Abstract
The existing deep learning-based denoising methods predicting standard-dose PET images (S-PET) from the low-dose versions (L-PET) solely rely on a single-dose level of PET images as the input of deep learning network. In this work, we exploited the prior knowledge in the form of multiple low-dose levels of PET images to estimate the S-PET images. To this end, a high-resolution ResNet architecture was utilized to predict S-PET images from 6 to 4% L-PET images. For the 6% L-PET imaging, two models were developed; the first and second models were trained using a single input of 6% L-PET and three inputs of 6%, 4%, and 2% L-PET as input to predict S-PET images, respectively. Similarly, for 4% L-PET imaging, a model was trained using a single input of 4% low-dose data, and a three-channel model was developed getting 4%, 3%, and 2% L-PET images. The performance of the four models was evaluated using structural similarity index (SSI), peak signal-to-noise ratio (PSNR), and root mean square error (RMSE) within the entire head regions and malignant lesions. The 4% multi-input model led to improved SSI and PSNR and a significant decrease in RMSE by 22.22% and 25.42% within the entire head region and malignant lesions, respectively. Furthermore, the 4% multi-input network remarkably decreased the lesions’ SUVmean bias and SUVmax bias by 64.58% and 37.12% comparing to single-input network. In addition, the 6% multi-input network decreased the RMSE within the entire head region, within the lesions, lesions’ SUVmean bias, and SUVmax bias by 37.5%, 39.58%, 86.99%, and 45.60%, respectively. This study demonstrated the significant benefits of using prior knowledge in the form of multiple L-PET images to predict S-PET images.
Keywords: Low-dose, Deep learning, Quantitative imaging, PET
Introduction
Positron emission tomography (PET) imaging is widely used as an essential tool for many clinical applications such as cancer diagnosis, tumor detection, evaluation of the lesion malignancy, staging of diseases, and treatment follow-up [1, 2]. Injection of a standard dose of radioactive tracer is normally required to achieve high-quality PET images in clinical settings. Though injection of high doses of radiopharmaceutical reduces the statistical noise and consequently leads to a better-quality PET image, it raises concerns due to the increased risk of radiation exposure to the patients and healthcare providers [3–5]. On the other hand, using a reduced dose of radioactive tracer would result in quality degradation in PET images due to the increased noise levels and loss of signal (low signal-to-noise ratio (SNR)) [6–8].
To address this issue, many efforts have been made to improve the quality of low-dose PET (L-PET) images through application of iterative reconstruction algorithms [9–12], image filtering and post-processing [13, 14], and machine learning (ML) methods [4, 15]. In iterative reconstruction algorithms, penalized reconstruction kernels and/or prior knowledge from the anatomical images (MR sequences) are exploited to regularize and/or guide PET image reconstruction to suppress the excessive noise induced by the reduce injected dose [12, 16]. There are two categories of post-reconstruction image denoising including image denoising techniques in the spatial domain and transform domain [13, 17]. These approaches may suffer from some drawbacks such as generating artifacts and/or pseudo signals, over smoothing, hallucinated structures, and high computational time. To address these issues, deep learning (DL) methods, as a special type of machine learning methods, have been dedicatedly developed in which a relationship between L-PET images and standard-dose PET (S-PET) images is learned to predict S-PET images from their low-dose counterparts [7, 18, 19]. Regarding the extraordinary performance of the deep learning methods [18], a number of deep learning-based solutions have been proposed for estimation of the high-quality PET from the low-dose versions with or without aid of anatomical images [20, 21].
Xu et al. [22] employed a U-Net model to predict S-PET images from 0.5% L-PET/MR images. This model exhibited superior performance in terms of noise suppression and signal recovery bias compared to conventional methods such as non-local means (NLM), block-matching and 3D filtering (BM3D), and auto-context network (AC-Net). Relying on the generative adversarial network (GAN), Wang et al. [23] introduced a 3D conditional GAN and compared it with 2D conditional GAN models, and some previously utilized approaches such as mapping-based sparse presentation method (m-SR), semi-supervised tripled dictionary learning method (t-DL), and convolutional neural networks (CNN). The GAN model, implemented in 3D mode, outperformed the aforementioned denoising approaches. Furthermore, Lei et al. [24] proposed a cycle-consistent GAN to estimate high-quality S-PET images from L-PET images (1/8th of S-PET). The results of this study showed a significant decrease in mean error (ME) values and a remarkable increase in PSNRs when using the cycle GAN model compared to L-PET and S-PET predicted by U-Net and conventional GAN models. Besides, Chen et al. [25] implemented a network similar to the model proposed by Xu et al. [22] to predict S-PET from a combination of L-PET (1% of S-PET) and MR images as well as L-PET alone. They claimed that the synthesized images generated from the combination of L-PET and MR bear higher image quality and lower SUV bias compared to the L-PET-alone model. All the above-mentioned studies merely consider the single-dose level of low-dose PET data as the input into the deep learning-based neural network, and they did not exploit the additional/auxiliary information that could be obtained from using multiple dose level of L-PET images (lower-dose levels of L-PET in addition to the original L-PET images).
