ABSTRACT
Over 100 human adenoviruses (HAdVs) have been isolated and allocated to seven species, A-G. Species F comprises two members-HAdV-F40 and HAdV-F41. As their primary site of infection is the gastrointestinal tract they have been termed, with species A, enteric adenoviruses. HAdV-F40 and HAdV-F41 are a common cause of gastroenteritis and diarrhoea in children. Partly because of difficulties in propagating the viruses in the laboratory, due to their restrictions on growth in many cell lines, our knowledge of the properties of individual viral proteins is limited. However, the structure of HAdV-F41 has recently been determined by cryo-electron microscopy. The overall structure is similar to those of HAdV-C5 and HAdV-D26 although with some differences. The sequence and arrangement of the hexon hypervariable region 1 (HVR1) and the arrangement of the C-terminal region of protein IX differ. Variations in the penton base and hexon HVR1 may play a role in facilitating infection of intestinal cells by HAdV-F41. A unique feature of HAdV-F40 and F41, among human adenoviruses, is the presence and expression of two fibre genes, giving long and short fibre proteins. This may also contribute to the tropism of these viruses. HAdV-F41 has been linked to a recent outbreak of severe acute hepatitis “of unknown origin” in young children. Further investigation has shown a very high prevalence of adeno-associated virus-2 in the liver and/or plasma of some cohorts of patients. These observations have proved controversial as HAdV-F41 had not been reported to infect the liver and AAV-2 has generally been considered harmless.
KEYWORDS: Adenovirus F41, adenovirus F40, HadV-F41, childhood hepatitis, hepatitis of unknown cause, adeno-associated virus 2
Introduction
Human adenoviruses (HAdVs), together with adenoviruses infecting other mammals, are members of the genus Mastadenovirus. They were first isolated in 1953 from human adenoids, following an outbreak of respiratory disease in a military facility [1]. Since then, over 100 different human adenoviruses have been characterized. They have been divided into seven species or groups, A to G. Viruses within a species show a marked similarity in nucleotide sequence and in tropism for site of infection. Initial subtyping was on the basis of haemagglutination inhibition and serum neutralization assays, but more recently genetic analysis has been used to assign viruses to a particular species [2–4]. Most HAdVs have been allocated to species D which has 75 members at the last count; species A comprises 4 viruses, species B 16 viruses, species C 5 viruses, species F 2 viruses, and E and G, 1 member each. In the family Adenoviridae, as well as mammalian viruses (Mastadenovirus), there are also adenoviruses which infect birds (Aviadenovirus), and fish (Ichtadenovirus). In addition, two other genera have been characterized Atadenovirus contains a mixture of adenoviruses infecting birds, mammals, and reptiles and Siadenovirus contains viruses which infect birds and amphibians [5].
The HAdVs are non-enveloped DNA viruses with a linear double stranded genome of approximately 30–40 kbp [6,7]. They tend to be restricted in the tissues and organs they infect, and this limitation results in specific adenovirus-associated diseases [8–10]. Thus, species B, C, and E target the respiratory tract and cause pneumonia and acute respiratory infection; species C also causes hepatitis and pharyngitis; species B and D infect the eye and cause keratoconjunctivitis and species B has also been linked to renal and urinary tract infections; species A, F, and G target the gastrointestinal tract and cause gastroenteritis and diarrhoea. Of the viruses causing gastrointestinal disease, the group F adenoviruses are most common. Species F has two members, HAdV-F40 and HAdV-F41. Together, they are known as the enteric adenoviruses and are the second most common cause of viral gastroenteritis and diarrhoea in children, worldwide.
Recently, HAdV-F41 has also been linked to a large number of cases of acute, severe hepatitis “of unknown origin” in young children. The nomenclature “hepatitis of unknown origin,” which is used throughout this review, is taken to mean that the patients are negative for hepatitis viruses (HAV, HBV, HCV, HDV, and HEV) and have serum aspartate aminotransferase or alanine aminotransferase level over 500 IU/L. This outbreak is considered, in detail, towards the end of this review (Section 8).
The enteric adenoviruses were first detected in stool samples from children suffering from acute gastroenteritis [11,12] and were later shown to comprise two closely related subtypes, HAdV-F40 and HAdV-F41 [13,14]. Complete nucleotide sequences are now available for several isolates of each virus: for example, HAdV-F40 Duggan (L19443) and HAdV-F41 Tak (DQ315364). HAdV-F40 and HAdV-F41 comprise 34,214 and 34,188 nucleotides, respectively [15,16]. Although the species F viruses are of considerable clinical importance, they have not been subject to the same detailed virological and biochemical analysis as the much more commonly studied species C (for example, HAdV-C2 and HAdV-C5) or species A (for example, HAdV-A12) viruses. This is partly because they are more difficult to propagate in the laboratory. Early studies showed that HAdV-F40 and F41 would grow well in certain commonly used cell lines, such as HT-29, but very poorly in others, such as HeLas. This led to them being termed “Fastidious viruses”; this is discussed in more detail in Section 4 of this review. The possible link of HAdV-F41 to hepatitis has stimulated scientific interest in the species F viruses, such that a cryo-EM structure has now been determined for the virus [17,18]. However, our knowledge of the properties and mode of action of individual HAdV-F41 proteins is very limited and, in most cases, non-existent. Therefore, some of the following discussion is based on extrapolation from our much more extensive knowledge of other HAdV species, in particular HAdV-C5. Several early reviews of HAdV-F species viruses contain useful information [16,19–23].
Species F adenovirus-associated disease
Infection with human adenoviruses is very common with most children infected with at least one species in early childhood. In immunocompetent individuals, symptoms are generally mild and disappear within one to two weeks. Simplistically, infection with adenoviruses from different groups is associated with particular clinical outcomes [9,10]. For example, respiratory disease, such as pneumonia, is mainly associated with species HAdV-B, C, and E, whereas keratoconjunctivitis is caused by species HAdV-B and D. Viral gastroenteritis and diarrhoea are predominantly caused species by HAdV-A, HAdV-F, and HAdV-G adenoviruses although it is notable that other adenovirus species can be shed in the gut (see Section 4).
The enteric, species F, adenoviruses cause gastroenteritis and diarrhoea in young children and infants throughout Europe, Africa, Asia, and the Americas, and are linked to 5–20% of the cases of diarrhoea [16]. They cause disease equally in industrialized and developing countries with little seasonal variation. An incubation time of about 1 week has been observed, after which infected children suffer from diarrhoea and vomiting but generally do not have fever. The two adenoviruses cause similar disease although HAdV-F40 infection has been suggested to result in milder diarrhoea symptoms [16]. In addition, both viruses tend to cause a milder disease than rotavirus, the primary cause of viral gastroenteritis in children. Transmission of HAdV-F viruses is probably by faecal-oral spread. Although most infected children recover quickly, enteric adenoviruses occasionally have been associated with fatal disease [24]. Infection with the HAdV-F species virus is common in children but is much rarer in adults. For example, in a wide-ranging study, in Brazil, of adenoviruses present in faecal samples from 5035 patients with gastroenteritis, the most common were enteric viruses (78% of the adenovirus-positive isolates, with 72% species F) [25]. The great majority of adenoviruses were present in the samples from children under 5 years of age (82%) with only 4.5% of the adenovirus-positive isolates from individuals over the age of 25 [25]. The mean and median ages of HAdV-positive patients in this study were 5.9 and 1 year, respectively.
In immunocompromised patients HAdV-F40 and 41 infections have not been commonly reported. For example, in a study of 289 immunocompromised patients only five were found to be infected with enteric adenoviruses [26]. In studies of AIDS patients, there was a weak correlation between increased shedding of enteric viruses and immunodeficiency although no specific virus was linked to AIDs-associated diarrhoea [27,28]. In a study of stem cell transplantation in children, adenovirus was isolated from 40% of the patients, but no species F viruses were observed [29]. Similarly, in a study of 153 transplant patients, adenovirus was isolated from the stool of 53 individuals. On genotyping the viruses, 76% were species C, 11% species A, 7% species D, 4% species B, and only 2% species F [30]. It has been concluded that in transplant patients there is “a very strong preponderance of species C in most instances” [31]. The corollary of this appears to be that immunosuppression makes very little difference to the incidence of HAdV-F40 or HAdV-F41 infection. In addition, transplantation tends not to occur in very young children who constitute most patients suffering from enteric adenovirus-associated gastroenteritis. It might be supposed that patients who are immunosuppressed will have passed the age when HAdV-F adenovirus infection is prevalent. A possible association of HAdV-F41 with severe, acute hepatitis in young children is considered in detail in Section 8.
Viral infection and persistence
Although most cases of adenovirus-attributable disease arise from de novo infection, it is now clear that the virus can persist in individuals in a latent form (reviewed [31,32]). Several sites of viral persistence have been identified. It was originally shown that group C adenoviruses persist in adenoids and tonsils [33] but adenoviruses have also been reported in other sites, such as lung epithelial cells and, importantly, the intestine [34–37]. Most studies of adenovirus persistence have concentrated on the species C types, such as HAdV-C2 and HAdV-C5. Adenoviruses were originally isolated from tonsils and adenoids [1]. Slightly later studies showed that small amounts of the group C viruses, which were able to replicate, could persist in these tissues [38–41]. However, it was also noted that in many cases adenoviral DNA could be isolated from tonsils, but this was not associated with infectious virus, leading to the suggestion that mucosal-associated lymphoid tissues were sites of adenovirus latent infection [42]. In a further study using material obtained from 203 patients undergoing tonsillectomies, it was shown that only a small proportion of samples (less than 15%) contained replicating virus; however, stimulation in vitro led to viral replication in most cases [41]. By the age of four, it appears that almost all children are positive for adenovirus DNA in their lymphoid cells, declining to around half by the age of 16 [41].
There is also strong evidence that the gastrointestinal (GI) tract can serve as a site of adenovirus persistence, particularly in children [32,35,43]. Shedding of adenovirus in the stool of adenovirus-infected individuals has been observed months after signs of infection had disappeared. Significantly, based on data obtained from screening paediatric transplant recipients, it was seen that those developing viraemia had detectable adenovirus in their stool samples before it could be observed in peripheral blood [44,45]. In a more detailed study of children receiving allogeneic stem cell transplantation, it was observed that adenoviremia was almost always preceded by the detection of adenovirus in stool samples [30]. However, many patients had adenovirus present in their stool who did not develop invasive disease [30]. These and other studies strongly suggest that the intestine probably serves as the primary site for general adenovirus persistence or latency. It is likely, particularly in view of their main sites of infection, that this is the case for the HAdV-F species viruses. However, when the relative prevalence of HAdV species in the GI tract has been analysed group C viruses have been found to predominate [30,35]. Similarly, in the analysis of stool samples from transplant patients, group C viruses were again the most common (species A, 11%; B, 4%; C, 76%; D, 7%; and F, 2% [43]). Although the group C viruses have been most commonly identified in association with the gastrointestinal tract, it seems reasonable to suggest that this is the site of persistence of all types of HAdV, including the species F, particularly as this is considered to be the primary site of infection by HAdV-F40 and HAdV-F41.
In a further study comparing HAdV present in the GI tracts of paediatric patients undergoing haematopoietic stem cell transplantation (HSCT) with a similar number of non-HSCT patients, HAdV was detected throughout the intestine but was most common in the ileum of about one-third of immunocompetent patients tested. Analysis of the biopsy material showed its presence in mucosal lymphocytes but not in epithelial cells. However, when the analysis was carried out on patients undergoing transplants, very large accumulations of HAdV were seen in epithelial cells [35]. This led the authors to propose that human adenoviruses persist in intestinal lymphocytes over long periods but cannot efficiently replicate in them. However, small numbers of viruses, leaking from there, can enter and replicate in proximal epithelial cells - these viruses are then detectable in stool. In immunosuppressed individuals, replication in epithelial cells increases to a very high level [43]. To what extent any or all of these observations apply to the HAdV-F40 and HAdV-F41 is not clear at present, but as this is the primary site of infection it would be surprising if the enteric viruses did not persist there.
Although the idea of adenovirus latency and persistence has been current for several decades, the mechanism responsible has received much less attention. However, in an elegant study by Zheng and colleagues using fibroblasts and human bronchial epithelial cells, it was shown that viral replication was appreciably attenuated in primary cells by interferons (IFN) [46]. IFNs block the binding of GA binding protein α/β (GABP) to the E1A enhancer region during the early stages of infection [46]. Mutational analysis showed that repression of E1A expression by IFNs requires an E2F binding site in the E1A enhancer; IFNs increased the level of E2F-associated Rb and p107 at the E1A enhancer. Addition of E1A 12S protein was sufficient to rescue HAdV replication by dissociating the E2F-Rb family protein interaction [46]. Although the study was carried out primarily using HAdV-C5 it was shown that the GABP and E2F/DP binding sites were all conserved in E1As from species A, B, D, and E viruses. Furthermore, IFNs were able to inhibit viral replication in human fibroblasts [46]. Comparison of the nucleotide sequences of HAdV-F40 and F41 E1A genes with those of HAdV-C5 and HAdV-A12 shows a high degree of homology in the first GA binding protein (GABP) binding sites and in the second E2F binding sites delineated by Zheng and colleagues (Figure 1). The second GABP site is also well conserved between the four viruses (Figure 1). However, there is less similarity when considering the 3’ binding site for E2F/DP (Figure 1 [46]). Indeed, it is not possible to identify an E2F/DP binding site 1 for HAdV-F40. It has been concluded that the enteric adenoviruses are also susceptible to IFN through binding to upstream sites in the E1A genes.
Figure 1.

Nucleotide sequence alignment of the GABP1 and E2F binding sites in the E1A enhancer regions from adenovirus species. The binding sites as designated in [46] are shown, together with the corresponding nucleotide sequences from HAdV-F40 and F41. Conserved nucleotides are in red.
