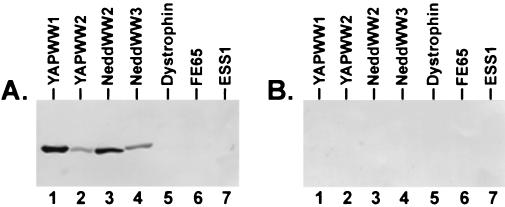
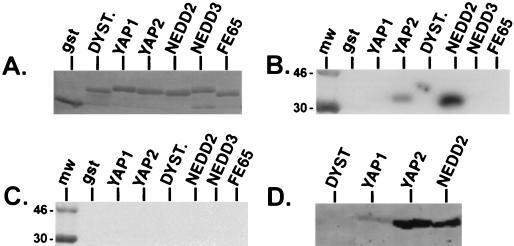
A second assay to detect protein-protein interactions was used to delineate further the minimal region of VSV M required to interact with WW domains and corroborate the specificity of the interactions demonstrated by far-Western analysis. Amino acids 17 to 33 of VSV M were joined in frame to BAP and expressed from constructs APVSVMWT (WT PPPY motif) and APVSVMA4 (PPPY was changed to AAAA; Fig. 2). Both the wild-type and mutant proteins were purified from E. coli, and equal amounts of protein were used to probe a panel of gstWW domain fusion proteins immobilized on nitrocellulose filters (Fig. 4). Amino acids 17 to 33 of VSV M containing the wild-type PY motif interacted with WW domains 1 and 2 from YAP as well as with WW domains 2 and 3 from the Nedd4 protein (Fig. 4A, lanes 1 to 4). APVSVMWT did not interact with the WW domains from dystrophin, FE65, and ESS1 (Fig. 4A, lanes 5, 6, and 7, respectively). Also, no interactions were observed when identical amounts of the gstWW domain fusion proteins were probed with APVSVMA4 (Fig. 4B, lanes 1 to 7). These results demonstrate that amino acids 17 to 33 of VSV M are sufficient to interact with specific WW domains and that the PY motif is essential for this interaction to occur. Last, the PY motif of VSV M not only interacts with select WW domains but also exhibits a preference for one domain over another (Fig. 3A, compare lanes 2 and 4; Fig. 4A, compare lanes 1 and 2).
FIG. 4.
BAP protein binding assay. Equivalent amounts (1.0 μg/lane) of the purified gstWW domain fusion proteins indicated were immobilized onto nitrocellulose filters and probed with the BAP fusion proteins APVSVMWT (panel A) or APVSVMA4 (panel B).
Rabies virus M protein interacts with cellular WW domains in vitro.
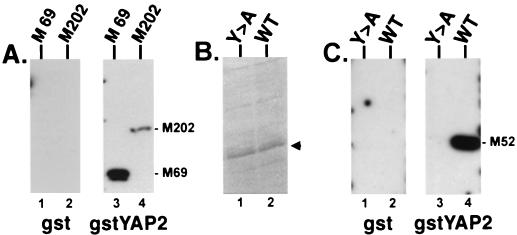
To determine whether a second rhabdoviral M protein could interact with WW domains, the full-length rabies virus M protein or various C-terminal truncations of rabies virus M protein were fused to GST (see Fig. 2). The fusion protein gstRabM52Y-A is identical to gstRabM52WT except for a single point mutation in the PY motif of the rabies virus M protein, which changes the tyrosine to an alanine (see Fig. 2). All four gstRabM fusion proteins were expressed in E. coli grown under inducing conditions and used in far-Western blotting assays (Fig. 5). Duplicate filters with gstRabM202 (containing full-length rabies virus M protein) and gstRabM69 were probed by using either GST alone or gstYAPWW2 (Fig. 5A). Both gstRabM202 and gstRabM69 fusion proteins interacted with the gstYAPWW2 probe (Fig. 5A, lanes 3 and 4) but not with GST alone (Fig. 5A, lanes 1 and 2). Identical amounts of gstRabM52WT and gstRabM52Y-A fusion proteins (as shown in Fig. 5B) were also probed with either GST alone or gstYAPWW2 (Fig. 5C). The gstRabM52WT fusion protein interacted with gstYAPWW2 (Fig. 5C, lane 4) but not with GST alone (lane 2). In contrast, a single point mutation within the PPxY motif in protein gstRabM52Y-A completely abolished the interaction with the YAP WW domain (Fig. 5C, lane 3).
