SUMMARY
Retinosomes are intracellular lipid bodies found in the retinal pigment epithelium (RPE). They contain retinyl esters (REs) and are thought to be involved in visual chromophore regeneration during dark adaptation and in case of chromophore depletion. However, key enzymes in chromophore regeneration, retinoid isomerase (RPE65), and lecithin:retinol acyltransferase (LRAT) are located in the endoplasmic reticulum (ER). The mechanism and the enzyme responsible for mobilizing REs from retinosomes remained unknown. Our study demonstrates that patatin-like phospholipase domain containing 2 (PNPLA2) mobilizes all-trans-REs from retinosomes. The absence of PNPLA2 in mouse eyes leads to a significant accumulation of lipid droplets in RPE cells, declined electroretinography (ERG) response, and delayed dark adaptation compared with those of WT control mouse. Our work suggests a function of PNPLA2 as an RE hydrolase in the RPE, mobilizing REs from lipid bodies and functioning as an essential component of the visual cycle.
Graphical Abstract

In brief
Hara et al. identify the PNPLA2 enzyme that mobilizes vitamin A from the retinosomes in the RPE to serve as the substrate for the isomerase RPE65, an essential enzyme for the regeneration of visual pigment chromophore and maintenance of vision.
INTRODUCTION
Vitamin A is essential for normal vision as an integral component of visual pigments, which consist of a chromophore, 11-cis-retinal linked via a Schiff base to the lysine residue of opsin.1,2 The initial event in the visual signal transduction is the photoisomerization of the 11-cis-retinal to all-trans-retinal, leading to the activation of G-protein coupled receptor (GPCR) opsin, which triggers the cascade of the signal transduction. The chromophore regeneration process after photobleaching is termed the visual cycle or retinoid cycle.3 All-trans-retinal dissociates from opsin, is reduced to all-trans-retinol by all-trans-retinol dehydrogenases (atRDHs) and transported to RPE cells, which generate the visual pigment chromophore 11-cis-retinal. All-trans-retinol is also delivered to RPE cells through signaling receptor and transporter of retinol stimulated by retinoic acid 6 (STRA6)4-6 from the circulation carried by a specific transport protein retinol-binding protein 4 (RBP4). After uptake of all-trans-retinol into RPE cells, lecithin:retinol acyltransferase (LRAT) esterifies all-trans-retinol by transferring a fatty acyl moiety from the membrane phosphatidylcholine to form retinyl esters.7,8 RPE65 hydrolyzes and isomerizes all-trans-RE to 11-cis-retinol,9-11 which is subsequently converted to 11-cis-retinal by 11-cis-retinol dehydrogenases (11cRDHs) in RPE cells. Finally, 11-cis-retinal is transported to photoreceptors and reattaches to apo-opsin to form light-sensitive rhodopsin and cone visual pigments.
RPE cells carry out multiple functions, including recycling, and storage of vitamin A in the form of RE. All-trans-REs are the substrate for RPE6512,13 to be hydrolyzed and isomerized to 11-cis-retinol, a critical step in the visual cycle. However, most of the RE pool does not directly participate in the isomerization.14 Excess REs are stored in the specialized storage particles termed retinosomes.15 Retinosomes contain a high level of REs and other lipids such as triglycerides, cholesterol, cholesteryl esters, and phospholipids.16 The proteome analysis of isolated lipid droplets from bovine RPE identified the presence of comparative gene identification 58 protein (CGI-58) and lipid droplet-associated proteins, perilipins (PLINs).16 REs are a highly hydrophobic compound insoluble in water; thus, REs in retinosomes cannot be released into the cytosol. Therefore, retinyl ester hydrolase (REH) activity is required to convert RE to all-trans-retinol, which is released from retinosomes and transported to the endoplasmic reticulum (ER) where RPE65 is located. RPE65 is the only known enzyme in the RPE that hydrolyzes and isomerizes RE to generate 11-cis-retinol.11 However, RPE65 resides in the ER and does not colocalize with retinosomes15; therefore RPE65 cannot access its substrate, REs stored in retinosomes. The mechanism for the release of retinoids from retinosomes and the enzyme responsible for mobilizing REs remained unknown.
Patatin-like phospholipase domain containing 2 (PNPLA2) is a key enzyme for lipolysis by catalyzing the first step of triglyceride hydrolysis in adipose, non-adipose tissues, and lipid droplets, yielding diacylglycerides (DAG) and fatty acids.17 Recently, PNPLA2 was found to be associated with lipid droplets in the hepatic stellate cells (HSCs) and to possess REH activity in vitro.18 However, its ablation did not increase RE levels in the liver, and thus, its function as REH in vivo was uncertain.
Four independent groups reported PNPLA2 under the different names, desnutrin, induced phospholipase A2ζ (iPLA2ζ)/transport secretion protein 2.2 (TTS-2.2), adipose triglyceride lipase (ATGL), and a receptor of pigment epithelium-derived factor (PEDF-R).17,19-21 Notari and colleagues showed that PNPLA2 is expressed in rodent ocular tissues and functions as a receptor of PEDF.21 PEDF is a neurotrophic and neuroprotective protein that protects photoreceptors from degeneration.22 It was reported that PEDF binding stimulates the phospholipase (PLA2) activity of PNPLA2, which releases the free fatty acid, linoleic acid, from the substrate, 1,2-dilinoleoyl-phosphatidylcholine.21 The function has been implicated in the degradation of rod outer segments by RPE cells during phagocytosis.23
In the present study, we report a function of PNPLA2 as an REH in the RPE. To better understand the role of PNPLA2 in the visual cycle, we employed a Pnpla2 knockout (KO) mouse model to analyze the subcellular localization of PNPLA2, and its role in the visual cycle, retinal function, and chromophore regeneration. We also analyzed retinoid profiles, focusing on REs and 11-cis-retinal regenerations in dark adaptation in Pnpla2 KO mouse eyes. In addition, we overexpressed PNPLA2 and its coactivator CGI-58 and used small interfering RNA (siRNA)-mediated knockdown of endogenous PNPLA2 gene in HEK293A cells to study the REH activity using in vitro assay. Moreover, liposome-based activity assay was used to verify REH activity of PNPLA2. The results demonstrated that expression of PNPLA2 promoted RPE65 isomerase activity for 11-cis-retinol production. These observations suggest that REH activity of PNPLA2 is essential for efficient mobilization of REs from lipid bodies in RPE cells and its possible contribution to the visual cycle.
RESULTS
PNPLA2 expression and distribution in ocular tissues
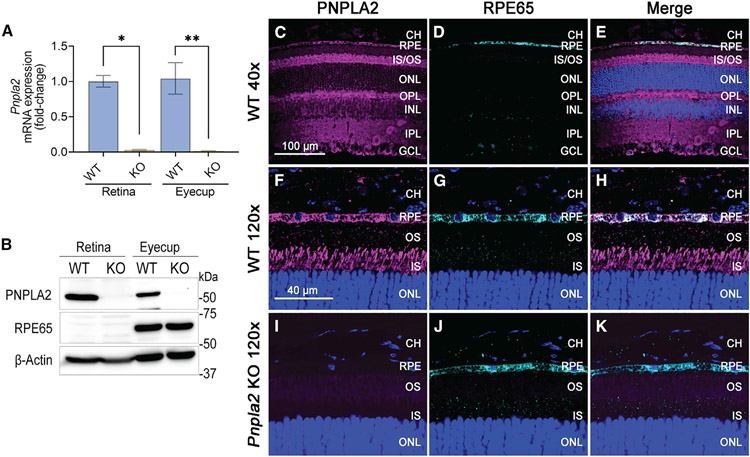
We first evaluated Pnpla2 mRNA and protein expression in the eyes of wild-type (WT) and Pnpla2 KO mice by qRT-PCR and western blot analysis. Figure 1A shows that prominent levels of the Pnpla2 mRNA were detected in the RPE and the retina of WT mice (Figure S1A). In Pnpla2 KO mice, we confirmed the absence of the Pnpla2 mRNA expression in both the retina and eyecups (Figures 1A and S1B). Western blot analysis detected PNPLA2 protein (54 kDa) in both the retinas and eyecups of WT mice, but not in those of Pnpla2 KO mice (Figure 1B). As expected, RPE65 (65 kDa), an RPE cell marker,24 was detected only in the eyecups in both WT and Pnpla2 KO mice (Figure 1B).
Figure 1. PNPLA2 expression and cellular localization in the retina and RPE.
(A) Pnpla2 mRNA expression in retina and eyecup (RPE and choroid) of WT and Pnpla2 KO mice at the age of 4 months was determined by qRT-PCR. Data are shown as relative fold to WT (mean ± SEM). Statistical analysis was performed using an unpaired two-tailed t test. *p < 0.05, **p < 0.01 vs. WT group (n = 3 per group).
(B) Western blot analysis of mouse retina and eyecup detecting PNPLA2 and RPE65.
(C–K) Immunofluorescence microscopy of mouse retina cross-sections from 3-month-old WT (C–H) and Pnpla2 KO (I–K) mice. Sections were immunostained with antibodies against PNPLA2 (magenta) and RPE65 (cyan), and nuclei counterstained with DAPI (blue). Immunolocalization of PNPLA2 and RPE65 in the WT (F–H) and Pnpla2 KO (I–K) mice retina and RPE. Merge indicates superimposed images of staining of PNPLA2, RPE65, and DAPI. CH, choroid; GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; IS/OS, photoreceptor inner and outer segments; ONL, outer nuclear layer; OPL, outer plexiform layer; RPE, retinal pigment epithelium.
Scale bars, 100 μm (C–E); 40 μm (F–K). See also Figure S1.
In addition, to analyze the cellular localization of PNPLA2 in the retina and RPE, we performed immunohistochemical analysis of mouse eyes. Immunostaining of cryosections of the retinal tissues from a WT mouse showed that PNPLA2 (magenta) was expressed throughout the retinal layers (Figure 1C). The immunostained RPE65 (cyan) was confined exclusively in the RPE layer (Figure 1D). PNPLA2 was also expressed in the RPE layer (Figure 1E). We examined the immunostained retinal sections between the choroid (CH) and the outer nuclear layer of photoreceptors (ONL) with higher magnification in WT (Figures 1F-1H) and Pnpla2 KO mice (Figures 1I-1K). The high intensity of the PNPLA2 signal was observed in the RPE and the inner segment of the photoreceptor (IS), but not in the outer segment (OS) or the outer nuclear layer of photoreceptors (ONL) (Figure 1F). RPE65 was observed in the RPE layer (Figure 1G). A merged image shows the signals of RPE65 overlapped with PNPLA2 observed as a white color in the RPE layer (Figure 1H). In contrast, PNPLA2 immunolabeling was virtually absent in retinal sections of Pnpla2 KO mice (Figure 1I). RPE65 was only observed in the RPE layer, the same as WT (Figures 1J and 1K). These observations indicate that PNPLA2 is expressed in RPE cells and throughout the retinal layers but not in the OS or ONL of photoreceptors.
