Abstract
We hypothesized that hepatitis C virus (HCV) persistence is related to the sequence variability of putative envelope genes. This hypothesis was tested by characterizing quasispecies in specimens collected every six months from a cohort of acutely HCV-infected subjects (mean duration of specimen collection, 72 months after seroconversion). We evaluated 5 individuals who spontaneously cleared viremia and 10 individuals with persistent viremia by cloning 33 1-kb amplicons that spanned E1 and the 5′ half of E2, including hypervariable region 1 (HVR1). To assess the quasispecies complexity and to detect variants for sequencing, the first PCR-positive sample was examined by using a previously described method that combines heteroduplex analysis and analysis of single-stranded conformational polymorphisms. The ratio of nonsynonymous to synonymous substitutions (dN/dS) within each sample was evaluated as an indicator of relative selective pressure. Amino acid sequences were analyzed for signature patterns, glycosylation signals, and charge. Quasispecies complexity was higher and E1 dN/dS ratios (selective pressure) were lower in those with persistent viremia; the association with persistence was strengthened by the presence of a combination of both characteristics. In contrast, a trend toward higher HVR1 dN/dS ratios was detected among those with persistent viremia. We did not detect any such association for factors that may affect complexity such as serum HCV RNA concentration. HVR1 had a lower positive charge in subjects with persistent viremia, although no consistent motifs were detected. Our data suggest that HCV persistence is associated with a complex quasispecies and immune response to HVR1.
An estimated 170 million people worldwide are infected with hepatitis C virus (HCV) (3), which may cause cirrhosis and hepatocellular carcinoma (2, 24, 35, 53). Viral persistence is central to HCV pathogenesis. Even though HCV-specific humoral and cellular immune responses are evident within months of exposure (7, 30, 37, 55), HCV RNA remains detectable for more than 20 years in the blood and livers of up to 85% of infected people.
It is plausible that HCV persistence relates to viral diversity during acute infection. Mathematical models of viral kinetics estimate that more than 1012 virions are produced each day in an infected person (39). Rapid replication and the absence of RNA polymerase proofreading result in accumulation of mutations at a rate of 0.4 × 10−3 to 1.2 × 10−3 base substitutions per site per year (1, 41, 42, 49). Consequently, many distinct but highly related variants coexist in the blood and liver of an individual, indicating that HCV exists as a quasispecies (23, 34, 51). Mutations may change an encoded amino acid (nonsynonymous) or result in the same amino acid (synonymous). Assuming that nonsynonymous mutations may allow immunologic escape (13, 59) and synonymous mutations have no direct immunological impact, the ratio of nonsynonymous to synonymous mutations may reflect the relative immune pressure at a locus (6, 47).
HCV diversity is greatest in the putative envelope genes, especially in a 27-amino-acid segment at the amino terminus of E2, designated hypervariable region 1 (HVR1) (21, 22, 28, 54). We hypothesized that individuals who clear viremia have an immune response directed against more conserved regions and that people who develop persistent infection have a more complex initial quasispecies. Hypotheses regarding acute HCV infection are difficult to test because acute HCV infection in humans is difficult to detect (patients are usually asymptomatic) and because experimental infection of chimpanzees, the only animal model, infrequently results in persistent viremia (4). In addition, the traditional method of examining viral complexity, namely, sequencing of viral clones, is too cumbersome to be applied to large numbers of individuals.
Two recent developments enabled us to test this hypothesis. First, we identified and characterized the long-term virologic outcomes for 43 individuals with acute HCV infection (55). Second, we developed a method for efficiently and accurately characterizing the HCV quasispecies (58). In this study, these resources were used to examine viral complexity and distortions in amino acid sequences of subjects with persistent viremia versus those with self-limited viremia. We also accounted for duration of infection and controlled for other factors (human immunodeficiency virus [HIV] infection, race, age, and frequency of drug use) that may affect HCV clearance.
MATERIALS AND METHODS
Study subjects.
Since 1988, approximately 3,000 former and current injection drug users, including 50 subjects who acquired HCV infection during follow-up, have been monitored in Baltimore, Md. In the principal cohort (ALIVE) (57), 43 HCV seroconverters were identified (56). In a second related cohort (REACH) (16), there were seven seroconverters. After a median of more than 6 years of semiannual follow-up subsequent to seroconversion, two distinct patterns of viremia were noted. For seven subjects HCV RNA was undetectable for a minimum of 2 years in at least four serum samples from each person. In contrast, for 43 subjects HCV RNA remained detectable in the last specimen tested. The viral load trajectories and temporal sequence of HCV RNA and levels of antibody detected for the 43 subjects from the ALIVE cohort are described elsewhere (55).
Of the seven subjects with self-limited viremia, HCV RNA was never recovered from one subject and was not amplified in E1 from a second, leaving five case subjects for further virologic study. HCV RNA characterizations for these 5 case subjects were compared with those for 10 control subjects chosen from 29 subjects exhibiting HCV seroconversion and having persistent viremia for at least 6 years (eight subjects did not have sufficient follow-up to be classified as persistently viremic). Case subjects and controls were matched for HIV-1 serostatus, race, age, and active versus inactive drug use, in that hierarchical order, based on the theoretical and empirically recognized effects of these factors on viral persistence. They are herein designated by letters of the alphabet.
Storage of serum and testing for anti-HCV.
All serum samples were centrifuged on site, stored for less than 1 week at −20°C, and subsequently stored at −70°C. They were tested for antibodies to HCV (HCV EIA 2.0; Ortho Diagnostics, Raritan, N.J.) and, if these results were positive, by a strip immunoblot assay (RIBA HCV 2.0; Chiron Corporation, Emeryville, Calif.), as previously described (56).
Generic detection of HCV RNA.
For all HCV seroconverters, we evaluated the presence of HCV RNA in sera collected 6 months before seroconversion, at seroconversion, and at a median of eight additional semiannual visits (55). HCV RNA was initially detected by a quantitative reverse transcriptase PCR (RT-PCR) assay (AMPLICOR HCV MONITOR; Roche Diagnostic Systems, Branchburg, N.J.), the linear range of which was determined to be 500 to 500,000 copies per ml of serum by our and other laboratories (18, 45). Results below the linear range of the quantitative assay were assigned a value of 250 copies per ml and, when additional sample was available, were tested again with one of two qualitative RT-PCR assays: an assay using an AMPLICOR HCV detection kit (Roche Diagnostic Systems) and an in-house nested-PCR assay using primers representing conserved sequences of the 5′ noncoding region (52). With the latter assays, the limit for detecting a subtype 1a reference strain (Hutchinson) (41) was approximately 100 copies per ml. In this study, data were analyzed for the first serum sample from which HCV cDNA was amplified.
Envelope region amplification.
