Abstract
Background
Brugada syndrome is a significant cause of sudden cardiac death (SCD), but the underlying mechanisms remain hypothetical.
Objectives
This study aimed to elucidate this knowledge gap through detailed ex vivo human heart studies.
Methods
A heart was obtained from a 15-year-old adolescent boy with normal electrocardiogram who experienced SCD. Postmortem genotyping was performed, and clinical examinations were done on first-degree relatives. The right ventricle was optically mapped, followed by high-field magnetic resonance imaging and histology. Connexin-43 and NaV1.5 were localized by immunofluorescence, and RNA and protein expression levels were studied. HEK-293 cell surface biotinylation assays were performed to examine NaV1.5 trafficking.
Results
A Brugada-related SCD diagnosis was established for the donor because of a SCN5A Brugada-related variant (p.D356N) inherited from his mother, together with a concomitant NKX2.5 variant of unknown significance. Optical mapping demonstrated a localized epicardial region of impaired conduction near the outflow tract, in the absence of repolarization alterations and microstructural defects, leading to conduction blocks and figure-of-8 patterns. NaV1.5 and connexin-43 localizations were normal in this region, consistent with the finding that the p.D356N variant does not affect the trafficking, nor the expression of NaV1.5. Trends of decreased NaV1.5, connexin-43, and desmoglein-2 protein levels were noted; however, the RT-qPCR results suggested that the NKX2-5 variant was unlikely to be involved.
Conclusions
This study demonstrates for the first time that SCD associated with a Brugada-SCN5A variant can be caused by localized functionally, not structurally, impaired conduction.
Key Words: Brugada syndrome, epicardial arrhythmogenic conduction substrate, right ventricle, SCN5A variant, sudden cardiac death
Central Illustration
The Brugada syndrome (BrS) is an inherited disease associated with a distinct electrocardiographic (ECG) phenotype associated with an increased risk of sudden cardiac death (SCD) due to ventricular fibrillation. Its prevalence is estimated at 1:2,000 with a first manifestation at an age of 41 ± 15 years. It represents a leading cause of death in young men, especially in Southeast Asia.1
Clinical studies on BrS patients have found electroanatomical abnormalities in the right ventricular outflow tract (RVOT) manifesting as low-voltage and fragmented electrograms. Their functional implication in SCD is proved by targeted catheter ablation suppressing ventricular fibrillation and normalizing the ECG.2, 3, 4, 5 The reasons for such localized abnormalities are not yet fully elucidated; RVOT-specific ion channels expression profile and microstructural defects are thought to contribute,5, 6, 7, 8, 9 but the current debate relates to the respective roles of depolarization versus repolarization disorders in the development of the BrS phenotype.9, 10, 11
In about 25% of BrS cases, there is a genetic basis involving SCN5A variants encoding for the cardiac sodium voltage-gated channel alpha subunit (NaV1.5). Numerous new genetic markers are continually identified, most of them as variants of uncertain significance,12,13 with genetic interactions being determinant for the extent and pathogenicity of electrophysiological abnormalities, potentially explaining the heterogeneous expression of the disease.4,13, 14, 15, 16, 17 These investigations demonstrate the critical role for the functional characterization of BrS patients, which remains challenging in the clinical context.
Here, we provide the first detailed functional, structural, and biochemical characterization of the impact of a BrS-associated SCN5A variant in a human explanted heart from a 15-year-old patient who experienced SCD.
Methods
More methodological details are provided in the Supplemental Appendix.
Human tissue samples
The study conformed to the Declaration of Helsinki and was approved by the Agence de la Biomédecine (PFS14-004). Ten human hearts were obtained through our organ donation program in collaboration with the University Hospital of Bordeaux from brain-dead patients who had not expressed their opposition to organ donation during their lifetimes, and whose hearts could not be used for transplantation. When possible, clinical examinations such as electrocardiogram (ECG) and echocardiography were performed on patients upon hospital admission. The hearts were flushed and immersed in precooled cardioplegic solution (CELSIOR, Institut Georges Lopez) after their excision before their transfer to the laboratory.
The SCD heart was from a 15-year-old adolescent boy who experienced SCD caused by a suspected ventricular fibrillation in the context of physical activity (warmup phase of rugby training) leading to irreversible neurologic damage and death. The results were compared with those from 9 human control hearts from donors with no cardiac diseases (control hearts 1 to 9); the reasons for the transplantation rejection of these hearts are listed in Supplemental Table 1 and do not constitute a significant limitation to their comparison with the SCD heart. Each control heart could not be used in all types of experiments because of the restricted availability of human tissues and the incompatibility between different experimental approaches on the same heart.
Ex vivo experiments
Functional, structural, and biochemical explorations were carried out: the right ventricle (RV) was optically mapped, followed by high-field magnetic resonance imaging (9.4-T) and histologic analysis. Connexin-43 and NaV1.5 were localized by immunofluorescence imaging. Conduction-related RNA and protein expression levels were studied through real-time qPCR and western blot analyses, and HEK-293 cell surface biotinylation assays were performed to examine NaV1.5 trafficking.
Clinical investigations and genetic analyses
In parallel, clinical and genetic analyses were carried out: 12-lead ECGs were measured in all of the donor’s first-degree relatives (parents, sister, half-brother, and 2 half-sisters), and ajmaline tests (1 mg/kg) were performed for the parents.
Next-generation sequencing was performed after informed consent. Whole genome sequencing of 8 BrS patients was also performed with a tagmentation-based DNA PCR-free library preparation kit; sequencing was performed on a NovaSeq 6000 (Illumina). Fastq files were mapped on the human reference (version hs37d5); the final VCF file containing all the samples was called using join-genotyping. Those steps were performed with the DRAGEN Bio-IT platform (Illumina). Details on annotation, filtering, classification, and pathogenicity prediction of variants are provided in the Supplemental Appendix.
Statistical analysis
Considering the particularity of this study, which compared the pathologic heart obtained from a single patient (SCD, n = 1) with a mean ± SEM of a population of control hearts (n = 4 for optical mapping experiments, n = 3-4 for western blot experiments), classic statistical tests could not be performed.
