Abstract
Skeletal muscle regeneration relies on muscle stem (satellite) cells. We previously demonstrated that satellite cells efficiently and accurately repair radiation-induced DNA double-strand breaks (DSBs) via the DNA-dependent kinase DNA-PKcs. We show here that DNA-PKcs affects myogenesis independently of its role in DSB repair. Consequently, this process does not require the accumulation of DSBs and it is also independent of caspase-induced DNA damage. We report that in myogenic cells DNA-PKcs is essential for the expression of the differentiation factor Myogenin in an Akt2-dependent manner. DNA-PKcs interacts with the p300-containing complex that activates Myogenin transcription. We show also that SCID mice that are deficient in DNA-PKcs, and are used for transplantation and muscle regeneration studies, display altered myofiber composition and delayed myogenesis upon injury. These defects are exacerbated after repeated injury/regeneration events resulting in reduced muscle size. We thus identify a novel, caspase-independent, regulation of myogenic differentiation, and define a differentiation phase that does not involve the DNA damage/repair process.
Subject terms: Cell biology, Molecular biology
Introduction
Double-strand breaks (DSBs) are dangerous DNA damages that can generate pathogenic mutations and genome rearrangements if incorrectly repaired, and compromise cell survival and cell fate if left unrepaired. The consequences of unrepaired DSBs are particularly critical in adult stem cells, which are responsible for tissue homeostasis and tissue regeneration throughout life. Unrepaired DNA damage may block proliferation, promote premature differentiation, or lead to cell death, thereby resulting in a reduction of the stem cell pool and decreased regenerative capacity of the tissue [1]. Muscle stem cells (satellite cells, SCs) are responsible for muscle regeneration and muscle homeostasis in the adult. Upon muscle damage, quiescent SCs are activated and give rise to myoblasts that differentiate and fuse into multinucleated myotubes and myofibers [2]. We previously showed that SCs repair DSBs more efficiently than their committed and differentiated progeny and that DSB repair by SCs is highly accurate, suggesting that the maintenance of genome stability is relevant for these cells [3].
The maintenance of genome stability is also important during myogenic differentiation, which is inhibited by induced genotoxic stress that blocks the activity of the myogenic determination factor MyoD [4]. Conversely, DNA damage induced by a specific DNase has been reported to promote myogenic differentiation [5]. Therefore, the role of DNA damage at different stages of myogenic differentiation needs to be clarified.
We previously demonstrated that efficient non-homologous end-joining (NHEJ) repair in SCs relies on the key repair enzyme DNA-PKcs (DNA-dependent protein kinase catalytic subunit) [3]. DNA-PKcs is recruited to DNA ends of DSBs in the presence of the Ku70/Ku80 heterodimer, then this protein kinase auto-transphosphorylates and becomes active by forming the DNA-PKcs/Ku70/ku80 complex called DNAPK [6]. Activated DNAPK plays a crucial role in tethering broken DNA ends [7], and acts as a signaling/mediator molecule by phosphorylating several proteins including the histone H2AX and Akt kinases thereby mediating the recruitment of these factors to the repair site, thus regulating the cellular response to the DNA lesion [8]. In addition to its direct involvement in NHEJ, upon DNA damage DNA-PKcs activates Akt kinases (Akts) prevalently located in the nucleus. Activated Akts phosphorylate substrates that participate in DNA repair and cell cycle arrest, i.e. the cellular response to DNA damage.
The Akt kinases Akt1 and Akt2 are expressed in muscle cells [9]. They impact on myogenesis upon activation by PI3Ks (phosphatidylinositol 3-kinases), which are stimulated by growth factors and cytokines [10]. PI3K/Akt1 play a role in the activation of SCs upon injury by inducing quiescence exit [11], whereas PI3K/Akt2 stimulates the expression of myogenic factors and thereby promotes muscle differentiation [12, 13]. Conversely, PIKKs (PI3K-like protein kinases) that are involved in DNA repair, like DNA-PKcs and the DNA damage sensors ATM and ATR are not known to be activated during myogenesis.
We show here that DNA-PKcs affects myogenesis independently of its role in DSB repair, via activation of Akts. We also show that chronic impairment of this pathway in vivo leads to alternative activation of Akts that results in compromised muscle regeneration.
Results
Differentiation of satellite cells is blocked upon inhibition of DNA-PKcs
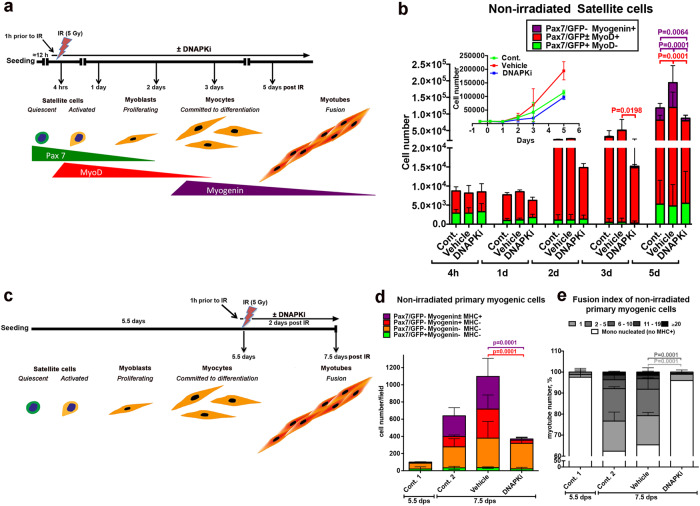
To assess the consequences of impaired DSB repair in survival and differentiation of muscle stem cells, GFP+ SCs isolated by FACS from Tg:Pax7-nGFP mice were treated for 1 h with 10 µM of the DNAPK inhibitor (DNAPKi) NU7441 then exposed to 5 Gy irradiation (IR) that induces DNA damage and DSBs. Cells were kept in culture in the presence and in the absence of the inhibitor, and examined at 4 h to 5 days post irradiation (dpIR) (scheme Fig. 1a). Proliferation and differentiation of SCs were assessed by the cell number, or the number of nuclei in multinucleated myofibers, and immunolabeling of myogenic markers. Upon activation, Pax7+ myoblasts express MyoD and progressively lose Pax7 expression as they differentiate to generate Myogenin+ (Myog+) myocytes and multinucleated myofibers [14]. Myog+ (differentiated) cells remain MyoD+ for some time, then both factors are downregulated in mature myotubes.
Fig. 1. Inhibition of DNA-PKcs blocks myogenic differentiation of satellite cells in the absence of irradiation (induced DSBs).
a Schematic representation of the experiment: GFP+ SCs isolated by FACS from Tg:Pax7-nGFP mice were seeded overnight (≈12 h) and pre-treated for 1 h with 10 µM of DNA-PKcs inhibitor (=DNAPKi= NU7441) or the corresponding volume of vehicle (0.2 % DMSO) before 5 Gy irradiation (IR), or were not irradiated. Cells were then kept in culture from 4 h to 5 days before analysis. The expected level of differentiation and the corresponding myogenic markers at these time points are schematized below. b Histograms of myogenic differentiation and growth curves of non-irradiated cells ± DNAPKi. Proliferation evaluated with the cell number, and differentiation with immunofluorescence of myogenic markers (Pax7, MyoD, Myogenin). Each condition was tested with SC derived from n = 3–7 mice, mean ± SD. Myogenic markers have been analyzed in 5–10 fields/condition (2,000-5,000 cells), and extrapolated to the total cell number. Significance evaluated by by two-way ANOVA (F = 7.37, DFn = 28, DFd = 162, p < 0.000001), with post-hoc Dunnett’s multiple comparisons test; significant P values are indicated in the histogram. P values and bars are of the same color as the category that is compared. c Schematic representation of the experiment: GFP+ SCs isolated by FACS from Tg:Pax7-nGFP mice were seeded overnight and let to proliferate exponentially. At 5.5 dps (days post-seeding) cells were pre-treated for 1 h with 10 µM of DNAPKi or the corresponding volume of vehicle, and analyzed 48 h later, i.e. 7.5 dps. d Histograms display the cell number and differentiation stage of non-irradiated SC-derived cells treated or not with DNAPKi, and defined by the indicated combinations of myogenic markers. Differentiation of MyoD+ myoblasts is characterized by the expression of Myogenin and later MHC. The fusion index, i.e. the number of nuclei per myotube, is a further parameter of differentiation at this stage. The color code indicates the differentiation state (from Pax7/GFP+ [not differentiated] to MHC+ [the most differentiated]). At 5.5 dps, before treatments, the SC-derived population was heterogeneous (e.g. up to 10% [differentating] Myogenin+ cells, in red in the histogram) in agreement with published data [71]. Myogenic markers have been analyzed in 5–10 fields/condition and extrapolated to the total cell number. Mean ± SD for each category. Significance by two-way ANOVA (F = 2.99, DFn = 9, DFd = 32, p = 0.0106), with post-hoc Dunnett’s multiple comparisons test; significant P values are indicated in histogram. P values and error bars are of the same color as the category that is compared. e Fusion indexes of non-irradiated SC-derived cells by enumeration of nuclei/myotube. Significance by two-way ANOVA (F = 22.68, DFn = 15, DFd = 35, p < 0.0001), with post-hoc Dunnett’s multiple comparisons test; significant P values are indicated in the histogram. P values and error bars are of the same color as the category that is compared. Significance bars are of the same color as the category that is compared. n = 3 experiments, n = 2500–5000 cells analyzed/condition, mean ± SD.
Irradiation decreased the number of SCs (Pax7+), committed (MyoD+) and differentiated (Myog+) cells, compared to non-irradiated cells (Fig. 1b and Fig. S1a, b), as expected [15]. This process was enhanced in the presence of DNAPKi, and in a dose-dependent manner (Fig. S1c, d). By 5dpIR, irradiation upon DNAPKi treatment resulted in no detectable Myog+ cells, and fully depleted reserve SCs (Pax7+/MyoD-/Myog-), that are an in vitro readout for self-renewal [16] (Fig. S1b). Impaired DSB repair upon DNAPKi treatment was verified by immunostaining and enumeration of γ-H2AX foci, a histone modification prevalently associated with DSBs, and 53BP1, a protein recruited at DSBs, (Fig. S1e–g).
Surprisingly, DNAPKi also affected proliferation and differentiation of non-irradiated SCs (50% reduction of the cell number at 5 days, and 87% reduction of the Myog+ population compared to vehicle, Fig. 1b, and Fig. S1a), with no apparent alteration of MyoD+ or reserve (Pax7+) cells. These perturbations were observed also with the immortalized myogenic cell line C2C7, which are cycling myoblasts with high proliferative capacity (Fig. S2). These results raise the possibility that DNA-PKcs blocks myogenic cells from expressing the differentiation marker Myogenin, displaying an activity that is independent from DSB repair.
To address the possibility that the effect of DNAPKi on myogenic differentiation is a consequence of the low cell number that results from slow proliferation and/or cell death upon treatment, GFP+ SCs were isolated by FACS and cultured in the absence of treatment until they reached confluence (>80 %) and were poised to differentiate (Fig. 1c). At 5.5 days post seeding (dps), cells were treated with DNAPKi (or vehicle) for 1 h, then irradiated or not, and analyzed two days later (i.e. 7.5 dps). In the absence of irradiation, treatment with DNAPKi reduced proliferation by about ≈1/3 compared to non-treated control and vehicle, where the number of Myog+ cells increased by sixfold to tenfold, respectively (Fig. 1d). Importantly, reduction in the cell number affected predominantly differentiated cells (Myog+, either MHC- or MHC+, a terminal differentiation marker), whereas Myogenin− cells were as numerous as in vehicle. Moreover, the fusion index was dramatically reduced compared to vehicle (80% of the few MHC+ cells were mononucleated in the presence of DNAPKi whereas 63% of MHC+ cells contained 2–20 nuclei in the absence of treatment, Fig. 1e). These experiments show that reduced differentiation of non-irradiated myogenic cells by DNAPKi is not due to reduced cell number. As expected, irradiation affected differentiation to a larger extent, in particular when DNA repair was inhibited, upon DNAPKi treatment (Fig. S3a–c).
These results were confirmed with C2C7 myoblasts using two distinct protocols that mimic either the conditions of highly proliferative SCs (Fig. S3d–g) or a later stage with myogenic cells ready to differentiate (Fig. S3h–k), before treatment (±DNAPKi and ± irradiation). In both cases, DNAPKi treatment inhibited Myog expression in non-irradiated cells. These data also reveal that DNAPKi affects mostly differentiating (Myog+) rather than committed (MyoD+) cells, and blocks further steps of differentiation in the absence of induced DNA damage.
