Abstract
The biota of Sulawesi is noted for its high degree of endemism and for its substantial levels of in situ biological diversification. While the island’s long period of isolation and dynamic tectonic history have been implicated as drivers of the regional diversification, this has rarely been tested in the context of an explicit geological framework. Here, we provide a tectonically informed biogeographical framework that we use to explore the diversification history of Sulawesi flying lizards (the Draco lineatus Group), a radiation that is endemic to Sulawesi and its surrounding islands. We employ a framework for inferring cryptic speciation that involves phylogeographic and genetic clustering analyses as a means of identifying potential species followed by population demographic assessment of divergence-timing and rates of bi-directional migration as means of confirming lineage independence (and thus species status). Using this approach, phylogenetic and population genetic analyses of mitochondrial sequence data obtained for 613 samples, a 50-SNP data set for 370 samples, and a 1249-locus exon-capture data set for 106 samples indicate that the current taxonomy substantially understates the true number of Sulawesi Draco species, that both cryptic and arrested speciations have taken place, and that ancient hybridization confounds phylogenetic analyses that do not explicitly account for reticulation. The Draco lineatus Group appears to comprise 15 species—9 on Sulawesi proper and 6 on peripheral islands. The common ancestor of this group colonized Sulawesi ~11 Ma when proto-Sulawesi was likely composed of two ancestral islands, and began to radiate ~6 Ma as new islands formed and were colonized via overwater dispersal. The enlargement and amalgamation of many of these proto-islands into modern Sulawesi, especially during the past 3 Ma, set in motion dynamic species interactions as once-isolated lineages came into secondary contact, some of which resulted in lineage merger, and others surviving to the present. [Genomics; Indonesia; introgression; mitochondria; phylogenetics; phylogeography; population genetics; reptiles.]
Graphical abstract
Graphical Abstract.
We now come to the island of Celebes, in many respects the most remarkable and interesting in the whole region, or perhaps on the globe, since no other island seems to present so many curious problems for solution.
Alfred Russel Wallace (1876) on the biogeography of Sulawesi.
At the interface between the Asian and Australo-Papuan realms, the Indonesian island of Sulawesi has long captivated researchers focused on biological diversification (Wallace 1860, 1869, 1876, 1880; Mayr 1944; Carlquist 1965; Lohman et al. 2011; Stelbrink et al. 2012). Whereas some islands provide simplified natural biological laboratories insulated from the evolutionary and ecological complexities of mainland systems, thereby offering opportunities to hone in on processes driving diversification (see MacArthur and Wilson 1967; Shaw and Gillespie 2016), Sulawesi resides on the opposite side of the insular complexity spectrum. In contrast with important model island archipelagos such as the Hawaiian, Galapagos, and Canary Islands that originated via the relatively straightforward process of hotspot island formation (Huppert et al. 2020), Sulawesi arose through an exceedingly complex combination of geological processes involving rifting, collision, subduction, and extension at the triple junction of the Australian, Sunda, and Philippine Sea plates (Hamilton 1979). Indeed, the dynamic tectonic processes that produced Sulawesi and its surrounding islands are considered to be among the most complex on Earth (Hall 1998; Leo et al. 2012). The complexity of Sulawesi’s tectonic setting suggests the potential for similarly complex processes underpinning biological diversification on this island and presents a daunting challenge to the biogeographer.
With the advent of High-Throughput Sequencing (HTS) and the collection of genome-scale data sets, it is now possible to interrogate in detail the processes underpinning even the complex biological diversification scenarios likely to be at play with Sulawesi. Such data are essential when the evolutionary processes responsible for the generation and maintenance of contemporary species diversity are a poor match to the simplifying assumptions of phylogenetic and population genetic analyses. For example, clades characterized by large effective population sizes and short internal branch lengths are notoriously difficult for phylogenetic inference and are expected to require large multi-locus data sets and coalescent-based inference methods to obtain reliable phylogenetic estimates (Edwards et al. 2007; Degnan and Rosenberg 2009; Xu and Yang 2016). A history involving interspecific hybridization further complicates matters because, until recently, the primary methods that have been developed to perform multispecies coalescent (MSC) phylogenetic analyses (e.g., Bouckaert et al. 2014; Mirabab et al. 2014; Yang 2015; Ogilvie et al. 2017) assumed that gene tree disagreement results strictly from incomplete lineage sorting and not from reticulation. New methods have appeared recently that account for both incomplete lineage sorting and gene flow in an MSC framework (e.g., Solís-Lemus et al. 2017; Wen et al. 2018; Jones 2019; Flouri et al. 2020), but these exciting approaches remain in their infancy and are computationally intensive. Cryptic speciation (Stuart et al. 2006; Bickford et al. 2007; Barley et al. 2013) and arrested speciation (lineage merger following substantial initial divergence) are additional complicating factors that may need to be accounted for in phylogenomic and population genomic analyses. Cryptic speciation is challenging to infer because there is substantial disagreement among researchers in terms of what constitutes sufficient evidence that genetically structured yet phenotypically homogeneous populations actually represent independent species (e.g., Jackson et al. 2017; Sukumaran and Knowles 2017; Leaché et al. 2019) which, of course, stems from the substantial disagreement regarding how species should be defined in the first place (see Coyne and Orr 2004; de Queiroz 2007 for discussion). Although not often discussed in the phylogenetics and biogeography literature, we argue here that arrested speciation events can also complicate matters, particularly when it comes to understanding the full complexity of the biogeographical story for a focal group, as well as for generating an accounting of the species diversity represented by the taxon in question.
In the present study, we explore the diversification history of the Draco lineatus Group of flying lizards of Sulawesi and adjacent islands in order to assess the biogeographical history of Sulawesi. A full understanding of the evolution of this assemblage required addressing the challenges outlined above. This study showcases an integrative multi-scale approach including 1) a densely sampled phylogeographic analysis, 2) multi-locus species delimitation based on exon-capture data, 3) population genomic analyses to further evaluate delimited species, 4) multispecies coalescent phylogenomic analyses that account for introgression, and 5) quantitative biogeographical evaluation. These analyses show that species diversity within the D. lineatus Group is underestimated, that successful and arrested cryptic speciation has occurred, that past hybridization confounds standard MSC phylogenetic analysis methods for this group, and that diversification within this clade has been driven by the dynamic tectonic history of Sulawesi.
History of Sulawesi Biogeography
Alfred Russel Wallace (1876, 1880) identified the massive tropical Southeast Asian island of Sulawesi (the 11th largest island in the world) as one of the most biogeographically interesting and perplexing islands on the planet based on its highly unusual and endemic fauna. Wallace (1859) recognized that Sulawesi was separated from Borneo and the contiguous Sunda Shelf by a deep channel, the Makassar Strait, and that this water barrier demarcated a major faunal transition that would come to be known as Wallace’s Line (Huxley 1868). An early appreciation of Sulawesi biogeography stemming from Wallace’s seminal work was largely focused on the fauna 1) being highly endemic, and 2) containing elements of both the Asian and Australo-Papuan biotas despite its close proximity to the Sunda Shelf island of Borneo (e.g., Mayr 1944; Darlington 1957). However, Sulawesi biogeography is more interesting than even Wallace and other early investigators appreciated, because substantial regional in situ diversification also characterizes the Sulawesi biota. Many studies have identified examples of regional in situ diversification among terrestrial taxa including cicadas (Duffels 1990; de Boer and Duffels 1996), Chitaura grasshoppers (Walton et al. 1997; Butlin et al. 1998; Bridle et al. 2001), cockroaches (Maekawa et al. 2001), Limnonectes fanged frogs (Evans et al. 2003a, Setiadi et al. 2011), Celebes toads (Evans et al. 2003c, 2008), Lamprolepis emerald skinks (Linkem et al. 2013), phalangers (Ruedas and Morales 2005), shrews (Esselstyn et al. 2009, 2021; Eldridge et al. 2018), rodents (Esselstyn et al. 2012; Achmadi et al. 2013; Rowe et al. 2016a, 2019; Giarla et al. 2018; Handika et al. 2021; Rowe et al. 2016b), tarsiers (Merker et al. 2009; Shekelle et al. 2010; Driller et al. 2015), anoas (Schreiber et al. 1999; Frantz et al. 2018), and forest pigs and babirusas (Frantz et al. 2018). In addition, there are impressive examples of in situ diversification in freshwater aquatic environments (von Rintelen et al. 2012), including in streams and the ancient deep-water rift lakes of central Sulawesi, as revealed by studies of aquatic snails (von Rintelen and Glaubrecht 2005; von Rintelen et al. 2007a, b, c; Glaubrecht and von Rintelen 2008), crabs (Schubart and Ng 2008), shrimps (von Rintelen et al. 2007a, 2010b), and fishes (Takehana et al. 2005; Mokodongan and Yamahira 2015; Mandagi et al. 2021; Utama et al. 2022).
The dominant biogeographical paradigm for in situ diversification of terrestrial organisms on Sulawesi involves seven areas of regional endemism (AOEs). This framework was established primarily via studies of Sulawesi’s endemic radiation of macaque monkeys, and on the general congruence of the monkey species boundaries with mitochondrial genetic breaks in the Celebes toad, Ingerophrynus celebensis (see Fooden 1969; Ciani et al. 1989; Evans et al. 1999, 2003a, 2003b; Evans 2012;). Many subsequent studies have found varying degrees of agreement with the macaque-based AOEs, suggesting a common underlying mechanism for Sulawesi regional diversification. Researchers have long suspected that the island’s dynamic tectonic history has likely played a pivotal role in this process, and recent studies have attempted to test this hypothesis more directly (e.g., Driller et al. 2015; Giarla et al. 2018; Frantz et al. 2018). These biogeographical studies have benefited from the ever more refined tectonic models describing Sulawesi’s complex paleo-island history, particularly those that have attempted to illustrate the presence of land and sea in the region over the past 25 million years (e.g., Hall 2009, 2012, 2013). Although progress has been made in recent years in generating and testing biogeographical hypotheses with reference to Sulawesi’s tectonic history, our goal was to formulate a more geologically and temporally explicit framework for Sulawesi biogeography and test it using phylogenomic and population genomic data obtained for the flying lizards of the Draco lineatus Group.
Geological and Biogeographic Framework
From a biogeographical standpoint, Sulawesi is effectively an isolated oceanic island. Since the Eocene formation of the Makassar Strait, Sulawesi had no land connections to the Sunda Shelf (Borneo, Java, Sumatra, and Malay Peninsula), nor to the more distant Sahul Shelf (New Guinea and Australia). Stelbrink et al. (2012) largely supported this argument using dated time trees in a reanalysis of 27 published phylogenetic data sets. Their analysis strongly rejected the hypothesis that Asian elements of the Sulawesi biota originated via trans-Makassar Strait vicariance, as the opening of the strait dates to ~45 Ma and the constituent Sulawesi fauna was demonstrably much younger in all but one case (mite harvestmen, Clouse and Giribet [2010]). Furthermore, despite a strong Asian signature, the composition of Sulawesi’s herpetofauna is very much consistent with the Makassar Strait having served as a very stringent barrier, with at least 98 Bornean amphibian and reptile genera absent from the Sulawesi fauna (Supplemental Material Appendix A). Stelbrink et al. (2012) were less definitive regarding the possibility that the eastern terranes might have served as vehicles for “tectonic dispersal” (Michaux 2010) from the Sahul Shelf. However, tectonic dispersal from the east also appears unlikely. The composition of the herpetofauna is nearly entirely Asian in origin, which is unlikely if the Asian representation arrived strictly via overwater dispersal while Australo-Papuan elements arrived as intact faunas on one or more paleo-islands. Thus, there is strong evidence supporting the view that the entirety of the native Sulawesi biota arrived via overwater dispersal rather than through vicariance.
Sulawesi is an ancient island, with land possibly present since West Sulawesi rifted from the Sunda Shelf margin in the Eocene (about 45 Ma) resulting in the formation of the Makassar Strait (Stelbrink et al. 2012). Whether the land was continuously present between 45 and 23 Ma is subject to debate (as discussed above), but the land has certainly been present at least since the collision of the Sula Spur micro-continent (an Australian continental promontory) with the North Sulawesi volcanic arc beginning approximately 23 Ma (Hall 2011; Nugraha and Hall 2018). The Sula Spur originated far to the southeast of West Sulawesi near the Bird’s Head of New Guinea and migrated a substantial distance with the Australian continent, of which it was part, prior to collision (Hall 2011). The Sula Spur collision and later subduction resulted in uplift and exhumation in Sulawesi’s Central Core, as well as the ultimate emergence of the Eastern and SE Peninsulas. Note that we use the term “Central Core” here in the biogeographical sense— as a convenient reference to the large land area in central Sulawesi from which the four peninsulas emanate—and not as an indication of the geological processes (such as metamorphic core complex formation) that may have contributed to the origin of that land area (see Fig. 1).
Figure 1.
Biogeographic framework for Sulawesi biogeography based on the paleo-island reconstructions of Nugraha and Hall (2018). Dashed lines represent potential contact points of the amalgamated paleo-islands, with the dates indicating the temporal duration of the paleo-island boundary. Gray shading indicates the Tempe Depression. Potentially biogeographically important fault zones (solid lines) overlay the map. The inset map color-codes the Central Core and four peninsulas of Sulawesi for ease of discussion in the text and demarcates with black lines the AOE boundaries as defined by Evans et al. (2003c).
Several islands were likely present within the current footprint of modern Sulawesi by 20 Ma, but some of these islands may have been submerged by 15 Ma according to paleo-island reconstructions of Nugraha and Hall (2018). Their reconstructions suggest the occurrence of two emergent land areas at 15 Ma, one in the Central Core and one spanning parts of the current Eastern and SE Peninsulas. Beginning at about this time, the conformation of modern Sulawesi was determined largely by extensional processes driven by rollback of the Banda subduction zone (Spakman and Hall 2010). Extension resulted in mountain building in emergent land areas of the developing Sulawesi archipelago, as well as in the formation of extremely deep inter-arm embayments, particularly during the relatively recent time interval of the past 1–2 Ma. From 10–2 Ma, the number of islands in the region is inferred to have increased systematically with three islands present at 10 Ma and as many as 13 present by 2 Ma. Perhaps more importantly, between 10 Ma and 4 Ma, the inferred process involves the sequential accumulation of additional relatively small islands within Sulawesi’s current geographical footprint, but beginning at 3 Ma these islands became progressively larger and began to amalgamate. By 1 Ma, Sulawesi had nearly achieved its current form with just the SW Peninsula separated from the remainder of the island by a submerged Tempe Depression (which would not become emergent until as recently as 4600 years ago; Gremmen 1990).
The key feature of the Nugraha and Hall (2018) model involves a series of relatively stationary paleo-islands that originated much closer to the Sunda Shelf than to the Sahul Shelf, existed for a considerable period of time as a disjunct archipelago, and then ultimately merged over the past 4 million years as additional uplift and exhumation filled the intervening inter-island gaps (Nugraha and Hall 2018). This model provides a potential explanation for Sulawesi’s regional endemism as early colonizing lineages may have initiated diversification on different paleo-islands prior to Sulawesi’s relatively recent unification. Notably, the tectonic history of Sulawesi suggests a biogeographical framework representing the inverse of typical vicariance biogeography scenarios. Rather than a once-unified land area fragmenting over time to produce allopatrically distributed descendent lineages, the history of Sulawesi reflects the relatively recent merger of once-isolated paleo-islands that apparently were never in prior contact with one another (Nugraha and Hall 2018). Thus, as was recently argued for Lesser Sundas fanged frogs and flying lizards (Reilly et al. 2019, 2022), a process involving dispersal to and among independent paleo-islands and subsequent diversification of these lineages in allopatry, followed by a merger of the paleo-islands bringing once-allopatric lineages into dynamic secondary contact is likely at the heart of the Sulawesi in situ diversification story.
