Abstract
We propose a deep learning approach to segment the skin lesion in dermoscopic images. The proposed network architecture uses a pretrained EfficientNet model in the encoder and squeeze-and-excitation residual structures in the decoder. We applied this approach on the publicly available International Skin Imaging Collaboration (ISIC) 2017 Challenge skin lesion segmentation dataset. This benchmark dataset has been widely used in previous studies. We observed many inaccurate or noisy ground truth labels. To reduce noisy data, we manually sorted all ground truth labels into three categories — good, mildly noisy, and noisy labels. Furthermore, we investigated the effect of such noisy labels in training and test sets. Our test results show that the proposed method achieved Jaccard scores of 0.807 on the official ISIC 2017 test set and 0.832 on the curated ISIC 2017 test set, exhibiting better performance than previously reported methods. Furthermore, the experimental results showed that the noisy labels in the training set did not lower the segmentation performance. However, the noisy labels in the test set adversely affected the evaluation scores. We recommend that the noisy labels should be avoided in the test set in future studies for accurate evaluation of the segmentation algorithms.
Keywords: Melanoma, Dermoscopy, Deep learning, Image segmentation, Noisy data
Introduction
An estimated 99,780 new cases of invasive melanoma and 97,920 in situ melanoma will be diagnosed in 2022 in the USA [1]. Dermoscopy is an imaging modality to aid dermatologists for the early detection of skin cancer and can improve diagnostic accuracy over clinical visual inspection by the experienced domain expert [2–4].
Computer vision techniques have improved appreciably in recent years [5–9] and have been successfully applied to many medical imaging problems [10–13]. In the skin cancer domain, deep learning techniques combined with dermoscopy have higher diagnostic accuracy than experienced dermatologists [10, 14–17]. Pathan et al. published a recent review detailing both handcrafted and deep learning (DL) techniques for computer-aided diagnosis of skin lesions [18]. Although deep learning eliminates a tedious feature engineering process, recent studies show that the fusion of deep learning and handcrafted features can improve accuracy in skin cancer diagnosis [17, 19–22]. Handcrafted features are not as straightforward as the deep learning method, and they require a lesion border to define the region of interest. Accurate calculation of handcrafted lesion features depends upon correct detection of the lesion border [22]. Thus, lesion segmentation is an important step in computer-aided diagnosis of skin cancer.
Traditional image processing methods were applied to segment the skin lesion in dermoscopic images [23–25]. These methods performed well on small sets but generated unsatisfactory results when applied to challenging conditions such as low contrast between lesion and background, lesions with different colors, and images with artifacts like hair, ruler marks, gel bubbles, and ink markers. Deep learning techniques have overcome these challenges to some extent and improved border detection in skin lesion images [26–30].
Al-Masni et al. [26] proposed a deep full-resolution convolutional network for skin lesion segmentation. Unlike U-Net [31], this method does not employ upsampling or downsampling operations so that the feature maps always have the same resolution from the input to the output. The deep learning methods require little preprocessing and the RGB color images are directly fed to the network. However, recent studies showed that adding more input color channels improves skin lesion segmentation. Yuan and Lo [28] combined three RGB channels, three HSV channels, and one L channel of CIELAB color space and input 7-channel images to their deep neural network model and showed improved results. Ozturk and Ozkaya [30] also used 7 channels in their deep learning method; however, their approach was slightly different. The first input layer took three RGB channels and four additional channels (S of HSV color space, I of YIQ color space, B of CBR color space, and Z of XYZ color space) were fed to deeper intermediate layers.
Xie et al. [29] created a high-resolution feature block (HRFB) having three branches — a normal convolutional branch, a spatial attention branch, and a channel attention branch. Tong et al. [32] used an extended U-Net architecture and proposed ASCU-Net by employing a triple attention mechanism of attention gate [33], spatial attention module, and channel attention module.
A transfer learning approach has also been applied to skin lesion segmentation problems. Kadry et al. [34] and Rajinikanth et al. [35] employed a pretrained VGG [7] network to encode the important features from the skin lesion image and then upsampled the feature maps repeatedly to generate the segmentation mask. Zafar et al. [36] employed a ResNet-50 [9] architecture pretrained on ImageNet [37] as the encoder network in their U-Net architecture. A similar method by Tschandl et al. [27] also used a ResNet-34 architecture as the encoding layers and investigated the effect of random weight initialization versus domain-specific or ImageNet pretraining. Nawaz et al. [38] presented a U-Net architecture using DenseNet [39] encoder to segment melanoma lesion of varying colors and sizes. Nguyen et al. [40] integrated a pretrained EfficientNet-B4 [41] and the residual blocks in their U-Net architecture. Despite the early success of deep learning methods on skin lesion segmentation, many current architectures still fail to produce satisfactory results on challenging conditions like low skin-versus-lesion contrast and presence of image artifacts like hair or ruler marks, ink markers, and gel bubbles. Another concern we found was the presence of inaccurate or noisy ground truth (GT) masks in the benchmark ISIC 2017 [42] skin lesion segmentation datasets used in previous studies. These noisy GTs in the benchmark dataset warrant investigation to determine their effect on skin lesion segmentation.
In this study, we propose a deep learning method to improve skin lesion segmentation in dermoscopic images. The proposed method uses a modified ChimeraNet [43] architecture that was used to detect hair and ruler marks in dermoscopic images. The main contributions of this paper are as follows:
-
(i)
The proposed method achieves state-of-the-art segmentation performance on the ISIC 2017 skin lesion segmentation dataset. This segmentation improvement can benefit conventional analysis of the lesion, which depends on accurate segmentation.
-
(ii)
We identify noisy or inaccurate ground truth labels in the benchmark public dataset.
-
(iii)
We investigate the effect of pruning the noisy or inaccurate ground truth labels from the dataset.
