Abstract
Converting tumor-associated macrophages (TAMs) from the M2 to the M1 phenotype is considered an effective strategy for cancer therapy. TRAF3 is known to regulate NF-κB signaling. However, the role of TRAF3 in TAM polarization has not yet been completely elucidated. Here, we found that ablation of TRAF3 increased M1 markers, iNOS, FGR and SLC4A7, while down-regulated M2 markers, CD206, CD36 and ABCC3, expression levels in macrophages. Moreover, TRAF3 deficiency enhanced LPS-induced M1 and abolished IL-4-induced macrophage polarization. Next, quantitative ubiquitomics assays demonstrated that among the quantitative 7618 ubiquitination modification sites on 2598 proteins, ubiquitination modification of IL-4 responding proteins was the most prominently reduced according to enrichment analysis. STAT6, a key factor of IL-4 responding protein, K450 and K129 residue ubiquitination levels were dramatically decreased in TRAF3-deficient macrophages. Ubiquitination assay and luciferase assay demonstrated that TRAF3 promotes STAT6 ubiquitination and transcriptional activity. Site mutation analysis revealed STAT6 K450 site ubiquitination played a vital role in TRAF3-mediated STAT6 activation. Finally, B16 melanoma mouse model demonstrated that myeloid TRAF3 deficiency suppressed tumor growth and lung metastasis in vivo. Taken together, TRAF3 plays a vital role in M2 polarization via regulating STAT6 K450 ubiquitination in macrophages.
Subject terms: Cancer microenvironment, Signal transduction, Ubiquitylation
Introduction
Macrophages are important components of innate immune cells and trigger a series of inflammatory and responses [1]. Tumor-associated macrophages (TAMs) are the most abundant immune cells in various tumor microenvironments [2, 3]. TAMs are highly plastic, which can be polarized into classically activated (M1) macrophages and alternatively activated (M2) macrophages [4–6]. M1 TAMs produce substantial inflammatory cytokines which facilitate anti-tumor immune responses and induce Th1 immunity, whereas alternatively activated M2 TAMs suppress tumor immune responds and stimulate tumor angiogenesis and progression [7, 8].
Emerging evidence demonstrates that targeting TAMs provides new weaponry to the immune arsenal. Technologies to disable the pro-tumorigenic M2 TAMs and increasing anti-tumorigenic M1 TAMs has been an obvious strategy for years. Several strategies have been proposed to block the recruitment of pro-tumor macrophages [9, 10], systemic activate anti-tumor macrophages [11] and target phagocytic signals to improve antibody therapy [12].
Tumor necrosis factor receptor (TNFR)-associated factor 3 (TRAF3) is an E3 ubiquitin ligase that participates in the activation and regulation of various protein kinases and phosphatases [13, 14]. TRAF3 can directly catalyze Lys (K) 63-linked polyubiquitination at the K174 site of apoptosis-associated speck-like protein containing a CARD (ASC), which is critical for speck formation and inflammasome activation [15]. TRAF3 can also mediate K48-linked ubiquitination degradation of NF-κB-inducing kinase (NIK) by recruiting two other E3 ubiquitin ligases, TRAF2 and cIAPs, to inhibit non-classical NF-κB and STING signal activation [16]. Our previous study confirmed that TRAF3 regulates Lys48-linked ubiquitination and degradation of Unc-51 like autophagy activating kinase 1 (ULK1) and activates NLRP3 inflammasome and pyroptosis in macrophages [17, 18]. TRAF3 catalyzes various types of polyubiquitinated chains, activating or degrading substrate molecules, and regulates signal transduction processes. However, the molecular mechanism of TRAF3 regulating TAMs remains unclear.
Signal transducer and activator of transcription 6 (STAT6) is a member of the STAT family of transcription factors and plays a central role in IL-4-mediated M2 TAM polarization [19–21]. STAT6 undergoes phosphorylation [22], acetylation [20], ubiquitination [19] and O-GlcNAcylation [23] under certain conditions and regulates various biological processes. Acetyltransferase CREB binding protein (CBP) acetylates K383 site of STAT6 and inhibits STAT6 transcriptional activity and macrophage M2 polarization [20]. Janus kinases (JAKs) recruit and phosphorylate STAT6 at Tyr641 and Ser756 sites, resulted in STAT6 dimer formation, nuclear translocation and transcriptional activation of M2 macrophage marker gene [24–26]. Several E3 ligases, such as RNF128 and RTA, mediate Lys48-linked polyubiquitination and degradation of STAT6, forming a negative feedback loop to inhibit the polarization of M2 TAMs [27, 28]. More recently, epithelial O-linked N-Acetylglucosamine modification (O-GlcNAcylation) of STAT6 was reported, which was demonstrated to activate the expression of GSDMC family genes and contribute to inflammation [23]. Although various forms of modifications activate or inhibit transcriptional activity of STAT6, how M2 TAM polarization is fine-turned by STAT6 remains unknown.
In the present study, we identified that E3 ubiquitin ligase TRAF3 regulates the ubiquitination of STAT6 K450 site in macrophages, and the ubiquitination level of STAT6 K450 positively correlates with the transcription activity of STAT6. Consequently, myeloid conditional TRAF3 deficiency dramatically suppresses STAT6 ubiquitination and M2 macrophage polarization and enhances the anti-tumor immune responds of macrophages.
Results
TRAF3 regulates M1/M2 macrophage polarization
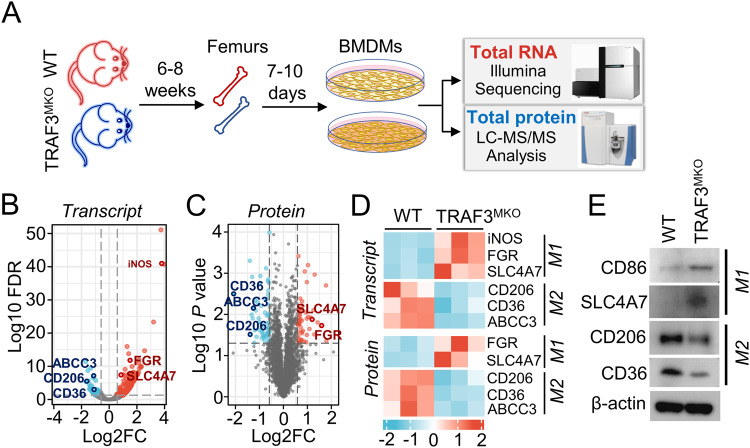
Previous studies indicated that TRAF3 regulates proinflammation cytokine production and innate immunity [29, 30]. To explore the potential role of TRAF3 in anti-tumor immunity, we analyzed cell type expression levels of TRAF3 in malignant solid tumor tissues using GEPIA2021 (http://gepia2021.cancer-pku.cn/) Sub-expression platform. As shown in Supplementary Fig. S1, TRAF3 expression levels were dramatically higher in M2 macrophages than other tumor infiltrated immune cell type, including CD4+ T cells, M1 macrophages, dendritic cells and natural killer (NK) cells. To further investigate the molecular mechanism of TRAF3 in myeloid cells, we constructed myeloid cell-conditional TRAF3 knockout C57BL/6 mice and the primary macrophages, BMDMs, were generated from WT and TRAF3MKO mice. Transcriptome and proteome expression profiles of BMDMs were analyzed by Illumina mRNA sequencing and LC-MS/MS protein peptides analysis (Fig. 1A). Compared with WT BMDMs, there are 325 different expression mRNAs (fold change >1.5, false discovery rate (FDR) < 0.05) and 110 different expression proteins (fold change >1.5, p < 0.05) in TRAF3MKO BMDMs (Fig. 1B, C). M1 macrophage marker genes, inducible NO Synthase (iNOS, also known as NOS2), significantly up-regulated in mRNA levels (Fig. 1D). Both protein and mRNA levels of M1 markers, FGR and SLC4A7, were up-regulated, and M2 markers, CD206 (also known as MRC1), CD36 and ABCC3 were down-regulated in TRAF3-deficient BMDMs (Fig. 1D). Moreover, the protein levels of M1 markers, CD86 and SLC4A7, and M2 markers, CD206 and CD36, were detected using immunoblotting, which showed that CD86 and SLC4A7 were significantly increased while CD206 and CD36 decreased in TRAF3MKO BMDMs (Fig. 1E). These results suggested the important role of TRAF3 in M1/M2 macrophage polarization.
Fig. 1. Combined analysis of transcriptome and proteome revealed TRAF3 regulates M1/M2 macrophage polarization.
A BMDMs were generated by cultivating bone marrow cells isolated from the femurs of 6–8 weeks old WT and TRAF3MKO mice. Total RNA was extracted and RNA-sequencing analysis was performed using Illumina Hiseq2500/X platform. Cell proteome expression profile was analyzed using LC-MS/MS analysis. B Volcano map of RNA-sequencing different expression genes. M1/M2 macrophage marker genes are provided. Fold change >1.5, FDR < 0.05, n = 3 for each group. C Volcano map of proteome expression profile. M1/M2 macrophage markers are provided. Fold change >1.5, p < 0.05, n = 3 for each group. D Heat map of M1/M2 macrophage marker gene mRNA and protein levels in WT and TRAF3MKO BMDMs. E BMDMs were harvested and lysed with RIPA buffer. Immunoblotting assays were performed to analyze M1 and M2 marker expression.