Sulawesi also has several major faults of unknown biogeographical importance (Fig. 1), which could add layers of complexity upon the paleo-island model proposed by Nugraha and Hall (2018). Several of these faults once appeared to represent potential paleo-island boundaries (see reconstructions in Moss and Wilson 1998; Hall 2009, 2012; Spakman and Hall 2010). However, there is no clear evidence that the faults represent boundaries between once-separated land areas despite that, in some instances, the faults serve as junctions between geological units with distinct soil and rock features (such as the Lawanopo Fault [see Fig. 1], a left-lateral strike-slip fault that separates ultramafic rocks to the north and metamorphic rocks to the south; Sukamto 1978, 1990; Natawidjaja and Daryono 2015). Some of these faults correlate closely with lineage/species boundaries detected via biogeographical analyses. For example, two species of tarsiers meet and form a narrow hybrid zone precisely at the Palu-Koro left-lateral strike-slip transform fault in the Palu Valley (Merker et al. 2009). In addition, many species exhibit morphological and/or genetic breaks at the position of the Gorontalo strike-slip fault, including macaque monkeys, Celebes toads, anoas, Sulawesi warty pigs, babirusas, and shrews (Evans et al. 2003c; Frantz et al. 2018; Esselstyn et al. 2021). Another example is provided by toads, monkeys, and shrews, all of which exhibit morphological and/or genetic breaks in close proximity to the geothermally active Kotomobagu Fault on the Northern Peninsula (Evans et al. 2003c; Esselstyn et al. 2021). Given these empirical findings, we believe that geological faults might be biogeographically important for Sulawesi even if the underlying mechanisms underpinning their functional significance in driving speciation are not yet understood. Therefore, our framework describing potentially important biogeographical boundaries on Sulawesi includes major faults as well as the inferred paleo-islands of Nugraha and Hall (2018). We summarize the biogeographical implications of the Nugraha and Hall (2018) model in Figure 1. We superimpose on a map of contemporary Sulawesi the geographical positions of biogeographical boundaries implied by the Nugraha and Hall (2018) reconstructions (with specific placements of the boundaries informed by the Sulawesi AOE framework of Evans et al. 2003c), as well as the temporal durations during which these boundaries may have existed. For example, the tectonic model suggests that two separate paleo-islands were present on either side of the Gorontalo boundary (which may coincide with the Gorontalo Fault) beginning 8 Ma and that this barrier ceased to exist about 1 Ma when this section of the Northern Peninsula became unified, and so on.
Species Delimitation
A phylogenetically informed analysis of the biogeographical history of Sulawesi flying lizards first requires an assessment of species boundaries in this assemblage. However, species delimitation is a controversial topic because researchers often disagree over what constitutes a species (Coyne and Orr 2004; de Queiroz 2007; Sukumaran and Knowles 2017; Leaché et al. 2019), and this is especially true for so-called “cryptic species” that differ little or not at all in external morphology. In our view, a species is a lineage on its own unique evolutionary trajectory, which can be identified as a temporally extended metapopulation lineage independent or largely independent from other such lineages. In other words, we advocate for the General Lineage Concept of species (de Queiroz 1998, 1999). When considering sympatric or parapatrically distributed lineages, our thinking also aligns well with the relaxed version of the Biological Species Concept advocated by Coyne and Orr (2004), a concept that emphasizes the evolution of reproductive isolating barriers but permits hybridization with limited gene flow. As Coyne and Orr (2004) have argued, this criterion results in a continuous distribution of possible degrees of reproductive isolation with no clear threshold to discretely demarcate the species boundary, thereby leaving the possibility of cases “where species status is more or less irresolvable.” In principle, we agree with this view, but in practice, we also see value in providing a testable taxonomic framework that can be reassessed with improved data or analytical methods. Therefore, to assess species status at putative Draco lineage boundaries, we have opted to assess lineage (and thus species) status by estimating bi-directional migration rates from a genomic data set, using as the threshold for lineage status a rate of 0.5 effective migrants per generation (see Shaffer and Thompson 2007; Burbrink and Ruane 2021; Reilly et al. 2022), the theoretical threshold rate expected to represent the tipping point between future genomic admixture versus ongoing genetic divergence under a pure drift model (Wright 1931; Slatkin 1987). We selected 0.5 migrants per generation as a conservative rule of thumb, suggesting a very limited rate of gene flow commensurate with lineage independence. We are not suggesting that such a value represents a discrete threshold separating species from non-species, and we furthermore appreciate the challenges of estimating migration rates with precision (Campbell et al. 2018). Despite these challenges, we suggest that taking migration into consideration in this manner is key to integrative taxonomic approaches (e.g., Fujita et al. 2012) that aim to delimit cryptic species using genetic data alone, as direct measurements inferring lack (or extreme limitation) of gene flow should be incorporated into species delimitation and taken as compelling evidence of lineage independence—just as traditional morphological character differences have been utilized in standard species descriptions.
The Draco lineatus Group
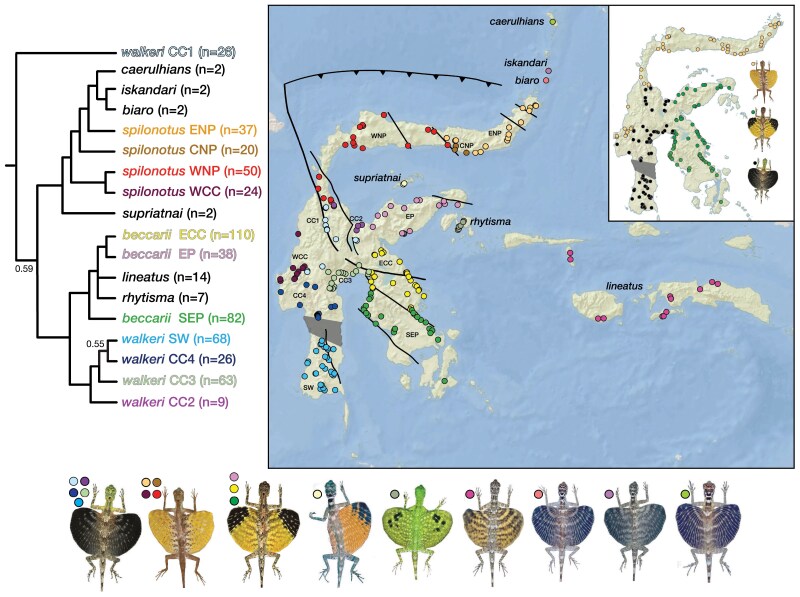
The flying lizards of the genus Draco are widespread across SE Asia and the Western Ghats region of southwestern India. Notably, this assemblage has crossed many oceanic barriers to independently colonize SE Asian archipelagos, including the Philippines (McGuire and Alacala 2000), the Lesser Sundas (Reilly et al. 2022), as well as Sulawesi and Maluku (McGuire et al. 2007). These arboreal lizards are famous for their wing-like patagial membranes supported by elongated thoracic ribs, which they utilize both for display and for gliding between trees (McGuire 2003; McGuire and Dudley 2005, 2011). The Draco lineatus Group (McGuire and Heang 2001; McGuire et al. 2007) is a monophyletic assemblage comprising nine described species inhabiting Sulawesi and its surrounding islands, reaching as far east as the Malukan island of Seram (McGuire et al. 2007). These are the only Draco species that occur on Sulawesi or on the Malukan islands that extend eastward from this island. Three of these species are parapatrically distributed Sulawesi endemics: 1) Draco beccarii on the Eastern and Southeastern Peninsulas and extreme eastern margin of the island’s Central Core, 2) D. spilonotus on the Northern Peninsula and western margin of the Central Core, and 3) D. walkeri on the remaining majority of the Central Core and on the Southwest Peninsula (Fig. 2 inset). The remaining six taxa are perhaps best characterized as “peripheral isolates” species (Mayr 1963), and five of these are confined to single islands or localized contiguous, offshore island archipelagos. The latter five species include three in the Sangir-Talaud Island Group, a string of volcanoes that lie between Sulawesi’s Northern Peninsula and the Philippine island of Mindanao (Morrice et al. 1983): the small-island Sangir-Talaud endemics include Draco biaro on Biaro Island, D. iskandari on Tahulandang Island, and D. caerulhians on Sangir Besar (McGuire et al. 2007). A fourth small-island endemic, D. supriatnai, is restricted to the Togian Islands, which are in Teluk Tomini (Tomini Bay) north of Sulawesi’s Eastern Peninsula. A fifth species, D. rhytisma, occurs on the Banggai Islands off the southeast coast of the Eastern Peninsula. Finally, a sixth peripheral isolates species, D. lineatus, has a much wider range than the other five non-Sulawesi members of the D. lineatus complex, and is known from the Sula Islands, Buru, and the cluster of islands that includes Ambon and Seram (Musters 1983; McGuire et al. 2007). These species are diagnosed primarily by differences in coloration among males. Given the complex tectonic history of Sulawesi and prior findings of substantial regional genetic divergence among otherwise morphologically homogeneous species (e.g., Evans et al. 2003c; Esselstyn et al. 2021), we assessed lineage boundaries and phylogenetic relationships of the Draco lineatus Group using a large-scale genetic data set. This effort showed that ancient hybridization, cryptic species, and arrested speciation must be accounted for to obtain a robust estimate of both the diversity and phylogenetic history of the Draco lineatus Group, and to provide a suitable framework for biogeographical inference.
Figure 2.
Bayesian phylogenetic estimate obtained via analysis of three mitochondrial genes (ND2, 12S ribosomal RNA, and 16S ribosomal RNA) for 593 Draco lineatus Group samples and 20 D. bimaculatus outgroup samples (D. bimaculatus not shown). The branches on the tree represent collapsed clades with the numbers following the clade names indicating the number of samples included in the analysis for that mitochondrial lineage. Posterior probabilities for all nodes including the 18 named clades are 1.0 unless otherwise indicated. Sulawesi sample localities (dots) are color-coded to reflect mitochondrial genetic structure. ENP = Eastern Northern Peninsula, CNP = Central Northern Peninsula, WNP = Western Northern Peninsula, WCC = Western Central Core, EP = Eastern Peninsula, SEP = Southeastern Peninsula, ECC = Eastern Central Core, SW = Southwestern Peninsula, CC1–CC4 = Central Core regions 1–4. The inset map shows the sampling localities color-coded to reflect the current taxonomy of the three Sulawesi Draco species (black dots for D. walkeri, orange dots for D. spilonotus, green dots for D. beccarii).
Materials and Methods
Sampling
All but 3 of the 613 Draco samples employed in this study were obtained by JAM and co-authors over the course of nearly two decades of fieldwork under Indonesian and Philippine research permits granted by LIPI and RISTEKDIKTI (now BRIN) and by the Philippine Department of the Environment and Natural Resources, Biodiversity Management Bureau (see Supplemental Materials Appendix B for a list of specimen voucher numbers and localities).
Genetic Data
The genetic data obtained for this study are nested in scope and were collected over two decades (see Table 1 for a summary of all genetic data sets analyzed for this study). We first generated a mitochondrial sequence data set which we have continued to add to over a 20-year interval. This data set now includes 593 individuals representing the nine currently recognized species in the Draco lineatus Group (McGuire et al. 2007) plus 20 additional individuals representing this clade’s sister taxon, Draco bimaculatus, from the Philippines (see McGuire and Alcala 2000). This approach allowed for many individuals to be screened for each species with particularly dense coverage of Sulawesi proper. Our mitochondrial data set is composed of three mitochondrial genes (ND2 and flanking tRNAs, 12S rRNA, and 16S rRNA). Standard Sanger sequencing laboratory methods were used and details can be found in Supplemental Materials Appendix C.
Table 1.
Analyses undertaken with data characteristics.
| Analysis | Sample # | # Loci | Base pairs |
|---|---|---|---|
| Bayesian phylogenetic analysis of mtDNA sequence data | 613 | 1 | 2337 |
| TESS analysis of Sequenom SNP data | 370 | 50 | 50 |
| STRUCTURE analysis of Draco beccarii | 29 | 1020 | 1020/23,313 |
| STRUCTURE analysis of D. spilonotus | 23 | 848 | 848/19,414 |
| STRUCTURE analysis of D. walkeri | 33 | 908 | 908/22,696 |
| STRUCTURE analysis of D. walkeri CC1-CC2 | 9 | 530 | 530/3761 |
| STRUCTURE analysis of D. walkeri CC3-CC4-SW | 24 | 858 | 858/18,356 |
| BFD* analysis of D. beccarii | 29 | 1020 | 1020 |
| BFD* analysis of D. spilonotus | 23 | 848 | 848 |
| BFD* analysis of D. walkeri | 33 | 908 | 908 |
| BPP version 3 analysis of D. beccarii | 29 | 500 | 696,314 |
| BPP version 3 analysis of D. spilonotus | 23 | 500 | 696,314 |
| BPP version 3 analysis of D. walkeri | 33 | 500 | 696,314 |
| Isolation-By-Distance Mantel test for D. beccarii | 29 | 1020 | 60,662 |
| Isolation-By-Distance Mantel test for D. spilonotus | 23 | 848 | 52,030 |
| Isolation-By-Distance Mantel test for D. walkeri | 33 | 908 | 60,840 |
| Isolation-By-Distance Mantel test for D. walkeri CC1-CC2 | 9 | 530 | 3761 |
| Isolation-By-Distance Mantel test for D. walkeri CC3-CC4-SW | 24 | 858 | 18,356 |
| G-PHOCS demographic analyses - D. beccarii EP | 10 | 667 | 696,314 |
| G-PHOCS demographic analyses - D. beccarii ECC | 11 | 667 | 696,314 |
| G-PHOCS demographic analyses - D. beccarii SEP | 8 | 667 | 696,314 |
| G-PHOCS demographic analysis of D. spilonotus NNP | 6 | 667 | 696,314 |
| G-PHOCS demographic analysis of D. spilonotus CNP | 2 | 667 | 696,314 |
| G-PHOCS demographic analysis of D. spilonotus WNP | 11 | 667 | 696,314 |
| G-PHOCS demographic analysis of D. spilonotus WCC | 4 | 667 | 696,314 |
| G-PHOCS demographic analysis of D. walkeri CC1 | 5 | 667 | 696,314 |
| G-PHOCS demographic analysis of D. walkeri CC2 | 4 | 667 | 696,314 |
| G-PHOCS demographic analysis of D. walkeri CC3 | 10 | 667 | 696,314 |
| G-PHOCS demographic analysis of D. walkeri CC4 | 6 | 667 | 696,314 |
| G-PHOCS demographic analysis of D. walkeri SW | 8 | 667 | 696,314 |
| Concatenated ML analysis of exome-capture and UCE sequence data | 108 | 1089 | 916,052 |
| ASTRAL-III summary MSC phylogenetic analysis | 108 | 1089 | 916,052 |
| StarBEAST2 MSC phylogenetic analysis all taxa included | 41 | 612 | 755,518 |
| StarBEAST2 MSC phylogenetic analysis w/out D. spilonotus WCC & WNP | 35 | 612 | 755,518 |
| StarBEAST2 MSC phylogenetic analysis w/out D. walkeri CC1-CC2 | 38 | 612 | 755,518 |
| ABBA-BABA (number of loci informative for ABBA-BABA comparison) | 28 | 1041 (267) | N/A |
| SNaQ summary MSci phylogenetic analysis | 41 | 999 | 737,452 |
| BPP version 4.3.8 full MSci phylogenetic analysis | 97 | 683 | 765,923 |
Second, we generated a 50-SNP dataset using the Sequenom platform for a subset of 370 Draco lineatus Group individuals informed by the mitochondrial results available at the time (2011) that again provides fine-scale geographic coverage but for a larger number of loci (see Fig. S1 for sampling map and Supplemental Appendix B for specimen information). We identified SNPs by first sequencing a reduced representation of the complete genome for the three Sulawesi Draco species on an Illumina sequencer. Enrichment for a subset of the genome was accomplished via restriction digestion of pooled genomic DNA for nine individuals prior to sequencing. The nine pooled individuals were not individually barcoded, so the SNPs selected for inclusion in the study were not known to be diagnostic for any one lineage or species. We initially identified 100 random SNPs that were surrounded by relatively invariant base positions that could serve as multiplex priming sites for screening (Gabriel et al. 2009). Sequenom then used their bioinformatics pipeline to identify batches of 30 and 20 SNPs, respectively, which were simultaneously screened in two successive runs.