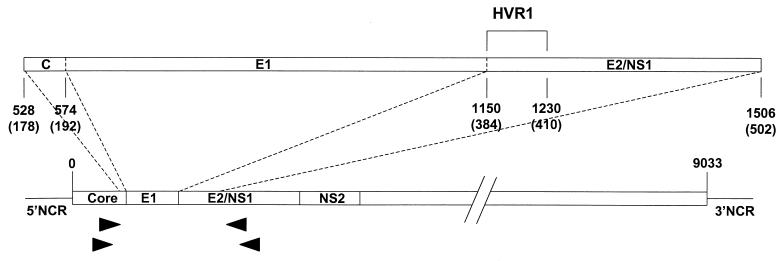
An HCV RNA characterization for each of 15 subjects was based on examination of 33 1,026-nucleotide cloned cDNAs spanning the region thought to encode envelope protein E1 and a segment of the E2 region, including HVR1 (Fig. 1). RNA was extracted from 100 μl of plasma or serum by using acid guanidinium thiocyanate (58). The RNA pellet was washed with 75% (vol/vol) ethanol, briefly air dried, and then redissolved in 50 μl of diethyl pyrocarbonate-treated water with 10 mM dithiothreitol (Promega, Madison, Wis.) and 5 U of RNasin ribonuclease inhibitor (Promega). After incubation at 65°C for 5 min, 5 μl of purified RNA was used to generate cDNA in a 20-μl reaction mixture at 37°C for 1 h with 20 U of Moloney murine leukemia virus RT (Perkin-Elmer, Foster City, Calif.) and the first-round PCR reverse primer. The entire 20-μl cDNA synthesis reaction mixture was used for the first-round PCR in a 25-μl reaction mixture containing 0.625 U of Taq polymerase (Life Technologies), 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, and 0.4 μM primers. The primers (and positions relative to the HCV-1 genome 5′ terminus [12]) were as follows: outer forward (positions 493 to 518), 5′-GCAACAGGGAACCTTCCTGGTTGCTC-3′; outer reverse (positions 1745 to 1723), 5′-GGGCAGDBCARRGTGTTGTTGCC-3′; inner forward (positions 502 to 527), 5′-AACCTTCCTGGTTGCTCTTTCTCTAT-3′; and inner reverse (positions 1527 to 1507), 5′-GAAGCAATAYTGYGGRCCACA-3′. Degenerate bases are indicated with standard codes of the International Union of Pure and Applied Chemistry. The forward primers are based on the work of Bukh et al. (8). Ten microliters of the first reaction product was used as the template for the inner nested PCR. Thermal-cycling conditions for both the inner and outer reactions were 10 cycles at 94°C for 10 s, 65°C for 30 s, and 72°C for 60 s, followed by 25 cycles at 94°C for 10 s, 65°C for 30 s, and 72°C for 90 s.
FIG. 1.
Diagram depicting the studied portion of the HCV genome and locations of the PCR primers (arrowheads) used in this study. Positions are based on the work of Choo et al. (12). 5′NCR and 3′NCR, 5′ and 3′ noncoding region, respectively.
Cloning of cDNA and complexity analysis of 33 cloned cDNAs by gel shift analysis.
The 1-kb HCV cDNA product was ligated into the vector pCR 2.1 and used to transform I F′ cells (TA cloning kit; Invitrogen, Carlsbad, Calif.). Transformants were detected according to the manufacturer’s protocol, and cloning efficiency was >90%.
For each subject, the gel shift patterns of 33 cloned cDNAs were examined by amplifying a 452-bp region spanning HVR1 and by using a nonradioactive method that detects distinct variants within a sample by a combination of heteroduplex analysis (HDA) and single-stranded conformational polymorphism analysis (SSCP) in a single gel (HDA+SSCP) (58). Clonotypes are defined as cloned cDNAs with indistinguishable patterns of electrophoretic migration by HDA+SSCP. In our earlier study, the mean (± standard deviation) genetic diversity of cloned cDNAs belonging to the same clonotype (intraclonotype diversity) was 0.6% (±0.9%), with 98.7% differing by less than 2%. The complexity of the quasispecies was characterized with the clonotype ratio, calculated as the number of clonotypes divided by 33, the number of cloned cDNAs examined. The clonotype ratio therefore varies from 0.03 (homogeneous) to 1 (highly complex).
Sequencing and signature pattern analysis.
To examine each subject’s quasispecies for signature sequences (motifs uniquely shared by a group of sequences) and for distortions in the ratio of nonsynonymous to synonymous substitutions (dN/dS), a subset of cloned cDNAs was identified for sequencing. For each subject, at least three cloned cDNAs were selected for sequencing based on gel shift patterns: two from the majority clonotype, one from each clonotype consisting of more than 10% of the 33 cloned cDNAs examined, and the cloned cDNA with the largest heteroduplex gel shift. Plasmid DNA was isolated from a 3.5-ml broth culture (High Pure plasmid isolation kit; Boehringer Mannheim) according to the manufacturer’s protocol. Sequences from this DNA and the forward and reverse primers were determined by using a PRISM version 2.1.1 automated sequencer (Applied Biosystems Inc., Foster City, Calif.). Sequences were assembled and edited with Sequencher (Gene Codes, Ann Arbor, Mich.) by a technician who was unaware of our hypotheses. Primer sequences were removed prior to analysis. Signature pattern analysis was performed with the Viral Envelope Signature Pattern Analysis (VESPA) program (26).
Variability analysis.
A software program (VarPlot for Windows) was developed by S. C. Ray to calculate values for dN, dS, or the dN/dS ratio in a “sliding window” of nucleotide sequence. A segment of defined length, in this case 60 bp (the window size), was used to determine the genetic distance, or number of mutations per site. This process was then repeated for an overlapping segment of 60 bp, which was shifted by 3 bp (the step size), and continued across the alignment. At each step all pairwise comparisons (up to 45) for a subject were performed and values were averaged. The mean values for all subjects were then averaged, ensuring that each subject was given equal weight. The method of Nei and Gojobori (38) was used to calculate the nonsynonymous genetic distance (number of nonsynonymous changes per nonsynonymous site) and the synonymous genetic distance. The Jukes-Cantor correction was used to correct for underestimation of distance due to multiple substitutions at the same site (20). To determine the dN/dS ratio, values for dN and dS, the dN/dS ratio for nonzero values of dS, and then the mean dN/dS ratio were calculated for each subject. In a similar manner dN minus dS was also determined, except that the calculation did not require discarding values when dN minus dS was equal to 0. VarPlot is available from S. C. Ray on request (sray@jhmi.edu).
Phylogenetic analysis.
The sequence alignment was randomly permuted 100 times by using the SEQBOOT program from the PHYLIP package, version 3.572c (14, 15). DNA distance matrices were calculated by using the DNADIST program, maximum-likelihood model, with a transition-to-transversion ratio of 4.25 (50). Permuted trees were generated by using the NEIGHBOR program with random addition, and bootstrap values were obtained by using CONSENSE. The indicated subtype reference sequences used for phylogenetic analysis had the following GenBank accession numbers: 1a, AF009606 and M62321; 1b, D90208; 1c, D14853; 2a, D00944; 2b, D10988; 3a, D17763; 4a, Y11604; 5a, Y13184; 6a, Y12083; “7a,” D84263; “8a,” D84264; “9a,” D84265; “10a,” D63821; and “11a,” D63822. Proposed subtype designations are in quotes.
