Abstract
NS3 protein of dengue virus type 2 has a serine protease domain within the N-terminal 180 residues. NS2B is required for NS3 to form an active protease involved in processing of the viral polyprotein precursor. The region carboxy terminal to the protease domain has conserved motifs present in several viral RNA-stimulated nucleoside triphosphatase (NTPase)/RNA helicases. To define the functional domains of protease and NTPase/RNA helicase activities of NS3, full-length and amino-terminal deletion mutants of NS3 were expressed in Escherichia coli and purified. Deletion of 160 N-terminal residues of NS3 (as in NS3del.2) had no detrimental effect on the basal and RNA-stimulated NTPase as well as RNA helicase activities. However, mutagenesis of the conserved P-loop motif of the RNA helicase domain (K199E) resulted in loss of ATPase activity. The RNA-stimulated NTPase activity was significantly affected by deletion of 20 amino acid residues from the N terminus or by substitutions of the cluster of basic residues, 184RKRK→QNGN, of NS3del.2, although both mutant proteins retained the conserved RNA helicase motifs. Furthermore, the minimal NS3 protease domain, required for cleavage of the 2B-3 site, was precisely defined to be 167 residues, using the in vitro processing of NS2B-NS3 precursors. Our results reveal that the functional domains required for serine protease and RNA-stimulated NTPase activities map within the region between amino acid residues 160 and 180 of NS3 protein and that a novel motif, the cluster of basic residues 184RKRK, plays an important role for the RNA-stimulated NTPase activity.
Dengue virus type 2 (DEN2), a member of the Flaviviridae, has a single-stranded RNA genome of positive-strand polarity which contains 10,723 nucleotides (nt) (in New Guinea C-strain [16]). Like other flavivirus RNA genomes, DEN2 RNA has a type I cap at the 5′ terminus, and a single open reading frame (10,173 nt in DEN2) encodes a polyprotein precursor (3,391 amino acid residues in DEN2) (for a review, see reference 6). The order of the polyprotein precursor is NH2-C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5-COOH, which is processed into three structural proteins (C, prM, and E) that are assembled into the virion and at least seven nonstructural proteins, NS1 to NS5, which are expressed in infected cells (6).
The processing of the amino-terminal region of the polyprotein precursor encoding the structural proteins is carried out by the host signal peptidase within the endoplasmic reticulum (22, 23, 32). Processing of the 2A-2B, 2B-3, 3-4A, and 4B-5 sites is catalyzed by a two-component viral proteinase, NS2B/NS3, of the serine proteinase family (3, 13). Previous studies established that the N-terminal region of about 180 amino acid residues of NS3 in the presence of NS2B is sufficient for processing at these junctions (4, 5, 7, 11, 26, 27, 40, 44). The viral proteinase-sensitive sites have in common two basic amino acid residues (such as Arg-Arg, Arg-Lys, and Lys-Arg), followed by a Gly, Ala, or Ser.
The region C-terminal to the proteinase domain of NS3 contains conserved motifs found in the nucleoside triphosphate (NTP)-binding proteins and DEXH family of RNA helicases (14, 17, 19). The role for a RNA helicase activity in flavivirus life cycle has been implicated in the genomic RNA replication in an unwinding step, because replication of flaviviruses is thought to proceed through a double-stranded RNA replicative form and replicative intermediate (for reviews, see references 6 and 42 and references therein). The unwinding of double-stranded RNA would require ATP hydrolysis; in support of this expectation, the RNA helicases have, in general, intrinsic enzymatic activity to catalyze hydrolysis of ATP or NTP. Many positive-strand RNA viruses have been shown to encode proteins possessing NTPase activities (10, 15, 20, 28, 29, 31, 34, 39, 41). Functional studies of a few NTPases in the virus life cycle have recently been reported (2, 35). In addition, viral NTPases are often stimulated by single-stranded RNA (15, 20, 28, 31, 34, 39, 41). NS3 protein of hepatitis C virus, a new member of the flavivirus family, and p80 protein of bovine viral diarrhea virus, a pestivirus, which are phylogenetically closer to each other but distant from arthropod-borne flaviviruses, have been demonstrated to possess RNA-stimulated NTPase and RNA helicase activities. However, among the arthropod-borne flaviviruses, the NS3 proteins of West Nile virus (41), yellow fever virus (39), and Japanese encephalitis virus (20) have been shown to have an RNA-stimulated NTPase activity but not RNA helicase activity. Moreover, the boundaries of the protease and RNA-stimulated NTPase functional domains of flavivirus NS3 proteins have not been clearly defined.
In this report, we show that a recombinant DEN2 NS3 protein expressed in Escherichia coli has an RNA-stimulated NTPase activity as well as RNA helicase activity. The kinetic and biochemical properties of the NTPase associated with DEN2 NS3 have been determined. Analyses of deletion mutants have revealed that the N-terminal 167 residues of NS3 are sufficient for the in vitro protease activity. Optimal RNA-stimulated NTPase activity, however, was obtained with a mutant NS3 with a N-terminal deletion of 160 residues. Deletion of 20 more residues from this mutant or mutagenesis of a novel cluster of basic amino acids, 184RKRK, significantly affected the RNA-stimulated NTPase activity. The results of this study indicate that the N-terminal region between residues 160 and 180 is shared by two distinct enzymatic activities of NS3 protein, the protease and the RNA-stimulated NTPase, and that the 184RKRK motif plays an important role in the NTPase activity stimulated by RNA.
MATERIALS AND METHODS
Materials.
Restriction enzymes, DNA-modifying enzymes, and SP6 RNA polymerase were purchased from New England Biolabs (Beverly, Mass.) or Promega (Madison, Wis.). E. coli DH5 and BL21(DE3) were from New England Biolabs. The rabbit reticulocyte coupled transcription-translation (TNT) system and canine pancreatic microsomal membranes were purchased from Promega. Ni2+-nitrilotriacetic acid (NTA) resin was from Qiagen (Chatsworth, Calif.). The Bradford protein assay kit was from Bio-Rad (Hercules, Calif.). Phosphoenolpyruvate and pyruvate kinase were purchased from Sigma. NADH, lactate dehydrogenase, and glycogen were purchased from Boehringer Mannheim (Indianapolis, Ind.). [α-32P]GTP and Tran[35S]-label (1,000 Ci/mmol) were purchased from Dupont-NEN (Boston, Mass.) and ICN Pharmaceuticals (Costa Mesa, Calif.). The pET-PFH-nef vector (46) was a gift from L. J. Zhao.
Construction of recombinant NS3 expression plasmids in the NTPase/RNA helicase domain.
Generally, all recombinant DNA and cloning procedures were carried out by standard methods (30). The pTM1-NS3 construct containing the full-length NS3 cDNA was previously described (44).
(i) pET-NS3AC-PFH.