Finally, we obtained a more comprehensive multi-locus data set comprising 1089 sequence loci for 104 individual Draco lineatus Group samples (see Fig. S2 for sampling map and Supplemental Appendix B for specimen information) plus two D. bimaculatus samples via exon-capture (Bi et al. 2012, 2013; Jones and Good 2016; Blom et al. 2017). Transcriptome-based exon-capture methods provide datasets ideal for phylogenetic and demographic inference, as they can screen hundreds to thousands of independent loci that can be quite long (>1000 bp) with approximately equal capture efficiency across samples characterized by considerable nuclear sequence divergence (Bragg et al. 2016; Portik et al. 2016). Targeted loci were identified initially via transcriptome sequencing of four individual Draco samples (representing two Draco boschmai, one D. timoriensis, and one D. beccarii/D. walkeri hybrid individual; see Reilly et al. 2022). Following the alignment of the transcriptome sequence data, genes that could be annotated via reference to the Anolis carolinensis genome (Alföldi et al. 2011) were considered for inclusion in our study. An initial chip-based sequence capture experiment undertaken for 95 Draco boschmai and D. timoriensis samples plus individual D. walkeri, D. volans, D. sumatranus, and D. modigliani samples (Reilly et al. 2022) had poor capture efficiency, inspiring a switch to the MYBaits in-solution capture approach (now Arbor Scientific; see Reilly et al. 2022). This transition improved our data set by allowing for the addition of flanking intronic and UTR sequences to the original exon targets. The transition to in-solution capture also allowed us to expand the scope of our capture array, and we added 540 lizard-specific UCE markers (Leaché et al. 2015), bringing the total number of targeted regions to 2040 and the number of targeted loci to 1249. Additional details describing laboratory methods for library preparation, capture reactions, and our bioinformatics pipeline are provided in Supplemental Appendix D.
Phylogenetic Analysis of Mitochondrial Data
The mitochondrial data set was subjected to partitioned Bayesian phylogenetic analysis using MrBayes 3.2.7 (Ronquist et al. 2012), with 5 a priori designated partitions (ND2 codon positions 1, 2, and 3, 12S rRNA+ND2 flanking tRNAs, and 16S rRNA). The most appropriate nucleotide substitution model for each partition was selected using likelihood ratio-tests implemented in MrModelTest (Nylander 2004). Four separate analyses were undertaken, each with Metropolis-coupling (four chains), and 50 million generations. Convergence was assessed by confirming that all four analyses settled on the same tree topology and parameter estimates and that all model parameters had ESS values over 200 using Tracer v1.7.2 (Rambaut et al. 2018). The tree was rooted using the outgroup Draco bimaculatus.
Population Structure
We first evaluated population structure using a Bayesian ancestry estimation clustering procedure implemented in TESS 2.3.1 (Chen et al. 2007), analyzing the Sequenom 50-SNP data set for 370 individual Draco lineatus Group samples with the goal of maximizing geographic coverage. Separate analyses with admixture were run for each of three groups of species (D. beccarii; D. walkeri; and D. spilonotus, D. biaro, D. iskandari, and D. caerulhians). Sample selection for these analyses reflects that we were mostly concerned about testing whether cryptic species might occur in the three Sulawesi species and the fact that the Sangir-Talaud species (D. biaro, D. caerulhians, and D. iskandari are deeply nested in the D. spilonotus clade). Each TESS analysis was undertaken with 100 replicates per K value, 25,000 burn-in cycles, and 50,000 post-burn-in cycles. The optimal K value was assessed by plotting the mean Deviation Information Criterion (DIC) outputs across the 100 runs performed for each K value and then visually inspecting the graph to identify the inflection point at which additional K increments did not improve the DIC as suggested in the program manual.
We then evaluated population structuring using a more extensive exon-capture data set for each of the three currently recognized Draco species on Sulawesi using a combination of approaches including the program STRUCTURE (Pritchard et al. 2000), BFD* (Bayes Factor Delimitation of Species (*with genomic data); Leaché et al. 2014a, b), and BPP (Bayesian Phylogenetics and Phylogeography) version 3 (Yang 2015). These analyses were undertaken as part of an integrative assessment of potential cryptic species implicated by the phylogeographic analyses of the mitochondrial data set.
Separate STRUCTURE analyses were undertaken using: 1) the linkage model with all informative SNPs and 2) using one SNP per locus. For D. beccarii, this corresponded to 23,313 informative SNPs and 1020 SNPs with one per locus, for D. spilonotus we analyzed 19,414 informative SNPs and 848 SNPs with one per locus, and for D. walkeri we analyzed 22,696 informative SNPs and 908 SNPs with one SNP per locus. For each data set, the program was run for 100,000 generations as burnin, followed by 100,000 generations for K = 2 through K = 5 populations with 10 replicates per K.
We performed a species delimitation analysis using BFD* (Leaché et al. 2014a, b), a program that makes use of the SNAPP (SNP and AFLP Package for Phylogenetic analysis) phylogenetic analysis computational machinery (Bryant et al 2012). As with SNAPP, the BFD* machinery requires SNP data, so we randomly selected one informative SNP per locus for analysis and default priors. The data sets were composed of 1020, 848, and 908 SNPs for D. beccarii, D. spilonotus, and D. walkeri, respectively, again reflecting the number of loci for which informative SNPs were detected for each species. We used the mitochondrial tree as a user-specified guide-tree to establish a priori species delimitation models, with up to three species compared for D. beccarii, up to four species for D. spilonotus, and up to five species for D. walkeri.
We also performed a species delimitation analysis using BPP version 3, a species delimitation/population structuring approach based on the multispecies coalescent (Rannala and yang 2003). The program allows for the analysis of aligned sequence data, and we ran a series of analyses for each species culminating in 500-locus analyses with all loci selected from the transcriptome-based exon capture fraction of our data set. Due to computational constraints, 500 was the largest number of loci for which we could efficiently run analyses, and we opted to use the top 500 loci ranked first by length and second by having the lowest percentage of missing data. Although BPP can simultaneously estimate the species tree and delimit species, we opted to fix the species tree so as to reduce the computational burden on the species delimitation component (thereby allowing inclusion of more loci). Ten separate 20,000-cycle analyses were run for each of the three focal species.
Isolation-By-Distance
We tested for IBD employing Mantel tests on matrices of Edwards’ genetic distances (Nei 1972) and Euclidean geographic distances (calculated in km using the PBSmapping R package; Schnute et al. 2003). Edwards’ genetic distances were calculated from data sets composed of all informative SNPs drawn from the exome and UCE data sets, with separate input files generated independently for each of five sample sets—1) D. beccarii (n = 29, number of SNPs = 60,662), 2) D. spilonotus (n = 23, number of SNPs = 52,030), 3) D. walkeri (n = 33, number of SNPs = 60,840), 4) D. walkeri CC1 and CC2 (Central Core 1, Central Core 2; n = 9, number of SNPs=3761), and 5) D. walkeri CC3, CC4, and SW (Central Core 3, Central Core 4, Southwest Peninsula; n = 24, number of SNPs = 18,356). Separate analyses were run for D. walkeri CC1 and CC2 and for D. walkeri CC3, CC4, and SW because of the substantial genetic divergence between these groups. Genetic distances were calculated for all possible two-sample comparisons using the mantel.randtest function in adegenet (Jombart 2008; Jombart and Ahmed 2011). We ran 100,000 permutations to assess the significance of each plot and to obtain p values. Geographic and genetic distances were then plotted against each other for each sample pair. Two-dimensional kernel density estimation of local point density was utilized to produce a color scale over the points using the R package MASS (Ripley and Venables 2002).
Demographic Analyses
We performed a series of demographic analyses to assess effective population sizes of extant and ancestral populations, timing of divergences, and rates of migration between putatively distinct lineages. These analyses were undertaken using the program g-phocs (Gronau et al. 2011), which is an isolation-with-migration program that can accommodate unphased genomic sequence data from unlinked neutrally evolving loci. We performed analyses on two different subsets of our larger genomic data set. For the first set of analyses, we utilized flanking sequences from each locus because these regions are more likely neutral and expected to be similarly influenced by the evolutionary history and demography of Draco populations (Luikart et al. 2003). After filtering, 518 flanking region (non-coding) loci were retained for analysis. We also performed analyses on a second data set composed of exons plus flanking sequences for 667 loci (the “allExons” data). We ran the second set of analyses despite likely violations of neutrality specifically for scaling the Theta, Tau, and migration estimates because we used a genome-wide mutation rate to convert unscaled parameter estimates to real-world values for the “allExons” markers (see below) and we had no firm basis for estimating the mutation rate for the flanking sequences. We performed test analyses using several priors for Theta and Tau parameters and because these had little impact on the resulting estimates settled on the broad prior of 1,10,000 for the tau-theta-alpha and tau-theta-beta gamma distribution attributes in the final analyses. Each population comparison was run for 500,000 generations discarding 10% as burnin after visually checking the parameter traces for convergence. Run outputs were visualized in tracer (Rambaut et al. 2018) to assess the posterior distributions of the parameters.
Demographic parameter estimates were converted to rough estimates of real-world values by applying a mutation rate of 1 × 10–9 (see Time Calibration below for justification). Estimates of effective population sizes (individuals) were obtained by dividing the Theta estimate by the mutation rate, then dividing that value by 4. The population divergence time in years was calculated by dividing the Tau estimate by the mutation rate. Migration rate estimates were converted to migrants per generation (2Nm) by multiplying the migration estimate by Theta and dividing by 4 (Gronau et al. 2011). Based on life history data from the related species Draco spilopterus (Alcala 1967), a generation time of one year was assumed for these conversions.
Phylogenomic Analyses
A concatenated alignment (916,052 bp) of exon-capture and UCE sequence data (683 exon-capture loci and 406 UCE loci that passed filtering) was subjected to an unpartitioned Maximum Likelihood phylogenetic analysis using IQ-TREE under the GTR + F + G4 model of sequence evolution (Minh et al. 2013). Nodal support was assessed with 1000 nonparametric bootstrap replicates. Individual gene trees for each of the 1089 loci were estimated using raxml (Stamatakis 2014) to serve as input files for the summary multispecies coalescent species tree approach implemented in astral-III (Mirarab et al. 2014). We opted to use ASTRAL-III for these analyses rather than an alternative summary method based on its good performance even with substantial incomplete lineage sorting in simulation studies (Shi and Yang 2018). For the astral-III analysis, a priori species were designated based on the results of population demographic analyses (see below).
We sought to obtain a calibrated time-tree estimate using full multispecies coalescent phylogenetic analyses, which can glean more signal from genomic data than can summary methods (Xu and Yang 2016; Jiao et al. 2021). We, therefore, performed exploratory 100-locus, time-calibrated species tree estimation on fixed species trees using BPP version 4. The results of these analyses (not shown) brought a number of important issues to our attention, including that two lineages included in these analyses were behaving as “rogue taxa” (Wilkinson 1996; Aberer et al. 2013) in a manner suggesting that ancient hybridization was confounding any analysis that simultaneously included both lineages. This result inspired several additional analyses, including full multispecies coalescent calibrated time-tree analyses of 612-locus data sets using StarBEAST2 version 2.5.2 (Ogilvie et al. 2017). The 612-locus data sets included all transcriptome-derived loci greater than 250 bp in length (but did not include any of the 406 retained UCE loci). We preferentially selected the transcriptome-derived markers over the UCE markers because they are on average ~3 times longer (1121 bp versus 367 bp) and substantially more variable (440 versus 52 SNPs per locus) in a dataset that includes all Draco species plus agamid outgroups (McGuire unpublished data). Because these analyses are computationally expensive, we reduced the scope of our data set to include 2 or 3 individuals per lineage (35, 38, and 40 exemplars in total depending on the analysis). StarBEAST2 analyses were based on the HKY+G nucleotide substitution model and an uncorrelated lognormal clock model with the clock rate set to 0.001 substitution/site/MY (Brandley et al. 2011; Blom et al. 2017; Allio et al. 2017).
To test for a signature of ancient hybridization we performed four sets of analyses. First, to test if including the “rogue” lineages discussed above influenced the overall structure of the inferred tree, we performed separate StarBEAST2 analyses with all species included, as well as with each of the two rogue lineages excluded. Second, we calculated D statistics (ABBA-BABA; Green et al. 2010; Durand et al. 2011) using modified versions of the scripts Pattern_By-Site.R and CalcD.R (Streicher et al. 2014; Blackmon and Adams 2015; scripts available here: https://github.com/libbybeckman/ABBA-BABA) and 10,000 bootstrap replicates for estimation of Z statistics and P values. ABBA-BABA analyses were run four times, each with a different set of individuals representing the species Draco spilonotus WCC (Western Central Core, pop1), D. spilonotus WNP (Western portion of Northern Peninsula, pop2), D. walkeri CC1+CC2 (pop3), and D. beccarii ECC-SEP (Eastern Central Core + SE Peninsula, pop4), with the D. spilonotus lineages (pops 1 and 2) representing sister lineages, and D. walkeri CC1+CC2 (pop3) the test lineage, and D. beccarii ECC-SEP (pop4) the outgroup. Third, we used SNaQ—a summary multispecies coalescent with introgression (MSci) method implemented in the PhyloNetworks package (Solís-Lemus et al. 2017)—to simultaneously estimate the species tree while detecting signatures of past hybridization. For this analysis, we first estimated 41-sample gene trees using RAxML for 999 sequence loci (639 exome and 360 UCE sequence loci that met RAxML requirements for the presence of outgroup sequences). ASTRAL-III was then used to calculate concordance factors for 101,271 species-level quartets obtained from the 999 gene trees. Finally, the concordance factors and mapping file indicating the a priori designated species for each of the 41 samples included in the analysis were analyzed using SNaQ (50 independent runs) to produce a species tree with an inferred hybridization band. Fourth, we used BPP version 4.3.8 (Flouri et al. 2020) to implement the full MSci model using a 683-locus transcriptome-derived data set for 97 individuals (excluding 13 individuals collected at or near a narrow D. walker–D. beccarii hybrid zone). These analyses were run with the species tree fixed to match the StarBEAST2 spliced tree (see Results below) and with three a priori designated alternative hybridization bands (one hybridization band per analysis; with one band connecting D. walkeri CC1 + CC2 and D. spilonotus WCC, one band connecting D. walkeri CC1 + CC2 and D. spilonotus WNP, and one band connecting D. walkeri CC1+CC2 and the common ancestor of D. spilonotus WCC + D. spilonotus WNP). The BPP 4.3.8 MSci analysis is a full (rather than summary) implementation of the MSci and provides time calibration of the a priori designated species tree with estimates of both the timing and extent of gene flow during the past hybridization event, as well as ancestral theta estimates for all nodes. These analyses were run under the HKY nucleotide substitution model, with two sets of priors. Two analyses were run for each of four prior combinations (BPP settings: thetaprior = 3 0.002 e and 3 0.004 e; tauprior = 3 0.002 and 3 0.004; phiprior = 1 1). Individual analyses were run for 1,000,000 generations with a sample frequency of 2 and a burn-in of 100,000 generations.
Time Calibration
Time calibration is often a thorny issue for both phylogenetic and population genetic analyses, particularly when appropriate fossils for calibration purposes are lacking (e.g., Schenk 2016). Unfortunately, this challenge applies to the present study as fossil calibrations are unavailable for the draconine agamids (the major clade that encompasses most of the diverse radiation of SE Asian agamids, including Draco). We first attempted to calibrate our time trees using secondary calibrations derived from a study of iguanian lizard phylogenetics by Townsend et al. (2011), a study which estimated an iguanian time tree using 29 nuclear loci and 11 fossil lizards for calibration purposes. Townsend et al. (2011) included a selection of agamids including Physignathus cocincinus, Pogona vitticeps, Chlamydosaurus kingi, and Ctenophorus isolepis, providing node age estimates for this assemblage. Consequently, we collected exon-capture data for these same taxa for inclusion as outgroups in our analyses. Initial 612-locus StarBEAST2 analyses fit truncated lognormal distributions with median node ages for this four-taxon outgroup assemblage as follows: 31.9 Ma, 18.7 Ma, and 15.0 Ma (taken from Townsend et al. 2011). However, the time-calibrated species trees inferred using these secondary calibrations returned trees with what appeared to be highly compressed branch lengths that were at odds with the inferred branch lengths obtained from uncalibrated analyses of the same data. Speculating that our analysis might have been confounded by interspecific gene flow, we next performed a similar set of analyses using a set of 17 Draco species that mostly reside outside of the Draco lineatus Group (just two D. lineatus Group species were included) which we deemed unlikely to be impacted by gene flow and for which phylogenetic estimation appears more straightforward (based on the observation that a diversity of analytical methods settle on the same topology, McGuire unpublished data). These analyses returned similarly compressed branch lengths that were again at odds with branch length estimates obtained in uncalibrated analyses of the same data. Finally, we opted to estimate a mutation rate for 612 exon-capture loci by calculating the mutation rate that would return the Townsend et al. (2011) node ages given the unscaled ultrametric StarBEAST2 tree. Utilizing a mutation rate to calibrate species time-tree estimates has the added benefit of also being suitable for converting unscaled effective population size and time-since-divergence estimates obtained in population demographic analyses into approximate real-world values. Separate mutation rates were estimated for each of the three nodes shared between our analysis and the Townsend et al. (2011) study, which returned median mutation rates of 0.867 × 10–9, 1.121 × 10–9, and 0.932 × 10–9 substitutions/site/year, respectively, with an average of the three equaling 0.973 × 10–9 substitutions/site/year, an estimate that is close to the nuclear DNA mutation rate of 1.0 × 10–9 substitutions/site/year used in a number of recent studies of squamate diversification (see Brandley et al. 2011; Allio et al. 2017; Blom et al. 2019; Reilly et al. 2022). Acknowledging the imprecision inherent in time calibration, we opted to calibrate time-tree and population demographic analyses using the standard mutation rate of 1.0 × 10–9 substitutions/site/year.
