Abstract
Abrupt aggregation of α-synuclein (α-Syn) leads to a formation of highly toxic protein oligomers. These aggregates are the underlying molecular cause of an onset of the irreversible degeneration of dopaminergic neurons in midbrain, hypothalamus, and thalamus, a pathology known as Parkinson’s disease. The transient nature of oligomers, as well as their structural and morphological heterogeneity, limits the use of cryo-electron microscopy and solid-state NMR, classical tools of structural biology, for elucidation of their secondary structure. Despite this limitation, numerous pieces of experimental evidence suggest that phospholipids can uniquely alter the structure and toxicity of oligomers. In this study, we utilize an innovative nano-infrared imaging technique, also known as atomic force microscopy infrared (AFM-IR) spectroscopy, to examine the structure of individual α-Syn oligomers grown in the presence of phosphatidylcholine (α-Syn:PC) and phosphatidylserine (α-Syn:PS). We determined the amount of the parallel and the antiparallel β-sheets, as well as the amount the α-helix and the unordered protein, in the secondary structure of α-Syn:PC and α-Syn:PS formed at day 2 (D2), 8 (D8), and 15 (D15) after initiation of protein aggregation. We found a gradual decrease in the amount of the parallel β-sheet in both α-Syn:PC and α-Syn:PS from D2 to D15 together with an increase in the α-helix and the unordered protein secondary structure. We infer that this is due to the presence of lipids in the structure of oligomers that prevent an expansion of the parallel β-sheet upon interaction of the oligomers with monomeric α-Syn.
Keywords: α-Synuclein aggregation, AFM-IR, protein−lipid interaction, oligomers, protein secondary structure, phosphatidylcholine and phosphatidylserine
Graphical Abstract

INTRODUCTION
Parkinson’s disease (PD) is the fastest growing neurodegenerative disease, projected to strike 12 million people by 2040 worldwide.1 PD is characterized by irreversible degeneration of dopaminergic neurons in substantia nigra pars compacta, as well as neurons in other regions of the brain, which progresses in the caudo-rostral axis. These neurodegenerative processes are linked to abrupt aggregation of α-synuclein (α-Syn), a small protein with an unordered secondary structure. α-Syn first forms soluble oligomers with a variety of structures that are observed in vitro and in vivo.2–8 Some of these oligomers can propagate into fibrils that are long unbranched β-sheet-rich assemblies. It should be noted that most fibrillary α-Syn is formed from monomers and not from oligomers.3,9 The use of cryo-EM and solid-state NMR allows for detailed elucidation of the correlation between molecular structures and the secondary structure of α-Syn fibrils.10–13 However, high structural heterogeneity of α-Syn oligomers, their transient nature, and low concentrations limit the use of both techniques. As a result, very little is known about the oligomeric structure of α-Syn. Despite this limitation, numerous pieces of experimental evidence point to a direct relationship between the crude shape of α-Syn oligomers and their propensity for exerting toxicity.14–17 Specifically, it has been found that small annular α-Syn oligomers induce Ca 2+ influx, caspase activation and cell death, whereas larger α-Syn oligomers exhibit no toxicity.15
