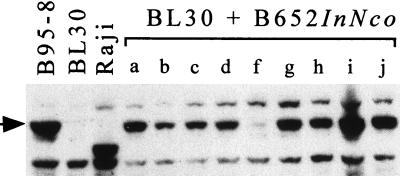Abstract
Replication and maintenance of the 170-kb circular chromosome of Epstein-Barr virus (EBV) during latent infection are generally believed to depend upon a single viral gene product, the nuclear protein EBNA-1. EBNA-1 binds to two clusters of sites at oriP, an 1,800-bp sequence on the EBV genome which can support replication and maintenance of artificial plasmids introduced into cell lines that contain EBNA-1. To investigate the importance of EBNA-1 to latent infection by EBV, we introduced a frameshift mutation into the EBNA-1 gene of EBV by recombination along with a flanking selectable marker. EBV genomes carrying the frameshift mutation could be isolated readily after superinfecting EBV-positive cell lines, but not if recombinant virus was used to infect EBV-negative B-cell lines or to immortalize peripheral blood B cells. EBV mutants lacking almost all of internal repeat 3, which encode a repetitive glycine and alanine domain of EBNA-1, were generated in the same way and found to immortalize B cells normally. An EBNA-1-deficient mutant of EBV was isolated and found to be incapable of establishing a latent infection of the cell line BL30 at a detectable frequency, indicating that the mutant was less than 1% as efficient as an isogenic, EBNA-1-positive strain in this assay. The data indicate that EBNA-1 is required for efficient and stable latent infection by EBV under the conditions tested. Evidence from other studies now indicates that autonomous maintenance of the EBV chromosome during latent infection does not depend on the replication initiation function of oriP. It is therefore likely that the viral chromosome maintenance (segregation) function of oriP and EBNA-1 is what is required.
The life cycle of Epstein-Barr virus (EBV) appears to depend upon a mechanism to replicate and maintain the viral genome in an autonomous state in expanding B-cell populations during the initial phase of latency (20–22). Maintenance of the viral chromosome in proliferating cells required that it be duplicated each cell cycle and passed on to daughter nuclei during mitosis. During latent infection, the circularized EBV genome replicates during the S phase of the cell cycle (14), apparently under a cellular control mechanism that limits each viral chromosome to one round of replication per S phase (1, 47). EBV chromosomes associate with the condensed cellular chromosomes during mitosis, ensuring that all copies of the EBV chromosome are enveloped within daughter nuclei as they form at the close of mitosis (15).
An 1,800-bp region of the EBV genome, oriP, was identified based on its ability to confer long-term maintenance of recombinant plasmids under selection in human cell lines (44). oriP requires only a single EBV-encoded protein, EBNA-1, for this activity (31, 48). oriP provides both replication and segregation functions to plasmids that carry it through two distinct and essential functional elements (4, 35), both of which bind EBNA-1 at multiple sites (34). A cluster of four EBNA-1 binding sites, usually called the DS for a dyad symmetry element contained within it, supports plasmid replication (42) and is the point from which replication forks diverge in both directions, at least within a few hundred base pairs, the resolution achieved by a two-dimensional gel analysis of replication intermediates (9).
The other essential element of oriP is a repetitive array of EBNA-1 binding sites, a family of 30-bp repeats called the FR, which is located nearly 1 kb away from the DS. The FR supports very little plasmid replication by itself, but in the presence of EBNA-1, it allows plasmids to be retained with their genes active for prolonged periods of time after being introduced into cells (23, 35). It was shown that oriP could confer mitotic stability to a 600-kb, circularized, cloned segment of a human chromosome in cells containing EBNA-1 and cause the artificial plasmid to associate with condensed human chromosomes during mitosis (36). EBNA-1 itself associates with human metaphase chromosomes (12) and also has the capacity to link together DNA molecules to which it is bound, by self-interaction (8, 32, 38). The mechanism behind the plasmid retention function of the FR and EBNA-1 is therefore likely to involve EBNA-1-mediated tethering of plasmids to human chromosomes during mitosis to prevent their loss to the cytoplasm.
It has long been assumed that oriP and EBNA-1 are together responsible for supporting the replication and maintenance of the circular EBV chromosome during latent infection of mitotically active cells, but a direct test of this has been lacking. However, in recent years, evidence has accumulated to suggest that the replication initiation function of oriP is dispensable to EBV. Two-dimensional gel analysis of replication intermediates has indicated that on the EBV chromosome, oriP is replicated passively from distant origins most of the time, with initiation occurring within oriP only a fraction of the time (29). An isolate of the cell line X50-7 was found to carry a variant EBV genome which had sustained a deletion that removed all of oriP except for the FR (43); when tested on plasmids, the FR does not support significant replication in the absence of the DS of oriP (4, 31, 35, 42, 44). Recent genetic studies in this laboratory have confirmed that the DS of oriP can be deleted from EBV without seriously compromising its ability to maintain its chromosome autonomously during latent infection (our unpublished data).
Could the viral chromosomal maintenance function, or segregation function, of EBNA-1 and oriP be redundant, too? EBNA-1 and oriP have been conserved during the evolution of the close relatives of EBV that infect Old World primates (6, 30, 46). However, the FR of oriP is also a potent EBNA-1-dependent enhancer of transcription, and EBNA-1 and the FR appear to be important for proper transcription from two EBV promoters that give rise to expression of genes required to immortalize B cells (10, 33, 39). Among the more distantly related gamma herpesviruses that are known to support autonomous replication of their viral genomes during latent infection, none appears to have a homolog of EBNA-1 or of oriP (2, 5, 24, 40). It therefore seemed possible that the plasmid maintenance property of EBNA-1 and oriP could be functionally redundant, like the replication initiation function.
To test the importance of EBNA-1 for latent infection by EBV, we introduced a frameshift mutation into the EBNA-1 gene of EBV by homologous recombination. Viruses carrying this mutation could not immortalize B cells nor stably infect an EBV-negative B-cell line. This suggests that the chromosome maintenance function of EBNA-1 and oriP is required for efficient and stable latent infection.
MATERIALS AND METHODS
Construction of plasmids.
p605 is a derivative of pUC12 which carries EBV strain B95-8 sequences between 107327 (HpaI) and 110760 (XbaI) and contains the CMVIE-neo selective marker inserted next to the EBNA-1 gene, as depicted in Fig. 1. Briefly, it was constructed as follows. Into a small plasmid (p277) carrying the EBNA-1 gene and 3′ sequences extending to a HindIII site at 110491, the CMVIE-neo marker was inserted as a 2.7-kb BamHI fragment (25) between the BstXI site near the middle of the EBNA-1 gene and the SstII site (109805) near the 3′ end. The BstXI-BamHI junction restored the BamHI site, which allowed the 5′ end of the EBNA-1 gene to be removed, and in its place was inserted the entire EBNA-1 gene and 3′ flanking sequences extending to the PvuII site. This resulted in duplication of 371 bp between the SstII and PvuII sites (109806 to 110176) so that it is present on both sides of the CMVIE-neo insertion. The 5′ and 3′ flanking sequences were then extended in three subsequent steps to create p605.