In a later report, it was shown that the HAdV-C5 E3-19K protein activates the unfolded protein response (UPR) sensor IRE1α in the endoplasmic reticulum (ER) [47]. This is specific as E3-19K has no effect on protein kinase R-like ER kinase or activating transcription factor 6 which are also UPR sensors. Activation of the IRE1α nuclease by E3-19K initiates splicing of X-box binding protein (XBP1) mRNA [47]. The association of XBP1 with the E1A enhancer/promoter stimulates E1A transcription, expression of E3-19K and viral infection. It has been concluded that the five components (E1A, E3-19K, IRE1α, XBP1s, and the E1A enhancer/promoter) constitute a feedforward loop sustaining persistent infection in the presence of IFN and lytic infection [47]. Whether a similar mechanism is active during species F virus infection is not known, but it seems reasonable to assume that it is. One point of interest is that the IRE1β ortholog is primarily expressed in the digestive tract, whereas IRE1α is expressed in most cell types. It is not clear whether IRE1β could function in the feedforward loop.
Viral entry
HAdV species A, C, D, E, and F gain entry to host cells by means of the widely expressed coxsackie-adenovirus receptor (CAR) [48,49]. This is recognised by the knob domain of the virus trimeric fibre capsid protein. The B group viruses are recognized by CD46 [50]; additionally, the glycans GD1a and polysialic acid, and desmoglein-2 can also act as receptors (reviewed [51]). CAR is an attachment receptor, but there is also a requirement for association with an entry receptor. For most adenoviruses, this is a member of the RGD-binding group of α5 integrins which bind to the viral penton base and act as secondary or accessory receptors, leading to internalisation of the virus. The group F viruses bind to the CAR receptor but, uniquely, they express two fibre proteins, encoded by adjacent genes (Figure 2). One is similar to the molecules present on the surface of group A, C, and D adenoviruses (termed long fibre) whereas the other (short fibre) is of lower molecular weight and does not bind CAR but probably interacts with heparan sulphate [53,20,54]. HAdV-F40 and HAdV-F41 make use of laminin-binding integrins, most probably α6β4, rather than the RGD-binding integrins [55]. The necessity for a different co-receptor is due to sequence variations in the penton base of these viruses. Thus, the RGD motif present in almost all human adenoviruses is replaced by an RGAD motif in HAdV-F40 and an IGDD motif in HAdV-F41 [55,56]. Interestingly, it was demonstrated that at low pH, as seen in the intestine, the interaction between the long fibre and CAR was inhibited, although the association of the short fibre with heparan sulphate was enhanced [54]. The possibility that the short fibre could act as the accessory receptor, rather than the penton base, has been largely discounted [49,55,57]. Thus, it has been suggested that, as small intestine epithelial cells do not express RGD binding α5 integrins, the enteric adenoviruses have evolved to make use of integrins which are expressed. Laminin-binding integrins are generally expressed on the basal surface of epithelial cells of the intestinal lining; when the cells migrate from the crypt they are shed from the villi, leaving gaps which may expose integrins and CAR molecules on the basal and lateral surfaces of the intestinal cells [55,58,59]. Other molecules on the cell surface have also been implicated in adenovirus attachment, such as desmoglein-2, sialic acid, heparan sulphate proteoglycans, GD1a glycans, MHC class I, CD80, and CD86, although it is not clear whether they are recognised by the enteric adenoviruses [60]. Although fibre and penton base are generally considered to be the major viral proteins involved in host cell recognition HAdV-C2/5 hexon interacts with the SR-A6 scavenger receptor [61,62:] [63]. However, this appears to occur in murine macrophages but not in human tumour cell lines such as A549.
Figure 2.
Locations of ORFs in the E4 genes of HAdV-F40. Locations of transcripts are shown as blocks. E4 transcripts are labelled with the number of constituent amino acids as in the original publication. The HAdV-F41 gene is arranged identically to that of HAdV-F40. Based on data from [16].
Specific evidence relevant to the uncoating and trafficking of HAdV-F40 and F41, within the infected cell, is very limited; therefore, it has been assumed here that the mechanism involved in dissociation of HAdV-F41 is broadly similar to that described for other adenovirus species, generally HAdV-C2/5 (reviewed [64,65] [66]). Once the HAdV-C species virus has gained entry into the host cell by endocytosis, a series of changes take place in which the capsid essentially breaks down [64,65,67]. Binding of the penton base to the integrin initiates endocytosis and eventually lysis of the endosome [66]. Simplistically, at the plasma membrane protein IIIa, protein VIII, fibre and some of the protein V and penton base proteins are dissociated from the virion, over the course of about 15 min [66–68]. Acidification of the endosome leads to the dissociation of penton base and protein IX. Lysis of the endosome membrane, mediated by upstream actomyosin-drifting motions of virus particles bound to CAR, results in release of the remains of the viral particle into the cytoplasm. Protein VI separates and is released on break down of the endosome membrane, together with a proportion of protein V [69]. The transport of the remaining virion to the nuclear envelope or to the vicinity of the centrosome (depending on cell type) is facilitated by microtubule motors [70–73]; reviewed [74]. In non-polarised cells, dynein/dynactin-based transport leads to virion concentration near the centrosome. On the other hand, in polarised epithelial cells the kinesin system transports virions to the nucleus [75] reviewed [66,76]. Once at the nuclear membrane the virion binds to Nup214 through hexon [77–79]. At the NPC, the capsids are disassembled through the action of the cellular E3 ubiquitin ligase MIB1 [80,81]. Partial disruption of the capsid results from the action of kinesins. MIB1 then ubiquitylates protein V, leading to its separation from the capsid and the viral DNA. The resulting complex of viral DNA, HAdV terminal protein (TP), and protein VII is a nuclear import substrate recognised by the nuclear import machinery [66]. Although this general outline applies to the C species viruses, it is not clear if it is the mechanism adopted by the enteric viruses. Indeed, the B group viruses, such as HAdV-B7 and HAdV-B35, accumulate in lysosomes rather than being rapidly trafficked to the nucleus like HAdV-C5 [82]. Similarly, differences in the structure of the viral capsid proteins, such as protein IX, may have effects on the fate of the HAdV-F41 once it has entered the host cell [17,18]. Also, interaction with different integrins by the enteric adenoviruses could have consequences for the way in which the virion is trafficked in the infected cell. Answers to these questions will have to await further investigation.
Species F adenoviruses as “fastidious viruses”
Historically, it has been reported that the enteric adenoviruses were restricted in their growth in human tumour cells, with notable differences between HAdV-F40 and F41 (reviewed [16,20,]). For example, in one report, appreciably lower titres of HAdV-F40, compared to HAdV-F41, were obtained after infection of WK conjunctiva cells, A549, HAdV-C5 E1HEK293 (293 cells) and KB cells [83]. Interestingly, HAdV-F40 did not replicate at all in HeLa cells in this study [83]. Other reports have indicated that neither virus can be grown in HeLa or KB cells, although this view is not universally held [13,20,83]. In other studies, it has been shown that both enteric viruses can be propagated in Chang conjunctival cells, 293 cells, and HT-29 and H1299 cells although generally less well than other adenovirus species (summarised in [16,20]). Again, conflicting reports have suggested that neither virus can be propagated in HT-29, PC, A431, or Tera-2 cells [16]. It is difficult to draw firm conclusions from these and many other studies although it is clear that the enteric viruses generally do not grow as well, under laboratory conditions, as adenoviruses from species A-E. One reasonably consistent result, however, is that HAdV-F41, and probably HAdV-F40, can be propagated in 293 cells. This is likely due to the helper function of the HAdV-C5 E1B55K protein, supplied in trans (reviewed [16]). In a detailed comparison of the infection of HeLa and 293 cells by the enteric viruses, it was observed that while most 293 cells could be infected, replication occurred in less than a fifth of the HeLa cells [84]. Furthermore, although the virus yield was similar for both cell lines, progeny viruses were only released from 293 cells. An extremely high particle-to-infectivity ratio was observed for the enteric viruses (100–1000-fold greater than HAdV-C5), although the yield of virus particles was similar for the three viruses [84]. It is likely that the very high ratio and the failure to release virus particles, seen with HeLa cells, may explain, at least partially, the difficulty in propagating the enteric viruses in the laboratory.
More recently, 293-based cell lines, which also express HAdV-C5 E4orf6, have been used to propagate HAdV-F viruses more successfully [85,86]. As both HAdV-C5 E1B55K and E4orf6 increase growth and propagation of fastidious adenoviruses, it is possible that the viruses lack, or have restricted, ability to recruit the cellular E3 ubiquitin ligases necessary for replication of almost all adenoviruses, although this has not been demonstrated [87]. Of course, both HAdV-F40 and F41 appear to have no difficulty in growing and replicating in the gastrointestinal tract of infected children. It might be assumed that a function supplied by the HAdV-C5 E1B55K and E4orf6 proteins is endogenously present, although there appears to be no evidence for that. Similarly, the apparent lack of an adenovirus death protein may reduce the release of progeny virus from infected cells in cell culture experiments (see section 6 and [52]). The rapid turnover of HAdV-F virus infected mucosal cells in the intestine might also facilitate the release of virus progeny without a highly developed mechanism for cell lysis.
HAdV-F40 and HAdV-F41 genomes and the roles of the viral early proteins
A complete nucleotide sequence for an enteric adenovirus, HAdV-F40, was first published in 1993 [15,16]. Since then, sequence data has become available for numerous strains of HAdV-F40 and HAdV-F41 and these have been deposited in appropriate data bases (for example, HAdV-F40 strain “Dugan,” L19443 and HAdV-F41 strain “Tak,” DQ315364.2). The notable increase in interest in HAdV-F41, initiated by the suggestion that it could be responsible for the recent outbreak of acute childhood hepatitis (discussed in detail in Section 8), has led to the sequencing of multiple viral isolates (for example [88]). However, to date, no consistent mutations in the HAdV-F41 genome have been detected, which are specifically associated with the childhood hepatitis (Section 8). The genomes of the two enteric adenoviruses are similar to each other with about 80% homology. Apart from that, the HAdV-F41 genome is more similar to the species HAdV-A than to any other species, with the possible exception of HAdV-G52 (the single member of that species). Comparisons of the sequences of several adenovirus genes (for example, hexon, L1, L2, penton base, and E4orf6) have been carried out, and in each case similarities of HAdV-F40 and F41 to species A have been appreciably greater than for species B, C, and D orthologs [89–93].
Detailed analysis of 65 available complete HAdV-F41 genomes has allowed the separation of three lineages [88]. Lineage 1, comprising seven members (one of which was the original “Tak” strain) had 99.3% average nucleotide identity and derived from several geographic locations. Lineage 2, with 53 members, again originated from multiple countries and had 99.8% nucleotide identity. Lineage 3 also had very high nucleotide identity but diverged from lineages 1 and 2 (98.2% identity). The most notable differences between the lineages were seen as a 45-nucleotide deletion in the lineage 3 long fibre gene, together with 21 SNPs. Four amino acid substitutions were seen in lineage 3 fibre knob region [88]. There was also divergence in the lineage 3 short fibre gene, which resulted in 20 amino acid substitutions. Considerable variations between the lineages were also reported for the E3 and E4 regions. Differences between the lineage 3 E3 region and a reference HAdV-F41 genome were greater than between the “Dugan” HAdV-F40 and “Tak” HAdV-F41 strains. Lineage 1 had an appreciable number of SNPs in the E4 region compared to the reference sequence [88]. Unfortunately, only one sequence of HAdV-F41 from a child suffering from severe hepatitis was available for this study and that was most similar to lineage 2.
The overall organisation of the genomes of the species F adenoviruses is similar to species C and A viruses, with a few notable exceptions. The E1 regions of HAdV-F40 and F41 are 80% homologous and have 52% homology to HAdV-C5 [16]. HAdV-F40 and HAdV-F41 E1A proteins have similar conserved regions (CRs) to other adenovirus E1As but have only about 50% homology to the HAdV-C5 protein [94,95]. Interestingly, differences between HAdV-F41 and a large number of other E1As have been noted in very highly conserved residues in the second Rb binding site (which overlaps the N-terminal region of CR1) and in a highly conserved CBP/p300 binding motif at the C-terminal end of CR1 [94]. It is not clear what effect these differences, which are not conserved in HAdV-F40 E1A, makes to the virus and its ability to replicate. The E1B55K proteins from the two enteric viruses are highly homologous (90%) with almost all differences localized to the N-terminal fifth of the protein. Comparison with the sequences of E1B55K from other species shows a reasonable level of homology, with most differences, again, occurring in the N-terminal quarter [87]. Although the binding sites on E1B55K for cellular proteins have not been mapped in detail, it seems likely that the enteric virus E1B55K proteins associate with many of the same cellular targets identified for species A and C adenoviruses. Thus, HAdV-F40 E1B55K and E4orf6 can degrade MRE11, p53, and DNA Ligase IV [96]. Presumably similar activities can be attributed to HAdV-F41 proteins, although this has not been demonstrated. HAdV-F40 E1B55K can, like the protein from most other species, interact with p53 and inhibit its transcriptional activity [97]. Interestingly, HAdV-F40 and HAdV-F41 E1B55K can complement HAdV-C5 E1A in transformation assays almost as well as HAdV-C5 E1B55K [16,97]. However, the enteric virus E1A genes are unable to co-operate with E1B genes from HAdV-C5, HAdV-A12, or HAdV-F41 to transform BRKs fully, although they can co-operate effectively with activated ras [98]. It is possible that this is due to the differences in the E1A binding sites, reducing the ability of E1A to interact with and inactivate Rb family proteins and CBP/p300, although this has not been confirmed.
