Summary
Osteocytes are the mechano-sensors of bones. Large gradient high-static magnetic fields (LG-HMFs) produce stable, high-precision, and non-attenuation mechanical forces. We discovered that magnetic forces opposite to gravity inhibited MLO-Y4 osteocyte proliferation and viability by inducing structural damage and apoptosis. In contrast, magnetic force loading in the same direction as that of gravity promoted the proliferation and inhibited apoptosis of MLO-Y4 osteocytes. Differentially expressed gene (DEG) analysis after magnetic force stimulation indicated that the ECM-integrin-CSK axis responded most significantly to mechanical signals. Wisp2 was the most significant DEG between the 12 T upward and downward groups, showing the highest correlation with the Wnt pathway according to the STRING protein interaction database. Explaining the cellular and molecular mechanisms by which mechanical stimuli influence bone remodeling is currently the focus of osteocyte-related research. Our findings provide insights into the effects of LG-HMFs on bone cells, which have further implications in clinical practice.
Subject areas: Cellular physiology, Molecular biology, Developmental biology
Graphical abstract

Highlights
-
•
Magnetic force direction affects the morphology and proliferation of MLO-Y4 osteocytes
-
•
Magnetic direction affects ECM-integrin-CSK axis-related proteins in MLO-Y4 cells
-
•
Cytoskeleton stabilization influences intracellular Wnt signaling in MLO-Y4 cells
-
•
Thus, magneto-mechanical stimulation affects the function of MLO-Y4 osteocytes
Cellular physiology; Molecular biology; Developmental biology.
Introduction
In 1936, Kolin1 first proposed the idea of employing electromagnetic fields in medical research. He demonstrated that, in general, biological systems are greatly influenced by the application of external magnetic fields. Permanent and man-made alternating magnetic fields of various strengths surround humans in daily life, including geomagnetic fields, stray fields from electromagnets and permanent magnets, as well as electromagnetic fields near electric devices and electric power lines.2 Magnetic resonance imaging (MRI) is undoubtedly a major success in the use of man-made high-magnetic fields in the clinical field. As a non-invasive, non-ionizing, and non-destructive medical diagnostic technique, MRI is widely used in clinical and animal research. The three distinct fields formed during the use of MRI include a static magnetic field (), time-dependent gradient fields, and the radiofrequency transmitter field ().3,4 High-strength magnets allow for greater signal-to-noise ratios, resulting in higher resolution or shorter scan times. The increasing demand for spatial resolution and a high signal-to-noise ratio drives the development of stronger magnetic field systems. The 10.5 T whole-body MRI scanner installed at the University of Minnesota’s Center for Magnetic Resonance Research (CMRR) is the most powerful whole-body MRI machine available for human use till date.5 The past few decades have seen the rapid development of a wide range of technologies for generating high and ultra-high magnetic fields.6,7,8 The increasing magnetic field strength is accompanied by an exacerbation of the field inhomogeneity problem, that is, the magnetic field gradient.9 The magnetic field gradient describes the change in external magnetic field strength with respect to distance (dB/dz). The spatial gradient is the key parameter of the field, which determines the attraction, a translation force exerted on an object. It is measured in Gauss per centimeter (G/cm) or Tesla per meter (T/m) and is represented by , with its magnitude as 10. Magnetic fields have different or even opposite biological effects at the cellular and whole-organism level. A major issue is that the magnetic induction intensity and the magnetic signal gradients that drive cells are too small and are below the significance threshold.11,12
Hall et al.13 reported that working ion channels generate weak magnetic fields of 10−9–10−5 T due to ion currents using a nitrogen-vacancy probe. In fact, various components within cells have certain magnetic properties. For example, magnetic correlation and magnetic moment formation are key electronic properties of oxygenated and deoxygenated hemoglobin. These properties, which arise from the strongly correlated electrons, affect hemoglobin function.14 The response of cells and intracellular structures to magnetic forces in an applied magnetic field can be elastic, viscous, or viscoelastic.15 Magnetic forces influence orientation and sample movement in gradient fields.16
Over the past 20 years, large gradient high-static magnetic fields (LG-HMFs) have undergone significant advances for ground-based simulations of weightlessness17 and cellular magnetism studies.18 Qiang et al.19 collaborated with Japan Superconductor Technology Inc. to design and manufacture superconducting magnets (JMTA-16T50MF) that can generate LG-HMFs. The specifications of the superconducting magnet are similar to those described by Hirose.20 If the diamagnetic material droplet is placed in a vertically oriented magnetic field gradient, the apparent gravity of the droplet can be attenuated or enhanced to different extents by the magnetic force. In a mouse safety study investigating the effect of 3.5–23.0 T static magnetic fields, the increase in spleen weight was more evident in groups with field intensity of 13.5 T and a gradient of 117.2 T/m, rather than in those with the highest field intensity (23.0 T).21 This observation further suggested that the magnetic field gradient is a key factor contributing to magnetic field-induced biological effects. Water and biological materials, such as DNA, proteins, and lipids are examples of diamagnetic materials.7,22 The magnetic susceptibility of diamagnetic materials is represented by a small negative value. For example, for water at 37°C, the diamagnetic susceptibility χ is ≈ −9.05 × 10−6 (SI unit).23 Most tissue constituents have a volume magnetic susceptibility that is similar to that of water.24 When placed in a gradient magnetic field, the direction of the magnetic force on the diamagnetic substance points from the stronger field to the weaker region.12,25 The repulsive diamagnetic force on diamagnetic materials can counteract and superimpose with gravity in the bore of a superconducting solenoid and powerful “bitter” electromagnet.19,25
The magnetic force of a unit volume of material is calculated according to the equation:
where B is the magnetic flux density, μ0 is the magnetic permeability of vacuum (=4π × 10−7 H/m), χ represents volume susceptibility, and z is the vertical site coordinate.26,27 The magnetic force is special as a mechanical stimulus as it is a function of the density of materials.28 Since the advent of the LG-HMF, it has been used for life science research, which has led to novel findings.27,29,30
Osteocytes are the most abundant resident cells in the bone, accounting for 90–95% of the total cellular component in the adult skeleton.31 As such, osteocytes are the master regulators of bone homeostasis.31 Furthermore, osteocytes are endocrine cells that regulate phosphate metabolism in multiple tissues. Most importantly, osteocytes act as primary mechanosensory cells.32,33 They are more responsive to mechanical stimulation than osteoblasts and trigger a greater calcium influx while releasing more prostaglandin (PGE2) and nitric oxide (NO), in addition to exhibiting more rapid β-catenin-mediated transcription.31 Osteocyte mechano-transduction is a complex but exquisite regulatory process occurring between different mechanosensory cells, between cells and their environment, as well as between neighboring cells.34 The cellular mechano-transduction process requires designated mechanotransducers to convert mechanical force into various fundamental biological signals.35 In general, the force applied to the cell-matrix adhesion first causes physical deformation of the ECM, with mechanical signals then transmitted from the ECM-integrin bond, to adapter proteins, and finally, to the cytoskeleton.36 Catch bonds have been mostly described for ECM-integrin interactions but were recently identified in adapter protein interactions, such as that between vinculin and actin.37 Huang et al. discovered that vinculin forms a force-dependent catch bond with F-actin through its tail domain, with lifetimes that depended on the direction of the applied force.37 The cytoskeleton typically moves mechanical signals from the site of assembly at the leading edge toward the cell centers.38 Osteocytes respond to mechanical stimuli by initiating intracellular signaling and producing factors that activate both osteoclasts and osteoblasts to remodel bone.39 The most researched and best described pathways induced by mechanical stimulations are those involving NO, ATP, PGEs, Ca2+, and Wnt, which are the first factors to be released by mechanically stressed osteocytes. Blocking one of these signaling pathways inhibits loading-induced bone anabolism.40,41 Several experimental settings have been developed to study cells under reduced mechanical cues, including the random positioning machine,42 slow-rotating wall vessels, the 3D clinostat, or antimagnetism levitation.43 Commercial and custom-made devices used in studies can load cells with a wide range of mechanical stimuli such as tension, compression, centrifugation, shear, and vibration.44 Different concepts and equipment designs produce mechanical stimuli of different amplitude and frequency that may lead to different strain axes and thus exhibit distinct effects on cellular reflection. Comparisons between studies should thus be made with caution. Therefore, it is of particular importance to study the effects of mechanical stimuli of the same type, size, and duration acting on cells.
When the mechanical state to which the bone is subjected changes, the internal structure of the bone changes and adapts accordingly while creating an optimal load-bearing structure. Explaining the cellular and molecular mechanisms through which this mechanical stimulus leads to bone reconstruction is the main current research task in the field of bone cell mechanics. In this study, magneto-mechanical stimuli of the same size and properties but in opposite directions were used to study the changes in cell structure and function of osteoblasts in response to mechanical loading and unloading.