TRAF3 deficiency decreased IL-4-induced M2 macrophage polarization and promotes LPS-induced M1 macrophage polarization
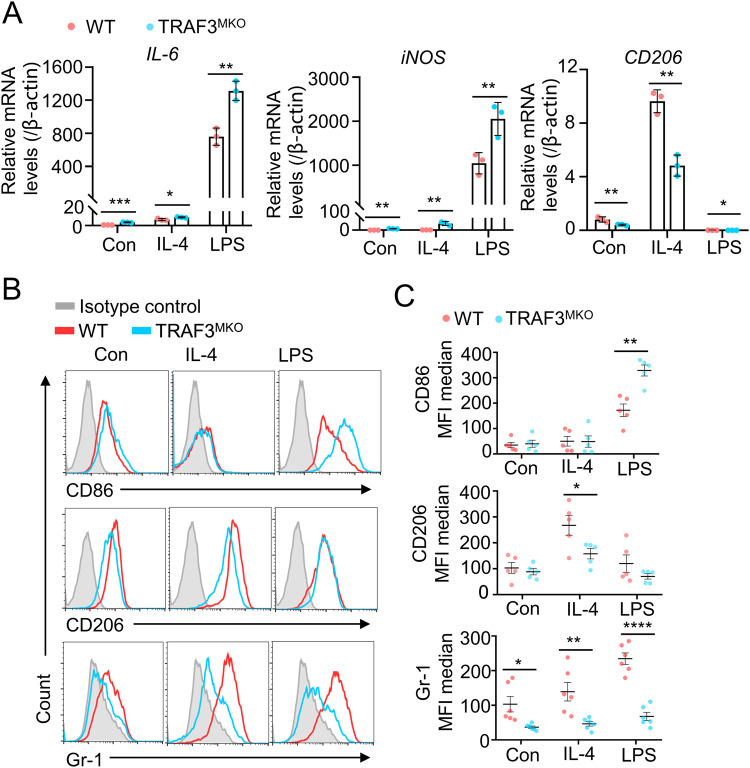
To further analyze the role of TRAF3 in macrophage polarization, BMDMs from WT and TRAF3MKO mice were treated with IL-4 and LPS, respectively, for 48 h to induce polarization, and M1/M2 marker genes were detected. In untreated cells, IL-6 and iNOS mRNA levels were significantly higher and CD206 mRNA level were reduced in TRAF3MKO BMDMs (Fig. 2A). LPS treatment significantly increased M1 marker genes, IL-6 and iNOS mRNA levels, which was 814.16 and 1773.52 fold higher than that of control group, respectively (Fig. 2A, left and middle panels). IL-4 treatment promotes mRNA expression of CD206, a M2 marker gene, which was 11.88 fold higher than that of untreated cells (Fig. 2A, right panel). Compared with WT BMDMs, IL-6 and iNOS expression levels significantly increased in LPS-treated TRAF3MKO BMDMs, and CD206 level decreased in IL-4-treated TRAF3MKO BMDMs (Fig. 2A). Furthermore, the protein levels of membrane M1/M2 markers were determined by flow cytometry analysis and immunoblotting. TRAF3 deficiency significantly promoted CD86 level in LPS-induced M1 macrophages and attenuated CD206 level in IL-4-induced M2 macrophages (Fig. 2B, C, upper and middle panels and Supplementary Fig. S3).
Fig. 2. TRAF3 deficiency decreased IL-4-induced M2 macrophage polarization and promotes LPS-induced M1 macrophage polarization.
A BMDMs from WT and TRAF3MKO mice were treated with IL-4 (10 ng/ml) or LPS (1 μg/ml) for 48 h. Total RNA was extracted and qRT-PCR analysis of IL-6, CD206 and iNOS were performed. Bar graphs with error bars are represented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001; n = 3 for each group. B, C BMDMs from WT and TRAF3MKO mice were treated with IL-4 (10 ng/ml) or LPS (1 μg/ml) for 48 h. Gr-1, CD206 and CD86 levels were analyzed by flow cytometric analysis. B Representative FACS plots are shown; C Summary graphs of MFI median based on multiple mice (n = 6/group) are shown. Scattered graphs with error bars are represented as mean ± SD. Each panel is a representative experiment of at least three independent biological replicates. *p < 0.05, **p < 0.01, ***p < 0.001.
In addition to macrophages, myeloid-derived suppressor cells (MDSCs) also derived from myeloid cells and play critical roles in tumor immunosuppression and metastasis [31, 32]. Several evidences demonstrated that MDSCs can differentiate to macrophages under certain conditions and alter tumor immunosuppressive microenvironment [33, 34]. Interestingly, we found that the expression levels of MDSC marker, GR-1, were dramatically down-regulated in untreated, IL-4- or LPS-stimulated TRAF3-deficient BMDMs (Fig. 2B, C, lower panels), which suggested TRAF3 may serve as a regulator MDSC differentiation. Taken together, these results demonstrated that TRAF3 plays vital roles in myeloid-derived M1/M2 macrophage polarization and MDSC differentiation.
Quantitative ubiquitomics demonstrated that TRAF3 regulates M2 macrophage programming via ubiquitinating IL-4 response proteins
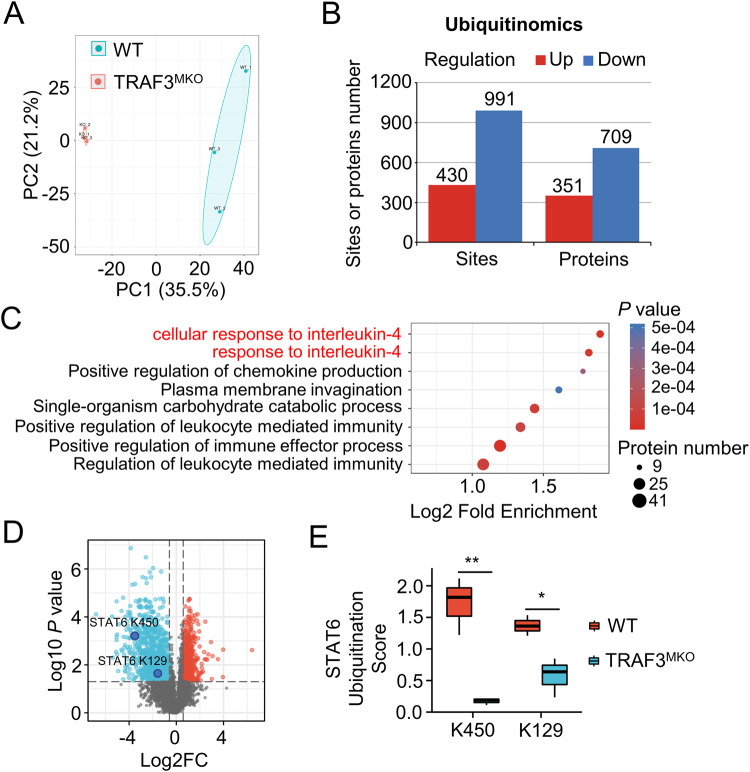
TRAF3 is known to be an E3 ubiquitin ligase, which catalyzes K48-linked [16] and K63-linked [15] ubiquitination of target molecules and drives activation or inhibition of downstream signaling [35]. To reveal the molecular mechanism of TRAF3 regulating M1/M2 macrophage polarization, quantitative ubiquitomics were analyzed in WT and TRAF3-deficient BMDMs. Based on enrichment of ubiquitin-modified peptides combined with mass spectrometry-based quantitative proteomics technology, 17,485 ubiquitination modification sites on 4326 proteins were detected. Among them, 7618 ubiquitination modification sites on 2598 proteins contain quantitative information, which was used for subsequent bioinformatics analysis (Supplementary Table S1). The repeatability of the quantitative ubiquitomics was assessed by principal component analysis (PCA) and the results showed good aggregation and quantitative repeatability (Fig. 3A). A total 1421 differential modified sites (TRAF3MKO/WT, fold change >1.5, p < 0.05, 430 up-regulated, 991 down-regulated) on 1060 proteins (351 up-regulated, 709 down-regulated) were found in macrophages (Fig. 3B). These results revealed the critical role of TRAF3 on ubiquitin modification.
Fig. 3. Quantitative ubiquitomics assays demonstrated that TRAF3 regulates IL-4 pathway.
BMDMs from WT and TRAF3MKO mice were treated with MG132 for 2 h before harvest and quantitative ubiquitomics assays were performed. A Principal component analysis (PCA) was done to evaluate the quantitative repeatability of ubiquitination modification. B Bar graph shows the significant different proteins or sites number of ubiquitin modification. Regulation means TRAF3MKO/WT up-regulated or down-regulated. Fold change >1.5, p < 0.05. C Functional enrichment of Gene Ontology (GO) analysis of biological process was done based on down-regulated ubiquitination modifying protein. For each category, a two-tailed Fisher’s exact test was employed to test the enrichment of the differentially modified protein against all identified proteins. The GO with a corrected p value < 0.05 is considered significant. D Volcano map of ubiquitin-modified sites profile. Blue, TRAF3MKO/WT down-regulated sites; red, TRAF3MKO/WT up-regulated sites. Fold change >1.5, p < 0.05, n = 3 for each group. E STAT6 K450 and K129 residue ubiquitination scores in WT and TRAF3MKO BMDMs. Student’s t test, *p < 0.05, **p < 0.01, n = 3 for each group.
Interestingly, functional enrichment analysis revealed that TRAF3 deficiency leaded to prominently reduced ubiquitination in IL-4 response proteins (Fig. 3C), which are key factors in programming M2 macrophages [36]. There were 13 down-regulated ubiquitinated sites on 10 IL-4 responding proteins in TRAF3-deficient BMDMs (Supplementary Table S2). Among the differential modified proteins, STAT6, the classical downstream factor of IL-4, K450 and K129 site ubiquitination was significantly attenuated in TRAF3-deficient macrophages (Fig. 3D–F). Taken together, these results suggested that TRAF3 regulates M2 macrophage programming via ubiquitinating IL-4 response proteins.