Inhibition of DNA-PKcs blocks Myogenin expression in Akt-dependent manner
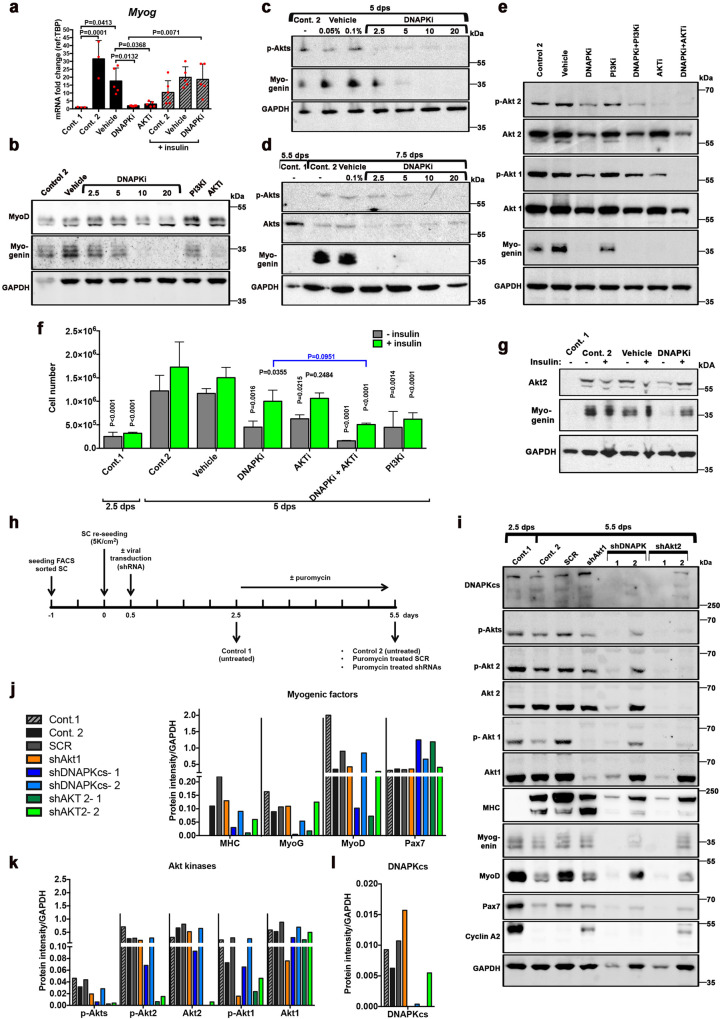
To verify that DNAPKi affects Myogenin expression, we treated C2C7 cells with increasing doses of DNAPKi (5–20 µM), following the previous scheme (Fig. S3d). We also examined the effect on Myogenin expression of inhibitors of PI3Ks (PI3Ki; ZSTK474, or LY294002) since DNA-PKcs is a PI3K-related kinase [17, 18], as well as an inhibitor of the downstream Akt kinases (AKTi; MK2206). Akt kinases are activated (phosphorylated at Ser473 and Thr308) by PI3Ks, as well as by PIKKs, upon DNA damage [19].
The transcript levels of Myog were reduced by 88% in cells treated with 10 µM DNAPKi compared to vehicle at 5 dps (Fig. 2a) and, accordingly, the Myogenin protein signal was depleted in a dose-dependent manner (Fig. 2b). The block in Myogenin expression was associated with reduced phosphorylation of Akts (phospho-Akts, or p-Akts) (Fig. 2c), indicating that DNAPKi also acts on Akts activation. Further, treatment with PI3Ki to some extent reduced, and AKTi depleted, Myogenin expression (Fig. 2e). Importantly, MyoD was not reduced with either of these three inhibitors (Fig. 2b). Depletion of Myogenin, reduction of p-Akts with increasing doses of DNAPKi, as well as reduced levels of Akts, were confirmed in SCs (Fig. 2d).
Fig. 2. DNA-PKcs is required for the expression of myogenin in a Akt-2 dependent manner.
a–g Myogenic cells treated with inhibitor of kinases. C2C7 cells were grown until 2.5 dps, following the scheme in Fig. S3d, and treated with inhibitors (alone or in combination, at the indicated doses) until 5 dps (panels a–c, e). SCs (d) were treated following the scheme in Fig. 1c, i.e. treated at 5.5 dps and collected at 7.5 dps. Control 1, untreated at 2.5 dps (or 5.5 dps, panel d); control 2, untreated at 5 dps (or 7.5 dps, panel d); vehicle, untreated (DMSO) at 5 dps (or 7.5 dps, d). a mRNA fold changes of Myog expression (n = 3–6) in C2C7 cells, mean ± SD normalized to the TBP-housekeeping gene, black columns, left part of the histogram. In the right part of the histogram, gray hatched columns indicate the same samples in the presence of 10 µg of insulin. Significance by ordinary one-way ANOVA (F = 9.943, DFn = 7, DFd = 30, p < 0.0001) with post-hoc Tukey’s multiple comparisons test, significant p values are indicated on the histogram. b Western blot (WB) of C2C7 cells treated with increasing doses of DNAPKi (NU7441), or 1 µM PI3Ki (ZSTK474), or 5 µM AKTi (MK2206); two bands of MyoD are present [72], and a band shift is observed at the highest doses of DNAPKi. c WB of Myogenin and p-Akts in C2C7 cells at increasing concentrations of DNAPKi. Two vehicle lanes (0.05% and 0.1%) correspond to the volume of DMSO used with 2.5 µM and 10 µM DNAPKi, respectively. d WB of Myogenin, Akts, and p-Akts of SCs in culture until 5.5 dps before treatment, with increasing concentrations of DNAPKi for two more days. e WB of C2C7 cells treated with 10 µM DNAPKi, PI3Ki (LY294002), and 5 µM AKTi, alone or in combination (left panels), and cultured in the presence or in the absence of inhibitor(s) until 5 dps; in panels b and e, vehicle contains 0.1% DMSO. In WBs, GAPDH was used as reference housekeeping protein. f C2C7 cell number and g WB of Myogenin and Akt2, upon growth and differentiation in the presence and in the absence of various inhibitors and insulin (green columns). As expected, insulin did not improve proliferation of cells treated with PI3Ki, because the PI3Ks are inhibited in this condition. In WB GAPDH was used as reference housekeeping protein. Significance by two-way ANOVA (F = 4.66, DFn = 18, DFd = 69, p < 0.0001), with post-hoc Tukey’s multiple comparisons of test vs. the corresponding vehicle conditions (vehicle or vehicle + insulin) when not otherwise indicated (P values shown on the histogram). h–l Analysis of myogenic factors and Akt kinases upon shRNA-dependent silencing of Akt1, Akt2, or DNA-PKcs. h Scheme and readouts of the experiment: SCs tested at 2.5 dps and 5.5 dps (control 1 and control 2, respectively) in the absence of puromycin (selection) treatment; SCR (scramble RNA) or shAkt1, shDNA-PKcs (2 independent clones), and shAkt2 (2 independent clones) at 5.5 dps upon puromycin selection. i WB of myogenic factors (differentiation markers: MHC (MF20) and Myogenin; myoblast marker MyoD, and stem cell marker Pax7), kinases (Akt1, Akt2, phospho-Akt1, phospho-Akt2, phospho-Akts, and DNA-PKcs), and the cell cycle marker cyclin A2. MyoD levels remained relatively high in SCR, perhaps as a consequence of the lentiviral and selection procedure. Normalization of protein levels with the reference protein GAPDH upon quantification of bands with Imagelab (BIORAD) for j myogenic factors, k individual Akt kinases and their phosphorylated forms, global p-Akts, and l DNA-PKcs. Uncropped gels are shown in “Original data file”.
Both members of the Akt family that are expressed in muscle (Akt1 and Akt2 [9]), and their respective phosphorylated forms, in particular p-Akt2, were affected by DNA-PKcs inhibition (Fig. 2e). Conversely, PI3Ki, which moderately reduced Myogenin levels, poorly reduced p-Akt2, and did not affect the levels of Akt1, p-Akt1, and Akt2, confirming a limited effect of PI3Ks on myogenesis. Double treatment with DNAPKi and PI3Ki resulted in a larger reduction of p-Akt2 than DNAPKi alone suggesting that activation of Akt2 by DNA-PKcs and PI3Ks are at least in part, non-overlapping processes. Conversely, this double treatment did not change the levels of Akt1/p-Akt1 and global Akt2 compared to DNAPKi alone, confirming the above-mentioned lack of effect of PI3Ki on these factors.
Finally, AKTi that inhibits Akts phosphorylation thereby reducing the levels of p-Akt1 and almost suppressing p-Akt2, inhibited Myogenin expression thus confirming that this effect depends on both Akts activation and DNA-PKcs (Fig. 2e). Combined AKTi and DNAPKi treatment further reduced the levels of Akt1 and Akt2, above that of each single inhibitor alone, confirming the notion that Akt1 and Akt2 phosphorylation during early myogenesis depends to a large extent, but not exclusively, on DNA-PKcs.
In summary, experiments with multiple inhibitors of kinases indicate that DNA-PKcs affects the fate of myogenic cells by promoting Myogenin expression, and this depends on activation of both Akt1 and Akt2. Conversely, PI3Ks have a minor effect and only result on moderate Akt2 activation.
Insulin-induced differentiation alleviates DNAPKi-blocked myogenesis through the PI3K/Akt pathway
Insulin and insulin-like growth factors (IGF) regulate and activate PI3Ks [20]. Insulin was added to the DNAPKi treatment, to assess whether insulin-dependent PI3K activation of Akts, and consequently inhibition of myogenesis, were rescued. C2C7 cells were treated with DNAPKi and/or inhibitors of the PI3K/Akt pathway (PI3Ki or AKTi), in the presence and in the absence of 10 µg/ml of insulin. Insulin (green columns, Fig. 2f) rescued to a large extent proliferation of cells treated with either DNAPKi or AKTi by doubling the cell number, and displayed its largest effect (threefold increase) in the presence of the two combined inhibitors, i.e. the condition in which proliferation was the most significantly reduced. Consistent with our model, insulin partially restored the levels of Myogenin transcripts (hatched columns Fig. 2a), and protein (Fig. 2g) that were significantly reduced upon DNAPKi treatment. Moreover, insulin increased the levels of Akt2 that was also depleted upon treatment with DNAPKi (Fig. 2g). Thus, insulin alleviates DNAPKi-dependent/AKTi-dependent depletion of Myogenin and the decline in cell proliferation, suggesting that Myogenin expression can be, at least partially, activated by PI3Ks, when DNA-PKcs is inhibited.
Myogenin expression is blocked by silencing DNA-PKcs and Akt2
To confirm that impairment of myogenic differentiation by DNAPKi and AKTi is due to bona fide inhibition of DNA-PKcs and Akt, the expression of DNA-PKcs, Akt1, and Akt2 was independently silenced in SCs by shRNAs lentivirus transduction (Fig. 2h). Control 1 (cells harvested 2.5 dps, in absence of treatment) displayed strong Pax7 and MyoD, and little Myogenin signal (Fig. 2i, j), confirming the prevalent myoblast state; the proliferation of these cells was verified by high levels of cyclin A2, a marker of dividing cells. At 5.5 dps, as expected, cyclin A2 levels dropped in control 2 (untreated) and the SCR sample (scramble shRNA). Advanced differentiation of both cells was evidenced by high levels of MHC, concomitantly with reduction of Myogenin and MyoD, as well as depletion of Pax7.
Silencing DNA-PKcs (by >95%) with two independent clones resulted in strongly reduced or undetectable MHC and Myogenin (with a larger effect with shDNAPK-1) compared to SCR, confirming that DNA-PKcs is necessary for myogenic differentiation (Fig. 2i, j). Silencing with shDNAPK-1 decreased MyoD levels, indicating that the committed myogenic population was also affected by DNA-PKcs knock-down. The more efficient silencer, shDNAPK-1, reduced the levels of globals p-Akts, and specifically Akt1, Akt2, and the respective phosphorylated forms (Fig. 2i, k).
Akt1silencing (by >90% with shAkt1) had a weak effect on committed myogenesis as it essentially affected myoblasts (Fig. 2i, j), in agreement with previous findings [9], and reduced MHC but not Myogenin levels. Conversely, Akt2 silencing with either shRNAs reduced the levels of MHC and MyoD as much as silencing of DNA-PKcs; the levels of Myogenin were reduced only by shAkt2–1. Of note, silencing of Akt2 also depleted DNA-PKcs (Fig. 2i, l).
Altogether, silencing experiments confirmed that DNA-PKcs is implicated in myogenesis, and showed that the DNA-PKcs downstream target Akt2 has similar effect on myogenesis as DNA-PKcs itself, whereas Akt1 only mildly affects this process.