Statistical analysis.
After examination of the distribution of data, statistical inference was made by using the nonparametric Mann-Whitney test of medians. A P value less than 0.05 was considered significant.
Nucleotide sequence accession numbers.
The sequences were submitted to GenBank and were assigned accession no. AF118570 through AF118632.
RESULTS
Subjects and initial sequence analysis.
No difference was detected between case and control subjects in the matching criteria (HIV status, race, age, and drug use activity) or the levels of HCV RNA in serum (Table 1).
TABLE 1.
Sociodemographic, virologic, serologic, and drug use characteristics of members of the study groupa
| Determinant | Value for:
|
|
|---|---|---|
| Case subjects (n = 5) | Control subjects (n = 10) | |
| Ageb | 27 (25–33) | 30 (23–50) |
| Male (%) | 80 | 80 |
| African-American (%) | 40 | 60 |
| Sampling intervalc | 3.3 (−3.3–40.9) | 3.0 (−13.9–41.4) |
| Drug used | 173 (1–917) | 377 (0–1180) |
| HIV seropositive (%) | 20 | 20 |
| Log10 [HCV RNA]e | 5.2 (3.3–7.6) | 4.8 (2.7–7.4) |
P > 0.05 for all comparisons. Case subjects showed clearance of viremia; control subjects showed persistence of viremia.
Median age at the time of HCV seroconversion in years (range).
Number of months from HCV seroconversion to HCV RNA detection. Negative values indicate RNA detected prior to seroconversion; median (range).
Self-reported drug use in episodes during the year following HCV seroconversion; median (range).
[HCV RNA], HCV RNA copies per milliliter of serum, at the time of sampling for this study; median (range).
From 15 serum samples, representing the first specimen from each subject in which HCV envelope RNA was detected, 63 sequences were obtained. All such sequences were 979 bp, except for the sequences of single variants from subjects F and BE, which had a 3-bp deletion. For case subject F, analysis was limited to 961 bp, due to a sequencing artifact at the 3′ end of the amplified region. There were a total of 67 sporadic nonsynonymous substitutions, defined as substitutions occurring in only 1 of the 63 clones sequenced, resulting in a sporadic substitution frequency of 2.4 × 10−5 per nonsynonymous site per PCR cycle. This frequency is consistent with that of expected artifacts of amplification (48) and is similar to the rate (3.5 × 10−5 per site per cycle) calculated for another cohort (36). Four of these sporadic substitutions resulted in termination codons. Sequences from case subjects and controls had the same rate of sporadic substitutions.
Phylogenetic analysis revealed that twelve subjects’ sequences clustered with subtype 1a, while those of the other three (D, AW, and BF) (Table 2) clustered with subtype 1b (data not shown). Both groups (subjects exhibiting clearance and those exhibiting persistence) had a 4:1 ratio of subtypes 1a and 1b. For all 15 pairs of sequences representing each majority clonotype, intraclonotype diversity was less than 1%, underscoring the sensitivity of the HDA+SSCP method.
TABLE 2.
Characteristics of subjects and samples and results of HDA+SSCP and sequence analysisa
| Subject | Genotype | Clonotype distributionb | No. of clonotypes | Clonotype ratio |
|---|---|---|---|---|
| Case subjects (clearance) | ||||
| A | 1a | 20, 5, 3, 2, 2, 1 | 6 | 0.18 |
| B | 1a | 32, 1 | 2 | 0.06 |
| D | 1b | 25, 4, 1, 1, 1, 1 | 6 | 0.18 |
| F | 1a | 20, 12, 1 | 3 | 0.09 |
| G | 1a | 14, 11, 2, 2, 1, 1, 1, 1 | 8 | 0.24 |
| Controls (persistence) | ||||
| AF | 1a | 30, 1, 1, 1 | 4 | 0.12 |
| AM | 1a | 21, 2, 2, 2, 1, 1, 1, 1, 1, 1 | 10 | 0.30 |
| AR | 1a | 5, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1 | 29 | 0.88 |
| AS | 1a | 23, 2, 1, 1, 1, 1, 1, 1, 1, 1 | 10 | 0.30 |
| AU | 1a | 11, 7, 4, 3, 1, 1, 1, 1, 1, 1, 1, 1 | 12 | 0.36 |
| AW | 1b | 24, 3, 2, 1, 1, 1, 1 | 7 | 0.21 |
| BC | 1a | 15, 6, 5, 1, 1, 1, 1, 1, 1, 1 | 10 | 0.30 |
| BE | 1a | 8, 5, 5, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1 | 17 | 0.52 |
| BF | 1b | 32, 1 | 2 | 0.06 |
| BH | 1a | 6, 2, 2, 2, 2, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1 | 23 | 0.70 |
Clonotype, a group of electrophoretically indistinguishable cloned cDNAs; clonotype ratio, the number of clonotypes divided by 33, the number of clones examined.
Each number represents a clonotype; the value indicates the number of cloned cDNAs assigned to that clonotype by the HDA + SSCP method.
Analysis of virologic determinants of viremia.
Because viral envelope proteins are important determinants of tropism and immunogenicity, we assessed the physicochemical properties of the protein sequences deduced from the amplified sequences by using a majority representative sequence for each subject (Fig. 2). HVR1 sequences from the subjects who cleared their viremia were significantly more positively charged than those who did not (median, +3 versus +1.5; P < 0.03).
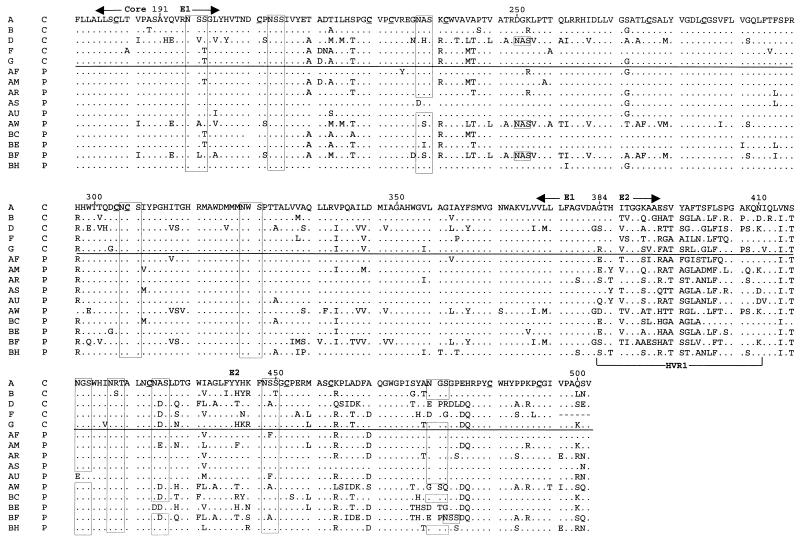
FIG. 2.