Biogeographic Model Comparison
We employed a Bayesian model-testing approach using the R package BioGeoBEARS (Matzke 2013) to select among alternative biogeographical inference models for Sulawesi and surrounding islands across a 12-million-year time window. The BPP time-tree was used for these analyses, which compared the DEC (dispersal, extinction, cladogenesis), DEC + J (dispersal, extinction, cladogenesis, plus jump dispersal), DIVA-like and DIVA-like +J, BAYAREA, and BAYAREA + J models. The analyses were time-stratified, with six included time slices (12, 6, 4, 3, 2, 1 Ma), and a series of areas-allowed matrices (one for each time slice) that indicated the presence or absence of available land that could be occupied by each species during that time window. We compared likelihoods for the different dispersal models using the AIC test implemented in BioGeoBEARS.
Results
Phylogeographic Analysis of Mitochondrial Sequence Data
The complete mitochondrial sequence alignment included 2337 bp for 613 samples. Bayesian phylogenetic analysis of these data returned a well-resolved and supported phylogenetic estimate for the Draco lineatus Group (Figs. 2 and S3). Several features of this phylogenetic estimate are noteworthy. First, whereas there are 9 currently recognized species in the D. lineatus Group, 18 well-supported geographically cohesive clades were recovered in the phylogenetic analysis (Figs. 2 and S3). All nine additional clades reflect geographic structuring within the three Sulawesi species, with D. beccarii composed of three deeply divergent clades, D. spilonotus composed of four, and D. walkeri composed of five. Each of these groupings is quite distinct, with uncorrected ND2 sequence divergences ranging from 5.4% between D. spilonotus CNP (Central Northern Peninsula) and D. spilonotus ENP (Eastern Northern Peninsula) to 15.8% between D. walkeri CC1 and D. walkeri CC4 (Central Core 4). Furthermore, each of the three described Sulawesi species are found to be paraphyletic relative to other species. For example, the three Sangir-Talaud species (D. biaro, D. caerulhians, and D. iskandari) were found to be nested within D. spilonotus; D. rhytisma and D. lineatus were nested within D. beccarii; and the D. walkeri CC1 clade is more closely related to D. supriatnai, D. spilonotus, and the Sangir-Talaud species than it is to the other four D. walkeri clades (Figs. 2 and S3). The combination of relatively deep genetic sub-structuring and lack of species monophyly indicated the possibility that one or more of these intraspecific clades represent cryptic species.
Sequenom
Our TESS analysis of the 50-SNP Sequenom data set for 370 Draco lineatus Group samples recovered most, but not all, of the mtDNA structure, while indicating some ancestral admixture near points of contact between genetically distinct clusters (Fig. S4). For example, whereas the mitochondrial data set suggests that Draco walkeri represents five divergent clades, the 50-SNP data set did not discriminate between the walkeri CC1 and CC2 groups. Furthermore, the samples comprising the CC3 cluster showed a signature of admixture in the easternmost samples in the vicinity of a known D. walkeri–D. beccarii hybrid zone (McGuire, unpublished). In the case of Draco beccarii, the mitochondrial tree includes three divergent clades but the 50-SNP data set did not differentiate the ECC (Eastern Central Core) and SEP (Southeastern Peninsula) clades, and admixture was detected in samples closest to the ECC-SEP versus EP (Eastern Peninsula) boundary. Both the mitochondrial and 50-SNP data sets agree that there are four D. spilonotus genetic clusters but with disagreement on the precise position of the CNP grouping. Furthermore, admixture was detected between the D. spilonotus WNP and ENP clusters.
Population Structure
Our STRUCTURE, BFD*, and BPP analyses of the exon-capture data are concordant in inferring that each of the three Sulawesi Draco lineatus Group species exhibits intraspecific genetic structuring. However, results were not entirely consistent across the three methods. BFD* returned the maximum number of clusters in each analysis (D. beccarii = 3 clusters, D. spilonotus = 4 clusters, D. walkeri = 5 clusters) and thus matched phylogeographic results obtained with the mitochondrial sequence data (Table S1). BPP v3 also found the maximum number of clusters in all 10 replicate runs for both D. spilonotus (4 clusters) and D. walkeri (5 clusters), and found the maximum number of clusters (3) in five of the 10 replicate runs for D. beccarii, with the remaining five runs supporting two clusters for this species (D. beccarii EP and D. beccarii ECC-SEP). Finally, STRUCTURE provided the most conservative results of the three methodologies (Fig. 3), returning K = 2 clusters for D. beccarii (D. beccarii EP and D. beccarii ECC-SEP), K = 2 clusters for D. spilonotus (D. spilonotus WNP + WCC [Western Central Core] and D. spilonotus CNP + ENP), and K = 3 or K = 4 clusters for D. walkeri (depending on interpretation). The first D. walkeri STRUCTURE run found strong support for K = 2 clusters, dividing the full set of samples into a D. walkeri CC1 + CC2 cluster and a D. walkeri CC3 + CC4 + SW [Southwest Peninsula] cluster. This reflected a very deep split in the genetic data and, therefore, independent analyses were run for D. walkeri CC1 + CC2 and for D. walkeri CC3+CC4+SW. The D. walkeri CC1 + CC2 analyses returned two well-supported clusters but the composition of the clusters did not match the mitochondrial CC1 and CC2 groupings. Draco walkeri CC3 + CC4 + SW analyses clearly identify D. walkeri SW as a distinct cluster, but return somewhat ambiguous results for CC3 and CC4 that are consistent with two clusters experiencing extensive admixture (Fig. 3).
Figure 3.
Maps depicting population demographic structure in (A) Draco spilonotus, (B) D. walkeri, and (C) D. beccarii with sampling localities color-coded by mitochondrial haplotype. The bar graphs reflect genetic clustering based on STRUCTURE analysis of the exon-capture data set under the linkage model (based on 19414, 22696, and 23313 SNPs, respectively). At putative cryptic species boundaries, bi-directional migration rates and time since divergence estimates based on G-PHocS demographic analyses are presented. The lines extending from the bar graphs to the sampling locations indicate which samples were included in both the STRUCTURE and G-PHocS analyses. Gray lines indicate samples that show little to no admixture and black lines point to more extensively admixed samples.
Isolation-By-Distance
Analyses of IBD showed mixed evidence of isolation-by-distance and population structure, indicating that IBD predominates within otherwise structured lineages (see Fig. S5). When all five D. walkeri mitochondrial groupings were analyzed together (Fig. S5 panel A), there was a little signature of IBD (p = 0.317), but the analysis restricted to D. walkeri CC1 and CC2 shows a signature of IBD (Fig. S5 panel B, p = 0.011) and the analysis restricted to D. walkeri CC3, CC4, and SW is indicative of two distinct clusters each with IBD (Fig. S5 panel C, p < 0.001). The results for D. spilonotus suggest IBD over shorter distances, and less IBD over longer distances consistent with the linear nature of the geographic distribution of this species that is divided into 2–4 distinct clusters (Fig. S5 panel D, p < 0.001). The results for D. beccarii are consistent with two distinct clusters each exhibiting limited internal IBD (Fig. S5 panel E, p < 0.001).
Population Demography
The g-phocs analyses returned results consistent with extensive intraspecific structuring of the three Sulawesi species, but do not support the maximum number of geographic clusters (Fig. 3). For Draco walkeri, we interpret the g-phocs results to be consistent with three distinct lineages (D. walkeri CC1-CC2, D. walkeri CC3-CC4, and D. walkeri SW). For D. spilonotus, the g-phocs results are consistent with four distinct lineages that match the mitochondrial groupings (D. spilonotus WCC, WNP, CNP, and ENP). For D. beccarii, the g-phocs results are consistent with two distinct lineages (D. beccarii EP and D. beccarii ECC-SEP). Our interpretation is based on the inferred migration rates between putative lineages and their times since divergence. For comparisons between each inferred distinct lineage, g-phocs detected limited gene flow representing less than one migrant per generation and divergence dates ranged from ~250,000 years (between D. biaro and D. iskandari) to 7.7 million years (between D. walkeri CC1-CC2 and D. walkeri CC3-CC4). The inferred effective population sizes are notably large for each lineage as well as for shared common ancestors (see Table S2).
Phylogenomic Analyses
Maximum likelihood phylogenetic analysis of the concatenated 1089-locus data set using IQ-TREE returned a well-supported, fully resolved tree topology (Fig. S6) that disagrees in several respects from the mitochondrial phylogenetic estimate. For example, the well-supported, highly divergent, non-sister Draco walkeri CC1 and CC2 clades obtained with the mitochondrial data set were recovered as sister clades in the concatenated ML tree. Similarly, the well-supported non-sister D. beccarii ECC and D. beccarii SEP clades recovered in analyses of the mitochondrial data set were not returned in the concatenated RAxML analyses of the phylogenomic data set, and D. beccarii ECC and SEP samples instead formed a single well-supported clade with limited internal structure. Although the RAxML phylogenetic estimate disagreed with the estimate obtained using mitochondrial data in these ways, D. walkeri and D. spilonotus were each found to be paraphyletic as was the case with the mitochondrial data, with the Sangir-Talaud species nested within D. spilonotus, and D. walkeri CC1-CC2 found to be more closely related to D. supriatnai, D. spilonotus, and the Sangir-Talaud species than to the other D. walkeri clades. The ASTRAL-III summary MSC analysis of the 1089-locus data set, which included a priori species designations for all samples, returned a species tree topology (Fig. S7) that agreed broadly with the RAxML tree. Notably, the ASTRAL-III species designations assumed D. walkeri CC1-CC2, D. walkeri CC3-CC4, and D. beccarii ECC-SEP as single lineages as indicated by the RAxML, STRUCTURE, and g-phocs analyses.
A series of BPP v. 3 phylogenetic analyses were undertaken with 100-locus subsets of the exon capture data on fixed tree topologies (beginning initially with the ASTRAL-III topology). These analyses, which were intended to provide the final divergence-dating estimates for this study, returned species tree estimates that exhibited peculiar “compressed” internal branches that we interpreted as reflecting possible disagreement between the phylogenetic signal in the data and the ASTRAL-III fixed tree topologies we were enforcing. Further exploration of the data using BPP indicated that the removal of select taxa (either Draco walkeri CC1-CC2 or D. spilonotus WCC) resulted in species tree estimates with regular (non-compressed) internodes. We suspected this reflected ancient hybridization between the geographically adjacent D. walkeri CC1-CC2 and D. spilonotus WCC lineages (see Fig. 3), which motivated a series of additional analyses. These analyses included application of the ABBA-BABA test, and analyses using SNaQ, which can accommodate both incomplete lineage sorting and reticulation. The ABBA-BABA tests showed a strong signature of past introgression between D. walkeri CC1-CC2 and D. spilonotus WCC with all four sets of samples (D statistics ranged from 0.21 to 0.35; Z scores ranged from 4.0 to 28.3; P values << 0.001). Analyses using SNaQ also inferred that the reticulation event to be between D. walkeri CC1-CC2 and D. spilonotus WCC (Fig. S8).
Full MSC StarBEAST2 analyses of our 612-locus data set were employed to infer the Draco lineatus Group species time tree. Given the inferred past hybridization between D. walkeri CC1-CC2 and D. spilonotus WCC (and possibly D. spilonotus WNP), we performed these analyses with three alternative sampling regimes: (1) complete lineage representation, (2) with D. walkeri CC1-CC2 excluded, and (3) with D. spilonotus WCC and D. spilonotus WNP excluded. The analysis with complete sampling returned a topology similar/identical to the concatenated RAxML and ASTRAL-III topologies, with the three D. walkeri lineages not forming a monophyletic group and D. supriatnai embedded within the D. spilonotus-Sangir-Talaud island group cluster of lineages. However, when D. walkeri CC1-CC2 was excluded from consideration, D. spilonotus WCC and WNP were returned as sister taxa with D. supriatnai placed at the base of the clade that includes the four D. spilonotus lineages plus the three species in the Sangir-Talaud island chain. When D. spilonotus WCC and WNP were excluded, the three D. walkeri lineages were placed as a monophyletic grouping for the first time, and together were placed as the sister group of the clade composed of the two D. beccarii lineages, plus D. lineatus and D. rhytisma.
Having identified a preferred species tree topology by splicing the results of the StarBEAST2 analyses that iteratively excluded Draco walkeri CC1-CC2 and D. spilonotus WCC and WNP, we then fixed this topology in an analysis employing BPP version 4.3.8 and the MSci framework (which requires an a priori fixed topology). BPP v 4.3.8 can provide an optimized species tree estimate with coalescent branch lengths and effective population sizes while also allowing a single a priori designated hybridization band. The program provides estimates of the timing and extent of bi-directional gene flow. We performed analyses with three alternative hybridization bands under a variety of prior settings, each of which involved a band connecting D. walkeri CC1-CC2 to another lineage (to D. spilonotus WCC, to D. spilonotus WNP, and to the branch subtending D. spilonotus WCC and WNP). All three sets of analyses suggested that ancient hybridization was primarily or exclusively between D. walkeri CC1-CC2 and D. spilonotus WCC. The hybridization band involving D. spilonotus WNP was placed immediately above the node representing the common ancestor of D. spilonotus WNP and D. spilonotus WCC, whereas the hybridization band involving the common ancestor of D. spilonotus WCC + D. spilonotus WNP was placed immediately below the node—in each case the inferred placement of the band was as close to the D. spilonotus WCC lineage as permitted by the constraints of the analysis). The preferred Draco lineatus Group time-tree estimate is presented in Figure 4. In this species tree estimate, hybridization between D. walkeri CC1–CC2 and D. spilonotus WCC is estimated to have taken place 1.85 Ma, with 37% of the alleles present in D. spilonotus WCC derived from D. walkeri CC1–CC2 and 26% of the alleles present in D. walkeri CC1–CC2 derived from D. spilonotus WCC.
Figure 4.
a) StarBEAST2 species time tree estimate with all lineages included (612 loci), b) a spliced StarBEAST2 tree combining the relationships inferred from two analyses, one with Draco walkeri CC1-CC2 excluded and one with D. spilonotus WCC excluded (612 loci), c) BPP v 4.3.8 MSci species time tree based on 683 exon-capture loci. The analysis includes a single a priori hybridization band that BPP inferred to have occurred 1.85 Ma. Photo credits: D. beccarii (Robin Moore), D. walkeri (Heidi Rockney), D. spilonotus (Coke Smith).
Biogeographic Model Comparison
Time-stratified BioGeoBears analyses strongly preferred the three models that included a jump-dispersal parameter (DEC + J, DIVA-like + J, BAYAREA + J) over those that did not. The three top models had very similar likelihood scores (LnL = –44.51, –44.18, and –44.99, respectively), and returned virtually identical dispersal histories (differing only in the state inferred for the root node). Figure 5 illustrates the sequence of dispersal and vicariance events inferred via BioGeoBears on the paleo-island reconstructions of Nugraha and Hall (2018). The timing of the inferred dispersal events is quite consistent with the hypothesized availability of land according to the paleo-island ages, with two exceptions. One exception is that the Togian Islands endemic, Draco supriatnai, is inferred to be older (~4.35 Ma; 95% HPD interval 4.24–4.47 Ma) than the Togian Islands (~3 Ma). The second exception is the split between D. beccarii EP and D. beccarii ECC-SEP, which is found to be slightly younger (2.5 Ma) than expected given the 3–8 Ma temporal extent of the hypothesized oceanic barrier.