FIG. 5.
Far-Western analysis of GST-rabies virus M fusion proteins. (A) Duplicate nitrocellulose filters with gstRabM69 (M 69; lanes 1 and 3) and gstRabM202 (containing the full-length M protein M202; lanes 2 and 4) were probed with either GST alone (lanes 1 and 2) or gstYAPWW2 (lanes 3 and 4). (B) Coomassie brilliant blue stain of bacterial cell extracts expressing approximately 1.0 μg of gstRabM52Y-A (Y>A; lane 1) or gstRabM52WT (WT [wild type]; lane 2) indicated by the arrowhead. (C) Identical amounts (1.0 μg/lane) of gstRabM52Y-A and gstRabM52WT to those seen in panel B were immobilized onto duplicate nitrocellulose filters and probed with either GST alone (lanes 1 and 2) or gstYAPWW2 (lanes 3 and 4).
To examine further the rabies virus M protein and WW domain interaction, the reverse of the experiment shown in Fig. 5C was performed. The rabies virus fusion proteins were now purified, quantitated, radiolabeled, and used to probe a panel of gstWW domain fusion proteins (Fig. 6). In addition to GST alone, equivalent amounts (as shown in Fig. 6A) of gstYAPWW1, gstYAPWW2, gstDystrophinWW, gstNedd4WW2, gstNedd4WW3, and gstFE65WW fusion proteins were probed with either gstRabM52WT (Fig. 6B) or gstRabM52Y-A (Fig. 6C). As expected, gstRabM52WT interacted with WW domain 2 and WW domain 1 (upon longer exposure of the filter) of YAP (Fig. 6B). Interestingly, gstRabM52WT also interacted strongly with WW domain 2 from the Nedd4 protein (Fig. 6B). The gstRabM52WT protein did not interact with the remaining gstWW domain fusion proteins, again demonstrating that there is specificity in this protein-protein interaction. As in the previous experiment, the interactions between the rabies virus M protein and the various WW domains observed (Fig. 6B) were completely abolished by the introduction of a single point mutation in the PPxY motif in the gstRab52MY-A protein (Fig. 6C).
FIG. 6.
Far-Western and BAP binding analyses of rabies virus M fusion proteins. Identical amounts (1.0 μg/lane; shown by Coomassie blue stain) (A) of GST, gstYAPWW1 (YAP1), gstYAPWW2 (YAP2), gstDystrophinWW (DYST), gstNedd4WW2 (NEDD2), gstNedd4WW3 (NEDD3), and gstFE65WW (FE65) were immobilized onto nitrocellulose and probed with either gstRabM52WT (B) or gstRabM52Y-A (C). Identical amounts (1.0 μg/lane) of the four gstWW domain fusion proteins indicated were immobilized on nitrocellulose and probed with APRabMWT (D). mw, radiolabeled protein standards.
To delineate further the minimal region of rabies virus M protein required for interaction with WW domains, amino acids 29 to 44 of rabies virus M protein were joined in frame to BAP to generate plasmids expressing the wild-type fusion protein (APRabMWT) or mutant fusion protein (APRabMY-A) (Fig. 2). The APRabMWT protein was able to interact strongly with WW domain 2 of YAP and Nedd4 and weakly with WW domain 1 of YAP (Fig. 6D). No interaction was observed with the WW domain of dystrophin (Fig. 6D). In contrast, the APRabMY-A protein was unable to interact with any of the WW domains (data not shown). These results demonstrate that 15 amino acids (29 to 44) of the rabies virus M protein containing the PY motif were sufficient to interact with specific WW domains.