The deep learning-based denoising approaches solely rely on a single-dose level of L-PET images as the input of models to predict the S-PET images. Given the PET raw data, any low-dose versions of the PET data could be generated. For instance, given a low-dose imaging at 10% of the standard dose, lower-dose levels of the current PET data, such as 8%, 6%, and 4%, could easily be generated/reconstructed from the 10% low-dose data. Due to the stochastic nature of PET acquisition, any of these low-dose versions of the PET data would bear complementary/additional information regarding the underlying signal in the standard PET image. In other words, all these lower-dose versions of the PET data contain the same or similar signals contaminated with different noise levels and/or distributions. In this light, lower-dose PET images could provide prior and/or additional knowledge for the prediction of the standard-dose PET images. This prior and/or additional knowledge could be exploited in a deep learning-based denoising framework to enhance the quality of standard dose prediction. To the best of our knowledge, this is the first study employing multiple low-dose levels of PET images as prior knowledge to develop a deep learning-based denoising model.
In this study, we investigate the benefits of utilizing additional information in the form of multiple low-dose PET images in a deep learning model. In this regard, we use 6% L-PET imaging data as the input of deep learning model, wherein lower-dose PET images with 4% and 2% of standard dose levels (extracted from the raw data of the 6% low-dose PET data) were employed as additional information.
Materials and Methods
Data Acquisition
This study was conducted on PET/CT brain images from 140 patients with head and neck malignant lesions (68 males and 72 females, 71 ± 9 years, mean age ± standard deviation (SD)) (100 subjects for training and 40 subjects for evaluation) acquired on a Biograph-6 scanner (Siemens Healthcare, Erlangen, Germany) with a standard dose of 210 ± 8 MBq of 18F-FDG. PET images were acquired for an acquisition time of 20 min, about 40 min after the injection. The PET raw data was registered in list-mode format, and then 6% low-dose PET data were extracted from the standard data. Then, lower-dose PET data, including 4%, 3%, and 2%, were generated from the 6% low-dose data. The low-dose data with the aforementioned percentages were reconstructed using ordered subsets-expectation maximization (OSEM) algorithm with 4 iterations and 18 subsets. The entire PET images were normalized to a range of between 0 and about 1 using a fixed normalization factor for the entire dataset.
The aim of low-dose and/or fast PET imaging is to reduce patient-absorbed dose in PET imaging (for instance in multiple follow-up PET scans) or acquire high-quality PET images in fast or short-time PET imaging (for instance in dynamic PET scans). To achieve clinically valid low-dose and/or fast PET imaging, the signal degradation due to low-count statistics should be retrievable to identify underlying signals and suppress statistical noise. In this regard, injected dose reduction in low-dose PET imaging or acquisition time reduction in fast PET imaging should not be too extreme that hinder/impair proper signal discrimination from the noise. Depending on the injected dose and the PET scanner sensitivity, low-dose or fast PET imaging could be conducted down to 1/30th of normal/standard PET imaging. In this light, noise suppression in 6% and 4% low-dose PET imaging was considered in this work that would greatly reduce the patient dose (equivalent to 20 × faster PET imaging), while the increased noise levels would not hinder the identification of the underlying signals. Beyond these levels, the quality of the PET images would be severely degraded (non-retrievable) with impaired clinical value.
Deep Neural Network Architecture
We adopted NiftyNet, an open-source convolutional neural networks (CNNs) platform for deep learning solutions in Python environment, to estimate S-PET images from L-PET images. The S-PET prediction was carried out using a high-resolution ResNet (HighResNet) model. This network consists of nineteen 3 × 3 × 3 convolution layers and one final 1 × 1 × 1 convolution layer. In the first seven layers with 16 kernels, the low-level features of the images such as corners and edges are extracted. The twelve subsequent convolution layers with 32 and 64 kernels and the final layer with 160 kernels are designed to capture medium and high-level features. Four different models were separately developed to compare/investigate the benefits of employing the multiple low-dose images as the additional information to the denoising network. The deep learning models were assessed to predict the standard-dose PET images from 6 to 4% low-dose data separately. For the 6% low-dose level, first, a model with a single input of 6% was first developed. Then, this model was compared with the model with 3-input channels getting 6%, 4%, and 2% low-dose data as input. In fact, the reference or output of these two models was identical (standard-dose PET images); only the model was once developed with a single-input channel (6% low dose) and once with three-input channels for 6%, 4%, and 2% low-dose data. Similarly, for 4% low-dose data, first, a model with only a single input for 4% low-dose data was developed. Then, the model was compared with a model with 3-input channels getting 4%, 3%, and 2% as input. The results for 4% and 6% low-dose PET images (each including single- and three-input models) were separately reported in two distinct tables. The same standard-dose PET images were considered as reference for the training of these four models.
