Abstract
Background
Chronic spontaneous urticaria (CSU) is a common and disabling disease. Assessments of IgE and C‐reactive protein (CRP) are recommended in the diagnostic work‐up, but the role and clinical relevance of these biomarkers are not well characterized. Moreover, it remains unknown if elevated levels of IgE or CRP are linked to CSU microRNA (miRNA) signatures or interleukin 31 (IL‐31).
Methods
We measured IgE and CRP serum levels in 47 CSU patients (and 45 healthy controls) and determined CSU disease activity using the urticaria activity score (UAS7). Expression levels of miR‐155 and miR‐221 were assessed by RT‐PCR, and IL‐31 levels were determined by ELISA.
Results
Total IgE and CRP levels were independently increased in CSU patients. IgE and CRP levels were highest and lowest in patients with high and mild disease activity. IgE levels correlated with miR‐155 levels, whereas CRP levels correlated with miR‐221 levels. miR‐155 and miR‐221 were significantly overexpressed in CSU patients. ROC analyses linked miRNA‐155 and CSU with a sensitivity of 79% and specificity of 87%, and miRNA‐221 and CSU with a sensitivity of 75% and specificity of 91%. High CRP and miR‐221 expression levels were linked to elevated levels of IgG anti‐TPO and IL‐31.
Conclusion
IgE and CRP are useful biomarkers for disease activity in CSU, with distinct miRNA profiles. High CRP and miR‐221 levels may point to autoimmune CSU and a role for IL‐31.
Keywords: CRP, IgE, interleukin‐31, micro‐RNA, urticaria
1. INTRODUCTION
Chronic spontaneous urticaria (CSU) is a common and disabling inflammatory disease characterized by itchy wheals, angioedema, or both. 1 These signs and symptoms are caused by the activation and degranulation of skin mast cells and their release of proinflammatory mediators including histamine. 2 In most patients with CSU, skin mast cells are activated by autoantibodies, either IgE antibodies against autoantigens, in autoallergic CSU, or IgG autoantibodies against IgE or its high‐affinity receptor, FceRI, in autoimmune CSU. 3 Autoallergic and autoimmune CSU differ in prevalence, disease activity and duration, and response to treatment. Autoimmune CSU is characterized by high disease activity, the presence of comorbid autoimmune diseases, elevated levels of IgG‐anti–thyroid peroxidase, basopenia, eosinopenia, and poor response to antihistamine and omalizumab treatment. 4 In routine clinical practice, it is important but difficult to assess patients for autoimmune CSU because three tests need to be performed and be positive, that is, the autologous serum skin test (ASST), an immunoassay for IgG anti‐IgE/FceRI, and a basophil activation test. 5
The most recent version of the international guideline for urticaria 6 provides recommendations for the diagnostic work‐up of CSU patients. These include the following blood tests, all of which are readily available for use in routine clinical practice: C‐reactive protein (CRP) and/or ESR, a differential blood count, total IgE, and IgG‐anti‐TPO. Some of these blood tests can help to screen CSU patients for the underlying cause of their disease, that is, autoallergy and autoimmunity. For example, elevated blood levels of total IgE, in some studies, were linked to autoallergic CSU, whereas low IgE levels point to autoimmune CSU. 7 Elevated levels of CRP/ESR have also been suggested to be linked to autoimmune CSU. 8 To date, however, the clinical profiles of CSU patients with high IgE or elevated CRP remain ill‐defined. Whether these two CSU biomarkers are linked to inflammation‐associated microRNAs (miRNA) or the itch cytokine interleukin‐31 (IL‐31) is currently unknown.
miRNAs, small noncoding RNAs, are believed to be involved in the pathogenesis of several chronic inflammatory skin disorders, 9 but little is known about their role in CSU. 10 Two miRNAs, miRNA‐155 (miR‐155) and miRNA‐221 (miR‐221), are of special interest in CSU as they have been implicated in the modulation of mast cell activation, IgE and FceRI, and autoimmunity. miR‐155 is needed for antibody production, B‐cell maturation, differentiation, and immunoglobulin class switching. 11 miR‐155 has also been reported to downregulate FceRI expression and FcεRI‐mediated mast cell degranulation, 12 , 13 and its expression is increased in human skin‐derived mast cells following FcεRI crosslinking with antigen. 14 CSU patients have not yet been assessed for miR‐155, but patients with atopic dermatitis exhibit increased expression in mast cells at lesional skin sites. 15 miR‐221 contributes to the regulation of the cell cycle and cytoskeleton of mast cells as well as their degranulation and cytokine production. 16 miR‐221 has also been linked to several autoimmune disorders, including psoriasis 17 and rheumatoid arthritis, 18 but, like miR‐155, has not been studied in CSU.
Interleukin 31 is involved in the pathogenesis of chronic inflammatory skin disorders, including atopic dermatitis, allergic contact dermatitis, and mastocytosis. 19 , 20 , 21 , 22 It is also believed to contribute to the pathogenesis of CSU, although there is little evidence for this. In a recent study, CSU patients had significantly higher mean serum IL‐31 levels as compared to healthy subjects. Interestingly, CSU patients with elevated ANA titers, a marker of autoimmune CSU, had significantly higher mean serum IL‐31 levels than those who were negative for ANA. 23 Also, successful omalizumab treatment of patients with CSU is associated with lowering serum IL‐31 levels. 24 Whether IL‐31, in CSU, is linked to IgE or CRP is currently unknown.
To address these gaps of knowledge, we investigated CSU patients for total IgE and CRP levels and their links to clinical features. We then assessed miR‐155 and miR‐221 levels and their association with IgE and CRP levels. Finally, we explored interleukin‐31 levels in CSU and how they are linked to IgE, CRP, and miRNAs.
2. METHODS
2.1. Study subjects and conduct
This study was conducted at the Dermatology outpatient clinic of Firat University Faculty of Medicine with a total of 100 adult participants, that is, 50 patients with CSU and 50 healthy individuals, and the sociodemographic characteristics of all the participants were recorded. Each patient underwent a detailed dermatological examination and completed the urticaria activity score (UAS). Laboratory parameters, that is, CRP, IgG anti‐thyroid peroxidase (anti‐TPO), and total IgE, were determined by enzyme‐linked immunosorbent assay (ELISA). The upper limit of normal for total IgE was 100 IU/ml. The study was initiated after obtaining the approval of the local ethics committee (Date: 29/09/2016, no: 02). All participants provided written informed consent.