The PY motif of VSV M facilitates exocytosis.
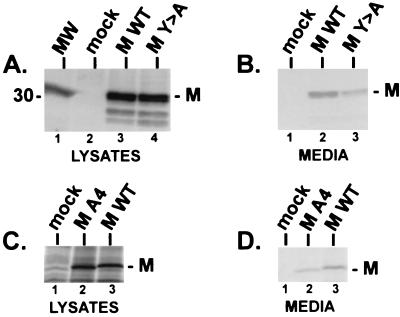
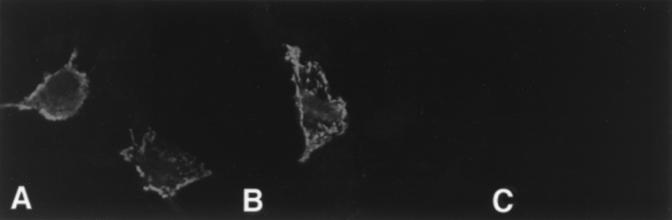
To determine whether the PY motif of VSV M plays a role in M-mediated release or budding, we took advantage of a previously described assay to measure membrane vesiculation and exocytosis of M protein from mammalian cells (20). Justice et al. (20) demonstrated that the VSV M protein, when expressed alone in mammalian cells, can induce the formation and release of lipid vesicles from the cell surface; the process may mimic virion assembly and budding from host cells but is clearly not identical to the formation and budding of the classic bullet-shaped virions. This phenomenon of M-mediated release was also observed when M protein of VSV was expressed from a baculovirus vector (29). The exocytosis assay in CV-1 cells was utilized to determine whether the PPxY motif of VSV M protein is important in M-mediated release. CV-1 cells were first infected with the recombinant vaccinia virus (VVT7) expressing the bacteriophage T7 polymerase and then transfected with plasmid pT7VSVMWT (expressing full-length, wild-type M protein), plasmid pT7VSVMY-A (identical to wild-type M except for a single point mutation within the PY motif resulting in a change from tyrosine to alanine), or no DNA (mock transfected). Both the cells and media were harvested and subjected to immunoprecipitation by using polyclonal anti-VSV M antiserum (Fig. 7). Identical amounts of both wild-type and mutant VSV M proteins were observed in the cell lysates (Fig. 7A, lanes 3 and 4), while no M protein was detected in mock-transfected cells (Fig. 7A, lane 2). In contrast, the amount of the mutant M protein (Fig. 7B, lane 3) released into the media was reduced in multiple independent experiments by about 70% (3.5-fold) as compared to the amount of wild-type VSV M protein in the media (Fig. 7B, lane 2). Similar results were obtained when a mutant M protein containing four alanines in place of PPxY was used in this assay (Fig. 7C and D). Again, identical amounts of wild-type and mutant M proteins were observed in the cell lysates (Fig. 7C, lanes 2 and 3); however, the amount of the A4 mutant protein present in the media was significantly reduced as compared to wild-type protein levels measured in multiple independent experiments (Fig. 7D, lanes 2 and 3). These results suggest that the PPxY motif of VSV M is important for M-mediated release in this assay and may reflect a similar role for the PPxY motif of VSV M in the release and budding of intact virions. To determine whether the defect in release of the mutant M proteins was due to their inability to localize to the cell membrane, indirect immunofluorescence and confocal microscopy were performed on cells expressing either the wild-type or mutant (Y-A) M protein (Fig. 8). Transfected cells were examined at 5, 8, and 10 h posttransfection (data not shown for 5- and 10-h time points). For all time points tested, the wild-type and mutant M proteins localized to the cytoplasmic membrane equally well (Fig. 8). These data indicate that the defect in budding of the mutant M protein (as shown in Fig. 7B) is not simply due to mislocalization within the cell and is likely due to a defect in a late step of the release process.