Figure 5.
Results of the BioGeoBEARS analysis superimposed on the paleo-island reconstructions from Nugraha and Hall (2018). The inferred biogeographical events are reported in temporal sequence across the six panels beginning in the upper left. Arrows represent dispersal events. Dashed bars represent diversification events that could involve either dispersal or vicariance. Numbers adjacent to arrows and dashed bars reflect the temporal order of the dispersal and/or vicariance events.
Discussion
The Draco lineatus complex of Sulawesi and surrounding islands exemplifies the manifold challenges that can impact species delimitation and phylogenomic analysis. Species delimitation efforts are likely to be controversial when cryptic species are involved (Chan et al. 2020, 2022), and this becomes even more problematic for cases such as this one in which independent, cryptic lineages may have subsequently merged or may be in the process of merging following secondary contact. Phylogenomic analyses are rendered suspect when hybridization has occurred (Leaché et al. 2014a, b) and it is clear that some lineages within the D. lineatus Group are not only experiencing contemporary gene flow (at a narrow hybrid zone between D. walkeri CC3–CC4 and D. beccarii ECC-SEP, McGuire unpublished) but also show signatures of ancient hybridization between non-sister lineages as well (e.g., between D. walkeri CC1–CC2 and D. spilonotus WCC). Untangling the diversification history under such a scenario is challenging. This study confronts each of these challenges by attempting to delimit species, estimate a time-calibrated phylogenomic tree under the MSci model that accommodates both incomplete lineage sorting and gene flow and using the recovered estimate to infer the biogeographical history of a clade occupying one of the most tectonically complex regions on Earth—Sulawesi and its surrounding islands.
Species Delimitation—Inferring Cryptic and Arrested Speciation
In this study, we first identified possible species using a phylogeographic analysis of a three-gene, mitochondrial sequence data set with dense sampling across the Draco lineatus Group. Whereas the current taxonomy includes 9 morphologically distinct species (McGuire et al. 2007), the mitochondrial phylogenetic estimate indicates that the D. lineatus Group is actually composed of 18 deeply divergent clades, including the 6 peripheral isolates Draco species on islands surrounding Sulawesi, along with 12 mitochondrial lineages divided among the 3 recognized Draco species on Sulawesi proper. Notably, none of the three currently recognized species on Sulawesi were found to be monophyletic in that analysis. This lack of monophyly along with deep divergences among the 12 Sulawesi clades (uncorrected patristic distances for the ND2 gene range from 5.4% to 15.8%) suggested that each could represent a separate species, with D. walkeri composed of as many as five cryptic species, D. spilonotus composed of as many as four cryptic species, and D. beccarii composed of as many as three cryptic species. Evaluation of the lineage status of these putative cryptic species in the context of our 50-SNP and (up to) 1089 locus genomic data sets using a combination of genetic structure, species delimitation, and population demographic analyses (and applying a 0.5 migrants per generation lineage boundary threshold) surprisingly indicated that most, but not all, of these mitochondrially distinct clades do appear to represent legitimate evolutionary species. Indeed, all six morphologically distinct “peripheral isolates” species (Mayr 1963) are supported as independent lineages, as are nine lineages on Sulawesi proper (six of which are inferred to be cryptic species).
Perhaps more interesting than the documentation that several cryptic Draco species exist on Sulawesi is our finding that three highly divergent mitochondrial boundaries do not seem to reflect cryptic species boundaries (i.e., the mitochondrial breaks between D. beccarii ECC and SEP, D. walkeri CC3 and CC4, and D. walkeri CC1 and CC2). What mechanisms might be responsible for deep mitochondrial divergences that are not indicative of lineage independence? One possibility is that these mitochondrial barriers simply appeared randomly, which has been shown to be plausible via simulation, particularly when migration distances and effective population sizes are small (Irwin 2002). Another possibility is sex-biased dispersal. Extreme female philopatry—a scenario in which females remain within their natal group while males disperse at reproductive maturity to join other groups—has been suggested as the most plausible explanation for mitochondrial breaks within rhesus and pig-tailed macaques (Tosi et al. 2003; Evans et al. 2021). We find both of these mechanisms to be implausible explanations for at least two of the three Draco intraspecific mitochondrial boundaries based on key deviations from model expectations. Both the random- and sex-biased dispersal models predict that mitochondrial breaks will occur in geographically random locations. However, the 10.6% mitochondrial boundary for D. beccarii ECC and SEP is tightly correlated with the Lawonopo Fault, which seems unlikely to be coincidental. Both models also predict that genetic divergence will be restricted to specific regions of the genome—to the mitochondrion for the random model, and to the mitochondrion plus sex chromosomes with a female-biased mode of inheritance (i.e., X or W chromosomes). However, the discrete mitochondrial break between Draco walkeri CC3 and CC4 is accompanied by a clear genomic signature of admixture consistent with two once-distinct lineages now in the process of merging (see Fig. 3). The STRUCTURE analysis, in particular, suggests a broad genomic cline at the junction of two once-separated lineages, with distinct, non-admixed samples remaining at the western and eastern limits of their joint range (Fig. 3). In addition, the remarkably large effective population sizes that we have estimated for all of these Draco lineatus Group lineages (Table S2) are inconsistent with the random model, and there is little ecological evidence for extreme female philopatry in flying lizards (though males may be more dispersive than females, see Alcala 1967; Mori and Hikida 1993).
Assuming the random- and sex-biased dispersal models are unlikely explanations for at least two of the three intraspecific mitochondrial breaks, we here propose a third possible mechanism that is consistent with all three, that of arrested speciation—the (perhaps temporary) merger of once-isolated lineages following secondary contact that nevertheless results in some genes being locked up at the point of secondary contact (e.g., those encoded on the mitochondrial genome). Mitochondrial divergences may have evolved in allopatry and then could be maintained following secondary contact and species merger through the formation and retention of mito-nuclear Dobzhansky–Muller Incompatibilities (DMIs; Dobzhansky 1936; Muller 1942; Orr and Turelli 2001; Burton et al. 2013) involving the mitochondrial genome plus one or a few genes in the nuclear genome that interact with mitochondrially encoded genes (e.g., genes involved in cellular respiration such as those composing the OXPHOS pathway; Rand et al. 2004; Lamb et al. 2018). Mito-nuclear DMIs have been increasingly implicated as a mechanism driving speciation (Burton and Barreto 2012; Burton et al. 2013; Hill 2017), but less stringent incompatibilities could also result in semi-permeable mito-nuclear boundaries where gene flow is limited but not entirely prevented, thereby resulting in the retention of deep mitochondrial boundaries within species (Lima et al. 2019). This hypothesis is testable, for example by assessing the fitness of offspring generated by crossing experiments involving individuals from opposite sides of the mitochondrial boundary, and is a promising direction for future work. We term this hypothetical process “arrested speciation” because the speciation process, which was initiated while the lineages were diverging in allopatry, was terminated when the lineages came back into secondary contact and experienced gene flow but could recommence since the mitochondrial genomes continue to diverge on either side of the zone of secondary contact and could ultimately spawn more DMIs that complete the speciation process.
Taken together, our species delimitation analyses suggest that the Draco lineatus Group is composed of 15 species, including the six currently recognized “peripheral isolates” species on Sulawesi’s surrounding islands, as well as nine species on Sulawesi proper. Two of the currently recognized Sulawesi species, D. beccarii and D. walkeri, are now shown to be morphologically diagnosable monophyletic assemblages composed of two and three cryptic species, respectively. The third currently recognized species, D. spilonotus, is also composed of multiple cryptic species (n = 4) but these four species do not form a monophyletic group. Rather, the two northernmost cryptic lineages, D. spilonotus CNP and ENP together form the sister clade of the Sangir-Talaud assemblage composed of the morphologically distinct D. biaro, D. iskandari, and D. caerulhians.
Phylogenomic Analyses and Ancient Hybridization
We performed a series of phylogenomic analyses of the Draco lineatus complex, which were ultimately geared toward estimating a time-calibrated species tree for the 15 species supported herein. These analyses included Maximum Likelihood inference of the concatenated data set using RAxML, summary multispecies coalescent analyses of the full data set using ASTRAL III, BPP v3 analyses of 100 and 500-locus subsets of the full data, and StarBEAST2 analyses of 612 exon-capture loci. Each of these analyses served its own purpose, with the RAxML analyses aimed at identifying phylogenomic structure across all samples, the ASTRAL-III analyses aimed at providing a summary MSC phylogenomic estimate that included all 105 samples in our exon-capture data set but with species designated a priori, and BPP v3 and StarBEAST2 analyses aimed at utilizing the power of a full MSC approach to estimate a time-calibrated species tree. By performing this diverse series of analyses, we were able to identify an underlying confounding process—ancient hybridization between D. walkeri CC1-CC2 and D. spilonotus WCC—that could have been easily overlooked and which we believe resulted in highly inaccurate phylogenomic estimates in all of the above analyses. Estimating a reliable multispecies coalescent time-tree under these circumstances is challenging. We opted for a two-stage approach involving first estimating two independent species trees using StarBEAST2, each with one of the two lineages involved in ancient hybridization deleted, followed by splicing these trees together. This provided a robust topology but with unreliable branch lengths. We then fixed this topology and employed BPP v 4.3.8 to obtain a more reliable time-tree estimate while accounting for, and indeed quantifying, the scale and timing of ancient hybridization between D. walkeri CC1-CC2 and D. spilonotus WCC. We believe that similar underlying processes could be at play when rogue taxa are found to be confounding phylogenomic analyses in other systems. The discovery that ancient hybridization between non-sister lineages could so dramatically alter the outcomes of our initial phylogenomic analyses using essentially any phylogenetic approach that could not account for that gene flow should serve as a cautionary tale for others engaged in species-level phylogenomic analyses.
Historical Biogeography
The comprehensive phylogenomic time tree for the Draco lineatus Group allows for a detailed biogeographical analysis of Sulawesi taking into consideration the complex tectonic history of the island and its immediate surroundings (Fig. 5). Our analysis indicates that the clade composed of D. bimaculatus and the D. lineatus Group split ~11.4 million years ago. Based on our analysis, it is not possible to infer whether the common ancestor of D. bimaculatus + the D. lineatus Group reached the southern Philippines first and then colonized Proto-Sulawesi or the reverse—either possibility is consistent with our phylogenetic estimate. After an apparent period of stasis, the crown Draco lineatus Group began to diversify ~6.3 Ma, and the series of subsequent branching events are very much consistent with the biogeographic framework that we proposed based on the Nugraha and Hall (2018) paleo-island reconstructions with superimposed fault zones. Indeed, we find that eight of the paleo-island boundaries are geographically concordant with species boundaries, and all but two of the inferred speciation events are temporally concordant as well (Fig. 6). The first temporally incongruent split involves Draco supriatnai on the Togian Islands, a species that appears to be older (~4.4 Ma ± 0.12 Ma) than the islands on which it resides today (~3 Ma), suggesting either that the Togian Islands are older than indicated by their model or that the time tree overestimates the age of this lineage. The second temporally incongruent split occurs between D. beccarii EP and ECC-SE, which is estimated to be slightly younger (2.5 + 0.07 Ma) than the age of the barrier (~3 Ma). We further find that the arrested speciation event involving the D. beccarii ECC and SEP mitochondrial groups very closely corresponds to the position of the Lawanopo Fault, a fault that separates ultramafic rocks to the north and metamorphic rocks to the south (Sukamto 1978, 1990; Natawidjaja and Daryono 2015) and may therefore represent a habitat boundary. It is difficult to assess whether there is a temporal congruence for that split because the mechanism by which the fault may have promoted divergence (and thus any assessment of timing) remains unknown. However, the 10.6% uncorrected ND2 divergence between the D. beccarii SEP and ECC mitochondrial haplotypes, and the mitochondrial gene tree estimate places D. beccarii SEP as the sister lineage of a clade composed of D. beccarii ECC, D. beccarii EP, D. rhytisma, and D. lineatus, together strongly suggest that whatever role the Lawanopo Fault may have played in the arrested speciation event between D. beccarii SEP and D. beccarii ECC must have been at least 3.8 million years ago (the node age for D. beccarii EP, D. beccarii ECC-SEP, D. rhytisma, and D. lineatus common ancestor). Notably, although the Lawanopo Fault has likely been inactive since the late Pleistocene, it appears to be an old system, with 120–250 km of movement along this fault and the parallel Matano-Palu-Koro fault system, which is estimated to have occurred over a duration of 3–8 million years (Natawidjaja and Daryono 2015).
Figure 6.
Biogeographical framework for Sulawesi based on paleo-island history, with temporally congruent Draco speciation events denoted with red dashed bars and the temporally incongruent speciation events represented by green dashed bars. Black arrows denote species boundaries not explained by the paleo-island model.
The framework that we have proposed here can also explain why lineage boundaries are often not perfectly geographically congruent across diverse taxa. For example, the boundaries between Draco spilonotus WNP and D. walkeri CC1-CC2, Macaca tonkeana and M. hecki, and the mitochondrial break in Ingerophrynus celebensis each occur near the base of the Northern Peninsula but are offset from one another (see Evans et al. 2003c). If lineages that began diverging in isolation following stepping-stone colonization of paleo-islands were brought into secondary contact following island merger, there are multiple possible outcomes including genetic homogenization, sympatric co-occurrence, and the formation of hybrid zones (including tension zones). In the case of tension zones, theory predicts that they are likely to settle in density troughs (Barton and Hewitt 1985), and the nature and position of such density troughs can vary between taxa and across time (Wielstra 2019). Given Sulawesi’s tectonic history, we suspect that a substantial component of in situ speciation on Sulawesi occurred on the paleo-islands, and that tension zone formation following paleo-island merger explains similar, but not entirely geographically congruent, parapatric species boundaries such as those that we see in Sulawesi flying lizards and macaque monkeys.
Although our geological model appears to explain many Draco lineatus Group divergence events, there nevertheless are several speciation events within this clade that remain unexplained. For example, the geographic distribution of D. spilonotus CNP corresponds on its western margin to the Gorontalo Fault and depression, but this lineage’s small range to the east of the Gorontalo Fault and its boundary with D. spilonotus ENP has no obvious geological explanation of which we are aware. The most prominent unexplained divergences are within Sulawesi’s Central Core, where we presently cannot offer geological explanations for the splits between D. spilonotus WCC and D. walkeri CC1-CC2, or between D. spilonotus WCC and D. walkeri CC3-CC4 (D. walkeri CC1-CC2 and D. walkeri CC3-CC4 are separated by a continuous mountainous ridge that is 1800 m or more in elevation for nearly its entire length). Furthermore, we cannot explain the initial divergences that resulted in the mitochondrially distinct D. walkeri CC1 and CC2 populations that merged or the D. walkeri CC3 and CC4 populations that are in the process of merging now. The biogeographical processes driving divergence in the Central Core remain enigmatic, and we consider it quite possible that the extremely dynamic tectonic processes that have driven extensive uplift and erosion in the Central Core have erased the geological signatures required to reconstruct the events that might have contributed to lineage divergences. Nevertheless, in this first analysis linking a well-supported phylogenomic time tree to an explicit geological model describing the paleo-island history of Sulawesi, we find it reassuring that there is substantial congruence between inferred speciation events and the tectonic history of Wallace’s “remarkable island.”
Conclusions
This study highlights many of the challenges likely to be faced when conducting species delimitation and phylogenetic analysis at and below the species level, particularly for complex assemblages likely to be characterized by secondary contact, gene flow, and cryptic speciation. Such studies are likely to require an integrative approach such as the one utilized here. Cryptic speciation is a controversial topic in part because of disagreement over what constitutes a species. But even among researchers who are like-minded in terms of their preferred species concept—for example, devotees of the Evolutionary or General Lineage Species Concepts—there is still plenty of room for disagreement in terms of what constitutes sufficient evidence of lineage status (e.g., Burbrink and Ruane 2021; Hillis et al. 2021; Burbrink et al. 2022; Chambers et al. in press). Here, we present evidence of potential cryptic lineages from phylogenetic analyses of mitochondrial and exome-capture sequence data and then attempt to test for lineage status using a combination of genetic clustering and population demographic analyses. We argue here that if a morphological character difference diagnosing two lineages can be assumed to be evidence of a barrier to gene flow, as is routinely assumed in traditional species descriptions, then a direct analysis of migration indicating a strong barrier to gene flow should also suffice. Based on population genetic theory, we opted for a threshold of 0.5 effective migrants per generation as providing sufficient evidence for a lineage boundary. Although others may opt for different threshold criteria, we believe that quantifying gene flow—even while acknowledging that obtaining reliable estimates of migration is challenging—is a path forward in resolving the cryptic species problem.