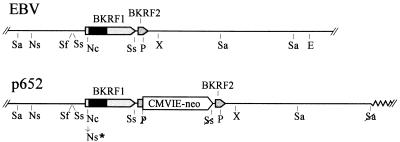
FIG. 1.
Insertion of a selective gene, CMVIE-neo, next to the EBNA-1 gene without disrupting the adjacent gene, BKRF2. (Upper) Part of the EBV genome, from the SalI site at 105,296 to the EcoRV site at 116,865. The EBNA-1 coding sequence (BKRF1) and an adjacent gene, BKRF2, are indicated. The dark portion of the EBNA-1 coding region is IR3, which encodes the Gly-Gly-Ala repeats. The indicated restriction enzyme sites are as follows: E, EcoRV; P, PvuII; Nc, NcoI; Ns, NsiI; Sa, SalI; Sf, SfiI; Ss, SstII; X, XbaI. Only the most relevant sites are shown for NcoI, PvuII, and SstII. (Lower) Relevant portion of the plasmid p652. The CMVIE-neo gene was inserted along with a duplication of nearly 400 bp of EBV sequence, from an SstII site to a PvuII site, placing the duplicated sequence on each side of the insertion. The duplicated sequence includes the 3′ end of the EBNA-1 gene (BKRF1) and continues through much of the adjacent gene, BKRF2. Thus a complete copy of each gene together with its 3′ and 5′ regulatory regions remains intact. Restriction sites that were destroyed where sequences were joined are indicated with a slash. Ns∗ represents filling in of an NcoI site to create an NsiI site and a frameshift mutation in the EBNA-1 gene. The zig-zag line at the right end indicates plasmid vector sequences.
p652 is a 39-kb cosmid which carries B95-8 sequences from a ClaI site at 93256 to a SalI site at 116209 inserted between the ClaI and SalI sites of the cosmid vector cos553, with the CMVIE-neo gene inserted next to the EBNA-1 gene (Fig. 1). cos553 is a 13-kb cosmid vector that carries oriP and oriLyt of EBV and fosters efficient homologous recombination with EBV during lytic replication (25). To construct p652, first the 3.1-kb SalI H fragment of B95-8 (113282 to 116206) was placed into cos553 between its pBR322-derived cloning sites, SalI and HindIII, such that both sites were lost, but with retention of the SalI site at the 113282 end, which was positioned close to the ClaI cloning site of the vector. Next, the adjacent EBV SalI fragment, F (105296 to 113282), was inserted in the proper orientation at the remaining SalI site. The sequences between the unique SfiI and XbaI sites (at 107430 and 110760) were then replaced with the corresponding region from p605, which carries the CMVIE-neo gene insertion, to give p629. Finally, flanking EBV sequences from ClaI (93256) to SalI (105296) were excised from pMH304 (obtained from Marie Hardwick) and inserted into p629, cut with ClaI and cut partially with SalI; p652 was isolated with the aid of Gigapack lambda packing extracts (Stratagene). The ClaI site at 99620 was not cleaved in pMH304 because of dam methylation.
The internal repeat 3 (IR3) deletion, dl7, was reported previously (48). It was sequenced and found to be a deletion of nucleotides 108253 to 108924, which removes 224 of the 237 codons within IR3. The frameshift mutant InNco was made by filling the central 4 bp of the NcoI site in the EBNA-1 gene of p277 by using the Klenow large fragment. Both mutations were transferred into p605 by using the AvrII and BstXI sites within the EBNA-1 gene, and from there, they were transferred into p652 as described above.
Recombinant EBV.
All cells were grown in RPMI 1640 medium supplemented with 9% fetal bovine serum and antibiotics, penicillin, and streptomycin (all from Gibco-BRL). Ten million B95-8 cells were suspended in 0.6 ml of complete medium and electroporated in a 0.4-cm-wide chamber with 3 to 6 μg of p652 (or p652 carrying the dl7 or InNco mutation) plus 3 μg of pCMV-BZLF1 (13) by using a BRL Cell-porator set at 800 μF and 400 V. The cells were then cultured in 10 ml of medium, which was collected after 4 days, passed through a 0.4-μm-pore-diameter filter, and stored at 4°C. The titer of recombinant, G418-resistant EBV, generally 25 to 80 per ml, was determined as described below.
Recombinant virus was isolated in immortalized B cells as follows. Peripheral blood leukocytes (1.0 × 106 to 1.5 × 106), depleted of T cells by rosette formation with sheep erythrocytes (19), were mixed with 1.25 ml of B95-8 supernatant and 10 ml of medium and then dispensed into a 24-well plate. The 24-well plates previously had been seeded with lethally gamma-irradiated (3,000 rad) human fibroblasts to serve as a feeder layer, split 1:4 from a confluent 10-cm-diameter dish. After 2 to 3 weeks, when transformed B cells had proliferated enough to cover most of the bottom of most wells, most of the medium in each well was removed by gentle suction and replaced with medium containing G418 at 650 μg/ml (active drug concentration). Medium was replaced in individual wells every few days when it became acidified, until either all the cells had died or a G418-resistant clone had emerged. After 2 to 4 weeks of selection, the G418 concentration was raised to 700 μg/ml to eliminate a few very slowly growing clones that were infected only with B95-8 virus and that did not carry the CMVIE-neo gene.
To render B95-8 cells infectable by EBV, cells were electroporated with pMEP4-CR2 (7), which carries oriP, an expressed cDNA for the EBV receptor, CD21, and which confers resistance to hygromycin B. A resistant population of B95-8 cells was selected in medium containing hygromycin B at 200 μg/ml. A total of 1.4 × 106 of these cells were mixed with 1 ml of supernatant from B95-8 containing B652InNco recombinants, described above, and then selected for clonal outgrowth in medium containing G418 at 840 μg/ml (active drug concentration) in a 48-well plate.
To determine the titer of G418-resistant EBV, generally 40,000 Raji or BL30 cells in 0.3 ml of medium were placed in each well of 48-well dishes, or half this number of cells and medium were used per well of 96-well dishes. Different amounts of virus stock were added to groups of 16 or more wells. After 2 days of incubation, the medium in the wells was gradually replaced with medium containing G418, at active concentrations of 900 μg/ml for Raji cells and 1,200 μg/ml for BL30 cells. Wells containing viable clones were scored after 2 to 3 weeks, and titers were determined by using Poisson statistics.
Antibodies against EBNA-1.
Affinity-purified rabbit antibodies specific for the carboxyl-terminal domain of EBNA-1 were prepared as described previously (37).
RESULTS
Introduction of EBNA-1 gene mutations into the EBV genome.