Alignment of inferred amino acid sequences for the majority sequences from each subject. In the first column, an alphabetical label is given for each subject, while in the second column, C indicates clearance of viremia and P indicates persistence. Periods indicate identity to the amino acid at that position in the first sequence. Positions of the Core and E1 and E2 regions are indicated above the alignment, whereas that of HVR1 is indicated below the alignment at the N terminus of E2. Boxes indicate predicted N-linked glycosylation sites. Cysteine residues in the first sequence are underlined.
There were 10 potential N-linked glycosylation sites (NXS or NXT) present in the amplified region based on the HCV-1 and Hutchinson sequences. These were highly conserved in all subtype 1a sequences (Fig. 2). Subtype 1b sequences (from subjects F, AW, and BF, as well as reference strain HCV-J) shared 9 of the 10 sites of HCV-1, with loss of the site at position 476 and addition of a site at position 250. Sequences from subject BF also carried an additional site at position 478. N-linked glycosylation sites were 100% conserved among the sequenced cloned cDNAs from each individual. Viremia persistence did not correlate with predicted N-linked glycosylation. Likewise, there were 14 cysteine residues in the amplified region, and all 14 were conserved in 61 of 63 sequences; the two isolated exceptions were consistent with sporadic substitution.
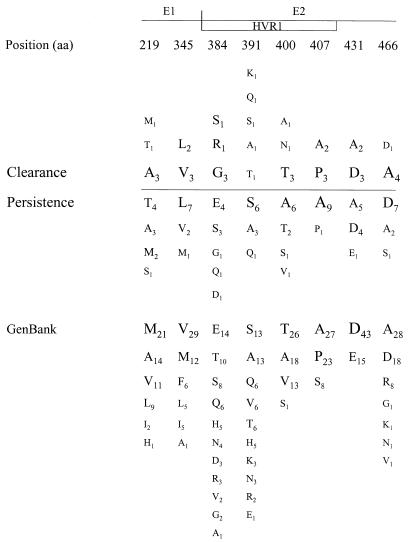
To test the hypothesis that a signature sequence within the amplified region is linked to clearance or persistence of viremia, signature pattern analysis was applied. One representative sequence was chosen for each sample. In all cases, one of the majority clonotype sequences also represented the consensus sequence at each amino acid position for that sample. Signature pattern analysis identified eight amino acid positions at which the majority amino acid differed between case and control subject sequences (Fig. 3). However, at none of these positions was a residue uniquely present in either outcome group and, in all but one case, the amino acids found in the 15 HCV seroconverters were well represented among 58 other HCV sequences from GenBank. The one exception was position 431, which contained alanine in 7 of our 15 sequences. The GenBank sequences uniformly had an acidic residue (aspartate or glutamate) at position 431; hence, none had an alanine at this position. This residue may be a feature of the regional (Baltimore) epidemic from which the subjects were enrolled; however, the proportions of samples containing this alanine were not different between the two outcome groups.
FIG. 3.
Comparison of the frequencies of amino acids in consensus sequences for the 5 case subjects (group showing clearance) and the 10 control subjects (group showing persistence of viremia). A subscript indicates the number of sequences having that residue at that position. Sites identified by using signature pattern analysis are displayed; for sites not shown, the most frequently observed amino acids (aa) did not differ between case subjects and control subjects. Positions are based on the work of Choo et al. (12). Also shown are the amino acids in these positions for 58 GenBank sequences spanning the same region.
Quasispecies complexity and the outcome of acute infection.
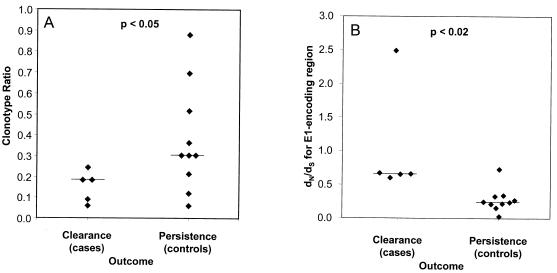
As hypothesized, case subjects (who cleared viremia) had lower median quasispecies complexity as measured by clonotype ratio than controls (whose viremia persisted) (P < 0.05) (Fig. 4A). While no one who cleared viremia had a quasispecies complexity value greater than 0.3, 3 of the 10 controls had levels of complexity as low as those of the five case subjects. Therefore, low quasispecies complexity may be necessary, but not sufficient, for clearance of hepatitis C viremia, suggesting that other factors may be important.
FIG. 4.
Virologic correlates of outcome. (A) Clonotype ratio, calculated as the ratio of the number of clonotypes detected to the number of cloned DNAs examined (Table 2), versus outcome. (B) E1 dN/dS ratio versus outcome. For each subject, all pairwise nonsynonymous and synonymous distances were calculated for the E1-coding region. These distances were averaged, and the dN/dS ratio was then calculated. Horizontal lines represent medians.
dN/dS ratios and outcome of acute infection.
Case subjects had significantly higher dN/dS ratios for E1 than controls (P < 0.02) (Fig. 4B). This difference was decreased when the E2 segment was added to the analysis (data not shown), suggesting that the patterns of nonsynonymous substitutions differed in the E1 and E2 regions.
Segmental differences in dN/dS ratios.
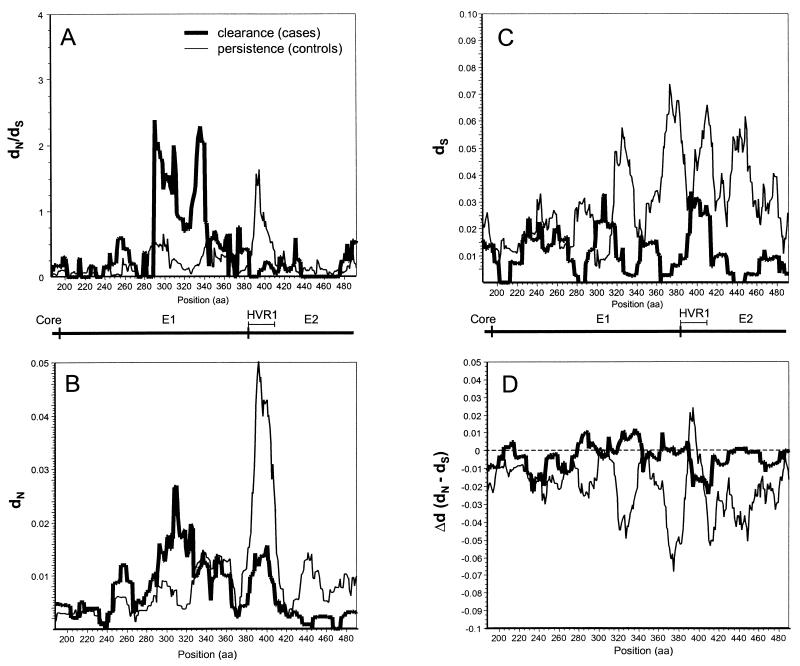
To test the hypothesis that the regions under the greatest selective pressure differed between case and control subjects, we performed a high-resolution analysis of differences in dN/dS ratios by using VarPlot. We found generally low dN/dS ratios as previously observed (50), with values of less than 1.0 throughout the envelope region studied. Two notable distortions in the dN/dS plots were observed: that in an E1 segment centered on amino acid 310 in sequences of the case subjects and that in a segment corresponding to HVR1 in sequences of the control subjects (Fig. 5A). These high-dN/dS-ratio segments corresponded to segments of high dN values (Fig. 5B) and were not due to differences in dS values (Fig. 5C). Results of an analysis based on the difference between dN and dS (Δd) was in agreement with results of the dN/dS analysis, with positive values being obtained for Δd in E1 segments of case subjects and in HVR1 of controls (Fig. 5D).
