Abstract
Circular RNAs (circRNAs) are a class of noncoding RNAs that have been found to be involved in intervertebral disc degeneration (IVDD) progression, and N6-methyladenosine (m6A) broadly exists in circRNAs. Here, we identified circGPATCH2L with a low m6A methylation level to be upregulated in degenerative nucleus pulposus tissues. Mechanistically, as a protein decoy for tripartite motif containing 28 (TRIM28) within aa 402–452 region, circGPATCH2L abrogates the phosphorylation of TRIM28 and inhibits P53 degradation, which contributes to DNA damage accumulation and cellular apoptosis and leads to IVDD progression. Moreover, m6A-methylated circGPATCH2L is recognised and endoribonucleolytically cleaved by a YTHDF2-RPL10-RNase P/MRP complex to maintain the physiological state of nucleus pulposus cells. Thus, our data show the physiological significance of m6A modification in regulating circRNA abundance and provide a potentially effective therapeutic target for the treatment of IVDD.
Subject terms: Epigenetics, Ageing, Cell biology
Introduction
Low back pain (LBP) is the most frequent cause of activity limitation in patients under the age of 45 years, and approximately four out of five adults will experience LBP in their lives, imposing huge socioeconomic burdens on society [1, 2]. Intervertebral disc degeneration (IVDD) is the major contributor to LBP in the clinic [3, 4]. Each intervertebral disc consists of a central nucleus pulposus (NP), an outer annulus fibrosus (AF), and superior/inferior cartilaginous endplates [5–7]. The secretion of collagen type II and aggrecan (ACAN) from nucleus pulposus cells (NPCs) decreases with age, which causes cellular metabolism switching from anabolism to catabolism [8]. Although research in the field of IVDD is continuously increasing, a consensus on the pathogenesis of IVDD has only been reached on multifactorial causes, such as senescence, DNA damage, apoptosis, abnormal loading, and nutrition deficiency [5, 7, 9]. Thus, the underlying molecular mechanism of IVDD is still under exploration.
As a type of noncoding RNA, circular RNAs (circRNAs) are produced by back-splicing of precursor mRNAs [10]. An increasing number of recent studies have been devoted to understanding the mechanisms of circRNAs in regulating NPC function and IVDD progression [11, 12]. It has been found that circRNAs localised in the cytoplasm can function as microRNA (miRNA) sponges [13]. For example, circVMA21 was demonstrated to reduce NPC apoptosis and extracellular matrix (ECM) degradation as a competing endogenous RNA (ceRNA) of miR-200c, releasing x linked inhibitor-of-apoptosis protein (XIAP) [14]. However, other functions of circRNAs remain to be explored in IVDD, such as as protein decoys. The regulatory mechanism of circRNA abundance in NPCs under physiological conditions has not yet been characterised.
N6-methyladenosine (m6A) is the most widely distributed RNA modification in eukaryotes, and it is related to every fundamental aspect of RNA metabolism, including splicing, nuclear exportation, translation, and degradation [15, 16]. Nevertheless, the function of m6A methylation depends on the specific transcript, the specific cell type, and whether m6A reader proteins are activated [17]. Thus, to understand the pathophysiological or physiological function of m6A, analysing its effects on a certain transcript under specific conditions is of great importance. A recent study showed the m6A methylation pattern of NP tissues in a mouse degenerated intervertebral disc model [18], but the specific m6A methylation pattern of circRNAs in IVDD pathogenesis is still unclear.
In the present study, we detected the m6A methylation of circRNA in NP tissues of IVDD patients and demonstrated that unmethylated circGPATCH2L could induce DNA damage accumulation and NPC apoptosis by binding to tripartite motif containing 28 (TRIM28), which accelerated IVDD progression. To secure a steady state in NPCs, m6A-containing circGPATCH2L was subjected to endoribonucleolytic cleavage via the YTHDF2-RPL10-RNase P/MRP complex. This study elaborates the mechanism of circRNA m6A methylation in maintaining the physiological state of NPCs and provides potential targets for IVDD therapy.
Results
circGPATCH2L, with a decreased m6A methylation level, is upregulated in degenerative NP tissues
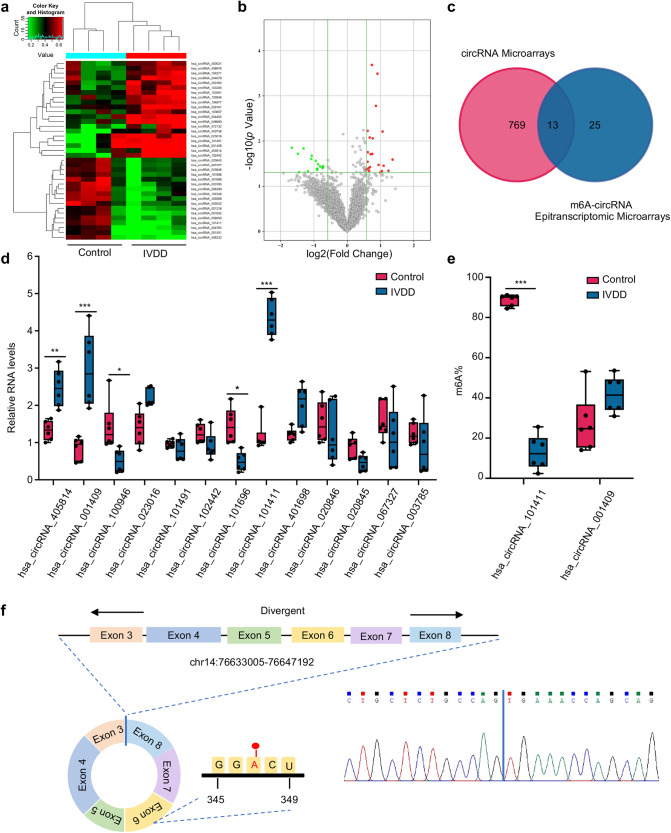
We performed m6A-circRNA epitranscriptomic microarray analysis using NP tissues obtained from four degenerative disc patients (IVDD group) and four nondegenerative disc patients (control group; Supplementary Table S1). Of the 4395 circRNAs detected by the epitranscriptomic microarray, 21 circRNAs were hypermethylated and 17 circRNAs were hypomethylated in the IVDD group compared with the control group when filtering with the fold change ≥ 1.5 and p ≤ 0.05 thresholds (Fig. 1a, b; Supplementary Fig. S1a, b; Supplementary Table S2). The identification of differentially methylated and expressed circRNAs was performed by overlapping m6A-circRNA epitranscriptomic microarray analysis and microarray analysis of circRNAs (Fig. 1c) [19]. A total of 13 candidate circRNAs were further validated using specific divergent primers in a quantitative real-time polymerase chain reaction (qRT–PCR) assay. hsa_circRNA_101411 and hsa_circRNA_001409 were found to be significantly upregulated in the IVDD group to an extent similar to that in the circRNA microarray data (Fig. 1d). Then, we adopted a T3 DNA ligase assay to test the m6A methylation status of hsa_circRNA_101411 and hsa_circRNA_001409 with specific DNA probes based on the sequence flanking the m6A site and nonm6A site, respectively [20]. The ligation efficiency of the m6A site was significantly decreased compared with that of the nonm6A site; thus, the m6A methylation ratio could be calculated using Ct values (Supplementary Fig. S2a). The methylation ratio of hsa_circRNA_101411 was significantly decreased in the IVDD group, which was consistent with the m6A-circRNA epitranscriptomic microarray analysis data. According to the circBase and circBank database annotation [21, 22], hsa_circRNA_101411, named circGPATCH2L (circBase ID: hsa_circ_0005267; conserved sequence: mmu_circ_0000404; Supplementary Table S3), is derived from exons 3 to 8 of the GPATCH2L gene (chr14:76633005–76647192) according to the human genome reference hg19/GRCh37 (Fig. 1f). The m6A site was predicted by the SRAMP prediction server with very high confidence (Supplementary Fig. S2b) [23]. circGPATCH2L was chosen for further study. The results of Sanger sequencing analysis of the PCR products of circGPATCH2L were in accordance with circBase database annotation (Fig. 1f).
Fig. 1. circGPATCH2L is hypomethylated and upregulated in IVDD.
a Hierarchical clustering for circRNAs with differential m6A methylation levels in the IVDD and control groups. The red–green gradient colour scheme represents the high and low m6A methylation relative levels as referenced in the Colour Key. b Volcano plot showing 38 differentially m6A-methylated circRNAs in NP tissues of the IVDD and control groups. The cut off is |fold change| ≥ 1.5, p ≤ 0.05. The red and green points in the plot indicate the upregulated and downregulated circRNAs with statistical significance, respectively. c Venn diagram identifying 13 differentially expressed circRNAs with altered methylation in IVDD samples based on the overlap of circRNA microarray and m6A-circRNA epitranscriptomic microarray data. d qRT–PCR analysis of the expression of 13 circRNAs in NP tissues. n = 6. e The m6A ratio of hsa_circRNA_101411 and hsa_circRNA_001409 was calculated using a T3 DNA ligase assay. n = 6. f Schematic of the genomic loci of circGPATCH2L and the m6A site. The back-splice junction site of circGPATCH2L was confirmed by Sanger sequencing. NP nucleus pulposus, m6A N6-methyladenosine, qRT–PCR quantitative real-time polymerase chain reaction, IVDD intervertebral disc degeneration. Data are shown as the mean ± S.D. ***p < 0.001; **p < 0.01; *p < 0.05.
Characteristics of circGPATCH2L in NP
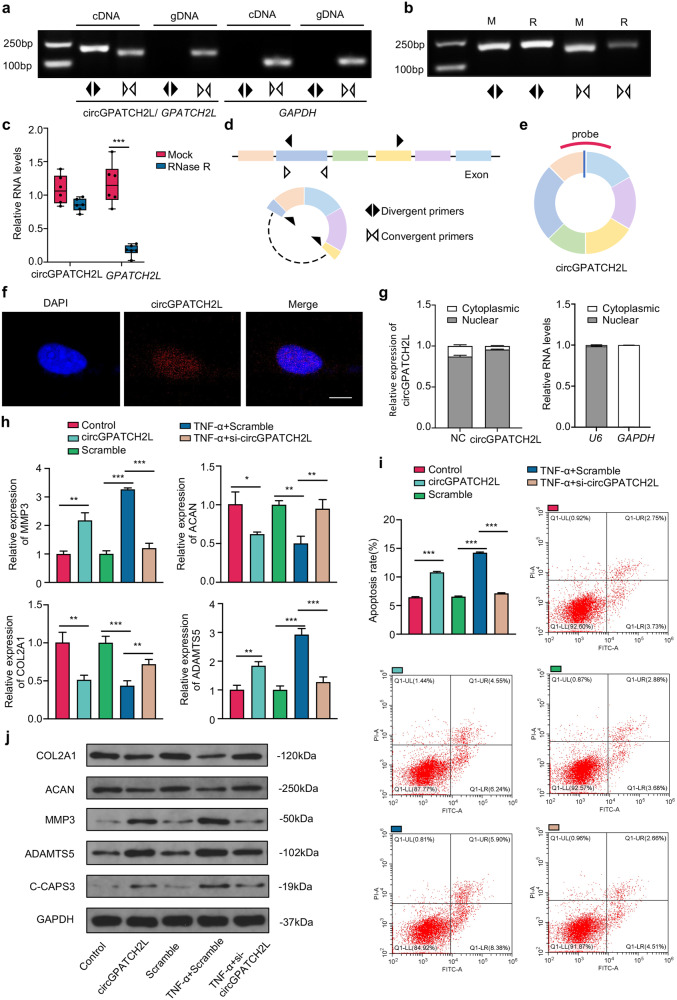
Prior to the functional study, a prerequisite was the identification of circGPATCH2L by different techniques as previously described [24]. PCR and qRT–PCR analyses showed that circGPATCH2L could be amplified by divergent primers and was resistant to ribonuclease R (RNase R) treatment, demonstrating that circGPATCH2L had greater stability than its linear transcript (Fig. 2a–d). Its homologous circRNA in mice, mmu_circ_0000404, was also confirmed (mmu_circ_0000404 shown in Supplementary Fig. S3b, c). We further analysed the subcellular localisation of circGPATCH2L expression in NPCs via nuclear and cytoplasmic fractionation and RNA fluorescent in situ hybridisation (FISH) examination using a specific probe targeting the back-spliced site of circGPATCH2L (Fig. 2e). The data demonstrated that circGPATCH2L was predominantly localised in the nucleus, and this tendency was more substantial when exogenous circGPATCH2L was overexpressed (Fig. 2f, g). According to reports, some circRNAs in the cytoplasm serve as ceRNAs through interacting with miRNAs or as translational templates [13, 14, 25], while circRNAs located in the nucleus can bind to proteins or be a competitor of DNA in protein binding [26, 27]. Therefore, we speculate that circGPATCH2L acts as a decoy for proteins or serves as a scaffold for protein complexes.