Results
Cells survive in 12 T LG-HMFs
The magnet we used (Figure 1A) provides a high-homogeneous static magnetic field (SMF) at its center. The magnetic field direction is vertically upward. MLO-Y4 cell colony numbers decreased under 12 T upward treatment, while increasing under 12 T downward treatment (Figure 2A). To observe the effects of LG-HMFs with opposite orientation on the MLO-Y4 relative cell number and viability, we used the blood count method and CCK-8 assays, respectively. Relative cell number and viability decreased under 12 T upward treatment, while increasing under 12 T downward treatment at 24, 48, and 72 h (Figures 2B and 2C). Flow cytometry revealed that 12 T upward treatment for 24 h significantly promoted cell apoptosis. Alternatively, 12 T downward treatment inhibited osteocyte apoptosis (Figures 2D and 2E). Western blotting analysis was performed to determine the changes in the expression of apoptosis-related proteins Bax and cleaved caspase 3. The 12 T upward treatment significantly promoted the expression of Bax and cleaved caspase3 in MLO-Y4 cells (Figure 2F), whereas the 12T downward treatment showed a decreasing trend, but this was not statistically significant (Figure 2F). The 12 T upward magnetic field applied for 24 h significantly altered osteocyte morphology, and the synapses became elongated. Conversely, the 12 T downward treatment had no significant effect on osteocyte morphology (Figure 2G). By counting the cell area and number of dendrites, we found that they decreased after 12T upward treatment, whereas no effect was observed after 12T downward treatment (Figure 2H).
Figure 1.
The LG-HMFs exposure system
(A) Superconducting magnet (JMTA-16T50MF) and the object stage of cell culture.
(B) Parameters of the superconducting magnet.
(C) Magnetic field distribution in the magnetic cavity.
(D) B(dB/dz) represents the variation of the magnetic force with position.
(E) Plot of radial apparent gravity fit at the 12 T upward position.
(F) Plot of radial apparent gravity fit at the 12 T downward position.
(G) Schematic diagram of the magnetic forces and gravity on the cell in the magnet chamber.
Figure 2.
The influence of a different direction of magnetic force on growth of MLO-Y4 cells
(A) The clonogenic ability of MLO-Y4 cells.
(B) The blood cell count method calculates the number of cells; n = 3.
(C) CCK8 assay for cell viability; n = 3.
(D and E) Flow cytometric analysis of osteocytes to examine cell apoptosis.
(F) Protein expression levels of Bax and cleaved caspase 3.
(G) Observation of cellular morphology by crystal violet staining. Scale bar: 100 μm.
(H) Analysis of the cellular area and the number of dendrites in osteocytes by using Image-Pro Plus software; n = 100.
Data shown as mean ± SD. ∗p < 0.05, ∗∗ <0.01, ∗∗∗ <0.001.
DEG analysis
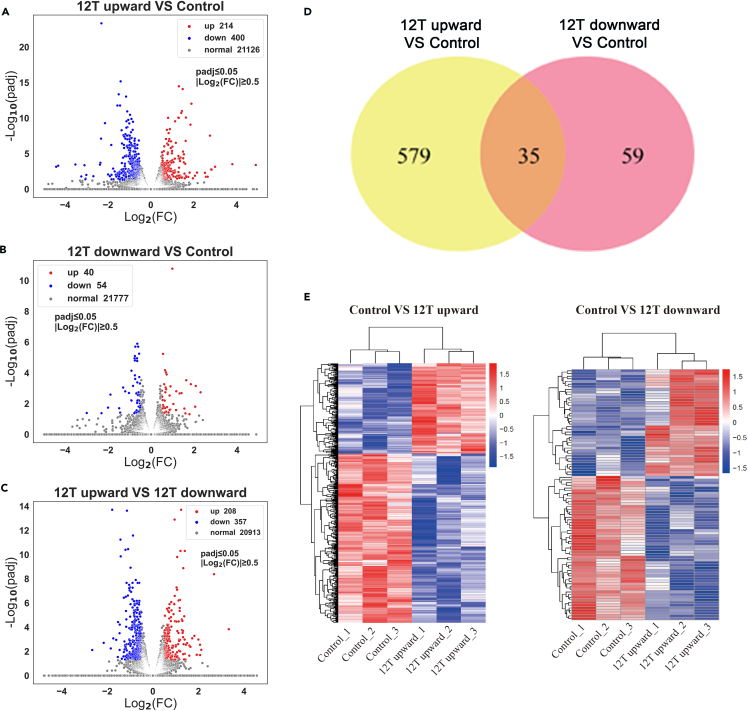
Control, 12 T upward, and 12 T downward-treated cells were subjected to RNA sequencing (Figure 9). Compared to the control, 12 T upward-treated MLO-Y4 cells exhibited 614 DEGs, of which 214 were upregulated and 400 were downregulated (Figure 3A). Alternatively, 12 T downward-treated MLO-Y4 cells exhibited 94 DEGs, of which 40 were upregulated and 54 were downregulated (Figure 3B). Furthermore, there were 565 DEGs in the 12 T upward group compared to the 12 T downward group. Upregulated and downregulated DEGs are shown in volcano plots (Figure 3C). The Venn diagram shows 35 common DEGs in the two treatment groups relative to the control (Figure 3D). These included Wisp2, Col5a3, Oas2, Plin4, Parp11, Sp100, Uba7, Trim30a, Stat1, Il4ra, Hist1h1c, Oas3, Gbp7, Cmpk2, Slfn8, Trim30d, Ifi47, Rbm3, Usp18, Tor3a, Uap1l1, Gm6136, Gm20559, Ccl2, Herc6, Cd80, Cirbp, Cbs, Ifi207, Rpl9-ps6, Gm9115, Ctso, Tnfaip3, Tlr3, and Gm7666. After identifying the DEGs, we performed a hierarchical cluster analysis with the upregulated and downregulated genes illustrated in a heatmap (Figure 3E). Gene counts were log10 transformed and normalized as Z score. In Figure 3E, two clusters were clearly displayed; all control groups were in one cluster and the LG-HMF groups in another cluster, which indicates high-intragroup consistency and high-intergroup variability.
Figure 9.

Library preparation for transcriptome sequencing.
Figure 3.
DEG expression profiles of osteocytes after different direction magnetic force treatment
(A) Volcano plot of DEGs between 12 T upward and control groups.
(B) Volcano plot of DEGs between 12 T downward and control groups.
(C) Volcano plot of DEGs between 12 T upward and 12 T downward groups.
(D) Venn diagram of DEGs.
(E) Heatmap of cluster analysis of DEGs. Red indicates DEGs with high expression, and blue indicates DEGs with low expression.
Gene ontology (GO) enrichment analysis of DEGs
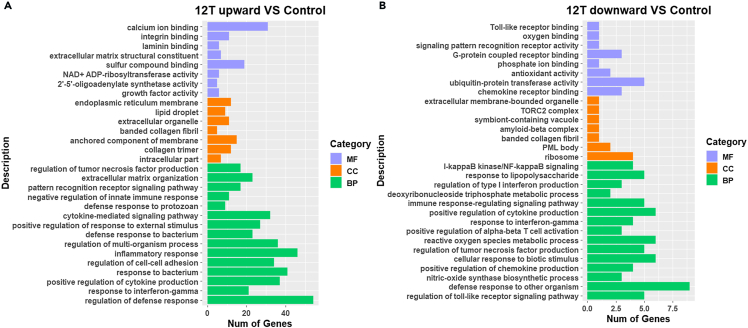
We performed GO enrichment analysis on DEGs, with p < 0.05 as the significance threshold. Genes were enriched in terms in the biological process (BP), cellular component (CC), and molecular function (MF) categories. GO enrichment classification maps of DEGs from the 12 T upward (Figure 4A) and 12 T downward (Figure 4B) treatment groups were generated. DEGs in the 12 T upward treatment group were enriched for defense response, regulation of cytokine production, and mitotic E in the BP category. For CC, the highest number of genes was enriched for ECM. Enriched MF terms included platelet-derived growth factor binding, growth factor binding, and receptor regulator activity (Figure 4A). For the 12 T downward treatment group, significant BP terms included toll-like receptor signaling pathway, defense response against other organisms, and positive regulation of nitric oxide synthase. Among CC terms, the ribosomal subunit had the highest number of enriched genes. Lastly, CCR chemokine receptor binding, sulfur compound binding, and glycosaminoglycan binding were the significant MF terms (Figure 4B).
Figure 4.
GO enrichment analysis of DEGs
(A) GO enrichment classification map of DEGs of 12 T upward with control.
(B) GO enrichment classification map of DEGs of 12 T downward with control.