To establish a method for transferring mutations within the EBNA-1 gene from plasmids to the EBV genome, a plasmid called p652 was constructed (Fig. 1). p652 carries 23 kb of EBV DNA spanning the EBNA-1 gene and was built using a cosmid vector, cos553, that carries oriLyt of EBV, which allows efficient homologous recombination with EBV during lytic viral replication (25). A G418 resistance gene, CMVIE-neo, was inserted close to the EBNA-1 gene to facilitate recovery of latently infected cell clones harboring a recombinant virus after infection. Because there is a size limit for packaging EBV genomes into viral capsids (3), the large size of p652, 39 kb, favors recovery of recombinant viral genomes that have recombined on both sides of the selective marker, resulting in replacement of wild-type EBV sequences with homologous sequences from the plasmid, rather than single recombination events which would insert the entire plasmid into the viral genome (25). Only 82 bp separates the EBNA-1 coding region, BKRF1, from the neighboring late open reading frame, BKRF2. In order to insert the CMVIE-neo gene here while minimizing the risk of disrupting regulatory signals for either gene, a 371-bp region spanning the last 70 bp of BKRF1 and most of BKRF2 was duplicated so as to be present on both sides of the CMVIE-neo insertion, as shown in Fig. 1.
Two mutations within the EBNA-1 gene were transferred from smaller plasmids to p652: a deletion of most of IR3 and a frameshift mutation. IR3 is a triplet repeat that encodes a repetitive array of glycine and alanine comprising over one-third of the EBNA-1 protein. The deletion dl7 removes 324 of the 337 IR3 codons and yet has no effect on the ability of EBNA-1 to support replication and maintenance of oriP-containing plasmids or to activate the FR (family of repeats) transcriptional enhancer of oriP (45, 48). The insertion mutation InNco is a four-base insertion at EBNA-1 codon 39, created by filling in an NcoI site, which should cause translation to terminate 29 out-of-frame codons later. The next in-frame methionine codon is located near the end of the EBNA-1 gene, so reinitiation of translation is not expected. Placement of InNco within the EBNA-1 gene on the oriP-containing plasmid p201 (48) abolished the ability of the plasmid to be maintained autonomously in cells (data not shown), so InNco is considered to be a null mutation.
Recombinant EBV strains that had acquired the CMVIE-neo gene from p652 were generated by electroporating B95-8 cells with p652 along with pCMV-BZLF1, which induces lytic EBV replication. We detected recombinant EBV in the culture medium 3 to 4 days after transfection, typically at 25 to 80 infectious units per ml, by infecting Raji cells and measuring the number of resistant clones that could emerge in medium containing G418. Analysis of the total cellular DNA from several clones showed that the CMVIE-neo gene was indeed linked to the EBV genome and that plasmid vector sequences from p652 were not present. We designate the B95-8-derived virus that has acquired the CMVIE-neo insertion from p652 by homologous recombination as B652.
Recombinant EBV strains were generated from B95-8 cells by using p652, p652dl7, and p652InNco, and the virus stocks were used to immortalize human peripheral blood B cells in 24-well culture dishes. At around 3 weeks after infection, when immortalized B cells, mostly infected with nonrecombinant B95-8 virus, had grown to a fairly high density, G418 was added to each well to select B-cell clones infected with recombinant EBV. B cells immortalized with B652 could be isolated with G418 at 700 μg/ml, but they were not fully resistant even at this concentration, which was the lowest concentration that was found to be sufficient for selection. It is not clear why B cells immortalized with B652 are not fully resistant to G418, since we have found that this virus can provide full resistance to G418 when infecting established Burkitt lymphoma cell lines or other human cell lines. However, probably for this reason, EBV recombinants generated with p652 could give rise to G418-resistant immortalized B-cell clones only about 1% as efficiently as they could confer G418 resistance to Raji cells, a far lower relative efficiency than was found previously with EBV carrying the same G418 resistance gene inserted at other locations (25). After several attempts, a few partially resistant B-cell clones were isolated by using recombinant EBV generated with p652 and with both derivatives of p652 carrying mutations in the EBNA-1 gene. Once partially G418-resistant B-cell clones were identified, they could be expanded in the absence of G418 for analysis.
Immortalization of B cells with EBV lacking most of IR3.
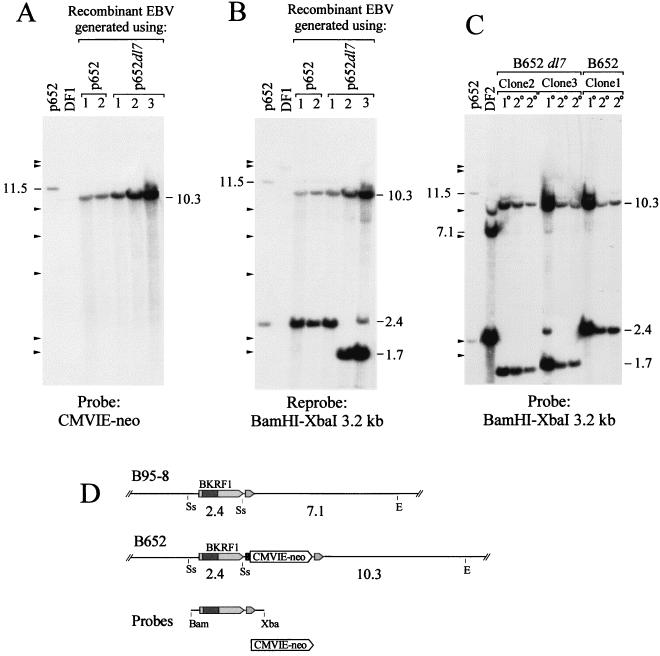
The structure of the viral genomes carried by the G418-resistant B-cell clones was investigated by Southern analysis. To verify that the CMVIE-neo gene had become incorporated into the viral genomes correctly by homologous recombination, total cellular DNA was cut with EcoRV plus SstII and probed with the CMVIE-neo gene. All clones contained the CMVIE-neo gene on a 10.3-kb EcoRV-SstII DNA fragment (Fig. 2A). Insertion of p652 into the EBV genome by a single recombination event would have placed the CMVIE-neo gene on either an 11.6- or an 8.8-kb fragment, depending on which side of the CMVIE-neo gene recombination occurred. Stripping the membrane and reprobing it with a BamHI-XbaI fragment containing the EBNA-1 gene revealed the presence of the IR3 deletion, dl7, in two of the three recombinants obtained by using p652dl7 (Fig. 2B). This deletion removes nearly 700 bp from the EBNA-1 gene, reducing the size of the SstII fragment from 2.4 kb to 1.7 kb. With one of these clones, clone 3, both the deletion-carrying 1.7-kb DNA and the normal-size 2.4-kb DNA were detected, the latter at only 1/10 the intensity of the former, indicating that this clone carried some parental B95-8 viral genomes in addition to more numerous B652dl7 mutant genomes. One of three clones that were analyzed, clone 1, appeared to have an EBNA-1 gene of normal size, indicating that, for this recombinant, recombination had occurred within the 1,250 bp that separates IR3 from the CMVIE-neo insertion.