Materials and Methods
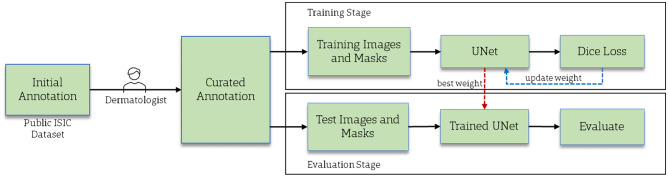
This section discusses the materials and methods used in this study. Our proposed method has three stages — annotation curation, training, and evaluation or inference. First, a dermatologist or specialist assesses the quality of ground truth annotations in a benchmark public dataset. Second, a UNet segmentation model is trained using the curated training set. Finally, the trained model is evaluated on the curated test set. The overall flow diagram of the proposed method is shown in Fig. 1.
Fig. 1.
The overall flow diagram of a proposed skin lesion segmentation method
Image Datasets
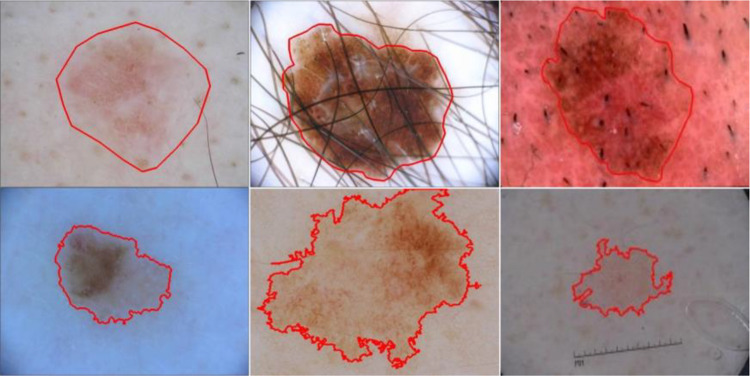
The dataset used in this study is the publicly available ISIC 2017 [42] skin lesion segmentation dataset. It is a large skin lesion segmentation dataset released as a part of the 2017 International Skin Imaging Collaboration (ISIC) Challenge. It provides 2750 dermoscopic skin lesion images with lesion boundary masks — 2000 training, 150 validation, and 600 test images. The ground truth (GT) lesion boundary masks were determined under the supervision of expert clinicians using both manual annotation and semi-automated process, as shown in Fig. 2. The images are 8-bit RGB images with varying height and width ranging from a few hundred pixels to a few thousand pixels. As the dataset provides a single train-validation split, we combined the official training and validation sets to create a single training set of 2150 images to run fivefold cross-validation experiments. The official 600 test images were used as a holdout test set to evaluate the performance of our proposed method against the state-of-the-art methods.
Fig. 2.
Skin lesion dermoscopy images with ground truth lesion boundary (red) from publicly available ISIC skin lesion datasets. The masks are manually drawn (first row) or generated using a semi-automated process (second row)
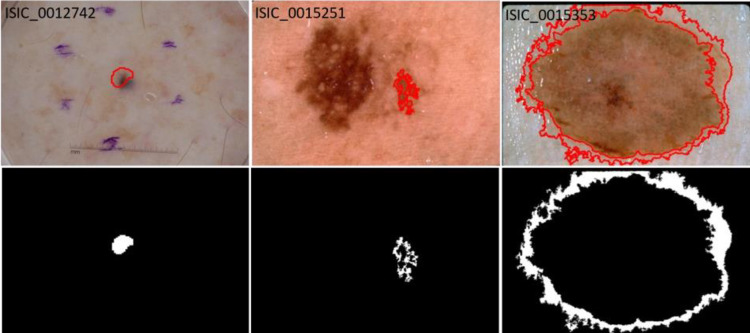
As the GT masks were created using both manual and semi-automated processes, we found some of the ground truth masks, especially those determined automatically, were inaccurate (Fig. 3). The noisy labels or inaccurate examples in the training set might affect the model adversely, reducing accuracy. Conversely, noisy labels might aid performance by increasing the number of training examples or regularizing the overparameterized deep learning model. Also, the noisy labels in the test set might not demonstrate a true evaluation of the model. Thus, all 2750 GT masks, including both train and test sets, were re-evaluated by a dermatologist and categorized into three categories — good, mildly noisy, and noisy. The number of GT masks in each category after reevaluation is shown in Table 1.
Fig. 3.
Examples of inaccurate or noisy ground truths on ISIC lesion segmentation dataset. Overlays show GT lesion boundaries on lesion images (top row) and ground truth lesion segmentation mask (bottom row). The lesion boundary (red) fails to cover the whole lesion in all examples
Table 1.
Number of images with good, mildly noisy, and noisy lesion boundary labels in ISIC 2017 train and test sets
| Image set | Good | Mildly noisy | Noisy |
|---|---|---|---|
| Train + validation (2150) | 1982 | 149 | 19 |
| Test (600) | 493 | 87 | 20 |
| Total (2750) | 2475 | 236 | 39 |
Data Augmentation
Data augmentation can be applied during the training of deep neural networks to increase the number of training images without adding new images. Augmentation will result in better generalization of deep network models and reduce the overfitting problem. Data augmentation performs different image transform methods on the original training images to generate more examples for training. In this study, we selected the following image transforms for data augmentation:
Height or width shift with a range of (− 0.15, + 0.15)
Horizontal or vertical flip
Rotation with a range between + 90° to − 90°
Zoom with a range (− 0.15, + 15)
Brightness with a range of (0.85, 1.15)
Contrast with a range of (0.85, 1.15)
Furthermore, all images were resized to 448 × 448, and the image pixel values were rescaled between 0 and 1. Finally, the images were normalized before feeding them to the deep network.
