Abstract
The retrovirus avian sarcoma and leukosis virus (ASLV) enters cells via pH-independent membrane fusion. This reaction is catalyzed by the viral glycoprotein Env, composed of a membrane-distal subunit, SU, and a membrane-anchored subunit, TM. Previous mutational analysis of a variable region, central within the SU subunit, indicates that this region constitutes part of the receptor-binding domain for subgroup A envelope (EnvA) and furthermore that basic residues (R210, R213, R223, R224, and K227) within this region are critical determinants of efficient ASLV infection. Substitutions of these basic residues exert effects on both receptor binding and postbinding events in EnvA-mediated entry. In this study, we performed biochemical analysis of the EnvA protein from three of the receptor-binding domain mutants (R213A/K227A, R213A/R223A/R224A, and R213S) to define the role of this domain in early molecular events in the entry pathway. Protease sensitivity assays demonstrated that receptor binding was sufficient to trigger conformational changes in the SU subunit of mutants R213A/K227A and R213S similar to those in the wild-type EnvA, while R213A/R223A/R224A was constitutively sensitive to protease. In contrast, all three receptor-binding domain mutants disrupted receptor-triggered conversion of EnvA to an active, membrane-binding conformation as assessed by liposome flotation assays. Our results demonstrate that mutations in the receptor-binding site can dissociate receptor-triggered conformational changes in the SU subunit from membrane binding. Furthermore, they suggest that communication between the receptor-binding subunit, SU, and the fusogenic subunit, TM, is crucial for efficient activation of the fusogenic state of EnvA. Analysis of these mutants continues earlier observations that binding to the cellular receptor provides the trigger for efficient activation of this pH-independent viral envelope protein.
Viral envelope glycoproteins play two critical roles in the entry of viruses into cells. They attach the virus to the cell surface through specific interactions with the host cell receptor, and they catalyze fusion of the viral and host cell membranes. The fusogenic capacity of these proteins is tightly regulated. Exposure to the appropriate environmental signals triggers the activation of the glycoproteins, converting them from the native, nonfusogenic state to an active, fusogenic state. Upon activation, the viral glycoproteins undergo structural rearrangements leading to exposure of the hydrophobic fusion peptide (6, 22, 57) and insertion into the host cell membrane (11, 27, 46, 47), beginning the process of membrane mixing. Viral glycoproteins can be categorized based on the nature of the activation signal. Glycoproteins of pH-dependent viruses, such as the hemagluttinin of influenza virus (HA) or the G protein of vesicular stomatitis virus, are activated by the low-pH environment of the endosome encountered following receptor-mediated endocytosis (6, 37, 55). In contrast, the glycoproteins of viruses such as avian sarcoma and leukosis virus (ASLV) and the human immunodeficiency virus (HIV) are fusion active at neutral pH (24, 35, 36, 48). The molecular mechanisms of pH-independent virus entry are poorly understood; however, current models for this class of viruses and recent supporting evidence suggest that activation is mediated by the interactions of the viral glycoprotein and the cellular receptor(s) (11, 26, 27).
The envelope glycoprotein of ASLV, EnvA, and its cellular receptor, Tva, provide an amenable and informative model system for elucidating early molecular events in pH-independent virus entry. EnvA, like numerous viral glycoproteins, is produced as an inactive precursor that is proteolytically processed into two subunits, the surface (SU) subunit and the transmembrane (TM) subunit (31, 39). These subunits are covalently bound and form stable trimers (17). The SU subunit of EnvA is responsible for the initial, high-affinity interaction between ASLV and Tva on the surfaces of host cells (2, 4, 5, 14, 15, 59), while the TM subunit is believed to mediate the fusogenic activity of the protein. Receptor binding induces conformational changes in EnvA and converts it to an activated, membrane-binding state (11, 22, 27). The receptor-triggered activation of EnvA is cooperative and appears to require binding of multiple receptor molecules by the trimeric EnvA protein (11). Once activated, the TM subunit of EnvA is believed to be tethered to two apposing membranes, the viral membrane through the membrane-spanning domain and the target cell membrane, most likely through the hydrophobic fusion peptide. It appears that during or immediately following activation, the SU subunit releases the receptor (11, 27), perhaps allowing for lateral mobility of the membrane-bound TM subunit within the plane of the membrane(s) and subsequent formation of a multimeric fusion pore. Although Tva is necessary for EnvA activation, it remains to be determined how receptor binding triggers this process.
Genetic and mutational analyses have partially defined the receptor-binding domain (RBD) within EnvA. Receptor specificity maps to two small, highly variable domains, hr1 and hr2, within the SU subunit (4, 5, 14, 15). Comparison of the sequences of the five major subgroups of ASLV reveals six conserved basic residues within the hr2 domain of subgroup A viruses. Mutational analysis of these residues indicates that five of the six basic residues (R210, R213, R223, R224, and K227) are critical for efficient EnvA-mediated entry (42). Two phenotypically distinct classes of RBD mutants were identified previously. The first class includes mutants impaired in the ability to mediate entry and to bind receptor, exemplified by the R213A/K227A (M20) and R213A/R223A/R224A (M21) mutants. A second class, represented by R213S (M28), is competent to bind receptor but is defective for infection, suggesting that this class represents a block in the entry pathway after receptor binding (Fig. 1). In order to determine how the receptor-binding site participates in the activation of pH-independent viral glycoproteins, we biochemically characterized the effects of mutations in the RBD on Tva-triggered activation of ASLV EnvA. We identified receptor-triggered conformational changes in EnvA by examining changes in the protease sensitivity of SU upon Tva binding and used a liposome association assay to analyze conversion of TM to a hydrophobic, membrane-binding conformation. Our analysis of these mutants appears to establish a sequence of events during receptor-triggered activation, with receptor-triggered conformational changes in SU preceding changes in TM. Mutations in the RBD physically and temporally uncoupled Tva-induced conformational changes in SU from fusogenic changes in TM, indicating that residues in the receptor-binding site of SU are crucial for transmitting an appropriate activation signal to TM. These results strengthen a model whereby the RBD is intimately involved in controlling the fusogenic activity of pH-independent viral glycoproteins and indicate that cross talk between the envelope subunits (SU and TM) is critical for efficient receptor-triggered activation.
FIG. 1.
Mutations within the RBD of EnvA. The sequence of the hr2 domain in the wild-type protein (wt) is aligned with the sequences of selected mutants, with the relative binding and infectivity of these mutants, as described previously (42), listed to the right. Basic residues conserved in ASLV (A) are in boldface.
MATERIALS AND METHODS
Protein production.
