Abstract
Background
Kdm6b, a specific histone 3 lysine 27 (H3K27) demethylase, has been reported to be implicated in a variety of developmental processes including cell differentiation and cell fate determination and multiple organogenesis. Here, we regulated the transcript level of kdm6bb to study the potential role in controlling the hearing organ development of zebrafish.
Methods
A morpholino antisense oligonucleotide (MO) strategy was used to induce Kdm6b deficiency; immunohistochemical staining and in situ hybridization analysis were conducted to figure out the morphologic alterations and embryonic mechanisms.
Results
Kdm6bb is expressed in the primordium and neuromasts at the early stage of zebrafish embryogenesis, suggesting a potential function of Kdm6b in the development of mechanosensory organs. Knockdown of kdm6bb severely influences the cell migration and proliferation in posterior lateral line primordium, abates the number of neuromasts along the trunk, and mRNA-mediated rescue test can partially renew the neuromasts. Loss of kdm6bb might be related to aberrant expressions of chemokine genes encompassing cxcl12a and cxcr4b/cxcr7b in the migrating primordium. Moreover, inhibition of kdm6bb reduces the expression of genes in Fgf signaling pathway, while it increases the axin2 and lef1 expression level of Wnt/β-catenin signaling during the migrating stage.
Conclusions
Collectively, our results revealed that Kdm6b plays an essential role in guiding the migration of primordium and in regulating the deposition of zebrafish neuromasts by mediating the gene expression of chemokines and Wnt and Fgf signaling pathway. Since histone methylation and demethylation are reversible, targeting Kdm6b may present as a novel therapeutic regimen for hearing disorders.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10565-021-09669-y.
Keywords: kdm6bb, Primordium migration, Cell proliferation, Chemokines, Fgf signaling
Background
The malformation of inner ear structure during the development process is a leading cause of congenital hearing loss. Multiple regulatory factors are involved in the developmental event of otic morphogenesis, but the underlying molecular mechanisms have not yet been entirely revealed. The lateral line (LL) system in zebrafish, an organ to sense water pressure and movements (Engelmann et al. 2000; Ghysen and Dambly-Chaudiere 2007; Ma and Raible 2009), is similar in structure and function with the vertebrate inner ear (Nicolson 2005; Whitfield 2002), which has been proven to be an excellent model to understand the fundamental principles in the development of sensory organs. As the mechanosensory organs of lateral line, neuromasts are divided into the cephalic anterior LL and the posterior LL which are distributed along the whole body of zebrafish (Aman and Piotrowski 2009; Dalle Nogare et al. 2014; Gallardo et al. 2010). Directional cell cluster migration and neuromast deposition are the fundamental behavior at the early stage of the development period of posterior lateral line (PLL), that the migration begins around 22 h post-fertilization (hpf) and quits at approximately 42 hpf in zebrafish, while the deposition starts once the fourth rosette of primordium is assembled, and ends in five to six trunk neuromasts and two or three terminal neuromasts between 25 and 54 hpf (Nechiporuk and Raible 2008). The microstructure of the migrating PLL is a series of rose-shaped organs, called rosettes, which deposits at regular intervals (Dambly-Chaudiere et al. 2003; Ghysen and Dambly-Chaudiere 2004). Mature neuromast consists of central hair cells (HCs) and surrounding supporting cells (SCs) as well as the periphery mantle cells. Previous studies have unmasked refined coordination involving the canonical Wnt pathway, Fgf family, Notch signaling, and the chemokine pathway during the posterior LL cluster cell migration and neuromast morphogenesis, which are the basics of normal mechanosensory behaviors (Aman and Piotrowski 2008; Lecaudey et al. 2008; Nechiporuk and Raible 2008). However, the dynamic and modifiable regulation tools in the development process are rarely reported.
The epigenetic mechanisms, characterized in changing gene expression but not DNA sequence, are indispensable to multiple development events. DNA methylation and histone modifications, including histone acetylation and histone demethylation, are the most important covalent modification forms catalyzed by a series of enzymes. By alterations of histone proteins at specific residues, lysine demethylases (KDMs) and lysine methyltransferases (KMTs) are widely involved in regulating gene expression during the developmental process (Clarke et al. 2020). The H3K27 demethylase Kdm6b (also known as Jmjd3) is considered to be an H3K27 demethylase that can specifically mediate demethylation of H3K27me2/3 peptides (Agger et al. 2007; Hong et al. 2007; Xiang et al. 2007; Yang et al. 2019). Through dynamic methylation and demethylation, epigenetic modification plays important roles in regulating many cellular processes; for example, trimethylated H3K27 is generally reported as a mark for gene repression, while monomethylated H3K27 is related to gene activation (Hong et al. 2007; Xiang et al. 2007). Recently, Kdm6b has been proposed to be involved in controlling differentiation of embryonic stem cells, endochondral and osteogenic organogenesis, and lung formation (Akerberg et al. 2017; Li et al. 2014; Ye et al. 2012; Zhang et al. 2015). In other species outside vertebrates, for example, oyster and mangrove rivulus, histone methylation was closely involved in their early development process (Fellous et al. 2019, 2015). Additionally, Kdm6b deficiency through knockdown or knockout strategy can inhibit the cell growth and survival of multiple myeloma cells, suggesting a central role in tumorigenesis (Ohguchi et al. 2017). Kdm6b has recently been described as a regulator in temperature-dependent sex determination by activating the male pathway (Ge et al. 2018; Weber et al. 2020). As reported, Jmjd3 functionality lost resulted in an inability to differentiate bipolar cell subpopulation, implying that the Jmjd3 is critical for the development and maturation of retinal cells (Iida et al. 2014). Taken together, growing evidence shows that Kdm6b is associated with many human diseases, for example, developmental diseases, cancer, immune systematic diseases, infection, and urinary systems disorders (Zhang et al. 2019). However, it remains unknown whether Kdm6b plays any role in the development of hearing organs.
In this study, we identified kdm6bb expression in the developing sensory organs of zebrafish. By specific morpholino targeting Kdm6b, we demonstrated that interference with Kdm6b expression resulted in a disarranged PLL pattern, destruction of cell proliferation, and a reduced number of neuromasts after deposition period in dose-dependent effect. In addition, the in situ hybridization analysis showed that Kdm6b regulated the posterior zebrafish lateral line by the chemokines and Fgf pathways and that the cxcl12a/cxcr4b/cxcr7b ligand-receptor system is significantly inhibited; the expression of Fgf family members fgf3, fgf10, pea3, and fgfr1 are all decreased; while the Wnt/β-catenin targets axin2 and lef1 restricted in the leading zone highly expressed into an expansion range. Our study uncovered that Kdm6b is essential for the development of the posterior LL system of zebrafish and its reversible modification might be a useful target in improving hearing level.
