Abstract
The tumor-associated latent membrane protein 1 (LMP1) gene in the Epstein-Barr virus (EBV) genome is activated by EBV-encoded proteins and cellular factors that are part of general signal transduction pathways. As previously demonstrated, the proximal region of the LMP1 promoter regulatory sequence (LRS) contains a negative cis element with a major role in EBNA2-mediated regulation of LMP1 gene expression in B cells. Here, we show that this silencing activity overlaps with a transcriptional enhancer in an LRS sequence that contains an E-box-homologous motif. Mutation of the putative repressor binding site relieved the repression both in a promoter-proximal context and in a complete LRS context, indicating a functional role of the repressor. Gel retardation assays showed that members of the basic helix-loop-helix transcription factor family, including Max, Mad1, USF, E12, and E47, and the corepressor mSin3A bound to the E-box-containing sequence. The enhancer activity correlated with the binding of USF. Moreover, the activity of the LMP1 promoter in reporter constructs was upregulated by overexpression of USF1 and USF2a, and the transactivation was inhibited by the concurrent expression of Max and Mad1. This suggests that Max-Mad1-mediated anchorage of a multiprotein complex including mSin3A and histone deacetylases to the E-box site constitutes the basis for the repression. Removal of acetyl moieties from histones H3 and H4 should result in a chromatin structure that is inaccessible to transcription factors. Accordingly, inhibition of deacetylase activity with trichostatin A induced expression of the endogenous LMP1 gene in EBV-transformed cells.
Epstein-Barr virus (EBV) is a ubiquitous herpesvirus in humans that is usually apathogenic but is also associated with a number of malignant diseases, including Burkitt’s lymphoma (BL), nasopharyngeal carcinoma, and Hodgkin’s disease (reviewed in reference 51). In healthy infected individuals, the virus persists for life as a latent infection of, probably, the B-lymphoid compartment. In vitro-infected primary B cells are induced to indefinite cell proliferation in which the virus persists in a latent state. These immortalized lymphoblastoid cell lines express six nuclear proteins (EBNA1 to -6), three integral membrane proteins (latent membrane protein 1 [LMP1], LMP2A, and LMP2B), and two small nuclear RNAs (EBER1 and -2) (reviewed in reference 33). Experiments with recombinant viruses have demonstrated that EBNA1, -2, -3, -5, and -6 and LMP1 are important for the growth transformation and immortalization of B lymphocytes (13, 25, 32, 42, 62, 67). LMP1 has a short hydrophilic amino-terminal domain and a long hydrophilic carboxy-terminal domain exposed at the cytosolic side of the cell membrane and six membrane-spanning hydrophobic domains (6, 23, 38). The attachment of LMP1 to the cytoskeleton, localization to patches in the plasma membrane, and rapid turnover correlate with the oncogenic activity of LMP1 and are properties that are reminiscent of activated growth factor receptors (43). It has recently been shown that LMP1, although presumably lacking ligands of its own, transmits signals to the B cells through the TRAF, TRADD/TRAF, and SEK/JNK-1 signalling pathways (for an overview, see reference 22).
The LMP1 gene regulatory sequence (LRS) is composed of both positive and negative transcriptional cis elements, and the gene is inactive in the absence of the inducers. It can be activated by signals reaching the promoter via the cellular protein kinase A (20) and protein kinase C (55) pathways. It is also activated by different EBV-encoded proteins: EBNA1 (24), EBNA6 (1), and EBNA2 acting alone (18, 58, 59) or in concert with EBNA5 (26, 48). The promoter regions of genes induced by EBNA2, including LMP1, generally contain one or several binding sites for the RBP-Jκ transcription factor, and EBNA2 has been shown to associate physically with this protein and block its repressor function (28, 64). Furthermore, it has been suggested that RBP-Jκ-mediated tethering of EBNA2 to the promoter is an essential step in EBNA2-induced transactivation (39, 66, 69). This does not seem to be the case, however, for the LMP1 gene, which retains EBNA2 responsiveness also when the RBP-Jκ site is deleted (18, 58, 59, 66). Reports from several groups, including ours, have implicated a purine-rich sequence (PU box) in the LRS and the PU.1 and Spi-B transcription factors in EBNA2-mediated transactivation of the LMP1 promoter (30, 37, 58, 59). Our results suggest that a POU domain protein, which binds to an octamer motif in LRS, may assist in the targeting of EBNA2 to the LMP1 promoter and that the POU domain protein and the PU box binding proteins cooperate in the transactivation of the promoter by EBNA2 (58). Furthermore, our studies indicate that the promoter-proximal −106 to +40 part of the LRS contains additional regulatory sequences that play an important role in EBNA2 responsiveness (18, 58). An ATF/CRE site in the region was shown to mediate both EBNA2-dependent and EBNA2-independent activation of the LMP1 promoter (60). In the present study we have focused on a sequence immediately upstream of the ATF/CRE site that contains a potential E-box site and which, according to previous results, is involved in silencing of the LMP1 gene. E-box sites bind proteins that belong to the basic helix-loop-helix (bHLH) family of transcription factors, which regulate the expression of differentiated cellular functions in various differentiated cell types (reviewed in reference 40). Protein-protein interactions can occur between different bHLH members, forming homo- or heterodimers, with the latter often being the biologically active species. The members of the large bHLH family have been categorized into higher-order groups, the A, B, and C classes of E-box binding proteins, based on distinct differences in the DNA binding specificities of the factors (15, 49).
The objective of the present study was to define the role of the E-box site in the EBNA2 responsiveness of the LMP1 promoter and to determine its relation to the previously identified negative element in the promoter-proximal LRS region. We demonstrate that the silencer sequence colocalizes with the E-box-homologous motif and that both overlap with an enhancer element. We have also obtained evidence indicating that the LMP1 promoter can be regulated via the recruitment of the mSin3A corepressor and histone deacetylase activity to the promoter.
MATERIALS AND METHODS
Plasmid constructions.
All constructs made were verified by dideoxy sequencing with the Sequenase system (United States Biochemical Corp.). The pSV2gpt, pEΔA6, pgCAT, pgLRS(−106)CAT, and pgLRS(−634)CAT constructs have been described earlier (19, 53). The LRS is defined as nucleotides 169477 to 170151 of B95-8 EBV DNA, which corresponds to positions −634 to +40 relative to the transcription initiation site.
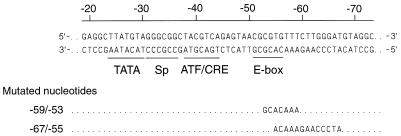
To make a series of 5′ deletion reporter plasmids, PCR amplifications were performed with the pgLRS(−214)CAT plasmid (19) as a template and primers that resulted in fragments with one end corresponding to position +40 in LRS and the other end corresponding to different 5′ positions. The pgLRS(−106)(mut−59/−53) plasmid was constructed by PCR amplification of the pgLRS(−214)CAT plasmid with one primer ending at position −106 which carried transverse mutations at position −59 to −53 in the E box (Fig. 1) and the other primer ending at position +40 in LRS. All of the PCR fragments were subcloned into the TA cloning vector (Invitrogen Corporation). Taking advantage of a synthetic HindIII site in one primer and a PstI site in the TA cloning vector, the PCR fragments were then subcloned between the HindIII and PstI sites in the pgCAT plasmid, resulting in pgLRS(−40)CAT, pgLRS(−50)CAT, pgLRS(−52)CAT, pgLRS(−54)CAT, pgLRS(−55)CAT, pgLRS(−56)CAT, pgLRS(−58)CAT, pgLRS(−63)CAT, pgLRS(−67)CAT, and pgLRS(−106)(mut−59/−53)CAT. The pgLRS(−106)(mut−67/−55)CAT construct was generated by ligation of a HindIII-MluI fragment containing −54/+40 of LRS and a −106/−55 oligonucleotide with MluI-HindIII ends containing transverse mutations in the −67/−55 part of LRS into the HindIII site in the pgCAT vector (Fig. 1). The pgLRS(−634)(mut−67/−55)CAT construct was made by ligating a HindIII-MluI fragment containing −54/+40 of the LRS, an oligonucleotide with MluI-RsaI ends comprising −112/−55 of the LRS carrying transverse mutations in the −67/−55 part of the LRS, and an RsaI-PstI fragment containing the −634/−113 part of the LRS into a HindIII-PstI-digested pgCAT vector (Fig. 1).
FIG. 1.
The promoter-proximal part of the LRS. The double-stranded DNA sequence is of B95-8 EBV DNA origin. The scale refers to the distance in base pairs from the transcription initiation site (+1). Transcription factor binding sites identified in a database search and possibly involved in the regulation of the LMP1 promoter are underlined. Two sets of mutations introduced in the LRS for a functional analysis of the E-box region are indicated below the sequence.
