Abstract
The retinoblastoma tumor suppressor protein (pRb) can associate with the transforming proteins of several DNA tumor viruses, including the large T antigen encoded by polyomavirus (Py T Ag). Although pRb function is critical for regulating progression from G1 to S phase, a role for pRb in S phase has not been demonstrated or excluded. To identify a potential effect of pRb on DNA replication, pRb protein was added to reaction mixtures containing Py T Ag, Py origin-containing DNA (Py ori-DNA), and murine FM3A cell extracts. We found that pRb strongly represses Py ori-DNA replication in vitro. Unexpectedly, however, this inhibition only partially depends on the interaction of pRb with Py T Ag, since a mutant Py T Ag (dl141) lacking the pRb interaction region was also significantly inhibited by pRb. This result suggests that pRb interferes with or alters one or more components of the murine cell replication extract. Furthermore, the ability of Py T Ag to be phosphorylated in such extracts is markedly reduced in the presence of pRb. Since cyclin-dependent kinase (CDK) phosphorylation of Py T Ag is required for its replication function, we hypothesize that pRb interferes with this phosphorylation event. Indeed, the S-phase CDK complex (cyclin A-CDK2), which phosphorylates both pRb and Py T Ag, alleviates inhibition caused by pRb. Moreover, hyperphosphorylated pRb is incapable of inhibiting replication of Py ori-DNA in vitro. We propose a new requirement for maintaining pRb phosphorylation in S phase, namely, to prevent deleterious effects on the cellular replication machinery.
Replication of DNA containing the polyomavirus (Py) origin in murine cells or cell extracts requires only one virally encoded product, the large T antigen (T Ag), with all other factors derived from the host cell. Based on the better-characterized simian virus 40 (SV40) replicon, the majority of cellular factors required for accomplishing viral DNA replication have now been identified from human cells (reviewed in reference 76). In many cases, these factors share functional and structural homology with analogous factors derived from organisms as far removed as budding yeast. Thus, SV40 and to a lesser extent Py have been useful models for gaining insight into the workings of putative cellular replicons. Less well understood are how viral replicons are regulated and how they have evolved to intersect or bypass the normal cellular controls imposed on the host replication origins.
The replication functions of the SV40 T Ag have been well characterized (reviewed in references 7 and 19). Although Py T Ag has not been as extensively studied as T Ag encoded by its SV40 counterpart, its replication-related activities are for the most part strikingly similar to those of SV40 T Ag. A cell-free replication system was developed in which Py T Ag mediates replication of DNA containing the Py replication origin (Py ori-DNA) in murine cell extracts (59, 65). Py T Ag binding to the Py origin palindrome is stimulated by nucleotides (47), and in the presence of nucleotides it can also form hexamers (85) and melt sites within the origin (4, 43). Additionally, Py T Ag can unwind origin DNA and display DNA helicase activity (71, 84). Like SV40 T Ag (13, 58, 70), purified Py T Ag binds to and requires specifically the murine DNA polymerase α-primase complex in order to mediate Py ori-DNA synthesis (9, 56, 59).
Although there has been extraordinary progress in elucidating genes and proteins involved in initiation of DNA synthesis in eukaryotic cells, cellular proteins which precisely fulfill the roles of the large T Ags have not yet been identified. Given that these viral proteins interact and function with the cellular factors that are highly likely to be involved in promoting replication from cellular origins, they serve as potentially important models for understanding not only how replication functions mechanistically but also how such replication is regulated during the cell cycle. Both SV40 (reviewed in references 19, 20, and 66) and Py (5, 6, 27, 83) T ags are phosphorylated at multiple sites in vivo. In each case as well, their ability to support viral ori-DNA replication depends on their state of phosphorylation. In particular, SV40 (references 51 and 55 and references therein) and Py (12, 42) T Ags each contain a single cyclin-dependent kinase (CDK) site within the vicinity of their origin binding domain, phosphorylation of which is essential for their ability to initiate DNA synthesis. Both T Ags are also negatively regulated by phosphorylation at other sites (11, 83), and it is likely that there are mechanisms for removal of repressing phosphates in each case.
Both SV40 (14, 31, 32; reviewed in reference 46) and Py (17, 30, 41, 63) T Ags possess an LXCXE motif within their N termini which is necessary for their interaction with the retinoblastoma tumor suppressor protein (pRb) and related family members. It is well documented that the pRb protein plays a critical role in controlling the progression of cells from G1 to S (reviewed in references 3, 28, 35, and 44). In its underphosphorylated state, pRb can bind to E2F family members and repress their ability to function as transcriptional activators (reviewed in references 60 and 61). However, after phosphorylation by G1-specific cyclin dependent kinases (CDKs), pRb can no longer bind E2Fs, which are then free to induce genes involved in initiation of DNA synthesis (61). Both SV40 (23, 48) and Py (38) T Ags bind specifically to the underphosphorylated form of pRb, although hyperphosphorylated forms of pRb can accumulate in cells expressing T Ag (48). Consistent with these observations, it has been shown that deletion of the pRb binding motifs of SV40 and Py T Ag (21, 41, 45, 57, 63, 73, 80) render them defective in immortalization function and transactivation of genes involved in initiation of S phase.
Whether pRb binding directly affects the replication functions of the T Ags is less clear. Although viral ori-DNA synthesis in cells expressing T Ag pRb-binding mutants is reduced, it is possible that this effect is indirect, resulting from the inability of mutant T Ag to counteract the negative regulation of passage into S phase by pRb (73, 81). A C-terminal fragment of Py T Ag which lacks the pRb interaction motif was shown to function as an autonomous mediator of viral ori-DNA synthesis in cycling cells, although both the pRb binding region and another, as yet unidentified function of the N terminus of Py T Ag are required for Py T Ag to mediate viral ori-DNA synthesis in resting cells (24). Based on the possibility of a regulatory loop between T Ag and pRb in which each negatively affects the function of the other, we set out to examine the effect(s) of pRb on the ability of T Ag to promote viral ori-DNA replication. Our results suggest that the presence of active underphosphorylated pRb can repress replication by Py T Ag, even without direct interaction with T Ag. By analogy with cellular processes, they provide a plausible explanation for the need to maintain hyperphosphorylated pRb in S phase.
MATERIALS AND METHODS
Recombinant baculoviruses.
Recombinant baculoviruses vEV55PyT (84) and vEV55Hp53 (22), expressing wild-type Py T Ag and wild-type human p53, respectively, were constructed previously; viruses expressing wild-type human pRb (vEV55HRB) and a mutant Py T Ag, dl141, which cannot bind to pRb (vEV55PyLTdl141) were generated as follows. To make vEV55HRB, a BamHI fragment from pJW3HRBc (50), containing the entire pRb coding region, was inserted into the BglII site of the polylinker of pEV55 (52), generating pEV55HRB. Sf21 insect cells were cotransfected with pEV55HRB and purified wild-type baculovirus DNA, and the recombinant pRb baculovirus was purified from an occlusion body-negative plaque. To construct vEV55PyLTdl141, a vector encoding Py T Ag with a deletion of nucleotides 978 to 995 that removes the pRb interaction motif DLXCXE (amino acids 141 to 146) (pPyLTdl141ori+ [41]; kindly provided by M. Bastin) was used to isolate a BstXI fragment (nucleotides 167 to 1695) and exchanged for the same fragment in the transplacement vector encoding wild-type Py T Ag, EV55PyT (84), to generate pEV55PyLTdl141. The pEV55PyLTdl141 plasmid was cotransfected with modified baculovirus DNA (BaculoGold; PharMingen) into Sf21 insect cells to generate the recombinant baculovirus vEV55PyLTdl141. Recombinant baculoviruses expressing cyclin A and a hemagglutinin influenza virus (HA) epitope-tagged CDK2 (HA-CDK2) were generously provided by D. Morgan (University of California, San Francisco).
Protein purification.