TRAF3 regulated STAT6 K450 ubiquitination and transcriptional activation
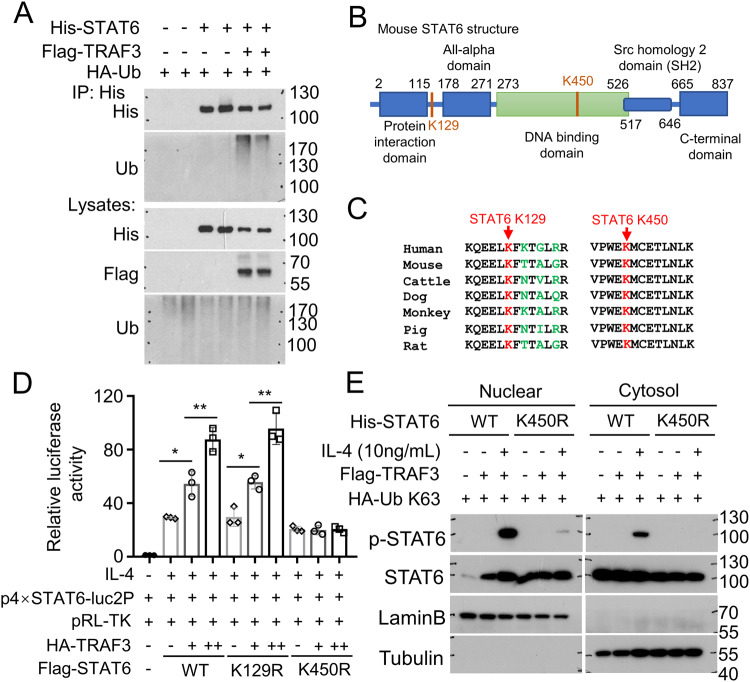
Our finding that ablation of TRAF3 in macrophages blocks the ubiquitination of STAT6 in K450 and K129 sites raise the question of whether TRAF3 directly catalyze STAT6 ubiquitination. His-STAT6, Flag-TRAF3 and HA-Ub were expressed in HEK293 cells and ubiquitination assays showed that overexpression of Flag-TRAF3 facilitates exogenous His-STAT6 ubiquitination (Fig. 4A). Both K450 and K129 sites ubiquitination levels were found down-regulated in TRAF3 ablated BMDMs. K129 residue locates between the Protein Interaction Domain and All-alpha Domain of STAT6, while K450 residue locates in DNA Binding Domain of STAT6 (Fig. 4B). Moreover, the K450 site is evolutionarily conserved in STAT6 proteins among different species (Fig. 4C), suggesting the functional specificity of this lysine in STAT6.
Fig. 4. TRAF3 promotes STAT6 ubiquitination and enhances IL-4 induced STAT6 transcriptional activity.
A HEK293 cells were co-transfected with His-STAT6, Flag-TRAF3 and HA-Ub expression plasmids. Ubiquitination assay of STAT6 was performed using anti-His antibody for IP and anti-Ub antibody for immunoblotting of ubiquitinated STAT6. B Schematic representation of the mouse STAT6 protein, showing the potential TRAF3-related ubiquinated lysine (K) residues (K129 and K450) highlighted in red. C Amino acid sequence alignment of STAT6 among the indicated species, showing K129 and K450 highlighted in red. D HEK293 cells were transfected with indicated plasmids and luciferase assay was performed to determine transcriptional activity of STAT6 or indicated STAT6 mutants (K129R and K450R) with or without TRAF3 expression. Cells were stimulated with IL-4 (10 ng/ml) for 24 h before harvest. Data with error bars are represented as mean ± SD. Each panel is a representative experiment of at least three independent biological replicates. One-way ANOVA, *p < 0.05, **p < 0.01, n = 3 for each group. E His-STAT6 WT or His-STAT6 K450R were transfected into HEK293 cells together with Flag-TRAF3 and HA-Ub K63. Cells were treated with (+) or without (−) IL-4 (10 ng/ml) for 2 h. Nuclear and cytosol fractions were extracted and p-STAT6 levels were detected by immunoblotting. LaminB shows nuclear fraction and Tubulin shows cytosol fraction.
Since STAT6 ubiquitination is strongly induced in TRAF3 overexpressed cells, we hypothesized that TRAF3 modulates IL-4-mediated STAT6 transcriptional activation. Luciferase assay showed that TRAF3 overexpression dramatically enhanced IL-4-induced STAT6 reporter gene transcription (Fig. 4D). To study the function of K450 and K129 ubiquitination, we then generate two STAT6 mutants by replacing K129 and K450 in STAT6 with Arginine (R) and verified their function. As shown in Fig. 4D, the presence of the K450R, not K129R mutants, dramatically abolished TRAF3-mediated STAT6-driven luciferase activity.
We next examined the effect of K450R mutant on the regulation of IL-4-induced STAT6 phosphorylation. IL-4 treatment for 2 h promotes only wild-type (WT) STAT6 phosphorylation but did not efficiently induce K450R mutant STAT6 phosphorylation (Fig. 4E). Taken together, these results suggested that K450 site on STAT6 play a critical role on TRAF3 mediated STAT6 ubiquitination and transcriptional activity.
Myeloid TRAF3 deficiency suppressed M2 TAM polarization in tumor context
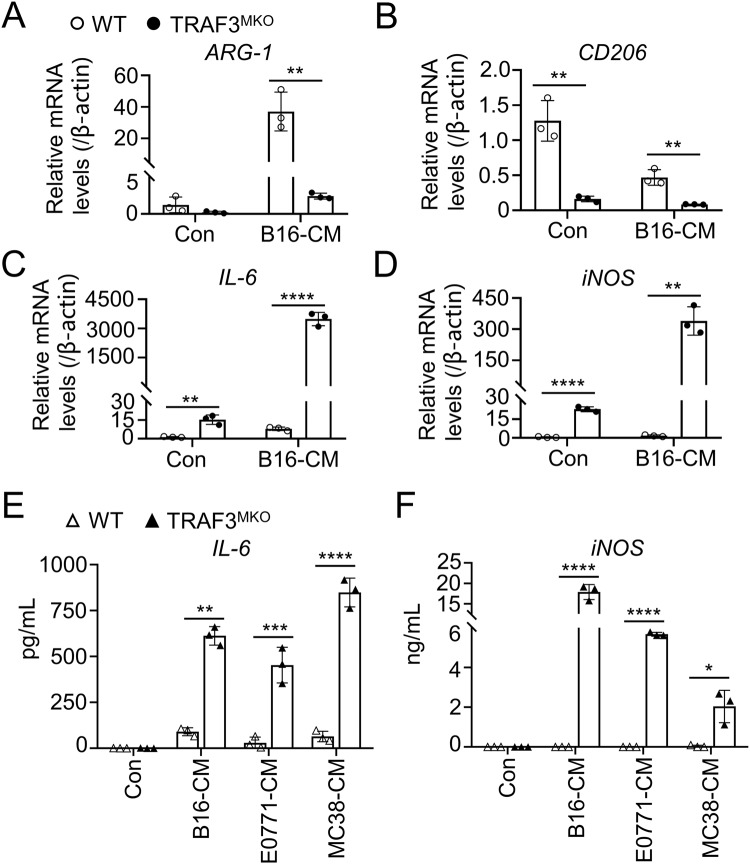
To assess the function of TRAF3 in regulating TAM polarization, BMDMs from WT and TRAF3MKO mice were co-cultured with B16 murine melanoma cell conditional media (B16-CM) for 48 h and M1/M2 marker genes were detected using qRT-PCR. TRAF3 deficiency markedly intercepted ARG-1 and CD206 expression (Fig. 5A, B) and facilitated IL-6 and iNOS expression in B16-CM-treated cells (Fig. 5C, D). When co-cultured with conditional media of two other cancer cell lines, MC38 murine colon carcinoma cells (MC38-CM) and E0771 murine breast cancer cells (E0711-CM), decreased ARG-1 and CD206 and increased IL-6 and iNOS expression levels were also observed in TRAF3 deficient BMDMs (Supplementary Fig. S4A–D). Moreover, the production of IL-6 and iNOS in the supernatant was examined using ELISA assay and the results showed that TRAF3 deficiency in BMDMs remarkably promoted IL-6 and iNOS production and secretion (Fig. 5E, F). These results suggested the potential role of TRAF3 in regulating TAM M1/M2 polarization balance in tumor context.
Fig. 5. TRAF3 deficiency suppressed tumor induced M2 TAM polarization.
A–D BMDMs were co-cultured with B16 conditional media (B16-CM) for 48 h. Total RNA was extracted and qRT-PCR analysis of ARG-1 (A), CD206 (B), IL-6 (C) and iNOS (D) was performed. E, F BMDMs were co-cultured with E0771, B16 or MC38 conditional media (E0771-CM, B16-CM or MC38-CM) for 48 h. The concentration of IL-6 (E) and iNOS (F) in the supernatant of cell culture medium was determined using ELISA assay. Bar graphs with error bars are represented as mean ± SD. Each panel is a representative experiment of at least three independent biological replicates. **p < 0.01, ***p < 0.001, ****p < 0.0001, n = 3 for each group.