Konno, Banerjee, and co-workers utilized high-speed atomic force microscopy to monitor the dynamics of changes in the protein secondary structure that is taken place upon propagation of amyloid oligomers into fibrils.18,19 Kurouski and co-workers characterized the surface of insulin protofilaments and fibrils by using tip-enhanced Raman spectroscopy (TERS).20 The authors found that the surface of amyloid fibrils had a very high content of the unordered protein secondary structure, whereas the core of the fibrils was primarily composed of the parallel β-sheet. It was also found that surfaces of twisted and tape-like fibril polymorphs have a distinctly different secondary structure. A TERS analogue, atomic force microscopy infrared (AFM-IR) spectroscopy, is a modern analytical technique that can be used to probe the secondary structure of individual amyloid oligomers.21–23 AFM-IR utilizes a metalized probe that is positioned at the protein aggregates and illuminated by pulsed tunable IR light.24 IR pulses cause thermal expansions in the sample that are recorded by the metalized scanning probe. It was recently demonstrated that AFM-IR possessed single-monolayer and even single-molecule sensitivity.21,22 It should be noted that conventional IR spectroscopy probes the changes in the bulk volume of the protein sample. Therefore, it cannot be used to reveal the structure of highly heterogeneous, from the perspective of the protein secondary structure, amyloid oligomers. Furthermore, concentrations of amyloid oligomers are too small for their structural characterization using conventional IR spectroscopy. Finally, AFM-IR and TERS provide different but complementary information about the structure of biological specimens.25 This high sensitivity and nanometer spatial resolution made AFM-IR highly attractive for structural analysis of amyloid fibrils,21,23,24,26–28 viruses,29 and plant epicuticular waxes.30,31 Using AFM-IR, Zhou and Kurouski were able to resolve structural changes that are taken place in the structure of a-Syn oligomers upon their propagation into fibrils and observed that α-Syn with similar height profiles could express a different protein secondary structure.32 Furthermore, the researchers revealed substantial structural heterogeneity in the structure of individual oligomers present at the same time of the protein aggregation. Dou and co-workers showed that an aggregation of α-Syn in the presence of large unilamellar vesicles (LUVs) of phosphatidylserine (PS) and phosphatidylcholine (PC) results in the formation of highly homogeneous, from the perspective of protein secondary structure, oligomers.33 These findings suggest that lipids facilitate protein aggregation. Furthermore, it was demonstrated that such oligomers contain lipids in their structure. Using AFM-IR, Rizevsky and co-workers recently showed that phospholipids uniquely alter the secondary structure and toxicity of insulin oligomers and fibrils.34 The researchers also demonstrated that early stage insulin oligomers exhibit strong lipid content, whereas the amount of lipids decreases as the oligomers propagate into filaments and fibrils, higher assembly structures of misfolded proteins. These findings are in good agreement with the experimental results previously reported by Galvagnion and co-workers showing that PS could facilitate the α-Syn aggregation by accelerating the primary nucleation.35 Furthermore, Galvagnion and Alza found that the rate of α-Syn aggregation directly depends on the lipid-to-protein ratio (P:L ratio).36–38 Specifically, with an increase in the concentration of PS, the researchers observed the increase in the rate of α-Syn aggregation. This suggested that lipids facilitated protein aggregation. However, with the subsequent increase in the amount of lipids, a decrease in rate of α-Syn aggregation was observed. This suggested that delocalization of monomeric α-Syn along surfaces of PS vesicles disfavored protein−protein interactions that were necessary for the aggregate formation.39–42