The plasmids pCB6 envA PI and pCB6 hr2 mutant envA have been described previously (23, 42). A fragment encoding a nine-amino-acid epitope tag from the myc gene was inserted at the amino terminus of envA PI and designated myc-EnvA PI (18). To generate mutant constructs encoding a glycosyl phosphatidylinositol (PI)-linked form of EnvA (EnvA PI), XbaI/EcoRI fragments encoding the mutant hr2 domains were subcloned into pCB6myc-envA PI vectors. These constructs were designated pCB6 m20 PI, pCB6 m21 PI, etc., denoting their PI linkage. Stable NIH 3T3 cell lines were established by CaPO4− transfection (58) and selection with G418 (300 μg/ml; Gibco-BRL). NIH 3T3 cells stably expressing wild-type EnvA PI were generously supplied by J. White of the University of Virginia. Soluble EnvA PI protein was produced as described previously, with modifications (11). Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% bovine calf serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and 300 μg of G418/ml. Subconfluent T150 flasks were induced with sodium butyrate (25 mM) for 16 h. The cells were incubated with serum-free Dulbecco’s modified Eagle’s medium for 1 h prior to harvest and then were washed twice with 2× phosphate-buffered saline (PBS) and one time with Ca2+- and Mg2+-free PBS. PI-linked protein was released in Ca2+- and Mg2+-free PBS plus protease inhibitors (aprotinin, leupeptin, pepstatin, and phenylmethyl sulfonyl fluoride [Sigma] with 50 mU of PI-phospholipase C (PLC) (Boehringer Mannheim Biochemica) at 37°C for 60 min. The supernatants were clarified by centrifugation, and samples were concentrated approximately 5-fold to 15-fold with a 100K Macrosep centrifugal concentrator (Filtron). The samples were stored at 4°C. The amounts of soluble envelope proteins were standardized within twofold of wild-type EnvA PI by quantitative Western blot analysis with rabbit anti-SU serum and 125I-protein A and by phosphorimaging. Soluble Tva (sTva) was produced in insect cells (Spodoptera frugiperda) and purified from cellular supernatants with a nickel affinity chromatography column (2). The proteins were stored at 4°C and diluted to the appropriate concentrations in Ca2+- and Mg2+-free PBS.
Analysis of oligomeric state.
PI-PLC-released protein was layered on a 10 to 30% sucrose gradient in PBS containing Triton X-114 (0.1%). Samples were centrifuged in an SW41 rotor (Beckman) at 41,000 rpm for 17 h at 4°C. Gradients were fractionated in 500-μl samples, and proteins were precipitated with trichloroacetic acid as described previously (11) followed by analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
Protease sensitivity assay.
Mutant and wild-type EnvA PI proteins were incubated with or without excess sTva (500 ng) in a total reaction volume of 50 μl of Ca2+- and Mg2+-free PBS at 4°C for 15 min, shifted to 37°C for 15 min, and then returned to 4°C. Samples were incubated on ice for 30 min in the presence of thermolysin (75 ng/μl; Boehringer Mannheim Biochemica) and CaCl2 (2 mM). The reactions were immediately terminated by the addition of Laemmli sample buffer, heated to 95°C for 10 min, and then analyzed by SDS-PAGE and Western blotting.
Lipids.
Phosphatidylcholine from eggs and cholesterol were purchased from Sigma Biochemicals. Lipids were stored under argon at −80°C as 100-mg/ml stock solutions in chloroform.
Preparation of liposomes.
Liposomes were produced by a modification of the protocol described previously (32). Briefly, phosphatidylcholine (13 μmol) and cholesterol (6.5 μmol) in chloroform were mixed at a 2:1 molar ratio and dried under argon in a round-bottom glass flask. Glass beads and absolute ethyl alcohol (preheated to 52°C) were added with vortexing. The lipids were dried to a thin film by heating them to 52°C under a vacuum. Liposomes were generated by the addition of 0.5 ml of Ca2+- and Mg2+-free PBS (preheated to 52°C) with vigorous mixing. The liposomes were sonicated for 60 s in a water bath sonicator (Heat Systems).
Liposome-binding assays.
The liposome-binding assay was a modification of the protocol described previously (11). EnvA PI was incubated with the indicated amount of sTva in a final volume of 40 μl of Ca2+- and Mg2+-free PBS on ice for 15 min. Liposomes (40 μl; preequilibrated to the indicated temperature) were added to the samples and incubated for an additional 15 min (unless otherwise indicated). Samples were placed on ice, and 73% (wt/vol) sucrose in PBS was added to the protein-liposome mixture to bring it to 50% sucrose. The samples were overlaid with 150 μl of 40% sucrose and 300 μl of 25% sucrose in a 700-μl Ultra-clear (Beckman) centrifuge tube. After centrifugation at 269,000 × g for 3 h at 4°C in a SW50 rotor, seven 100-μl fractions were drawn from the air-fluid interface. Proteins were precipitated from the fractions with the addition of an equal volume of Triton lysis buffer (150 mM NaCl, 1% Triton X-100, 50 mM Tris [pH 8.0], 5 mM EDTA), casein (50 ng/μl), and trichloroacetic acid to 10% (wt/vol). The samples were incubated on ice for 30 min, pelleted, and washed twice with ice-cold acetone. The dried pellets were resuspended in Triton lysis buffer and then subjected to SDS-PAGE and Western blot analysis.
Detection of proteins.
Western blots were probed with polyclonal rabbit antisera against SU generously provided by T. Matthews of Duke University. Rabbit antibodies were detected by incubation with goat anti-rabbit secondary antibodies conjugated to horseradish peroxidase from Boehringer Mannheim Biochemica. Horseradish peroxidase was identified by enhanced chemiluminescence (ECL) as instructed by the manufacturer (Pierce) and the blots were exposed to X-ray film. Alternatively, primary antibody was detected with 125I-protein A (0.1 μCi/μl; Dupont) and quantitated with a PhosphorImager and ImageQuant software (Molecular Dynamics).
RESULTS
Generation of aqueous, trimeric RBD mutant protein.