Pnpla2 KO mice showed impaired visual function compared with WT mice
To evaluate whether the absence of PNPLA2 affects retinal function in Pnpla2 KO mice, we performed electroretinography (ERG) recording and compared it with that of age-matched WT mice at the age of postnatal day 65 (PD65). Figure 2A shows the representative scotopic ERG waveforms of a fully dark-adapted WT (left column) and a Pnpla2 KO mouse (right column) elicited by increasing intensities of light flashes. Figures 2B and 2C summarize light intensity responses of ERG scotopic a-wave and b-wave, respectively. In both groups, the amplitudes of a-wave responses were proportional to the increasing light stimulus intensity. The amplitude of a-wave of Pnpla2 KO was significantly lower than that of age-matched WT mice at all tested intensities. The b-wave amplitudes of WT mice reached the maximum at the intensity of 1.3 log (cd·s/m2) and did not increase further following higher intensity light stimulations. In contrast, Pnpla2 KO mice showed significantly lower responses relative to WT mice, which steadily increased with increasing light intensities. For photopic ERGs, mice were light-adapted by the steady, rod-saturating background light of 2.8 log (cd/m2) for 7 min. Figure 2D shows the representative photopic ERG waveforms of a WT mouse and a Pnpla2 KO mouse. Figure 2E shows that the photopic b-wave amplitudes of Pnpla2 KO mice were significantly lower than that of WT. ERG data suggest that rod and cone functions in Pnpla2 KO mice are significantly depressed compared with those of age-matched WT.
Figure 2. ERG response of dark-adapted WT and Pnpla2 KO mice.
(A) Representative scotopic responses were obtained from a WT (left column) and a Pnpla2 KO (right column) mouse at PD65. Scotopic responses were elicited by a series of single flashes with increasing light intensities indicated with the log in the panel. Vertical lines crossing each trace at 0 msec are the timing of the stimulus flash. a- and b-waves are marked in the panel.
(B and C) Comparison of scotopic a- and b-wave amplitudes with increasing light stimulation in WT and Pnpla2 KO mice. The amplitude of a-wave is converted to positive values.
(D) Representative photopic waves of a WT and a Pnpla2 KO mouse.
(E) Comparison of photopic b-wave amplitudes of WT and Pnpla2 KO mice. All data are presented as mean ± SEM. Statistical analysis was performed using an unpaired two-tailed t test. *p < 0.05, **p < 0.01, ***p < 0.001 (n = 7) vs. WT group.
Lipid accumulation in RPE cells of Pnpla2 KO mice
To determine the function of PNPLA2 in RPE cells, we examined lipid accumulation and their subcellular distribution in the RPE of WT and Pnpla2 KO mice at the age of PD100. We first detected the uptake of BODIPY493/503 by the lipid droplets (green) in the RPE flat mount. Zonula occludens (ZO)-1 (blue) was immunostained to view RPE cell boundary with the nuclei counterstained with DAPI (blue) (Figures 3Aa-3a′ and 3Ab-3b′). We observed a significant accumulation of lipid droplets and increased size of lipid droplets in RPE cells in Pnpla2 KO mice (Figure 3A) as compared with that of age-matched WT mice. The merged images showed that lipid droplet accumulation in Pnpla2 KO mice was prominent along the lateral cell boundary of RPE cells (Figures 3Ab-3b′).
Figure 3. Pnpla2 KO mice have impaired visual chromophore regeneration.
(A) Confocal microscopy of RPE flatmount from WT and Pnpla2 KO mice at the age of PD100 (a and b) with lipid droplets stained with BODIPY (green) and the nuclei counterstained with DAPI (blue) and ZO-1 (blue) (a’ and b’). Scale bar, 40 μm.
(B) Scotopic ERG a-wave recoveries after photobleaching with 1,000 cd/m2 light for 2 min. Recovery data were acquired at every 5-min intervals with a scotopic single flash of 10 cd s/m2 stimuli for up to 60 min. A-wave recoveries are presented as percentage amplitudes. WT (n = 6). Pnpla2 KO (n = 5). (C and D) Comparison of retinoid profiles in mouse eyes during the recovery. The amounts of RE and 11-cis-retinol (picomole/eye) were quantified in WT and Pnpla2 KO mice with dark-adapted overnight (n = 4), light-adapted (n = 4), 15-min recovery after light bleach (n = 8), and 40-min recovery (n = 8) groups by HPLC. All data are presented as mean ± SEM. Statistical analysis was performed using an unpaired t test with Welch’s correction. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. WT group. See also Figures S2 and S3.
We also examined the RE accumulation in the fully dark-adapted and light-adapted RPE of the WT and Pnpla2 KO mice. REs-specific autofluorescence was compared among Rpe65 KO, WT, and Pnpla2 KO mice. An Rpe65 KO mouse at the age of 8 months was used as a positive RE accumulation model based on previous reports.25-27 Figures S2A-S2C show representative fluorescence images of the RPE of those mice. REs-specific autofluorescence clearly increased in the light-adapted mice as expected (Figures S2B and S2C). Relative fluorescence was measured and compared with WT. We observed significant fluorescence increase in the dark-adapted Pnpla2 KO mice compared with WT (Figure S2D), indicating that the dark-adapted Pnpla2 KO mice accumulated REs even more than WT. As expected, the light-adapted animals had higher fluorescence than those dark-adapted.
To examine the localization of PNPLA2 in RPE, we used immunostaining of the ex vivo retinol-oleate-treated RPE flatmounts of Rpe65 KO and WT mice (Figures S3A and S3B). PNPLA2 (green) signals were observed surrounding the surface of the retinosomes (blue). The overlapping of lipid droplets and retinosomes appeared in pink in the merged images in Figure S3A and S3B. In WT RPE flatmount, smaller lipid droplets and retinosomes were produced (Figure S3B). An enlarged image from the Rpe65 KO RPE demonstrated the PNPLA2 localization on the retinosomes (Figure S3C).
Delayed dark adaptation of visual sensitivity in Pnpla2 KO mice following visual pigment photobleach
To determine if PNPLA2 deletion affects the recovery of photoreceptor function after the photobleach, we measured the time course of a-wave recovery trace in the dark adaptation and compared it with that in WT mice. All mice were fully dark-adapted before ERG recording and then exposed to the rhodopsin-bleaching light (1,000 cd/m2) for 2 min and returned to the dark, followed by single flash (10 cd s/m2) responses every 5 min for up to 1 h. Figure 3B shows the recoveries of a-wave amplitudes plotted as percentage of maximum amplitudes, and the mean of each time point was calculated relative to the mean of the initial dark-adapted a-wave value (mean, WT = 216.5 μV, KO = 161.5 μV). The a-wave of Pnpla2 KO mice showed 66% recovery while the a-wave of WT mice reached 100% complete recovery at 60 min dark adaptation. At all the time points between 20 min and 60 min, the rates of a-wave recovery were significantly different between genotypes. The a-wave tracing of Pnpla2 KO mice revealed a significantly slower a-wave recovery relative to the WT mice (Figure 3B).
Impaired visual chromophore regeneration in Pnpla2 KO mice
Next, we investigated if the rate of 11-cis-retinal synthesis is impacted in Pnpla2 KO mice because the deficient synthesis of 11-cis-retinal is known to contribute to delayed dark adaptation. WT and Pnpla2 KO mice were subjected to a bleaching condition (5,000 lux for 30 min), followed by 15 min and 40 min of dark adaptation. We compared the amount of REs and 11-cis-retinal in the eyes between WT and Pnpla2 KO mice. HPLC quantification revealed that the level of REs was significantly higher in Pnpla2 KO mice in all the tested groups (Figure 3C). The amount of 11-cis-retinal in the eyes was similar in both fully dark- and light-adapted groups of Pnpla2 KO and WT mice (Figure 3D). However, levels of 11-cis-retinal after 15 min and 40 min of post-bleach dark adaptation were significantly lower in Pnpla2 KO mice compared with WT mice (Figure 3D). This result suggests that delayed dark adaptation of ERG a-wave in Pnpla2 KO mice is most likely due to the lower rate of 11-cis-retinal regeneration.
PNPLA2 and CGI-58 are associated with cellular lipid droplets
REs are mostly stored in lipid storage particles, retinosomes, in RPE cells. It is thought that they serve to replenish the chromophore.15,16 Because PNPLA2 is a lipid droplet-associated protein in hepatocytes, it is likely to localize in the retinosomes. To examine the expression of PNPLA2 in relation to the lipid droplets, HEK293A cells were treated with 400 μM oleic acid overnight to increase the formation of the lipid droplets, followed by transient plasmid transfection of PNPLA2, CGI-58, and RPE65 for another 24 h. Under this condition, the cells displayed lipid droplet (red) accumulation in their cytosols (Figures 4A-4D). We examined the subcellular localization of PNPLA2, PLIN2, CGI-58, and RPE65 using immunofluorescence. Cells were costained with LipidTox Red dye for neutral lipid droplet and 488-Alexa-flour fluorescent-labeled antibodies for respective proteins. First, we detected PLIN2, an abundant lipid droplet-associated protein28,29 primarily in the large lipid droplets (yellow) (Figure 4A). RPE65 showed a subcellular localization distinct from PLIN2. RPE65 is an ER-associated protein30,31; as expected, RPE65 staining was detected around the nucleus and formed small and irregular shapes spreading outward from the nucleus. The merged image revealed that no RPE65 signal overlaps with lipid droplets (Figure 4B). In contrast, the immunostaining of PNPLA2 showed a ring pattern overlapping with PLIN2 signal, suggesting colocalization of PNPLA2 with lipid droplets (Figure 4C). Multiple studies have shown that PNPLA2 is associated with a coactivator CGI-58 in the lipid droplets.32-35 As expected, CGI-58 and PNPLA2 were both detected predominantly in lipid droplets (Figures 4C and 4D). The intensity profiles were plotted by the distance between fluorescent signals of proteins of interest and lipid droplets on the z axis along the dotted line depicted in the merged image. The enlarged regions used for the profile analysis are shown in the upper right corner of each plot (Figures 4E-4H). Immunostaining of PLIN2, PNPLA2, and CGI-58 showed colocalization with lipid droplets (Figures 4E, 4G, and 4H). In contrast, RPE65 immunostaining showed no overlap with lipid droplets (Figure 4F).
Figure 4. PNPLA2 and CGI-58 are associated with cellular lipid droplets.
(A–D) Immunofluorescence microscopy of HEK293A cells treated with 400 μM oleic acid for 24 h followed by infection with indicated adenoviral expression vectors. Lipid droplets were stained with LipidTox Red (red). Cells were stained with indicated antibodies for the PLIN2 (A), RPE65 (B), PNPLA2 (C), or CGI-58 (D). Scale bar, 5 μm.
(E–H) RGB profile plot of IF microscopy images from (A–D). Dotted line in merged panels indicates the scan position selected for a plot. Lipid droplets (red); lipid droplet-associated proteins (green).