Expanding upon these findings, we investigate the changes in the secondary structure of α-Syn oligomers in the presence of PC and PS LUVs. To slow down α-Syn aggregation and understand the correlation between lipid and protein secondary structure, we collect the samples at day 2 (D2), day 8 (D8), and day 15 (D15) at room temperature with no agitation after initiation of protein aggregation in the presence of PC and PS.
RESULTS AND DISCUSSION
Microscopic examination of α-Syn aggregates grown in the presence of PC (α-Syn:PC) revealed the presence of spherical oligomers that had a height of ~ 1–3 nm (D2), Figure 1 and Figure S1. The height of these aggregates gradually increased during the course of protein aggregation reaching 2–4 nm (D8) and 3–7 nm (D15). Morphologically similar aggregates were observed for α-Syn grown in the presence of PS (α-Syn:PS). At D2, their height ranged within 1–3 nm. At D8, two populations of these oligomers were observed. The first population had a height of 1–5 nm, whereas the second one exhibited the height of 7–10 nm. α-Syn:PS oligomers observed at D15 exhibited a broad range of sizes ranging from 2 to 17 nm, Figure 1. It should be noted that the appearance of morphologically similar aggregates was observed under the lipid-free conditions, Figure S1 and S3.
Figure 1.
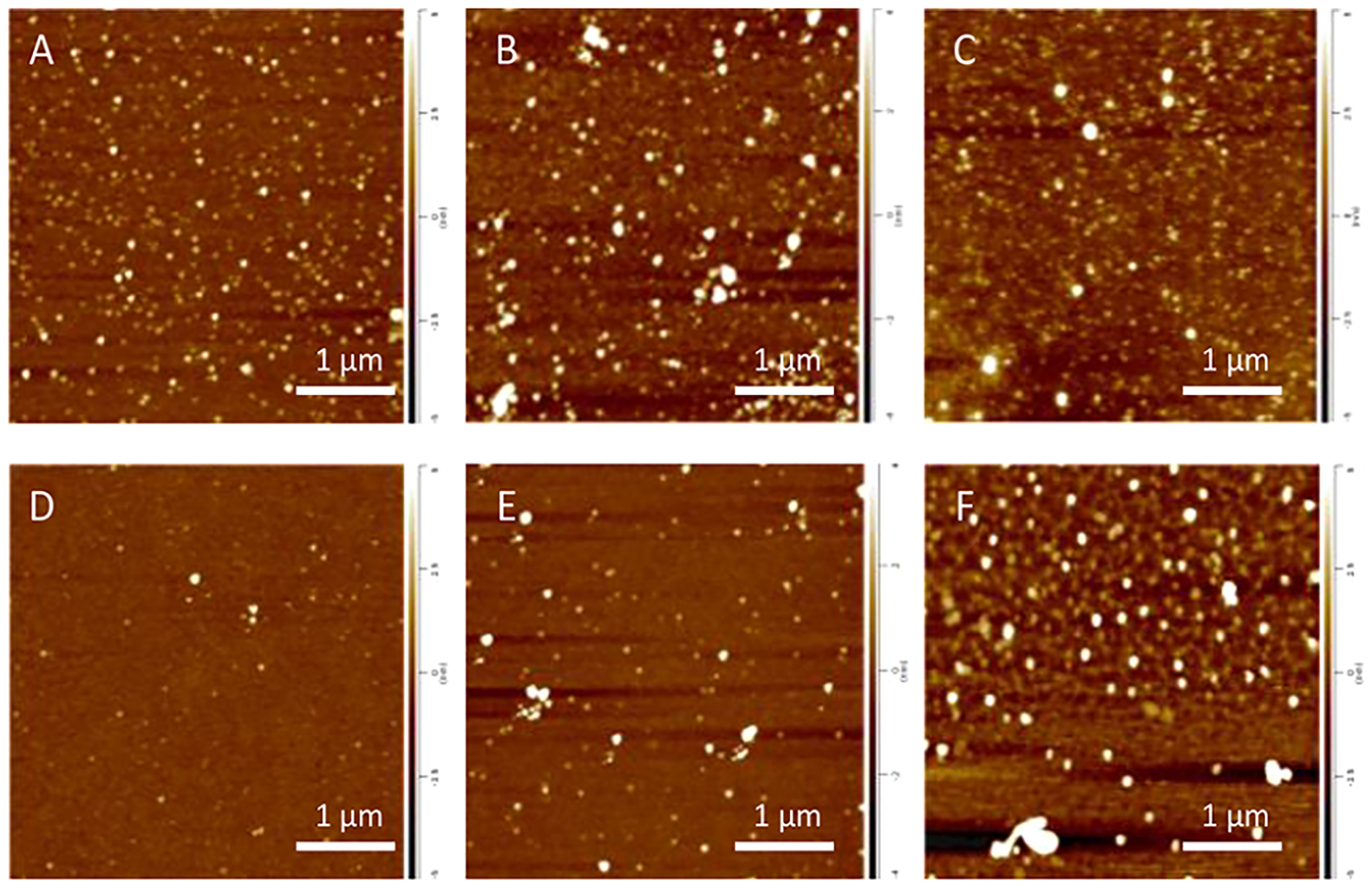
AFM images of α-Syn:PC (top row) and α-Syn:PS (bottom row) aggregates formed at D2 (A and D), D8 (B and E), and D15 (C and F) at a 1:2 lipid-to-protein (L:P) ratio.
Nanoscale infrared imaging of α-Syn:PC and α-Syn:PS oligomers formed at D2, D8, and D15 revealed their high parallel β-sheet content, as well as the presence of the antiparallel β-sheet in their structure, Figure 2. It should be noted that all observed oligomers present in each of the analyzed samples exhibited low structural heterogeneity (Figure S8), which confirms the previously reported hypothesis that lipids facilitate α-Syn aggregation. Thus, in the presence of a phospholipids, aggregation of α-Syn yields structurally similar oligomers that were not observed upon α-Syn aggregation in the lipid-free environment, Figure S3 and S4.
Figure 2.

AFM-IR maps of α-Syn:PC (A–D, H–K, and P–S) and α-Syn:PS (E–G, L–O, and T–W) formed at D2 (A–H), D8 (I–P), and D15 (Q–X) at a 1:2 lipid-to-protein (L:P) ratio. AFM height images (A, E, I, M, Q, U), IR absorption maps at 1100 cm−1 (ν(PO2−) of phospholipids) (B, F, J, N, R, V), IR ratio overlaying map of 1624 cm−1 (parallel β-sheet) (green) and 1655 cm−1 (α-helix/unordered protein secondary structure) (red) (C, G, K, O, S, W), and IR absorption map at 1694 cm−1 (antiparallel β-sheet) (D, H, L, P, T,X). Scale bar, 100 nm.
We observed a substantial amount of the unordered protein in the secondary structure of α-Syn:PS oligomers grown at D2.The amount of the unordered protein slightly decreased in these oligomers at D8 and D15 with an increase in the amount of the parallel β-sheet (Figure 2). Finally, we found that all analyzed α-Syn:PC and α-Syn:PS aggregates have high amount of lipids in their structure (Figure 2 and Figure S5). This experimental evidence demonstrates that lipids present in the α-Syn oligomers were grown in their presence.
Next, we collected more than 50 spectra from α-Syn:PC and α-Syn:PS oligomers present at D2, D8, and D15, as well as from the oligomers grown in the lipid-free environment at the same time points, Figure 3. We fitted the amide I band in these spectra to quantify the amount of the parallel and the antiparallel β-sheets, as well as the amount of the α-helix and the unordered protein, in these aggregates Figure S6.
Figure 3.