FIG. 7.
VSV M budding assay. (A) Radiolabeled lysates from CV-1 cells receiving no DNA (mock, lane 2), T7VSVMWT DNA (MWT, lane 3), and T7VSVMY-A DNA (MY>A, lane 4) were immunoprecipitated with polyclonal antiserum against the M protein of VSV and fractioned by SDS-PAGE. The position of the M protein of VSV is indicated. MW, 14C-labeled protein standards. (B) Radiolabeled proteins released into the media covering cells transfected with no DNA (mock, lane 1), T7VSVMWT DNA (lane 2), and T7VSVMY-A DNA (lane 3) were immunoprecipitated with polyclonal antiserum against the M protein of VSV and fractionated by SDS-PAGE. The position of the M protein of VSV is indicated. (C) Radiolabeled lysates from CV-1 cells receiving no DNA (mock, lane 1), T7VSVMA4 DNA (lane 2), and T7VSVMWT DNA (lane 3) were immunoprecipitated with polyclonal antiserum raised against VSV virions (ATCC) and fractionated by SDS-PAGE. (D) Radiolabeled proteins released into the media covering cells transfected with no DNA (mock, lane 1), T7VSVMA4 (lane 2), and T7VSVMWT (lane 3) were immunoprecipitated with polyclonal antiserum raised against VSV virions (ATCC) and fractionated by SDS-PAGE.
FIG. 8.
Indirect immunofluorescence and confocal microscopy of transfected CV-1 cells. (A) CV-1 cells expressing wild-type VSV M protein at 8 h posttransfection. (B) CV-1 cells expressing the VSV M protein containing a tyrosine (Y) to alanine (A) mutation within the PY motif at 8 h posttransfection. (C) Untransfected CV-1 cells. Primary polyclonal antiserum (identical to that used in the experiment shown in Fig. 7A and B) was directed against the M protein of VSV.
DISCUSSION
Much progress has been made in studying the assembly and budding pathways of negative-sense RNA viruses in general; however, many questions remain concerning the role of both viral and host proteins in the late stages of the viral life cycle. Toward a better understanding of the role of M protein in rhabdoviral assembly and egress, we have demonstrated in this report that: (i) a highly conserved PY motif within the M protein of VSV and rabies virus can function as a ligand which interacts in vitro with WW domains of specific cellular proteins, and (ii) this same PY motif of VSV M protein may be functionally important for the budding process. Taken together, these findings suggest that rhabdoviral budding mediated by the M protein may be facilitated by virus-host interactions.
The polyproline ligand which interacts with the WW domain has been identified and well characterized as having the core consensus sequence PPxY. While the M proteins of many rhabdoviruses maintain the PY motif at their amino termini (Table 1), it should be noted that the M proteins from several rhabdoviruses of fish possess a PPxH (where H denotes histidine) motif rather than PPxY (2). Although the aromatic nature of the amino acid position occupied by H rather than Y is maintained, it is not clear whether this PPxH motif can interact with either WW domains or perhaps a WW-like domain. Nevertheless, WW domains from YAP and the Nedd4 protein interacted strongly and specifically with the PY motifs of both VSV and rabies virus M proteins, whereas WW domains from other cellular proteins (dystrophin, FE65, and ESS1; for a review see reference 48) did not interact with either viral protein. Single point mutations within the PPxY motifs of VSV M protein and rabies virus M protein were sufficient to either significantly reduce or abolish interactions with cellular WW domains. Attempts were made to determine whether full-length YAP can interact with VSV M protein and whether YAP was present in mature virions. Results from these experiments were negative, suggesting that YAP is an unlikely interacting agent. However, the authentic in vivo interacting agent may possess a “YAP-like” class of WW domain.
