Abstract
Background
The spread of extended-spectrum β-lactamases (ESBL) producing E. coli from food animals and the environment to humans has become a significant public health concern. The objectives of this study were to determine the occurrence, pathotypes, virulotypes, genotypes, and antimicrobial resistance patterns of ESBL-producing E. coli in retail meat samples and workers in retail meat shops in Egypt and to evaluate the bactericidal efficacy of silver nanoparticles (AgNPs-H2O2) against multidrug resistant (MDR) ESBL-producing E. coli.
Results
A total of 250 retail meat samples and 100 human worker samples (hand swabs and stool) were examined for the presence of ESBL- producing E. coli. Duck meat and workers’ hand swabs were the highest proportion of ESBL- producing E. coli isolates (81.1%), followed by camel meat (61.5%). Pathotyping revealed that the isolates belonged to groups A and B1. Virulotyping showed that the most prevalent virulence gene was Shiga toxin 2 (stx2) associated gene (36.9%), while none of the isolates harbored stx1 gene. Genotyping of the identified isolates from human and meat sources by REP-PCR showed 100% similarity within the same cluster between human and meat isolates. All isolates were classified as MDR with an average multiple antibiotic resistance (MAR) index of 0.7. AgNPs-H2O2 at concentrations of 0.625, 1.25, 2.5 and 5 μg/mL showed complete bacterial growth inhibition.
Conclusions
Virulent MDR ESBL-producing E. coli were identified in retail meat products in Egypt, posing significant public health threats. Regular monitoring of ESBL-producing E. coli frequency and antimicrobial resistance profile in retail meat products is crucial to enhance their safety. AgNPs-H2O2 is a promising alternative for treating MDR ESBL-producing E. coli infections and reducing antimicrobial resistance risks.
Keywords: Retail meat, ESBL, E. coli, Antimicrobial resistance, Multidrug resistant, Silver nanoparticles
Background
For several decades, β-lactam antibiotics have been considered the antimicrobial agents of choice in humans and veterinary medicine. However, the effectiveness of these drugs has been diminished due to the emergence of β-lactamase-producing bacteria, particularly within the Enterobacterales [1]. Recently, as a result of the increased production of extended-spectrum β-lactamases (ESBL) a rise in β-lactam resistance in E. coli has increased [2]. ESBL-producing E. coli or their resistance genes can potentially be transmitted through direct contact, the food chain, or environmental sources [3]. The emergence of ESBL-producing bacteria linked to cattle, poultry, and pigs may be linked to the gradual increase in the usage of third-generation cephalosporins in food animal production [4, 6].
The genome of E. coli consists of a mobile gene core that determines the strain pathotypes. Various virulence factors determinants have been attributed to E. coli pathogenicity, including Shiga toxin-associated genes (stx1 and stx2), toxin production genes such as hemolysin (hly) and the astA gene encoding enteroaggregative E. coli heat-stable enterotoxin, intimin encoding gene (eae), and fimbrial H gene (fimH). E. coli are classified into eight phylotypes (A, B1, B2, C, D, E, F, clade 1) based on the presence or absence of ChuA, yjaA, TspE4.C2, and arpA genes [5, 6]. Phylotypes B2 and D are frequently found in humans, with phylotype B2 being associated with extraintestinal disease in both animals and humans [7]. Different properties of isolates are indicated by their typing, which is helpful for identifying the circulation pattern across various sources and hosts [8]. The presence of similarities between clinical and foodborne ESBL isolates suggests that food products can serve as a reservoir for ESBL-producing bacteria and their genes [9, 10, 13].
ESBL enzymes hydrolyze broad-spectrum cephalosporins, including ceftazidime, cefotaxime, cefuroxime, ceftriaxone and cefepime. Most ESBLs are classified as Ambler class A enzymes, which includes the sulfhydryl variable (SHV), Temoneria (TEM) and cefotaxime (CTX-M) types [11]. Unlike other bacterial families, Enterobacterales encode SHV, TEM and CTX-M genes on plasmids rather than the chromosome [12], ESBL-producing E. coli is becoming more resistant to fluoroquinolones and aminoglycosides, leading to the evaluation of colistin as an important alternative antimicrobial for human therapy [13, 17].However, colistin has been extensively used in veterinary medicine for years to prevent and treat gastrointestinal infections caused by Enterobacterales in food producing animals [14]. The mcr-1 gene, responsible for mobile colistin resistance, has been detected in Enterobacterales isolates from humans, food, companion animals, meat, and the environment in various studies [15].
Cephalosporins are widely used to treat various infections caused by both Gram-negative and Gram-positive bacteria [16]. They possess a broader activity range and are less susceptible to inactivation by β-lactamase enzymes compared to other β-lactam antibiotics, with successive generations of cephalosporins having a wider spectrum of activity [17]. The World Health Organization has prioritized third-generation cephalosporins (such as cefotaxime and ceftriaxone) in monitoring and stewardship programs for antibiotic resistance due to their significance to human health [18, 23].ESBLs, which hydrolyze third-generation cephalosporins, can transmit acquired resistance to other bacterial populations, leading to the spread of antibiotic resistance [12, 19].
In the past, the most common ESBL genes found in K. pneumoniae were the allelic variants of TEM-1, TEM-2, and SHV-1 that are the traditional TEM- and SHV-types, exhibit enhanced activity against extended-spectrum cephalosporines, and are primarily found in the hospital environment. However, in recent times, ESBL genes have been more frequently identified in E. coli, with the majority belonging to the CTX-M- group [20, 21].
The increasing prevalence of infections caused by MDR bacteria, coupled with the rising problem of acquired resistance resulting from improper antibiotic use, highlights the urgent need to reduce antibiotic consumption by exploring alternative approaches [22]. Among the potential alternatives, nanoparticles have emerged as a promising alternative to antibiotics for controlling infectious agents. Silver nanoparticles (AgNPs) exhibit biocidal effects against various foodborne bacteria [23]. They can interact with the cell surface of Gram-negative bacteria, causing damage and structural changes that enhance bacterial permeability [24]. Therefore, this study aims to (i) determine the occurrence, pathotypes, virulotypes, genotypes, and antimicrobial resistance patterns of ESBL-producing E. coli in retail meat samples and workers in contact in Egypt, and (ii) evaluate the bactericidal efficacy of silver nanoparticles (AgNPs-H2O2) against MDR ESBL-producing E. coli.
Materials and methods
Sample collection
The study received approval from the Zagazig University Institutional Animal Care and Use Committee (ZU-IACUC) under approval number ZU-IACUC/2/F/214/2022. Animal procedures were conducted following the ARRIVE guidelines. A total of 250 meat samples (chicken breast, duck breast, turkey breast, beef meat, and camel meat) were collected from retail meat shops in Zagazig City, Sharkia Governorate, Egypt. Samples were collected aseptically in sterile plastic bags between January to March 2022. Additionally, a total of 100 hand and stool swabs (50, each) were collected from workers at the retail shops where meat samples were collected. Hand swabs were collected by rolling a sterile swab moistened in buffered peptone water over the palmar surface of the workers’ hand. The swab was then placed in the collection tube containing buffered peptone water. Stool samples were collected in clean sterile cups, and a swab was taken from each sample and inserted in a collection tube containing buffered peptone water. All samples and swabs were transported in an icebox at 4 °C to the laboratory and processed within 12–24 h.
E. coli isolation and identification
From each meat sample, 25 g were aseptically homogenized in 1:10 buffered peptone water and incubated for 24 h at 37 °C. Hand and stool swabs were also incubated in buffered peptone water. A loopful of incubated samples was then streaked onto Chromocult® Tryptone Bile X-glucuronide agar (Sigma Aldrich, Millipore, ISO 16,649) supplemented with 2 μg/mL of cefotaxime (CTX) and incubated for 24 h at 37 °C. Green blue colonies were picked and purified by streaking into Tryptone Soy Agar (TSA) (Merck, Darmstadt, Germany). The purified colonies were then identified by biochemical tests for indole, oxidase, catalase, Methlye-Red (MR), Voges-Proskauer (VP), citrate, urease, nitrate reduction and sugar fermentation.
To confirm the presence of E. coli, bacterial DNA from presumptive E. coli colonies was extracted using the QIAamp DNA Mini kit according to the manufacturer’s instructions (Qiagen GmbH, Hilden, Germany, Catalogue no. 51,304). Specific primers targeting the phoA gene (Table 1) were employed for amplification to confirm E. coli isolates [25].
Table 1.