Effects of LG-HMFs on the ECM-integrin-CSK
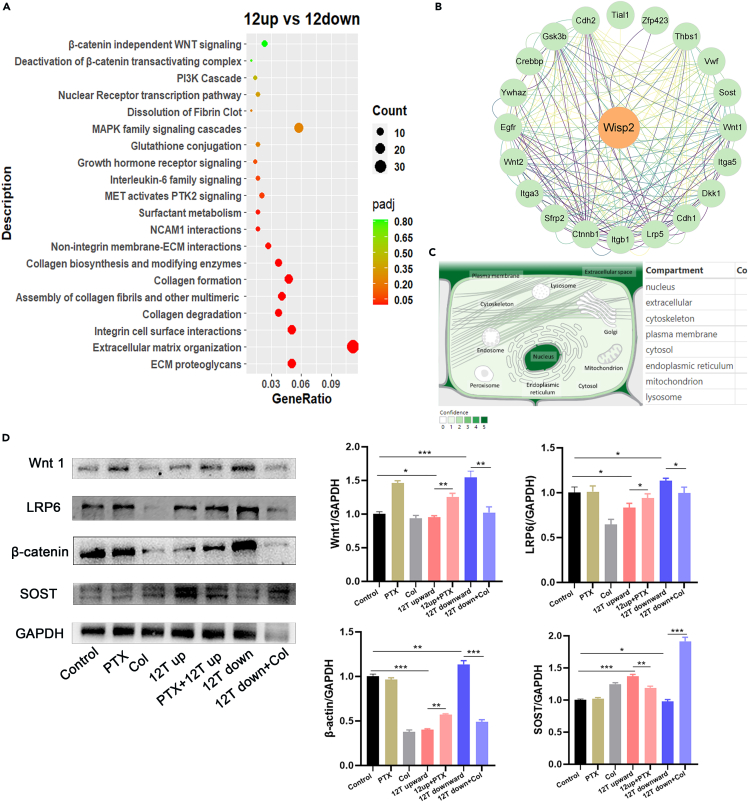
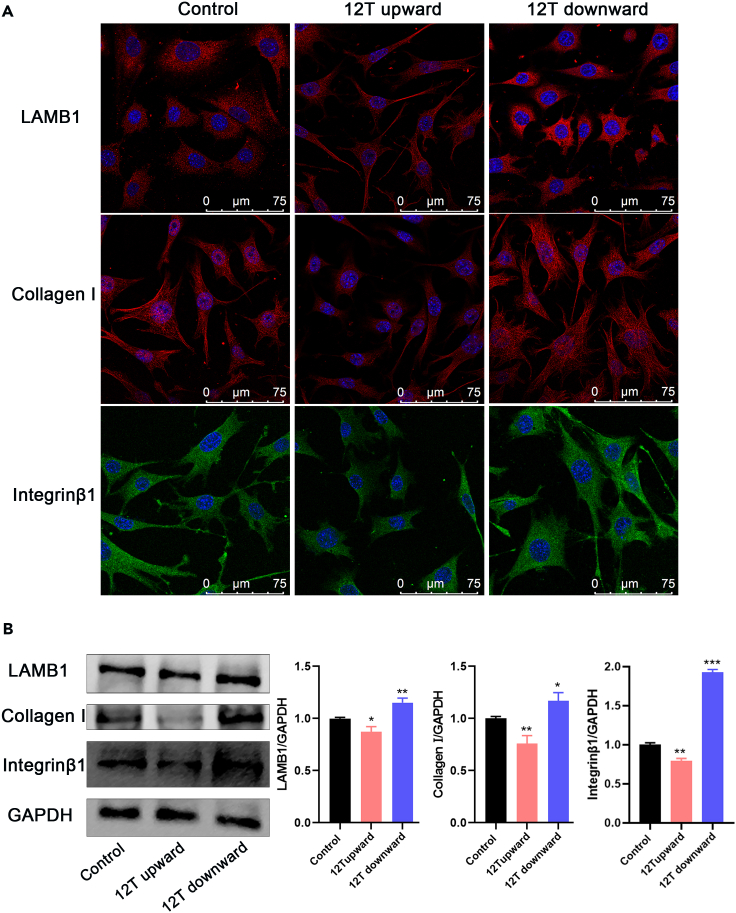
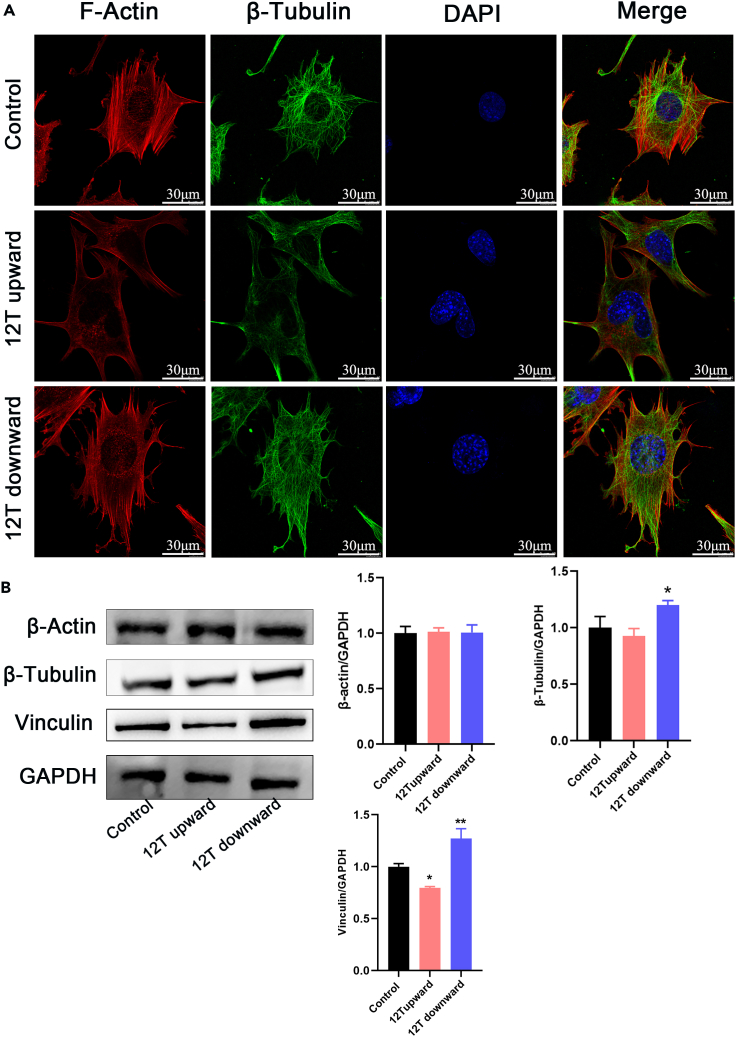
We subjected DEGs between the 12 T upward and 12 T downward treatment groups to Reactome pathway analysis, with p < 0.05 as a significance threshold. In Figure 7A, significant pathways were plotted using a bubble diagram and result indicated that DEGs were mainly associated with ECM proteoglycans, ECM organization, and integrin cell surface interactions. Further, 12 T upward treatment significantly inhibited the expression of ECM proteins LAMB1 and integrin β1. In contrast, 12 T downward treatment significantly promoted the expression of ECM proteins LAMB1 and integrin β1 (Figures 5A and 5B). Under control and 12 T downward conditions, MLO-Y4 cells showed star-like shape, and their actin fibers were evenly distributed across the cell bodies. The cells are abundant in actin fibers, which branch or rebranch in different planes of the cell body, forming intercellular connections with surrounding bone cells. However, under the 12 T upward conditions, MLO-Y4 cells exhibited a relatively long cell body shape, with a discontinuous and loose cytoskeleton (Figure 6A). We also examined the expression of microfilament protein β-actin and microtubulin protein β-tubulin, observing no significant differences except that 12 T downward treatment could promote β-tubulin expression. However, 12 T upward treatment significantly downregulated the expression of skeleton-binding protein vinculin, while 12 T downward treatment had the opposite effect (Figure 6B).
Figure 7.
Effect of magnetic force on the Wnt signaling pathway in MLO-Y4 cells
(A) The Reactome enrichment analysis of 12 T upward and 12 T downward; significant pathways were plotted in the bubble diagram.
(B) Protein interactions network map centered on wisp2.
(C) Subcellular localizations of wisp2.
(D) Protein expression levels of the Wnt signaling pathway (Wnt1, LRP6, β-catenin, and SOST).
Data shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; vs. control.
Figure 5.
The influence of different direction of magnetic force on ECM and integrin
(A) Immunoluminescent staining of ECM proteins (LAMB1 and Collagen Ι) and integrin ligand proteins (integrin β1). Scale bar: 75 μm.
(B) Protein expression levels of LAMB1, Collagen Ι, and integrin β1. Data shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 vs. control.
Figure 6.
The influence of different direction of magnetic force on the cytoskeleton in osteocytes
(A) Immunoluminescent staining of F-actin and β-tubulin. Scale bar: 30 μm.
(B) Protein expression levels of β-actin, β-tubulin, and Vinculin.
Data shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01.