To clone the cDNA encoding all of the conserved motifs of the NS3 helicase domain into the pET-PFH vector, a 1.3-kb cDNA (nt 5062 to 6375) was amplified by PCR using pTM1-NS3 as a template and primers A (5′-aagataccatgGACATTTTTCGA-3′; containing an engineered NcoI site [underlined] which includes an in-frame translational start codon; DEN2 sequence is in uppercase) and C (5′-agatacaggcctCTT TCTTCCAGC-3′, containing an StuI site [underlined] and the C-terminal sequence of NS3). The PCR product was double digested with NcoI and StuI and cloned into the pET-PFH vector between the NcoI and StuI sites to yield pET-NS3AC-PFH.
(ii) pET-NS3BC-PFH.
To clone pET-NS3BC, a 0.48-kb fragment (nt 5896 to 6375) was amplified using primers B (5′-aagataccatGGGGAGAATAGGA-3′) and C (see above). The PCR product was double digested with NcoI and StuI and cloned into the pET-PFH vector between NcoI and StuI sites to yield pET-NS3BC-PFH. The PCR-amplified region was verified by sequencing.
(iii) pET-NS3-PFH and pET-NS3-PH.
Plasmid pET-NS3-PFH was obtained by replacing the NcoI-to-EcoRI fragment of pET-NS3AC-PFH with the corresponding fragment obtained from pTM1-NS3. The pET-NS3-PH was obtained by replacing the StuI-to-BamHI (located in the vector) fragment of pET-NS3-PFH with a synthetic DNA fragment encoding a protein kinase A site and a hexahistidine tag with the deletion of the FLAG epitope.
(iv) pET-NS3del.1-PH and pET-NS3del.2-PH.
Plasmid pET-NS3-PH was digested partially with NdeI and completely with NcoI and then treated with Klenow DNA polymerase in the presence of dNTPs to create blunt ends. The large DNA fragments were purified and religated to give rise to the N-terminal deletion constructs pET-NS3del.1-PH and pET-NS3del.2-PH.
(v) pET-NS3mt.1-PH and pET-NS3mt.2-PH.
Site-directed mutagenesis in the NS3 P-loop region was carried out by using a PCR-based point mutagenesis protocol (47). Briefly, we synthesized five PCR primers with convenient cloning sites (under lined): D (5′-GCT GCC ATG GGA GCA TAT GTG AGT GCT-3′), E (5′-CTT CGT CTC TCC CGC TCC-3′), F (5′-CA TGC TCG AGT TGA GAT GTA TCC TCT AGC-3′), G1 (5′-CAA AAC GGA AAC TTG ACC ATC ATG GAC CTC-3′), and G2 (5′-GTT TCC GTT TTG AAA AAT GTC ATC TTC GAT CTC-3′). Using pTM1-NS3 as the template, fragments NS3-DE, NS3-DG2, and NS3-G1F were first amplified and purified. NS3-DE was then used as a primer along with the primer F for PCR to generate the PCR fragment DF. This fragment was digested with NcoI and XhoI and cloned into NcoI-XhoI sites of pET-NS3-PH to give rise to pET-NS3mt.1-PH (198GKT→GET). For construction of pET-NS3mt.2-PH, PCR fragments NS3-DG2 and NS3-G1F were mixed and PCR was carried out in the presence of primers D and F. The desired product was digested with NcoI and XhoI and was used to replace the NcoI-XhoI fragment from plasmid pET-NS3-PH to generate pET-NS3mt.2-PH (184RKRK→QNGN). The NdeI-to-XhoI fragment of pET-NS3mt.2-PH was replaced with that of pTM1-NS3 to obtain the wild-type control construct pET-NS3wt-PH. The integrity of PCR-amplified regions was confirmed by sequencing.
Construction of NS3 expression plasmids in the protease domain.
Expression plasmids encoding the NS2B-NS3 protease domain containing the N-terminal 183, 176, 170, 169, 168, 167, 166, 165, and 164 amino acids (aa) of NS3 [designated NS2B-NS3(183aa), etc.] were constructed by PCR. The 5′-upstream primer, CATGCCATGGAACAAACACTGACCATACTCATC-3′ (nt 4405 to 4422), was used for all PCRs. The 3′-downstream primer was different for each of the constructs; the sequences are shown in Table 1. The template for PCR was pLZ-NS2B3(pro) (45). The resulting PCR product was digested with NsiI (nt 4696) and BamHI and cloned into plasmid pTM1-NS2B-NS3(50%)-PFH to give rise to several plasmids containing different lengths (from 164 to 183 amino acid residues) of the NS3 protease domain.
TABLE 1.
Oligonucleotide primers used for PCR to construct NS2B-NS3 proteins containing a protease domaina
| Precursor protease | Nucleotide position | Sequence of PCR primer |
|---|---|---|
| NS2B-NS3 upstream primer | 4405–4422 (+) | 5′-CATGCCATGGAACAAACACTGACCATACTCATC |
| NS2B-NS3(183aa) | 5050–5067 (−) | 5′-TGATGGATCCCTATTAAATGTCATCTTCGATCTC |
| NS2B-NS3(176aa) | 5032–5049 (−) | 5′-TGATGGATCCCTATTATGGATTGTCTTCAATACT |
| NS2B-NS3(170aa) | 5014–5031 (−) | 5′-TGATGGATCCCTATTATTTTTCAGTCTGGGCTAT |
| NS2B-NS3(169aa) | 5011–5028 (−) | 5′-TGATGGATCCCTATTATTCAGTCTGGGCTATAGC |
| NS2B-NS3(168aa) | 5008–5025 (−) | 5′-TGATGGATCCCTATTAAAGTCTGGGCTATAGCAC |
| NS2B-NS3(167aa) | 5005–5022 (−) | 5′-TGATGGATCCCTATTACTGGGCTATAGCACTCA |
| NS2B-NS3(166aa) | 5003–5019 (−) | 5′-TGATGGATCCTATTAGGCTATAGCACTCACATA |
| NS2B-NS3(165aa) | 4999–5016 (−) | 5′-TGATGGATCCTATTATATAGCACTCACATATGC |
| NS2B-NS3(164aa) | 4996–5013 (−) | 5′-TGATGGATCCCTATTAAGCACTCACATATGCTCC |
The upstream primer was kept constant, and the downstream primer was varied to make sequential deletions. (+) and (−) refer to the sequences from sense and antisense strands, respectively.
Expression and purification of NS3 proteins containing the NTPase/RNA helicase domain.