Results
Detection of Kdm6b in the developing sensory organs of zebrafish
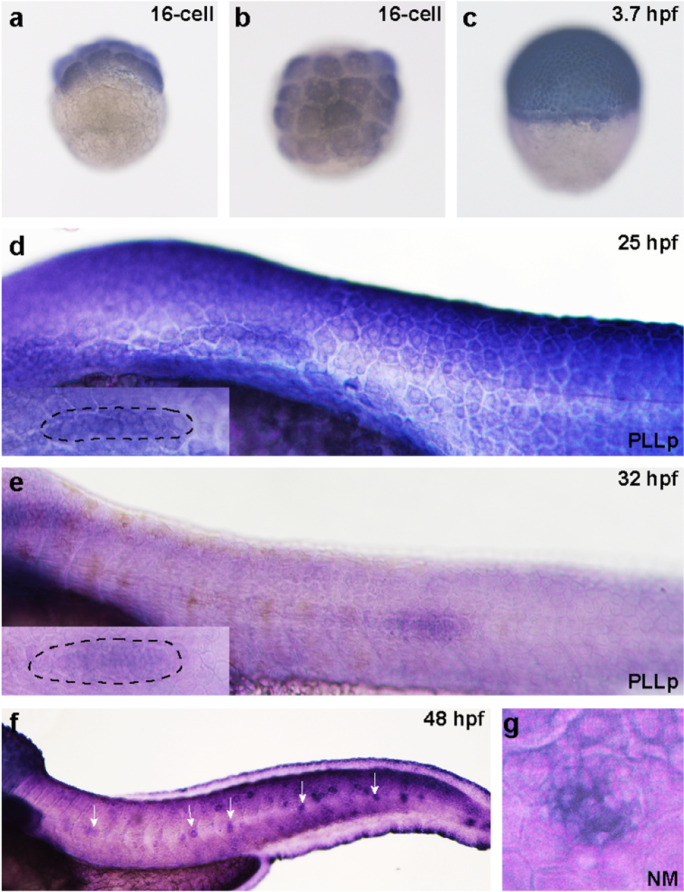
We first used WISH analysis to detect whether Kdm6b is expressed in the developing zebrafish embryo and to figure out its expression pattern. We collected the embryo in chronological order from 16-cell stage to 48 hpf according to the stage series by Kimmel CB et al. (Kimmel et al. 1995). We conducted both the sense and antisense mRNA probe for kdm6bb. However, we failed to detect any expression of Kdm6b in the whole body of zebrafish, and the representative AS and S pictures were shown in Figure S1. We found Kdm6b was expressed in the blastomeres at 16-cell stage from lateral and top view (Fig. 1a–1b). At 3.7 hpf, Kdm6b was detected in the elliptical shape (Fig. 1c). At 25–32 hpf, Kdm6b was found prominently expressed in the migrating primordium (Fig. 1d–1f). By 48 hpf, Kdm6b staining was found in the deposited posterior lateral line (PLL) neuromasts and mainly focused on the central hair cell region (Figs. 1f and 1g).
Fig. 1.
The expression pattern of kdm6bb during the early development stage of zebrafish. a, b Kdm6bb is detected at the early embryonic stage at 16-cell from the lateral and top view (a, b; n = 14 embryos) and blastula period (3.7 hpf) (c; n = 12 embryos) by whole-mount in situ hybridization. d–f Kdm6bb is expressed in the migrating primordium (d, e) and deposited neuromasts (f). The dotted lines of (d; n = 8 embryos) and (e; n = 10 embryos) outline the migrating primordia at higher magnification. White arrows in (f; n = 16 embryos) mark the neuromasts stained with kdm6bb along the trunk. The details of kdm6bb staining in magnified neuromasts are presented in (g). The experiment was duplicated for two times
Kdm6b is required for the normal depositing pattern in the lateral line system
Since the whole-mount ISH data showed evident expression of Kdm6b in the PLL neuromasts, we studied whether Kdm6b is required during neuromasts deposition. We chose the transgenic line Tg (cldnb: lynGFP) zebrafish as experimental models for more direct observation on the primordium migration and neuromast deposition since green fluorescent protein (GFP) is expressed in the cell membrane of primordium and neuromast.
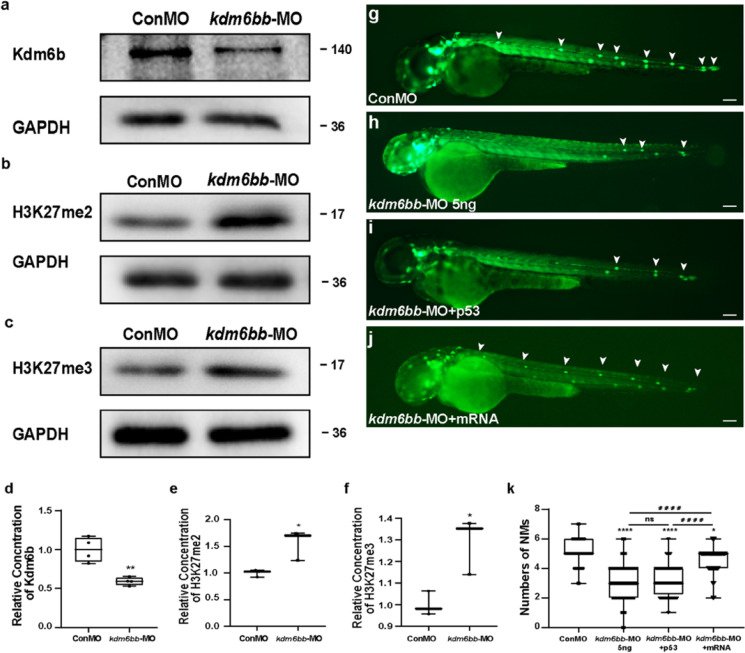
To investigate whether Kdm6b has role on the development of zebrafish, we first introduced a translational antisense morpholino (MO) strategy to downregulate the Kdm6b level. To exclude the effect of injection operation, the control group was injected with control-MO. We used Western blotting analysis to detect the efficacy of knockdown strategy and found a significant reduction of Kdm6b level after injection with kdm6bb-MO compared to that injected with Con-MO (Fig. 2a and 2d). Besides, we also detected the protein levels of H3K27me2 and H3K27me3 and confirmed that H3K27me2/3 were both upregulated when the level of Kdm6b was knockdown by kdm6bb-MO (Figs. 2b-2c, and 2e-2f).
Fig. 2.
Kdm6b is required for cell migration and neuromast deposition in zebrafish posterior lateral lines. a–f Kdm6b level is effectively downregulated using the morpholino technology. The protein blotting of Kdm6b is significantly decreased (a), and the protein blottings of H3K27me2 (b) and H3k27me3 (c) were both upregulated by the special antisense morpholino injection both in the band intensity. d–f The quantification analysis of relative concentration of Kdm6b, H3K27me2, and H3K27me3. Data are recorded as mean (minimum and maximum values). *p < 0.05 and **p < 0.01. g–j At 48 hpf, the deposited neuromasts are labeled in green along the posterior body of control embryos (g), Kdm6b-deficient mutants (h), co-injection of kdm6bb-MO + p53 (i), and co-injection of kdm6bb-MO + kdm6bb mRNA specimens (j). The neuromasts of PLL are labeled in green fluorescence in the transgenic cldnb:lynGFP embryos, and a severe reduction in number of neuromasts is found (g–h). The decreased number of neuromasts was confirmed when co-injected with p53 and kdm6bb-MO (i). The decreased number of neuromasts by Kdm6b-defect is partially rescued by the combined injection of kdm6bb mRNA (j). White arrowheads label the neuromast along the trunk and terminal of the posterior LL (g–j). k Statistical analysis of the number of posterior LL neuromasts at 48 hpf in controls (n = 185), Kdm6b-deficient embryos (n = 129), and kdm6bb-MO + mRNA members (n = 42). ****p < 0.0001 (the contrast between kdm6bb-MO group with Con-MO group), ####p < 0.0001 (the contrast between kdm6bb-MO group with kdm6bb-MO + mRNA group), *p < 0.05 (the contrast between kdm6bb-MO + mRNA group with Con-MO group). Scale bars mark a distance of 100 μm
At 48 hpf, a stage when PLL primordium stops to migrate and finishes deposition, an average of 5.065 ± 0.056 neuromasts (n = 185) was distributed periodically over the trunk in the embryos injected with control-MO (Fig. 2g and 2k). In contrast, the average number of neuromasts was significantly reduced to 3.116 ± 0.091 (n = 129) after injection with kdm6bb-MO (Fig. 2h and 2k). Interestingly, we did not examine any significant difference in the number of terminal neuromasts between the experiment and the control group. According to the previous study, up to 18% of morpholinos are in the possibility of off-target, resulting in cell death or central nervous system (CNS) destruction mainly on the cause of p53-mediated apoptosis (Ekker and Larson 2001; Robu et al. 2007). To avoid the non-specific effect of morpholinos, we co-injected p53 with kdm6bb morpholinos and found a severe reduction in the number of neuromasts, similar to those injected with KDM6B-MO only, confirming a powerful efficacy of kdm6bb-MO (Fig. 2h and 2i).