The Details of Implementation
The training of the four models was performed using a L2-norm loss function (the best performance was observed with this loss function) with Adam optimizer and leaky ReLU as activation function. The models were developed in a 2-dimensional mode with batch size of 20, weight decay of 10−4, and max iteration of 104. The training was continued to reach the plateau of the training loss. Five percent of the dataset was considered as the evaluation set within the training process. No significant overfitting was observed during the training of the denoising models. The starting learning rate was set at 0.1 which was multiplied by 0.05 every 100 iterations following the recommendation by [26].
Evaluation Strategy
To evaluate the prediction performance of the different denoising models and the benefits of applying the prior knowledge in the form of multiple L-PET images, we utilized five quantitative metrics including structural similarity index (SSI), peak signal-to-noise ratio (PSNR), and root mean square error (RMSE) based on the standard uptake value (SUV) within the entire head region and the malignant lesions. Manual segmentation of the lesions, drawn by specialist, in the test dataset is available; thus, quantitative metrics were calculated separately for the malignant lesions. The lesions were segmented by nuclear medicine specialists as a part of the clinical routine for diagnosis, lesion assessment, and treatment. The lesion contouring was performed on PET images; however, the structures in the CT images were considered within lesion identification. In addition to the abovementioned metrics, we calculated the bias of mean SUV (SUVmean bias) and max SUV (SUVmax bias) on the predicated S-PET images versus the reference S-PET images for the malignant lesions.
Results
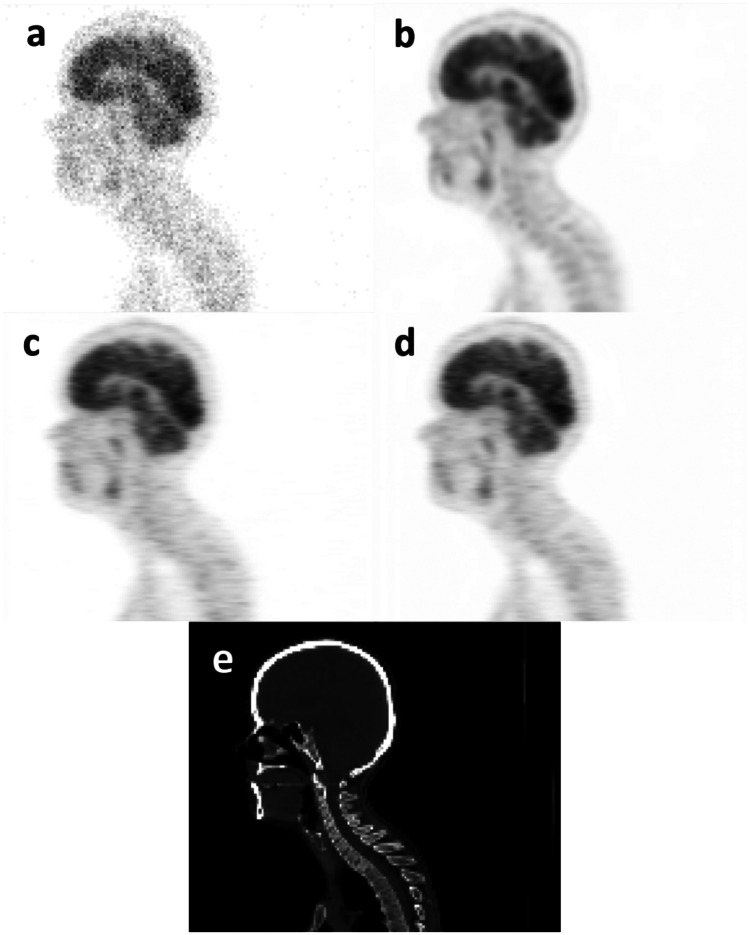
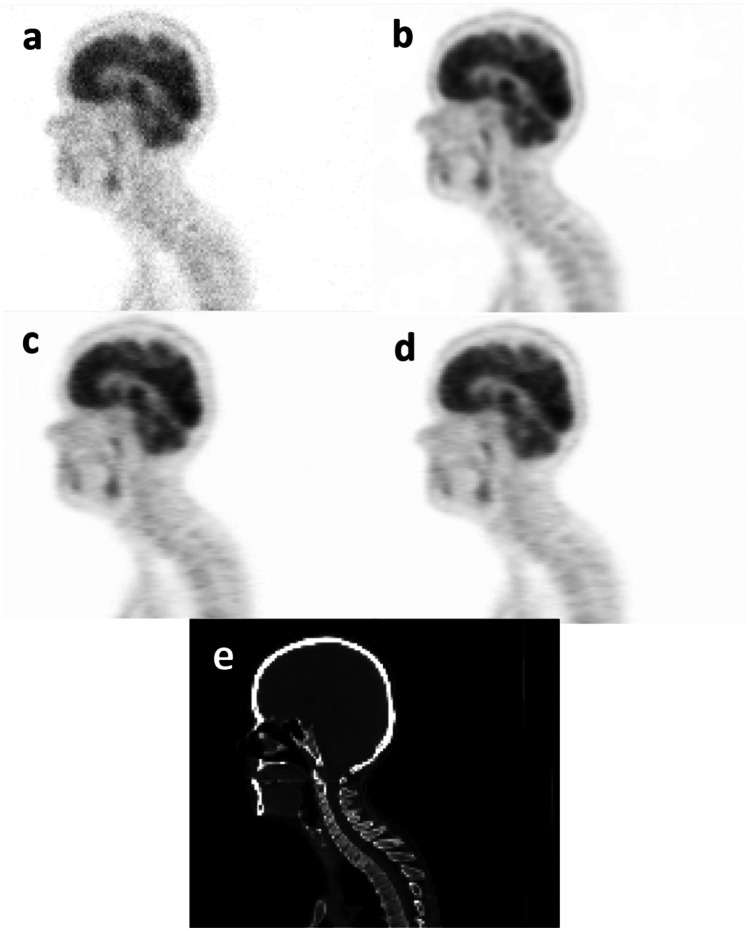
In order to visually compare the quality of the predicted PET images, sagittal views of the S-PET images predicted by the single-input and the multi-input networks together with the ground truth S-PET image, the corresponding CT image, and the L-PET images of a representative patient are shown in Figs. 1 and 2. The increased levels of signal-to-noise ratio (SNR) were observed when using multi-input models compared to the single-input models.
Fig. 1.
Sagittal views of a 4% L-PET image, b ground-truth S-PET image, c S-PET predicted by the single-input network (4% L-PET image), d S-PET predicted by the multi-input network (3% and 2% L-PET images as prior knowledge, in addition to 4% L-PET image), and e CT image