Competent E. coli DE3(BL21) cells, transformed with the expression constructs, were grown in LB medium containing ampicillin (100 μg/ml) at 37°C until the optical density at 600 nm reached 0.6. The cells were induced with isopropyl-β-d-thiogalactopyranoside (IPTG; 0.4 mM) for 4 to 6 h, harvested by centrifugation, and stored at −70°C until use. NS3 polypeptides of expected sizes were produced in E. coli predominantly as inclusion bodies. These were fractionated from the inclusion bodies into a soluble supernatant fraction and an insoluble pellet fraction essentially as described previously (39). The majority of the NS3 polypeptide was associated with the pellet fraction as inclusion bodies. The pellet fraction was resuspended in 20 ml of solubilization buffer (6 M guanidine-HCl, 50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10 mM β-mercaptoethanol) and kept in ice for 1 h. The extract was centrifuged at 30,000 × g for 30 min at 4°C. The supernatant solution was purified by passage through a Ni2+-NTA affinity column. A Bio-Rad Econo column (1.5 cm in diameter) containing 5 ml of Ni2+-NTA resin was preequilibrated with 20 ml of lysis buffer at room temperature. The column was preequilibrated with 3 bed volumes of buffer A (8 M urea, 150 mM NaCl, 10 mM β-mercaptoethanol, 50 mM Tris-HCl [pH 8.0]) and then with buffer B (buffer A, with Tris-HCl [pH 8.0] replaced with by morpholineethanesulfonic acid [MES]-HCl, [pH 6.5]). Proteins were eluted from the column in buffer A containing 400 mM imidazole. The elution profile was monitored by using the Bio-Rad protein estimation kit. The peak fractions were pooled, and the proteins were precipitated by the addition of 3 volumes of 100% acetone kept at −20°C. After a brief spin in a tabletop clinical centrifuge (1,000 × g), the supernatant was discarded, and the protein pellet was dissolved in 2 ml of buffer C (6 M guanidine-HCl, 50 mM Tris-HCl [pH 8.0], 5 mM dithiothreitol). This fraction was subjected to Sephadex G-75 gel filtration chromatography through a column of 1.75-cm diameter (200-ml bed volume) and equibrated with elution buffer D (6 M urea, 150 mM NaCl, 20 mM β-mercaptoethanol, 50 mM Tris-HCl [pH 8.0], 1 mM EDTA). The elution was monitored at A280, and the samples from peak fractions were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). The protein concentration of the pooled fractions was diluted to less than 0.5 mg/ml in buffer D, and the refolding of the denatured protein was carried out by dialysis against buffer E (0.5 M guanidine-HCl [pH 8.0], 1 mM EDTA, 1 mM dithiothreitol, 20% glycerol) for 16 h at 4°C. The supernatant was dialyzed against buffer F (25 mM HEPES-K+ [pH 7.4], 50 mM KCl, 1 mM EDTA, 0.02% β-mercaptoethanol, 0.01% Triton-X-100, 50% glycerol) for 10 h at 4°C. The final dialysate was clarified by centrifugation at 30,000 × g for 30 min at 4°C. The supernatant fraction was stored as aliquots at −70°C.
NTPase assay.
In the standard NTPase assay, hydrolysis of NTP was coupled to the oxidation of NADH, which was followed spectrophotometrically at 340 nm and was carried out essentially as described previously (39), with slight modifications. Briefly, the reaction mixture (200 μl) contained 50 mM HEPES-K+ (pH 7.5), 2.5 mM MgCl2, 0.5 mM NTP, 2 mM phosphoenolpyruvate, NADH (100 μg/ml), pyruvate kinase (100 μg/ml), and lactate dehydrogenase (25 μg/ml) with or without 0.5 mM poly(A) (measured as mononucleotide equivalents) and 1 to 3 μg of purified protein. The A340 was measured continuously with a Spectronic 3000 array spectrophotometer (5 s per cycle, 2 s per exposure) for 3 to 5 min. The blank controls contained all components except the enzyme. For assays in the absence of divalent cations, the reactions were first carried out as described above, stopped by the addition of 20 μl of a freshly prepared solution of 1.0 N HCl (12), and after 3 min neutralized with 1.0 N KOH. The ADP formed in the coupled system was assayed as described above.
RNA helicase assay.
Plasmid pSP65 was linearized with BstNI and was used as a template for SP6 RNA polymerase to generate an unlabeled single-stranded RNA of 154 nt. pSP64 was linearized by digestion with BamHI and was transcribed in the presence of [α-32P]GTP (800 Ci/mmol) and 10-fold molar excess of unlabeled GTP and three NTPs. The conditions for in vitro transcription were those recommended by the manufacturer (Promega). After incubation for 1 h at 37°C, the reaction mixtures were treated with RNase-free DNase I and extracted with phenol-CHCl3. The unincorporated NTPs were separated by a Sephadex G-25 spin column, and the RNAs were precipitated with ethanol. The unlabeled and radiolabeled RNAs were mixed in a molar ratio of 5:1 and annealed (0.5 M NaCl, 25 mM HEPES-K+ [pH 7.5], 1 mM EDTA, 0.1% SDS) in a thermocycler (95°C for 5 min, 55°C for 30 min, and 25°C overnight). After ethanol precipitation, the RNAs were resolved by PAGE (8% gel) using a buffer containing 0.045 M Tris-borate, 0.001 M EDTA, and 0.1% SDS. The duplex substrate band was detected by autoradiography, excised, and eluted (0.5 M ammonium acetate, 2 mM EDTA, 0.1% SDS) for 2 h at 37°C. The eluates were precipitated with ethanol and redissolved in 10 mM Tris-HCl (pH 7.5)–1 mM EDTA. The substrate was stored at −70°C until use.
The reaction mixture (10 μl) for the RNA helicase assay contained 25 mM HEPES (pH 7.5), 5 mM ATP, 3 mM MnCl2, 2 mM dithiothreitol, 100 μg of bovine serum albumin per ml, 5 U of RNasin and the RNA substrate (3,000 cpm containing approximately 0.25 pmol/assay), and NS3del.2 protein (54 pmol). The reaction mixture was incubated for 30 min at 37°C, and the reaction was terminated by adding 2.5 μl of 5× buffer (100 mM Tris-HCl [pH 7.5], 50 mM EDTA, 0.1% Triton X-100, 0.5% SDS, 50% glycerol, 0.1% bromophenol blue). The helicase assay mixtures were analyzed by SDS-PAGE (75% gel; acrylamide/bisacrylamide at 30:0.8, 0.5× Tris-borate-EDTA, 0.1% SDS; 15 mA of constant current) and autoradiography.
In vitro processing of NS2B-NS3(Pro) in a TNT system.
To define the minimal protease domain of NS3, the NS2B-NS3 protease domain precursors containing variable lengths of the N-terminal amino acids of NS3 were analyzed for in vitro processing using the Promega TNT system in the presence of rabbit reticulocyte lysate. Plasmid DNAs were purified by CsCl-ethidium bromide equilibrium density gradients (30). The TNT reactions for in vitro processing were carried out essentially as specified by the manufacturer. Briefly, the reaction mixtures contained the plasmid DNA [1 μg of NS2B-NS3(Pro)], T7 RNA polymerase, Trans-label (20 μCi of [35S]methionine/cysteine), and canine pancreatic microsomal membranes (9). Reaction mixtures were incubated at 30°C for 90 min. Microsomal membrane pellet fractions were isolated by centrifugation of TNT reaction mixtures at 15,000 × g for 15 min, washed with phosphate-buffered saline, and analyzed by SDS-PAGE followed by fluorography.
RESULTS
Expression and purification of DEN2 NS3 polypeptides.