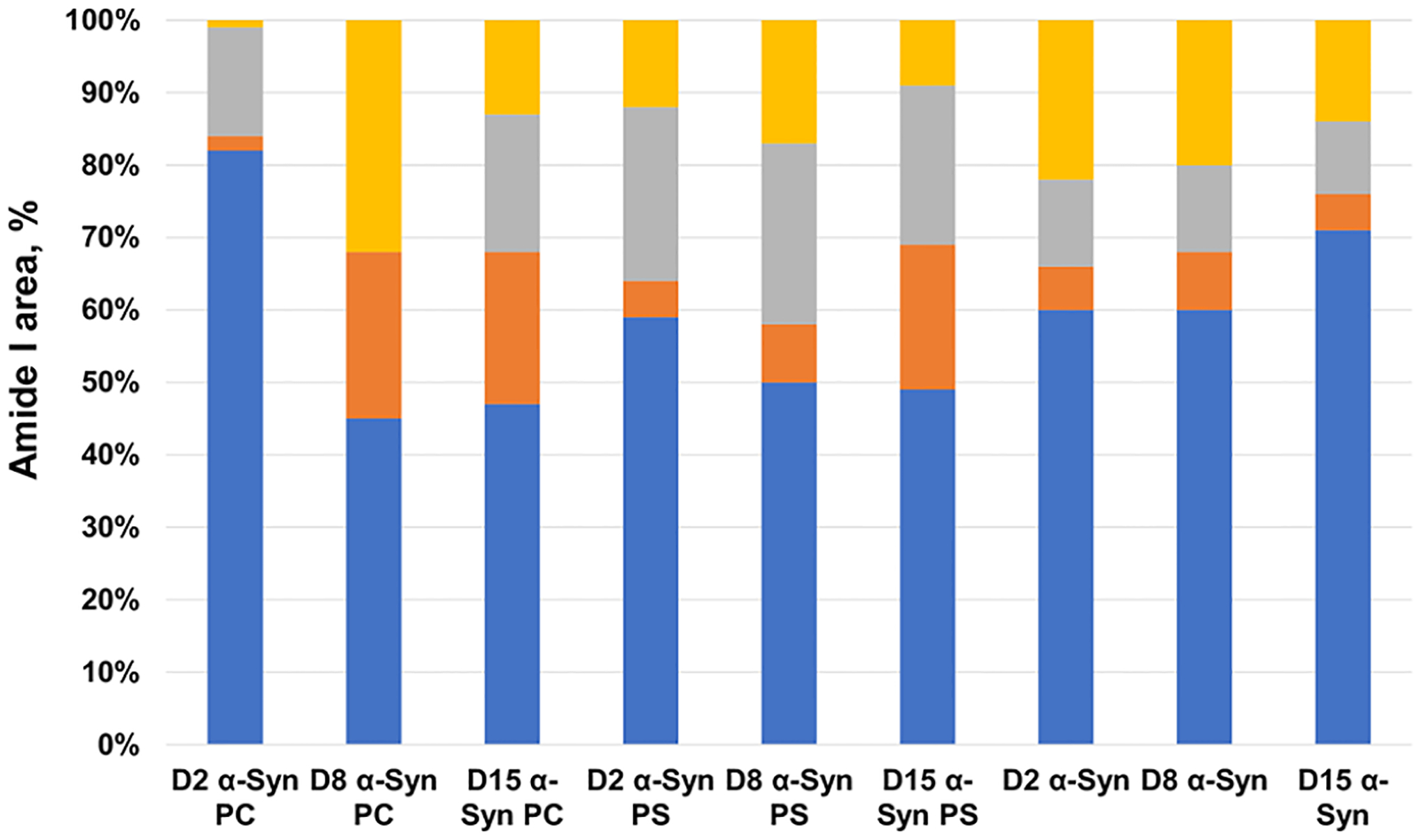
Histograms of changes in the protein secondary structure in α-Syn:PC, α-Syn:PS, and α-Syn oligomers present at D2, D8, and D15 at 1:2 lipid-to-protein (L:P) ratio. The percentage of the parallel β-sheet is shown in blue, the antiparallel β-sheet in yellow, the α-helix and the random coil in orange, and the β-turn in gray in the secondary structure of α-Syn:PC, α-Syn:PS, and α-Syn oligomers.
Our results show that α-Syn:PC oligomers present at D2 have ~82% of the parallel β-sheet, ~18 with the unordered protein secondary structure and small 1–2% α-helix and the unordered protein. AFM-IR analysis of D8 α-Syn:PC oligomers revealed a drastic increase in the amount of the α-helix (22%) and the antiparallel β-sheet (31%) in their structure relative to the amount of the parallel β-sheet (44%). These findings show that initial interactions between α-Syn and PC result in the formation of β-sheet-rich nuclei that further facilitate protein aggregation. However, the presence of PC on the surface of these oligomers prevents the expansion of their parallel β-sheet content. Instead, monomeric proteins that bind to these nuclei adopt an α-helical structure, as was previously reported for a monomeric α-Syn that transitions from unordered into α-helix in the presence of lipid bilayers.43,44 The increase of an α-helix structure can be related to the recruitment of free monomer to the aggregates. Further oligomerization and elongation are inhibited under no agitation condition because free monomers are not accessible to nucleation sites in this condition. We also observed an increase in the amount of the antiparallel β-sheet as the oligomers matured from D2 to D8. We also observed high structural similarity between the α-Syn:PC oligomers present at D8 and D15. Specifically, the content of the parallel β-sheet and the α-helix remained nearly constant, whereas an increase in the amount of the β-turn secondary structure increased with a simultaneously decrease in the amount of the antiparallel β-sheet. This observation is interesting because both the antiparallel β-sheet and the β-turn structure have been known to inhibit the aggregation of amyloid protein. We observed a gradually increase in the amount of the antiparallel β-sheet and the β-turn structure from D2 to D15 of α-Syn aggregates formed in the presence of PS and PC LUVs. The β-turn structure could facilitate the nucleation of a β-hairpin-turn structure, which is known to occur between two antiparallel β-sheets.45 It has been previously reported that PC will inhibit the α-Syn aggregation. Our results suggested that the interaction between α-Syn and PC LUVs could result in an increase of the antiparallel β-sheet and the β-turn structure. However, it is not clear whether the inhibition triggers the formation of those secondary structure or if the lipid and protein interaction shapes the secondary structure and inhibits the aggregation.