FIG. 2.
Southern analysis of EBV genomes carrying the inserted CMVIE-neo gene and a deletion of IR3 in immortalized B-cell clones. (A) B cells were immortalized with recombinant EBV that was generated by using each indicated plasmid. Five micrograms of total cellular DNA was digested with EcoRV plus SstII. The blot was probed with the CMVIE-neo gene. To the left, 294 pg of digested p652 DNA was used, corresponding to 9 DNA molecules per cell. For p652, the CMVIE-neo gene is on an 11.5-kb SstII-EcoRV fragment; its position is indicated. (B) The blot was stripped and reprobed with a BamHI-XbaI fragment spanning the EBNA-1 gene. DF1 is an EBV-immortalized B-cell line that carries between 5 and 10 EBV genomes per cell. Cytosine methylation at several SstII sites can account for the large size of the major band and for the partial digestion seen with DF1. (C) Southern analysis of two secondary B-cell clones (2°) that were obtained by using virus released from each of the primary isolates (1°), clones 1, 2, and 3, that were analyzed for panels A and B. DNA was digested with EcoRV plus SstII as for panels A and B and probed with the BamHI-XbaI 3.2-kb fragment. DF2 is an EBV-immortalized B-cell line that contains approximately 100 EBV genomes per cell. Marks to the left of A, B, and C indicate size markers (bacteriophage lambda DNA cut with HindIII). (D) Restriction maps of the relevant portion of the EBV genome for strain B95-8 and for B652. Cleavage sites for SstII (Ss) and EcoRV (E) and the sizes of fragments from the double digest, in kilobase pairs, are indicated; see the legend to Fig. 1 for details. DNAs used to make hybridization probes are shown below.
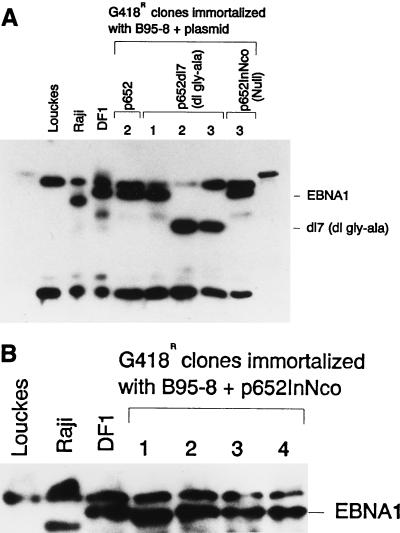
The presence of the dl7 deletion in the recombinant virus in the two B-cell clones was verified by immune blotting of whole-cell extracts by using EBNA-1-specific rabbit antibodies. The EBNA-1 detected in clones 2 and 3 had a mobility consistent with a size of 47 kDa as expected for the loss of almost all of the glycine-alanine repeats (Fig. 3A).
FIG. 3.
Detection of EBNA-1 in B-cell clones carrying recombinant EBV. (A and B) Immunoblot analysis of protein extracted from 100,000 cells of different B-cell clones that were immortalized with EBV that had recombined with the indicated plasmids. The numbers above the lanes of the gels designate specific clones, most of which were also analyzed for viral DNA in Fig. 2 and 4. EBNA-1 of the B95-8 size and of the dl7 size are indicated by EBNA1 and dl7, respectively, to the left. Louckes is an EBV-negative Burkitt’s lymphoma cell line. EBV of Raji has fewer IR3 repeats than B95-8 EBV, so its EBNA-1 is correspondingly smaller. DF1 is a human B-cell clone immortalized with B95-8 EBV.
To verify that IR3-deleted virus could immortalize B cells in the absence of wild-type EBNA-1, clones 2 and 3, carrying B652dl7 virus, were gamma irradiated, cocultivated with peripheral blood leukocytes, and cultured in multiwell plates. Transformed B-cell clones emerged at frequencies close to 1 per 20,000 donor cells for both clones, a frequency that is typical of the release of EBV from immortalized B cells, and this compared to 1 per 40,000 donor cells obtained with clone 1 carrying B652 virus. Five transformants obtained with B652dl7 virus were examined by Southern analysis and shown to be infected only with virus carrying the IR3 deletion. The analysis of four such secondary transformants, two from each of the primary isolates, clones 2 and 3, is shown in Fig. 2C. These results indicate that the IR3 repeats, which comprise over one-third of the coding sequence of EBNA-1, are dispensable for immortalization of B cells and for virus production.
All of the secondary B-cell transformants obtained in this experiment exhibited the same partial G418 resistance as observed with the primary isolates. The secondary transformants, which were isolated and expanded in the absence of G418, tended to have fewer EBV genome copies per cell than the primary transformants that were grown under drug selection (Fig. 2C). The results indicate that the low frequencies with which the primary isolates were obtained most likely reflect inadequate expression of the CMVIE-neo gene in EBV-immortalized B cells.
Inability of EBNA-1-deficient EBV to immortalize B cells.
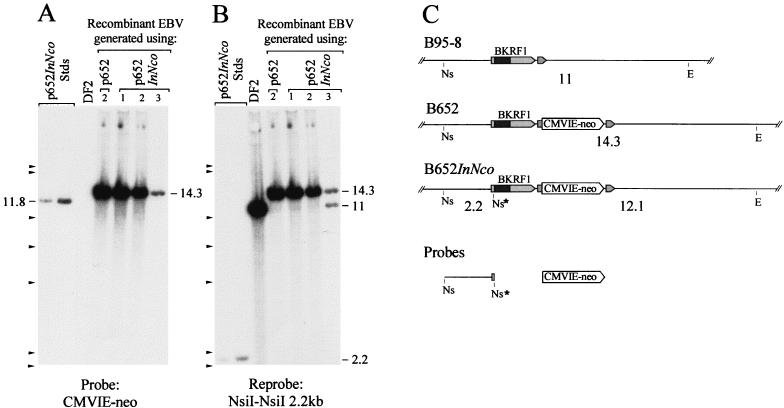
Three G418-resistant human B-cell clones that were obtained by using recombinant EBV generated with p652InNco were analyzed for the structure of the viral genomes they carried. The insertion of 4 bp at the NcoI site in the EBNA-1 gene created an NsiI site. If this NsiI site were present on the viral chromosomes containing the CMVIE-neo gene, Southern analysis would reveal a 12.1-kb EcoRV-NsiI DNA fragment hybridizing to a labeled CMVIE-neo probe. What was detected instead in DNA from all three clones was the 14.3-kb fragment expected in B652 genomes lacking the frameshift mutation (Fig. 4A), thus indicating that the CMVIE-neo gene was linked to the wild-type EBNA-1 gene on the EBV genomes in these clones. When the blot was stripped and reprobed with the 2.2-kb of DNA extending upstream from the frameshift mutation to the nearest NsiI site, again the 14.3-kb band was detected while the 2.2-kb NsiI fragment was not, confirming the absence of the frameshift insertion within the EBNA-1 gene (Fig. 4B). One clone, clone 3, also appeared to be infected with nonrecombinant B95-8 virus, as indicated by the 11-kb fragment. By Western blotting, it was shown that these three clones, and a fourth G418-resistant B-cell clone that was also obtained with p652InNco, all contained EBNA-1 protein similar in size to EBNA-1 of strain B95-8 (Fig. 3B).