Primers, product size and annealing temperatures used for identification, virulence and antimicrobial resistance genes reported in the present study
| Gene | Primer sequences (5’-3’) | Product size (bp) | Annealing (°C) | References |
|---|---|---|---|---|
| hlyA | fw: AACAAGGATAAGCACTGTTCTGGCT | 1177 | 60˚C | [28] |
| rev: ACCATATAAGCGGTCATTCCCGTCA | ||||
| phoA | fw: CGATTCTGGAAATGGCAAAAG | 720 bp | 60˚C | [25] |
| rev: CGTGATCAGCGGTGACTATGAC | ||||
| stx1 | fw: ACACTGGATGATCTCAGTGG | 614 | 58˚C | [30] |
| rev: CTGAATCCCCCTCCATTATG | ||||
| stx2 | fw: CCATGACAACGGACAGCAGTT | 779 | 58˚C | |
| rev: CCTGTCAACTGAGCAGCACTTTG | ||||
| astA | fw: CCATCAACACAGTATATCCGA | 110 | 55˚C | [29] |
| rev: GGTCGCGAGTGACGGCTTTGT | ||||
| stx2f | fw: AGA TTG GGC GTC ATT CAC TGG TTG | 428 | 57˚C | [31] |
| rev: TAC TTT AAT GGC CGC CCT GTC TCC | ||||
| fimH | fw: TGCAGAACGGATAAGCCGTGG | 508 | 50˚C | [32] |
| rev: GCAGTCACCTGCCCTCCGGTA | ||||
| eaeA | fw: ATG CTT AGT GCT GGT TTA GG | 248 | 51˚C | [33] |
| rev: GCC TTC ATC ATT TCG CTT TC | ||||
| blaIMP | fw: CATGGTTTGGTGGTTCTTGT | 488 | 53˚C | [39] |
| rev: ATAATTTGGCGGACTTTGGC | ||||
| blaVIM | fw: AGTGGTGAGTATCCGACA | 280 | 53˚C | |
| rev: ATGAAAGTGCGTGGAGAC | ||||
| blaNDM | fw: GGCGGAATGGCTCATCACGA | 287 | 55˚C | |
| rev: CGCAACACAGCCTGACTTTC | ||||
| blaTEM | fw: ATCAGCAATAAACCAGC | 516 | 54˚C | [40] |
| rev: CCCCGAAGAACGTTTTC | ||||
| blaOXA-1 | fw: ATATCTCTACTGTTGCATCTCC | 619 | 54˚C | |
| rev: AAACCCTTCAAACCATCC | ||||
| blaSHV | fw: AGGATTGACTGCCTTTTTG | 392 | 54˚C | |
| rev: ATTTGCTGATTTCGCTCG | ||||
| blaCMY | fw: GACAGCCTCTTTCTCCACA | 1143 | 60˚C | [41] |
| rev: TGGAACGAAGGCTACGTA | ||||
| blaCTX-M-1 | fw: GTTACAATGTGTGAGAAGCAG | 1041 | 60˚C | [42] |
| rev: CCGTTTCCGCTATTACAAAC | ||||
| tetA(A) | fw: GGTTCACTCGAACGACGTCA | 576 | 50˚C | [43] |
| rev: CTGTCCGACAAGTTGCATGA | ||||
| tetA(B) | fw: CCTCAGCTTCTCAACGCGTG | 633 | 50˚C | |
| rev: GCACCTTGCTCATGACTCTT | ||||
| sul | fw: CGGCGTGGGCTACCTGAACG | 60˚C | [44] | |
| rev: GCCGATCGCGTGAAGTTCCG | ||||
| cmlA | fw: CCGCCACGGTGTTGTTGTTATC | 698 | 50˚C | [45] |
| rev: CACCTTGCCTGCCCATCATTAG | ||||
| floR | fw: TTTGGWCCGCTMTCRGAC | 494 | 50˚C | [46] |
| rev: SGAGAARAAGACGAAGAAG | ||||
| mcr-1 | fw: AGTCCGTTTGTTCTTGTGGC | 320 | 58 °C | [47] |
| rev: AGATCCTTGGTCTCGGCTTG | ||||
| mcr-5 | fw: ATGCGGTTGTCTGCATTTATC | 1644 | 58 °C | |
| rev: TCATTGTGGTTGTCCTTTTCTG | ||||
| chuA | fw: GAC GAA CCA ACG GTC AGG AT | 279 | 55˚C | [27] |
| rev: TGC CGC CAG TAC CAA AGA CA | ||||
| yjaA | fw: TGA AGT GTC AGG AGA YGC TG | 211 | ||
| rev: ATG RAG AAT GCG TTC CTC AAC | ||||
| tspE4C2 | fw: GAG TAA TGT CGG GGC ATT CA | 152 | ||
| rev: CGC GYC AAC AAA GTA TTR CG |
Characterization of ESBL-producing E. coli
The Double Disc Synergy Test (DDST) was used to verify the confirmed E. coli strains for phenotypic ESBL expression [26].
ESBL-producing E. coli phylotyping, virulotyping and genotyping
The confirmed strains were phylotyped using primers for the amplification of chuA, yjaA, and, tspE4C2 genes (Table 1), according to the phylotype classification scheme previously described [27].
Virulotyping was also performed using specific primers for amplification of the virulence associated genes (Table 1), including hly [28],, astA [29], stx1 and stx2 [30], stx2f [31],, fimH [32], eaeA [33].
Genotyping was performed on extracted DNA through fingerprinting PCR using REP-primers synthesized by Metabion (Germany) with the following sequences: Rep1R-I 5’- III ICG ICG ICA TCI GGC-3’ and Rep2-I 5’- ICG ICT TAT CIG GCC TAC-3’ [34]. The primers were included in a 25- μL reaction mixture containing 12.5 μL of EmeraldAmp Max PCR Master Mix (Takara, Japan), 1 μL of each primer ( 20 pmol), 4.5 μL of water, and 6 μL of DNA template. The PCR procedure was carried out using an Applied Biosystem 2720 thermal cycler.
To assess the discriminatory power of the REP-PCR fingerprinting data, the Simpson’s index of diversity (D) was utilized. The fingerprinting data was converted into a binary code, indicating the presence or absence of each band. A D value greater than 0.9 indicated good differentiation [35].
Antimicrobial susceptibility testing
The Kirby-Bauer disc diffusion method was used to evaluate the isolates’ antibiotic susceptibility in accordance with the standards established by the National Committee for Clinical Laboratory Standards (NCCLS). Nineteen different antimicrobial agents were tested, and the zones of inhibition were measured and interpreted based on the guidelines provided by the Clinical and Laboratory Standards Institute (CLSI) [26]. In accordance with CLSI recommendations, the double fold dilution procedure (0.125-256 g/mL) was used to establish the minimum inhibitory concentration (MIC) for colistin [36]. The antimicrobial agents used included penicillin (PEN), ampicillin (AMP), amoxicillin (AMX), streptomycin (STR), erythromycin (ERY), nalidixic acid (NAL), amikacin (AMK), trimethoprim-sulfamethoxazole (SXT), kanamycin (KAN), neomycin (NEO), gentamicin (GEN), ciprofloxacin (CIP), tetracycline (TET), colistin (CST), imipenem (IPM), chloramphenicol (CHL), cefotaxime (CTX), ceftriaxone (CRO), ceftazidime (CAZ). E. coli ATCC 25,922 and Staphylococcus aureus ATCC 25,923 served as the microorganisms for quality control.
Multiple antibiotic resistance (MAR) index was calculated by dividing the number of antibiotics to which E. coli isolates showed resistance by the total number of drugs tested [37]. Multidrug resistance (MDR) was defined as the resistance of an isolate to at least one agent in three or more antibiotic classes [38].
Antimicrobial resistance genes
Bacterial DNA from the E. coli confirmed isolates was also screened for ESBL encoding genes (Table 1); blaIMP, blaVIM, and blaNDM [39], blaTEM, blaOXA-1, and blaSHV [40], blaCMY [41], blaCTX-M-1 [42], tetracycline; tetA(A) and tetA(B) [43], sulfonamides; sul [44], chloramphenicol; cmlA [45], and florquinolones; floR [46]. The presence of colistin resistance genes mcr-1 to mcr-5 was also examined [47].
Antimicrobial effect of silver nanoparticles on MDR ESBL-producing E. coli
AgNPs-H2O2 (Top Superpower-vision) was obtained as a commercial product from El-Delta Center for Nanosilver Technology Company, Mansoura, Egypt. The stock solution of the product contained 45-nm silver nanoparticles (0.00004467 mL/liter), hydrogen peroxide (50% per liter) and natural herb mint (1 mL/liter) at a concentration of 5 mL/liter of water. The particles size was previously determined to be 30.17–67.92 nm with a zeta potential estimation of − 0.192 mV [48]. The AgNPs-H2O2 mixture was prepared by diluting the stock solution in sterile distilled water to achieve the desired commercial concentration. The minimum inhibitory concentrations (MIC50 and MIC90) of AgNPs-H2O2 were determined against MDR ESBL-E. coli by the broth microdilution method [36]. Briefly, microtiter plate wells were supplemented with various concentrations of AgNPs-H2O2 ranging from 100, 50, 25, 10, 5, 2.5 1.25, 0.625, 0.312, 0.156 and 0.078 μg/mL. MDR- ESBL-producing colonies were added in Muller Hinton broth and adjusted to the density of a 0.5 McFarland standard (1 × 108 cfu/ml). Each well received a final inoculum of 5 × 105 cfu/mL, and the plates were incubated for 24 h at 37 °C. A well with MHB alone and another well included MHB with AgNPs-H2O2 were used as reference control. The lowest agent concentration that entirely prevents an organism’s observable growth is known as the MIC endpoint. The MIC50 and MIC90 were calculated using an orderly array method [49], where the middle value was selected as MIC50. The MIC90 was determined in the same way by selecting the appropriate value from the orderly array.
Data analysis
For statistical analysis and data visualization, R software was used (R Core Team, 2022; version 4.2.0). The E. coli isolation rate was calculated by dividing the number of E. coli positive samples by the total number of samples tested for each source. Pearson’s chi-square test was employed to assess any variations in the E. coli isolation rates among different sample types. The heatmap was created using the “Complex heatmap” package [50], and the dendrogram was created using the “hclust” function of the stats package. Furthermore, one-way analysis of variance was conducted to compare the MAR index of isolates from different sources. P-values < 0.05 were considered statistically significant.
Results
E. coli isolation and identification
Out of the 350 samples tested, 112 (32%) were E. coli positive with 68.75% (77/112) were isolated from retail meat samples and the remaining 31.25% (35/112) were recovered from retail market workers. E. coli was identified in various types of retail meat and market workers samples. The E. coli isolation rates exhibited significant variation (P = 0.0003) among the different sample sources. The highest isolation rate (54%) was observed in beef meat samples followed by workers stool samples (48%) (Table 2).
Table 2.
Proportion of E. coli and ESBL-producing E. coli isolated from retail meat samples and retail shop workers
| Source | Sampling site | Number examined | No. of E. coli positive (%) | No. of ESBL positive E. coli (%) | Phylogenetic group | |
|---|---|---|---|---|---|---|
| A | B1 | |||||
| Chickens | Breast meat | 50 | 17 (34%) | 10 (58.8%) | 0 | 10 (100%) |
| Ducks | Breast meat | 50 | 11 (22%) | 9 (81.8%) | 0 | 9 (100%) |
| Turkey | Breast meat | 50 | 9 (18%) | 5 (55.6%) | 0 | 5 (100%) |
| Beef | Cube meat | 50 | 27 (54%) | 13 (48.1%) | 0 | 13 (100%) |
| Camel | Cube meat | 50 | 13 (26%) | 8 (61.5%) | 0 | 8 (100%) |
| Workers | Hand swabs | 50 | 11 (22%) | 9 (81.8%) | 0 | 9 (100%) |
| Stool swabs | 50 | 24 (48%) | 11 (45.8%) | 4 (36.4%) | 7 (63.6%) | |
| Total | 350 | 112 (32%) | 65 (58%) | 4 (6.2%) | 61 (93.8%) | |
ESBL-producing E. coli
Overall, 58% (65/112) of E. coli isolates were ESBL producers, with 45 (69.2%) and 20 (30.8%) isolates recovered from retail meat and market workers samples, respectively (Table 2). The highest proportion of ESBL-producing E. coli was observed in duck meat and worker hand swabs (81.8%), followed by camel meat samples (61.5%).