Fig. 2.
Sagittal views of a 6% L-PET image, b ground-truth S-PET image, c S-PET predicted by the single-input network (6% L-PET image), d S-PET predicted by the multi-input network (4% and 2% L-PET images as prior knowledge, in addition to 6% L-PET image), and e CT image
To quantitatively assess the quality of predicted PET images, SSI, PSNR, and RMSE metrics were calculated within the entire head region as well as the malignant lesions. Furthermore, the SUV bias for the lesions were measured on the S-PET images predicted from single-input and multi-input models versus the ground-truth S-PET images. Among the 140 patient datasets, 40 cases were not used within the training/development of the models, and they were regarded as the external dataset for the evaluation of different models. Table 1 summarizes the average and the standard deviation of the abovementioned metrics calculated across 40 patients in the external test dataset for 4% low-dose PET imaging, wherein 3% and 2% low-dose PET data were used as prior knowledge. Besides, Table 2 reports the same metrics calculated for low-dose PET imaging with 6% of S-PET data. The differences between the predicted S-PET images by the single-input and multi-input models were assessed with the paired t-test analysis (considering P-values of smaller than 0.05 statistically significant). The paired t-test analysis demonstrated statistically significant differences between single-input and multi-input models and the benefits of employing the prior knowledge in the form of lower-dose PET images.
Table 1.
Quantitative metrics calculated for the S-PET images predicted by the single-input (4% L-PET image) and multi-input (4%, 3%, and 2% L-PET images) deep learning models. P-values are calculated between the single-input and multi-input denoising models
| L-PET (4%) | Single-input (4%) | Multi-input (4%, 3%, 2%) | P-value | ||
|---|---|---|---|---|---|
| PSNR ± SD | 29.59 ± 1.26 | 39.73 ± 2.75 | 41.87 ± 2.81 | 0.01 | |
| SSI ± SD | 0.86 ± 0.033 | 0.97 ± 0.005 | 0.98 ± 0.003 | 0.04 | |
| RMSE ± SD (head) | 0.30 ± 0.04 | 0.09 ± 0.03 | 0.07 ± 0.02 | 0.01 | |
| RMSE ± SD (lesion) | 1.19 ± 0.23 | 0.59 ± 0.24 | 0.44 ± 0.18 | < 0.01 | |
| SUVmean bias ± SD(%) (lesion) | − 0.01 ± 0.93 | − 1.92 ± 1.53 | −0.68 ± 0.77 | < 0.01 | |
| SUVmax bias ± SD(%) (lesion) | 20.20 ± 5.29 | − 2.29 ± 6.02 | 1.44 ± 3.95 | < 0.01 | |
Table 2.