FIG. 4.
Failure to transfer the InNco frameshift mutation to recombinant EBV genomes recovered in immortalized B cells. Five micrograms of DNA from each clone was digested with EcoRV plus NsiI and probed with the CMVIE-neo gene in panel A. For panel B, the membrane was stripped and reprobed with the 2.2 kb of DNA extending from the NsiI site at the frameshift mutation (InNco) leftward to the next NsiI site (Fig. 1). DF2 is a human B-cell clone that carries approximately 100 EBV genomes (strain B95-8) per cell. To the left, digested p652 DNA was loaded at 294 and 589 pg per lane, corresponding to 9 and 18 copies per cell. An 11.8-kb NsiI-EcoRV fragment from the plasmid is indicated. The other indicated bands are described in the text. (C) Restriction maps of the relevant portion of the EBV genome for strains B95-8, B652, and B652InNco. Cleavage sites for NsiI (Ns) and EcoRV (E) and the sizes of fragments from the double digest, in kilobase pairs, are indicated; see Fig. 1 for details. DNAs used to make hybridization probes are shown below.
The recombinant EBV genomes all acquired the CMVIE-neo gene by recombination on both sides of the selective marker. For the clones obtained with p652InNco, the position where recombination occurred on the “left” side of the CMVIE-neo gene was in all four cases within the 2 kb of DNA separating the frameshift mutation and the CMVIE-neo gene and was never within the 15 kb of homologous EBV sequence that extends from the left of the frameshift mutation on the plasmid, which would have introduced the EBNA-1 mutation into the viral genome. It seems likely that most recombinant viruses that acquired the CMVIE-neo gene also acquired the mutation in the EBNA-1 gene but were not isolated because the frameshift mutation within the EBNA-1 gene prevented efficient immortalization of B cells.
To determine at what frequency the frameshift mutation remained linked to the selective marker among the recombinant viruses that were generated, stocks of B652InNco were obtained by transient transfection of B95-8 cells and used to infect EBV-positive Raji cells, where the endogenous EBV would complement a superinfecting EBNA-1-defective virus. Of 13 G418-resistant clones that were expanded and examined by Southern analysis, the CMVIE-neo gene was linked to the frameshift mutation in the recombinant EBV genomes of 11 clones, while for two clones, the recombinant EBV contained a wild-type EBNA-1 gene. A recombinant virus stock was also used to superinfect B95-8 cells which had been made to express the CR2 gene, as described below. Of five superinfected clones that were analyzed, recombinant EBV chromosomes had acquired the frameshift mutation in four clones and retained the wild-type EBNA-1 gene in one clone. When B652dl7 was isolated by immortalizing B cells as discussed above, the IR3 deletion remained linked to the selective marker in two of the three clones that were isolated. Considering all of these results together, of 21 cases in which the mutation in the EBNA-1 gene was harmless (dl7) or else EBNA-1 function was supplied in trans, the CMVIE-neo gene remained linked to the mutant EBNA-1 gene 17 times and became linked to the wild-type EBNA-1 gene 4 times. In contrast, the InNco mutation did not remain linked to the selective marker in any of four B-cell clones that were immortalized with recombinant virus generated from p652InNco. By comparing the two distributions, 17:4 to 0:4, the likelihood of recovering only the wild-type EBNA-1 gene on recombinant EBV in the immortalized B-cell clones by chance alone can be calculated to be 0.007 by Fisher’s exact test (Table 1). This is evidence that EBNA-1 is required for efficient immortalization of B cells by EBV.
TABLE 1.
Failure to recover EBNA-1-deficient EBV in immortalized B cells
| Type of cells infected | Recombining plasmid | No. of clones with G418-resistant EBV carrying:
|
|
|---|---|---|---|
| Mutant EBNA-1 | Wild-type EBNA-1 | ||
| Raji | p652InNco | 11 | 2 |
| B95-8/CR2 | p652InNco | 4 | 1 |
| Human B cells | p652dl7 | 2 | 1 |
| Total | 17 | 4 | |
| Human B cells | p652InNco | 0 | 4 (P = 0.007)a |
Fisher exact test for consistency in a two-by-two table, comparing distributions of 17:4 and 0:4.
Inability of EBNA-1-deficient EBV to establish a latent infection in BL30 cells.
EBNA-1 might be required for the control of EBV gene transcription in immortalized B cells (10, 33, 39), so it was important to test whether EBNA-1-deficient EBV could establish a latent infection in an established cell line that does not depend on EBV. For this study, it was advantageous to generate a high-titer stock of B652InNco virus. This was accomplished by generating a low-titer stock, by transiently transfecting B95-8 cells with p652InNco plus pCMV-BZLF1, and using this to superinfect B95-8 cells that carried a plasmid expressing the EBV receptor, CD21 (7). After expansion of several G418-resistant, superinfected clones, Southern analysis showed that four clones contained recombinant EBV genomes in which the frameshift mutation remained linked to the CMVIE-neo gene, while in one clone the recombinant EBV genomes contained the wild-type EBNA-1 gene. Interestingly, the clones that carried the EBNA-1-deficient EBV genomes grew noticeably more slowly than the clone that contained only the wild-type EBNA-1 gene, although after several weeks of propagation, their growth rates improved.
The Southern analysis indicated that the superinfecting recombinant EBV genomes were present at high copy and similar in number to the preexisting B95-8 viral genomes (see Fig. 6). This indicated that the superinfecting viral genomes were being amplified in the fraction of cells undergoing lytic development and that they should be released efficiently as infectious virus. One superinfected clone was chosen as a source of B652InNco virus, and the clone in which the recombinant EBV genome had regained the wild-type EBNA-1 gene provided a source of B652 virus. The titer of EBV that could confer clonal resistance to G418 was determined by infecting Raji cells, which are EBV positive, and by infecting BL30 cells, which are EBV negative. B652 virus was detected at high titers when used to infect either cell line (63,000 and 120,000 per ml for Raji and BL30 cells, respectively). In contrast, B652InNco stably infected Raji cells 20 times more efficiently than it could infect BL30 cells (15,000/ml versus 750/ml). These data, which are presented in Table 2, indicated that, without EBNA-1, EBV was at most 5% as effective as wild-type virus in establishing a latent infection that could provide G418 resistance. If the data are normalized for the relative efficiency with which the EBNA-1-proficient virus, B652, conferred G418 resistance to the two cell lines, then the relative efficiency of establishing a latent infection in the absence of EBNA-1 was no more than 2.6%.