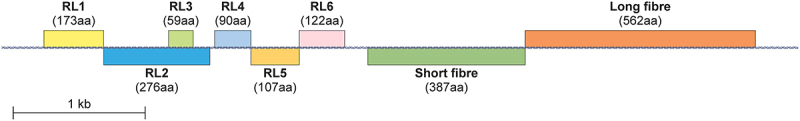
The E2 regions encode the viral DNA binding protein (DBP/E2A), the pre-terminal protein (pTP), and the viral polymerase (pol), and these are thought to be highly conserved in HAdV-F40 and F41. It is assumed that they function in the same way as their HAdV-C5 and HAdV-A12 counterparts. The E3 regions of the two enteric viruses are very similar but have differences to HAdV-C5 E3 (Figure 3) [16,52]. HAdV-F41 E3 encodes six open reading frames, RL1–6 [52]. RL1, RL2, and RL3 appear to have no equivalent in the species C adenoviruses and encode proteins of 173, 276, and 59 amino acids, respectively. RL3 is read in a different reading frame to the others. The three other ORFs are similar to the well-characterized HAdV-C2 proteins; thus, RL4 is homologous to HAdV-C2 10.4-kDa, RL5 is homologous to HAdV-C2 14.5-kDa, and RL6 is considered to be analogous to HAdV-C2 14.7-kDa protein (Figure 3). Comparable proteins are also expressed by species B and E adenoviruses [52]. Although RL1 is equivalent in location on the viral genome to HAdV-C2 gp19 it shares no obvious homology; however, it is approximately 30% homologous to the HAdV-A12 E3 264 residue protein. Similarly, RL2 appears to have no equivalent in HAdV-C2, HAdV-C5, HAdV-B3, or HAdV-E4 although it has approximately 34% homology to HAdV-A12 268 amino acid protein [16,52,99]. Based on a structural prediction, it has been suggested that HAdV-F41 RL3 is equivalent to HAdV-C2 E3 6.7-kDa protein although it has no sequence homology [52]. In HAdV-C5 proteins encoded by E3 protect adenovirus infected cells from killing mediated by cytotoxic T cells and toxic cytokines (see, for example, review by [100] [In the latter review a more modern nomenclature for the E3 proteins has been used, but in the present article we have retained the nomenclatures used in the original publications]). E3gp19K blocks MHC class I-restricted antigen presentation. The receptor internalization and degradation complex (RID) comprises E3–10.4kDa and E3–14.4kDa and controls degradation of surface receptors, such as Fas. E3 6.7-kDa acts with RID and may help to inhibit apoptosis. E3–14.7kDa is an also an inhibitor of apoptosis [100]. Adenovirus death protein (ADP) is encoded by the E3 region in HAdV-C2 and C5 viruses but appears to be absent from HAdV-F virus E3 regions. Similarly, no evidence of a “death like function” was found for HAdV-B1 virus [101]. Interestingly, the incorporation of the HAdV-C5 ADP gene into recombinant HAdV-F41 increased virus yield substantially through enhancing the spread of the progeny virus among packaging cells [102]. It has been suggested that the similarity of the enteric virus E3 regions to that of HAdV-A12 rather than the group C viruses, such as HAdV-C2 or 5, may be linked to a shared site of infection, the gastrointestinal rather than the respiratory tract [16].
Figure 3.
Locations of ORFs in the E3 and fibre genes of HAdV-F41. Locations of transcripts are shown as blocks. E3 transcripts are labelled RL1–6. The constituent amino acid numbers are shown. Based on data from [52].
The E4 regions of HAdV-F40 and HAdV-F41 are generally similar to those of HAdV-C2 and HAdV-A12 except that there is no equivalent of E4orf1 (Figure 2) [16]. However, it has been shown that HAdV-F40 can complement the HAdV-C2 deletion mutant dl808, which lacks most of the E4 region [103]. A major role of the adenovirus E4orf6 protein is, together with the viral E1B55K protein, to recruit a cellular ubiquitin E3 ligase, to ubiquitylate host proteins and, in many cases, target them for degradation by the proteasome [104] reviewed [87; 105]. HAdV-F40 E4orf6 forms a complex with the complementary E1B55K protein although some differences have been noted in their mode of action. The HAdV-F40 E4orf6 forms complexes containing Elongins B and C and E1B55K as is the case with HAdV-C5 and this is able to cause degradation of p53 and Mre11, as is the case for species A and C but not B, D, and E, adenovirus protein complexes [96]. A central component of the HAdV-C5 E1B55K/E4orf6 E3 ligase is cullin 5 (Cul5) [104]. However, it appears that HAdV-F40, and presumably HAdV-F41, recruit cullin 2, rather than Cul5, as is the case for species A adenoviruses [96,106]. The interaction of HAdV-F40 E4orf6 with Cul2 is due to the presence of a sequence resembling the cellular consensus Cul2 box; a similar sequence is present in HAdV-A12 E4orf6 [107]. As cullin 2 is part of the E3 ubiquitin ligase recruited by species A, F, and G adenoviruses, it has been suggested that this may be linked to their tropism for the gastrointestinal tract; thus, other human adenoviruses which target the respiratory tract or the eye do not have a Cul2 box and almost entirely recruit Cul5 [90]. Interestingly, in an extensive study, it was shown that all adenoviruses isolated from great apes encode E4orf6 proteins which bind Cul5, whereas those E4orf6 proteins isolated from monkey adenoviruses have a Cul2 box, suggesting that the ability to bind Cul5 coincided with the evolution of apes and hominids from monkeys [90]. It is not clear what advantage the use of Cul2 over Cul5 gives to the viruses. It has been shown, however, that cullin 2 is far more abundant in the intestine of the mouse than in lung tissue, whereas cullin 5 is present in both to an equal extent [90]. In humans, cullin 2 seems to be expressed at a reasonably high level in both respiratory and gastrointestinal tracts (https://www.proteinatlas.org/ENSG00000108094-CUL2/tissue) although cullin 5 appears to be particularly abundant in respiratory tract tissues (https://www.proteinatlas.org/ENSG00000166266-CUL5/tissue).
The structure of adenovirus F41 and the roles of the viral late proteins
The overall structures have been determined for three human adenovirus types: HAdV-C5, HAdV-D26, and HAdV-F41; structures have also been determined for bovine BAdV-3, snake SnAdV-1, and lizard LAdV-2 [17,18,108–114]. All three HAdVs are broadly similar, with large, non-enveloped capsids of approximately 950 Å in diameter pseudo-T = 25 icosahedral particle with 12 trimeric hexons per facet (giving a total of 240 trimers) and pentameric pentons at the vertices. The hexons are stabilized by the minor capsid proteins-IIIa, and VIII. Proteins IIIa, VIII, fragments of VI and core protein VII are present in the interior of the capsid. Penton base subunits are organized into 12 homopentamers at the vertices of the capsid. These associate with the N-terminal regions of the trimeric fibre proteins. The N-terminal region of protein IX is present on the outer surface of the capsid between hexon trimers. The structure of HAdV-F41 has been solved, using cryo-electron microscopy to a resolution of 3.8 Å or 4 Å [17,18]. This was determined at both pH7.4 and pH4.0 [18], the latter to take into account the fact that the virus normally infects the GI tract; however, there appears to be little difference in the structure of the capsid at the lower pH [18]. On examination of the surface charge of the three human viruses at pH7.4 HAdV-D26 has been found to have a highly negatively charged surface, whereas HAdV-C5 only has negatively charged areas at the tops of the hexons; HAdV-F41 capsid is generally uncharged at pH7.4 and has two uncharged surface regions at pH 4.0. It was concluded that the icosahedral part of the capsid is unaffected by low pH, the virus having evolved so there are fewer charged amino acids on its surface compared to species C and D viruses, presumably as an adaptation to the acidic GI tract [17,18].
The major late genes present in the enteric viruses are comparable to those in other virus species. Hexon is 88% identical between the two viruses, which is similar to that seen within other species, such as HAdV-C. HAdV-F hexon most closely resembles that present in species A viruses. The major sequence difference between HAdV-F40 and HAdV-F41 hexons and HAdV-C5 hexon is in the loop regions exposed on the virus surface [115,116]. The overall structural fold of HAdV-F41 hexon is similar to that seen with other adenovirus species, although differences were observed in the seven hypervariable regions (HVRs), when comparing it to HAdV-C5 and HAdV-D26 [18]. In particular, HVR1 is much shorter and less charged in HAdV-F41 giving it a rigid structure, compared to HAdV-C5 [18]. Again, this may be an adaptation to the low pH found in the intestinal tract.
The L4 regions of HAdV-F40 and F41 resemble those seen for most other adenoviruses with the major similarity observed in the L4-100K protein. L4-33K and −22K are also suggested to be expressed by the enteric viruses [16]. As noted in Section 3 the penton base is involved in virus entry and binding to the accessory receptor. The penton base forms multiple interactions in the virion binding to hexon, fibre and IIIa protein [18]. Comparison of structures for the penton base determined in free solution and in the virion has indicated that the presence of other viral proteins causes four disordered regions to adopt defined conformations [18]. For example, the conserved random coil region between amino acids T33 and G49 is stabilized in the virion through association with two loop regions from protein IIIa [18]. Other regions of HAdV-F41 penton base which become structured in the virion are located between Y419 and L429 which becomes largely α-helical possibly through interactions in the capsid or binding to fibre. The two loop regions within the penton base V70 to N110 become structured through interaction with hexon molecules in the HAdV-F41 virion [18]. As mentioned in Section 3 the integrin binding RGD motif, present on the penton base of most adenoviruses, is not conserved in the enteric viruses but is replaced by IGDD in HAdV-F41 and RGAD in HAdV-F40. The loops containing the RGD sequence are disordered in HAdV-C5 and HAdV-D26 as is the IGDD sequence in HAdV-F41, presumably giving it some flexibility to allow binding to the host cell surface [17,18,109,114,117]. Overall, the structure of HAdV-F41 penton base is similar to that of the protein from species C and D [17,18]. However, it has been noted that the binding of penton to fibre may be different in HAdV-F41 and HAdV-C5 with less hydrogen-bridging and more hydrophobic interactions in the latter virus [17].
Two fibre proteins are expressed by the enteric adenoviruses-a “long fibre,” which binds to the CAR receptor and is similar to that present in all other human adenoviruses, and a “short” fibre which does not bind to CAR but to heparan sulphate [53,118,54,119]. This short fibre is considered to help protect the virus from the low pH found in the intestine. The major difference between the short and long fibres lies in the length of the shaft region between the tail and knob regions. In HAdV-F41 long fibre there are twenty-two 16 amino acid repeats, whereas there are only twelve repeats in the short form [119]. In the structural studies of the short fibre “head region” it was shown that there are distinct differences to the head regions of long fibres occurring in both HAdV-F41 and other adenovirus species, possibly explaining why there is no binding to CAR [120]. Whilst they are superficially similar, there is a specific deletion in the AB loop in the short fibre head, resulting in an appreciable conformational change, incompatible with interaction with CAR [120]. Other structural differences between the heads of the short and long fibres have also been reported [120].
A notable difference between HAdV-F41 and HAdV-C5 and HAdV-D26 is the arrangement of minor capsid protein IX on the surface of the capsid. In the case of the latter two viruses, it forms a highly ordered, tight triskelion-shaped network across the virion surface, locating in the valleys between the hexons; it also interacts and binds the hexons together imparting thermostability to the virus [109,110]; [17; 18]. In HAdV-F41, however, the structure of protein IX on the capsid surface is quite distinct and not so ordered [17,18]. It is probable that changes in the amino acid sequence of the protein are responsible for reorganization of protein IX. A model has been suggested in which HAdV-F41 protein IX forms four triskelions and three mobile helix bundles per facet [17,121]. In HAdV-C5 and D26, the helix bundles are not mobile [17]. Consistent with these data, in another investigation, it was observed that the C-terminal half of protein IX is flexible in HAdV-F41, exposing its C-terminus to the capsid exterior, unlike HAdV-C5 and HAdV-D26 [18].
Information for the minor capsid proteins IIIa, V, VI, VII, and VIII indicates that their structure and distribution in the virus particle is comparable for HAdV-F41 and HAdV-C5 [17,18]. Proteins IIIa and VIII line the internal capsid surface in HAdV-F41. Some VIII proteins interact with IIIa, stabilizing the vertices, while others maintain nonperipentonal hexons in the central plate of the facet [17]. In the cryo-EM study of HAdV-F41 it has been shown that the core protein V probably links the viral genome to the capsid; similar electron density was observed in the study of HAdV-C5 and HAdV-D26 [18,114,122].
Differences in the amino acid sequences and consequent structure of components of the adenovirus capsid determine differences in the abilities of various species to resist neutralization by human α-defensins (HDs). In most cases binding of defensin to the infecting adenovirus particle stabilizes the capsid and inhibits release of protein VI, blocking uncoating and exposure of the viral genome [123–126]. However, this is not the case for species D, and F viruses, which are resistant to HD5 and neutrophil peptide 1 (NP1) [126]. The ability of defensins to bind to the viral capsid is determined by a short four amino acid sequence in the N-terminal region of the fibre protein which is acidic in species A, B, and C HAdVs but positively charged and hydrophobic in HAdV-D and F viruses. The defensin also binds the penton base in the susceptible species, associating it with a disordered region at the top of the base. It was suggested that stabilization of the HAdV vertex region comprising the penton base and fibre can be achieved by a three-fold increase in intermolecular nonbonded interactions [126,127].
Adenovirus F41 as a vector
Thousands of gene therapy trials have been or are being carried out with human adenovirus vectors (see for example [128] [129,130]). Most are for the treatment of cancer (oncolytic viruses). Others are for the use of adenovirus vectors as vaccines in which the vector expresses an antigenic protein or for gene therapy in which the vector expresses a protein to correct a genetic defect. It might be assumed that appreciable use would have been made of the species F adenoviruses to target intestinal cells, based on their tropism. This does not appear to be the case as most studies have used vectors based on HAdV-C5 (reviewed [131]). However, it has been shown that an HAdV-F41 vector binds to differentiated and undifferentiated enterocytes much better than a HAdV-C5 vector. In addition, the uptake of HAdV-F41 was much more efficient [132]. Unfortunately, these experiments do not appear to have been followed up in any systematic way.