Both the WW domain and the related SH3 domain have been implicated in mediating virus-host protein-protein interactions. The Nef protein of human immunodeficiency virus type 1 (HIV-1) possesses a polyproline ligand which has been shown to interact with Src-family SH3 domains of cellular proteins and to be important for optimal viral replication (25, 26). The LMP2 protein of Epstein-Barr virus contains two polyproline motifs that have been postulated to mediate an interaction between LMP2 and the Src-family tyrosine kinases, FYN and LYN (30). Interestingly, the PY motif is highly conserved in the Gag proteins of many animal and human retroviruses (55, 58, 59). One of the well-characterized Gag proteins in terms of functional domains important for gag-mediated budding is that of RSV (3, 4, 13, 52, 53, 55, 56, 57). Elegant studies have demonstrated that the PY motif present within the p2b protein of RSV Gag not only interacts with WW domains in vitro but also functions as a late budding domain (L domain) which is essential for a late stage in retroviral assembly and release (15, 55, 58). The Gag polyprotein of RSV, like the M protein of VSV, can bud from cells in the absence of any other viral protein. RSV Gag proteins with mutations in the PY motif exhibit a decreased ability to bud from cells, yet budding was not completely abolished (55, 58). We observed similar results with the VSV M budding assay, whereby PY mutants of VSV M were released from cells at a level significantly lower than that observed for the wild-type M protein (Fig. 8). This low level of budding of the mutant M proteins may be attributed to redundancy in the signal for budding at either the cellular (through multiple proteins and/or pathways involved in exocytosis) or viral (through multiple budding signals in the M protein) level. Indeed, the sequence PSAP at amino acids 37 to 40 of VSV M closely resembles the functional budding domain identified in the p6 protein of HIV-1 (for which the sequence is PT/SAPP) (18, 19).
In addition to the RSV Gag protein, late budding domains have been identified in the Gag proteins of HIV-1, equine infectious anemia virus, and recently Mason-Pfizer monkey virus (17, 19, 41, 59). Whether these retroviral L domains actually mediate their function by interacting with cellular proteins remains to be determined. Our findings obtained with the M protein of rhabdoviruses are the first to demonstrate that the PY motifs of VSV and rabies virus M can interact with WW domains and suggest that they may function as L domains in the budding process of rhabdoviruses. Taken together, these results provide further evidence that the M protein of rhabdoviruses and the Gag protein of retroviruses are in many ways functionally equivalent. Although not presented here, it should be noted that the N-terminal 74 amino acids of VSV M protein containing the PY motif were shown recently to be capable of functionally replacing the L domain of the p2b protein of RSV in a retroviral budding assay (12). Moreover, point mutations that altered the PY motif of VSV M in these chimeric M-Gag proteins resulted in a protein that was defective in budding (12).
Should the interaction between WW domains and viral PY motifs prove to be a specific one in vivo, then this virus-host interaction could serve as a potential target for antiviral agents. Since the WW domain and the core motif of its ligand are relatively short, one could speculate that such antiviral agents could be easily selected from chemical libraries of low-molecular-weight compounds (49). Also, if indeed the PY-WW domain interaction is a common step in the assembly pathways of rhabdoviruses, retroviruses, and filoviruses, it is tempting to speculate that antivirals which target this interaction may be effective against a variety of viral pathogens, including Ebola and Marburg viruses.
The effects of mutations within the rhabdoviral PY motif on budding and release of virions are currently being tested with a reverse-genetics approach (24, 54). Also, several cellular proteins that interact with the N-terminal portions of both VSV and rabies virus M proteins are being analyzed and may provide useful information as to the identity of host proteins that are involved in rhabdoviral budding (18). It is our hope that these studies will enhance our understanding of the role of both viral and host proteins in enveloped RNA virus assembly and release.
ACKNOWLEDGMENTS
We acknowledge the generosity of W. Wunner for the rabies virus M clone, R. R. Wagner and Z. Ye for antisera against VSV M, and M. Schubert for the Indiana serotype of VSV. We also thank J. Wills and R. Craven for fruitful discussions, B. Kay for the BAP vector, and J. Lenard for critical review of the manuscript.