LG-HMFs regulate the Wnt pathway in MLO-Y4
DEGs between the 12 T upward and 12 T downward treatment groups were subjected to pathway enrichment analysis, with significant terms including the Wnt/β-catenin pathway (Figure 7A). Wisp2 (Wnt1-inducible signaling pathway protein 2) was the most significant DEG between the 12 T upward and 12 T downward groups. We analyzed Wisp2 using the STRING protein interaction database to obtain the protein networks that interact with Wisp2 (Figure 7B). The GeneCards database indicated that Wisp2 localized to the nucleus, ECM, and CSK (Figure 7C). The STRING database highlighted the Wnt signaling pathway as the most relevant to Wisp2. Wnt signaling is mechanosensitive and acts downstream of mechanical stimuli during skeletogenesis.45 Here, 12 T upward treatment significantly inhibited the expression of Wnt pathway-related proteins, including Wnt1, β-catenin, and LRP6, in MLO-Y4 cells (Figure 7D). Sclerosteosis (SOST) has an inhibitory effect on the Wnt pathway. Furthermore, 12 T upward treatment for 48 h significantly promoted the protein expression of SOST, while inhibiting that of Wnt pathway members (Figure 7D). To verify whether the LG-HMF affects Wnt pathway factor expression in light of its effect on the skeleton, we pretreated MLO-Y4 cells with a low concentration of paclitaxel (0.5 μM) for 24 h before placing them under 12 T upward magnetic field conditions. We discovered that cytoskeleton stabilization in the LG-HMF stimulated Wnt signaling (Figure 7D). At the same time, colchicine (0.5 μM) pretreatment at a low concentration significantly inhibited the effect of 12 T downward treatment on Wnt signaling in MLO-Y4 cells (Figure 7D).
Conditioned medium (CM) from LG-HMF-treated osteocytes promotes osteoclasts differentiation
Osteocytes are the main regulators of bone homeostasis, exerting their effects through paracrine signaling to coordinate the differentiation and function of osteoclasts and osteoblasts.46 To determine whether LG-HMFs influence osteocyte function, we examined the effect of CM on bone differentiation in pre-osteoblast RAW264.7 cells. Osteoclast differentiation was evaluated based on F-actin rings and TRAP staining. Compared with those in control CM-treated group, the area of F-actin rings (Figure 8A) and number of TRAP-positive multinucleated cells (Figures 8B and 8C) were increased after treatment with CM from 12 T upward-treated osteocytes, while they decreased after treatment with the CM from 12 T downward-treated osteocytes. OPG and NO are factors released by osteoblasts to inhibit osteoclast differentiation and activity.47 The 12 T upward treatment inhibited OPG (osteoclastogenesis inhibitory factor) and NO synthesis, whereas 12 T downward treatment promoted it (Figures 8D and 8E).
Figure 8.
LG-HMFs affect the function of MLO-Y4 osteocytes
(A) Osteoclast differentiation was evaluated by the F-actin ring staining.
(B and C) Osteoclast differentiation was evaluated by the TRAP staining.
(D) Detection of NO secretion in CM.
(E) Protein expression levels of OPG.
Data shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; vs. control. #p < 0.05, ##p < 0.01, ###p < 0.001, ####p < 0.0001; 12 T upward vs. 12 T downward. Scale bar: 200 μm.
Discussion
Magnetic forces generated by magnetic fields act on cellular components, including the cell membrane, cytoplasm, cytoskeleton, nucleus, and intracellular motors, thus, triggering cellular signaling pathways. We have previously shown that simulated microgravity generated by LG-HMF influences the fate of MC3T3-E1 osteoblasts.27,48 In the current study, we used LG-HMF-generated magnetic forces with the same magnitude and opposite direction to investigate the effects of magnetic force loading in different directions on cell fate and the underlying mechanisms. We observed that the viability and relative number of osteocytes cultured under 12 T downward treatment for 24 h were enhanced, while the same parameters were decreased following 12 T upward treatment. There was no significant change in cell morphology, cell area, and number of cell dendrites in the 12 T downward treatment group compared with those in the geomagnetic group, and apoptosis tended to decrease, which may account for the increase viability of MLO-Y4 cells. Alternatively, the 12 T upward treatment significantly promoted cell apoptosis, with changes in osteocyte morphology, dendrites elongation and reduced dendrites number, which directly led to a decrease in cell number and viability.
Compared to controls, 12 T upward-treated MLO-Y4 cells exhibited 614 DEGs, while 94 DEGs were obtained for 12 T downward-treated cells. This suggests that the upward magnetic force may cause more biological effects. We further performed GO functional enrichment analysis of the DEGs. GO enrichment results showed that the biological processes that changed after both 12 T upward and 12 T downward treatment were associated with the regulation of defense responses, collagen polymerization, extracellular matrix protein binding, integrin binding, and ion binding. Interestingly, the biological processes that change in MLO-Y4 cells after 12 T upward treatment include regulation of cell-cell adhesion, while the biological processes that change after 12 T downward treatment include nitric oxide synthase biosynthesis processes. At the same time, DEGs between the 12 T upward and 12 T downward groups were subjected to analysis based on the Reactome database. These were associated with ECM proteoglycans, ECM organization, and integrin cell surface interactions. To further validate the transcriptome-sequencing results, we examined the expression of extracellular matrix proteins and integrin-related proteins using immunofluorescence and Western blot assays. The 12 T upward treatment inhibited the expression of LAMB1, collagen I, and integrin β1 in MLO-Y4 cells. Mechanical homeostasis involves ECM constituents such as collagens and laminin, which support and transmit loads. Integrins bridge extracellular and intracellular structures, while their associated linker proteins (such as talin and vinculin) connect receptors to the cytoskeleton.49 The microfilament skeleton of osteocytes was clustered at the edges of cells treated with 12 T upward, indicative of altered microfilament distribution. Alternatively, 12 T downward promoted the skeletal arrangement of osteocytes. Our Western blot results showed that the expression of β-tubulin and β-actin was less affected by magnetism. However, vinculin, which is involved in cytochemical signaling in combination with F-actin, showed opposite expression trends in the 12 T upward and 12 T downward treatment groups.
Wisp2, the most significant DEG between 12 T upward- and 12 T downward-treated cells, localized to the nucleus, ECM, and cytoskeleton. Wisp2 was most strongly correlated with the Wnt pathway based on the STRING protein interactions database. Wisp2 is both a secretory and cytosolic protein that activates the classical Wnt pathway and increases β-catenin levels.50 Herein, we discovered that the 12 T upward treatment inhibited the Wnt1/LRP6/β-catenin pathway. In combination with our experimental results, cytoskeleton stabilizer pretreatment of MLO-Y4 cells attenuated the inhibitory effect of 12 T upward on the Wnt signaling pathway. Cytoskeleton inhibitor pretreatment of MLO-Y4 cells inhibited the effect of the 12 T downward treatment of the Wnt signaling pathway. Thus, cytoskeletal stability is important for the transduction of the Wnt signaling pathway in MLO-Y4 cells. Furthermore, the 12 T upward treatment decreased NO production and increased SOST expression, further synergistically inhibiting the Wnt pathway in MLO-Y4 cells. Wnt pathway inhibition further reduced the expression of downstream bone function protein OPG. NO and OPG are typical cytokines that inhibit osteoclast differentiation.47 Alternatively, osteoclast formation was significantly enhanced following the 12 T upward treatment due to the significantly lower NO and OPG secreted by cells. Conversely, the 12 T downward treatment increased NO production and decreased SOST expression. Higher secretion of NO and OPG suppressed osteoclast differentiation in the 12 T downward treatment group relative to the control group.
Overall, the parameters associated with the mechanical stimulation to which cells were subjected in this study are more standard and accurate. In MLO-Y4 osteocytes, the ECM-integrin-CSK axis and Wnt1/LRP6/β-catenin/OPG pathway were altered by magnetic force. Osteocyte fate and function were altered as a result. We further clarified the effect of osteocytes on bone reconstruction in response to different mechanical stimuli. We provide a basis for further research on this issue in terms of cellular-molecular mechanisms that are of great importance to clinical practice.