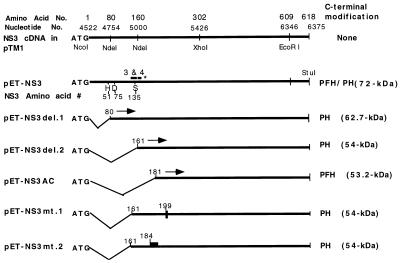
To study the biochemical properties of the putative NTPase/RNA helicase domain of the DEN2 NS3, full-length (618-aa) and mutant polypeptides containing the C-terminal 540, 458, 438, and 160 amino acid residues of NS3 were expressed in E. coli by using the pET-PFH vector (46) or its derivative, pET-PH, between NcoI (which provided the translational start codon) and StuI (to which C-terminal Lys of NS3 was fused) (Fig. 1). These NS3 polypeptides were also engineered to express a C-terminal fusion of a protein kinase A phosphorylation site, synthetic FLAG epitope, and a hexahistidine tag (PFH) (26 amino acid residues) or the protein kinase A site and the His tag (PH). The calculated molecular sizes of the polypeptides are shown in Fig. 1. To study the function of the P loop in the NTPase/RNA helicase region of NS3, we constructed a substitution mutant, NS3mt.1-PH, in which the conserved Gly-199Lys-Thr in the P-loop motif was mutated to Gly-199Glu-Thr. A second substitution mutant, NS3mt.2-PH, was constructed such that a stretch of basic amino acid residues of NS3 protein, 184Arg-Lys-Arg-Lys, was mutated to Gln-Asn-Gly-Asn. The wild-type full-length NS3-PH was also constructed as a control.
FIG. 1.
DEN2 NS3 expression constructs. The full-length and N-terminal deletion constructs of NS3 were cloned into the pET-PFH or pET-PH vector as described in Materials and Methods. These constructs encode recombinant NS3 proteins with a Met residue at the N terminus and His tags (PFH or PH) at the C terminus. The filled boxes in pET-NS3mt.1 and -2 refer to the substitution mutants of NS3 in which the P-loop motif 199GKT is changed to GET and the stretch of basic residues 184RKRK is changed to QNGN, respectively. Conserved boxes 3 and 4 of 10 flavivirus NS3 sequences, indicated as 3 & 4, are from a previous report (36) based on the model of Bazan and Fletterick (3). The asterisk indicates the endpoint of the minimal protease domain.
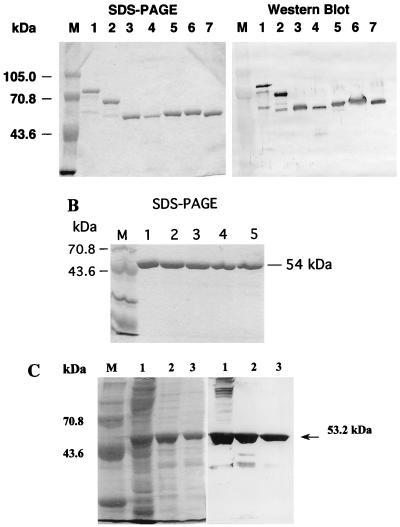
The expression of all of these recombinant NS3 proteins in E. coli BL21(DE3) cells subsequent to induction with IPTG was confirmed by Western blot analyses using rabbit polyclonal anti-NS3 antibody. The desired proteins were expressed at high levels but formed insoluble inclusion bodies. The NS3 polypeptides from the inclusion bodies were purified by Ni2+ affinity chromatography under denaturing conditions as described in Materials and Methods. Purified polypeptides were analyzed by SDS-PAGE and Western blotting as shown for full-length NS3, NS3del.1, NS3del.2, NS3AC, NS3mut.1, and NS3mut.2 in Fig. 2A, as well as NS3BC (data not shown). In addition to the proteins with the predicted molecular weights, some specific protein bands with lower molecular weights were detected and copurified with the desired proteins in the Ni2+ column chromatography. Smaller polypeptides were possibly generated from either proteolytic degradation or internal initiation of translation of the mRNAs. The number of lower-molecular-weight bands increased in proportion to the size of the expressed proteins. The NS3 polypeptides were further purified by gel filtration chromatography, and the eluates were refolded by dialysis as described in Materials and Methods. The purity of the refolded proteins NS3del.2 and NS3AC subsequent to gel filtration column chromatography as determined by SDS-PAGE and Coomassie blue staining is shown in Fig. 2B and C.
FIG. 2.
Expression of NS3 polypeptides in E. coli. (A) Recombinant DEN2 NS3 proteins were expressed and purified from E. coli BL21(DE3) cells by using Ni-NTA affinity column chromatography as described in Materials and Methods. Left panel, Coomassie blue-stained gel of Ni 2+-NTA-purified proteins separated by SDS-PAGE. Right panel, Western blot prepared with anti-DEN2 NS3 polyclonal antibodies. Lanes: M, protein molecular weight marker; 1, NS3-PFH; 2, NS3del.1-PH; 3, NS3del.2-PH; 4, NS3AC-PFH; 5, NS3del.2-PH; 6, NS3mt.2-PH; 7, NS3mt.1-PH. (B) NS3del.2 protein was further purified by Sephadex G-75 column chromatography under denaturing conditions, and eluates were refolded described in Materials and Methods. The refolded fractions were analyzed by SDS-PAGE and stained with Coomassie blue. Lanes: M, molecular weight standards; 1 to 5, pooled fractions 21 to 26, 27, 28, 29, and 30, respectively. (C) SDS-PAGE followed by Coomassie blue staining (left panel) and Western blot analysis (right panel) of the refolded NS3AC polypeptide.
Biochemical and kinetic properties of NTPase activity.
The ATPase activities of DEN2 NS3 polypeptides were determined using a coupled enzymatic assay in which oxidation of NADH was followed spectrophotometrically as described in Materials and Methods. The rate of nonenzymatic hydrolysis of ATP or oxidation of NADH was insignificant under the assay conditions (data not shown). Although full-length NS3 and NS3del.1 had ATPase activities (data not shown), the 54-kDa NS3del.2 polypeptide was used for all subsequent kinetic and biochemical analyses of NTPase/RNA helicase since it was more soluble and purer than the full-length NS3 and NS3del.1 proteins (Fig. 2A, lanes 1 to 3).
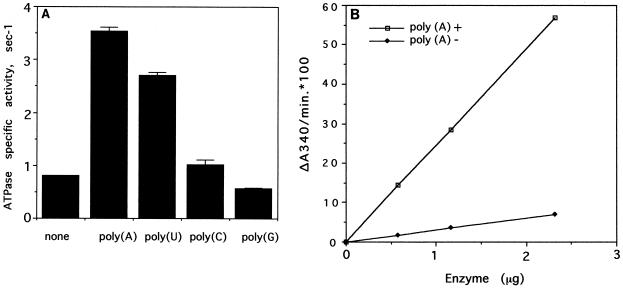
The effects of various polyribonucleotides on the ATPase activity associated with NS3del.2 were determined. The ATPase activity was stimulated 4.4-fold by poly(A) and 3.4-fold by poly(U). Poly(C) had almost no effect (1.3-fold), and poly(G) slightly inhibited the ATPase activity (70% of the basal activity) under the reaction conditions used (Fig. 3A).
FIG. 3.