Sf21 insect cells were seeded at a density of 2.5 × 107 cells per 150-mm-diameter dish prior to infection with recombinant baculoviruses expressing wild-type or mutant Py T Ag, human pRb, or human p53. Infected cells were extracted with lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 10 mM β-mercaptoethanol, 1% Nonidet P-40 [NP-40], 35 mM phenylmethylsulfonyl fluoride, 0.1% [vol/vol] aprotinin [Sigma], 0.5 mM sodium vanadate) 48 h postinfection, and Py T Ag (84) or p53 (82) was immunopurified on PAb F5 or PAb 421 columns, respectively, as previously described. pRb was immunopurified by passing the infected insect cell extract over a Sepharose column cross-linked with a pRb-specific antibody, IF-8 (2). pRb was eluted from the immunoaffinity column in an alkaline buffer (20 mM triethylamine [pH 10.8], 200 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, 10% glycerol), followed by rapid neutralization in buffer containing 1/20 fraction volume of 1 M Tris-HCl (pH 7.5). In some cases, pRb was purified from cells infected with a recombinant His-tagged pRb baculovirus generously provided by S. Dowdy (Washington University). To purify His-pRb, insect cell extracts were incubated with Ni-nitrilotriacetic acid (NTA) beads (Sigma) for 1 h at 4°C followed by two successive washes in NP-40 (+) buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol, 40 mM imidazole, 10% [vol/vol] glycerol, 1% [vol/vol] NP-40). After two washes in NP-40 (−) buffer (same as above but lacking NP-40), beads were packed into a column and washed with NP-40 (−) buffer containing 250 mM NaCl. pRb was eluted in NP-40 (−) buffer containing 250 mM NaCl and 300 mM imidazole. All baculovirus-expressed and purified proteins were dialyzed into buffer D (10 mM HEPES [pH 7.5], 5 mM NaCl, 0.1 mM EDTA, 50% glycerol, 1 mM DTT) after release from the affinity columns.
To prepare 32P-labeled Py T Ag, insect cells were switched to a phosphate-free medium at 43 h postinfection, and then 0.5 mCi of radioactive orthophosphate (NEN) was added for the last hour before extraction. 32P-labeled Py T Ag was purified as described previously (84).
Cyclin A-CDK2 complexes were isolated from Sf21 insect cells coinfected with baculoviruses expressing cyclin A and HA-CDK2. After harvesting as described above, the complex was purified from the insect cell extract on an anti-HA antibody column as previously published (86), using a synthetic HA peptide to elute the proteins from the immunoaffinity column. Pooled fractions were dialyzed into buffer D containing 10% glycerol. The kinase activity of cyclin A-CDK2 was standardized by using histone H1 (Sigma) as the substrate. To prepare hyperphosphorylated pRb, High 5 insect cells (Invitrogen) were infected with His-tagged pRb-, cyclin A-, and HA-CDK2-expressing baculoviruses, and pRb protein was purified on a Ni-NTA column as described above.
Purified replication proteins RP-A (generously provided by E. Ferrari and U. Hubscher, University of Zurich-Irchel) and PCNA (a generous gift from B. Stillman, Cold Spring Harbor) were isolated from calf thymus and from HeLa cells, respectively.
Specific complex formation between Py T Ag and pRb.
Sf21 insect cells were coinfected with recombinant baculoviruses expressing pRb and either wild-type or mutant Py T Ag. The cells were extracted with lysis buffer (84) 48 h postinfection, and proteins were immunoprecipitated from the extracts by using monoclonal antibodies PAb F5 (specific for Py T Ag) and IF-8 (2) (specific for pRb) cross-linked to protein G- and protein A-Sepharose, respectively. The Sepharose beads were washed four times with radioimmunoprecipitation assay buffer prior to addition of a denaturing protein sample buffer, and polypeptides were resolved by sodium dodecyl sulfate–7.5% polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting with antibody PAb F5 or IF-8.
In vitro DNA replication.
Standard Py ori-DNA replication mixtures (50 μl) and the preparation of murine FM3A cell extracts have been described elsewhere (59). Reaction mixtures containing FM3A extract (60 μg), wild-type or mutant Py T Ag, and Py ori-DNA (pBE102) (65) were incubated at 33°C for 3 h. A molar ratio of Py T Ag to pBE102 of from 50:1 to 200:1 was used. This ratio has been observed to give maximum levels of incorporation of deoxynucleotides (65).
To analyze the phosphorylation of Py T Ag in the replication extract, [γ-32P]ATP (25 μCi) was included in replication reaction mixtures as described above. Subsequently, Py T Ag was immunoprecipitated from the reactions with monoclonal antibody PAb F5 cross-linked to protein G-Sepharose. The levels of protein and phosphorylation were examined by Western blotting and autoradiography.
Protein kinase assay.
Reaction mixtures contained 50 mM HEPES (pH 7.5), 10 mM MgCl2, 1 mM DTT, Py T Ag, pRb, and cyclin A-CDK2. After a 30-min incubation period at 20°C, the reactions were terminated by the addition of protein sample buffer, and samples were analyzed by SDS-PAGE followed by autoradiography.
RESULTS
pRb inhibits Py ori-DNA replication supported by both wild-type Py T Ag and a mutant Py T Ag (dl141) which lacks the pRb interaction motif.
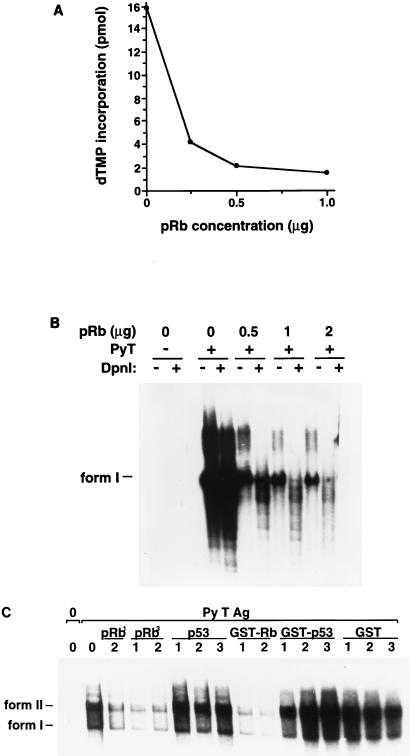
Both human and murine pRb proteins were shown previously to bind similarly to T Ags derived from rodent and simian polyomaviruses (17). To analyze the effect that pRb may have on in vitro DNA replication supported by Py T Ag, human pRb was isolated from recombinant baculovirus-infected insect cells. While initially pRb preparations were immunopurified by using IF-8 antibody columns, in some later experiments His-tagged pRb protein was purified on Ni-NTA beads. The two sources of affinity-purified pRb were found to provide essentially identical results whenever they were compared. Increasing concentrations of pRb protein were added to the replication reaction mixtures such that the molar ratio of pRb to Py T Ag increased from 0.5:1 to 2:1. DNA synthesis supported by Py T Ag was analyzed by acid precipitation of the total [α-32P]dTMP incorporated into replicated DNA (Fig. 1A), or the DNA was purified from the reaction mixtures, linearized, and then digested with DpnI to remove the unreplicated DNA and resolved on 1% agarose gels (Fig. 1B). In the absence of pRb, as expected, Py T Ag mediated the replication of a Py ori-DNA containing plasmid (Fig. 1). Both methods revealed that pRb markedly inhibited ori-DNA synthesis supported by Py T Ag. The pRb inhibitory effect was specific since, confirming our previous observations (53, 82), addition of immunopurified p53 protein did not inhibit Py ori-DNA replication (Fig. 1C). Additionally, bacterially expressed glutathione S-transferase (GST)–pRb inhibited Py ori-DNA replication, while GST-p53 or GST alone did not.
FIG. 1.
pRb, but not p53, inhibits the replication of Py T Ag in vitro. (A) Increasing concentrations of pRb were added to replication mixtures containing FM3A extract (300 μg), Py ori-DNA (0.2 μg), and Py T Ag (0.6 μg). After a 3-h incubation at 33°C, 5-μl aliquots were acid precipitated and counted by scintillation. A background of 0.5 pmol has been subtracted. (B) DNA replication products were purified from the replication mixtures, linearized, digested with DpnI, and analyzed on 1% agarose gels followed by autoradiography. Linear DNA (form I) is indicated on the left. (C) Replication mixtures containing FM3A extract (300 μg), Py ori-DNA (0.2 μg), and Py T Ag (0.6 μg) received increasing concentrations of pRb or p53 purified from insect cells or from bacteria (GST-Rb, GST-p53, or GST). pRb1 and pRb3 are different preparations of pRb. 1 (0.5:1), 2 (1:1), and 3 (2:1) denote the ratios of pRb or p53 to Py T Ag. After a 3-h incubation at 33°C, the DNA products were analyzed as described for panel B. Form I and form II DNAs are indicated on the left.