2.2. Assessment of disease activity by the use of the urticaria activity score
The disease activity of CSU patients was assessed with the urticaria activity score (UAS) as recommended using the EAACI/GA2LEN/EuroGuiDerm/APAAACI guideline for urticaria. 6 Patients recorded daily, for 7 days, the number of wheals (no wheals = 0 points; <20 wheals = 1 point; 20–50 wheals = 2 points; >50 wheals = 3 points) and the intensity of itch (no itch = 0 points; mild itch = 1 point; moderate itch = 2 points; severe itch = 3 points). The weekly UAS (UAS7) was calculated by adding the UAS values of 7 consecutive days, with a minimum score of 0 and a maximum score of 42. Patients with UAS7 values of 1–15, 16–27, and 28–42 were considered to have minimal/mild, moderate, and severe disease activity, respectively. 25
2.3. RT‐PCR analysis of plasma samples
Total RNA was isolated from serum samples (200 μL per sample) of 50 CSU patients and 50 controls using a miRNeasy Serum/Plasma Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Qiagen miScript Reverse Transcription (RT) Kit II (Hilden, Germany) was used for cDNA extraction. The cDNAs were amplified using the Qiagen miScript PreAMP PCR kit (Hilden, Germany). Following the cDNA extraction, the real‐time polymerase chain reaction (RT‐PCR) stage began to analyze the expression levels of miR‐155 and miR‐221. The reaction components for RT‐PCR were prepared using the Qiagen miScript SYBR Green PCR kit. SNORD61 was used for the normalization of RT‐PCR. RT‐PCR Rotor‐Gene Q (Qiagen) was used to identify SNORD61 expression levels as a reference along with the hsa‐miR‐155‐3p and hsa‐miR‐221‐3p miRNA primers.
2.4. Calculation of fold changes by 2−ΔΔCt analysis
Changes in miR‐155 and miR‐221 gene expression were calculated. First, the delta CT values of SNORD61 were calculated considering it as the reference miRNA. The Ct values provided by the Real‐Time analysis were entered into the Excel program, and the statistical 2−ΔΔCt analyzed of all miRNAs using the GeneGlobe Data Analysis Center (Qiagen) online analysis program.
2.5. IL‐31 analysis
Serum samples obtained by centrifugation were stored at −80°C and concentrations of IL‐31 in serum samples were measured in 47 CSU patients and 45 control subjects by ELISA using commercially available kits (Elabscience Biotechnology Co. Ltd).
2.6. Statistical analysis
Statistical analysis was performed using the Statistical Package for Social Sciences for Windows (SPSS) 22.0. The results of categorical measurements were expressed as numbers and percentages. The chi‐square test was used for the analysis of two categorical variables. Normally distributed numeric measurements were presented as mean ± standard deviation, and non‐normally distributed numeric measurements were given as mean ± standard deviation, median (percentiles 25th–75th). The Mann‐Whitney U test was used to compare non‐normally distributed numerical data between two groups and the Kruskal‐Wallis test was used to compare the means among three or more groups (Significance values have been adjusted by the Bonferroni correction for multiple tests). Independent sample t‐test was used to compare normally distributed data between two groups, whereas one‐way ANOVA was used to compare the means among three or more groups. Spearman and Pearson correlation tests were used for correlation analysis. A p‐value of <0.05 was considered statistically significant.
3. RESULTS
In the patient group (32 females), the mean age was 35.6 ± 6.8; and in the control group (34 females), the mean age was 33.5 ± 9.9 years. Based on their UAS7 values, 6 (12.8%), 20 (42.6%), and 21 (44.7%) patients had minimal/mild, moderate, and severe disease activity, respectively.
3.1. Blood levels of total IgE and CRP are independently increased and linked with disease activity in patients with CSU
Mean total IgE levels were significantly higher in CSU patients (129 ± 14 IU/L) as compared to healthy controls (53 ± 9 IU/L, p < 0.01; Figure 1A). More than half of the CSU patients, 24 of 47 (51.4%), had elevated total IgE levels as compared to 8 of 45 healthy controls (17.7%, p < 0.05). Across all CSU patients, IgE levels were highest [138 ± 105 IU/L, median = 110 IU/L (65.0, 197.5 IU/L)] in patients with high disease activity, followed by moderate disease activity [127 ± 89 IU/L, median = 91.7 IU/L (60.62, 187.0 IU/L)], and they were lowest [104 ± 100 IU/L, median = 82.3 IU/L (41.8, 152.2 IU/L)] in patients with mild disease activity (Figure 1B), as assessed by UAS7.
FIGURE 1.
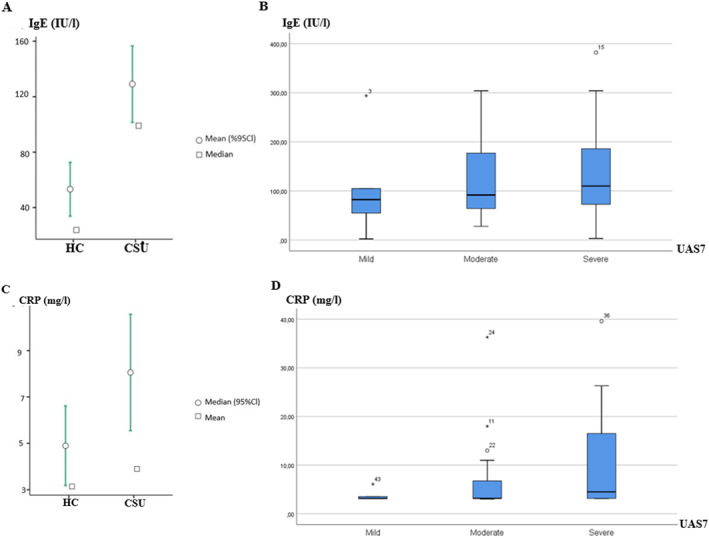
IgE and CRP levels increased independently and were linked to disease activity in CSU patients. (A) Total IgE levels were significantly higher in CSU patients (129 ± 14 IU/L) than in HCs (53 ± 9 IU/L, p < 0.01). (B) IgE levels were highest (138 ± 105 IU/L) in patients with high disease activity, intermediate in patients with moderate disease activity (127 ± 89 IU/L), and lowest (104 ± 100 IU/L) in patients with mild disease activity. (C) CRP levels were higher in CSU patients (8.1 ± 1.3 mg/L) than in HCs (4.8 ± 0.8 mg/L, p < 0.05). (D) CRP levels were highest (10.3 ± 10.2 mg/L) in patients with high disease activity, intermediate in patients with moderate disease activity (6.9 ± 8.0 mg/L), and lowest in patients with mild disease activity (3.7 ± 1.2 mg/L). CRP, C‐reactive protein; CSU, chronic spontaneous urticaria; HC, heathy controls, disease activity assessed by UAS7; UAS, urticaria activity score.