Quantitative metrics calculated for the S-PET images predicted by the single-input (6% L-PET image) and multi-input (6%, 4%, and 2% L-PET images) deep learning models. P-values are calculated between the single-input and multi-input denoising models
| L-PET (6%) | Single-input (6%) | Multi-input (6%, 4%, 2%) | P-value | |
|---|---|---|---|---|
| PSNR ± SD | 35.33 ± 1.27 | 41.44 ± 2.90 | 44.89 ± 2.89 | 0.01 |
| SSI ± SD | 0.93 ± 0.016 | 0.98 ± 0.002 | 0.99 ± 0.001 | 0.04 |
| RMSE ± SD (head) | 0.15 ± 0.02 | 0.08 ± 0.02 | 0.05 ± 0.02 | < 0.01 |
| RMSE ± SD (lesion) | 0.61 ± 0.11 | 0.48 ± 0.20 | 0.29 ± 0.13 | < 0.01 |
| SUVmean bias ± SD(%) (lesion) | 0.09 ± 0.45 | − 5.46 ± 0.65 | −0.71 ± 0.49 | < 0.01 |
| SUVmax bias ± SD(%) (lesion) | 9.08 ± 1.34 | − 4.43 ± 2.05 | 2.41 ± 1.55 | < 0.01 |
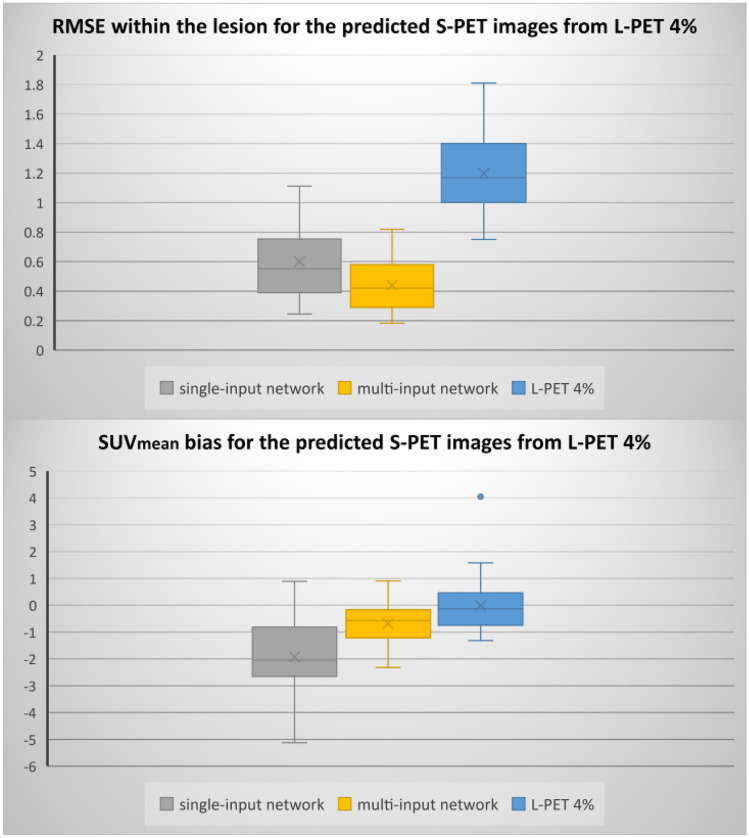
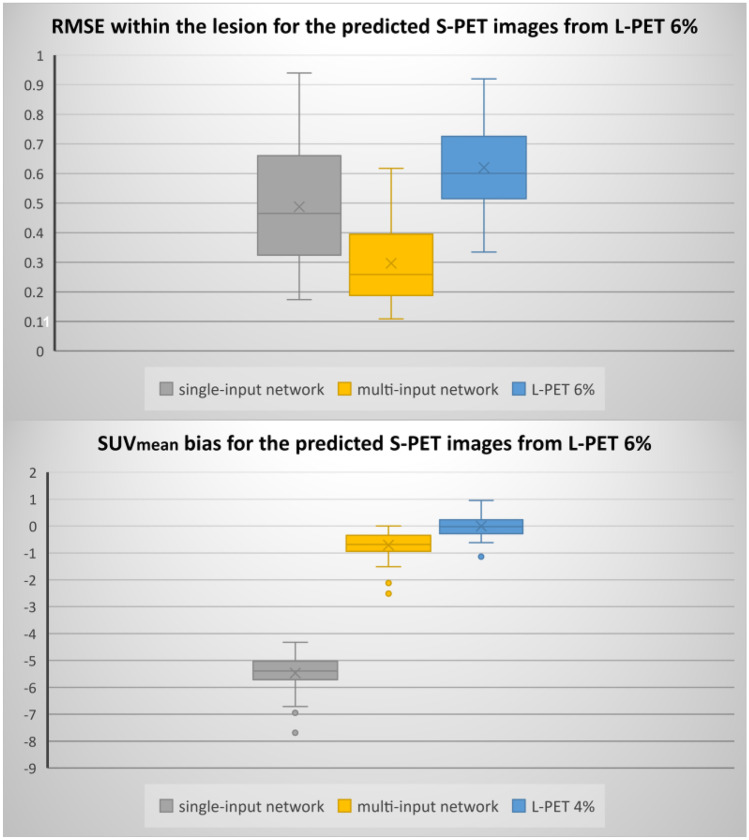
Moreover, to meticulously investigate the performance of the models, the boxplots of RMSE and SUVmean bias within the malignant lesions are presented in Figs. 3 and 4.