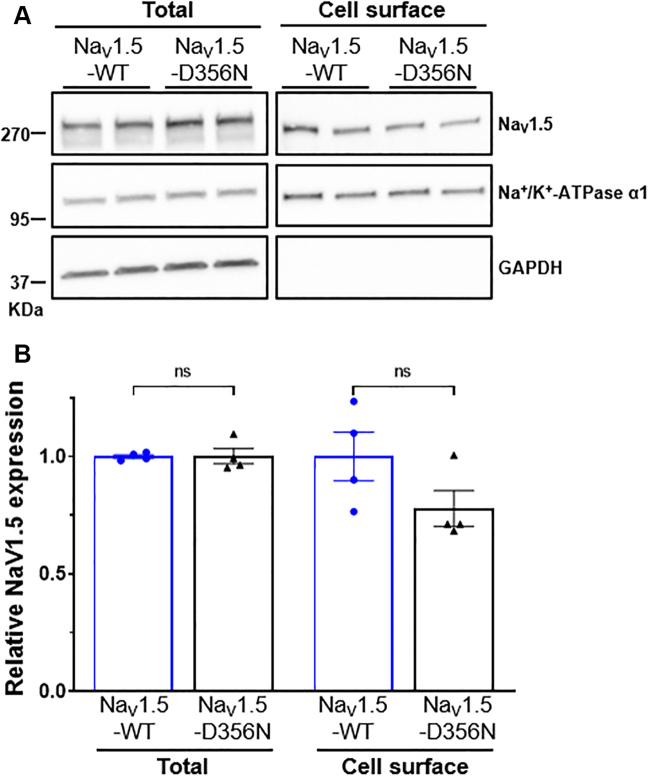
For histologic analyses, the fibrosis percentages of myocardium were represented as mean ± SEM of 8 values (8 images per section) for each region of each individual, allowing us to determine the differences between groups using Kruskall-Wallis tests. For NaV1.5-D356N variant trafficking assessment in HEK-293 cells, the expression of either total or cell surface NaV1.5 was compared between cells transfected with NaV1.5-WT (wild type) + NaVβ1 (n = 4) and those transfected with NaV1.5-D356N + NaVβ1 (n = 4) using Mann-Whitney tests.
The fixed P value was 0.05, and all statistical analyses were performed on Prism software (GraphPad Software).
Results
Clinical phenotype and genetic variations
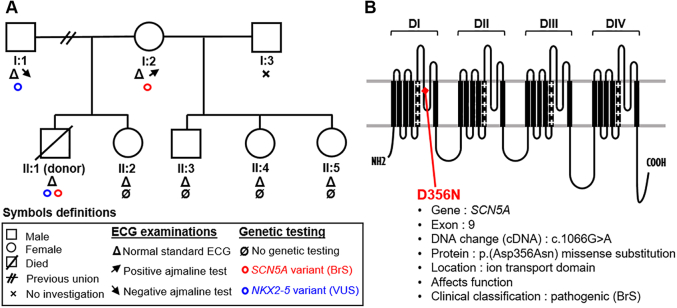
The mechanisms underlying SCD in this case were unexplained at the time of inclusion in our research protocol because the heart donor’s ECG and echocardiography at hospital admission before death were normal, and no cardiovascular antecedents or family history were known (Supplemental Figure 1, Supplemental Table 1). The suspected cause was a rhythm disorder, although no recordings are available to confirm this. Sinus rhythm was recovered after administration of 1 mg adrenaline, with an estimated low-flow period of 15 minutes and no flow of <5 minutes. The patient was then intubated and spent 14 days in the intensive care unit, during which time he showed progressive neurologic deterioration leading to encephalic death. However, postmortem panel sequencing of the donor identified rare variants in SCN5A and NKX2-5 genes (Supplemental Tables 2 and 3; case 1). The p.D356N variant is a pathogenic and BrS-associated single-nucleotide variation (c.1066G>A) in exon 9 of SCN5A, causing a missense substitution (p.Asp356Asn), which leads to a loss of function of the ion transport domain of NaV1.5.18 The variant in the gene encoding the Nkx2-5 transcription factor is a variant of unknown significance reported as NM_004387.3(NKX2.5):c.595G>C(p.Asp199His). The mother of the victim carried the same SCN5A variant, and the NKX2-5 variant was found in his father. Twelve-lead ECGs of the donor’s first-degree relatives were normal. Sodium blocker (ajmaline) test results were negative for the father but positive for BrS for the mother (Figure 1).
Figure 1.
Clinical and Genotyping Data of Heart Donor and His Family
(A) SCD heart donor’s family tree summarizing ECG and genetic testing. (B) Location of the p.D356N variant in the ion transport domain of NaV1.5. ECG = electrocardiogram; SCD = sudden cardiac death.
In addition, 8 additional individuals carrying the SCN5A c.1066G>A disease-causing variant (Supplemental Tables 2 and 3) were identified and sequenced. Filtering for additional rare variants among 109 genes including the cardiomyopathy-arrhythmia gene panel were applied. This screening revealed 11 rare variants in TTN, DSP, FLNC, NEXN, JUP, PRDM16, HCN4, SCN1B, CDH2, RYR2, and SLC8A1, considered as variants of unknown significance or likely benign by the American College of Medical Genetics classification.
Baseline electrophysiological properties of the RV
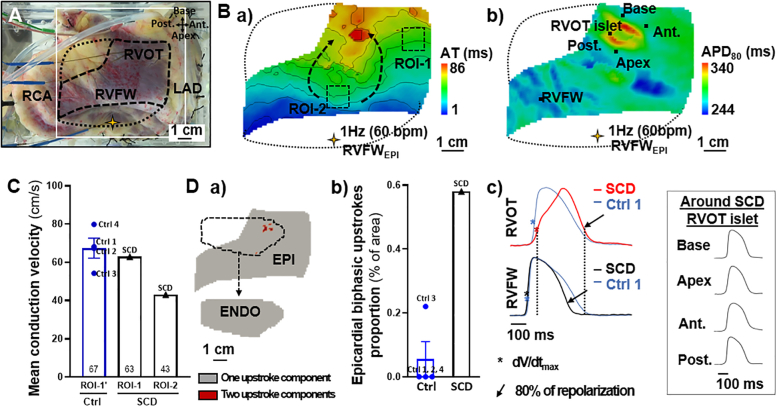
Upon stimulation of the RV preparation at 1 Hz (60 beats/min), optical mapping showed a distinct area of slow conduction in the epicardial anterior RV (Figure 2B.a, dotted arrows). In this region (ROI-2), the local mean conduction velocity was decreased by a third compared with ROI-1 (43.0 vs 62.9 cm/s) and those at similar locations in control human hearts (ROI-1’; 67.4 ± 5.2 cm/s) (Figure 2C, Supplemental Figure 2). The transmural conduction velocities in the RVOT and right ventricular free wall (RVFW) were similar (20.6 vs 23.1 cm/s; data not shown).
Figure 2.
Right Ventricular Conduction and Upstroke Abnormalities of the SCD Heart
(A) Epicardial view of the RV preparation (yellow star: stimulation; open square: recorded optical window; solid dotted line: noise-free optical signals mask). (B) Epicardial a) activation time and b) APD80 maps. (C) Epicardial mean conduction velocities in a “normal” conduction area (ROI-1) compared with an “impaired” conduction area (ROI-2). ROI-1′ is equivalent to the ROI-1 in control hearts (ROIs localizations in B.a and Supplemental Figure 2). (D) a) Binary maps for optical upstroke morphology from SCD heart (epicardial and endocardial masks detailed in Supplemental Figure 3), and b) epicardial biphasic upstroke proportions in SCD and control hearts.; c) optical action potential traces from SCD and control 1 hearts’ RVFW and RVOT (other control hearts shown in Supplemental Figure 2), and from areas bordering the SCD RVOT islet (pixels indicated in B.b). Data presented for 1 Hz (60 beats/min) pacing in epicardial RVFW (RVFWEPI). Data from control hearts (n = 4) are presented as mean ± SEM.