DNA-PKcs interacts with the complex that activates Myogenin expression
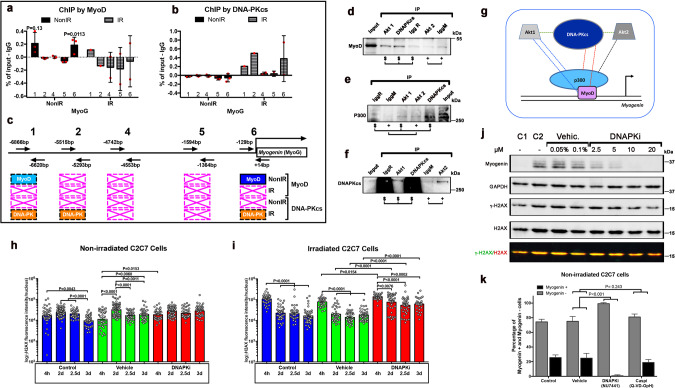
DNA-PKcs may regulate Myogenin expression by direct intervention on its promoter or activating one of the effectors of the Myogenin promoter. Chromatin immunoprecipitation (ChIP) experiments on the Myogenin promoter region confirmed the expected presence of MyoD [21, 22] but not DNA-PKcs, which is instead present on the locus only upon DNA damage (positive control; Fig. 3a–c). These experiments suggest that DNA-PKcs does not directly regulate the Myogenin promoter. Conversely, co-immunoprecipitation experiments with DNA-PKcs pulldown revealed the presence of MyoD and the acetyltransferase p300 (Fig. 3d, e), two key components of the complex that activates myogenic genes including Myogenin [21], suggesting that DNA-PKcs is part of the same complex as these Myogenin activators. P300 and MyoD were also present upon Akt1 pulldown (Fig. 3d), and p300 upon Akt2 pulldown, indicating that p300 and MyoD containing complexes interact with Akts as well, consistent with Akts phosphorylating p300 to promote the p300/MyoD interaction [21]. Finally, Akt1 and Akt2 were present upon DNA-PKcs pulldown (Fig. 3f).
Fig. 3. DNA-PKcs interacts with the complex that activates Myogenin expression, and activates Myogenin/myogenic differentiation without acting in DNA repair.
Quantitative PCR (histogram) analyses of several DNA fragments in the Myogenin promoter [73] from ChIP assays with either a anti-MyoD or b anti-DNA-PKcs antibodies in differentiating cells. Significance by unpaired t-test (F = 2.9, DFn = 2, DFd = 2). c Scheme (not at scale) of primers positions on the the Myog promoter with the respective distances from the initiation of transcription site (+1), and summary of positive amplifications in light blue, and statistically significant in dark blue (MyoD) or orange (DNA-PKcs), whereas crossed boxes in pink indicate no amplification, in the absence and in the presence of irradiation: n = 3, unless differently indicated. Immunostaining with d MyoD, e p300, and f DNA-PKcs of immunoprecipitation with DNA-PKcs and Akt1 (IgG rabbit, IgR), and Akt2 (IgG Mouse, IgM) pulldown. Input, empty IgG rabbit (IgGR, for control of DNA-PKcs and Akt1) and IgGM (IgG Mouse, IgM, for control of Akt2) in C2C7 cells at 5 dps. g Schematic representation of the interactions (dotted lines) observed in immunoprecipiatation experiments; dotted lines of the same color indicate interactions detected with pulldown of the same protein. Protein sizes are not at scale. Histone γH2AX immunostaining evaluated per cell in the absence [control, vehicle (0.2% DMSO)] and in the presence of 10 µM DNAPKi at different time points h in non-irradiated cells and i irradiated C2C7 cells. Total γH2AX immunofluorescence intensity was assessed with ImageJ of single cells after acquisition with Cell Voyager CV1000, confocal scanner box (Yokogawa). N = 40–80 cells from 3 independent experiments. Significance by one-way ANOVA (Non-irradiated: F = 16.37, DFn = 11, DFd = 588, p < 0.0001. Irradiated: F = 53.68, DFn = 11, DFd = 457, p < 0.0001) with post-hoc Tukey’s multiple comparisons test. P values are indicated on the histogram. j WB of Myogenin, global histone H2AX, γH2AX, and the housekeeping GAPDH protein in C2C7 cells at 4.5 dps. The γH2AX/H2AX ratio is shown in the bottom line after superimposition of the 700 nm (Goat α-mouse Starbright blue 700) and 800 nm (goat α-rabbit CF770) signal with the ImageLab program of the ChemiDoc MP imaging system (BioRad). The highest doses of DNAPKi result in slightly reduced levels of γ-H2AX, compatibly with H2AX being also phosphorylated by DNA-PKcs. k Percentage of Myogenin+ and Myogenin- cells upon immunostaining per image in the presence and in the absence of inhibitors of DNA-PKcs (10 µM NU7441) and caspases (30 µM Q-VD-OpH) (2–4 fields/condition, 150–300 cells, analyzed, n = 3). Significance by two-way ANOVA (F = 114.8, DFn = 3, DFd = 44, p < 0.0001) with post-hoc Tukey’s multiple comparisons test vs. vehicle (P values shown on the histogram). Uncropped gels are shown in “Original data file”.
Altogether, these data indicate that DNA-PKcs interacts with p300 and MyoD that mediate Myogenin expression at its promoter, as well as its regulators Akt1 and Akt2 (recapitulative scheme in Fig. 3g). Whether DNA-PKcs participates in these complexes in the context of DNA repair remained unclear.
DNA-PKcs is required to activate Myogenin-dependent differentiation without acting on DNA repair
Since DNA-PKcs is normally activated by DNA damage, DNA-PKcs function in promoting myogenesis could be triggered by endogenous DNA damage, for instance generated during DNA replication or, alternatively, by caspase-induced DNase (CAD). CAD-generated DNA damage at promoters has been reported to induce the expression of genes such as p21, which blocks the cell cycle, to promote late muscle differentiation [5]. In the same study, direct DNA damage on the Myogenin locus was not observed. We reported above DNA damage in the absence of irradiation in SCs (Fig. S1e–g) and activated C2C7 myoblasts (not shown) up to 24 and 48 h in culture, respectively, which is compatible with DNA lesions induced during DNA replication [23, 24]. Analysis of DNA damage at later time points showed that in the absence of irradiation, the γ-H2AX signal/cell remained essentially constant after 3 days in culture (Fig. 3h), a time point when Myogenin starts to be expressed in C2C7 cells and DNAPKi affects myogenesis (see above Fig. S2c). The γ-H2AX signal was not affected by DNAPKi (Fig. 3h). This result indicates a nearly constant load of most likely replication-induced DNA damage that is not repaired by DNA-PKcs, as expected for this type of DNA lesion [23, 24]. As a control, DNAPKi largely impaired the repair of IR-induced DNA damage in the same culture conditions (Fig. 3i). Consistent with this result, WB analysis confirmed that γ-H2AX is present in C2C7 myoblasts at 2.5 dps (control 1) as well as during differentiation (control 2), and did not increase in the presence of DNAPKi (Fig. 3j). These data suggest that DNA-PKcs is not activated for the repair of endogenous DNA damage during differentiation. Also, DNA-PKcs is not involved in phosphorylation of H2AX in response to replication-induced DNA damage, which substantially does not vary during differentiation.
DNA-PKcs may then be involved in myogenesis through processing DNA damage produced by other sources, e. g. CAD, where DNA-PKcs has been previously reported to phosphorylate H2AX [25]. However, differently from that study, inhibition of caspases by CASPi (Q-VD-OpH) did not reduce the number of Myogenin expressing cells nor the cell fusion marked by MHC, whereas 1 µM and 10 µM [optimal dose in growth medium, see Fig. S1c, d] of DNAPKi reduced, and fully blocked, respectively, the appearance of these cells in growth medium (serum-rich) used in our experimental paradigm (Figs. 3k and S4). This was also the case for experiments performed in differentiation medium (DM, serum-depleted, as in ref [25]) where 1 µM of DNAPKi blocked the expression of the differentiation factor Myogenin (Fig. S4). However, in DM, caspase inhibition reduced myogenic fusion whereas Myogenin expression was only poorly affected, in marked contrast with experiments in growth medium. These paired experiments show that caspases do play a role (i.e. induce myogenesis) in the differentiation medium but not in growth medium, whereas DNA-PKcs inhibition fully blocks myogenesis in both conditions (Fig. S4).
These data suggest that in our model DNA-PKcs impairment affects muscle differentiation independent of caspases, and possibly acts at an earlier stage of myogenesis than caspases.
Satellite cells from SCID mice have reduced proliferation and differentiation
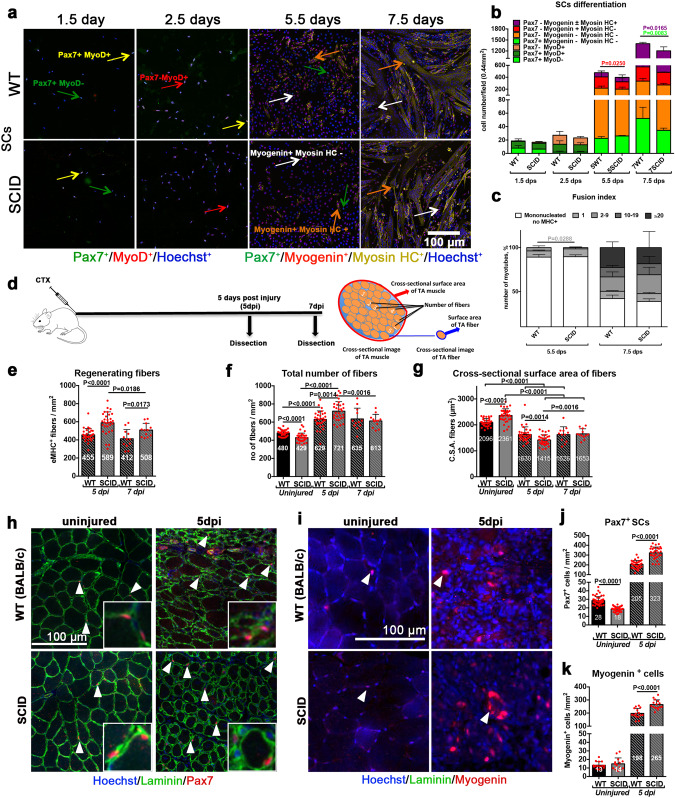
To evaluate whether altered myogenesis affects muscle regeneration in the context of chronic DNA-PKcs depletion, we analyzed SCs derived from severe combined immunodeficiency (SCID) mice that are naturally DNA-PKcs-deficient, and control mice on the same genetic background (BALB/c) [26]. Freshly isolated SCs that were plated and assessed from 1.5 to 7.5 dps displayed comparable proliferation rates in WT and SCID mice until 5.5 dps, although at this time point about 10% less Myog+ cells were present in SCID-derived cells (Fig. 4a, b). Moreover, the fusion index (MHC staining) showed about 80% mononucleated MHC+ cells in SCID versus 60% of mononucleated and 40% multinucleated cells in WT (Fig. 4c). No delay in myofusion in mutant cells was observed at 7.5 dps (Fig. 4c), but we detected 30 % less reserve SCs in SCID compared to WT (light green, Fig. 4b), indicating that not only differentiation but also self-renewal of SCs was perturbed in SCID-derived cells. Thus, chronic DNA-PKcs deficiency in mice affects myogenesis, but to a lower extent than the DNAPKi treatment or DNA-PKcs silencing, suggesting a compensatory mechanism as these mice develop muscles in the absence of DNA-PKcs.