To further validate the role of Kdm6b in embryonic PLL morphogenesis, kdm6bb mRNA and kdm6bb-MO were injected together. Co-injection of kdm6bb mRNA and morpholino could partially rescue the defects of posterior LL, and the reduced number of neuromasts on the body trunk was restored to 4.667 ± 0.111 (n = 42) (Fig. 2j and 2k). Taken together, these conclusions can be illustrated that the deletion of kdm6bb affects the normal PLL morphogenesis during the development of zebrafish embryos.
Kdm6b knockdown disrupts normal rosette assembly and cell proliferation of the primordium
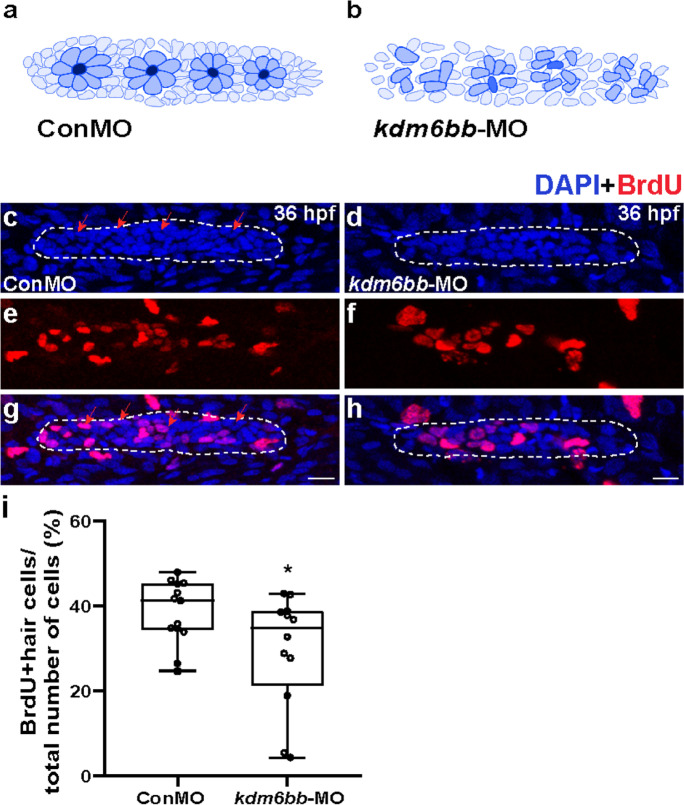
At the early stage of lateral line development, the primordium is migrating forward to the tail region and is organized into rosettes in the trailing zone. Neuromast deposition occurs when the fourth rosette is assembled. However, in the kdm6bb morphants, we did not find a normal rosette pattern at 36 hpf when compared with the corresponding control primordia, suggesting obvious damage of rosette assembly by inhibition of kdm6bb (Figs. 3a-3d).
Fig. 3.
The downregulation of Kdm6b represses cell proliferation and disrupts rosette assembly during primordium migration. a, b The diagram of rosette organization in controls and kdm6bb-MO embryos. c–h Representative images show the comparison in number of BrdU labeled proliferating cells between controls embryos (c, e, g) and Kdm6b-deficient mutants (d, f, h) at 36 hpf. Red arrows indicate the rosette-shaped clusters of the primordium (c). Scale bars mark the 10 μm scale. i Significant difference in quantification of BrdU index in control embryos and kdm6bb-MO embryos. Data are recorded as mean (minimum and maximum values). *p < 0.05
Cell proliferation has been reported to be widely involved in many processes such as the size of primordium, the capacity of migration, and the deposition cycle of proneuromasts in the migrating primordium (Aman et al. 2011). In our previous study, we found GSK-J4 (a selective inhibitor of H3K27me3 histone demethylase) treatment prevented both hair cell and supporting cell regeneration through reducing proliferation (Bao et al. 2017). Here, we investigated whether cell proliferation behavior is vitiated in Kdm6b-deficient mutants during the zebrafish embryo development. BrdU staining was used to capture the proliferating cells in the primordia of kdm6bb mutants and control embryos at 36 hpf. The BrdU index (the ratio of BrdU-marked nuclei to total cell nuclei) was significantly decreased in the kdm6bb-MO primordia compared to the parallel controls, while the BrdU index was 29.56% ± 0.04 (n = 13) in kdm6bb mutants and 38.45% ± 0.02 (n = 12) in control siblings, respectively (Fig. 3e–3i). Collectively, our data manifested that Kdm6b knockdown induces inhibited cell proliferation and apoptosis during PLL development, resulting an abnormal assembly of classical rosettes.
Kdm6b downregulation impacts primordium migration by restriction of chemokine signaling
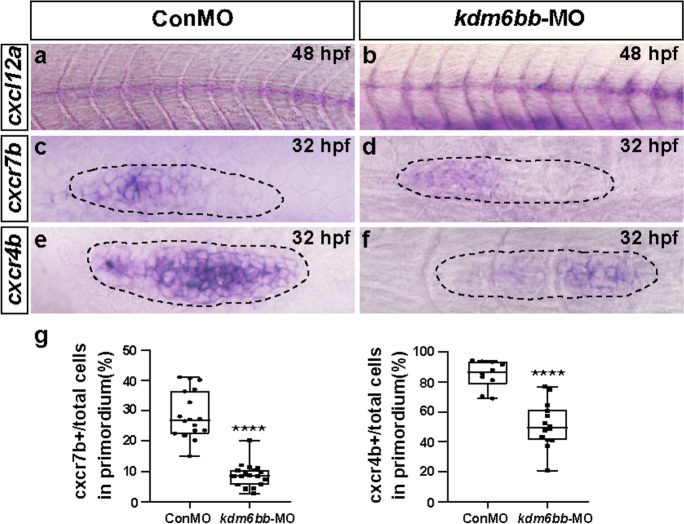
Chemokines and coupled receptors are active in promoting cell proliferation and in guiding the direction of migrating cells (Wang and Knaut 2014). Of the detected chemokine ligands and receptors in humans and mouse, cxcl12-cxcr4 together with cxcl12-cxcr7 pathway is one of the most famous in guidance of primordium migration. Previous studies have shown that cxcl12a, cxcr4b, and cxcr7b are asymmetrically expressed in zebrafish during embryonic development and is crucial for posterior LL cell migration (Aman and Piotrowski 2008; Valentin et al. 2007). We asked whether the expression pattern of chemokines is restricted because of Kdm6b defects. At 48 hpf, cxcl12a was found expressed like a narrow stripe along the horizontal myoseptum through in situ hybridization test in control-MO embryos. However, in the kdm6bb morphant embryos, the staining of cxcl12a was inconsecutive and partially missed (Fig. 4a and 4b). At 32 hpf, cxcr7b was strongly recognized in the primordium mainly in the trailing region but absent in the leading zone in control siblings while the expression level of cxcr7b was impressively downsized in the kdm6bb morphant primordia. In addition, the apparent rosette-shaped staining of cxcr7b was completely invisible in kdm6bb mutants compared with the controls (Fig. 4c, 4d, and 4g). We further detected a similar expression reduction of cxcr4b in the disordered KDM6B morphant primordia. Consistent with the previous data, cxcr4b was verified broadly and highly expressed in most regions of the primordium, especially in the leading part in the controls. However, after Kdm6b knockdown, cxcr4b expression was dramatically decreased both from the trailing zone to the leading zone (Fig. 4e–4g). To verify our results, we conducted real-time qPCR experiments to examine the mRNA level of cxcl12a, cxcr4b, and cxcr7b. Consistent with the ISH data, the qPCR analysis revealed significant reduction of relative cxcr7b and cxcr4b level in kdm6bb-MO embryos compared to the ConMO group while the change of cxcl12a was not significant between the experimental and control group (Figure S3). Taken together, the WISH data showed that Kdm6b might be a regulator of PLL morphogenesis during primordium migration by modulating chemokine signaling.
Fig. 4.