Network Architecture
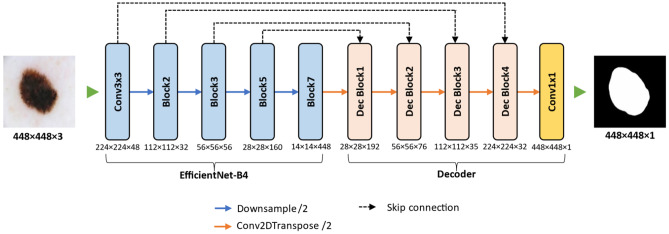
In this study, we used a modified U-Net [31] convolutional neural network (CNN) architecture for skin lesion segmentation by Lama et al. [43]. The proposed encoder-decoder based image segmentation model, named ChimeraNet, uses a pretrained EfficientNet [41] model in the encoder and squeeze-and-excitation [44] structures in the decoder. Furthermore, we applied a dilated convolution [45] operation in place of a regular convolution operation in these squeeze-and-excitation residual blocks. As artifacts like hair, ruler marks, and purple marks hinder the detection of important features from skin lesion images [23, 46], we adopted the CNN architecture that was already successful in segmenting fine structures like hair and ruler marks from the skin lesion images. However, a few minor modifications were performed on the original ChimeraNet [43] model to accommodate skin lesion segmentation task. The overall pipeline of the proposed UNet architecture is shown in Fig. 4.
Fig. 4.
Proposed architecture for skin lesion segmentation. An encoder-decoder architecture with pretrained EfficientNet model as the encoder network, and the decoder network comprised four squeeze-and-excitation residual blocks
Encoder
In the encoder part, we used the EfficientNet [41] model pretrained on the ImageNet [37] image classification challenge dataset. EfficientNets are composed of mobile inverted bottleneck convolution (MBConv) structures and have 8 network variants from EfficientNet-B0 to EfficientNet-B7. These networks use multiple MBConv blocks grouped together to form seven larger blocks named Block1 to Block7, as given in Table 2. EfficientNetB0 is the baseline architecture, and other variants are scaled up by employing a compound scaling method that uniformly scales network depth, width, and resolution with a fixed set of scaling factors. In the proposed model, we used the pretrained EfficientNet-B4 variant of EfficientNet models as the encoder network. Like many CNN architectures, the EfficientNet model downsamples the feature map repeatedly while extracting the most useful features from the image. The spatial dimension of the final feature map gets much smaller than the original dimension of an input image. The dimensions of feature maps at different levels are given in Table 2.
Table 2.
Different blocks of EfficientNet-B4 model and their output feature map sizes and the number of channels
| Block name | Feature map size (W × H) | #Feature map (C) |
|---|---|---|
| Input layer | 448 × 448 | 3 |
| Conv3 × 3 | 224 × 224 | 48 |
| Block 1 | 224 × 224 | 24 |
| Block 2 | 112 × 112 | 32 |
| Block 3 | 56 × 56 | 56 |
| Block 4 | 28 × 28 | 112 |
| Block 5 | 28 × 28 | 160 |
| Block 6 | 14 × 14 | 272 |
| Block 7 | 14 × 14 | 448 |
Conversely, the decoder network needs to expand these low-resolution feature maps to generate the segmentation map with spatial dimensions equal to those of the input image. The U-Net architecture uses the skip-connections to recover the spatial information lost in the encoder due to downsampling process. For precise localization of features, the skip-connection feeds the high-resolution output feature maps at various levels in the encoder to the decoder by skipping some blocks, as shown in Fig. 4. In the proposed method, we used the outputs of Conv3 × 3, Block2, Block3, and Block5 as sources of the skip-connections. These blocks are selected for skip connections because the size of output feature maps is downsampled by a factor of 2 in the subsequent block. The output dimensions of each block corresponding to the skip connections and the final output of the encoder are given in Table 2.
Decoder
The decoder network is constructed using a squeeze-and-excitation residual (SERes) structure [44], as shown in Fig. 5. The SERes block has a better feature representation capability than the plain convolution block, as it emphasizes the informative features and suppresses the weaker ones by modeling the interdependencies between channels of convolutional features. The decoder network has 4 blocks named Dec Block1 to Dec Block4, as shown in Fig. 4. Each decoder block is composed of a SERes block and gets two feature maps as inputs — an output feature map from the previous stage and a low-level feature map via a skip-connection from the encoder. For example, the first block (Dec Block1) of the decoder gets 14 × 14 × 448 feature input from the previous stage, the final output of the encoder, and 28 × 28 × 160 low-level feature input from Block5 via a skip-connection. Here, three dimensions of the feature map represent width (W), height (H), and number of feature map or channel (C). Both feature inputs are concatenated before feeding to the SERes block. However, the dimensions of both inputs are not the same. To combine both inputs, first, the 14 × 14 × 448 feature map from the previous stage is upsampled using a transposed convolution, also called deconvolution. The transposed convolution performs 2 × 2 upsampling followed by a 3 × 3 convolution operation. We selected the number of filters for transposed convolution as half of the number of input channels, i.e., 224 (= 448/2), thus generating a 28 × 28 × 224 feature map. Then, the two inputs are concatenated along the channel axis to form 28 × 28 × 384 feature map before feeding to the SERes block. The SERes block combines an SE block with a residual structure [44], as shown in Fig. 5. The residual unit in the SERes block is a double convolution block, which applies two sets of 3 × 3 dilated convolutions (dilation rate = 2), batch normalization [47], and rectified linear unit (ReLU) operations. Again, we selected the number of filters for two convolution layers in the residual unit as half of the input channels, i.e., 192 (= 384/2). The residual unit outputs a 28 × 28 × 192 feature map, and then the squeeze-and-excitation operation is performed to scale the features along the channel axis. To find the weights for each channel of the feature map, SE first applies global average pooling to reduce the feature map to 1 × 1 × 192 and then applies non-linear operations like FC, ReLU, FC, and sigmoid. The number of neurons in two FC layers are C/r and C, respectively, where r is a feature reduction factor and empirically selected as r = 8. The SE generates a 1 × 1 × 192 weight vector with each value in the range of 0 to 1. Then the residual feature is multiplied with a weight vector to scale the features and generate a 28 × 28 × 192 scaled feature map.
Fig. 5.
