Abstract
The 2010 Deepwater Horizon disaster remains one of the largest oil spills in history. This event caused significant damage to coastal ecosystems, the full extent of which has yet to be fully determined. Crude oil contains toxic heavy metals and substances such as polycyclic aromatic hydrocarbons that are detrimental to some microbial species and may be used as food and energy resources by others. As a result, oil spills have the potential to cause significant shifts in microbial communities. This study assessed the impact of oil contamination on the function of endophytic microbial communities associated with saltmarsh cordgrass (Spartina alterniflora). Soil samples were collected from two locations in coastal Louisiana, USA: one severely affected by the Deepwater Horizon oil spill and one relatively unaffected location. Spartina alterniflora seedlings were grown in both soil samples in greenhouses, and GeoChip 5.0 was used to evaluate the endophytic microbial metatranscriptome shifts in response to host plant oil exposure. Oil exposure was associated with significant shifts in microbial gene expression in functional categories related to carbon cycling, virulence, metal homeostasis, organic remediation, and phosphorus utilization. Notably, significant increases in expression were observed in genes related to metal detoxification with the exception of chromium, and both significant increases and decreases in expression were observed in functional gene subcategories related to hydrocarbon metabolism. These findings show that host oil exposure elicits multiple changes in gene expression from their endophytic microbial communities, producing effects that may potentially impact host plant fitness.
Keywords: Deepwater Horizon, GeoChip 5.0, Metatranscriptome, Saltmarsh cordgrass, PAH, Heavy metals
Introduction
Between 4 and 5 million barrels of oil were estimated to have escaped into the Gulf of Mexico following the catastrophic explosion on April 20, 2010 and subsequent sinking of the Deepwater Horizon (DWH) drilling unit (McNutt et al. 2011; U.S. Coast Guard 2011). The unprecedented scale of the spill marked the DWH disaster as the largest offshore oil spill in U.S. history. Remedial actions involved the application of chemical dispersants, controlled in situ burns, and physical removal by skimming (U.S. Coast Guard 2011). Such efforts were extensive but did not prevent oil contamination from reaching the Gulf Coast of the United States, where nearly 2100 km of coastline were affected (Deepwater Horizon Natural Resource Damage Assessment Trustees 2016).
The resulting exposure of marine and terrestrial ecosystems to crude oil, oil residues, and chemical dispersants was catastrophic. Thousands of shore-birds, turtles, and individuals of other marine and coastal species were recorded to have been directly killed by oiling (Deepwater Horizon Natural Resource Damage Assessment Trustees 2016). Beyond those species visibly affected by oiling, thousands of kilometers of coastal soils were contaminated with toxic compounds. Several studies have determined the composition of the crude oil released by the DWH spill included polycyclic aromatic hydrocarbons (PAHs), low molecular weight hydrocarbons, and heavy metals such as As, Pb, Co, Ni, Zn, Fe, and Cr, representing a significant toxicological hazard to exposed organisms (Groudeva et al. 2001; Liu et al. 2012).
Unlike animal species, floral and microbial communities cannot relocate from contaminated sites. Marine microbial communities have shown significant shifts in response to crude oil contamination, including enrichment of those species capable of degrading hydrocarbons (Doyle et al. 2018). While such species may utilize hydrocarbons as an energy source, detrimental effects to microbes from exposure to toxic metals also occur and have been reported for decades (Bååth 1989; Giller et al. 1998). Furthermore, resulting overgrowth of hydrocarbon degraders may lead to proportionate suppression of other microbial species. Community shifts may lead to far-reaching ecological disturbances should those outcompeted species be beneficial to more complex organisms. Such effects have the potential to be especially detrimental in microbe-plant interactions due to the importance microbial symbionts can represent to host plant health (Brundrett 2004; Liu et al. 2017).
Microbial species residing within plant tissues may be beneficial, parasitic, or have little to no effect on their host. Endophytes are those microbial species which neither damage plant tissue nor form membrane-bound or intracellular structures, as seen in endosymbiotic species (Reinhold-Hurek and Hurek 2011). They have been observed to increase host resistance to both pathogens and abiotic stressors, indicating that endophytic communities play a crucial role in maintaining host health (Khare et al. 2018). Further study of endophytes and their effects on host plants has the potential to reveal further benefits.
Plant tissues serve as the external environment for endophytic communities. Therefore, host plant responses to biotic and abiotic stressors have the potential to induce shifts in these communities. The DWH spill contaminated thousands of kilometers of coastal soils, affecting countless plants and their associated endophytes and representing a significant abiotic environmental stressor (Deepwater Horizon Natural Resource Damage Assessment Trustees 2016; Odukoya et al. 2019). The purpose of this study was to assess whether shifts occurred in endophytic community function arising from host exposure to DWH oil spill contamination, and, if such shifts were present, which functional gene categories were affected. A gene array, the GeoChip 5.0 (Shi et al. 2019), which targets functional genes across divergent taxonomic groups including bacteria, fungi, archaea, eukaryotes, and viruses, was used. Saltmarsh cordgrass (Spartina alterniflora) was selected as a model organism as it is a coastal marsh grass commonly found in the regions of Louisiana contaminated by the DWH spill, where it often comprises the bulk of marshland vegetation (Edwards and Mills 2005). A two-year experiment was conducted to investigate the potential effects of oil exposure on the endophyte communities within S. alterniflora host plants. Specimens of S. alterniflora were exposed to DWH crude oil residue after inoculation with microbial communities from two salt marsh locations with differing oil exposures and grown inside greenhouses at Tulane University to reduce the impact of environmental variability. While prior studies involving the impacts of the DWH on marsh plants have focused on plant transcriptomes (Alvarez et al. 2018) and endophyte community structure (Kandalepas et al. 2015), this study will be the first to utilize a comprehensive environmental metatranscriptome gene array to reveal shifts in endophyte community function.