NTPase assays in the absence or presence of polyribonucleotides. (A) ATPase assays were carried out in the absence or presence of different homopolymers at 0.5 mM (concentration measured as mononucleotides). The specific activities of the NS3del.2 ATPase under each reaction condition were calculated based on the protein used (1.45 μg or 27 pmol/assay) and measured ATP hydrolysis rates. Specific activity is defined as moles of ADP generated per mole of protein/per second. Each point on the plot represents the mean value of triplicate assays, and each error bar represents the standard deviation. (B) ATPase assays of the purified NS3del.2 protein were carried out under standard reaction conditions with or without 0.5 mM poly(A). The initial velocity, ΔA340, was proportional to the amount of protein (18.62 pmol/μg of protein) used in the assay. Each point is the mean value of two measurements.
Poly(A) was used in subsequent studies since it had the highest stimulation of NTPase activity. The initial velocity, ΔA340, was proportional to the amount of enzyme added and was linear in either the presence or the absence of poly(A) (Fig. 3B). The optimum pH for the enzyme was determined to be between 5.5 and 9.0 in the absence or presence of poly(A). The pH profile in the absence of poly(A) was relatively flat, although there was an optimum at about pH 6.5. In the presence of poly(A), the pH profile was a typical bell-shaped curve with a distinct optimum at pH 7.5 (data not shown). These results indicated that the poly(A)-dependent ATPase activity was more sensitive to pH than the basal ATPase activity.
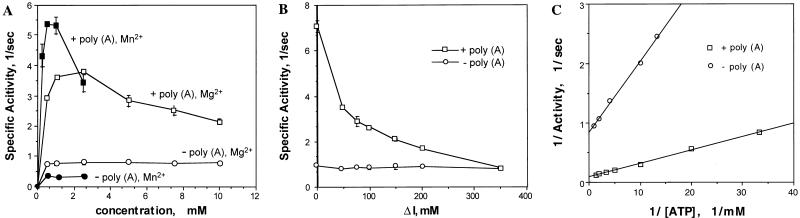
The optimum Mg2+ concentration for the ATPase activity of NS3del.2 was determined. In the absence of poly(A), the ATPase activity was not affected by Mg2+ up to 10 mM once the Mg2+ concentration exceeded the ATP concentration, which was 0.5 mM. In the presence of poly(A), the optimum Mg2+ concentration was between 1 and 2.5 mM for the ATPase activity, and further increases of Mg2+ caused a gradual decrease in ATPase activity (Fig. 4A). The optimum Mn2+ concentration was between 0.5 and 1 mM under the reaction conditions described, and higher concentrations were inhibitory.
FIG. 4.
Biochemical and kinetic analysis of ATPase activity of NS3. (A) Effect of divalent cation. The effects of various Mg2+ or Mn2+ concentrations on the ATPase were determined in 50 mM HEPES-K+ (pH 7.5)–16 mM (NH4)2SO4–0.5 mM ATP with or without 0.5 mM poly(A), as indicated. (B) Effect of ionic strength. The reactions were carried out in 50 mM HEPES-K+ (pH 7.5)–2.5 mM MgCl2–0.5 mM ATP and increasing concentration of KCl in the absence or presence of 0.5 mM poly(A). The increased ionic strength [I = 1/2 · (Ci · Zi2)] is indicated. (C) Effect of ATP concentration. The ATPase activity of NS3del.2 was measured at increasing concentrations of ATP but constant Mg2+ concentration (2.5 mM), with the rest of the components the same. The ATP concentrations used in the assays are 0.03, 0.05, 0.1, 0.2, 0.3, 0.5, and 0.75 mM. Km and Kcat constants for NS3del.2 protein in the absence (○) or presence (□) of 0.5 mM poly(A) were determined from Lineweaver-Burk plots.
The poly(A)-dependent ATPase activity of NS3del.2 was very sensitive to high ionic strengths, as increasing the KCl concentration decreased the poly(A)-stimulated ATPase activity (Fig. 4B). Fifty percent inhibition occurred at 50 mM KCl, and at 300 mM KCl the poly(A)-dependent ATPase activity was almost abolished. In contrast, the ATPase activity in the absence of poly(A) remained unchanged at this ionic strength (Fig. 4B).
The kinetics of ATP hydrolysis by the NS3del.2 protein at various ATP concentrations were determined at 2.5 mM Mg2+ ion concentration in the absence or presence of 0.5 mM poly(A). The ATP concentrations used, 0.03, 0.05, 0.1, 0.2, 0.3, 0.5, and 0.75 mM, were below the Mg2+ ion concentration. The specific activity of ATPase increased proportional to ATP concentrations with or without poly(A), and both curves were rectangular hyperbolas. The Lineweaver-Burk plots of these data were linear over the range of the ATP concentration used (Fig. 4C). In the presence of poly(A), the apparent Km of NS3del.2 for ATP was 0.25 mM and the apparent Vmax was 10.8 s−1. In the absence of poly(A), the apparent Km for ATP was 0.14 mM and the apparent Vmax was 1.178 s−1. Under these experimental conditions, the apparent Km for ATP was increased only slightly (1.8-fold) by poly(A) but the apparent Vmax was increased dramatically (9.7-fold).
The apparent kcat for Mg2+-ATP in the presence of poly(A) for the NS3del.2 protein was about 10 pmol of ATP hydrolyzed/s/pmol of protein, close to the reported kcat values for Mg2+-ATP of the recombinant hepatitis C virus NS3 and yellow fever virus NS3 proteins expressed in E. coli at their optimum reaction conditions (3.5 and 14.7 s−1, respectively [31, 39]). These kcat values are also close to that of the 50-kDa proteolytic product of West Nile virus NS3 purified from virus infected cells by treatment with subtilisin (3.2 s−1) (41) but are lower than that of recombinant baculovirus-expressed and partially purified p80 protein of bovine viral diarrhea virus (70 s−1) (34).
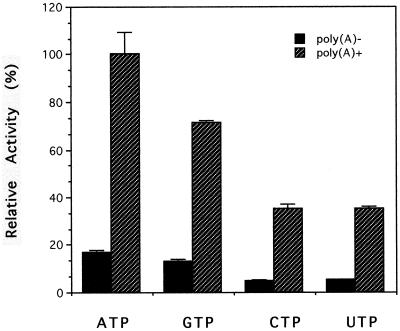
In addition to ATP, DEN2 NS3 can utilize other NTPs. Among all of the NTP substrates, ATP and GTP were preferred over pyrimidines for both the basal and RNA-dependent NTPase activity (Fig. 5). In this regard, DEN2 NS3 is similar to yellow fever virus NS3 but differs from West Nile virus NS3, which utilizes ATP, CTP, and UTP equally well but hydrolyzes GTP poorly.
FIG. 5.
Enzyme-coupled NTPase assays. The NTPase assays were carried out under standard conditions as described in Materials and Methods in the absence or presence of 0.5 mM poly(A) and with 0.5 mM each NTP (ATP, GTP, CTP, and UTP) as substrates. Relative activities in triplicates were plotted in a bar graph; each error bar represents the standard deviation.