In this study, we not only found evidence of limited gene flow between contemporary cryptic species but also found evidence of ancient hybridization that severely confounded our phylogenomic estimates. The underlying problem was not an easy one to identify or recognize, but its effect on our phylogenomic analyses brought to mind the effect of so-called “rogue taxa” (Wilkinson 1996; Aberer et al. 2013), as removal of either D. walkeri CC1-CC2 or D. spilonotus WCC radically altered the recovered topology. Because multispecies coalescent analyses are computationally challenging, it is perhaps unreasonable to expect researchers to perform a thorough set of analyses in which individual taxa are systematically deleted in an effort to root out the phenomenon uncovered here, but a signature of ancient hybridization is certainly an appropriate thing to look for when the inferred tree provides surprising results—such as when Draco walkeri CC1-CC2 was returned as distantly related to D. walkeri CC3-CC4 and D. walkeri SW despite being morphologically indistinguishable.
Finally, this study provides the first biogeographical analysis of Sulawesi using phylogenomic and population genomic data sets paired with a tectonically explicit biogeographical framework. Our analyses found strong congruence between Draco lineatus Group diversification on Sulawesi and its surrounding islands with the paleo-island history of Sulawesi, and also found additional evidence that faults on Sulawesi are likely important in the history of Sulawesi biological diversification as did Merker et al. (2009) in their study of tarsiers. The framework we have proposed is likely to explain patterns of speciation more generally on Sulawesi—an island characterized by substantial in situ diversification. We note, however, that future researchers should expect general agreement with the framework even if species contact zones are not perfectly congruent. The paleo-island history of Sulawesi is such that many lineages likely diverged in allopatry following stepping-stone colonization of what was once an archipelago of islands, and the subsequent amalgamation of those paleo-islands initiated dynamic species interactions. Only those lineages that failed to achieve sympatry and avoided a process of competitive replacement following contact are likely to show the parapatric species distribution pattern seen in flying lizards, monkeys, and numerous other lineages. These parapatric lineage boundaries are likely to represent tension zones which are expected to settle in density troughs, the geographic positions of which may very well differ between species based on their ecological characteristics. Thus, we predict that many species boundaries across diverse taxa will be found in the same region, but they are unlikely to be perfectly congruent because the mechanism that drove initial diversification likely has long since ceased to exist.
Supplementary Material
Data available from the Dryad Digital Repository: http://dx.doi.org/10.6078/dryad.D1S99X.
Acknowledgments
Three anonymous reviewers, the Editor, and the Associate Editor provided suggestions that substantially improved the work. The authors thank Umilaela Arifin, Jarot Arizona, David Bickford, Luke Bloch, Boy, Frederick Chain, Tom Devitt, Ari Ekapratama, Rosita Elianur, CJ Hayden, Mistar Kamsi, Ben Karin, Tri Wahyu Laksono, Ruud de Lang, Adam Leaché, Mumpuni, Gilang Ramadhan, Ferdi Rangkuti, Heidi Rockney, Abdul Rosyid, Dan Scantlebury, Iqbal Setiadi, and Ted Townsend for their help with the field collection and preparation of specimens and tissues. The authors thank CJ Hayden and Charles Linkem for assistance with molecular lab work, Lydia Smith for laboratory support, Ke Bi for bioinformatics support, and Carol Spencer for accession and curation of specimens. Fieldwork was performed under research permits issued by the Indonesian Institute of Sciences (LIPI) and RISTEKDIKTI, the Philippine Department of Environment and Natural Resources Biodiversity Management Bureau, as well as AUP #R279 issued by the U.C. Berkeley IACUC.
Contributor Information
Jimmy A Mcguire, Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720, USA; Department of Integrative Biology, University of California, Berkeley, CA 94720, USA.
Xiaoting Huang, College of Marine Life Sciences, Ocean University of China, No. 5 Yushan Road, Qindao, Shandong, 266003, PR China.
Sean B Reilly, Department of Ecology and Evolutionary Biology, University of California, Santa Cruz, CA 95060, USA.
Djoko T Iskandar, School of Life Sciences and Technology, Institut Teknologi Bandung, Bandung, Indonesia.
Cynthia Y Wang-Claypool, Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720, USA; Department of Integrative Biology, University of California, Berkeley, CA 94720, USA.
Sarah Werning, Department of Anatomy, Des Moines University, 3200 Grand Avenue, Des Moines, IA 50312-4198, USA.
Rebecca A Chong, Department of Biology, University of Hawaii at Manoa, Honolulu, HI 96822, USA.
Shobi Z S Lawalata, Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720, USA; Department of Integrative Biology, University of California, Berkeley, CA 94720, USA; United in Diversity Foundation, Jalan Hayam Wuruk, Jakarta, Indonesia.
Alexander L Stubbs, Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720, USA; Department of Integrative Biology, University of California, Berkeley, CA 94720, USA.
Jeffrey H Frederick, Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720, USA; Department of Integrative Biology, University of California, Berkeley, CA 94720, USA.
Rafe M Brown, Biodiversity Institute and Department of Ecology and Evolutionary Biology, 1345 Jayhawk Blvd., University of Kansas, Lawrence, KS 66045, USA.
Ben J Evans, Biology Department, McMaster University, Hamilton, Ontario, Canada.
Umilaela Arifin, Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720, USA; School of Life Sciences and Technology, Institut Teknologi Bandung, Bandung, Indonesia; Center for Taxonomy and Morphology, Zoologisches Museum Hamburg, Leibniz Institute for the Analysis of Biodiversity Change, Martin-Luther-King-Platz 3, R230 20146 Hamburg, Germany.
Awal Riyanto, Laboratory of Herpetology, Museum Zoologicum Bogoriense, Research Center for Biosystematics and Evolution, National Research and Innovation Agency of Indonesia (BRIN), Cibinong 16911, Indonesia.
Amir Hamidy, Laboratory of Herpetology, Museum Zoologicum Bogoriense, Research Center for Biosystematics and Evolution, National Research and Innovation Agency of Indonesia (BRIN), Cibinong 16911, Indonesia.
Evy Arida, Research Center for Applied Zoology, National Research and Innovation Agency of Indonesia (BRIN), Cibinong 16911, Indonesia.
Michelle S Koo, Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720, USA.
Jatna Supriatna, Department of Biology, Institute for Sustainable Earth and Resources (I-SER), Gedung Laboratorium Multidisiplin, and Research Center for Climate Change (RCCC-UI), Gedung Laboratorium Multidisiplin, Faculty of Mathematics and Natural Sciences, Universitas Indonesia, Depok 16424, Indonesia.
Noviar Andayani, Department of Biology, Institute for Sustainable Earth and Resources (I-SER), Gedung Laboratorium Multidisiplin, and Research Center for Climate Change (RCCC-UI), Gedung Laboratorium Multidisiplin, Faculty of Mathematics and Natural Sciences, Universitas Indonesia, Depok 16424, Indonesia.
Robert Hall, SE Asia Research Group (SEARG), Department of Earth Sciences, Royal Holloway University of London, Egham, Surrey TW20 0EX, UK.
Conflict of Interest statement
The authors declare no conflict of interest.
Funding
This work was supported by the Museum of Vertebrate Zoology, the Department of Integrative Biology at the University of California at Berkeley, and the National Science Foundation (DEB-1258185, DEB-1652988, DEB-1457845, DEB-0640967, DEB-0328700 awarded to J.A.M. and DEB 0640737 issued to RMB). This study utilized the Vincent J. Coates Genomic Sequencing Laboratory at U.C. Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303.
DATA AVAILABILITY STATEMENT
Sequence data for the ND2, 12S, and 16S mitochondrial genes have been submitted to the GenBank database under accession numbers OQ598699–OQ599306 (ND2), OQ628522-OQ629047 (12S), and OQ624360-OQ624885 (16S). Sequence data from the exon-capture experiment were submitted to the NCBI sequence read archive under BioSample accession numbers SAMN33714970-SAMN33715073. Software input files for the mtDNA data set and multiple versions of the exome-capture data (full data nexus file, StarBEAST2 data sets for 35, 38, and 41 samples, and BPP 4.3.8 input files for 683 loci and 97 individuals) are available from the Dryad data repository (doi:10.6078/D1S99X).
References
- Aberer A.J., Krompass D., Stamatakis A.. 2013. Pruning rogue taxa improves phylogenetic accuracy: an efficient algorithm and webservice. Syst. Biol. 62:162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achmadi A.S., Esselstyn J.A., Rowe K.C., Maryanto I., Abdullah M.T.. 2013. Phylogeny, diversity, and biogeography of Southeast Asian spiny rats (Maxomys). J. Mammal. 94:1412–1423. [Google Scholar]
- Alcala A.C. 1967. Population biology of the “flying” lizard, Draco volans, on Negros Island, Philippines. Nat. Appl. Sci. Bull. 20:335–372. [Google Scholar]
- Alföldi J., Di Palma F., Grabherr M., Williams C., Kong L., Mauceli E., Russell P., Lowe C.B., Glor R.E., Jaffe J.D., Ray D.A., Boissinot S., Shedlock A.M., Botka C., Castoe T.A., Colbourne J.K., Fujita M.K., Moreno R.G., ten Hallers B.F., Haussler D., Heger A., Heiman D., Janes D.E., Johnson J., de Jong P.J., Koriabine M.Y., Lara M., Novick P.A., Organ C.L., Peach S.E., Poe S., Pollock D.D., de Queiroz K., Sanger T., Searle S., Smith J.D., Smith Z., Swofford R., Turner-Maier J., Wade J., Young S., Zadissa A., Edwards S.V., Glenn T.C., Schneider C.J., Losos J.B., Lander E.S., Breen M., Ponting C.P., Lindblad-Toh K.. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477:587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allio R., Donega S., Galtier N., Nabholz B.. 2017. Large variation in the ratio of mitochondrial to nuclear mutation rate across animals: implications for genetic diversity and the use of mitochondrial DNA as a molecular marker. Mol. Biol. Evol. 34:2762–2772. [DOI] [PubMed] [Google Scholar]
- Barley A.J., White J., Diesmos A.C., Brown R.M.. 2013. The challenge of species delimitation at the extremes: diversification without morphological change in Philippine sun skinks. Evolution 67:3556–3572. [DOI] [PubMed] [Google Scholar]
- Barton N.H., Hewitt G.M.. 1985. Analysis of hybrid zones. Annu. Rev. Ecol. Evol. Syst. 16:113–148. [Google Scholar]
- Bi K., Linderoth T., Vanderpool D., Good J.M., Nielsen R., Moritz C.. 2013. Unlocking the vault: next-generation museum population genomics. Mol. Ecol. 22:6018–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi K., Vanderpool D., Singhal S., Linderoth T., Moritz C., Good J.M.. 2012. Transcriptome-based exon capture enables highly cost-effective comparative genomic data collection at moderate evolutionary scales. BMC Genom. 13:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickford D., Lohman D.J., Sodhi N.S., Ng P.K.L., Meier R., Winker K., Ingram K.K., Das I.. 2007. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22:148–155. [DOI] [PubMed] [Google Scholar]
- Blackmon, H., Adams R. H... 2015. evobiR: comparative and population genetic analyses. R Package version 1. [Google Scholar]
- Blom M.P.K., Bragg J.G., Potter S., Moritz C.. 2017. Accounting for uncertainty in gene tree estimation: summary-coalescent species tree inference in a challenging radiation of Australian lizards. Syst. Biol. 66:352–366. [DOI] [PubMed] [Google Scholar]
- Blom M.P.K., Matzke N.J., Bragg J.G., Arida E., Austin C.C., Backlin A.R., Carretero M.A., Fisher R.N., Glaw F., Hathaway S.A., Iskandar D.T., McGuire J.A., Karin B.R., Reilly S.B., Rittmeyer E.N., Rocha S., Sanchez M., Stubbs A.L., Vences M., Moritz C.. 2019. Habitat preference modulates trans-oceanic dispersal in a terrestrial vertebrate. Proc. R. Soc. B. 286:20182575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer A.J., Duffels J.P.. 1996. Historical biogeography of the cicadas of Wallacea, New Guinea and the West Pacific: a geotectonic explanation. Palaeogeogr. Palaeoclimatol. Palaeoecol. 124:153–177. [Google Scholar]
- Bouckaert R., Heled J., Kühnert D., Vaughan T., Wu C.-H., Xie D., Suchard M.A., Rambaut A., Drummond A.J.. 2014. BEAST 2: A Software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10:e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg J.G., Potter S., Bi K., Moritz C.. 2016. Exon capture phylogenomics: efficacy across scales of divergence. Mol. Ecol. Resour. 16:1059–1068. [DOI] [PubMed] [Google Scholar]
- Brandley M.C., Wang Y., Guo X., Montes de Oca A.N., Fería-Ortíz M., Hikida T., Ota H.. 2011. Accommodating heterogenous rates of evolution in molecular divergence dating methods: an example using intercontinental dispersal of plestiodon (eumeces) lizards. Syst. Biol. 60:3–15. [DOI] [PubMed] [Google Scholar]
- Bridle J.R., Garn A-K., Monk K.A., Butlin R.K.. 2001. Speciation in Chitaura grasshoppers (Acridae: Oxyinae) on the island of Sulawesi: colour patterns, morphology and contact zones. Biol. J. Linn. Soc. 72:373–390. [Google Scholar]
- Bryant D., Bouckaert R., Felsenstein J., Rosenberg N.A., RoyChoudhury A.. 2012. Inferring species trees directly from biallelic genetic markers: bypassing gene trees in a full coalescent analysis. Mol. Biol. Evol. 29:1917–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbrink F.T., Bernstein J.M., Kuhn A., Gehara M., Ruane S.. 2022. Ecological divergence and the history of gene flow in the Nearctic milksnakes (Lampropeltis Triangulum complex). Syst. Biol. 71:839–858. [DOI] [PubMed] [Google Scholar]
- Burbrink F.T., Ruane S.. 2021. Contemporary philosophy and methods for studying speciation and delimiting species. Ichthyol. Herpetol. 109:874–894. [Google Scholar]
- Burton R.S., Barreto F.S.. 2012. A disproportionate role mtDNA in Dobzhansky-Muller incompatibilities? Mol. Ecol. 21:4942–4957. [DOI] [PubMed] [Google Scholar]
- Burton R.S., Pereira R.J., Barreto F.S.. 2013. Cytonuclear genomic interactions and hybrid breakdown. Annu. Rev. Ecol. Evol. Syst. 44:281–302. [Google Scholar]
- Butlin R.K., Walton C., Monk K.A., Bridle J.R.. 1998. Biogeography of Sulawesi grasshoppers, genus Chitaura, using DNA sequence data. In: Hall R., Holloway J.D., editors. Biogeography and Geological Evolution of SE Asia. Leiden: Backhuys Publishers. p. 355–359. [Google Scholar]
- Campbell C.R., Poelstra J.W., Yoder A.D.. 2018. What is speciation genomics? The roles of ecology, gene flow, and genomic architecture in the formation of species. Biol. J. Linn. Soc. 124:561–583. [Google Scholar]
- Carlquist, S. 1965. Island life: a natural history of the islands of the world. Dargen City: The Natural History Press. [Google Scholar]
- Chambers E.A., Marshall T.L., Hillis D.M.. in press. The importance of contact zones for distinguishing interspecific from intraspecific geographic variation. Syst. Biol. 72. [DOI] [PubMed] [Google Scholar]
- Chan K.O., Hutter C.R., Wood P.L., Grismer L.L., Das I., Brown R.M.. 2020. Gene flow creates a mirage of cryptic species in a Southeast Asian spotted stream frog complex. Mol. Ecol. 29:3970–3987. [DOI] [PubMed] [Google Scholar]
- Chan K.O., Hutter C.R., Wood P.L., Su Y.-C., Brown R.M.. 2022. Gene flow increases phylogenetic structure and inflates cryptic species estimations: a case study on widespread philippine puddle frogs (Occidozyga laevis). Syst. Biol. 71:40–57. [DOI] [PubMed] [Google Scholar]
- Chen C., Durand E., Forbes F., François O.. 2007. Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol. Ecol. Notes 7:747–756. [Google Scholar]
- Ciani A.C., Stanyon R., Scheffrahn W., Sampurno B.. 1989. Evidence of gene flow between Sulawesi macaques. Am. J. Primatol. 17:257–270. [DOI] [PubMed] [Google Scholar]
- Clouse R.M., Giribet G.. 2010. When Thailand was an island—the phylogeny and biogeography of mite harvestmen (Opiliones, Cyphophthalmi, Stylocellidae) in Southeast Asia: the biogeography of Southeast Asian mite harvestmen. J. Biogeogr. 37:1114–1130. [Google Scholar]
- Coyne J.A., Orr H.A.. 2004. Speciation. Sunderland, Mass: Sinauer Associates. [Google Scholar]
- Darlington P.J. 1957. Zoogeography: the geographical distribution of animals. New York: Wiley. [Google Scholar]
- de Queiroz K. 1998. The general lineage concept of species, species criteria, and the process of speciation. In: Howard D.J., Berlocher S.H.. Endless Forms. Species and Speciation. Oxford: Oxford University Press. p. 57–75. [Google Scholar]