CSU patients also had higher CRP levels than healthy controls, that is, 8.1 ± 1.3 and 4.8 ± 0.8 mg/L, respectively (p < 0.05; Figure 1C), and higher rates of elevated CRP (36.2% vs. 13.3%). CRP levels were highest [10.3 ± 10.2 mg/L, median = 4.5 mg/L (3.15, 16.55 mg/L)] in patients with high disease activity, lowest in patients with mild disease activity [3.7 ± 1.2 mg/L, median = 3.1 mg/L (3.01, 3.35 mg/L)] and intermediate in patients with moderate disease activity [6.9 ± 8.0 mg/L, median = 3.2 mg/L (3.14, 6.7 mg/L); Figure 1D]. There was no correlation between IgE and CRP levels in CSU patients.
3.2. IgE and CRP are independently linked to miR‐155 and miR‐221, respectively, both of which are overexpressed in CSU
Across all study subjects, IgE levels, but not CRP levels, were correlated with levels of miR‐155 (r = 0.28, p < 0.05; Figure 2A), whereas levels of CRP, but not IgE, were correlated with levels of miR‐221 (r = 0.25, p < 0.05; Figure 2B). In CSU, the expression levels of miR‐221 and miR‐155 were correlated and significantly higher as compared to the control group (>2.2‐fold, p < 0.01; Figure 2C). ROC analyses linked miR‐155 and CSU with a sensitivity of 79% and specificity of 87%, and miR‐221 and CSU with a sensitivity of 75% and specificity of 91% (Figure 2D).
FIGURE 2.
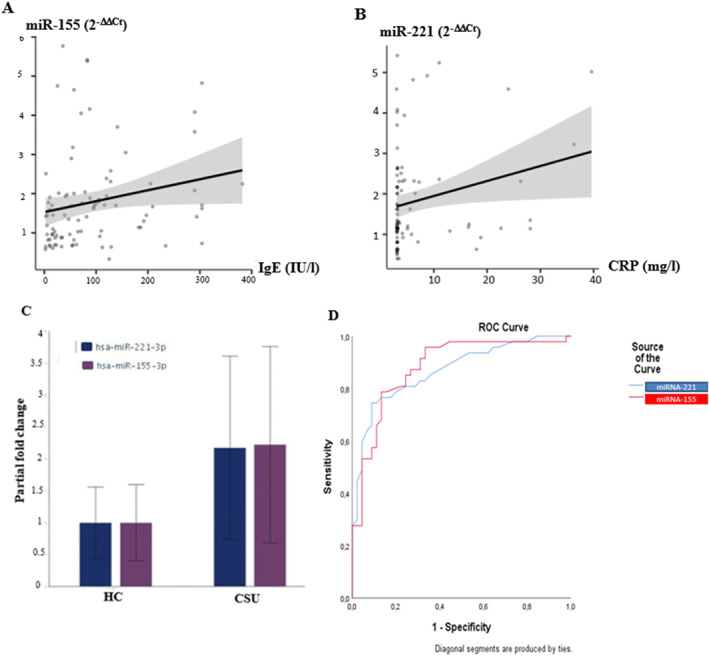
miR‐155 and miR‐221 are upregulated in CSU and linked to total IgE and CRP, respectively. (A) IgE levels correlated with miR‐155 levels in all study participants (r = 0.28, p < 0.05). (B) CRP levels correlated with miR‐155 levels in all study participants (r = 0.25, p < 0.05). (C) miR‐221 and miR‐155 were correlated and significantly higher in CSU patients than in HCs (fold change>2.2, p < 0.01). (D) ROC analyses linked miR‐155 and CSU with a sensitivity of 79% and specificity of 87%, and miR‐221 and CSU with a sensitivity of 75% and specificity of 91%. CRP, C‐reactive protein; CSU, chronic spontaneous urticaria; HC, heathy controls.
3.3. CRP and miR‐221 expression are linked to features of autoimmune CSU and interleukin‐31
In CSU patients, levels of CRP correlated with those of anti‐TPO (r = 0.33, p < 0.05), and anti‐TPO‐positive patients had higher CRP levels as compared to anti‐TPO‐negative patients [16.5 ± 15.9 mg/L, median = 8.74 mg/L (3.93, 32.95 mg/L) versus 7.0 ± 7.2 mg/L, median = 3.37 mg/L (3.14, 6.69 mg/L), p < 0.05]. High CRP was also linked to elevated blood levels of IL‐31 (r = 0.36 p < 0.05) (Figure 3A). The mean IL‐31 level in patients in the high CRP group was markedly higher than the mean IL‐31 level in the low CRP group [12.8 ± 19.3 pg/mL, median = 7.14 pg/mL (4.43, 10.0 pg/mL) versus 5.0 ± 2.3 pg/mL, median = 5.06 pg/mL (3.41, 6.46 pg/mL), p = 0.01] (Table 1).
FIGURE 3.
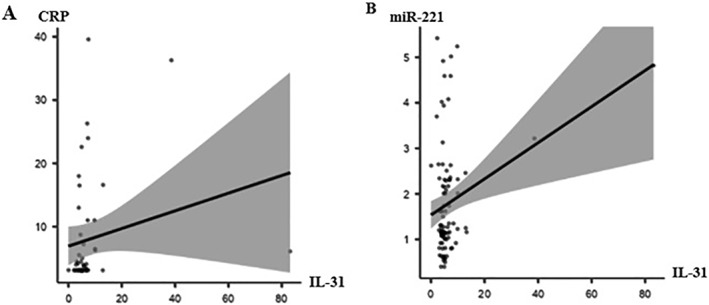
CRP and miR‐221 expression are linked to IL‐31. A. In CSU patients, there was a positive correlation between CRP levels and blood levels of IL‐31 (r = 0.36, p < 0.05). B. IL‐31 and miR‐221 expression levels showed a weak but significant correlation (r = 0.29, p < 0.05) in the entire study group. CRP, C‐reactive protein; CSU, chronic spontaneous urticaria.
TABLE 1.
Laboratory findings (a) and clinical characteristics (b) in CSU patients with low and high CRP levels.
| (a) Laboratory findings | Low CRP | High CRP | value |
|---|---|---|---|
| Mean ± SD | Mean ± SD | ||
| Median, percentiles a | Median, percentiles a | ||
| miR‐221 (2−ΔΔCT) | 2.42 ± 1.15 | 2.61 ± 1.59 | 0.651 |
| miR‐155 (2−ΔΔCT) | 2.48 ± 1.43 | 2.35 ± 1.19 | 0.756 |
| IL‐31 (pg/dL) a | 5.02 ± 2.28 | 12.77 ± 19.25 | 0.01 |
| 5.06 (3.41, 6.46) | 7.14 (4.43, 10.0) | ||
| Total IgE (IU/L) a | 125.4 ± 93.8 | 135.1 ± 102.9 | 0.948 |
| 95.8 (72.0, 171.5) | 112.5 (54.25, 229.25) |
| (b) Clinical characteristic | Low CRP n (%) | High CRP n (%) | value | |
|---|---|---|---|---|
| Duration | <1 year | 19 (65.5) | 7 (38.9) | <0.05 |
| >1 year | 10 (34.5) | 11 (61.1) | ||
| Atopic status | Yes | 1 (3.4) | 5 (27.8) | <0.05 |
| No | 28 (96.6) | 13 (72.2) | ||
| Angioedema | Yes | 10 (34.5) | 9 (50) | >0.05 |
| No | 19 (65.5) | 9 (50) | ||
| ASST | Negative | 23 (79.3) | 15 (83.3) | >0.05 |
| Positive | 6 (20.7) | 3 (16.7) | ||
| UAS7 | Mild | 5 (17.2) | 1 (5.6) | >0.05 |
| Moderate | 13 (44.8) | 7 (38.9) | ||
| Severe | 11 (37.9) | 10 (55.6) | ||
Note: Low CRP <5 mg/L; high CRP >5 mg/L. Significant results indicating p‐values below 0.05 are reported in bold.