(I) Western blot analysis of sucrose density gradient fractionation of oleate-treated HEK293A cells with transiently expressed proteins of interest and endogenous lipid droplet-associated proteins. Fractions: Input (total lysate); pellet (cell pellet); Cyto, cytosol; LD, lipid droplet.
Next, to further examine the association of proteins to the lipid droplets and other organelles, we isolated lipid droplets from the transfected cells. Cell lysates were fractionated by density gradient centrifugation, collected by the layer, and analyzed by western blotting to compare total lysate (Input), cell pellet (Pellet), cytosol (Cyto), and purified lipid droplets (LDs) (Figure 4I). These gradient fractions were immunoblotted for lysosomal-associated membrane protein 1 (LAMP1),36 Calnexin, an integral membrane protein of the ER,37 and PLIN2 and PLIN3, LD marker proteins,38 and transiently expressed proteins of interest, PNPLA2, CGI-58, and RPE65. Among three cellular fractions and the total lysate (input), the LD fraction was distinct from others and showed all lipid droplet-associated proteins PLIN2, PLIN3,39 PNPLA2, and CGI-58. PLIN2 was found exclusively in the LD fraction. PLIN3, PNPLA2, and CGI-58 were abundant in the LD fraction. In contrast, RPE65, LAMP1, and calnexin were not detected in the LD fraction. RPE65 was predominantly detected in the cytosol and pellet fractions containing ER (Figure 4I). These results confirmed that PNPLA2 and GCI-58 are lipid droplet-associated proteins, whereas RPE65 was not associated with LDs.
REH activity of PNPLA2 is enhanced by CGI-58
Several studies showed that CGI-58 is a coactivator of PNPLA2 and specifically enhances its activity in different tissues and cells.17,32,40 Therefore, we examined the effect of CGI-58 on the REH activity of PNPLA2. We generated adenoviruses expressing human CGI-58 (Ad-CGI-58) and human PNPLA2 (Ad-PNPLA2) and introduced them into HEK293A cells pretreated with all-trans-retinol and oleic acid for 24 h. REH activity was measured by HPLC analysis of the RE (substrate of PNPLA2) levels.
Figure 5A shows representative retinoid profiles extracted from HEK293A cells infected with null adenovirus (Adv) as a negative control, Ad-CGI-58 alone, and Ad-PNPLA2 alone, and coinfected with Ad-CGI-58 and Ad-PNPLA2. Western blot analysis of the examined groups demonstrated that all the infected cells expressed the proteins of interest (Figure 5B). The retinoid quantification showed the highest RE level (629 pmol/h) in the cells infected by control Adv. Lysates from cells infected with Ad-CGI-58 had a 26.3% decrease (464 pmol/mg of protein) of RE relative to Adv control. Lysates from cells infected with Ad-PNPLA2 showed a reduction by 31.3% (433 pmol/mg of protein) because PNPLA2 alone without CGI-58 has relatively weak REH activity. The last group of lysates from coinfection of Ad-CGI-58 and Ad-PNPLA2 showed a further decrease in RE by approximately 66.7% (210 pmol/mg of protein) compared with that of Adv control (Figure 5C). These results indicate that RNPLA2 acts as REH, and that CGI-58 enhanced the REH activity of PNPLA2.
Figure 5. REH activity of PNPLA2 is enhanced by CGI-58.
(A) Retinoid profiles comparing RE levels in HEK293A cells infected with adenovirus expressing CGI-58, PNPLA2 (multiplicity of infection [MOI] = 50 each) adjusted for total MOI 100 with null Adv for control (MOI = 50), and coexpressing of CGI-58 and PNPLA2 (MOI = 50 each)with null Adv alone as control (MOI = 100). The products of REH activity by PNPLA2 were detected at 320 nm and quantified by HPLC. Peak 1: REs; Peak 2: all-trans-retinol.
(B) Western blot analysis using the same cell lysates used in retinoid profile assay.
(C) Comparison of the amounts of REs detected in the cells. REs were quantified by HPLC (n = 4 per group). Data are presented as mean ± SEM. Statistical analysis was performed using one-way ANOVA with a two-stage linear step-up procedure. ns: not significant, *p < 0.05, ***p < 0.001, and ****p < 0.0001.
(D) Western blot analysis showing expression of endogenous PNPLA2 inhibited by the PNPLA2 siRNA. Left lane: HEK293A cells transfected with 100 nM scrambled siRNA (scr-siRNA). Right lane: HEK293A cells transfected with 100 nM PNPLA2 siRNA.
(E) Comparison of the amounts of REs in scrambled and PNPLA2 siRNA-transfected cells (n = 4 per group). The amounts of REs were quantified by HPLC. Data are presented as mean ± SEM. Statistical analysis was performed using an unpaired two-tailed t test. *p < 0.05.
(F and G) Measurement of REH activity of PNPLA2 using liposome assay. All-trans-retinyl palmitate (RP) containing liposomes (250 μM lipids, 3.3 μM all-trans-RP) were incubated in 200 μL of 1% BSA in 0.1 M Tris-HCl for 2 h at 37°C with 250 μg of total cellular protein. Cells were infected by either Ad-mCherry (MOI = 100) (control) or by Ad-PNPLA2 (MOI = 100) and Ad-CGI (MOI = 100). (F) Representative retinoid profile for the liposome-based assay. Peak 1: REs; Peak 2: all-trans-retinol. (G) Comparison of the amount of all-trans-retinol produced by HEK293A cell homogenates that were separately infected with Ad-mCherry (control), Ad-CGI-58 (CGI-58), or Ad-PNPLA2 (PNPLA2) (MOI = 100 each) for 24 h. An equal amount of cell homogenates (125 μg of total protein) of PNPLA2 and CGI-58 lysates were mixed with liposomes to examine the effect of the coactivator (n = 3 per group). Data are presented as mean ± SEM. Statistical analysis was performed using one-way ANOVA followed by Tukey’s test. ns: not significant. ****p < 0.0001.
siRNA-mediated knockdown of endogenous PNPLA2 decreased endogenous REH activity
We further confirmed the REH activity of PNPLA2 by siRNA-mediated knockdown of endogenous PNPLA2 expression in HEK293 cells treated with all-trans-retinol and oleic acid. Western blot analysis confirmed an approximately 63% reduction of endogenous PNPLA2 protein in HEK293A cells 48 h after siRNA transfection (Figure 5D). HPLC analysis showed that REs were significantly elevated by approximately 42% (control, 156 pmol/mg of protein vs. PNPLA2 siRNA, 221 pmol/mg of protein) in the PNPLA2 siRNA-transfected cells, indicating a negative correlation with the levels of PNPLA2 protein (Figure 5E). These results suggest that endogenous PNPLA2 contributes significantly to the endogenous REH activity.
Verification of REH activity of PNPLA2 in vitro using liposome-based REH activity assay
Finally, to confirm the enzymatic activity of PNPLA2 and the role of CGI-58 as a coactivator, we performed cell-free liposome-based REH activity assay.41 An equal amount of cell lysate from HEK293A cells (without preincubation with all-trans-retinol) singly infected with adenovirus expressing mCherry protein (Ad-mCherry [negative control]) or a mixture of lysates from cells infected by Ad-PNPLA2 and that infected by Ad-CGI-58 was incubated with liposomes containing RP (all-trans-retinyl palmitate)41 as a substrate for PNPLA2 activity assay. Figure 5F shows the representative retinoid profiles of HPLC analysis. A mixture of lysates from the cells infected with Ad-PNPLA2 and that with Ad-CGI-58 decreased the level of REs (peak 1) and generated significant amounts of all-trans-retinol (peak 2) (Figure 5F, lower panel, and Figure 5G). Retinoid quantification demonstrated that lysates from cells infected with Ad-mCherry and Ad-CGI-58 alone (Figure 5G) showed no differences in levels of all-trans-retinol. Lysates from cells infected with Ad-PNPLA2 alone showed production of all-trans-retinol (Figure 5G). The mixture of cell lysates from cells infected with Ad-CGI-58 and Ad-PNPLA2 produced the highest amount of all-trans-retinol (Figure 5G). These results confirmed that CGI-58 enhanced the REH activity of PNPLA2 not only when they are coexpressed in the same cell but also when they are expressed separately and then mixed in a cell-free system.
PNPLA2 enhances the production of 11-cis-retinol by RPE65
To determine if REH activity of PNPLA2 facilitates the production of 11-cis-retinol, we employed HEK293A cells stably expressing LRAT (HEK293A-LRAT)42 preincubated with 25 μM all-trans-retinol to induce RE and LD accumulation in the cells. The cells were then infected with Ad-RPE65 alone or coinfected with Ad-RPE65 and Ad-PNPLA2 and incubated for another 24 h. The expression of PNPLA2 and RPE65 was confirmed by western blotting (Figure 6B), and retinoid profiles were analyzed by HPLC. As both added all-trans-retinol and 11-cis-retinol produced by RPE65 are converted by LRAT to their corresponding REs inside HEK293A-LRAT cells, and these REs are not separated by HPLC, we used the saponification procedure to quantify the level of total 11-cis-retinol generated during the retinol isomerase reaction.
Figure 6. PNPLA2 enhanced the production of 11-cis-retinol by RPE65.
(A–C) PNPLA2 enhances intracellular 11-cis-retinol production by RPE65 in a cell culture assay. (A) HPLC profiles of retinol isomers from saponified REs extracted from HEK293A-LRAT cells infected with Adv (null, control) (MOI = 50), Ad-RPE65 (RPE65) (MOI = 30) plus Adv (MOI = 20), and RPE65 (MOI = 30) plus Ad-PNPLA2 (PNPLA2) (MOI = 20). The inset shows the absorbance spectra of peak 2, 11-cis-ROL at 318.5 nm. HEK293A-LRAT cells were supplemented with 25 μM all-trans-retinol for 48 h before infection with the adenoviruses. Peak 1, REs; Peak 2, 11-cis-retinol (blue dotted line); Peak 3, 13-cis-retinol; Peak 4, all-trans-retinol. (B) Western blot analysis using the samples for retinoid profile analysis (40 μg of protein per lane). (C) Comparison of the amount of 11-cis-retinol production in HEK293A-LRAT cells expressing RPE65 alone (tan) and that coexpressing RPE65 and PNPLA2 (teal). Data are shown as mean ± SEM. Statistical analysis was performed using an unpaired two-tailed t test. ***p = 0.00023 (n = 9 per group).
(D and E) PNPLA2 increased 11-cis-retinol production in vitro retinol isomerase assay. Retinoids generated after incubation of HEK293A-LRAT cell homogenates expressing RPE65 alone or together with PNPLA2 and CGI-58 (250 μg of total cellular protein) with 0.3 μM of all-trans-retinol for 2 h were analyzed by HPLC.
(D) Representative retinoid profiles are shown for the samples with and without PNPLA2 and CGI-58 expression. Peak 1: REs; Peak 2: 11-cis-retinol.