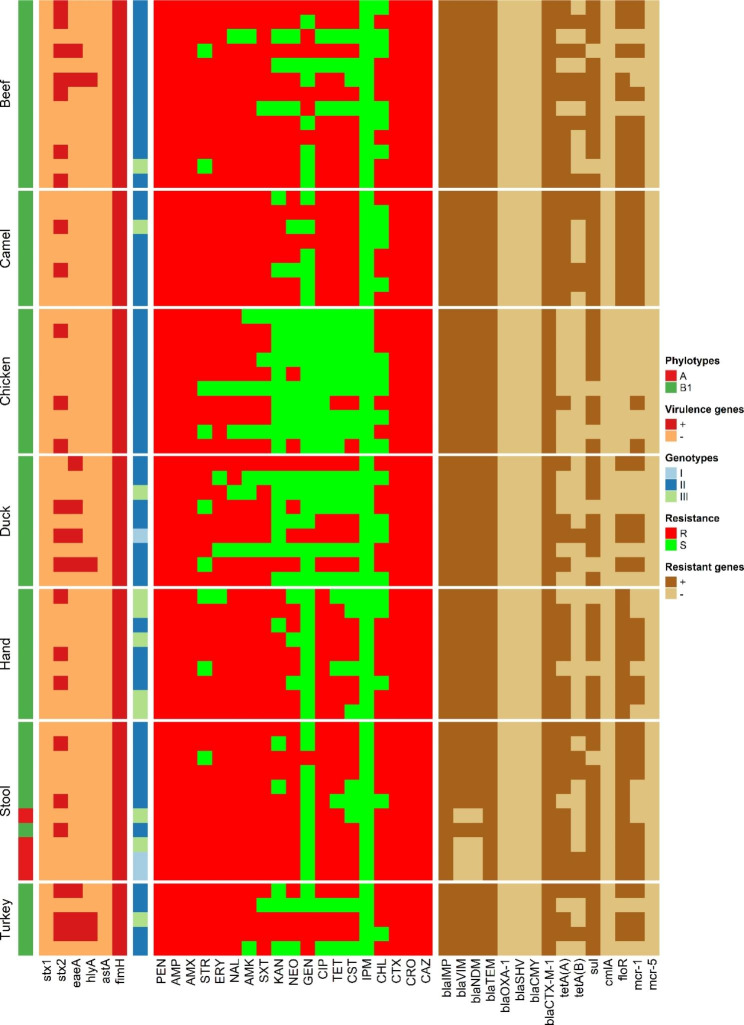
Phylotyping, virulotyping and genotyping
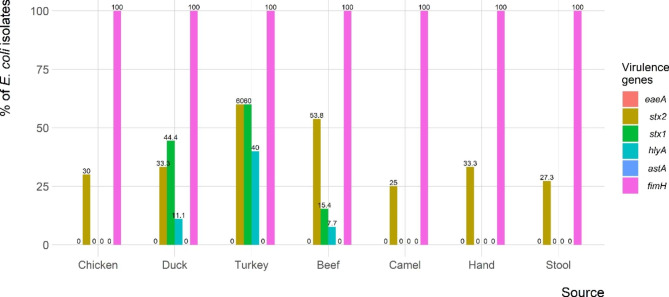
The phylogenetic grouping of the 65 ESBL-producing E. coli showed that 4 (6.2%) isolates belonged to group A and 61 (93.8%) to group B1 (Table 2). All group A isolates were recovered from workers stool samples (Fig. 1). None of the isolates belonged to group B2 or D. All the ESBL-producing E. coli isolates (100%) harbored at least one virulence gene, but only 25 (38.5%) of the isolates harbored two or more virulence genes (Fig. 2). The stx2 gene was amplified in 24 (36.9%) of the isolates, followed by eaeA and hlyA genes in 9 (13.8%) and 4 (6.2%) isolates, respectively. The stx2f gene was identified in only four isolates that tested positive for stx2 gene (one each from chicken, turkey, duck, and beef meat samples). None of the isolates carried the astA or stx1genes, however, all of them tested positive for the fimH gene (Fig. 2).
Fig. 1.
Heatmap representation of ESBL-producing E. coli phylotypes, virulotypes, genotypes, antimicrobial resistance patterns and genes
Fig. 2.
Frequency of virulence genes of ESBL-producing E. coli isolated from retail meat samples and retail shop workers
A single amplification profile was used to analyze the REP-PCR patterns of the 65 ESBL-producing E. coli isolates. The profiles were distinguished based on the number and position of the amplified fragments, which ranged in size from 290 to 1600 bp. Visual examination of the banding patterns revealed that five profiles (E1 to E5) were generated (Fig. 3). Simpson’s index of diversity was used to evaluate the discriminatory power of the REP-PCR, and the results showed that it had relatively low discriminatory power with a D value of 0.42. There were three main clusters revealed by the dendrogram analysis of the 65 analyzed isolates. (Figures 1 and 3). Notably, isolates from workers and meat within the same cluster exhibited 100% similarity.
Fig. 3.
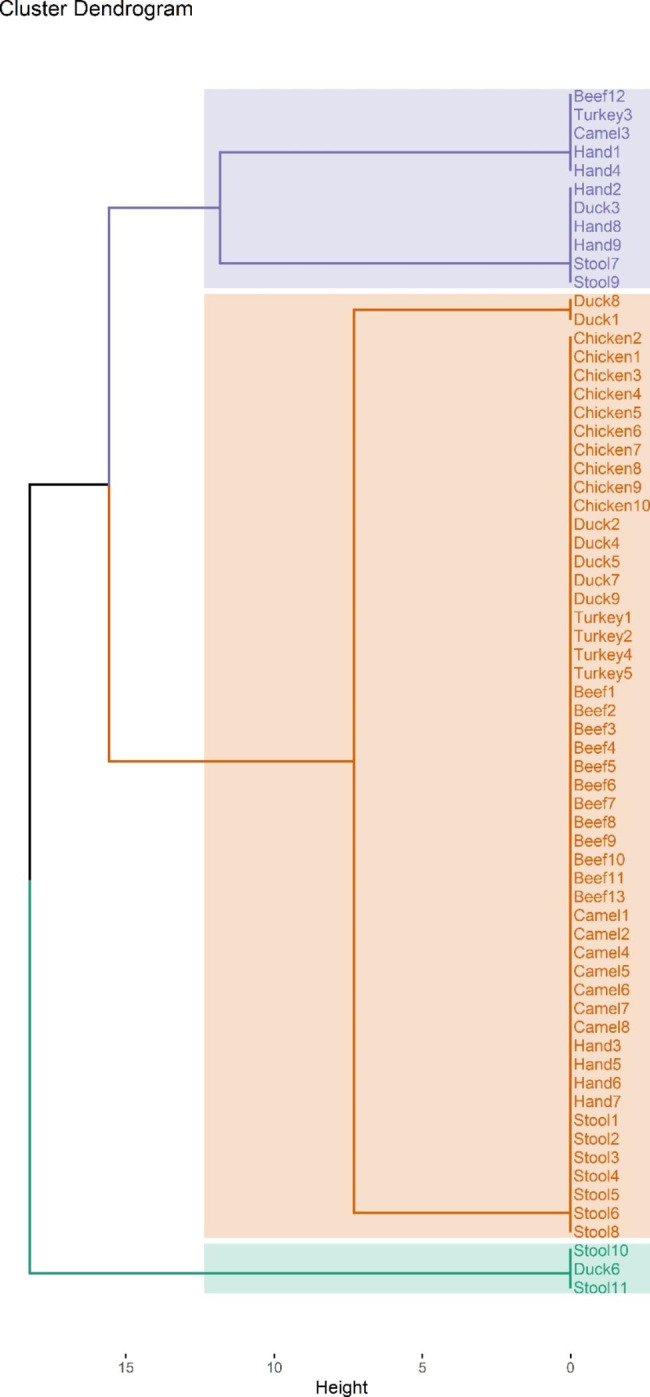
REP-PCR based dendrogram for ESBL-producing E. coli isolated from retail meat samples and retail shop workers
Antimicrobial susceptibility testing
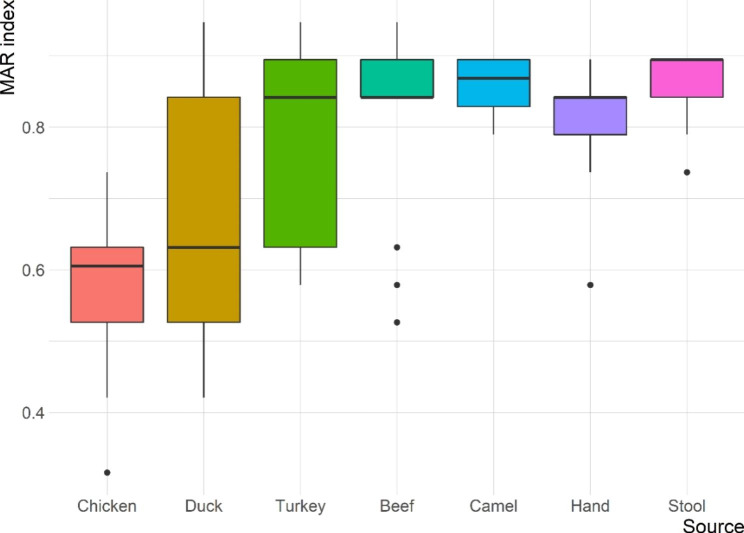
Table 3 presents the antimicrobial susceptibility profiles of the 65 ESBL-producing E. coli isolates. The isolates showed high frequencies of resistance to PEN, AMP, AMX, CTX, CAZ and CRO (100%, each), while the lowest resistance rate (21.5%) was observed for GEN. All isolates were susceptible to IMP. Figure 4 displays the frequency of antimicrobial resistance by source of isolation. However, no significant difference (P > 0.05) was found between resistance rates of ESBL-producing E. coli isolated from retail meat and market workers. All isolates were classified as MDR with MAR index ranging from 0.32 to 0.95 and an average of 0.70 (Fig. 5). The average MAR index of isolates recovered from various sources differed significantly, with workers’ stool having the highest MAR index (0.86) followed by camel meat (0.85).