FIG. 5.
Variability plots of the envelope region. For each subject, the intrasample dN/dS ratio (A), dN value (B), dS value (C), or Δd value (dN − dS) (D) was calculated for overlapping windows of 20 amino acids (aa; 60 nucleotides), sliding in increments of 1 amino acid across E1 and the first 119 amino acids of E2. The mean values for each group (clearance or persistence) were then plotted. Positions are based on the work of Choo et al. (12).
DISCUSSION
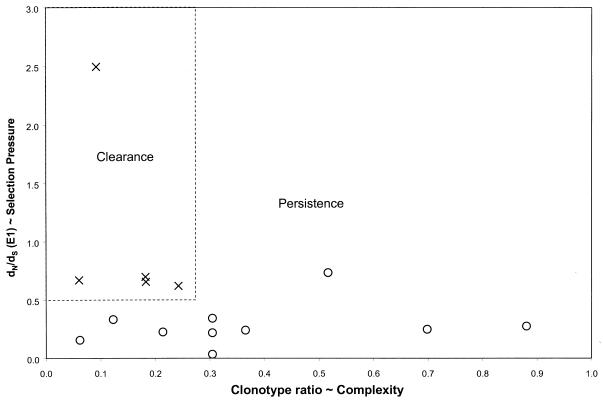
In this prospective study of subjects with acute HCV infection, clearance of viremia was associated with lower early quasispecies complexity and a higher ratio of nonsynonymous to synonymous mutations in E1. In addition, subjects with clearance of viremia were segregated from those with persistent viremia by combining these two measures (Fig. 6). By using high-resolution sequence analysis, we were able to demonstrate that the correlation between clearance and nonsynonymous change in E1 was complemented by a similar correlation between persistence and nonsynonymous change in HVR1 (in E2), suggesting that HVR1 may act as an immunologic decoy during acute infection.
FIG. 6.
Ratio of nonsynonymous to synonymous distances versus clonotype ratio. Values from Fig. 4A and B were plotted on the same graph, and a box (dotted line) is drawn around the points representing values for the case subjects (with clearance of viremia).
During acute infection, each individual develops a quasispecies, or swarm of highly related viral sequences. While this may involve random mutation with certain functional constraints, current evidence suggests a more directed process. In a longitudinal study of three subjects, HVR1 variation during the first 12 months of infection did not reveal a common pattern of increasing diversity within each sample, though later sequences did diverge from earlier ones, indicating the action of selective forces (33). The direction of these changes does not appear to be programmed by the viral sequence, because in a cohort of persons infected from the same homogeneous source, each developed a distinct quasispecies (36). While diversification may be dependent on the characteristics of the virus, selection is a function of the environment in which the virus replicates.
Diversification does not ensure evolution.
The genetic sequences of HCV variants are very heterogeneous, varying by more than 30% across the entire genome among the six major genotypes, 20% among subtypes, and up to 10% within a subtype (50). Within a single infected individual, the diversity of viral variants varies greatly, depending on the stage of disease and the genomic region assessed, but even in acute infection it may be as high as 6% (58). This profound variability is generally attributed to the combination of three factors: an error-prone RNA-directed RNA polymerase, a high rate of viral replication, and persistent infection.
Despite generating a large number of diverse progeny, a quasispecies in a constant environment may not appear to evolve over time (51). This predicted equilibrium has been demonstrated in HCV-infected chimpanzees, in which extremely limited change in the quasispecies was observed (5, 9, 32); the lack of genetic drift appears to correlate with weak immune responses in chimpanzees (54). Reduced evolution of the quasispecies has also been observed in immunocompromised humans (27, 40). Thus, the progressive change of the distribution of variants in a quasispecies requires an additional factor: selective pressure.
Sequence variation as a result of selection.
We attempted to reduce the number of variables, particularly those that would lead to a bias suggesting selective pressure. We did this by controlling for duration of infection and for the genomic region assessed and by separately examining dN and dS values. Case and control subjects had similar durations of infection and concentrations of HCV RNA in sera (Table 1). Because sequence analysis was restricted to intrasample comparisons of the E1-E2 region, differential effects of RNA secondary structure (on dN and dS) and protein function (on dN) were minimized.
In addition, because results based on dN, the dN/dS ratio, and Δd (dN minus dS) led to the same conclusions, we have addressed concerns regarding which indicator should have been used to indicate selective pressure. While many researchers have used the dN/dS ratio or Δd as surrogate indicators for immune pressure on RNA viruses (for example, see references 6, 43, and 60), there is disagreement over which calculation should be used and how to interpret the results (44). In protein-coding regions, multiple forces affect the balance between fixation of silent (synonymous) mutations versus those that alter amino acid sequence (nonsynonymous). Synonymous changes are often thought to represent a “molecular clock,” independent of external pressures and expected to occur at a rate proportional to the organism’s reproductive rate, whereas nonsynonymous changes are selected by immune pressure. It may be difficult to interpret comparisons of values of dN/dS or Δd for different genomic regions, due to unrecognized differences in RNA secondary structures (restricting dS) or protein functions (restricting dN). We controlled for these effects by comparing the same regions in different groups of individuals and by demonstrating the same findings for both the dN/dS ratio and Δd.
Had we demonstrated a correlation between clearance of viremia and higher dN/dS ratios for the entire region (E1 and 5′ segment of E2) that we analyzed, we might simply have concluded that stronger antienvelope immune pressure was advantageous for preventing persistent viremia. However, dN/dS ratios were similar among case subjects (those exhibiting clearance) and controls (those exhibiting persistence of viremia).
While we found that case subjects had higher dN/dS ratios for E1 alone (Fig. 4A and 5A), controls exhibited a trend toward higher HVR1 dN/dS ratios (Fig. 5A). These reciprocal findings are compelling and may indicate segmental differences in the effects of selective pressure. The latter finding suggests that HVR1 can function as an immunologic decoy, stimulating a strong immune response that is ineffective for clearing viremia.
A curious result shown in Fig. 5C is the trend, most pronounced in the control group, toward lower dS values in the 5′ portion of E1 than in a 3′ segment of E1 (just preceding E2) and E2. This trend has also been observed with a cohort of women who received HCV-contaminated anti-D immunoglobulin (36) and by cross-sectional analysis of complete genome sequences (50). Lower dS values may indicate that the 5′ portion of E1 has some constraints on synonymous variation, such as RNA secondary structure or binding sites for factors that regulate replication or translation.