Materials and methods
Soil sample collection and germination of plant specimens
Seeds of wild S. alterniflora were collected in southern Louisiana in November 2015 and cold stratified at 4 °C for 3 months. Stratified seeds were surface-sterilized via subsequent immersion baths of 95% ethanol for 3 min and 0.825% NaOCl for 30 min before being rinsed in sterile deionized water for 10 s. The seeds were germinated in sterile deionized water and transplanted into trays containing the prepared substrate (equal volumes of organic humus and vermiculite, which was autoclaved three times at 121 °C for 60 min). Planted trays were placed into a Conviron Model GR48 Plant Grow Room (Controlled Environments Ltd., Winnipeg, Canada) and grown under lighting and moisture conditions designed to replicate the climate of southern Louisiana (Krauss et al. 1998). Seedlings were fertilized with approximately 9 g of Scott’s Osmocote® (Marysville, Ohio) fertilizer (14:14:14 NPK) and watered with deionized water. After approximately 1 month, plant trays were relocated to a greenhouse environment and allowed to acclimate for 3 weeks. Air temperature and humidity within the greenhouse were monitored with an iButton, model DS1923 (Maxim Integrated, San Jose, CA, USA), and soil temperature was monitored with an iButton, model DS1922L. Greenhouse air temperature averaged 4 °C higher per month than those reported at the New Orleans Airport (MSY), while experimental pot soil temperatures averaged <1 °C cooler. The average greenhouse monthly humidity was 67.5%.
Soil collection and initial transplanting methods
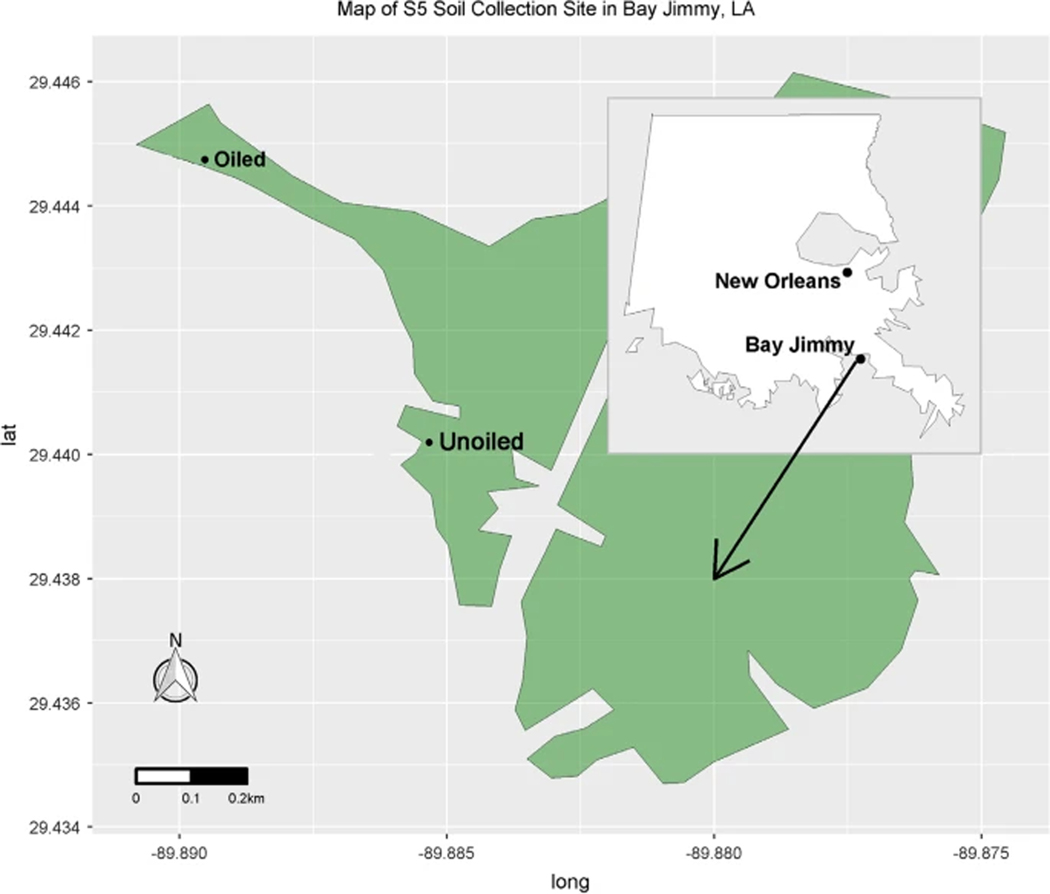
In order to inoculate the plants with endophyte communities potentially altered by oil exposure, soil samples were collected from two locations within Bay Jimmy, LA (Fig. 1) from a site reported to have been relatively unaffected by oil contamination (29.44006° N, 89.88583 W) and a site reported to have been heavily impacted (29.44464° N, 89.88959° W) by the 2010 Deepwater Horizon oil spill (Zengel et al. 2015). Each soil sample was sieved with a 1.27 cm screen constructed from steel hardware cloth (Blue Hawk, Largo, FL) and used to fill 3.8 L plastic nursery trade pots. On May 1, 2016, acclimatized S. alterniflora greenhouse specimens were transplanted into the prepared trade pots no later than 60 h after the soil samples were collected. Pot placement in the greenhouse was randomized; pots were divided into five sections oriented approximately East–West on four tables oriented approximately North–South. Plants were grown in the Bay Jimmy soil for 1 month before being transplanted into 19 L mesocosms.
Fig. 1.