Table 3.
Frequency of antimicrobial resistance profiles of ESBL-producing E. coli isolated from retail meat samples and retail shop workers
| Antimicrobial resistance profiles1 | Frequency of E. coli isolates from each source | Total | ||||||
|---|---|---|---|---|---|---|---|---|
| Chicken | Duck | Turkey | Beef | Camel | Hand | Stool | ||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, KAN, NEO, CIP, TET, CST, CHL, CTX, CRO, CAZ | 2 | 2 | 2 | 6 | 12 | |||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, KAN, NEO, GEN, CIP, TET, CST, CHL, CTX, CRO, CAZ | 1 | 1 | 2 | 4 | ||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, KAN, NEO, GEN, CIP, TET, CST, CTX, CRO, CAZ | 1 | 1 | 2 | 4 | ||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, NEO, CIP, TET, CST, CHL, CTX, CRO, CAZ | 1 | 1 | 1 | 1 | 4 | |||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, KAN, NEO, CIP, TET, CST, CTX, CRO, CAZ | 2 | 1 | 3 | |||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, KAN, NEO, CIP, TET, CHL, CTX, CRO, CAZ | 1 | 1 | 1 | 3 | ||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, CHL, CTX, CRO, CAZ | 2 | 1 | 3 | |||||
| PEN, AMP, AMX, ERY, NAL, AMK, SXT, KAN, NEO, CIP, TET, CST, CHL, CTX, CRO, CAZ | 1 | 1 | 2 | |||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, KAN, CIP, TET, CST, CTX, CRO, CAZ | 1 | 1 | 2 | |||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, CTX, CRO, CAZ | 1 | 1 | 2 | |||||
| PEN, AMP, AMX, ERY, NAL, AMK, SXT, KAN, NEO, GEN, CIP, TET, CST, CHL, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, ERY, NAL, AMK, SXT, KAN, NEO, GEN, CIP, TET, CST, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, KAN, CIP, TET, CST, CHL, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, NEO, GEN, CIP, TET, CST, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, CIP, TET, CST, CHL, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, KAN, NEO, CIP, TET, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, NEO, CIP, TET, CHL, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, ERY, NAL, AMK, SXT, KAN, NEO, CIP, CHL, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, CIP, TET, CST, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, KAN, NEO, CIP, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, TET, CST, CHL, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, NEO, CST, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, ERY, NAL, AMK, SXT, NEO, CHL, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, SXT, NEO, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, SXT, GEN, CHL, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, NAL, AMK, SXT, KAN, CIP, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, CHL, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, GEN, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, AMK, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, NAL, CHL, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, SXT, CHL, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, ERY, SXT, GEN, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, ERY, CHL, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, CHL, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, STR, NAL, CTX, CRO, CAZ | 1 | 1 | ||||||
| PEN, AMP, AMX, CTX, CRO, CAZ | 1 | 1 | ||||||
1 PEN, penicillin; AMP, ampicillin; AMX, amoxicillin; STR, streptomycin; AMK, amikacin; KAN, kanamycin; NEO, neomycin; GEN, gentamicin; SXT, trimethoprim-sulfamethoxazole; ERY, erythromycin; NAL, nalidixic acid; CIP, ciprofloxacin; TET, tetracycline; CST, colistin; IPM, imipenem; CHL, chloramphenicol; CTX, cefotaxime; CRO, ceftriaxone; CAZ, ceftazidime
Fig. 4.
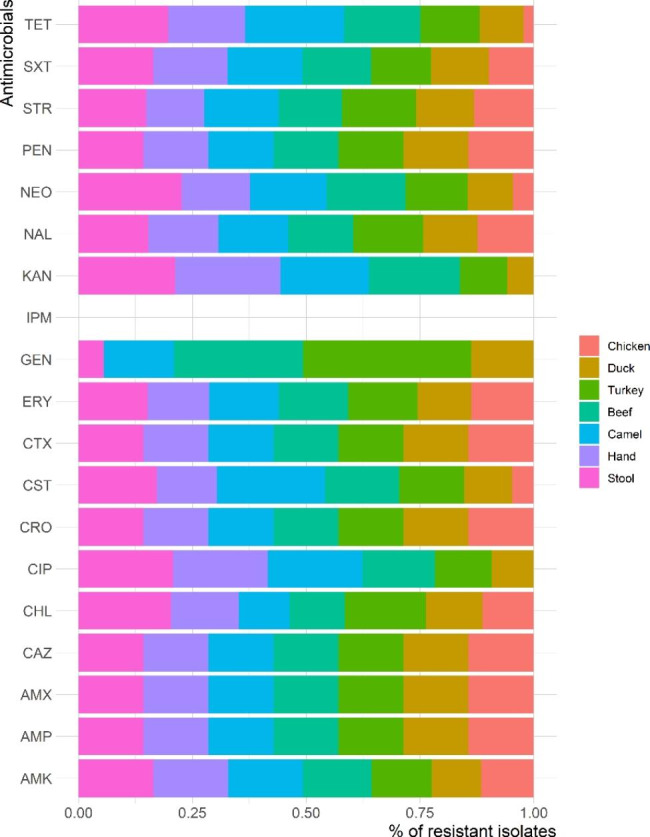
Frequency of antimicrobial resistance of ESBL-producing E. coli isolated from retail meat samples and retail shop workers
Fig. 5.
Multiple antibiotic resistance (MAR) index box plot of ESBL-producing E. coli isolated from retail meat samples and retail shop workers
Antimicrobial resistance genes
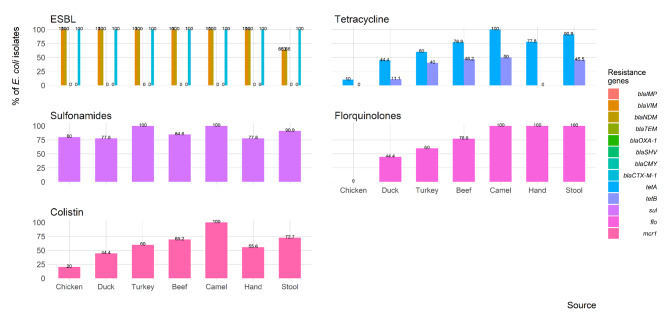
The frequency of antimicrobial resistance genes detected in ESBL-producing E. coli isolates obtained from retail meat and market workers is shown in Fig. 6. The ESBL encoding genes (blaIMP, blaTEM, and blaCTX-M-1) were found in 100% of the isolates, while blaVIM, and blaNDM were detected in 93.8% of the isolates. However, none of the isolates were positive for blaOXA-1, blaSHV, or blaCMY. Tetracycline resistance genes (tetA and tetB) were detected in 66.2% and 27.7% of the tested isolates, respectively The resistance genes for sulfonamides (sul), and florquinolones (floR) were identified in 86.2% and 69.2% of the isolates, respectively. Only mcr-1 was detected among the tested colistin resistance genes (mcr-1 and mcr-5), and it was found in 60% of the isolates.
Fig. 6.
Frequency of antimicrobial resistance genes of ESBL-producing E. coli isolated from retail meat samples and retail shop workers
Antimicrobial effect of silver nanoparticles
The antimicrobial activity of AgNPs-H2O2 against ESBL-producing E. coli was evaluated using the broth microdilution method. The MIC values of different concentrations of AgNPs-H2O2 against ESBL-producing E. coli isolated from retail meat and market workers are illustrated in Table 4. AgNPs-H2O2 concentrations of 0.625, 1.25, 2.5 and 5 μg/mL showed complete bacterial growth inhibition (no turbidity). The MIC50 and MIC90 were 0.625 and 2.5 μg/mL, respectively.
Table 4.
The distribution of minimum inhibition concentration (MIC) values of AgNPs concentrations against ESBL-producing E. coli isolated from retail meat samples and retail shop workers
| Source | No. of sensitive E. coli isolate at different AgNPs concentrations (μg/mL) | ||||||
|---|---|---|---|---|---|---|---|
| 5 | 2.5 | 1.25 | 0.625 | 0.312 | 0.156 | 0.078 | |
| Chickens | 2 | 2 | 0 | 6 | 0 | 0 | 0 |
| Ducks | 0 | 3 | 1 | 5 | 0 | 0 | 0 |
| Turkey | 0 | 2 | 0 | 3 | 0 | 0 | 0 |
| Beef | 3 | 1 | 0 | 9 | 0 | 0 | 0 |
| Camel | 0 | 3 | 1 | 4 | 0 | 0 | 0 |
| Worker’s hand | 0 | 1 | 2 | 6 | 0 | 0 | 0 |
| Worker’s stool | 0 | 0 | 2 | 9 | 0 | 0 | 0 |
| Total | 5 | 12 | 6 | 42 | 0 | 0 | 0 |
Discussion
Unhygienic food handling and processing procedures facilitate the dissemination of MDR bacteria, including ESBL-E. coli to human consumers. Thus, enforcing strict monitoring measures and promoting sanitary procedures for meat distribution are necessary to prevent the proliferation of antibiotic-resistant bacteria [4].
In this study, ESBL-E. coli prevalence in duck meat was higher compared to the other sources (81.8%), followed by camel meat (61.5%). Similar results were reported in other studies in China [51] and Thailand [52], where MDR E. coli isolates were more common in ducks than in chicken. The higher isolation rate of ESBL-E. coli from ducks could be attributed to their nature as waterfowl, which excretes feces in water, thus enhancing the spread of the pathogens within the duck population [52].
A high ESBL-producing E. coli isolation rate of 86.7 from chicken meat in Turkey was reported by Kürekci, et al. [53]. The authors attributed the high isolation rate from chicken meat due to the extensive use of fluoroquinolones, cephalosporines and aminoglycosides in poultry industry which resulted in selection pressure for the high carriage rate of ESBL-producing E. coli in chickens [54].