Adenovirus F41 and acute, severe childhood hepatitis
During the first six months of 2022 there were many reports of acute, severe hepatitis in young (under the age of five), otherwise healthy, children (reviewed, for example [133;] [134,135;] [136] [137; 138]; and references therein). Although most patients appeared to be in the U.K. and U.S., cases were seen in at least 30 countries, including Canada, Indonesia, Spain, Israel, Denmark, Ireland, France, Romania, and Belgium. The children presented with vomiting, jaundice, and diarrhoea. Overall, at least 1000 hospitalized children have been reported worldwide, some requiring liver transplants and a few dying. Although there was no obvious cause for the hepatitis that is not particularly unusual, as a direct cause for many cases of acute liver failure is often not found (see, for example [139,140]). However, for this cohort of children with acute hepatitis, almost all tested negative for hepatitis viruses (HAV, HBV, HCV, often HDV, HEV), Epstein–Barr virus (EBV), Cytomegalovirus (CMV), and Human Immunodeficiency Virus (HIV). Most patients had not been infected with SARS-CoV-2, nor had they been vaccinated against COVID-19. However, many of the children were positive for adenovirus. For example, in the UK Health Security Agency technical report published at the end of July 2022, summarizing cases in the U.K. up to that time, there had been 270 confirmed cases of severe hepatitis in children under the age of 10. Of these, 15 received liver transplants but none died. Almost all were tested for adenovirus and 65.9% were positive. Similarly, in a National Center for Immunization and Respiratory Diseases (NCIRD) report published in August 2022 (https://www.cdc.gov/ncird/investigation/hepatitis-unknown-cause/technical-report.html), summarizing the then current state of knowledge in 43 states of the U.S., it was reported that there were 358 patients under investigation with a median age of 2 years. At that time 6% had required liver transplants and 4% had died; 299 patients were tested for adenovirus and 45% were positive. A more detailed analysis showed that HAdV-F41 was, by far, the most common adenovirus. In one study from the U.K. 44 children (median age 4 years) with acute severe hepatitis were treated, of whom 6 required liver transplants. Of 29 patients who were tested, 93% were positive for adenovirus [141]. In another study of 15 children with acute severe hepatitis (median age 2 years 11 months), 9 had no obvious cause. Of these, eight tested positive for adenovirus, of whom five were positive for HAdV-F41 [142]. Thus, by late summer of 2022, HAdV-F41 was thought to be a likely cause of the hepatitis, even though it had not been detected in all the patients nor was there evidence of adenovirus-mediated tissue damage in many cases.
As the enteric adenoviruses show a marked tropism for the gastrointestinal tract and not the liver, this led to considerable confusion and debate. A further complication was added, towards the end of 2022 and in 2023, when three carefully performed, specific studies of young children, presenting with severe hepatitis with no known cause, showed the presence of adeno-associated virus-2 (AAV-2) in almost all members of the cohorts examined [143–145]. HAdV-F41 was detected in an appreciable proportion of patients but was not ubiquitous unlike AAV-2. Adeno-associated viruses (AAVs) require a “helper virus” to replicate; in these reports, it was suggested that the helper function was provided in some patients by adenovirus (generally HAdV-F41) but in others by Human herpesvirus 6B (HHV-6B). The results of these later studies are summarized in Table 1.
Table 1.
AAV-2 and HAdV detected in cases of hepatitis of unknown origin. Summary of data from studies by [143; 145] and [144].
| Ho et al. 2023[143] | Servellita et al. 2023[145] | Morfopoulou et al. 2023[144] | |
|---|---|---|---|
| Number of cases | 32 | 16 | 38 |
| Median age | 4.1 years | 3 years | <10 years |
| Number of controls | 25 immunocompetent Including 12 HAdV positive controls |
113 | 66 immunocompetent 21 immunocompromised |
| AAV-2 detected | 9/9 | 13/14 | 27/28 |
| HAdV detected | 6/9 | 14/14 (10/14 HAdV-F41). | 22/23 |
| HHV-6B detected | 3/9 | 7/14 (11/14 positive for EBV) | 16/23 |
| Viruses detected in the livers of patients | AAV-2 4/4 | AAV-2 0/8 HAdV 3/8 |
AAV-2 5/5 HHV-6B 5/5 |
| AAV-2 in controls | 0/25 | 4/113 | 6/65 in blood of immunocompetent cases. 6/17 of immunocompromised cases. |
| HAdV in controls | 0/13 in healthy controls 6/12 in HAdV-positive controls |
9/113 | 1/65 |
| HHV-6B in controls | 0/13 in healthy controls 9/12 in HAdV positive controls. |
1/113 | 0/2 livers |
| COVID-19 vaccination | 0 | Not known | Not known |
| SARS-Cov-2 infection | 3/31 but 12/23 had evidence of past infection. | Not detected in cases or controls | 15/20 seropositive |
| DRB1 × 04:01 allele | 25/27 compared with 10/64 in controls | Not determined | 12/13 |
Adeno-associated viruses are members of the Parvovirus family, of the genus Dependovirus [146–149]. They are small non-enveloped viruses, with a single stranded DNA genome comprising about 4.6K nucleotides. Approximately 100 genomic isolates and 13 serotypes have been identified. AAV is widespread in humans, with AAV-2 the most commonly identified and the most studied [150]. More than 90% of the adult population is naturally infected with AAV, with antibodies to the different AAV serotypes detected in most individuals [151–153]. Antibodies against AAV-2, for example, are present in 50–80% of the individuals although only about a third tend to be neutralizing antibodies [151,154]. The incidence of AAV-2 increases to approximately 90% in immunosuppressed individuals. Antibodies against AAV can be detected at birth, suggesting maternal transmission. Over the first year of life, antibody levels tend to fall and then rise until late adolescence [155]. It seems likely that AAV infection occurs again later in life, giving rise to increased seropositivity after the age of about 30 [156]. In studies of AAV distribution in human tissues, AAV cap gene sequences have been detected in brain, colon, liver, lung, kidney, spleen, and bone marrow, with the highest level in the liver and bone marrow [157]. Other reports have indicated the presence of AAV-2 in cervix, penis, uterus, abortion material, and blood [158–161]. High levels of AAV (predominantly AAV-2) are present in blood from about a third of healthy donors, with AAV residing in CD3+ T lymphocytes. This was thought to be a possible site for AAV persistence [162].
The consensus is that AAV is not associated with any disease and does not produce any clinical symptoms in the great majority of cases, although there is evidence for hepatitis following AAV gene therapy [163–169]. However, there have been suggestions of a link between AAV and cervical carcinoma, reproductive system disorders, and hepatocellular carcinoma (reviewed [170]). While some studies have indicated that AAV can have a protective role in cervical tumorigenesis, others have concluded that there is neither a positive nor negative link. Similarly, limited evidence has shown that AAV infection may contribute to placental complications and spontaneous abortions, whereas others have found no statistically significant difference between levels of AAV in spontaneous and therapeutic abortions [159,170]. There have also been suggestions that AAV could contribute to hepatocellular carcinoma by integration into the genomes of patients [171–174]. Insertion of AAV DNA was mainly in the CCNA2, CCNE1, TERT, TNFSF10, and KMT2B cancer driver genes, generally leading to protein over-expression. The original study by Nault and colleagues was the subject of strong criticism at the time, but a later widespread screen produced results consistent with the earlier investigation [171,172,175,176]. However, in studies of liver carcinomas from Korean, Thai, and Mongolian patients, the integration of AAV was at a very low level, leading the authors to conclude that AAV posed a minimal risk of causing hepatocarcinogenesis [177,178]. In immunocompromised individuals, levels of AAV tend to be higher (see [162] for example) but the virus still does not produce clinical symptoms.
Adeno-associated viruses require the presence of a co-infecting “helper” virus which is usually adenovirus or a herpesvirus although other viruses, such as HPV, can fulfil this role. Presumably to overcome the inefficiency of the requirement for infection of a host cell by two viruses at the same time, AAVs can establish a latent infection, which may, but often does not, involve integration of its genome into that of the host. The integration site has been mapped to chromosome 19q13.4, a site termed AAVS1 [179,180]. Commonly, the AAV-2 genome is present in human tissue samples, in the absence of a helper virus, as an extrachromosomal rolling circle. Activation of the virus requires expression of the viral Rep proteins, which are needed for replication, transcription, integration, and encapsulation. The cap gene encodes the three VP1, VP2, and VP3 proteins, which constitute the viral capsid and the membrane-associated accessory protein (MAAP) which aids AAV replication and has a role in controlling HAdV infection. The assembly of the capsid is facilitated by the assembly activating protein (AAP) [148,181–183]. Many host cell surface molecules are recognised by infecting AAV-2 (reviewed [184]). Glycans, which were once thought to be primary receptors, may now be considered low specificity attachment factors. Proteins constitute the most likely candidates for AAV receptors with knock-out of more than 50 genes affecting AAV resistance [184]. Adeno-associated virus receptor (AAVR, also known as KIAA0319L) is a widely expressed membrane protein, which is important for trafficking and likely serves as a viral entry receptor for AAV-2. The capsid spikes surrounding the threefold axes bind to the PKD2 motif in the ectodomain of AAVR [185–187].
Whilst the popular press and social media largely considered the mystery solved, with AAV-2 the cause of the hepatitis, several important unanswered questions remain. Firstly, although AAV-2 can infect liver cells, it is normally considered to be harmless and there are virtually no clinical conditions associated with “normal” AAV infection. Why should AAV-2 suddenly cause severe liver disease? Secondly, HAdV-F41 is well-known to infect the gut but not the liver; as discussed throughout this review, it is widely associated with gastroenteritis but not associated with hepatitis, even in immunocompromised patients. On a more general level, it is interesting to note that there have been virtually no reports of HAdV-F40 in the patients with hepatitis, which is remarkable as it is very similar to HAdV-F41 and is equally prevalent in the gut.
In an MMWR report, published 14 June 2022, based on results from four data sources in the U.S., it was concluded that there had been no significant increase in hospital admissions of children suffering from hepatitis over the period from January 2017 to March 2022. Nor had there been an increase in the frequency of liver transplants [188]. However, it was noted that there was a marked decrease in HAdV-F41 in stool specimens from children over the period May 2020 to August 2021, presumably corresponding to COVID-19 lockdown. From October 2021 the number of children positive for HAdV-F41 returned to the pre-pandemic level, slightly preceding the reports of the hepatitis [188]. This could imply that there has always been a cohort of children with severe hepatitis, of unknown origin, who were infected with HAdV-F41 and AAV-2 but went unobserved. However, in a large retrospective study of non-A-E-hepatitis in children covering the period from 1990 to 2022, very few cases of adenovirus infection were seen, although there was a marked increase in hepatitis cases from 2019 [189]. It was suggested that there is, indeed, an increased incidence of acute, severe hepatitis of unknown origin in children but several factors could trigger it, one of which could be HAdV-F41 and/or AAV-2 [189]. For example, an increase in HAdV-F41 and AAV-2 occurrences has been observed in wastewater in Northern Ireland, coincident with an increase in severe hepatitis in young children [190,191]. Although there was an increase in HAdV-F41 at the same time as the increase in hepatitis, the levels of other adenoviruses in the wastewater stayed approximately constant [190].
It is possible that a lack of exposure to micro-organisms in general and viruses in particular, such as HAdV-F41 and AAV-2, during COVID-19 lock-down could have meant that young children developed “an immunity gap” which has been defined as “a group of susceptible individuals who avoided infection and therefore lack pathogen-specific immunity to protect against future infection” [192]. Obviously, this could have contributed to the hepatitis. It is notable that an increase in childhood infection, generally, has occurred after the end of COVID-19 restrictions (for example [192,193] [194–196]). This has been most pronounced among young children.
In one of the latest studies, showing the presence of AAV-2 in the patients with hepatitis, it was noted that the patients had a high frequency of the MHC alleles HLA-DRB *04:01 (25/27 cases compared to 10/64 in controls), DQA1 × 03:03 (23/27 compared to 11/64 controls), and DRB × 01:03 (23/27 compared to 21/64 controls) [143]. In another study HLA-DRB *04:01 was also seen in 12/13 cases [144]. HLA-DRB *04:01 is known to be associated with autoimmune disease, and it was suggested that its presence could increase susceptibility to viral infection [143]. Further studies are required to examine links between viral infection, MHC alleles, and hepatitis.
Whether the hepatitis is caused by direct viral infection or an indirect immune-mediated mechanism (summarized in Figure 4) it appears that the behaviour of HAdV-F41 and AAV-2 is abnormal, in that species F adenoviruses do not infect liver cells and AAV-2 is not considered to be associated with a disease or produce clinical symptoms. It is possible, therefore, that mutations could have occurred to account for the changes in properties of the viruses. Several investigations have shown that mutation in the HAdV fibre gene can alter the tropism of adenoviruses [197–199, 200,201]. For example, the addition of a 7-lysine-residue motif at the C-terminal end of HAdV-F40 short fibre (which does not bind CAR) facilitated efficient transduction of target cells, which were not previously susceptible to infection, via the heparan-containing receptor [202]. In the studies of hepatitis of unknown origin, full or partial sequencing of the HAdV-F41 genome was carried out in many cases. For example, in one reportseveral early region proteins were found not to have significant mutations compared to control HAdV-F41 [144]. When the entire HAdV-F41 genome was sequenced, it was concluded either that SNPs were shared between cases and controls [144] or the patient HAdV-F41 sequence was similar to a previously reported virus [143]. Other partial HAdV-F41 sequences appear to vary slightly from previously published data but not to such an extent that it is likely to account for the virus’s change in tropism and/or its ability to cause novel disease. However, it is clear from the example of the omicron variant of SARS-Cov-2 that a single amino acid substitution can radically alter virus infectivity and severity of disease caused.
Figure 4.