Myeloid TRAF3 deficiency suppressed B16 murine melanoma tumorigenesis and lung metastasis
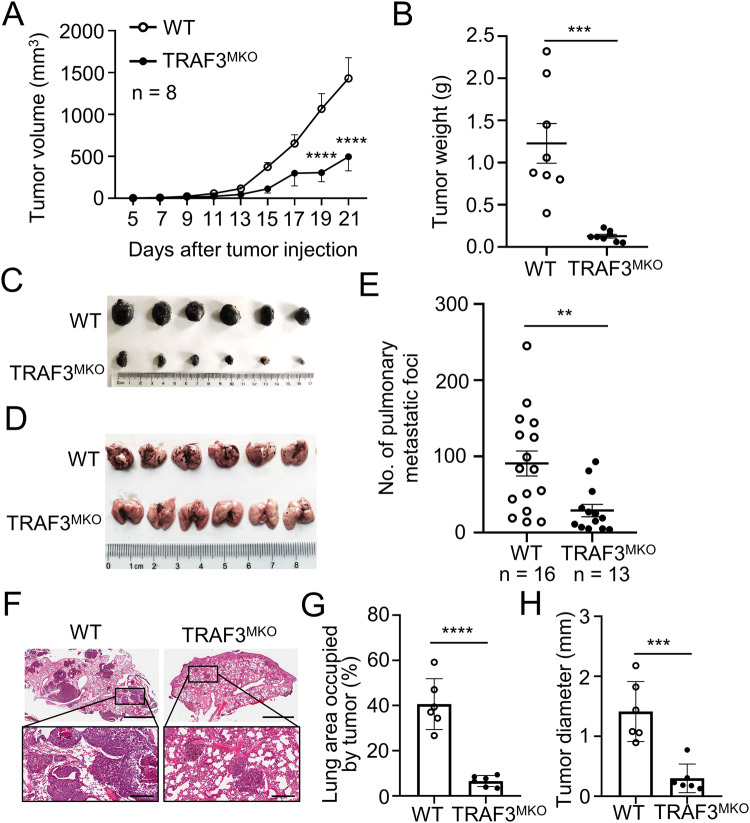
Next, we generated murine B16 melanoma tumor bearing mice to investigate the role of TRAF3 on tumor progression in vivo. WT and TRAF3MKO mice were injected s.c. with B16 cells and tumor growth curve were observed. Although B16 tumors grew vigorously in WT mice, TRAF3 conditional knockout in myeloid cells resulted in smaller tumor formation in TRAF3MKO mice (Fig. 6A–C).
Fig. 6. Myeloid TRAF3 deficiency suppressed B16 melanoma tumorigenesis and lung metastasis in mouse model.
A–C WT and TRAF3MKO mice were injected s.c. with B16 melanoma cells at 5 × 105 cells per site. Tumor volume was calculated by the formula: V = 1/2 × a (length) × b2 (width). Tumors were harvested from the mice 21 days after tumor injection. Tumor growth curve (A), tumor weight (B) and presentative photograph of tumors (C) are shown. n = 8 for each group. D–H WT and TRAF3MKO mice were dosed by tail-veil injection with 0.5 × 105 B16 cells. D Presentative photograph of lungs are shown; E Summary graphs of the number of pulmonary metastases at day 14 are shown (n = 16 for WT group and n = 13 for TRAF3MKO group); F H&E staining of lung tissue (Upper, magnification ×20, bar = 2 mm; Lower, magnification ×100, bar = 300 μm); G Lung area occupied by tumor, n = 6 for each group; H Tumor size, n = 6 for each group. Data are represented as mean ± SD. Each panel is a representative experiment of at least three independent biological replicates. Two-way ANOVA (A), Student’s t test (B, E, G, H). **p < 0.01, ***p < 0.001, ****p < 0.0001.
To further determine the role of TRAF3-deficient myeloid cells in tumor metastasis, a pulmonary metastasis mouse model was established by tail-veil injection with B16 cells in WT and TRAF3MKO mice. Pulmonary metastatic foci numbers and tumor sizes were significantly reduced in TRAF3MKO mice than that in WT mice (Fig. 6D–H). These data indicated that TRAF3 deficiency in myeloid cells promotes tumor progression and metastasis in mice model.
TRAF3 regulates TAMs and MDSCs infiltration in B16 tumor microenvironment
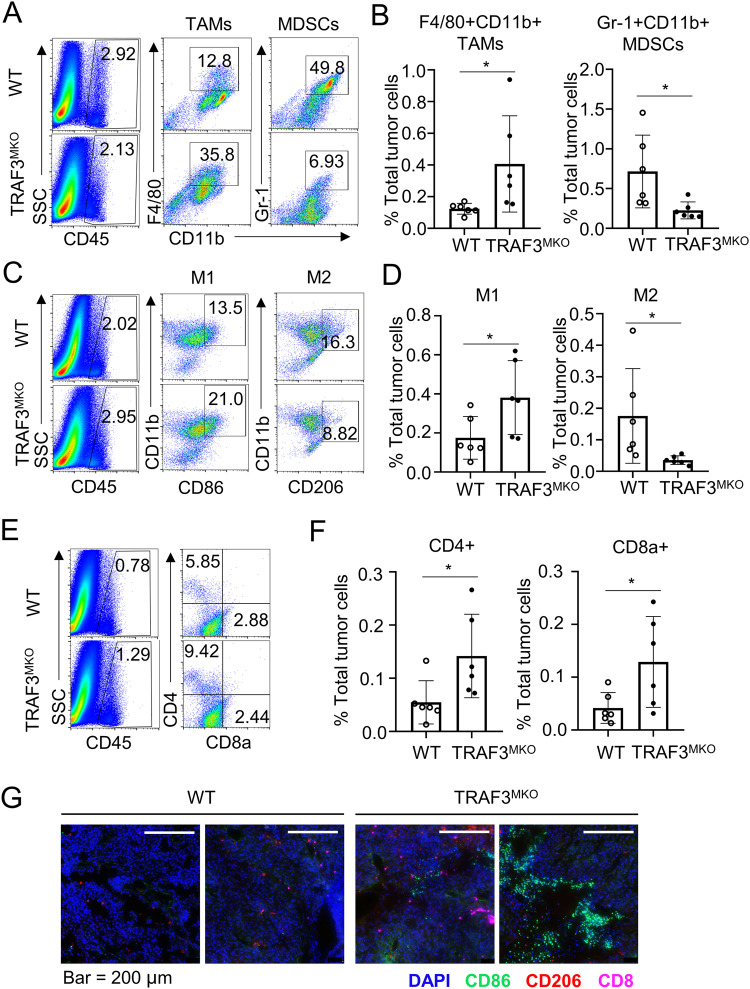
Macrophages are one of the major populations of tumor-infiltrating immune cells and play important roles in tumor growth and metastasis [3, 37]. As TRAF3 suppresses M1 TAM marker and facilitates M2 marker expression, we supposed that TRAF3 might play a vital role in regulating tumor-infiltrating TAMs. B16 cell-challenged WT and TRAF3MKO mice were sacrificed and tumor-infiltrating immune cells (CD45+) were assessed by flow cytometry analysis. Among the CD45+ immune cells, the B16 cell-challenged TRAF3MKO mice had increased frequency of F4/80+CD11b+ TAMs and decreased frequency of Gr-1+CD11b+ MDSCs (Fig. 7A, B). The frequency and number of CD86+CD11b+ M1 TAMs significantly increased and CD206+CD11b+ M2 TAMs decreased (Fig. 7C, D). Moreover, there was a considerable increase in the frequency and number of CD4+ and CD8a+ effector T cells (Fig. 7E, F). Multiplexed immunofluorescence staining of tumor tissue demonstrated that CD86 positive M1 TAM and CD8+ T cell infiltration in B16 tumor tissue was significantly increased in TRAF3MKO mice (Fig. 7G). Besides, we also detected tumor infiltrated DC, NK and B cells, the frequency of these cells didn’t change in TRAF3MKO mice bearing tumor (Supplementary Fig. S5). These results revealed cellular and molecular mechanisms of tumor rejection ability of the TRAF3MKO mice.
Fig. 7. TRAF3 regulates immune cell infiltration in B16 tumor model.
Flow cytometric analysis of immune cells (CD45+) infiltrated to day 21 tumor tissues. A Frequency of tumor infiltrated F4/80+CD11b+ TAMs and Gr-1+CD11b+ MDSCs. B Summary of flow cytometry data of (A) based on multiple mice, showing the frequency of the indicated cell populations within total tumor cells. C Frequency of tumor infiltrated CD86+CD11b+ M1 TAMs and CD206+CD11b+ M2 TAMs. D Summary of flow cytometry data of (C) based on multiple mice. E Frequency of tumor infiltrated CD4+ T cells and CD8a+ T cells. F Summary of flow cytometry data of (E) based on multiple mice. G Multiplexed immunofluorescence staining of infiltrating immune cells in B16 tumor tissue. Presentative images are shown. Bar = 200 μm. CD8, pink; CD86, green; CD206, red. Data are represented as mean ± SD. Each panel is a representative experiment of at least three independent biological replicates. Student’s t test, *p < 0.05, n = 6 for each group.
Discussion
Macrophage infiltration in tumor microenvironment is associated with enhanced tumor progression and poor clinical outcome. Emerging evidence indicate that strategies targeting TAMs display excellent anti-tumor potency [10–12], suggesting the vital role of macrophages in anti-tumor immunotherapy.