The cDNAs for human USF1 and mouse USF2a were kindly provided by M. Sawadogo, in the pSG5(USF1) and pSG5(USF2a) vectors. The reference plasmid pSG5 was constructed by the removal of USF2a from the pSG5(USF2a) vector by EcoRI and BglII cleavage, followed by blunt-end filling of the 5′ ends and ligation. Mouse USF2a was used because a cDNA for the corresponding full-length human protein was not available at the time. The vectors pSP(Max p21) and pSP(Mad1) were kindly provided by R. N. Eisenman and B. Blackwood. The cDNAs corresponding to Max and Mad1 were isolated by EcoRI digestion of the pSP(Max p21) and pSP(Mad1) vectors and subcloning in the pCI vector (Promega), resulting in the pCI(Max) and pCI(Mad1) vectors, respectively.
Cell culture, DNA transfections, and CAT assays.
DG75 is an EBV genome-negative BL cell line (7). Rael (35), P3HR-1 (27), and Daudi (34) are EBV-positive BL cell lines. The lymphoid cells were maintained as suspension cultures in RPMI 1640 medium (Life Technologies Inc.) supplemented with 10% fetal calf serum (Life Technologies Inc.), penicillin, and streptomycin. The transfections were carried out by electroporation as described previously (60). DG75 cells (5 × 106) were cotransfected with 8 μg of DNA of the reporter construct and, for Fig. 2, with 1.4 pmol of DNA of the EBNA2 expression vector pEΔA6 or 1.4 pmol of DNA of the pSV2gpt vector. For Fig. 3 only pSV2gpt was added. In the pSG5(USF1) and pSG5(USF2a) transfections, 1.2 μg of either expression vector, the corresponding amount of the control vector pSG5, or half of each of the pSG5(USF1) and pSG5(USF2a) expression vectors was utilized. In the pSG5(USF2a)-pCI(Max)-pCI(Mad1) transfections, 1 μg of the pSG5(USF2a) expression vector and either 7.5 μg of pCI(Max) and 7.5 μg of pCI(Mad1) or the corresponding amount of pCI expression vector were used. Cells were harvested after 72 h, and aliquots of the cell lysates were assayed for chloramphenicol acetyltransferase (CAT) activity (52).
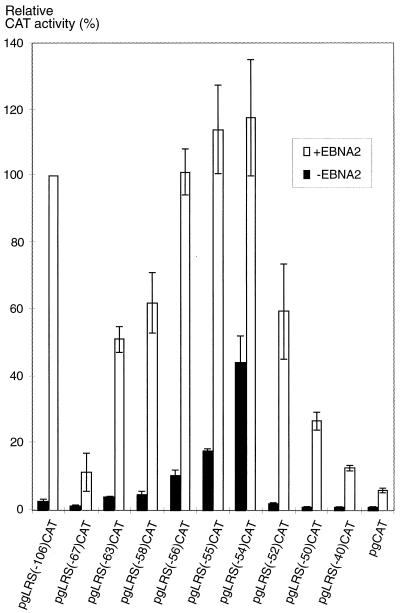
FIG. 2.
Deletion mutation analysis of transcriptional cis elements in the promoter-proximal part of the LRS. Reporter plasmids carrying LRS inserts with 5′ deletions covering the −106 to −40 region (as detailed in Materials and Methods) were transfected together with the EBNA2 expression vector pEΔA6 (+EBNA2) or with an equivalent amount of the empty vector pSV2gpt (−EBNA2) into the EBV-negative B-cell line DG75. The CAT activity is given as relative chloramphenicol acetylation expressed as a percentage of the activity obtained with pgLRS(−106)CAT in the presence of EBNA2. The 100% value corresponded to acetylation of 38% of the substrate in the assay. The values are the means from four independent transfections. Error bars indicate standard errors of the means.
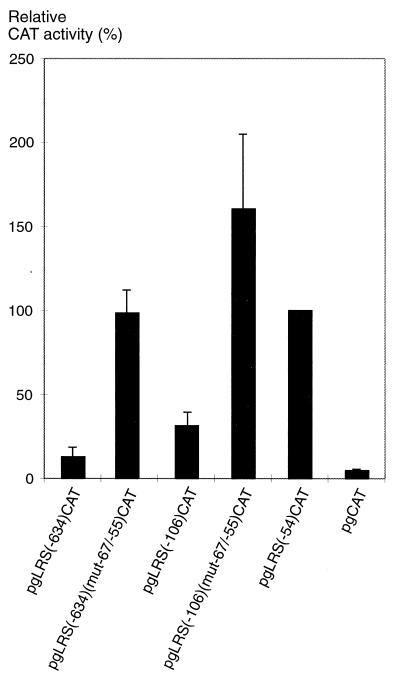
FIG. 3.
A repressor element is present in the −67 to −55 region of the LRS. Mutations in the putative repressor site were introduced in the pgLRS(−106)CAT and pgLRS(−634)CAT plasmids, as indicated in Fig. 1 and in Materials and Methods. The reporter plasmids were transfected into the EBV-negative B-cell line DG75. The CAT activity is given as relative chloramphenicol acetylation expressed as a percentage of the activity obtained with the pgLRS(−54)CAT plasmid. The 100% value corresponded to acetylation of 19% of the substrate in the assay. The values are the means from three independent transfections with double samples. Error bars indicate standard errors of the means.
EMSAs.
Nuclear extracts were prepared as described previously (60). Electrophoretic mobility shift assays (EMSAs) were performed with two double-stranded oligonucleotides corresponding to the −73 to −29 and the −66 to −41 segments of the LRS. The blunt-ended double-stranded oligonucleotides were labelled with [γ-32P]ATP, and the binding reactions were performed as previously described (60). In the competition experiments, a 500-fold (for Fig. 4A) or 150-fold (for Fig. 5A) excess of competing oligonucleotide was added before the 32P-labelled probe. After incubation at room temperature for 20 min, the samples were separated by electrophoresis in 5% polyacrylamide gels (acrylamide-bisacrylamide, 29:1) in 0.5× Tris-borate-EDTA for 2 h at 300 V. The oligonucleotides used in the EMSA experiments are shown in Fig. 4B and 5B.
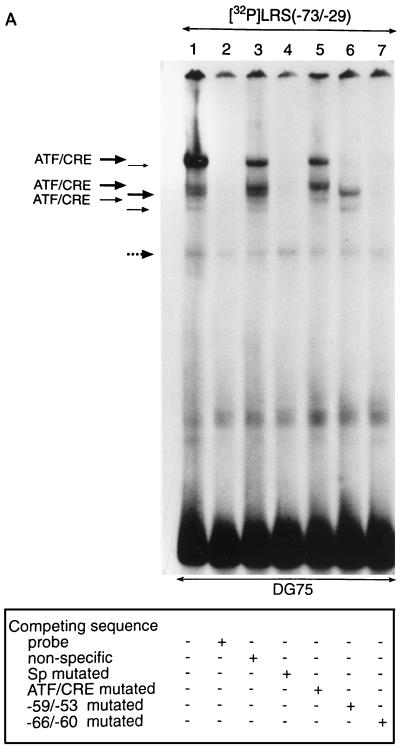
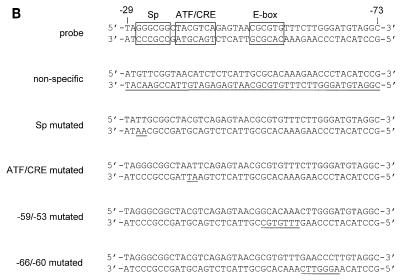
FIG. 4.
Transcription factors in B-lymphoid cells bind to the E-box-containing −59/−53 region of the LRS. (A) A 32P-labelled double-stranded oligonucleotide corresponding to the −73 to −29 LRS region was incubated with nuclear extracts from DG75 cells and subjected to EMSA. Lane 1 shows the binding pattern obtained with the nuclear extract. Competition reactions was carried out as indicated below the autoradiogram and described in Materials and Methods. Six complexes (indicated by solid arrows) are considered specific. Three complexes were shown to interact with the ATF/CRE site in the LRS and are designated ATF/CRE (bands remaining after competition with an LRS fragment that contained a mutated ATF/CRE site). The other three complexes interacted with the −59/−53 sequence in the LRS (bands remaining after competition with an LRS fragment that contained a mutated −59/−53 sequence). One unspecific band that was not abolished by competition with unlabelled probe is indicated by a dotted arrow. (B) Nucleotide sequences of the double-stranded oligonucleotides used in the competition experiment. Binding sites conforming to Sp, ATF/CRE, and E-box consensus sequences are boxed, and mutated nucleotides are underlined.