Limitations of the study
This study had some limitations. The first is the short exposure time of 24 h in LG-HMFs for the cells used for transcriptome sequencing to ensure that the cells are in optimal condition. Second, only the differentiation of osteoclasts based on osteocyte secretory factors and not that based on osteocyte secretory factors was tested for osteoblastes function. Finally, only the effect of magnetic force on the expression of ECM-integrin-CSK was examined. The information transfer between ECM-integrin-CSK is based on previous studies. Further experiments are needed to verify the transmission of magneto-mechanical signals in cells.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| GAPDH Monoclonal antibody | Proteintech | RRID:AB_210743; Cat#60004-1-Ig |
| Anti-Bax antibody | Abcam | RRID:AB_2938987; Cat#ab182733 |
| Cleaved Caspase-3 (Asp175) Antibody | Cell Signaling Technologies | RRID:AB_2341188; Cat# 9661 |
| Laminin beta 1 Polyclonal antibody | Proteintech | RRID:AB_2879288; Cat#23498-1-AP |
| Collagen Type I Polyclonal antibody | Proteintech | RRID:AB_2082037; Cat#14695-1-AP |
| Integrin Beta 1 Polyclonal antibody | Proteintech | RRID:AB_2130085; Cat#12594-1-AP |
| β-Actin (13E5) Rabbit mAb | Cell Signaling Technologies | RRID:AB_2223172; Cat#4970 |
| Anti β-Tubulin Mouse Monoclonal Antibody | Proteintech | RRID:AB_2881629; Cat#66240-1-Ig |
| WNT1 Polyclonal antibody | Proteintech | RRID:AB_2881013; Cat#27935-1-AP |
| LRP6 (C47E12) Rabbit mAb | Cell Signaling Technologies | RRID:AB_1950408; Cat#3395 |
| Beta Catenin Polyclonal antibody | Proteintech | RRID:AB_2086128; Cat#51067-2-AP |
| Anti-Sclerostin antibody | Abcam | RRID:AB_956321; Cat#ab63097 |
| Anti-Osteoprotegerin antibody | Abcam | RRID:AB_2934183; Cat#ab183910 |
| Cy3-labeled Goat Anti-Rabbit IgG (H + L) | Beyotime Biotechnology | RRID:AB_2893015; Cat#A0516 |
| Goat Anti-Mouse IgG H&L (Alexa Fluor® 488) | Abcam | RRID:AB_2576208; Cat#ab150113 |
| HRP-labeled Goat Anti-Rabbit IgG(H + L) | Beyotime Biotechnology | RRID:AB_2892644; Cat#A0208 |
| HRP-labeled Goat Anti-Mouse IgG(H + L) | Beyotime Biotechnology | RRID:AB_2860575; Cat#A0216 |
| Chemicals, peptides, and recombinant proteins | ||
| Murine sRANK-Ligand | Peprotech | Cat# 315-11-10UG |
| Paclitaxel | TopScience | Cat# T0968 |
| Colchicine | TopScience | Cat# T0320 |
| Deposited data | ||
| RNA-seq transcriptome data | This paper | NCBI-based platforms: NCBI Sequence Read Archive (SRA): BioProject ID PRJNA932050 |
| Experimental models: Cell lines | ||
| MLO-Y4 | University of Texas Health Science Center | RRID:CVCL_M098 |
| RAW 264.7 | ATCC | RRID:CVCL_0493; Cat#TIB-71 |
| Software and algorithms | ||
| GraphPad Prism 8.2.1 | https://www.graphpad.com/ | |
| Python 3.8 | https://www.python.org/ | |
| R-project | https://www.r-project.org/ | |
| ImageJ | https://ImageJ.net/software/fiji/downloads | |
| Image-Pro Plus6.0 | https://www.epixinc.com/vision_archive/imagepro.htm | |
| Other | ||
| Collagen Type Ι | BD Corning | Cat#354236 |
| Crystal Violet | Amersco | Cat#0528-25G |
| Triton X-100 | Sinopharm Chemical ReagentCo., Ltd | Cat#30188928 |
| Cell Counting Kit-8 | Beyotime Biotechnology | Cat#C0038 |
| Annexin V-FITC Apoptosis Detection Kit | Beyotime Biotechnology | Cat#C1062M |
| ActinRed™ 555 ReadyProbes™ | Thermo Fisher Scientific | Cat#R37112 |
| DAPI dihydrochloride | Beyotime Biotechnology | Cat#C1002 |
| NO assay kit | Beyotime Biotechnology | Cat#S0021S |
| Leukocyte acid phosphatase (TRAP) Kit | Sigma-Aldrich | Cat# 387A-1KT |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Peng Shang, Northwestern Polytechnical University, China (shangpeng@nwpu.edu.cn).
Materials availability
This study did not generate new unique reagents. Further information and request for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Peng Shang, Northwestern Polytechnical University, China (shangpeng@nwpu.edu.cn).
Experiment methods and subject details
Cell lines
The mouse osteocyte-like cell line (MLO-Y4) used in this study was gifted by Prof. Jean X. Jiang (University of Texas Health Science Center, San Antonio, TX, USA). MLO-Y4 cells were cultured on collagen-coated (Collagen Type Ι, BD Corning, Cat#354236) petri dishes. The cells were cultured in α-MEM medium (Gibco) supplemented with 5% fetal bovine serum (FBS, Gibco), 5% calf serum (Every Green), 2 mM L-glutamine, and 1% penicillin/streptomycin (Beyotime Biotechnology, Cat#C0222) in a humidified 5% CO2 and 37°C controlled by an incubator system (Thermo Fisher Scientific).
Pre-osteoclast RAW 264.7 cells were purchased from the Cell Bank of Chinese Academy of Science (CAS; Shanghai, China), ATCC (TIB-71). RAW 264.7 cells were cultured in α-MEM medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco), 2 mM L-glutamine, and 1% penicillin/streptomycin (Beyotime Biotechnology, Cat#C0222). The cells were cultured in a humidified 5% CO2 and 37°C controlled incubator system (Thermo Fisher Scientific).
Method details
Large gradient high magnetic fields exposure system
In this study, LG-HMFs were produced by a superconducting magnet (JMTA-16T50MF) (Figure 1A). The LG-HMF consisted of the NbTi and Nb3Sn superconductors. To generate the 16 T magnetic field in a region with a diameter of 50 mm, the superconducting magnet was combined with reverse and main coils. The main coils consisted of two NbTi coils and one Nb3Sn coil. The main coil was designed to generate a high field gradient. The reverse coil consisted of a Nb3Sn coil that generates a reverse field for future high magnetic field gradients of approximately 150 T/m.27,51 In addition to the space occupied by the temperature control unit in the magnet hole, the diameter of the space used for the experiment was approximately 44 mm, while the total length was 400 mm. We independently developed an experimental platform to study the cell biology effect of superconducting magnets, including object stages (Figure 1B), temperature control systems, and gas control systems. The constant temperature circulation system (MPG-13A, Bluepard instruments) was composed of a temperature controller, temperature sensor, heater, circulation pump, and low water lever alarm device. The temperature of the magnetic chamber was controlled through heat conduction of temperature-controlled water via the magnet chamber at 37°C–37.5°C. We designed our original gas mixing device to mix CO2 and air proportionally by adjusting the gas flow meters and then humidifying them through the humidity controller and passing them into the magnet chamber to ensure the humidity as well as the 5% CO2 concentration during cell culture. The direction of the magnetic field inside the magnet chamber was vertically upward (Figure 1C). In this study, cells were placed in the positions of 12.56 T, −109 T/m and 12.50 T, 105 T/m.
The object stage (Figure 1B) contained two 12 T cell culture platforms that were corresponding to different magnetic field gradients of −109, and 109 T/m in a 5 cm diameter room temperature bore, respectively. The change in magnetic field strength and gradient within the LG-HMF exhibited continuity (Figures 1D and 1E). Schematic diagram of the magnetic and gravity on the cell in the magnet chamber are illustrated in Figure 1F.
Cell proliferation and viability
For cell colony formation assays, MLO-Y4 cells were seeded in 35 mm petri dishes at 1000 cells/dish. After 12 h of cell adhesion on the plates, cells were cultured separately in a geomagnetic incubator, 12 T upward and 12 T downward LG-HMFs. The medium was refreshed every two days and colonies were observed after 10 days. Next, cells were washed with 1×PBS and fixed with 4% paraformaldehyde for 20 min. Subsequently, 1 mL of 0.1% crystal violet staining solution was added to each dish for 30 min. The excess crystal violet staining solution was washed with distilled water and air-dried. Finally, we observed the results using the body vision microscope (SZ61, Olympus) and acquired images using a DSC-H5 Sony camera.
For cell counting, MLO-Y4 cells were seeded into 35 mm petri dishes at 1 × 105 cells/dish. After adherence to the plates, cells were treated with 12 T upward and 12 T downward LG-HMFs for 24, 48, and 72 h. After trypsin digestion, we counted the total number of cells using a hemocytometer.
MLO-Y4 cell viability was measured using a Cell Counting Kit-8 (Beyotime Biotechnology, Cat#C0038). MLO-Y4 cells were inoculated in removable 96-well plates at a density of 5 × 103 cells/dish. After adherence to the plates, cells were treated with 12 T upward and 12 T downward LG-HMFs for 24, 48, and 72 h. Next, 10% CCK-8 was added to the culture medium and incubated for 2 h at 37°C. The absorbance values were measured at 490 nm using an enzyme marker (Synergy HT, BioTek).
Cell apoptosis assay
MLO-Y4 cell were seeded into 35-mm petri dishes at 1.5 × 105 cells/dish. After the MLO-Y4 cells were treated with 12 T upward and 12 T downward LG-HMFs for 24 h, cells were stained using Annexin V-FITC apoptosis Detection Kit (Beyotime Biotechnology, Cat#C1062M). The cell culture solution was aspirated into a 2-mL centrifuge tube and the apoptotic cells were washed once with 1 × PBS; then, an appropriate amount of trypsin solution was added to digest the cells. The cells were incubated at room temperature until they could be blown off by a gentle blow, the collected cell culture medium was added, cells were gently blown off, transferred to a centrifuge tube, centrifuged at 1000 × g for 5 min, and the supernatant was discarded. The cells were washed once by gently resuspending in 1 × PBS, centrifugation at 1000 × g for 5 min, and ultimately, the supernatant was discarded. Next, we added 195 μL Annexin V-FITC conjugate, 5 μL Annexin V-FITC, as well as 10 μL propidium iodide staining solution and mix gently. Subsequently, we incubated the solution for 10–20 min at room temperature (20°C–25°C), protected from light. The detection was performed using a flow cytometer (FACSCalibur, BD Bioscience). Annexin V+/PI− were considered early apoptotic cells. Annexin V+/PI+ were considered late apoptotic cells.