Ant = anterior; AT = activation time; EPI = epicardial; ENDO = endocardial; LAD = left anterior descending artery; Post = posterior; RCA = right coronary artery; ROI = region of interest; RVFWEPI = pacing site in epicardial right ventricular free wall.
The conduction delay in ROI-2 led to a delayed activation of the epicardial RVOT with a localized “islet” that was associated with an area of prolonged action potential duration at 80% of repolarization (APD80) (Figure 2B.b), caused by a biphasic morphology of the upstrokes of optical action potential appearing in red on the binary maps (Figure 2D.a) and action potential traces (Figure 2D.c). Such morphologies were absent from the remaining epicardium and from the endocardium (Figure 2D.a, Supplemental Figure 3). The surface area with epicardial biphasic upstrokes (0.58%) was significantly larger in the SCD heart than in the control hearts (Figure 2D.b).
Rate dependence of the epicardial conduction properties
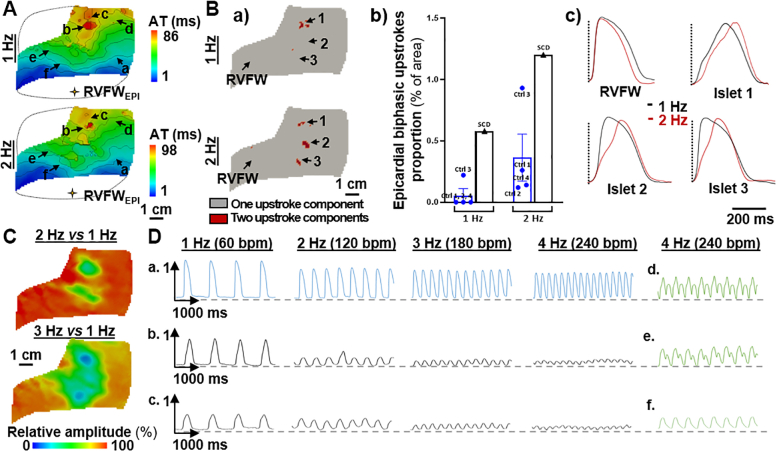
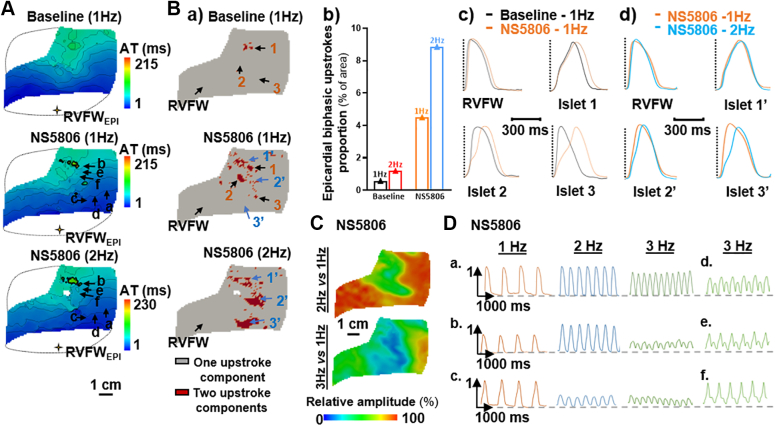
Increasing the pacing frequency had a significant impact on conduction properties of the SCD RV (Figure 3). At 2 Hz (120 beats/min) we observed a prolongation of total activation time (98 ms versus 86 ms at 1 Hz) (Figure 3A) associated with a prolongation of the biphasic upstrokes in the RVOT islet (islet 1) as well as the emergence of new regions with such abnormal upstrokes (islets 2 and 3). The surface area of pixels showing biphasic upstrokes was doubled and remained higher than in control hearts (Figure 3B).
Figure 3.
Impact of Increasing Pacing Frequency
RV conduction parameters in the SCD heart in response to epicardial RVFW pacing at 1, 2, 3, and 4 Hz. (A) Activation time maps. (B) a) Binary maps for optical upstroke morphology with b) corresponding biphasic upstroke proportions for SCD and control hearts (1 and 2 Hz only; maps of control hearts are available in Supplemental Figure 2) and c) averaged action potential traces from 4 areas. (C) Relative optical signal amplitude maps and (D) amplitude variations of nonaveraged optical action potential traces from different pixels (localizations in A). Data from control hearts (n = 4) are presented as mean ± SEM. Abbreviations as in Figure 2.
Important variations in optical action potential amplitude were additionally observed: the signal amplitude decreased at 2 Hz pacing (120 beats/min) in the abnormal RVOT islet by a factor >2, and the low-amplitude area further widened as the frequency increased (Figure 3C). This effect can be seen qualitatively on the action potential traces (Figure 3D): the signal amplitude showed only a modest decrease in a “normal” site a, whereas in initially slower conduction sites b and c, it decreased dramatically as the pacing frequency increased. At 4 Hz pacing (240 beats/min), amplitude alternans was observed in areas d, e, and f with previously observed slower conduction (ROI-2) (Figure 2B.a), and an epicardial area (close to ROI-2) showed 2:1 conduction failure under stimulation (f in Figure 3, Supplemental Figure 4), resulting in figure-of-eight activation patterns, although they did not result in sustained re-entries (Supplemental Figure 4). Strikingly these abnormalities were not observed on the endocardium.
Impact of Ito activation
Pharmacologic activation of Ito under NS5806 perfusion led to an increase in epicardial total activation time (86 vs 215 ms) (Figure 4A) and in biphasic upstrokes surface area (0.58 vs 4.50%) (Figure 4B). The frequency effect on the spatial extent of areas showing biphasic upstrokes persisted under NS5806 perfusion (Figure 4B). Activation of Ito currents also led to an increased reduction in amplitude at higher frequencies and to the appearance of amplitude alternans (3 Hz, 180 beats/min) (Figures 3C, 3D, 4C, and 4D).
Figure 4.
Impact of Ito Current Activation
Epicardial conduction parameters in baseline versus NS5806 perfusion (10 µM) when pacing in epicardial RVFW at 1 Hz, and impact of increasing pacing frequency under NS5806 perfusion (1, 2, and 3 Hz pacing). (A) Activation time maps. (B) a) Binary maps of optical upstroke morphology with b) associated epicardial biphasic upstroke proportions; averaged optical action potential traces from different regions; localizations in B.a are represented in c) for baseline versus NS5806 (1 Hz) and in d) for NS5806 1 Hz versus 2 Hz. (C) Relative optical signal amplitude maps and (D) amplitude variations of nonaveraged optical action potential traces from different regions at 1, 2, and 3 Hz under NS5806 perfusion (localizations of pixels in A). Abbreviations as in Figure 2.
Interestingly, applying long pauses in the stimulation sequence (≤2.5 s) in baseline conditions, which allows for full reactivation of Ito currents, had little effect on activation time and upstroke morphologies (Supplemental Figure 5).