TRAF3 is broadly involved in receptor-mediated signaling pathways, including tumor necrosis factor receptor (TNFR) or Toll-like receptor (TLR) trigged macrophage activation [14, 38]. Acting alone or in combination, TRAF3 is known to control many biological processes, including cytokine production [16, 39], autophagy [40] and pyroptosis [15, 18]. TRAF3 positively controls type I interferon production [41, 42], and negatively regulates non-canonical NF-κB signaling [43]. As an E3 ligase, TRAF3 mediates integration of different signaling cascades in macrophages remains unclear. Sub-expression data of TRAF3 in tumor infiltrated immune cells (GEPIA2021, http://gepia2021.cancer-pku.cn/) revealed that TRAF3 expression levels markedly higher in M2 macrophages, which indicates the critical role of TRAF3 in TAM polarization. Next, our present data of transcriptome-proteome combined analysis of BMDMs revealed that TRAF3 ablation facilitated the transcriptional activity of M1 genes and compromised the mRNA and protein expression of M2 genes and thus restrained M2 macrophage polarization and potentiated anti-tumor immunity.
Considering the critical role of TAMs in regulating tumor microenvironments, the identification of TRAF3 ubiquitin modulation in TAM polarization may provide therapeutic targets for anti-tumor immunotherapy. The present study showed that the Th2 cytokine IL-4 induced upregulation of M2 TAM marker gene, CD206, was significantly blocked in TRAF3-deficient macrophages. Meanwhile, LPS promoted M1 TAM marker gene, iNOS, IL-6 and CD86, dramatically enhanced in TRAF3-deficient macrophages.
E3 ligases usually catalyze substrate proteins to form 8 types of polyubiquitin chains, M1-, K6-, K11-, K27-, K29-, K33-, K48- and K63-linked polyubiquitin chains, which participates in a series of biological processes, such as autophagy [44], immune response [45] and tumorigenesis [46, 47]. K48-linked polyubiquitin chains often cause substrate molecules to enter proteasome mediated degradation system [46]. K63- and M1-linked polyubiquitin chains can promote the activation of substrate molecules and intracellular signal transduction [15, 48, 49]. TRAF3 catalyzes K63- and K48-linked polyubiquitination of target molecules. TRAF3 is known to mediate K48-linked ubiquitination degradation of NIK via forming complex with E3 ligases TRAF2 and cIAPs [16, 43, 50]. TFAF3 can also catalyze K63-linked polyubiquitination activation of substrate protein ASC and enhance NLRP3 inflammasome [15]. However, the catalytic activity and mechanism of TRAF3 in regulating M2 TAM polarization are still unclear. Our previous study confirmed that TRAF3 regulates ULK1 ubiquitination and degradation and promotes LPS/Nigericin induced pyroptotic cell death in macrophages [17].
To reveal the new mechanism of TRAF3 regulating M1/M2 TAM marker expression, we performed quantitative ubiquitomics assays to analyze TRAF3 targeting molecules in wild-type and TRAF3-deficient primary murine macrophages. Functional enrichment of differentially ubiquitination sites demonstrated that TRAF3 mediated ubiquitination substrates were particularly enriched in IL-4 response pathway proteins, which was aligned with the phenotype of M2 TAM polarization. Among the differentially ubiquitination proteins, the ubiquitination levels of transcriptional factor STAT6 were found significantly reduced in TRAF3-deficient macrophages. LC-MS/MS analysis identified K450 and K129 as predominant ubiquitination sites in STAT6.
The ubiquitination of STAT family proteins has been widely studied, and ubiquitination modifications in STAT1, STAT2 and STAT3 have been identified [51–54]. STAT1 has linear ubiquitination at K511 and K652 sites, which inhibits STAT1 activation and type-I interferon signaling homeostasis [54]. Phosphorylation at tyrosine 701 site of STAT1 (pY701-STAT1) is the major form of ubiquitinated-STAT1, which undergoes rapid degradation by ubiquitin-proteasome system [55]. Tripartite motif-containing protein 21 (TRIM21) was identified as an E3 ligase of STAT1 and mediates STAT1 stability in gastric cancer [51]. TRAF6 mediates K63 ubiquitination within the Src homology 2 (SH2) domain of STAT3, which is an essential step for STAT3 membrane recruitment and subsequent phosphorylation in response to S Typhimurium infection [52]. Recent studies reported that STAT6 is ubiquitination substrate of E3 ligases RNF128 and RTA [27, 28]. The present results demonstrated that TRAF3 catalyzes STAT6 ubiquitination at K450 site. Site-mutation analysis of STAT6 K450 revealed that TRAF3-mediated K450 site of STAT6 ubiquitination facilitates IL-4-induced STAT6 phosphorylation and transcriptional activity.
TAMs are the most abundant immune cells in the tumor microenvironment and function as key regulators of the complex interplay between tumor cells and the immune microenvironment [3]. STAT6 is a critical signal molecule that regulates M2 polarization of TAMs [20]. However, the role of TRAF3/STAT6 axis on M2 TAM polarization, tumorigenesis and tumor metastasis still needs to be solved urgently. Our present data showed that myeloid conditional knockout of TRAF3 suppressed B16 murine melanoma tumorigenesis and lung metastasis in C57BL/6 mouse model. Ablation of TRAF3 in myeloid cells significantly reduced tumor infiltrated CD206+ M2 TAMs and Gr-1+ MDSCs in B16 tumors.
Moreover, tumor-infiltrating M2 TAMs directly promote cancer progression by suppressing anti-tumor T cell function through secreting regulatory cytokines, including IL-10, ARG-1 [56, 57]. TAMs reprogramming to an anti-tumor M1 phenotype can increase their antigen cross-presentation ability and in turn enhance CD8+ T cell-mediated anti-tumor immunity [58]. Our study showed that myeloid cell-specific deletion of TRAF3 greatly enhanced the frequency of tumor-infiltrating T cells, which provided explanation for the stronger tumor rejection ability of the TRAF3MKO mice. TRAF3 deficient TAMs produced decreased levels of ARG-1 and elevated levels of IL-6 and iNOS, suggesting an M1-like phenotype that is known to promote anti-tumor T cell responds [59].
In summary, the present study identified the essential positively regulatory role of STAT6 K450 ubiquitination mediated by TRAF3 in M2 TAM polarization. Due to the critical role of M2 TAMs in tumorigenesis and metastasis, we propose that targeting TRAF3 or downstream STAT6 K450 ubiquitination has clinical potential in cancer immunotherapy.
Methods
Mice
C57BL/6 background TRAF3-flox mice were kindly provided by Dr Robert Brink (Garvan Institute of Medical Research) [60]. Age-matched myeloid cell-conditional TRAF3 knockout mice (TRAF3fl/fllyz2Cre/wt, termed TRAF3MKO) and wild-type control mice (TRAF3fl/fl-lyz2wt/wt, termed WT) were produced as previously [17]. Briefly, TRAF3-flox mice were crossed with lysozyme 2-Cre (Lyz2-Cre) mice. The mice were maintained in specific pathogen-free facility of Hebei University and all animal experiments were conducted in accordance with guidelines and regulations approved by the Animal Ethical and Welfare Committee (AEWC) of Hebei University.
Antibodies and reagents
Antibodies for TRAF3 (66310-1-Ig, 1:1000), CD206(18704-1-AP, 1:1000) CD36 (18836-1-AP, 1:1000), CD86 (26903-1-AP, 1:1000) were from ProteinTech (Wuhan, China). Antibody for ubiquitin (ET1609-21, 1:1000) was from HuaAn Biotechnology (Hangzhou, Zhejiang, China). Antibodies for STAT6 (5397, 1:1000), p-STAT6 (Y641, 9362, 1:1000) were purchased from Cell Signaling Technology (Boston, MA, USA). Antibody for SLC4A7 (82335, 1:1000) was purchased from Abcam (Boston, MA, USA). HA (12CA5, 1:1000 for IP) and HA-HRP (3F10, 1:5000) antibodies were from Roche Life Science (Basel, Switzerland). Flag-HRP (A8592, 1:5000) and β-actin (A2228, 1:10,000) antibodies were from Sigma-Aldrich (St. Louis, MO, USA). His-tag antibody (D291-3, 1:1000) was from MBL (Tokyo, Japan). The fluorochrome-labeled antibodies specific to mouse proteins PE-conjugated anti-mouse Ly-6g/Ly-6C (Gr-1) (108407), APC-conjugated anti-mouse CD206 (141708), APC-conjugated anti-mouse CD86 (105012), PE-conjugated anti-mouse CD4 (100511), APC-conjugated anti-mouse CD8a (100711), PE anti-mouse CD49b (108907), PerCP-Cy5.5 anti-mouse NK1.1 (108727), PE anti-mouse CD19 (115507), PerCP-Cy5.5 anti-mouse CD45R/B220 (103236) and their corresponding isotype controls were from Biolegend (San Diego, CA, USA). FITC-conjugated anti-mouse CD45 (553080), Alexa Fluor 647 rat anti-mouse F4/80 (565853), PerCP-Cy5.5-conjugated rat anti-mouse CD11b (550993), PE anti-mouse CD11c (561044) and their corresponding isotype controls were from BD Bioscience (New York, USA). LPS (derived from Escherichia coli strain 0127: SB8), Z-Leu-Leu-Leu-al (MG132) were from Sigma-Aldrich (St. Louis, MO, USA). Recombinant Murine IL-4 (214-14) were from Pepro Tech (Hangzhou, Zhejiang, China).
Cell culture
Human embryo kidney 293 (HEK293), B16 murine melanoma cells and MC38 murine colon carcinoma cells were obtained from Cell Resource Center (IBMS, CAMS/PUMC, Beijing, China). E0771 murine breast cancer cells were obtained from BeNa Culture Collection (BNCC, Beijing, China). HEK293, B16 and E0771 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% of FBS (Gemini Bio), 100 U/ml of penicillin, and 100 μg/ml of streptomycin. MC38 cells were cultured in RPMI-1640 containing 10% of FBS, 100 U/ml of penicillin, and 100 μg/ml of streptomycin. Murine primary bone marrow-derived macrophages (BMDMs) were prepared as previously described [17]. Briefly, bone marrow cells isolated from the femurs of indicated young adult WT or TRAF3MKO mice were cultured in a DMEM medium supplemented with M-CSF conditional medium for 7 days. BMDMs were starved overnight in DMEM medium supplemented with 0.5% FBS before being stimulated with LPS (1 μg/ml) and recombinant murine IL-4 (10 ng/ml). Total cell lysates were prepared for immunoblotting assays, and total RNA was prepared for RT-PCR assays.