FIG. 6.
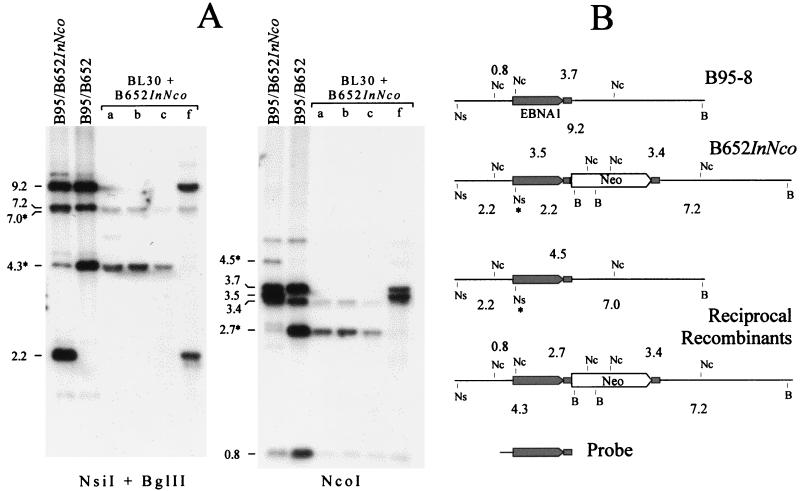
Stable infection of BL30 cells with B652InNco EBV selects for recombinants with the EBNA-1 gene restored. (A) Southern analysis of 0.5 μg of DNA from superinfected B95-8 clones carrying either B652InNco virus or B652 virus and of 5 μg of DNA from four G418-resistant clones of BL30 infected with virus released from B95/B652InNco superinfected cells. The DNA was cut with a combination of NsiI and BglII (left) or with NcoI (right). Approximate sizes are given for bands of interest. DNA fragments that resulted from recombination between the two viruses, B652InNco and B95-8, are marked with an asterisk. (B) Restriction site maps of the relevant part of the B95-8 and B652InNco viral genomes and their recombination products. B, BglII; Nc, NcoI; Ns, NsiI. An asterisk marks the NsiI site created by the frameshift mutation. The probe that was used contained the sequences 3′ of the EBNA-1 gene that were duplicated (small filled rectangle) and are present on both sides of the selective marker. Note that one of the reciprocal recombinants is identical to B652. (The probe contained, in addition to the region shown, about 100 bp of EBV sequence to the left of oriP, so EBV DNA fragments of 5.1 kb in the NcoI digestion and 0.6 kb in the NsiI-BglII digestion [not shown] were also detected weakly.)
TABLE 2.
Inability of EBNA-1-deficient EBV to stably infect BL30 cells
| Virus | Titer (G418-resistant CFU/ml) when infecting cell linea:
|
Ratio of titers infecting BL30/Raji cells | |
|---|---|---|---|
| BL30 | Raji | ||
| B-652 | 120,000 | 63,000 | 1.9 |
| B-652InNco | 720 | 15,000 | 0.05 |
See Materials and Methods for details.
Nine G418-resistant clones of BL30 that had been infected with the B652InNco stock of virus were examined by using in situ-lysis gels (11), and all were found to contain circular EBV genomes (data not shown). Western analysis revealed that all nine clones contained EBNA-1 protein of the B95-8 size (Fig. 5). One clone, designated clone f, expressed EBNA-1 at a level that was abnormally low but still detectable. DNA from four clones, including clone f, was investigated by Southern analysis (Fig. 6). The analysis is explained below. In short, the EBV genomes in three of the clones had the same structure as B652; that is, they had acquired the wild-type EBNA-1 gene by recombination with B95-8 virus in the superinfected B95-8 cells. Clone f appeared to be doubly infected, carrying genomes of both B652InNco and B95-8 viruses.
FIG. 5.
BL30 clones, obtained by selecting for stable infection by EBNA-1-deficient virus, all contain EBNA-1 protein. Immunoblot of protein extracted from 100,000 cells. Nine G418-resistant clones of BL30, labeled a through j, were obtained by infection with EBV released from B95-8 cells superinfected with B652InNco. The arrow indicates B95-8 EBNA-1.
The Southern analysis shown in Fig. 6 helped to account for the relative efficiency with which B652InNco virus, as released from superinfected B95-8 cells, could stably infect EBV-negative BL30 cells. A probe containing the EBNA-1 gene detects (primarily) a 2.2-kb NsiI-BglII fragment when the NsiI site, created by the frameshift mutation, is present in the EBNA-1 gene and linked to the selective marker; this probe detects a 4.3-kb NsiI-BglII fragment when the wild-type EBNA-1 gene is linked to the selective marker, and it detects a 9.2-kb NsiI-BglII fragment in B95-8 DNA (Fig. 6A, left blot, and B). Similarly, with NcoI-digested DNA, the EBNA-1 gene probe detects (primarily) a 3.5-kb fragment if InNco is linked to the selective marker, a 2.7-kb fragment if wild-type EBNA-1 is linked to the selective marker, and a 3.7-kb fragment in B95-8 EBV (right blot). It is clear from both blots that some of the viral genomes in B95-8 cells superinfected with B652InNco (indicated as B95/B652InNco) have the wild-type EBNA-1 gene linked to the selective marker (4.3-kb DNA in the NsiI-BglII digest, 2.7-kb DNA in the NcoI digest). These, it appears, were generated by homologous recombination between the mutant EBV genome and the B95-8 EBV genome. The reciprocal product of this recombination would place the frameshift mutation on a B95-8 EBV chromosome lacking the selective marker, as the diagram indicates in Fig. 6B. A 7.0-kb NsiI-BglII DNA fragment and a 4.5-kb NcoI fragment that would be cleaved from the reciprocal recombination product were detected at the expected intensities.
The products of recombination made up about 6% of EBV chromosomes present in DNA from B95-8 cells superinfected with B652InNco, based on the strengths of hybridization signals measured with a PhosphorImager and averaged for the two blots in Fig. 6A. This agrees well with the 2.6 to 5% relative efficiency with which virus released from these cells could confer stable G418 resistance to EBV-negative BL30 cells, as determined in Table 2. Of the G418-resistant clones that emerged from the infection—at the frequency of 2.6 to 5%—nine out of nine that were tested were EBNA-1-positive. If EBNA-1-deficient EBV could establish a latent infection of BL30 cells with 1% the efficiency of wild-type EBV, then it would be expected that two to four of the nine clones would have carried only EBNA-1-deficient EBV, presumably integrated into a host chromosome. Therefore, an upper estimate of the relative efficiency by which EBNA-1-deficient EBV might stably infect BL30 cells under these conditions would be, conservatively, less than 1%.