Fig. 3.
Boxplots of SUVmean bias and RMSE within the lesions for the different models and 4% L-PET
Fig. 4.
Boxplots of SUVmean bias and RMSE within the lesion regions for the different models and 6% L-PET
Discussion
In this study, we set out to examine the benefits of employing the prior knowledge in the form of lower-dose PET images for the task of normal PET prediction from low-dose PET data. In this regard, the quality of the predicted S-PET images from the L-PET images at different 4% and 6% dose levels of the standard dose was assessed using two models with single-input and multi-input architectures. As shown in Figs. 1 and 2, the quality of predicted PET image improved for both 6% and 4% low-dose imaging when multi-input models were employed getting lower-dose PET images as complementary knowledge. Regarding Figs. 1 and 2, the multi-input networks resulted in better image quality compared to single-input networks.
The analysis of the results in Table 1 indicates the superiority of the network trained with multi-input L-PET images over the single input. The results in Table 1 demonstrated that the 4% multi-input network (using 2% and 3% L-PET as the input in addition to 4% L-PET) would lead to a significant decrease in RMSE by 22.22% (from 0.09 to 0.07) and by 25.42% (from 0.59 to 0.44) within the entire head region and lesions, respectively, compared to the single-input model (using only 4% L-PET images as input). Furthermore, the network trained with multiple L-PET images produces higher values for the SSI and PSNR metrics. It should be emphasized that utilizing prior knowledge in the form of multi-input network remarkably decreases the SUVmean bias and SUVmax bias within the lesions by 64.58% (from 1.92 to 0.68%) and by 37.12% (from 2.29 to 1.44%), respectively.
Regarding Table 2, a similar trend was observed in the analysis of the predicted image by the network trained with 6% L-PET images. SSI and RMSE metrics significantly improved with multi-input network (taking 4% and 2% L-PETs in addition to 6% L-PET). In addition, utilizing multi-input network reduced RMSE within the head region, RMSE within the malignant lesions, and SUVmean bias in the lesions by 37.5%, 39.58%, and 86.99%, respectively. Furthermore, using multi-input model led to a noticeable decrease in the bias of SUVmax by 45.60% (from 4.43 to 2.41%).
Figures 3 and 4 compared the distributions of the SUVmean bias and RMSE within the lesions for both single-input and multi-input models. Quantitative results in these figures demonstrate the superior performance of the multi-input network over the single-input network. Regarding Fig. 3, the median of RMSE and SUVmean bias for single-input and multi-input networks within the lesion are 0.55 and 0.42 (SUV) (for RMSE) and − 2.04 and − 0.56 (for SUVmean bias), respectively. Regarding Fig. 4, the multi-input network (with three-input channels for 6%, 4%, and 2% low-dose PET images) yielded a median RMSE of 0.25 (SUV) SUVmean bias of − 0.68, compared to 0.46 and − 5.39 achieved by the single-input network. In addition, both Figs. 3 and 4 indicated higher variation of errors in the single-input models in comparison with the multi-input models.
To determine the statistical significance of the differences between these metrics, P-values calculated for SUVmean bias, SUVmax bias, and RMSE (within the lesion) demonstrated significant improvement achieved through employing low-dose prior knowledge in the denoising models.
The primary aim of this work was to investigate the benefits of employing lower-dose PET data in the denoising models for prediction of the standard PET images. Owing to the stochastic nature of PET acquisition and signal formation, reconstruction of the PET data at different low-dose (or low-count) levels would provide different distributions of noise and presentations of signal-to-noise ratio. Since the underlying signals/uptake patterns in these images are the same, and they are contaminated with different noise levels and/or distributions, they would provide valuable knowledge to the model to distinguish between the noise and underlying signals. The benefits of lower-dose prior knowledge were demonstrated for two low-dose levels of 6% and 4%, wherein significant improvement was observed in both models. The proposed framework could be used in any denoising models where the raw PET (or SPECT) data are available, and the image reconstruction could be performed with different count/dose levels. Moreover, this framework is applicable to the models implemented in the sinogram or projection domain [7, 27–29].