(E) Quantification of 11-cis-retinol produced by HEK293A-LRAT cells infected with Ad-RPE65 (MOI = 50) that were additionally infected with either Adv (null) (MOI = 100) (control), or Ad-PNPLA2 and Ad-CGI-58 (MOI = 50 each) for 24 h. An equal amount of cell homogenates was mixed with all-trans-retinol added in 2 μL of dimethyl formamide and incubated for 2 h at 37°C. Data are presented as mean ± SEM. Statistical analysis was performed using an unpaired two-tailed t test. **p < 0.0027 (n = 4 per group).
First, cells infected by control Adv (null) showed no 11-cis-retinol production since they do not contain any endogenous RPE65 (Figure 6A, top panel). Next, the cells expressing RPE65 alone showed the production of 11-cis-retinol as expected (Figure 6A, middle panel). Coexpression of RPE65 with PNPLA2 showed an increase in the output of 11-cis-retinol compared with the cells expressing RPE65 alone (Figure 6A, bottom panel). Quantification of 11-cis-retinol showed that the cells infected with Ad-RPE65 alone produced 11-cis-retinol at a level of 1,660 pmol/mg of protein. Coexpression of RPE65 with PNPLA2 generated 11-cis-retinol at 2,008 pmol/mg of protein (Figure 6C). These data suggest that PNPLA2 facilitates the production of 11-cis-retinol catalyzed by RPE65.
Alternatively, we studied the effect of PNPLA2 and CGI-58 overexpression on the 11-cis-retinol production by RPE65 using in vitro retinol isomerase assay. In this case, the retinol isomerase reaction was started by mixing all-trans-retinol with the homogenates of HEK293A-LRAT cells expressing either RPE65 alone or coexpressing RPE65, PNPLA2, and CGI-58. As shown in Figures 6D and 6E, the 11-cis-retinol amount produced by RPE65 was 50% higher in the presence of PNPLA2 and CGI-58 than in control (Adv).
DISCUSSION
In the RPE, all-trans REs serve as the substrate for RPE65 to produce 11-cis-retinol, a precursor of 11-cis-retinal, the visual pigment chromophore.11,13 REs are mainly stored in morphologically unique LD structures termed retinosomes in the RPE.15,16 Protein seipin43 and fat storage-inducing transmembrane protein 244 drive the LD biogenesis. The breakdown of LDs is regulated by several proteins of the PAT family (perilipins).45 Degradation of triglycerides in LDs occurs by the sequential action of PNPLA2, hormone-sensitive lipase, and monoglyceride lipase.46
It has been shown that retinosomes participate in the regeneration of 11-cis-retinal through the visual cycle.15 Retinosomes rapidly increase in size after the photobleach while decreasing in size during the dark adaptation.15 However, it was unclear how the retinosomes contribute to the visual cycle. RPE65 is localized in the ER, while retinosomes are located in peripheral cytosol close to the plasma membrane15; these two distinct subcellular localizations suggest that RPE65 cannot access and cleave REs stored in retinosomes. REs are hydrophobic water-insoluble compounds, as such they cannot be transported to the ER through the water milieu of cytoplasm. Therefore, we hypothesized that an enzyme REH might be present in retinosomes and hydrolyze all-trans-REs from retinosomes to produce all-trans-retinol, which can be transported to ER to provide substrate for LRAT and RPE65. However, no such REH has ever been identified in the RPE. The present study showed that PNPLA2 is expressed in the RPE and colocalized with LDs (Figures S3A-S3C). Further, Pnpla2 KO mice displayed declined ERG responses, delayed regeneration of 11-cis-retinal, and increased levels of all-trans-RE in the RPE, suggesting that PNPLA2 functions as an REH in the RPE and is essential for the visual cycle and normal vision.
In line with the previous studies,21 PNPLA2 protein was detected in the isolated eyecups (containing RPE and choroid) and in the retinas as a single band with the expected molecular weight by western blot analysis. The results showed that the antibody for PNPLA2 only recognized a band in the eyecups and retinas of WT mice in western blot, but not in that of Pnpla2 KO mice (Figure 1B), confirming the specificity of the anti-PNPLA2 antibody. We also measured RPE65 levels in the same eyecups in WT and Pnpla2 KO mice. Western blot analysis showed that RPE65 levels were unchanged in the RPE of the Pnpla2 KO mice (Figure 1B), suggesting that the declined ERG (Figures 2B and 2C) and delayed regeneration of the chromophore (Figure 3B) are not due to the downregulation of RPE65. As shown by immunohistochemistry using the same anti-PNPLA2 antibody, PNPLA2 was detected in the RPE and retina (Figures 1C and 1E). Although PNPLA2 was originally found in the RPE as a receptor for PEDF,21 its function in the visual cycle and its role in vision have not been established.
As shown by scotopic ERG, both a- and b-wave amplitudes were significantly declined in Pnpla2 KO mice relative to their WT littermates (Figures 2B, 2C, and 2E), suggesting an important role of PNPLA2 in visual function. We did not observe a decrease in opsin level (Figures S4A and S4B), photoreceptors (Figures S5A-S5F), retina degeneration by TUNEL assay (Figures S6A-S6D), or decreased retinal thickness by optical coherence tomography (Figures S7A and S7B) in Pnpla2 KO mice; therefore, we proposed that the diminished visual response in these mice might be due to an impaired retinoid cycle and impaired visual pigment regeneration. Indeed, our results demonstrated a significant increase of LD accumulation in the RPE of Pnpla2 KO mice relative to WT mice (Figure 3A), suggesting that REs and other lipids may accumulate as a component of these LDs. Retinoid profile analysis and autofluorescence measurement showed that Pnpla2 KO mouse RPE contains significantly higher RE levels than WT mice (Figures 3C and S2B-S2D), suggesting a deficient RE utilization. Dark adaptation after photobleaching showed that ERG response recovery and 11-cis-retinal chromophore regeneration were delayed in Pnpla2 KO mice (Figures 3B and 3D). Similarly, a delayed dark adaptation and decrease in 11-cis chromophore regeneration were observed previously in RPE65 D477G mutant mice.47 However, in the case of Pnpla2 KO mice, the RPE65 level is similar to that of WT mice, as shown by western blot analysis (Figure 1B). Therefore, the diminished 11-cis-retinol regeneration rate cannot be ascribed to the RPE65 level. Moreover, since RE levels are increased in Pnpla2 KO mice, one might expect that production of 11-cis-retinol would be even higher in Pnpla2 KO mice due to the higher level of substrate for RPE65. This seeming contradiction can be explained by different subcellular localizations of RPE65 (in ER) and the majority of REs (accumulated in retinosomes after the photobleach). The results of immunostaining showed that PNPLA2 is colocalized with LDs (Figure 4C). In contrast, RPE65 did not show colocalization with LDs (Figure 4B). In line with previous studies, CGI-58, a coactivator of PNPLA2,18 was also colocalized with LDs (Figure 4D), confirming the involvement of PNPLA2 in the LD metabolism. Gradient density fractionation also showed that PNPLA2 and CGI-58 cofractionated with LDs in contrast to RPE65 (Figure 4I). This colocalization of PNPLA2 with retinosomes (Figures 4B, 4C, and S3A-S3C) suggests that PNPLA2 has a direct access to REs stored in LDs, while RPE65 has no such access. This finding agrees with the previously established notion that most of the RE pool in the RPE does not participate in the isomerization reaction catalyzed by RPE65.14
PNPLA2 (also known as ATGL) was originally identified as a triglyceride lipase in the liver and adipose tissue.17,18 In the HSCs, PNPLA2 is associated with LDs.18 It has been shown that PNPLA2 can hydrolyze REs in vitro.18 However, surprisingly, the ablation of PNPLA2 in mice did not result in the accumulation of REs in the liver.18 Therefore, it was previously uncertain if PNPLA2 functions as REH in vivo. To determine if PNPLA2 has REH activity, we expressed PNPLA2 alone or together with its coactivator CGI-58 in HEK293A cells loaded with all-trans-retinol and oleic acid to promote intracellular LDs formation. In HEK293A cells with adenovirus-mediated overexpression of PNPLA2, we observed a robust decrease of RE levels (Figure 5A). We did not quantify the increase of all-trans-retinol production since there was a significant amount of all-trans-retinol (Figure 5A, peak 2) absorbed by the cells during preincubation of HEK293A cells with all-trans-retinol before the adenovirus infection. As expected, the PNPLA2 REH activity was increased in the presence of CGI-58 (Figures 5A and 5C). Further, we confirmed this result using the cell-free liposome-based REH activity assay (Figures 5F and 5G). In cells with the knockdown of endogenous PNPLA2, we observed increased RE accumulation, which is in line with the function of PNPLA2 as REH (Figure 5E). Our results established that PNPLA2 is an efficient REH in the RPE cells that can mobilize REs from LDs to provide substrate for RPE65 localized in ER.
Next, we were interested if the overexpression of PNPLA2 in HEK293A-LRAT cells would affect the RPE65-mediated production of 11-cis-retinol. As shown in Figures 6A and 6C-6E, the amount of 11-cis-retinol produced by RPE65 in the presence of overexpressed PNPLA2 is significantly higher than that in RPE65 expression alone. The apparent activity of an enzyme can be changed either by increasing its expression level or by increasing its substrate concentration (if the substrate level did not reach saturation). We found that the RPE65 level in the presence of PNPLA2 was similar to that in the absence of PNPLA2 (Figure 6B). Therefore, the increased production of 11-cis-retinol was not due to the changed RPE65 expression level, suggesting that PNPLA2 contributes to increased 11-cis-retinol production by providing additional substrate for RPE65. It is likely that all-trans-retinol, which was generated by PNPLA2 in retinosomes, is released and reaches ER where LRAT and RPE65 reside. All-trans-retinol produced from retinosomes may be bound to cellular retinol-binding protein 1 (CRBP1)48 when it is transported to the ER. In the ER, all-trans-retinol is re-esterified by LRAT to provide a substrate for RPE65 (Figure 7). Retinosomes contain not only REs but also triglycerides and cholesteryl esters, which are surrounded by a phospholipid monolayer.16 Mobilization of these stored lipids requires the hydrolysis of respective ester bonds by lipases and ester hydrolases. PNPLA2 is ideally suited for its role since it catalyzes the hydrolysis of all these lipids in retinosomes.
Figure 7. Schematic of a proposed mechanism of the visual cycle.
PNPLA2 plays a role in visual pigment regeneration in RPE cells. All-trans-REs (AtREs) accumulate and form retinosomes in RPE cells. PNPLA2 hydrolyzes and mobilizes AtREs from the retinosomes. The released all-trans-retinol (AtROL) was transferred back to the ER to regenerate the visual pigment chromophore, 11-cis-retinal (11cRAL), through the visual cycle.
It is worth noting that the deletion of PNPLA2 does not completely abolish the production of 11-cis-retinol in Pnpla2 KO mice. There are two possible reasons for it. First, part of all-trans-retinol is delivered to the ER not from retinosome storage but from the circulation. Second, RPE may contain some other, yet unidentified, REHs which act on retinosomes.