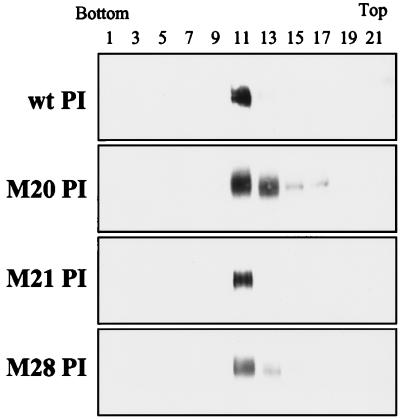
To evaluate the effects of RBD mutations on postbinding events in Tva-triggered activation, including membrane association, water-soluble forms of the RBD mutants were generated by replacing the proteinaceous membrane anchor of TM with the glycosyl PI signal of decay-accelerating factor. Mutants from two phenotypic classes of RBD mutations were chosen for this biochemical analysis. The first class of mutants exhibits defects in both receptor binding and viral entry (M20 and M21), whereas the second class is defective for entry but retains wild-type receptor-binding activity (M28) (Fig. 1) (42). These mutations were cloned into an EnvA PI-expressing plasmid and were given designations with the suffix PI. NIH 3T3 lines stably expressing these RBD envelope proteins were generated, and the PI-linked proteins were released from the cell surface with PI-PLC. Clarified and concentrated PI-PLC-released supernatants were evaluated by SDS-PAGE and Western blotting with polyclonal antiserum against the SU subunit of EnvA. The three EnvA RBD mutants had mobilities on reducing SDS-PAGE similar to that of wild-type EnvA PI (Fig. 2, lanes 1, 4, 7, and 10). The mobilities of the three RBD mutants compared to that of the uncleaved form of EnvA PI suggested that the proteins released from the cell surface were fully cleaved and processed (Fig. 2, lane 13). This is consistent with previous observations that these RBD mutations do not appear to alter the processing or cleavage of EnvA (42). The oligomeric status of the RBD mutant EnvA PI glycoproteins was also evaluated by sucrose density gradient analysis (Fig. 3). This analysis demonstrated that, similar to wild-type EnvA PI, the majority of the mutant aqueous protein migrated to a position in the gradient consistent with an oligomeric structure (Fig. 3, fractions 11 and 13) as determined previously for the trimeric ASLV (A) envelope protein (17, 23). A minimal amount of protein was detected in fractions 15 and 17 for M20 PI. This spread of M20 PI within the gradient may reflect dissociation of the protein during ultracentrifugation. We have occasionally observed similar patterns of migration of wild-type EnvA PI (data not shown). From this, it appears that mutations in the RBD have no appreciable effect on the processing, cleavage, or stability of the EnvA PI trimer.
FIG. 2.
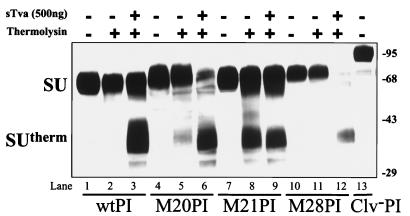
Protease sensitivity of SU in the absence and presence of sTva. Soluble EnvA PI was incubated with (+) or without (−) excess sTva and subjected to thermolysin digestion before analysis by SDS-PAGE. The western blot was probed with antiserum against SU, which was detected by ECL. The SU subunit and proteolytic product, SUtherm, migrated as indicated on the left, and molecular weight markers are on the right. The uncleaved form of EnvA PI (Clv− PI) was included as a size reference. wt, wild-type.
FIG. 3.
Sucrose density centrifugation analysis of PI-PLC-released envelope proteins. Wild-type (wt) and RBD mutant proteins were subjected to ultracentrifugation on a 10 to 30% sucrose gradient as described in Materials and Methods. Odd-numbered fractions were precipitated and analyzed by SDS–12.5% PAGE, followed by Western blotting with antiserum against the SU subunit and detection by ECL.
Effects of RBD mutations on receptor-induced structural rearrangements in EnvA.
In the native, nonfusogenic conformation, EnvA is relatively insensitive to digestion with the protease thermolysin. Receptor binding induces conformational changes in EnvA, exposing previously inaccessible protease sites and rendering the SU subunit thermolysin sensitive (22). This enhanced thermolysin sensitivity results in the formation of a proteolyzed fragment of SU, called SUtherm, that can be detected by Western blot analysis with polyclonal antiserum against SU. To assess whether receptor binding was capable of inducing conformational changes in the RBD mutants, protease sensitivity assays were performed. Soluble EnvA PI was incubated with or without excess soluble receptor (sTva) at 37°C for 15 min and then treated with thermolysin prior to analysis by SDS-PAGE and Western blotting. Similar to wild-type EnvA PI, two other RBD mutants, M20 PI and M28 PI, exhibited enhanced sensitivity to thermolysin in the presence of sTva (Fig. 2). The resulting proteolytic products had mobilities comparable to that of wild-type SUtherm, indicating that the structural alterations induced in M20 PI and M28 PI were similar to those observed in the SU subunit of wild-type EnvA following receptor binding. These results suggested that these two mutants bound receptor sufficiently and were competent to undergo receptor-induced conformational rearrangements in SU. We occasionally observed minimal proteolytic cleavage of M20 PI in the absence of sTva (Fig. 2, lane 5). However, this mutant demonstrated a clear increase in sensitivity to thermolysin following the addition of receptor (Fig. 2, lanes 5 and 6), consistent with sTva-triggered conformational changes. Thus, under conditions where receptor was not limiting, mutant M20 was competent to undergo structural rearrangements, suggesting that despite quantitative differences in its ability to bind Tva, it can respond appropriately to receptor binding. This is in contrast to the behavior of M21 PI, which was sensitive to thermolysin independent of sTva and did not demonstrate an appreciable increase in sensitivity following the addition of receptor (Fig. 2, lanes 8 and 9). This inherent increased sensitivity to protease and unresponsiveness to sTva suggested that M21 PI was in an unstable or open conformation or that it existed in a “preactivated” form that no longer required receptor binding.
Effects of RBD mutations on conversion of EnvA to a fusogenic state.
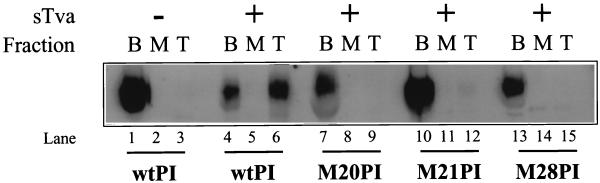
In addition to the structural alterations in the SU subunit detected by protease digestion, Tva binding also induces conformational changes in the TM subunit of EnvA that expose the previously buried hydrophobic fusion peptide domain (22) and convert the protein to an activated, membrane-binding state (11, 27). Receptor-triggered activation is detected in vitro by a liposome flotation assay. In the native state, EnvA PI is water soluble and does not bind a target liposomal membrane. However, when activated by receptor binding, EnvA forms a stable protein-membrane complex, presumably by insertion of the hydrophobic fusion peptide of TM into the liposome membrane. The buoyancy of the liposomes causes this complex to float to the top of a sucrose step gradient during ultracentrifugation, while the inactive protein remains at the bottom of the gradient. Thus, receptor-triggered activation of EnvA is detectable as a shift in position in the step gradient from the bottom to the top, liposome-containing fraction. In order to determine whether Tva binding could convert the RBD mutants to an active, membrane-binding conformation, liposome flotation assays were performed with excess sTva. As described previously, wild-type EnvA PI was activated by receptor binding and colocalized with liposomes in the presence of receptor (11, 27). In contrast, none of the three RBD mutants colocalized with liposomes under comparable conditions (Fig. 4, lanes 9, 12, and 15). Experiments utilizing a Myc epitope-tagged form of wild-type EnvA PI showed that, similar to the untagged EnvA PI, this protein was able to efficiently associate with liposomes, indicating that the amino-terminal tag present on the RBD mutants was not responsible for their failure to convert to a liposome-binding state (data not shown). Therefore, the apparent inability of the RBD mutants to bind liposomes suggests a defect in their conversion to a membrane-binding state by sTva.