There are only minor transformations in the secondary structure content between D2 and D8 oligomers formed in the presence of PS. We found that the amount of the parallel β-sheet slightly decreased from D2 (58%) to D8 (50%), whereas the small increase in the antiparallel β-sheet took place (from 12% at D2 to 16% at D8). We also observed an increase in the amount of the α-helix as the oligomers propagated from D8 to D15 with a significant reduction in the amount of the antiparallel β-sheet. It should be noted that observed structural changes for α-Syn:PC and α-Syn:PS were not evident for α-Syn oligomers grown in the lipid-free environment, Figure 3. Specifically, in the absence of lipids, we observed a gradual increase in the amount of the parallel β-sheet from D2 to D15, followed by a decrease in the amount of the antiparallel β-sheet, which is in good agreement with the previously reported by Zhou and Kurouski results.32 We also found that, under the lipid-free environment, the amount of the α-helix, random coil, and β-turns remains nearly the same in D2, D8, and D15 oligomers. Thus, lipids not only drastically alter the secondary structure of α-Syn oligomers that were grown in their presence but also significantly change the evolution of the protein secondary structure that takes place upon oligomer propagation into fibrils.
CONCLUSIONS
Our experimental findings show that all examined α-Syn:PC and α-Syn:PS oligomers present at the lag phase of protein aggregation (D2-D15) possess corresponding phospholipids in their structure. We also found that the presence of the lipid in the oligomer structure lowers the amount of the parallel β-sheet in their secondary structure as the oligomers propagate from D2 to D15. At the same time, the amount of the α-helix and the unordered protein secondary structure gradually increase in these oligomers. We infer that the changes in the secondary structures are due to the presence of lipids in the oligomers that modify the assembly of monomeric proteins upon their assembly onto the very early stage (D2) oligomers. It should be noted that the opposite transformations in the corresponding secondary structures have been observed upon α-Syn aggregation under a lipid-free environment. One can expect that α-Syn:PC and α-Syn:PS oligomers grown in the presence of lipids would exert different cell toxicities in comparison to the oligomers grown in the absence of lipids.
EXPERIMENTAL METHODS
Protein and Lipid Preparation.
α-Syn is purchased from AnaSpec, CA, USA. The preparation of α-Synuclein follows a group protocol by Zhou and Kurouski.32 Briefly, α-Syn was dissolved to final concentration of 150 μM in 1X PBS buffer, pH at 7.5 stock. Next the stock was mixed with DMPC or DMPS lipid unilamellar vesicles (LUVs) and reach final protein concentration at 45 μM. For early stage oligomers study, the solution was kept under room temperature without any agitation.
The preparation of lipid unilamellar vesicles (LUVs) was repeated by using previous protocol by Dou and Kurouski.33 After LUVs were collected through the extruder, the sizes of lipid LUVs were checked by using dynamic light scattering (DLS). In this experiment, we investigate the lipid to protein ratio 1:2. Samples for AFM-IR, CD, and FT-IR measurements were collected on day 2, day 8, and day 15, denoted as D2, D8, and D15.
AFM-IR Imaging and Spectroscopy.
The solution (3–6 μL) of aggregate samples was deposited on a silicon wafer, exposed for 5–10 min for drying. Next, remove the excess samples, rinsed with DI water and dried under N2 flow. AFM-IR imaging was conducted by using a Nano-IR3 system (Bruker, Santa Barbara, CA, USA). The IR source was a QCL laser. Contact-mode AFM tips (ContGB-G AFM probe, NanoAndMore) were used to obtain all spectra and maps. No evidence of the sample distortion was observed upon contact-mode AFM imaging. IR maps at 1624, 1655, and 1694 cm−1 wavenumber values were obtained to study the secondary structure of α-Syn:PS and α-Syn:PC oligomers and at 1100 cm−1 to study the presence of these phospholipids in α-Syn aggregates. The phase loop lock was enabled during the mapping with a 0.03 V threshold. The AFM height and deflection images were acquired simultaneously with IR maps. Totally, 20 point spectra were taken from every analyzed oligomer and individual aggregates. The spectra were zapped from 1648 to 1652 cm−1 due to the chip-to-chip transition artifact from the instrument. Savitzky−Golay smoothing was applied to all spectra with 2 polynomial order by using MATLAB.