In summary, we demonstrated that PNPLA2 affects the visual cycle by promoting RPE65 isomerase activity, likely by increasing the supply of its substrate in the ER. Consequently, the absence or dysfunction of PNPLA2 may impair vision. Furthermore, we identified a function of PNPLA2 as an RE mobilizing enzyme of the visual cycle, which plays a significant role in the retinoid homeostasis in the RPE and in the regeneration of 11-cis-retinal chromophore for normal vision.
Limitations of the study
This study has some limitations. We used the Pnpla2 global knockout mouse model in this work to study its role as REH in the visual cycle. It functions as REH in RPE and the deletion of Pnpla2 in RPE may affect retina function through the deficient visual cycle. However, since PNPLA2 is also expressed in neural retina, the ERG signal decrease may be a possible result of ablation of Pnpla2 in the photoreceptors. Currently, the role of PNPLA2 in the neural retina is unknown. However, PNPLA2 cannot function as REH in the photoreceptors since they do not contain retinyl esters and LDs. Finally, Pnpla2 knockout mice die prematurely at the age of 3 months from heart complications. Therefore, this study was unable to evaluate the phenotype in aged animals.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Gennadiy Moiseyev (gmoiseye@wakehealth.edu).
Materials availability
Adenoviruses and plasmids generated in this study will be made available from the lead contact upon request.
Data and code availability
This paper does not report new or original code.
The raw data and original images reported in this paper will be shared by the lead contact upon request.
Any additional information required to reanalyze the data reported in this paper will be available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethical approval
All protocols were approved by the Institutional Animal Use and Care Committee of the University of Oklahoma Health Sciences Center (OUHSC). All animal experiments were performed following the guideline of the animal care and use program with the division of comparative medicine (DCM) of the University of Oklahoma.
Animals
Heterozygous of Pnpla2−/− (Knockout, KO) mice (C57BL/6J background) were purchased from Jackson Laboratory (Bar Harbor, ME). Breeding colonies were established in the Rodent Barrier Facility at the OUHSC. All animals were group-housed in the pathogen-free animal facility and maintained in a temperature-controlled environment with 12-h light/12-h dark cycle and were fed a standard diet (PicoLab Rodent Diet 20 5053, LabDiet) and water ad libitum. The 8–16 week-old mice of both sexes were used in this study. All animals were euthanized by CO2 asphyxiation followed by cervical dislocation, and ocular tissue collection was performed under dim red or ambient room light, depending on the experiment. Littermates were used as controls.
Cell lines and cell cultures
Human embryonic kidney (HEK) 293A was originally purchased from QBiogene (Irvine, CA), and HEK293A-LRAT stable cell line was generated as described previously.42 HEK 293A cells were maintained in complete media containing Dulbecco’s modified Eagle’s medium (DMEM) (Corning) supplemented with 10% fetal bovine serum (FBS) (R&D Systems) and 1x Antibiotic-Antimycotic (Corning). HEK293A-LRAT cell line was maintained in the complete media supplemented with 10 μg/mL blasticidin (Gibco). Cells were maintained at 37°C in 5% CO2. For lipid droplet formation, cells were cultured 24–48 h in the complete medium supplemented with 10–25 μM retinol (Sigma-Aldrich) and 300 μM oleic acid (Sigma-Aldrich) conjugated with BSA.
METHOD DETAILS
RNA extraction, qRT-PCR and mRNA expression analysis
Total RNAs were isolated from the eyecups and retinas of WT and Pnpla2 KO mice using a Quick DNA/RNA Miniprep Plus Kit (Zymo Research) following the manufacturer’s instructions. The RNA concentration was determined by NanoDrop ND-1000 UV-Vis spectrophotometer (Thermo Scientific). The cDNA was synthesized from 1 μg of the total RNAs using TaqMan Reverse Transcription Reagents (Applied Biosystems). Quantitative Reverse Transcription-PCR (qRT-PCR) was performed using a CFX96 real-time PCR system (Bio-Rad) with a pair of gene-specific primers for the mouse Pnpla2 and 18s RNA as a reference gene (qRT-PCR primer information are shown in Table S1). Bio-Rad CFX Manager 3.1 (Bio-Rad) was used for relative mRNA expression analysis. Relative expression of Pnpla2 was calculated using the comparative 2−ΔΔCT method of the gene-expression with the 18s RNA as an internal reference for normalization.54
Protein extraction and Western blot analysis
Mouse eye tissues and cultured cell pellets were sonicated in RIPA buffer with 1x Halt protease inhibitor cocktail (Thermo Scientific). Lysates of total cellular proteins were collected by centrifugation at 21,000 × g for 15 min at 4°C. The total protein concentration of eye tissue and cell lysates were determined using Bradford Reagent55 or Pierce BCA Protein Assay Kit (Thermo Scientific). The equal amount of protein (20 or 40 μg) was resolved in 10% SDS-PAGE and transferred onto the nitrocellulose membrane. Western blot analysis was performed with primary antibodies: anti-PNPLA2 (1:1000, 2183S, Cell Signaling Technology), anti-CGI-58 (1:2000, sc-100468, Santa Cruz Biotechnology, for Western blot; 1:2000, ab183739, Abcam, for immunostaining), anti-RPE65 (1:1000, house-made antibody),49 anti-LAMP1 (1:1000, 21997-1-AP, Proteintech), anti-Calnexin (1:1000, ab10286, Abcam), Anti-PLIN2 (1:1000, ab108323, Abcam), Anti-PLIN3 (1:1000, 106941-1-AP, Proteintech), and anti-β-Actin (C4) HRP (1:1000, sc-47778 HRP, Santa Cruz Biotechnology). HRP-conjugated secondary antibodies, anti-mouse IgG, anti-rabbit IgG, and anti-goat IgG (1:5000, Vector Laboratories) were used. Western blot signals were acquired using a ChemiDoc system (Bio-Rad), and the signal intensities were quantified using a densitometry application, Image Lab software version 6.1.0 (Bio-Rad).
Immunohistochemistry of mouse retina
Immunofluorescence chemistry (IFC) of the mouse retina/RPE was performed following a published method.56 Briefly, mouse eyes were enucleated by dissection, and the superior side of the cornea was demarcated with green tattoo dye (Ketchum Mfg). The eyes were fixed in Davidson’s fixative (2% Formaldehyde, 35% Ethanol, 10% Glacial Acetic Acid) for 48 h, and stored in 70% ethanol until paraffin processing. Sagittal sections along the superior-inferior retinal axis were cut at 5 μm thickness. Prior to IFC, sections were deparaffinized and rehydrated in graded ethanol. Sections were blocked in the blocking buffer 2 h (5% BSA, 0.3% Triton X-100, in PBS). Sections were incubated at 4°C with the primary antibody (anti-PNPLA2 and anti-RPE65) diluted in blocking solution 1:100 for ON. Sections were then washed three times in PBS and incubated with the fluorophore-conjugated secondary antibodies (PNPLA2, 1:400, Donkey anti-rabbit Alexa 594; RPE65, 1:400, Donkey anti-mouse Alexa 488) and the nucleus dye (1:1000, 4′,6-diamidino-2-phenylindole (DAPI), Life Technologies) for 2 h at RT.
Electroretinography
Animals were fully dark-adapted overnight (ON, 16 h) in the darkroom before ERG recording. Animals were handled under dim red light and anesthetized with ketamine hydrochloride(K)/xylazine hydrochloride solution(X) (2 μL/g of body weight, final concentration of 45 mg/mL (K), 5 mg/mL (X)) diluted with saline and mice were allowed to be fully sedated. Pupils were dilated with 1% tropicamide ophthalmic solution (Bausch + Lomb). The mouse was placed on the heated stage, and platinum ground and reference needle electrodes were placed on the base of the tail and on the mouse’s cheek, respectively. Mouse eyes were coated with GONIOVISC ophthalmic lubricant (HUB Pharmaceuticals) before the gold-wire loop electrodes (Diagnosys) made contacting with the cornea. The recording was made using the Espion E3 system with a Ganzfeld ColorDome system (Diagnosys) with a standard ERG protocol.57 For the retinal function test, responses were recorded with 8-step scotopic protocols starting with a single flash of −3.7 log cd·s/m2 intensity followed by a step each increasing light intensity to −1.7, −0.7, 0.3, 1.3, 2.3, 2.6 and 2.8 log cd·s/m2. In the last step, the animal was exposed to a steady rod suppressing 2.8 log cd/m2 light for 2 min to elicit a cone response. For dark adaptation rhodopsin recovery, ERG was performed with a modified timing and photobleaching light intensity described in the previously published method.47 Responses of the fully dark-adapted mice were acquired by applying a single flash 10 cd s/m2 stimulus before and after complete photobleaching of rods by 1,000 cd/m2 for 2 min. Then mice were allowed to recover in the dark for 60 min while acquiring a response every 5 min with a single flash of 10 cd s/m2 stimulus. The a-waves of rhodopsin recovery data were presented as percentage amplitudes. The a-wave values were first converted to positive values, then the mean amplitudes at each time point were calculated with the mean of the initial dark-adapted a-wave values (WT = 216.5 μV, n = 6, KO = 161.5 μV, n = 5).
Immunostaining of mouse eyecup flatmount
For immunostaining of eyecup flatmounts, mouse eyeballs were fixed in phosphate-buffered 4% paraformaldehyde (PFA) for 30 min on ice before dissection. Muscle and fat surrounding the eye globe were cleaned off. The anterior portion of the cornea and the lens were dissected away, and the retina was removed to expose underlying RPE. The dissected eyecup (sclera, choroid, and RPE) was washed three times in PBS-T (PBS with 0.2% Triton X-100), and permeabilized and blocked for 1 h in PBS containing 10% FBS and 0.1% Triton X-100. Following overnight incubation at 4°C with a primary antibody diluted in the blocking solution, the eyecups were washed three times with PBS containing 0.1% Triton X-100, and incubated with anti-ZO1 (1:200, MABT11, Millipore), the secondary antibody (1:200, DyLight405, 712-475-450, Jackson ImmunoResearch), BODIPY493/503 (Invitrogen) for lipid labeling, and the nuclei were counterstained with DAPI (1:1000, D9542, Sigma-Aldrich) for 2 h. The eyecup was flattened on a slide, and a total of 4 straight cuts from the limbal edge were made to form 4-petaL flower-like structure and mounted in Vectashield mounting media (Vector Laboratories).
Visual chromophore regeneration in mouse eyes
All mice were dark-adapted ON before the experiment. All procedures were conducted under dim red light unless otherwise noted. Animals were euthanized by CO2 asphyxiation. WT and Pnpla2 KO mice (n = 24 per group) were divided into four groups. Group 1 (n = 4) was subjected to the ON dark adaptation only. Group 2 (n = 4) was exposed to the 5,000 lux white light for 30 min and then immediately euthanized. Group 3 (n = 8) was subjected to 15 min recovery in the complete dark after the photobleach under 5,000 flux white light for 30 min. Group 4 (n = 8) was subjected to 40 min recovery in the dark after the same photobleach. The 1% tropicamide ophthalmic solution (Bausch + Lomb) was applied in both eyes 30 min before photobleaching in groups 2 to 4. Each mouse was exposed to 5,000 lux in the open-top container (112 mm dia. x 151 mm ht., 1000 mL Nalgene straight-sided wide-mouth polycarbonate jars, Nalgene). Dark recovery groups 3 and 4 were transferred back to the cages and then dark-adapted for indicated periods. All animals were euthanized at the end of the experiment, and eyes were collected to snap-freeze in liquid nitrogen. They were stored in the dark at −80°C until they were used for retinoid quantification.