The prevalence of ESBL-producing E. coli in beef was lower than that in chicken, which is consistent with the findings of Randall, et al. [55] who found rates of 20% in beef samples compared to 63% in chicken samples. Rao, et al. [56], also reported that only 1.9% of beef samples were positive for ESBL-producing E. coli, whereas 65.4% of chicken samples tested positive. Meanwhile, the prevalence of ESBL-producing E. coli in beef samples in the current study (48.1%) was much higher than the average (7.1%) of ESBL/AmpC-producing E. coli found in beef samples purchased at retail in the EU [57].
Meat is highly susceptible to microbial contamination due to multiple contacts from the slaughterhouse until consumption as food [4]. The prevalence of ESBL-E. coli was found to be high in camel (61.5%) and beef meat (48.1%) samples, this is in line with another study [4]. Similarly, El-Ghareeb, et al. [58] reported the isolation of ESBL-producing E. coli from 11.3% camel minced meat samples in Saudi Arabia. The high water and protein content of red meat provide a favorable environment for bacterial growth and may contribute to the high contamination rates [59].
In the present study, ESBL-E. coli was found to be prevalent in workers hand swabs (81.8%) and stool samples (45.8%). Similar findings were reported in studies conducted in Thailand [52, 60]. In Ethiopia, E. coli was isolated from 20% of hand swabs, with ESBL-E. coli accounting for 25% of the isolates [4]. Variations in the hygienic procedures of meat handlers and the sanitary standards of meat retail shops may be the cause of the observed differences in E. coli prevalence. Inadequate hygienic procedures of meat handlers and insufficient sanitation standards in meat retailer stores may lead to cross-contamination of meat with E. coli [4].
E. coli can be classified into four main phylogenetic groups named A1, B1, B2, and D, which can be identified by PCR of four genes [6]. Generally, commensal strains belong to groups A1 and B1, while most virulent strains belong to groups B2 and D [6, 61]. In the present study, all the isolates were of the commensal groups A (6.2%) and B1 (93.8). Previous studies have also reported that the majority of the E. coli phylotypes from different sources were of the commensal groups A1 and B1 [61–63]. In Egypt, Abdallah, et al. [64] reported that 80% of ESBL-producing E. coli isolated from chicken meat in the same study area were also of the commensal groups. The high proportion of commensal strains highlight their critical silent role in the spread and dissemination of ESBL-resistance genes [61, 64].
Virulotyping of the isolates revealed that all had at least one virulence gene, with stx2 (36.9%) being the most frequently identified gene, and four isolates being positive for stx2f gene. The existence of genes related to virulence such as stx1, stx2, and eae have been found to be crucial factors for the pathogenicity of E. coli strains [65]. Stx2-encoding strains of Shiga toxin producing E. coli (STEC) have been linked to more severe infections than those only possessing stx1 [66]. In our study, stx2 was the most common virulence gene (36.9%), consistent with reports from Germany, Argentina, and China [67–69]. In line with our findings, a study conducted in Egypt reported that 36% of MDR E. coli isolates obtained from chicken meat carried the stx2 gene [70].
In China, 28.57% and 51.02% of E. coli isolates had stx1 and stx2 genes, respectively [69]. Although Shiga toxin is necessary for STEC pathogenicity, it is not sufficient. Therefore, we examined four additional virulence-associated genes: hly, ast, eae, and fimH, that are linked to bacterial virulence. The combination of the eae and stx genes has been associated with increased virulence [71]. None of the isolates in our investigation carried the astA or stx1genes, while Nong, et al. [69] found astA in 20.41% and Ali, et al. [70] detected stx1 gene in 4% of the E.coli strains from chicken meat. All tested isolates in our study were positive for fimH gene, which is responsible for adhesion (the first step in the colonization process). Similar results were reported in poultry meat from Brazil [53, 72]. This is expected because this gene has been reported from both clinical and commensal E. coli isolates from different sources [53, 57]. The eaeA and hlyA genes were found in 13.8% and 6.2% of our isolates, respectively. A lower proportion of eaeA and hlyA encoding genes (5 and 4%, respectively) among E. coli isolates was reported previously in Egypt by Ali, et al. [70]. However, a higher percentage of eae -encoding genes (34.69%) among the E. coli isolates was observed in China [69]. In contrast, none of ESBL-producing E. coli recovered from retail meat in Mexico were positive of stx1, stx2, hlyA, or eae, virulence genes [61].
Recent studies highlight the significance of comparing isolates from various sources to evaluate the relevance of the foodborne pathway in human infection [73]. This study utilized REP-PCR to investigate the genetic relatedness between the isolates from different sources. The isolates were grouped into five profiles in three clusters. The 100% similarity between human and meat isolates within the same cluster indicates genetic relatedness and the possibility of transmission of strains from meat to humans. This is supported by the isolation of the same ESBL-associated genes from human and meat isolates. A similar study conducted in the Netherlands also reported a partially close genetic relationship between strains obtained from human carriers and chicken meat samples [74]. A Japanese study also reported 80% similarity index between E. coli isolates harboring β-lactamase obtained from domestic and imported chicken meat samples [75]. Similarly, a study in Sweden revealed that less than 0.1% of the population carried ESBL-producing isolates associated with poultry, but 5% of individuals were colonized with ESBL-encoding plasmids that were identical to those found in chicken meat and poultry isolates [76]. Additionally, a study conducted in rural Ghana demonstrated genetic links between ESBL-producing E. coli, suggesting possible transmission between poultry and human populations [77].
However, Belmar Campos, et al. [21] reported that ESBL genes produced by E. coli from chicken meat are different from those found in human stool samples and their data do not support the notion that ESBL strains from chicken meat significantly contribute to human colonization. The authors attributed this discrepancy to the fact that the chicken meat samples were collected more than six months after the human feces samples were obtained. Similarly, other studies have reported no genetic relatedness between ESBL-producing isolates from food animals and humans in close contact [73, 76]. In a study conducted in the Netherlands, no evidence was found to support the hypothesis of clonal transmission of ESBL-producing E. coli isolates between humans and poultry [78].
All isolates tested in our study were found to be MDR, with an average MAR index ranging from 0.32 to 0.95. The isolates showed 100% resistance to penicillins, beta-lactams and cephems. The MDR isolates are considered reservoirs for both resistance and virulence genes and can be transferred to other strains of the same species and other species, thereby increasing the source of antibiotic resistance [61]. Similar results were reported by other studies [52, 55]. Abayneh, et al. [4] reported that the majority of E. coli isolates (74.3%) exhibited resistance to three or more classes of antibiotics, including TET, ERY and cotrimoxazole. Additionally, 85.7% of ESBL-producers were resistant to CTX and CRO, while 71.4% were resistant to CAZ. Another study reported high resistance of ESBL-E. coli to AMP (69.4%), SXT (66.7%), TET (88.9%) and Sulfonamide (75%) [79]. In China, ESBL-producing E. coli were found to be resistant to different antibiotics such as AMP (98.9%) and TET (97.6%) [80]. Another study in Egypt TET (80.9%), STR (61.9%) and SXT (61.9%) [81]. All our isolates were susceptible to imipenem, this is consistent with another study conducted in Turkey on chicken meat samples [53]. The difference between the resistance rates of ESBL-producing E. coli to antimicrobials could be attributed to antibiotic administration practices in the veterinary field as growth promoters or for therapy, also due to geographic distribution [3].
Colistin resistance has been observed in E. coli isolates from food-producing animals, especially poultry [82]. In this study, 60% of the isolates were resistant to colistin. The resistance to colistin is mediated by chromosomal mutations in pmrA/B, phoP/Q, and mgrB genes. However, an acquired colistin-resistance gene mcr-1 has also been identified in E. coli [83, 84]. To date, nine variants of mcr have been identified in humans and different animals [85]. The existence of mcr genes in mobile genetic elements raises concerns about their potential horizontal transfer in the food chain, posing a risk to public health [14]. Since 2006, the coexistence of mcr genes with other resistance determinants, such as ESBL and/or carbapenemase genes, has been reported in Enterobacterales. Notably, an increase in the prevalence of mcr-1 genes has been observed among ESBL-producing E. coli strains in animals, while their occurrence remains low in non-ESBL-producing E. coli strains. This suggests that the use of extended-spectrum cephalosporins may have contributed to the dissemination of mcr-1 [86, 87]. A study conducted in Turkey revealed a close genetic relationship between mcr-1 genes found in chicken meat and isolates of human origin, indicating the emergence and spread of mcr-mediated colistin resistance in E. coli across various sources with zoonotic potential in the food chain [88].
In this study, none β-lactamase resistance genes including tetA, tetB, sul and flor were identified with high isolation rates ranging from 27.7% to 86.2%. In the same line, a previous study has reported high prevalence of tetA (72.58%), and sul1 (44.67%) [89]. The investigation of resistance determinants in our study indicated that ESBL-encoding genes were highly prevalent in the isolates. TEM, SHV, OXA, CMY, and CTX-M beta-lactamases are the most prevalent beta-lactamases in Gram-negative bacteria. In accordance, a study in Egypt reported that 57.55%, 46.23%, and 23.58% of the isolates had TEM, CTX-M, and SHV genes, respectively [64], In Bangladesh, Rahman, et al. [89] reported only blaSHV gene from ESBL-E. coli isolates, while in the Netherlands, blaCTX-M-1(58.1%) is the most prevalent gene found in chicken meat, followed by blaTEM-52 (14%) and blaSHV-12 (14%) [90]. In Singapore, Guo, et al. [91] found that out of 225 ESBL-producing E. coli isolates, 76.4% carried blaCTX-M genes, 45.3% carried blaTEM genes and 23.1% carried blaSHV genes. Additionally, Lim, et al. [92] emphasized the prevalence of CTX-M genes as ubiquitous ESBL genes in ESBL-producing E. coli.
Although our isolates were sensitive to imipenem by phenotypic test, carbapenem resistance genes were detected in the isolates by PCR. This indicates that not all carbapenemase-producing isolates exhibit phenotypic resistance to carbapenems due to either lack of expression or the level of gene expression is less than the required to exhibit phenotypic resistance [93, 94].