RNA-sequencing analysis
BMDMs from WT and TRAF3MKO mice were applied for total RNA extraction and RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). A total amount of 3 μg RNA per sample was used and sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA). The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) and the library preparations were sequenced on an Illumina Hiseq2500/X platform. The raw transcriptomic reads were mapped to a reference genome (GRCm39, http://www.ensembl.org/Mus_musculus/Info/Index) by using Bowtie v2.2.3. For quantification of gene expression level, Cuffquant and cuffnorm (v2.2.1) was used to calculate FPKMs (Fragments Per Kilobase of exon model per Million mapped fragments) of genes in each sample.
Mass spectrometry
BMDMs from WT and TRAF3MKO mice were treated with MG132 for 2 h before harvest. Cells were sonicated for three times on ice and lysed in a lysis buffer (8 M urea, 1% Protease Inhibitor Cocktail, 3 μM TSA, 50 mM NAM, 2 mM EDTA). The protein solution was then applied with trypsin digestion. The tryptic peptides were proceeded with LC-MS/MS analysis by using EASY-nLC 1000 UPLC system and tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo Fisher Scientific, Waltham, MA, USA) coupled online to the UPLC. The resulting MS/MS data were processed using Maxquant (v1.6.6.0).
Plasmids
Expression plasmids encoding HA-TRAF2, HA-TRAF3, HA-Ub, HA-Ub K63 were from Addgene (Watertown, MA, USA). Expression plasmids encoding murine Flag-STAT6 and STAT6 luciferase plasmid p4 × STAT6-Luc2P were provided by Dr. Yichuan Xiao [20]. The cDNA encoding mouse STAT6 (NM_009284.2) was subcloned into HA-tagged expression vector to generate His-STAT6 expression plasmid. His-STAT6 mutants (His-STAT6 K450R and His-STAT6 K129R) were constructed by site-directed mutagenesis.
Luciferase assay
HEK293 cells were seeded in a 24-well plate and grown for 24 h before transfection with Stat6 luciferase plasmid p4 × Stat6-Luc2P, control reporter plasmid pRL-TK and indicated gene expression plasmids using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). Twenty-four hours later, the cells were lysed and luciferase assays were performed using Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Specific promoter activity was expressed as the relative activity ratio of firefly luciferase to Renilla luciferase.
Flow cytometry
Single cell suspensions of BMDMs and B16 melanoma were stained and subjected to flow cytometry using FACS Calibur (BD Bioscience, New York, USA). Cells were stained with the following fluorescence-labeled antibodies were used: PE-conjugated anti-mouse Ly-6g/Ly-6C (Gr-1), APC-conjugated anti-mouse CD206, APC-conjugated anti-mouse CD86, PE-conjugated anti-mouse CD4, APC-conjugated anti-mouse CD8a, FITC-conjugated anti-mouse CD45, Alexa Fluor 647 rat anti-mouse F4/80, PerCP-Cy5.5-conjugated rat anti-mouse CD11b and their corresponding isotype controls. Cells were then washed with PBS solution and re-suspended for Flow cytometry.
Peripheral blood mononuclear cells (PBMCs) isolation
PBMCs were isolated from heparinized blood samples via Percoll density-gradient centrifugation (MLSM1092, Muti science, China) according to the manufacturer’s instructions. Briefly, blood was obtained from 6–8 weeks old mice using heparinized tubes and was diluted with an equal volume of phosphate-buffered saline, pH 7.4 (PBS), containing 0.05 M ethylenediaminetetraacetic acid (EDTA; Invitrogen). The cell suspension was layered over a Ficoll-Paque gradient and centrifuged at 400 × g for 15 min at room temperature. The PBMC interface was carefully removed by pipetting and washed with PBS. Cell number and viability were determined using a Countstar Automated Cell Counter.
Real-time quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from BMDMs using TRIzol Reagent (15596026, Invitrogen, Carlsbad, CA, USA). HiScript III RT SuperMix (Vazyme Biotech, Nanjing, China) were used for cDNA synthesis. Real-time PCR was performed using and ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech, Nanjing, China). The expression of individual genes was calculated and normalized to the expression of β-Actin. The gene-specific PCR primers are shown in Table 1.
Table 1.
Gene-specific primer sequences used in quantitative RT-PCR.
| Gene | Primer | Sequence |
|---|---|---|
| IL-6 | Forward | CACAGAGGATACCACTCCCAACA |
| Reverse | TCCACGATTTCCCAGAGAACA | |
| CD206 | Forward | CAGGTGTGGGCTCAGGTAGT |
| Reverse | TGGCATGTCCTGGAATGAT | |
| iNOS | Forward | GTTCTCAGCCCAACAATACAAGA |
| Reverse | GTGGACGGGTCGATGTCAC | |
| ARG-1 | Forward | TTTTTCCAGCAGACCAGCTT |
| Reverse | AGAGATTATCGGAGCGCCTT | |
| β-actin | Forward | GGCTGTATTCCCCTCCATCG |
| Reverse | CCAGTTGGTAACAATGCCATGT |
Enzyme-linked immunosorbent assay (ELISA)
Cell culture supernatants were harvested and the concentration of IL-6 and iNOS in the supernatant of the cell culture medium was examined using mouse IL-6 (ABclonal, RK00008) and iNOS (Mabtech, 3421-1H-6) ELISA kits according to the manufacturer’s instructions.
Immunoblotting
Cells were harvest and lysed with RIPA buffer (Solarbio, Beijing China). Cell lysates were subjected to immunoblotting and coIP assays. In brief, cell lysates were separated by SDS-PAGE and transferred to PVDF membranes. The membranes were blocked with 5% non-fat milk and incubated with a specific primary antibody. Horseradish peroxidase-conjugated secondary antibody was applied and membranes were visualized by ECL detection.
Ubiquitination and quantitative ubiquitinomics assays
Cells were pretreated with MG132 for 2 h before harvest and then lysed with RIPA lysis buffer containing 6 M urea and protease inhibitors. The indicated proteins were isolated by immunoprecipitation (IP) with indicated antibody for 1 h at 4 °C, followed by incubation with protein A-agarose (Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight at 4 °C. Then, the protein A-agarose-antigen-antibody complex pellet were washed five times and the ubiquitinated proteins was detected by immunoblotting using anti-ubiquitin antibody.
For quantitative ubiquitinomics assay, BMDMs from WT and TRAF3MKO mice were treated with MG132 for 2 h before harvest. Cells were sonicated and lysed in a lysis buffer (8 M urea, 1% Protease Inhibitor Cocktail, 3 μM TSA, 50 mM NAM, 2 mM EDTA) and the protein solution was then applied with trypsin digestion. To enrich ubiquitin-modified peptides, the tryptic peptides dissolved in NETN buffer (100 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl, 0.5% NP-40, pH 8.0) were incubated with pre-washed anti-ubiquitin antibody beads (PTM Bio, Hangzhou, Zhejiang, China) at 4 °C overnight. The beads were washed and the bound peptides were eluted with 0.1% trifluoroacetic acid. The eluted peptides were subjected to LC-MS/MS analysis.
Tumor models
Murine B16 melanoma cells (5 × 105) were injected s.c. into 6–8-weeks old, age- and sex-matched WT and TRAF3MKO mice. Tumor growth were monitored as tumor volume, which was estimated according to the formula: Volume = 1/2 × a × b2, where (a) and (b) represents the largest and smallest diameters of the tumor, respectively. Mice were sacrificed at day 21 after injection according to the protocol approved by the Animal Ethical and Welfare Committee (AEWC) of Hebei University. The xenograft tumors were dissected, weighted, and photographed. All the experiments were carried out in double blind manner.
Multiplexed immunofluorescence staining
Multiplexed immunofluorescence staining of B16 xenograft tumor tissues were performed using the OpalTM chemistry (PerkinElmer, Waltham, USA) with antibodies against CD3 (78588T, CST), CD4 (25229T, CST), CD8 (98941T, CST), CD86 (26903-1-AP, Proteintech) and CD206 (18704-1-AP, Proteintech). Briefly, the tissue slide baked at 63 °C for 1 h, after deparaffinization by automatic dyeing machine (LEICAST5020, Leica), the slides in antigen retrieval buffer were processed with microwave (3 min 100% power, 15–20 min 20% power), then blocked with blocking buffer for 10 min at room temperature. Slides were incubated with the primary antibody for 60 min, and subsequently incubated with HRP-conjugated secondary antibody for 10 min. Thereafter, slides were incubated with Opal 520 working buffer for 10 min at room temperature and then washed in TBST buffer. Above procedures were repeated until all markers (Opal 520, Opal 570, Opal 620, Opal 650, and Opal 690) were assessed. And antibodies complexes were removed by microwave (3 min 100% power, 15–20 min 20% power) before another marker was counterstained. Finally, the slides were counterstained with DAPI for 5 min. Immunofluorescence imagings were acquired by Tissue-FAXS system (TissueFAXS Spectra, TissueGnostics).