Retinoid analysis
The procedure of retinoid extraction was performed as described previously.58 Briefly, the eyes were individually homogenized in lysis buffer [10 mM NH2OH, 50% ethanol, 50% 2-(N-morpholino) ethanesulfonic acid, pH 6.5] with a glass grinder, and the retinoids were extracted with hexane and dried under argon. Dried retinoid samples were resuspended in 200 μL of HPLC mobile phase (11.2% ethyl acetate, 2.0% dioxane, 1.4% octanol, 85.4% hexane) and analyzed by HPLC (515 HPLC pump; Waters Corp) with a normal phase LiChrosphere SI-60 (Alltech) 5 μm column and isocratic mobile phase (1 mL/min). Absorbance was monitored at 320 nm or 360 nm, and the peak of each retinoid isomer in HPLC profile was identified by retention time and its absorption spectra of the retinoid standards. All data were presented as mean ± SEM from 8 independent measurements. For cell studies, cells were sonicated in 200 μL PBS (3 times for 20 s) and extracted by 300 μL of ethanol and 10 mL of hexanes. The top organic phase was collected and dried under argon. The dried samples were processed in the same procedure described above. Absorbance was monitored at 320 nm, and the peak of each retinoid in the HPLC profile was identified by comparison to the retinoid standard. For 11-cis-retinol quantification, HEK293A-LRAT cells containing REs were saponified with 0.35 N KOH as described previously,59 and the products were analyzed by HPLC as described above. The PNPLA2 REH activity was calculated based on the amounts of the remaining all-trans-REs, and the RPE65 isomerase activity was calculated from the area of the generated 11-cis-retinol peak. The amounts of retinoids were normalized by the total protein concentration. All data were presented as mean ± SEM from 4 independent measurements.
Immunostaining of lipid droplet and associated proteins
HEK293A cells grown on coverslips were treated with 400 μM oleic acid for 24 h, followed by Polyethylenimine (PEI) MAX (Polysciences) mediated transfection of indicated expression vectors at 2 μg/well in a 6 well plate (1:2 ratio, DNA 2 μg: PEI 4 μL).60,61 At 24 h post-transfection, the cells were fixed with 3% PFA and 0.02% glutaraldehyde diluted in PBS for 15 min at RT. The cells were washed with PBS three times and permeabilized with 0.2% digitonin and 10% FBS diluted in PBS for 1 h. The cells were incubated with primary antibodies against indicated proteins diluted at 1:200 in blocking buffer overnight. Secondary antibodies Alexa Fluor 488 (indicated proteins) (1:500, 715-545-151, Jackson ImmunoResearch), DAPI (1:1000, Sigma-Aldrich), and lipid droplet stain were applied together for 2 h. The cells were washed 3 times in PBS and mounted in Prolong Gold antifade reagent (Thermo Scientific). Lipid droplets were stained with HCS LipidTox Red neural lipid stain (1:500, Invitrogen).
Microscopy and image analysis
Fluorescence images were captured under a laser scanning confocal microscope (FV 1000 or FV 1200, Olympus) using a 60x or 100× oil-immersion objective. A z stack was applied in most images collected during a single session. Two- or three-channel images were taken consecutively at the same location to detect the association of protein and lipid droplets. The fluorescent intensity was measured using ImageJ graphic RGB profiler module.53
Subcellular fractionation of lipid droplets
Purification of lipid droplets from cells treated with oleic acid and all-trans-retinol overnight was performed using the Lipid Droplet Isolation Kit (Cell Biolabs) according to the manufacturer’s instructions. Cells were treated with 400 μM oleic acid overnight, followed by transient transfection of plasmids expressing RPE65,52 PNPLA2, and CGI-58 for 24 h with 1 μg DNA/100-mm plate. Briefly, 5 100-mm plates of confluent HEK293A cells were pooled for each prep. The cells were washed with PBS three times and collected. The cell pellet was resuspended with 200 μL of reagent A. After incubation for 10 min, 800 μL of reagent B was added to resuspended reagent A, and the cells were homogenized by passing the cells five times through a 27-gauge needle. Reagent B (600 μL) was layer gently onto the cell homogenate and was centrifuged for 5 h (24,000 × g, 4°C). The lipid droplets in the top fraction of 200 μL were collected; the next 500 μL of supernatant was discarded, then 200 μL was collected as the cytosol fraction. Next, all supernatant was discarded, then the pellet portion was solubilized with 200 μL of PBS and collected. Equal amounts of the total lipid droplet, cytosol, and pellet (membrane cytoskeleton) fractions were analyzed by immunoblotting. The purity of the isolated lipid droplets was assessed through blotting for PLIN. ER and lysosome associations were indicated by calnexin and LAMP1, respectively.
Adenovirus and plasmid construction
For the construction of Ad-PNPLA2 and Ad-CGI-58, the full length of human PNPLA2 (hPNPLA2) cDNA (HsCD00334354) and human CGI-58 (hCGI-58) cDNA (HsCD00043192) were purchased from Harvard PlasmID Repository (Harvard Medical School). The primers were designed to amplify with restriction enzyme sites of HindIII and EcoRV for hPNPLA2, and KpnI and NotI for hCGI-58 gene (DNA sequence of primers are shown in Table S1). PCR products of hPNPLA2 and hCGI-58 were first cloned into a cloning vector, pGEM-T Easy (Promega) and then subcloned into the pShuttle-CMV vector (Agilent) at compatible restriction sites. The recombinant adenoviruses were generated and propagated using the same procedure described in the methods.11 The titer of adenovirus was determined with Adeno-X Rapid Titer Kit (Clontech Laboratories) following the manufacturers’ instructions. Null adenovirus (Adv) was purchased from Vector Biolabs (Malvern, PA). The same procedure described above was used to generate the plasmids for hPNPLA2 and hCGI-58 gene into pcDNA6 V5-HisA vector (Invitrogen). Briefly, the pairs of primers were designed with same restriction sites (DNA sequence of primers are shown in Table S1). Plasmids were amplified and extracted from E. coli JM109 cells (Promega) using Plasmid Spin Miniprep or Maxi (QIAGEN) kits as required. Sequences of the plasmids were confirmed by the Laboratory for Molecular Biology and Cytometry Research (LMBCR) Core DNA Sequencing and Genomics Facility at the OUHSC.
In vitro retinyl ester hydrolysis assay
To test the REH activity of PNPLA2, we used HEK293A cells or HEK293A-LRAT stable cell lines.42 First, the cells (1 × 106 cell/plate) were seeded in 100-mm culture plates with the complete growth medium and incubated for 24 h at 37°C with 5% CO2. After the change to a fresh growth medium, all cell culture manipulation was performed under dim red light. Cell culture media were supplemented with 10–25 μM all-trans-retinol and 300 mM oleic acid conjugated with fatty acid-free BSA and incubated for 48 h. The cells were counted for multiplicity of infection (MOI) calculation prior to adenovirus infection. The cells were gently washed with 1x PBS, pH 7.4 twice to remove excess supplemented all-trans-retinol and oleic acid. Then, the fresh growth media were added to each cell culture plate. Cells were infected with indicated MOI of Adv (null), Ad-PNPLA2, Ad-CGI-58, Ad-RPE65, or coinfected Ad-PNPLA2 with either Ad-CGI-58 or Ad-RPE65. To adjust MOI to the same in each coinfected cell group, we infected all other cell cultures with the same total virus titer. After 24 h, the cells were harvested and washed with 1x PBS, pH 7.4 twice. Cell pellets were stored at −80°C and protected from light until used for retinoid quantification and analysis.
In vitro PNPLA2 siRNA-mediated gene silencing assay
HEK293A cells were seeded (2.5 × 106 cells/plate) in 100-mm culture plates with the complete growth medium and cultured overnight. All the following work was performed under dim red light. After replacing growth media, cell cultures were supplemented with 10 μM retinol and 300 μM Oleic acid/plate and incubated for another 48 h. Then, the cells were rinsed with 1x PBS twice. The medium was replaced again. Next, cells were transfected with 100 μM gene-specific or control siRNAs (ON_TARGETplus Human PNPLA2 siRNA SMARTPool, and ON-TARGETplus Non-targeting Control Pool, Horizon Discovery) using the HiPerFect Transfection Reagent (QIAGEN) according to the manufacturer’s instruction. After incubation for 48 h, the cells were collected in PBS buffer, protein expression was analyzed by Western blot analysis, and intracellular retinoid contents were analyzed by HPLC as described above.
In vitro isomerase assay
All-trans-retinol (Sigma-Aldrich) dissolved in N,N-dimethyl formamide (DMF) was used as the substrate for the isomerase assay. The 293A-LRAT cells expressing RPE65, PNPLA2, and CGI-58 were homogenized by sonication in the reaction buffer (10 mM 1,3-bis [tris(hydroxymethyl)-methylamino]propane (BTP), pH 8.0, 100 mM NaCl). For each reaction, 250 μg of total proteins from the cell homogenates expressed either RPE65 only or RPE65, PNPLA2 and CGI-58 were added into 200 μL of reaction buffer containing 0.3 μM of all-trans-retinol, 1% BSA and 25 μM of cellular retinaldehyde-binding protein (CRALBP). The reaction mixture was incubated at 37°C for 2 h and then stopped. Retinoids were extracted with 300 μL of cold methanol and 300 μL of hexane and centrifuged at 10, 000 × g for 5 min. The upper layer was collected, and the generated retinoids were analyzed by normal phase HPLC as described.13 The peak of each retinoid isomer was identified based on its characteristic retention time of retinoid standards and spectrum. The isomerase activity was calculated from the area of the 11-cis-retinol peak using Waters Empower software (Waters) with synthetic 11-cis-retinol as a standard.
Illustration
Figure 7 illustration was created using Adobe Illustrator (www.adobe.com) by Y.T. and was modified by M.H.
Data extraction
Data for absorption spectrum of 11-cis-retinol (Figure 6A, inset) generated by HPLC software was extracted from the image using a data extraction tool Plotdigitizer Version 3.0.0 (PlotDigitizer, https://plotdigitizer.com/).