FIG. 4.
Conversion of RBD mutants to an active conformation. Liposome flotation assays were performed on wild-type (wt) and RBD mutant EnvA PI proteins in the presence (+) or absence (−) of excess sTva. EnvA PI, with and without sTva, and liposomes were incubated at 37°C for 15 min before ultracentrifugation. The fractions were collected, acid precipitated, and analyzed by Western blotting with antiserum against SU detected by ECL, as described in Materials and Methods. B, M, and T denote the bottom, middle, and top fractions, respectively. Greater that 90% of the liposomes recovered are within the top fraction (data not shown).
Membrane-bound EnvA PI is protease sensitive.
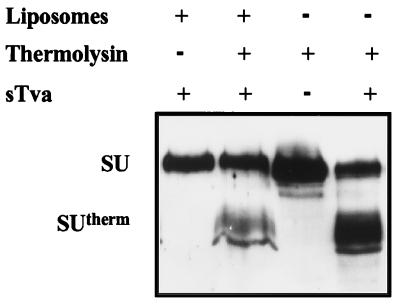
To determine the relationship between induction of a the protease-sensitive structure in SU and the active, membrane-binding conformation of EnvA PI and to further clarify the discrepant behavior of M20 PI and M28 PI in the two assays described above (i.e., their ability to undergo Tva-induced conformational changes and their failure to bind membranes), thermolysin sensitivity assays were performed on the wild-type EnvA PI isolated from the liposomes at the tops of the gradients. As with the aqueous protein, the SU subunit of membrane-bound EnvA PI was sensitive to thermolysin and the proteolytic product had a mobility on SDS-PAGE comparable to that of SUtherm (Fig. 5). The fact that EnvA PI that is associated with target membranes is thermolysin sensitive strongly suggests that the forms of envelope biochemically identified in the two assays above do not represent distinct populations of EnvA PI. Furthermore, this result indicates that the protease sensitivity and liposome-binding assays reflect coincident or sequential receptor-induced changes in the structure of EnvA PI. Thus, the ability of the M20 PI and M28 PI RBD mutants to undergo conformational changes in SU while failing to bind membranes is most consistent with the TM subunit failing to acquire a fusogenic, membrane-binding conformation.
FIG. 5.
Membrane-bound wild-type EnvA PI is sensitive to protease. A liposome flotation assay was performed as described in Materials and Methods. The top 100-μl fractions were subjected to thermolysin digestion. Soluble EnvA PI proteins with (+) and without (−) sTva were treated with thermolysin in parallel. The products were analyzed by reducing SDS-PAGE, and Western blots were probed with antiserum against SU detected by ECL. The SU subunit and the proteolytic product, SUtherm, migrated as indicated on the left. The distortion of the SUtherm band in the lanes containing liposomes is the result of lipids remaining in the sample.
Thermal profile of membrane association.
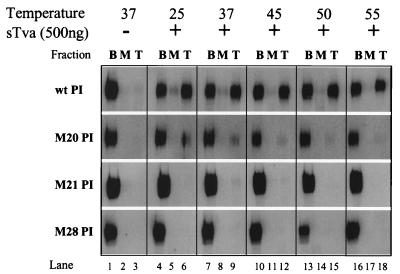
Fusion of ASLV with cells and the conversion of wild-type EnvA PI to the active, liposome-binding form are temperature dependent and are inefficient at temperatures below 20°C (11, 24, 27). This suggests that, in addition to receptor binding, a thermodynamic barrier must be overcome during the transition of EnvA to a membrane-binding state. Therefore, we postulated that the block to activation observed with the RBD mutants may represent an alteration in this thermodynamic barrier. To address this question, we performed liposome flotation assays at temperatures ranging from 25 to 55°C (Fig. 6). In the presence of sTva, wild-type EnvA PI was converted to the membrane-binding conformation throughout the temperature range evaluated with no appreciable increase at temperatures above 37°C. In contrast, mutants M21 PI and M28 PI were not detectable in the top of the gradient and thus failed to associate with liposomal membranes at any of the temperatures tested. The inability to overcome the block to activation with increasing energy suggested that these RBD mutations are unlikely to act by stabilizing an intermediate structure in the entry pathway. Minimal amounts of M20 PI (less than 10%) were detected in the liposome-containing top fraction when the Western blots were exposed for a prolonged time (Fig. 6, lanes 6, 9, and 12), indicating that M20 PI is inefficiently converted to the active conformation. No increase in conversion of M20 PI to the liposome-associated form was seen at elevated temperatures, suggesting that, similar to the M21 PI and M28 PI mutants, increasing energy was unable to alleviate the block and that the inefficient activation was likely not caused by an alteration in the thermodynamic barrier.
FIG. 6.
Thermodynamic profiles of sTva-triggered activation. EnvA PI was prebound to sTva (500 ng) (+) by incubation at 4°C. Liposomes preequilibrated to the indicated temperatures were added, and the samples were shifted to these temperatures for an additional 15 min. Flotation and SDS-PAGE–Western blotting determined the extent of binding of EnvA PI to the liposomes. The blots of gradient fractions were probed with antiserum against SU and I125-protein A detected by phosphorimaging. B, bottom fraction; M, middle fraction; T, top fraction; wt, wild-type; −, no sTva.
Kinetics of membrane association.