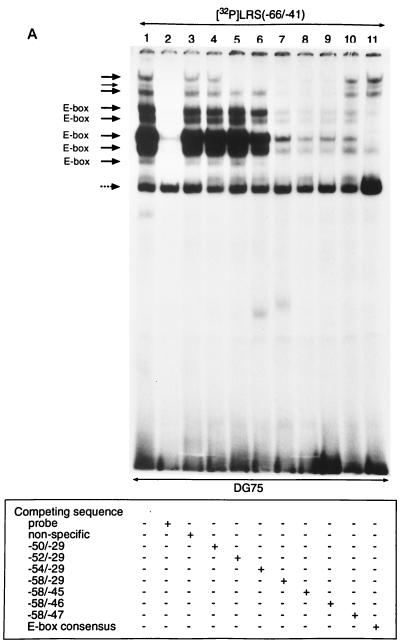
FIG. 5.
Mapping of transcription factor binding sites in the E-box containing region of the LRS. (A) A 32P-labelled double-stranded oligonucleotide corresponding to the −66 to −41 LRS region was incubated with nuclear extracts from DG75 cells and subjected to EMSA. Lane 1 shows the binding pattern obtained with the nuclear extract. Competition reactions was performed as indicated below the autoradiogram and described in Materials and Methods. Eight complexes (designated by solid arrows) were considered specific. The two slowest-migrating complexes required the LRS nucleotides between positions −52 and −46 (bands competed by LRS oligonucleotides containing either the −52/−29 part or the −58/−46 part of the LRS). The remaining six specific complexes interacted with the LRS nucleotides between positions −58 and −46 (bands competed by an LRS oligonucleotide consisting of the −58/−46 region). Five of the specific complexes were competed by an E-box consensus oligonucleotide (designated E-box). One nonspecific band that was not abolished by competition is indicated by a dotted arrow. (B) Nucleotide sequences of the double-stranded oligonucleotides used in the competition experiment. The potential E-box binding site in the LRS and the E-box class B consensus sequence are boxed, while mutated nucleotides are underlined.
The EMSA supershift analysis were performed as previously described (60). The following antibodies were used: anti-USF (sc-229X), anti-c-Myc (sc-42X), anti-Max (sc-197X), anti-Mad1 (sc-222X), anti-mSin3A (sc-767X), anti-mSin3B (sc-768X), anti-E47 (sc-763X), anti-E12 (sc-762X), anti-c-Fos/FosB/Fra-1/Fra-2 (sc-253X), and anti-c-Jun/JunB/JunD (sc-44X) (Santa Cruz Biotechnology).
Inductions and immunoblot analysis.
Induction of the viral lytic cycle in P3HR-1, Rael, and Daudi cells was performed by the addition of 12-O-tetradecanoylphorbol-13-acetate (TPA) to a concentration of 70 ng/ml (for P3HR-1 cells), 5-azacytidine to a concentration of 5 mM (for Rael cells) (44), and n-butyrate to a concentration of 4 mM (for Daudi cells) (41) and incubation for 48 h. Inhibition of deacetylation was carried out by the addition of trichostatin A to a concentration of 100 ng/ml followed by incubation for 24 h (36). The cells were harvested, sonicated in Western sample buffer, and cleared by centrifugation as described previously (60). The samples were boiled, and 10 μl of each extract (corresponding to 500,000 cells) was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% polyacrylamide gel) and blotted onto a Hybond C-extra nitrocellulose membrane (Amersham Life Science). The membranes were blocked in phosphate-buffered saline (PBS) (180 mM NaCl, 3.6 mM KCl, 11 mM Na2HPO4, 2.0 mM KH2PO4) containing 5% milk and 0.1% Tween 20, followed by a wash in PBS containing 0.1% Tween 20. The membranes were incubated for 1 h with the mouse anti-LMP1 antibody CS 1-4 (DAKO A/S) or anti-BZLF1 antibody (DAKO A/S) diluted 1:2,000 in PBS containing 0.2% milk and 0.1% Tween 20, followed by repeated washings in PBS containing 0.1% Tween 20. The membranes were incubated for 1 h with alkaline phosphatase-conjugated goat antimouse antibody (DAKO A/S) diluted 1:3,000 or 1:1,500 in PBS containing 0.2% milk and 0.1% Tween 20, followed by repeated washings in PBS containing 0.1% Tween 20. The proteins were visualized with the Immune-Star chemiluminescent protein detection system as described by the manufacturer of the reagents (Bio-Rad Laboratories).
RESULTS
A transcriptional silencer and an EBNA2-independent enhancer overlap with an E-box site in the LRS.
We have previously shown that a negative regulatory element is localized to the −106/−54 part of the LRS, the silencing activity of which is overridden by EBNA2 via an undefined mechanism (18, 20). A database search for potential transcription factor binding sites in the promoter-proximal part of the LRS revealed the presence of possible Sp, ATF/CRE, and E-box regulatory motifs in the −60 to +40 region, as indicated in Fig. 1. In a recent study, we have presented evidence showing that the ATF/CRE site is important for both the EBNA2-dependent and the EBNA2-independent regulation of the LMP1 promoter and that the Sp site may also contribute to the activation (60). To analyze the role of the E-box-containing region in the regulation of LMP1 promoter activity, we have now generated a series of CAT reporter plasmids containing 5′ deletion mutations of the LRS from position −106 to −40 (Fig. 2) and have introduced them into the EBV-negative B-cell line DG75 together with an EBNA2 expression vector or a control vector. The results indicated that the reporter plasmids could operationally be divided into three categories according to the pattern of CAT expression and the length of the LRS insert. The first group of plasmids, which contained short LRS fragments from position +40 up to and including −52, were inactive in the absence but activated in the presence of EBNA2, with the maximal response obtained with the pgLRS(−52)CAT plasmid. The level of activation corresponded to an inducibility (defined as the ratio between CAT activities induced by the reporter in the presence and in the absence of EBNA2) of about 30. The second category of reporter plasmids had inserts of intermediate length that contained the additional LRS sequences from position −54 to −67. These plasmids were active to various degrees both in the presence and in the absence of EBNA2. Maximal activity was obtained with the pgLRS(−54)CAT plasmid, and the activity then gradually decreased to close to baseline levels in constructs in which 1, 2, 4, 9, and 13 bp was added to the upstream end of the LRS insert. The plasmids assigned to this group could be activated by EBNA2, but the inducibility was significantly reduced compared with that for the plasmids in category 1. The third category of mutants is represented by the pgLRS(−106)CAT plasmid. This construct lacked significant EBNA2-independent activity but was highly responsive to activation by EBNA2, with an inducibility similar to that of plasmids in category 1. We suggest the following interpretation of the results. The properties of the reporter plasmids in category 1 are due to the effect of the stepwise inclusion of an EBNA2-dependent positive regulatory element. As demonstrated in a previous report (60), this element is an ATF/CRE site, and the activating effect is mediated by an ATF-2–c-Jun heterodimer. In plasmids belonging to category 2, an EBNA2-independent positive element and a negative element are included in the constructs, with the positive effect dominating in the shorter [pgLRS(−54)CAT] and the negative effect dominating in the longer LRS inserts. The pgLRS(−67)CAT plasmid represents the situation where the putative repressor has almost completely silenced both the EBNA2-dependent and the EBNA2-independent activities of the plasmid. The addition of the LRS sequence between positions −67 and −106 [pgLRS(−106)CAT] resulted in the reconstitution of EBNA2 responsiveness of the reporter plasmid without adding to the absolute level of activation compared with pgLRS(−50)CAT. This suggests to us that the −67/−106 region contains elements that participate in the EBNA2-induced alleviation of the repressor effect on the LMP1 promoter but that may not be conventional enhancer elements. Taken together, our results show that a repressor element and an EBNA2-independent enhancer element overlap with an E-box-homologous motif at position −56 to −51 in the LRS.
In order to confirm the presence of the negative element and to assess the importance of this element in a complete LRS context, the sequence between positions −67 and −55 in the LRS was mutated in pgLRS(−106)CAT and pgLRS(−634)CAT and the resulting reporter constructs were subjected to the transfection assay in DG75 cells (Fig. 3). Mutation of the −67/−55 region relieved the repression of the LRS in both constructs to a level of activity corresponding to that of pgLRS(−54)CAT. Thus, our results demonstrate that the negative element present in the −67 to −55 part of the LRS plays a functionally important role in the regulation of LMP1 promoter activity. Other negative elements, previously shown to be present in upstream regions of the LRS (19, 58, 59), cannot substitute for the −67/−55 element in silencing the EBNA2-independent enhancer element at position −54 of the LRS.
Members of the bHLH family of transcription factors interact with the E-box motif in the LRS.