Attenuated Total Reflectance Fourier-Transform Infrared (ATR-FTIR) Spectroscopy.
The solution (3–6 μL) of aggregates samples was deposited on an ATR crystal and dried at room temperature. Spectra were measured by using a Spectrum 100 FTIR spectrometer (PerkinElmer, Waltham, MA, USA). Three spectra were collected from each sample and averaged by using MATLAB.
Cicular Dichroism Spectroscopy (CD).
The circular dichroism spectra were recored on a JASCO J-1500 CD instrument at room temperature. Aliquots of 300 μL of the α-synuclein, α-syn:PC, and α-syn:PS solution after incubation for 2 days (D2), 8 days (D8), and 15 days (D15) were loaded. Spectra were recorded after averaging five accumulations in the range 190–260 nm using a 1.0 length quartz cuvette (Starna Cells, CA, USA). The data step size is 0.2 nm with a speed of 50 nm/min and a digital integration time of 2 s. Three spectra were collected and averaged by using MATLAB.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the National Institute of Health for the provided financial support (R35GM142869).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschemneuro.2c00355.
AFM image and height profiles of α-Syn aggregates formed at D2, D8, and D15 (Figure S1), height profiles of α-Syn:PC and α-Syn:PS oligomers formed at D2, D8, and D15 (Figure S2), AFM and AFM-IR maps of α-Syn oligomers formed at D2-D15 (Figure S3), averaged AFM-IR spectra of the protein aggregates and results of amide I fit (Figures S4–S6), Bar plot of protein secondary structure (Figure S7), Mean and standard deviation AFM-IR spectra (Figure S8), CD spectra (Figure S9), and FT-IR spectra (Figure S10) (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acschemneuro.2c00355
The authors declare no competing financial interest.
Contributor Information
Tianyi Dou, Department of Biochemistry and Biophysics, Texas A&M University, College Station, Texas 77843, United States.
Dmitry Kurouski, Department of Biochemistry and Biophysics and Department of Biomedical Engineering, Texas A&M University, College Station, Texas 77843, United States.
REFERENCES
- (1).Dorsey ER; Sherer T; Okun MS; Bloem BR The Emerging Evidence of the Parkinson Pandemic. J. Parkinsons Dis 2018, 8 (s1), S3–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Pieri L; Madiona K; Melki R Structural and functional properties of prefibrillar α-synuclein oligomers. Sci. Rep 2016, 6, 24526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Chen SW; Drakulic S; Deas E; Ouberai M; Aprile FA; Arranz R; Ness S; Roodveldt C; Guilliams T; De-Genst EJ; Klenerman D; Wood NW; Knowles TP; Alfonso C; Rivas G; Abramov AY; Valpuesta JM; Dobson CM; Cremades N Structural characterization of toxic oligomers that are kinetically trapped during alpha-synuclein fibril formation. Proc. Natl. Acad. Sci. U. S. A 2015, 112 (16), E1994–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Cremades N; Cohen SI; Deas E; Abramov AY; Chen AY; Orte A; Sandal M; Clarke RW; Dunne P; Aprile FA; Bertoncini CW; Wood NW; Knowles TP; Dobson CM; Klenerman D Direct observation of the interconversion of normal and toxic forms of alpha-synuclein. Cell 2012, 149 (5), 1048–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Apetri MM; Maiti NC; Zagorski MG; Carey PR; Anderson VE Secondary structure of alpha-synuclein oligomers: characterization by raman and atomic force microscopy. J. Mol. Biol 2006, 355 (1), 63–71. [DOI] [PubMed] [Google Scholar]
- (6).O’Leary EI; Lee JC Interplay between alpha-synuclein amyloid formation and membrane structure. Biochim. Biophys. Acta Proteins. Proteom 2019, 1867 (5), 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kurouski D; Van Duyne RP; Lednev IK Exploring the structure and formation mechanism of amyloid fibrils by Raman spectroscopy: a review. Analyst 2015, 140 (15), 4967–80. [DOI] [PubMed] [Google Scholar]
- (8).Hong DP; Han S; Fink AL; Uversky VN Characterization of the non-fibrillar alpha-synuclein oligomers. Prot. Pept. Lett 2011, 18 (3), 230–240. [DOI] [PubMed] [Google Scholar]
- (9).Knowles TP; Vendruscolo M; Dobson CM The amyloid state and its association with protein misfolding diseases. Nat. Rev 2014, 15 (6), 384–96. [DOI] [PubMed] [Google Scholar]