Fig. 2. Characterisation of circGPATCH2L and its function in NPCs.
a PCR analysis of circGPATCH2L using divergent and convergent primers in cDNA and gDNA. GAPDH was used as a control. b PCR analysis of circGPATCH2L using divergent and convergent primers after treatment with RNase R. c qRT–PCR showing resistance of circGPATCH2L to RNase R digestion. n = 6. d A schematic diagram representing divergent (black arrow) and convergent (white arrow) primers. e A schematic diagram of a probe targeting the back-spliced site of circGPATCH2L. f RNA-FISH analysis of circGPATCH2L in NPCs. The circGPATCH2L probe was labelled with Cy3. Cell nuclei were stained with DAPI. Scale bar = 10 μm. g Relative RNA levels of circGPATCH2L in the nuclear and cytoplasmic fractions of NPCs. circGPATCH2L was mainly localised in the nucleus. Relative RNA levels of U6 and GAPDH were tested as quality controls. n = 3. h qRT–PCR analysis of COL2A1, ACAN, MMP3, and ADAMTS5 mRNA expression in each group. The control group was transfected with a blank vector with flanking introns containing complementary Alu elements. NPCs transfected with sequence-scrambled DNA oligos were named Scramble. TNF-α (10 ng/ml) was used to induce an in vitroIVDD model. The circGPATCH2L overexpression plasmid successfully upregulated circGPATCH2L expression and siRNA for circGPATCH2L downregulated circGPATCH2L expression. n = 3. i Representative flow cytometry dot plots of apoptosis after Annexin V-FITC/PI dual staining. Histograms showing the apoptosis rate in the control, circGAPTCH2L (circGAPTCH2L overexpression), Scramble, TNF-α + Scramble, and TNF-α + si-circGAPTCH2L-2 groups. n = 3. j Western blot analysis of COL2, ACAN, MMP3, ADAMTS5, and C-CASP3 protein levels. circGPATCH2L overexpression induced degeneration of NPCs, and circGPATCH2L knockdown rescued the degeneration phenotype created by TNF-α (10 ng/ml) treatment. NPCs nucleus pulposus cells, M mock, R RNase R, RNase R ribonuclease R, PCR polymerase chain reaction, cDNA complementary DNA, gDNA genomic DNA, FISH fluorescent in situ hybridisation, DAPI 4,6-diamidino-2-phenylindole, qRT–PCR quantitative real-time PCR, GAPDH glyceraldehyde 3-phosphate dehydrogenase, COL2A1 collagen type II alpha 1 chain, ACAN aggrecan, MMP3 matrix metallopeptidase 3, ADAMTS5 a disintegrin and metalloproteinase with thrombospondin type 1 motif 5, IVDD intervertebral disc degeneration, C-CASP3 cleaved Caspase 3, TNF-α tumour necrosis factor-α. Data are shown as the mean ± S.D. ***p < 0.001; **p < 0.01; *p < 0.05.
circGPATCH2L regulates apoptosis and degeneration of NPCs
To determine the role of circGPATCH2L in IVDD, we performed loss- and gain-of-function experiments in an in vitro IVDD model using tumour necrosis factor-α (TNF-α, 10 ng/ml) [14]. First, NPCs isolated from nondegenerative disc patients were transfected with a circGPATCH2L overexpression plasmid, and the overexpression efficiency was confirmed by qRT–PCR (Supplementary Fig. S3d). For circGPATCH2L knockdown, two small interfering RNAs (siRNAs) specifically targeting the back-splice junction region were designed. si-circGPATCH2L-1 and si-circGPATCH2L-2 successfully silenced circGPATCH2L, and si-circGPATCH2L-2 was more efficient than si-circGPATCH2L-1 and thus was selected for the following loss-of-function assays (Supplementary Fig. S3d). Subsequently, qRT–PCR analysis (Fig. 2h) and Annexin V-FITC/propidium iodide (PI) dual staining (Fig. 2i) showed that circGPATCH2L overexpression markedly increased catabolic degeneration and apoptosis in NPCs and that knockdown of circGPATCH2L significantly rescued these phenotypes after TNF-α treatment. Moreover, western blot analysis confirmed the procatabolic and proapoptotic effects of circGPATCH2L (Fig. 2j). These data indicate a critical role of circGPATCH2L in regulating NPC functions and survival.
circGPATCH2L adjusts DNA damage repair and apoptosis in NPCs through TRIM28
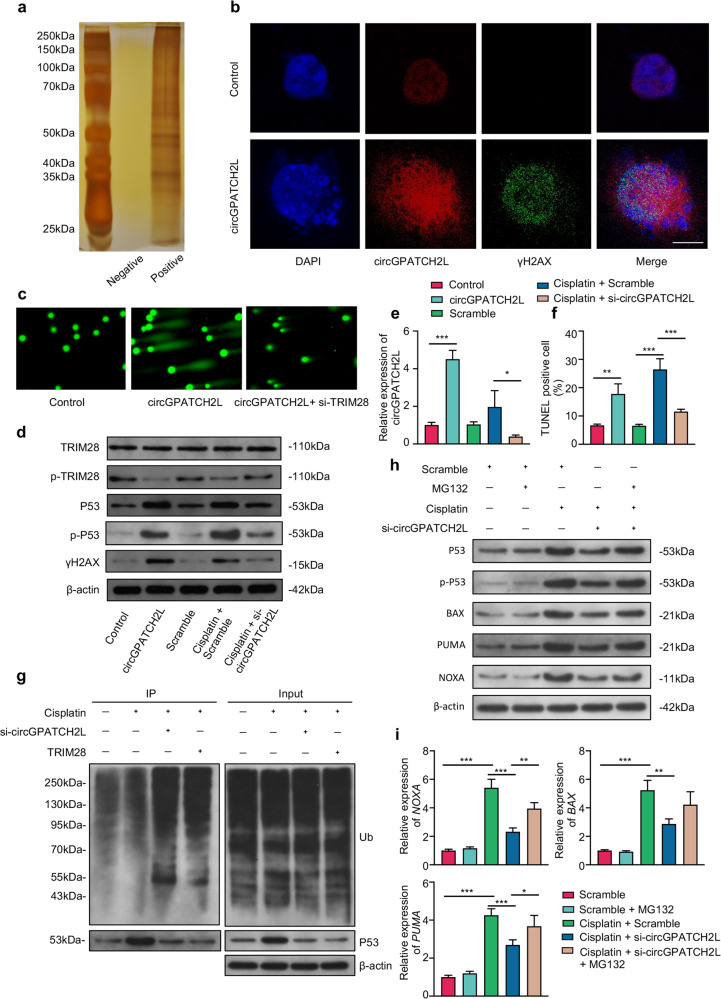
To better understand the molecular mechanism(s) involved in NP degeneration, we performed an RNA pulldown assay of circGPATCH2L using biotinylated probes targeting the circGPATCH2L back-spliced sequence in circGPATCH2L-overexpressing primary NPCs, and subsequently, the precipitates were subjected to proteomic screening (Fig. 3a). A large number of proteins were identified by liquid chromatography-tandem mass spectrometry (LC–MS/MS) analysis, among which tripartite motif containing 28 (TRIM28) was the one of most abundant engaging in the DNA damage response (DDR) pathway (Supplementary Fig. S4a, c) [28]. Notably, the Argonaute-2 (AGO2) protein was not detected in the precipitates (Supplementary Table S4), which further confirmed that circGPATCH2L did not act as a miRNA sponge [29]. Ataxia telangiectasia-mutated (ATM) kinase phosphorylates TRIM28 protein on Ser824 within the C-terminus, halting heterochromatin compaction and providing sufficient elasticity to facilitate DNA repair [30–32]. Thus, we performed RNA-FISH and γH2AX immunofluorescence staining. The imaging showed that circGPATCH2L overexpression increased the fluorescence intensity of H2AX phosphorylation (γH2AX), indicating that circGPATCH2L elicits DNA double-strand breaks (DSBs) (Fig. 3b). Then, a comet assay, which is commonly used to assess DSBs [33], was used to assess the critical role of TRIM28 in circGPATCH2L-induced DNA damage. Figure 3c showed that, in circGPATCH2L-overexpressed cells, an obvious smeared pattern of nuclear DNA was observed, and this phenomenon was attenuated after the treatment of siRNA targeting TRIM28 (si-TRIM28). TRIM28 has also been shown to suppress P53-mediated apoptosis by interacting directly with the E3 ubiquitin-protein ligase Mdm2 (MDM2) and promoting P53 polyubiquitination and degradation in the proteasome [34]. Afterwards, we performed loss- and gain-of-function experiments in an in vitro DNA damage model induced by cisplatin (50 μM) treatment [35]. In accordance with the comet assay, Western blot analysis showed that circGPATCH2L overexpression inhibited TRIM28 phosphorylation and increased γH2AX, while circGPATCH2L knockdown significantly rescued these phenotypes after cisplatin treatment (Fig. 3d), which confirmed the significant role of TRIM28 in the circGPATCH2L-mediated DNA damage response. In addition, P53 and its phosphorylation (Ser392, p-P53) were upregulated after circGPATCH2L overexpression and downregulated after circGPATCH2L knockdown (Fig. 3d), suggesting that circGPATCH2L was involved in both the degradation and phosphorylation of P53. qRT–PCR analysis was used to confirm the expression of circGPATCH2L (Fig. 3e). TUNEL assay confirmed the role of circGPATCH2L in the regulation of DNA damage repair and cellular apoptosis (Fig. 3f; Supplementary Fig. S4b).