Potential limitations.
The strength of our conclusions may be limited by the small size and heterogeneity of the cohort, by restricting our focus to a segment comprising approximately 10% of the viral genome, and by current methods for assessing HCV replication. However, we performed careful matching and followed up our subjects for a long period to ensure that clearance was durable. Although our results may not apply to all genotypes of HCV, because every subject in this study was infected with genotype 1, heterogeneity among infecting viruses may make our results more generally applicable than those from a single inoculum (36). We cannot exclude the possibility that our findings were due to interactions between mutations that we characterized and those that occurred in another genomic region.
By matching our case subjects and controls for similar durations of infection and finding similar concentrations of HCV RNA in sera, we hoped to have limited differences between the two groups in viral replicative cycles. Figure 5C shows, however, a trend toward higher dS values in E2 and a 3′ segment of E1 in the control (persistence) group. There is evidence to suggest that this trend (P > 0.05) indicates that more replicative cycles occurred in the control group, namely, our finding of greater quasispecies complexity among controls and, from our study of a larger portion of the same cohort, an association between higher levels of HCV RNA in sera and persistent viremia (55). Therefore, despite early sampling (median, 3 months after seroconversion), the persistence group may already have experienced more replicative cycles than the clearance group. If so, our conclusions would not have been affected, because of the reciprocal nature of our findings (as discussed in the preceding section): each group had high dN/dS ratios in different genome segments.
Artifactual substitutions and template resampling.
Sequences generated from a quasispecies after PCR amplification may include errors due to nucleotide misincorporation as well as template resampling. Nucleotide misincorporation was estimated by calculating the rate of sporadic nonsynonymous substitutions, which was remarkably similar to those of previous reports (36) and predictions (48). It is unlikely that nucleotide misincorporation substantially affected the results of this investigation, since the rates of sporadic nonsynonymous substitutions were similar for case subjects and controls, who were examined by the same methods. In addition, the use of the sporadic substitution rate as an index of nucleotide misincorporation overestimates this error because it also includes mutations genuinely present in the quasispecies but observed only once.
Template resampling may result in underestimation of quasispecies complexity when a small number of distinct genome templates is used in a PCR to generate sequence data. To evaluate the likelihood of resampling, the average number of distinct clones among r sampled clones can be estimated by the equation N[1−(1−1/N)r], where N is the number of molecules used as PCR templates (31). The average and smallest numbers of templates (N) in our study were 1,000 and 5, respectively, and the number of sampled clones (r) was 33. The estimated number of distinct templates among the 33 cloned cDNAs we examined was therefore 32 in the average sample and as low as 5 in the specimens with the lowest concentrations of RNA. Because we used our HDA+SSCP method to identify three to five distinct cloned cDNAs for sequencing, it is unlikely that the sequences analyzed were affected by resampling. In addition, because this source of error relates directly to the template numbers, which were similar between the two groups, the comparisons on which our conclusions were based were not affected. Our finding that there was no relationship between complexity and HCV RNA concentration supported these theoretical considerations (data not shown).
Lack of power to detect differences in levels of viremia.
The finding that the level of early viremia did not predict later clearance was probably due to the small number of subjects. As noted above, a larger study of the same cohort did demonstrate such a correlation (55). In cross-sectional studies, high-level viremia has also correlated with advanced liver disease (17) and failure of interferon therapy (29). Importantly, our current and previous findings (55) suggest that there is not a threshold of viremia above which persistent infection is a certainty.
Implications.
Our results offer some new insights into the elusive mechanisms and parameters of HCV persistence. While previous studies have linked HCV diversity with persistent infection, the question of whether this diversity was the cause or the result of persistent infection could not be addressed. In our cohort, higher quasispecies complexity was apparent within months of infection in those who developed persistent viremia. If abundant early replication is a major contributor to this higher complexity, then it may be possible to prevent persistence by using early measures to limit replication such as antiviral medications. In contrast, if segmental targeting of the immune response is a major determinant of persistence, this may offer hope for an effective vaccine, because a vaccine that reduces replication may be more achievable for HCV than one that provides sterilizing immunity. A similar argument could be applied to occupational exposures and other situations of known acute HCV infection, such that therapy directed toward shifting immune specificity or limiting replication might not prevent infection but might alter its natural history.
The proposed role of HVR1 as an immunologic decoy is not easily reconciled with prior evidence of an association between self-limited viremia and early expression of antibodies directed against HVR1 (25, 61). High-titer antibodies to HVR1 have been demonstrated to prevent HCV infection after in vitro neutralization, but protection was incomplete, possibly because of a minor population of neutralization escape mutants (13). The role of a highly variable domain as a major immunologic target and neutralization determinant would be advantageous for HCV, like the putative role of the HIV-1 hypervariable domains. The V1, V2, and V3 hypervariable loops of HIV-1 Env, which contain neutralization epitopes, may protect other more conserved neutralization epitopes (10) and determinants of coreceptor usage (19).
An additional finding was a higher positive charge in HVR1 among case subjects (who cleared viremia). While provocative, there is not sufficient information about an HCV receptor to place this finding in proper perspective. An association between pathogenetic outcome and HVR1 charge is reminiscent of the link between positive charge in the HIV-1 V3 loop and disease progression (46). Because too little is known to suggest a biologically plausible role for HVR1 charge, this finding should be confirmed with a similar, independent cohort.
Using a well-characterized cohort, analysis of a large number of HCV variants, and high-resolution analysis of nonsynonymous and synonymous substitutions, we were unable to identify an envelope sequence motif that predicts clearance or persistence of viremia. We did find differences between the two outcomes in quasispecies complexity and in the segmental patterns of selection pressure.
ACKNOWLEDGMENTS
This study was supported in part by National Institutes of Health grant IU19 AI-40035.
We thank the participants in the ALIVE and REACH cohorts for contributing the samples used in this study. J.R.T. thanks his Microbiology Branch (DCLD, ODE, CDRH, FDA) colleagues for their support.