Cell morphology observation
MLO-Y4 cells were seeded into 35 mm petri dishes at 1 × 105 cells/dish. After adhering to the plates, cells were immediately placed in the LG-HMFs for 24 h. After treatment, cells were washed with 1 × PBS and fixed with 4% paraformaldehyde for 20 min. Next, the fixed cells were washed with 1 × PBS, stained with 0.1% crystal violet solution for 30 min, and washed cleanly using ultrapure water. Finally, light microscopy (MD IL HC, Leica) was used to observe and image the cells. The cell area and number of dendrites were determined using the Image-Pro Plus 6.0 software.
Library construction and sequencing
First, total RNA of MLO-Y4 cells was obtained using the MagZol reagent (Magen, China). The mRNA was enriched using poly-T oligo-attached magnetic beads. Subsequently, the obtained mRNA was randomly interrupted in fragmentation buffer, and the first strand of cDNA was synthesized using the fragmented mRNA as the template. RNaseH was used to degrade the RNA strand, and the second cDNA strand was synthesized from dNTPs as raw material under the DNA polymerase I system. The purified double-stranded cDNA underwent terminal repair, had an A tail added, and was attached to the sequencing linker. Next, we screened approximately 370–420 bp of cDNA with AMPureXP beads, amplified the sequences using PCR, and purified the PCR product again using AMPureXP beads to finally obtained a library. Fluorescently labeled polymerase was added to the sequencing flow cell, and the sequencing instrument passes through to capture the fluorescence signal. Data collection and statistical analysis were performed using a specific computer software (Novogene, China).52,53,54 (Figure 9).
RNA-seq data processing and function gene annotation
DEGs analysis between the two comparative combinations was performed using DESeq2 software (1.20.0). DEG analysis of two conditions were performed using the edgeR software package (3.22.5). p-values were adjusted using the Benjamini & Hochberg method. After correction, P-values and log2 fold change (|log2 FC|) were used as broad values for significant differential expression. DEGs were screened using adjusted P-values <0.05 and |log2 FC| > 0.5 as screening criteria. The acquired differentially expressed genes were imported into Python 3.8, data were read using numpy and pandas, and then the volcanos were plotted using matplotlib and seaborn. We used gene set enrichment analysis (GSEA) tools (http://www.broadinstitute.org/gsea/index.jsp) to analyze the GO and Reactome datasets. GSEA results were plotted using the ggplot2 plotting package in R-project 4.2.2. PPI analysis of DEGs was based on the STRING database (https://cn.string-db.org/) of known and predicted protein-protein interactions. The GeneCards database (https://www.genecards.org/) was used to obtain the localization of genes in cells.
Immunofluorescence staining
MLO-Y4 cells were seeded into confocal petri dishes at a density of 1 × 105 cells/dish. After adhering to the plates, cells were placed under the LG-HMFs for 48 h. The supernatant was removed, and 1× PBS was used to wash the cells once. Next, cells were fixed with 4% paraformaldehyde for 20 min and permeabilized with 0.2% Triton X-100 for 5 min (Sinopharm Chemical ReagentCo., Ltd, Cat#30188928). Subsequently, cells were blocked in 2% BSA (2% BSA-PBS) for 30 min at room temperature. Next, after 3 washes with 1× PBST, cells were cultured overnight at 4°C with primary antibodies, including actinRed 555 ReadyProbes Reagent (1:400 dilution in 1× PBS, Thermo Fisher Scientific, Cat#R37112), mouse anti-β-Tubulin antibody (1:200 dilution in 1× PBS, Proteintech, Cat#66240-1-Ig), rabbit anti-LAMB 1 antibody (1:200 dilution in 1× PBS, Proteintech, Cat#23498-1-AP), rabbit anti-collagen I antibody (1:200 dilution in 1× PBS, Proteintech, Cat#14695-1-AP), and rabbit anti-Integrin β1 antibody (1:200 dilution in 1× PBS, Proteintech, Cat#12594-1-AP). The cells were washed 5 times with 1× PBS, then stained with Goat anti-Rabbit Alexa Fluor550-conjugated antibody (1:500 dilution in 1× PBS, Beyotime Biotechnology, Cat#A0516) and Goat anti-Mouse Alexa Fluor488-conjugated antibody (1:500 dilution in 1× PBS, Abcam, Cat#ab150113) for 60 min at room temperature in the dark. Nuclei were stained with DAPI (1:1000 dilution in 1× PBS, Beyotime Biotechnology, Cat#C1002), followed by 5 washing steps. The cells were imaged with using a laser scanning confocal microscope (SP8, Leica).
Protein expression assay
MLO-Y4 cells were subjected to LG-HMFs for 48 h and washed with 1× PBS. The RIPA Lysis Buffer (Beyotime Biotechnology, Cat#P0013B) containing 2 mM Protease inhibitor cocktail (Beyotime Biotechnology, Cat#P1005) and 5 mM Phosphatase inhibitor cocktail (Beyotime Biotechnology, Cat#P1081) was used to lyse the cells. A commercial BCA Protein Assay Kit (Beyotime Biotechnology, Cat# P0009) was used to examine the protein content. Proteins were loaded into the SDS-PAGE gels, separated through electrophoresis, and then transferred onto a polyvinylidene fluoride membranes. The polyvinylidene fluoride membranes were blocked with 5% nonfat-dried milk and then incubated with specific primary antibodies, including mouse anti-GAPDH (1:5000 dilution in 1× TBST, Proteintech, Cat#60004-1-Ig), Anti-Bax antibody (1:2000 dilution in 1× TBST, abcam, Cat# ab182733), Cleaved Caspase-3 (Asp175) Antibody (1:1000 dilution in 1× TBST, Cell Signaling Technologies, Cat# 9661), rabbit anti-LAMB 1 antibody (1:1000 dilution in 1× TBST, Proteintech, Cat#23498-1-AP), rabbit anti-collagen I antibody (1:2000 dilution in 1× TBST, Proteintech, Cat#14695-1-AP), rabbit anti-Integrin β1 antibody (1:4000 dilution in 1× TBST, Proteintech, Cat#12594-1-AP), rabbit anti-β actin antibody (1:1000 dilution in 1× TBST, Cell Signaling Technologies, Cat#4970), mouse anti-β-Tubulin antibody (1:20000 dilution in 1× TBST, Proteintech, Cat#66240-1-Ig), rabbit anti-Wnt 1 antibody (1:1000 dilution in 1× TBST, Proteintech, Cat#27935-1-AP), rabbit anti-LRP6 antibody (1:1000 dilution in 1× TBST, Cell Signaling Technology, Cat#3395), rabbit anti-β catenin antibody (1:1000 dilution in 1× TBST, Proteintech, Cat#51067-2-AP), rabbit anti-Sclerostin antibody (1:1000 dilution in 1× TBST, Abcam, Cat#ab63097), and rabbit anti-Osteoprotegerin antibody (1:1000 dilution in 1× TBST, Abcam, Cat#ab183910) overnight at 4°C. After washing the membranes five times with 1× TBST, they were incubated with species-specific secondary antibody conjugated to horseradish peroxidase (1:2000 dilution in 1× TBST, Beyotime Biotechnology, Cat#A0208 and Cat#A0216) for 2 h at room temperature. An ECL Plus Western Blotting Detection System (JS-M6P, P&Q Science Technology) was used to image the immunoreactive bands. The grayscale bands were determined using the ImageJ 1.8.0 software.
Detection of osteoclast differentiation
MLO-Y4 cells were collected in the medium treated with control, 12 T upward, and 12 T downward LG-HMFs for 48 h. The medium was filtered through a 0.22-μm membrane to remove bacteria and mixed with α-MEM containing 10% fetal bovine serum at a ratio of 1:2 to obtain conditioned medium (CM). To induce osteoclast differentiation, Raw264.7 cells were cultured in α-MEM containing 10% fetal bovine serum and 50 ng/mL soluble RANKL as previously described.55 After 24 h, the medium was replaced with fresh CM. This procedure was repeated every day.
For F-actin rings staining, until day 4, cells were stained with actinRed 555 ReadyProbes Reagent (1:400 dilution in 1× PBS, Thermo Fisher Scientific, Cat#R37112) and DAPI (1:1000 dilution in 1× PBS, Beyotime Biotechnology, Cat#C1002). The procedure was similar to that of immunofluorescence staining. The osteoclast F-actin rings were observed and imaged using an inverted fluorescence microscope (Observer 3, ZEISS).
For tartrate-resistant acid phosphate (TRAP) staining, on day 4, cells were washed with 1× PBS and fixed with 4% paraformaldehyde for 20 min. Next, cells were stained with Leukocyte Acid Phosphatase Kit (Sigma-Aldrich, Cat#387A-1KT) according to the manufacturer’s protocol. The results were observed and imaged using a light microscope (CKX53, Olympus). The cells with a burgundy color or more than three nuclei were recognized as osteoclasts. The number or area of formed osteoclasts was calculated using the Image-Pro Plus 6.0 software.