Fig. 3. circGPATCH2L promotes DNA damage and apoptosis in NPCs through TRIM28.
a Silver staining of the precipitates from circGPATCH2L pulldown with positive and negative probes after SDS–PAGE. b RNA-FISH assay of circGPATCH2L (red) and immunofluorescence staining of γH2AX (green) in NPCs with and without circGPATCH2L overexpression. The circGPATCH2L probe was labelled with Cy3. Cell nuclei were stained with DAPI (blue). Scale bar = 10 μm. c The comet assay was performed to detect cellular DNA damage (original magnification, 200×). The circGPATCH2L overexpression plasmid and siTRIM28 were used. d Western blot analysis of TRIM28, p-TRIM28, γH2AX, P53, p-P53 (Ser392) and β-actin in NPCs. Gain- and loss-of-function experiments showed that circGPATCH2L abrogated TRIM28 phosphorylation and induced DNA damage, P53 phosphorylation and accumulation. The control group was transfected with a blank vector with flanking introns containing complementary Alu elements. NPCs transfected with sequence-scrambled DNA oligos were named Scramble. Cisplatin (50 μM) was used to induce DNA damage in NPCs in vitro. The circGPATCH2L overexpression plasmid and si-circGPATCH2L were used to regulate circGPATCH2L expression. e qRT–PCR analysis of circGPATCH2L expression in different groups. n = 3. f Quantification of TUNEL-positive cells. n = 3. g Ubiquitination of p53 was analysed by immunoprecipitation with p53 antibody and followed by western blot analysis in NPCs with circGPATCH2L or TRIM28 depletion in the presence of Cisplatin (50 μM). h Western blot analysis of P53, p-P53 (Ser392), BAX, PUMA, NOXA and β-actin expression in NPCs. Scramble and Scramble with MG132 (5 µM, 24 h) were used as control. Cisplatin (50 μM) was used to induce DNA damage in NPCs. MG132 and si-circGPATCH2L were used to regulate total P53. i qRT–PCR analysis confirmed the expression of BAX, PUMA and NOXA mRNA in different groups. n = 3. NPCs nucleus pulposus cells, qRT–PCR quantitative real-time polymerase chain reaction, TRIM28 tripartite motif containing 28, p-TRIM28 TRIM28 phosphorylation, MDM2 E3 ubiquitin-protein ligase Mdm2, γH2AX H2A.X variant histone phosphorylation, P53 cellular tumour antigen p53, p-P53 phosphorylation of p53, β-actin actin beta. Data are shown as the mean ± S.D. ***p < 0.001; **p < 0.01; *p < 0.05.
To investigate the involvement of circGPATCH2L and TRIM28 in the ubiquitination and degradation of P53, we first transfected si-circGPATCH2L and a TRIM28 overexpression plasmid into cisplatin-pretreated NPCs. The ubiquitination analysis revealed that knocking down circGPATCH2L and overexpressing TRIM28 resulted in enhanced ubiquitination of P53 and decreased total P53 (Fig. 3g). Additionally, a proteasome inhibitor MG132 (5 µM, 24 h) [36] was used in the in vitro DNA damage model to test the total P53. Western blot showed that P53, p-P53 and P53-dependent apoptosis-related proteins (BAX, PUMA and NOXA) [37] were all downregulated after circGPATCH2L knockdown in the cisplatin-pretreated NPCs and these phenotypes were practically rescued when MG132 was added. Following that, qRT-PCR analysis indicated that apoptosis-related genes expression was increased after MG132 was introduced into NPCs (Fig. 3i). Taken together, we conclude that circGPATCH2L participates in DNA damage repair and subsequent apoptosis in NPCs, in which TRIM28 and subsequent P53 may play a crucial role.
circGPATCH2L binds to the aa 402–452 region of TRIM28
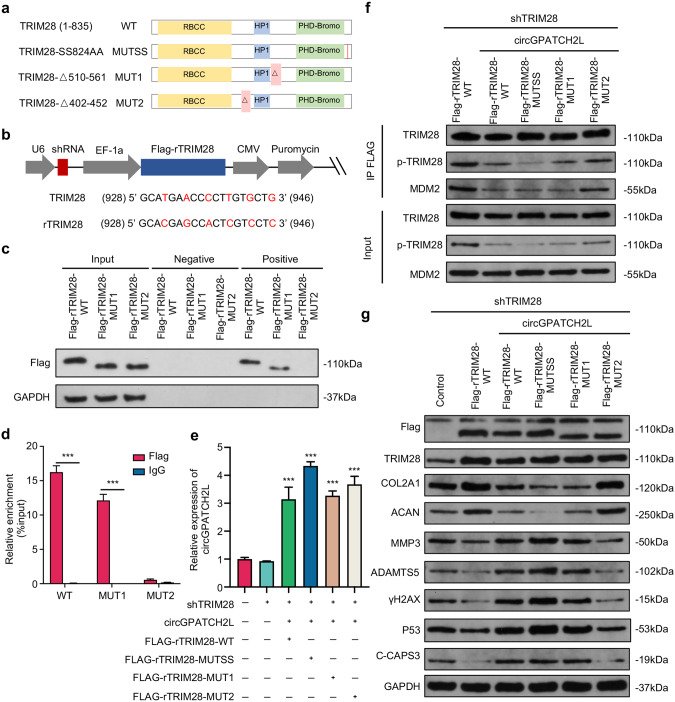
The catRAPID algorithm [38] was used to analyse the interaction propensities of circGPATCH2L with TRIM28 and predicted that circGPATCH2L-TRIM28 binding was mediated through TRIM28 residues 510–561 and 402–452 with the highest interaction capacity (Supplementary Table S5). Considering that Ser824 phosphorylation of TRIM28 plays a decisive role in the DDR [31, 39] and that circGPATCH2L overexpression inhibited TRIM28 phosphorylation, we next examined whether the phosphorylation of TRIM28 at Ser824 was dependent on the direct binding of circGPATCH2L with TRIM28. To address this question, we constructed short hairpin RNA (shRNA)-resistant 3×FLAG-TRIM28 (Flag-rTRIM28) plasmids containing TRIM28 shRNA to avoid interference of endogenous TRIM28 protein and reconstituted them with FLAG-tagged TRIM28 [40]. The plasmids were then packed into lentivirus. Three mutant shRNA-resistant cDNAs were generated (Fig. 4a, b): one point mutant, Flag-rTRIM28-SS823/824AA (Flag-rTRIM28-MUTSS), and two truncated mutants, Flag-rTRIM28-△510–561 (Flag-rTRIM28-MUT1) and Flag-rTRIM28-△402–452 (Flag-rTRIM28-MUT2). Cells were transfected with the relevant expression vectors mentioned above and selected in medium containing puromycin for 6 days. The surviving cells with stable knockdown of TRIM28 and exogenous expression of Flag-rTRIM28, confirmed by western blot (Supplementary Fig. S4d), were further transfected with circGPATCH2L plasmid. RNA-pulldown and RNA immunoprecipitation (RIP) assays showed that Flag-rTRIM28 wild type (Flag-rTRIM28-WT) and Flag-rTRIM28-MUT1 bound to circGPATCH2L, indicating that the interaction only occurred when aa 402–452 of TRIM28 were present (Fig. 4c, d). The expression of circGPATCH2L was confirmed by qRT–PCR (Fig. 4e). Furthermore, coimmunoprecipitation (Co-IP) assays confirmed that the phosphorylation of Ser824 was successfully inhibited with SS823/824AA and that the binding of circGPATCH2L-TRIM28 not only suppressed the phosphorylation of Ser824 but also hindered the TRIM28-MDM2 interaction (Fig. 4f).
Fig. 4. circGPATCH2L functions in NPCs by binding the aa 402–452 region of TRIM28.
a Schematic representation of the full-length, truncated or point mutant TRIM28. TRIM28 wild type (TRIM28-WT) was full-length and contained three main TRIM28 domains. TRIM28-MUT1 and TRIM28-MUT2 lacked the aa 510–561 and aa 402–452 regions, respectively. Each serine was mutated to alanine at aa 823/824 (TRIM28-MUTSS) to avoid the possibility that Ser823 becomes phosphorylated when Ser824 is mutated. b Schematic diagrams of the lentiviral plasmids used in this study. The modified plasmid was designed for simultaneous expression of shTRIM28, Flag-tagged shTRIM28-resistant wild-type or mutant TRIM28 cDNA (rTRIM28), and the puromycin resistance gene. The shRNA-targeted sequences and the silencing mutations in shTRIM28-resistant cDNAs are shown below. The numbers correspond to the nt positions in the open reading frames of TRIM28, and the changed bases are marked in red. c, d RNA pulldown and RIP assays confirmed the interaction between circGPATCH2L and TRIM28 within the site at residues 402–452. c Coprecipitation of circGPATCH2L and wild-type or mutant TRIM28 in an RNA pulldown assay. Total protein (Input), precipitates with biotinylated probes targeting the circGPATCH2L back-spliced site (positive) or negative biotinylated probes (negative) were analysed via western blot with Flag antibody. d RIP experiments were performed using Flag or negative IgG antibody. Purified RNA was used for qRT–PCR assays. n = 3. e qRT–PCR analysis confirmed the overexpression efficiency of circGPATCH2L in NPCs after treatment with the circGPATCH2L overexpression plasmid in different groups. n = 3. f Cell lysates were immunoprecipitated with an antibody against Flag and analysed via western blot with TRIM28 antibody, p-TRIM28 antibody, or MDM2 antibody (top). Total protein from cell lysates was analysed as input (bottom). g Western blot analysis of Flag, TRIM28, COL2, ACAN, MMP3, ADAMTS5, γH2AX, P53, C-CAPS3, and GAPDH expression in NPCs. The control group was transfected with a blank vector with 3×FLAG. NPCs nucleus pulposus cells, qRT–PCR quantitative real-time polymerase chain reaction, TRIM28 tripartite motif containing 28, p-TRIM28 TRIM28 phosphorylation, MDM2 E3 ubiquitin-protein ligase Mdm2, γH2AX H2A.X variant histone phosphorylation, P53 cellular tumour antigen p53, COL2A1 collagen type II alpha 1 chain, ACAN aggrecan, MMP3 matrix metallopeptidase 3, ADAMTS5 a disintegrin and metalloproteinase with thrombospondin type 1 motif 5, C-CASP3 cleaved Caspase 3, GAPDH glyceraldehyde 3-phosphate dehydrogenase. Data are shown as the mean ± S.D. ***p < 0.001.
To determine whether the interaction of circGPATCH2L and TRIM28 affects the function of NPCs, western blot assays were performed. As shown in Fig. 4g, the overexpression of circGPATCH2L markedly counteracted the antiapoptotic and DNA damage repair effects of TRIM28 and promoted the catabolic effects of NPCs. Importantly, MUT2 but not WT, MUTSS, or MUT1 TRIM28 restored normal metabolism of ECM in circGPATCH2L-overexpressing NPCs and reduced DNA damage, apoptosis, and P53 accumulation (Fig. 4g). Together, these data establish that circGPATCH2L, which acts as a molecular decoy by binding to aa 402–452 of TRIM28, induces DNA damage and apoptosis in NPCs, accelerating IVDD progression.