Treatment of infections caused by ESBL-producing E. coli requires high doses of antibiotics, which can lead to antibiotic resistance and adverse effects on patients [95]. Nanoparticles, such as silver nanoparticles, are considered alternatives to antibiotics for treating various infections. Silver nanoparticles have large surface area to volume ratio, allowing for increased contact with bacteria and resulting in direct interaction with the bacterial cell wall to produce antibacterial activity [96]. Our results showed that the MIC50 of AgNPs-H2O2 was 0.625 μg/mL against ESBL-producing E. coli. Another study in Egypt reported that the average MIC value of AgNPs against ESBL-producing E. coli was 27 μg/ml [95]. In India, an average MIC values of 11.25-45 μg/mL was reported [97],[103] while in Mexico, an MIC of 10 μg/mL was demonstrated for AgNPs [98]. The discrepancies in MIC values could be attributed to differences in the particle size of AgNPs [95]. Shafreen, et al. [99] argued that silver nanoparticles suspensions prepared by biological methods and with concentrations higher than 100 μg/mL may lose their antibacterial effect on microorganisms.
Conclusions
Results of present study showed high prevalence of virulent MDR ESBL-producing E. coli in retail meat products and workers in retail meat shops in Egypt. Therefore, regular monitoring of retail meat and application of hygienic food safety practices by food handlers are required for protecting consumers. Silver nanoparticles are considered a promising alternative for treating MDR ESBL-producing E. coli infections and reducing the risk of antimicrobial resistance.
Acknowledgements
The authors thank the retail shop workers who agreed to participate in this study. The authors are also thankful to Prof. Dr. Fatma A. El-Gohary, Department of Hygiene and Zoonoses, Faculty of Veterinary Medicine, Mansoura University, for kindly providing us with the commercial stock of nanoparticles.
Authors’ contributions
H.A. and R.E. participated in the study design. A.E., A.A., G.E., M.E., R.M., and T.E. performed the sampling, bacterial isolation and identification. H.A. and R.E. performed the antimicrobial susceptibility test and silver nanoparticle experiment. I.E. performed data analysis and visualization. H.A. and I.E. wrote and drafted the manuscript. All authors have read and approved the final manuscript.
Funding
This study did not receive any specific funding.
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Data Availability
The entirety of the data generated or analyzed during this study has been included in the published article.
Declarations
Ethics approval and consent to participate
The study received approval from the Zagazig University Institutional Animal Care and Use Committee (ZU-IACUC) under approval number ZU-IACUC/2/F/214/2022. Animal procedures were conducted following the ARRIVE guidelines. Procedures involving human samples were reviewed and approved by the Research Ethical Committee of Faculty of Medicine, Zagazig University. The study adhered to the principles outlined in the Declaration of Helsinki. Written informed consent was obtained from the retail meat shop workers participating in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8(4):251–9. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 2.Gutkind GO, Di Conza J, Power P, Radice M. β-lactamase-mediated resistance: a biochemical, epidemiological and genetic overview. Curr Pharm Design. 2013;19(2):164–208. doi: 10.2174/138161213804070320. [DOI] [PubMed] [Google Scholar]
- 3.Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infect. 2012;18(7):646–55. doi: 10.1111/j.1469-0691.2012.03850.x. [DOI] [PubMed] [Google Scholar]
- 4.Abayneh M, Tesfaw G, Woldemichael K, Yohannis M, Abdissa A. Assessment of extended-spectrum β-lactamase (ESBLs) – producing Escherichia coli from minced meat of cattle and swab samples and hygienic status of meat retailer shops in Jimma town, Southwest Ethiopia. BMC Infect Dis. 2019;19(1):897. doi: 10.1186/s12879-019-4554-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fulham M, Power M, Gray R. Diversity and distribution of Escherichia coli in three species of free-ranging australian Pinniped pups. Front Mar Sci. 2020;7. 10.3389/fmars.2020.571171
- 6.Gordon DM, Clermont O, Tolley H, Denamur E. Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environ Microbiol. 2008;10(10):2484–96. doi: 10.1111/j.1462-2920.2008.01669.x. [DOI] [PubMed] [Google Scholar]
- 7.Clermont O, Christenson JK, Daubié A-S, Gordon DM, Denamur E. Development of an allele-specific PCR for Escherichia coli B2 sub-typing, a rapid and easy to perform substitute of multilocus sequence typing. J Microbiol Methods. 2014;101:24–7. doi: 10.1016/j.mimet.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Sabat AJ, Budimir A, Nashev D, Sá-Leão R, van Dijl J, Laurent F, et al. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill. 2013;18(4):20380. doi: 10.2807/ese.18.04.20380-en. [DOI] [PubMed] [Google Scholar]
- 9.Franco A, Leekitcharoenphon P, Feltrin F, Alba P, Cordaro G, Iurescia M, et al. Emergence of a clonal lineage of Multidrug-Resistant ESBL-Producing Salmonella Infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS ONE. 2016;10(12):e0144802. doi: 10.1371/journal.pone.0144802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyrrell JM, Wootton M, Toleman MA, Howe RA, Woodward M, Walsh TR. Genetic & virulence profiling of ESBL-positive E. coli from nosocomial & veterinary sources. Vet Microbiol. 2016;186:37–43. doi: 10.1016/j.vetmic.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Peirano G, Pitout JDD. Extended-spectrum β-Lactamase-producing Enterobacteriaceae: update on Molecular Epidemiology and Treatment Options. Drugs. 2019;79(14):1529–41. doi: 10.1007/s40265-019-01180-3. [DOI] [PubMed] [Google Scholar]
- 12.Brolund A, Sandegren L. Characterization of ESBL disseminating plasmids. Infect Dis. 2016;48(1):18–25. doi: 10.3109/23744235.2015.1062536. [DOI] [PubMed] [Google Scholar]
- 13.Kempf I, Jouy E, Chauvin C. Colistin use and colistin resistance in bacteria from animals. Int J Antimicrob Agents. 2016;48(6):598–606. doi: 10.1016/j.ijantimicag.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 14.Clemente L, Manageiro V, Correia I, Amaro A, Albuquerque T, Themudo P, et al. Revealing mcr-1-positive ESBL-producing Escherichia coli strains among Enterobacteriaceae from food-producing animals (bovine, swine and poultry) and meat (bovine and swine), Portugal, 2010–2015. Int J Food Microbiol. 2019;296:37–42. doi: 10.1016/j.ijfoodmicro.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Poirel L, Jayol A, Nordmanna P, Polymyxins Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30(2):557–96. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein NC, Cunha BA. Third-generation cephalosporins. Med Clin North Am. 1995;79(4):705–19. doi: 10.1016/S0025-7125(16)30034-7. [DOI] [PubMed] [Google Scholar]
- 17.Prescott JF. Beta-lactam antibiotics: cephalosporins. Antimicrobial therapy in veterinary medicine. 2013:153 – 73.
- 18.WHO. World Health Organization releases the 2019 AWaRe classification antibiotics. World Health Organization: New York, NY, USA.; 2019.
- 19.Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657–86. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cantón R, Novais A, Valverde A, Machado E, Peixe L, Baquero F, et al. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Europe. Clinical Microbiology and Infection. 2008;14:144 − 53; 10.1111/j.1469-0691.2007.01850.x [DOI] [PubMed]
- 21.Belmar Campos C, Fenner I, Wiese N, Lensing C, Christner M, Rohde H, et al. Prevalence and genotypes of extended spectrum beta-lactamases in Enterobacteriaceae isolated from human stool and chicken meat in Hamburg. Ger Int J Med microbiology: IJMM. 2014;304(5–6):678–84. doi: 10.1016/j.ijmm.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 22.Franci G, Falanga A, Galdiero S, Palomba L, Rai M, Morelli G, et al. Silver nanoparticles as potential antibacterial agents. Molecules. 2015;20(5):8856–74. doi: 10.3390/molecules20058856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones N, Ray B, Ranjit KT, Manna AC. Antibacterial activity of ZnO nanoparticle suspensions on a broad spectrum of microorganisms. FEMS Microbiol Lett. 2008;279(1):71–6. doi: 10.1111/j.1574-6968.2007.01012.x. [DOI] [PubMed] [Google Scholar]
- 24.Lazar V. Quorum sensing in biofilms – how to destroy the bacterial citadels or their cohesion/power? Anaerobe. 2011;17(6):280–5. doi: 10.1016/j.anaerobe.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 25.Hu Q, Tu J, Han X, Zhu Y, Ding C, Yu S. Development of multiplex PCR assay for rapid detection of Riemerella anatipestifer, Escherichia coli, and Salmonella enterica simultaneously from ducks. Journal of Microbiological Methods. 2011;87(1):64 – 9; 10.1016/j.mimet.2011.07.007 [DOI] [PubMed]
- 26.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. 2019.