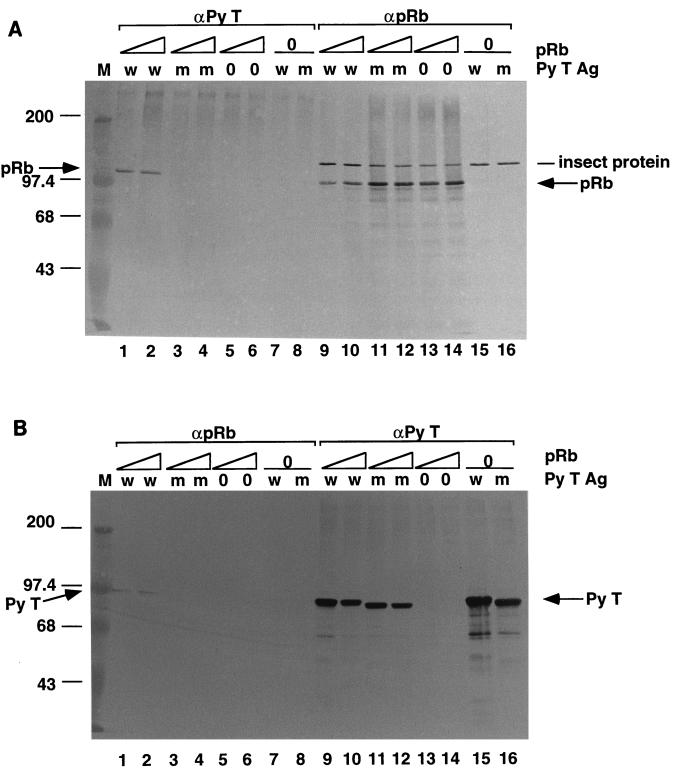
We then examined whether complex formation between pRb and Py T Ag was responsible for the observed inhibition. To address that question, we generated a recombinant baculovirus that expresses a mutant Py T Ag, dl141, which lacks the pRb interaction motif, DLXCXE (41). The importance of this sequence was verified by analyzing complex formation in insect cells that had been coinfected with recombinant baculoviruses expressing human pRb and either wild-type or the mutant dl141 Py T Ag (Fig. 2A and B). The recombinant baculoviruses expressing the Py large T Ags yielded similar quantities of wild-type and mutant proteins (Fig. 2B). Immunoprecipitation-Western blotting was performed to determine the relative abilities of the wild-type and mutant forms of Py T Ag to bind to pRb. First the extracts were immunoprecipitated with the Py T Ag-specific antibody (PAb F5), and then the supernatant (the unbound pRb) was immunoprecipitated with the pRb-specific antibody IF 8 (Fig. 2A). The presence of pRb in the complex was detected by immunoblotting with IF-8. The reverse order of immunoprecipitations was performed starting with IF-8 and immunoblotting with the Py T Ag-specific antibody, PAb F5 (Fig. 2B). As expected, pRb formed a complex with wild-type Py T Ag but not with the dl141 mutant Py T Ag. Approximately 50% of the pRb protein entered the complex (Fig. 2A; compare lanes 1 and 2 with lanes 13 and 14), whereas the majority of the wild-type Py T Ag was free (Fig. 2B; compare lanes 1 and 2 to lane 15). An unknown protein of insect cell origin cross-reacted with the pRb specific antibody (Fig. 2A). Wild-type but not mutant Py T Ag-pRb complexes could also be detected when the immunopurified proteins were mixed together in vitro. However, the binding in vitro was inefficient compared to the interaction in coinfected insect cells (data not shown).
FIG. 2.
pRb inhibits the replication activities of wild-type Py T Ag and a mutant Py T Ag (dl141) that does not bind to pRb. (A and B) Insect cells were coinfected with recombinant baculoviruses expressing either wild-type Py T Ag (w; lanes 1, 2, 7, 9, 10, and 15) or mutant Py T Ag dl141 (m; lanes 3, 4, 8, 11, 12, and 16) and pRb (lanes 1 to 6 and 9 to 14). Extracts from the coinfected insect cells were immunoprecipitated with Py T Ag-specific antibody PAb F5 (A, lanes 1 to 8). The supernatants from those immunoprecipitates were reimmunoprecipitated with a pRb-specific antibody IF-8 (lanes 9 to 16). In panel B, the extracts were first immunoprecipitated with the pRb-specific antibody (lanes 1 to 8) and then reimmunoprecipitated with the Py T Ag-specific antibody (lanes 9 to 16). The immunoprecipitates were analyzed by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with IF-8 (A) or PAbF5 (B). Sizes are indicated in kilodaltons. (C) Increasing concentrations of pRb were added to mixtures containing FM3A extract (300 μg), Py ori-DNA (0.2 μg), and wild-type (wt) or mutant (mt; dl141) Py T Ag (0.6 μg). After a 3-h incubation at 33°C, 5-μl aliquots were acid precipitated and counted by scintillation. A background of 0.5 pmol has been subtracted.
To determine whether the inhibition by pRb requires its interaction with Py T Ag, the effect of pRb on the ability of the dl141 mutant Py T Ag to support ori-DNA synthesis was tested. Importantly, the wild-type and mutant Py T Ags supported replication of Py ori-DNA in vitro to the same extent when used in equal quantities (Fig. 2C). We were surprised to observe that pRb inhibited replication mediated by both Py T Ags (Fig. 2C). However, ori-DNA replication supported by the mutant Py T Ag was inhibited overall to a lesser extent by pRb than was the wild-type protein, which was nearly maximally inhibited by the lowest concentration of pRb. This finding suggests that there are two modes of pRb inhibition: one exerted through its association with Py T Ag, and another which is independent of this interaction.
Despite the fact that binding to Py T Ag was not a requirement for inhibition by pRb, it is still possible that Py T Ag is somehow affected by the presence of pRb. We reasoned that if this is the case, increasing the concentration of Py T Ag in the replication reaction mixtures would at least partly overcome the pRb inhibition. To be able to add Py T Ag in excess to pRb, we set up initial conditions with a lesser concentration of Py T Ag that supported low but detectable DNA synthesis and then added increasing amounts of pRb to the reaction mixtures such that strong inhibition of Py ori-DNA replication was reached. Under these conditions, increasing the concentration of Py T Ag partially overcame the replication inhibition by pRb, suggesting that T Ag itself is a target of pRb in vitro (data not shown).
Our data suggested that the inhibition by pRb was at least partially mediated through effects on the FM3A replication extract. By analogy to the SV40 system, T Ag, single-stranded DNA-binding protein (RP-A), and DNA polymerase α-primase are required for the initial steps of replication (reviewed in references 19 and 76). However, neither human RP-A nor murine polymerase α purified from FM3A cells reversed the pRb inhibition of Py DNA replication (data not shown). Similarly, PCNA, which is a processivity factor for polymerase δ (76) did not relieve the inhibition of Py replication (data not shown). Despite these negative results, it is possible that other factors in the murine cell extract are involved in the effect of pRb. To address this potentiality, conditions were then established such that the amount of FM3A extract that was added to the replication mixtures was varied. In this case, increasing concentrations of FM3A extract were added to reaction mixtures which contained a fixed concentration of Py T Ag in the presence or absence of a given amount of pRb. Under these conditions, increasing the concentration of the replication extract partially overcame the replication block by pRb (data not shown). A suggested explanation for these results, namely, that Py T Ag and pRb compete for a limiting component of the replication extract, is supported by the experiments described below.
pRb changes the phosphorylation state of Py T Ag in murine extracts.