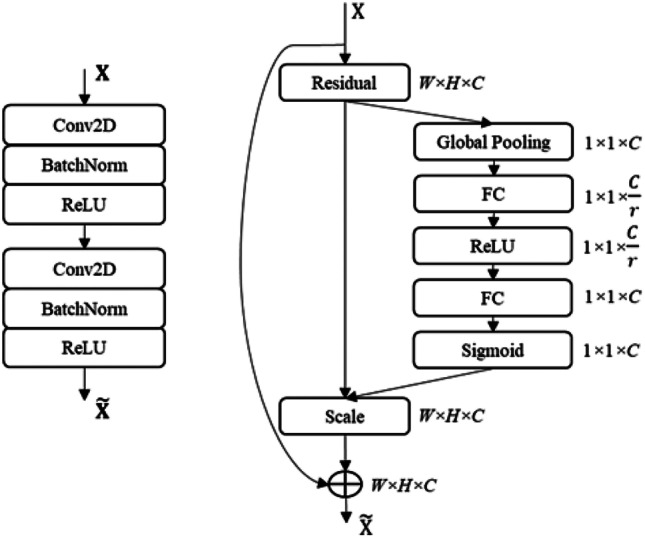
Structures of convolution blocks in the decoder network. Double convolution block (left) and squeeze-and-excitation residual block (right)
Furthermore, the SERes block combines this scaled residual feature map with the original input feature map. However, the channels in the original input feature map (28 × 28 × 224) and residual feature output (28 × 28 × 192) are not the same, so a 1 × 1 convolution operation with 192 filters followed by a batch normalization operation are performed on the original input feature map. Then, the SERes block adds two feature maps together and applies the ReLU operation to generate the final 28 × 28 × 192 feature output.
Similarly, the remaining decoder blocks (Dec Block2 to Dec Block4) apply the same set of operations as Dec Block1. Only the size and the number of feature maps are different, as shown in Table 3. The number of feature maps (C) corresponds to the number of convolutional filters applied in each SERes block. Also, the dropout operation with 0.4 probability was applied after each decoder block to regularize the network from the overfitting during the training. The output resolution of the final decoder block, Dec Block4, is still smaller than the original input resolution so it is upsampled by a factor of 2. Finally, 1 × 1 convolution and a sigmoid function are applied to generate the final segmentation map of size 448 × 448 × 1. The 1 × 1 convolution reduces the number of channels to the desired number of classes, and the sigmoid operation converts all pixel values to the range between 0 and 1. Each pixel value in the segmentation map represents the probability score of that pixel belonging to the skin lesion.
Table 3.
SERes blocks in the decoder and their output sizes
| Block name | Size (W × H) | Feature map (C) |
|---|---|---|
| Dec Block1 | 28 × 28 | 192 |
| Dec Block2 | 56 × 56 | 76 |
| Dec Block3 | 112 × 112 | 35 |
| Dec Block4 | 224 × 224 | 32 |
During inference, we give five different augmented versions of an input image to the trained deep network: an original image, a horizontally flipped image, a vertically flipped image, a 90° clockwise rotated image, and a 90° counterclockwise rotated image. The deep network generates the segmentation output for each image, and the final segmentation mask is generated by aggregating these five outputs using the unweighted average of the five predicted masks. The mask is binarized using the threshold of 0.5 to generate the final segmentation mask.
Training Details
All models were built using Keras with a Tensorflow backend in Python 3 and trained using a single 32 GB Nvidia V100 graphics card. We used a fivefold cross-validation method to tune the hyperparameters, which are shown in Table 4. The networks were trained using a Dice [48] loss function and Kingma and Adam [49] optimization algorithm. To reduce overfitting of a deep neural network model, we used data augmentation (see details in section “Data Augmentation”), a dropout layer, and an early stopping technique. The dropout probability of 0.4 was selected for the dropout layers in each decoder block. All images were resized to 448 × 448 using bilinear interpolation.
Table 4.
Training hyperparameters
| Parameter | Value |
|---|---|
| image size | 448 × 448 |
| learning rate | 0.0001 |
| batch size | 10 |
| epoch | 200 |
| dropout probability | 0.4 |
| optimizer | Adam |
| loss method | dice |
| early stopping patience | 30 |
Experimental Results
We evaluated the performance of the proposed method by comparing the predicted lesion segmentation masks with the provided ground truth masks on the official ISIC 2017 [42] skin lesion segmentation dataset having 600 test images. In addition, the proposed method was also evaluated on curated ISIC 2017 test sets. The evaluation metrics used are Jaccard index (Jac), Dice similarity coefficient (Dsc), and accuracy (Acc).
Segmentation Performance of the Proposed and State-of-the-Art Methods on ISIC 2017 Test Images
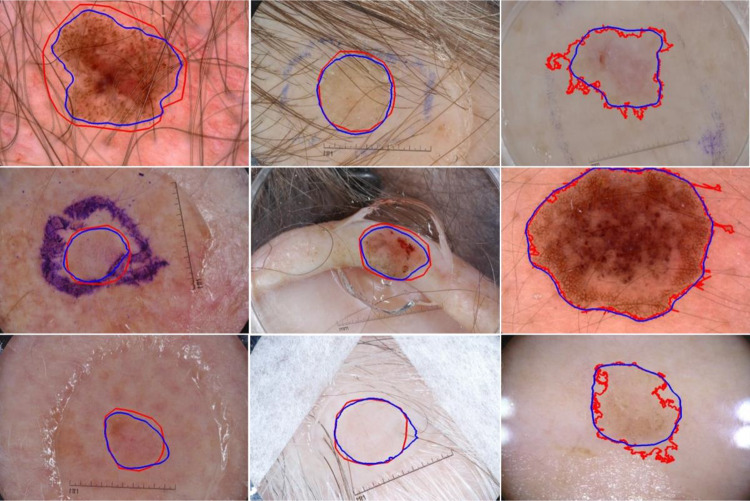
In this section, we compared the lesion segmentation performance of the proposed method on 600 ISIC 2017 test images with the previously reported methods, as shown in Table 5. The proposed method achieved the highest Jaccard score of 0.807, compared to the state-of-the-art methods [53]. In Fig. 6, we show the segmentation results of the proposed method on ISIC 2017 test images. The segmentation results showed that the proposed method successfully finds the lesion border despite the presence of hair, ruler marks, ink marker, and sticker artifacts. Also, the proposed method accurately segments the skin lesion from the background in challenging images having low contrast between the skin and the lesion (see row 3). The predicted masks have smooth lesion borders (blue) compared to the jagged ground truth lesion borders (red) generated by semi-automated processes (see third column).