Tissue microstructure
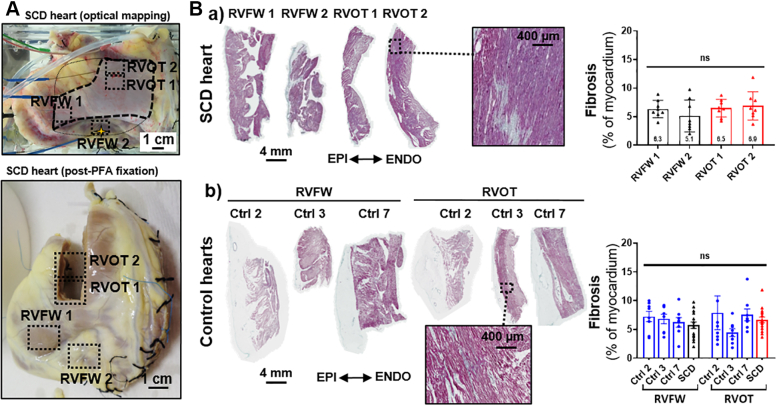
High-field magnetic resonance imaging did not show any structural abnormalities or fatty infiltrations in the anterior RV regions that showed conduction delays and biphasic upstrokes (Supplemental Figure 6). The quantification of collagen content in histologic analyses showed no significant difference in fibrosis levels between the impaired conduction islet sections and those from other areas (Figure 5B.a). In addition, fibrosis levels were similar to those found in both RVOT and RVFW sections of control hearts (Figure 5B.b).
Figure 5.
Histologic Examination of Fibrosis Levels
(A) RV sampling from the SCD heart in relation to optical mapping results (RVOT 1 and RVOT 2 were collected in RVOT biphasic upstrokes islet and RVFW 1 and RVFW 2 in areas without conduction abnormalities; dashed squares). (B) Masson’s trichrome stained transmural sections (6 μm) from a) SCD heart and b) 3 control hearts (for control hearts, 1 sample from each RVOT and RVFW) and associated quantification of fibrosis as percentage of myocardium (each section divided in 8 areas, so n = 8 for each area of each individual; mean ± SEM; ns indicates a nonsignificant difference; fixed P = 0.05; Kruskall-Wallis test followed by the Dunn’s posthoc test). Abbreviations as in Figure 2.
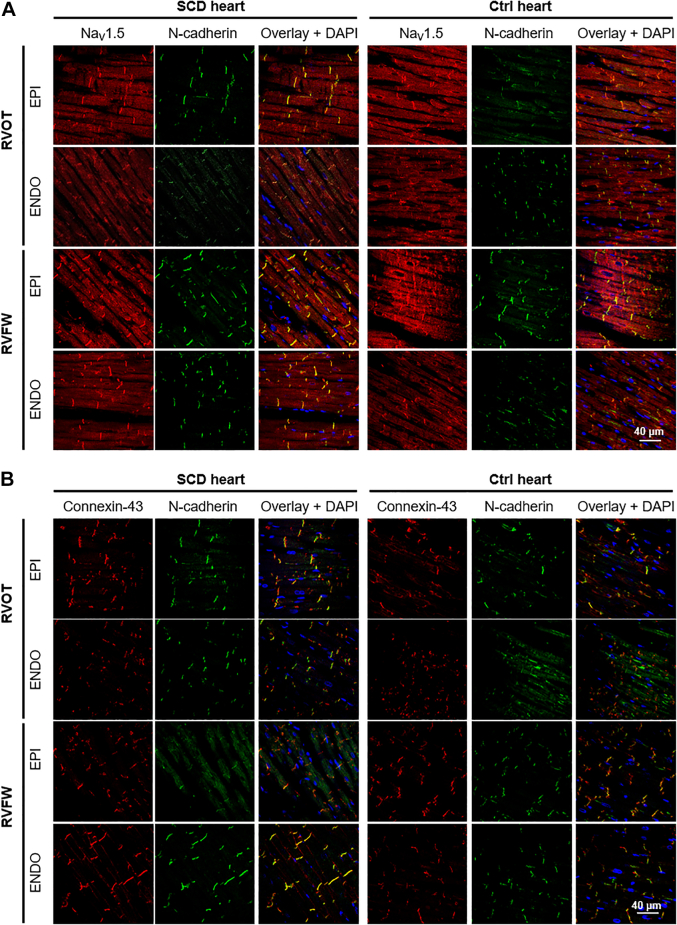
Subcellular localization of NaV1.5, Connexin-43, and N-cadherin
In Figure 6, the immunofluorescent labeling of tissue sections from different epicardial or endocardial regions of the SCD and control hearts’ RV demonstrated that NaV1.5, connexin-43, and N-cadherin were correctly localized at the intercalated discs, even in regions of impaired conduction identified during optical mapping experiments (RVOT EPI). NaV1.5 was also found at the lateral membrane in transverse tubules.
Figure 6.
Subcellular Localization of NaV1.5 and Connexin-43 in SCD Heart
Representative confocal fluorescent images (objective ×20, oil immersion) of 6-μm-thick longitudinal sections of myocardial paraffin pre-embedded blocks from RVOT epicardium (EPI), RVOT endocardium (ENDO); RVFW EPI and RVFW ENDO of SCD and control 3 hearts immunolabeled for (A) NaV1.5 and N-cadherin or for (B) connexin-43 and N-cadherin. Abbreviations as in Figure 2.
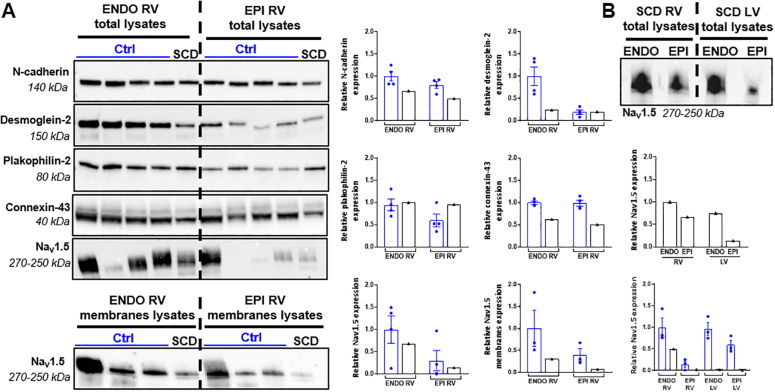
Changes in conduction-related proteins and gene expression levels
Western blot analyses showed that the endocardial desmoglein-2 and epicardial and endocardial connexin-43 and N-cadherin were clearly underexpressed in the RV of the SCD heart compared with the control hearts (Figure 7A), whereas the differences are smaller in the left ventricle (Supplemental Figure 7). In addition, the expression level of NaV1.5 was lower in the right and left ventricles (in both total or membranes lysates). Similar to control hearts, a gradient of NaV1.5 expression was detected from the epicardium to the endocardium in the SCD heart (Figure 7B).
Figure 7.
Cardiac Conduction Related–Protein Expression in Tissue Samples
(A) Relative expression of N-cadherin, desmoglein-2, plakophilin-2, connexin-43, and NaV1.5 in SCD and control hearts endocardial and epicardial RV total and membrane lysates. (B) NaV1.5 relative expression in RV versus LV ENDO and EPI total lysates of SCD (black) and control (blue) hearts. Relative protein expression was normalized by stain free technology. Data from control hearts (n = 3 to 4) are presented as mean ± SEM. LV = left ventricular; other abbreviations as in Figure 2.