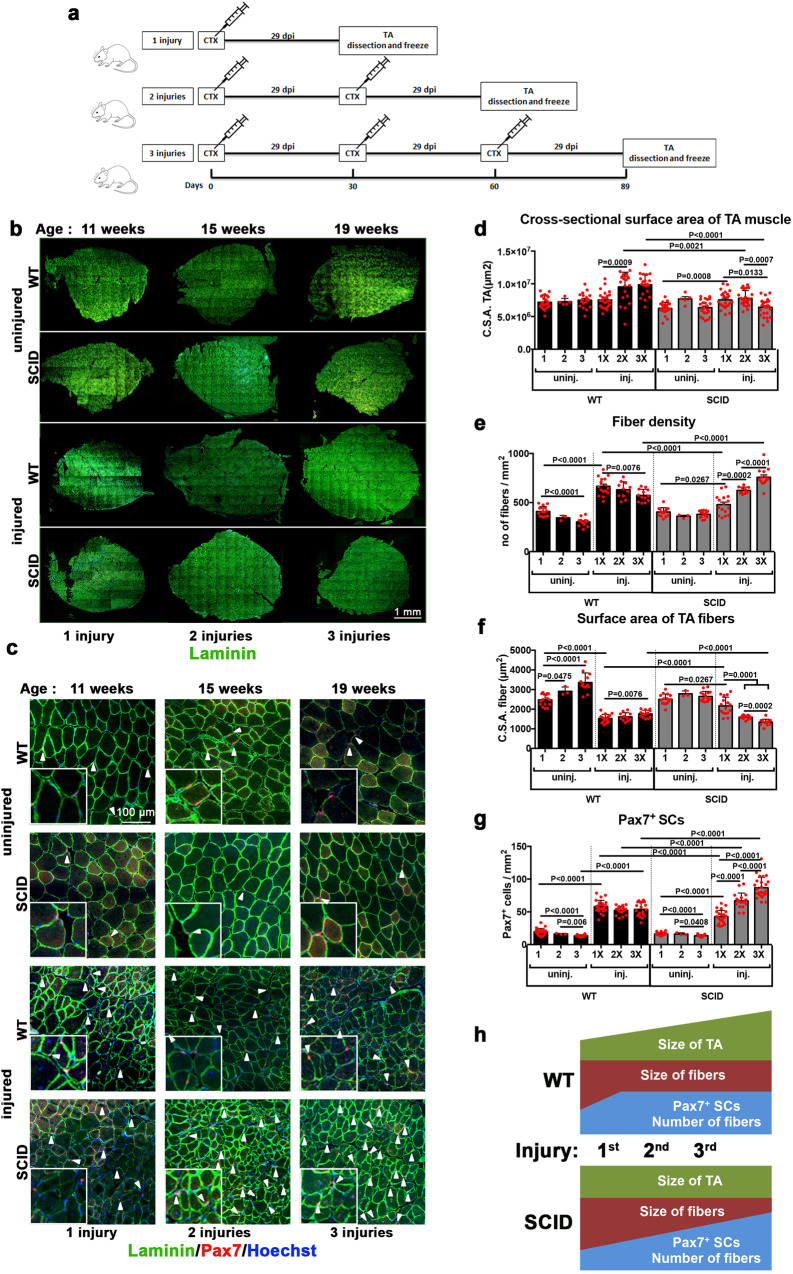
Fig. 4. Delayed SC differentiation in SCID mice results in altered muscle composition during regeneration.
a–c SCs isolated from SCID and the corresponding WT mice. a Immunofluorescent labeling of WT or SCID SC-derived myogenic cells with: Pax7 (green), MyoD (red), Myogenin (red), and MHC (yellow); nuclei are counterstained with Hoechst (blue). Pax7+ cells are indicated with a green arrow, MyoD+ cells with a red arrow, Myogenin+ cells with a white arrow, and MHC+ cells with an orange arrow. b Histograms of myogenic differentiation of SC derived from WT and SCID mice. Combination of myogenic markers reveals several myogenic or differentiated subpopulations, indicated in the legend of the histogram, and displayed with increasing differentiation level from bottom to top. n = 3–4 mice, 5–10 fields/condition, mean ± SD. Significance by Mann–Whitney test, P values indicated on the histogram. c Fusion index 5.5 and 7.5 days post seeding. The number of nuclei/structures have been arbitrarily defined and vary from 0 to >20. Sixty-seven then 789 MHC+ cells/0.44 mm2 in SCID mice, and 73 then 870 MHC+ cells/0.44 mm2 in WT mice at 5.5 dps and 7.5 dps, respectively. n = 3–4 mice, 5000 nuclei/condition, mean ± SD. Significance by Mann–Whitney test, P values are indicated on the histogram. d–h In vivo TA muscle regeneration in WT and SCID mice 5 and 7 dpi. d Experimental scheme: 10 µM of cardiotoxin injury followed by TA dissection 5 and 7 dpi, along with uninjured TA from the same mouse. Cross-sectional scheme of TA muscle and parameters measured are shown. Histograms of e number of regenerating eMHC+ fibers/mm2, f number of total fibers/mm2, and g cross-sectional surface area (CSA) of fibers on TA sections. The CSA of the fibers is a measure of their size (thickness), obtained by dividing the TA surface by the number of fibers. Representative images of e, f and g are shown in Fig. S5a. Representative images of h Pax7+ SCs (in red) and i Myogenin+ differentiating cells (in red), indicated with arrowheads on uninjured and 5 dpi TA sections. Quantification of j Pax7+ SCs/mm2 and k Myogenin+ cells/mm2 on uninjured and 5 dpi TA sections. n = 3 mice, 10 sections/condition, mean ± SD. Significant P values between conditions by Mann–Whitney test are indicated on the histograms.
Altered myofiber composition and structure after regeneration in SCID mice
To assess regeneration of DNA-PKcs deficient SCs in vivo, SCID and the corresponding WT control mice were injured with cardiotoxin (CTX) in the tibialis anterior (TA) muscle. TA muscles were dissected and analyzed 5 and 7 days post injury (dpi) (Fig. 4d). Regeneration was assessed by quantification of embryonic eMyHC+ fibers (Fig. S5a), a marker that is expressed transiently during muscle regeneration [27]. We observed a higher density of eMyHC+ fibers in injured SCID compared to WT TA muscle (589 and 455 eMHC+ fibers/mm2 at 5 dpi respectively, Fig. 4e), and consequently a higher density of total fibers (enumerated after Laminin+ labeling; 721/mm2 in SCID and 628/mm2 in WT; Fig. 4f). Although the number of eMyHC+ regenerating fibers decreased from 5 dpi to 7dpi, they remained at higher density in injured SCID than WT TA muscle (508 and 412 eMyHC+ fibers/mm2, respectively at 7dpi; Fig. 4e), and the total number of fibers was not significantly different at 7dpi (613 fibers/mm2 in SCID TA and 635 fibers/mm2 in WT TA; Fig. 4f). In contrast, in uninjured TA muscle the density of total fibers was higher in WT than SCID mice (480/mm2 and 429/mm2, respectively, Fig. 4f). To uncouple the regeneration defects due to DNA-PKcs deficiency from immunodeficiency [28], we also injured Rag2−/−γc−/− immunodeficient mice and the corresponding control (C57BL/6), and observed no difference in the density of regenerating fibers at 5 dpi (Fig. S5b).
As immunodeficiency may also affect the innate immunity, we analyzed whether SCID mice had altered composition of the M1 and M2 macrophages that have a regulatory role in myogenesis at the proliferative and differentiation steps, respectively [29, 30]. M1 (CD68+, CD206−) and M2 (CD68+, CD206+) populations, identified by combinations of the pan-macrophage marker CD68 and anti-inflammatory marker CD206, were assessed in uninjured and injured TA muscle of WT and SCID mice at 7 dpi (Fig. S5c–e). As expected, we observed a marked increase of both populations in WT as well as SCID mice upon injury, but no significant differences in number between WT and SCID mice (Fig. S5e), indicating that M1and M2 were present during muscle regeneration in both mice.
SCID fibers displayed a smaller (16%) cross-sectional area (CSA) than WT at 5 dpi, and this difference was lost at 7dpi (Fig. 4g). In contrast, in uninjured mice the CSA of fibers was 10% larger in SCID than WT mice. In summary, SCID mice have slightly less fibers, but these fibers cover a larger surface than in WT. Upon injury, the situation is reversed and fibers increase in number and cover a smaller surface in SCID than in WT mice during early regeneration, then recover later, pointing to a delay in muscle regeneration in SCID mice.
Cross-sectional area of the entire TA muscle showed that uninjured SCID have less Pax7+ SCs than WT (18 cells/mm2 and 28 cells/mm2, respectively) (Fig. 4h, j). This situation was reversed after CTX injury (323 Pax7+ SCs cells/mm2 in SCID vs. 205 Pax7+ SCs cells/mm2 in WT TA). Similarly, differentiating (Myogenin+) cells were more numerous in injured SCID (265/mm2) than WT TA muscle (198/mm2) (Fig. 4i, k). Therefore, SCID mice show hallmarks of delayed regeneration that are compensated at 7 dpi.
Repetitive injuries accentuate regeneration defects in SCID mice
We then asked if SCID mice can sustain multiple cycles of regeneration, as is the case for WT mice [26, 31]. To do so, TA muscles were injured with CTX from 1 to 3 consecutive times, allowing 30 days for regeneration for each cycle [26]. Three sets of mice were used for one, two, and three rounds of injury, respectively (Fig. 5a).
Fig. 5. Marked decrease of muscle mass and increased SC population in SCID mice after multiple regeneration events.
a Experimental planning of repetitive cycles of muscle injury and regeneration. WT and SCID mice were injected 1 or 2 or 3 times with cardiotoxin, at intervals of 29 days between injections. TA muscles were dissected and frozen either at 29 days post injury (1 injury), or 59 days post injury (2 injuries), or 89 days post injury (3 injuries). b Complete sections and c zoomed images of cross-sectional TA, immunostained for Laminin (green) and Pax7 (red), and counterstained for Hoechst (blue, nuclei). Histograms of d cross-sectional surface area of the entire TA (measured from sections as in (4d); n = 4–5 TA (mice)/condition), e number of fibers per mm2 (i.e. fiber density), f cross-sectional surface area of fibers g number of Pax7+ SCs per mm2. n = 4–5 mice/condition, 5 sections/condition, mean ± SD. Significant P values between conditions according to the Mann–Whitney test are indicated on the histograms. h schematic representation (not at scale) of changes in the parameters measured in d–g upon each injury/regeneration event per mm2 of a TA muscle.
The total body weight of uninjured WT and SCID mice increased from the beginning to the end of the experiment (from 7 to 19 weeks; not shown), but the weight of their TA did not (Table S1). In contrast, after the third regeneration event, TA muscles increased by 34% and 21% in weight in WT and SCID mice, respectively. Overall, the CSA of the TA was smaller in SCID compared to WT mice after the second and third regeneration cycles (Fig. 5b-d). Thus, despite the increasing in body weight, the CSA of the TA muscle was essentially unchanged in SCID mice after 3 regeneration cycles.
The number of fibers (and thereby fiber density), which increased in both mice in the acute phase of the first regeneration (at 5 dpi) (see Fig. 4f), remained high in WT mice at the end of the first regeneration cycle, and slightly decreased in the successive ones (Fig. 5e). Conversely, in SCID mice the fiber density increased slowly at the end of the first regeneration cycle, and progressively increased during the successive regeneration cycles. Thus, after the third regeneration event the fiber density was higher in SCID than WT mice. The fiber size also decreased at 5 dpi (Fig. 4g), and remained low until the third regeneration cycle in all mice, in particular SCID (Fig. 5f). Finally, the density of Pax7+ SCs increased from 19/mm2 in uninjured mice to 58/mm2 after the first regeneration cycle, and remained stable until the third regeneration cycle in WT mice. Conversely, the density of Pax7+ SCs kept increasing in SCID mice, and reached 87/mm2 after the third regeneration cycle, thus exceeding the number in the WT (Fig. 5g).
It was reported that increased DNAPK, that inhibits the AMP-activated protein kinase (AMPK), has a regulatory role in mitochondrial homeostasis and metabolism, and thus on the mass and fiber composition (oxidative or glycolytic) of the muscle [32, 33]. We wondered whether alterations in the structure of the muscle fibers upon regeneration was associated with changes in the composition of myofibers in SCID mice. Thus, we assessed the fiber compositions by immunofluorescence of MHC isoforms (MHC type I, slow oxidative; MHC type IIa, fast oxidative; MHC type IIb, fast glycolytic; MHC type IIx, fast oxidative/glycolytic intermediate) of the TA muscle [34] after 1 and 3 successive regeneration events, and in uninjured TA muscle (Fig. S6a). The aim was to assess whether DNA-PKcs deficiency in SCID mice has an impact on the fiber composition, in addition to the observed differences in the fiber size and density. After consecutive injury and successive regeneration, we observed a slight reduction of MHC type IIa+ fibers (oxidative fibers) in the TA muscle of WT mice, whereas this was not the case in SCID mice. Despite SCID mice experienced slight changes in MHC type IIx and MHC type IIa+ fibers after the first regeneration event, these differences were resumed after the third regeneration event (Fig. S6b), indicating that differences in the myofibers upon regeneration in SCID mice were not associated with changes in fiber composition.