The chemokine signaling pathway is disrupted by kdm6bb-MO. a–f Representative in situ staining of chemokine members cxcl12a, cxcr4b, and cxcr7b are compared between the controls and kdm6bb morphants at 32–48 hpf. (a, b)The consecutive expression of cxcl12a is interrupted in Kdm6b knockdown embryos, both (c, d) cxcr4b and (e, f) cxcr7b expressions are downregulated in kdm6bb morpholino mutants in comparison with the control-MO-injected embryos. g The ratio of cxcr7b and cxcr4b-positive cells to total cells in the primordia of controls (ConMO; n = 18 embryos for cxcr7b, and n = 10 embryos for cxcr4b) and experiment group (kdm6bb-MO; n = 19 embryos for cxcr7b, and n = 14 embryos for cxcr4b). The black dotted lines outline the primordium appearance. The leading region is positioned to the right and the trailing region is to the left. Data are recorded as mean (minimum and maximum values). ****p < 0.0001
Kdm6b controls the migrating primordium through Wnt and Fgf signaling
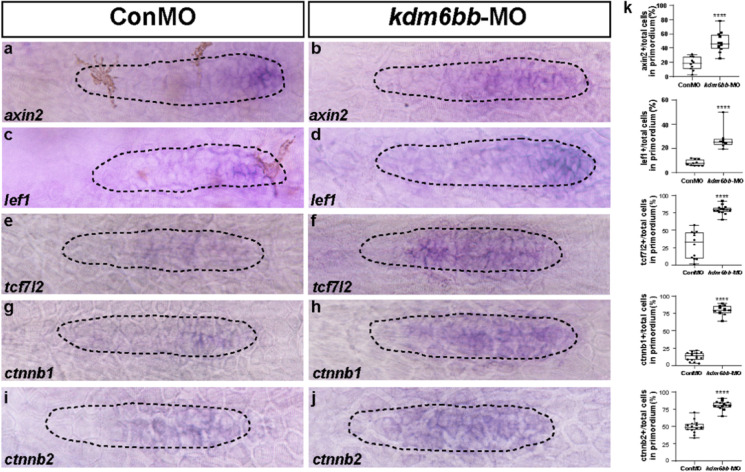
Previous studies concerning the function of canonical Wnt/β-catenin and Fgf signaling have demonstrated that their coordination plays crucial roles in controlling the cell migration and proliferation during early stage of primordia and proneuromast development(Aman and Piotrowski 2008). We therefore asked whether Kdm6b has interactions with the two associated signaling pathways in lateral line primordium migration process. Firstly, we examined the expression level of Wnt target genes axin2 and lef1 and found that both genes were restricted in the right and leading edge of the migrating primordium as labeled by dotted lines in control embryos between 32 hpf. In contrast, in kdm6bb-MO morphants, expression of both genes, especial the area of axin2 staining significantly expanded, crossing the leading tip to the trailing region (Fig. 5a, 5b, and 5k), which indicates a negative regulation of kdm6bb on Wnt signaling. To confirm our findings, we further detected the expression levels of other Wnt target genes, such as tcf7l2, ctnnb1, and ctnnb2. Our results demonstrated significant upregulation of tcf7l2, ctnnb1, and ctnnb2 transcript levels in kdm6bb-MO embryos compared with the controls (Fig. 5e–5k).
Fig. 5.
Knockdown of Kdm6b upregulates the Wnt signaling pathway. a–j In situ staining of the Wnt signaling target genes, axin2, lef1, tcf7l2, ctnnb1, and ctnnb2 are all upregulated in kdm6bb-MO embryos compared to the controls by in situ hybridization at 32 hpf. k The ratio of axin2, lef1, tcf7l2, ctnnb1, and ctnnb2-positive cells to total cells in the primordia of controls (ConMO; n = 10 embryos for axin2 and lef1, respectively; n = 12, 15, and 15 embryos for tcf7l2, ctnnb1, and ctnnb2, respectively) and experiment group (kdm6bb-MO; n = 12 embryos for axin2, n = 8 embryos for lef1, n = 15 embryos for tcf7l2, n = 12 embryos for ctnnb1, and n = 14 embryos for ctnnb2).The black dotted lines are drawn to shape the primordium. Data are recorded as mean (minimum and maximum values). ****p < 0.0001
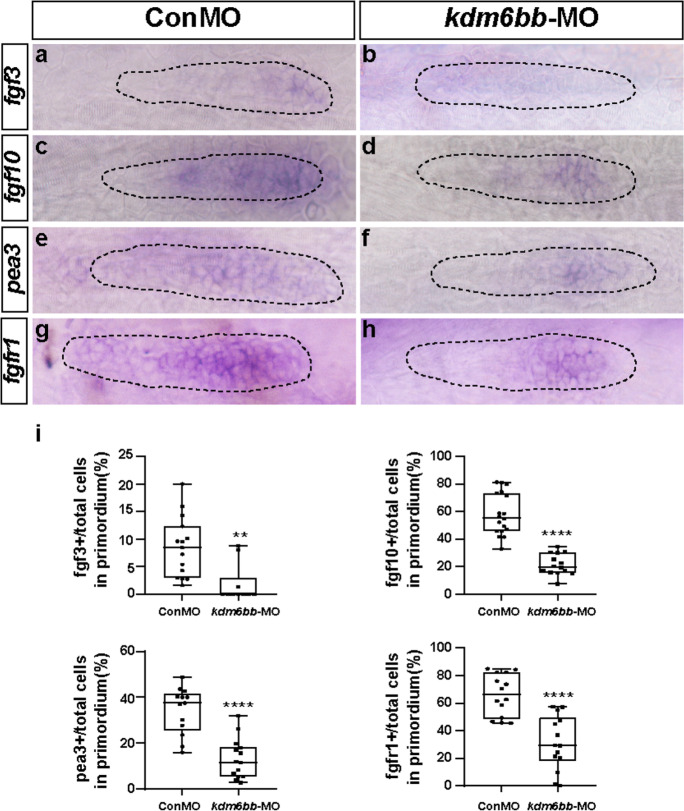
The Fgf signaling pathway members fgf3 and fgf10 have been described to be the only detectable FGF ligands in the migrating primordia (Nechiporuk and Raible 2008). We next examined the expression level of fgf3 and fgf10 in the lateral line system. We collected 30–32 hpf embryos, using WISH staining, we found fgf3 expression of the controls was restricted in the leading zone, and fgf10 had a wider expression than fgf3. However, lower distributions of fgf3 and fgf10 were detected after loss of Kdm6b function in posterior LL primordia (Fig. 6a–6d, and 6i). There was a transcript reduction of pea3, a Fgf signaling target downstream gene, in the PLL primordia of the kdm6bb variants compared to that of controls (Fig. 6e, 6f, and 6i). Furthermore, the transcript encoding FGF receptor, named fgfr1, was found strongly expressed in the control posterior LL. However, in the primordium of kdm6bb morphants, fgfr1 gene expression was inhibited to a minor area (Fig. 6g–6i).
Fig. 6.
Kdm6b-depletion represses Fgf signaling in zebrafish primordium. a–h Corresponding decreased expression of Fgf signaling components including fgf3, fgf10, pea3, and fgfr1 are presented in kdm6bb morphants compared to the control siblings at 32 hpf. i The ratio of fgf3, fgf10, pea3, and fgfr1-positive cells to total cells in the primordia of controls (ConMO; n = 15 embryos for fgf3, n = 18 embryos for fgf10, n = 13 embryos for pea3, and n = 14 embryos for fgfr1) and experiment group (kdm6bb-MO; n = 10 embryos for fgf3, n = 14 embryos for fgf10, n = 14 embryos for pea3, and n = 14 embryos for fgfr1).The primordium appearance is shaped by dotted lines. Data are recorded as mean (minimum and maximum values). **p < 0.01, and ****p < 0.0001
In addition, our real-time quantitative PCR analysis demonstrated the same variation tendency with the ISH semi-quantitative analysis results, especially in the changes of fgf10, pea3, and fgfr1 mRNA levels (Figure S3). Collectively, these results indicated that the loss of Kdm6b reduced the expression of Fgf signaling pathway but increased the Wnt/β-catenin signaling in PLL primordia, which might be the reason of disturbed cell migration and proliferation.