Regarding the limitation of this work, it should be noticed that the data utilized in this work were obtained from a Biograph 6 PET/CT scanner which does not have time-of-flight (TOF) imaging capability. TOF imaging would result in an enhanced signal-to-noise ratio. In this light, in modern PET scanners, lower levels of low-dose imaging could be conducted with similar image quality. Thus, the results presented in this work could be valid for this PET scanner, and further investigation is required for TOF PET imaging. Moreover, previous studies have shown that the incorporation of structural information (for instance, in the form of MR images) would improve the quality and quantitative accuracy of low-dose imaging [25]. In this work, no structural information was considered within the model development which could further improve the outcome of these models. Moreover, only a ResNet model was employed to train the different models; however, novel deep learning architectures such as GAN models may lead to improved outcomes.
Conclusion
In this paper, we applied prior knowledge/additional information to the deep learning-based denoising models via utilizing multiple dose levels of L-PET data as the extra input channels to network to estimate the S-PET images. The quantitative evaluation of the proposed framework demonstrated the benefits of employing lower-dose PET data in the denoising models for prediction of the standard PET images. The proposed framework was examined for 6% and 4% low-dose imaging levels. This study recommends using of multiple levels of low-dose PET imaging as prior knowledge to predict the standard-dose PET images.
Author Contribution
All authors contributed to the study conception and design. All authors read and approved the final manuscript.
Data Availability
Data supporting the findings of this study are available from the corresponding author upon journal request.
Declarations
Ethics Approval
The patients involved in this study gave their informed consent for data collection.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Behnoush Sanaei, Email: behnoush.sanaeii@gmail.com.
Reza Faghihi, Email: faghihir@shirazu.ac.ir.
Hossein Arabi, Email: hossein.arabi@unige.ch.
References
- 1.Basu S, Hess S, Braad PE, Olsen BB, Inglev S, Høilund-Carlsen PF. The basic principles of FDG-PET/CT imaging. PET clinics. 2014;9(4):355–370. doi: 10.1016/j.cpet.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Zimmer L. PET imaging for better understanding of normal and pathological neurotransmission. Biologie aujourd'hui. 2019;213(3–4):109–120. doi: 10.1051/jbio/2019025. [DOI] [PubMed] [Google Scholar]
- 3.Khoshyari-morad Z, Jahangir R, Miri-Hakimabad H, Mohammadi N, Arabi H: Monte Carlo-based estimation of patient absorbed dose in 99mTc-DMSA,-MAG3, and-DTPA SPECT imaging using the University of Florida (UF) phantoms. arXiv preprint arXiv:2103.00619. 2021 Feb 28.
- 4.Sanaat A, Shiri I, Arabi H, Mainta I, Nkoulou R, Zaidi H. Deep learning-assisted ultra-fast/low-dose whole-body PET/CT imaging. European journal of nuclear medicine and molecular imaging. 2021;48(8):2405–2415. doi: 10.1007/s00259-020-05167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahey FH. Dosimetry of pediatric PET/CT. Journal of Nuclear Medicine. 2009;50(9):1483–1491. doi: 10.2967/jnumed.108.054130. [DOI] [PubMed] [Google Scholar]
- 6.Sanaei B, Faghihi R, Arabi H: Quantitative investigation of low-dose PET imaging and post-reconstruction smoothing. arXiv preprint arXiv:2103.10541. 2021 Mar 18.
- 7.Sanaat A, Arabi H, Mainta I, Garibotto V, Zaidi H. Projection space implementation of deep learning–guided low-dose brain PET imaging improves performance over implementation in image space. Journal of Nuclear Medicine. 2020;61(9):1388–1396. doi: 10.2967/jnumed.119.239327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aghakhan Olia N, Kamali-Asl A, Hariri Tabrizi S, Geramifar P, Sheikhzadeh P, Farzanefar S, Arabi H, Zaidi H. Deep learning–based denoising of low-dose SPECT myocardial perfusion images: quantitative assessment and clinical performance. European journal of nuclear medicine and molecular imaging. 2022;49(5):1508–1522. doi: 10.1007/s00259-021-05614-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Case JA. 3D iterative reconstruction can do so much more than reduce dose. Journal of Nuclear Cardiology. 2019;2:1–5. doi: 10.1007/s12350-019-01827-4. [DOI] [PubMed] [Google Scholar]