- (10).Li B; Ge P; Murray KA; Sheth P; Zhang M; Nair G; Sawaya MR; Shin WS; Boyer DR; Ye S; Eisenberg DS; Zhou ZH; Jiang L Cryo-EM of full-length alpha-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun 2018, 9 (1), 3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Guerrero-Ferreira R; Taylor NM; Mona D; Ringler P; Lauer ME; Riek R; Britschgi M; Stahlberg H Cryo-EM structure of alpha-synuclein fibrils. Elife 2018, 7, e36402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Tuttle MD; Comellas G; Nieuwkoop AJ; Covell DJ; Berthold DA; Kloepper KD; Courtney JM; Kim JK; Barclay AM; Kendall A; Wan W; Stubbs G; Schwieters CD; Lee VMY; George JM; Rienstra CM Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol 2016, 23 (5), 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Heise H; Hoyer W; Becker S; Andronesi OC; Riedel D; Baldus M Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc. Natl. Acad. Sci. U. S. A 2005, 102 (44), 15871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Chen L; Jin J; Davis J; Zhou Y; Wang Y; Liu J; Lockhart PJ; Zhang J Oligomeric alpha-synuclein inhibits tubulin polymerization. Biochem. Biophys. Res. Commun 2007, 356 (3), 548–53. [DOI] [PubMed] [Google Scholar]
- (15).Danzer KM; Haasen D; Karow AR; Moussaud S; Habeck M; Giese A; Kretzschmar H; Hengerer B; Kostka M Different species of alpha-synuclein oligomers induce calcium influx and seeding. J. Neurosci 2007, 27 (34), 9220–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Nasstrom T; Fagerqvist T; Barbu M; Karlsson M; Nikolajeff F; Kasrayan A; Ekberg M; Lannfelt L; Ingelsson M; Bergstrom J The lipid peroxidation products 4-oxo-2-nonenal and 4-hydroxy-2-nonenal promote the formation of alpha-synuclein oligomers with distinct biochemical, morphological, and functional properties. Free. Radic. Biol. Med 2011, 50 (3), 428–37. [DOI] [PubMed] [Google Scholar]
- (17).Sharon R; Bar-Joseph I; Frosch MP; Walsh DM; Hamilton JA; Selkoe DJ The formation of highly soluble oligomers of alpha-synuclein is regulated by fatty acids and enhanced in Parkinson’s disease. Neuron 2003, 37 (4), 583–95. [DOI] [PubMed] [Google Scholar]
- (18).Konno H; Watanabe-Nakayama T; Uchihashi T; Okuda M; Zhu L; Kodera N; Kikuchi Y; Ando T; Taguchi H Dynamics of oligomer and amyloid fibril formation by yeast prion Sup35 observed by high-speed atomic force microscopy. Proc. Natl. Acad. Sci. U. S. A 2020, 117 (14), 7831–7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Banerjee S; Sun Z; Hayden EY; Teplow DB; Lyubchenko YL Nanoscale Dynamics of Amyloid β−42 Oligomers As Revealed by High-Speed Atomic Force Microscopy. ACS Nano 2017, 11 (12), 12202–12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Kurouski D; Deckert-Gaudig T; Deckert V; Lednev IK Surface characterization of insulin protofilaments and fibril polymorphs using tip-enhanced Raman spectroscopy (TERS). Biophys. J 2014, 106 (1), 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Ruggeri FS; Longo G; Faggiano S; Lipiec E; Pastore A; Dietler G Infrared nanospectroscopy characterization of oligomeric and fibrillar aggregates during amyloid formation. Nat. Commun 2015, 6, 7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ruggeri FS; Mannini B; Schmid R; Vendruscolo M; Knowles TPJ Single molecule secondary structure determination of proteins through infrared absorption nanospectroscopy. Nat. Commun 2020, 11 (1), 2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Rizevsky S; Kurouski D Nanoscale Structural Organization of Insulin Fibril Polymorphs Revealed by Atomic Force Microscopy-Infrared Spectroscopy (AFM-IR). Chembiochem 2020, 21 (4), 481–485. [DOI] [PubMed] [Google Scholar]
- (24).Ramer G; Ruggeri FS; Levin A; Knowles TPJ; Centrone A Determination of Polypeptide Conformation with Nanoscale Resolution in Water. ACS Nano 2018, 12 (7), 6612–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Dou T; Li Z; Zhang J; Evilevitch A; Kurouski D Nanoscale Structural Characterization of Individual Viral Particles Using Atomic Force Microscopy Infrared Spectroscopy (AFM-IR) and Tip-Enhanced Raman Spectroscopy (TERS). Anal. Chem 2020, 92 (16), 11297–11304. [DOI] [PubMed] [Google Scholar]
- (26).Ruggeri FS; Benedetti F; Knowles TPJ; Lashuel HA; Sekatskii S; Dietler G Identification and nanomechanical characterization of the fundamental single-strand protofilaments of amyloid alpha-synuclein fibrils. Proc. Natl. Acad. Sci. U. S. A 2018, 115 (28), 7230–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Ruggeri FS; Flagmeier P; Kumita JR; Meisl G; Chirgadze DY; Bongiovanni MN; Knowles TPJ; Dobson CM The Influence of Pathogenic Mutations in alpha-Synuclein on Biophysical and Structural Characteristics of Amyloid Fibrils. ACS Nano 2020, 14 (5), 5213–5222. [DOI] [PubMed] [Google Scholar]