RNase P/MRP elicits YTHDF2-mediated RNA decay on m6A-methylated circGPATCH2L in NPCs
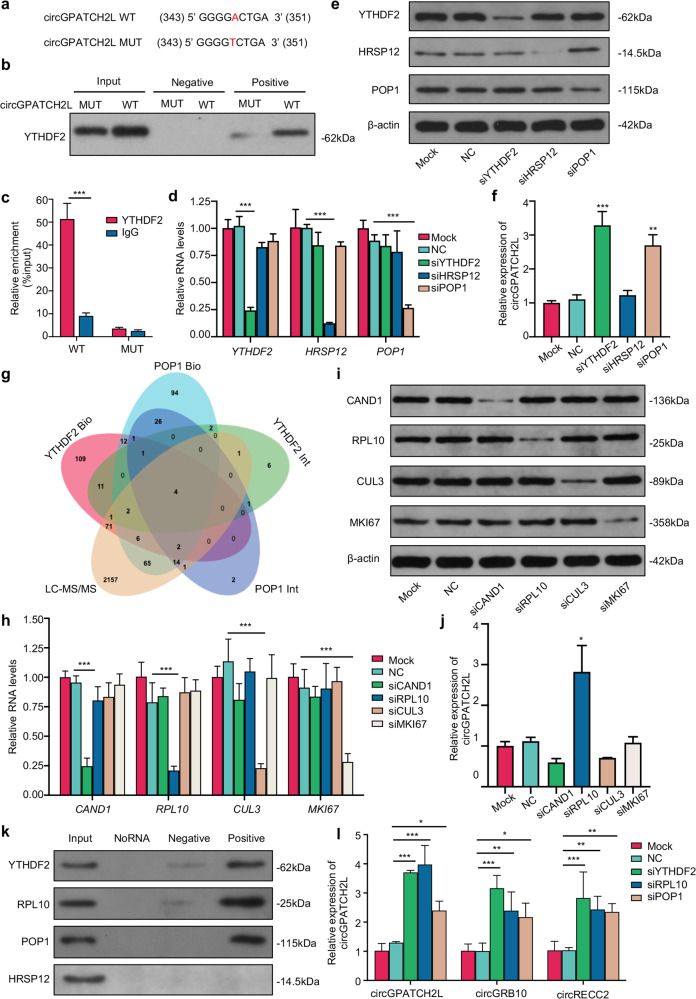
Since circGPATCH2L was upregulated with a decreased m6A level in degenerative NP tissues, we speculated that the m6A level of circGPATCH2L might influence its degradation. m6A-containing circRNAs were experimentally found to be susceptible to endoribonucleolytic cleavage via the YTHDF2 (YTH N6-methyladenosine RNA binding protein 2, an m6A reader protein), HRSP12 (2-iminopropanoate deaminase, an adaptor protein), and RNase P/MRP (ribonuclease P/MRP, an endoribonuclease) complexes [41]. Thus, to first analyse the m6A site in circGPATCH2L, we constructed wild-type circGPATCH2L (circGPATCH2L WT) and point mutant circGPATCH2L (circGPATCH2L MUT) vectors, in which the adenine residue embedded within the m6A motif was replaced by thymine (A-T transversion mutation, Fig. 5a). After transfection of NPCs with circGPATCH2L WT or MUT, RNA pulldown and RIP assays were performed to confirm the binding of circGPATCH2L with YTHDF2 through the m6A site (Fig. 5b, c). To further investigate a possible function of the complex mentioned above in NPCs, we designed siRNAs targeting these three proteins, and the knockdown efficiency was identified by qRT–PCR (Fig. 5d; Supplementary Fig. S5a) and western blot (Fig. 5e). Notably, circGPATCH2L significantly increased in abundance only after downregulation of YTHDF2 and RNase P/MRP protein subunit POP1 (POP1), indicating that m6A-methylated circGPATCH2L associated with YTHDF2 could be endoribonucleolytically cleaved by RNase P/MRP but not in an HRSP12-dependent manner (Fig. 5f).
Fig. 5. m6A methylated circGPATCH2L was subject to endoribonucleolytic cleavage via YTHDF2-RPL10-RNase P/MRP complex.
a The m6A motif sequence and the silencing mutation of circGPATCH2L. The numbers correspond to the nt positions in the open reading frames. A-T transversion mutations are indicated in red. b Western blot analysis of YTHDF2 in precipitates from a circGPATCH2L pulldown assay in NPCs with circGPATCH2L-WT or circGPATCH2L-MUT overexpression. The input was cell lysates with total protein; the positive and negative groups were precipitates with positive and negative biotinylated probes, respectively. c circGPATCH2L was detected by qRT–PCR in a RIP assay with YTHDF2 antibody-conjugated beads. IgG antibody was used as a negative control. n = 3. d qRT–PCR analysis of YTHDF2, HRSP12, and POP1 showed that the expression of each gene was downregulated by the indicated siRNAs in an independent manner. The mock group did not receive any treatment, and the NC group was treated with transfection reagent only. n = 3. e Western blot analysis identifying the knockdown efficiency of YTHDF2, HRSP12, POP1, and β-actin at the protein level. f qRT–PCR analysis of circGPATCH2L expression after transfection with siRNA. Knockdown of YTHDF2 and POP1 caused a significant increase in circGPATCH2L. n = 3. g Venn diagram showing the intersection of proteins among two PPI databases (BioGRID and IntAct) and our mass spectrometry data. Four candidate proteins were identified. h qRT–PCR analysis of CAND1, CUL3, RPL10, and MKI67 showing that the expression of each gene was downregulated by the indicated siRNAs in an independent manner. The mock group did not receive any treatment, and the NC group was treated with transfection reagent only. n = 3. i Western blot confirming the knockdown efficiency of CAND1, RPL10, CUL3, MKI67, and β-actin at the protein level; β-actin was used as an internal reference. j qRT–PCR analysis of circGPATCH2L expression after knockdown of each candidate protein. Knockdown of RPL10 increased circGPATCH2L abundance. n = 3. k Western blot analysis of YTHDF2, RPL10, POP1 and HRSP12 in precipitates from a circGPATCH2L pulldown assay in NPCs overexpressing circGPATCH2L. The input group was cell lysates with total protein; the NoRNA group with no probe was used as a control; biotinylated probes targeting the circGPATCH2L back-spliced site (positive) or negative biotinylated probes (negative) were also used. l qRT–PCR analysis of circGPATCH2L, circGRB10, and circRECC2 in NPCs after knockdown of each protein confirmed that the YTHDF2-RPL10-RNase P/MRP complex could mediate endoribonucleolytic cleavage of m6A-methylated circRNAs. n = 3. qRT–PCR quantitative real-time polymerase chain reaction, NC negative control, NPCs nucleus pulposus cells, YTHDF2 YTH N6-methyladenosine RNA binding protein 2, HRSP12 2-iminopropanoate deaminase, POP1 ribonuclease P/MRP protein subunit POP1, β-actin actin beta, WT wild type, MUT mutant, PPI Protein–Protein Interaction, Bio BioGrid, Int IntAct, LC–MS/MS liquid chromatography-tandem mass spectrometry, CAND1 cullin-associated and neddylation-dissociated 1, RPL10 ribosomal protein L10, CUL3 cullin 3, MKI67 marker of proliferation Ki-67. Data are shown as the mean ± S.D. ***p < 0.001; **p < 0.01; *p < 0.05.
To further explore a scaffold protein bridging YTHDF2 and POP1, two PPI (Protein–Protein Interaction) databases, BioGrid [42] and IntAct [43], together with our LC–MS/MS data, were used for candidate protein prediction (Fig. 5g). Four candidate proteins were identified as candidate bridge proteins and subjected to further tests. siRNAs targeting CAND1, RPL10, CUL3, and MKI67 were designed, and the knockdown efficiency was identified by qRT–PCR (Fig. 5h; Supplementary Fig. S5b) and western blot (Fig. 5i). After transfection of NPCs with relevant siRNAs, circGPATCH2L markedly increased in abundance only after downregulation of RPL10 (Fig. 5j). We then performed an RNA pulldown assay of circGPATCH2L using a biotinylated probe in circGPATCH2L-overexpressing NPCs. YTHDF2, RPL10, and POP1, but not HRSP12, were found in the precipitate of circGPATCH2L-positive probes (Fig. 5k), indicating that RPL10 might act as a scaffold protein bridging YTHDF2 and POP1 to perform endoribonucleolytic cleavage of circGPATCH2L. In support of this assumption, other circRNAs reported in NPCs (circGRB10 and circERCC2) [19, 44] increased remarkably in abundance when YTHDF2, RPL10, and POP1 were downregulated separately by siRNAs (Fig. 5l). Consistent with these results, HRSP12 was not identified in LC–MS/MS analysis of circGPATCH2L pulldown precipitates either. Collectively, our data indicate that m6A-methylated circGPATCH2L can be endoribonucleolytically cleaved by a YTHDF2-RPL10-RNase P/MRP complex in NPCs.
Adenovirus (Adv) delivery of shRNA against circGPATCH2L alleviates IVDD in vivo
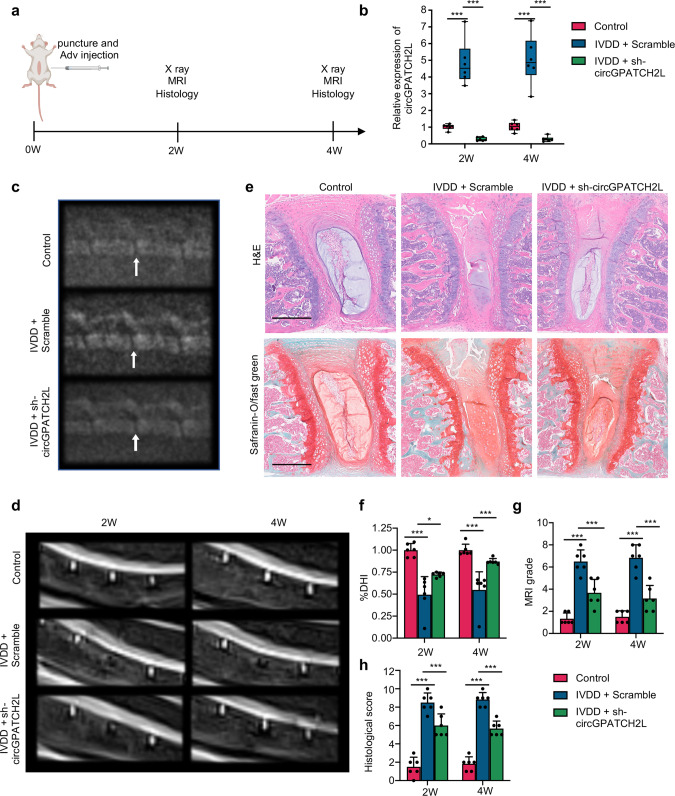
We next investigated circGPATCH2L function in an IVDD mouse model. A total of 36 C57BL/6J mice (8 weeks old, male) were operated under general anaesthesia. The control group received a sham operation, while the IVDD + Scramble group and IVDD +sh-circGPATCH2L group were punctured and injected at the lumbar 4/5 segment with Adv-sh-Scramble and adenoviral mouse sh-circGPATCH2L (mmu_circ_0000404), respectively, on Day 0. The dose of the adenovirus was 2 ×108 viral genome particles using a 33 G Hamilton syringe. X-ray imaging, magnetic resonance imaging (MRI), and histological staining were conducted at 2 and 4 weeks (Fig. 6a). High transfection and knockdown efficiencies were observed in the IVDD + sh-circGPATCH2L group (Fig. 6b). As shown in Fig. 6c, disc height decreased in the IVDD + Scramble group, and knockdown of circGPATCH2L restored disc height, which was confirmed by the percentage of disc height index (%DHI) [45] (Fig. 6f). Meanwhile, at each time point after operation, modified Pfirrmann grades based on T2-weighted MRI showed a significant difference between the IVDD + Scramble group and the IVDD + sh-circGPATCH2L group (Fig. 6d, g). The Pfirrmann grading system is the most widely known classification for intervertebral disc degeneration based on MR signal intensity, disc structure, distinction between nucleus and anulus, and disc height [46, 47]. And the modified Pfirrmann grading system with stronger discriminatory power for IVDD is necessary for IVDD research [46]. In addition, safranin-O/fast green and haematoxylin-eosin (H&E) staining showed abundant fibrous cartilaginous tissue and ruptured AF in the IVDD + Scramble group, while the IVDD + sh-circGPATCH2L group showed more proteoglycans and fewer ruptures than the IVDD + Scramble group (Fig. 6e). The histological classification score system according to a reported mouse IVDD histological classification was used to detect degenerative progression [48]. As it was showed in Fig. 6h, the histological score was significantly lower in the IVDD + sh-circGPATCH2L group than in the IVDD + Scramble group. Taken together, these results reveal the positive effects of circGPATCH2L knockdown on inhibiting ECM degradation, restoring NP functions, and alleviating IVDD in a mouse model.