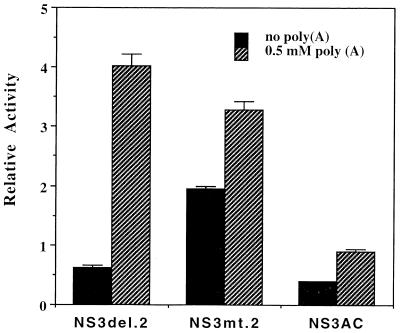
Next, we sought to determine the minimum functional domain for the ATPase activity. We assayed the NTPase activity associated with the purified NS3AC polypeptide, which contains all of the conserved motifs of NTPase/RNA helicase domain between 180 and 618 amino acid residues of NS3 (Fig. 1). The results showed that the basal ATPase activity was reduced about twofold, but the poly(A)-stimulated activity was reduced dramatically compared with that of NS3del.2, being only twofold higher than its basal activity (Fig. 6).
FIG. 6.
ATPase activities of NS3 mutants. The standard enzyme-coupled ATPase assays were carried out in triplicate in the presence or absence of 0.5 mM poly(A) and 0.5 mM ATP. Relative activity denotes the specific activity units as defined for Fig. 3.
The NS3AC polypeptide is 20 amino acid residues shorter than the NS3.del.2 protein at the N terminus, which indicated that this 20-aa region, between amino acid residues 160 and 180 from the N terminus of NS3, is crucial for the protein’s RNA-stimulated NTPase activity or proper folding of this domain. This 20-amino-acid region is N terminal to the first conserved motif of the DEXH family of RNA helicases and the P-loop motif (Gly-199Lys-Thr). The Lys-199→Glu substitution in the P-loop motif abolished the ATPase activity (data not shown), consistent with the involvement of this motif in ATP binding (37). This result suggests that there is only one ATP binding site in the NS3 protein. Interestingly, replacement of a stretch of four basic amino acids, RKRK at positions 184 to 187, with QNGN increased the basal activity about 2-fold, whereas the poly(A)-stimulated activity was less than 1.5-fold, compared with the 8- to 10-fold stimulation of ATPase activity observed with the parent NS3del.2 protein (Fig. 6). These results indicate that this stretch of basic amino acid residues plays an important role in RNA-stimulated NTPase activity.
Mapping the minimal protease domain required for processing of the 2B-3 site.
Since the results obtained thus far indicated that the region between 160 and 180 amino acid residues of NS3 plays a role in the basal as well as the RNA-stimulated ATPase activity, we sought to determine whether this region is also required for processing of the 2B-3 site by the protease domain of NS3. The catalytic triad of the serine protease domain of DEN2 NS3 consists of His-51, Asp-75, and Ser-135. The work of Bazan and Fletterick has determined the amino acid residues which constitute the substrate binding pocket to be within two conserved regions of boxes 3 and 4 (3). In DEN2 these residues have been identified to be Asp-129 and Phe-130 in box 3 and Tyr-150, Asn-152, and G-153 in box 4, and the homology region spans the 39 residues up to residue 158 (36). Furthermore, Gly-153 and Ser-163 of DEN2 NS3 are the analogs of Gly-216 and Gly-226, which are located at the entry to the substrate binding pocket and thus play an important role in substrate selection for trypsin and chymotrypsin (24). These residues are highly conserved in all flavivirus NS3 proteins. Mutation of Gly-153 had a detrimental effect on the protease activity. However, the importance of residues C terminal to Gly-153 had not been established, and the minimal protease domain required for activity had not been mapped.
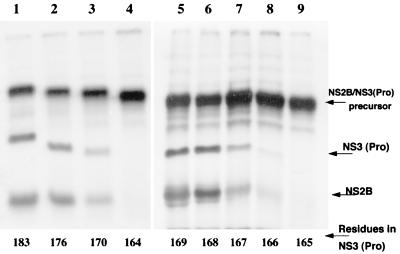
To map precisely the minimal protease domain, we generated a series of NS2B-NS3(Pro) precursors by PCR in which the protease domain of NS3 was progressively deleted from 183 to 164 amino acid residues. The 5′-upstream primer from the N terminus of NS2B was kept constant, and the 3′-downstream primers complementary to NS3 protease domain were designed to generate progressive deletions from amino acid positions 183 to 164. The resulting plasmids were used for the TNT procedure as described in Materials and Methods, in the presence of 35S-labeled methionine and canine microsomal membranes, which enhanced the cleavage efficiency of 2B-3 site (9). The precursor products were analyzed for the ability to undergo processing by cis cleavage of the 2B-3 site. Using four plasmid constructs, NS2B-NS3(183aa), NS2B-NS3(176aa), NS2B-NS3(170aa), and NS2B-NS3(164aa), we mapped the C-terminal boundary of the protease domain to a region between residues 170 and 164 as processing occurred with all precursors except for NS2B-NS3(164aa) (Fig. 7, lanes 1 to 4). To precisely map the C-terminal boundary, we generated five additional constructs, NS2B-NS3(169aa) to NS2B-NS3(165aa), and analyzed their abilities to undergo processing (Fig. 7, lanes 5 to 9). The results shown in Fig. 7 indicate that the precursor NS2B-NS3(167aa) but not NS2B-NS3(166aa) underwent processing in vitro. Taken together, these data suggest that the N-terminal protease and the C-terminal NTPase/RNA helicase represent two distinct domains and at most share a 6-aa region for distinct enzymatic activities.
FIG. 7.
Mapping the boundary of the NS3 protease domain. NS2B-NS3 precursor constructs containing successive C-terminal deletions of the protease domain were expressed in TNT system in the presence of canine microsomal membranes as described in Materials and Methods. The lysates were centrifuged to isolate the microsomal membrane fraction, and the processing reactions were analyzed by SDS-PAGE and autoradiography. Lanes: 1, NS2B-NS3(183aa); 2, NS2B-NS3(176aa); 3, NS2B-NS3(170aa); 4, NS2B-NS3(164aa); 5 to 9: NS2B-NS3(169aa) to NS2B-NS3(165aa).
DEN2 NS3 has an intrinsic RNA helicase activity.
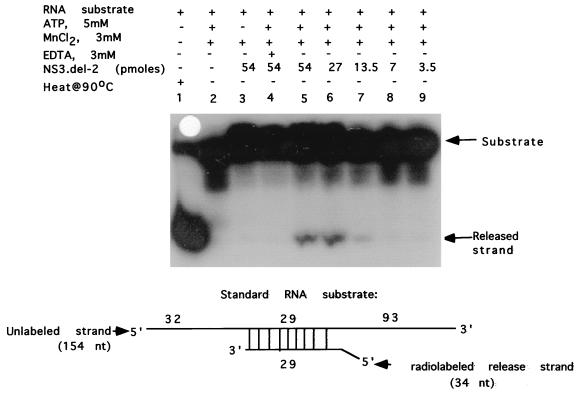
Since the NS3del.2 protein has the RNA-stimulated ATPase activity which is an intrinsic property of RNA helicases, we examined whether it also had any RNA helicase activity. A partially duplex RNA substrate containing both 5′ and 3′ single-stranded regions was prepared and tested in the RNA helicase assay as described in Materials and Methods. As shown in Fig. 8, the NS3del.2 protein had an intrinsic RNA helicase activity in the presence of Mn2+ and ATP (lanes 5 to 7). In control reactions, in the absence of the NS3del.2 protein (lane 2) or ATP (lane 3) or in the presence of EDTA (lane 4), the enzyme was inactive in releasing the radiolabeled strand. Under these conditions, the full-length NS3 had a lower RNA helicase activity but the NS3AC polypeptide lacked any detectable RNA helicase activity (data not shown).