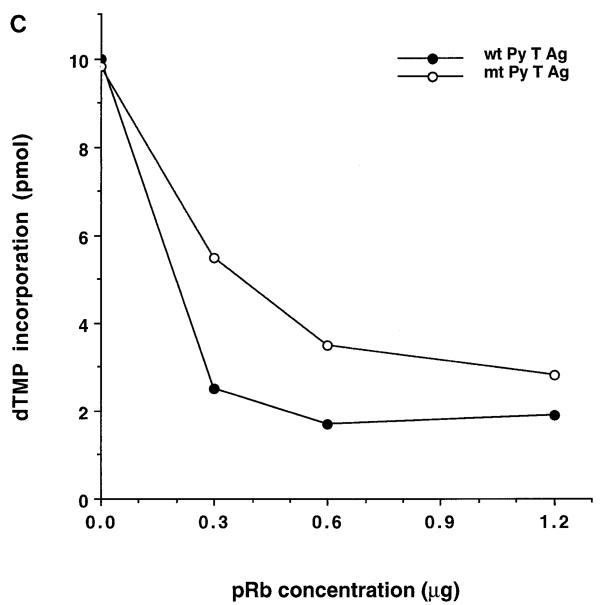
Our results did not exclude the possibility that pRb causes degradation of Py T Ag in FM3A extracts, hence leading to reduced ori-DNA synthesis. This possibility was examined by direct Western blotting of the amount of Py T Ag remaining after a 3-h incubation with or without pRb in complete reaction mixtures (Fig. 3). While it was clear that the levels of Py T Ag protein were not reduced in the presence of inhibitory concentrations of pRb, we made the unexpected observation that Py T Ag migrated slightly faster after incubation in mixtures containing pRb than ones lacking pRb (Fig. 3A). This could be explained either by specific (but highly controlled) proteolytic cleavage of Py T Ag or by an alteration in posttranslational modification causing altered gel mobility. A change in electrophoretic mobility of Py T Ag, attributed to differences in phosphorylation, has been observed previously in extracts from lytically infected cells (5, 33), as well as after phosphorylation by S- and G2-phase CDKs in vitro (42). This issue was pursued by examining the ability of Py T Ag to be phosphorylated in the DNA replication reaction mixtures. Murine extracts containing Py T Ag were incubated with [γ-32P]ATP and either no pRb or inhibitory concentrations of pRb. We reproducibly found that while Py T Ag can be labeled by 32P under these conditions, Py T Ag was markedly less phosphorylated in the presence of pRb than in its absence (Fig. 3B). The pRb-induced change in the phosphorylation of Py T Ag did not depend on DNA synthesis: when 0.1 mM ATP was used instead of 4 mM ATP, which is required to support replication, Py T Ag still acquired less 32P in the presence of pRb than in its absence (data not shown). Importantly, similar results were obtained with the dl141 mutant Py T Ag (data not shown), leading to the conclusion that interactions with pRb are not required for this effect.
FIG. 3.
pRb changes the phosphorylation state of Py T Ag in extracts of FM3A cells. (A) Py T Ag (0.6 μg) was incubated in complete Py ori-DNA replication reaction mixtures at 33°C for 3 h in the presence (+) or absence (−) of pRb (1.2 μg). Two different preparations of FM3A extract (1 and 2, 300 μg each) were used. The total reaction volume (50 μl) was analyzed by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with Py T Ag-specific antibody PAb F5. (B) Py T Ag (0.6 μg) was incubated in a complete replication reaction including [γ-32P]ATP at 33°C for 3 h with (+) or without (−) pRb (1.2 μg). Py T Ag was immunoprecipitated with Py T Ag-specific antibody PAb F5 and analyzed by SDS-PAGE. The Western blot (Western) was probed with PAb F5 and IF-8 (specific for pRb) before being subjected to autoradiography (Autorad). Molecular masses of marker proteins are indicated in kilodaltons on the left; the positions of Py T Ag and pRb are indicated on the right.
The reduced phosphorylation of Py T Ag in the presence of pRb could be due to an induction of a phosphatase or an inhibition of a kinase. pRb has been shown to associate with the protein phosphatase type I catalytic subunit (16), D-type cyclins (15, 18), and the c-Abl tyrosine kinase (87). Although each of these reported interactions would be unlikely to directly affect the phosphorylation state of Py T Ag, it is possible that they may do so indirectly. Alternately, as yet unidentified kinases or phosphatases might interact with pRb. To further explore these possibilities, Py T Ag was purified from infected insect Sf9 cells that had been labeled with 32Pi prior to extraction and immunopurification. The prelabeled T Ag was then incubated in complete replication reaction mixtures in the presence or absence of pRb. If pRb stimulates a Py T Ag phosphatase, a significant loss of phosphate might be expected. However, while Py ori-DNA replication supported by the pre-labeled Py T Ag, was inhibited by pRb (Fig. 4B), there was no reduction in the phosphorylation of this source of Py T Ag in the presence of pRb (Fig. 4A). As discussed in greater detail later, while these data might appear to contradict the results shown in Fig. 3, it is important to stress that Py T Ag can be phosphorylated on multiple sites in mammalian cells as well as in baculovirus-infected insect cells (5, 6, 27, 83). These results suggest that pRb affects a kinase in the FM3A replication extract rather than a phosphatase. Furthermore, incubating the 32P-labeled Py T Ag and pRb together in the absence of FM3A extract did not change the phosphorylation state of Py T Ag, indicating that a phosphatase was not associated with the immunopurified pRb preparations (data not shown).
FIG. 4.
pRb does not affect the phosphorylation state of a prelabeled Py T Ag. Increasing concentrations of pRb were added to complete replication reactions containing 32P-labeled Py T Ag. After 3 h of incubation at 33°C, the mixtures were split to two. (A) One half was analyzed by SDS-PAGE, transferred to nitrocellulose, and immunoblotted by PAb F5 (Western) prior to autoradiography (Autorad). (B) From the other half (25 μl), DNA was purified from the replication mixtures, linearized, digested with DpnI, and examined on 1% agarose gel and autoradiography. Form I DNA is indicated on the right; sizes of markers are indicated in kilodaltons on the left.
Cyclin A-CDK2 kinase reverses the inhibition of Py replication caused by pRb.
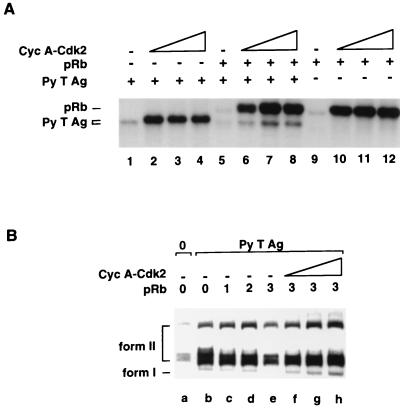
Our data show that pRb affects the ability of one or more protein kinases in the FM3A replication extract to phosphorylate Py T Ag. We recently demonstrated that cyclin A-CDK2, the predominant CDK complex during S phase (reviewed in references 25, 62, and 64), is capable of phosphorylating Py T Ag in vitro on T278 (42). Since pRb itself is phosphorylated extensively by CDKs (reviewed in references 40, 54, and 79), we considered the possibility that at least part of the inhibitory effect of pRb on Py T Ag is through competition for phosphorylation by a CDK complex, in this case cyclin A-CDK2. To test this possibility, cyclin A-CDK2 was purified from insect cells which had been coinfected with baculoviruses expressing cyclin A and HA-CDK2 and was incubated along Py T Ag and pRb, individually and together (Fig. 5A). Confirming our previous observations, Py T Ag alone was efficiently phosphorylated by the cyclin A-CDK2 complex, leading both to incorporation of 32P and to a shift in its electrophoretic mobility (Fig. 5A, lanes 1 to 4). pRb was also phosphorylated by cyclin A-CDK2, although not sufficiently to engender its characteristic mobility shift after extensive phosphorylation. In the presence of pRb, however, phosphorylation of Py T Ag by cyclin A-CDK2 was markedly reduced (lanes 5 to 8). By contrast, phosphorylation of pRb by cyclin A-CDK2 was not affected by the presence of Py T Ag (compare lanes 4 to 8 to lanes 9 to 12). pRb, therefore, efficiently competes with Py T Ag for phosphorylation by A-CDK2 in vitro.
FIG. 5.
pRb inhibits phosphorylation of Py T Ag by cyclin A-CDK2. (A) Py T Ag (0.1 μg; lanes 1 to 8), pRb (0.8 μg; lanes 5 to 12), and increasing concentrations of lysates of Sf9 cells coinfected with cyclin (Cyc) A- and CDK2-expressing baculoviruses (0 μl [lanes 1, 5, and 9], 0.5 μl [lanes 2, 6, and 10], 1 μl [lanes 3, 7, and 11], and 2 μl [lanes 4, 8, and 12]) were incubated in kinase buffer at room temperature for 30 min with 1 mM ATP and [γ-32P]ATP. The phosphorylated proteins were analyzed by SDS-PAGE. (B) Py T Ag (0.15 μg) was incubated with increasing concentrations of pRb (1 [0.35 μg], 2 [0.7 μg], and 3 [1.4 μg]) in complete replication reactions at 33°C for 3 h. Then increasing concentrations of cyclin A-CDK2 (0 μl [lanes a to e], 0.5 μl [lane f], 1 μl [lane g], and 2 μl [lane h]) were added to the reactions containing the most inhibitory levels of pRb. The DNA products were purified from the reactions, linearized, digested with DpnI, and analyzed on 1% agarose gels and by autoradiography. Form I and form II DNAs are indicated on the left.