QUANTIFICATION AND STATISTICAL ANALYSIS
All graphs were generated by GraphPad Prism 9 (GraphPad Software, www.graphpad.com/) or Igor Pro 8.04 (WaveMetrics, www.wavemetrics.com). Statistical analysis were performed with GraphPad Prism 9.0 software. Statistical significance was determined by two-tailed unpaired Student t-test or Welch’s t test. For statistical comparisons of more than two groups were determined by one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc tests or two-state linear step-up procedure of Benjamini, Kreiger and Yekutieli as indicated in figure legends. All data were presented as mean ± SEM unless otherwise noted. A p value of less than 0.05 was considered significant. Statistical significance was defined as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. ns, not significant.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-ATGL | Cell Signaling Technology | Cat# 2138S; RRID: AB_2167955 |
| Rabbit polyclonal anti-ATGL | Thermo Fisher Scientific | Cat# PA5-17436; RRID:AB_10982805 |
| Mouse monoclonal anti-ABHD5 (36A) | Santa Cruz Biotechnology | Cat# sc-100468; RRID:AB_2220720 |
| Recombinant anti-Abhd5/CGI-58 | Abcam | Cat# ab183739; RRID:AB_2909444 |
| Rabbit anti-RPE65 | Ma et al.49 | N/A |
| Mouse anti-rhodopsin (1D4) | Molday and MacKenzie.50 | N/A |
| Horse anti-Mouse IgG (H + L), Peroxidase | Vector Laboratories | Cat# PI-2000; RRID:AB_2336177 |
| Rabbit polyclonal anti-LAMP1 | Proteintech | Cat# 21997-1-AP; RRID:AB_2878966 |
| Rabbit polyclonal anti-Calnexin | Abcam | Cat# ab10286; RRID:AB_2069009 |
| Rabbit monoclonal ADFP (Perilipin2/PLIN2) | Abcam | Cat# ab108323; RRID:AB_1086346 |
| Rabbit polyclonal anti-TIP47 (Perilipin3/PLIN3) | Proteintech | Cat# 10694-1-AP; RRID:AB_2297252 |
| Goat anti-Rabbit IgG (H + L), Peroxidase | Vector Laboratories | Cat# PI-1000; RRID:AB_2336198 |
| Horse anti-Goat IgG (H + L), Peroxidase | Vector Laboratories | Cat# PI-9500; RRID:AB_2336124 |
| Mouse monoclonal anti-β -Actin (C4) | Santa Cruz Biotechnology | Cat# sc-47778 HRP; RRID:AB_2714189 |
| Mouse monoclonal anti-β-Actin | Sigma-Aldrich | Cat# A1978; RRID:AB_476692 |
| Alexa Fluor 488 AffiniPure Donkey Anti-Mouse IgG (H + L) | Jackson ImmunoResearch Labs | Cat# 715-545-151; RRID:AB_2341099 |
| Alexa Fluor 594 AffiniPure Donkey Anti-Rabbit IgG (H + L) | Jackson ImmunoResearch Labs | Cat# 711-585-152; RRID:AB_2340621 |
| Bacterial and virus strains | ||
| Ad-hPNPLA2 | This manuscript | N/A |
| Ad-hCGI-58 | This manuscript | N/A |
| Ad-hRPE65 | Moiseyev et al.11 | N/A |
| Ad-cRPE65 | Moiseyev et al.51 | N/A |
| Ad-mCherry | This manuscript | N/A |
| Adv (Ad-CMV-Null) | Vector Biolabs | Cat# 1300 |
| Biological samples | ||
| Liposomes | Nikolaeva et al.41 | N/A |
| Critical commercial assays | ||
| Quick DNA/RNA Miniprep Plus Kit | Zymo Research | Cat# D7003 |
| HSC LipidTox Red Neutral Lipid Stain | Thermo Fisher | Cat# H34476 |
| DAB Substrate Kit | BD Pharmingen | Cat# 550880 |
| HiPerFect Transfection Reagent | QIAGEN | Cat# 301705 |
| PEI MAX | Polysciences | Cat# 24765 |
| Experimental models: Cell lines | ||
| HEK293A (QBI-293A) | Qbiogene (MP Biomedicals) | Cat# AES0503, RRID:CVCL_6910 |
| HEK293A-LRAT stable cell line | Takahashi et al.42 | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: B6;129P2-Pnpla2tm1Rze/J | The Jackson Laboratory | RRID:IMSR_JAX:019003 |
| Oligonucleotides | ||
| Oligonucleotides used in this study | This manuscript | Table S1 |
| ON-TARGETplus Non-Targeting Control Pool | Horizon Discovery | Cat# D-001810-10-50 |
| ON-TARGETplus Human PNPLA2 siRNA SMART Pool | Horizon Discovery | Cat# L-009003-01-0050 |
| Recombinant DNA | ||
| PNPLA2-pOTB7 | PlasmID | HsCD00334354 |
| ABHD5-pDONR221 | PlasmID | HsCD00043192 |
| Plasmid: hPNPLA2-pcDNA6 V5-His A | This manuscript | N/A |
| Plasmid: hCGI-58-pcDNA6 V5-His A | This manuscript | N/A |
| Plasmid: hRPE65-pcDNA3.1(−) | Takahashi et al.52 | N/A |
| Software and algorithms | ||
| CFX Manager 3.1 | Bio-Rad | RRID:SCR_017251 |
| Image Lab 6.1 | Bio-Rad | RRID:SCR_014210 |
| GraphPad Prism 9 | GraphPad software | RRID:SCR_002798 |
| IGOR Pro 8 | WaveMetrics | RRID:SCR_000325 |
| Olympus Fluoview FV10-ASW | Olympus | RRID:SCR_014215 |
| ImageJ | Schneider et al.53 | RRID:SCR_003070 |
| PlotDigitizer Version 3.0 | PlotDigitizer | RRID:SCR_014484 |
| Other | ||
| 515 HPLC Pump | Waters | https://www.waters.com/waters/en_US/515-HPLC-Pump/nav.htm?cid=534312&locale=en_US |
| LiChrosphere SI-60 5μm | Alltech | N/A |
| Espion E3 Electroretinography Console, ColorDome with Lab Cradle | Diagnosys | https://www.diagnosysllc.com/research/ |
Highlights.
PNPLA2 deficiency delays 11-cis-retinal regeneration in the visual cycle
Pnpla2 KO mice accumulate REs in lipid droplets in RPE
PNPLA2 mobilizes REs from lipid droplets via REH activity
PNPLA2 provides the source of substrate for RPE65 in the visual cycle
ACKNOWLEDGMENTS
This work was supported by NIH grants (EY019309, EY033330, EY030472, EY012231, EY028949, EY032930, EY032931). This study was also supported by the Diabetic Animal Core and Histology and Image Core of Diabetes Research COBRE in Oklahoma (grant no. GM122744) and the Vision Core supported by NEI P30 (grant no. EY021725).
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112091.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Wald G (1968). Molecular basis of visual excitation. Science 162, 230–239. 10.1126/science.162.3850.230. [DOI] [PubMed] [Google Scholar]
- 2.Honig B, and Ebrey TG (1974). The structure and spectra of the chromophore of the visual pigments. Annu. Rev. Biophys. Bioeng 3, 151–177. 10.1146/annurev.bb.03.060174.001055. [DOI] [PubMed] [Google Scholar]
- 3.Kiser PD, Golczak M, and Palczewski K (2014). Chemistry of the retinoid (visual) cycle. Chem. Rev 114, 194–232. 10.1021/cr400107q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D, and Sun H (2007). A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science 315, 820–825. 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- 5.Blaner WS (2007). STRA6, a cell-surface receptor for retinol-binding protein: the plot thickens. Cell Metab. 5, 164–166. 10.1016/j.cmet.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Isken A, Golczak M, Oberhauser V, Hunzelmann S, Driever W, Imanishi Y, Palczewski K, and von Lintig J (2008). RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome. Cell Metab. 7, 258–268. 10.1016/j.cmet.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacDonald PN, and Ong DE (1988). Evidence for a lecithin-retinol acyltransferase activity in the rat small intestine. J. Biol. Chem 263, 12478–12482. 10.1016/S0021-9258(18)37779-2. [DOI] [PubMed] [Google Scholar]
- 8.Saari JC, and Bredberg DL (1989). Lecithin:Retinol acyltransferase in retinal pigment epithelial microsomes. J. Biol. Chem 264, 8636–8640. 10.1016/S0021-9258(18)81839-7. [DOI] [PubMed] [Google Scholar]
- 9.Redmond TM, Poliakov E, Yu S, Tsai J-Y, Lu Z, and Gentleman S (2005). Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc. Natl. Acad. Sci. USA 102, 13658–13663. 10.1073/pnas.0504167102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin M, Li S, Moghrabi WN, Sun H, and Travis GH (2005). Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell 122, 449–459. 10.1016/j.cell.2005.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moiseyev G, Chen Y, Takahashi Y, Wu BX, and Ma J.-x. (2005). RPE65 is the isomerohydrolase in the retinoid visual cycle. Proc. Natl. Acad. Sci. USA 102, 12413–12418. 10.1073/pnas.0503460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gollapalli DR, and Rando RR (2003). All-trans-retinyl esters are the substrates for isomerization in the vertebrate visual cycle. Biochemistry 42, 5809–5818. 10.1021/bi0341004. [DOI] [PubMed] [Google Scholar]
- 13.Moiseyev G, Crouch RK, Goletz P, Oatis J Jr., Redmond TM, and Ma JX (2003). Retinyl esters are the substrate for isomerohydrolase. Biochemistry 42, 2229–2238. 10.1021/bi026911y. [DOI] [PubMed] [Google Scholar]
- 14.Stecher H, Gelb MH, Saari JC, and Palczewski K (1999). Preferential release of 11-cis-retinol from retinal pigment epithelial cells in the presence of cellular retinaldehyde-binding protein. J. Biol. Chem 274, 8577–8585. 10.1074/jbc.274.13.8577. [DOI] [PubMed] [Google Scholar]
- 15.Imanishi Y, Batten ML, Piston DW, Baehr W, and Palczewski K (2004). Noninvasive two-photon imaging reveals retinyl ester storage structures in the eye. J. Cell Biol 164, 373–383. 10.1083/jcb.200311079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orban T, Palczewska G, and Palczewski K (2011). Retinyl ester storage particles (retinosomes) from the retinal pigmented epithelium resemble lipid droplets in other tissues. J. Biol. Chem 286, 17248–17258. 10.1074/jbc.M110.195198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, and Zechner R (2004). Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306, 1383–1386. 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 18.Taschler U, Schreiber R, Chitraju C, Grabner GF, Romauch M, Wolinski H, Haemmerle G, Breinbauer R, Zechner R, Lass A, and Zimmermann R (2015). Adipose triglyceride lipase is involved in the mobilization of triglyceride and retinoid stores of hepatic stellate cells. Biochim. Biophys. Acta 1851, 937–945. 10.1016/j.bbalip.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villena JA, Roy S, Sarkadi-Nagy E, Kim K-H, and Sul HS (2004). Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J. Biol. Chem 279, 47066–47075. 10.1074/jbc.M403855200. [DOI] [PubMed] [Google Scholar]
- 20.Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, and Gross RW (2004). Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J. Biol. Chem 279, 48968–48975. 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 21.Notari L, Baladron V, Aroca-Aguilar JD, Balko N, Heredia R, Meyer C, Notario PM, Saravanamuthu S, Nueda M-L, Sanchez-Sanchez F, et al. (2006). Identification of a lipase-linked cell membrane receptor for pigment epithelium–derived factor. J. Biol. Chem. 281, 38022–38037. 10.1074/jbc.M600353200. [DOI] [PubMed] [Google Scholar]