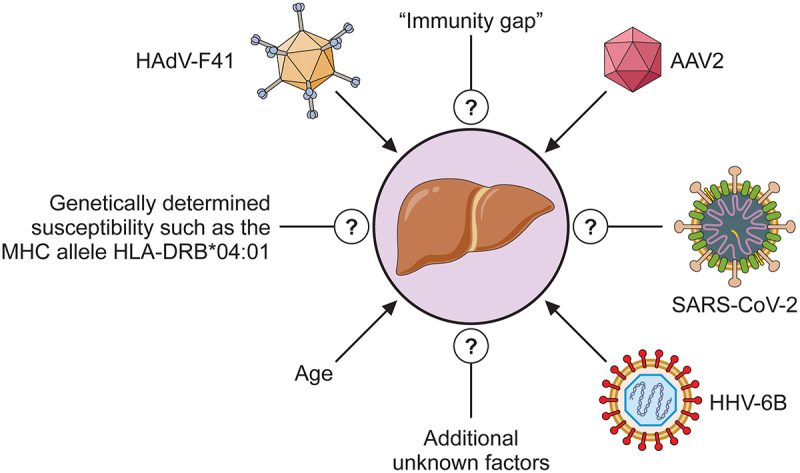
Cartoon showing possible factors contributing to severe acute childhood hepatitis “of unknown origin.”
Possible mutations in AAV-2 could also change its ability to produce a deleterious infection. Sequencing of AAV-2 has also been carried out in detail. In one report, the genomes of AAV-2 were sequenced from 13 hepatitis patients [145]. Of 35 mutations, 43% were in the AAP gene, 40% in VP1, and 17% in Rep78. Perhaps, it is significant that over 70% of these mutations were the same as those identified in a second study [143]. In this latter investigation, it was noted that nine of the capsid gene mutations seen in the AAV-2 from hepatitis patients were associated with an AAV-2 variant (AAVv66) which has an altered phenotype [143,203]. AAVv66 has increased tissue spread, increased virion stability, and production of progeny, as well as an ability to evade neutralizing antibody [203]. In the third study identifying AAV-2 in the hepatitis patients, extensive sequencing of AAV-2 genomes showed little or no difference between cases and controls although it was noted that there were changes in the capsid gene in all contemporary samples which were absent from historic AAV-2s [144].
Concluding remarks
Historically, interest in the enteric adenoviruses has stemmed from their ability to cause gastroenteritis and diarrhoea in children. This probably represents the most serious clinical condition due to any adenovirus species, in immunocompetent individuals. Specific studies of the properties of the viruses have been partially limited by difficulties in growing them in the laboratory because replication is restricted in several commonly used cell lines. Additionally, adenovirologists, when investigating viral replication, for example, or the biochemistry of particular adenovirus proteins, have tended to concentrate on those species for which most information is already available, such as HAdV-C5 and, to a lesser extent, HAdV-A12. Therefore, much of our knowledge of the life cycle and properties of HAdV-F41 has been based on the assumption that they are similar to those of HAdV-C5. This has been aided by the availability of nucleotide sequences for HAdV-F40 and HAdV-F41, allowing a comparison of theoretical protein primary structures with HAdV-C5 orthologs. Even so, the identity between HAdV-F40 and HAdV-C5 or HAdV-A12 for most early genes is less than 60%, suggesting possible marked differences in the mode of action [16].
The recent reports linking HAdV-F41 to severe, acute hepatitis in children has stimulated interest in the virus, to such an extent that cryo-EM structures are now available [17,18]. Some notable differences to HAdV-C5 have been observed, and it has been suggested that these may be responsible for the tropism of the enteric adenoviruses and their ability to withstand the low pH found in the gut. It would be beneficial if this renewed interest in HAdV-F41 could also be extended to a biochemical analysis of the properties of the early region proteins.
Regarding the causes of the severe hepatitis, there is no unequivocal explanation (Figure 4). The number of cases has decreased since the late summer of 2021 to pre-pandemic levels. However, it is notable that there have always been cases of childhood hepatitis, which could not be attributed to any of the common hepatitis viruses [139,140]. It is possible that they share an aetiology with the latest outbreak, although in one retrospective study there was little evidence of HAdV infection in the pre-2019 cases [189]. It seems reasonable to suppose that the COVID-19 pandemic may have contributed to the outbreak. Although most of the patients tested negative for SARS-CoV-2, at the time of hospitalization, it is possible that previous infections could have occurred. There is extensive evidence that SARS-CoV-2 can cause acute hepatitis (see, for example, [204] [205,206] [207]). In addition, it is clear that the prolonged lock-down and isolation have meant that very young children have reduced immunity. These factors, as well as a possible change in tropism of HAdV-F41 and perhaps alteration in responses to AAV infection, could, taken together, have led to an increase in acute hepatitis in children but without a single factor being responsible. Further investigation is required for a definitive explanation.
Funding Statement
The author(s) reported that there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability statement
All data mentioned and referenced in this review is available and open to use without restriction.
References
- [1].Rowe WP, Huebner RJ, Gilmore LK, et al. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc Soc Exp Biol Med. 1953;84(3):570–21. doi: 10.3181/00379727-84-20714 [DOI] [PubMed] [Google Scholar]
- [2].Aoki K, Benkö M, Davison AJ, et al. Toward an integrated human adenovirus designation system that utilizes molecular and serological data and serves both clinical and fundamental virology.J Virol. 2011;85(11):5703–5704. doi: 10.1128/JVI.00491-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kajon AE, Echavarria M, de Jong JC.. Designation of human adenovirus types based on sequence data: an unfinished debate. J Clin Virol. 2013;58(4):743–744. doi: 10.1016/j.jcv.2013.10.014 [DOI] [PubMed] [Google Scholar]
- [4].Seto D, Chodosh J, Brister JR, et al. Using the whole-genome sequence to characterize and name human adenoviruses. J Virol. 2011;85(11):5701–5702. doi: 10.1128/JVI.00354-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Harrach B, Tarján ZL, Benkő M. Adenoviruses across the animal kingdom: a walk in the zoo. FEBS Lett. 2019;593(24):3660–3673. doi: 10.1002/1873-3468.13687 [DOI] [PubMed] [Google Scholar]
- [6].Hearing P. Adenoviridae: the viruses and their replication. In: Howley P, Knipe D, Cohen J, and Damania B, editors. Fields Virology: DNA Viruses. Netherlands: Walters Kluwer; 2021. p. 98–128. [Google Scholar]
- [7].Lion T, Wold WSM. Adenoviruses. In: PM H, DM K, JL C, and BA D, editors. Fields Virology: DNA Viruses. Netherlands: Walters Kluwer; 2021. p. 129–171. [Google Scholar]
- [8].Khanal S, Ghimire P, Dhamoon AS. The repertoire of adenovirus in human disease: the innocuous to the deadly. Biomedicines. 2018;6(1):30. doi: 10.3390/biomedicines6010030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lynch JP 3rd, Kajon AE. Adenovirus: epidemiology, global spread of novel serotypes, and advances in treatment and prevention. Semin Respir Crit Care Med. 2016;37(4):586–602. doi: 10.1055/s-0036-1584923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lynch JP 3rd, Kajon AE. Adenovirus: epidemiology, global spread of novel types, and approach to treatment. Semin Respir Crit Care Med. 2021;42(6):800–821. doi: 10.1055/s-0041-1733802 [DOI] [PubMed] [Google Scholar]
- [11].Flewett TH, Morris CA, Bryden AS, et al. Epidemic viral enteritis in a long-stay children’s ward. Lancet. 1975;1(7897):4–5. doi: 10.1016/s0140-6736(75)92370-3 [DOI] [PubMed] [Google Scholar]
- [12].Schoub BD, Koornhof HJ, Lecatsas G, et al. Letter: viruses in acute summer gastroenteritis in black infants. Lancet. 1975;1(7915):1093–1094. doi: 10.1016/s0140-6736(75)91869-3 [DOI] [PubMed] [Google Scholar]
- [13].de Jong JC, Wigand R, Kidd AH, et al. Candidate adenoviruses 40 and 41: fastidious adenoviruses from human infant stool. J Med Virol. 1983;11(3):215–231. doi: 10.1002/jmv.1890110305 [DOI] [PubMed] [Google Scholar]
- [14].Uhnoo I, Wadell G, Svensson L, et al. Two new serotypes of enteric adenovirus causing infantile diarrhoea. Dev Biol Stand. 1983;53:311–318. [PubMed] [Google Scholar]
- [15].Davison AJ, Telford EA, Watson MS, et al. The DNA sequence of adenovirus type 40. J Mol Biol. 1993;234(4):1308–1316. doi: 10.1006/jmbi.1993.1687 [DOI] [PubMed] [Google Scholar]
- [16].Mautner V, Steinthorsdottir V, Bailey A. Enteric adenoviruses. Curr Top Microbiol Immunol. 1995;199(Pt 3):229–282. doi: 10.1007/978-3-642-79586-2_12 [DOI] [PubMed] [Google Scholar]
- [17].Pérez-Illana M, Martínez M, Condezo GN, et al. Cryo-EM structure of enteric adenovirus HAdV-F41 highlights structural variations among human adenoviruses. Sci Adv. 2021;7(9):eabd9421. 2021 Feb 24. 10.1126/sciadv.abd9421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rafie K, Lenman A, Fuchs J, et al. The structure of enteric human adenovirus 41-A leading cause of diarrhea in children. Sci Adv. 2021;7(2):eabe0974. doi: 10.1126/sciadv.abe0974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Albert MJ. Enteric adenoviruses. Brief review. Arch Virol. 1986;88(1–2):1–17. doi: 10.1007/BF01310885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tiemessen CT, Kidd AH. The subgroup F adenoviruses. J Gen Virol. 1995;76(Pt 3):481–497. doi: 10.1099/0022-1317-76-3-481 [DOI] [PubMed] [Google Scholar]
- [21].Uhnoo I, Svensson L, Wadell G. Enteric adenoviruses. Baillieres Clin Gastroenterol. 1990;4(3):627–642. doi: 10.1016/0950-3528(90)90053-j [DOI] [PubMed] [Google Scholar]
- [22].Wadell G, Allard A, Johannson M, et al. Enteric adenoviruses. In: Kapikian A, editor. Viral infections of the gastrointestinal tract. New York: Marcel Dekker; 1994. p. 57–74. [Google Scholar]
- [23].Wadell G, Allard A, Johansson M, et al. Enteric adenoviruses. Ciba Found Symp. 1987;128:63–91. doi: 10.1002/9780470513460.ch5 [DOI] [PubMed] [Google Scholar]
- [24].Whitelaw A, Davies H, Parry J. Electron microscopy of fatal adenovirus gastroenteritis. Lancet. 1977;1(8007):361. doi: 10.1016/s0140-6736(77)91158-8 [DOI] [PubMed] [Google Scholar]
- [25].de Souza EV, de Souza YFVP, Medeiros RS, et al. Diversity of enteric and non-enteric human adenovirus strains in Brazil, 2006-2011. Arch Virol. 2021;166(3):897–903. doi: 10.1007/s00705-020-04946-3 [DOI] [PubMed] [Google Scholar]
- [26].Hierholzer JC. Adenoviruses in the immunocompromised host. Clin Microbiol Rev. 1992;5(3):262–274. doi: 10.1128/CMR.5.3.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Thea DM, Glass R, Grohmann GS, et al. Prevalence of enteric viruses among hospital patients with AIDS in Kinshasa, Zaire. Trans R Soc Trop Med Hyg. 1993b;87(3):263–266. doi: 10.1016/0035-9203(93)90119-b PMID: 8236386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Thea DM, St Louis ME, Atido U, et al. A prospective study of diarrhea and HIV-1 infection among 429 Zairian infants. N Engl J Med. 1993a;329(23):1696–1702. doi: 10.1056/NEJM199312023292304 [DOI] [PubMed] [Google Scholar]
- [29].Kroes AC, de Klerk EP, Lankester AC, et al. Sequential emergence of multiple adenovirus serotypes after pediatric stem cell transplantation.J Clin Virol. J Clin Virol. 2007;38(4):341–347. doi: 10.1016/j.jcv.2007.01.001 [DOI] [PubMed] [Google Scholar]
- [30].Lion T, Kosulin K, Landlinger C, et al. Monitoring of adenovirus load in stool by real-time PCR permits early detection of impending invasive infection in patients after allogeneic stem cell transplantation. Leukemia. 2010;24(4):706–714. doi: 10.1038/leu.2010.4 [DOI] [PubMed] [Google Scholar]
- [31].Lion T. Adenovirus infections in immunocompetent and immunocompromised patients. Clin Microbiol Rev. 2014;27(3):441–462. doi: 10.1128/CMR.00116-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kosulin K. Intestinal HAdV infection: tissue specificity, persistence, and implications for antiviral therapy. Viruses. 2019;11(9):804. doi: 10.3390/v11090804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Green M, Wold WS, Mackey JK, et al. Analysis of human tonsil and cancer DNAs and RNAs for DNA sequences of group C (serotypes 1, 2, 5, and 6) human adenoviruses. Proc Natl Acad Sci U S A. 1979;76(12):6606–6610. doi: 10.1073/pnas.76.12.6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hogg JC. Role of latent viral infections in chronic obstructive pulmonary disease and asthma. Am J Respir Crit Care Med. 2001;164(10 Pt 2):S71–5. doi: 10.1164/ajrccm.164.supplement_2.2106063 [DOI] [PubMed] [Google Scholar]
- [35].Kosulin K, Geiger E, Vécsei A, et al. Persistence and reactivation of human adenoviruses in the gastrointestinal tract. Clin Microbiol Infect. 2016;22(4):.e381.1–.e381.8. doi: 10.1016/j.cmi.2015.12.013 [DOI] [PubMed] [Google Scholar]
- [36].Leung AY, Chan M, Cheng VC, et al. Quantification of adenovirus in the lower respiratory tract of patients without clinical adenovirus-related respiratory disease. Clin Infect Dis. 2005;40(10):1541–1544. doi: 10.1086/429627 [DOI] [PubMed] [Google Scholar]
- [37].Roy S, Calcedo R, Medina-Jaszek A, et al. Adenoviruses in lymphocytes of the human gastro-intestinal tract. PLoS One. 2011;6(9):e24859. doi: 10.1371/journal.pone.0024859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fox JP, Brandt CD, Wassermann FE, et al. The virus watch program: a continuing surveillance of viral infections in metropolitan New York families. VI. Observations of adenovirus infections: virus excretion patterns, antibody response, efficiency of surveillance, patterns of infections, and relation to illness. Am J Epidemiol. 1969. Jan;89(1):25–50. [DOI] [PubMed] [Google Scholar]