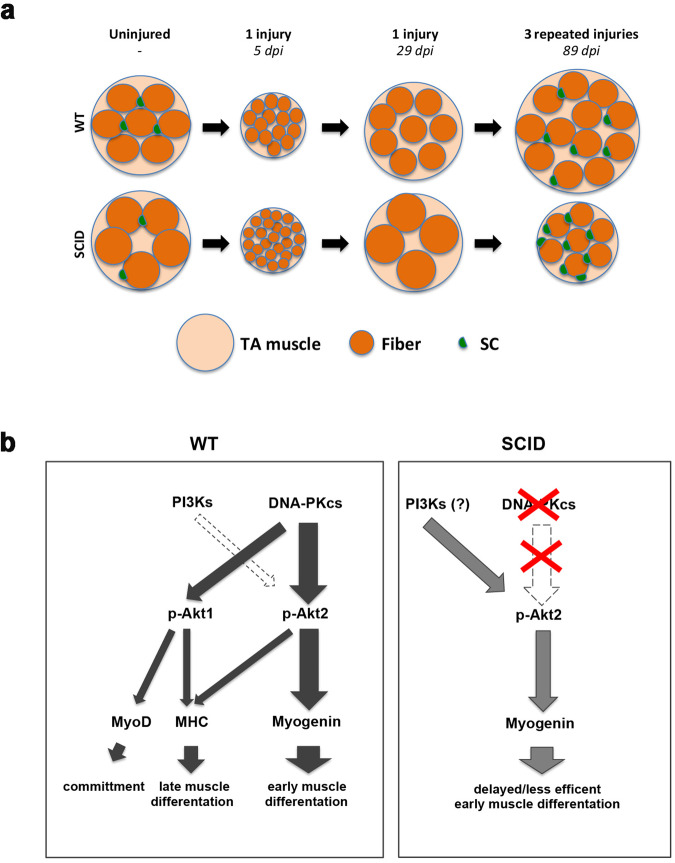
In summary, following multiple rounds of injuries, the TA muscle of WT mice was larger in size, and SCID mice of smaller size, compared to uninjured muscle (see Figs. 5h and 6a). Despite these differences, upon repetitive injuries the TA of either mice contained more fibers, which were of smaller size, and had a higher number of SCs compared to the corresponding uninjured muscle, pointing to different regeneration dynamics in mice mutant for DNAPK.
Fig. 6. Scheme activation of Myogenin and muscle fibers before and after regeneration in WT vs SCID.
a Scheme of cross-sectional area of uninjured (the comparable cross-sectional surface area of uninjured TA muscle as WT: 5.4 ± 1.2 mm2 and SCID: 5.5 ± 1.1 mm2), one-time injured TA at 5 dpi and 29 dpi, and 3 times injured (every 29 days) TA muscle at 89 dpi from WT and SCID mice. Big cantaloupe circles represent TA muscle, orange circles represent individual fibers and green half circles represents SCs. The scheme, which is not at scale, aims to summarize the difference in SC number, fiber number, and size in conditions tested in this study. b Left panel: schematic representation of the activation of Myogenin by DNA-PKcs through phosphorylation of Akt2. The activation of other myogenic and differentiation factors (MyoD and MHC) by DNA-PKcs through Akt2 and/or Akt1 is also shown. The larger the arrow the stronger the effect. The minor role of PI3Ks is indicated with a white dotted arrow. In the absence of DNA-PKcs, PI3Ks in large part activates Myogenin (right panel).
Discussion
Muscle stem cell differentiation undergoes multiple regulatory states. We identify a new, caspase-independent, regulatory function for DNA-PKcs kinase in this process where it acts independently of its role in DSB repair and via activation of Akts. We also show that chronic impairment of this pathway in vivo leads to alternative activation of Akts that results in compromised muscle regeneration.
DNA-PKcs activates the expression of Myogenin in an Akt2-dependent manner
We report here that differentiation of myogenic cells requires the protein kinase DNA-PKcs to promote expression of the key differentiation factor Myogenin. Inhibition or silencing of DNA-PKcs blocks the expression of Myogenin and myogenic differentiation in an Akt2-dependent manner (Fig. 6b), and reduces phosphorylation of Akt. Further, inhibition of Akts as well as Akt2 silencing blocks Myogenin expression. Consistently with these findings, Myogenin expression has been reported to correlate with the expression and activation of Akt2 [35, 36]. Importantly, our findings show that DNA-PKcs acts upstream of Akt2, and is directly responsible for Akt2 activation.
Akts have been shown to play a role in myogenesis upon activation by PI3Ks, specifically Akt1 which promotes SC activation, whereas Akt2 is implicated in differentiation [11,12, 13, 37]. These studies mostly focused on activated or committed myogenic cells (MyoD+). Our findings focus on the pivotal role of regulating the expression of Myogenin and differentiation, which we show requires Akt2 phosphorylation by DNA-PKcs. Conversely, PI3Ks, which are major effectors of Akt activation [38], and were hypothesized to act also during this phase of myogenesis [13], do play a minor role.
Notably, we show that DNA-PKcs intervenes at multiple phases of muscle differentiation, since it acts also upstream and downstream of its major target Myogenin. Indeed, by phosphorylating Akt1, DNA-PKcs promotes expression of MyoD, and by phosphorylating Akt1 and Akt2, it leads to the expression of the late differentiation factor MHC (Fig. 6b). However, inhibition or silencing of DNA-PKcs reduces but does not block the expression of MyoD and MHC, in contrast to the effect on Myogenin.
DNA-PKcs activates Myogenin expression in the absence of induced DNA damage
DNA-PKcs has been reported to activate Akts essentially at DSB sites where it regulates the DNA repair mechanism of NHEJ [39, 40]. DNA-PKcs is a multifunctional kinase implicated in several processes linked to the repair of DNA damage, in addition to its direct function in NHEJ [41]. Namely, this kinase modulates transcription through interaction with the transcriptional machinery (in particular RNAPII), and phosphorylation of transcription factors including master regulator genes [42], and histones [43]. In these processes, DNA-PKcs is essentially activated by DNA damage [44]. In other cases, for instance activation of the hypoxia factor HIF1α, the action of DNA-PKcs depends on histone acetylation rather than DNA damage [45, 46].
We identify a novel role of DNA-PKcs in muscle differentiation that we propose to be independent of DNA damage, either induced (e.g. caspase-dependent) or of endogenous origin. To activate Myogenin expression, DNA-PKcs does not require irradiation, and this activation is not associated with increased DNA damage or γH2AX levels. Accordingly, the Myogenin promoter does not undergo DNA damage during muscle differentiation [5]. DNA-PKcs-dependent activation of Myogenin appears also to be independent of DNA damage specifically associated with muscle differentiation, and which is induced by a caspase-dependent nuclease (CAD) [5]. In that context caspase-3 and caspase-9 are involved in muscle differentiation [47, 48], and DNA-PKcs phosphorylates the histone H2AX in response to caspase-dependent DNA damage [25]. Transient DNA damage has been proposed to play a role in muscle differentiation, but this process has not been fully elucidated. As such, caspase-induced DNA damage is repaired, likely by nucleotide excision repair (NER), and it is thought to regulate the expression of genes that are relevant for myogenesis, for instance p21 that is necessary for cell cycle withdrawal and thus myogenic differentiation [5, 49]. In our experiments inhibition of caspases did not reduce the number of Myog+ cells, whereas inhibition of DNA-PKcs fully depleted this myogenic population. DNA-PKcs inhibition blocked Myogenin expression and myogenesis also in the conditions used by Connolly et al. [25], i.e. (serum-depleted) DM, whereas in these conditions caspase inhibition only poorly affected Myogenin expression and myogenesis, and rather affected myogenic fusion. These data show that DNA-PKcs-dependent myogenesis through activation of Myogenin is well distinct from DNA-PKcs phosphorylation of H2AX in response to caspase-induced DNA damage.
DNA-PKcs interacts with the complex that activates the Myog promoter
DNA-PKcs-dependent expression of Myogenin does not require the presence of the kinase directly on the Myog promoter, compatible with the notion that activation is not triggered by DNA damage. Instead, DNA-PKcs interacts with components of the complex that activates Myog expression and includes the transcription activator p300 and the transcription factor MyoD [21]. Co-immunoprecipitation experiments showed that this complex also includes Akt1, and Akt2 (at least in complex with p300), in analogy with Akts phosphorylating p300 to promote the interaction with MyoD [21]. Our experiments also indicate that DNA-PKcs itself interacts with Akt1 and Akt2. It is thus likely that DNA-PKcs phosphorylates Akt2 that in turn activates p300 and MyoD at the Myogenin promoter. This mechanism appears similar to activation of the fatty acid synthase promoter, although in that case DNA-PKcs activation largely relies on DNA damage at the target promoter [50]. It is possible that DNA-PKcs also activates Akt1 at the MyoD promoter, and Akt1/Akt2 at the MHC promoter in a similar way, although we have not dissected these two mechanisms where DNA-PKcs has a lesser impact.
Delayed muscle differentiation in SCID mice
Although inhibition and silencing of DNA-PKcs activity severely affects myogenesis in WT SCs in vitro, DNA-PKcs-deficient (SCID) mice do develop muscles. Muscle is normally formed also in engineered DNA-PKcs knock-out mice that are deleted for the kinase domain [51, 52]. We showed that nevertheless, SCID mice exhibited detectable defects in differentiation in vivo and in vitro (e.g. delayed myotube formation and reduced number of MHC+ cells). This was accompanied by a higher number of renewing SCs, as well as MHC+ and Myogenin+ cells in SCID compared to WT during early muscle regeneration. An increased number of myogenic and differentiated cells in regenerating SCID may result from hyperactivity or, alternatively, delayed regeneration. The latter possibility is in agreement with in vitro data reported in the present study, which indicate that muscle regeneration in SCID mice is delayed according to the scheme provided by Murphy et al. [53]. These alterations are not associated with changes in the composition of glycolytic and oxidative myofibers.
Notably, regeneration deficits were exacerbated after multiple rounds of muscle injury and regeneration, resulting in smaller TA muscles in SCID mice. SCID TA also contained less myofibers than WT. These alterations suggest that regeneration defects in SCID mice may affect long-term muscle function. Observations that SCID mice regenerate muscle after multiple rounds of injury suggest that DNA-PKcs-deficient mice rely on alternative mechanisms, probably resulting from adaptation to chronic deficiency of DNA-PKcs. These mechanisms would ensure activation of Akt kinases and Myogenin expression. PI3K kinases are good candidates for this task since insulin, that activates PI3Ks, partially rescued the pAkt signal and Myogenin expression in DNA-PKcs-inhibited cells. Moreover, inhibition of PI3Ks to some extent reduced Akt2 phosphorylation and Myogenin levels.
DNA-PKcs deficiency in SCID mice [54] is due to a point mutation (Tyr-4046) that generates an early termination codon, abolishing the kinase activity [55]. The missing domain is thought to be responsible for the stability and/or the folding of the protein, and indeed DNA-PKcs protein levels are negligible in SCID mice [56, 57]. It is unlikely that the residual DNA-PKcs (if any) would trigger myogenesis in SCID mice, since the myogenic function of DNA-PKcs depends on its kinase activity that is missing in these mutants [55]. The kinase domain is also depleted in engineered DNA-PKcs mutants, that also do form muscle [51, 52]. Thus, alternative, PI3K-dependent mechanisms likely promote myogenesis when DNA-PKcs is inactive.
SCID mice are a common model for muscle dystrophy and transplantation studies. For example, SCID/mdx mice are immunodeficient due to the DNA-PKcs mutation (NHEJ is required for immunoglobulin V(D)J recombination) and a model of muscular dystrophy due to mutation in the dystrophin gene (DMD, Duchenne muscular dystrophy). Immunodeficiency makes these mice good recipients for transplantation experiments, thereafter most studies are concentrated on the effects of immunodeficiency over muscle regeneration, by assessing transplanted cells derived from WT mice or human [58], or focusing on inflammation [59] or fibrosis during regeneration of the injured muscle [60]. Although SCID mice remain a good model for many studies, we show here that impairment of DNA-PKcs results in different muscle regeneration dynamics and muscle composition compared to WT, which occur upon unaltered composition of anti-inflammatory macrophages, and these differences should be considered in future studies with these mice.
Conclusions
We have demonstrated that DNA-PKcs is essential for myogenesis in vitro where it activates Myogenin expression through phosphorylation of Akt2. To a minor extent DNA-PKcs also activates the upstream myogenic factor MyoD and the downstream marker MHC, thereby playing multiple roles during muscle commitment and differentiation. In the absence of DNA-PKcs, Myogenin is activated by PI3Ks, which however results in delayed dynamics of regeneration. These alterations are exacerbated following repeated rounds of muscle injury. SCID mice that are naturally mutated in DNA-PKcs may have developed compensatory mechanism that may not necessarily operate in WT mice. Since SCID mice are currently used for stem cell transplantation studies, it should be considered that these mice regenerate muscle through an adaptative and essentially DNA-PKcs-independent mechanism.
Materials and methods
Animal models
For most in vitro experiments and drug treatments SCs were isolated from the limbs of Tg:Pax7-nGFP mice [61]. DNAPK-deficient SCs and TA cryosections were obtained from CB17/lcr-Prkdcscid/lcrlcoCrl (SCID) mice and Balb/cAnN (wild-type, WT, control) purchased from Charles River laboratories. For regeneration analysis of injured TA muscles, also Rag2−/− γc−/− and C57BL/B6 mice as control were used, provided by Shahragim Tajbakhsh, Paris. The genotype, phenotype and genetic background of each mouse strain are indicated in Table S2.