REFERENCES
- 1.Abe K, Inchauspe G, Fujisawa K. Genomic characterization and mutation rate of hepatitis C virus isolated from a patient who contracted hepatitis during an epidemic of non-A, non-B hepatitis in Japan. J Gen Virol. 1992;73:2725–2729. doi: 10.1099/0022-1317-73-10-2725. [DOI] [PubMed] [Google Scholar]
- 2.Alter M J, Margolis H S, Krawczynski K, Judson F N, Mares A, Alexander W J, Hu P Y, Miller J K, Gerber M A, Sampliner R E, Meeks E, Beach M J. The natural history of community acquired hepatitis C in the United States. N Engl J Med. 1992;327:1899–1905. doi: 10.1056/NEJM199212313272702. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. Hepatitis C: global prevalence. Weekly Epidemiol Rec. 1997;72:341–348. [PubMed] [Google Scholar]
- 4.Bassett S E, Brasky K M, Lanford R E. Analysis of hepatitis C virus-inoculated chimpanzees reveals unexpected clinical profiles. J Virol. 1998;72:2589–2599. doi: 10.1128/jvi.72.4.2589-2599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassett S E, Thomas D L, Brasky K M, Lanford R E. Viral persistence, antibody to E1 and E2, and hypervariable region 1 sequence stability in hepatitis C virus-inoculated chimpanzees. J Virol. 1999;73:1118–1126. doi: 10.1128/jvi.73.2.1118-1126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonhoeffer S, Holmes E C, Nowak M A. Causes of HIV diversity. Nature. 1995;376:125. doi: 10.1038/376125a0. [DOI] [PubMed] [Google Scholar]
- 7.Bradley D W, Krawczynski K, Ebert J W, McCaustland K A, Choo Q L, Houghton M A, Kuo G. Parenterally transmitted non-A, non-B hepatitis: virus-specific antibody response patterns in hepatitis C virus-infected chimpanzees. Gastroenterology. 1990;99:1054–1060. doi: 10.1016/0016-5085(90)90626-c. [DOI] [PubMed] [Google Scholar]
- 8.Bukh J, Purcell R H, Miller R H. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc Natl Acad Sci USA. 1993;90:8234–8238. doi: 10.1073/pnas.90.17.8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukh J, Yanagi M, Emerson S U, Purcell R H. Presented at the Fifth International Meeting on Hepatitis C Virus and Related Viruses: Molecular Virology and Pathogenesis, 25–28 June 1998, Venice, Italy. 1998. Course of infection and evolution of monoclonal hepatitis C virus (HCV) in chimpanzees transfected with a cDNA clone of genotype 1a, abstr. 131. [Google Scholar]
- 10.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reference deleted.
- 12.Choo Q L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby A, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghton M. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter H J, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci USA. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein J. PHYLIP-phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 16.Garfein R S, Doherty M C, Brown D, Thomas D L, Villano S A, Monterroso E, Vlahov D. Hepatitis C virus infection among short-term injection drug users. J Acquired Immune Defic Syndr. 1998;18:S11–S19. doi: 10.1097/00042560-199802001-00004. [DOI] [PubMed] [Google Scholar]
- 17.Gretch D, Corey L, Wilson J, Dela Rosa C, Willson R, Carithers R, Busch M, Hart J, Sayers M, Han J. Assessment of hepatitis C virus RNA levels by quantitative competitive RNA polymerase chain reaction: high titer viremia correlates with advanced stage of disease. J Infect Dis. 1994;169:1219–1225. doi: 10.1093/infdis/169.6.1219. [DOI] [PubMed] [Google Scholar]
- 18.Hadziyannis E, Fried M W, Nolte F S. Evaluation of two methods for quantitation of hepatitis C virus RNA. Mol Diagn. 1997;2:39–46. doi: 10.1054/MODI00200039. [DOI] [PubMed] [Google Scholar]
- 19.Hoffman T L, Stephens E B, Narayan O, Doms R W. HIV type I envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors. Proc Natl Acad Sci USA. 1998;95:11360–11365. doi: 10.1073/pnas.95.19.11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jukes T H, Cantor T R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 21.Kao J-H, Chen P-J, Lai M-Y, Wang T-H, Chen D-S. Quasispecies of hepatitis C virus and genetic drift of the hypervariable region in chronic type C hepatitis. J Infect Dis. 1995;172:261–264. doi: 10.1093/infdis/172.1.261. [DOI] [PubMed] [Google Scholar]
- 22.Kato N, Ootsuyama Y, Sekiya H, Ohkoshi S, Nakazawa T, Hijikata M, Shimotohno K. Genetic drift in hypervariable region 1 of the viral genome in persistent hepatitis C virus infection. J Virol. 1994;68:4776–4784. doi: 10.1128/jvi.68.8.4776-4784.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato N, Ootsuyama Y, Tanaka T, Nakagawa M, Nakazawa T, Muraiso K, Ohkoshi S, Hijikata M, Shimotohno K. Marked sequence diversity in the putative envelope proteins of hepatitis C viruses. Virus Res. 1992;22:107–123. doi: 10.1016/0168-1702(92)90038-b. [DOI] [PubMed] [Google Scholar]
- 24.Kiyosawa K, Sodeyama T, Tanaka E, Gibo Y, Yoshizawa K, Nakano Y, Furuta S, Akahane Y, Nishioka K, Purcell R H. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology. 1990;12:671–675. doi: 10.1002/hep.1840120409. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi M, Tanaka E, Matsumoto A, Ichijo T, Kiyosawa K. Antibody response to E2/NS1 hepatitis C virus protein in patients with acute hepatitis C. J Gastroenterol Hepatol. 1997;12:73–76. doi: 10.1111/j.1440-1746.1997.tb00350.x. [DOI] [PubMed] [Google Scholar]
- 26.Korber B, Myers G. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992;8:1549–1560. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- 27.Kumar U, Monjardino J, Thomas H C. Hypervariable region of hepatitis C virus envelope glycoprotein (E2/NS1) in an agammaglobulinemic patient. Gastroenterology. 1994;106:1072–1075. doi: 10.1016/0016-5085(94)90770-6. [DOI] [PubMed] [Google Scholar]
- 28.Kurosaki M, Enomoto N, Marumo F, Sato C. Rapid sequence variation in the hypervariable region of hepatitis C virus during the course of chronic infection. Hepatology. 1993;18:1293–1299. [PubMed] [Google Scholar]
- 29.Lau J Y, Davis G L, Kniffen J, Qian K P, Urdea M S, Chan C S, Mizokami M, Neuwald P D, Wilber J C. Significance of serum hepatitis C virus RNA levels in chronic hepatitis C. Lancet. 1993;341:1501–1504. doi: 10.1016/0140-6736(93)90635-t. [DOI] [PubMed] [Google Scholar]
- 30.Lechmann M, Ihlenfeldt H G, Braunschweiger I, Giers G, Jung G, Matz B, Kaiser R, Sauerbruch T, Spengler U. T- and B-cell responses to different hepatitis C virus antigens in patients with chronic hepatitis C infection and in healthy anti-hepatitis C virus-positive blood donors without viremia. Hepatology. 1996;24:790–795. doi: 10.1002/hep.510240406. [DOI] [PubMed] [Google Scholar]