- 27.Jeong YW, Kim TE, Kim JH, Kwon HJ. Pathotyping avian pathogenic Escherichia coli strains in Korea. J Vet Sci. 2012;13(2):145–52. doi: 10.4142/jvs.2012.13.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol. 1995;12(2):85–90. doi: 10.1111/j.1574-695X.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto T, Echeverria P. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect Immun. 1996;64(4):1441–5. doi: 10.1128/iai.64.4.1441-1445.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dipineto L, Santaniello A, Fontanella M, Lagos K, Fioretti A, Menna LF. Presence of Shiga toxin-producing Escherichia coli O157:H7 in living layer hens. Lett Appl Microbiol. 2006;43(3):293–5. doi: 10.1111/j.1472-765X.2006.01954.x. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt H, Scheef J, Morabito S, Caprioli A, Wieler LH, Karch H. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Applied and environmental microbiology. 2000;66(3):1205-8; 10.1128/AEM.66.3.1205-1208.2000 [DOI] [PMC free article] [PubMed]
- 32.Siqueira AK, Ribeiro MG, Leite Dda S, Tiba MR, Moura C, Lopes MD, et al. Virulence factors in Escherichia coli strains isolated from urinary tract infection and pyometra cases and from feces of healthy dogs. Res Vet Sci. 2009;86(2):206–10. doi: 10.1016/j.rvsc.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 33.Wang G, Clark CG, Rodgers FG. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J Clin Microbiol. 2002;40(10):3613–9. doi: 10.1128/JCM.40.10.3613-3619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohapatra BR, Broersma K, Mazumder A. Comparison of five rep-PCR genomic fingerprinting methods for differentiation of fecal Escherichia coli from humans, poultry and wild birds. FEMS Microbiol Lett. 2007;277(1):98–106. doi: 10.1111/j.1574-6968.2007.00948.x. [DOI] [PubMed] [Google Scholar]
- 35.Hunter PR. Reproducibility and indices of discriminatory power of microbial typing methods. J Clin Microbiol. 1990;28(9):1903–5. doi: 10.1128/jcm.28.9.1903-1905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CLSI . Methods for Dilution Antimicrobial susceptibility tests for Bacteria that grow aerobically; approved Standard—Ninth Edition. Wayne, PA: Clinical and Laboratory Standards Institute; 2012. [Google Scholar]
- 37.Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol. 1983;46(1):165–70. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol infection: official publication Eur Soc Clin Microbiol Infect Dis. 2012;18(3):268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 39.Xia Y, Liang Z, Su X, Xiong Y. Characterization of carbapenemase genes in Enterobacteriaceae species exhibiting decreased susceptibility to carbapenems in a university hospital in Chongqing, China. Ann Lab Med. 2012;32(4):270–5. doi: 10.3343/alm.2012.32.4.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colom K, Pérez J, Alonso R, Fernández-Aranguiz A, Lariño E, Cisterna R. Simple and reliable multiplex PCR assay for detection of blaTEM, bla(SHV) and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol Lett. 2003;223(2):147–51. doi: 10.1016/s0378-1097(03)00306-9. [DOI] [PubMed] [Google Scholar]
- 41.Gray JT, Hungerford LL, Fedorka-Cray PJ, Headrick ML. Extended-spectrum-cephalosporin resistance in Salmonella enterica isolates of animal origin. Antimicrob Agents Chemother. 2004;48(8):3179–81. doi: 10.1128/aac.48.8.3179-3181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jouini A, Vinué L, Slama KB, Sáenz Y, Klibi N, Hammami S, et al. Characterization of CTX-M and SHV extended-spectrum beta-lactamases and associated resistance genes in Escherichia coli strains of food samples in Tunisia. J Antimicrob Chemother. 2007;60(5):1137–41. doi: 10.1093/jac/dkm316. [DOI] [PubMed] [Google Scholar]
- 43.Randall LP, Cooles SW, Osborn MK, Piddock LJ, Woodward MJ. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J Antimicrob Chemother. 2004;53(2):208–16. doi: 10.1093/jac/dkh070. [DOI] [PubMed] [Google Scholar]
- 44.Ibekwe AM, Murinda SE, Graves AK. Genetic diversity and Antimicrobial Resistance of Escherichia coli from Human and Animal sources uncovers multiple resistances from human sources. PLoS ONE. 2011;6(6):e20819. doi: 10.1371/journal.pone.0020819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keyes K, Hudson C, Maurer JJ, Thayer S, White DG, Lee MD. Detection of florfenicol resistance genes in Escherichia coli isolated from sick chickens. Antimicrob Agents Chemother. 2000;44(2):421–4. doi: 10.1128/AAC.44.2.421-424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doublet B, Lailler R, Meunier D, Brisabois A, Boyd D, Mulvey MR, et al. Variant Salmonella genomic island 1 antibiotic resistance gene cluster in Salmonella enterica serovar Albany. Emerg Infect Dis. 2003;9(5):585–91. doi: 10.3201/eid0905.020609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rebelo AR, Bortolaia V, Kjeldgaard JS, Pedersen SK, Leekitcharoenphon P, Hansen IM, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018;23(6):17–00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El-Gohary FA, Abdel-Hafez LJM, Zakaria AI, Shata RR, Tahoun A, El-Mleeh A, et al. Enhanced antibacterial activity of silver nanoparticles combined with hydrogen peroxide against Multidrug-Resistant pathogens isolated from dairy farms and beef slaughterhouses in Egypt. Infect Drug Resist. 2020;13:3485–99. doi: 10.2147/idr.S271261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamilton-Miller JM. Calculating MIC50. J Antimicrob Chemother. 1991;27(6):863–4. doi: 10.1093/jac/27.6.863. [DOI] [PubMed] [Google Scholar]
- 50.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–9. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 51.Yassin AK, Gong J, Kelly P, Lu G, Guardabassi L, Wei L, et al. Antimicrobial resistance in clinical Escherichia coli isolates from poultry and livestock, China. PLoS ONE. 2017;12(9):e0185326. doi: 10.1371/journal.pone.0185326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tansawai U, Walsh TR, Niumsup PR. Extended spectrum ß-lactamase-producing Escherichia coli among backyard poultry farms, farmers, and environments in Thailand. Poult Sci. 2019;98(6):2622–31. doi: 10.3382/ps/pez009. [DOI] [PubMed] [Google Scholar]
- 53.Kürekci C, Osek J, Aydın M, Tekeli İO, Kurpas M, Wieczorek K, et al. Evaluation of bulk tank raw milk and raw chicken meat samples as source of ESBL producing Escherichia coli in Turkey: recent insights. J Food Saf. 2019;39(2):e12605. doi: 10.1111/jfs.12605. [DOI] [Google Scholar]
- 54.Cantón R, Novais A, Valverde A, Machado E, Peixe L, Baquero F, et al. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect. 2008;14(s1):144–53. doi: 10.1111/j.1469-0691.2007.01850.x. [DOI] [PubMed] [Google Scholar]
- 55.Randall LP, Lodge MP, Elviss NC, Lemma FL, Hopkins KL, Teale CJ, et al. Evaluation of meat, fruit and vegetables from retail stores in five United Kingdom regions as sources of extended-spectrum beta-lactamase (ESBL)-producing and carbapenem-resistant Escherichia coli. Int J Food Microbiol. 2017;241:283–90. doi: 10.1016/j.ijfoodmicro.2016.10.036. [DOI] [PubMed] [Google Scholar]
- 56.Rao M, Laidlaw A, Li L, Young K, Tamber S. Isolation of third generation cephalosporin resistant Enterobacteriaceae from retail meats and detection of extended spectrum beta-lactamase activity. J Microbiol Methods. 2021;189:106314. doi: 10.1016/j.mimet.2021.106314. [DOI] [PubMed] [Google Scholar]
- 57.EFSA ECDC. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 2020;18(3):e06007. doi: 10.2903/j.efsa.2020.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.El-Ghareeb WR, Abdel-Raheem M, Al-Marri SM, Alaql TA, Fayez FM. Isolation and identification of extended spectrum β-lactamases (ESBLs) Escherichia coli from minced camel meat in Eastern province, Saudi Arabia. Thai J Veterinary Med. 2020;50(2):155–61. doi: 10.56808/2985-1130.3013. [DOI] [Google Scholar]
- 59.I.C.M.S. Micro-organisms in Foods 6: Microbial Ecology of Food Commodities. Springer US; 2000.
- 60.Niumsup PR, Tansawai U, Na-udom A, Jantapalaboon D, Assawatheptawee K, Kiddee A, et al. Prevalence and risk factors for intestinal carriage of CTX-M-type ESBLs in Enterobacteriaceae from a thai community. Eur J Clin Microbiol Infect Dis. 2018;37(1):69–75. doi: 10.1007/s10096-017-3102-9. [DOI] [PubMed] [Google Scholar]
- 61.Martínez-Vázquez AV, Mandujano A, Cruz-Gonzalez E, Guerrero A, Vazquez J, Cruz-Pulido WL, et al. Evaluation of Retail Meat as a source of ESBL Escherichia coli in Tamaulipas. Mexico Antibiot. 2022 doi: 10.3390/antibiotics11121795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barrios-Villa E, Cortés-Cortés G, Lozano Zarain P, Romero-Romero S, Lara Flores N, Estepa V, et al. Characterization of extended-spectrum and CMY-2 -lactamases, and associated virulence genes in from food of animal origin in México. Br Food J. 2018;120(7):1457–73. doi: 10.1108/BFJ-02-2018-0104. [DOI] [Google Scholar]
- 63.Baran A, Adıgüzel M, Yüksel M. Prevalence of antibiotic-resistant and extended-spectrum beta-lactamase-producing Escherichia coli in chicken meat from eastern Turkey. Pakistan Veterinary Journal. 2020;40(3).