Fig. 6. Knockdown of circGPATCH2L in NPCs represses IVDD in a mouse model.
a Diagram showing the in vivo experimental procedure. A total of 36 mice (8 weeks old, male) were randomly divided into three groups: the sham operation group (control), Adv-sh-Scramble injection with puncture group (IVDD + Scramble), and Adv-sh-circGPATCH2L injection with puncture group (IVDD + sh-circGPATCH2L). b The knockdown efficiency of circGPATCH2L after Adv-sh-circGPATCH2L transfection was confirmed by qRT–PCR in NP tissues. n = 6. c Representative radiographs of mice in the three groups at 4 weeks after the operation. L4/5 were punctured. The white arrow indicates the punctured segment. d Representative T2-weighted MRI images of the mouse lumbar spine with punctured segments at 2 and 4 weeks after the operation. e H&E staining and safranin-O/fast green staining of the punctured discs in each group at 4 weeks after the operation. Scale bar = 500 μm. f The disc height index (%DHI) decreased significantly in the IVDD + Scramble group and was resumed after Adv-sh-circGPATCH2L injection at 2 and 4 weeks after the operation. n = 6. g The MRI grade based on a modified Pfirrmann grading system was significantly lower in the IVDD + sh-circGPATCH2L group than in the IVDD + Scramble group. n = 6. h A significantly decreased histological score was noted in the IVDD + sh-circGPATCH2L group compared with the IVDD + Scramble group. The histological score was calculated using a mouse intervertebral disc histological classification. n = 6. W week, IVDD intervertebral disc degeneration, Adv adenovirus, qRT–PCR quantitative real-time polymerase chain reaction, L lumbar, H&E haematoxylin-eosin, MRI magnetic resonance imaging. Data are shown as the mean ± S.D. ***p < 0.001, *p < 0.05.
Discussion
Accumulating evidence has demonstrated that circRNAs play a critical role in IVDD by regulating signalling pathways that are fundamental to NPC functions, such as the WNT pathway [12, 49]. However, most studies assume that circRNAs act as ceRNAs and have focused on the pathological states of NPCs. In this study, we identified that circGPATCH2L participates in the DNA damage response and apoptosis in IVDD by binding to TRIM28 as a protein decoy. We then demonstrated that m6A-methylated circGPATCH2L is endoribonucleolytically cleaved by the YTHDF2-RPL10-RNase P/MRP complex in normal NPCs. To the best of our knowledge, this is not only the first study on the influence of m6A epigenetic modification of circRNAs in the progression of IVDD but also the first exploration of the regulatory mechanism of circRNA abundance under physiological conditions in NPCs.
Similar to many linear mRNAs, most circRNAs composed of exons are located in the cytoplasm, while circRNAs composed of introns and exons tend to be located in the nucleus and participate in transcriptional regulation [50, 51]. A recent study showed that depletion of DDX39A/B (the two human homologues of Hel25E) induced the accumulation of long (>1,300 nucleotides) or short (<400 nucleotides) circRNAs in the nucleus [52]. However, Holdt et al. recently reported that circANRIL (409 nucleotides), located in the nucleus, bound to intranuclear protein pescadillo homologue 1 (PES1) and impaired ribosome biogenesis [26]. Likewise, our results showed that circGPATCH2L (531 nucleotides) consisted of exons 3 to 8 of the GPATCH2L gene and was predominantly localised in the nucleus. According to studies, m6A methylation occurs primarily in the nucleus, and YTHDC1 (YTH domain containing 1, an m6A reader protein) may facilitates nuclear export of the m6A methylated circGPATCH2L to the cytoplasm for further degradation [53, 54]. Even so, further research is necessary to fully understand any unidentified processes governing circRNA nuclear export or retention.
In this study, shTRIM28-resistant lentiviral TRIM28 overexpression vectors were constructed to simultaneously knockdown endogenous TRIM28 and express exogenous TRIM28. Our data showed that after circGPATCH2L bound to the 402–452 aa region of TRIM28, the phosphorylation of Ser824 TRIM28 was abrogated, which hindered the downstream DNA damage response and simultaneously impeded the binding of MDM2 and TRIM28, leading to P53 accumulation and consequent NPC apoptosis. Unrepaired DNA damage triggers cell cycle arrest/senescence or programmed cell death [55]. Consequently, these senescent cells secrete proinflammatory cytokines, chemokines, and growth factors, known as the senescence-associated secretory phenotype (SASP), leading to ECM degradation and IVDD [56]. Moreover, P53 is a critical tumour suppressor factor controlling almost 500 target genes involved in cell cycle arrest, senescence, DNA damage repair, and apoptosis, among other processes [57, 58]. In addition, ATM kinase activated by DNA damage phosphorylates P53 [59], and phosphorylation of Ser392 enhanced the binding of p53 and target DNA as well as the transcriptional activity of p53 [60]. Our results showed that circGPATCH2L could lead to the phosphorylation of P53 and promote the transcription of its downstream apoptosis-related proteins, which further strengthens NPCs apoptosis and leads to consequent cellular catabolism caused by the binding of circGPATCH2L and TRIM28.
Dynamic reversible m6A modification is the most common type of internal RNA posttranscriptional modification and exhibits unprecedented spatiotemporal specificity [61]. A previous study showed that m6A-containing mRNAs and certain m6A-methylated circRNAs could be degraded by the YTHDF2-HRSP12-RNase P/MRP complex [41]. HRSP12 and YTHDF2 are bound to each other and work cooperatively to bind to m6A-methylated mRNAs. Meanwhile, RNase P/MRP-mediated cleavage was elicited by binding to HRSP12. The HRSP12 binding site and RNase P/MRP cleavage site was located upstream and downstream of the m6A site, respectively [41]. Nevertheless, the abundance of circGPATCH2L was not affected by HRSP12 in our data, likely because the “GGUUC” motif of HRSP12 is located downstream of the m6A site in circGPATCH2L. Furthermore, it is possible that conformational differences between circRNAs might account for their differential recruitment of protein complexes, together with other undefined factors, for degradation [13]. Given that m6A-containing circGPATCH2L could be endoribonucleolytically cleaved by RNase P/MRP in NPCs, we speculated that there might be a new protein bridging YTHDF2 and RNase P/MRP. After we reviewed PPI databases and our mass spectrometry data, we focused on RPL10, which we finally found to bind YTHDF2 and POP1 through subsequent experiments. Then, we demonstrated that m6A-methylated circRNAs were degraded by the YTHDF2-RPL10-RNase P/MRP complex. Our study provides groundbreaking data on a detailed mechanism of circRNA degradation in NPCs. However, how m6A methylation is regulated in the pathogenesis of IVDD requires further study.
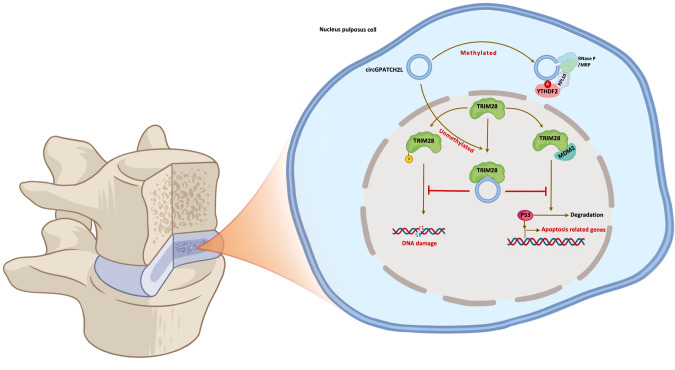
In summary, the findings presented here demonstrate that circGPATCH2L suppresses DNA damage repair and induces apoptosis by binding to TRIM28 and serves as a potentially effective therapeutic target for the treatment of IVDD (Fig. 7). Our study also highlights the functional significance of the m6A modification in regulating circGPATCH2L degradation, which provides important insights into the underlying regulation mechanism by which circRNAs maintain a steady state in NPCs.
Fig. 7. Schematic illustration for the working model of circGPATCH2L.
circGPATCH2L regulates intervertebral disc degeneration via TRIM28 and m6A-mediated circGPATCH2L degradation is elicited by the YTHDF2-RPL10-RNase P/MRP complex.
Methods and materials
Patient tissue sample collection
Degenerative intervertebral disc tissues were obtained from 10 patients with degenerative disc disease who underwent discectomy for intervertebral discs. Control intervertebral disc tissues were taken from 10 patients undergoing surgery due to thoracolumbar fracture or scoliosis. The detailed information for each patient is listed in Supplementary Table S1.
Isolation and culture of NPCs
Nucleus pulposus tissue was digested with 0.2% Pronase and 65 U/ml collagenase type 2 solution (Worthington, cat. #LS004177) in succession. The released cells were filtered through a 100 μm cell strainer and collected by centrifugation for 5 min at 500 g. Cells were then plated into culture flasks and cultured in Minimum Essential Medium α (α-MEM; Gibco, cat. #11900-073) with 10% foetal bovine serum (Corning, cat. #35-010-CV) and maintained in a humidified incubator at 37 °C with 2% O2. When grown to 80% confluence, the cells were digested with 0.05% trypsin/0.48 mM EDTA (Gibco, cat. #15400054) and passed for expansion. Cells from passages 3 to 5 were used for further experiments. To induce NP degeneration, cells were treated with TNF-α (10 ng/ml; Abcam, cat. #ab259420) for 12 h. To induce DNA damage, cells were treated with cisplatin (50 μM; Sigma, cat. #15663-27-1) for 12 h. To reduce the degradation of ubiquitin-conjugated proteins, cells were pre-treated with MG132 (5 μM; Sigma, cat. #474790) for 24 h. NPCs were identified by COL2A1 immunofluorescence staining (Supplementary Fig. S3a).