Abbreviations: ASST, autologous serum skin test; CRP, C‐reactive protein; IgE, immunoglobulin E; IL‐31, interleukin 31; miR, microRNA; UAS, urticaria activity score.
Results of non‐parametric tests are shown as mean ± standard deviation and median and 25th and 75th percentiles.
Expression levels of miR‐221 were higher in anti‐TPO‐positive patients than in negative patients (3.5 ± 1.4 vs. 2.4 ± 1.3, p < 0.05). MiR‐221 expression levels were highest (2.7 ± 1.4) in patients with high disease activity, lowest in patients with mild disease activity (2.1 ± 1.2), and intermediate in patients with moderate disease activity (2.4 ± 1.3). Across the entire study population, expression levels of miR‐221 were significantly correlated with levels of interleukin‐31 (r = 0.29, p < 0.05), albeit weakly (Figure 3B). Both, miR‐221 and CRP levels, were significantly higher in patients who had their CSU for longer than 1 year (p < 0.01, p < 0.05, respectively).
4. DISCUSSION
This study identified several novel features of CSU, most importantly that total IgE and CRP are independently increased and linked to disease activity, that IgE and CRP are independently linked to miR‐155 and miR‐221, respectively, and that CRP and miR‐221 expression are linked to features of autoimmune CSU and interleukin‐31.
It is well known that CRP and total IgE levels are elevated on average in CSU, and it has also been suggested that they are potential biomarkers for disease activity. 7 , 8 , 26 , 27 Although the rates reported in studies vary, IgE is elevated in approximately half of the CSU patients 7 , 28 and CRP in about one‐third of CSU patients. 8 In our study, IgE elevation was found in 51.4% of the patients and CRP in 36.2%, confirming previous reports.
Total IgE has been the subject of numerous studies in CSU over the past years, but there are few studies on its association with disease activity, with contradictory results. 29 , 30 , 31 Some studies found IgE levels to be correlated with disease activity, 30 , 31 whereas others did not. 27 , 29 The reasons for this may include differences in study populations, including rates of comorbid atopic diseases.
Most studies on CRP levels in CSU found them to be linked with disease activity, 8 , 27 , 32 , 33 , 34 but two did not. 26 , 29 In our study, total IgE and CRP levels were highest in severe disease and lowest in mild disease activity. However, they were not correlated with each other, which supports their association with different CSU endotypes, that is, autoallergic and autoimmune CSU. 7 , 8 IgE antibodies against interleukin (IL)‐24, 35 thyroid peroxidase, 36 , 37 double‐stranded DNA, 38 tissue factor, 39 and thyroglobulin 40 in CSU patients are seen as drivers of the pathogenesis of autoallergic CSU. Many but not all studies have linked the presence of IgE autoantibodies to elevated total IgE levels in CSU patients. 37 , 41 Clinical features such as a higher frequency of atopy 37 and a rapid response to omalizumab 42 have been reported in CSU patients with high IgE levels. In a recent study, similar to our results, total IgE and CRP values were also not correlated with each other. 27
Remarkably, CSU patients had higher levels of miR‐155 and miR‐221. This has not previously been reported but does not come completely unexpected. miR‐155 and miR‐221 have been shown to have effects on mast cell functions. 14 , 16 , 43 , 44 Furthermore, overexpression of miR‐155 has been detected in mast cell‐driven diseases such as atopic dermatitis, 15 , 45 allergic contact dermatitis, 46 asthma, 47 and allergic rhinitis, 48 while overexpression of miR‐221 has been detected in chronic inflammatory diseases such as psoriasis, 17 psoriatic arthritis, 49 rheumatoid arthritis, 18 and ankylosing spondylitis. 50
Even more remarkably, miR‐155 was linked to IgE and miR‐221 to CRP. Why IgE and miR‐155 are associated is currently unclear, but they are also linked in atopic dermatitis (AD), where total IgE levels tend to be markedly higher than in CSU. 7 In a previous study, patients with atopic dermatitis had 4.6‐fold higher miR‐155, 15 as compared to 2.2‐fold elevated levels in our CSU patients. Interestingly, miR‐155 suppresses CTLA4, an inhibitory molecule of T cell response, in activated T cells. 15 In animal models, CTLA‐4 blockade has been shown to maintain or increase allergic responses and inflammation with increased IgE levels. 51 Furthermore, miR‐155 was upregulated in type 2 native lymphoid cells in response to IL‐33, 52 which can be increased in CSU 53 , 54 and causes elevated serum IgE. 55 Further studies are needed to better characterize the relationship between miR‐155, IgE, and IL‐33 in the pathogenesis of CSU.
Of note, miR‐155 is encoded within a region known as the B cell integration cluster (Bic) gene located on chromosome 21q21, 56 which has been linked with IgE‐mediated diseases. 57 In fact, miR‐155 is implicated in the pathogenesis of allergic diseases such as asthma, 47 allergic rhinitis, 58 and atopic dermatitis, 15 and many studies show that it plays an important role in IgE‐mediated immune responses. 14 , 43 Also, miR‐155 expression was increased in human skin‐derived mast cells and mouse bone marrow‐derived mast cells following FcεRI crosslinking with IgE antigen. 14 On the other hand, miR‐155 increased IgE‐dependent cytokine production by targeting the suppressor of cytokine signaling 1. 43 It was shown that miR‐155 specifically targets the FcεRI signaling pathway leading to prostaglandin biosynthesis and cytokine production but not the leukotriene production or degranulation pathways. 14 These results suggest positive feedback between IgE‐mediated cytokine release and miR‐155 expression. It has been reported that miR‐155 expression is higher in patients with allergic rhinitis than in those with non‐allergic rhinitis. In the same study, miR‐155 expression was found to be higher in patients with a positive skin prick test, which detects type I reactions related to allergens, compared with patients with negative prick test. 58 In our study, miR‐155 was overexpressed in CSU patients and had a specificity of 87% by ROC analysis. Taken together, these results suggest that miR‐155 is a biomarker for CSU, especially for autoallergic CSU.