Bioassay for NO production
MLO-Y4 cell were seeded into 35 mm petri dishes at 1.5 × 105 cells/dish. After the treatment with LG-HMFs for 48 h, the MLO-Y4 cells supernatant was collected. First, we diluted the standard with the solution used for the sample to be tested and made a standard curve according to manufacturer’s protocol (Beyotime Biotechnology, Cat#S0021S). We added standards and samples in 96-well plates at a volume of 50 μL/well. Next we added Griess Reagent I to each well (50 μL/well) followed by Griess Reagent II (50 μL/well). The absorbance values were measured at 540 nm using an enzyme marker (Synergy HT, BioTek).
Quantification and statistical analysis
All experimental data are expressed as mean ± SD, and statistical analysis was performed using GraphPad Prism version 8.2.1 software for Windows (GraphPad Software, San Diego, California USA). Data obtained from western blots are representative for 3 independent experiments. One-way ANOVA with Tukey’s multiple-comparison method was used to evaluate the differences between the control, 12 T upward, and 12 T downward. P-values <0.05 were considered statistically significant. Significance was defined as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Differential expression analysis was compared between the two combinations using DESeq2 1.20.0 software. The method of Benjamini and Hochberg was used to adjust the resulting P-values to control for false discovery rates (FDRs). Genes with an FDR-corrected P-value <0.05 by DESeq2 were considered differentially expressed. Significance for GO and Reactome enrichment analysis was determined using an FDR-corrected P-value (<0.05).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (52037007); the Innovation Foundation for Doctor Dissertation of Northwestern Polytechnical University (CX2021102), and the Chongming Project of the Heye Health Technology Co., Ltd. (HYCMP-2023016).
Author contributions
Conceptualization, P.S. and B.Z.; methodology & investigation, B.Z., G.Z., and D.Y.; validation, C.L., formal analysis, X.L. and X.Z., resources, P.S. and Y.F.; data curation, P.S. and B.Z.; writing-original draft, B.Z., X.L., and X.Z.; writing-review & editing, B.Z., S.W., H.L., G.Z., and P.S.; project administration, P.S. and B.Z.; funding acquisition, P.S., Y.F., and B.Z. supervision, P.S.
Declaration of interests
The authors declare no competing interests.
Published: July 13, 2023
Data and code availability
-
•
All standardized datasets have been deposited within the NCBI Sequence Read Archive (SRA): BioProject ID PRJNA932050, and are publicly available as of the data of publication.
-
•
This study does not report original code.
-
•
Any additional information required to reanalyze the data reported in this study is available from the lead contact upon request.
References
- 1.Kolin A. An electromagnetic flowmeter - Principle of the method and its application to bloodflow measurements. P Soc Exp Biol Med. 1936;35:53–56. doi: 10.3181/00379727-35-8854P. [DOI] [Google Scholar]
- 2.Gayathri K., Shailendhra K. MRI and Blood Flow in Human Arteries: Are There Any Adverse Effects? Cardiovasc. Eng. Technol. 2019;10:242–256. doi: 10.1007/s13239-019-00400-x. [DOI] [PubMed] [Google Scholar]
- 3.Schenck J.F. Safety of strong, static magnetic fields. J. Magn. Reson. Imag. 2000;12:2–19. doi: 10.1002/1522-2586(200007)12:1<2::Aid-Jmri2>3.0.Co;2-V. [DOI] [PubMed] [Google Scholar]
- 4.Wosik J., Villagran M., Uosef A., Ghobrial R.M., Miller J.H., Kloc M. Effects of Magnetic Fields and Field Gradients on Living Cells. Emagres. 2019;8:371–385. doi: 10.1002/9780470034590.emrstm1609. [DOI] [Google Scholar]
- 5.Nowogrodzki A. The world's strongest MRI machines are pushing human imaging to new limits. Nature. 2018;563:24–26. doi: 10.1038/d41586-018-07182-7. [DOI] [PubMed] [Google Scholar]
- 6.Hahn S., Kim K., Kim K., Hu X., Painter T., Dixon I., Kim S., Bhattarai K.R., Noguchi S., Jaroszynski J., Larbalestier D.C. 45.5-tesla direct-current magnetic field generated with a high-temperature superconducting magnet. Nature. 2019;570:496–499. doi: 10.1038/s41586-019-1293-1. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L., Hou Y., Li Z., Ji X., Wang Z., Wang H., Tian X., Yu F., Yang Z., Pi L., et al. 27 T ultra-high static magnetic field changes orientation and morphology of mitotic spindles in human cells. Elife. 2017;6 doi: 10.7554/eLife.22911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian X., Lv Y., Fan Y., Wang Z., Yu B., Song C., Lu Q., Xi C., Pi L., Zhang X. Safety evaluation of mice exposed to 7.0-33.0 T high-static magnetic fields. J. Magn. Reson. Imag. 2021;53:1872–1884. doi: 10.1002/jmri.27496. [DOI] [PubMed] [Google Scholar]
- 9.Mullen M., Garwood M. Contemporary approaches to high-field magnetic resonance imaging with large field inhomogeneity. Prog. Nucl. Magn. Reson. Spectrosc. 2020;120–121:95–108. doi: 10.1016/j.pnmrs.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panych L.P., Kimbrell V.K., Mukundan S., Jr., Madore B. Relative Magnetic Force Measures and Their Potential Role in MRI Safety Practice. J. Magn. Reson. Imag. 2020;51:1260–1271. doi: 10.1002/jmri.26924. [DOI] [PubMed] [Google Scholar]
- 11.Zablotskii V., Polyakova T., Dejneka A. Modulation of the Cell Membrane Potential and Intracellular Protein Transport by High Magnetic Fields. Bioelectromagnetics. 2021;42:27–36. doi: 10.1002/bem.22309. [DOI] [PubMed] [Google Scholar]
- 12.Zhang B., Yuan X., Lv H., Che J., Wang S., Shang P. Biophysical mechanisms underlying the effects of static magnetic fields on biological systems. Prog. Biophys. Mol. Biol. 2023;177:14–23. doi: 10.1016/j.pbiomolbio.2022.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Hall L.T., Hill C.D., Cole J.H., Städler B., Caruso F., Mulvaney P., Wrachtrup J., Hollenberg L.C.L. Monitoring ion-channel function in real time through quantum decoherence. Proc. Natl. Acad. Sci. USA. 2010;107:18777–18782. doi: 10.1073/pnas.1002562107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayda S., Kandemir Z., Bulut N., Maekawa S. Magnetic mechanism for the biological functioning of hemoglobin. Sci. Rep. 2020;10:8569. doi: 10.1038/s41598-020-64364-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., Law J., Luo M., Gong Z., Yu J., Tang W., Zhang Z., Mei X., Huang Z., You L., Sun Y. Magnetic Measurement and Stimulation of Cellular and Intracellular Structures. ACS Nano. 2020;14:3805–3821. doi: 10.1021/acsnano.0c00959. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J., He X., Chen X., Wu Y., Dong L., Cheng K., Lin J., Wang H., Weng W. Enhancing osteogenic differentiation of BMSCs on high magnetoelectric response films. Mater. Sci. Eng. C Mater. Biol. Appl. 2020;113 doi: 10.1016/j.msec.2020.110970. [DOI] [PubMed] [Google Scholar]
- 17.Hammer B.E., Kidder L.S., Williams P.C., Xu W.W. Magnetic Levitation of MC3T3 Osteoblast Cells as a Ground-Based Simulation of Microgravity. Microgravity Sci. Technol. 2009;21:311–318. doi: 10.1007/s12217-008-9092-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao Q., Zhang L., Han X., Chen H., Ji X., Zhang X. Magnetic Susceptibility Difference-Induced Nucleus Positioning in Gradient Ultrahigh Magnetic Field. Biophys. J. 2020;118:578–585. doi: 10.1016/j.bpj.2019.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian A.R., Yin D.C., Yang P.F., Jia B., Zhang W., Shang P. Development of a Ground-Based Simulated Experimental Platform for Gravitational Biology. IEEE Trans. Appl. Supercond. 2009;19:42–46. doi: 10.1109/TASC.2009.2012422. [DOI] [Google Scholar]
- 20.Hirose R., Hayashi S., Watanabe Y., Yokota Y., Takeda M., Kurahashi H., Kosaka K., Shibutani K. Development of a superconducting magnet for high magnetic force field application. IEEE Trans. Appl. Supercond. 2007;17:2299–2302. doi: 10.1109/Tasc.2007.898496. [DOI] [Google Scholar]
- 21.Tian X., Wang D., Feng S., Zhang L., Ji X., Wang Z., Lu Q., Xi C., Pi L., Zhang X. Effects of 3.5-23.0T static magnetic fields on mice: A safety study. Neuroimage. 2019;199:273–280. doi: 10.1016/j.neuroimage.2019.05.070. [DOI] [PubMed] [Google Scholar]
- 22.Parfenov V.A., Mironov V.A., van Kampen K.A., Karalkin P.A., Koudan E.V., Pereira F.D., Petrov S.V., Nezhurina E.K., Petrov O.F., Myasnikov M.I., et al. Scaffold-free and label-free biofabrication technology using levitational assembly in a high magnetic field. Biofabrication. 2020;12 doi: 10.1088/1758-5090/ab7554. [DOI] [PubMed] [Google Scholar]