- 10.Yu X, Wang C, Hu H, Liu H. Low dose PET image reconstruction with total variation using alternating direction method. PloS one. 2016;11(12):e0166871. doi: 10.1371/journal.pone.0166871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeraatkar N, Sajedi S, Farahani MH, Arabi H, Sarkar S, Ghafarian P, Rahmim A, Ay MR. Resolution-recovery-embedded image reconstruction for a high-resolution animal SPECT system. Physica Medica. 2014;30(7):774–781. doi: 10.1016/j.ejmp.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 12.Mehranian A, Reader AJ. Model-based deep learning PET image reconstruction using forward–backward splitting expectation–maximization. IEEE transactions on radiation and plasma medical sciences. 2020;5(1):54–64. doi: 10.1109/TRPMS.2020.3004408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arabi H, Zaidi H. Improvement of image quality in PET using post-reconstruction hybrid spatial-frequency domain filtering. Physics in Medicine & Biology. 2018;63(21):215010. doi: 10.1088/1361-6560/aae573. [DOI] [PubMed] [Google Scholar]
- 14.Arabi H, Zaidi H. Non-local mean denoising using multiple PET reconstructions. Annals of nuclear medicine. 2021;35(2):176–186. doi: 10.1007/s12149-020-01550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou L, Schaefferkoetter JD, Tham IW, Huang G, Yan J. Supervised learning with cyclegan for low-dose FDG PET image denoising. Medical image analysis. 2020;65:101770. doi: 10.1016/j.media.2020.101770. [DOI] [PubMed] [Google Scholar]
- 16.Bland J, Mehranian A, Belzunce MA, Ellis S, McGinnity CJ, Hammers A, Reader AJ. MR-guided kernel EM reconstruction for reduced dose PET imaging. IEEE transactions on radiation and plasma medical sciences. 2017;2(3):235–243. doi: 10.1109/TRPMS.2017.2771490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arabi H, Zaidi H. Spatially guided nonlocal mean approach for denoising of PET images. Medical physics. 2020;47(4):1656–1669. doi: 10.1002/mp.14024. [DOI] [PubMed] [Google Scholar]
- 18.Arabi H, Zaidi H. Applications of artificial intelligence and deep learning in molecular imaging and radiotherapy. European Journal of Hybrid Imaging. 2020;4(1):1–23. doi: 10.1186/s41824-020-00086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arabi H, AkhavanAllaf A, Sanaat A, Shiri I, Zaidi H. The promise of artificial intelligence and deep learning in PET and SPECT imaging. Physica Medica. 2021;83:122–137. doi: 10.1016/j.ejmp.2021.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Chen KT, Gong E, de Carvalho Macruz FB, Xu J, Boumis A, Khalighi M, Poston KL, Sha SJ, Greicius MD, Mormino E, Pauly JM. Ultra–low-dose 18F-florbetaben amyloid PET imaging using deep learning with multi-contrast MRI inputs. Radiology. 2019;290(3):649–656. doi: 10.1148/radiol.2018180940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Wu J, Lu W, Onofrey JA, Liu YH, Liu C. Noise reduction with cross-tracer and cross-protocol deep transfer learning for low-dose PET. Physics in Medicine & Biology. 2020;65(18):185006. doi: 10.1088/1361-6560/abae08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu J, Gong E, Pauly J, Zaharchuk G: 200x low-dose PET reconstruction using deep learning. arXiv preprint arXiv:1712.04119, 2017.
- 23.Wang Y, Yu B, Wang L, Zu C, Lalush DS, Lin W. … Zhou L: 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage. 2018;174:550–562. doi: 10.1016/j.neuroimage.2018.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei Y, Dong X, Wang T, Higgins K, Liu T, Curran W. J, … Yang X: Whole-body PET estimation from low count statistics using cycle-consistent generative adversarial networks. Physics in Medicine & Biology. 2019;64(21):215017. doi: 10.1088/1361-6560/ab4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen KT, Gong E, de Carvalho Macruz F. B, Xu J, Boumis A, Khalighi M, … Zaharchuk G: Ultra–low-dose 18F-florbetaben amyloid PET imaging using deep learning with multi-contrast MRI inputs. Radiology. 2019;290(3):649–656. doi: 10.1148/radiol.2018180940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith LN: A disciplined approach to neural network hyper-parameters: Part 1--learning rate, batch size, momentum, and weight decay. arXiv preprint arXiv:1803.09820. 2018 Mar 26.
- 27.Arabi H, Zaidi H. Assessment of deep learning-based PET attenuation correction frameworks in the sinogram domain. Physics in Medicine & Biology. 2021;66(14):145001. doi: 10.1088/1361-6560/ac0e79. [DOI] [PubMed] [Google Scholar]
- 28.Olia NA, Kamali-Asl A, Tabrizi SH, Geramifar P, Sheikhzadeh P, Farzanefar S, Arabi H: Deep learning-based noise reduction in low dose SPECT Myocardial Perfusion Imaging: Quantitative assessment and clinical performance. arXiv preprint arXiv:2103.11974. 2021 Mar 22.
- 29.Olia NA, Kamali-Asl A, Tabrizi SH, Geramifar P, Sheikhzadeh P, Arabi H, Zaidi H: Deep Learning-based Low-dose Cardiac Gated SPECT: Implementation in Projection Space vs. Image Space. In2021 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC) 2021 Oct 16 (pp. 1–3). IEEE.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data supporting the findings of this study are available from the corresponding author upon journal request.