SCN5A transcript levels of the SCD endocardial RV samples were lower than those measured in both control hearts; however, they were considerably higher in the other regions (≤4 times in the endocardial left ventricle) (Figure 8A). Important trends of increased RNA levels (SCD vs control hearts) were also found in epicardial RV and endocardial left ventricle for CDH2, DSG2, PKP2, GAJ1, and NKX2.5 and in epicardial left ventricle only for DSG2, GAJ1, and NKX2.5 (Supplemental Figure 8).
Figure 8.
Total and Cell Surface Expression of NaV1.5-D356N in HEK-293 Cells
(A) Representative western blots of total and cell surface NaV1.5 from HEK-293 cells transfected with NaV1.5-WT (wild type) + NaVβ1 or NaV1.5-D356N + NaVβ1. Samples were probed in parallel with the anti–Na+/K+-ATPase α1 and anti-GAPDH (glyceraldehyde 3-phosphate dehydrogenase) antibodies. (B) Quantitative data (mean ± SEM) of total and cell surface NaV1.5 protein relative expression (n = 4 in 2 different experiments). The expression of NaV1.5 in each sample was first normalized to the Na+/K+-ATPase α1 in the same blot and then expressed relative to NaV1.5 protein expression (total or cell surface) in cells transfected with NaV1.5-WT + NaVβ1 (ns indicates nonsignificant differences; Mann-Whitney tests with fixed P = 0.05).
Impact of the SCN5A c.1066G>A disease-causing variant on NaV1.5 channel trafficking
Surface biotinylation experiments in HEK-293 cells revealed no differences in the expression of NaV1.5 both at the cell and in total cell lysates between cells expressing the D356N mutant and the WT channels (Figure 8).
Discussion
A localized epicardial and purely functional arrhythmogenic conduction substrate
This study is the first to investigate the functional mechanisms associated with a SCN5A disease-causing variant in human cardiac tissues in the context of BrS. Detailed optical mapping of the RV allowed us to identify specific epicardial regions showing abnormal conduction and altered optical upstroke morphologies (Figure 2). These conduction abnormalities were rate dependent and increased with pacing frequency, leading to large epicardial areas showing abnormal upstroke morphologies and amplitude alternans at higher rates (Figure 3). Interestingly, high-resolution cardiac magnetic resonance and histologic analysis did not reveal any microstructural abnormalities in these specific regions (Figure 5, Supplemental Figure 6). Our results therefore indicate that localized functional conduction alterations were the likely cause of arrhythmogenesis in this case.
Several studies have previously highlighted the presence of localized RV epicardial abnormalities in BrS patients and idiopathic ventricular fibrillation survivors, manifested by distinct areas of abnormal signals and fibrotic tissue.2, 3, 4, 5,19 The precise mechanisms underlying these specific features in BrS, and more widely in J-wave syndromes, are still uncertain; they have been attributed to slow and tortuous conduction as the consequence of a structural defect,6, 7, 8, 9,12 whereas experimental studies have highlighted a role for repolarization heterogeneity and regions with action potentials showing prominent phase 1 repolarization or abbreviated duration.20,21
In our experiments, we observed an epicardial region of slowed conduction and areas displaying biphasic upstroke morphologies (Figure 2). These abnormal optical upstrokes could in principle be associated with either conduction or repolarization abnormalities: spatial averaging effects of optical signals may produce biphasic upstrokes through slow conduction layers in subepicardium, as described previously,22,23 or near epicardial sites of conduction blocks and colliding wave fronts. By contrast, a prominent phase 1 repolarization could also result in altered upstrokes, with a loss of the overshoot and delay in reaching peak fluorescence during the plateau phase. Our experiments provide 2 key observations in favor of conduction abnormalities. First, the abnormal upstroke morphologies are enhanced (both in amplitude and in spatial extent) at increasing pacing frequencies (Figure 3B), which is inconsistent with the phase 1 repolarization hypothesis, inasmuch as Ito, the main current responsible for this phase, is gradually inactivated at higher pacing rates. Second, the drop in signal amplitude at high frequencies (Figures 3C and 3D) indicates reduced electrical activity or, in other words, partial conduction failure in subepicardial layers, which ultimately results in conduction alternans manifesting as amplitude alternans. Our results obtained with the Ito activator (Figure 4) indicate that this current may play a role at rest, but that in this case it merely further aggravated the conduction disorder that was already present at baseline, by opposing already impaired sodium currents.
Implications of the Brugada-associated SCN5A variant
Our observations are in line with the genotyping (Figure 1, Supplemental Table 2), which revealed a c.1066G>A variant in SCN5A causing a pathogenic missense substitution (p.D356N) that affected the ion transport domain of NaV1.5 and had been previously identified in BrS and progressive cardiac conduction disorder patients, including young people (<16 years).18,24,25 A biophysical assay in HEK293 cells had already revealed, before this study, that the p.D356N variant results in total NaV1.5 loss of function.18 At the tissular level, it may thus be associated with strong conduction disturbances prone to appear first in the RVOT epicardium because of its lower conduction reserve (physiological low expression of NaV1.5) compared with other ventricular regions,8,9 as also confirmed by our western blotting (Figure 7, Supplemental Figure 7). This functional conduction heterogeneity, which led to figure-of-eight activation patterns at high frequencies (Supplemental Figure 4), could thus provide the substrate for arrhythmias in this adolescent boy who experienced cardiac arrest in a context of physical activity.
Consistently with the correct subcellular localization of NaV1.5 in the SCD RV (Figure 6), the cell surface biotinylation assays indicate that the c.1066G>A disease-causing variant in SCN5A does not affect the trafficking, nor the expression and stability of the sodium channels in HEK-293 cells (Figure 8). However, the results of our biochemical experiments from tissue samples suggest trends to reduction in NaV1.5 expression compared with control hearts in both total and membrane lysates (Figure 7, Supplemental Figure 7), which could be caused by perturbations during transcription in the endocardial RV, where SCN5A RNA levels have also been found lower than in control hearts (Supplemental Figure 8), but it seems to be more due to post-transcriptional disturbances in the other regions where RNA levels are instead higher than in control hearts. Whatever its cause, the NaV1.5 under expression could contribute to a synergistic effect, together with the sodium current reduction, more readily leading to the depletion of the low conduction reserves, first in the epicardial RVOT, providing the arrhythmogenic substrate.