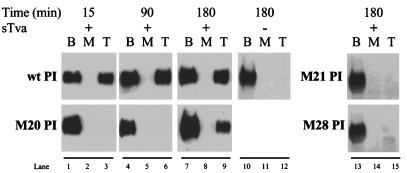
We hypothesized that the observed block to membrane insertion seen with M20 PI, M21 PI, and M28 PI might represent changes in the rate of sTva-induced activation of the glycoproteins. Previous analysis indicated that the temperature-dependent, receptor-triggered activation of wild-type EnvA PI occurs rapidly when receptor is in excess and plateaus within 5 min (11, 27). In order to determine whether these RBD mutants were kinetically delayed compared to wild-type EnvA PI, we performed liposome flotation assays with increasing incubation periods at 37°C (Fig. 7). M20 PI was found in the liposome-containing top fraction of the gradient at the 3-h time point (Fig. 7, lane 9). At the early time points of 15 and 90 min, membrane association of M20 PI was only detectable when the Western blots were overexposed (data not shown). Receptor-triggered activation of M20 PI, therefore, was relatively inefficient and demonstrated delayed kinetics compared to the wild type, which had reached a plateau within 15 min (Fig. 7, lane 3). In contrast, M21 PI and M28 PI were not found in the top fraction after a 3-h incubation with liposomes and excess receptor, indicating that they failed to be activated by sTva within this period (Fig. 7, lane 15). The failure of M21 PI to associate with the membrane, despite extended incubations and increased energy, is less consistent with the protein existing in a “presprung,” active conformation. Rather, these results favor the hypothesis that this triple mutation (R213A/R223A/R224A) destabilizes the SU protein, resulting in constitutive sensitivity to thermolysin, but does not lead to TM activation. The inability to detect membrane association of M28 PI despite prolonged incubation is consistent with a model in which there is a physical block during the conversion of the protein to an active state. An alternative and compatible interpretation would be that this mutation severely retarded the rate of conversion of the TM subunit to the membrane-binding conformation beyond the 180 min tested. This analysis suggests that the signal for activation was not transmitted appropriately from the SU subunit of M28 PI to the TM subunit.
FIG. 7.
Kinetics of sTva-triggered activation. EnvA PI was incubated in the absence (−) or presence (+) of sTva (500 ng) and liposomes at 37°C for the length of time indicated, shifted to 4°C, and immediately processed on sucrose step gradients as described in Materials and Methods. Following flotation, the extent of binding of EnvA PI to the liposomes was determined by SDS-PAGE, and Western blots were probed with antiserum against SU detected by ECL. B, bottom fraction; M, middle fraction; T, top fraction; wt, wild-type; −, no sTva.
DISCUSSION
Entry into a host cell is a critical initiating event in the infectious cycle of a virus. Our understanding of this event is crucial both for future hopes of developing therapeutics to block the entry of viral pathogens and for the ability to exploit viruses as vectors for targeted gene delivery. It is clear that enveloped viruses have evolved to use a variety of host molecules to mediate their attachment to the cell and use different physical clues to activate the fusogenic capacities of their glycoproteins. Despite these differences, there is ever-growing evidence that many viral glycoproteins from highly divergent viruses share common architectural and structural motifs (8, 19, 21, 52–54, 56), suggesting that these viruses may exploit common mechanisms to mediate membrane fusion. EnvA, the envelope glycoprotein of ASLV, a prototypical retrovirus, contains many of these motifs (21, 56). Thus, our increased appreciation of the dynamic interactions and structural changes that occur during EnvA-mediated entry may have implications for our understanding of entry by other retroviruses, such as HIV.
Here, we examined the effects of mutations within the RBD on early events in ASLV entry to delineate the role of the RBD in receptor-triggered activation of EnvA. Mutations within the RBD do not appear to disrupt the global structure or oligomeric status of EnvA. Previous analysis of these three RBD mutants had demonstrated that they are processed and incorporated into viral particles similarly to wild-type EnvA (42), indicating that there are no gross disruptions in the overall structure of the glycoprotein. Density gradient centrifugation demonstrates that all three mutants sediment at a position comparable to that of wild-type EnvA PI, indicating that these mutations did not alter the formation or stability of the glycoprotein trimer. Therefore, the biochemical and functional properties of these RBD mutants do not appear to be due to altered stability of the glycoprotein oligomer, and as might be expected, the RBD does not appear to play a role in EnvA oligomerization.
The abilities of the RBD mutants to respond to receptor binding appropriately, inducing structural rearrangements in the SU and TM subunits, were variable and did not correlate with the abilities of the proteins to bind receptor. Both M20 PI and M28 PI demonstrated enhanced sensitivity to protease in the presence of high levels of sTva (Table 1), consistent with receptor-induced structural rearrangements within the SU subunits of these envelope proteins. There were no detectable differences in the rate or thermal profile of Tva-induced protease sensitivity for M20 PI or M28 PI compared to those of wild-type EnvA PI (data not shown.) These two RBD mutants, therefore, were competent to undergo the initial structural changes in SU induced by Tva binding.
TABLE 1.
Phenotypes of RBD mutants
| Name | Mutation | Relative binding (%)a | Receptor-triggered protease sensitivityb | Receptor-triggered membrane associationb | Relative infectivity (%)a |
|---|---|---|---|---|---|
| wtc | 100 | + | + | 100 | |
| M20 | R213A/K227A | 14 | + | Delayede | 7 |
| M21 | R213A/R223A/R234A | 11 | −d | − | 5 |
| M28 | R213S | 91 | + | − | 5 |
From reference 42.
+, present; −, absent.
wt, wild type.
Sensitivity to thermolysin was independent of sTva.
Membrane association was delayed.
In contrast, biochemical analysis of M21 PI established that this mutant was sensitive to proteolytic digestion independent of Tva binding, suggesting an alteration in the native structure of SU. This phenotype initially suggested the possibility that the protein no longer required Tva in order to undergo structural rearrangements and existed in a preactivated state. However, analysis by liposome flotation assays demonstrated that M21 PI failed to bind the target membrane despite prolonged incubation with or without excess receptor (data not shown). The inability of M21 PI to associate with liposomes favors the hypothesis that the basic residues at positions 223 and 224 (Table 1) played a role in stabilizing the structure of SU or of EnvA as a whole. This is consistent with previous mutagenesis studies, which indicated that mutations at only positions 223 and 224 of SU had negligible effects on receptor binding but appeared to accentuate the effects of mutations at residues 213 and 227 (42). These results suggested that residues 223 and 224 participate in receptor binding by maintaining the tertiary structure of the RBD and/or by directly stabilizing the interactions with receptor. The constitutive sensitivity of M21 PI to thermolysin, coupled with the inability of this mutant to form a complex with target membranes, is consistent with these mutations destabilizing the structure of SU. Analysis of the protease-sensitive phenotype of additional mutants containing alterations at residues 223 and 224 is required to confirm this hypothesis.
The pH-dependent glycoprotein of influenza, HA, can be converted to a fusogenic structure at neutral pH through destabilization of the protein with heat or chemicals, supporting a “metastable model” for the native conformation of the fusion protein (7, 43). The recent work of Carr et al. suggests that low pH is not a specific requirement for fusion activity and that activation of HA involves general destabilization of the native, metastable conformation and conversion to a more thermodynamically stable, active state (7). Receptor-induced changes in EnvA are biochemically distinct from the changes induced by general destabilization. M21 PI appears to represent a destabilized form of the SU subunit of EnvA PI, yet under all conditions analyzed this protein was unable to convert to an active, membrane-binding state. Unlike the effects of mutant M21 PI on protease sensitivity, heat destabilization of wild-type EnvA PI does not result in the formation of SUtherm in protease sensitivity assays. Rather, a smaller proteolytic product of approximately 19 kDa is formed when EnvA PI is exposed to temperatures greater than 50°C (data not shown). In the absence of sTva, this heat-destabilized EnvA PI protein does not bind liposomal membranes to an appreciable degree (unpublished data), further suggesting that it is not in an active conformation. While the native state of EnvA may also represent a metastable conformation, destabilization through mutagenesis or with heat appears insufficient for conversion to an active conformation. Coupled with the findings for M21 PI, these results suggest that specific interactions with the viral receptor are required for activation of EnvA, and this may represent another distinction between the activation of pH-independent and pH-dependent glycoproteins.