CRP levels have previously been reported to be higher in autoimmune CSU. 8 , 32 Our finding of a correlation between CRP and anti‐TPO, a marker of autoimmune CSU, supports this notion. As a remarkable and new finding, one important parameter we found to be correlated with CRP was miR‐221. miR‐221 has been studied in autoimmune diseases such as psoriasis, 17 psoriatic arthritis, 49 and rheumatoid arthritis 54 and was suggested to be a biomarker for disease activity, early diagnosis, or response to treatment. For example, miR‐221 expression was increased in psoriasis and correlated with disease activity and inflammatory cytokine levels (tumor necrosis factor‐α, IL‐17A, and IL‐22). 17 Also, miR‐221 expression was increased in patients with psoriatic arthritis, and low miR‐221 levels were associated with poor treatment response. 49 In rheumatoid arthritis, miR‐221 was higher in patients with high disease activity, and the reduction of inflammatory cytokines (TNFa, IL‐6, and IL‐1β) by inhibition of miR‐221 suggests that it may be a target for treatment. 18 , 59 The fact that miR‐221 is more strongly stimulated by non‐IgE antibodies in previous studies suggests that the association between miR‐221 and anti‐TPO or CRP is not accidental and indicates that miR‐221 is a biomarker for autoimmune CSU.
We also observed that both CRP and miR‐221 are linked to IL‐31. IL‐31, a member of the IL‐6 cytokine family, is a pruritic proinflammatory cytokine. 60 Not surprisingly, IL‐31 levels are elevated in pruritic diseases such as atopic dermatitis, 19 prurigo nodularis, 61 lichen planus, 62 and uremic pruritus. 63 Furthermore, elevated IL‐31 levels have been reported in autoimmune diseases such as psoriasis, 64 systemic lupus erythematosus, 65 dermatomyositis, 66 bullous pemphigoid, 67 and subtypes of pemphigus including the non‐pruritic subtype pemphigus vegetans. 68
IL‐31 levels were previously reported to be elevated in CSU, but its role in the pathogenesis has not been clarified yet. 20 , 23 , 69 The decrease in IL‐31 levels after omalizumab treatment in CSU patients suggests that IL‐31 is a pathogenic driver of CSU. 24 In line with our results, IL‐31 was not associated with disease activity as measured by UAS7 in previous studies, 23 , 24 , 54 although results regarding the correlation with itch scores are conflicting. 23 , 54 The detection of higher IL‐31 levels in patients with high ANA titers 23 prompts us to consider whether IL‐31 contributes to the pathogenesis of autoimmune CSU, although there is insufficient evidence for this. IL‐31 levels were higher in CSU patients with thyroid autoimmunity (assessed by anti‐TPO and/or anti‐TG) than in patients without thyroid autoimmunity, but the difference was not statistically significant. 23 In previous studies, there was no difference in IL‐31 plasma levels in ASST‐positive or negative CSU patients, 69 , 70 and IL‐31 was not linked to CRP. 54 In contrast, in our study, IL‐31 was linked to miR‐221 and CRP. The mean IL‐31 level in patients in the high CRP group was higher than the mean IL‐31 level in the low CRP group. A previous CSU study suggested that IL‐31 is not the primary mediator of pruritus, as some patients with high disease activity had undetectable levels of IL‐31, and the highest IL‐31 level was lower than what is required to induce itch sensation. 24 The fact that IL‐31 is not only elevated in pruritic skin diseases but also in autoimmune diseases suggests that it plays an important role in inflammatory responses and autoimmunity beyond itch.
Our study has two limitations: (i) a relatively small number of patients and (ii) the absence of post‐treatment levels of the biomarkers assessed. However, the strength of the study is that it is a prospective study in which the clinical and laboratory findings of the patients were evaluated comprehensively.
5. CONCLUSİON
Total IgE and CRP appear to be drivers in different subtypes of CSU and may be biomarkers of disease activity. Upregulation of miR‐155 and miR‐221 with high specificity in CSU patients makes their role in CSU pathogenesis worth investigating. The association of miR‐221 with CRP, anti‐TPO, IL‐31, and disease activity indicates that it is a potent biomarker for CSU, especially for autoimmune CSU. Further studies should explore the role of miRNAs and interleukin‐31 signatures in CSU.
AUTHOR CONTRIBUTIONS
Ozge Sevil Karstarli Bakay and Betül Demir designed the study. Ozge Sevil Karstarli Bakay, Betül Demir, and Demet Cicek coordinated the study. Deniz Erol and Zulal Aşçı Toraman established and performed the laboratory tests. Demet Cicek and Yunus Gural performed statistical analyses. Ozge Sevil Karstarli Bakay and Marcus Maurer were involved in data interpretation. Ozge Sevil Karstarli Bakay and Marcus Maurer drafted the manuscript. All authors proof‐read and approved the final version of the manuscript.
CONFLICT OF INTEREST STATEMENT
None, in relation to this work. Outside of it, MM is or recently was a speaker and/or advisor for and/or has received research funding from Allakos, Amgen, Aralez, ArgenX, AstraZeneca, Celldex, Centogene, CSL Behring, FAES, Genentech, GIInnovation, GSK, Innate Pharma, Kyowa Kirin, Leo Pharma, Lilly, Menarini, Moxie, Novartis, Pfizer, Roche, Sanofi/Regeneron, Third Harmonic Bio, UCB, and Uriach.
ACKNOWLEDGMENTS
This work was supported by the Firat University Coordinatorship of Scientific Research Projects (No: TF.16.36).
Open Access funding enabled and organized by Projekt DEAL.
Karstarli Bakay OS, Demir B, Cicek D., et al. In chronic spontaneous urticaria, IgE and C‐reactive protein are linked to distinct microRNAs and interleukin‐31. Clin Transl Allergy. 2023;e12290. 10.1002/clt2.12290
DATA AVAILABILITY STATEMENT
Data can be requested by the corresponding authors.