This work was supported by grants from the National Institutes of Health (P.P.). Confocal laser scanning microscopy was performed at the MSSM-CLSM core facility, with the support of funding from an NIH shared instrumentation grant and an NSF Major Research Instrumentation grant.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1992. [Google Scholar]
- 2.Benmansour A, Paubert G, Bernard J, DeKinkelin P. The polymerase-associated protein (M1) and the matrix protein (M2) from a virulent and an avirulent strain of viral hemorrhagic septicemia virus (VHSV), a fish rhabdovirus. Virology. 1994;198:602–612. doi: 10.1006/viro.1994.1072. [DOI] [PubMed] [Google Scholar]
- 3.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett R P, Rhee S, Craven R C, Hunter E, Wills J W. Amino acids encoded downstream of gag are not required by Rous sarcoma virus protease during Gag-mediated assembly. J Virol. 1991;65:272–280. doi: 10.1128/jvi.65.1.272-280.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bork P, Sudol M. The WW domain: a signaling site in dystrophin. Trends Biochem Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 6.Bukreyev A A, Volchkov V E, Blinou V M, Dryga S A, Netesov S V. The complete nucleotide sequence of the Popp (1967) strain of Marburg virus: a comparison with the musoke (1980) strain. Arch Virol. 1995;140:1589–1600. doi: 10.1007/BF01322532. [DOI] [PubMed] [Google Scholar]
- 7.Chen H I, Sudol M. The WW domain of YES-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H I, Sudol M. Identification and characterization of protein ligands to the WW domain by Western ligand blotting. Tech Protein Chem VII. 1996;7:3–12. [Google Scholar]
- 9.Chen H I, Einbond A, Kwak S-J, Linn H, Koepf E, Peterson S, Kelley J W, Sudol M. Characterization of the WW domain of human Yes-associated protein and its polyproline-containing ligands. J Biol Chem. 1997;272:17070–17077. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- 10.Chong L D, Rose J K. Membrane association of functional vesicular stomatitis virus matrix protein in vivo. J Virol. 1993;67:407–414. doi: 10.1128/jvi.67.1.407-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chong L D, Rose J K. Interaction of normal and mutant vesicular stomatitis virus matrix proteins with the plasma membrane and nucleocapsids. J Virol. 1994;68:441–447. doi: 10.1128/jvi.68.1.441-447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craven R C, Harty R N, Paragas J, Palese P, Wills J W. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J Virol. 1999;73:3359–3365. doi: 10.1128/jvi.73.4.3359-3365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craven R C, Leure-duPree A E, Erdie C R, Wilson C B, Wills J W. Necessity of the spacer peptide between CA and NC in the Rous sarcoma virus Gag protein. J Virol. 1993;67:6246–6252. doi: 10.1128/jvi.67.10.6246-6252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Einbond A, Sudol M. Towards prediction of cognate complexes between the WW domain and proline-rich ligands. FEBS Lett. 1996;384:1–8. doi: 10.1016/0014-5793(96)00263-3. [DOI] [PubMed] [Google Scholar]
- 15.Garnier L, Wills J W, Verderame M F, Sudol M. WW domains and retrovirus budding. Nature. 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 16.Gill D S, Banerjee A K. Complete nucleotide sequence of the matrix protein mRNA of vesicular stomatitis virus (New Jersey serotype) Virology. 1986;150:308–312. doi: 10.1016/0042-6822(86)90293-x. [DOI] [PubMed] [Google Scholar]
- 17.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harty, R. N., J. P. Paragas, and P. Palese. Unpublished data.