Discussion
The development and morphogenesis of auditory organs involve complicated interplay of multiple genetic and epigenetic factors. Our previous study showed that using GSK-J4 to inhibit H3K27me3 demethylase reduced the proliferative hair cell regeneration in posterior LL neuromasts of zebrafish (Bao et al. 2017). As the specific demethylase of H3K27me3, we hypothesized that Kdm6b would have an impact on the development of zebrafish lateral line. In this study, we investigated the function of Kdm6b during zebrafish posterior LL development. WISH results solidly displayed that Kdm6b was expressed in the migrating primordium and deposited neuromasts of zebrafish in a temporally and spatially controlled manner. Using the antisense MO knockdown technology, we found that the primordium migration was disrupted, the proportion of cell proliferation was severely decreased, and the number of PLL neuromasts in kdm6bb morphants was significantly reduced. Moreover, the result of partially restored number of neuromasts in RNA rescue test confirmed the important role of Kdm6b in PLL morphogenesis. Furthermore, the kdm6bb variants showed a consistent decreased distribution of cxcr4b and cxcr7b, and the intact staining of cxcl12a was broken off as shown in the in situ hybridization data. In addition, Wnt signaling was upregulated while Fgf signaling was downregulated in the kdm6bb-MO embryos. To make our data more convincing, we also conducted the ISH analysis using sense mRNA probe of indicated genes and failed to find any transcript expression, indicating a negative control of sense probe for ISH (Figure S2). Therefore, our research data suggests that Kdm6b might be a novel and hopeful target for hearing loss.
Epigenetic modifications including histone methylation and demethylation and histone acetylation and deacetylation have been found to play dual regulation roles in gene activation or repression, thus active in multiple biological processes by regulating chromatin structure and protein function. For example, the regulation and level of H3K9me2 has been reported to be involved in age-related hearing loss (rat model) (Li et al. 2021), in protection of hair cell from death (mouse damage model) (Yu et al. 2013), and in the process of hair cell regeneration after neomycin damage (zebrafish model) (Tang et al. 2016). The methylated H3K9 and H3K4 are classic examples of antagonistic effects concerning the dynamic modification of histone methylation, which usually the methylated H3K9 is related to negative transcription, but hypermethylation of H3K4 is usually correlated with active gene expression (Briggs et al. 2001; Völkel and Angrand 2007). However, the statement is not absolute. In our previous study, when treatment with the inhibitor of LSD1/KDM1A, the upregulated H3K4me2 level was related to a reduced number of both HCs and SCs in zebrafish larvae (He et al. 2013). In addition, the LSD1 inhibitor 2-PCPA has been confirmed to induce suppression of HC proliferative regeneration after neomycin induced HC damage (He et al. 2016). Similarly, inhibition of H3K9me2 with BIX01294 led to decreased number of HCs during regeneration events following neomycin-induced hair cell loss (Tang et al. 2016). As previously referred, Kdm6b, as an H3K27me3 demethylase, is involved in embryonic development and the formation of the endoderm, mesoderm, and ectoderm (Jiang et al. 2013; Li et al. 2014; Morales Torres et al. 2013; Tang et al. 2014; Zhang et al. 2015). In addition, Kdm6b is also involved in maintaining the pluripotency of mouse embryonic stem cells, assisting the self-renewal and differentiation of cardiovascular cells, and skin repairment (Agger et al. 2009; Apostolou and Hochedlinger 2013; Ohtani et al. 2013; Shaw and Martin 2009), immune system (De Santa et al. 2007; Liu et al. 2015; Salminen et al. 2014; Yan et al. 2014), neurodegenerative disease (Gomez-Sanchez et al. 2013; Tang et al. 2014), and cancer (Anderton et al. 2011; Ramadoss et al. 2012; Wei et al. 2013). The trimethylation of histone H3K27 is related to repressive transcription and gene silence. The reduced H3K27me3 level has been delineated as a biomarker of poor prognosis and invasive tumors, suggesting inhibition of gene expression of H3K27me3 (Bayliss et al. 2016). Loss of H3K27me3 has also been reported to mark aggressiveness of breast cancer, and increased H3K27me3 led to decreased tumor cell growth and migration (Hsieh et al. 2020). Here, our study detected the upregulation of H3K27me3 demethylase by kdm6bb knockdown severely disrupted the migration and deposition behavior of zebrafish posterior lateral line, suggesting a similar inactivation role of H3K27me3 in auditory organs compared with cancer. The multiple roles of KDM6B in various organs or systems make it a potential therapeutic target for some diseases; for example, KDM6B is upregulated in blood disorders, and inhibition of its negative regulator AP-1 in Kdm6b-deficient hematopoietic stem cells restores the functional defects (Mallaney et al. 2019). In patients with myelodysplastic syndromes and chronic myelomonocytic leukemia, overexpression of KDM6B has been reported. By generating a Vav-Kdm6b mouse model, KDM6B is overexpressed in the hematopoietic compartment, resulting in significant hematopoietic defects. However, using the KDM6B inhibitor GSK-J4 can ameliorate the ineffective hematopoiesis observed in Vav-Kdm6b mice, suggesting a therapeutic potential for myeloid disorders through targeting KDM6B (Wei et al. 2018). In epithelial ovarian cancer cells, overexpression of KDM6B promoted proliferation, migration, and invasion, while silencing KDM6B inhibited these processes and served as a therapeutic target (Liang et al. 2019). In this study, considering the knockdown strategy by morpholino, we chose co-injection of kdm6bb mRNA and morpholino as the complementary strategy, and we found it could partially rescue the defects of PLL.
During migration, the PLL primordial cells are clustered and organized into a radial arrangement called rosettes, which are deposited repeatedly in a stereotyped manner, and then the deposited proneuromasts differentiate into mature neuromasts (Chitnis et al. 2012; Ghysen and Dambly-Chaudiere 2004). The chemokine network involving the broad bindings between ligands and corresponding receptors is well studied for the function of promoting cell migration, particularly in leukocytes, inflammation, heart and vascular system, and lateral line (Hughes and Nibbs 2018). The chemokine ligand-receptor (cxcl12a-cxcr4b-cxcr7b) system has been reported to be an important guidance signal of directional migration in zebrafish (David et al. 2002; Li et al. 2004). Cxcl12a-cxcr4b signal is mostly restricted in the posterior and leading region of the zebrafish primordium, while cxcr7b is also tethered to cxcl12a but asymmetrically distributed in the opposite and trailing zone (Aman and Piotrowski 2008). Cxcl12 is the most elemental ligand that is expressed consecutively along the intermediate segment; however, the in situ staining of cxcl12a completely ceased in KDM6B-deficient embryos, resulting in disturbed migration. Similarly, the cxcl12a receptors cxcr7b and cxcr4b were detected in lower expression levels in kdm6bb morphants contrary to the controls. This finding uncovers that kdm6bb regulates chemokine network to induce the posterior primordium migration of zebrafish.
The Fgf signaling is essential for rosette formation and primordium migration, while the Wnt/β-catenin signal activates the expression of FGF ligands fgf3 and fgf10, and Wnt activation or Fgf reduction both inhibit proper primordium developing patterns (Aman and Piotrowski 2008; Nechiporuk and Raible 2008). To investigate the mechanism of Kdm6b in controlling primordium migration, we performed ISH experiments with probes of the Fgf and Wnt signaling pathways. The results showed that Kdm6b-depletion significantly induced hyper-expression of axin2 and lef1, but hypo-expression of Fgf components (fgf3, fgf10, pea3, and fgfr1). The enhancement expression in the region of Wnt signal target genes in KDM6B-deficient primordium might be related to the blocking of Fgf signal in the trailing region.