While mutants M20 PI and M28 PI exhibited wild-type responses to sTva binding in protease sensitivity assays, the mutants demonstrated minimal or no detectable conversion to a membrane-binding conformation in liposome flotation assays. Membrane association is mediated through the TM subunit (27); therefore, these results suggest that the mutants M20 PI and M28 PI are blocked at a step or steps prior to exposure of hydrophobic regions within TM. The observed block to activation was not alleviated by increasing the temperature of the reaction, suggesting it was not a result of alterations in the thermodynamic activation profile of EnvA and could not be overcome by destabilizing the protein. The inefficient and slow rate of membrane binding seen with M20 PI indicates that this mutant dissociates Tva-triggered conformational changes in SU from changes in TM. The decreased sTva-binding capacity of M20 PI may contribute in part to the observed delay in membrane association. However, the significant increase in the sensitivity of SU to protease suggests that sTva binding is sufficiently avid to induce a relatively rapid conformational change in M20 PI. We cannot formally exclude the possibility that suboptimal binding is sufficient for the initial conformational changes in SU while wild-type receptor-binding affinity is necessary for acquisition of a fusogenic state. However, it is clear from studies of retroviral envelope proteins that high-affinity binding is not a prerequisite for viral entry. There are numerous examples of mutations within ASLV, murine leukemia virus, and HIV envelope proteins that diminish their capacities to bind their respective host cell receptors, yet these mutations have no demonstrable effects on envelope-mediated membrane fusion or infection (33, 42, 50). Therefore, we favor a model in which the RBD of ASLV envelope is intimately involved in transducing the activation signal from the receptor-binding site to the TM subunit.
The requirement for an intact RBD during the activation of EnvA is more dramatically illustrated by biochemical analysis of mutant M28 PI. Despite the ability of M28 PI to efficiently bind to sTva and undergo conformational rearrangements, this RDB mutant failed to bind to target membranes under any circumstances tested. In vitro, M28 PI demonstrates a complete uncoupling of receptor-triggered changes in the SU and TM subunits, again supporting the role of the RBD in transduction of the activation signal to the fusogenic TM subunit. The biochemical properties of M28 PI suggest that a basic residue at position 213 of SU is critical for transmission of the activation signal to TM. Thus, these two RBD mutants, M20 and M28, clearly demonstrate dissociation of the Tva-induced changes in the receptor-binding subunit from changes in the fusogenic subunit and confirm an essential role of the RBD for full activation of this pH-independent virus.
Acquisition of protease sensitivity and membrane association represent changes on the same pathway (Fig. 5); therefore, identification of mutants that uncouple these events favors a sequential model in which receptor binding induces changes in the structure of SU prior to membrane binding via TM (Fig. 8). The sequential nature of the activation of EnvA is analogous, in part, to the early events believed to occur when the glycoprotein of HIV type 1 (HIV-1) interacts with the host cell. For HIV-1, the SU subunit (gp120) initially binds the primary receptor, CD4 (1, 34), leading to structural changes that enable the glycoprotein to utilize a coreceptor molecule, CXCR4 or CCR5 (9, 12, 13, 16, 20). Similar to sTva-triggered changes in EnvA, CD4 binding exposes cryptic protease sites in gp120 (44). However, CD4-induced changes, which also include structural rearrangements within SU and frequently shedding of this subunit (3, 25, 38, 45), are insufficient for virus entry (34). Secondary conformational changes are required for the glycoprotein to expose hydrophobic residues and acquire a fusogenic potential, and these changes occur following coreceptor binding (29). In contrast to HIV-1 Env, EnvA requires a single cellular receptor, Tva, to achieve an active conformation. Here, we have demonstrated that mutations within the RBD are able to dissociate the first Tva-induced changes in SU from the secondary changes needed to insert into the target cell membrane (Fig. 8). The dissociation of early steps in the activation pathway supports a role for the RBD in coupling the activation signal induced by receptor binding from the SU to the TM subunit. This suggests that communication between the two subunits may be critical for efficient receptor-triggered activation of pH-independent viral entry.
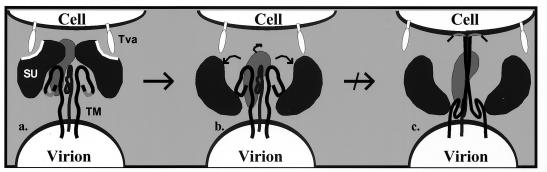
FIG. 8.
Model of early events in EnvA-mediated entry. Through the RBD in SU, trimeric EnvA binds multiple Tva molecules on the surfaces of avian cells (a). This binding triggers structural rearrangements in the SU subunit, which likely include lateral dissociation of the globular head groups (curved arrows) (b). This precedes transformation of EnvA to an active state, which occurs following exposure of the fusion peptide domain and insertion into the host cell membrane (c). Mutations in the RBD disrupt the conversion of EnvA to an active state (slashed arrow) by uncoupling receptor-triggered changes in SU and TM, resulting in a block in the entry pathway after the formation of an early intermediate (b).
The functional defect defined by the RBD mutant M28 suggests that there is cross talk between the SU and TM subunits during receptor-triggered activation of EnvA, with the RBD in SU playing a central role in coupling the activation signal. It appears that mutations in the viral envelope proteins which produce phenotypes similar to that of M28 (i.e., thus mainten wild-type receptor binding yet are defective for entry) are very rare. Obvious exceptions are the HIV-1 gp120 mutants, which maintain wild-type CD4 binding but have a restricted cellular tropism, likely representing changes in the interaction of gp120 with the coreceptor molecules (CXCR4 and CCR5) (10). Nonetheless, there is indirect evidence of communication between the RBD within gp120 and the TM subunit of HIV-1, gp41. Resistance to neutralization by antibodies to the CD4-binding site maps to a single substitution in gp41 (582 A/T) (30, 40, 41, 50). This substitution dramatically reduces the ability of antibodies to bind the CD4-binding site within gp120 compared to their ability to bind that of the parental virus, HXB2 (49). The neutralization-resistant mutant, HXB2thr582, has a greater propensity to form syncytia in tissue culture than HXB2, suggesting that the mutant envelope protein may be more fusogenic (49). This indicates that the TM subunit of HIV can modulate the structure of the receptor-binding site; moreover, it suggests that interactions between the receptor-binding site in gp120 and the TM subunit are functionally important for initiating HIV entry into cells. Analysis of M28 PI demonstrates the first biochemical characterization of a receptor-binding site mutant which uncouples these early events in viral entry and further supports the importance of functional interactions between the receptor-binding site in SU and the TM subunit during pH-independent retrovirus entry.