- [39].Fox JP, Hall CE, Cooney MK. The Seattle virus watch. VII. Observations of adenovirus infections. Am J Epidemiol. 1977;105(4):362–386. doi: 10.1093/oxfordjournals.aje.a112394 [DOI] [PubMed] [Google Scholar]
- [40].Garnett CT, Erdman D, Xu W, et al. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J Virol. 2002;76(21):10608–10616. doi: 10.1128/jvi.76.21.10608-10616.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Garnett CT, Talekar G, Mahr JA, et al. Latent species C adenoviruses in human tonsil tissues. J Virol. 2009;83(6):2417–2428. doi: 10.1128/JVI.02392-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Neumann R, Genersch E, Eggers HJ. Detection of adenovirus nucleic acid sequences in human tonsils in the absence of infectious virus. Virus Res. 1987;7(1):93–97. doi: 10.1016/0168-1702(87)90060-8 [DOI] [PubMed] [Google Scholar]
- [43].Lion T. Adenovirus persistence, reactivation, and clinical management. FEBS Lett. 2019;593(24):3571–3582. doi: 10.1002/1873-3468.13576 [DOI] [PubMed] [Google Scholar]
- [44].Hoffman JA. Adenovirus infections in solid organ transplant recipients. Curr Opin Organ Transplant. 2009;14(6):625–633. doi: 10.1097/MOT.0b013e3283324e1b [DOI] [PubMed] [Google Scholar]
- [45].Lion T, Baumgartinger R, Watzinger F, et al. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood. 2003;102(3):1114–1120. doi: 10.1182/blood-2002-07-2152 [DOI] [PubMed] [Google Scholar]
- [46].Zheng Y, Stamminger T, Hearing P. E2F/Rb family proteins mediate interferon induced repression of adenovirus immediate early transcription to promote persistent viral infection. PLOS Pathog. 2016;12(1):e1005415. doi: 10.1371/journal.ppat.1005415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Prasad V, Suomalainen M, Jasiqi Y, et al. The UPR sensor IRE1α and the adenovirus E3-19K glycoprotein sustain persistent and lytic infections. Nat Commun. 2020;11(1):1997. doi: 10.1038/s41467-020-15844-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Bergelson JM, Cunningham JA, Droguett G, et al. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275(5304):1320–1323. doi: 10.1126/science.275.5304.1320 [DOI] [PubMed] [Google Scholar]
- [49].Roelvink PW, Lizonova A, Lee JG, et al. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol. 1998;72(10):7909–7915. doi: 10.1128/JVI.72.10.7909-7915.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Gaggar A, Shayakhmetov DM, Lieber A. CD46 is a cellular receptor for group B adenoviruses.Nat med. 2003;9(11):1408–1412.doi: 10.1038/nm952 [DOI] [PubMed] [Google Scholar]
- [51].Stasiak AC, Stehle T. Human adenovirus binding to host cell receptors: a structural view. Med Microbiol Immunol. 2020;209(3):325–333. doi: 10.1007/s00430-019-00645-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yeh HY, Pieniazek N, Pieniazek D, et al. Genetic organization, size, and complete sequence of early region 3 genes of human adenovirus type 41. J Virol. 1996;70(4):2658–2663. doi: 10.1128/jvi.70.4.2658-2663.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kidd AH, Chroboczek J, Cusack S, et al. Adenovirus type 40 virions contain two distinct fibers. Virology. 1993;192(1):73–84. doi: 10.1006/viro.1993.1009 [DOI] [PubMed] [Google Scholar]
- [54].Rajan A, Palm E, Trulsson F, et al. Heparan sulfate is a cellular receptor for enteric human adenoviruses. Viruses. 2021;13(2):298. doi: 10.3390/v13020298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rajan A, Persson BD, Frängsmyr L, et al. Enteric species F human adenoviruses use laminin-binding integrins as co-receptors for infection of Ht-29 cells. Sci Rep. 2018;8(1):10019. doi: 10.1038/s41598-018-28255-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Albinsson B, Kidd AH. Adenovirus type 41 lacks an RGD alpha(v)-integrin binding motif on the penton base and undergoes delayed uptake in A549 cells. Virus Res. 1999;64(2):125–136. doi: 10.1016/s0168-1702(99)00087-8 [DOI] [PubMed] [Google Scholar]
- [57].Favier AL, Burmeister WP, Chroboczek J. Unique physicochemical properties of human enteric Ad41 responsible for its survival and replication in the gastrointestinal tract. Virology. 2004;322(1):93–104. doi: 10.1016/j.virol.2004.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Stewart RL, O’Connor KL. Clinical significance of the integrin α6β4 in human malignancies. Lab Invest. 2015;95(9):976–986. doi: 10.1038/labinvest.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Williams JM, Duckworth CA, Burkitt MD, et al. Epithelial cell shedding and barrier function: a matter of life and death at the small intestinal villus tip. Vet Pathol. 2015;52(3):445–455. doi: 10.1177/0300985814559404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Arnberg N. Adenovirus receptors: implications for targeting of viral vectors. Trends Pharmacol Sci. 2012;33(8):442–448. doi: 10.1016/j.tips.2012.04.005 [DOI] [PubMed] [Google Scholar]
- [61].Khare R, Reddy VS, Nemerow GR, et al. Identification of adenovirus serotype 5 hexon regions that interact with scavenger receptors.J Virol. 2012;86(4):2293–2301.doi: 10.1128/JVI.05760-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Maler MD, Nielsen PJ, Stichling N, et al. Key role of the scavenger receptor MARCO in mediating adenovirus infection and subsequent innate responses of macrophages. MBio. 2017;8(4):e00670–17. doi: 10.1128/mBio.00670-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Stichling N, Suomalainen M, Flatt JW, et al. Lung macrophage scavenger receptor SR-A6 (MARCO) is an adenovirus type-specific virus entry receptor. PLOS Pathog. 2018;14(3):e1006914. doi: 10.1371/journal.ppat.1006914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Greber UF, Flatt JW. Adenovirus entry: from infection to immunity. Annu Rev Virol. 2019;6(1):177–197. doi: 10.1146/annurev-virology-092818-015550 [DOI] [PubMed] [Google Scholar]
- [65].Greber UF, Singh I, Helenius A. Mechanisms of viral un coating. Trends Microbiol. 1994;2(2):52–56. doi: 10.1016/0966-842x(94)90126-0 [DOI] [PubMed] [Google Scholar]
- [66].Greber UF, Suomalainen M. Adenovirus entry: stability, uncoating, and nuclear import. Mol Microbiol. 2022;118(4):309–320. doi: 10.1111/mmi.14909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Greber UF, Willetts M, Webster P, et al. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75(3):477–486. doi: 10.1016/0092-8674(93)90382-z [DOI] [PubMed] [Google Scholar]
- [68].Nakano MY, Boucke K, Suomalainen M, et al. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J Virol. 2000;74(15):7085–7095. doi: 10.1128/jvi.74.15.7085-7095.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Puntener D, Engelke MF, Ruzsics Z, et al. Stepwise loss of fluorescent core protein V from human adenovirus during entry into cells. J Virol. 2011;85(1):481–496. doi: 10.1128/JVI.01571-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bremner KH, Scherer J, Yi J, et al. Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe. 2009;6(6):523–535. doi: 10.1016/j.chom.2009.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Dales S, Chardonnet Y. Early events in the interaction of adenoviruses with HeLa cells. IV. Association with microtubules and the nuclear pore complex during vectorial movement of the inoculum. Virology. 1973;56(2):465–483. doi: 10.1016/0042-6822(73)90050-0 [DOI] [PubMed] [Google Scholar]
- [72].Miles BD, Luftig RB, Weatherbee JA, et al. Quantitation of the interaction between adenovirus types 2 and 5 and microtubules inside infected cells. Virology. 1980;105(1):265–269. doi: 10.1016/0042-6822(80)90177-4 [DOI] [PubMed] [Google Scholar]
- [73].Wang IH, Burckhardt CJ, Yakimovich A, et al. Imaging, tracking and computational analyses of virus entry and egress with the cytoskeleton. Viruses. 2018;10(4):166. doi: 10.3390/v10040166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Scherer J, Vallee RB. Adenovirus recruits dynein by an evolutionary novel mechanism involving direct binding to Ph-primed hexon. Viruses. 2011;3(8):1417–1431. doi: 10.3390/v3081417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Zhou J, Scherer J, Yi J, et al. Role of kinesins in directed adenovirus transport and cytoplasmic exploration. PLOS Pathog. 2018;14(5):e1007055. doi: 10.1371/journal.ppat.1007055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Scherer J, Yi J, Vallee RB. Role of cytoplasmic dynein and kinesins in adenovirus transport. FEBS Lett. 2020;594(12):1838–1847. doi: 10.1002/1873-3468.13777 [DOI] [PubMed] [Google Scholar]
- [77].Cassany A, Ragues J, Guan T, et al. Nuclear import of adenovirus DNA involves direct interaction of hexon with an N-terminal domain of the nucleoporin Nup214. J Virol. 2015;89(3):1719–1730. doi: 10.1128/JVI.02639-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Trotman LC, Mosberger N, Fornerod M, et al. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat Cell Biol. 2001;3(12):1092–1100. doi: 10.1038/ncb1201-1092 [DOI] [PubMed] [Google Scholar]
- [79].Wang IH, Burckhardt CJ, Yakimovich A, et al. The nuclear export factor CRM1 controls juxta-nuclear microtubule-dependent virus transport. J Cell Sci. 2017;130(13):2185–2195. doi: 10.1242/jcs.203794 [DOI] [PubMed] [Google Scholar]
- [80].Bauer M, Flatt JW, Seiler D, et al. The E3 ubiquitin ligase mind bomb 1 controls adenovirus genome release at the nuclear pore complex. Cell Rep. 2019;29(12):3785–3795.e8. doi: 10.1016/j.celrep.2019.11.064 [DOI] [PubMed] [Google Scholar]
- [81].Sarbanes SL, Blomen VA, Lam E, et al. E3 ubiquitin ligase mindbomb 1 facilitates nuclear delivery of adenovirus genomes. Proc Natl Acad Sci U S A. 2021;118(1):e2015794118. doi: 10.1073/pnas.2015794118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Shayakhmetov DM, Li ZY, Ternovoi V, et al. The interaction between the fiber knob domain and the cellular attachment receptor determines the intracellular trafficking route of adenoviruses. J Virol. 2003;77(6):3712–3723. doi: 10.1128/jvi.77.6.3712-3723.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Witt DJ, Bousquet EB. Comparison of enteric adenovirus infection in various human cell lines. J Virol Methods. 1988;20(4):295–308. doi: 10.1016/0166-0934(88)90133-4 [DOI] [PubMed] [Google Scholar]
- [84].Brown M, Wilson-Friesen HL, Doane F. A block in release of progeny virus and a high particle-to-infectious unit ratio contribute to poor growth of enteric adenovirus types 40 and 41 in cell culture. J Virol. 1992;66(5):3198–3205. doi: 10.1128/JVI.66.5.3198-3205.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Lemiale F, Haddada H, Nabel GJ, et al. Novel adenovirus vaccine vectors based on the enteric-tropic serotype 41. Vaccine. 2007;25(11):2074–2084. doi: 10.1016/j.vaccine.2006.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Yamasaki S, Miura Y, Brown E, et al. Development of a method for effective amplification of human adenovirus 40. Arch Virol. 2010;155(7):1059–1068. doi: 10.1007/s00705-010-0683-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Blackford AN, Grand RJ. Adenovirus E1B 55-kilodalton protein: multiple roles in viral infection and cell transformation. J Virol. 2009;83(9):4000–4012. doi: 10.1128/JVI.02417-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Götting J, Cordes AK, Steinbrück L, et al. Molecular phylogeny of human adenovirus type 41 lineages. eCollection 2022 Virus Evol. 2022;8(2):veac098. doi: 10.1093/ve/veac098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ebner K, Pinsker W, Lion T. Comparative sequence analysis of the hexon gene in the entire spectrum of human adenovirus serotypes: phylogenetic, taxonomic, and clinical implications. J Virol. 2005;79(20):12635–12642. doi: 10.1128/JVI.79.20.12635-12642.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Gilson T, Blanchette P, Ballmann MZ, et al. Using the E4orf6-based E3 ubiquitin ligase as a tool to analyze the evolution of adenoviruses. J Virol. 2016;90(16):7350–7367. doi: 10.1128/JVI.00420-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Madisch I, Harste G, Pommer H, et al. Phylogenetic analysis of the main neutralization and hemagglutination determinants of all human adenovirus prototypes as a basis for molecular classification and taxonomy.J. Virol. 2005;79(24):15265–15276. doi: 10.1128/JVI.79.24.15265-15276.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Madisch I, Hofmayer S, Moritz C, et al. Phylogenetic analysis and structural predictions of human adenovirus penton proteins as a basis for tissue-specific adenovirus vector design. J Virol. 2007;81(15):8270–8281. doi: 10.1128/JVI.00048-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Madisch I, Wölfel R, Harste G, et al. Molecular identification of adenovirus sequences: a rapid scheme for early typing of human adenoviruses in diagnostic samples of immunocompetent and immunodeficient patients. J Med Virol. 2006;78(9):1210–1217. doi: 10.1002/jmv.20683 [DOI] [PubMed] [Google Scholar]