The above experiment led us to examine whether cyclin A-CDK2 could reverse the inhibition of Py ori-DNA replication by pRb. Cyclin A-CDK2 was added into replication reaction mixtures at a point where partial inhibition by pRb was reached (Fig. 5B; compare lanes b and e). Under such conditions, addition of the CDK complex indeed completely reversed the inhibition by pRb (compare lane e with lanes f to h). Note that under conditions where sufficient pRb was added to cause a greater inhibition, we were unable to add sufficient cyclin A-CDK2 to reverse the inhibitory effect (data not shown). Furthermore, the ability of the CDK complex to block the inhibitory effect of pRb was reversed when additional pRb was added to the reaction mixtures (data not shown). Nevertheless, this result is consistent with the observation that phosphorylation of Py T Ag by cyclin A-CDK2 in vitro is greatly reduced in the presence of pRb protein.
Hyperphosphorylated pRb does not inhibit Py ori-DNA replication in vitro.
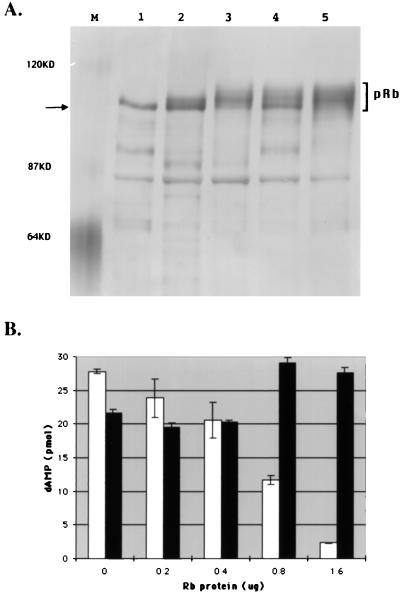
If one mode by which pRb protein can inhibit Py T Ag is through competition for available CDK complexes, it follows that hyperphosphorylated pRb might not be able to inhibit Py ori-DNA replication. To test this, pRb protein was purified from insect cells which had been infected solely with the pRb baculovirus or which had been coinfected with recombinant baculoviruses expressing cyclin A and CDK2 in addition to pRb. As seen in Fig. 6A, when different preparations of singly (lanes 1 and 2) and triply (lanes 3 to 5) infected insect cells were compared, the pRb from cells coinfected with the cyclin A- and CDK2-expressing viruses consisted of a heterogeneous group of electrophoretic species with retarded electrophoretic mobility. The fact that baculovirus-expressed pRb is more efficiently phosphorylated by CDKs in vivo than in vitro has been previously demonstrated (37). When two preparations of pRb, underphosphorylated pRb (from lane 1) and hyperphosphorylated pRb (from lane 5), were tested for their effects on Py ori-DNA synthesis in vitro, only the latter was inert and did not inhibit T Ag mediated ori-DNA synthesis (Fig. 6B). It should be mentioned here that when each of the five samples shown in Fig. 6A was tested for its effect on Py ori-DNA synthesis, as expected, both hypophosphorylated samples shown in lanes 1 and 2 repressed replication. However, there was variation in the abilities of the hyperphosphorylated pRb proteins to affect replication. Specifically, while the pRb proteins shown in lanes 3 and 5 failed to repress DNA synthesis, that shown in lane 4 was inhibitory. However, pRb preparations shown in lanes 1, 2, and 4 had markedly greater amounts of the most underphosphorylated (rapidly migrating) form of pRb (indicated by the arrow), while the two noninhibitory samples in lanes 3 and 5 had the highest ratio of hyperphosphorylated forms. This finding is consistent with the possibility that the ability of underphosphorylated pRb to serve as an effective substrate for CDKs may be at least partially responsible for its ability to block both the phosphorylation and the ability to support ori-DNA replication of Py T Ag.
FIG. 6.
Hyperphosphorylated pRb does not inhibit Py ori-DNA replication. (A) High 5 (lanes 1, 3, 4, and 5) or Sf9 (lane 2) cells were infected with baculoviruses expressing His-pRb (lanes 1 to 5) either alone or along with cyclin A- and HA-CDK2-expressing baculoviruses (lanes 3 to 5). pRb proteins were purified on Ni-NTA columns and analyzed by SDS-PAGE. The molecular masses of marker proteins are indicated at the left. The arrow at the left points to the underphosphorylated form of pRb. (B) Underphosphorylated (from lane 1 in panel A; white bars) and hyperphosphorylated (from lane 5 in panel A; black bars) pRb proteins at the indicated amounts were added to replication mixtures (as described in the legend to Fig. 1 and in Materials and Methods). The incorporation of [α-32P]dAMP into acid-insoluble DNA products was measured by scintillation counting.
DISCUSSION
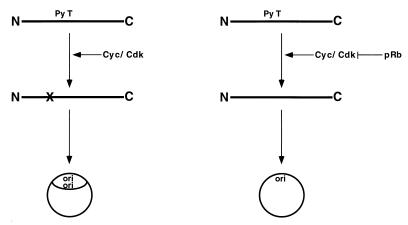
Our initial goal was to determine the consequences of the interaction of Py T Ag with the pRb protein for the replication function of T Ag. We found that the pRb protein can strongly repress Py ori-DNA synthesis in vitro. Unexpectedly, however, pRb also significantly inhibits replication supported by a mutant Py T Ag which is incapable of binding to pRb. This result suggests an effect of pRb on one or more processes occurring in the replication extract in addition to its association with T Ag. Based on the following observations made either previously or in this study, we have developed a hypothesis (Fig. 7) which may explain how pRb can inhibit ori-DNA replication supported by Py T Ag without detectably associating with T Ag: (i) Py T Ag can be phosphorylated by the S-phase CDK complex, cyclin A-CDK2, at a site, T278, which is critical for its replication function; (ii) Py T Ag is significantly less phosphorylated in FM3A extracts when pRb protein is present; (iii) pRb competes for or inhibits phosphorylation of Py T Ag by a cyclin A-CDK2 complex in vitro, (iv) adding excess cyclin A-CDK2 reverses the inhibition of Py ori-DNA synthesis caused by pRb; (v) the net phosphorylation on Py T Ag which was prelabeled with 32P prior to adding to reaction mixtures is not detectably altered by the presence of pRb protein; and (vi) hyperphosphorylated pRb does not inhibit Py ori-DNA synthesis in vitro.
FIG. 7.
pRb inhibits Py ori-DNA replication by interfering with CDK phosphorylation of Py T Ag. Cyc, cyclin.
To expand on the model shown in Fig. 7, we suggest that Py T Ag isolated from insect cells is relatively underphosphorylated at T278 and needs to be more efficiently phosphorylated in order to effectively mediate ori-DNA replication. This is consistent with our observation that Py T Ag can be efficiently phosphorylated by cyclin A-CDK2 in vitro and, in fact, undergoes a quantitative electrophoretic shift by SDS-PAGE after such phosphorylation. It is assumed that CDK complex(es) present in FM3A extracts can phosphorylate Py T Ag at this site, thus converting it to an active form for DNA replication. This critical phosphorylation of T Ag is inhibited in the presence of underphosphorylated pRb, possibly because pRb, which can be phosphorylated by CDKs on several sites, effectively outcompetes the Py T Ag for the available CDK activity in the reaction mixtures. Hyperphosphorylated pRb cannot compete and is therefore not inhibitory. By contrast, it was reported that SV40 T Ag is hyperphosphorylated at its CDK site in infected insect cells (29), and it should be mentioned here that we observed that pRb was markedly less inhibitory to SV40 T Ag-supported SV40 ori-DNA replication in vitro (data not shown). Furthering the hypothesis is the experiment shown in Fig. 4, in which Py T Ag that was prelabeled in insect cells prior to extraction showed no change in phosphorylation in the replication mixtures with or without pRb. This is best explained by the fact that T Ag is multiply phosphorylated at a number of different sites in insect cells as it is in mammalian cells, although, as suggested above, only weakly so at its CDK site. If its major (if not sole) site of phosphorylation in FM3A extracts were at T278 (possibly because the other sites are already relatively well phosphorylated), then there would not be a significant change in overall phosphorylation of T Ag after incubation. Indeed, we demonstrated previously that the replication activity of Py T Ag is stimulated by low levels of phosphatase (83), suggesting that at least one or more repressing phosphates are stoichiometrically present on insect cell-derived Py T Ag. An excellent test of this model would be to determine whether Py T Ag prephosphorylated with a CDK complex was more refractory to inhibition by pRb. Unfortunately, this experiment has been technically challenging because attempts to efficiently phosphorylate Py T Ag by immobilized cyclin A-CDK2 have been unsuccessful.