Membrane fusion is an important and ubiquitous cellular process that plays a role in diverse biological events ranging from fertilization to neurotransmission. Protein-protein interactions appear to play a critical role in the regulation of vesicle fusion (28, 51). Recent work by Weber et al. suggests that interactions between two proteins, vesicle SNARE and target SNARE, are sufficient to mediate membrane fusion in vitro and, thus, that these SNARE proteins represent a minimal machinery of intracellular vesicle fusion (51). The regulation of protein fusogenicity by specific protein-protein interactions may therefore represent a common means of modulating membrane fusion. The interactions between EnvA and Tva represent a simple viral model of a protein-regulated fusion machine. Thus, a detailed understanding of this system may further our understanding of virus entry, as well as cellular membrane fusion in general.
ACKNOWLEDGMENTS
We thank Robert Doms at the University of Pennsylvania and members of the Bates laboratory for helpful discussions.
This work was supported by grants to P.B. from the National Institutes of Health (CA63531 and CA76256) and the American Heart Association (95015200). R.D. was supported by T32 GM07229 from the NIH.
REFERENCES
- 1.Ashorn P A, Berger E A, Moss B. Human immunodeficiency virus envelope glycoprotein/CD4-mediated fusion of nonprimate cells with human cells. J Virol. 1990;64:2149–2156. doi: 10.1128/jvi.64.5.2149-2156.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balliet J W, Berson J, D’Cruz C M, Huang J, Crane J, Gilbert J, Bates P. Production and characterization of a soluble, active form of Tva, the subgroup A avian sarcoma and leukosis virus receptor. J Virol. 1999;73:3054–3061. doi: 10.1128/jvi.73.4.3054-3061.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger E A, Lifson J D, Eiden L E. Stimulation of glycoprotein gp120 dissociation from the envelope glycoprotein complex of human immunodeficiency virus type 1 by soluble CD4 and CD4 peptide derivatives: implications for the role of the complementarity-determining region 3-like region in membrane fusion. Proc Natl Acad Sci USA. 1991;88:8082–8086. doi: 10.1073/pnas.88.18.8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bova C A, Manfredi J P, Swanstrom R. env genes of avian retroviruses: nucleotide sequence and molecular recombinants define host range determinants. Virology. 1986;152:343–354. doi: 10.1016/0042-6822(86)90137-6. [DOI] [PubMed] [Google Scholar]
- 5.Bova C A, Olsen J C, Swanstrom R. The avian retrovirus env gene family: molecular analysis of host range and antigenic variants. J Virol. 1988;62:75–83. doi: 10.1128/jvi.62.1.75-83.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 7.Carr C M, Chaudhry C, Kim P S. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc Natl Acad Sci USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 9.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 10.Cordonnier A, Montagnier L, Emerman M. Single amino-acid changes in HIV envelope affect viral tropism and receptor binding. Nature. 1989;340:571–574. doi: 10.1038/340571a0. [DOI] [PubMed] [Google Scholar]
- 11.Damico R L, Crane J, Bates P. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc Natl Acad Sci USA. 1998;95:2580–2585. doi: 10.1073/pnas.95.5.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 13.Doranz B J, Berson J F, Rucker J, Doms R W. Chemokine receptors as fusion cofactors for human immunodeficiency virus type 1 (HIV-1) Immunol Res. 1997;16:15–28. doi: 10.1007/BF02786321. [DOI] [PubMed] [Google Scholar]
- 14.Dorner A J, Coffin J M. Determinants for receptor interaction and cell killing on the avian retrovirus glycoprotein gp85. Cell. 1986;45:365–374. doi: 10.1016/0092-8674(86)90322-3. [DOI] [PubMed] [Google Scholar]
- 15.Dorner A J, Stoye J P, Coffin J M. Molecular basis of host range variation in avian retroviruses. J Virol. 1985;53:32–39. doi: 10.1128/jvi.53.1.32-39.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 17.Einfeld D, Hunter E. Oligomeric structure of a prototype retrovirus glycoprotein. Proc Natl Acad Sci USA. 1988;85:8688–8692. doi: 10.1073/pnas.85.22.8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fass D, Kim P S. Dissection of a retrovirus envelope protein reveals structural similarity to influenza hemagglutinin. Curr Biol. 1995;5:1377–1383. doi: 10.1016/s0960-9822(95)00275-2. [DOI] [PubMed] [Google Scholar]
- 20.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 21.Gallaher W R, Ball J M, Garry R F, Griffin M C, Montelaro R C. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Hum Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert J M, Hernandez L D, Balliet J W, Bates P, White J M. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J Virol. 1995;69:7410–7415. doi: 10.1128/jvi.69.12.7410-7415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert J M, Hernandez L D, Chernov-Rogan T, White J M. Generation of a water-soluble oligomeric ectodomain of the Rous sarcoma virus envelope glycoprotein. J Virol. 1993;67:6889–6892. doi: 10.1128/jvi.67.11.6889-6892.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert J M, Mason D, White J M. Fusion of Rous sarcoma virus with host cells does not require exposure to low pH. J Virol. 1990;64:5106–5113. doi: 10.1128/jvi.64.10.5106-5113.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hart T K, Kirsh R, Ellens H, Sweet R W, Lambert D M, Petteway S R, Jr, Leary J, Bugelski P J. Binding of soluble CD4 proteins to human immunodeficiency virus type 1 and infected cells induces release of envelope glycoprotein gp120. Proc Natl Acad Sci USA. 1991;88:2189–2193. doi: 10.1073/pnas.88.6.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 27.Hernandez L D, Peters R J, Delos S E, Young J A, Agard D A, White J M. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J Cell Biol. 1997;139:1455–1464. doi: 10.1083/jcb.139.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jahn R, Hanson P I. Membrane fusion. SNAREs line up in new environment. Nature. 1998;393:14–15. doi: 10.1038/29871. [DOI] [PubMed] [Google Scholar]
- 29.Jones P L, Korte T, Blumenthal R. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J Biol Chem. 1998;273:404–409. doi: 10.1074/jbc.273.1.404. [DOI] [PubMed] [Google Scholar]