- [94].Avvakumov N, Kajon AE, Hoeben RC, et al. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology. 2004;329(2):477–492. doi: 10.1016/j.virol.2004.08.007 [DOI] [PubMed] [Google Scholar]
- [95].van Loon AE, Ligtenberg M, Reemst AM, et al. Structure and organization of the left-terminal DNA regions of fastidious adenovirus types 40 and 41. Gene. 1987;58(1):109–126. doi: 10.1016/0378-1119(87)90034-5 [DOI] [PubMed] [Google Scholar]
- [96].Cheng CY, Gilson T, Dallaire F, et al. The E4orf6/E1B55K E3 ubiquitin ligase complexes of human adenoviruses exhibit heterogeneity in composition and substrate specificity. J Virol. 2011;85(2):765–775. doi: 10.1128/JVI.01890-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Cheng CY, Gilson T, Wimmer P, et al. Role of E1B55K in E4orf6/E1B55K E3 ligase complexes formed by different human adenovirus serotypes. J Virol. 2013;87(11):6232–6245. doi: 10.1128/JVI.00384-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Cousin C, Winter N, Gomes SA, et al. Cellular transformation by E1 genes of enteric adenoviruses. Virology. 1991;181(1):277–287. doi: 10.1016/0042-6822(91)90493-u [DOI] [PubMed] [Google Scholar]
- [99].Bailey A, Mautner V. Phylogenetic relationships among adenovirus serotypes. Virology. 1994;205(2):438–452. doi: 10.1006/viro.1994.1664 [DOI] [PubMed] [Google Scholar]
- [100].Lichtenstein DL, Toth K, Doronin K, et al. Functions and mechanisms of action of the adenovirus E3 proteins. Int Rev Immunol. 2004;23(1–2):75–111. doi: 10.1080/08830180490265556 [DOI] [PubMed] [Google Scholar]
- [101].Frietze KM, Campos SK, Kajon AE. No evidence of a death-like function for species B1 human adenovirus type 3 E3-9K during A549 cell line infection. BMC Res Notes. 2012;5(1):429. doi: 10.1186/1756-0500-5-429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Lu ZZ, Zou XH, Lastinger K, et al. Enhanced growth of recombinant human adenovirus type 41 (HAdV-41) carrying ADP gene. Virus Res. 2013;176(1–2):61–68. doi: 10.1016/j.virusres.2013.05.003 [DOI] [PubMed] [Google Scholar]
- [103].Mautner V, Mackay N. Enteric adenovirus type 40: complementation of the E4 defect in Ad2 dl808. Virology. 1991;183(1):433–436. doi: 10.1016/0042-6822(91)90161-4 [DOI] [PubMed] [Google Scholar]
- [104].Querido E, Blanchette P, Yan Q, et al. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 2001;15(23):3104–3117. doi: 10.1101/gad.926401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Schreiner S, Wimmer P, Dobner T. Adenovirus degradation of cellular proteins. Future Microbiol. 2012;7(2):211–225. doi: 10.2217/fmb.11.153 [DOI] [PubMed] [Google Scholar]
- [106].Blackford AN, Patel RN, Forrester NA, et al. Adenovirus 12 E4orf6 inhibits ATR activation by promoting TOPBP1 degradation. Proc Natl Acad Sci U S A. 2010;107(27):12251–12256. doi: 10.1073/pnas.0914605107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Gilson T, Cheng CY, Hur WS, et al. Analysis of the Cullin binding sites of the E4orf6 proteins of human adenovirus E3 ubiquitin ligases. J Virol. 2014;88(7):3885–3897. doi: 10.1128/JVI.03579-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Cheng L, Huang X, Li X, et al. Cryo-EM structures of two bovine adenovirus type 3 intermediates. Virology. 2014;450-451:174–181. doi: 10.1016/j.virol.2013.12.012 [DOI] [PubMed] [Google Scholar]
- [109].Kundhavai Natchiar S, Venkataraman S, Mullen TM, et al. Revised crystal structure of human adenovirus reveals the limits on protein IX quasi-equivalence and on analyzing large macromolecular complexes. J Mol Biol. 2018;430(21):4132–4141. doi: 10.1016/j.jmb.2018.08.011 [DOI] [PubMed] [Google Scholar]
- [110].Liu H, Jin L, Koh SB, et al. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329(5995):1038–1043. doi: 10.1126/science.1187433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Marabini R, Condezo GN, Krupovic M, et al. Near-atomic structure of an atadenovirus reveals a conserved capsid-binding motif and intergenera variations in cementing proteins. Sci Adv. 2021;7(14):eabe6008. doi: 10.1126/sciadv.abe6008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Menéndez-Conejero R, Nguyen TH, Singh AK, et al. Structure of a reptilian adenovirus reveals a phage tailspike fold stabilizing a vertebrate virus capsid. Structure. 2017;25(10):1562–1573.e5. doi: 10.1016/j.str.2017.08.007 [DOI] [PubMed] [Google Scholar]
- [113].Reddy VS, Natchiar SK, Stewart PL, et al. Crystal structure of human adenovirus at 3.5 a resolution. Science. 2010;329(5995):1071–1075. doi: 10.1126/science.1187292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Yu X, Veesler D, Campbell MG, et al. Cryo-EM structure of human adenovirus D26 reveals the conservation of structural organization among human adenoviruses. Sci Adv. 2017;3(5):e1602670. doi: 10.1126/sciadv.1602670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Toogood CI, Hay RT. DNA sequence of the adenovirus type 41 hexon gene and predicted structure of the protein. J Gen Virol. 1988;69(Pt 9):2291–2301. doi: 10.1099/0022-1317-69-9-2291 [DOI] [PubMed] [Google Scholar]
- [116].Toogood CI, Murali R, Burnett RM, et al. The adenovirus type 40 hexon: sequence, predicted structure and relationship to other adenovirus hexons. J Gen Virol. 1989;70(Pt 12):3203–3214. doi: 10.1099/0022-1317-70-12-3203 [DOI] [PubMed] [Google Scholar]
- [117].Yu X, Mullen TM, Abrishami V, et al. Structure of a cell entry defective human adenovirus provides insights into precursor proteins and capsid maturation. J Mol Biol. 2022;434(2):167350. doi: 10.1016/j.jmb.2021.167350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Pieniazek NJ, Slemenda SB, Pieniazek D, et al. Human enteric adenovirus type 41 (Tak) contains a second fiber protein gene. Nucleic Acids Res. 1990;18(7):1901. doi: 10.1093/nar/18.7.1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Yeh HY, Pieniazek N, Pieniazek D, et al. Human adenovirus type 41 contains two fibers. Virus Res. 1994;33(2):179–198. doi: 10.1016/0168-1702(94)90054-x [DOI] [PubMed] [Google Scholar]
- [120].Seiradake E, Cusack S. Crystal structure of enteric adenovirus serotype 41 short fiber head. J Virol. 2005;79(22):14088–14094. doi: 10.1128/JVI.79.22.14088-14094.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Gallardo J, Pérez-Illana M, Martín-González N, et al. Adenovirus structure: what is new? Int J Mol Sci. 2021;22(10):5240. doi: 10.3390/ijms22105240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Dai X, Wu L, Sun R, et al. Atomic structures of minor proteins VI and VII in human adenovirus. J Virol. 2017;91(24):e00850–17. doi: 10.1128/JVI.00850-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Nemerow GR, Stewart PL. Insights into adenovirus uncoating from interactions with integrins and mediators of host immunity. Viruses. 2016;8(12):337. doi: 10.3390/v8120337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Nguyen EK, Nemerow GR, Smith JG. Direct evidence from single-cell analysis that human {alpha}-defensins block adenovirus uncoating to neutralize infection. J Virol. 2010;84(8):4041–4049. doi: 10.1128/JVI.02471-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Smith JG, Nemerow GR. Mechanism of adenovirus neutralization by human alpha-defensins. Cell Host Microbe. 2008;3(1):11–19. doi: 10.1016/j.chom.2007.12.001 [DOI] [PubMed] [Google Scholar]
- [126].Smith JG, Silvestry M, Lindert S, et al. Insight into the mechanisms of adenovirus capsid disassembly from studies of defensin neutralization. PLOS Pathog. 2010;6(6):e1000959. doi: 10.1371/journal.ppat.1000959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Flatt JW, Kim R, Smith JG, et al. An intrinsically disordered region of the adenovirus capsid is implicated in neutralization by human alpha defensin 5. PLoS One. 2013;8(4):e61571. Print 2013. doi: 10.1371/journal.pone.0061571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Beatty MS, Curiel DT. Chapter two–adenovirus strategies for tissue-specific targeting. Adv Cancer Res. 2012;115:39–67. doi: 10.1016/B978-0-12-398342-8.00002-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Watanabe M, Nishikawaji Y, Kawakami H, et al. Adenovirus biology, recombinant adenovirus, and adenovirus usage in gene therapy. Viruses. 2021;13(12):2502. doi: 10.3390/v13122502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Wold WS, Toth K. Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr Gene Ther. 2013;13(6):421–433. doi: 10.2174/1566523213666131125095046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Buckinx R, Timmermans JP. Targeting the gastrointestinal tract with viral vectors: state of the art and possible applications in research and therapy. Histochem Cell Biol. 2016;146(6):709–720. doi: 10.1007/s00418-016-1496-6 [DOI] [PubMed] [Google Scholar]
- [132].Croyle MA, Stone M, Amidon GL, et al. In vitro and in vivo assessment of adenovirus 41 as a vector for gene delivery to the intestine. Gene Ther. 1998;5(5):645–654. doi: 10.1038/sj.gt.3300645 [DOI] [PubMed] [Google Scholar]
- [133].Grand RJ. A link between severe hepatitis in children and adenovirus 41 and adeno-associated virus 2 infections. J Gen Virol. 2022;103(11). doi: 10.1099/jgv.0.001783 [DOI] [PubMed] [Google Scholar]
- [134].Ilic I, Ilic M. Multi-country outbreak of severe acute hepatitis of unknown origin in children, 2022. Acta Paediatr. 2023;112(6):1148–1156. doi: 10.1111/apa.16685 [DOI] [PubMed] [Google Scholar]
- [135].Karpen SJ. Acute hepatitis in children in 2022 - human adenovirus 41? N Engl J Med. 2022;387(7):656–657. doi: 10.1056/NEJMe2208409 [DOI] [PubMed] [Google Scholar]
- [136].Matthews PC, Campbell C, Săndulescu O, et al. Acute severe hepatitis outbreak in children: A perfect storm. What do we know, and what questions remain? Front Pharmacol. 2022;13:1062408. doi: 10.3389/fphar.2022.1062408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Uwishema O, Mahmoud A, Wellington J, et al. A review on acute, severe hepatitis of unknown origin in children: A call for concern. Ann Med Surg (Lond). 2022;81:104457. doi: 10.1016/j.amsu.2022.104457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Wang C, Gao ZY, Walsh N, et al. Acute hepatitis of unknown aetiology among children around the world. Infect Dis Poverty. 2022;11(1):112. doi: 10.1186/s40249-022-01035-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Bucuvalas J, Yazigi N, RH S Jr. Acute liver failure in children. vii Clin Liver Dis. 2006;10(1):149–168. doi: 10.1016/j.cld.2005.10.006 [DOI] [PubMed] [Google Scholar]
- [140].Squires RH Jr, Shneider BL, Bucuvalas J, et al. Acute liver failure in children: the first 348 patients in the pediatric acute liver failure study group. J Pediatr. 2006;148(5):652–658. doi: 10.1016/j.jpeds.2005.12.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Kelgeri C, Couper M, Gupte GL, et al. Clinical spectrum of children with acute hepatitis of unknown cause. N Engl J Med. 2022;387(7):611–619. doi: 10.1056/NEJMoa2206704 [DOI] [PubMed] [Google Scholar]
- [142].Gutierrez Sanchez LH, Shiau H, Baker JM, et al. A case series of children with acute hepatitis and human adenovirus infection. N Engl J Med. 2022. 2022 Aug 18;387(7):620–630. doi: 10.1056/NEJMoa2206294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Ho A, Orton R, Tayler R, et al. Nature. Adeno-associated virus 2 infection in children with non-A-E hepatitis. Online ahead of print;2023. doi: 10.1038/s41586-023-05948-2 [DOI] [PubMed] [Google Scholar]
- [144].Morfopoulou S, Buddle S, Montaguth OET, et al. Genomic investigations of unexplained acute hepatitis in children. Nature. 2023. Mar 30;617(7961):564–573. Online ahead of print. doi: 10.1038/s41586-023-06003-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Servellita V, Gonzalez AS, Lamson DM, et al. Adeno-associated virus type 2 in US children with acute severe hepatitis. Nature. 2023;617(7961):574–580. doi: 10.1038/s41586-023-05949-1 Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Samulski RJ, Muzyczka N. AAV-Mediated gene therapy for research and therapeutic purposes. Annu Rev Virol. 2014;1(1):427–451. doi: 10.1146/annurev-virology-031413-085355 [DOI] [PubMed] [Google Scholar]
- [147].Kerr JR, Cotmore SF, Bloom R, editors et al. Parvoviruses. London. Hodder Arnold Publication; 2006. doi: 10.1201/b13393 [DOI] [Google Scholar]
- [148].Muzyczka N, Berns K. Parvoviridae: the viruses and their replication. Fields Virology. 2001;2:2327–2359. [Google Scholar]
- [149].Weitzman MD, Linden RM. Adeno-associated virus biology. Methods Mol Biol. 2011;807:1–23. doi: 10.1007/978-1-61779-370-7_1 [DOI] [PubMed] [Google Scholar]
- [150].Moullier P, Snyder RO. International efforts for recombinant adeno-associated viral vector reference standards. Mol Ther. 2008;16(7):1185–1188. doi: 10.1038/mt.2008.125 [DOI] [PubMed] [Google Scholar]
- [151].Boutin S, Monteilhet V, Veron P, et al. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther. 2010;21(6):704–712. doi: 10.1089/hum.2009.182 [DOI] [PubMed] [Google Scholar]