FIG. 8.
RNA helicase assay of NS3del.2 protein. RNA helicase assays were performed as described in Materials and Methods. The double-stranded RNA substrate used for helicase assay is shown. Lanes: 1, RNA substrate heated at 90°C before loading; 2, no protein added (negative control); 3, standard RNA helicase reaction mixture omitting ATP but containing 54 pmol of NS3del.2; 4, standard RNA helicase reaction mixture except containing 3 mM EDTA and 54 pmol of NS3del.2; 5 to 9, standard RNA helicase reaction mixtures containing 54, 27, 14, 7, and 3.5 pmol of NS3del.2, respectively. Autoradiography of the polyacrylamide gel is shown.
DISCUSSION
The viral NTPase/RNA helicases have been classified into three superfamilies (SF1 to SF3) (for a review, see reference 17). The NS3-like proteins encoded by the potyvirus-flavivirus-pestivirus groups and the helicase of vaccinia virus are classified in SF2. DEN2 NTPase/RNA helicase has some characteristics in common with RNA-stimulated NTPases/RNA helicases, members of SF2 family (17) of positive-strand RNA viruses such as West Nile virus (41), yellow fever virus (39), hepatitis C virus (31), bovine viral diarrhea virus (a pestivirus) (34), tamarillo mosaic potyvirus (10), and plum pox potyvirus (21). One common property of all established viral RNA helicases is the Mg2+ ion-dependent basal NTPase activity. This activity is stimulated by single-stranded homopolymeric RNA. The ATPase activity of DEN2 NS3 was stimulated maximally by poly(A) and poly(U) (10- to 16-fold) and minimally by poly(C) (1.5-fold), similar to the West Nile virus and yellow fever virus NS3 proteins, whereas hepatitis C virus NS3, bovine viral diarrhea virus p80, and Japanese encephalitis virus NS3 were maximally stimulated by poly(U) and poly(C) (20, 31, 34) but minimally by poly(A). Poly(G) inhibited the activity of DEN2 NS3, as has been reported for other NTPases.
The stimulation of NTPases by single-stranded RNA is anticipated, as these proteins bind to a partially single-stranded region of duplex RNA and unwind the base-paired region by using the energy from ATP hydrolysis. The model proposed based on the crystal structure of hepatitis C virus NS3 RNA helicase (8) (see also references 18a and 43) is also consistent with this property of RNA helicases. It is thought that RNA binding induces a conformational change in the protein resulting in a more kinetically favorable active site–Mg2+-ATP substrate interaction (25), consistent with the change in the Vmax of DEN2 NS3 for Mg2+-ATP from 1.2 to 10.8 s−1 in the presence of poly(A). This basal ATPase activity associated with DEN2 NS3 was not sensitive to the change of pH or ionic strength, whereas the RNA-stimulated ATPase was inhibited by high ionic strengths. This result suggests that high-ionic-strength conditions preclude the conformational change resulting from interaction between NS3 and RNA that is necessary to provide kinetically favorable fit between the active site and ATP.
DEN2 NS3del.2 protein has a dose-dependent RNA unwinding activity (Fig. 8). To date none of the arthropod-borne flavivirus NS3 proteins, which have a distant phylogenetic relationship to hepatitis C virus and pestiviruses proteins, have been demonstrated to have RNA helicase activity. However, we noticed that this activity is lower than that for hepatitis C virus NS3 or bovine viral diarrhea virus (a pestivirus) p80 RNA helicase (18, 33, 38). The bovine viral diarrhea virus p80 and hepatitis C virus NS3 RNA helicases exhibited significant RNA helicase activities with 0.1 to 1 pmol of purified proteins (18, 38). By comparison, we had to use 27 pmol of the purified NS3del.2 protein for detection of helicase activity in the presence of approximately similar substrate concentration (Fig. 8). One possible explanation for this low activity is that the denaturation and refolding conditions used in this study for the E. coli-expressed NS3del.2 proteins were not optimum. Alternatively, the RNA helicase activity of arthropod-borne flaviviruses may be stimulated by interaction with other viral replicase components. In this regard, it is worth noting that the RNA helicase activity of the cellular eukaryotic translation initiation factor 4A (eIF-4A) is dependent on heterodimerization between eIF-4A, which has a single RNA binding domain, and eIF-4B, which has two RNA binding domains (for a review, see reference 17).
The N-terminal region of NS3 interacts with NS2B and functions as a two-component serine protease involved in processing the viral polyprotein precursor. The N-terminal 184 residues of NS3 were shown to be sufficient to form a bimolecular complex with NS2B (1). Thus, NS3 has two distinct enzymatic activities carried out by two independent domains. Our data indicate that the N-terminal 160 aa are not required for NTPase activity. However, when 20 additional aa were deleted from the N terminus of NS3 (NS3AC), or the stretch of basic amino acid residues 184RKRK was mutated to neutral amino acid residues (NS3mut.2 protein), the mutant proteins had altered RNA-stimulated ATPase activities. For example, NS3AC had reduced basal activity compared with NS3del.2. This activity was stimulated only about twofold by poly(A) and was inhibited about threefold by poly(U) (data not shown). These properties were quite different from those of NS3del.2 protein. Interestingly, 184RKRK→QNGN substitution mutations increased the basal activity but reduced the level of stimulation in the presence of poly(A) to less than twofold.
In this study, we have precisely mapped the minimal protease domain of NS3 to the N-terminal 167 residues. Recent extensive mutational analyses of the NS3 protease domain in conserved boxes 3 and 4 (36) revealed critical residues within these conserved regions required for protease activity. The C-terminal boundary of the minimal protease domain, Gln-167, is outside conserved box 4; this residue is conserved among 8 of 10 flavivirus NS3 proteins listed (36).
The conserved motif A at the N-terminal region of all NTPase/RNA helicases is GK(S/T), which, from X-ray crystallographic data, is involved in binding the γ- and β-phosphates of ATP. The importance of this P-loop motif (GxxxxGK[S/T] [37]) for the ATPase activity of flavivirus NS3 is first demonstrated in this study, as the GKT→GET mutation abolished the ATPase activity. In addition, we have identified a novel motif, a stretch of four basic amino acid residues (184RKRK) which is located close to the P-loop motif A (196GAGKT) of NS3; this motif seems to be an important determinant for the RNA-stimulated ATPase activity. We propose that this basic cluster of amino acid residues is involved in an RNA-protein interaction sensitive to high ionic strength which affects the conformation of the substrate-binding P loop.
In addition to NTPase/RNA helicase function, DEN2 NS3 is likely to function also as a 5′-RNA triphosphatase, as observed for the West Nile virus NS3. The 5′-RNA triphosphatase activity is involved in the first step in formation of the 5′-cap structure. Since the loss of the ATPase activity associated with the mutation (K199E) in the P-loop motif supports the presence of only one ATP binding site in NS3, this site may also be involved in 5′-RNA triphosphatase activity. Further work is necessary to characterize the 5′-RNA triphosphatase activity and the functional domain of NS3 involved in this function.