- 19.Huang M, Orenstein J M, Martin M, Freed E O. p6 Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Justice P A, Sun W, Li Y, Ye Z, Grigera P R, Wagner R R. Membrane vesiculation function and exocytosis of wild-type and mutant matrix proteins of vesicular stomatitis virus. J Virol. 1995;69:3156–3160. doi: 10.1128/jvi.69.5.3156-3160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaelin W G, Jr, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Yue L, Farnham P J, Blanar M A, Livingston D M, Flemington E K. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2f-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 22.Kaptur P E, McKenzie M O, Wertz G W, Lyles D S. Assembly functions of vesicular stomatitis virus matrix protein are not disrupted by mutations at major sites of phosphorylation. Virology. 1995;206:894–903. doi: 10.1006/viro.1995.1012. [DOI] [PubMed] [Google Scholar]
- 23.Kiuchi A, Roy P. Comparison of the primary sequence of spring viremia of carp virus M protein with that of vesicular stomatitis virus. Virology. 1984;134:238–243. doi: 10.1016/0042-6822(84)90290-3. [DOI] [PubMed] [Google Scholar]
- 24.Lawson N, Stillman E A, Whitt M A, Rose J K. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C-H, Leung B, Lemmon M A, Zheng J, Cowburn D, Kuriyan J, Saksela K. A single amino acid in the SH3 domain of Hck determines its high affinity and specificity in binding to HIV-1 Nef protein. EMBO J. 1995;14:5006–5015. doi: 10.1002/j.1460-2075.1995.tb00183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee C-H, Saksela K, Mirza U A, Chait B T, Kuriyan J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 1996;85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- 27.Lenard J. Negative-strand virus M and retrovirus MA proteins: all in a family? Virology. 1996;216:289–298. doi: 10.1006/viro.1996.0064. [DOI] [PubMed] [Google Scholar]
- 28.Lenard J, Vanderoef R. Localization of the membrane-associated region of vesicular stomatitis virus M protein at the N terminus, using the hydrophobic, photoreactive probe 125I-TID. J Virol. 1990;64:3486–3491. doi: 10.1128/jvi.64.7.3486-3491.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Luo L, Schubert M, Wagner R R, Kang C Y. Viral liposomes released from insect cells infected with recombinant baculovirus expressing the matrix protein of vesicular stomatitis virus. J Virol. 1993;67:4415–4420. doi: 10.1128/jvi.67.7.4415-4420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longnecker R, Druker B, Roberts T M, Kieff E. An Epstein-Barr virus protein associated with cell growth transformation interacts with a tyrosine kinase. J Virol. 1991;65:3681–3692. doi: 10.1128/jvi.65.7.3681-3692.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyles D S, McKensie M, Parce J W. Subunit interactions of vesicular stomatitis virus envelope glycoprotein stabilized by binding to viral matrix protein. J Virol. 1992;66:349–358. doi: 10.1128/jvi.66.1.349-358.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macias M J, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Structure of the WW-domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 33.McCreedy B J, Jr, Lyles D S. Distribution of M protein and nucleocapsid protein of vesicular stomatitis virus in infected cell plasma membranes. Virus Res. 1989;14:189–205. doi: 10.1016/0168-1702(89)90001-4. [DOI] [PubMed] [Google Scholar]
- 34.Mebatsion T, Konig M, Conzelmann K-K. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- 35.Newcomb W W, Tobin G J, McGowan J J, Brown J C. In vitro reassembly of vesicular stomatitis virus skeletons. J Virol. 1982;41:1055–1062. doi: 10.1128/jvi.41.3.1055-1062.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pal R, Wagner R R. Rhabdovirus membrane and maturation. In: Wagner R R, editor. The rhabdoviruses. New York, N.Y: Plenum; 1987. pp. 75–128. [Google Scholar]
- 37.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peeples M E. Paramyxovirus M proteins: pulling it all together and taking it on the road. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 427–456. [Google Scholar]