- 23.Schenck J.F. The role of magnetic susceptibility in magnetic resonance imaging: MRI magnetic compatibility of the first and second kinds. Med. Phys. 1996;23:815–850. doi: 10.1118/1.597854. [DOI] [PubMed] [Google Scholar]
- 24.Duyn J.H. Studying brain microstructure with magnetic susceptibility contrast at high-field. Neuroimage. 2018;168:152–161. doi: 10.1016/j.neuroimage.2017.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaugnon E., Tournier R. Levitation of Organic Materials. Nature. 1991;349:470. doi: 10.1038/349470a0. [DOI] [Google Scholar]
- 26.Catherall A.T., Eaves L., King P.J., Booth S.R. Magnetic levitation: Floating gold in cryogenic oxygen. Nature. 2003;422:579. doi: 10.1038/422579a. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y., Yin D.C., Liu Y.M., Shi J.Z., Lu H.M., Shi Z.H., Qian A.R., Shang P. Design of shared instruments to utilize simulated gravities generated by a large-gradient, high-field superconducting magnet. Rev. Sci. Instrum. 2011;82 doi: 10.1063/1.3557402. [DOI] [PubMed] [Google Scholar]
- 28.Guevorkian K., Valles J.M., Jr. Swimming Paramecium in magnetically simulated enhanced, reduced, and inverted gravity environments. Proc. Natl. Acad. Sci. USA. 2006;103:13051–13056. doi: 10.1073/pnas.0601839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bras W., Torbet J., Diakun G.P., Rikken G.L.J.A., Diaz J.F. The diamagnetic susceptibility of the tubulin dimer. J. Biophys. 2014;2014 doi: 10.1155/2014/985082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L., Wang J., Wang H., Wang W., Li Z., Liu J., Yang X., Ji X., Luo Y., Hu C., et al. Moderate and strong static magnetic fields directly affect EGFR kinase domain orientation to inhibit cancer cell proliferation. Oncotarget. 2016;7:41527–41539. doi: 10.18632/oncotarget.9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robling A.G., Bonewald L.F. The Osteocyte: New Insights. Annu. Rev. Physiol. 2020;82:485–506. doi: 10.1146/annurev-physiol-021119-034332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riquelme M.A., Cardenas E.R., Xu H., Jiang J.X. The Role of Connexin Channels in the Response of Mechanical Loading and Unloading of Bone. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21031146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin L., Liu W., Cao H., Xiao G. Molecular mechanosensors in osteocytes. Bone Res. 2020;8:23. doi: 10.1038/s41413-020-0099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh A.V., Romeo A., Scott K., Wagener S., Leibrock L., Laux P., Luch A., Kerkar P., Balakrishnan S., Dakua S.P., Park B.W. Emerging Technologies for In Vitro Inhalation Toxicology. Adv. Healthc. Mater. 2021;10 doi: 10.1002/adhm.202100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh A.V., Chandrasekar V., Laux P., Luch A., Dakua S.P., Zamboni P., Shelar A., Yang Y., Pandit V., Tisato V., Gemmati D. Micropatterned Neurovascular Interface to Mimic the Blood-Brain Barrier's Neurophysiology and Micromechanical Function: A BBB-on-CHIP Model. Cells. 2022;11 doi: 10.3390/cells11182801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gauthier N.C., Roca-Cusachs P. Mechanosensing at integrin-mediated cell-matrix adhesions: from molecular to integrated mechanisms. Curr. Opin. Cell Biol. 2018;50:20–26. doi: 10.1016/j.ceb.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 37.Huang D.L., Bax N.A., Buckley C.D., Weis W.I., Dunn A.R. Vinculin forms a directionally asymmetric catch bond with F-actin. Science. 2017;357:703–706. doi: 10.1126/science.aan2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puklin-Faucher E., Sheetz M.P. The mechanical integrin cycle. J. Cell Sci. 2009;122:179–186. doi: 10.1242/jcs.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isomursu A., Lerche M., Taskinen M.E., Ivaska J., Peuhu E. Integrin signaling and mechanotransduction in regulation of somatic stem cells. Exp. Cell Res. 2019;378:217–225. doi: 10.1016/j.yexcr.2019.01.027. [DOI] [PubMed] [Google Scholar]
- 40.Uda Y., Azab E., Sun N., Shi C., Pajevic P.D. Osteocyte Mechanobiology. Curr. Osteoporos. Rep. 2017;15:318–325. doi: 10.1007/s11914-017-0373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson E., Lara-Castillo N., Akhter M.P., Dallas M., Scott J.M., Ganesh T., Johnson M.L. Osteocyte Wnt/beta-catenin pathway activation upon mechanical loading is altered in ovariectomized mice. BoneKEy Rep. 2021;15 doi: 10.1016/j.bonr.2021.101129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hauslage J., Cevik V., Hemmersbach R. Pyrocystis noctiluca represents an excellent bioassay for shear forces induced in ground-based microgravity simulators (clinostat and random positioning machine) NPJ Microgravity. 2017;3:12. doi: 10.1038/s41526-017-0016-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mellor L.F., Steward A.J., Nordberg R.C., Taylor M.A., Loboa E.G. Comparison of Simulated Microgravity and Hydrostatic Pressure for Chondrogenesis of hASC. Aerosp. Med. Hum. Perform. 2017;88:377–384. doi: 10.3357/AMHP.4743.2017. [DOI] [PubMed] [Google Scholar]
- 44.Manokawinchoke J., Pavasant P., Limjeerajarus C.N., Limjeerajarus N., Osathanon T., Egusa H. Mechanical loading and the control of stem cell behavior. Arch. Oral Biol. 2021;125 doi: 10.1016/j.archoralbio.2021.105092. [DOI] [PubMed] [Google Scholar]
- 45.Brunt L.H., Begg K., Kague E., Cross S., Hammond C.L. Wnt signalling controls the response to mechanical loading during zebrafish joint development. Development. 2017;144:2798–2809. doi: 10.1242/dev.153528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delgado-Calle J., Bellido T. The osteocyte as a signaling cell. Physiol. Rev. 2022;102:379–410. doi: 10.1152/physrev.00043.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dallas S.L., Prideaux M., Bonewald L.F. The osteocyte: an endocrine cell . and more. Endocr. Rev. 2013;34:658–690. doi: 10.1210/er.2012-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qian A.R., Hu L.F., Gao X., Zhang W., Di S.M., Tian Z.C., Yang P.F., Yin D.C., Weng Y.Y., Shang P. Large gradient high magnetic field affects the association of MACF1 with actin and microtubule cytoskeleton. Bioelectromagnetics. 2009;30:545–555. doi: 10.1002/bem.20511. [DOI] [PubMed] [Google Scholar]
- 49.Humphrey J.D., Dufresne E.R., Schwartz M.A. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 2014;15:802–812. doi: 10.1038/nrm3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grünberg J.R., Hammarstedt A., Hedjazifar S., Smith U. The Novel Secreted Adipokine WNT1-inducible Signaling Pathway Protein 2 (WISP2) Is a Mesenchymal Cell Activator of Canonical WNT. J. Biol. Chem. 2014;289:6899–6907. doi: 10.1074/jbc.M113.511964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirose R., Saito K., Watanabe Y., Tanimoto Y. Development of a superconducting magnet for high magnetic force application. IEEE Trans. Appl. Supercond. 2004;14:1693–1695. doi: 10.1109/Tasc.2004.831037. [DOI] [Google Scholar]
- 52.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garber M., Grabherr M.G., Guttman M., Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods. 2011;8:469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- 54.Maharjan R.S., Singh A.V., Hanif J., Rosenkranz D., Haidar R., Shelar A., Singh S.P., Dey A., Patil R., Zamboni P., et al. Investigation of the Associations between a Nanomaterial's Microrheology and Toxicology. ACS Omega. 2022;7:13985–13997. doi: 10.1021/acsomega.2c00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang P., Tang C., Wu J., Yang Y., Yan Z., Liu X., Shao X., Zhai M., Gao J., Liang S., et al. Pulsed electromagnetic fields regulate osteocyte apoptosis, RANKL/OPG expression, and its control of osteoclastogenesis depending on the presence of primary cilia. J. Cell. Physiol. 2019;234:10588–10601. doi: 10.1002/jcp.27734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
-
•
All standardized datasets have been deposited within the NCBI Sequence Read Archive (SRA): BioProject ID PRJNA932050, and are publicly available as of the data of publication.
-
•
This study does not report original code.
-
•
Any additional information required to reanalyze the data reported in this study is available from the lead contact upon request.