Although our results can be explained by the above model, we cannot exclude alternative hypotheses. For example, it remains possible that the results seen are caused not by pRb protein itself but by another component of the pRb preparation. We have noted that the pRb protein fractions contain significantly more additional polypeptides than do other proteins immunopurified from insect cells. Based on the well-documented ability of pRb to bind to a number of cellular proteins (reviewed in reference 79) we would postulate that such a protein(s) need not necessarily be viewed as an artifactual contamination but rather considered a possibly physiologically relevant association. However, as mentioned above, the preparations of pRb that we have used do not contain detectable contaminating kinases or phosphatases.
More recently another conserved sequence (HPDKGG) within the N termini of BK virus, SV40, and Py T Ags (10, 26, 68, 72, 74, 89) has been shown to function as a DnaJ domain. This region, in a fashion analogous to the Escherichia coli J domain, serves to recruit cellular DnaK-like chaperons (reviewed in reference 8). While SV40 deleted of its J domain is defective in mediating viral ori-DNA replication in vivo (10), the corresponding region on Py T Ag may not be required since Gjørup et al. (24) have shown that the N terminus is dispensable for mediating viral DNA replication. Strikingly, an intact J domain is required for the ability of SV40 T Ag to regulate pRb family phosphorylation through an as yet unidentified mechanism (80).
The role of the pRb protein as the central gatekeeper which regulates progression from G1 into S phase has been extensively studied (28, 35, 44, 46, 60, 67). A wealth of evidence suggests that a critical function of unphosphorylated pRb is to bind E2F proteins leading to the repression of promoters bearing E2F response elements. Whether pRb functions during other phases of the cell cycle has not yet been firmly established. However, a number of experiments have suggested that pRb may be directly or indirectly involved in regulating viral DNA replication. Wilcock and Lane (88) reported the colocalization of pRb to sites of viral replication in herpesvirus-infected cells. Amin et al. (1) showed that the pocket domain of the pRb-related protein p107, when bound to SV40 T Ag, prevents assembly of T Ag hexamers at the core origin and also interferes with the interaction of T Ag with DNA polymerase α. Since we observed that pRb is more inhibitory to wild-type Py T Ag than to the Rb-binding-defective form of T Ag, it is likely that the tumor suppressor can repress Py T Ag through at least two ways, one producing direct effects similar to those noted by Amin et al. (1) and the other involving down-regulation of Py T Ag phosphorylation in the replication reaction mixtures. Savoysky et al. (69) reported that the ability of SV40 T Ag to stimulate DNA polymerase α was reduced if T Ag was preincubated with pRb. However, a stimulation of DNA polymerase α by hyperphosphorylated pRb was also reported (78). Cellular replicons can be affected as well, since pRb, through its N terminus, was recently reported to interact with MCM7 in vitro, as well as to inhibit Xenopus laevis DNA replication in vitro (75). Furthermore, Knudson et al. (39) have provided evidence that pRb mutated at a number of its phosphorylation sites can block progression of cells through S phase. By contrast, however, Karantza et al. (36) reported that ectopically expressed wild-type or T Ag-binding-defective pRb proteins lead to accumulation of cells in G2 but not in S.
With few exceptions, the vast majority of studies to date support the likelihood that the functional unphosphorylated or underphosphorylated pRb presides in G1 and that the S-phase hyperphosphorylated form of pRb is inert. pRb possess 16 CDK sites and can be phosphorylated by many if not all of the known CDK complexes (reviewed in reference 79). The early G1 cyclin D1-CDK4 complex phosphorylates different sites on pRb than does the late G1 cyclin E-CDK2 complex (79), and D- and E cyclin-CDK complexes together result in sufficient pRb phosphorylation to allow progression to S phase (49; reviewed in reference 67). Interestingly, these G1 CDK complexes are not capable of phosphorylating Py T Ag in vitro (reference 42 and unpublished data). Our data thus suggest the possibility of a regulatory loop between Py T Ag and pRb: while underphosphorylated pRb blocks ori-DNA replication mediated by Py T Ag in G1, binding of T Ag to pRb results in pRb inactivation and hyperphosphorylation. The net result is progression through G1 to S phase where viral ori-DNA synthesis can initiate in appropriately timed fashion, through availability of cellular replication factors as well as critical phosphorylation at the regulatory CDK site on T Ag.
While it is assumed that phosphorylation of pRb renders it incapable of binding to its cellular targets such as E2F, our results suggest an additional requirement for hyperphosphorylation of pRb in S phase. Although full understanding of the roles of S phase CDK complexes in regulation of DNA replication is not yet available, there is ample and increasing evidence supporting the probability that CDK functions are critical for the correct function and regulation of cellular origins (reviewed in references 34 and 77). If the inhibitory effects that we have noted for under- but not hyperphosphorylated pRb were to be documented as well for processes necessary for cellular DNA replication, we can propose that pRb phosphorylation is a multiply determined requirement for successful passage of cells through S phase.
ACKNOWLEDGMENTS
We are most grateful for the expert technical assistance of E. Freulich. J. Manfredi is thanked for his contribution to preparing the pRb-expressing baculovirus.
This work was supported by NIH grant CA26905.
REFERENCES
- 1.Amin A A, Murakami Y, Hurwitz J. Initiation of DNA replication by simian virus 40 T antigen is inhibited by the p107 protein. J Biol Chem. 1994;269:7735–7743. [PubMed] [Google Scholar]
- 2.Bártek J, Vojtesek B, Grand R J A, Gallimore P H, Lane D P. Cellular localization and T antigen binding of the retinoblastoma protein. Oncogene. 1992;7:101–108. [PubMed] [Google Scholar]
- 3.Bártek J, Bartkova J, Lukas J. The retinoblastoma protein pathway in cell cycle control and cancer. Exp Cell Res. 1997;237:1–6. doi: 10.1006/excr.1997.3776. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharyya S, Lorimer H E, Prives C. Murine polyomavirus and simian virus 40 large T antigens produce different structural alterations in viral origin DNA. J Virol. 1995;69:7579–7585. doi: 10.1128/jvi.69.12.7579-7585.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bockus B J, Schaffhausen B. Phosphorylation of polyomavirus large T antigen: effects of viral mutations and cell growth state. J Virol. 1987;61:1147–1154. doi: 10.1128/jvi.61.4.1147-1154.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bockus B J, Schaffhausen B. Localization of the phosphorylations of polyomavirus large T antigen. J Virol. 1987;61:1155–1163. doi: 10.1128/jvi.61.4.1155-1163.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boroweic J A, Dean F B, Bullock P A, Hurwitz J. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell. 1990;60:181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- 8.Brodsky J L, Pipas J M. Polyomavirus T antigens: molecular chaperones for multiprotein complexes. J Virol. 1998;72:5329–5334. doi: 10.1128/jvi.72.7.5329-5334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruckner A, Stadlbauer F, Guarino L A, Brunahl A, Schneider C, Rehfuess C, Previes C, Fanning E, Nasheuer H P. The mouse DNA polymerase alpha-primase subunit p48 mediates species-specific replication of polyomavirus DNA in vitro. Mol Cell Biol. 1995;15:1716–1724. doi: 10.1128/mcb.15.3.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, K. S., K. P. Mullane, I. A. Aksoy, H. Stubdal, J. Zalvide, J. M. Pipas, P. A. Silver, T. M. Roberts, B. S. Schaffhausen, and J. A. DeCaprio. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 11:1098–1110. [DOI] [PubMed]