Table 5.
Performance comparison with other lesion segmentation methods on the original ISIC 2017 test dataset
| Methods | Year | Jac | Dsc | Acc |
|---|---|---|---|---|
| Al-Masni et al. [26] | 2018 | 0.771 | 0.871 | 0.940 |
| Tschandl et al. [27] | 2019 | 0.768 | 0.851 | |
| Yuan and Lo [28] | 2019 | 0.765 | 0.849 | 0.934 |
| Navarro et al. [50] | 2019 | 0.769 | 0.854 | 0.955 |
| Xieet al. [29] | 2020 | 0.783 | 0.862 | 0.938 |
| Ozturk and Ozkaya [30] | 2020 | 0.783 | 0.886 | 0.953 |
| Shan et al. [51] | 2020 | 0.763 | 0.846 | 0.937 |
| Kaymak et al. [52] | 2020 | 0.725 | 0.841 | 0.939 |
| Nguyen et al. [40] | 2020 | 0.781 | 0.861 | |
| Zafar et al. [36] | 2020 | 0.772 | 0.858 | |
| Goyal et al. [53] | 2020 | 0.793 | 0.871 | |
| Tong et al. [32] | 2021 | 0.742 | 0.926 | |
| Chen et al. [54] | 2022 | 0.8036 | 0.8704 | 0.9471 |
| Ashraf et al. [55] | 2022 | 0.8005 | ||
| Our method | 0.807 | 0.880 | 0.948 |
The bold values emphasize the highest values
Fig. 6.
Segmentation results of the proposed method on ISIC 2017 test set. Overlays of ground truth lesion boundary (red) and predicted lesion boundary (blue) on skin lesion images. Lesion border predictions are accurate even in the presence of artifacts like hair, ruler marks, and ink markers
Effect of Pruning the Noisy GT Labels from ISIC 2017 Dataset
In this section, we investigated the effect of pruning the noisy ground truth (GT) labels from both training and test sets of the ISIC 2017 lesion segmentation dataset. Table 6 shows the segmentation performance of the proposed method on 600 test images before and after pruning the noisy labels from the dataset.
Table 6.
Segmentation performance comparison of the proposed method before and after pruning noisy and mildly noisy GT labels from ISIC 2017 train and test sets
| Train pruned (Ntrain) | Test pruned (Ntest) | Jac | Dsc | Acc |
|---|---|---|---|---|
| None (2150) | None (600) | 0.807 | 0.880 | 94.779 |
| Noisy (580) | 0.817 | 0.889 | 95.536 | |
| Noisy + Mildly noisy (493) | 0.832 | 0.900 | 96.393 | |
| Noisy (2131) | None (600) | 0.806 | 0.878 | 94.723 |
| Noisy (580) | 0.815 | 0.887 | 95.518 | |
| Noisy + Mildly noisy (493) | 0.827 | 0.895 | 96.210 | |
| Noisy + Mildly noisy (1982) | None (600) | 0.802 | 0.875 | 94.658 |
| Noisy (580) | 0.812 | 0.885 | 95.398 | |
| Noisy + Mildly noisy (493) | 0.824 | 0.893 | 96.073 |
The values in bold are the highest scores
First, we removed the noisy GT labels from the training set. When 19 noisy labels were removed from the training set of 2150 images, there was no significant change in the performance per Jaccard scores (0.807 vs. 0.806) on 600 test images. However, when both mildly noisy and noisy labels (168 images) were removed, the performance slightly decreased from a Jaccard score of 0.807 to 0.802 on 600 test images. Larger training sets provide more examples, advantageous for training the deep learning model even if the labels are noisy or mildly noisy.
Second, we removed the noisy GT labels from 600 ISIC 2017 test images. The model trained on the full training set (2150 images) improved the Jaccard score by 0.01 from 0.807 to 0.817 (a 1% improvement) when 20 noisy labels were removed. Furthermore, when both noisy (= 20) and mildly noisy (= 87) GT labels were removed, we achieved the highest Jaccard score of 0.832, which is 2.5% improvement from 0.807.
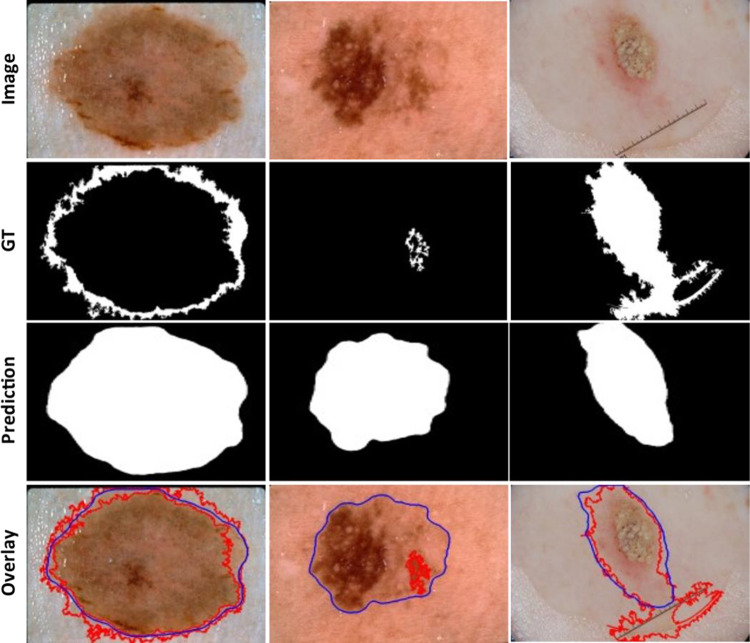
In Fig. 7, we showed the segmentation results of the proposed method on the ISIC2017 test images having noisy or inaccurate ground truth masks. The overlays of the predicted lesion boundary (indicated by blue line) and the ground truth lesion boundary (indicated by red line) on the third-row show that the predicted segmentation covers the lesion area more accurately than the ground truth lesion mask.
Fig. 7.