DISCUSSION
This report provides strong genetic evidence that EBNA-1 is required in order for EBV to establish a stable latent infection in proliferating cells. It has long been understood that EBNA-1 is required in order for oriP of EBV to be active on artificial plasmids, to allow such plasmids to be replicated and maintained stably in cells (31, 48). It could not be assumed that this would be the case for the EBV chromosome, however. As discussed in the introduction, much evidence now suggests that EBV does not require the replication initiation function of oriP, and other gamma herpesviruses that maintain their chromosomes autonomously in proliferating cells do not have a genetic homolog of EBNA-1.
The implication of this work is that EBNA-1 is essential for maintenance of the circular EBV chromosome in proliferating cells, almost certainly in conjunction with the extrachromosomal maintenance function of oriP, if not its replication function. Replication appears to initiate most of the time at sites distant from oriP on EBV latent chromosomes (29), and EBNA-1 is unlikely to be involved in replication that initiates away from oriP, since specific binding sites for EBNA-1 have been found only at oriP and at the Qp promoter (34). This reasoning leads to the conclusion that it is the extrachromosomal maintenance function of EBNA-1 and of oriP, rather than their replication function, that is required. Thus, replication of the EBV chromosome would appear to be insufficient for its stable maintenance.
Studies that have employed the FR of oriP and EBNA-1 to provide a plasmid segregation function have shown this to be the case for artificial plasmids that can otherwise replicate (16, 23), even for plasmids much larger than the EBV genome (36). It should be expected that other viruses that maintain their chromosomes autonomously in proliferating cells will also have mechanisms to ensure mitotic stability. Recent work has shown that bovine papillomavirus plasmids achieve this by attachment to cellular chromosomes during mitosis (26), as does EBV through oriP and EBNA-1 (see introduction), so this mechanism might turn out to be common.
The failure of EBV carrying the InNco frameshift mutation in the EBNA-1 gene to be recovered among EBV-transformed B-cell clones was statistically significant, despite the fact only a few clones were obtained for this analysis (Table 1). This is evidence that EBNA-1 contributes to the efficiency with which EBV immortalizes B cells, but it does not necessarily mean that EBNA-1 is essential for B-cell immortalization. A rigorous test of this will require obtaining a stock of EBNA-1-null EBV that contains little or no virus carrying a functional EBNA-1 gene. It might be expected that at some frequency EBV could sustain the transformed state in a B-cell clone by having integrated its genome into a human chromosome. However, in only one documented case has an EBV-immortalized B-cell line been found to carry only integrated EBV genomes in the absence of circular copies, and in this case, the EBV genome became integrated after it had circularized and perhaps after the clone had been established in culture (18).
More than one-third of the EBNA-1 protein sequence is a repetitive array of alanine and glycine repeats, which were previously shown to be dispensable for the plasmid replication, plasmid maintenance, and transcription-activating functions of EBNA-1 (45, 48). Subsequent work has revealed that the Gly-Ala repeats of EBNA-1 become essential for these functions when a neighboring arginine-rich region of the protein has been deleted, suggesting that the Gly-Ala repeats might contribute to these functions normally in some way (our unpublished data). More recently, this region of the EBNA-1 protein was shown to prevent protein processing for display and recognition by cytotoxic T cells (27, 28). The demonstration here that the Gly-Ala repeats are also dispensable for the stable latent infection and immortalization of B cells by EBV reinforces the idea that the primary function of this unusual sequence could indeed be to prevent a cellular immune response against latently infected cells.
In contrast to the situation with immortalized B cells, when EBV-negative Burkitt’s lymphoma cell lines have been stably infected with EBV in culture, the resulting cell lines have been found frequently to carry only an integrated EBV genome (17, 41). For this reason, it was somewhat surprising that EBV lacking a functional EBNA-1 gene could not stably infect BL30, an EBV-negative Burkitt’s lymphoma cell line, at even 1% the frequency at which EBV carrying a wild-type EBNA-1 gene could stably infect these cells. One possible explanation is that EBV integrates into BL30 less frequently than it does when infecting other Burkitt’s lymphoma cell lines, which is consistent with the limited available data (17, 41). It is also conceivable that the selection for expression of a G418 resistance gene present on the EBV chromosome in our study favored an episomal configuration over an integrated one, although this could not have been for reasons of gene copy number, since several of the infected BL30 clones that we analyzed appeared to carry only one or two circular viral chromosomes per cell on average (data not shown). Another possibility is that expression of the selective marker was to some extent dependent on the FR enhancer of oriP, which requires EBNA-1, despite being tens of kilobases from oriP on the EBV chromosome. A final possibility is that EBNA-1 might be important to stabilize the EBV chromosome soon after infection or for some other function upon which integration might depend.
The results of this study confirm the central importance of EBNA-1, and of oriP, presumably, to EBV in that they allow the virus to maintain its genome autonomously in mitotically active cells. This knowledge increases the incentive to discover the molecular details of this function.
ACKNOWLEDGMENTS
We thank Sarah M. Camiolo for expert help in constructing the large plasmids and Jackie M. Bashaw for composing the figures.
This work was supported by grant CA4312212 from the NIH.