- 22.Cao W, Tombran-Tink J, Elias R, Sezate S, Mrazek D, and McGinnis JF (2001). In vivo protection of photoreceptors from light damage by pigment epithelium-derived factor. Invest. Ophthalmol. Vis. Sci 42, 1646–1652. [PubMed] [Google Scholar]
- 23.Bullock J, Polato F, Abu-Asab M, Bernardo-Colón A, Aflaki E, Agbaga M-P, and Becerra SP (2021). Degradation of photoreceptor outer segments by the retinal pigment epithelium requires pigment epithelium-derived factor receptor (PEDF-R). Invest. Ophthalmol. Vis. Sci 62, 30. 10.1167/iovs.62.2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamel CP, Tsilou E, Pfeffer BA, Hooks JJ, Detrick B, and Redmond TM (1993). Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J. Biol. Chem 268, 15751–15757. 10.1016/S0021-9258(18)82319-5. [DOI] [PubMed] [Google Scholar]
- 25.Redmond TM, Yu S, Lee E, Bok D, Hamasaki D, Chen N, Goletz P, Ma J-X, Crouch RK, and Pfeifer K (1998). Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat. Genet 20, 344–351. 10.1038/3813. [DOI] [PubMed] [Google Scholar]
- 26.Katz ML, and Redmond TM (2001). Effect of Rpe65 knockout on accumulation of lipofuscin fluorophores in the retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci 42, 3023–3030. [PubMed] [Google Scholar]
- 27.Sheridan C, Boyer NP, Crouch RK, and Koutalos Y (2017). RPE65 and the accumulation of retinyl esters in mouse retinal pigment epithelium. Photochem. Photobiol 93, 844–848. 10.1111/php.12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolins NE, Rubin B, and Brasaemle DL (2001). TIP47 associates with lipid droplets. J. Biol. Chem 276, 5101–5108. 10.1074/jbc.M006775200. [DOI] [PubMed] [Google Scholar]
- 29.Hickenbottom SJ, Kimmel AR, Londos C, and Hurley JH (2004). Structure of a lipid droplet protein: the PAT family member TIP47. Structure 12, 1199–1207. 10.1016/j.str.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 30.Båvik CO, Busch C, and Eriksson U (1992). Characterization of a plasma retinol-binding protein membrane receptor expressed in the retinal pigment epithelium. J. Biol. Chem 267, 23035–23042. 10.1016/S0021-9258(18)50052-1. [DOI] [PubMed] [Google Scholar]
- 31.Hamel CP, Tsilou E, Harris E, Pfeffer BA, Hooks JJ, Detrick B, and Redmond TM (1993). A developmentally regulated microsomal protein specific for the pigment epithelium of the vertebrate retina. J. Neurosci. Res 34, 414–25. 10.1002/jnr.490340406. [DOI] [PubMed] [Google Scholar]
- 32.Lass A, Zimmermann R, Haemmerle G, Riederer M, Schoiswohl G, Schweiger M, Kienesberger P, Strauss JG, Gorkiewicz G, and Zechner R (2006). Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 3, 309–319. 10.1016/j.cmet.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 33.Granneman JG, Moore H-PH, Granneman RL, Greenberg AS, Obin MS, and Zhu Z (2007). Analysis of lipolytic protein trafficking and interactions in adipocytes. J. Biol. Chem 282, 5726–5735. 10.1074/jbc.M610580200. [DOI] [PubMed] [Google Scholar]
- 34.Gruber A, Cornaciu I, Lass A, Schweiger M, Poeschl M, Eder C, Kumari M, Schoiswohl G, Wolinski H, Kohlwein SD, et al. (2010). The N-terminal region of comparative gene identification-58 (CGI-58) is important for lipid droplet binding and activation of adipose triglyceride lipase. J. Biol. Chem 285, 12289–12298. 10.1074/jbc.M109.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eichmann TO, Grumet L, Taschler U, Hartler J, Heier C, Woblistin A, Pajed L, Kollroser M, Rechberger G, Thallinger GG, et al. (2015). ATGL and CGI-58 are lipid droplet proteins of the hepatic stellate cell line HSC-T6. J. Lipid Res 56, 1972–1984. 10.1194/jlr.M062372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen JW, Cha Y, Yuksel KU, Gracy RW, and August JT (1988). Isolation and sequencing of a cDNA clone encoding lysosomal membrane glycoprotein mouse LAMP-1. Sequence similarity to proteins bearing onco-differentiation antigens. J. Biol. Chem 263, 8754–8758. 10.1016/S0021-9258(18)68370-X. [DOI] [PubMed] [Google Scholar]
- 37.Wada I, Rindress D, Cameron PH, Ou WJ, Doherty JJ, Louvard D, Bell AW, Dignard D, Thomas DY, and Bergeron JJ (1991). SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J. Biol. Chem 266, 19599–19610. 10.1016/S0021-9258(18)55036-5. [DOI] [PubMed] [Google Scholar]
- 38.Sztalryd C, and Brasaemle DL (2017). The perilipin family of lipid droplet proteins: gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta. Mol. Cell Biol. Lipids 1862, 1221–1232. 10.1016/j.bbalip.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenberg AS, Egan JJ, Wek SA, Garty NB, Blanchette-Mackie EJ, and Londos C (1991). Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem 266, 11341–11346. 10.1016/S0021-9258(18)99168-4. [DOI] [PubMed] [Google Scholar]
- 40.Korbelius M, Vujic N, Sachdev V, Obrowsky S, Rainer S, Gottschalk B, Graier WF, and Kratky D (2019). ATGL/CGI-58-Dependent hydrolysis of a lipid storage pool in murine enterocytes. Cell Rep. 28, 1923–1934.e4. 10.1016/j.celrep.2019.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikolaeva O, Takahashi Y, Moiseyev G, and Ma J.-x. (2009). Purified RPE65 shows isomerohydrolase activity after reassociation with a phospholipid membrane. FEBS J. 276, 3020–3030. 10.1111/j.1742-4658.2009.07021.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi Y, Moiseyev G, Chen Y, and Ma J.-x. (2005). Identification of conserved histidines and glutamic acid as key residues for isomerohydrolase activity of RPE65, an enzyme of the visual cycle in the retinal pigment epithelium. FEBS Lett. 579, 5414–5418. 10.1016/j.febslet.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Wang H, Becuwe M, Housden BE, Chitraju C, Porras AJ, Graham MM, Liu XN, Thiam AR, Savage DB, Agarwal AK, et al. (2016). Seipin is required for converting nascent to mature lipid droplets. Elife 5, e16582. 10.7554/eLife.16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen F, Yan B, Ren J, Lyu R, Wu Y, Guo Y, Li D, Zhang H, and Hu J (2021). FIT2 organizes lipid droplet biogenesis with ER tubule-forming proteins and septins. J. Cell Biol 220, e201907183. 10.1083/jcb.201907183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tansey JT, Sztalryd C, Hlavin EM, Kimmel AR, and Londos C (2004). The central role of perilipin A in lipid metabolism and adipocyte lipolysis. IUBMB Life 56, 379–385. 10.1080/15216540400009968. [DOI] [PubMed] [Google Scholar]
- 46.Fader Kaiser CM, Romano PS, Vanrell MC, Pocognoni CA, Jacob J, Caruso B, and Delgui LR (2021). Biogenesis and breakdown of lipid droplets in pathological conditions. Front. Cell Dev. Biol 9, 826248. 10.3389/fcell.2021.826248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin Y, Moiseyev G, Chakraborty D, and Ma J.-x. (2017). A dominant mutation in Rpe65, D477G, delays dark adaptation and disturbs the visual cycle in the mutant knock-in mice. Am. J. Pathol 187, 517–527. 10.1016/j.ajpath.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghyselinck NB, Båvik C, Sapin V, Mark M, Bonnier D, Hindelang C, Dierich A, Nilsson CB, Håkansson H, Sauvant P, et al. (1999). Cellular retinol-binding protein I is essential for vitamin A homeostasis. EMBOJ. 18, 4903–4914. 10.1093/emboj/18.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma J, Zhang J, Othersen KL, Moiseyev G, Ablonczy Z, Redmond TM, Chen Y, and Crouch RK (2001). Expression, purification, and MALDI analysis of RPE65. Invest. Ophthalmol. Vis. Sci 42, 1429–1435. [PubMed] [Google Scholar]
- 50.Molday RS, and MacKenzie D (1983). Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry 22, 653–660. 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 51.Moiseyev G, Takahashi Y, Chen Y, Kim S, and Ma J.-x. (2008). RPE65 from cone-dominant chicken is a more efficient isomerohydrolase compared with that from rod-dominant species. J. Biol. Chem 283, 8110–8117. 10.1074/jbc.M703654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi Y, Moiseyev G, Ablonczy Z, Chen Y, Crouch RK, and Ma J.-x. (2009). Identification of a novel palmitylation site essential for membrane association and isomerohydrolase activity of RPE65. J. Biol. Chem 284, 3211–3218. 10.1074/jbc.M807248200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Livak KJ, and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 55.Bradford MM (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 72, 248–254. 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 56.Qiu F, Ma X, Shin Y-H, Chen J, Chen Q, Zhou K, Wu W, Liang W, Wu Y, Song Q, and Ma J-X (2020). Pathogenic role of human C-reactive protein in diabetic retinopathy. Clin. Sci 134, 1613–1629. 10.1042/cs20200085. [DOI] [PubMed] [Google Scholar]
- 57.Malechka VV, Moiseyev G, Takahashi Y, Shin Y, and Ma J.-x. (2017). Impaired rhodopsin generation in the rat model of diabetic retinopathy. Am. J. Pathol 187, 2222–2231. 10.1016/j.ajpath.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shin Y, Moiseyev G, Petrukhin K, Cioffi CL, Muthuraman P, Takahashi Y, and Ma J-X (2018). A novel RPE65 inhibitor CU239 suppresses visual cycle and prevents retinal degeneration. Biochim. Biophys. Acta, Mol. Basis Dis 1864, 2420–2429. 10.1016/j.bbadis.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garwin GG, and Saari JC (2000). [19] High-performance liquid chromatography analysis of visual cycle retinoids. In Methods Enzymol. (Academic Press; ), pp. 313–324. 10.1016/S0076-6879(00)16731-X. [DOI] [PubMed] [Google Scholar]
- 60.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, and Behr JP (1995). A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. USA 92, 7297–7301. 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sonawane ND, Szoka FC, and Verkman AS (2003). Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J. Biol. Chem 278, 44826–44831. 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper does not report new or original code.
The raw data and original images reported in this paper will be shared by the lead contact upon request.
Any additional information required to reanalyze the data reported in this paper will be available from the lead contact upon request.