RNase R treatment
RNase R treatment was performed using Ribonuclease R (RNase R, Epicentre, cat. #RNR07250) according to the manufacturer’s recommendations. In particular, the total RNA and DNA were treated with 3 U/μg RNase R (20 U/μl) at 37 °C for 15 min [62]. The untreated RNA was incubated with DEPC-treated water without RNase R in the control group.
DNA and RNA isolation
For each tissue or cell sample, 1 ml TRI reagent (Molecular Research Center, cat. #TR-118) and 5 μL polyacryl carrier (Molecular Research Center, cat. #PC-152) were added for isolation of DNA or RNA. Total RNA or DNA was extracted using a RNeasy Mini Kit (Qiagen, cat. #74106) or DNeasy Blood & Tissue Kit (Qiagen, cat. #69504) according to the manufacturer’s instructions. Nuclear and cytoplasmic RNA were isolated with a cytoplasmic and nuclear RNA purification kit (Norgen Biotek, cat. #21000) strictly following the manufacturer’s protocol. A NanoDrop™ One spectrophotometer (Thermo Scientific) was used to monitor nucleic acid quality and quantity.
PCR and qRT–PCR
Vilo Superscript reagents (Invitrogen, cat. #11754050) were used to generate cDNA from total RNA. AmpliTaq™ DNA Polymerase (Applied Biosystems, cat. #N8080157) was used for PCR. The products were then detected by agarose gel electrophoresis and Sanger sequencing. SYBR Green PCR master mix (Applied Biosystems, cat. #4309155) was used to perform qRT–PCR. GAPDH or β-actin served as a housekeeping gene, and U6 was used as a reference for nuclear genes. Relative expression was calculated by a comparative threshold cycle (Ct) method using the formula 2−(ΔΔct) [63]. All primers are listed in Supplementary Table S7.
T3 DNA ligase assay for m6A detection
A refined single-based T3 ligase–based method was used to validate m6A sites [20]. The unmethylated adenosine site was used as a reference. DNA probe L1 and probe R1 were designed to match the flanking sequences of the m6A site, and probe L2 and probe R2 were designed to match the flanking sequences of the reference site. Ligation reaction mixture A consisted of 20 nM probe L1 or probe L2, 20 nM probe R1 or probe R2, 1× T3 ligation buffer (New England Biolabs, cat. #M0317S) and 1 mg RNase R-treated RNA. Ligation reaction mixture B consisted of an appropriate amount of T3 DNA ligase with ligation buffer. Mixture A was heated at 85 °C for 3 min and then incubated at 35 °C for 10 min, and then, ligation reaction mixture B was added to a final volume of 20 μl. This mixture was incubated at room temperature for the ligation reaction for 10 min and chilled on ice immediately. DNA probe ligation efficiency was quantified using qRT–PCR as described above. A schematic of the procedure is shown in Supplementary Fig. S2a. The specific probes and primers are listed in online Supplementary Table S7. The m6A ratio was calculated by employing the following equation:
Comet assay
Comet assay was performed according to the manufacturer’s instructions of a Comet Assay Kit (Abcam, cat. #ab238544). The Comet Assay is a single cell gel electrophoresis assay (SCGE) for the measurement of cellular DNA damage. Briefly, lysis buffer (pH 10.0), alkaline solution, and electrophoresis running solution were prepared prior to performing the assay and chilled to 4 °C thoroughly. The 37 °C liquefied comet agarose was added per well onto the Comet Slide and cooled to 4 °C for 15 min. NPCs was collected and washed once with ice-cold PBS and resuspended at 1 ×105 cells/mL in ice-cold PBS (without Mg2+ and Ca2+). The cell samples were combined with comet agarose at 1/10 ratio (v/v) and immediately transferred onto the top of the comet agarose layer for 15 min, 4 °C in the dark. For assay procedure, the agarose was electrophoresed after having been substituted to a pre-chilled alkaline electrophoresis solution for 30 mins. The treated slide was then washed three times with distilled water, substituted with 70% ethanol and dried. Diluted Vista Green DNA Dye (1:10,000) was used for staining DNA at room temperature for 15 min. A fluorescent microscope (Leica, M205 FA) was used for observation.
Western blot
Samples were lysed using RIPA buffer (Sigma, cat. #R0278) with Protease and Phosphatase Inhibitor Cocktail (Sigma, cat. #P8340, P0044) and then quantified using a Pierce™ BCA Protein Assay Kit (Thermo Scientific, cat. #23225). The total proteins isolated from cells or IP precipitates were separated by SDS–PAGE and then transferred to PVDF membranes (Millipore, cat. #IPVH00010). The membranes were blocked and incubated with the primary antibodies and HRP-conjugated secondary antibodies successively. Bands were visualised with SuperSignal West Dura Extended Duration Substrate (Thermo Scientific, cat. #34075) and were scanned using an Epson Perfection V19 Photo scanner (Epson, Japan). All antibodies are listed in Supplementary Table S6.
RNA pulldown assay
The RNA pulldown assay was modified from previous studies [64]. Briefly, 1 × 107 NPCs from the circGPATCH2L overexpression group were washed with ice-cold phosphate-buffered saline (PBS) and lysed in 500 µl RIP buffer for 5 min on ice. Cell lysates were centrifuged at 13,000 g for 10 min at 4 °C, and the supernatant was obtained. Biotinylated RNA probes were prepared using Large Scale RNA Production Systems-T7 (Promega, cat. #P1300) and a Pierce™ RNA 3′ End Desthiobiotinylation Kit (Invitrogen, cat. #20163) according to the manufacturers’ instructions. Then, the previous supernatant was incubated with probes for RNA pulldown assays using a Pierce™ Magnetic RNA–Protein Pull-Down Kit (Invitrogen, cat. #20164). Finally, the eluents were denatured in loading buffer and resolved by SDS–PAGE either for silver staining or immunodetection. Probe sequences are listed in Supplementary Table S7.
Silver staining
Equal amounts of the retrieved proteins obtained from the RNA pulldown assay were loaded in a 10% SDS–PAGE gel and separated by electrophoresis. The gel was further stained using a Pierce Silver Stain for Mass Spectrometry kit (Thermo Scientific, cat. #24600) in accordance with the manufacturer’s instructions. The specific bands were cut for mass spectrometry analysis. Proteins detected by LC–MS/MS are presented in Supplementary Table S4.
Co-IP assay
Co-IP was performed according to the manufacturer’s instructions for a Pierce™ Classic Magnetic IP/Co-IP Kit (Thermo Scientific, cat. #88804). Following transfection with exogenous expression vectors of TRIM28 and circGAPTCH2L overexpression plasmid, NPCs were harvested and homogenised in ice-cold lysis buffer (25 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% NP40 and 5% glycerol; pH 7.4) with Protease and Phosphatase Inhibitor Cocktail (Sigma, cat. #P8340, P0044). After centrifugation at 13,000 g for 10 min, the supernatant was collected, and the protein concentration was quantified. The same amount of protein in each group was mixed with 10 μg of anti-Flag IP antibody and incubated at 4 °C overnight. IgG was utilised as a negative control. Then, the mixture was added to prewashed magnetic beads and incubated at 4 °C for 6 h. The precipitates were then washed and denatured in 120 μL of Lane Marker Sample Buffer and incubated at 95 °C for 5 min. The precipitates were separated by SDS–PAGE and analysed by western blotting with TRIM28, p-TRIM28, and MDM2 antibodies. All antibodies are listed in Supplementary Table S6.
RNA immunoprecipitation (RIP) assay
RNA immunoprecipitation was performed using an Imprint® RNA Immunoprecipitation Kit (Sigma, cat. #RIP) according to the manufacturer’s instructions. YTHDF2 antibody and FLAG antibody were used to precipitate RNA. The binding RNA was detected by qRT–PCR with specific primers. All antibodies are listed in Supplementary Table S6.
Ubiquitination assay
Cisplatin pre-treated NPCs were transfected with si-circGPATCH2L or a TRIM28 overexpression plasmid before harvesting. Cells were lysed by boiling in buffer (50 mM Tris, 1% NP40, 1 mM EDTA, 150 mM NaCl, 0.5% sodium deoxycholate, 1% SDS, 1 mM PMSF, and 10 mM NaF) for 10 mins. Cell lysates were diluted ten-fold with lysis buffer without SDS and subjected to immunoprecipitation with p53 antibody. The ubiquitination of endogenous p53 was detected by western blot. All antibodies are listed in Supplementary Table S6.
LC–MS/MS
Protein bands cut from the gel were washed and then dehydrated in acetonitrile. After destaining with 50% acetonitrile and 50% 100 mM ammonium bicarbonate, the bands were digested in trypsin (13 ng/μL, 10 mM ammonium bicarbonate solution containing 10% acetonitrile) overnight at room temperature. Samples were analysed via LC–MS/MS on a Q-ExactiveTM HF Hybrid Quadrupole-Orbitrap mass spectrometer coupled to an EASY-nanoLC 1000 system (Thermo Scientific). The chromatography column used for separation was an AcclaimTM PepMapTM 100 C18 column (Thermo Scientific, 100 Å, 75 μm × 25 cm). Raw data were analysed using PEAKS studio X+ software (Bioinformatics Solutions Inc., Waterloo, Canada) based on the human Swiss-Prot database containing 20414 sequences (ver. 201907). For high confidence, the false discovery rate (FDR) was controlled below 1%, and protein identifications were accepted with a confidence score (−10lgP) > 20 for peptides and (−10lgP) > 20 for proteins, with at least 1 unique peptide per protein.
m6A-circRNA epitranscriptomic microarray and bioinformatics analysis
The sample preparation and microarray hybridisation were performed based on Arraystar’s standard protocols. Briefly, total RNA (2 μg) from the control/degenerative group was added to a mixture of 300 μL IP buffer and 2 μg anti-m6A rabbit polyclonal antibody (Synaptic Systems, Goettingen, Germany). Then, the enriched RNA was eluted with 200 μL of elution buffer at 50 °C for 1 h. The m6A-modified RNAs eluted from the magnetic beads were marked as “IP”, and unmodified RNAs in the supernatant were marked as “Sup”. The “IP” and “Sup” RNAs were treated with RNase R to eliminate linear RNAs. The remaining RNA was subsequently purified using a RNeasy MinElute Cleanup Kit (Qiagen, cat. #74204). Then, the enriched “IP” and “Sup” RNAs were labelled with Cy3 (for “Sup”) or Cy5 (for “IP”) using a Super RNA Labelling Kit (Arraystar) and purified with a RNeasy Mini Kit. The labelled RNAs were hybridised onto an Arraystar Human circRNA Epitranscriptomic Microarray (8 ×15 K, Arraystar). The microarrays were scanned with an Agilent Scanner G2505C (Agilent, Beijing, China) and analysed using Agilent Feature Extraction software (version 11.0.1.1). Differentially m6A-methylated circRNAs between the two comparison groups were identified by filtering with the fold change (|FC| ≥ 1.5) and statistical significance (p ≤ 0.05) thresholds.