Segmentation results of the proposed method on examples having noisy (or inaccurate) ground truth (GT) on an official ISIC 2017 test set. The predicted lesion borders (blue) cover the lesion area more accurately than the GT lesion border (red)
Discussion
In this study, we demonstrated that our proposed deep learning method successfully detects the skin lesion boundary on most dermoscopic skin lesion images. We scored segmentation performance using the Jaccard index, dice similarity coefficient, and accuracy. As the accuracy metric counts true-negative pixels and true-positive pixels equally, accuracy overstates the actual performance when positive (lesion) and negative (background) pixels are highly imbalanced. Accordingly, accuracy is less useful than the other methods in assessing segmentation performance for lesions which occupy a small area of the image.
The proposed network architecture in this study was the same model, ChimeraNet, used in detecting hair and ruler marks in dermoscopic images [43]. The encoder-decoder architecture uses a pre-trained EfficientNet [41] model as the encoder network and a squeeze-and-excitation residual [44] structure as the convolutional block to construct the decoder network. The proposed method performed better than the state-of-the-art methods on the skin lesion segmentation task, with the highest Jaccard scores of 0.807 on the official ISIC2017 test set and 0.832 on the curated ISIC2017 test set. A very similar method was employed by Nguyen et al. [40], with a pretrained EfficientNet [41] model as an encoder network. Our proposed model improved the Jaccard score of Nguyen by 0.026, from 0.781 to 0.807, (a 3.2% improvement) on the ISIC 2017 test set. The main difference was the use of a different decoder network which employs a squeeze-and-excitation residual structure and dilated convolution operations. The squeeze-and-excitation convolutional structure improved the segmentation performance by focusing on more critical channels of the feature maps [44], resulting in a better feature representation and generalization than the basic convolutional blocks. The dilated convolution operations give the larger receptive field without increasing the number of filter parameters. Lama et al. [43] compared loss functions and various U-Net architectures and found that the U-Net architecture presented here was best in the dermoscopy domain. Thus, ablation studies comparing various architectures are not repeated here.
Although the ISIC 2017 skin lesion segmentation dataset is the largest publicly available and most-used dataset in skin lesion segmentation studies in the deep learning era, we found many inaccurate ground truth masks in the dataset. These inaccurate GT masks might affect the segmentation performance of the deep learning model. Thus, our dermatologist manually reevaluated all ground truth masks and graded them into three categories — good, mildly noisy, and noisy. Then we conducted multiple experiments to analyze the effect of removing the noisy or inaccurate ground truth masks from both training and test sets. The results in Table 6 show that the model trained on the complete training set performed slightly better than the model trained on the curated training set (after removing noisy and mildly noisy examples). The full training set model had 0.807 Jaccard and 0.880 Dice scores on 600 ISIC2017 test images, while the curated training set model only achieved 0.802 Jaccard and 0.875 Dice scores. These experimental results show that the presence of noisy or inaccurate labels in the training set does not reduce the model’s performance. Instead, some noisy or inaccurate labels in the training set might provide a regularization effect for the overparameterized deep learning model and thus generalize better, aside from the beneficial effect of a more extensive training set. Conversely, the noisy or inaccurate GTs in the test set adversely affected the evaluation scores. The Jaccard and Dice similarity scores were improved from 0.807 to 0.832 and 0.88 to 0.90, respectively when the noisy and mildly noisy GT labels were removed from the official ISIC 2017 lesion segmentation test set. This result shows that the segmentation performance is significantly underestimated when evaluated on the test set having noisy or inaccurate GT labels. As many previous studies have used the official test set to evaluate their method against the state-of-the-art methods, comparisons might not be fair and accurate.
Image segmentations created by ChimeraNet deep learning are subjectively improved, compared to both automatic and manual borders. The excessive jaggedness of the automatic borders is remedied with the new technique. The manual borders, characterized by straight lines joined at points, are smoothed. Both types of distortion in the ground truth segmentations, excessive jaggedness, and straight-line junctions, are non-physiologic and may lead to error in handcrafted feature analysis which depends upon an accurate border.
There are limitations to this study. Only one dermatologist scored the accuracy of the segmentations. No new noisy segmentations were added to the benchmark ISIC 2017 dataset to see the effect of noisy data at different proportions in the training set. The experiments were conducted using only the available noisy data in the original dataset. Furthermore, we did not create new ground truths for the noisy or inaccurate ground truths.
Conclusion
In this study, we employed a novel deep learning technique to segment skin lesions in dermoscopic images. The proposed method performed better than the previous state-of-the-art methods. We observed the presence of noisy or inaccurate ground truth labels in a large benchmark dataset. With help of a dermatologist, we manually re-evaluated the ground truth masks. Furthermore, we investigated the effect of noisy ground truth labels in the benchmark dataset. Our experimental results show that more training data, including noisy data, yields better performance than the condensed curated data. However, the noisy data adversely affects the evaluation scores when present in the test data. The test scores were improved when the noisy or inaccurate labels were removed from the official test set. We recommend that future researchers avoid the noisy data in the test set for a fair and accurate evaluation of their lesion segmentation algorithms.
This manuscript has NOT been copyrighted, published, submitted, or accepted for publication elsewhere.
Author Contribution
We confirm that all the authors contributed to the study conception and design. The experiments are conducted by Norsang Lama. Data analysis and validation are performed by all the authors. The first draft of the manuscript was written by Norsang Lama, and all the authors commented on the previous versions of the manuscript. All the authors read and approved the final manuscript.
Data Availability
The datasets used in this study are publicly available benchmark datasets.
Declarations
Ethics Approval
NA.
Consent to Participate
NA.