- 11.Cegielska A, Moarefi I, Fanning E, Virshup D M. T-antigen kinase inhibits simian virus 40 DNA replication by phosphorylation of intact T antigen on serines 120 and 123. J Virol. 1994;68:269–275. doi: 10.1128/jvi.68.1.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee A, Bockus B J, Gjørup O V, Schaffhausen B S. Phosphorylation sites in polyomavirus large T antigen that regulate its function in viral, but not cellular, DNA synthesis. J Virol. 1997;71:6472–6478. doi: 10.1128/jvi.71.9.6472-6478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins K L, Russo A R, Tseng B Y, Kelly T J. The role of the 70 KDa subunit of human DNA polymerase α in DNA replication. EMBO J. 1993;12:4555–4566. doi: 10.1002/j.1460-2075.1993.tb06144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeCaprio J A, Ludlow J W, Figge J, Shew J Y, Huang C M, Lee W H, Marsilio E, Paucha E, Livingston D M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 15.Dowdy S F, Hinds P W, Louie K, Reed S I, Arnold A, Weinberg R A. Physical interaction of the retinoblastoma protein with human cyclins. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- 16.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 17.Dyson N, Bernards R, Friend S H, Gooding L R, Hassell J A, Major E O, Pipas J M, Vandyke T, Harlow E. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J Virol. 1990;64:1354–1356. doi: 10.1128/jvi.64.3.1353-1356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ewen M E, Sluss H K, Sherr C J, Matsushime H, Kato J Y, Livingston D M. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 19.Fanning E, Knippers R. Structure and function of simian virus 40 large T antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 20.Fanning E. Control of SV40 DNA replication by protein phosphorylation: a model for cellular DNA replication? Trends Cell Biol. 1994;4:250–255. doi: 10.1016/0962-8924(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 21.Freund R, Bauer P H, Crissman H A, Bradbury E M, Benjamin T L. Host range and cell cycle activation properties of polyomavirus large T-antigen mutants defective in pRB binding. J Virol. 1994;68:7227–7234. doi: 10.1128/jvi.68.11.7227-7234.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman P N, Kern S E, Vogelstein B, Prives C. Wild type, but not mutant, human p53 proteins inhibit the replication activities of SV40 large T antigen. Proc Natl Acad Sci USA. 1990;87:9275–9279. doi: 10.1073/pnas.87.23.9275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedrich T D, Laffin J, Lehman J M. Hypophosphorylated retinoblastoma gene product accumulates in SV40-infected cells acquiring a tetraploid DNA content. Oncogene. 1993;8:1673–1677. [PubMed] [Google Scholar]
- 24.Gjørup O V, Rose P E, Holman P S, Bockus B J, Schaffhausen B S. Protein domains connect cell cycle stimulation directly to initiation of DNA replication. Proc Natl Acad Sci USA. 1994;91:12125–12129. doi: 10.1073/pnas.91.25.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grana X, Reddy E P. Cell cycle control in mammalian cells: role of cyclins, cyclin dependent kinases (CDKs), growth suppressor genes and cyclin-dependent kinase inhibitors (CKIs) Oncogene. 1995;11:211–219. [PubMed] [Google Scholar]
- 26.Harris K F, Christensen J B, Radany E H, Imperiale M J. Novel mechanisms of E2F induction by BK virus large-T antigen: requirement of both the pRb-binding and the J domains. Mol Cell Biol. 1998;18:1746–1756. doi: 10.1128/mcb.18.3.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassauer M, Scheidtmann K H, Walter G. Mapping of phosphorylation sites in polyomavirus large T antigen. J Virol. 1986;58:805–816. doi: 10.1128/jvi.58.3.805-816.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatakeyama M, Weinberg R A. The role of RB in cell cycle control. Prog Cell Cycle Res. 1995;1:9–19. doi: 10.1007/978-1-4615-1809-9_2. [DOI] [PubMed] [Google Scholar]
- 29.Hoss A, Moarefi I, Scheidtmann K H, Cisek L J, Corden J L, Dornreiter I, Arthur A K, Fanning E. Altered phosphorylation pattern of simian virus 40 T antigen expressed in insect cells by using a baculovirus vector. J Virol. 1990;64:4799–4807. doi: 10.1128/jvi.64.10.4799-4807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howes S H, Bockus B J, Schaffhausen B S. Genetic analysis of polyomavirus large T nuclear localization: nuclear localization is required for productive association with pRb family members. J Virol. 1996;70:3581–3588. doi: 10.1128/jvi.70.6.3581-3588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Q J, Dyson N, Harlow E. The regions of the retinoblastoma protein needed for binding to adenovirus E1A or SV40 large T antigen are common sites for mutations. EMBO J. 1990;9:1147–1155. doi: 10.1002/j.1460-2075.1990.tb08221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang S, Wang N P, Tseng B Y, Lee W H, Lee E H. Two distinct and frequently mutated regions of retinoblastoma protein are required for binding to SV40 T antigen. EMBO J. 1990;9:1815–1822. doi: 10.1002/j.1460-2075.1990.tb08306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hutchinson M, Hunter T, Eckhart W. Characterization of T antigens in polyoma-infected and transformed cells. Cell. 1978;17:65–77. doi: 10.1016/0092-8674(78)90083-1. [DOI] [PubMed] [Google Scholar]
- 34.Jallepalli P V, Kelly T J. Cyclin-dependent kinase and initiation at eukaryotic origins: a replication switch? Curr Opin Cell Biol. 1997;9:358–363. doi: 10.1016/s0955-0674(97)80008-7. [DOI] [PubMed] [Google Scholar]
- 35.Kaelin W G. Recent insights into the functions of the retinoblastoma susceptibility gene product. Cancer Investig. 1997;15:243–254. doi: 10.3109/07357909709039722. [DOI] [PubMed] [Google Scholar]
- 36.Karantza V, Maroo A, Fay D, Sedivy J M. Overproduction of Rb protein after the G1/S boundary causes G2 arrest. Mol Cell Biol. 1993;13:6640–6652. doi: 10.1128/mcb.13.11.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato J, Matsushime H, Hiebert S W, Ewen M E, Sherr C J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 38.Khandjian E W, Tremblay S. Phosphorylation of the retinoblastoma protein is modulated in mouse kidney cells infected with polyomavirus. Oncogene. 1992;7:909–917. [PubMed] [Google Scholar]
- 39.Knudsen E S, Buckmaster C, Chen T T, Feramisco J, Wang J Y. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 1998;12:2278–2292. doi: 10.1101/gad.12.15.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knudsen E S, Wang J Y. Differential regulation of retinoblastoma protein function by specific Cdk phosphorylation sites. J Biol Chem. 1996;271:8313–8320. doi: 10.1074/jbc.271.14.8313. [DOI] [PubMed] [Google Scholar]
- 41.Larose A, Dyson N, Sullivan M, Harlow E, Bastin M. Polyomavirus large T antigen mutants affected in retinoblastoma protein binding are defective in immortalization. J Virol. 1991;65:2308–2313. doi: 10.1128/jvi.65.5.2308-2313.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Bhattacharyya S, Prives C. Regulation of the replication functions of polyoma large T antigen by S- and G2-phase cyclin-dependent kinases. J Virol. 1997;71:6469–6485. doi: 10.1128/jvi.71.9.6479-6485.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, Li B L, Hock M, Wang E, Folk W R. Sequences flanking the pentanucleotide T-antigen binding sites in the polyomavirus core origin help determine selectivity of DNA replication. J Virol. 1995;69:7570–7578. doi: 10.1128/jvi.69.12.7570-7578.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin B T, Wang J Y. Cell cycle regulation of retinoblastoma protein phosphorylation. Ciba Found Symp. 1992;170:227–241. [PubMed] [Google Scholar]
- 45.Linder S, Nilsson M, Martens I, Magnusson G. A viable mouse polyomavirus mutant without immortalizing or transforming activities. Virology. 1990;179:78–86. doi: 10.1016/0042-6822(90)90276-w. [DOI] [PubMed] [Google Scholar]
- 46.Livingston D M. Functional analysis of the retinoblastoma gene product and of RB-SV40 T antigen complexes. Cancer Surv. 1992;12:153–160. [PubMed] [Google Scholar]
- 47.Lorimer H E, Wang E H, Prives C. The DNA binding properties of polyomavirus large T antigen are altered by ATP and other nucleotides. J Virol. 1991;65:687–699. doi: 10.1128/jvi.65.2.687-699.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ludlow J W, DeCaprio J A, Huang C M, Lee W H, Paucha E, Livingston D M. SV40 large T antigen binds preferentially to an underphosphorylated member of the retinoblastoma susceptibility gene product family. Cell. 1989;56:57–65. doi: 10.1016/0092-8674(89)90983-5. [DOI] [PubMed] [Google Scholar]