- (28).Ruggeri FS; Vieweg S; Cendrowska U; Longo G; Chiki A; Lashuel HA; Dietler G Nanoscale studies link amyloid maturity with polyglutamine diseases onset. Sci. Rep 2016, 6, 31155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Dou T; Li Z; Zhang J; Evilevitch A; Kurouski D Nanoscale Structural Characterization of Individual Viral Particles Using Atomic Force Microscopy Infrared Spectroscopy (AFM-IR) and Tip-Enhanced Raman Spectroscopy (TERS). Anal. Chem 2020, 92 (16), 11297–11304. [DOI] [PubMed] [Google Scholar]
- (30).Farber C; Li J; Hager E; Chemelewski R; Mullet J; Rogachev AY; Kurouski D Complementarity of Raman and Infrared Spectroscopy for Structural Characterization of Plant Epicuticular Waxes. ACS Omega 2019, 4, 3700–3707. [Google Scholar]
- (31).Farber C; Wang R; Chemelewski R; Mullet J; Kurouski D Nanoscale Structural Organization of Plant Epicuticular Wax Probed by Atomic Force Microscope Infrared Spectroscopy. Anal. Chem 2019, 91 (3), 2472–2479. [DOI] [PubMed] [Google Scholar]
- (32).Zhou L; Kurouski D Structural Characterization of Individual alpha-Synuclein Oligomers Formed at Different Stages of Protein Aggregation by Atomic Force Microscopy-Infrared Spectroscopy. Anal. Chem 2020, 92 (10), 6806–6810. [DOI] [PubMed] [Google Scholar]
- (33).Dou T; Zhou L; Kurouski D Unravelling the Structural Organization of Individual alpha-Synuclein Oligomers Grown in the Presence of Phospholipids. J. Phys. Chem. Lett 2021, 12 (18), 4407–4414. [DOI] [PubMed] [Google Scholar]
- (34).Rizevsky S; Matveyenka M; Kurouski D Nanoscale Structural Analysis of a Lipid-Driven Aggregation of Insulin. J. Phys. Chem. Lett 2022, 13 (10), 2467–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Galvagnion C; Buell AK; Meisl G; Michaels TCT; Vendruscolo M; Knowles TPJ; Dobson CM Lipid vesicles trigger α-synuclein aggregation by stimulating primary nucleation. Nat. Chem. Biol 2015, 11 (3), 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Alza NP; Iglesias Gonzalez PA; Conde MA; Uranga RM; Salvador GA Lipids at the Crossroad of alpha-Synuclein Function and Dysfunction: Biological and Pathological Implications. Front. Cell Neurosci 2019, 13, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Galvagnion C The Role of Lipids Interacting with -Synuclein in the Pathogenesis of Parkinson’s Disease. J. Parkins. Dis 2017, 7, 433–450. [DOI] [PubMed] [Google Scholar]
- (38).Galvagnion C; Brown JW; Ouberai MM; Flagmeier P; Vendruscolo M; Buell AK; Sparr E; Dobson CM Chemical properties of lipids strongly affect the kinetics of the membrane induced aggregation of alpha-synuclein. Proc. Natl. Acad. Sci. U. S. A 2016, 113 (26), 7065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Ysselstein D; Joshi M; Mishra V; Griggs AM; Asiago JM; McCabe GP; Stanciu LA; Post CB; Rochet J-C Effects of impaired membrane interactions on α-synuclein aggregation and neurotoxicity. Neurobiol. Dis 2015, 79, 150–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Bodner CR; Dobson CM; Bax A Multiple Tight Phospholipid-Binding Modes of α-Synuclein Revealed by Solution NMR Spectroscopy. J. Mol. Biol 2009, 390 (4), 775–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Maltsev AS; Chen J; Levine RL; Bax A Site-Specific Interaction between α-Synuclein and Membranes Probed by NMRObserved Methionine Oxidation Rates. J. Am. Chem. Soc 2013, 135(8), 2943–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bodner CR; Maltsev AS; Dobson CM; Bax A Differential Phospholipid Binding of α-Synuclein Variants Implicated in Parkinson’s Disease Revealed by Solution NMR Spectroscopy. Biochemistry 2010, 49 (5), 862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Viennet T; Wordehoff MM; Uluca B; Poojari C; Shaykhalishahi H; Willbold D; Strodel B; Heise H; Buell AK; Hoyer W; Etzkorn M Structural insights from lipid-bilayer nanodiscs link alpha-Synuclein membrane-binding modes to amyloid fibril formation. Commun. Biol 2018, 1, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Pfefferkorn CM; Jiang Z; Lee JC Biophysics of alpha-synuclein membrane interactions. Biochim. Biophys. Acta 2012, 1818 (2), 162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Ahn JM; Kassees K; Lee TK; Manandhar B; Yousif AM 6.03 - Strategy and Tactics for Designing Analogs: Biochemical Characterization of the Large Molecules. In Comprehensive Medicinal Chemistry III; Chackalamannil S, Rotella D, Ward SE, Eds.; Elsevier: Oxford, 2017; pp 66–115. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