Ethics statement
Animals housing, husbandry and handling were performed in the animal facility of Institut Pasteur in accordance with the national and European community guidelines. SC isolations were performed from healthy 7–10 weeks old male mice, in vivo injury experiments were performed on healthy 7–19 weeks old male mice according to national and European guidelines, and protocols were approved by the ethics committee at Institut Pasteur and the French Ministry.
Satellite cell isolation by FACS and cell culture
SCs were freshly isolated from fore and hind limbs of ad hoc mice that express the Pax7 marker linked to the fluorescent label GFP (Tg:Pax7-nGFP mice), following the isolation protocol, as previoulsy described [62], and in more detail in [63]. Briefly, initially the limbs were chopped off, and precipitated in DMEM Glutamax (GIBCO) with 1% Penicillin Streptomycin (Pen/Strep). The floating lipid particles were successively removed by discarding the excess medium. This step was followed by digestion step with 0.1% Collagenase D (Roche), 0.25% Trpysin (GIBCO) and 0.1 mg/ml DNase I (Roche) in DMEM Glutamax (containing 1% Pen/Strep) for 30 min at 37 °C, which was repeated 5 times until full digestion of the tissues. After each digestion step, where the tissue was placed in a fetal bovine serum (FBS) solution to inactivate trypsin, the digested material (suspension) was filtered on ice through subsequently 100 µm, 70 µm, and 40 µm cell strainers respectively. The filtered digests were then pre-spinned for 10 min at 50 × g and the supernatant was collected. The supernatant was then centrifuged for 15 min at 550 × g at 4 °C. The supernatant was discarded and the pellet resuspended in fresh DMEM Glutamax with 1% of Pen/Strep. Centrifugation was repeated 3–4 times until the resuspension was sufficiently transparent to proceed further. After the final spin, the pellet was resuspended in 600 µl of DMEM Glutamax (with 1% Pen/Strep) and 2% FBS. Finally, the samples were processed by FACS (ARIA III, BD Biosciences) to sort SCs, which are Pax7-nGFP+, using GFP as marker.
In the case of SCID mice and the corresponding Balb/cAnN control mice, as well as Rag2−/−− γc−/− and the control C57BL/B6 mice, which SCs are not Pax7-nGFP+, the muscle bulk was digested with 2.4 U/ml of Dispase II (Roche), 0.2% of Collagenase A (Roche) and 0.1 mg/ml DNase I (Roche) in DMEM Glutamax (+1% Pen/Strep) for 2 h at 37 °C. Then filtration and centrifugation were performed as indicated above. At the final step, cells were labeled with receptor markers (conjugated antibodies) for 30 min at 4 °C. Cells were then sorted with FACS ARIA III, gating the SC population for Sca1−, CD45− and CD31−, CD34+, and Itgα-7+, as indicated in the published protocol [63].
Cell culture
Immediately after sorting, collected SCs were seeded at a density of 5000 cells/cm2 in Lab-TekTM chamber slides (Nunc) or cell dishes pre-coated with 0.1 mg/ml of poly-D Lysine (SIGMA) and Matrigel (Corning). The (growth) medium used consisted of: 40% of DMEM Glutamax with 1% Pen/Strep, 40% of MCDB (SIGMA), 20% of FBS, 2% of Ultraser G (Pall), and 10 µg/ml of insulin when indicated. Cells were incubated at 37 °C with 5% CO2 and 20% O2. With this medium, during differentiation serum reduction is gradual, due to the metabolism of dividing cells in culture, and differentiation is not induced.
C2C7 cells (immortalized myoblast cell line) [64, 65] were incubated in the same condition as SCs without pre-coating in DMEM Glutamax with 1% Pen/Strep and 20% FBS. When indicated, C2C7 cells were incubated in differentiation medium (DM) which consisted of DMEM Glutamax with 1% Pen/Strep and 2% horse serum (serum depletion induces differentiation).
Inhibitor treatment and irradiation
Cells were pre-treated by addition of inhibitors to the culture medium for 1 h before irradiation, at a final concentration of 10 µM of the DNAPK inhibitor (=DNAPKi: NU7441) (Axon Medchem), 5 µM of the Akt inhibitor (AKTi, MK2206) (APExBIO), 10 µM of the PI3K inhibitor (LY294002), or 1 µM of the PI3K inhibitor (ZSTK474) (Selleckchem), 30 µM of caspase inhibitor Q-VD-OPh (A1901) (APExBIO) or the corresponding volume of vehicle (DMSO, diluent of the drugs). After 1 h treatment, cells were irradiated at 5 Gy (with the Xstrahl RS320 Irradiator for X-ray), which is expected to induce approximately 200 DSBs [66], or were not exposed to irradiation (control and vehicle). Cells were incubated for different periods (from 1 h to 7 days) post irradiation before assessing SCs/C2C7 cells differentiation, viability, and cell number. Two experimental plans were used (see Figs. 1, S2, and S3). In the first plan, cells were treated with inhibitor (or vehicle) 12 h after seeding then analyzed at 1–5 or 7 days post seeding (dps). In the second plan, cells were treated with inhibitor (or vehicle) 2.5 or 4.5 days after seeding then analyzed at 5 or 7 dps. In experiment in Fig.1, cells were treated after overnight seeding and analyzed at the indicated times; in the following experiments SCs were treated 5.5 dps and analyzed at 7.5 dps.
shRNA cloning, transfection, and transduction
Lentiviruses were produced from the shRNA plasmids or scrambled (SCR) shRNA for control (Mission shRNA library, Sigma) cloned in Escherichia coli (Table S3). ShRNA plasmids, that carry ampicillin and puromycin resistance genes, were amplified in E. coli grown overnight in Luria Bertani broth medium with 100 µg/ml Ampicillin. ShRNA plasmids were extracted and concentrated with NulceoBond® Xtra Maxi Plus kit, Plasmid DNA purification kit (Macherey-Nagel, ref.: 740416.10) following the manufacturer’s protocol. Purified shRNA plasmids and plasmid coding for the viral particle (psPAX2-packaging, pMD2.G-envelope) were co-transfected in HEK-293T cells treated with 25 µM of chloroquine diphosphate (Sigma), a DNA intercalator that increases the transfection efficiency, using the calcium phosphate transfection method [67]. Two days post transfection, the HEK-293T cell culture medium was harvested, filtered through 0.45 µm filters, and concentrated by ultra-centrifugation at 19,000 rpm for 90 min at 4 °C. After centrifugation the supernatant was discarded and the pellet resuspended in phosphate buffered saline (PBS) in a final volume of 250 µl. For titration purposes, p24 viral particles were quantified by the Virus and Immunity laboratory at Institut Pasteur.
Freshly FACS-sorted SCs were put in culture for 12 h then re-seeded at a density of 5000 cells/cm2, and 12 h later transduced with shRNAs or SCR (scrambled non-targeting shRNA) concentrated viral particles. The viral particles were added to the cell culture medium with 8 µg/ml of the transduction enhancer Polybrene (hexadimethrine bromide) (Sigma). Six to 8 h post transduction the culture medium was refreshed (without viral particles and Polybrene), and 48 h post transduction (or 2.5 dps, considering re-seeding as the starting point), was again refreshed with 1 µg/ml puromycin containing medium for selection of cells stably expressing the shRNA. After 3 days of puromycin selection, i.e. 5 days post transduction (corresponding to 5.5 dps), surviving cells were harvested. In parallel, a first sample of cells was harvested 2.5dps and a second sample 5.5 dps, in the absence of puromycin treatment and shRNA transduction (control 1 and control 2, respectively).
Muscle injury
For regeneration analysis, mice were anesthesised with 0.5% Imalgene + 2% Rompun, and tibialis anterior (TA) muscles were injected with 50 µl of 10 µM protein kinase C specific inhibitor, cardiotoxin (Latoxan), as previously described [68]. The TA muscles were harvested for analysis 5 days post injury. For repetitive injury, the mice were injected with cardiotoxin for 3 times during 89 days (the second and third injections were done 30 and 60 days after the first injection, respectively). For these experiments, TA muscles were dissected and frozen 29 days post injury (1 injury), or 59 days post injury (2 injuries), or 89 days post injury (3 injuries), and analyzed all together. The first injury was performed on 7 weeks old mice and TA muscles injured 3 times were dissected from 19 weeks old mice.
Immunostaining and histology
Satellite cell-derived cultures were fixed in 4% PFA for 5 min and permeabilized with 0.5 % Triton X-100 for 5 min as previously described [62]. Upon permeabilization, samples were treated for antigen blocking with 2 % BSA and 5% serum in PBS for 1 h at room temperature. Primary antibodies were incubated overnight at 4 °C, and secondary antibodies (conjugated with fluorophores), and 5 µg/ml Hoechst 33342 (Sigma, ref #14533) (nuclear counterstaining) for 2 h at room temperature.
Freshly dissected TA muscles were frozen in liquid nitrogen cooled isopentane for 30 s. The frozen TA muscles were cryo-sectioned into 10 µm slices. These sections were fixed with 4% PFA for 15 min at room temperature and antigen unmasking was performed by incubating sections in boiling 10 mM citrate buffer pH 6 for 20 mins prior to immunostaining, as described [61]. For regeneration analysis with embryonic Myosin heavy chain (eMHC) specifically, the TA sections were left unfixed and treated with acetone for 10 min at -20 °C prior to immunostaining. All sections were permeabilized with 0.5% Triton X-100 for 5 min and antigen blocking performed with 10% goat serum (inactivated) or 2% goat serum for eMHC staining in PBS and 0.5% Triton X-100 containing PBS for 1 h at room temperature. Incubation with primary and secondary antibodies was performed as for cell immunostaining, see above.
Antibodies
The following antibodies have been used in this study: mouse monoclonal α-Pax7 (AB_528428), mouse monoclonal α−Myogenin (F5D, AB_2146602), mouse monoclonal α-Myosin Heavy chain, MHC (MF20, AB_2147781) and mouse monoclonal α-embryonic MyHC (F1.652, AB_528358), α-Myosin Heavy chain type I (BA-D5), α-Myosin Heavy chain type IIa (SC-71), α-Myosin Heavy chain type IIb (BF-F3) antibodies from Developmental Studies Hybridoma Bank; rabbit polyclonal α-Myogenin (sc-576), rabbit polyclonal α-MyoD (sc-304), Rabbit polyclonal α-GAPDH (sc-25778) from Santa Cruz; mouse monoclonal α-Akt2 (5239), rabbit polyclonal α-phospho-Akt2 (Ser474) (8599), rabbit polyclonal α-Akt1 (C73H10), rabbit polyclonal α-phospho-Akt1 (Ser473) (9018), rabbit polyclonal α-phospho-Akt (9271), rabbit polyclonal α-H2AX (2595) from Cell Signaling technology; mouse monoclonal α-phospho-H2AX(Ser139) (05-636) from Millipore; rabbit polyclonal α-53BP1 (NB100–304) from Novusbio; chicken polyclonal-α-GFP (ab13970), rabbit polyclonal-α-DNA-PKcs (ab70250) rabbit monoclonal α-Cyclin A2 (ab181591) from abcam; Rabbit α-Laminin (L9393), Propodium Iodide (P4864) from Sigma; goat α-rabbit fluor 555 (A21428), goat α-rabbit fluor 488 (A11034), goat α-mouse fluor 488 (A11029), goat α-mouse fluor 555 (A21425), goat α-mouse fluor cy5 (A10524), goat α-chicken fluor 488 (A11039), goat α-mouse-HRP (31430), goat α-rabbit-HRP (31460), goat α-mouse IgG1 fluor 488 (A-21121), goat α-mouse IgGM fluor 555 (A-21426), goat α-mouse IgG2b fluor 647 (A-21242), goat α-rabbit fluor 405 (A-31556) from Invitrogen; mouse α-CD68 (MCA1957), hFAB Rhodamine Anti-GAPDH IgG (12004168), Goat α-mouse Starbright blue 700 (12004159) from BioRad; goat α-rabbit CF770 (10078-1) from Biotium; mouse α-CD206-alexa fluor 647 (565250), FITC Annexin V Apoptosis Detection kit I (556547) from BD Biosciences.