- 49.Lundberg A S, Weinberg R A. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manfredi J J, Prives C. Binding of p53 and p105-Rb is not sufficient for oncogenic transformation by a hybrid polyoma simian virus 40 large T antigen. J Virol. 1990;69:5250–5259. doi: 10.1128/jvi.64.11.5250-5259.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McVey D, Woelker B, Tegtmeyer P. Mechanisms of simian virus 40 T antigen activation by phosphorylation of threonine 124. J Virol. 1996;70:3887–3893. doi: 10.1128/jvi.70.6.3887-3893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller D W, Safer P, Miller L K. An insect baculovirus host-vector system for high-level expression of foreign genes. In: Setlow J K, Hollander A, editors. Genetic engineering. 8. Principles and methods. New York, N.Y: Plenum Publishing Corp.; 1986. pp. 277–298. [Google Scholar]
- 53.Miller S, Farmer G, Prives C. p53 inhibits DNA replication in vitro in a DNA-binding-dependent manner. Mol Cell Biol. 1995;15:6554–6560. doi: 10.1128/mcb.15.12.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mittnacht S. Control of pRB phosphorylation. Curr Opin Genet Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 55.Moarefi I F, Small D, Gilbert I, Höpfner M, Randall S K, Schneider C, Russo A A R, Ramsperger U, Arthur A K, Stahl H, Kelly T J, Fanning E. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus 40 (SV40) large T antigen specifically blocks SV40 origin DNA unwinding. J Virol. 1993;67:4992–5002. doi: 10.1128/jvi.67.8.4992-5002.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moses K, Prives C. A unique subpopulation of DNA polymerase α interacts with polyoma T antigen and stimulates DNA replication. Mol Cell Biol. 1994;14:2767–2776. doi: 10.1128/mcb.14.4.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mudrak I, Ogris E, Rotheneder H, Wintersberger E. Coordinated trans activation of DNA synthesis- and precursor-producing enzymes by polyomavirus large T antigen through interaction with the retinoblastoma protein. Mol Cell Biol. 1994;14:1886–1892. doi: 10.1128/mcb.14.3.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murakami Y, Hurwitz J. DNA polymerase alpha stimulates the ATP-dependent binding of simian virus tumor T antigen to the SV40 origin of replication. J Biol Chem. 1993;268:11018–11027. [PubMed] [Google Scholar]
- 59.Murakami Y, Eki T, Tamada M-A, Prives C, Hurwitz J. Species specific in vitro synthesis of DNA containing the polyomavirus origin of replication. Proc Natl Acad Sci USA. 1986;83:6347–6351. doi: 10.1073/pnas.83.17.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nevins J R, Leone G, DeGregori J, Jakoi L. Role of the Rb/E2F pathway in cell growth control. J Cell Physiol. 1997;173:233–236. doi: 10.1002/(SICI)1097-4652(199711)173:2<233::AID-JCP27>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 61.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 62.Nigg E A. Cyclin dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays. 1995;6:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 63.Pilon A A, Desjardins P, Hassell J A, Mes-Masson A M. Functional implications of mutations within polyomavirus large T antigen Rb-binding domain: effects on pRb and p107 binding in vitro and immortalization activity in vivo. J Virol. 1996;70:4457–4465. doi: 10.1128/jvi.70.7.4457-4465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pines J. The cell cycle kinases. Semin Cancer Biol. 1994;5:305–313. [PubMed] [Google Scholar]
- 65.Prives C, Murakami Y, Kern F, Folk W, Basilico C, Hurwitz J. DNA sequence requirements of polyoma replication in vivo and in vitro. Mol Cell Biol. 1987;7:3694–3704. doi: 10.1128/mcb.7.10.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prives C. The replication functions of SV40 T antigen are regulated by phosphorylation. Cell. 1990;61:735–738. doi: 10.1016/0092-8674(90)90179-i. [DOI] [PubMed] [Google Scholar]
- 67.Reed, S. I. Control of the G1/S transition. Cancer Surv. 29:7–23. [PubMed]
- 68.Riley M I, Yoo W, Mda N Y, Folk W R. Tiny T antigen: an autonomous polyomavirus T antigen amino-terminal domain. J Virol. 1997;71:6068–6074. doi: 10.1128/jvi.71.8.6068-6074.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Savoysky E, Suzuki M, Simbulan C, Tamai K, Ohuchi T, Akiyama T, Yoshida S. Immunopurified Rb protein inhibits SV40 T antigen-dependent stimulation of DNA polymerase alpha. Oncogene. 1993;8:319–325. [PubMed] [Google Scholar]
- 70.Schneider C, Weisshart K, Guarino L A, Dornreiter I, Fanning E. Species-specific functional interactions of DNA polymerase alpha-primase with simian virus 40 (SV40) T antigen require SV40 origin DNA. Mol Cell Biol. 1994;14:3176–3185. doi: 10.1128/mcb.14.5.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seki M, Enomoto T, Eki T, Miyahima A, Murakami Y, Hanaoka R, Ui M. DNA helicase and 5′-triphosphate activities of polyoma virus large tumor antigen. Biochemistry. 1990;29:1003–1009. doi: 10.1021/bi00456a024. [DOI] [PubMed] [Google Scholar]
- 72.Sheng Q, Denis D, Ratnofsky M, Roberts T M, DeCaprio J A, Schaffhausen B. The DnaJ domain of polyomavirus large T antigen is required to regulate Rb family tumor suppressor function. J Virol. 1997;71:9410–9416. doi: 10.1128/jvi.71.12.9410-9416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Soderbarg K, Magnusson G. Lytic functions of mutant polyomavirus large T-antigen with deletion of retinoblastoma protein-binding motif. Virology. 1993;193:281–288. doi: 10.1006/viro.1993.1123. [DOI] [PubMed] [Google Scholar]
- 74.Srinivasan A, McClellan A J, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky J L, Pipas J M. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. J Virol. 1997;71:1888–1896. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sterner J M, Dew-Knight S, Musahl C, Kornbluth S, Horowitz J M. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol Cell Biol. 1998;18:2748–2757. doi: 10.1128/mcb.18.5.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stillman B. Smart machines at the DNA replication fork. Cell. 1994;78:725–728. doi: 10.1016/s0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 77.Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 78.Takemura M, Kitagawa T, Izuta S, Wasa J, Takai A, Akiyama T, Yoshida S. Phosphorylated retinoblastoma protein stimulates DNA polymerase alpha. Oncogene. 1997;15:2483–2492. doi: 10.1038/sj.onc.1201431. [DOI] [PubMed] [Google Scholar]
- 79.Taya Y. RB kinases and RB-binding proteins: new points of view. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- 80.Tevethia M J, Lacko H A, Kierstead T D, Thompson D L. Adding an Rb-binding site to an N-terminally truncated simian virus 40 T antigen restores growth to high cell density, and the T common region in trans provides anchorage-independent growth and rapid growth in low serum concentrations. J Virol. 1997;71:1888–1896. doi: 10.1128/jvi.71.3.1888-1896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Uzvolgyi E, Classon M, Henriksson M, Huang H J, Szekely L, Lee W H, Klein G, Sumegi J. Reintroduction of a normal retinoblastoma gene into retinoblastoma and osteosarcoma cells inhibits the replication associated function of SV40 large T antigen. Cell Growth Differ. 1991;2:297–303. [PubMed] [Google Scholar]
- 82.Wang E, Friedman P, Prives C. The murine p53 protein blocks replication of SV40 DNA in vitro by inhibiting the initiation function of SV40 large T antigen. Cell. 1989;57:379–392. doi: 10.1016/0092-8674(89)90913-6. [DOI] [PubMed] [Google Scholar]
- 83.Wang E H, Bhattacharyya S, Prives C. The replication functions of polyomavirus large tumor antigen are regulated by phosphorylation. J Virol. 1993;67:6788–6796. doi: 10.1128/jvi.67.11.6788-6796.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang E H, Prives C. DNA helicase and duplex DNA fragment unwinding activities of polyoma and simian virus 40 large T antigens display similarities and differences. J Biol Chem. 1991;266:12668–12675. [PubMed] [Google Scholar]
- 85.Wang E H, Prives C. ATP induces the assembly of polyoma large tumor antigen into hexamers. Virology. 1991;184:399–403. doi: 10.1016/0042-6822(91)90858-9. [DOI] [PubMed] [Google Scholar]
- 86.Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin dependent kinases. Nature. 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- 87.Welch P J, Wang J Y. A C-terminal protein-binding domain in the retinoblastoma protein regulates nuclear c-Abl tyrosine kinase in the cell cycle. Cell. 1993;75:779–790. doi: 10.1016/0092-8674(93)90497-e. [DOI] [PubMed] [Google Scholar]
- 88.Wilcock D, Lane D P. Localization of p53, retinoblastoma, and host replication proteins at sites of viral replication in herpes infected cells. Nature (London) 1991;349:429–431. doi: 10.1038/349429a0. [DOI] [PubMed] [Google Scholar]
- 89.Zalvide J, Stubdal H, DeCaprio J A. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol Cell Biol. 1998;18:1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]