Locations of soil sample collection. The landmass shape allowed for little oil contamination to reach the “unoiled” site despite close proximity. Crude oil residue was still visible at the “oiled” site at the time of sample collection
Mesocosm setup
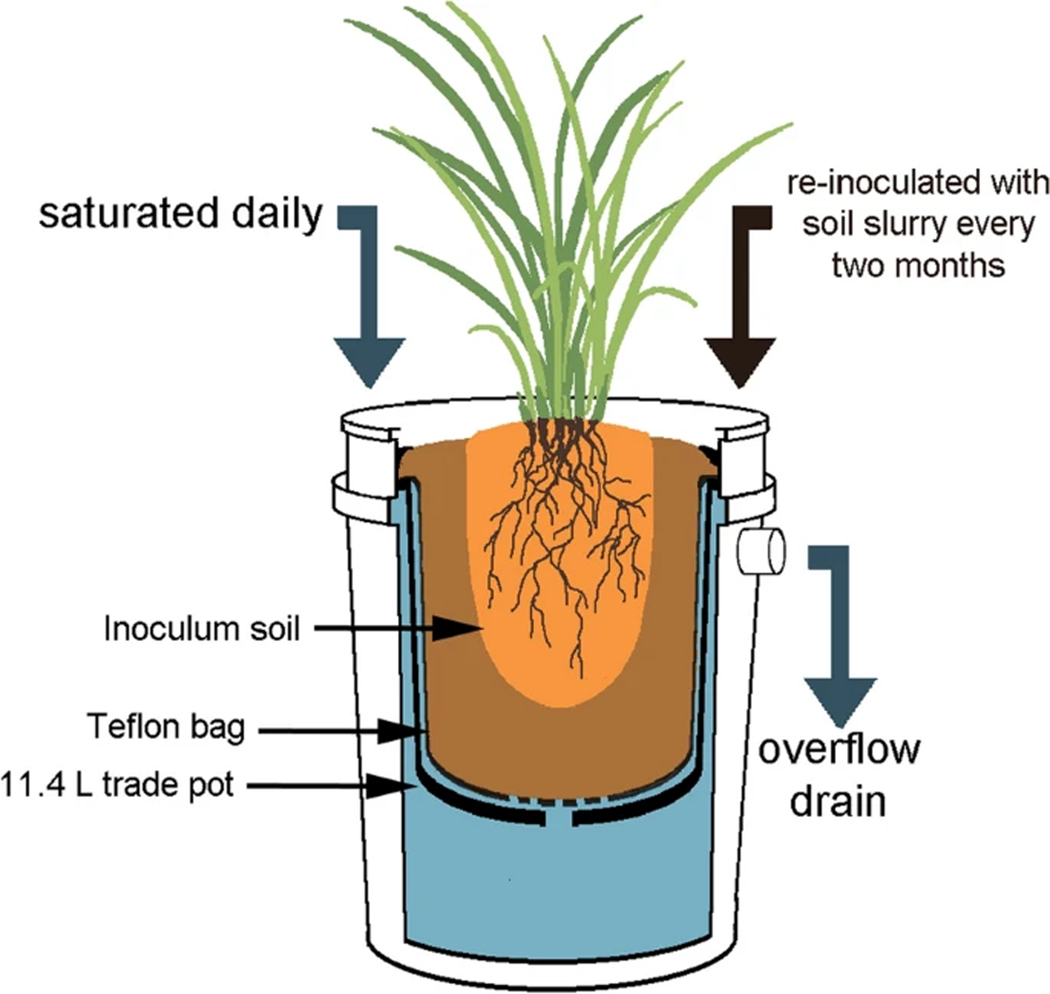
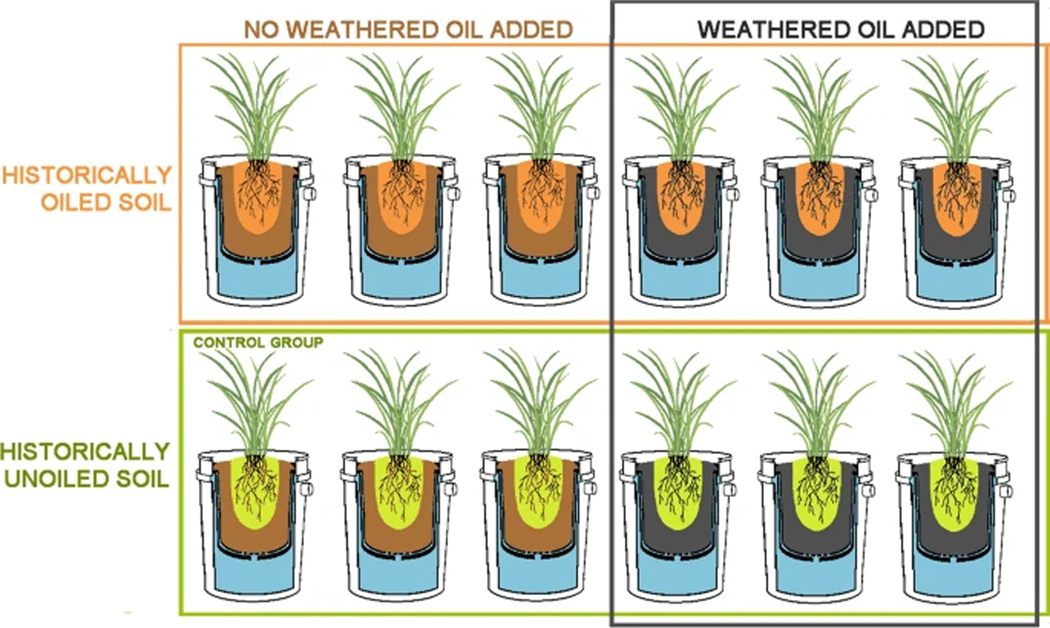
A total of 12 mesocosms were constructed to mimic estuarine conditions (Fig. 2). Each consisted of an 11.4 L plastic trade pot nested within a water-filled 19 L plastic bucket with a drainage tube installed near the top. The interiors of the trade pots were lined with Teflon bags (P-00113, Welch Fluorocarbon, Inc, Dover, NH, USA) to minimize reactions between the pot and the oil treatment. The bottom of each bag was punctured to allow drainage. The 11.4 L trade pots were filled with 7.6 L of a growth substrate consisting of 2:3 organic humus to sand (Quikrete® Play Sand, Quikrete, Atlanta, GA). Naturally-weathered oil skimmed initially from the ocean during the DWH spill was obtained from the Gulf of Mexico Research Initiative (GoMRI). All plants were transferred into mesocosms, with half of the plants in each soil group having 1.6 L of weathered oil added to each pot and mixed into the soil before the transfer. To reduce the likelihood of airborne contamination, 1.9 L of sand were then added in an even layer to the top of each 11.4 L pot. Thus, there were 3 plants per treatment, and treatments were as follows: (1) non-oiled soil, (2) non-oiled soil with weathered oil added, (3) oiled soil, and (4) oiled soil with weathered oil added (Fig. 3). Three weeks after being transplanted into the mesocosms, plants were fertilized with 8 g of Scott’s Osmocote® Plus (Marysville, Ohio), and each pot was covered with an additional 0.95 L of sand. Plants were irrigated on top of each pot with a timer-controlled drip irrigation system. The irrigation system was tested to confirm uniform flow to each pot, and the frequency of irrigation was adjusted as necessary to maintain moisture levels in the mesocosm. Each mesocosm was watered with an additional 1 L of 5 g L−1 Instant Ocean salt (Instant Ocean Spectrum Brands, Blacksburg, VA) solution once per month to simulate brackish water influx. Mesocosms were maintained by adding 4 g of fertilizer and reinoculating with 1 L of a soil slurry obtained from the original Bay Jimmy sites every 2 months. The soil slurry was sieved in the same manner as the initial soil samples and diluted to a concentration of approximately 1:10 with water from the same tap as the irrigation system before being applied to the mesocosms. Plants were occasionally misted with water to prevent salt buildup on the leaves.