- de Queiroz K. 1999. The general lineage concept of species and the defining properties of the species category. In: Wilson R. A., editor. Species, new interdisciplinary essays. Cambridge, UK: The MIT Press. p. 49–89. [Google Scholar]
- de Queiroz K. 2007. Species concepts and species delimitation. Syst. Biol. 56:879–886. [DOI] [PubMed] [Google Scholar]
- Degnan J.H., Rosenberg N.A.. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24:332–340. [DOI] [PubMed] [Google Scholar]
- Di Leo J.F., Wookey J., Hammond J.O.S., Kendall J.-M., Kaneshima S., Inoue H., Yamashina T., Harjadi P.. 2012. Mantle flow in regions of complex tectonics: insights from Indonesia. Geochem. Geophys. Geosyst. 13, Q12008. [Google Scholar]
- Dobzhansky T. 1936. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21:113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driller C., Merker S., Perwitasari-Farajallah D., Sinaga W., Anggraeni N., Zischler H.. 2015. Stop and go—waves of tarsier dispersal mirror the genesis of Sulawesi Island. PLoS One 10:e0141212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffels J.P. 1990. Biogeography of Sulawesi cicadas (Homoptera, Cicadoidea). In: Knight W.J., Holloway J.D., editors. Insects and the rain forest of South East Asia (Wallacea). London: Royal Entomological Society. p. 63–72. [Google Scholar]
- Durand E.Y., Patterson N., Reich D., Slatkin M.. 2011. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28:2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S.V., Liu L., Pearl D.K.. 2007. High-resolution species trees without concatenation. Proc. Natl. Acad. Sci. USA 104:5936–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge R.A., Achmadi A.S., Giarla T.C., Rowe K.C., Esselstyn J.A.. 2018. Geographic isolation and elevational gradients promote diversification in an endemic shrew on Sulawesi. Mol. Phylogenet. Evol. 118:306–317. [DOI] [PubMed] [Google Scholar]
- Esselstyn J.A., Achmadi A.S., Handika H., Swanson M.T., Giarla T.C., Rowe K.C.. 2021. Fourteen new, endemic species of shrew (genus Crocidura) from Sulawesi reveal a spectacular island radiation. Bull. Amer. Mus. Nat. Hist 454:1–108. [Google Scholar]
- Esselstyn J.A., Achmadi A.S., Rowe K.C.. 2012. Evolutionary novelty in a rat with no molars. Biol. Lett. 8:990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esselstyn J.A., Timm R.M., Brown R.M.. 2009. Do geological or climatic processes drive speciation in dynamic archipelagos? The tempo and mode of diversification in Southeast Asian shrews. Evolution 63:2595–2610. [DOI] [PubMed] [Google Scholar]
- Evans B.J. 2012. Coalescent-based analysis of demography. In: Gower D., Johnson K., Richardson J., Rosen B., Ruber L., Williams S., editors. Biotic Evolution and Environmental Change in Southeast Asia. Cambridge: Cambridge University Press. p. 270–289. [Google Scholar]
- Evans B.J., Brown R.M., McGuire J.A., Supriatna J., Andayani N., Diesmos A., Iskandar D., Melnick D.J., Cannatella D.C.. 2003a. Phylogenetics of fanged frogs: testing biogeographical hypotheses at the interface of the Asian and Australian faunal zones. Syst. Biol. 52:794–819. [PubMed] [Google Scholar]
- Evans B.J., McGuire J.A., Brown R.M., Andayani N., Supriatna J.. 2008. A coalescent framework for comparing alternative models of population structure with genetic data: evolution of Celebes toads. Biol. Lett. 4:430–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans B.J., Morales J.C., Supriatna J., Melnick D.J.. 1999. Origin of the Sulawesi macaques (Cercopithecidae: Macaca) as suggested by mitochondrial DNA phylogeny. Biol. J. Linn. Soc. 66:539–560. [Google Scholar]
- Evans B.J., Peter B.M., Melnick D.J., Andayani N., Supriatna J., Zhu J., Tosi A.J.. 2021. Mitonuclear interactions and introgression genomics of macaque monkeys (Macaca) highlight the influence of behaviour on genome evolution. Proc. R. Soc. B. 288:20211756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans B.J., Supriatna J., Andayani N., Melnick D.J.. 2003b. Diversification of Sulawesi macaque monkeys: decoupled evolution of mitochondrial and autosomal DNA. Evolution 57:1931–1946. [DOI] [PubMed] [Google Scholar]
- Evans B.J., Supriatna J., Andayani N., Setiadi M.I., Cannatella D.C., Melnick D.J.. 2003c. Monkeys and toads define areas of endemism on Sulawesi. Evolution 57:1436–1443. [DOI] [PubMed] [Google Scholar]
- Flouri T., Jiao X., Rannala B., Yang Z.. 2020. A Bayesian implementation of the multispecies coalescent model with introgression for phylogenomic analysis. Mol. Biol. Evol. 37:1211–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fooden J. 1969. Taxonomy and evolution of monkeys of Celebes (primates: Cercopithecidae). Bibl. Primatol. 10:1–148. [Google Scholar]
- Frantz L.A.F., Rudzinski A., Nugraha A.M.S., Evin A., Burton J., Hulme-Beaman A., Linderholm A., Barnett R., Vega R., Irving-Pease E.K., Haile J., Allen R., Leus K., Shephard J., Hillyer M., Gillemot S., van den Hurk J., Ogle S., Atofanei C., Thomas M.G., Johansson F., Mustari A.H., Williams J., Mohamad K., Damayanti C.S., Wiryadi I.D., Obbles D., Mona S., Day H., Yasin M., Meker S., McGuire J.A., Evans B.J., Rintelen T., Ho S.Y.W., Searle J.B., Kitchener A.C., Macdonald A.A., Shaw D.J., Hall R., Galbusera P., Larson G.. 2018. Synchronous diversification of Sulawesi’s iconic artiodactyls driven by recent geological events. Proc. R. Soc. B. 285:20172566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M.K., Leaché A.D., Burbrink F.T., McGuire J.A., Moritz C.. 2012. Coalescent-based species delimitation in an integrative taxonomy. Trends Ecol. Evol. 27:480–488. [DOI] [PubMed] [Google Scholar]
- Gabriel S., Ziaugra L., Tabbaa D.. 2009. SNP genotyping using the sequenom MassARRAY iPLEX platform. Curr. Prot. Human Genet. 60:2–12. [DOI] [PubMed] [Google Scholar]
- Giarla T.C., Maher S.P., Achmadi A.S., Moore M.K., Swanson M.T., Rowe K.C., Esselstyn J.A.. 2018. Isolation by marine barriers and climate explain areas of endemism in an island rodent. J. Biogeogr. 45:2053–2066. [Google Scholar]
- Glaubrecht M., von Rintelen T.. 2008. The species flocks of lacustrine gastropods: Tylomelania on Sulawesi as models in speciation and adaptive radiation. Hydrobiologia 615:181–199. [Google Scholar]
- Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M., Patterson N., Li H., Zhai W., Fritz M.H.-Y., Hansen N.F., Durand E.Y., Malaspinas A.-S., Jensen J.D., Marques-Bonet T., Alkan C., Prüfer K., Meyer M., Burbano H.A., Good J.M., Schultz R., Aximu-Petri A., Butthof A., Höber B., Höffner B., Siegemund M., Weihmann A., Nusbaum C., Lander E.S., Russ C., Novod N., Affourtit J., Egholm M., Verna C., Rudan P., Brajkovic D., Kucan Z., Gušic I., Doronichev V.B., Golovanova L.V., Lalueza-Fox C., de la Rasilla M., Fortea J., Rosas A., Schmitz R.W., Johnson P.L.F., Eichler E.E., Falush D., Birney E., Mullikin J.C., Slatkin M., Nielsen R., Kelso J., Lachmann M., Reich D., Pääbo S.. 2010. A draft sequence of the Neandertal genome. Science 328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremmen W.H.E. 1990. Palynological investigations in the Danau Tempe depression, southwest Sulawesi (Celebes). Mod. Quat. Res. Southeast Asia 11:123–134. [Google Scholar]
- Gronau I., Hubisz M.J., Gulko B., Danko C.G., Siepel A.. 2011. Bayesian inference of ancient human demography from individual genome sequences. Nat. Genet. 43:1031–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R. 1998. The plate tectonics of Cenozoic SE Asia and the distribution of land and sea. In: Hall R., Holloway J.D., editors, Biogeography and Geological Evolution of SE Asia. Leiden: Backhuys Publishers; p. 99–131. [Google Scholar]
- Hall R. 2009. Southeast Asia’s changing palaeogeography. Blumea 54:148–161. [Google Scholar]
- Hall R. 2011. Australia–SE Asia collision: plate tectonics and crustal flow. In: Hall R., Cottam M.A., Wilson M.E.J., editors. The SE Asian Gateway: history and tectonics of the Australia-Asia collision. London: Special Publications, Geological Society, 355:75–109. [Google Scholar]
- Hall R. 2012. Sundaland and Wallacea. In: Gower D., Johnson K., Richardson J., Rosen B., Ruber L., Williams S., editors. Biotic evolution and environmental change in Southeast Asia. Cambridge: Cambridge University Press. p. 32–78. [Google Scholar]
- Hall R. 2013. The palaeogeography of Sundaland and Wallacea since the late Jurassic. J Limnol 72:e1. [Google Scholar]
- Hamilton W. 1979. Tectonics of the Indonesian region. U.S. Geol. Surv. Prof. Pap. 1078(345):1–308. [Google Scholar]
- Handika H., Achmadi A.S., Esselstyn J.A., Rowe K.C.. 2021. Molecular and morphological systematics of the Bunomys division (Rodentia: Muridae), an endemic radiation on Sulawesi. Zool. Scr. 50:141–154. [Google Scholar]
- Hill G.E. 2017. The mitonuclear compatibility species concept. Auk 134:393–409. [DOI] [PubMed] [Google Scholar]
- Hillis D.M., Chambers E.A., Devitt T.J.. 2021. Challenges in species delimitation and the role of population models. Ichthyol. Herpetol. 109:895–903. [Google Scholar]
- Huppert K.L., Perron J.T., Royden L.H.. 2020. Hotspot swells and the lifespan of volcanic ocean islands. Sci. Adv. 6:eaaw6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley T.H. 1868. On the classification and distribution of the Alectoromorphae and Heteromorphae. Proc. Zool. Soc. London. 1868:294–319. [Google Scholar]
- Irwin D.E. 2002. Phylogeographic breaks without geographic barriers to gene flow. Evolution 56:2383–2394. [DOI] [PubMed] [Google Scholar]
- Jackson N.D., Carstens B.C., Morales A.E., O’Meara B.C.. 2017. Species delimitation with gene flow. Syst. Biol. 66:799–812. [DOI] [PubMed] [Google Scholar]
- Jiao X., Flouri T., Yang Z.. 2021. Multispecies coalescent and its applications to infer species phylogenies and cross-species gene flow. Natl. Sci. 8:nwab127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioformatics 24:1403–1405. [DOI] [PubMed] [Google Scholar]
- Jombart T., Ahmed I.. 2011. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioformatics 27:3070–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G.R. 2019. Divergence estimation in the presence of incomplete lineage sorting and migration. Syst. Biol. 68:19–31. [DOI] [PubMed] [Google Scholar]
- Jones M.R., Good J.M.. 2016. Targeted capture in evolutionary and ecological genomics. Mol. Ecol. 25:185–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb A.M., Gan H.M., Greening C., Joseph L., Lee Y.P., Morán-Ordóñez A., Sunnocks P., Pavlova A.. 2018. Climate-driven mitochondrial selection: a test in Australian songbirds. Mol. Ecol. 27:898–918. [DOI] [PubMed] [Google Scholar]
- Leaché A.D., Chavez A.S., Jones L.N., Grummer J.A., Gottscho A.D., Linkem C.W.. 2015. Phylogenomics of phrynosomatid lizards: conflicting signals from sequence capture versus restriction site associated DNA sequencing. Genome Biol. Evol. 7:706–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaché A.D., Fujita M.K., Minin V.N., Bouckaert R.R.. 2014a. Species delimitation using genome-wide SNP data. Syst. Biol. 63:534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaché A.D., Harris R.B., Rannala B., Yang Z.. 2014b. The influence of gene flow on species tree estimation: a simulation study. Syst. Biol. 63:17–30. [DOI] [PubMed] [Google Scholar]
- Leaché A.D., Zhu T., Rannala B., Yang Z.. 2019. The spectre of too many species. Syst. Biol. 68:168–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima T.G., Burton R.S., Willett C.S.. 2019. Genomic scans reveal multiple mito-nuclear incompatibilities in population crosses of the copepod Tigriopus californicus. Evolution 73:609–620. [DOI] [PubMed] [Google Scholar]
- Linkem C.W., Brown R.M., Siler C.D., Austin C.C., Iskandar D.T., Diesmos A.C., Supriatna J., Andayani N., McGuire J.A.. 2013. Stochastic faunal exchanges drive diversification in widespread Wallacean and Pacific Island lizards, (Squamata: Scincidae: Lamprolepis smaragdina). J. Biogeogr. 40:507–520. [Google Scholar]
- Lohman D.J., de Bruyn M., Page T., von Rintelen K., Hall R., Ng P.K.L., Shih H.-T., Carvalho G.R., von Rintelen T.. 2011. Biogeography of the Indo-Australian Archipelago. Annu. Rev. Ecol. Evol. Syst. 42:205–226. [Google Scholar]
- Luikart G., England P.R., Tallmon D., Jordan S., Taberlet P.. 2003. The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 4:981–994. [DOI] [PubMed] [Google Scholar]
- MacArthur R.H., Wilson E.O.. 1967. The theory of island biogeography. Princeton: Princeton University Press. [Google Scholar]
- Maekawa K., Kon M., Araya K., Matsumoto T.. 2001. Phylogeny and biogeography of wood-feeding cockroaches, genus Salganea Stål (Blaberidae: Panesthiinae), in Southeast Asia based on mitochondrial DNA sequences. J. Mol. Evol. 53:651–659. [DOI] [PubMed] [Google Scholar]
- Mandagi I.F., Kakioka R., Montenegro J., Kobayashi H., Masengi K.W.A., Inomata N., Nagano A.J., Toyoda A., Ansai S., Matsunami M., Kimura R., Kitano J., Kusumi J., Yamahira K.. 2021. Species divergence and repeated ancient hybridization in a Sulawesian lake system. J. Evol. Biol. 34:1767–1780. [DOI] [PubMed] [Google Scholar]
- Matzke N.J. 2013. BioGeoBEARS: BioGeography with Bayesian (and likelihood) evolutionary analysis in R Scripts. R Package, version 0.2.1. [Google Scholar]
- Mayr E. 1944. Wallace’s line in the light of recent zoogeographic studies. Q. Rev. Biol. 19:1–14. [Google Scholar]
- Mayr, E. 1963. Animal species and evolution. Cambridge: Harvard University Press. [Google Scholar]
- McGuire J.A. 2003. Allometric prediction of locomotor performance: an example from Southeast Asian flying lizards. Am. Nat. 161:337–339. [DOI] [PubMed] [Google Scholar]
- McGuire J.A., Alcala A.C.. 2000. A taxonomic revision of the flying lizards (Iguania: Agamidae: Draco) of the Philippine Islands, with a description of a new species. Herpetolological Monogr. 14:81–138. [Google Scholar]
- McGuire J.A., Brown R.M., Mumpuni R.A., Andayani N.. 2007. The flying lizards of theDraco lineatusgroup (Squamata: Iguania: Agamidae): a taxonomic revision with descriptions of two new species. Herpetological Monogr. 21:179–212. [Google Scholar]
- McGuire J.A., Dudley R.. 2005. The cost of living large: comparative gliding performance in flying lizards (Agamidae: Draco). Am. Nat. 166:93–106. [DOI] [PubMed] [Google Scholar]
- McGuire J.A., Dudley R.. 2011. The biology of gliding in flying lizards (Genus Draco) and their fossil and extant analogs. Integr. Comp. Biol. 51:983–990. [DOI] [PubMed] [Google Scholar]
- McGuire J.A., Heang K.B.. 2001. Phylogenetic systematics of Southeast Asian flying lizards (Iguania: Agamidae Draco) as inferred from mitochondrial DNA sequence data. Biol. J. Linn. Soc. 72:203–229. [Google Scholar]
- Merker S., Driller C., Perwitasari-Farajallah D., Pamungkas J., Zischler H.. 2009. Elucidating geological and biological processes underlying the diversification of Sulawesi tarsiers. Proc. Natl. Acad. Sci. USA 106:8459–8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaux B. 2010. Biogeology of Wallacea: geotectonic models, areas of endemism, and natural biogeographical units. Biol. J. Linn. Soc. 101:193–212. [Google Scholar]
- Minh B.Q., Nguyen M.A.T., von Haeseler A.. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30:1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirarab S., Reaz R., Bayzid M.S., Zimmermann T., Swenson M.S., Warnow T.. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30:i541–i548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokodongan D.F., Yamahira K.. 2015. Origin and intra-island diversification of Sulawesi endemic Adrianichthyidae. Mol. Phylogenet. Evol. 93:150–160. [DOI] [PubMed] [Google Scholar]
- Mori A., Hikida T.. 1993. Natural history observations of the flying lizard, Draco volans sumatranus (Agamidae, Squamata) from Sarawak, Malaysia. Raffles Bull. Zool. 41:83–94. [Google Scholar]
- Morrice M.G., Jezek P.A., Gill J.B., Whitford D.J., Monoarfa M.. 1983. An introduction to the Sangihe arc: volcanism accompanying arc—arc collision in the Molucca Sea, Indonesia. J. Volcanol. Geotherm. Res. 19:135–165. [Google Scholar]
- Moss S.J., Wilson M.E.. 1998. Biogeographic implications of the Tertiary paleogeographic evolution of Sulawesi and Borneo. In: Hall R., Holloway J.D., editors. Biogeography and Geological Evolution of SE Asia. Leiden: Backhuys Publishers. p. 133–164. [Google Scholar]
- Muller H.J. 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6:71–125. [Google Scholar]
- Musters C.J.M. 1983. Taxonomy of the genus Draco L. (Agamidae, Lacertilia, Reptilia). Zool. Verh. 199:1–132. [Google Scholar]
- Natawidjaja D.H., Daryono M.R.. 2015. The Lawanopo Fault, central Sulawesi, East Indonesia. 4th International Symposium on Earthquake and Disaster Mitigation 2014 (ISEDM 2014) AIP Conf. Proc. 1658, 030001-1-23.