Vector construction and siRNAs
A circGPATCH2L expression vector was constructed with amplified DNA fragments, including the sequence of exons 3–8, of the GPATCH2L gene, with flanking introns containing complementary Alu elements [65]. The amplified fragments were inserted into a pEX-3 (pGCMV/MCS/Neo) vector between the EcoRI and BamHI sites (GenePharma, Shanghai, China). The TRIM28 expression vector was constructed in the same way. For the mutant version of circGPATCH2L, the sequence of GGGGACTGA was replaced by GGGGTCTGA, which was verified by Sanger sequencing. For the control vector, the flanking intron sequence was synthesised and cloned into the EcoRI/BamHI-digested pEX-3 vector.
siRNAs for YTHDF2 (si-YTHDF2), HRSP12 (si-HRSP12), POP1 (si-POP1), CAND1 (si-CAND1), RPL10 (si-RPL10), CUL3 (si-CUL3), MKI67 (si-MKI67), TRIM28 (si-TRIM28) and circGPATCH2L (si-circGPATCH2L) and nonspecific control siRNA (NC) and scrambled siRNA (scramble) were purchased from GenePharma. si-circGPATCH2L targeted the back-splice junction of circGPATCH2L. NPCs were seeded in 6-well plates for 24 h and transfected with plasmids or siRNAs using Lipofectamine 3000 (Invitrogen, cat. #L3000015) according to the manufacturer’s instructions.
The lentiviral plasmid LV6 (EF1a-Puro-Amp) from GenePharma was used as a backbone for TRIM28 knockdown and complementation experiments. Briefly, U6-shRNA against TRIM28 was cloned into the XbaI/XbaI sites to create LV6-U6-shTRIM28 lentiviral vectors. Then, relevant shTRIM28-resistant 3×FLAG-TRIM28 cDNA containing silent point mutations (t931c, a934g, c937a, t940c, g943c, g946c) was ligated into the LV6-U6-shTRIM28 lentiviral vectors at the NotI/NsiI site, including a point mutation [3×FLAG-TRIM28-SS824AA(Flag-rTRIM28-MUT1)] and truncated mutations [3×FLAG-TRIM28-△510-561(Flag-rTRIM28-MUT2) and 3×FLAG-TRIM28-△402-452(Flag-rTRIM28-MUT3)]. The 3×FLAG sequence was cloned into the Notl/BamHI site of LV6 as a control. To package lentiviral particles, 293T (ATCC, cat. # CRL-3216) cells were plated at 80–90% confluence and transfected with the transfer plasmids pGag/Pol, pRev, and pVSV-G using RNAi-Mate (GenePharma, cat. #G04001). Viruses were harvested at 72 h post-transfection.
For the murine circGPATCH2L in vivo knockdown assay, the adenovirus1 (ADV1) vector from GenePharma was used. shRNA against mmu_circ_0000404 was cloned into the ADV1 vector at the EcoRI/BamHI sites. To package ADV1, 293A (Invitrogen, cat. #R70507) cells were plated at 80–90% confluence and transfected with transfer plasmid and pGP-Ad-pac vector using RNAi-Mate. Viruses were harvested 15 days post-transfection. All sequences are listed in Supplementary Table S7.
Cell transfection
For plasmid or siRNA transfection, NPCs were plated into 6-well plates for 24 h before transfection. Cells were then transfected with the indicated plasmids (4 μg per well) or siRNA (100 pmol per well) using Lipofectamine 3000 for 6 h. Two days after transfection, the cells were subjected to RNA isolation, protein isolation, or immunofluorescence analysis as described above. Lentiviral vectors were used for exogenous TRIM28 expression. NPCs were infected with lentiviral particles at a multiplicity of infection (MOI) of 50 pfu/cell for 48 h. The transduced cells were selected for at least 6 days in 1 μg/mL puromycin (Sangon Biotech, cat. #A610593) and then subjected to further study.
Immunofluorescence staining and RNA-FISH
Cy3-labelled RNA probes against the circGPATCH2L back-splice sequence were used in RNA-FISH assays. NPCs grown on glass coverslips to approximately 50% confluence were treated with plasmids for 48 h. The coverslips were then fixed, permeabilized, and prehybridized. Cells were incubated with RNA probes at 42 °C overnight. After being rinsed with saline sodium citrate (Servicebio, cat. #G3015) three times and blocked for 30 min with 1% BSA, the coverslips were incubated with relevant primary antibodies at 4 °C overnight. Subsequently, the coverslips were washed and incubated with secondary antibodies for 1 h, followed by 30 min of DAPI (Servicebio, cat. #G1012) staining. Image acquisition was performed using a confocal laser scanning microscope (Leica, TCS SP8). The probe sequences and antibodies are listed in Supplementary Tables S7 and S6.
Apoptosis assays
The rates of apoptosis were evaluated using an Annexin V-FITC/PI Apoptosis Detection Kit (KeyGEN BioTECH, cat. #KGA108) following the manufacturer’s instructions. Stained cells were analysed using flow cytometry (Beckman Coulter, CytoFLEX), and data were analysed using CytExpert software (Beckman Coulter). TUNEL assays were performed using a TUNEL BrightGreen Apoptosis Detection Kit (Vazyme, cat. #A112) according to the manufacturer’s protocol. Three random microscopic fields per slide were calculated.
Mouse model of IVDD and in vivo transduction
A total of 36 C57BL/6J mice (8 weeks old, male) from GemPharmatech Co., Ltd. were used for the experiment. Resource equation approach was used to sample size estimate [66]. The mice were randomly divided into three groups, with 12 mice in each group: the control group, IVDD + Scramble (Adv-sh-Scramble injection) group, and IVDD + sh-circGPATCH2L group. All mice were anaesthetised by gas anaesthesia using isoflurane (3% isoflurane for induction, 2% isoflurane for maintenance). Six mice in each group were sacrificed at each time point (2 and 4 weeks after the operation). The model of IVDD was established as described previously [67]. Briefly, the spine was exposed through an anterior midline transperitoneal approach, and L4/5 were punctured by a 33-gauge needle (Hamilton, Reno, NV). A total of 2 ×108 viral genome particles per mouse of Adv-sh-Scramble (5 ×1011 pfu/ml) or Adv-sh-circGPATCH2L (1 ×1011 pfu/ml) were injected into the indicated discs. After the operation, the incision was closed in a layered fashion, and the animals were allowed to move freely around their cages.
Radiography and MRI examination
All mice received radiographic and MRI examinations at the 2-week and 4-week time points post-operation. Radiography was conducted using a small animal digital X-ray scanner (Medsinglong, China). The heights of vertebral bodies and discs were measured with imaging software (ImageJ, NIH), and the disc height index (DHI) was calculated using a previously described method [45]. MRI was performed using a Magnetom Prisma 3.0T scanner (Siemens, Germany). T2-weighted sections in the median sagittal plane were obtained, and modified Pfirrmann grades were calculated for analysis [46].
Histological evaluation
All mice were euthanized via pentobarbital sodium intraperitoneal injection. The operated segments were harvested en bloc, fixed in 4% paraformaldehyde for 48 h, decalcified in 10% EDTA in PBS for 2 weeks, and then embedded in paraffin. Sections (5 μm) along the midsagittal plane were used for staining. Haematoxylin–eosin (H&E) and safranin-O/fast green staining were conducted according to standard protocols [14]. The grading score of histological staining was calculated following the criteria established by Takashi et al. [48].
Statistical analysis
Data are expressed as the mean ± S.D. Data distribution was tested using a Shapiro–Wilk test. The equality of variances was examined with a Brown–Forsythe test. To analyse normally distributed and equal variance data, ordinary one-way ANOVA with Tukey’s multiple comparison test was used for multiple groups, and an unpaired two-tailed Student’s t test was used between two groups. Ranked data were analysed using a Kruskal–Wallis test with Dunn’s multiple comparisons for multiple groups and a Mann–Whitney U test for two groups. All statistical analyses were performed with Prism version 8.0 (GraphPad, USA). Differences were considered significant at p < 0.05. Unless otherwise indicated, all in vitro experiments were performed in triplicate.
Supplementary information
Supplementary Table S1 Detailed information for each patient
Supplementary Table S2 Differentially m6A-methylated circRNAs identified by m6A-circRNA epitranscriptomic microarray
Supplementary Table S3 Differentially m6A methylated and expressed circRNAs
Supplementary Table S4 The results of liquid chromatography-tandem mass spectrometry
Supplementary Table S5 Interaction propensity of TRIM28 and circGPATCH2L predicted by catRAPID algorithm
Supplementary Table S6 Antibodies used in this study
Supplementary Table S7 All Oligonucleotides used in the study
Acknowledgements
We thank all voluntary intervertebral disc donors. We are grateful to the Central-Lab of Huashan Hospital for their excellent technical help.
Author contributions
HW, JY, and ZC designed the experiments. ZC, JS, and LX conducted the majority of the assays, acquired the data and wrote the manuscript. FZ, LX, and SM performed the animal experiments. ZC, JS, LX, and FZ analysed the data. FL, XX, GX, and CZ collected human samples and provided technical assistance. HW, JY, and FZ arranged the results and revised the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Numbers 82172490, 82102620, 81972109, 82072488) and Shanghai Science and Technology Development Funds (Numbers 23YF1404200).
Data availability
The raw data from the m6A-circRNA epitranscriptomic microarray of NPCs have been deposited in the Gene Expression Omnibus database under the accession code (GSE199993). All of the full-length original western blots are provided as supplementary material. Data supporting the present study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval
The study protocols were approved by the Ethics Committee of Fudan University, Huashan Hospital, and informed consent was obtained from each donor. The Animal Care and Use Committee of Fudan University approved the surgical interventions, treatments, and postoperative animal care procedures in this study.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhenhao Chen, Jian Song, Lin Xie.
Contributor Information
Fei Zou, Email: zoufei@huashan.org.cn.
Jianyuan Jiang, Email: jjy@fudanspine.com.
Hongli Wang, Email: wanghongli@huashan.org.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01190-5.