- 39.Pirozzi G, McConell S J, Uveges A J, Carter J M, Sparks A B, Kay B K, Fowlkes D M. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J Biol Chem. 1997;272:14611–14616. doi: 10.1074/jbc.272.23.14611. [DOI] [PubMed] [Google Scholar]
- 40.Plant P J, Yeger H, Staub O, Howard P, Rotin D. The C2 domain of the ubiquitin protein ligase Nedd4 mediates Ca-dependent plasma membrane localization. J Biol Chem. 1997;272:32329–32336. doi: 10.1074/jbc.272.51.32329. [DOI] [PubMed] [Google Scholar]
- 41.Puffer B A, Parent L J, Wills J W, Montelaro R C. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rayssiguier C, Cioe L, Withers E, Wunner W H, Curtis P J. Cloning of the rabies virus matrix protein mRNA and determination of its amino acid sequence. Virus Res. 1986;5:177–190. doi: 10.1016/0168-1702(86)90016-x. [DOI] [PubMed] [Google Scholar]
- 43.Rose J K, Gallione C J. Nucleotide sequences of the mRNAs encoding the vesicular stomatitis virus G and M proteins determined from cDNA clones containing the complete coding regions. J Virol. 1981;39:519–528. doi: 10.1128/jvi.39.2.519-528.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez A, Kiley M P, Holloway B P, Auperin D D. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 1993;29:215–240. doi: 10.1016/0168-1702(93)90063-s. [DOI] [PubMed] [Google Scholar]
- 45.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staub O, Dho S, Henry P C, Correa J, Ishikawa T, McGlade J, Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle’s syndrome. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 47.Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the YES proto-oncogene product. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- 48.Sudol M. Structure and function of the WW domain. Prog Biophys Mol Biol. 1996;65:113–132. doi: 10.1016/s0079-6107(96)00008-9. [DOI] [PubMed] [Google Scholar]
- 49.Sudol M. The WW domain and its proline-rich ligand in Alzheimer’s disease and muscular dystrophy. In: Anand R, Smith P, Warne P, editors. Emerging therapeutic targets. Vol. 1. London, United Kingdom: Ashley Pub. Ltd.; 1997. pp. 81–84. [Google Scholar]
- 50.Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, Huebner K, Lehman D. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module—the WW domain. J Biol Chem. 1995;270:14733–14741. doi: 10.1074/jbc.270.24.14733. [DOI] [PubMed] [Google Scholar]
- 51.Wagner R R, Rose J K. Rhabdoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1997. pp. 1121–1135. [Google Scholar]
- 52.Weldon R A, Jr, Erdie C R, Oliver M G, Wills J W. Incorporation of chimeric Gag protein into retroviral particles. J Virol. 1990;64:4169–4179. doi: 10.1128/jvi.64.9.4169-4179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weldon R A, Jr, Wills J W. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whelan S P J, Ball L A, Barr J N, Wertz G W. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wills J W, Craven R C. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 57.Wills J W, Craven R C, Weldon R A, Jr, Nelle T D, Erdie C R. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J Virol. 1991;65:3804–3812. doi: 10.1128/jvi.65.7.3804-3812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiang Y, Cameron C E, Wills J W, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yasuda J, Hunter E. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J Virol. 1998;72:4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye Z, Sun W, Suryanarayana K, Justice P, Robinson D, Wagner R R. Membrane-binding domains and cytopathogenesis of the matrix protein of vesicular stomatitis virus. J Virol. 1994;68:7386–7396. doi: 10.1128/jvi.68.11.7386-7396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zakowski J J, Petri W A, Jr, Wagner R R. Role of the matrix protein in assembling the membrane of vesicular stomatitis virus: reconstitution of matrix proteins with negatively charged phospholipid vesicles. Biochemistry. 1981;20:3902–3907. doi: 10.1021/bi00516a037. [DOI] [PubMed] [Google Scholar]
- 62.Zakowski J J, Wagner R R. Localization of membrane-associated proteins in vesicular stomatitis virus by use of hydrophobic membrane probes and cross-linking reagents. J Virol. 1980;36:93–102. doi: 10.1128/jvi.36.1.93-102.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]