In conclusion, we found certain functions of Kdm6b in the developing lateral line system, a mechanosensory organ of zebrafish. Kdm6b loss of function led to impaired PLL formation, interrupted migration of primordium, reduced cell proliferation, and depressed deposition of mature neuromasts. We demonstrated that Kdm6b is a decisive element in controlling normal PLL patterns by regulating Fgf and Wnt signaling and cxcl12a-cxcr7b/cxcr4b chemokine pathway, but further studies are needed to check the direct or indirect causality within the phenomenon. In addition, previous studies have shown that the H3K27me3 demethylase Kdm6b is associated with inflammation (Burchfield et al. 2015; Kruidenier et al. 2012; Neele et al. 2017; Satoh et al. 2010), so KDM6B may contribute to the treatment and prevention of otitis media related diseases. Our findings provide noteworthy links of epigenetic histone methylation to the development of hearing organ using zebrafish as an experimental model, enrich the theoretical mechanisms of Kdm6b in controlling the cell migration and proliferation behavior, and contribute to a potential therapeutic target of Kdm6b for hearing loss.
Materials and methods
Transgenic zebrafish line
All animal experiments of zebrafish were approved by the Institutional Animal Care and Use Committee of Fudan University in Shanghai. The PLL primordia and neuromasts were observed by the Tg (cldnb: lynGFP) line (Haas and Gilmour 2006). The embryos were maintained and raised in embryo medium followed standard recipe after spawning in the constant temperature incubator at 28.5 °C (Westerfield 2000). To avoid pigmentation, embryos were incubated with 1-phenyl-2-thiourea (Sigma-Aldrich) from 10 h after birth. The age and stage of embryos during development was recorded as hours post-fertilization (hpf).
Morpholino injections and mRNA rescue test
Kdm6bb-MO, with a sequence of 5′-AAAAGAACATGACTGACCTGGTGTG-3′ and a solution of 5 ng was injected into Tg (cldnb: lynGFP) embryos for kdm6bb knockdown at one or two cell stage. To avoid side effects by injection handling, control-MO, with a sequence of 5′-CCTCTTACCTCAGTTACAATTTATA-3′ was injected. For mRNA rescue test, 150 pg of kdm6bb mRNA generated by the mMESSAGE mMachine Sp6 Kit (Ambion, Austin, TX, US) was co-injected with kdm6bb morpholino.
Whole-mount in situ hybridization
The whole-mount in situ hybridization (WISH) experiment was manipulated as previously recited (He et al. 2014; Thisse and Thisse 2014). Briefly, after fixation and gradient dehydration, the embryos were warehoused in pure methanol at − 20 °C. Embryos were rehydrated and then digested in 20 μg/ml proteinase K for preparation of hybridization, then embryos were incubated with the probes at 65 °C overnight. After labeling, larvae were rinsed four times in a graded SSC-PBST series and then put in the blocking reagent at room temperature for 1 h. After bonding to the anti-digoxigenin-AP Fab fragment (Roche) overnight at 4 °C, the embryos were washed sufficiently and then incubated with BM purple AP substrate (Roche) for visualized staining in a dark place. The color reaction was discontinued by NTMT and washed thoroughly in PBST. For whole-mount neuromast imaging, specimens were mounted in 100% glycerol, with a Nikon fluorescence stereomicroscope. All image processing was performed with Photoshop and Illustrator software (2018, Adobe).
Western blot analysis
The extraction of total protein was operated using AllPrep DNA/RNA/Protein Mini Kit (QIAGEN, Hilden, Germany), and the protein concentrations were detected via A BCA protein kit (Thermo Fisher Scientific, Rockford, IL). Then, proteins were separated on SDS–polyacrylamide gels and transferred onto PVDF membranes (Immobilon-P; Millipore, Bedford, MA, USA). The membranes were incubated overnight with rabbit anti-KDM6B/JMJD3 polyclonal antibody (1:500 dilution, Abcam, USA), rabbit anti-Histone H3K27me3 polyclonal antibody (MEMD Millipore Corp, USA), rabbit anti-Histone H3K27me2 polyclonal antibody (MEMD Millipore Corp, USA), or mouse anti-GAPDH monoclonal antibody as reference (1:1000 dilution, Sigma) at 4 °C. The ECL kit (Pierce) was used to visualize the immunoreactive bands and the intensities of the bands were quantified with Fiji (National Institutes of Health).
BrdU incorporation and analysis
We manually dechorionated the embryos at first, and the embryos were incubated in 15 mM bromodeoxyuridine (BrdU) (Sigma–Aldrich) for 1.5 h. The embryos were collected individually, anesthetized in 0.02% MS-222 (Sigma–Aldrich), and fixed at 4 °C overnight in 4% PFA. After rinsing several times in PBT-2, the control and kdm6bb-deficient embryos were immersed in 2 N HCl for 30 min at 37 °C for blocking of non-specific binding domains. Immunocytochemistry staining was manipulated following the standard procedure after incubation with the primary mouse monoclonal anti-BrdU antibody (1:300 dilution, Santa Cruz Biotechnology) for 1 h at 37 °C and then 4 °C overnight. After washing 3 times in PBT-2, the secondary Cy3 donkey anti-mouse polyclonal antibody (1:300 dilution, Jackson) was added and co-incubated for 1 h at 37 °C successively. The proliferation analysis was conducted by detection of BrdU-positive cells. The fluorescent images were photographed with a multichannel confocal microscope (TCS SP8; Leica), and the further procession of pictures was realized with Photoshop and Illustrator software (2018, Adobe).
Real-time PCR quantification
Total RNAs were extracted from the experimental and control embryos at 48 hpf after deletion of yolk sac using TRIzol reagent (Thermo Fisher Scientific) and reverse transcription into cDNA was performed with the Transcriptor First Strand cDNA Synthesis Kit (Roche). To monitor the mRNA levels of desirable genes, quantitative real-time PCR system (LightCycler® 480, Roche) was constructed using SYBR Premix Ex Taq II (Takara Biomedical Technology). Results were analyzed using the ΔΔCt method, and primer sequences were described in Supplementary table 1.
Statistical analysis
The software GraphPad Prism (version, 8.0c) espoused all statistical analyses. Multiple comparisons were performed using one-way ANOVA, while two-group comparisons were illustrated with a two-tailed Student t-test. Statistics were all presented as mean (minimum and maximum values) with all the event plotting labeled, and p value < 0.05 was treated as statistically significant.
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations
- BrdU
Bromodeoxyuridine
- GFP
Green fluorescent protein
- HCs
Hair cells
- Hpf
Hours post-fertilization
- KDMs
Lysine demethylases
- KMTs
Lysine methyltransferases
- LL
Lateral line
- MO
Morpholino antisense oligonucleotide
- PLL
Posterior lateral line
- SCs
Supporting cells
- WISH
Whole-mount in situ hybridization
Author contribution
DT and YL performed the experiment and drafted the manuscript. YL NZ, RY, CW, and LW analyzed the data and approved the final manuscript. YH and SL conceived and designed the study, interpreted and analyzed the data, and approved the final manuscript.
YH and SL: conceptualization, methodology, writing—review and editing—project administration. DT, YL NZ, RY, CW, and LW: methodology, formal analysis. YH and SL: validation, investigation, formal analysis. DT and YL: validation, investigation. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 82071045, 81870728, and 81800912) and Shanghai Rising-Star Program (19QA1401800).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dongmei Tang and Yitong Lu contributed equally to this work.
Contributor Information
Shaofeng Liu, Email: liusf_cn@163.com.