To correlate the activity data from the mutational analysis with the potential binding of transcription factors, we performed EMSAs with nuclear extracts of DG75 cells and an oligonucleotide probe corresponding to the −73/−29 part of LRS (Fig. 4). Sequences involved in the protein-DNA interactions were defined further by competition experiments with unlabelled oligonucleotides. Six specific complexes were recognized (Fig. 4A, lanes 2 and 3). A seventh band not removed by competition with unlabelled probe was assumed to represent nonspecific complex formation. An LRS competitor oligonucleotide with a mutated Sp site removed all specific bands, showing that the Sp site in the probe was not involved in complex formation (Fig. 4A, lane 4). Competition with an LRS oligonucleotide with a mutated ATF/CRE site removed three of the specific bands; the remaining three were assumed to represent binding to the ATF/CRE site (Fig. 4A, lane 5). Competition with an LRS oligonucleotide with a mutation involving the −59/−53 region removed three bands, and the remaining three bands were assumed to represent binding to the mutated region (Fig. 4A, lane 6). Finally, competition with an LRS oligonucleotide with a mutated −66/−60 sequence removed all specific bands, indicating that this region was not involved in the formation of any of the complexes identified in our EMSA (Fig. 4A, lane 7). This might seem inconsistent with the results of the deletion mutation analysis described above (Fig. 2), which indicated that the −67/−60 region was part of a negative cis element. We suggest, however, that this apparent discrepancy is due to quantitative rather than qualitative reasons in the sense that the putative repressor can bind, albeit with a lower affinity, to the LRS probe even if the −66/−60 sequence is mutated. In conclusion, the ATF/CRE motif and the −59/−53 sequence seem to be the major protein binding sites in the −73/−29 part of the LRS.
To characterize the pattern of transcription factor binding to the E-box-containing region in further detail, we performed EMSAs with DG75 nuclear extracts and an oligonucleotide probe corresponding to positions −66 to −41 of the LRS (Fig. 5). Competition experiments with unlabelled oligonucleotides identified eight specific complexes (Fig. 5A, lanes 2 and 3). One band that was not abolished by competition with unlabelled probe was assumed to represent nonspecific complex formation. A similar factor binding pattern was observed with both EBV-negative and EBV-positive B cells and with epithelial cells and T cells (data not shown). To define the 5′ ends of the protein binding sites, a set of competitor oligonucleotides that contained the −58/−29, −56/−29, −54/−29, −52/−29, and −50/−29 sequences of LRS, respectively, in a mutated context was used (Fig. 5B). Competition with the −50/−29 region did not remove any of the bands, while the −52/−29 region competed for the two slowest-migrating bands (Fig. 5A, lanes 4 and 5). The six other bands were partly removed by competition with the −54/−29 region and completely removed by competition with the −58/−29 region (Fig. 5A, lanes 6 and 7). The 3′ ends of the factor binding sites were characterized in an analogous manner by using a set of competitor oligonucleotides that contained the −58/−45, −58/−46, or −58/−47 sequences of the LRS in a mutated context (Fig. 5B). Competition with the −58/−45 and −58/−46 sequences removed all of the specific bands, while the −58/−47 region was a less efficient competitor (Fig. 5A, lanes 8, 9, and 10). The results thus indicated that two sets of factor binding sites were present in this region of the LRS, one centered around nucleotides −58/−46 and the other centered around positions −52/−46. This notion was strengthened by competition experiments with an oligonucleotide that contained an E-box class B consensus motif (Fig. 5A, lane 11), which has a five-of-six nucleotide sequence identity with the E-box site in the LRS. The competitor removed five bands (marked E-box in Fig. 5A) but left three bands largely unaffected, indicating that members of several subfamilies of the bHLH group are involved in complex formation.
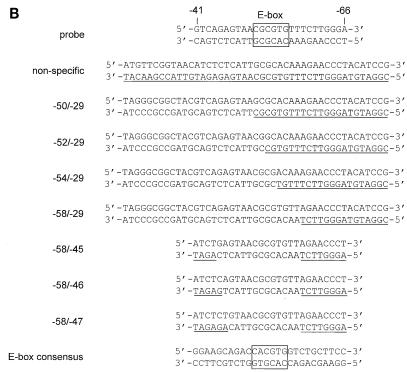
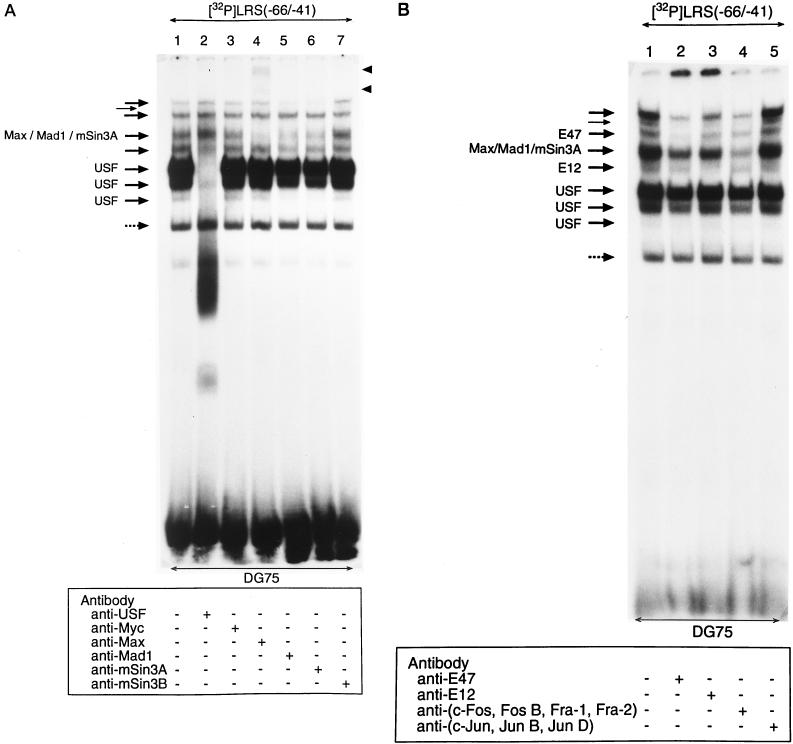
To identify the factors binding to the E-box region in the LRS, antibody supershift analysis was performed with the EMSA −66/−41 probe and a panel of commercially available antibodies against transcription factors that could conceivably be involved in this type of interaction (Fig. 6). Three of the eight bands of the EMSA pattern were abolished with an anti-USF antibody (Fig. 6A, lane 2), and one band was diminished with either of three different antibodies: anti-Max antibody (lane 4), anti-Mad1 antibody (lane 5), or anti-mSin3A antibody (lane 6). Of the remaining four unidentified EMSA bands, two were removed by anti-E47 or anti-E12 antibodies, respectively (Fig. 6B, lanes 2 and 3). The band shifted by the anti-E12 antibody was competed out by the E-box class B consensus oligonucleotide (Fig. 5A, lane 11), although E12 and E47 conventionally are classified as class A proteins. The complex removed by the anti-E47 antibody was not competed out by the same oligonucleotide (Fig. 5A, lane 11). In summary, our results demonstrated that the USF, Max, Mad1, mSin3A, E12, and E47 transcription factors are present in DG75 cells and interact with the E-box motif-containing sequence in the promoter-proximal part of the LRS.
FIG. 6.
Identification of the transcription factors interacting with the E-box site in the LRS. Nuclear extract from DG75 cells was incubated under binding conditions with a 32P-labelled double-stranded oligonucleotide corresponding to the −66 to −41 LRS region. Antibody supershifts were carried out by incubation with antibodies as indicated below the autoradiograms. The reaction mixtures were analyzed by EMSA. One nonspecific band that was not abolished by competition with unlabelled probe is indicated by a dotted arrow. (A) Eight specific complexes are indicated by solid arrows; three are designated USF and one is designated Max/Mad1/mSin3A, since it contains these three factors. The positions of the immunologically shifted complexes are shown by the solid arrowheads for the anti-Max shifts. (B) Eight specific complexes are indicated by solid arrows: three designated USF, one designated E12, one designated Max/Mad1/mSin3A, and one designated E47. Two complexes are not designated due to the fact that the protein components were not identified. It should be noted that the addition of anti-E12 and anti-E47 antibodies to the reaction mixtures shifted the respective protein complexes to the top of the gel.
USF-mediated activation of the LMP1 promoter is inhibited by the Max-Mad1 repressor.