Consent to for Publication
NA
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Pehamberger H, Binder M, Steiner A, Wolff K. In vivo epiluminescence microscopy: improvement of early diagnosis of melanoma. Journal of Investigative Dermatology. 1993;100(3 SUPPL.):S356–S362. doi: 10.1038/jid.1993.63. [DOI] [PubMed] [Google Scholar]
- 3.Soyer HP, Argenziano G, Talamini R, Chimenti S. Is dermoscopy useful for the diagnosis of melanoma? Arch Dermatol. 2001;137(10):1361–1363. doi: 10.1001/archderm.137.10.1361. [DOI] [PubMed] [Google Scholar]
- 4.Braun RP, Rabinovitz HS, Oliviero M, Kopf AW, Saurat JH. Pattern analysis: a two-step procedure for the dermoscopic diagnosis of melanoma. Clin Dermatol. 2002;20(3):236–239. doi: 10.1016/S0738-081X(02)00216-X. [DOI] [PubMed] [Google Scholar]
- 5.A. Krizhevsky, I. Sutskever, and G. Hinton, ImageNet classification with deep convolutional neural networks, in Advances in Neural Information and Processing Systems (NIPS), vol. 25, 2012, pp. 1097–1105.
- 6.C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens, and Z. Wojna, Rethinking the inception architecture for computer vision, in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 2818–2826.
- 7.K. Simonyan and A. Zisserman, Very deep convolutional networks for large-scale image recognition, arXiv preprint arXiv:1409.1556, 2014.
- 8.Goodfellow I, et al. Generative adversarial networks. Commun ACM. 2020;63(11):139–144. doi: 10.1145/3422622. [DOI] [Google Scholar]
- 9.K. He, X. Zhang, S. Ren, and J. Sun, Deep residual learning for image recognition, in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 770–778.
- 10.Esteva A, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulshan V, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 12.Sornapudi S, et al. Deep learning nuclei detection in digitized histology images by superpixels. J Pathol Inform. 2018;9(1):5. doi: 10.4103/jpi.jpi_74_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Litjens G, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Ferris LK, et al. Computer-aided classification of melanocytic lesions using dermoscopic images. J Am Acad Dermatol. 2015;73(5):769–776. doi: 10.1016/J.JAAD.2015.07.028. [DOI] [PubMed] [Google Scholar]
- 15.Marchetti MA, et al. Results of the 2016 International Skin Imaging Collaboration International Symposium on biomedical imaging challenge: comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J Am Acad Dermatol. 2018;78(2):270–277.e1. doi: 10.1016/j.jaad.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haenssle HA, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Annals of Oncology. 2018;29(8):1836–1842. doi: 10.1093/annonc/mdy166. [DOI] [PubMed] [Google Scholar]
- 17.N. C. F. Codella et al., Deep learning ensembles for melanoma recognition in dermoscopy images, IBM J. Res. Dev., vol. 61, no. 4–5, pp. 5:1–5:15, Jul. 2017, 10.1147/JRD.2017.2708299.
- 18.Pathan S, Prabhu KG, Siddalingaswamy PC. Techniques and algorithms for computer aided diagnosis of pigmented skin lesions—a review. Biomed Signal Process Control. 2018;39:237–262. doi: 10.1016/J.BSPC.2017.07.010. [DOI] [Google Scholar]
- 19.T. Majtner, S. Yildirim-Yayilgan, and J. Y. Hardeberg, Combining deep learning and hand-crafted features for skin lesion classification, 2016 6th International Conference on Image Processing Theory, Tools and Applications, IPTA 2016, 2017, 10.1109/IPTA.2016.7821017.
- 20.N. Codella, J. Cai, M. Abedini, R. Garnavi, A. Halpern, and J. R. Smith, Deep learning, sparse coding, and SVM for melanoma recognition in dermoscopy images BT - Machine Learning in Medical Imaging, 2015, pp. 118–126.
- 21.González-Díaz I. DermaKNet: incorporating the knowledge of dermatologists to convolutional neural networks for skin lesion diagnosis. IEEE J Biomed Health Inform. 2019;23(2):547–559. doi: 10.1109/JBHI.2018.2806962. [DOI] [PubMed] [Google Scholar]
- 22.Hagerty JR, et al. Deep learning and handcrafted method fusion: higher diagnostic accuracy for melanoma dermoscopy images. IEEE J Biomed Health Inform. 2019;23(4):1385–1391. doi: 10.1109/JBHI.2019.2891049. [DOI] [PubMed] [Google Scholar]
- 23.G. Celebi, Emre M.; Wen, Quan; Iyatomi, Hitoshi; Shimizu, Kouhei; Zhou, Huiyu; Schaefer, A state-of-the-art on lesion border detection in dermoscopy images, in Dermoscopy Image Analysis, J. S. Celebi, M. Emre; Mendonca, Teresa; Marques, Ed. Boca Raton: CRC Press, 2015, pp. 97–129. [Online]. Available: 10.1201/b19107
- 24.Mishra NK, et al. Automatic lesion border selection in dermoscopy images using morphology and color features. Skin Research and Technology. 2019;25(4):544–552. doi: 10.1111/srt.12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.M. E. Celebi, H. Iyatomi, G. Schaefer, and W. v Stoecker, Lesion border detection in dermoscopy images, Computerized Medical Imaging and Graphics, vol. 33, no. 2, pp. 148–153, 2009, 10.1016/j.compmedimag.2008.11.002. [DOI] [PMC free article] [PubMed]
- 26.Al-masni MA, Al-antari MA, Choi MT, Han SM, Kim TS. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput Methods Programs Biomed. 2018;162:221–231. doi: 10.1016/j.cmpb.2018.05.027. [DOI] [PubMed] [Google Scholar]
- 27.Tschandl P, Sinz C, Kittler H. Domain-specific classification-pretrained fully convolutional network encoders for skin lesion segmentation. Comput Biol Med. 2019;104:111–116. doi: 10.1016/j.compbiomed.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Yuan Y, Lo YC. Improving dermoscopic image segmentation with enhanced convolutional-deconvolutional networks. IEEE J Biomed Health Inform. 2019;23(2):519–526. doi: 10.1109/JBHI.2017.2787487. [DOI] [PubMed] [Google Scholar]
- 29.F. Xie, J. Yang, J. Liu, Z. Jiang, Y. Zheng, and Y. Wang, Skin lesion segmentation using high-resolution convolutional neural network, Comput Methods Programs Biomed, vol. 186, p. 105241, 2020, 10.1016/j.cmpb.2019.105241. [DOI] [PubMed]
- 30.Öztürk Ş, Özkaya U. Skin lesion segmentation with improved convolutional neural network. J Digit Imaging. 2020;33(4):958–970. doi: 10.1007/s10278-020-00343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O. Ronneberger, P. Fischer, and T. Brox, U-Net: convolutional networks for biomedical image segmentation. [Online]. Available: http://lmb.informatik.uni-freiburg.de/
- 32.X. Tong, J. Wei, B. Sun, S. Su, Z. Zuo, and P. Wu, Ascu-net: attention gate, spatial and channel attention U-net for skin lesion segmentation, Diagnostics, vol. 11, no. 3, 2021, 10.3390/diagnostics11030501. [DOI] [PMC free article] [PubMed]
- 33.O. Oktay et al., Attention u-net: learning where to look for the pancreas, arXiv preprint arXiv:1804.03999, 2018.