Western blotting
Cells were lysed in lysis buffer (5 mM EDTA, 50 mM Tris pH 8, 150 mM NaCl, 0.5 % Triton X-100, 0.1 % SDS) with 2× of cOmpleteTM protease inhibitor cocktail (Roche), 1× PhosSTOP (phosphotase inhibitor, Roche) for 30 min on ice. In order to get rid of viscosity due to DNA, the lysed samples were sonicated 5 times for 10 s (with 10 sec intervals) at medium frequency in the sonicator bath. Samples were denatured in 1× Laemmli buffer with 1/10 β-mercaptoethanol for 5 min at 95 °C and seperated in NuPAGE 4–12% Bis-Tris Protein Gels (Invitrogen). The separated proteins were transferred and bound on nitrocellulose membrane by Trans-Blot Turbo system (Bio-Rad) or classical electro-transfer in 1× NuPAGE transfer buffer (with 20% ethanol) bound on an Amersham Protran nitrocellulose membrane (0.45 µm porosity) (GE Healthcare, Life Sciences). The membranes were blocked with 5% nonfat dry milk for detection of non-phosphorylated residues, or 5% BSA for detection of phosphorylated proteins in 1× PBS with 0.1% Tween 20 for 1 h at room temperature. Primary antibodies were incubated in the corresponding blocking buffer (milk or BSA in 1× PBS with 0.1% Tween 20) overnight at 4 °C followed by 1 h incubation with the secondary antibody (conjugated with either peroxidase [HRP] or fluorophore) at room temperature.
Imaging and enumeration of immunomarkers
Cells were analyzed with a fluorescence microscopy Zeiss Axioplan equipped with an Apotome and Axiovision software or Cell Voyager CV1000, confocal scanner box (Yokogawa). γ-H2AX and 53BP1 foci (DSB markers) were enumerated in at least 30–50 cells/condition, from 3 independent experiments. For differentiation analysis, cells positive for one or more myogenic markers were enumerated per condition, and the percentage was evaluated on the total number of nuclei (Hoechst staining), by assessing 2000–5000 nuclei/condition and 5–10 fields/condition. The fusion index was calculated with the number of nuclei/myosin heavy chain positive myotubes, by assessing 2000–5000 nuclei/condition and 5–10 fields/condition. Image analyses were performed with the ImageJ 2.0 software.
For the histology analysis, 4–10 complete muscle section per condition were analyzed. The complete muscle section images were taken by Cell Voyager CV1000, confocal scanner box (Yokogawa) and SP8 resonant scanner (Leica) as 5 stacks and maximum projection were analyzed. Image analyses were performed by ImageJ 2.0 software. The actual numbers of eMHC- and eMHC+ fibers, Pax7+ and Myogenin+ nuclei per section were counted. Upon measurement of cross-sectional surface area (CSA) of the TA muscle sections, average cross-sectional surface areas of fibers for each section were calculated by (CSA of TA muscle section/total number of fibers).
Western blots were either exposed on films and scanned, followed by analysis of protein signal intensity by ImageJ 2.0 software when needed, or exposed by Chemidoc (BioRad) and in this case protein signals were analyzed with the Imagelab software. The ratio of proteins were calculated by dividing the the signal intensity of the proteins analyzed by Image J, and protein intensities were normalized by dividing the protein of interest by the intensity of GAPDH in the same condition.
RNA extraction and RT-qPCR
Total RNA was isolated from cells using the RNAeasy extraction kits (Qiagen) and reverse transcribed with Superscript IV reverse transcriptase (Invitrogen ref #18090010), following the manufacturer’s instructions. The reaction mix consisted of 2.5 µM of Oligo d(T)20 primer, 0.5 mM dNTPs, up to 1 µg template RNA, 1× SSIV buffer, 5 mM of DTT and 10 U/µl of SuperscriptTM IV Reverse Transcriptase. The cDNAs were quantified by real-time RT-qPCR using the Power SYBR Green Master mix (Applied Biosystems) on a StepOne Plus RealTime PCR system (Applied Biosystems). Myogenin transcripts were amplified using the forward (5′GTGAATGCAACTCCCACAGC) and reverse (5′CGCGAGCAAATGATCTCCTG) primers, and Tbp transcripts using the forward (5′ATCCCAAGCGATTTGCTG) and reverse (5′CCTGTGCACACCATTTTTCC) primers [69].
Cell counting to assess proliferation
Cells in culture were washed with PBS, trypsinised with 0.05% Trypsin in EDTA for 5 min at 37 °C. Cell counting was performed either (1) with Scepter, Handheld-automated Cell counter (Millipore), which gives the cell count/ml, detected by 60 μm sensors, or (2) manually with the cell counting chambers (KOVA international, ref #87144) and counting under light microscope. When the Scepter was used, at the beginning of each experiment a triple counting was performed to validate the reproducibility of the measurement, then counting was done once/sample. In several cases counting was performed with both methods to validate the cell number.
CHIP (chromatin immunoprecipitation) and IP (Immunoprecipitation) assays
For ChIP, 10 × 106 C2C7 cells at 2.5 dps were cross-linked in 1% formaldehyde (Sigma-Aldrich, 252549) for 10 min, followed by addition of 125 mM glycine to stop the reaction (5 min). Cells were then washed in PBS, resuspended in lysis buffer (10 mM Tris-HCl (pH 8), 10 mM EDTA, 0.5 mM EGTA, 0.25% (v/v) Triton X-100, and protease and RNase inhibitors) for 5 min on ice. The pellets were then resuspended in 250 mM NaCl, 50 mM Tris-HCl (pH 8), 1 mM EDTA, 0.5 mM EGTA, for 30 min on ice to wash out non-cross-linked material. The resulting pellets were resuspended in 10 mM Tris-HCl (pH 8), 1 mM EDTA, 0.5 mM EGTA, 1% (w/v) SDS and chromatin was sheared by sonication (final average size of 200–500 base pairs, (bp)). The chromatin was diluted to obtain the following buffer composition: 0.1% (w/v) SDS, 1% (v/v) Triton, 0.1% (w/v) sodium deoxycholate, 10 mM Tris-HCl (pH 8), 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and protease and RNase inhibitors.
ChIPs were carried out by incubating 10–20 μg of chromatin with 1 μg of either antibody: rabbit polyclonal-α-DNA-PKcs (ab70250), rabbit polyclonal α-MyoD (sc-304), or α-rabbit IgG (Santa Cruz Biotechnology, sc-2027) as a control. After overnight incubation at 4 °C, 40 μl of protein G-Dynabeads™ (Thermo-Fisher Scientific, 10004D) were added on lysate for 2 h at 4 °C. After extensive washing, DNA was isolated from the beads by successive boiling for 10 min in the presence of 10% (w/v) Chelex 100 Resin (Bio-Rad, 1421253), incubating at 55 °C for 30 min in the presence of 100 µg ml−1 of Proteinase K (Eurobio Ingen, GEXPRK01-E), and boiling again for 10 min. After centrifugation, the supernatant was used as a direct template for qPCR detection of 5 different regions of the Myogenin promoter (Fig. 3c) (primers listed in Table S4). Immunoprecipitated chromatin with the indicated antibodies was calculated as a percentage of the input DNA after normalization with control ChIP performed with rabbit IgGs (Mock) [70].
For the IP assays, 10 × 106 C2C7 cells were lysed at 2.5 dps by lysis buffer for 30 min on ice as for WB. The 40 µl of protein G-Dynabeads™ (Thermo-Fisher Scientific, 10004D) were incubated with primary antibodies (α-mouse IgG (Santa Cruz Biotechnology, sc-2025) and α-rabbit IgG (Santa Cruz Biotechnology, sc-2027) were used as controls) for 2 h at 4 °C followed by incubation of antibody-conjugated beads with 10 µg of cell lysates overnight at 4 °C. The immunoprecipitated proteins were analyzed by Western blot.
Statistical analyses
Statistical analyses were performed using the Graphpad Prism (version 6–7) software. For multiple grouped data was applied a multiple comparison analysis: two-way ANOVA coupled with Dunnett’s multiple comparisons test; for the other experiments one-way ANOVA with post-hoc Tukey’s multiple comparisons test, non-parametric Mann–Whitney test or Unpaired Student T test were performed, as indicated.
Supplementary information
Acknowledgements
We thank the lab of Shahragim Tajbakhsh for the material provided to perform in vivo muscle injuries and antibodies targeting of myogenic factors, Hiroshi Sakai for guidance to initate the in vivo injury experiments, and Shahragim Tajbakhsh for helpful discussion, Sebastien Mella for his advice on statistical analysis. We thank the Virus and Immunity Laboratory at Institut Pasteur (director Dr. Schwartz) for quantification of p24 viral particles, the components of Elisa Perdiguero-Gomez lab at Institut Pasteur, in particular Alina Sommer, for the technical help with the analysis of macrophage immunolabeling, and the center for Translational Science (CRT)- Cytometry and Biomarkers Unit and Photonic BioImaging Unit of Technology and Service (CBUTechS and PBI UTechS) at Institut Pasteur for technical support in this study.
Author contributions
HHS planned, analyzed, and performed all experiments, except the immunostaining of macrophages, fiber composition (slow and fast fibers), and some experiments with the caspase inhibitor, which have been performed by BM. HHS also contributed to writing the manuscript. MR supervised the study, analyzed the data, and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by AFM (research grant (16580), and thesis grant (18425)).
Data availability
The data analyzed during this study are included in this published article and the supplemental data files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01177-2.