- 64.Abdallah HM, Reuland EA, Wintermans BB, al Naiemi N, Koek A, Abdelwahab AM, et al. Extended-spectrum β-Lactamases and/or Carbapenemases-Producing Enterobacteriaceae isolated from Retail Chicken meat in Zagazig, Egypt. PLoS ONE. 2015;10(8):e0136052. doi: 10.1371/journal.pone.0136052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson RP, Clarke RC, Wilson JB, Read SC, Rahn K, Renwick SA, et al. Growing concerns and recent outbreaks involving Non-O157:H7 serotypes of Verotoxigenic Escherichia coli. J Food Prot. 1996;59(10):1112–22. doi: 10.4315/0362-028x-59.10.1112. [DOI] [PubMed] [Google Scholar]
- 66.Johannes L, Römer W. Shiga toxins–from cell biology to biomedical applications. Nat Rev Microbiol. 2010;8(2):105–16. doi: 10.1038/nrmicro2279. [DOI] [PubMed] [Google Scholar]
- 67.Brusa V, Aliverti V, Aliverti F, Ortega E, de la Torre J, Linares L, et al. Shiga toxin-producing Escherichia coli in beef retail markets from Argentina. Front Cell Infect Microbiol. 2013;2. 10.3389/fcimb.2012.00171 [DOI] [PMC free article] [PubMed]
- 68.Hoang Minh S, Kimura E, Hoang Minh D, Honjoh K, Miyamoto T. Virulence characteristics of Shiga toxin-producing Escherichia coli from raw meats and clinical samples. Microbiol Immunol. 2015;59(3):114–22. doi: 10.1111/1348-0421.12235. [DOI] [PubMed] [Google Scholar]
- 69.Nong F, Zhang P, Meng J, Xie Q, Li Y, Pan Y, et al. Characterization of shiga-toxin producing Escherichia coli (STEC) isolated from retail raw meats in Southeast China. Food Control. 2021;126:108061. doi: 10.1016/j.foodcont.2021.108061. [DOI] [Google Scholar]
- 70.Ali SS, Sonbol FI, Sun J, Hussein MA, Hafez AE, Abdelkarim EA, et al. Molecular characterization of virulence and drug resistance genes-producing Escherichia coli isolated from chicken meat: metal oxide nanoparticles as novel antibacterial agents. Microb Pathog. 2020;143:104164. doi: 10.1016/j.micpath.2020.104164. [DOI] [PubMed] [Google Scholar]
- 71.Werber D, Fruth A, Buchholz U, Prager R, Kramer MH, Ammon A, et al. Strong association between shiga toxin-producing Escherichia coli O157 and virulence genes stx2 and eae as possible explanation for predominance of serogroup O157 in patients with haemolytic uraemic syndrome. Eur J Clin Microbiol Infect Dis. 2003;22(12):726–30. doi: 10.1007/s10096-003-1025-0. [DOI] [PubMed] [Google Scholar]
- 72.Ferreira JC, Penha Filho RAC, Kuaye APY, Andrade LN, Chang Y-F, Darini ALC. Virulence potential of commensal multidrug resistant Escherichia coli isolated from poultry in Brazil. Infect Genet Evol. 2018;65:251–6. doi: 10.1016/j.meegid.2018.07.037. [DOI] [PubMed] [Google Scholar]
- 73.Dorado-García A, Smid JH, Van Pelt W, Bonten MJ, Fluit AC, van den Bunt G, et al. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: a pooled analysis. J Antimicrob Chemother. 2018;73(2):339–47. doi: 10.1093/jac/dkx397. [DOI] [PubMed] [Google Scholar]
- 74.Kluytmans JAJW, Overdevest ITMA, Willemsen I, Kluytmans-van den Bergh MFQ, van der Zwaluw K, Heck M, et al. Extended-spectrum β-Lactamase–producing Escherichia coli from Retail Chicken meat and humans: comparison of strains, plasmids, resistance genes, and virulence factors. Clin Infect Dis. 2012;56(4):478–87. doi: 10.1093/cid/cis929. [DOI] [PubMed] [Google Scholar]
- 75.Kawamura K, Goto K, Nakane K, Arakawa Y. Molecular epidemiology of extended-spectrum β-lactamases and Escherichia coli isolated from retail foods including chicken meat in Japan. Foodborne Pathog Dis. 2014;11(2):104–10. doi: 10.1089/fpd.2013.1608. [DOI] [PubMed] [Google Scholar]
- 76.Börjesson S, Ny S, Egervärn M, Bergström J, Rosengren Ã, Englund S, et al. Limited dissemination of extended-spectrum β-Lactamase– and plasmid-encoded AmpC–Producing < em > Escherichia coli from Food and Farm Animals, Sweden. Emerg Infect Disease J. 2016;22(4):634. doi: 10.3201/eid2204.151142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Falgenhauer L, Imirzalioglu C, Oppong K, Akenten CW, Hogan B, Krumkamp R, et al. Detection and characterization of ESBL-Producing Escherichia coli from humans and poultry in Ghana. Front Microbiol. 2019;9. 10.3389/fmicb.2018.03358 [DOI] [PMC free article] [PubMed]
- 78.de Been M, Lanza VF, de Toro M, Scharringa J, Dohmen W, Du Y, et al. Dissemination of Cephalosporin Resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet. 2014;10(12):e1004776. doi: 10.1371/journal.pgen.1004776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang H, McEntire JC, Zhang L, Li X, Doyle M. The transfer of antibiotic resistance from food to humans: facts, implications and future directions. Rev Sci Tech. 2012;31(1):249–60. doi: 10.20506/rst.31.1.2117. [DOI] [PubMed] [Google Scholar]
- 80.Grave K, Torren-Edo J, Mackay D. Comparison of the sales of veterinary antibacterial agents between 10 european countries. J Antimicrob Chemother. 2010;65(9):2037–40. doi: 10.1093/jac/dkq247. [DOI] [PubMed] [Google Scholar]
- 81.Moawad AA, Hotzel H, Awad O, Tomaso H, Neubauer H, Hafez HM, et al. Occurrence of Salmonella enterica and Escherichia coli in raw chicken and beef meat in northern Egypt and dissemination of their antibiotic resistance markers. Gut Pathog. 2017;9:57. doi: 10.1186/s13099-017-0206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hassen B, Abbassi MS, Ruiz-Ripa L, Mama OM, Hassen A, Torres C, et al. High prevalence of mcr-1 encoding colistin resistance and first identification of blaCTX-M-55 in ESBL/CMY-2-producing Escherichia coli isolated from chicken faeces and retail meat in Tunisia. Int J Food Microbiol. 2020;318:108478. doi: 10.1016/j.ijfoodmicro.2019.108478. [DOI] [PubMed] [Google Scholar]
- 83.Uz Zaman T, Albladi M, Siddique MI, Aljohani SM, Balkhy HH. Insertion element mediated mgrB disruption and presence of ISKpn28 in colistin-resistant Klebsiella pneumoniae isolates from Saudi Arabia. Infect Drug Resist. 2018;11:1183–7. doi: 10.2147/IDR.S161146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cao L, Li X, Xu Y, Shen J. Prevalence and molecular characteristics of mcr-1 colistin resistance in Escherichia coli: isolates of clinical infection from a Chinese University Hospital. Infect Drug Resist. 2018;11:1597–603. doi: 10.2147/IDR.S166726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium isolate. mBio. 2019;10(3):e00853–19. doi: 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haenni M, Métayer V, Gay E, Madec JY. Increasing trends in mcr-1 prevalence among extended-spectrum-β-lactamase-producing Escherichia coli isolates from french calves despite decreasing exposure to colistin. Antimicrob Agents Chemother. 2016;60(10):6433–4. doi: 10.1128/AAC.01147-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perrin-Guyomard A, Bruneau M, Houée P, Deleurme K, Legrandois P, Poirier C, et al. Prevalence of mcr-1 in commensal Escherichia coli from French livestock, 2007 to 2014. Eurosurveillance. 2016;21(6):30135. doi: 10.2807/1560-7917.ES.2016.21.6.30135. [DOI] [PubMed] [Google Scholar]
- 88.Adiguzel MC, Baran A, Wu Z, Cengiz S, Dai L, Oz C, et al. Prevalence of Colistin Resistance in Escherichia coli in Eastern Turkey and genomic characterization of an mcr-1 positive strain from Retail Chicken meat. Microb Drug Resist. 2020;27(3):424–32. doi: 10.1089/mdr.2020.0209. [DOI] [PubMed] [Google Scholar]
- 89.Rahman MM, Husna A, Elshabrawy HA, Alam J, Runa NY, Badruzzaman ATM, et al. Isolation and molecular characterization of multidrug-resistant Escherichia coli from chicken meat. Sci Rep. 2020;10(1):21999. doi: 10.1038/s41598-020-78367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, et al. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, the Netherlands. Emerg Infect Dis. 2011;17(7):1216–22. doi: 10.3201/eid1707.110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo S, Aung KT, Leekitcharoenphon P, Tay MYF, Seow KLG, Zhong Y, et al. Prevalence and genomic analysis of ESBL-producing Escherichia coli in retail raw meats in Singapore. J Antimicrob Chemother. 2020;76(3):601–5. doi: 10.1093/jac/dkaa461. [DOI] [PubMed] [Google Scholar]
- 92.Lim EJ, Ho SX, Cao DY, Lau QC, Koh TH, Hsu LY. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in Retail Chicken meat in Singapore. Ann Acad Med Singapore. 2016;45(12):557–9. doi: 10.47102/annals-acadmedsg.V45N12p557. [DOI] [PubMed] [Google Scholar]
- 93.Oteo J, Hernández JM, Espasa M, Fleites A, Sáez D, Bautista V, et al. Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. J Antimicrob Chemother. 2013;68(2):317–21. doi: 10.1093/jac/dks383. [DOI] [PubMed] [Google Scholar]
- 94.van der Bij AK, Pitout JD. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J Antimicrob Chemother. 2012;67(9):2090–100. doi: 10.1093/jac/dks214. [DOI] [PubMed] [Google Scholar]
- 95.Mohammed ASA, Mourad MI, Alsewy FZ, Azzam NFAEM. Combination of silver nanoparticles with ineffective antibiotics against extended spectrum beta-lactamases producing isolates at Alexandria Main University Hospital, Egypt. Beni-Suef Univ J Basic Appl Sci. 2021;10(1):58. doi: 10.1186/s43088-021-00147-2. [DOI] [Google Scholar]
- 96.Castillo RR, Lozano D, González B, Manzano M, Izquierdo-Barba I, Vallet-Regí M. Advances in mesoporous silica nanoparticles for targeted stimuli-responsive drug delivery: an update. Expert Opin Drug Deliv. 2019;16(4):415–39. doi: 10.1080/17425247.2019.1598375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ansari MA, Khan HM, Khan AA, Cameotra SS, Pal R. Antibiofilm efficacy of silver nanoparticles against biofilm of extended spectrum β-lactamase isolates of Escherichia coli and Klebsiella pneumoniae. Appl Nanosci. 2014;4(7):859–68. doi: 10.1007/s13204-013-0266-1. [DOI] [Google Scholar]
- 98.Vazquez-Muñoz R, Meza-Villezcas A, Fournier PGJ, Soria-Castro E, Juarez-Moreno K, Gallego-Hernández AL, et al. Enhancement of antibiotics antimicrobial activity due to the silver nanoparticles impact on the cell membrane. PLoS ONE. 2019;14(11):e0224904. doi: 10.1371/journal.pone.0224904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shafreen RB, Seema S, Ahamed AP, Thajuddin N, Ali Alharbi S. Inhibitory effect of Biosynthesized Silver Nanoparticles from Extract of Nitzschia palea against curli-mediated biofilm of Escherichia coli. Appl Biochem Biotechnol. 2017;183(4):1351–61. doi: 10.1007/s12010-017-2503-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The entirety of the data generated or analyzed during this study has been included in the published article.