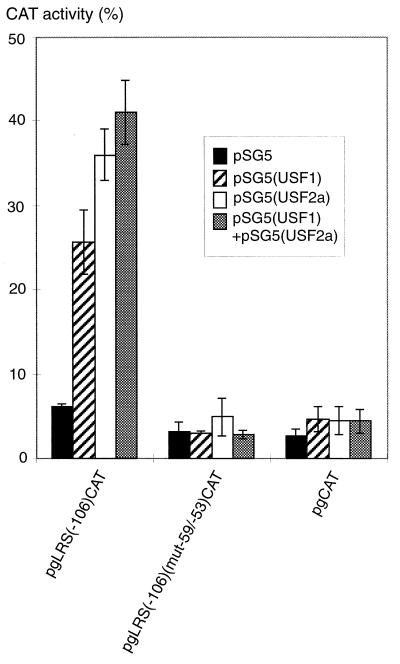
EMSA experiments using a −54/−41 fragment of the LRS resulted in the predominant formation of three major bands, all of which were recognized by the anti-USF antibody (data not shown). In the light of the previous observation that a positive element is included with the 5′ addition of nucleotide −54 in the deletion mutation analysis of the LRS (Fig. 2), it seems reasonable to assume that USF constitutes the corresponding EBNA2-independent transactivating factor. The occurrence of several EMSA bands fits with the fact that the different forms of USF are ubiquitously expressed and bind as homo- and heterodimers to an E-box site. To assess whether USF transcription factors can activate the LMP1 promoter in an EBNA2-independent manner, reporter vectors containing the −106/+40 LRS region with or without mutation of the −59/−53 region were cotransfected with expression vectors for human USF1 and/or mouse USF2a into DG75 cells. The −59/−53 mutation of the E box was tested in EMSA experiments, which showed that the binding of all of the E-box binding proteins was abolished (data not shown). Transfections of either USF1 or USF2a, resulting in the dominant generation of homodimeric forms, transactivated the LMP1 promoter (Fig. 7). When half of the amount of each vector was transfected together, favoring the formation of heterodimeric forms (63), the same level of induction was obtained as with the USF2a vector only.
FIG. 7.
USF1 and USF2a transactivate the LMP1 promoter independently of EBNA2. The pSG5(USF1) and pSG5(USF2a) expression vectors, separately or mixed, or the pSG5 control vector was cotransfected with the reporter plasmid pgLRS(−106)CAT or pgLRS(−106)(mut−59/−53)CAT or the pgCAT control plasmid into DG75 cells, as detailed in Materials and Methods. The CAT activity is given as percent chloramphenicol acetylation. The values shown are the means from three independent transfections. Error bars indicate standard errors of the means.
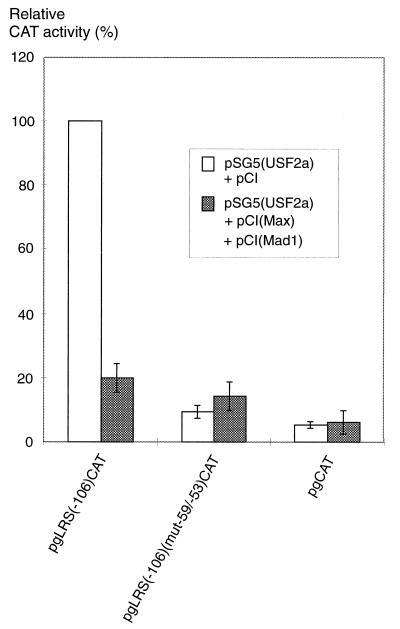
Since the enhancer activity in the LRS was shown to be localized very close to a repressor element, the repressor might be identical to the putative ternary complex Max-Mad1-mSin3A observed in our EMSA experiments. Transfection of the reporter plasmid carrying the promoter-proximal −106/+40 LRS region with or without a mutated E box together with expression vectors for USF2a, Max, and Mad1 into DG75 cells showed that Max-Mad1 repressed the activity of the LMP1 promoter in an E-box-dependent manner (Fig. 8). Cotransfection with the mSin3A expression vector was not necessary because of the abundance of this protein in the cells (5). Protein levels in the transfected cells were analyzed with immunoblotting analysis (data not shown). Cotransfection with the corresponding expression vectors increased the levels of Max, Mad1, and USF in the cells severalfold from a basal level, ruling out the possibility that Max-Mad1 downregulated the expression of USF. Taken together, the results suggested that USF proteins confer EBNA2-independent activity to the LMP1 promoter via the E-box region and that this activation can be downregulated by the Max-Mad1-mSin3A factors.
FIG. 8.
USF2a-mediated transactivation of the LMP1 promoter is repressed by the Max-Mad1 heterodimer. The pSG5(USF2a) expression vector was cotransfected with the pCI(Max) and pCI(Mad1) expression vectors or an equivalent amount of the pCI control vector and with the reporter plasmid pgLRS(−106)CAT or pgLRS(−106)(mut−59/−53)CAT or the pgCAT control plasmid into DG75 cells, as detailed in Materials and Methods. Cotransfection with the mSin3A expression vector was not performed because the cells express this protein constitutively at a high level. The CAT activity is given as relative chloramphenicol acetylation expressed as a percentage of the activity obtained with the pgLRS(−106)CAT plasmid in the presence of the pSG5(USF2a) expression vector. The 100% value corresponded to acetylation of 17% of the substrate in the assay. The values are the means from three independent transfections. Error bars indicate standard errors of the means.
Expression of the LMP1 gene is upregulated by inhibition of deacetylation.
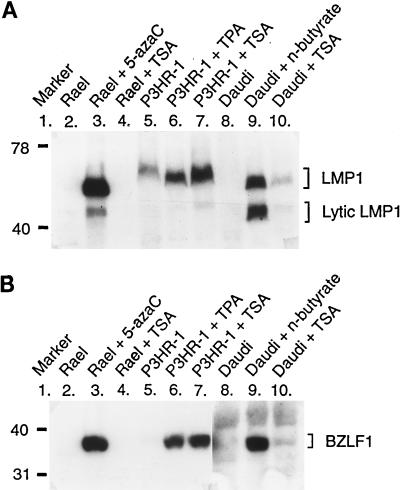
The experiments described above strongly suggested the involvement of the Max-Mad1-mSin3A complex in the regulation of the LMP1 gene. It has been postulated that Max-Mad1-mSin3A functions as a repressor by recruiting deacetylases to the promoter, thereby lowering the level of acetylated histones in the surrounding chromatin and creating a more compact chromatin structure. To analyze whether the expression of LMP1 from the endogenous EBV genome was affected by an increase of the level of histone acetylation, three EBV-positive cell lines, Rael, P3HR-1, and Daudi, were treated with the deacetylase inhibitor trichostatin A. The expression of LMP1 in the cells was monitored by immunoblotting (Fig. 9A). Under normal conditions, these cell lines express LMP1 only at very low levels or not at all, and they do not express EBNA2. The effect of the addition of trichostatin A to the culture medium varied between the analyzed cell lines. Trichostatin A did not induce LMP1 expression in Rael cells, whereas both full-length LMP1 and the truncated variant of this protein found in lytic infection were expressed in P3HR-1 and Daudi cells. To determine whether inhibition of deacetylation induced the lytic cycle in P3HR-1 and Daudi cells, the expression of BZLF1 in the trichostatin A-treated cells was analyzed by immunoblotting (Fig. 9B). BZLF1 is an immediate-early EBV protein expressed in the lytic cell cycle. The results revealed the appearance of significant levels of BZLF1 in P3HR-1 and Daudi cells, indicating that the lytic cycle was induced in these cells but not in Rael cells. Thus, the results are compatible with the hypothesis that core histone acetylation, presumably with secondary effects on chromatin structure, plays a role in the relief of LMP1 gene repression in the endogenous EBV genome, in addition to inducing the lytic cell cycle in transformed B cells. Obviously, other regulatory mechanisms also exist, as in Rael cells, with the power to override these effects.
FIG. 9.
Treatment with the deacetylase inhibitor trichostatin A upregulates the expression of the LMP1 gene and induces the lytic cycle in some EBV-transformed B-cell lines. Trichostatin A (TSA) was added to the culture media of three EBV-positive cell lines, Rael, P3HR-1, and Daudi, and the expression of the LMP1 and BZLF1 proteins was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting. For control purposes, the expression of LMP1 and BZLF1 was induced in parallel cultures by using 5-azacytidine (5-azaC), TPA, or n-butyrate, depending on the cell line, as indicated above the lanes and described in Materials and Methods. (A) The mouse anti-LMP1 antibody CS 1-4 was used. The positions of the full-length LMP1 and the truncated form found in lytically infected cells are indicated on the right. The sizes of the LMP1 proteins differ between the cell lines due to varying numbers of a specific repeat in the proteins. (B) The mouse anti-BZLF1 antibody was used. It should be noted that the BZLF1 protein was expressed at low levels in uninduced P3HR-1 cells. This cell line is known to contain lytic cell subpopulations. A longer exposure of the autoradiogram for the Daudi cell extracts was required in order to detect BZLF1 protein expression. Numbers on the left are molecular masses in kilodaltons.