Fig. 2.
Mesocosm construction and maintenance
Fig. 3.
All experimental groups
Sample collection and processing
Samples for GeoChip 5.0 analysis were collected in June 2018, approximately two years after the start of the experiment. Leaves were inspected for overall health, and approximately 5 g of whole leaves and 5 g of root pieces were collected from three plants per treatment group, for a total of twelve 5 g samples of each tissue type. Samples were immediately sealed in Ziploc bags and refrigerated. The edges of each leaf were removed, and the remaining portion was chopped into approximately 3 cm pieces using a sterilized blade. All downstream processing was conducted in a biosafety cabinet using sterile techniques. Leaf samples were submerged in subsequent baths of 95% ethanol for 10 s, 0.525% NaOCl for 2 min, and 70% ethanol for 2 min to reduce microbial contamination on leaf exteriors (Kandalepas et al. 2015). Root samples were submerged in subsequent baths of 70% ethanol for 10 s and 2.6% NaOCl for 2 min before being rinsed three times in sterile deionized water. Disinfected samples were then frozen in CTAB buffer (2% CTAB, 0.02 M EDTA, 0.1 M Tris, 1.4 M NaCl) at −20 °C until RNA extraction. All processing was completed within 12 h of initial collection. Approximately 10 g of soil were collected from each mesocosm, prepared as described in Rodrigue et al. (2020), and four groups of alkylated PAHs (C1- to C4-naphthalenes, C1- to C4-phenanthrenes, C1- to C3-dibenzothiophenes and C1- to C3-chrysenes) were quantified through analysis with a 6890 N gas chromatograph (Agilent Technologies, Santa Clara, USA) equipped with a 5973 N mass selective detector.
RNA extraction and GeoChip 5.0S analysis
Samples were thawed at room temperature, and CTAB buffer was removed. Samples were then dried at 55 °C for up to 1.5 h on aluminum dishes to evaporate CTAB buffer completely. Both leaf and root samples were pulverized by submerging each sample and PowerBeads from PowerBead tubes of RNeasy PowerSoil Total RNA kit (Qiagen, Germantown, MD) in liquid nitrogen and grinding with a mortar and a pestle. RNA was then extracted using the RNeasy PowerSoil Total RNA kit (Qiagen, Germantown, MD) according to manufacturer’s instructions. Extracted RNA was quantified on Qubit 2.0 Fluorometer (ThermoFisher, Waltham, MA, USA) and immediately converted into cDNA using a High Capacity cDNA Reverse Transcription Kit (ThermoFisher, Waltham, MA) according to manufacturer’s instructions, and sent to Glomics, Inc. (Norman, OK) for analysis via GeoChip 5.0. Hybridization was carried out at 68 °C for 22 h using hybridization buffer (Agilent, New Castle, DE) and 10% formamide. Reads were scanned and quantified as described in Cong et al. (2015).
Computational analysis
GeoChip data received from Glomics, Inc. were normalized using the O.U. Microarray Data Manager (http://ieg.ou.edu/microarray/). Only the reads with a signal-to-noise ratio greater than 2.0 were retained for further downstream analyses. The resulting GeoChip read data were subdivided into 9 major functional group categories: carbon cycling, metal homeostasis, nitrogen cycling, organic remediation, phosphorus, secondary metabolism, sulfur, virulence, or other. Each functional group was visualized for trends using heatmaps and nonmetric multidimensional scaling (NMDS). For NMDS visualization, the data were transformed to a Bray–Curtis dissimilarity index and assessed using the envfit and adonis functions (permutational distance-based multivariate analysis of variance (PERMANOVA)). The Bray–Curtis dissimilarity index was chosen because it is one of the least vulnerable indices to several types of errors (Schroeder and Jenkins 2018). Functional gene categories displaying a p-value of <0.1 from adonis were considered indicative of an overall shift and subsequent subcategory analyses were completed. Treatment effects per subcategory of these functional categories were analyzed via ANOVA, one-sample t-test, and t-test depending on the number of conditions being examined. Due to low sample size and variation between replicates, both samples with historical oil exposure and recent amendment with weathered oil were considered to have oil exposure and combined during analysis to provide a less variable dataset to understand the overall effects of oil exposure on gene function.. Heatmaps were produced in R 4.0.0 statistical software (R Core Team 2020), and Bray–Curtis, NMDS, envfit, adonis, and permutest were calculated using vegan package (version 2.5–6) in R 4.0.0 statistical software (Oksanen et al. 2019). One-sample t test and unpaired t-test with Welch’s correction were completed using GraphPad Prism version 9.1.2 for Windows (GraphPad Software, San Diego, CA). For unpaired t-test in GraphPad Prism, the model comparison option was used to provide additional confirmation that the two means were different due to low sample size and variation between replicates. Gene target diversity metrics were calculated using the O.U. Microarray Data Manager GeoChip data analysis pipeline.
Results
Both visualization techniques (heatmaps and NMDS) displayed some visual clustering between samples of each tissue type (leaf or root) and treatment (oiled or not) for secondary metabolism, carbon, and virulence functional categories. Categories with visual clustering were partially supported by a low p-value (p<0.1) from adonis, though this was not considered statistically significant. The ANOVA analyses did not indicate statistical significance (p ≤ 0.05) for any subcategories with visual clustering. The following subcategories (Supplemental 1) were found to have significant (p ≤ 0.05) changes associated with oil exposure from one-sample t test, but not unpaired t test: antibiotic resistance, aromatics, arsenic detoxification, chlorinated solvents, chromium detoxification, herbicide-related compounds, methane, mercury detoxification, mercury transport, other hydrocarbons, pesticide-related compounds, phytic acid hydrolysis, polyphosphate synthesis, silicon biosynthesis, and tellurium detoxification. Unpaired t test provided additional support for identifying statistically significant shifts, and both one-sample t test and unpaired t test analyses indicated statistical significance for the 8 subcategories reviewed below.