REFERENCES
- 1. Kolkhir P, Giménez‐Arnau AM, Kulthanan K, Peter J, Metz M, Maurer M. Urticaria. Nat Rev Dis Prim. 2022;8(1):61. 10.1038/s41572-022-00389-z [DOI] [PubMed] [Google Scholar]
- 2. Church MK, Kolkhir P, Metz M, Maurer M. The role and relevance of mast cells in urticaria. Immunol Rev. 2018;282(1):232‐247. 10.1111/imr.12632 [DOI] [PubMed] [Google Scholar]
- 3. Bracken SJ, Abraham S, MacLeod AS. Autoimmune theories of chronic spontaneous urticaria. Front Immunol. 2019;29(10):627. 10.3389/fimmu.2019.00627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kolkhir P, Muñoz M, Asero R, et al. Autoimmune chronic spontaneous urticaria. J Allergy Clin Immunol. 2022;149(6):1819‐1831. 10.1016/j.jaci.2022.04.010 [DOI] [PubMed] [Google Scholar]
- 5. Schoepke N, Asero R, Ellrich A, et al. Biomarkers and clinical characteristics of autoimmune chronic spontaneous urticaria: results of the PURIST Study. Allergy. 2019;74(12):2427‐2436. 10.1111/all.13949 [DOI] [PubMed] [Google Scholar]
- 6. Zuberbier T, Abdul Latiff AH, Abuzakouk M, et al. The international EAACI/GA2LEN/EuroGuiDerm/APAAACI guideline for the definition, classification, diagnosis, and management of urticaria. Allergy. 2022;77(3):734‐766. 10.1111/all.15090 [DOI] [PubMed] [Google Scholar]
- 7. Altrichter S, Fok JS, Jiao Q, et al. Total IgE as a marker for chronic spontaneous urticaria. Allergy Asthma Immunol Res. 2021;13(2):206‐218. 10.4168/aair.2021.13.2.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kolkhir P, Altrichter S, Hawro T, Maurer M. C‐reactive protein is linked to disease activity, impact, and response to treatment in patients with chronic spontaneous urticaria. Allergy. 2018;73(4):940‐948. 10.1111/all.13352 [DOI] [PubMed] [Google Scholar]
- 9. Lin CE, Kaptein JS, Sheikh J. Differential expression of microRNAs and their possible roles in patients with chronic idiopathic urticaria and active hives. Allergy Rhinol. 2017;8(2):67‐80. 10.2500/ar.2017.8.0199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang L, Qi R, Yang Y, Gao X, Chen H, Xiao T. Serum miR‐125a‐5p and CCL17 upregulated in chronic spontaneous urticaria and correlated with treatment response. Acta Derm Venereol. 2019;99(6):571‐578. 10.2340/00015555-3149 [DOI] [PubMed] [Google Scholar]
- 11. Calame K. MicroRNA‐155 function in B cells. Immunity. 2007;27(6):825‐827. 10.1016/j.immuni.2007.11.010 [DOI] [PubMed] [Google Scholar]
- 12. Biethahn K, Orinska Z, Vigorito E, et al. miRNA‐155 controls mast cell activation by regulating the PI3Kc pathway and anaphylaxis in a mouse model. Allergy. 2014;69(6):752‐762. 10.1111/all.12407 [DOI] [PubMed] [Google Scholar]
- 13. Leib N, Herrmann N, Koch S, et al. MicroRNA‐155 mediates downregulation of the high‐affinity receptor for IgE through toll‐like receptor signaling. J Allergy Clin Immunol. 2018;141(1):425‐429. 10.1016/j.jaci.2017.08.014 [DOI] [PubMed] [Google Scholar]
- 14. Mohammed Z, McHale C, Kubinak JL, Dryer S, Gomez G. miR‐155 is a positive regulator of FcεRI‐induced cyclooxygenase‐2 expression and cytokine production in mast cells. Front Allergy. 2022;3:835776. 10.3389/falgy.2022.835776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sonkoly E, Janson P, Majuri ML, et al. MiR‐155 is overexpressed in patients with atopic dermatitis and modulates T‐cell proliferative responses by targeting cytotoxic T lymphocyte‐associated antigen 4. J Allergy Clin Immunol. 2010;126(3):581‐589. 10.1016/j.jaci.2010.05.045 [DOI] [PubMed] [Google Scholar]
- 16. Mayoral RJ, Deho L, Rusca N, et al. MiR‐221 influences effector functions and actin cytoskeleton in mast cells. PLoS One. 2011;6(10):26133. 10.1371/journal.pone.0026133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meng Z, Qiu J, Zhang H. MiR‐221‐3p as a potential biomarker for patients with psoriasis and its role in inflammatory responses in keratinocytes. Skin Pharmacol Physiol. 2021;34(5):300‐306. 10.1159/000515114 [DOI] [PubMed] [Google Scholar]
- 18. Abo ElAtta AS, Ali YBM, Bassyouni IH, Talaat RM. Upregulation of miR‐221/222 expression in rheumatoid arthritis (RA) patients: correlation with disease activity. Clin Exp Med. 2019;19(1):47‐53. 10.1007/s10238-018-0524-3 [DOI] [PubMed] [Google Scholar]
- 19. Sokołowska‐Wojdyło M, Gleń J, Zabłotna M, et al. Association of distinct IL‐31 polymorphisms with pruritus and severity of atopic dermatitis. J Eur Acad Dermatol Venereol. 2013;27(5):662‐664. 10.1111/j.1468-3083.2012.04649.x [DOI] [PubMed] [Google Scholar]
- 20. Raap U, Wieczorek D, Gehring M, et al. Increased levels of serum IL‐31 in chronic spontaneous urticaria. Exp Dermatol. 2010;19(5):464‐466. 10.1111/j.1600-0625.2010.01067.x [DOI] [PubMed] [Google Scholar]
- 21. Schulz F, Marenholz I, Fölster‐Holst R, et al. A common haplotype of the IL‐31 gene influencing gene expression is associated with nonatopic eczema. J Allergy Clin Immunol. 2007;120(5):1097‐1102. 10.1016/j.jaci.2007.07.065 [DOI] [PubMed] [Google Scholar]
- 22. Hartmann K, Wagner N, Rabenhorst A, et al. Serum IL‐31 levels are increased in a subset of patients with mastocytosis and correlate with disease severity in adult patients. J Allergy Clin Immunol. 2013;132(1):232‐235. 10.1016/j.jaci.2012.11.008 [DOI] [PubMed] [Google Scholar]
- 23. Chaowattanapanit S, Choonhakarn C, Salao K, et al. Increased serum IL‐31 levels in chronic spontaneous urticaria and psoriasis with pruritic symptoms. Heliyon. 2020;6(12):e05621. 10.1016/j.heliyon.2020.e05621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Altrichter S, Hawro T, Hänel K, et al. Successful omalizumab treatment in chronic spontaneous urticaria is associated with lowering of serum IL‐31 levels. J Eur Acad Dermatol Venereol. 2016;30(3):454‐455. 10.1111/jdv.12831 [DOI] [PubMed] [Google Scholar]