Yingzi He, Email: yingzihe09611@126.com.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23(10):1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerberg AA, Henner A, Stewart S, Stankunas K. Histone demethylases Kdm6ba and Kdm6bb redundantly promote cardiomyocyte proliferation during zebrafish heart ventricle maturation. Dev Biol. 2017;426(1):84–96. doi: 10.1016/j.ydbio.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman A, Nguyen M, Piotrowski T. Wnt/beta-catenin dependent cell proliferation underlies segmented lateral line morphogenesis. Dev Biol. 2011;349(2):470–482. doi: 10.1016/j.ydbio.2010.10.022. [DOI] [PubMed] [Google Scholar]
- Aman A, Piotrowski T. Wnt/beta-catenin and Fgf signaling control collective cell migration by restricting chemokine receptor expression. Dev Cell. 2008;15(5):749–761. doi: 10.1016/j.devcel.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Aman A, Piotrowski T. Multiple signaling interactions coordinate collective cell migration of the posterior lateral line primordium. Cell Adh Migr. 2009;3(4):365–368. doi: 10.4161/cam.3.4.9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderton JA, Bose S, Vockerodt M, Vrzalikova K, Wei W, Kuo M, et al. The H3K27me3 demethylase, KDM6B, is induced by Epstein-Barr virus and over-expressed in Hodgkin's Lymphoma. Oncogene. 2011;30(17):2037–2043. doi: 10.1038/onc.2010.579. [DOI] [PubMed] [Google Scholar]
- Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502(7472):462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao B, He Y, Tang D, Li W, Li H. Inhibition of H3K27me3 histone demethylase activity prevents the proliferative regeneration of zebrafish lateral line neuromasts. Front Mol Neurosci. 2017;10:51. doi: 10.3389/fnmol.2017.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss J, Mukherjee P, Lu C, Jain SU, Chung C, Martinez D, et al. Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci Transl Med. 2016;8(366):366ra161. doi: 10.1126/scitranslmed.aah6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15(24):3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchfield JS, Li Q, Wang HY, Wang RF. JMJD3 as an epigenetic regulator in development and disease. Int J Biochem Cell Biol. 2015;67:148–157. doi: 10.1016/j.biocel.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis AB, Nogare DD, Matsuda M. Building the posterior lateral line system in zebrafish. Dev Neurobiol. 2012;72(3):234–255. doi: 10.1002/dneu.20962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TL, Tang R, Chakraborty D, Van Rechem C, Ji F, Mishra S, et al. Histone lysine methylation dynamics control EGFR DNA copy-number amplification. Cancer Discov. 2020;10(2):306–325. doi: 10.1158/2159-8290.Cd-19-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle Nogare D, Somers K, Rao S, Matsuda M, Reichman-Fried M, Raz E, et al. Leading and trailing cells cooperate in collective migration of the zebrafish posterior lateral line primordium. Development. 2014;141(16):3188–3196. doi: 10.1242/dev.106690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambly-Chaudiere C, Sapede D, Soubiran F, Decorde K, Gompel N, Ghysen A. The lateral line of zebrafish: a model system for the analysis of morphogenesis and neural development in vertebrates. Biol Cell. 2003;95(9):579–587. doi: 10.1016/j.biolcel.2003.10.005. [DOI] [PubMed] [Google Scholar]
- David NB, Sapede D, Saint-Etienne L, Thisse C, Thisse B, Dambly-Chaudiere C, et al. Molecular basis of cell migration in the fish lateral line: role of the chemokine receptor CXCR4 and of its ligand, SDF1. Proc Natl Acad Sci U S A. 2002;99(25):16297–16302. doi: 10.1073/pnas.252339399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130(6):1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Ekker SC, Larson JD. Morphant technology in model developmental systems. Genesis. 2001;30(3):89–93. doi: 10.1002/gene.1038. [DOI] [PubMed] [Google Scholar]
- Engelmann J, Hanke W, Mogdans J, Bleckmann H. Hydrodynamic stimuli and the fish lateral line. Nature. 2000;408(6808):51–52. doi: 10.1038/35040706. [DOI] [PubMed] [Google Scholar]
- Fellous A, Earley RL, Silvestre F. Identification and expression of mangrove rivulus (Kryptolebias marmoratus) histone deacetylase (HDAC) and lysine acetyltransferase (KAT) genes. Gene. 2019;691:56–69. doi: 10.1016/j.gene.2018.12.057. [DOI] [PubMed] [Google Scholar]
- Fellous A, Favrel P, Riviere G. Temperature influences histone methylation and mRNA expression of the Jmj-C histone-demethylase orthologues during the early development of the oyster Crassostrea gigas. Mar Genomics. 2015;19:23–30. doi: 10.1016/j.margen.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Gallardo VE, Liang J, Behra M, Elkahloun A, Villablanca EJ, Russo V, et al. Molecular dissection of the migrating posterior lateral line primordium during early development in zebrafish. BMC Dev Biol. 2010;10:120. doi: 10.1186/1471-213X-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C, Ye J, Weber C, Sun W, Zhang H, Zhou Y, et al. The histone demethylase KDM6B regulates temperature-dependent sex determination in a turtle species. Science. 2018;360(6389):645–648. doi: 10.1126/science.aap8328. [DOI] [PubMed] [Google Scholar]
- Ghysen A, Dambly-Chaudiere C. Development of the zebrafish lateral line. Curr Opin Neurobiol. 2004;14(1):67–73. doi: 10.1016/j.conb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Ghysen A, Dambly-Chaudiere C. The lateral line microcosmos. Genes Dev. 2007;21(17):2118–2130. doi: 10.1101/gad.1568407. [DOI] [PubMed] [Google Scholar]
- Gomez-Sanchez JA, Gomis-Coloma C, Morenilla-Palao C, Peiro G, Serra E, Serrano M, et al. Epigenetic induction of the Ink4a/Arf locus prevents Schwann cell overproliferation during nerve regeneration and after tumorigenic challenge. Brain. 2013;136(Pt 7):2262–2278. doi: 10.1093/brain/awt130. [DOI] [PubMed] [Google Scholar]
- Haas P, Gilmour D. Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line. Dev Cell. 2006;10(5):673–680. doi: 10.1016/j.devcel.2006.02.019. [DOI] [PubMed] [Google Scholar]
- He Y, Tang D, Cai C, Chai R, Li H. LSD1 is Required for Hair Cell Regeneration in Zebrafish. Mol Neurobiol. 2016;53(4):2421–2434. doi: 10.1007/s12035-015-9206-2. [DOI] [PubMed] [Google Scholar]
- He Y, Wu J, Mei H, Yu H, Sun S, Shou J, et al. Histone deacetylase activity is required for embryonic posterior lateral line development. Cell Prolif. 2014;47(1):91–104. doi: 10.1111/cpr.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Yu H, Sun S, Wang Y, Liu I, Chen Z, et al. Trans-2-phenylcyclopropylamine regulates zebrafish lateral line neuromast development mediated by depression of LSD1 activity. Int J Dev Biol. 2013;57(5):365–373. doi: 10.1387/ijdb.120227hl. [DOI] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104(47):18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh IY, He J, Wang L, Lin B, Liang Z, Lu B, et al. H3K27me3 loss plays a vital role in CEMIP mediated carcinogenesis and progression of breast cancer with poor prognosis. Biomed Pharmacother. 2020;123:109728. doi: 10.1016/j.biopha.2019.109728. [DOI] [PubMed] [Google Scholar]
- Hughes CE, Nibbs RJB. A guide to chemokines and their receptors. FEBS J. 2018;285(16):2944–2971. doi: 10.1111/febs.14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida A, Iwagawa T, Kuribayashi H, Satoh S, Mochizuki Y, Baba Y, et al. Histone demethylase Jmjd3 is required for the development of subsets of retinal bipolar cells. Proc Natl Acad Sci U S A. 2014;111(10):3751–3756. doi: 10.1073/pnas.1311480111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wang J, Zhang Y. Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway. Cell Res. 2013;23(1):122–130. doi: 10.1038/cr.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488(7411):404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecaudey V, Cakan-Akdogan G, Norton WH, Gilmour D. Dynamic Fgf signaling couples morphogenesis and migration in the zebrafish lateral line primordium. Development. 2008;135(16):2695–2705. doi: 10.1242/dev.025981. [DOI] [PubMed] [Google Scholar]