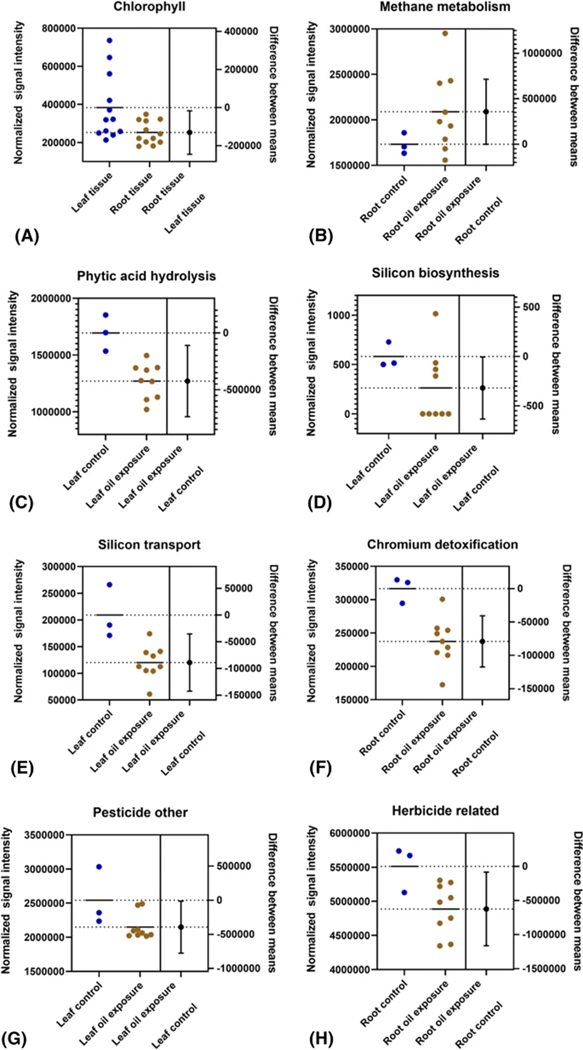
For microbial gene targets in the secondary metabolism category, oil exposure was not found to exert an effect on the expression of either rhodopsin or microbial chlorophyll-related genes. However, the difference in microbial gene expression for the chlorophyll subcategory was found to differ significantly (p = 0.0279, t = 2.461, df = 13.59, 83% confidence in distinct population means) between tissue types, with leaf tissue having higher expression levels (Fig. 4A). The expression levels of microbial functional genes related to methane metabolism were found to increase significantly (p = 0.05, t = 2.234, df = 9.95, 31% confidence in distinct population means) in root samples exposed to oil, however, there was no such effect observed in leaves (Fig. 4B). Oil exposure was associated with significant shifts in gene expression levels for the phosphorus category, though these shifts were restricted to leaf samples and involved only the phytic acid hydrolysis subcategory (p = 0.0216, t = 4.017, df = 3.423, 98% confidence in distinct population means) which was lower in samples exposed to oil (Fig. 4C). For metal homeostasis, oil exposure was associated with a significant decrease in expression levels for silicon biosynthesis (p = 0.047, t = 2.270, df = 9.653, 34% confidence in distinct population means) and silicon transport (p = 0.0041, t = 3.704, df = 10, 97% confidence in distinct population means) subcategories in leaf samples (Fig. 4D, E), while root samples displayed a significant decrease in expression levels in the chromium detoxification (p = 0.002, t = 4.915, df = 6.789, 96% confidence in distinct population means) subcategory (Fig. 4F). A shift associated with oil exposure in the organic remediation functional category for leaf tissue samples indicated decreased expression levels of genes in the pesticide-related compounds subcategory (p = 0.04, t = 2.305, df = 10, 67% confidence in distinct population means) (Fig. 4G), while root tissue samples displayed decreased expression levels in the herbicide-related compounds (p = 0.0273, t = 2.58, df = 10, 77% confidence in distinct population means) subcategory (Fig. 4H). Shannon diversity values for all samples ranged from 8.94 to 9.90 (σ = 0.25) and Simpson diversity values ranged from 0.09 to 0.24 (σ = 0.05). Total soil concentrations of the four groups of alkylated PAH congeners averaged 1.884 μg/g for control samples and 1.894 μg/g for oiled samples, with one outlier reading disregarded.
Fig. 4.
Estimation plots of GeoChip functional subcategories that displayed consistent statistically significant changes related to tissue type or overall oil exposure. Functional gene categories: A secondary metabolism, B carbon, C phosphorus, D–F metal homeostasis, and G, H organic remediation. Dots represent the total sum of each sample, solid and dotted horizontal lines indicate the mean of each group of samples. Note that the plot for Chlorophyll (A) compares expression between root and leaf samples and thus includes all samples of each tissue (both control and oiled), while plots B–H compare the control samples of one tissue type against oiled samples of the same tissue type
Discussion
Because endophytic communities establish an intimate relationship with their host, they must respond to changes in an internal environment governed by host health and responses to environmental stimuli and stressors. Previous studies have shown that the presence or absence of toxicants such as PAHs and heavy metals can elicit biochemical responses from plants, and such alterations to the internal chemistry of the host plant represent a change in habitat to which endophytic communities must adapt (Maliszewska-Kordybach and Smreczak 2000; Sethy and Ghosh 2013; Seneviratne et al. 2017; Udo and Fayemi 1975).
Clustering was observed in heatmaps between both samples with the same soil inoculant collection location and samples obtained from the same tissue type, suggesting similar trends in microbial gene expression between these experimental cohorts. This was confirmed through t test analyses of the subcategories in question. For example, the expression of microbial genes related to chlorophyll were expected to differ between leaf and root tissue types regardless of oil exposure. Leaves, which are exposed to light, had a high expression level, while roots, which would normally not be exposed to sunlight, had low expression levels. These findings are in line with other studies, as it has been previously demonstrated that microbial communities can differ vastly between different organ systems and tissue types of their host (Ugarelli et al. 2019; Wallace et al. 2018; Wei and Ashman 2018).