- 30.Klasse P J, McKeating J A, Schutten M, Reitz M S, Jr, Robert-Guroff M. An immune-selected point mutation in the transmembrane protein of human immunodeficiency virus type 1 (HXB2-Env:Ala 582(→Thr)) decreases viral neutralization by monoclonal antibodies to the CD4-binding site. Virology. 1993;196:332–337. doi: 10.1006/viro.1993.1484. [DOI] [PubMed] [Google Scholar]
- 31.Leamnson R N, Halpern M S. Subunit structure of the glycoprotein complex of avian tumor virus. J Virol. 1976;18:956–968. doi: 10.1128/jvi.18.3.956-968.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long D, Berson J F, Cook D, Doms R W. Characterization of human immunodeficiency virus type 1 gp120 binding to liposomes containing galactosylceramide. J Virol. 1994;68:5890–5898. doi: 10.1128/jvi.68.9.5890-5898.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacKrell A J, Soong N W, Curtis C M, Anderson W F. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J Virol. 1996;70:1768–1774. doi: 10.1128/jvi.70.3.1768-1774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 35.McClure M O, Marsh M, Weiss R A. Human immunodeficiency virus infection of CD4-bearing cells occurs by a pH-independent mechanism. EMBO J. 1988;7:513–518. doi: 10.1002/j.1460-2075.1988.tb02839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McClure M O, Sommerfelt M A, Marsh M, Weiss R A. The pH independence of mammalian retrovirus infection. J Gen Virol. 1990;71:767–773. doi: 10.1099/0022-1317-71-4-767. [DOI] [PubMed] [Google Scholar]
- 37.Miller D K, Lenard J. Inhibition of vesicular stomatitis virus infection by spike glycoprotein. Evidence for an intracellular, G protein-requiring step. J Cell Biol. 1980;84:430–437. doi: 10.1083/jcb.84.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore J P, McKeating J A, Weiss R A, Sattentau Q J. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 39.Perez L G, Hunter E. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein that block processing to gp85 and gp37. J Virol. 1987;61:1609–1614. doi: 10.1128/jvi.61.5.1609-1614.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reitz M S, Jr, Wilson C, Naugle C, Gallo R C, Robert-Guroff M. Generation of a neutralization-resistant variant of HIV-1 is due to selection for a point mutation in the envelope gene. Cell. 1988;54:57–63. doi: 10.1016/0092-8674(88)90179-1. [DOI] [PubMed] [Google Scholar]
- 41.Robert-Guroff M, Reitz M S, Jr, Robey W G, Gallo R C. In vitro generation of an HTLV-III variant by neutralizing antibody. J Immunol. 1986;137:3306–3309. [PubMed] [Google Scholar]
- 42.Rong L, Edinger A, Bates P. Role of basic residues in the subgroup-determining region of the subgroup A avian sarcoma and leukosis virus envelope in receptor binding and infection. J Virol. 1997;71:3458–3465. doi: 10.1128/jvi.71.5.3458-3465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruigrok R W, Martin S R, Wharton S A, Skehel J J, Bayley P M, Wiley D C. Conformational changes in the hemagglutinin of influenza virus which accompany heat-induced fusion of virus with liposomes. Virology. 1986;155:484–497. doi: 10.1016/0042-6822(86)90210-2. [DOI] [PubMed] [Google Scholar]
- 44.Sattentau Q J, Moore J P. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sattentau Q J, Moore J P, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stegmann T, Delfino J M, Richards F M, Helenius A. The HA2 subunit of influenza hemagglutinin inserts into the target membrane prior to fusion. J Biol Chem. 1991;266:18404–18410. [PubMed] [Google Scholar]
- 47.Stegmann T, White J M, Helenius A. Intermediates in influenza induced membrane fusion. EMBO J. 1990;9:4231–4241. doi: 10.1002/j.1460-2075.1990.tb07871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stein B S, Gowda S D, Lifson J D, Penhallow R C, Bensch K G, Engleman E G. pH-independent HIV entry into CD4-positive T cells via virus envelope fusion to the plasma membrane. Cell. 1987;49:659–668. doi: 10.1016/0092-8674(87)90542-3. [DOI] [PubMed] [Google Scholar]
- 49.Stern T L, Reitz M S, Jr, Robert-Guroff M. Spontaneous reversion of human immunodeficiency virus type 1 neutralization-resistant variant HXB2thr582: in vitro selection against cytopathicity highlights gp120-gp41 interactive regions. J Virol. 1995;69:1860–1867. doi: 10.1128/jvi.69.3.1860-1867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thali M, Olshevsky U, Furman C, Gabuzda D, Li J, Sodroski J. Effects of changes in gp120-CD4 binding affinity on human immunodeficiency virus type 1 envelope glycoprotein function and soluble CD4 sensitivity. J Virol. 1991;65:5007–5012. doi: 10.1128/jvi.65.9.5007-5012.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber T, Zemelman B V, McNew J A, Westermann B, Gmachl M, Parlati F, Sollner T H, Rothman J E. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 52.Weissenhorn W, Calder L J, Wharton S A, Skehel J J, Wiley D C. The central structural feature of the membrane fusion protein subunit from the Ebola virus glycoprotein is a long triple-stranded coiled coil. Proc Natl Acad Sci USA. 1998;95:6032–6036. doi: 10.1073/pnas.95.11.6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 54.Weissenhorn W, Wharton S A, Calder L J, Earl P L, Moss B, Aliprandis E, Skehel J J, Wiley D C. The ectodomain of HIV-1 env subunit gp41 forms a soluble, alpha-helical, rod-like oligomer in the absence of gp120 and the N-terminal fusion peptide. EMBO J. 1996;15:1507–1514. [PMC free article] [PubMed] [Google Scholar]
- 55.White J, Matlin K, Helenius A. Cell fusion by Semliki Forest, influenza, and vesicular stomatitis viruses. J Cell Biol. 1981;89:674–679. doi: 10.1083/jcb.89.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 57.White J M, Wilson I A. Anti-peptide antibodies detect steps in a protein conformational change: low-pH activation of the influenza virus hemagglutinin. J Cell Biol. 1987;105:2887–2896. doi: 10.1083/jcb.105.6.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wigler M, Silverstein S, Lee L S, Pellicer A, Cheng Y, Axel R. Transfer of purified herpes virus thymidine kinase gene to cultured mouse cells. Cell. 1977;11:223–232. doi: 10.1016/0092-8674(77)90333-6. [DOI] [PubMed] [Google Scholar]
- 59.Zingler K, Young J A T. Residue Trp-48 of Tva is critical for viral entry but not for high-affinity binding to the SU glycoprotein of subgroup A avian leukosis and sarcoma viruses. J Virol. 1996;70:7510–7516. doi: 10.1128/jvi.70.11.7510-7516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]