- 25. Stull D, McBride D, Tian H, et al. Analysis of disease activity categories in chronic spontaneous/idiopathic urticaria. Br J Dermatol. 2017;177(4):1093‐1101. 10.1111/bjd.15454 [DOI] [PubMed] [Google Scholar]
- 26. Plavsic A, Tomic‐Spiric V, Arandjelovic S, Miskovic R, Dimitrijevic M, Peric‐Popadic A. Biomarkers of disease activity in patients with chronic spontaneous urticaria. Postepy Dermatol Alergol. 2021;38(6):1017‐1022. 10.5114/ada.2021.112276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Montjoye L, Darrigade AS, Giménez‐Arnau A, Herman A, Dumoutier L, Baeck M. Correlations between disease activity, autoimmunity, and biological parameters in patients with chronic spontaneous urticaria. Eur Ann Allergy Clin Immunol. 2021;53(2):55‐66. 10.23822/eurannaci.1764-1489.132 [DOI] [PubMed] [Google Scholar]
- 28. Staubach P, Vonend A, Burow G, Metz M, Magerl M, Maurer M. Patients with chronic urticaria exhibit increased rates of sensitisation to Candida albicans, but not to common moulds. Mycoses. 2009;52(4):334‐338. 10.1111/j.1439-0507.2008.01601.x [DOI] [PubMed] [Google Scholar]
- 29. Baek YS, Jeon J, Kim JH, Oh CH. Severity of acute and chronic urticaria correlates with D‐dimer level, but not C‐reactive protein or total IgE. Clin Exp Dermatol. 2014;39(7):795‐800. 10.1111/ced.12413 [DOI] [PubMed] [Google Scholar]
- 30. Altrichter S, Hawro T, Liedtke M, et al. In chronic spontaneous urticaria, IgE against staphylococcal enterotoxins is common and functional. Allergy. 2018;73(7):1497‐1504. 10.1111/all.13381 [DOI] [PubMed] [Google Scholar]
- 31. Kessel A, Helou W, Bamberger E, et al. Elevated serum total IgE – a potential marker for severe chronic urticaria. Int Arch Allergy Immunol. 2010;153(3):288‐293. 10.1159/000314370 [DOI] [PubMed] [Google Scholar]
- 32. Czarnecka‐Operacz M, Szulczyńska‐Gabor J, Leśniewska K, et al. Acute‐phase response and its biomarkers in acute and chronic urticaria. Postepy Dermatol Alergol. 2018;35(4):400‐407. 10.5114/ada.2018.77672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tedeschi A, Asero R, Lorini M, Marzano AV, Cugno M. Plasma levels of matrix metalloproteinase‐9 in chronic urticaria patients correlate with disease severity and C‐reactive protein but not with circulating histamine‐releasing factors. Clin Exp Allergy. 2010;40(6):875‐881. 10.1111/j.1365-2222.2010.03473.x [DOI] [PubMed] [Google Scholar]
- 34. Deza G, Ricketti PA, Giménez‐Arnau AM, Casale TB. Emerging biomarkers and therapeutic pipelines for chronic spontaneous urticaria. J Allergy Clin Immunol Pract. 2018;6(4):1108‐1117. 10.1016/j.jaip.2018.02.024 [DOI] [PubMed] [Google Scholar]
- 35. Schmetzer O, Lakin E, Topal FA, et al. IL‐24 is a common and specific autoantigen of IgE in patients with chronic spontaneous urticaria. J Allergy Clin Immunol. 2018;142(3):876‐882. 10.1016/j.jaci.2017.10.035 [DOI] [PubMed] [Google Scholar]
- 36. Altrichter S, Peter HJ, Pisarevskaja D, Metz M, Martus P, Maurer M. IgE mediated autoallergy against thyroid peroxidase–a novel pathomechanism of chronic spontaneous urticaria? PLoS One. 2011;6(4):e14794. 10.1371/journal.pone.0014794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sánchez J, Sánchez A, Cardona R. Clinical characterization of patients with chronic spontaneous urticaria according to anti‐TPO IgE levels. J Immunol Res. 2019;2019:4202145. 10.1155/2019/4202145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hatada Y, Kashiwakura J, Hayama K, et al. Significantly high levels of anti‐dsDNA immunoglobulin E in sera an the ability of dsDNA to induce the degranulation of basophils from chronic urticaria patients. Int Arch Allergy Immunol. 2013;161(Suppl 2):154‐158. 10.1159/000350388 [DOI] [PubMed] [Google Scholar]
- 39. Cugno M, Asero R, Ferrucci S, et al. Elevated IgE to tissue factor and thyroglobulin are abated by omalizumab in chronic spontaneous urticaria. Allergy. 2018;73(12):2408‐2411. 10.1111/all.13587 [DOI] [PubMed] [Google Scholar]
- 40. Kolkhir P, Metz M, Altrichter S, Maurer M. Comorbidity of chronic spontaneous urticaria and autoimmune thyroid diseases: a systematic review. Allergy. 2017;72(10):1440‐1460. 10.1111/all.13182 [DOI] [PubMed] [Google Scholar]
- 41. Sánchez J, Sánchez A, Cardona R. Causal relationship between anti‐TPO IgE and chronic urticaria by in vitro and in vivo tests. Allergy Asthma Immunol Res. 2019;11(1):29‐42. 10.4168/aair.2019.11.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marzano AV, Genovese G, Casazza G, et al. Predictors of response to omalizumab and relapse in chronic spontaneous urticaria: a study of 470 patients. J Eur Acad Dermatol Venereol. 2019;33(5):918‐924. 10.1111/jdv.15350 [DOI] [PubMed] [Google Scholar]
- 43. Qayum AA, Paranjape A, Abebayehu D, et al. IL‐10‐Induced miR‐155 targets SOCS1 to enhance IgE‐mediated mast cell function. J Immunol. 2016;196(11):4457‐4467. 10.4049/jimmunol.1502240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mayoral RJ, Pipkin ME, Pachkov M, van Nimwegen E, Rao A, Monticelli S. MicroRNA‐221‐222 regulate the cell cycle in mast cells. J Immunol. 2009;182(1):433‐445. 10.4049/jimmunol.182.1.433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ma L, Xue H‐B, Wang F, Shu C‐M, Zhang J‐H. MiRNA‐155 may be involved in the pathogenesis of atopic dermatitis by modulating the differentiation and function of T helper type 17 (Th17) cells. Clin Exp Immunol. 2015;181(1):142‐149. 10.1111/cei.12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Z, Yi T, Long M, Ding F, Ouyang L, Chen Z. Involvement of the negative feedback of IL‐33 signaling in the anti‐inflammatory effect of electro‐acupuncture on allergic contact dermatitis via targeting MicroRNA‐155 in mast cells. Inflammation. 2018;41(3):859‐869. 10.1007/s10753-018-0740-8 [DOI] [PubMed] [Google Scholar]