- Nei M. 1972. Genetic distance between populations. Am. Nat. 106:283–292. [Google Scholar]
- Nugraha A.M.S., Hall R.. 2018. Late Cenozoic palaeogeography of Sulawesi, Indonesia. Palaeogeogr. Palaeoclimatol. Palaeoecol. 490:191–209. [Google Scholar]
- Nylander J.A.A. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- Ogilvie H.A., Bouckaert R.R., Drummond A.J.. 2017. StarBEAST2 brings faster species tree inference and accurate estimates of substitution rates. Mol. Biol. Evol. 34:2101–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H.A., Turelli M.. 2001. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller Incompatibilities. Evolution 55:1085–1094. [DOI] [PubMed] [Google Scholar]
- Portik D.M., Smith L.L., Bi K.. 2016. An evaluation of transcriptome-based exon capture for frog phylogenomics across multiple scales of divergence (Class: Amphibia, Order: Anura). Mol. Ecol. Resour. 16:1069–1083. [DOI] [PubMed] [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P.. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A.. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67:901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand D.M., Haney R.A., Fry A.J.. 2004. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 19:645–653. [DOI] [PubMed] [Google Scholar]
- Rannala B., Yang Z.. 2003. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164:1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly S.B., Stubbs A.L., Arida E., Karin B.R., Arifin U., Kaiser H., Bi K., Iskandar D.T., McGuire J.A.. 2022. Phylogenomic analysis reveals dispersal-driven speciation and divergence with gene flow in Lesser Sunda flying lizards (Genus Draco). Syst. Biol. 71:221–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly S.B., Stubbs A.L., Karin B.R., Bi K., Arida E., Iskandar D.T., McGuire J.A.. 2019. Leap-frog dispersal and mitochondrial introgression: phylogenomics and biogeography of Limnonectes fanged frogs in the Lesser Sundas Archipelago of Wallacea. J. Biogeogr. 46:757–769. [Google Scholar]
- von Rintelen K., Glaubrecht M., Schubart C.D., Wessel A., Rintelen T.. 2010b. Adaptive radiation and ecological diversification of Sulawesi’s ancient lake shrimps: adaptive radiation of ancient lake shrimps. Evolution 64:3287–3299. [DOI] [PubMed] [Google Scholar]
- von Rintelen K., von Rintelen T., Glaubrecht M.. 2007a. Molecular phylogeny and diversification of freshwater shrimps (Decapoda, Atyidae, Caridina) from ancient Lake Poso (Sulawesi, Indonesia)—The importance of being colourful. Mol. Phylogenet. Evol. 45:1033–1041. [DOI] [PubMed] [Google Scholar]
- von Rintelen K., von Rintelen T., Meixner M., Lüter C., Cai Y., Glaubrecht M.. 2007c. Freshwater shrimp–sponge association from an ancient lake. Biol. Lett. 3:262–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Rintelen T., Bouchet P., Glaubrecht M.. 2007b. Ancient lakes as hotspots of diversity: a morphological review of an endemic species flock of Tylomelania (Gastropoda: Cerithioidea: Pachychilidae) in the Malili lake system on Sulawesi, Indonesia. Hydrobiologia 592:11–94. [Google Scholar]
- von Rintelen T., Glaubrecht M.. 2005. Anatomy of an adaptive radiation: a unique reproductive strategy in the endemic freshwater gastropod Tylomelania (Cerithioidea: Pachychilidae) on Sulawesi, Indonesia and its biogeographical implications: anatomy of Tylomelania. Biol. J. Linn. Soc. 85:513–542. [Google Scholar]
- von Rintelen T., von Rintelen K., Glaubrecht M., Schubart C.D., Herder F.. 2012. Aquatic biodiversity hotspots in Wallacea: the species flocks in the ancient lakes of Sulawesi, Indonesia. In: Gower D., Johnson K., Richardson J., Rosen B., Ruber L., Williams S., editors. Biotic evolution and environmental change in Southeast Asia. Cambridge, UK: Cambridge University Press. p. 290–315. [Google Scholar]
- Ripley B.D., Venables W.N.. 2002. Modern Applied Statistics with S. New York: Springer. [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P.. 2012. MRBAYES 3.2: efficient Bayesian phylogenetic inference and model selection across a large model space. Syst. Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe K.C., Achmadi A.S., Esselstyn J.A.. 2016a. Repeated evolution of carnivory among Indo-Australian rodents: carnivorous rodents of Indo-Australia. Evolution 70:653–665. [DOI] [PubMed] [Google Scholar]
- Rowe K.C., Achmadi A.S., Esselstyn J.A.. 2016b. A new genus and species of omnivorous rodent (Muridae: Murinae) from Sulawesi, nested within a clade of endemic carnivores. J. Mammal. 97:978–991. [Google Scholar]
- Rowe K.C., Achmadi A.S., Fabre P., Schenk J.J., Steppan S.J., Esselstyn J.A.. 2019. Oceanic islands of Wallacea as a source for dispersal and diversification of murine rodents. J. Biogeogr. 46:2752–2768. [Google Scholar]
- Ruedas L.A., Morales J.C.. 2005. Evolutionary relationships among genera of Phalangeridae (Metatheria: Diprotodontia) inferred from mitochondrial DNA. J. Mammal. 86:353–365. [Google Scholar]
- Schenk J.J. 2016. Consequences of secondary calibrations on divergence time estimates. PLoS One 11:e0148228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnute J.T., Boers N.M., Haigh R.. 2003. PBS Software: maps, spatial analysis, and other utilities. Can. Tech. Rep. Fish. Aquat. Sci. 2496:1–82. [Google Scholar]
- Schreiber A., Seibold I., Nötzold G., Wink M.. 1999. Cytochrome b gene haplotypes characterize chromosomal lineages of Anoa, the Sulawesi dwarf buffalo (Bovidae: Bubalus sp.). Heredity 90:165–176. [DOI] [PubMed] [Google Scholar]
- Schubart C.D., Ng P.K.L.. 2008. A new molluscivore crab from Lake Poso confirms multiple colonization of ancient lakes in Sulawesi by freshwater crabs (Decapoda: Brachyura). Zool. J. Linn. Soc. 154:211–221. [Google Scholar]
- Setiadi M.I., McGuire J.A., Brown R.M., Zubairi M., Iskandar D.T., Andayani N., Supriatna J., Evans B.J.. 2011. Adaptive radiation and ecological opportunity in Sulawesi and Philippine fanged frog (Limnonectes) communities. Am. Nat. 178:221–240. [DOI] [PubMed] [Google Scholar]
- Shaffer H.B., Thomson R.C.. 2007. Delimiting species in recent radiations. Syst. Biol. 56:896–906. [DOI] [PubMed] [Google Scholar]
- Shaw K.L., Gillespie R.G.. 2016. Comparative phylogeography of oceanic archipelagos: hotspots for inferences of evolutionary process. Proc. Natl. Acad. Sci. USA 113:7986–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekelle M., Meier R., Wahyu I., Wirdateti T.N.. 2010. Molecular phylogenetics and chronometrics of Tarsiidae based on 12S mtDNA haplotypes: evidence for Miocene origins of crown tarsiers and numerous species within the Sulawesian clade. Int. J. Primatol. 31:1083–1106. [Google Scholar]
- Shi C.-M., Yang Z.. 2018. Coalescent-based analyses of genomic sequence data provide a robust resolution of phylogenetic relationships among major groups of gibbons. Mol. Biol. Evol. 35:159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. 1987. Gene flow and the geographic structure of natural populations. Science 236:787–792. [DOI] [PubMed] [Google Scholar]
- Solís-Lemus C., Bastide P., Ané C.. 2017. PhyloNetworks: a package for phylogenetic networks. Mol. Biol. Evol. 34:3292–3298. [DOI] [PubMed] [Google Scholar]
- Spakman W., Hall R.. 2010. Surface deformation and slab–mantle interaction during Banda arc subduction rollback. Nature Geosci. 3:562–566. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelbrink B., Albrecht C., Hall R., Rintelen T.. 2012. The biogeography of Sulawesi revisited: is there evidence for a vicariant origin of taxa on Wallace’s “anomalous island?”: biogeography of Sulawesi. Evolution 66:2252–2271. [DOI] [PubMed] [Google Scholar]
- Streicher J.W., Devitt T.J., Goldberg C.S., Malone J.H., Blackmon H., Fujita M.K.. 2014. Diversification and asymmetrical gene flow across time and space: lineage sorting and hybridization in polytypic barking frogs. Mol. Ecol. 23:3273–3291. [DOI] [PubMed] [Google Scholar]
- Stuart B.L., Inger R.F., Voris H.K.. 2006. High level of cryptic species diversity revealed by sympatric lineages of Southeast Asian forest frogs. Biol. Lett. 2:470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukamto, R. 1978. The structure of Sulawesi in the light of plate tectonics. Proceedings of the Regional Conference on the Geology and Mineral Resources of Southeast Asia, Wiryosujono, S., Sudradjat, A., editors, Jakarta: Indonesian Association of Geologists, pp 121–141. [Google Scholar]
- Sukamto, R. 1990. Geology of the Ujung Pandang Sheet, South Sulawesi. Scale 1:1,000,000. Bandung: Geological Survey of Indonesia, Directorate of Mineral Resources, Geological Research and Development Centre, 12pp. [Google Scholar]
- Sukumaran J., Knowles L.L.. 2017. Multispecies coalescent delimits structure, not species. Proc. Natl. Acad. Sci. USA 114:1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana Y., Naruse K., Sakaizumi M.. 2005. Molecular phylogeny of the medaka fishes genus Oryzias (Beloniformes: Adrianichthyidae) based on nuclear and mitochondrial DNA sequences. Mol. Phylogenet. Evol. 36:417–428. [DOI] [PubMed] [Google Scholar]
- Tosi A.J., Morales J.C., Melnick D.J.. 2003. Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution 57:1419–1435. [DOI] [PubMed] [Google Scholar]
- Townsend T.M., Mulcahy D.G., Noonan B.P., Sites J.W. Jr., Kuczynski C.A., Wiens J.J., Reeder T.W.. 2011. Phylogeny of iguanian lizards inferred from 29 nuclear loci, and comparison of concatenated and species tree approaches for an ancient, rapid radiation. Mol. Phylogenet. Evol. 61:363–380. [DOI] [PubMed] [Google Scholar]
- Utama I.V., Mandagi I.F., Lawelle S.A., Masengi K.W.A., Watanabe K., Sawada N., Nagano A.J., Kusumi J., Yamahira K.. 2022. Deeply divergent freshwater fish species within a single river system in central Sulawesi. Mol. Phylogenet. Evol. 173:107519. [DOI] [PubMed] [Google Scholar]
- Wallace A.R. 1859. The geographical distribution of birds. Ibis 1:449–454. [Google Scholar]
- Wallace A.R. 1860. On the zoological geography of the Malay Archipelago. J. Linn. Soc. 14:172–184. [Google Scholar]
- Wallace, AR. 1869. The Malay Archipelago: the land of the orangutan and the bird of paradise; a narrative of travel with studies of man and nature. London: Macmillan and Co. [Google Scholar]
- Wallace A.R. 1876. The geographical distribution of animals, with a study of the relations of living and extinct faunas as elucidating the past changes of the earth’s surface. London: Macmillan and Co. [Google Scholar]
- Wallace A.R. 1880. Island life; or, the phenomena and causes of insular faunas and floras, including a revision and attempted solution of the problem of geological climates. London: Macmillan and Co. [Google Scholar]
- Walton C., Butlin R.K., Monk K.A.. 1997. A phylogeny for grasshoppers of the genus Chitaura (Orthoptera: Acridae) from Sulawesi, Indonesia, based on mitochondrial DNA sequence data. Biol. J. Linn. Soc. 62:365–382. [Google Scholar]
- Wen D., Yu Y., Zhu J., Nakhleh L.. 2018. Inferring phylogenetic networks using PhyloNet. Syst. Biol. 67:735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielstra B. 2019. Hybrid zone movement: more pervasive than appreciated. J. Biogeog. 46:1300–1305. [Google Scholar]
- Wilkinson M. 1996. Majority-rule reduced consensus trees and their use in bootstrapping. Mol. Biol. Evol. 13:437–444. [DOI] [PubMed] [Google Scholar]
- Wright S. 1931. Evolution in Mendelian populations. Genetics 16:97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Yang Z.. 2016. Challenges in species tree estimation under the multispecies coalescent model. Genetics 204:1353–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2015. The BPP program for species tree estimation and species delimitation. Curr. Zool. 61:854–865. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequence data for the ND2, 12S, and 16S mitochondrial genes have been submitted to the GenBank database under accession numbers OQ598699–OQ599306 (ND2), OQ628522-OQ629047 (12S), and OQ624360-OQ624885 (16S). Sequence data from the exon-capture experiment were submitted to the NCBI sequence read archive under BioSample accession numbers SAMN33714970-SAMN33715073. Software input files for the mtDNA data set and multiple versions of the exome-capture data (full data nexus file, StarBEAST2 data sets for 35, 38, and 41 samples, and BPP 4.3.8 input files for 683 loci and 97 individuals) are available from the Dryad data repository (doi:10.6078/D1S99X).