- [152].Calcedo R, Vandenberghe LH, Gao G, et al. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis. 2009;199(3):381–390. doi: 10.1086/595830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Louis Jeune V, Joergensen JA, Hajjar RJ, et al. Pre-existing anti-adeno-associated virus antibodies as a challenge in AAV gene therapy. Hum Gene Ther Methods. 2013;24(2):59–67. doi: 10.1089/hgtb.2012.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Chirmule N, Propert K, Magosin S, et al. Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 1999;6(9):1574–1583. doi: 10.1038/sj.gt.3300994 [DOI] [PubMed] [Google Scholar]
- [155].Calcedo R, Morizono H, Wang L, et al. Adeno-associated virus antibody profiles in newborns, children, and adolescents. Clin Vaccine Immunol. 2011;18(9):1586–1588. doi: 10.1128/CVI.05107-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Erles K, Sebökovà P, Schlehofer JR. Update on the prevalence of serum antibodies (IgG and IgM) to adeno-associated virus (AAV). J Med Virol. 1999;59(3):406–411. doi: [DOI] [PubMed] [Google Scholar]
- [157].Gao G, Vandenberghe LH, Alvira MR, et al. Clades of adeno-associated viruses are widely disseminated in human tissues. J Virol. 2004;78(12):6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Grossman Z, Mendelson E, Brok-Simoni F, et al. Detection of adeno-associated virus type 2 in human peripheral blood cells. J Gen Virol. 1992;73(Pt 4):961–966. doi: 10.1099/0022-1317-73-4-961 [DOI] [PubMed] [Google Scholar]
- [159].Sayyadi-Dehno Z, Seyed Khorrami SM, Ghavami N, et al. Molecular detection of adeno-associated virus DNA in cases of spontaneous and therapeutic abortion. Fetal Pediatr Pathol. 2019;38(3):206–214. doi: 10.1080/15513815.2019.1576817 [DOI] [PubMed] [Google Scholar]
- [160].Tobiasch E, Rabreau M, Geletneky K, et al. Detection of adeno-associated virus DNA in human genital tissue and in material from spontaneous abortion. J Med Virol. 1994;44(2):215–222. doi: 10.1002/jmv.1890440218 [DOI] [PubMed] [Google Scholar]
- [161].Walz CM, Anisi TR, Schlehofer JR, et al. Detection of infectious adeno-associated virus particles in human cervical biopsies. Virology. 1998;247(1):97–105. doi: 10.1006/viro.1998.9226 [DOI] [PubMed] [Google Scholar]
- [162].Hüser D, Khalid D, Lutter T, et al. High prevalence of infectious adeno-associated virus (AAV) in human peripheral blood mononuclear cells indicative of t lymphocytes as sites of AAV persistence. J Virol. 2017;91(4):e02137–16. doi: 10.1128/JVI.02137-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Berns KI, Giraud C. Biology of adeno-associated virus. Curr Top Microbiol Immunol. 1996;218:1–23. doi: 10.1007/978-3-642-80207-2_1 [DOI] [PubMed] [Google Scholar]
- [164].Blacklow NR, Hoggan MD, Kapikian AZ, et al. Epidemiology of adenovirus-associated virus infection in a nursery population. Am J Epidemiol. 1968a;88(3):368–378. doi: 10.1093/oxfordjournals.aje.a120897 [DOI] [PubMed] [Google Scholar]
- [165].Blacklow NR, Hoggan MD, Rowe WP. Serologic evidence for human infection with adenovirus-associated viruses. J Natl Cancer Inst. 1968b;40(2):319–327. [PubMed] [Google Scholar]
- [166].Chand D, Mohr F, McMillan H, et al. Hepatotoxicity following administration of onasemnogene abeparvovec (AVXS-101) for the treatment of spinal muscular atrophy. J Hepatol. 2021;74(3):560–566. doi: 10.1016/j.jhep.2020.11.001 [DOI] [PubMed] [Google Scholar]
- [167].Chowdary P, Shapiro S, Makris M, et al. Phase 1-2 trial of AAVS3 gene therapy in patients with hemophilia B. N Engl J Med. 2022;387(3):237–247. doi: 10.1056/NEJMoa2119913 [DOI] [PubMed] [Google Scholar]
- [168].Mullard A. Gene therapy community grapples with toxicity issues, as pipeline matures. Nat Rev Drug Discov. 2021;20(11):804–805. doi: 10.1038/d41573-021-00164-x [DOI] [PubMed] [Google Scholar]
- [169].Muzyczka N, Berns KI. AAV’s Golden Jubilee. Mol Ther. 2015;23(5):807–808. doi: 10.1038/mt.2015.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Sant’anna TB, Araujo NM. Adeno-associated virus infection and its impact in human health: an overview. Virol J. 2022;19(1):173. doi: 10.1186/s12985-022-01900-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].La Bella T, Imbeaud S, Peneau C, et al. Adeno-associated virus in the liver: natural history and consequences in tumour development. Gut. 2020;69(4):737–747. doi: 10.1136/gutjnl-2019-318281 [DOI] [PubMed] [Google Scholar]
- [172].Nault JC, Datta S, Imbeaud S, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas.Nat Genet. 2015;47(10):1187–1193. doi: 10.1038/ng.3389 [DOI] [PubMed] [Google Scholar]
- [173].Nault JC, Datta S, Imbeaud S, et al. Adeno-associated virus type 2 as an oncogenic virus in human hepatocellular carcinoma. Mol Cell Oncol. 2016;3(2):e1095271. doi: 10.1080/23723556.2015.1095271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Tatsuno K, Midorikawa Y, Takayama T, et al. Impact of AAV2 and hepatitis B virus integration into genome on development of hepatocellular carcinoma in patients with prior hepatitis B virus infection. Clin Cancer Res. 2019;25(20):6217–6227. doi: 10.1158/1078-0432.CCR-18-4041 [DOI] [PubMed] [Google Scholar]
- [175].Berns KI, Byrne BJ, Flotte TR, et al. Adeno-Associated virus type 2 and hepatocellular carcinoma? Hum Gene Ther. 2015;26(12):779–781. doi: 10.1089/hum.2015.29014.kib [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Büning H, Schmidt M. Adeno-associated vector toxicity-to be or not to be? Mol Ther. 2015;23(11):1673–1675. doi: 10.1038/mt.2015.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Park KJ, Lee J, Park JH, et al. Adeno-associated virus 2-mediated hepatocellular carcinoma is very rare in Korean patients. Ann Lab Med. 2016;36(5):469–474. doi: 10.3343/alm.2016.36.5.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Schäffer AA, Dominguez DA, Chapman LM, et al. Integration of adeno-associated virus (AAV) into the genomes of most Thai and Mongolian liver cancer patients does not induce oncogenesis. BMC Genomics. 2021;22(1):814. doi: 10.1186/s12864-021-08098-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Kotin RM, Siniscalco M, Samulski RJ, et al. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci, USA. 1990;87(6):2211–2215. doi: 10.1073/pnas.87.6.2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Samulski RJ, Zhu X, Xiao X, et al. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. Embo J. 1991;10(12):3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Galibert L, Hyvönen A, Eriksson RAE, et al. Functional roles of the membrane associated AAV protein MAAP. Sci Rep. 2021;11(1):21698. doi: 10.1038/s41598-021-01220-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Mietzsch M, Pénzes JJ, Agbandje-McKenna M. Twenty-five years of structural parvovirology. Viruses. 2019;11(4):362. doi: 10.3390/v11040362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Qiu J, Pintel DJ. The adeno-associated virus type 2 rep protein regulates RNA processing via interaction with the transcription template. Mol Cell Biol. 2002;22(11):3639–3652. doi: 10.1128/MCB.22.11.3639-3652.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Meyer NL, Chapman MS. Adeno-associated virus (AAV) cell entry: structural insights.Trends microbiol. 2022;30(5):432–451.doi: 10.1016/j.tim.2021.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Meyer NL, Hu G, Davulcu O, et al. Structure of the gene therapy vector, adeno-associated virus with its cell receptor. AAVR Elife. 2019;8:e44707. doi: 10.7554/eLife.44707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Zhang R, Cao L, Cui M, et al. Adeno-associated virus 2 bound to its cellular receptor AAVR. Nat Microbiol. 2019a;4(4):675–682. doi: 10.1038/s41564-018-0356-7 [DOI] [PubMed] [Google Scholar]
- [187].Zhang R, Xu G, Cao L, et al. Divergent engagements between adeno-associated viruses with their cellular receptor AAVR. Nat Commun. 2019b;10(1):3760. doi: 10.1038/s41467-019-11668-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Kambhampati AK, Burke RM, Dietz S, et al. Trends in acute hepatitis of unspecified etiology and adenovirus stool testing results in children - United States, 2017-2022. MMWR Morb Mortal Wkly Rep. 2022;71(24):797–802. doi: 10.15585/mmwr.mm7124e1 [DOI] [PubMed] [Google Scholar]
- [189].Leiskau C, Tsaka S, Meyer-Ruhnke L, et al. Acute severe non-A-E-hepatitis of unknown origin in children - a 30-year retrospective observational study from north-west Germany. Hepatol. 2023;78(5):971–978. doi: 10.1016/j.jhep.2022.12.012 [DOI] [PubMed] [Google Scholar]
- [190].Martin NA, Gonzalez G, Reynolds LJ, et al. Adeno-associated virus 2 and human adenovirus F41 in wastewater during outbreak of severe acute hepatitis in children, Ireland. Emerg Infect Dis. 2023;29(4):751–760. doi: 10.3201/eid2904.221878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Reyne MI, Allen DM, Levickas A, et al. Detection of human adenovirus F41 in wastewater and its relationship to clinical cases of acute hepatitis of unknown aetiology. Sci Total Environ. 2023;857(Pt 2):159579. doi: 10.1016/j.scitotenv.2022.159579 [DOI] [PubMed] [Google Scholar]
- [192].Messacar K, Baker RE, Park SW, et al. Preparing for uncertainty: endemic paediatric viral illnesses after COVID-19 pandemic disruption. Lancet. 2022;400(10364):1663–1665. doi: 10.1016/S0140-6736(22)01277-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Al-Jwadi RF, Mills EHA, Torp-Pedersen C, et al. Consequences of COVID-19-related lockdowns and reopenings on emergency hospitalizations in pediatric patients in Denmark during 2020-2021. Eur J Pediatr. 2023;182(1):285–293. doi: 10.1007/s00431-022-04682-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Amar S, Avni YS, O’Rourke N, et al. Prevalence of common infectious diseases after COVID-19 vaccination and easing of pandemic restrictions in Israel. JAMA Netw Open. 2022;5(2):e2146175. doi: 10.1001/jamanetworkopen.2021.46175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Benschop KS, Albert J, Anton A, et al. Re-emergence of enterovirus D68 in Europe after easing the COVID-19 lockdown, September 2021. Euro Surveill. 2021;26(45):2100998. doi: 10.2807/1560-7917.ES.2021.26.45.2100998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Eden JS, Sikazwe C, Xie R, et al. Off-season RSV epidemics in Australia after easing of COVID-19 restrictions. Nat Commun; 13(1):2884. doi: 10.1038/s41467-022-30485-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Korokhov N, Mikheeva G, Krendelshchikov A, et al. Targeting of adenovirus via genetic modification of the viral capsid combined with a protein bridge. J Virol. 2003;77(24):12931–12940. doi: 10.1128/jvi.77.24.12931-12940.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Krasnykh VN, Douglas JT, van Beusechem VW. Genetic targeting of adenoviral vectors. Mol Ther. 2000;1(5 Pt 1):391–405. doi: 10.1006/mthe.2000.0062 [DOI] [PubMed] [Google Scholar]
- [199].Krasnykh VN, Mikheeva GV, Douglas JT, et al. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J Virol. 1996;70(10):6839–6846. doi: 10.1128/JVI.70.10.6839-6846.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Noureddini SC, Curiel DT. Genetic targeting strategies for adenovirus. Mol Pharm. 2005;2(5):341–347. doi: 10.1021/mp050045c [DOI] [PubMed] [Google Scholar]
- [201].Wickham TJ, Tzeng E, Shears LL 2nd, et al. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J Virol. 1997;71(11):8221–8229. doi: 10.1128/JVI.71.11.8221-8229.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Nakamura T, Sato K, Hamada H. Reduction of natural adenovirus tropism to the liver by both ablation of fiber-coxsackievirus and adenovirus receptor interaction and use of replaceable short fiber. J Virol. 2003;77(4):2512–2521. doi: 10.1128/jvi.77.4.2512-2521.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Hsu HL, Brown A, Loveland AB, et al. Structural characterization of a novel human adeno-associated virus capsid with neurotropic properties. Nat Commun. 2020;11(1):3279. doi: 10.1038/s41467-020-17047-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Brisca G, Mallamaci M, Tardini G, et al. Brisca-CoV-2 infection may present as acute hepatitis in children. Pediatr Infect Dis J. 2021;40(5):e214–e215. doi: 10.1097/INF.0000000000003098 [DOI] [PubMed] [Google Scholar]
- [205].Mameli S, Marcialis MA, Bassareo PP, et al. COVID-19 and hepatic damage: what we know? Panminerva Med. 2023;65(1):58–64. doi: 10.23736/S0031-0808.21.04239-7 [DOI] [PubMed] [Google Scholar]
- [206].Sun J, Aghemo A, Forner A, et al. COVID-19 and liver disease. Liver Int. 2020;40:1278–1281. doi: 10.1111/liv.14470 [DOI] [PubMed] [Google Scholar]
- [207].Wander P, Epstein M, Bernstein D. COVID-19 presenting as acute hepatitis. Am J Gastroenterol. 2020;115(6):941–942. doi: 10.14309/ajg.0000000000000660 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data mentioned and referenced in this review is available and open to use without restriction.