- 31.Liu S L, Rodrigo A G, Shankarappa R, Learn G H, Hsu L, Davidov O, Zhao L P, Mullins J I. HIV quasispecies and resampling. Science. 1996;273:415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- 32.Major M E, Mihalik K, Kolykhalov A A, Kleiner D, Rice C M, Feinstone S M. Presented at the Fifth International Meeting on Hepatitis C Virus and Related Viruses: Molecular Virology and Pathogenesis, 25–28 June 1998, Venice, Italy. 1998. Long term follow-up of chimpanzees inoculated with the first HCV infectious clone: immune responses, disease progression, and sequence evolution, abstr. 27. [Google Scholar]
- 33.Manzin A, Solforosi L, Petrelli E, Macarri G, Tosone G, Piazza M, Clementi M. Evolution of hypervariable region 1 of hepatitis C virus in primary infection. J Virol. 1998;72:6271–6276. doi: 10.1128/jvi.72.7.6271-6276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martell M, Esteban J I, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattsson L, Sonnerborg A, Weiland O. Outcome of acute symptomatic non-A, non-B hepatitis: a 13-year follow-up study of hepatitis C virus markers. Liver. 1993;13:274–278. doi: 10.1111/j.1600-0676.1993.tb00644.x. [DOI] [PubMed] [Google Scholar]
- 36.McAllister J, Casino C, Davidson F, Power J, Lawlor E, Yap P L, Simmonds P, Smith D B. Long-term evolution of the hypervariable region of hepatitis C virus in a common-source-infected cohort. J Virol. 1998;72:4893–4905. doi: 10.1128/jvi.72.6.4893-4905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Missale G, Bertoni R, Lamonaca V, Valli A, Massari M, Mori C, Rumi M G, Houghton M, Fiaccadori F, Ferrari C. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J Clin Investig. 1996;98:706–714. doi: 10.1172/JCI118842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 39.Neumann A U, Lam N P, Dahari H, Gretch D R, Wiley T E, Layden T J, Perelson A S. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 40.Odeberg J, Yun Z B, Sönnerborg A, Bjoro K, Uhlén M, Lundeberg J. Variation of hepatitis C virus hypervariable region 1 in immunocompromised patients. J Infect Dis. 1997;175:938–943. doi: 10.1086/513995. [DOI] [PubMed] [Google Scholar]
- 41.Ogata N, Alter H J, Miller R H, Purcell R H. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:3392–3396. doi: 10.1073/pnas.88.8.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto H, Kojima M, Okada S, Yoshizawa H, Iizuka H, Tanaka T, Muchmore E E, Peterson D A, Ito Y, Mishiro S. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology. 1992;190:894–899. doi: 10.1016/0042-6822(92)90933-g. [DOI] [PubMed] [Google Scholar]
- 43.Pawlotsky J M, Germanidis G, Neumann A U, Pellerin M, Frainais P O, Dhumeaux D. Interferon resistance of hepatitis C virus genotype 1b: relationship to nonstructural 5A gene quasispecies mutations. J Virol. 1998;72:2795–2805. doi: 10.1128/jvi.72.4.2795-2805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodrigo A G, Mullins J I. Human immunodeficiency virus type 1 molecular evolution and the measure of selection. AIDS Res Hum Retroviruses. 1996;12:1681–1685. doi: 10.1089/aid.1996.12.1681. [DOI] [PubMed] [Google Scholar]
- 45.Roth W K, Lee J H, Rüster B, Zeuzem S. Comparison of two quantitative hepatitis C virus reverse transcriptase PCR assays. J Clin Microbiol. 1996;34:261–264. doi: 10.1128/jcm.34.2.261-264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shankarappa R, Gupta P, Learn G H J, Rodrigo A G, Rinaldo C R J, Gorry M C, Mullins J I, Nara P L, Ehrlich G D. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology. 1998;241:251–259. doi: 10.1006/viro.1997.8996. [DOI] [PubMed] [Google Scholar]
- 47.Simmonds P, Balfe P, Ludlam C A, Bishop J O, Brown A J. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990;64:5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith D B, McAllister J, Casino C, Simmonds P. Virus ‘quasispecies’: making a mountain out of a molehill? J Gen Virol. 1997;78:1511–1519. doi: 10.1099/0022-1317-78-7-1511. [DOI] [PubMed] [Google Scholar]
- 49.Smith D B, Pathirana S, Davidson F, Lawlor E, Power J, Yap P L, Simmonds P. The origin of hepatitis C virus genotypes. J Gen Virol. 1997;78:321–328. doi: 10.1099/0022-1317-78-2-321. [DOI] [PubMed] [Google Scholar]
- 50.Smith D B, Simmonds P. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J Mol Evol. 1997;45:238–246. doi: 10.1007/pl00006226. [DOI] [PubMed] [Google Scholar]
- 51.Steinhauer D A, Holland J J. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409–433. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]
- 52.Thomas D L, Zenilman J Z, Alter H J, Shih J W, Galai N, Quinn T C. Sexual transmission of hepatitis C virus among patients attending Baltimore sexually transmitted diseases clinics—an analysis of 309 sexual partnerships. J Infect Dis. 1995;171:768–775. doi: 10.1093/infdis/171.4.768. [DOI] [PubMed] [Google Scholar]
- 53.Tong M J, El-Farra N S, Reikes A R, Co R L. Clinical outcomes after transfusion-associated hepatitis C. N Engl J Med. 1995;332:1463–1466. doi: 10.1056/NEJM199506013322202. [DOI] [PubMed] [Google Scholar]
- 54.van Doorn L J, Capriles I, Maertens G, DeLeys R, Murray K, Kos T, Schellekens H, Quint W. Sequence evolution of the hypervariable region in the putative envelope region E2/NS1 of hepatitis C virus is correlated with specific humoral immune responses. J Virol. 1995;69:773–778. doi: 10.1128/jvi.69.2.773-778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Villano S A, Vlahov D, Nelson K E, Cohn S, Thomas D L. Persistence of viremia and the importance of long-term follow-up after acute hepatitis C infection. Hepatology. 1999;29:908–914. doi: 10.1002/hep.510290311. [DOI] [PubMed] [Google Scholar]
- 56.Villano S A, Vlahov D, Nelson K E, Lyles C M, Cohn S, Thomas D L. Incidence and risk factors for hepatitis C among injection drug users in Baltimore, Maryland. J Clin Microbiol. 1997;35:3274–3277. doi: 10.1128/jcm.35.12.3274-3277.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vlahov D, Anthony J C, Muñoz A, Margolik J, Celentano D D, Solomon L, Polk B F. The ALIVE Study: a longitudinal study of HIV-1 infection in intravenous drug users: description of methods. J Drug Issues. 1991;21:759–776. [PubMed] [Google Scholar]
- 58.Wang Y, Ray S C, Laeyendecker O, Ticehurst J R, Thomas D L. Assessment of hepatitis C virus sequence complexity by the electrophoretic mobility of both single- and double-stranded DNA. J Clin Microbiol. 1998;36:2982–2989. doi: 10.1128/jcm.36.10.2982-2989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, et al. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang L, Diaz R S, Ho D D, Mosley J W, Busch M P, Mayer A. Host-specific driving force in human immunodeficiency virus type 1 evolution in vivo. J Virol. 1997;71:2555–2561. doi: 10.1128/jvi.71.3.2555-2561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zibert A, Meisel H, Kraas W, Schulz A, Jung G, Roggendorf M. Early antibody response against hypervariable region 1 is associated with acute self-limiting infections of hepatitis C virus. Hepatology. 1997;25:1245–1249. doi: 10.1002/hep.510250530. [DOI] [PubMed] [Google Scholar]