References
- 1.Knezevic NN, Candido KD, Vlaeyen JWS, Van Zundert J, Cohen SP. Low back pain. Lancet. 2021;398:78–92. doi: 10.1016/S0140-6736(21)00733-9. [DOI] [PubMed] [Google Scholar]
- 2.Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol. 2014;10:44–56. doi: 10.1038/nrrheum.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartvigsen J, Hancock MJ, Kongsted A, Louw Q, Ferreira ML, Genevay S, et al. What low back pain is and why we need to pay attention. Lancet. 2018;391:2356–67. doi: 10.1016/S0140-6736(18)30480-X. [DOI] [PubMed] [Google Scholar]
- 4.Zhou M, Wang H, Zeng X, Yin P, Zhu J, Chen W, et al. Mortality, morbidity, and risk factors in China and its provinces, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2019;394:1145–58. doi: 10.1016/S0140-6736(19)30427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams MA, Freeman BJ, Morrison HP, Nelson IW, Dolan P. Mechanical initiation of intervertebral disc degeneration. Spine. 2000;25:1625–36. doi: 10.1097/00007632-200007010-00005. [DOI] [PubMed] [Google Scholar]
- 6.Vergroesen PP, Kingma I, Emanuel KS, Hoogendoorn RJ, Welting TJ, van Royen BJ, et al. Mechanics and biology in intervertebral disc degeneration: a vicious circle. Osteoarthr Cartil. 2015;23:1057–70. doi: 10.1016/j.joca.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Nasto LA, Roughley P, Leme AS, Houghton AM, Usas A, et al. Spine degeneration in a murine model of chronic human tobacco smokers. Osteoarthr Cartil. 2012;20:896–905. doi: 10.1016/j.joca.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakai D, Nakamura Y, Nakai T, Mishima T, Kato S, Grad S, et al. Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nat Commun. 2012;3:1264. doi: 10.1038/ncomms2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–47. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Yujiao W, Fang W, Linhui Y, Ziqi G, Zhichen W, et al. The roles of miRNA, lncRNA and circRNA in the development of osteoporosis. Biol Res. 2020;53:40. doi: 10.1186/s40659-020-00309-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Tian L, Li J, Li Y, Du L, Huo Z, et al. The roles of circRNAs in intervertebral disc degeneration: inflammation, extracellular matrix metabolism, and apoptosis. Anal Cell Pathol. 2022;2022:9550499. doi: 10.1155/2022/9550499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Chen X, Xu D, Li S, Chan MTV, Wu WKK. Circular RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Prolif. 2019;52:e12704. doi: 10.1111/cpr.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–90. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 14.Cheng X, Zhang L, Zhang K, Zhang G, Hu Y, Sun X, et al. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis. 2018;77:770–9. doi: 10.1136/annrheumdis-2017-212056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74. doi: 10.1038/s41392-020-00450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Gong W, Shao X, Shi T, Zhang L, Dong J, et al. METTL3-mediated m(6)A modification of ATG7 regulates autophagy-GATA4 axis to promote cellular senescence and osteoarthritis progression. Ann Rheum Dis. 2022;81:87–99. doi: 10.1136/annrheumdis-2021-221091. [DOI] [PubMed] [Google Scholar]
- 17.Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608–24. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 18.Zhu B, Chen HX, Li S, Tan JH, Xie Y, Zou MX, et al. Comprehensive analysis of N6-methyladenosine (m(6)A) modification during the degeneration of lumbar intervertebral disc in mice. J Orthop Translat. 2021;31:126–38. doi: 10.1016/j.jot.2021.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie L, Huang W, Fang Z, Ding F, Zou F, Ma X, et al. CircERCC2 ameliorated intervertebral disc degeneration by regulating mitophagy and apoptosis through miR-182-5p/SIRT1 axis. Cell Death Dis. 2019;10:751. doi: 10.1038/s41419-019-1978-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W, Yan J, Zhang Z, Pian H, Liu C, Li Z. Identification of a selective DNA ligase for accurate recognition and ultrasensitive quantification of N(6)-methyladenosine in RNA at one-nucleotide resolution. Chem Sci. 2018;9:3354–9. doi: 10.1039/C7SC05233B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu M, Wang Q, Shen J, Yang BB, Ding X. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019;16:899–905. doi: 10.1080/15476286.2019.1600395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–70. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia P, Wang S, Ye B, Du Y, Li C, Xiong Z, et al. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity. 2018;48:688–701.e687. doi: 10.1016/j.immuni.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Czerwińska P, Mazurek S, Wiznerowicz M. The complexity of TRIM28 contribution to cancer. J Biomed Sci. 2017;24:63. doi: 10.1186/s12929-017-0374-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misir S, Wu N, Yang BB. Specific expression and functions of circular RNAs. Cell Death Differ. 2022;29:481–91. doi: 10.1038/s41418-022-00948-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Lu H, Chen C, Lyu Y, Cole RN, Semenza GL. HIF-1 Interacts with TRIM28 and DNA-PK to release paused RNA polymerase II and activate target gene transcription in response to hypoxia. Nat Commun. 2022;13:316. doi: 10.1038/s41467-021-27944-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White D, Rafalska-Metcalf IU, Ivanov AV, Corsinotti A, Peng H, Lee SC, et al. The ATM substrate KAP1 controls DNA repair in heterochromatin: regulation by HP1 proteins and serine 473/824 phosphorylation. Mol Cancer Res. 2012;10:401–14. doi: 10.1158/1541-7786.MCR-11-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Burton EM, Bhaduri-McIntosh S. Chloroquine triggers Epstein-Barr virus replication through phosphorylation of KAP1/TRIM28 in Burkitt lymphoma cells. PLoS Pathog. 2017;13:e1006249. doi: 10.1371/journal.ppat.1006249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Copp ME, Chubinskaya S, Bracey DN, Shine J, Sessions G, Loeser RF, et al. Comet assay for quantification of the increased DNA damage burden in primary human chondrocytes with aging and osteoarthritis. Aging Cell. 2022;21:e13698. doi: 10.1111/acel.13698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin JO, Lee GD, Nam SH, Lee TH, Kang DH, Yun JK, et al. Sequential ubiquitination of p53 by TRIM28, RLIM, and MDM2 in lung tumorigenesis. Cell Death Differ. 2021;28:1790–803. doi: 10.1038/s41418-020-00701-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han Y, Zhou CM, Shen H, Tan J, Dong Q, Zhang L, et al. Attenuation of ataxia telangiectasia mutated signalling mitigates age-associated intervertebral disc degeneration. Aging Cell. 2020;19:e13162. doi: 10.1111/acel.13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan G, Yang S, Yang S. Macrophage RGS12 contributes to osteoarthritis pathogenesis through enhancing the ubiquitination. Genes Dis. 2022;9:1357–67. doi: 10.1016/j.gendis.2021.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yee KS, Wilkinson S, James J, Ryan KM, Vousden KH. PUMA- and Bax-induced autophagy contributes to apoptosis. Cell Death Differ. 2009;16:1135–45. doi: 10.1038/cdd.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livi CM, Klus P, Delli Ponti R, Tartaglia GG. catRAPID signature: identification of ribonucleoproteins and RNA-binding regions. Bioinformatics. 2016;32:773–5. doi: 10.1093/bioinformatics/btv629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McAvera RM, Crawford LJ. TIF1 proteins in genome stability and cancer. Cancers. 2020;12:2094. [DOI] [PMC free article] [PubMed]
- 40.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–6. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 41.Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol Cell. 2019;74:494–507.e498. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 42.Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47:D529–d541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Del Toro N, Shrivastava A, Ragueneau E, Meldal B, Combe C, Barrera E, et al. The IntAct database: efficient access to fine-grained molecular interaction data. Nucleic Acids Res. 2022;50:D648–d653. doi: 10.1093/nar/gkab1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo W, Mu K, Zhang B, Sun C, Zhao L, Li HR, et al. The circular RNA circ-GRB10 participates in the molecular circuitry inhibiting human intervertebral disc degeneration. Cell Death Dis. 2020;11:612. doi: 10.1038/s41419-020-02882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masuda K, Aota Y, Muehleman C, Imai Y, Okuma M, Thonar EJ, et al. A novel rabbit model of mild, reproducible disc degeneration by an anulus needle puncture: correlation between the degree of disc injury and radiological and histological appearances of disc degeneration. Spine. 2005;30:5–14. doi: 10.1097/01.brs.0000148152.04401.20. [DOI] [PubMed] [Google Scholar]
- 46.Griffith JF, Wang YX, Antonio GE, Choi KC, Yu A, Ahuja AT, et al. Modified Pfirrmann grading system for lumbar intervertebral disc degeneration. Spine. 2007;32:E708–712. doi: 10.1097/BRS.0b013e31815a59a0. [DOI] [PubMed] [Google Scholar]
- 47.Pfirrmann CW, Metzdorf A, Zanetti M, Hodler J, Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine. 2001;26:1873–8. doi: 10.1097/00007632-200109010-00011. [DOI] [PubMed] [Google Scholar]
- 48.Ohnishi T, Sudo H, Iwasaki K, Tsujimoto T, Ito YM, Iwasaki N. In vivo mouse intervertebral disc degeneration model based on a new histological classification. PLoS One. 2016;11:e0160486. doi: 10.1371/journal.pone.0160486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang F, Lin F, Xu Z, Huang Z. Circular RNA ITCH promotes extracellular matrix degradation via activating Wnt/β-catenin signaling in intervertebral disc degeneration. Aging. 2021;13:14185–97. doi: 10.18632/aging.203036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188–206. doi: 10.1038/s41571-021-00585-y. [DOI] [PubMed] [Google Scholar]
- 51.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 52.Huang C, Liang D, Tatomer DC, Wilusz JE. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639–44. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 54.Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017;6:e31311. [DOI] [PMC free article] [PubMed]
- 55.Ou HL, Schumacher B. DNA damage responses and p53 in the aging process. Blood. 2018;131:488–95. doi: 10.1182/blood-2017-07-746396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang S, Zhang F, Ma J, Ding W. Intervertebral disc ageing and degeneration: The antiapoptotic effect of oestrogen. Ageing Res Rev. 2020;57:100978. doi: 10.1016/j.arr.2019.100978. [DOI] [PubMed] [Google Scholar]
- 57.Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018;25:104–13. doi: 10.1038/cdd.2017.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han CW, Lee HN, Jeong MS, Park SY, Jang SB. Structural basis of the p53 DNA binding domain and PUMA complex. Biochem Biophys Res Commun. 2021;548:39–46. doi: 10.1016/j.bbrc.2021.02.049. [DOI] [PubMed] [Google Scholar]
- 59.Reinhardt HC, Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet. 2012;28:128–36. doi: 10.1016/j.tig.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dai Z, Li G, Chen Q, Yang X. Ser392 phosphorylation modulated a switch between p53 and transcriptional condensates. Biochim Biophys Acta Gene Regul Mech. 2022;1865:194827. doi: 10.1016/j.bbagrm.2022.194827. [DOI] [PubMed] [Google Scholar]
- 61.Anders M, Chelysheva I, Goebel I, Trenkner T, Zhou J, Mao Y, et al. Dynamic m(6)A methylation facilitates mRNA triaging to stress granules. Life Sci Alliance. 2018;1:e201800113. doi: 10.26508/lsa.201800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verduci L, Ferraiuolo M, Sacconi A, Ganci F, Vitale J, Colombo T, et al. The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription-competent complex. Genome Biol. 2017;18:237. doi: 10.1186/s13059-017-1368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 64.Ghosh A, Pandey SP, Ansari AH, Sundar JS, Singh P, Khan Y, et al. Alternative splicing modulation mediated by G-quadruplex structures in MALAT1 lncRNA. Nucleic Acids Res. 2022;50:378–96. doi: 10.1093/nar/gkab1066. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–47. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 66.Arifin WN, Zahiruddin WM. Sample size calculation in animal studies using resource equation approach. Malays J Med Sci. 2017;24:101–15. doi: 10.21315/mjms2017.24.5.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang H, Ma SY, Feng G, Shen FH, Joshua Li X. Therapeutic effects of adenovirus-mediated growth and differentiation factor-5 in a mice disc degeneration model induced by annulus needle puncture. Spine J. 2010;10:32–41. doi: 10.1016/j.spinee.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1 Detailed information for each patient
Supplementary Table S2 Differentially m6A-methylated circRNAs identified by m6A-circRNA epitranscriptomic microarray
Supplementary Table S3 Differentially m6A methylated and expressed circRNAs
Supplementary Table S4 The results of liquid chromatography-tandem mass spectrometry
Supplementary Table S5 Interaction propensity of TRIM28 and circGPATCH2L predicted by catRAPID algorithm
Supplementary Table S6 Antibodies used in this study
Supplementary Table S7 All Oligonucleotides used in the study
Data Availability Statement
The raw data from the m6A-circRNA epitranscriptomic microarray of NPCs have been deposited in the Gene Expression Omnibus database under the accession code (GSE199993). All of the full-length original western blots are provided as supplementary material. Data supporting the present study are available from the corresponding author upon reasonable request.