Tumor-infiltrating immune cell analysis
Tumor tissues were dissected and ground in RPMI 1640 medium containing 10% FBS followed by treatment with DNaseI (0.2 mg/ml, DN25, Sigma-Aldrich, St. Louis, MO, USA) and collagenase IV (0.5 mg/ml, V900893, Sigma-Aldrich, St. Louis, MO, USA) for 30 min at 37 °C. The cells were then passed through a 70 μm mesh. The resulting dissociated cells were collected and resuspended in Red Blood Cell Lysis Buffer for 3 min and washed twice in PBS. The cells were subjected to flow cytometry analysis.
Tumor metastasis assay
Experimental metastasis assay was performed using tail-veil injection method. Briefly, WT and TRAF3MKO mice were dosed by tail-veil injection with 0.5 × 105 B16 cells. Mice were sacrificed at day 14 after injection according to the protocol approved by the Animal Ethical and Welfare Committee (AEWC) of Hebei University. The lungs were dissected and photographed. The numbers of pulmonary metastatic foci were counted.
Hematoxylin and eosin (H&E) staining
The lung tissues were fixed with 4% paraformaldehyde overnight, embedded in paraffin, and sectioned into 4 μm thick slices using automatic tissue microtome (TP1020, Leica, USA). Slices were stained with H&E and scanned using digital pathology scanner (Aperio CS2, Leica, USA). The tumor diameters and the lung area occupied by tumor were analyzed using ImageScopex64 software (Media Cybernetics, Silver Spring, MD). For each section, six random non-contiguous microscopic fields were analyzed.
Statistical analysis
The data are shown as mean ± SD, and all the presented data are the representative results of at least three independent repeats. Statistical analysis was performed using GraphPad Prism 8 software. Two-tailed unpaired Student’s t tests or one-way or two-way ANOVA analysis with Tukey’s multiple comparisons were used according to the number of groups compared. p < 0.05 were considered significant.
Supplementary information
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by J-HS, L-NL, D-DS, W-WL, CL, F-XW and T-TW. BL, N-PC, YQ and J-HS analyzed and interpreted the data. The first draft of the manuscript was written by J-HS and Z-YN and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by National Natural Science Foundation of China (Nos. 82273463, 82103181, 31971304), Natural Science Foundation of Hebei Province (Nos. H2022201065, H2022201067, C2020201052, H2021201028), Hebei Province Foundation for Returned Overseas Scholars (No. C20200305), Government Foundation of Clinical Medicine Talents Training Program of Hebei Province (No. 361007), Foreign Intelligence Introduction Project of Hebei Province (No. 360601), CAMS Innovation Fund for Medical Sciences (No. 2019-I2M-5-055), Science and Technology Research Project of Colleges in Hebei Province (No. QN2023008), Foundation of President of Hebei University under grant (No.202204) and Tumor Microecological Metabolism Regulation Research Innovation Team of Hebei University.
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Ethical approval
The animal experiments were conducted according to the protocol approved by the Animal Ethical and Welfare Committee (AEWC) of Hebei University.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jian-Hong Shi, Email: shijianhong@hbu.edu.cn.
Yan Qin, Email: qinyan@hbu.edu.cn.
Zhi-Yu Ni, Email: nizhiyu@hbu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01194-1.
References
- 1.Cassetta L, Pollard JW. A timeline of tumour-associated macrophage biology. Nat Rev Cancer. 2023;23:238–57. doi: 10.1038/s41568-022-00547-1. [DOI] [PubMed] [Google Scholar]
- 2.Onkar S, Cui J, Zou J, Cardello C, Cillo AR, Uddin MR, et al. Immune landscape in invasive ductal and lobular breast cancer reveals a divergent macrophage-driven microenvironment. Nat Cancer. 2023;4:516–34. doi: 10.1038/s43018-023-00527-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao X, Li B, Chen J, Dang J, Chen S, Gunes EG, et al. Effect of cabazitaxel on macrophages improves CD47-targeted immunotherapy for triple-negative breast cancer. J Immunother Cancer. 2021;9:e002022. doi: 10.1136/jitc-2020-002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H, et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell. 2019;35:588–602.e10. doi: 10.1016/j.ccell.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei YT, Wang XR, Yan C, Huang F, Zhang Y, Liu X, et al. Thymosin alpha-1 reverses M2 polarization of tumor-associated macrophages during efferocytosis. Cancer Res. 2022;82:1991–2002. doi: 10.1158/0008-5472.CAN-21-4260. [DOI] [PubMed] [Google Scholar]
- 6.Vadevoo SMP, Gunassekaran GR, Lee C, Lee N, Lee J, Chae S, et al. The macrophage odorant receptor Olfr78 mediates the lactate-induced M2 phenotype of tumor-associated macrophages. Proc Natl Acad Sci USA. 2021;118:e2102434118. doi: 10.1073/pnas.2102434118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mantovani A, Locati M. Macrophage metabolism shapes angiogenesis in tumors. Cell Metab. 2016;24:887–8. doi: 10.1016/j.cmet.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Tang K, Ma J, Huang B. Macrophages' M1 or M2 by tumor microparticles: lysosome makes decision. Cell Mol Immunol. 2022;19:1196–7. doi: 10.1038/s41423-022-00892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barreira da Silva R, Leitao RM, Pechuan-Jorge X, Werneke S, Oeh J, Javinal V, et al. Loss of the intracellular enzyme QPCTL limits chemokine function and reshapes myeloid infiltration to augment tumor immunity. Nat Immunol. 2022;23:568–80. doi: 10.1038/s41590-022-01153-x. [DOI] [PubMed] [Google Scholar]
- 10.Muller AK, Kohler UA, Trzebanski S, Vinik Y, Raj HM, Girault JA, et al. Mouse modeling dissecting macrophage-breast cancer communication uncovered roles of PYK2 in macrophage recruitment and breast tumorigenesis. Adv Sci. 2022;9:e2105696. doi: 10.1002/advs.202105696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishiga Y, Drainas AP, Baron M, Bhattacharya D, Barkal AA, Ahrari Y, et al. Radiotherapy in combination with CD47 blockade elicits a macrophage-mediated abscopal effect. Nat Cancer. 2022;3:1351–66. doi: 10.1038/s43018-022-00456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Ye Y, Liu Z, Zhang G, Dai H, Li J, et al. Macrophage mitochondrial fission improves cancer cell phagocytosis induced by therapeutic antibodies and is impaired by glutamine competition. Nat Cancer. 2022;3:453–70. doi: 10.1038/s43018-022-00354-5. [DOI] [PubMed] [Google Scholar]
- 13.Arkee T, Hostager BS, Houtman JCD, Bishop GA. TRAF3 in T cells restrains negative regulators of LAT to promote TCR/CD28 signaling. J Immunol. 2021;207:322–32. doi: 10.4049/jimmunol.2001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi JH, Sun SC. Tumor necrosis factor receptor-associated factor regulation of nuclear factor kappaB and mitogen-activated protein kinase pathways. Front Immunol. 2018;9:1849. doi: 10.3389/fimmu.2018.01849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan K, Wei C, Zheng Z, Song T, Wu F, Zhang Y, et al. MAVS promotes inflammasome activation by targeting ASC for K63-linked ubiquitination via the E3 ligase TRAF3. J Immunol. 2015;194:4880–90. doi: 10.4049/jimmunol.1402851. [DOI] [PubMed] [Google Scholar]
- 16.Parvatiyar K, Pindado J, Dev A, Aliyari SR, Zaver SA, Gerami H, et al. A TRAF3-NIK module differentially regulates DNA vs RNA pathways in innate immune signaling. Nat Commun. 2018;9:2770. doi: 10.1038/s41467-018-05168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen Y, Liu WW, Zhang X, Shi JG, Jiang S, Zheng L, et al. TRAF3 promotes ROS production and pyroptosis by targeting ULK1 ubiquitination in macrophages. FASEB J. 2020;34:7144–59. doi: 10.1096/fj.201903073R. [DOI] [PubMed] [Google Scholar]
- 18.Shi JH, Ling C, Wang TT, Zhang LN, Liu WW, Qin Y, et al. TRK-fused gene (TFG) regulates ULK1 stability via TRAF3-mediated ubiquitination and protects macrophages from LPS-induced pyroptosis. Cell Death Dis. 2022;13:93. doi: 10.1038/s41419-022-04539-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou C, Lu C, Pu H, Li D, Zhang L. TRAF6 promotes IL-4-induced M2 macrophage activation by stabilizing STAT6. Mol Immunol. 2020;127:223–9. doi: 10.1016/j.molimm.2020.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Yu T, Gan S, Zhu Q, Dai D, Li N, Wang H, et al. Modulation of M2 macrophage polarization by the crosstalk between Stat6 and Trim24. Nat Commun. 2019;10:4353. doi: 10.1038/s41467-019-12384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamerkar S, Leng C, Burenkova O, Jang SC, McCoy C, Zhang K, et al. Exosome-mediated genetic reprogramming of tumor-associated macrophages by exoASO-STAT6 leads to potent monotherapy antitumor activity. Sci Adv. 2022;8:eabj7002. doi: 10.1126/sciadv.abj7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong X, Yang C, He WQ, Yu J, Xin Y, Zhang X, et al. Sirtuin 6 maintains epithelial STAT6 activity to support intestinal tuft cell development and type 2 immunity. Nat Commun. 2022;13:5192. doi: 10.1038/s41467-022-32846-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao M, Ren K, Xiong X, Xin Y, Zou Y, Maynard JC, et al. Epithelial STAT6 O-GlcNAcylation drives a concerted anti-helminth alarmin response dependent on tuft cell hyperplasia and Gasdermin C. Immunity. 2022;55:623–38.e5. doi: 10.1016/j.immuni.2022.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang CL, Jiang H, Feng W, Liu H, Han L, Chen Y, et al. Total glucosides of paeony ameliorate pristane-induced lupus nephritis by inducing PD-1 ligands(+) macrophages via activating IL-4/STAT6/PD-L2 signaling. Front Immunol. 2021;12:683249. doi: 10.3389/fimmu.2021.683249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahal OM, Wolfe AR, Mandal PK, Larson R, Tin S, Jimenez C, et al. Blocking interleukin (IL)4- and IL13-mediated phosphorylation of STAT6 (Tyr641) decreases M2 polarization of macrophages and protects against macrophage-mediated radioresistance of inflammatory breast cancer. Int J Radiat Oncol Biol Phys. 2018;100:1034–43. doi: 10.1016/j.ijrobp.2017.11.043. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Malabarba MG, Nagy ZS, Kirken RA. Interleukin 4 regulates phosphorylation of serine 756 in the transactivation domain of Stat6. Roles for multiple phosphorylation sites and Stat6 function. J Biol Chem. 2004;279:25196–203. doi: 10.1074/jbc.M313668200. [DOI] [PubMed] [Google Scholar]