DISCUSSION
In previous reports we have presented evidence demonstrating that the proximal region of the LMP1 promoter contains a negative cis element with a major role in EBNA2-mediated regulation of LMP1 gene expression in B-lymphoid cells. Here, we show that this silencing activity overlaps with a transcriptional enhancer and is localized in a sequence that contains an E-box-homologous motif. Mutation of the putative repressor binding site relieved the repression both in a promoter-proximal and a complete LRS context, indicating a functional role of the repressor in LMP1 gene regulation. A number of proteins belonging to the bHLH family of transcription factors, including Max, Mad1, USF, E12, and E47, and the transcriptional corepressor mSin3A bound in a sequence-specific manner to the E-box-containing sequence. The activity of the LMP1 promoter in reporter constructs was downregulated by the concurrent expression of Max, Mad1, and mSin3A, consistent with the notion that a ternary complex between these factors constitutes the previously postulated repressor. Moreover, the promoter was upregulated in an EBNA2-independent manner by USF, and the activation correlated with the binding of USF proteins to the E-box site. Interestingly, inhibition of deacetylase activity with trichostatin A induced expression of the endogenous LMP1 gene in EBV-transformed cells, suggesting that the LMP1 promoter can be regulated via Max-Mad1-mSin3A-mediated recruitment of deacetylases to the promoter, leading to core histone deacetylation and modulation of chromatin structure.
Our DNA binding studies revealed the formation of a number of specific complexes with the E-box-containing region of the LRS. The majority of the EMSA bands contained protein components identified with specific antibodies and shown to belong to the bHLH family of transcription factors. Two of the complexes, however, remained unidentified. The binding sites for the latter proteins seemed to be somewhat displaced towards the transcription initiation site relative to the bHLH factor binding site and did not completely encompass the E-box motif (see the results of the competition experiments in Fig. 5A). Judging from the competition experiments, the nucleotides in the −52 to −46 sequence were essential for binding (Fig. 5A). Thus, the observation that the inducibility of pgLRS(−52)CAT and pgLRS(−50)CAT by EBNA2 was largely the same in spite of the fact that nucleotides −52 and −51 are important for binding showed that the unidentified proteins are not involved in the E-box-independent, EBNA2-mediated transactivation of the LRS. We cannot exclude, however, the possibility that the unidentified proteins may play a role in the E-box-mediated regulation of the LMP1 promoter. No obvious candidates emerged in a database search for potential transcription factor binding sites corresponding to the −52 to −46 sequence in the LRS. On the other hand, all of the other factors that formed complexes with the E-box-containing sequence were identified as known members of the bHLH family. Nucleotides −58 to −46 of the LRS were required for the binding. Thus, nucleotides in the E-box-flanking regions were involved in the interaction with the factors, in accordance with previous investigations of E-box-containing promoters (61). The members of the bHLH transcription factor family have been divided into two classes depending on the sequence of the canonical bHLH binding site CANNTG. Class A proteins, which include AP-4, E-12, E-47, E2-2, and others, bind to the sequence CACCTG or CAGCTG. Class B proteins, which include c-Myc, Max, MyoD, myogenin, and USF, bind to CACGTG or CATGTG. Class A proteins do not bind to class B sites and vice versa. In addition, some proline-containing bHLH repressor proteins, although recognizing the class B canonical sites, have been shown to prefer the noncanonical CACGCG and CACGAG sites (15, 49). This group of proteins, which has only a few members, including the Drosophila hairy factor, has been designated class C bHLH factors. Recently, a new class, class D, has been defined, the members of which lack the basic region (3). It should be noted that the E-box site in the LRS, CACGCG, is a noncanonical class C sequence, although the proteins which in the present study have been found to bind to this site belong to class A and class B. However, experiments employing the strategy of sequential selection and amplification of oligonucleotides have demonstrated that at least some class B factors, including c-Myc, Max, and USF, can bind to the CACGCG sequence, albeit with a lower affinity than to a class B site (8, 49). The class A proteins E12 and E47 also bound to the E-box-containing sequence in the LRS, although they are regarded as class A factors. It is, however, well established that each of the two subunits in a heterodimeric bHLH protein recognizes different parts of the asymetric CANNTG palindromic sequence (9). Conceivably, the E2A factors bind to the LRS as a part of a heterodimeric complex in which the partner is a bHLH protein recognizing the other half of the E-box sequence, i.e., the GCG part of the CACGCG motif. The functional role of these factors in the LMP1 gene context remains to be established.
The USF proteins represent the larger part of the LRS E-box DNA binding activity in the B cells investigated in the present study. Interestingly, this group of transcription factors, while being ubiquitously expressed, is involved in the expression of several tissue-specific or developmentally regulated genes (40). The factors are encoded by two distinct genes (the USF1 and USF2 genes) and exist in the form of homomeric and heteromeric dimers able to bind to specific E-box sites. In vivo, four combinations of the different USF proteins are prevalent, with the most common species being heterodimers between the USF1 and USF2a isoforms (63). In the present study it is shown that the E-box site in the LRS is a transcriptional enhancer of the LMP1 promoter and that transactivation of the promoter is mediated by the USF proteins in an EBNA2-independent way. We have so far not identified the specific members of the USF factor family that interact with the E-box motif in the LRS. However, the quantitative dominance of the most slow-moving USF complex in the EMSA suggests that it corresponds to the USF1-USF2a heterodimer. Transfections under conditions that favor the formation of the homomeric or heteromeric forms of USF1 and USF2a suggested that all dimer combinations were equally effective in the transactivation of the LMP1 promoter.
Our EMSA supershift analysis indicated that a complex consisting of the Max and Mad1 factors in association with the mSin3A protein interacts with the E-box sequence. The Max protein is thought to play an essential role in the function of this biologically important group of transcription factors by being a partner in complex formation with Myc or Mad1 to -4 or with itself (4, 10, 11, 29, 68). Dimerization with Max is necessary for these proteins to be able to bind to DNA and exert their effects on transcription. Myc-Max heterodimers, regarded as the biologically active form of Myc, transactivate genes involved in cell proliferation and apoptosis which contain the specific E-box sequence. Max itself is thought to be transcriptionally inert (31). Myc-Max heterodimers are favored over homodimers when the two proteins are at equilibrium, since both the Myc and Max proteins preferentially heterodimerize. The Max-Mad dimeric molecules are repressors of Myc-Max-mediated transcriptional activation through competition for the same E-box site (reviewed in reference 2). Max-Mad forms ternary complexes in solution with mSin3A and mSin3B that recognize the E-box site (5). It has been postulated that transcriptional repression by the Mad-Max-mSin3 complex involves deacetylation of core histones via recruitment of deacetylases to the promoter region (50). Deacetylation will increase the net positive charge of the histone proteins, resulting in a higher affinity for the DNA and a more compact chromatin structure. Hence, transcriptional cis elements will become less accessible for transcription factors and components of the basal transcriptional machinery. It is well established that the EBV genome is packaged into a nucleosomal structure in the cell (16, 56). The finding that Max-Mad1-mSin3A bound to the promoter region and that the binding was associated with a repression of promoter activity in reporter plasmids therefore suggested that protein deacetylation plays an important role in the regulation of LMP1 gene expression. Our observation that treatment of the cells with the deacetylase inhibitor trichostatin A induced LMP1 expression in Daudi and P3HR-1 cells is consistent with this hypothesis. The difference between Rael and the other cell lines regarding the sensitivity to trichostatin A might be due to differences in the methylation pattern of the LMP1 promoter region. It has been shown by transfection of in vitro-methylated LRS reporter plasmids into Raji cells that the activity of the promoter is downregulated by sequence-specific methylation (45). It is also known that the LMP1 promoter is only partially methylated in Daudi cells but is fully methylated in Rael cells (17, 21, 46). It was recently shown that the methyl-CpG binding protein MeCP2 associates with a corepressor complex containing mSin3A and histone deacetylases (47). Transcriptional repression was relieved by trichostatin A, indicating that deacetylation of histones is an essential component of this type of methylation-mediated repression. It should be noted, however, that repression by MeCP2 was not completely alleviated by trichostatin A, suggesting that part of the repression was deacetylase independent. Furthermore, our experiments showed that demethylation of the heavily methylated endogenous EBV genome in Rael cells by 5-azacytidine induced the expression of LMP1, while treatment with trichostatin A had no measurable activating effect on the gene (Fig. 9A). Taken together, the data suggest that transcriptional repression by methylation can be attained through several mechanisms, at least one of which does not involve the recruitment of the mSin3A-deacetylase corepressor complex to the promoter region. A possibility which still cannot be ruled out is that methylation at a specific CpG site in certain promoters blocks transcription by interfering with the binding of a transcription factor even under the conditions of inhibition of deacetylation.