- Li Q, Shirabe K, Kuwada JY. Chemokine signaling regulates sensory cell migration in zebrafish. Dev Biol. 2004;269(1):123–136. doi: 10.1016/j.ydbio.2004.01.020. [DOI] [PubMed] [Google Scholar]
- Li Q, Wang HY, Chepelev I, Zhu Q, Wei G, Zhao K, et al. Stage-dependent and locus-specific role of histone demethylase Jumonji D3 (JMJD3) in the embryonic stages of lung development. PLoS Genet. 2014;10(7):e1004524. doi: 10.1371/journal.pgen.1004524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Xiang YH, Liang XJ, Zhang Y, Zhao PP, Wang M, et al. Expression of G9a in auditory cortex is downregulated in a rat model of age-related hearing loss. Journal of Molecular Neuroscience : MN. 2021;71(2):409–418. doi: 10.1007/s12031-020-01663-z. [DOI] [PubMed] [Google Scholar]
- Liang S, Yao Q, Wei D, Liu M, Geng F, Wang Q, et al. KDM6B promotes ovarian cancer cell migration and invasion by induced transforming growth factor-β1 expression. J Cell Biochem. 2019;120(1):493–506. doi: 10.1002/jcb.27405. [DOI] [PubMed] [Google Scholar]
- Liu Z, Cao W, Xu L, Chen X, Zhan Y, Yang Q, et al. The histone H3 lysine-27 demethylase Jmjd3 plays a critical role in specific regulation of Th17 cell differentiation. J Mol Cell Biol. 2015;7(6):505–516. doi: 10.1093/jmcb/mjv022. [DOI] [PubMed] [Google Scholar]
- Ma EY, Raible DW. Signaling pathways regulating zebrafish lateral line development. Curr Biol. 2009;19(9):R381–386. doi: 10.1016/j.cub.2009.03.057. [DOI] [PubMed] [Google Scholar]
- Mallaney C, Ostrander EL, Celik H, Kramer AC, Martens A, Kothari A, et al. Kdm6b regulates context-dependent hematopoietic stem cell self-renewal and leukemogenesis. Leukemia. 2019;33(10):2506–2521. doi: 10.1038/s41375-019-0462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales Torres C, Laugesen A, Helin K. Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLoS ONE. 2013;8(4):e60020. doi: 10.1371/journal.pone.0060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechiporuk A, Raible DW. FGF-dependent mechanosensory organ patterning in zebrafish. Science. 2008;320(5884):1774–1777. doi: 10.1126/science.1156547. [DOI] [PubMed] [Google Scholar]
- Neele AE, Prange KH, Hoeksema MA, van der Velden S, Lucas T, Dimmeler S, et al. Macrophage Kdm6b controls the pro-fibrotic transcriptome signature of foam cells. Epigenomics. 2017;9(4):383–391. doi: 10.2217/epi-2016-0152. [DOI] [PubMed] [Google Scholar]
- Nicolson T. The genetics of hearing and balance in zebrafish. Annu Rev Genet. 2005;39:9–22. doi: 10.1146/annurev.genet.39.073003.105049. [DOI] [PubMed] [Google Scholar]
- Ohguchi H, Harada T, Sagawa M, Kikuchi S, Tai YT, Richardson PG, et al. KDM6B modulates MAPK pathway mediating multiple myeloma cell growth and survival. Leukemia. 2017;31(12):2661–2669. doi: 10.1038/leu.2017.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K, Zhao C, Dobreva G, Manavski Y, Kluge B, Braun T, et al. Jmjd3 controls mesodermal and cardiovascular differentiation of embryonic stem cells. Circ Res. 2013;113(7):856–862. doi: 10.1161/CIRCRESAHA.113.302035. [DOI] [PubMed] [Google Scholar]
- Ramadoss S, Chen X, Wang CY. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J Biol Chem. 2012;287(53):44508–44517. doi: 10.1074/jbc.M112.424903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, et al. p53 activation by knockdown technologies. PLoS Genet. 2007;3(5):e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K, Hiltunen M, Kauppinen A. Histone demethylase Jumonji D3 (JMJD3/KDM6B) at the nexus of epigenetic regulation of inflammation and the aging process. J Mol Med (berl) 2014;92(10):1035–1043. doi: 10.1007/s00109-014-1182-x. [DOI] [PubMed] [Google Scholar]
- Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11(10):936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10(8):881–886. doi: 10.1038/embor.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Lin Q, He Y, Chai R, Li H. Inhibition of H3K9me2 reduces hair cell regeneration after hair cell loss in the zebrafish lateral line by down-regulating the Wnt and Fgf signaling pathways. Front Mol Neurosci. 2016;9:39. doi: 10.3389/fnmol.2016.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Li T, Li J, Yang J, Liu H, Zhang XJ, et al. Jmjd3 is essential for the epigenetic modulation of microglia phenotypes in the immune pathogenesis of Parkinson's disease. Cell Death Differ. 2014;21(3):369–380. doi: 10.1038/cdd.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Thisse C. In situ hybridization on whole-mount zebrafish embryos and young larvae. Methods in molecular biology (Clifton, N.J.) 2014;1211:53–67. doi: 10.1007/978-1-4939-1459-3_5. [DOI] [PubMed] [Google Scholar]
- Valentin G, Haas P, Gilmour D. The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b. Curr Biol. 2007;17(12):1026–1031. doi: 10.1016/j.cub.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Völkel P, Angrand PO. The control of histone lysine methylation in epigenetic regulation. Biochimie. 2007;89(1):1–20. doi: 10.1016/j.biochi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Wang J, Knaut H. Chemokine signaling in development and disease. Development (cambridge, England) 2014;141(22):4199–4205. doi: 10.1242/dev.101071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C, Zhou Y, Lee JG, Looger LL, Qian G, Ge C, et al. Temperature-dependent sex determination is mediated by pSTAT3 repression of Kdm6b. Science. 2020;368(6488):303–306. doi: 10.1126/science.aaz4165. [DOI] [PubMed] [Google Scholar]
- Wei Y, Chen R, Dimicoli S, Bueso-Ramos C, Neuberg D, Pierce S, et al. Global H3K4me3 genome mapping reveals alterations of innate immunity signaling and overexpression of JMJD3 in human myelodysplastic syndrome CD34+ cells. Leukemia. 2013;27(11):2177–2186. doi: 10.1038/leu.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zheng H, Bao N, Jiang S, Bueso-Ramos CE, Khoury J, et al. KDM6B overexpression activates innate immune signaling and impairs hematopoiesis in mice. Blood Adv. 2018;2(19):2491–2504. doi: 10.1182/bloodadvances.2018024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. A guide for the laboratory use of zebrafish (Danio rerio). zebrafish book a guide for the laboratory use of zebrafish. 2000.
- Whitfield TT. Zebrafish as a model for hearing and deafness. J Neurobiol. 2002;53(2):157–171. doi: 10.1002/neu.10123. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007;17(10):850–857. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]
- Yan Q, Sun L, Zhu Z, Wang L, Li S, Ye RD. Jmjd3-mediated epigenetic regulation of inflammatory cytokine gene expression in serum amyloid A-stimulated macrophages. Cell Signal. 2014;26(9):1783–1791. doi: 10.1016/j.cellsig.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zha Y, Ding J, Ye B, Liu M, Yan C, et al. Histone demethylase KDM6B has an anti-tumorigenic function in neuroblastoma by promoting differentiation. Oncogenesis. 2019;8(1):3. doi: 10.1038/s41389-018-0112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Fan Z, Yu B, Chang J, Al Hezaimi K, Zhou X, et al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell. 2012;11(1):50–61. doi: 10.1016/j.stem.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Lin Q, Wang Y, He Y, Fu S, Jiang H, et al. Inhibition of H3K9 methyltransferases G9a/GLP prevents ototoxicity and ongoing hair cell death. Cell Death Dis. 2013;4(2):e506. doi: 10.1038/cddis.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Xu L, Xu L, Xu Q, Li D, Yang Y, et al. JMJD3 promotes chondrocyte proliferation and hypertrophy during endochondral bone formation in mice. J Mol Cell Biol. 2015;7(1):23–34. doi: 10.1093/jmcb/mjv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu L, Yuan X, Wei Y, Wei X. JMJD3 in the regulation of human diseases. Protein Cell. 2019;10(12):864–882. doi: 10.1007/s13238-019-0653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.