Crude oil has been demonstrated to contain heavy metals and can contaminate areas affected by spills with significantly elevated concentrations (Liu et al. 2012; Osuji and Onojake 2004). The uptake of such metals into plant tissue from contaminated sediment is well-documented, to the extent that hyperaccumulator species play critical roles in phytoremediation (Leitenmaier and Küpper 2013). The GeoChip 5.0 array contains microbial gene targets for arsenic, chromium, copper, mercury, and tellurium detoxification. Significant shifts were detected in the expression of chromium detoxification genes, and shifts in the arsenic, mercury, and tellurium detoxification subcategories were partially supported, suggesting that the presence of crude oil residue may have altered the environmental concentration of one or more of these metals. We suspect that increased exposure of the host plants to crude oil residue resulted in increased uptake of heavy metals, which in turn may have induced endophyte cellular defenses against metal toxicity as demonstrated in previous studies (Luo et al. 2011). While the increase in gene expression observed in most of the metal detoxification subcategories was consistent with our expectation, the decrease observed in the chromium detoxification subcategory was not consistent with our expectations. Further study will be necessary to elucidate the mechanisms behind this finding.
GeoChip 5.0 gene targets in the organic remediation category include genes associated with the degradation of PAHs, including genes identified explicitly with the degradation of benzene, toluene, ethylbenzene, and xylene, and chlorinated aromatics. On average, crude oil is comprised of between 0.2 and 7% PAH by volume, most of which are low molecular weight 2-ring or 3-ring molecules (Neff et al. 2005). The DWH well falls on the higher end of this range, with a PAH concentration estimated at 3.9% (Reddy et al. 2012; Tidwell et al. 2016). It was expected that shifts would be observed in these categories in response to oil exposure. However, only partial support for such shifts was observed in our study. This may indicate that concentrations of PAHs and other hydrocarbons in the inoculant soil and weathered oil amendments have decreased substantially over the 8 years between the DWH spill and the conclusion of this experiment, which is supported by the similarity in PAH concentrations between oiled and control samples. Low molecular weight PAHs are more volatile, water-soluble, and easier for microorganisms to degrade than high molecular weight PAHs, and as such, are removed from the environment much faster (Ghosal et al. 2016). However, the overall lack of response in this category may indicate that even higher molecular weight PAHs have substantially declined in concentration. Alternatively, although PAHs have been found to infiltrate plant tissue, both via translocation following root uptake and through their stomata as a result of atmospheric deposition (Liu et al. 2017), there exists the possibility that effects were limited to the external rhizosphere microbiome or that the cellular defenses of S. alterniflora or its associated external microbial communities exerted a prohibitive effect on hydrocarbon uptake (Liu et al. 2018). Should the latter have occurred, a link may exist to the shifts observed in the category of functional genes related to phosphorus. Phosphorus has been observed to be a limiting factor in the degradation of hydrocarbons by known degraders (Cunliffe and Kertesz 2006; Vyas and Dave 2010). Therefore, the activation of hydrocarbon degradation pathways may require increased phosphorus uptake, resulting in the observed shifts in phosphorus-related gene expression (phytic acid hydrolysis subcategory) as endophytes responded to a decrease in available phosphorus.
This study suggests that endophytic community function is impacted by and responds to host exposure to crude oil residue. Subsequent alterations in the expression of functional genes related to secondary metabolism, carbon, phosphorus, metal homeostasis, and organic remediation suggest a change in the internal chemistry of exposed S. alterniflora. These biochemical shifts within host tissue may represent a response to compounds present within the crude oil residue or a direct adaptation by the microbial communities to crude oil compounds taken up by host tissues. Despite these shifts, diversity was not observed to change significantly across treatments. This may be due in part to the fact that the GeoChip 5.0 array is not designed to focus on identifying community composition. Further study is necessary to determine such changes and those related to community density and spatial distribution throughout host tissue.
The interaction between host plant development and plant-associated microbial communities is a developing research area with much importance to the broad field of phytoremediation (Redfern and Gunsch 2016). The plant species used in this study, S. alterniflora, is of particular interest due to its role as a foundational ecosystem engineer and its tolerance to the presence of elevated levels of crude oil contaminants. Should associated endophyte species or the plants themselves prove efficient at remediating these contaminants, S. alterniflora may present a viable option for stabilizing and remediating marsh environments affected by oil spills. However, such treatment would require careful study before implementation. Although S. alterniflora has displayed resistance to crude oil-associated toxicants, it can readily hybridize with other Spartina species and as such has been labeled an invasive species in several regions (Liu et al. 2020; Xia et al. 2020).
Herein, efforts were made to ensure seedlings for this study were germinated in sterile conditions. Nonetheless, while the seeds themselves may have contained endophytes that were unaffected by external sterilization, the potential lack of endophytic and symbiotic microbial communities may have affected host seedling development and led to alterations in mature host immune response that shifted experimental communities away from those that would have been observed in natural ecosystems. Thus, subsequent studies may benefit from an earlier exposure of host seedlings to inoculant substrate or the incorporation of field trials to establish target microbial associations more firmly. Sampling more replicates per treatment will be beneficial and serve to clarify smaller-scale shifts potentially masked by the sensitivity of the GeoChip microarray. Finally, the bimonthly additions of a soil amendment from sample locations allowed for experimental soil communities to track the seasonal variation of natural systems potentially absent in greenhouse conditions, but such exposures may have contributed to some of the variation in the GeoChip results seen in this study. Research gaps remain to fully untangle the exact relationships between the external environment, host plant function, and endophyte community health. However, further research into the influence of endophytic communities on the health and resiliency of their host plants holds excellent potential for advances in agriculture, the conservation of natural ecosystems, and phytoremediation.
Supplementary Material
Funding
This research was made possible in part by a grant from The Gulf of Mexico Research Initiative, and in part by Duke’s Superfund Research Center, the National Institute for Environmental Health Sciences under the Grant NIEHS P42-ES010356, and the National Science Foundation’s Graduate Research Fellowship Program (Grant No. DGE-1644868).
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s10532-021-09968-5.
Declarations
Conflict of interest Not applicable.
Contributor Information
Samantha D. Addis, Department of Civil and Environmental Engineering, Duke University, Durham, NC, USA
Stephen K. Formel, Ecology and Evolutional Biology, Tulane University, New Orleans, LA, USA
Yeon Ji Kim, Department of Civil and Environmental Engineering, Duke University, Durham, NC, USA.
Paige B. Varner, Department of Civil and Environmental Engineering, Duke University, Durham, NC, USA
Daniel B. Raudabaugh, Department of Civil and Environmental Engineering, Duke University, Durham, NC, USA
Emilie Lefevre, Department of Civil and Environmental Engineering, Duke University, Durham, NC, USA.
Brittany M. Bernik, Ecology and Evolutional Biology, Tulane University, New Orleans, LA, USA
Vijaikrishnah Elango, Civil and Environmental Engineering, Louisiana State University, Baton Rouge, LA, USA.