- 27.Gu F, Wang C, Wei F, Wang Y, Zhu Q, Ding L, et al. STAT6 degradation and ubiquitylated TRIML2 are essential for activation of human oncogenic herpesvirus. PLoS Pathog. 2018;14:e1007416. doi: 10.1371/journal.ppat.1007416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waqas SFH, Hoang AC, Lin YT, Ampem G, Azegrouz H, Balogh L, et al. Neuropeptide FF increases M2 activation and self-renewal of adipose tissue macrophages. J Clin Invest. 2017;127:2842–54. doi: 10.1172/JCI90152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao P, Ma X, Yuan M, Yi Y, Liu G, Wen M, et al. E3 ligase Nedd4l promotes antiviral innate immunity by catalyzing K29-linked cysteine ubiquitination of TRAF3. Nat Commun. 2021;12:1194. doi: 10.1038/s41467-021-21456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang N, de la Rosa X, Libreros S, Pan H, Dreyfuss JM, Serhan CN. Cysteinyl-specialized proresolving mediators link resolution of infectious inflammation and tissue regeneration via TRAF3 activation. Proc Natl Acad Sci USA. 2021;118:e2013374118. doi: 10.1073/pnas.2013374118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sui H, Dongye S, Liu X, Xu X, Wang L, Jin CQ, et al. Immunotherapy of targeting MDSCs in tumor microenvironment. Front Immunol. 2022;13:990463. doi: 10.3389/fimmu.2022.990463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hegde S, Leader AM, Merad M. MDSC: markers, development, states, and unaddressed complexity. Immunity. 2021;54:875–84. doi: 10.1016/j.immuni.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar S, Torres MP, Kaur S, Rachagani S, Joshi S, Johansson SL, et al. Smoking accelerates pancreatic cancer progression by promoting differentiation of MDSCs and inducing HB-EGF expression in macrophages. Oncogene. 2015;34:2052–60. doi: 10.1038/onc.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang N, Gao X, Zhang W, Xiong J, Cao X, Fu ZF, et al. JEV infection induces M-MDSC differentiation into CD3(+) macrophages in the brain. Front Immunol. 2022;13:838990. doi: 10.3389/fimmu.2022.838990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bishop GA, Stunz LL, Hostager BS. TRAF3 as a multifaceted regulator of B lymphocyte survival and activation. Front Immunol. 2018;9:2161. doi: 10.3389/fimmu.2018.02161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czimmerer Z, Halasz L, Daniel B, Varga Z, Bene K, Domokos A, et al. The epigenetic state of IL-4-polarized macrophages enables inflammatory cistromic expansion and extended synergistic response to TLR ligands. Immunity. 2022;55:2006–26.e6. doi: 10.1016/j.immuni.2022.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordon SR, Maute RL, Dulken BW, Hutter G, George BM, McCracken MN, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545:495–9. doi: 10.1038/nature22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin J, Xiao Y, Hu H, Zou Q, Li Y, Gao Y, et al. Proinflammatory TLR signalling is regulated by a TRAF2-dependent proteolysis mechanism in macrophages. Nat Commun. 2015;6:5930. doi: 10.1038/ncomms6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng PH, Keats JJ, Wang H, et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol. 2008;9:1364–70. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman AC, Kemp AJ, Drabsch Y, Behrends C, Wilkinson S. Autophagy acts through TRAF3 and RELB to regulate gene expression via antagonism of SMAD proteins. Nat Commun. 2017;8:1537. doi: 10.1038/s41467-017-00859-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hornick EL, Wallis AM, Bishop GA. TRAF3 enhances type I interferon receptor signaling in T cells by modulating the phosphatase PTPN22. Sci Signal. 2022;15:eabn5507. doi: 10.1126/scisignal.abn5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perkins DJ, Polumuri SK, Pennini ME, Lai W, Xie P, Vogel SN. Reprogramming of murine macrophages through TLR2 confers viral resistance via TRAF3-mediated, enhanced interferon production. PLoS Pathog. 2013;9:e1003479. doi: 10.1371/journal.ppat.1003479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 2008;9:1371–8. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pohl C, Dikic I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 2019;366:818–22. doi: 10.1126/science.aax3769. [DOI] [PubMed] [Google Scholar]
- 45.Hu H, Sun SC. Ubiquitin signaling in immune responses. Cell Res. 2016;26:457–83. doi: 10.1038/cr.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Q, Aminu B, Roscow O, Zhang W. Targeting the ubiquitin signaling cascade in tumor microenvironment for cancer therapy. Int J Mol Sci. 2021;22:791. doi: 10.3390/ijms22020791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Senft D, Qi J, Ronai ZA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 2018;18:69–88. doi: 10.1038/nrc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu G, Li D, Liang W, Sun W, Xie X, Tong Y, et al. PP6 negatively modulates LUBAC-mediated M1-ubiquitination of RIPK1 and c-FLIP(L) to promote TNFalpha-mediated cell death. Cell Death Dis. 2022;13:773. doi: 10.1038/s41419-022-05206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aalto A, Martinez-Chacon G, Kietz C, Tsyganova N, Kreutzer J, Kallio P, et al. M1-linked ubiquitination facilitates NF-kappaB activation and survival during sterile inflammation. FEBS J. 2022;289:5180–97. doi: 10.1111/febs.16425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanjo H, Zajonc DM, Braden R, Norris PS, Ware CF. Allosteric regulation of the ubiquitin:NIK and ubiquitin:TRAF3 E3 ligases by the lymphotoxin-beta receptor. J Biol Chem. 2010;285:17148–55. doi: 10.1074/jbc.M110.105874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L, Li Q, Yang J, Xu P, Xuan Z, Xu J, et al. Cytosolic TGM2 promotes malignant progression in gastric cancer by suppressing the TRIM21-mediated ubiquitination/degradation of STAT1 in a GTP binding-dependent modality. Cancer Commun. 2022;43:123–49. doi: 10.1002/cac2.12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruan HH, Zhang Z, Wang SY, Nickels LM, Tian L, Qiao JJ, et al. Tumor necrosis factor receptor-associated factor 6 (TRAF6) mediates ubiquitination-dependent STAT3 activation upon salmonella enterica serovar typhimurium infection. Infect Immun. 2017;85:e00081–17. doi: 10.1128/IAI.00081-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Precious B, Young DF, Andrejeva L, Goodbourn S, Randall RE. In vitro and in vivo specificity of ubiquitination and degradation of STAT1 and STAT2 by the V proteins of the paramyxoviruses simian virus 5 and human parainfluenza virus type 2. J Gen Virol. 2005;86:151–8. doi: 10.1099/vir.0.80263-0. [DOI] [PubMed] [Google Scholar]
- 54.Zuo Y, Feng Q, Jin L, Huang F, Miao Y, Liu J, et al. Regulation of the linear ubiquitination of STAT1 controls antiviral interferon signaling. Nat Commun. 2020;11:1146. doi: 10.1038/s41467-020-14948-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren Y, Zhao P, Liu J, Yuan Y, Cheng Q, Zuo Y, et al. Deubiquitinase USP2a sustains interferons antiviral activity by restricting ubiquitination of activated STAT1 in the nucleus. PLoS Pathog. 2016;12:e1005764. doi: 10.1371/journal.ppat.1005764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engblom C, Pfirschke C, Pittet MJ. The role of myeloid cells in cancer therapies. Nat Rev Cancer. 2016;16:447–62. doi: 10.1038/nrc.2016.54. [DOI] [PubMed] [Google Scholar]
- 57.Ruffell B, Chang-Strachan D, Chan V, Rosenbusch A, Ho CM, Pryer N, et al. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 2014;26:623–37. doi: 10.1016/j.ccell.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma S, Sun B, Duan S, Han J, Barr T, Zhang J, et al. YTHDF2 orchestrates tumor-associated macrophage reprogramming and controls antitumor immunity through CD8(+) T cells. Nat Immunol. 2023;24:255–66. doi: 10.1038/s41590-022-01398-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klug F, Prakash H, Huber PE, Seibel T, Bender N, Halama N, et al. Low-dose irradiation programs macrophage differentiation to an iNOS(+)/M1 phenotype that orchestrates effective T cell immunotherapy. Cancer Cell. 2013;24:589–602. doi: 10.1016/j.ccr.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 60.Gardam S, Sierro F, Basten A, Mackay F, Brink R. TRAF2 and TRAF3 signal adapters act cooperatively to control the maturation and survival signals delivered to B cells by the BAFF receptor. Immunity. 2008;28:391–401. doi: 10.1016/j.immuni.2008.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.