References
- 1.Vitale I, Manic G, De Maria R, Kroemer G, Galluzzi L. DNA damage in stem cells. Mol Cell. 2017;66:306–19. doi: 10.1016/j.molcel.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Zammit PS, Heslop L, Hudon V, Rosenblatt JD, Tajbakhsh S, Buckingham ME, et al. Kinetics of myoblast proliferation show that resident satellite cells are competent to fully regenerate skeletal muscle fibers. Exp Cell Res. 2002;281:39–49. doi: 10.1006/excr.2002.5653. [DOI] [PubMed] [Google Scholar]
- 3.Vahidi Ferdousi L, Rocheteau P, Chayot R, Montagne B, Chaker Z, Flamant P, et al. More efficient repair of DNA double-strand breaks in skeletal muscle stem cells compared to their committed progeny. Stem Cell Res. 2014;13:492–507. doi: 10.1016/j.scr.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Simonatto M, Marullo F, Chiacchiera F, Musaró A, Wang JY, Latella L, et al. DNA damage-activated ABL-MyoD signaling contributes to DNA repair in skeletal myoblasts. Cell Death Differ. 2013;20:1664–74. doi: 10.1038/cdd.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc Natl Acad Sci USA. 2010;107:4230–5. doi: 10.1073/pnas.0913089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Xu X, Chen Y, Cheung JC, Wang H, Jiang J, et al. Structure of an activated DNA-PK and its implications for NHEJ. Mol Cell. 2021;81:801–10.e803. doi: 10.1016/j.molcel.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell. 2017;66:801–17. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Kotula E, Faigle W, Berthault N, Dingli F, Loew D, Sun JS, et al. DNA-PK target identification reveals novel links between DNA repair signaling and cytoskeletal regulation. PLoS ONE. 2013;8:e80313. doi: 10.1371/journal.pone.0080313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matheny RW, Jr., Geddis AV, Abdalla MN, Leandry LA, Ford M, McClung HL, et al. AKT2 is the predominant AKT isoform expressed in human skeletal muscle. Physiol Rep. 2018;6:e13652. doi: 10.14814/phy2.13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian J, Wang Q, Dose M, Pruett N, Kieffer-Kwon KR, Resch W, et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–37. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, Zhu H, Situ C, Han L, Yu Y, Cheung TH, et al. p110alpha of PI3K is necessary and sufficient for quiescence exit in adult muscle satellite cells. EMBO J. 2018;37. 10.15252/embj.201798239. [DOI] [PMC free article] [PubMed]
- 12.Chen J, Wang Y, Hamed M, Lacroix N, Li Q. Molecular basis for the regulation of transcriptional coactivator p300 in myogenic differentiation. Sci Rep. 2015;5:13727. doi: 10.1038/srep13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knight JD, Kothary R. The myogenic kinome: protein kinases critical to mammalian skeletal myogenesis. Skelet Muscle. 2011;1:29. doi: 10.1186/2044-5040-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida N, Yoshida S, Koishi K, Masuda K, Nabeshima Y. Cell heterogeneity upon myogenic differentiation: down-regulation of MyoD and Myf-5 generates ‘reserve cells’. J Cell Sci. 1998;111:769–79. doi: 10.1242/jcs.111.6.769. [DOI] [PubMed] [Google Scholar]
- 15.Puri PL, Bhakta K, Wood LD, Costanzo A, Zhu J, Wang JY. A myogenic differentiation checkpoint activated by genotoxic stress. Nat Genet. 2002;32:585–93. doi: 10.1038/ng1023. [DOI] [PubMed] [Google Scholar]
- 16.Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166:347–57. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abraham RT. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair. 2004;3:883–7. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Hunter T. When is a lipid kinase not a lipid kinase? When it is a protein kinase. Cell. 1995;83:1–4. doi: 10.1016/0092-8674(95)90225-2. [DOI] [PubMed] [Google Scholar]
- 19.Bozulic L, Hemmings BA. PIKKing on PKB: regulation of PKB activity by phosphorylation. Curr Opin Cell Biol. 2009;21:256–61. doi: 10.1016/j.ceb.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–51. doi: 10.1002/j.1460-2075.1996.tb01045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serra C, Palacios D, Mozzetta C, Forcales SV, Morantte I, Ripani M, et al. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol Cell. 2007;28:200–13. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faralli H, Dilworth FJ. Turning on myogenin in muscle: a paradigm for understanding mechanisms of tissue-specific gene expression. Comp Funct Genomics. 2012;2012:836374. doi: 10.1155/2012/836374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017;168:644–56. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connolly PF, Fearnhead HO. DNA-PK activity is associated with caspase-dependent myogenic differentiation. FEBS J. 2016;283:3626–36. doi: 10.1111/febs.13832. [DOI] [PubMed] [Google Scholar]
- 26.Fukada S, Morikawa D, Yamamoto Y, Yoshida T, Sumie N, Yamaguchi M, et al. Genetic background affects properties of satellite cells and mdx phenotypes. Am J Pathol. 2010;176:2414–24. doi: 10.2353/ajpath.2010.090887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carraro U, Dalla Libera L, Catani C. Myosin light and heavy chains in muscle regenerating in absence of the nerve: transient appearance of the embryonic light chain. Exp Neurol. 1983;79:106–17. doi: 10.1016/0014-4886(83)90382-5. [DOI] [PubMed] [Google Scholar]
- 28.Brzoska E, Ciemerych MA, Przewozniak M, Zimowska M. Regulation of muscle stem cells activation: the role of growth factors and extracellular matrix. Vitam Horm. 2011;87:239–76. doi: 10.1016/b978-0-12-386015-6.00031-7. [DOI] [PubMed] [Google Scholar]
- 29.Bencze M, Negroni E, Vallese D, Yacoub-Youssef H, Chaouch S, Wolff A, et al. Proinflammatory macrophages enhance the regenerative capacity of human myoblasts by modifying their kinetics of proliferation and differentiation. Mol Ther. 2012;20:2168–79. doi: 10.1038/mt.2012.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panci G, Chazaud B. Inflammation during post-injury skeletal muscle regeneration. Semin Cell Dev Biol. 2021;119:32–38. doi: 10.1016/j.semcdb.2021.05.031. [DOI] [PubMed] [Google Scholar]
- 31.Hardy D, Besnard A, Latil M, Jouvion G, Briand D, Thepenier C, et al. Comparative study of injury models for studying muscle regeneration in mice. PLoS ONE. 2016;11:e0147198. doi: 10.1371/journal.pone.0147198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Röckl KS, Hirshman MF, Brandauer J, Fujii N, Witters LA, Goodyear LJ. Skeletal muscle adaptation to exercise training: AMP-activated protein kinase mediates muscle fiber type shift. Diabetes. 2007;56:2062–9. doi: 10.2337/db07-0255. [DOI] [PubMed] [Google Scholar]
- 33.Egawa T, Ohno Y, Goto A, Yokoyama S, Hayashi T, Goto K. AMPK mediates muscle mass change but not the transition of myosin heavy chain isoforms during unloading and reloading of skeletal muscles in mice. Int J Mol Sci. 2018;19. 10.3390/ijms19102954. [DOI] [PMC free article] [PubMed]
- 34.Bloemberg D, Quadrilatero J. Rapid determination of myosin heavy chain expression in rat, mouse, and human skeletal muscle using multicolor immunofluorescence analysis. PLoS ONE. 2012;7:e35273. doi: 10.1371/journal.pone.0035273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneko S, Feldman RI, Yu L, Wu Z, Gritsko T, Shelley SA, et al. Positive feedback regulation between Akt2 and MyoD during muscle differentiation. Cloning of Akt2 promoter. J Biol Chem. 2002;277:23230–5. doi: 10.1074/jbc.M201733200. [DOI] [PubMed] [Google Scholar]
- 36.Vandromme M, Rochat A, Meier R, Carnac G, Besser D, Hemmings BA, et al. Protein kinase B beta/Akt2 plays a specific role in muscle differentiation. J Biol Chem. 2001;276:8173–9. doi: 10.1074/jbc.M005587200. [DOI] [PubMed] [Google Scholar]
- 37.Wilson EM, Tureckova J, Rotwein P. Permissive roles of phosphatidyl inositol 3-kinase and Akt in skeletal myocyte maturation. Mol Biol Cell. 2004;15:497–505. doi: 10.1091/mbc.E03-05-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer. 2020;20:74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol Cell. 2008;30:203–13. doi: 10.1016/j.molcel.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 40.Liu P, Gan W, Guo C, Xie A, Gao D, Guo J, et al. Akt-mediated phosphorylation of XLF impairs non-homologous end-joining DNA repair. Mol Cell. 2015;57:648–61. doi: 10.1016/j.molcel.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yue X, Bai C, Xie D, Ma T, Zhou PK. DNA-PKcs: a multi-faceted player in DNA damage response. Front Genet. 2020;11:607428. doi: 10.3389/fgene.2020.607428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Damia G. Targeting DNA-PK in cancer. Mutat Res. 2020;821:111692. doi: 10.1016/j.mrfmmm.2020.111692. [DOI] [PubMed] [Google Scholar]
- 43.Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–20. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 44.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 45.Bouquet F, Ousset M, Biard D, Fallone F, Dauvillier S, Frit P, et al. A DNA-dependent stress response involving DNA-PK occurs in hypoxic cells and contributes to cellular adaptation to hypoxia. J Cell Sci. 2011;124:1943–51. doi: 10.1242/jcs.078030. [DOI] [PubMed] [Google Scholar]
- 46.Wrann S, Kaufmann MR, Wirthner R, Stiehl DP, Wenger RH. HIF mediated and DNA damage independent histone H2AX phosphorylation in chronic hypoxia. Biol Chem. 2013;394:519–28. doi: 10.1515/hsz-2012-0311. [DOI] [PubMed] [Google Scholar]
- 47.Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA. Caspase 3 activity is required for skeletal muscle differentiation. Proc Natl Acad Sci USA. 2002;99:11025–30. doi: 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murray TV, McMahon JM, Howley BA, Stanley A, Ritter T, Mohr A, et al. A non-apoptotic role for caspase-9 in muscle differentiation. J Cell Sci. 2008;121:3786–93. doi: 10.1242/jcs.024547. [DOI] [PubMed] [Google Scholar]
- 49.Al-Khalaf MH, Blake LE, Larsen BD, Bell RA, Brunette S, Parks RJ, et al. Temporal activation of XRCC1-mediated DNA repair is essential for muscle differentiation. Cell Discov. 2016;2:15041. doi: 10.1038/celldisc.2015.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong RH, Chang I, Hudak CS, Hyun S, Kwan HY, Sul HS. A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell. 2009;136:1056–72. doi: 10.1016/j.cell.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taccioli GE, Amatucci AG, Beamish HJ, Gell D, Xiang XH, Torres Arzayus MI, et al. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity. 1998;9:355–66. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]
- 52.Anne Esguerra Z, Watanabe G, Okitsu CY, Hsieh CL, Lieber MR. DNA-PKcs chemical inhibition versus genetic mutation: Impact on the junctional repair steps of V(D)J recombination. Mol Immunol. 2020;120:93–100. doi: 10.1016/j.molimm.2020.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murphy MM, Keefe AC, Lawson JA, Flygare SD, Yandell M, Kardon G. Transiently active Wnt/beta-catenin signaling is not required but must be silenced for stem cell function during muscle regeneration. Stem Cell Rep. 2014;3:475–88. doi: 10.1016/j.stemcr.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bosma GC, Custer RP, Bosma MJ. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;301:527–30. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 55.Beamish HJ, Jessberger R, Riballo E, Priestley A, Blunt T, Kysela B, et al. The C-terminal conserved domain of DNA-PKcs, missing in the SCID mouse, is required for kinase activity. Nucleic Acids Res. 2000;28:1506–13. doi: 10.1093/nar/28.7.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Araki R, Fujimori A, Hamatani K, Mita K, Saito T, Mori M, et al. Nonsense mutation at Tyr-4046 in the DNA-dependent protein kinase catalytic subunit of severe combined immune deficiency mice. Proc Natl Acad Sci USA. 1997;94:2438–43. doi: 10.1073/pnas.94.6.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blunt T, Gell D, Fox M, Taccioli GE, Lehmann AR, Jackson SP, et al. Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc Natl Acad Sci USA. 1996;93:10285–90. doi: 10.1073/pnas.93.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pisciotta A, Riccio M, Carnevale G, Lu A, De Biasi S, Gibellini L, et al. Stem cells isolated from human dental pulp and amniotic fluid improve skeletal muscle histopathology in mdx/SCID mice. Stem Cell Res Ther. 2015;6:156. doi: 10.1186/s13287-015-0141-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grabowska I, Mazur MA, Kowalski K, Helinska A, Moraczewski J, Streminska W, et al. Progression of inflammation during immunodeficient mouse skeletal muscle regeneration. J Muscle Res Cell Motil. 2015;36:395–404. doi: 10.1007/s10974-015-9433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farini A, Meregalli M, Belicchi M, Battistelli M, Parolini D, D’Antona G, et al. T and B lymphocyte depletion has a marked effect on the fibrosis of dystrophic skeletal muscles in the scid/mdx mouse. J Pathol. 2007;213:229–38. doi: 10.1002/path.2213. [DOI] [PubMed] [Google Scholar]
- 61.Gayraud-Morel B, Chretien F, Jory A, Sambasivan R, Negroni E, Flamant P, et al. Myf5 haploinsufficiency reveals distinct cell fate potentials for adult skeletal muscle stem cells. J Cell Sci. 2012;125:1738–49. doi: 10.1242/jcs.097006. [DOI] [PubMed] [Google Scholar]
- 62.Gayraud-Morel B, Chretien F, Flamant P, Gomes D, Zammit PS, Tajbakhsh S. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev Biol. 2007;312:13–28. doi: 10.1016/j.ydbio.2007.08.059. [DOI] [PubMed] [Google Scholar]
- 63.Gayraud-Morel B, Pala F, Sakai H, Tajbakhsh S. Isolation of muscle stem cells from mouse skeletal muscle. Methods Mol Biol. 2017;1556:23–39. doi: 10.1007/978-1-4939-6771-1_2. [DOI] [PubMed] [Google Scholar]
- 64.Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–7. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 65.Pinset C, Montarras D, Chenevert J, Minty A, Barton P, Laurent C, et al. Control of myogenesis in the mouse myogenic C2 cell line by medium composition and by insulin: characterization of permissive and inducible C2 myoblasts. Differentiation. 1988;38:28–34. doi: 10.1111/j.1432-0436.1988.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 66.Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci USA. 2003;100:5057–62. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwon M, Firestein BL. DNA transfection: calcium phosphate method. Methods Mol Biol. 2013;1018:107–10. doi: 10.1007/978-1-62703-444-9_10. [DOI] [PubMed] [Google Scholar]
- 68.Dentice M, Ambrosio R, Damiano V, Sibilio A, Luongo C, Guardiola O, et al. Intracellular inactivation of thyroid hormone is a survival mechanism for muscle stem cell proliferation and lineage progression. Cell Metab. 2014;20:1038–48. doi: 10.1016/j.cmet.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baghdadi MB, Castel D, Machado L, Fukada SI, Birk DE, Relaix F, et al. Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche. Nature. 2018;557:714–8. doi: 10.1038/s41586-018-0144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crochemore C, Fernandez-Molina C, Montagne B, Salles A, Ricchetti M. CSB promoter downregulation via histone H3 hypoacetylation is an early determinant of replicative senescence. Nat Commun. 2019;10:5576. doi: 10.1038/s41467-019-13314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Urbani L, Piccoli M, Franzin C, Pozzobon M, De Coppi P. Hypoxia increases mouse satellite cell clone proliferation maintaining both in vitro and in vivo heterogeneity and myogenic potential. PLoS ONE. 2012;7:e49860. doi: 10.1371/journal.pone.0049860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tintignac LA, Sirri V, Leibovitch MP, Lecluse Y, Castedo M, Metivier D, et al. Mutant MyoD lacking Cdc2 phosphorylation sites delays M-phase entry. Mol Cell Biol. 2004;24:1809–21. doi: 10.1128/MCB.24.4.1809-1821.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Londhe P, Davie JK. Sequential association of myogenic regulatory factors and E proteins at muscle-specific genes. Skelet Muscle. 2011;1:14. doi: 10.1186/2044-5040-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed during this study are included in this published article and the supplemental data files.