Overexpression of Max and Mad1 in EBV-negative DG75 lymphoid cells repressed the USF2a-mediated transactivation of the LMP1 promoter in reporter plasmids. Cotransfection of Myc and Max expression vectors in the same cell line did not reveal any stimulatory effect on the promoter by this factor combination (unpublished data). We were also unable to demonstrate binding of Myc to the LRS E-box site by supershift experiments with specific antibodies (Fig. 6A, lane 3). Thus, we conclude that the well-known mechanism for the repressor function of Max-Mad, i.e., a competition between the transactivating Myc-Max and the repressive Max-Mad complex for a specific E-box site, is not valid for the LMP1 promoter. Instead, the enhancement of the LMP1 promoter activity is mediated by several factors, including ATF-1/CREB-1, ATF-2/c-Jun, USF, and possibly other factors binding further upstream, and this activation is counteracted by the binding of the Max-Mad1-mSin3A repressor. LMP1 gene silencing might then occur via the recruitment of a deacetylase to the promoter-proximal region and a modulation of chromatin structure.
It is known that induction of demethylation by 5-azacytidine or activation of the protein kinase C signalling pathway by TPA triggers activation of the lytic cell cycle and expression of LMP1 in EBV-transformed cells (12, 14, 54). The induction of expression of the truncated LMP1 variant in the Daudi and P3HR-1 cell lines by trichostatin A suggested that the lytic cycle might have been activated in these cells. This was confirmed by the observation that expression of the BZLF1 protein occurred concomitantly with the induction of LMP1 by trichostatin A in the cells. Furthermore, previous investigations involving n-butyrate treatment of EBV-transformed cells, which also inhibits deacetylation, have demonstrated that the lytic cycle and expression of LMP1 are induced by this substance (14). This raises the question whether trichostatin A-induced expression of LMP1 is a direct effect on core histones in the LMP1 promoter region or a phenomenon secondary to a general induction of the lytic cycle. Speaking against the latter interpretation is the observation that treatment with trichostatin A activated the LMP1 promoter in reporter plasmids transfected into DG75 cells (data not shown).
We have previously shown that one important element of EBNA2-induced transactivation of the LMP1 promoter is the overriding of the effect of a negative element in the promoter-proximal region, but the mechanism for this action was not clarified (18, 58). The identification of Max-Mad1-mSin3A as the likely repressor and the assumption that repression occurs via deacetylation open up a number of possible options for EBNA2-induced reversal of the repression. In one model, the balance between the binding of the Max-Mad1-mSin3A complex and USF to the LRS E box is influenced by EBNA2 in favor of USF. This could be achieved via several conceivable mechanisms. In this way the recruitment of deacetylases to the promoter would be impeded. However, DNA binding studies of proteins in EBV-negative and EBV-positive cells reveal factor binding patterns in the E-box region that are indistinguishable from each other, which would be an argument against this hypothesis. In a second model, EBNA2 abolishes the repressive effect of Max-Mad1-mSin3A by affecting histone acetylation in a more direct manner. Several transcription factors, including Gcn5, CBP/p300, and TAFII250, have been found to possess histone acetyltransferase activity (57). EBNA2 might have the same catalytic activity or in some indirect way be able to recruit histone acetyltransferase activity to the LMP1 promoter. Under the assumption that EBNA2 confers acetyltransferase activity, one might also postulate that acetylation of nonhistone proteins, such as high-mobility-group proteins or transcription factors, is important for transcriptional regulation and contributes to the transcriptional effects of EBNA2. Another possible way for EBNA2 to counteract deacetylation and overcome Max-Mad repression would be to recruit the SNF-SWI complex to the promoter. The SNF-SWI complex removes surrounding histones by an ATP-dependent mechanism, creating a chromatin structure that is more accessible for protein interactions and thereby for induction of transcription. It has, in fact, been shown that EBNA2 can interact with the hSNF5/Ini1 component of the SNF-SWI complex (65).
ACKNOWLEDGMENTS
We gratefully acknowledge Carina Ström and Jane Löfvenmark for skillful technical assistance. We thank M. Sawadogo for the pSG5(USF1) and pSG5(USF2a) plasmids and R. N. Eisenman and B. Blackwood for the pSP(Max) and pSP(Mad1) plasmids.
This study was supported by grants from the Swedish Medical Research Council, the Swedish Cancer Society, and the Sahlgrenska University Hospital.
REFERENCES
- 1.Allday M J, Crawford D H, Thomas J A. Epstein-Barr virus (EBV) nuclear antigen 6 induces expression of the EBV latent membrane protein and an activated phenotype in Raji cells. J Gen Virol. 1993;74:361–369. doi: 10.1099/0022-1317-74-3-361. [DOI] [PubMed] [Google Scholar]
- 2.Amati B, Land H. Myc-Max-Mad: a transcriptional factor network controlling cell cycle progression, differentiation and death. Curr Biol. 1994;4:102–108. doi: 10.1016/0959-437x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 3.Atchley W R, Fitch W M. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayer D E, Kretzner L, Eisenman R N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 5.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 6.Bankier A T, Deininger P L, Satchwell S C, Baer R, Farrell P J, Barrell B G. DNA sequence analysis of the EcoRI Dhet fragment of B95-8 Epstein-Barr virus containing the terminal repeat sequences. Mol Biol Med. 1983;1:425–445. [PubMed] [Google Scholar]
- 7.Ben-Bassat H, Goldblum N, Mitrani S, Goldblum T, Yoffey J M, Cohen M M, Bentwich Z, Ramot B, Klein E, Klein G. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt-like” malignant lymphoma (line D.G.-75) Int J Cancer. 1977;19:27–33. doi: 10.1002/ijc.2910190105. [DOI] [PubMed] [Google Scholar]
- 8.Blackwell T K, Huang J, Ma A, Kretzner L, Alt F W, Eisenman R N, Weintraub H. Binding of Myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackwell T K, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1109. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 10.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 11.Blackwood E M, Lüscher B, Eisenman R N. Myc and Max associate in vivo. Genes Dev. 1992;6:71–80. doi: 10.1101/gad.6.1.71. [DOI] [PubMed] [Google Scholar]
- 12.Boos H, Berger R, Kuklik-Roos C, Iftner T, Mueller-Lantzsch N. Enhancement of Epstein-Barr virus membrane protein (LMP) expression by serum, TPA, or n-butyrate in latently infected Raji cells. Virology. 1987;159:161–165. doi: 10.1016/0042-6822(87)90360-6. [DOI] [PubMed] [Google Scholar]
- 13.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Contreras-Salazar B, Ehlin-Henriksson B, Klein G, Masucci M G. Up regulation of the Epstein-Barr virus (EBV)-encoded membrane protein LMP in the Burkitt’s lymphoma line Daudi after exposure to n-butyrate and after EBV superinfection. J Virol. 1990;64:5441–5447. doi: 10.1128/jvi.64.11.5441-5447.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dang C V, Dolde C, Gillison M L, Kato G J. Discrimination between related DNA sites by a single amino acid residue of Myc-related basic-helix-loop-helix proteins. Proc Natl Acad Sci USA. 1992;89:599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyson P, Farrell P J. Chromatin structure of Epstein-Barr virus. J Gen Virol. 1985;66:1931–1940. doi: 10.1099/0022-1317-66-9-1931. [DOI] [PubMed] [Google Scholar]
- 17.Ernberg I, Falk K, Minarovits J, Busson P, Tursz T, Masucci M G, Klein G. The role of methylation in the phenotype-dependent modulation of Epstein-Barr nuclear antigen 2 and latent membrane protein genes in cells latently infected with Epstein-Barr virus. J Gen Virol. 1989;70:2989–3002. doi: 10.1099/0022-1317-70-11-2989. [DOI] [PubMed] [Google Scholar]
- 18.Fåhraeus R, Jansson A, Ricksten A, Sjöblom A, Rymo L. Epstein-Barr virus-encoded nuclear antigen 2 activates the viral latent membrane protein promoter by modulating the activity of a negative regulatory element. Proc Natl Acad Sci USA. 1990;87:7390–7394. doi: 10.1073/pnas.87.19.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fåhraeus R, Jansson A, Sjöblom A, Nilsson T, Klein G, Rymo L. Cell phenotype dependent control of Epstein-Barr virus latent membrane protein 1 (LMP1) gene regulatory sequences. Virology. 1993;195:71–80. doi: 10.1006/viro.1993.1347. [DOI] [PubMed] [Google Scholar]