REFERENCES
- 1.Adams A. Replication of latent Epstein-Barr virus genomes in Raji cells. J Virol. 1987;61:1743–1746. doi: 10.1128/jvi.61.5.1743-1746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloss T A, Sugden B. Optimal lengths for DNAs encapsidated by Epstein-Barr virus. J Virol. 1994;68:8217–8222. doi: 10.1128/jvi.68.12.8217-8222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chittenden T, Lupton S, Levine A J. Functional limits of oriP, the Epstein-Barr virus plasmid origin of replication. J Virol. 1989;63:3016–3025. doi: 10.1128/jvi.63.7.3016-3025.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decker L L, Shankar P, Khan G, Freeman R B, Dezube B J, Lieberman J, Thorley-Lawson D A. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J Exp Med. 1996;184:283–288. doi: 10.1084/jem.184.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dillner J, Rabin H, Letvin N, Henle W, Henle G, Klein G. Nuclear DNA-binding proteins determined by the Epstein-Barr virus-related simian lymphotropic herpesviruses H. gorilla, H. pan, H. pongo and H. papio. J Gen Virol. 1987;68:1587–1596. doi: 10.1099/0022-1317-68-6-1587. [DOI] [PubMed] [Google Scholar]
- 7.Evans T J, Jacquemin M G, Farrell P J. Efficient EBV superinfection of group I Burkitt’s lymphoma cells distinguishes requirements for expression of the Cp viral promoter and can activate the EBV productive cycle. Virology. 1995;206:866–877. doi: 10.1006/viro.1995.1009. [DOI] [PubMed] [Google Scholar]
- 8.Frappier L, O’Donnell M. Epstein-Barr nuclear antigen 1 mediates a DNA loop within the latent replication origin of Epstein-Barr virus. Proc Natl Acad Sci USA. 1991;88:10875–10879. doi: 10.1073/pnas.88.23.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gahn T A, Schildkraut C L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 10.Gahn T A, Sugden B. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J Virol. 1995;69:2633–2636. doi: 10.1128/jvi.69.4.2633-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardella T, Medveczsky P, Sairenji T, Mulder C. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J Virol. 1984;50:248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grogan E A, Summers W P, Dowling S, Shedd D, Gradoville L, Miller G. Two Epstein-Barr viral nuclear neoantigens distinguished by gene transfer, serology, and chromosome binding. Proc Natl Acad Sci USA. 1983;80:7650–7653. doi: 10.1073/pnas.80.24.7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 14.Hampar B, Tanaka A, Nonoyama M, Derge J G. Replication of the resident repressed Epstein-Barr virus genome during the early S phase (S-1 period) of nonproducer Raji cells. Proc Natl Acad Sci USA. 1974;71:631–633. doi: 10.1073/pnas.71.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris A, Young B D, Griffin B E. Random association of Epstein-Barr virus genomes with host cell metaphase chromosomes in Burkitt’s lymphoma-derived cell lines. J Virol. 1985;56:328–332. doi: 10.1128/jvi.56.1.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinzel S S, Krysan P J, Tran C T, Calos M P. Autonomous DNA replication in human cells is affected by the size and the source of the DNA. Mol Cell Biol. 1991;11:2263–2272. doi: 10.1128/mcb.11.4.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurley E A, Agger S, McNeil J A, Lawrence J B, Calendar A, Lenoir G, Thorley-Lawson D A. When Epstein-Barr virus persistently infects B-cell lines, it frequently integrates. J Virol. 1991;65:1245–1254. doi: 10.1128/jvi.65.3.1245-1254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurley E A, Klaman L D, Agger S, Lawrence J B, Thorley-Lawson D A. The prototypical Epstein-Barr virus-transformed lymphoblastoid cell line IB4 is an unusual variant containing integrated but no episomal viral DNA. J Virol. 1991;65:3958–3963. doi: 10.1128/jvi.65.7.3958-3963.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan M E, Clark C. An improved rosetting assay for detection of human T lymphocytes. J Immunol Methods. 1974;5:131–135. doi: 10.1016/0022-1759(74)90003-9. [DOI] [PubMed] [Google Scholar]
- 20.Kieff E, Rickinson A B. Epstein-Barr virus. In: Knipe D M, Fields B N, Howley P M, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 21.Kieff E. Epstein-Barr virus and its replication. In: Knipe D M, Fields B N, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2324–2396. [Google Scholar]
- 22.Klein G. Epstein-Barr virus strategy in normal and neoplastic B cells. Cell. 1994;77:791–793. doi: 10.1016/0092-8674(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 23.Krysan P J, Haase S B, Calos M P. Isolation of human sequences that replicate autonomously in human cells. Mol Cell Biol. 1989;9:1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kung S-H, Medveczky P G. Identification of a herpesvirus saimiri cis-acting DNA fragment that permits stable replication of episomes in transformed T cells. J Virol. 1996;70:1738–1744. doi: 10.1128/jvi.70.3.1738-1744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee M A, Kim O J, Yates J L. Targeted gene disruption in Epstein-Barr virus. Virology. 1992;189:253–265. doi: 10.1016/0042-6822(92)90701-p. [DOI] [PubMed] [Google Scholar]
- 26.Lehman C W, Botchan M R. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc Natl Acad Sci USA. 1998;95:4338–4343. doi: 10.1073/pnas.95.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen P M, Klein G, Kurilla M G, Masucci M G. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 28.Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci M G. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc Natl Acad Sci USA. 1997;94:12616–12621. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Little R D, Schildkraut C L. Initiation of latent DNA replication in the Epstein-Barr virus genome can occur at sites other than the genetically defined origin. Mol Cell Biol. 1995;15:2893–2903. doi: 10.1128/mcb.15.5.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loeb D D, Sung N S, Pesano R L, Sexton C J, Hutchison III C D, Pagano J S. Plasmid origin of replication of herpesvirus papio: DNA sequence and enhancer function. J Virol. 1990;64:2876–2883. doi: 10.1128/jvi.64.6.2876-2883.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lupton S, Levine A J. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985;5:2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Middleton T, Sugden B. EBNA1 can link the enhancer element to the initiator element of the Epstein-Barr virus plasmid origin of DNA replication. J Virol. 1992;66:489–495. doi: 10.1128/jvi.66.1.489-495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puglielli M T, Woisetschlaeger M, Speck S H. oriP is essential for EBNA gene promoter activity in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J Virol. 1996;70:5758–5768. doi: 10.1128/jvi.70.9.5758-5768.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rawlins D R, Milman G, Hayward S D, Hayward G S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 35.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simpson K, McGuigan A, Huxley C. Stable episomal maintenance of yeast artificial chromosomes in human cells. Mol Cell Biol. 1996;16:5117–5126. doi: 10.1128/mcb.16.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sternås L, Middleton T, Sugden B. The average number of molecules of Epstein-Barr nuclear antigen 1 per cell does not correlate with the average number of Epstein-Barr virus (EBV) DNA molecules per cell among different clones of EBV–immortalized cells. J Virol. 1990;64:2407–2410. doi: 10.1128/jvi.64.5.2407-2410.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su W, Middleton T, Sugden B, Echols H. DNA looping between the origin of replication of Epstein-Barr virus and its enhancer site: stabilization of an origin complex with Epstein-Barr nuclear antigen 1. Proc Natl Acad Sci USA. 1991;88:10870–10874. doi: 10.1073/pnas.88.23.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugden B, Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989;63:2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virgin H W, IV, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang F, Marchini A, Kieff E. Epstein-Barr virus (EBV) recombinants: use of positive selection markers to rescue mutants in EBV-negative B-lymphoma cells. J Virol. 1991;65:1701–1709. doi: 10.1128/jvi.65.4.1701-1709.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wysokenski D A, Yates J L. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J Virol. 1989;63:2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yandava C N, Speck S H. Characterization of the deletion and rearrangement in the BamHI C region of the X50-7 Epstein-Barr virus genome, a mutant viral strain which exhibits constitutive BamHI W promoter activity. J Virol. 1992;66:5646–5650. doi: 10.1128/jvi.66.9.5646-5650.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yates J L, Camiolo S M. Dissection of DNA replication and enhancer activation functions of Epstein-Barr virus nuclear antigen 1. In: Stillman B, Kelley T, editors. Cancer cells. Vol. 6. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. pp. 197–205. [Google Scholar]
- 46.Yates J L, Camiolo S M, Ali S, Ying A. Comparison of the EBNA1 proteins of Epstein-Barr virus and herpesvirus papio in sequence and function. Virology. 1996;222:1–13. doi: 10.1006/viro.1996.0392. [DOI] [PubMed] [Google Scholar]
- 47.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]