Sunshine A. Van Bael, Ecology and Evolutional Biology, Tulane University, New Orleans, LA, USA
John H. Pardue, Civil and Environmental Engineering, Louisiana State University, Baton Rouge, LA, USA
Claudia K. Gunsch, Department of Civil and Environmental Engineering, Duke University, Durham, NC, USA
Data availability
Data are publicly available through the Gulf of Mexico Research Initiative Information & Data Cooperative (GRIIDC) at https://data.gulfresearchinitiative.org (https://doi.org/10.7266/n7-rzxc-4x96). All raw data are publicly available on the NCBI SRA database (Accession Number: GSE148431) and upon request.
Code availability
Not applicable.
References
- Alvarez M, Ferreira de Carvalho J, Salmon A, Ainouche ML, Cavé-Radet A, El Amrani A, Foster TE, Moyer S, Richards CL (2018) Transcriptome response of the foundation plant Spartina alterniflora to the Deepwater Horizon oil spill. Mol Ecol 27(14):2986–3000. 10.1111/mec.14736 [DOI] [PubMed] [Google Scholar]
- Bååth E (1989) Effects of heavy metals in soil on microbial processes and populations (a review). Water Air Soil Pollut 47:335–379. 10.1007/BF00279331 [DOI] [Google Scholar]
- Brundrett M (2004) Diversity and classification of mycorrhizal associations. Biol Rev 79:473–495. 10.1017/S1464793103006316 [DOI] [PubMed] [Google Scholar]
- Cong J, Liu X, Lu H, Xu H, Li Y, Deng Y, Li D, Zhang Y (2015) Analyses of the influencing factors of soil microbial functional gene diversity in tropical rainforest based on GeoChip 5.0. Genom Data 5:397–398. 10.1016/j.gdata.2015.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe M, Kertesz MA (2006) Effect of Sphingobium yanoikuyae B1 inoculation on bacterial community dynamics and polycyclic aromatic hydrocarbon degradation in aged and freshly PAH-contaminated soils. Environ Pollution 144(1):228–237. 10.1016/j.envpol.2005.12.026 [DOI] [PubMed] [Google Scholar]
- Deepwater Horizon Natural Resource Damage Assessment Trustees (2016). Deepwater horizon oil spill: final programmatic damage assessment and restoration plan and final programmatic environmental impact statement. http://www.gulfspillrestoration.noaa.gov/restoration-planning/gulf-plan
- Doyle SM, Whitaker EA, De Pascuale V, Wade TL, Knap AH, Santschi PH, Quigg A, and Sylvan JB (2018) Rapid formation of microbe-oil aggregates and changes in community composition in coastal surface water following exposure to oil and the dispersant corexit. Front Microbio 9:689. 10.3389/fmicb.2018.00689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KR, Mills KP (2005) Aboveground and belowground productivity of Spartina alterniflora (smooth cordgrass) in natural and created Louisiana salt marshes. Estuaries 28(2):252–265. 10.1007/BF02732859 [DOI] [Google Scholar]
- Ghosal D, Ghosh S, Dutta T, Ahn Y (2016) Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHS): a review. Front Microbiol 7:1369. 10.3389/fmicb.2016.01369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giller KE, Witter E, Mcgrath SP (1998) Toxicity of heavy metals to microorganisms and microbial processes in agricultural soils: a review. Soil Biol Biochem 30(10–11):1389–1414. 10.1016/S0038-0717(97)00270-8 [DOI] [Google Scholar]
- Groudeva VI, Groudev SN, Doycheva AS (2001) Bioremediation of waters contaminated with crude oil and toxic heavy metals. Int J Miner Process 62:293–299. 10.1016/S0301-7516(00)00060-0 [DOI] [Google Scholar]
- Kandalepas D, Blum MJ, Van Bael SA (2015) Shifts in symbiotic endophyte communities of a foundational salt marsh grass following oil exposure from the Deepwater Horizon oil spill. PLoS ONE. 10.1371/journal.pone.0122378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare E, Mishra J, Arora NK (2018) Multifaceted interactions between endophytes and plant: developments and prospects. Front Microbiol. 10.3389/fmicb.2018.02732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss KW, Chambers JL, Allen JA (1998) Salinity effects and differential germination of several half-sib families of bald cypress from different seed sources. New For 15:53–68. 10.1023/A:1006572609171 [DOI] [Google Scholar]
- Leitenmaier B, Küpper H (2013) Compartmentation and complexation of metals in hyperaccumulator plants. Front Plant Sci. 10.3389/fpls.2013.00374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Tao S, Zuo Q, Coveney RM (2007) Uptake of polycyclic aromatic hydrocarbons by maize plants. Environ Pollut 148(2):614–619. 10.1016/j.envpol.2006.11.026 [DOI] [PubMed] [Google Scholar]
- Liu Z, Liu J, Zhu Q, Wu W (2012) The weathering of oil after the Deepwater Horizon oil spill: insights from the chemical composition of the oil from the sea surface, salt marshes and sediments. Environ Res Lett 7:035302. 10.1088/1748-9326/7/3/035302 [DOI] [Google Scholar]
- Liu H, Carvalhais LC, Crawford M, Singh E, Dennis PG, Pieterse CMJ, Schenk PM (2017) Inner plant values: diversity, colonization and benefits from endophytic bacteria. Front Microbiol 8:2552. 10.3389/fmicb.2017.02552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang Z, Sheng Y, Gao Y, Zhao Z (2018) Phenanthrenedegrading bacteria on root surfaces: a natural defense that protects plants from phenanthrene contamination. Plant Soil 425:335–350. 10.1007/s11104-018-3575-z [DOI] [Google Scholar]
- Liu W, Zhang Y, Chen X, Maung-Douglass K, Strong DR, Pennings SC (2020) Contrasting plant adaptation strategies to latitude in the native and invasive range of Spartina alterniflora. New Phytol 226:623–634. 10.1111/nph.16371 [DOI] [PubMed] [Google Scholar]
- Luo S, Wan Y, Xiao X, Guo H, Chen L, Xi Q, Zeng G, Liu C, Chen J (2011) Isolation and characterization of endophytic bacterium LRE07 from cadmium hyperaccumulator Solanum nigrum L. and its potential for remediation. Appl Microbiol Biotechnol 89:1637–1644. 10.1007/s00253-010-2927-2 [DOI] [PubMed] [Google Scholar]