- 20.Fåhraeus R, Palmqvist L, Nerstedt A, Farzad S, Rymo L, Laín S. Response to cAMP levels of the Epstein-Barr virus EBNA2-inducible LMP1 oncogene and EBNA2 inhibition of a PP1-like activity. EMBO J. 1994;13:6041–6051. doi: 10.1002/j.1460-2075.1994.tb06950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falk K I, Szekely L, Aleman A, Ernberg I. Specific methylation pattern in two control regions of Epstein-Barr virus latency: the LMP-1-coding upstream regulatory region and an origin of DNA replication (oriP) J Virol. 1998;72:2969–2974. doi: 10.1128/jvi.72.4.2969-2974.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farrell P J. Signal transduction from the Epstein-Barr virus LMP-1 transforming protein. Trends Microbiol. 1998;6:175–177. doi: 10.1016/s0966-842x(98)01262-1. [DOI] [PubMed] [Google Scholar]
- 23.Fennewald S, van Santen V, Kieff E. Nucleotide sequence of an mRNA transcribed in latent growth-transforming virus infection indicates that it may encode a membrane protein. J Virol. 1984;51:411–419. doi: 10.1128/jvi.51.2.411-419.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gahn T A, Sugden B. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J Virol. 1995;69:2633–2636. doi: 10.1128/jvi.69.4.2633-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 26.Harada S, Kieff E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J Virol. 1997;71:6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinuma Y, Grace J T. Cloning of immunoglobulin-producing human leukemic and lymphoma cells in long-term cultures. Proc Soc Exp Biol Med. 1967;124:107–111. doi: 10.3181/00379727-124-31677. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh J J-D, Hayward S D. Masking of the CBF1/RBPJκ transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 29.Hurlin P J, Quéva C, Koskinen P J, Steingrímsson E, Ayer D E, Copeland N G, Jenkins N A, Eisenman R N. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johannsen E, Koh E, Mosialos G, Tong X, Kieff E, Grossman S R. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato G J, Lee W M F, Chen L, Dang C V. Max: functional domains and interaction with c-Myc. Genes Dev. 1992;6:81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- 32.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 34.Klein E, Klein G, Nadkarni J S, Nadkarni J J, Wigzell H, Clifford P. Surface IgM-kappa specificity of a Burkitt lymphoma cell in vivo and in derived culture lines. Cancer Res. 1968;28:1300–1310. [PubMed] [Google Scholar]
- 35.Klein G, Dombos L, Gothoskar B. Sensitivity of Epstein-Barr virus (EBV) producer and non-producer human lymphoblastoid cell lines to superinfection with EB-virus. Int J Cancer. 1972;10:44–57. doi: 10.1002/ijc.2910100108. [DOI] [PubMed] [Google Scholar]
- 36.Laherty C D, Yang W-M, Sun J-M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 37.Laux G, Adam B, Strobl L J, Moreau-Gachelin F. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-Jκ interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 1994;13:5624–5632. doi: 10.1002/j.1460-2075.1994.tb06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liebowitz D, Wang D, Kieff E. Orientation and patching of the latent infection membrane protein encoded by Epstein-Barr virus. J Virol. 1986;58:233–237. doi: 10.1128/jvi.58.1.233-237.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Littlewood T, Evan G. Transcription factors 2. Helix-loop-helix. Protein Profile. 1995;2:621–702. [PubMed] [Google Scholar]
- 41.Luka J, Kallin B, Klein G. Induction of the Epstein-Barr virus (EBV) cycle in latently infected cells by n-butyrate. Virology. 1979;94:228–231. doi: 10.1016/0042-6822(79)90455-0. [DOI] [PubMed] [Google Scholar]
- 42.Mannick J B, Cohen J I, Birkenbach M, Marchini A, Kieff E. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J Virol. 1991;65:6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin J, Sugden B. The latent membrane protein oncoprotein resembles growth factor receptors in the properties of its turnover. Cell Growth Differ. 1991;2:653–660. [PubMed] [Google Scholar]
- 44.Masucci M G, Contreras-Salazar B, Ragnar E, Falk K, Minarovits J, Ernberg I, Klein G. 5-Azacytidine up-regulates the expression of Epstein-Barr virus nuclear antigen 2 (EBNA2) through EBNA-6 and latent membrane protein in the Burkitt’s lymphoma line Rael. J Virol. 1989;63:3135–3141. doi: 10.1128/jvi.63.7.3135-3141.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minarovits J, Hu L-F, Minarovits-Kormuta S, Klein G, Ernberg I. Sequence-specific methylation inhibits the activity of the Epstein-Barr virus LMP 1 and BCR2 enhancer-promoter regions. Virology. 1994;200:661–667. doi: 10.1006/viro.1994.1229. [DOI] [PubMed] [Google Scholar]
- 46.Minarovits J, Minarovits-Kormuta S, Ehlin-Henriksson B, Falk K, Klein G, Ernberg I. Host cell phenotype-dependent methylation patterns of Epstein-Barr virus DNA. J Gen Virol. 1991;72:1591–1599. doi: 10.1099/0022-1317-72-7-1591. [DOI] [PubMed] [Google Scholar]
- 47.Nan X, Ng H-H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 48.Nitsche F, Bell A, Rickinson A. Epstein-Barr virus leader protein enhances EBNA-2 mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J Virol. 1997;71:6619–6628. doi: 10.1128/jvi.71.9.6619-6628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohsako S, Hyer J, Panganiban G, Oliver I, Caudy M. Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev. 1994;8:2743–2755. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
- 50.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 51.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 52.Ricksten A, Olsson A, Andersson T, Rymo L. The 5′ flanking region of the gene for the Epstein-Barr virus-encoded nuclear antigen 2 contains a cell type specific cis-acting regulatory element that activates transcription in transfected B-cells. Nucleic Acids Res. 1988;16:8391–8410. doi: 10.1093/nar/16.17.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ricksten A, Svensson C, Welinder C, Rymo L. Identification of sequences in Epstein-Barr virus DNA required for the expression of the second Epstein-Barr virus-determined nuclear antigen in COS-1 cells. J Gen Virol. 1987;68:2407–2418. doi: 10.1099/0022-1317-68-9-2407. [DOI] [PubMed] [Google Scholar]
- 54.Rowe M, Evans H S, Young L S, Hennessy K, Kieff E, Rickinson A B. Monoclonal antibodies to the latent membrane protein of Epstein-Barr virus reveal heterogeneity of the protein and inducible expression in virus-transformed cells. J Gen Virol. 1987;68:1575–1586. doi: 10.1099/0022-1317-68-6-1575. [DOI] [PubMed] [Google Scholar]
- 55.Rowe M, Lear A L, Croom-Carter D, Davies A H, Rickinson A B. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J Virol. 1992;66:122–131. doi: 10.1128/jvi.66.1.122-131.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaw J E, Levinger L F, Carter C W. Nucleosomal structure of Epstein-Barr virus DNA in transformed cell lines. J Virol. 1979;29:657–665. doi: 10.1128/jvi.29.2.657-665.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shikama N, Lyon J, La Thangue N B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 58.Sjöblom A, Jansson A, Yang W, Laín S, Nilsson T, Rymo L. PU box-binding transcription factors and a POU domain protein cooperate in the Epstein-Barr virus (EBV) nuclear antigen 2-induced transactivation of the EBV latent membrane protein 1 promoter. J Gen Virol. 1995;76:2679–2692. doi: 10.1099/0022-1317-76-11-2679. [DOI] [PubMed] [Google Scholar]
- 59.Sjöblom A, Nerstedt A, Jansson A, Rymo L. Domains of the Epstein-Barr virus nuclear antigen 2 (EBNA2) involved in the transactivation of the latent membrane protein 1 and the EBNA Cp promoters. J Gen Virol. 1995;76:2669–2678. doi: 10.1099/0022-1317-76-11-2669. [DOI] [PubMed] [Google Scholar]
- 60.Sjöblom A, Yang W, Palmqvist L, Jansson A, Rymo L. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J Virol. 1998;72:1365–1376. doi: 10.1128/jvi.72.2.1365-1376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Solomon D L C, Amati B, Land H. Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers. Nucleic Acids Res. 1993;21:5372–5376. doi: 10.1093/nar/21.23.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Viollet B, Lefrançois-Martinez A-M, Henrion A, Kahn A, Raymondjean M, Martinez A. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J Biol Chem. 1996;271:1405–1415. doi: 10.1074/jbc.271.3.1405. [DOI] [PubMed] [Google Scholar]
- 64.Waltzer L, Bourillot P Y, Sergeant A, Manet E. RBP-Jκ repression activity is mediated by a co-repressor and antagonized by the Epstein-Barr virus transcription factor EBNA2. Nucleic Acids Res. 1995;23:4939–4945. doi: 10.1093/nar/23.24.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu D Y, Kalpana G V, Goff S P, Schubach W H. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J Virol. 1996;70:6020–6028. doi: 10.1128/jvi.70.9.6020-6028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yalamanchili R, Tong X, Grossman S, E. J, Mosialos G, Kieff E. Genetic and biochemical evidence that EBNA2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology. 1994;204:634–641. doi: 10.1006/viro.1994.1578. [DOI] [PubMed] [Google Scholar]
- 67.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in a variety of mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 68.Zervos A S, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 69.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-JK, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]