- 34.S. Kadry, D. Taniar, R. Damaševičius, V. Rajinikanth, and I. A. Lawal, Extraction of abnormal skin lesion from dermoscopy image using VGG-SegNet, in 2021 Seventh International Conference on Bio Signals, Images, and Instrumentation (ICBSII), 2021, pp. 1–5.
- 35.Rajinikanth V, Kadry S, Damaševičius R, Sankaran D, Mohammed MA, Chander S. Skin melanoma segmentation using VGG-UNet with Adam/SGD optimizer: a study, in. Third International Conference on Intelligent Computing Instrumentation and Control Technologies (ICICICT) 2022;2022:982–986. [Google Scholar]
- 36.Zafar K, et al. Skin lesion segmentation from dermoscopic images using convolutional neural network. Sensors (Switzerland) 2020;20(6):1–14. doi: 10.3390/s20061601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L. Imagenet: a large-scale hierarchical image database, in. IEEE Conference on Computer Vision and Pattern Recognition. 2009;2009:248–255. [Google Scholar]
- 38.Nawaz M, et al. Melanoma segmentation: a framework of improved DenseNet77 and UNET convolutional neural network. Int J Imaging Syst Technol. 2022;32(6):2137–2153. doi: 10.1002/ima.22750. [DOI] [Google Scholar]
- 39.G. Huang, Z. Liu, L. Van Der Maaten, and K. Q. Weinberger, Densely connected convolutional networks, in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 4700–4708.
- 40.D. K. Nguyen, T. T. Tran, C. P. Nguyen, and V. T. Pham, Skin Lesion segmentation based on integrating EfficientNet and residual block into U-Net neural network, Proceedings of 2020 5th International Conference on Green Technology and Sustainable Development, GTSD 2020, pp. 366–371, 2020, 10.1109/GTSD50082.2020.9303084.
- 41.M. Tan and Q. Le, Efficientnet: rethinking model scaling for convolutional neural networks, in International Conference on Machine Learning, 2019, pp. 6105–6114.
- 42.N. C. F. Codella et al., Skin lesion analysis toward melanoma detection: a challenge at the 2017 International symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC), Proceedings - International Symposium on Biomedical Imaging, vol. 2018-April, pp. 168–172, 2018, 10.1109/ISBI.2018.8363547.
- 43.N. Lama et al., ChimeraNet: U-Net for hair detection in dermoscopic skin lesion images, J Digit Imaging, no. 0123456789, 2022, 10.1007/s10278-022-00740-6. [DOI] [PMC free article] [PubMed]
- 44.J. Hu, L. Shen, and G. Sun, Squeeze-and-excitation networks, in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2018, pp. 7132–7141.
- 45.F. Yu and V. Koltun, Multi-scale context aggregation by dilated convolutions, arXiv preprint arXiv:1511.07122, 2015.
- 46.Q. Abbas, M. E. Celebi, and I. F. Garc\’\ia, Hair removal methods: a comparative study for dermoscopy images, Biomed Signal Process Control, vol. 6, no. 4, pp. 395–404, 2011.
- 47.S. Ioffe and C. Szegedy, Batch normalization: accelerating deep network training by reducing internal covariate shift.
- 48.C. H. Sudre, W. Li, T. Vercauteren, S. Ourselin, and M. Jorge Cardoso, Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations, in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Springer, 2017, pp. 240–248. [DOI] [PMC free article] [PubMed]
- 49.D. P. Kingma and J. Ba, Adam: A method for stochastic optimization, arXiv preprint arXiv:1412.6980, 2014.
- 50.Navarro F, Escudero-Viñolo M, Bescós J. Accurate segmentation and registration of skin lesion images to evaluate lesion change. IEEE J Biomed Health Inform. 2019;23(2):501–508. doi: 10.1109/JBHI.2018.2825251. [DOI] [PubMed] [Google Scholar]
- 51.P. Shan, Y. Wang, C. Fu, W. Song, and J. Chen, Automatic skin lesion segmentation based on FC-DPN, Comput Biol Med, vol. 123, no. April, p. 103762, 2020, 10.1016/j.compbiomed.2020.103762. [DOI] [PubMed]
- 52.R. Kaymak, C. Kaymak, and A. Ucar, Skin lesion segmentation using fully convolutional networks: a comparative experimental study, Expert Syst Appl, vol. 161, p. 113742, 2020, 10.1016/j.eswa.2020.113742.
- 53.Goyal M, Oakley A, Bansal P, Dancey D, Yap MH. Skin lesion segmentation in dermoscopic images with ensemble deep learning methods. IEEE Access. 2020;8:4171–4181. doi: 10.1109/ACCESS.2019.2960504. [DOI] [Google Scholar]
- 54.Chen P, Huang S, Yue Q. Skin lesion segmentation using recurrent attentional convolutional networks. IEEE Access. 2022;10(September):94007–94018. doi: 10.1109/ACCESS.2022.3204280. [DOI] [Google Scholar]
- 55.Ashraf H, Waris A, Ghafoor MF, Gilani SO, Niazi IK. Melanoma segmentation using deep learning with test-time augmentations and conditional random fields. Sci Rep. 2022;12(1):1–16. doi: 10.1038/s41598-022-07885-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used in this study are publicly available benchmark datasets.