- Maliszewska-Kordybach B, Smreczak B (2000) Ecotoxicological activity of soils polluted with polycyclic aromatic hydrocarbons (PAHs): effect on plants. Environ Tech 21:1099–1110. 10.1080/09593330.2000.9618996 [DOI] [Google Scholar]
- McNutt MK, Camilli R, Crone TJ, Guthrie GD, Hsieh PA, Ryerson TB, Savas O, Shaffer F (2011) Review of flow rate estimates of the Deepwater Horizon oil spill. Proc Natl Acad Sci USA 109:20260–20267. 10.1073/pnas.1112139108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff JM, Stout SA, Gunster DG (2005) Ecological risk assessment of polycyclic aromatic hydrocarbons in sediments: Identifying sources and ecological hazard. Integr Environ Assess Manag 1:22–33. 10.1897/IEAM_2004a-016.1 [DOI] [PubMed] [Google Scholar]
- Odukoya J, Lambert R, Sakrabani R (2019) Understanding the impacts of crude oil and its induced abiotic stresses on agrifood production: a review. Horticulturae 5(2):47. 10.3390/horticulturae5020047 [DOI] [Google Scholar]
- Oksanen J, Guillaume BF, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, and Wagner H (2019) vegan: community ecology package. R package version 2.5–6. https://CRAN.R-project.org/package=vegan [Google Scholar]
- Osuji LC, Onojake CM (2004) Trace heavy metals associated with crude oil: a case study of Ebocha-8 oil-spill-polluted site in Niger delta, Nigeria. Chem Biodivers 1:1708–1715. 10.1002/cbdv.200490129 [DOI] [PubMed] [Google Scholar]
- R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/ [Google Scholar]
- Reddy CM, Arey JS, Seewald JS, Sylva SP, Lemkau KL, Nelson RK, Carmichael CA, McIntyre CP, Fenwick J, Ventura GT, Van Mooy BA, Camilli R (2012) Composition and fate of gas and oil released to the water column during the Deepwater Horizon oil spill. Proc Natl Acad Sci USA 109(50):20229–20234. 10.1073/pnas.1101242108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfern LK, Gunsch CK (2016) Endophytic phytoaugmentation: treating wastewater and runoff through augmented phytoremediation. Ind Biotechnol 12(2):83–90. 10.1089/ind.2015.0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Hurek T (2011) Living inside plants: bacterial endophytes. Curr Opin Plant Biol 14(4):435–443. 10.1016/j.pbi.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Rodrigue M, Elango V, Curtis D, Collins A, Pardue JH (2020) Biodegradation of MC252 polycyclic aromatic hydrocarbons and alkenes in two coastal wetlands. Mar Pollut Bull 157:111319. 10.1016/j.marpolbul.2020.111319 [DOI] [PubMed] [Google Scholar]
- Seneviratne M, Rajakaruna N, Rizwan M, Madawala HMSP, Ok YS, Vithanage M (2017) Heavy metal-induced oxidative stress on seed germination and seedling development: a critical review. Environ Geochem Health 41(4):1813–1831. 10.1007/s10653-0170005-8 [DOI] [PubMed] [Google Scholar]
- Sethy SK, Ghosh S (2013) Effect of heavy metals on germination of seeds. J Nat Sci Biol Med 4(2):272–275. 10.4103/0976-9668.116964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder PJ, Jenkins DG (2018) How robust are popular beta diversity indices to sampling error? Ecosphere 9(2):e02100. 10.1002/ecs2.2100 [DOI] [Google Scholar]
- Shi Z, Yin H, Van Nostrand JD, Voordeckers JW, Tu Q, Deng Y, Yuan M, Zhou A, Zhang P, Xiao N, Ning D, He Z, Wu L, Zhou J (2019) Functional gene array-based ultrasensitive and quantitative detection of microbial populations in complex communities. mSystems. 10.1128/mSystems.00296-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidwell LG, Allan SE, O’Connell SG, Hobbie KA, Smith BW, Anderson KA (2016) PAH and OPAH flux during the deepwater horizon incident. Environ Sci Technol 50(14):7489–7497. 10.1021/acs.est.6b02784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Coast Guard (2011) B.P. Deepwater Horizon Oil Spill: Incident Specific Preparedness Review (ISPR), Final Report. Department of Homeland Security, Washington, DC [Google Scholar]
- Udo EJ, Fayemi AAA (1975) The effect of oil pollution of soil on germination, growth and nutrient uptake of corn. J Environ Qual 4(4):537–540. 10.2134/jeq1975.00472425000400040023x [DOI] [Google Scholar]
- Ugarelli K, Laas P, Stingl U (2019) The microbial communities of leaves and roots associated with turtle grass (Thalassia testudinum) and manatee grass (Syringodiumfilliforme) are distinct from seawater and sediment communities, but are similar between species and sampling sites. Microorg 7(1):4. 10.3390/microorganisms7010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas TK, Dave BP (2010) Effect of addition of nitrogen, phosphorus, and potassium fertilizers on biodegradation of crude oil by marine bacteria. Indian J Mar Sci 39(10):143–150 [Google Scholar]
- Wallace J, Laforest-Lapointe I, Kembel S (2018) Variation in the leaf and root microbiome of sugar maple (Acer saccharum) at an elevational range limit. PeerJ 6:e5293. 10.7717/peerj.5293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Ashman TL (2018) The effects of host species and sexual dimorphism differ among root, leaf and flower microbiomes of wild strawberries in situ. Sci Rep 8:5195. 10.1038/s41598-018-23518-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Geng Q, An S (2020) Rapid genetic divergence of an invasive species, Spartina alterniflora, in China. Front Genet 11:284. 10.3389/fgene.2020.00284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel S, Bernik BM, Rutherford N, Nixon Z, Michel J (2015) Heavily oiled salt marsh following the Deepwater Horizon oil spill, ecological comparisons of shoreline cleanup treatments and recovery. PLoS ONE 10:e0132324. 10.1371/journal.pone.0132324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are publicly available through the Gulf of Mexico Research Initiative Information & Data Cooperative (GRIIDC) at https://data.gulfresearchinitiative.org (https://doi.org/10.7266/n7-rzxc-4x96). All raw data are publicly available on the NCBI SRA database (Accession Number: GSE148431) and upon request.
Not applicable.