Possible implications of the variant of uncertain significance in NKX2-5
A variant of unknown significance in NKX2-5 was detected in the SCD donor and his father, who has a normal phenotype with a negative ajmaline test (Figure 1, Supplemental Table 2). Although not reported before, its location close to the homeobox (Asp199His) could have an additional impact because the transcription factor Nkx2-5 is a key regulator of cardiac development.26 Interestingly, adult Nkx2-5+/- mice display major anatomical and functional disturbances in the His-Purkinje system.27 Although these disorders cannot elucidate our observations from the RVOT epicardium (Figures 2, 3, and 4), it has been shown that Nkx2-5 null cardiomyocytes have abnormal physiology with asynchronous contractions, prolonged APD, and slowed initial upstroke velocity,28 and that Nkx2-5 regulates the expression of several conduction-related genes, like those coding for NaV1.5 or connexin-43, which were underexpressed in NKX2-5 mutant models.29,30 Our RT-qPCR showed trends to overexpression of NKX2-5 gene in the epicardial RV and epicardial and endocardial left ventricular samples from the SCD patient (probable consequence of the NKX2-5 variant), which could explain why the RNAs of all the conduction-related genes studied trended to be present in increased amounts in these regions of the SCD heart (Supplemental Figure 8). Nevertheless, this upregulation of genes transcription could not explain the observed decrease in protein levels of NaV1.5, connexin-43, N-cadherin, and desmoglein-2 (Figure 7, Supplemental Figure 7), which suggests that posttranscriptional downregulations may be responsible. Some of these could result from the SCN5A disease-causing variant, inasmuch as it is known that the sodium channel is strongly coregulated with these 3 proteins (and others) in the intercalated-disc microdomaines.31 Despite this limited information, and given that the impact of the NKX2-5 c.595G>C variant on the transcription factor function has never been assessed, we cannot fully exclude a synergistic effect between the NKX2-5 and SCN5A variants in this specific case, even though the genotyping and the ajmaline test results in the parents would be in favor of a more prominent role of the SCN5A variant in arrhythmogenesis.
Role of SCN5A variants in severity of BrS and SCD risk stratification
These results are consistent with recent studies that have shown the major role of SCN5A variants in BrS pathogenesis and SCD risk stratification.4,13,14,17,24,32 In our study, 2 family members (mother and son) carrying the same disease-causing SCN5A variant presented different degrees of arrhythmogenicity, indicating that SCN5A variants are probably not the sole determinant of disease severity. It is well known that BrS has a higher incidence rate and severity in male individuals, probably caused by a limited depolarization reserve of male cells resulting from their high Ito density versus female individuals33 and not from the effects of the sex hormones on NaV1.5.34 Nevertheless, there is probably a complex interplay between sex, age, genetic factors, and other precipitating or modulating factors like exercise, body temperature and its impact on sodium channels,35 and sympathovagal unbalance,36 which could all apply to our case. In addition, common genetic variations could induce variable severity of BrS in these 2 individuals,13,15,16,32 and we cannot exclude a synergistic effect of the NKX2.5 variant of uncertain significance in the SCD victim, which was not found in his mother.
Study limitations
This study is subject to limitations. 1) We cannot confirm that the young donor had a BrS phenotype, because only few ECGs were obtained at hospital admission before his death, and no sodium-blocker testing was performed during his lifetime. However, the fact that the same variant was detected in his mother, who also had a normal baseline ECG but a positive sodium-blocker test result, strongly supports an arrhythmic SCD associated with BrS, in the presence of a known SCN5A mutant. This variant was detected in 8 BrS patients followed up in our centers, providing an additional argument in favor of BrS-related SCD, here likely at an early stage. 2) The experiments were performed in ex vivo perfused human hearts, which is different from in vivo conditions. This limitation is mitigated by the fact that optical mapping allows a much more detailed and spatially resolved electrophysiological characterization than is currently feasible in clinical investigations. 3) The genotyping results were unknown at the time of the functional ex vivo characterization, which prevented us from designing a more patient-specific protocol. 4) The availability of human hearts for ex vivo functional characterization from donors having experienced SCD is exceptional. Although our results are based on a single case, we believe they provide unique advancements in our mechanistic insights into this cause of SCD in the young. 5) The control hearts used in this study were not strictly healthy hearts because they were rejected for transplantation; however, the reasons for nontransplantation (age or hypokinesia) are in our view unlikely to have significantly affected the conclusions of our study.
Conclusions
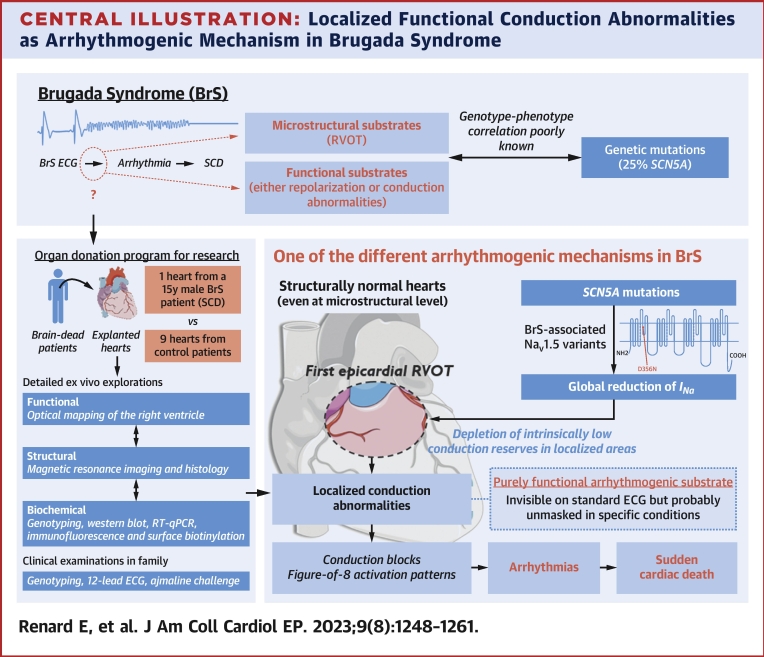
In line with recent studies that have demonstrated the importance of a better phenotype-genotype correlation to improve the prediction of lethal events in BrS patients,3,14 we have for the first time performed an extensive characterization of the functional, structural, and biochemical impacts of a p.D356N variant of the SCN5A, a gene with pleomorphic clinical impact. Our data demonstrate that loss-of-function sodium channel disease–causing variants will lead to impaired conduction in localized regions with intrinsic low conduction reserve and can cause SCD even in the absence of microstructural abnormalities (Central Illustration).
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: Medical knowledge in BrS as well as SCN5A mutations: BrS can be a purely functional disorder without scar or fibrotic substrate.
TRANSLATIONAL OUTLOOK: Subtle conduction disturbances may be the cause of sudden death in the young. Mapping protocol should be optimized to identify localized functional conduction dysfunction.