- 47. Zhou H, Li J, Gao P, Wang Q, Zhang J. miR‐155: a novel target in allergic asthma. Int J Mol Sci. 2016;17(10):1773. 10.3390/ijms17101773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhu Y, Liu Y, Zhu X, Wang Z, Wang M. Upregulation of miR‐155 regulates group 2 innate lymphoid cells by targeting c‐maf in allergic rhinitis. Eur J Pharmacol. 2020;887:173564. 10.1016/j.ejphar.2020.173564 [DOI] [PubMed] [Google Scholar]
- 49. Wade SM, McGarry T, Wade SC, Fearon U, Veale DJ. Serum microRNA signature as a diagnostic and therapeutic marker in patients with psoriatic arthritis. J Rheumatol. 2020;47(12):1760‐1767. 10.3899/jrheum.190602 [DOI] [PubMed] [Google Scholar]
- 50. Lai NS, Yu HC, Chen HC, Yu CL, Huang HB, Lu MC. Aberrant expression of microRNAs in T cells from patients with ankylosing spondylitis contributes to the immunopathogenesis. Clin Exp Immunol. 2013;173(1):47‐57. 10.1111/cei.12089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jen KY, Campo M, He H, et al. CD45RB ligation inhibits allergic pulmonary inflammation by inducing CTLA4 transcription. J Immunol. 2007;179(6):4212‐4218. 10.4049/jimmunol.179.6.4212 [DOI] [PubMed] [Google Scholar]
- 52. Johansson K, Malmhäll C, Ramos‐Ramírez P, Rådinger M. MicroRNA‐155 is a critical regulator of type 2 innate lymphoid cells and IL‐33 signaling in experimental models of allergic airway inflammation. J Allergy Clin Immunol. 2017;139(3):1007‐1016. 10.1016/j.jaci.2016.06.035 [DOI] [PubMed] [Google Scholar]
- 53. Kay AB, Clark P, Maurer M, Ying S. Elevations in T‐helper‐2‐initiating cytokines (interleukin‐33, interleukin‐25 and thymic stromal lymphopoietin) in lesional skin from chronic spontaneous ('idiopathic') urticaria. Br J Dermatol. 2015;172(5):1294‐1302. 10.1111/bjd.13621 [DOI] [PubMed] [Google Scholar]
- 54. Lin W, Zhou Q, Liu C, Ying M, Xu S. Increased plasma IL‐17, IL‐31, and IL‐33 levels in chronic spontaneous urticaria. Sci Rep. 2017;7(1):17797. 10.1038/s41598-017-18187-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schmitz J, Owyang A, Oldham E, et al. IL‐33, an interleukin‐1‐like cytokine that signals via the IL‐1 receptor‐related protein ST2 and induces T helper type 2‐associated cytokines. Immunity. 2005;23(5):479‐490. 10.1016/j.immuni.2005.09.015 [DOI] [PubMed] [Google Scholar]
- 56. Leng RX, Pan HF, Qin WZ, Chen GM, Ye DQ. Role of microRNA‐155 in autoimmunity. Cytokine Growth Factor Rev. 2011;22(3):141‐147. 10.1016/j.cytogfr.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 57. Blumenthal MN, Langefeld CD, Barnes KC, et al. A genome‐wide search for quantitative trait loci contributing to variation in seasonal pollen reactivity. J Allergy Clin Immunol. 2006;117(1):79‐85. 10.1016/j.jaci.2005.09.038 [DOI] [PubMed] [Google Scholar]
- 58. Suojalehto H, Toskala E, Kilpeläinen M, et al. MicroRNA profiles in nasal mucosa of patients with allergic and nonallergic rhinitis and asthma. Int Forum Allergy Rhinol. 2013;3(8):612‐620. 10.1002/alr.21179 [DOI] [PubMed] [Google Scholar]
- 59. Yang S, Yang Y. Downregulation of microRNA221 decreases migration and invasion in fbroblast‐like synoviocytes in rheumatoid arthritis. Mol Med Rep. 2015;12(2):2395‐2401. 10.3892/mmr.2015.3642 [DOI] [PubMed] [Google Scholar]
- 60. Murdaca G, Greco M, Tonacci A, et al. IL‐33/IL‐31 axis in immune‐mediated and allergic diseases. Int J Mol Sci. 2019;20(23):5856. 10.3390/ijms20235856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chaowattanapanit S, Wongjirattikarn R, Chaisuriya N, et al. Increased IL‐31 expression in serum and tissue protein in prurigo nodularis. Ther Adv Chronic Dis. 2022;13:20406223221112561. 10.1177/20406223221112561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Welz‐Kubiak K, Kobuszewska A, Reich A. IL‐31 is overexpressed in lichen planus but its level does not correlate with pruritus severity. J Immunol Res. 2015;2015:854747. 10.1155/2015/854747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Oweis AO, Al‐Qarqaz F, Bodoor K, et al. Elevated interleukin 31 serum levels in hemodialysis patients are associated with uremic pruritus. Cytokine. 2021;138:155369. 10.1016/j.cyto.2020.155369 [DOI] [PubMed] [Google Scholar]
- 64. Purzycka‐Bohdan D, Gleñ J, Zabłotna M, et al. Significance of interleukin‐31 (IL‐31) gene polymorphisms and IL‐31 serum level in psoriasis in correlation with pruritus. Postepy Dermatol Alergol. 2021;38(4):657‐664. 10.5114/ada.2021.108926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang HT, Chen JM, Guo J, Lan Y, Wei YS. The association of interleukin‐31 polymorphisms with interleukin‐31 serum levels and risk of systemic lupus erythematosus. Rheumatol Int. 2016;36(6):799‐805. 10.1007/s00296-016-3422-6 [DOI] [PubMed] [Google Scholar]
- 66. Kim HJ, Zeidi M, Bonciani D, et al. Itch in dermatomyositis: the role of increased skin interleukin‐31. Br J Dermatol. 2018;179(3):669‐678. 10.1111/bjd.16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Salz M, Haeberle S, Hoffmann J, Enk AH, Hadaschik EN. Elevated IL‐31 serum levels in bullous pemphigoid patients correlate with eosinophil numbers and are associated with BP180‐IgE. J Dermatol Sci. 2017;87(3):309‐311. 10.1016/j.jdermsci.2017.07.019 [DOI] [PubMed] [Google Scholar]
- 68. Okuno S, Hashimoto T, Yamazaki Y, Okuzawa M, Satoh T. IL‐31 and IL‐31 receptor alpha in pemphigus: contributors to more than just itch? J Dermatol. 2023;50(7):927‐930. 10.1111/1346-8138.16730 [DOI] [PubMed] [Google Scholar]
- 69. Crisan IG, Bocsan CI, Vesa SC, Cristea V. Correlations between serum levels of IL‐17, IL‐4, IL‐31, IFN‐ϒ and etiological factors in patients with CSU. HVM Bioflux. 2014;6:25‐29. [Google Scholar]
- 70. Raap U, Gehring M, Kleiner S, et al. Human basophils are a source of ‐ and are differentially activated by ‐ IL‐31. Clin Exp Allergy. 2017;47(4):499‐508. 10.1111/cea.12875 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data can be requested by the corresponding authors.