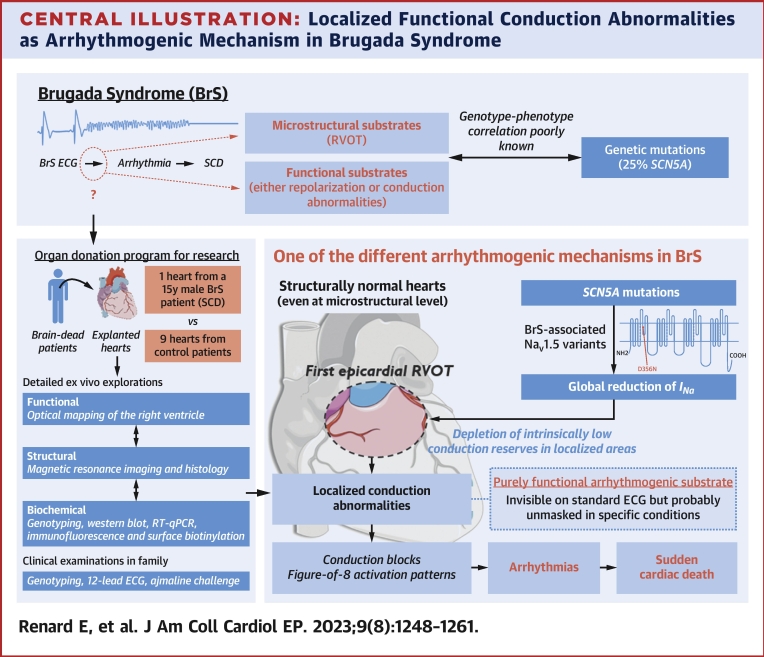
Central Illustration.
Localized Functional Conduction Abnormalities as Arrhythmogenic Mechanism in Brugada Syndrome
Aiming to help understand the arrhythmogenic mechanisms of Brugada syndrome (BrS), this study provides detailed ex vivo functional, structural, and biochemical explorations of a human heart with a BrS-associated SCN5A variant. The functional consequences of this variant were optically mapped in a human heart, showing for the first time that the arrhythmogenic substrate can be of purely functional origin and involve localized conduction abnormalities prone to appear in the epicardial right ventricular outflow tract.
INa = sodium current; NaV1.5 = sodium channel protein type 5 subunit alpha; RVOT = right ventricular outflow tract; SCD = sudden cardiac death; SCN5A = NaV1.5-encoding gene.
Funding Support and Author Disclosures
This work received financial support from the French Government as part of the “Investments of the Future” program managed by the National Research Agency (ANR-10-IAHU04-LIRYC), the Leducq-Foundation (RHYTHM network, 16CVD02), and the Fondation Coeur et Artères (FC17T2). Dr Barc is supported by the ANR JCJC LEARN (R21006NN, RPV21014NNA). Dr Schott is supported by IRP-, an I-SITE NExT health and engineering initiative (Ecole Centrale & Nantes University) and by the IRP- GAINES funded by INSERM and CNRS. Dr Marionneau is supported by the ANR-16-CE92-0013-01 and the National Institutes of Health (R01-HL148803). All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
The authors thank all the researchers and clinicians involved in our organ donation program, Bruce Smaill for providing the NaV1.5 antibody, the Genomics and Bioinformatics Core Facility of Nantes GenoBiRD (ANR-11-INBS-0013) for the use of its resources and for its technical support, the Biological Resource Centre for biobanking (CHU Nantes, Nantes Université, Centre de ressources biologiques [BB-0033-00040]), and Valentin Crusson for his technical support.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental figures, tables, and an expanded Methods section, please see the online version of this paper.
Appendix
References
- 1.Brugada J., Campuzano O., Arbelo E., Sarquella-Brugada G., Brugada R. Present status of Brugada syndrome: JACC state-of-the-art review. J Am Coll Cardiol. 2018;72(9):1046–1059. doi: 10.1016/j.jacc.2018.06.037. [DOI] [PubMed] [Google Scholar]
- 2.Ten Sande J.N., Coronel R., Conrath C.E., Driessen A.H.G., de Groot J.R., Tan H.L., et al. ST-segment elevation and fractionated electrograms in Brugada syndrome patients arise from the same structurally abnormal subepicardial RVOT area but have a different mechanism. Circ Arrhythm Electrophysiol. 2015;8(6) doi: 10.1161/CIRCEP.115.003366. [DOI] [PubMed] [Google Scholar]
- 3.Nademanee K., Veerakul G., Chandanamattha P., Chaothawee L., Ariyachaipanich A., Jirasiriojanakorn K., et al. Prevention of ventricular fibrillation episodes in Brugada syndrome by catheter ablation over the anterior right ventricular outflow tract epicardium. Circulation. 2011;123(12):1270–12794. doi: 10.1161/CIRCULATIONAHA.110.972612. [DOI] [PubMed] [Google Scholar]
- 4.Ciconte G., Monasky M.M., Santinelli V., Micaglio E., Vicedomini G., Anastasia L., et al. Brugada syndrome genetics is associated with phenotype severity. Eur Heart J. 2021;42(11):1082–1090. doi: 10.1093/eurheartj/ehaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pieroni M., Notarstefano P., Oliva A., Campuzano O., Santangeli P., Coll M., et al. Electroanatomic and pathologic right ventricular outflow tract abnormalities in patients with Brugada syndrome. J Am Coll Cardiol. 2018;72 doi: 10.1016/j.jacc.2018.09.037. [DOI] [PubMed] [Google Scholar]
- 6.Nademanee K., Raju H., de Noronha S.V., Papadakis M., Robinson L., Rothery S., et al. Fibrosis, connexin-43, and conduction abnormalities in the Brugada syndrome. J Am Coll Cardiol. 2015;66(18):1976–1986. doi: 10.1016/j.jacc.2015.08.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coronel R., Casini S., Koopmann T.T., Wilms-Schopman F.J.G., Verkerk A.O., de Groot J.R., et al. Right ventricular fibrosis and conduction delay in a patient with clinical signs of Brugada syndrome. Circulation. 2005;112(18):2769–2777. doi: 10.1161/CIRCULATIONAHA.105.532614. [DOI] [PubMed] [Google Scholar]
- 8.Boukens B.J., Sylva M., de Gier-de Vries C., Remme C.A., Bezzina C.R., Christoffels V.M., et al. Reduced sodium channel function unmasks residual embryonic slow conduction in the adult right ventricular outflow tract. Circ Res. 2013;113(2):137–141. doi: 10.1161/CIRCRESAHA.113.301565. [DOI] [PubMed] [Google Scholar]
- 9.Behr E.R., Ben-Haim Y., Ackerman M.J., Krahn A.D., Wilde A.A.M. Brugada syndrome and reduced right ventricular outflow tract conduction reserve: a final common pathway? Eur Heart J. 2021;42(11):1073–1181. doi: 10.1093/eurheartj/ehaa1051. [DOI] [PubMed] [Google Scholar]
- 10.Haïssaguerre M., Nademanee K., Hocini M., Cheniti G., Duchateau J., Frontera A., et al. Depolarization versus repolarization abnormality underlying inferolateral J-wave syndromes: new concepts in sudden cardiac death with apparently normal hearts. Heart Rhythm. 2018;16:781–790. doi: 10.1016/j.hrthm.2018.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilde A.A.M., Postema P.G., Di Diego J.M., Viskin S., Morita H., Fish J.M., et al. The pathophysiological mechanism underlying Brugada syndrome: depolarization versus repolarization. J Mol Cell Cardiol. 2010;49:543–553. doi: 10.1016/j.yjmcc.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Priori S.G., Napolitano C. J-wave syndromes: electrocardiographic and clinical aspects. Card Electrophysiol Clin. 2018;10:355–369. doi: 10.1016/j.ccep.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Campuzano O., Sarquella-Brugada G., Cesar S., Arbelo E., Brugada J., Brugada R. Update on genetic basis of Brugada syndrome: monogenic, polygenic or oligogenic? Int J Mil Ssi. 2020;21(19):7155. doi: 10.3390/ijms21197155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa T., Kimoto H., Mishima H., Yamagata K., Ogata S., Aizawa Y., et al. Functionally validated SCN5A variants allow interpretation of pathogenicity and prediction of lethal events in Brugada syndrome. Eur Heart J. 2021;42(29):2854–2863. doi: 10.1093/eurheartj/ehab254. [DOI] [PubMed] [Google Scholar]
- 15.Barc J., Tadros R., Glinge C., Chiang D.Y., Jouni M., Simonet F., et al. Genome-wide association analyses identify new Brugada syndrome risk loci and highlight a new mechanism of sodium channel regulation in disease susceptibility. Nat Genet. 2022;54:232–239. doi: 10.1038/s41588-021-01007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bezzina C.R., Barc J., Mizusawa Y., Remme C.A., Gourraud J.B., Simonet F., et al. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet. 2013;45:1044–1049. doi: 10.1038/ng.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Postema P.G., Walsh R., Bezzina C.R. Illuminating the path from genetics to clinical outcome in Brugada syndrome. Eur Heart J. 2021;42(11):1091–1093. doi: 10.1093/eurheartj/ehaa994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makiyama T., Akao M., Tsuji K., Doi T., Ohno S., Takenaka K., et al. High risk for bradyarrhythmic complications in patients with Brugada syndrome caused by SCN5A gene mutations. J Am Coll Cardiol. 2005;46(11):2100–2106. doi: 10.1016/j.jacc.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 19.Haïssaguerre M., Hocini M., Cheniti G., Duchateau J., Sacher F., Puyo S., et al. Localized structural alterations underlying a subset of unexplained sudden cardiac death. Circ Arrhythm Electrophysiol. 2018;11(7) doi: 10.1161/CIRCEP.117.006120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szél T., Antzelevitch C. Abnormal repolarization as the basis for late potentials and fractionated electrograms recorded from epicardium in experimental models of Brugada syndrome. J Am Coll Cardiol. 2014;63(19):2037–2045. doi: 10.1016/j.jacc.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patocskai B., Yoon N., Antzelevitch C. Mechanisms underlying epicardial radiofrequency ablation to suppress arrhythmogenesis in experimental models of Brugada syndrome. J Am Col Cardiol EP. 2017;3(4):353–363. doi: 10.1016/j.jacep.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pertsov A., Walton R.D., Bernus O. Optical imaging of cardiac action potential. Adv Exp Med Biol. 2015;859:299–i311. doi: 10.1007/978-3-319-17641-3_12. [DOI] [PubMed] [Google Scholar]
- 23.Efimov I.R., Mazgalev T.N. High-resolution, three-dimensional fluorescent imaging reveals multilayer conduction pattern in the atrioventricular node. Circulation. 1998;98(1):54–57. doi: 10.1161/01.cir.98.1.54. [DOI] [PubMed] [Google Scholar]
- 24.Baruteau A.E., Kyndt F., Behr E.R., Vink A.S., Lachaud M., Joong A., et al. SCN5A mutations in 442 neonates and children: genotype-phenotype correlation and identification of higher-risk subgroups. Eur Heart J. 2018;39(31) doi: 10.1093/eurheartj/ehy412. [DOI] [PubMed] [Google Scholar]
- 25.Chockalingam P., Clur S.A.B., Breur J.M.P.J., Kriebel T., Paul T., Rammeloo L.A., et al. The diagnostic and therapeutic aspects of loss-of-function cardiac sodium channelopathies in children. Heart Rhythm. 2012;9(12):1986–1992. doi: 10.1016/j.hrthm.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Schott J.J., Benson D.W., Basson C.T., Pease W., Silberbach G.M., Moak J.P., et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281(5373):108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 27.Choquet C., Kelly R.G., Miquerol L. Nkx2-5 defines distinct scaffold and recruitment phases during formation of the murine cardiac Purkinje fiber network. Nat Commun. 2020;11(1):5300. doi: 10.1038/s41467-020-19150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson D.J., Kaplan D.I., Bell K.M., Koutsis K., Haynes J.M., Mills R.J., et al. NKX2-5 regulates human cardiomyogenesis via a HEY2 dependent transcriptional network. Nat Commun. 2018;9(1):1373. doi: 10.1038/s41467-018-03714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briggs L.E., Takeda M., Cuadra A.E., Wakimoto H., Marks M.H., Walker A.J., et al. Perinatal loss of Nkx2-5 results in rapid conduction and contraction defects. Circ Res. 2008;103(6):580–590. doi: 10.1161/CIRCRESAHA.108.171835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teunissen B.E.J., Jansen A.T., van Amersfoorth S.C.M., O’Brien T.X., Jongsma H.J., Bierhuizen M.F.A. Analysis of the rat connexin 43 proximal promoter in neonatal cardiomyocytes. Gene. 2003;322:123–136. doi: 10.1016/j.gene.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 31.Rivaud M.R., Delmar M., Remme C.A. Heritable arrhythmia syndromes associated with abnormal cardiac sodium channel function: ionic and non-ionic mechanisms. Cardiovasc Res. 2020;116(9):1557–1570. doi: 10.1093/cvr/cvaa082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wijeyeratne Y.D., Tanck M.W., Mizusawa Y., Batchvarov V., Barc J., Crotti L., et al. SCN5A mutation type and a genetic risk score associate variably with Brugada syndrome phenotype in SCN5A families. Circ Genom Precis Med. 2020;13(6) doi: 10.1161/CIRCGEN.120.002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Diego J.M., Cordeiro J.M., Goodrow R.J., Fish J.M., Zygmunt A.C., Pérez G.J., et al. Ionic and cellular basis for the predominance of the Brugada syndrome phenotype in males. Circulation. 2002;106(15):2004–2011. doi: 10.1161/01.cir.0000032002.22105.7a. [DOI] [PubMed] [Google Scholar]
- 34.Yang G., Liu J., Wang Y., Du Y., Ma A., Wang T. Lack of influence of sex hormones on Brugada syndrome-associated mutant Nav1.5 sodium channel. J Electrocardiol. 2019;52:82–87. doi: 10.1016/j.jelectrocard.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 35.Dumaine R., Towbin J.A., Brugada P., Vatta M., Nesterenko D.V., Nesterenko V.V., et al. Ionic mechanisms responsible for the electrocardiographic phenotype of the Brugada syndrome are temperature dependent. Circ Res. 1999;85(9):803–809. doi: 10.1161/01.res.85.9.803. [DOI] [PubMed] [Google Scholar]
- 36.Liu M., Yang K.C., Dudley S.C. Cardiac sodium channel mutations: why so many phenotypes? Nat Rev Cardiol. 2014;11(10):607–615. doi: 10.1038/nrcardio.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.