ACKNOWLEDGMENTS
This research was supported by a grant from the National Institutes of Health (AI 32078) and partly by a Focused Giving Grant from the Johnson & Johnson Foundation (to R.P.). S.C. was partly supported by a predoctoral fellowship from Kansas Health Foundation.
REFERENCES
- 1.Arias C F, Preugschat F, Strauss J H. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993;193:888–899. doi: 10.1006/viro.1993.1198. [DOI] [PubMed] [Google Scholar]
- 2.Barton D J, Flanegan J B. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J Virol. 1997;71:8482–8489. doi: 10.1128/jvi.71.11.8482-8489.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazan J F, Fletterick R J. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology. 1989;171:637–639. doi: 10.1016/0042-6822(89)90639-9. [DOI] [PubMed] [Google Scholar]
- 4.Cahour A, Falgout B, Lai C-J. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J Virol. 1992;66:1535–1542. doi: 10.1128/jvi.66.3.1535-1542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers T J, Grakoui A, Rice C M. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991;65:6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers T J, Hahn C S, Galler R, Rice C M. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 7.Chambers T J, Weir R C, Grakoui A, McCourt D W, Bazan J F, Fletterick R J, Rice C M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci USA. 1990;87:8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho H S, Ha N C, Kang L W, Chung K M, Back S H, Jang S K, Oh B H. Crystal structure of RNA helicase from genotype 1b hepatitis C virus. J Biol Chem. 1998;273:15045–15052. doi: 10.1074/jbc.273.24.15045. [DOI] [PubMed] [Google Scholar]
- 9.Clum S, Ebner K E, Padmanabhan R. Cotranslational membrane insertion of dengue virus type 2 NS2B/NS3 serine proteinase precursor is required for efficient processing in vitro. J Biol Chem. 1997;272:30715–30723. doi: 10.1074/jbc.272.49.30715. [DOI] [PubMed] [Google Scholar]
- 10.Eagles R M, Balmori M E, Beck D L, Gardner R C, Forster R L. Characterization of NTPase, RNA-binding and RNA-helicase activities of the cytoplasmic inclusion protein of tamarillo mosaic potyvirus. Eur J Biochem. 1994;224:677–684. doi: 10.1111/j.1432-1033.1994.t01-1-00677.x. [DOI] [PubMed] [Google Scholar]
- 11.Falgout B, Pethel M, Zhang Y M, Lai C J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald D K, McKenzie L, Ebner K E. Galactosyl transferase activity in a variety of sources. Biochim Biophys Acta. 1971;235:425–428. doi: 10.1016/0005-2744(71)90282-8. [DOI] [PubMed] [Google Scholar]
- 13.Gorbalenya A E, Donchenko A P, Koonin E V, Blinov V M. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 1989;17:3889–3897. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gros C, Wengler G. Identification of an RNA-stimulated NTPase in the predicted helicase sequence of the rubella virus nonstructural polyprotein. Virology. 1996;217:367–372. doi: 10.1006/viro.1996.0125. [DOI] [PubMed] [Google Scholar]
- 16.Irie K, Mohan P M, Sasaguri Y, Putnak R, Padmanabhan R. Sequence analysis of cloned dengue virus type 2 genome (New Guinea-C strain) Gene. 1989;75:197–211. doi: 10.1016/0378-1119(89)90266-7. [DOI] [PubMed] [Google Scholar]
- 17.Kadare G, Haenni A L. Virus-encoded RNA helicases. J Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D W, Gwack Y, Han J H, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 18a.Kim J L, Morgenstern K A, Griffith J P, Dwyer M D, Thomson J A, Murcko M A, Lin C, Caron P R. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 19.Koonin E V. A new group of putative RNA helicases. Trends Biochem Sci. 1992;17:495–497. doi: 10.1016/0968-0004(92)90338-a. [DOI] [PubMed] [Google Scholar]
- 20.Kuo M D, Chin C, Hsu S L, Shiao J Y, Wang T M, Lin J H. Characterization of the NTPase activity of Japanese encephalitis virus NS3 protein. J Gen Virol. 1996;77:2077–2084. doi: 10.1099/0022-1317-77-9-2077. [DOI] [PubMed] [Google Scholar]
- 21.Lain S, Martin M T, Riechmann J L, Garcia J A. Novel catalytic activity associated with positive-strand RNA virus infection: nucleic acid-stimulated ATPase activity of the plum pox potyvirus helicaselike protein. J Virol. 1991;65:1–6. doi: 10.1128/jvi.65.1.1-6.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markoff L. In vitro processing of dengue virus structural proteins: cleavage of the pre-membrane protein. J Virol. 1989;63:3345–3352. doi: 10.1128/jvi.63.8.3345-3352.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nowak T, Farber P M, Wengler G, Wengler G. Analyses of the terminal sequences of West Nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology. 1989;169:365–376. doi: 10.1016/0042-6822(89)90162-1. [DOI] [PubMed] [Google Scholar]
- 24.Polgar L. Structure and function of serine proteases. Amsterdam, The Netherlands: Elsevier Science Publishers; 1987. [Google Scholar]
- 25.Preugschat F, Averett D R, Clarke B E, Porter D J T. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J Biol Chem. 1996;271:24449–24457. doi: 10.1074/jbc.271.40.24449. [DOI] [PubMed] [Google Scholar]
- 26.Preugschat F, Lenches E M, Strauss J H. Flavivirus enzyme-substrate interactions studied with chimeric proteinases: identification of an intragenic locus important for substrate recognition. J Virol. 1991;65:4749–4758. doi: 10.1128/jvi.65.9.4749-4758.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preugschat F, Yao C W, Strauss J H. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J Virol. 1990;64:4364–4374. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rikkonen M, Peranen J, Kaariainen L. ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsP2. J Virol. 1994;68:5804–5810. doi: 10.1128/jvi.68.9.5804-5810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez P L, Carrasco L. Poliovirus protein 2C has ATPase and GTPase activities. J Biol Chem. 1993;268:8105–8110. [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Suzich J A, Tamura J K, Palmer Hill F, Warrener P, Grakoui A, Rice C M, Feinstone S M, Collett M S. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svitkin Y V, Lyapustin V N, Lashkevich V A, Agol V I. Differences between translation products of tick-borne encephalitis virus RNA in cell-free systems from Krebs-2 cells and rabbit reticulocytes: involvement of membranes in the processing of nascent precursors of flavivirus structural proteins. Virology. 1984;135:536–541. doi: 10.1016/0042-6822(84)90207-1. [DOI] [PubMed] [Google Scholar]
- 33.Tai C L, Chi W K, Chen D S, Hwang L H. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3) J Virol. 1996;70:8477–8484. doi: 10.1128/jvi.70.12.8477-8484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura J K, Warrener P, Collett M S. RNA-stimulated NTPase activity associated with the p80 protein of the pestivirus bovine viral diarrhea virus. Virology. 1993;193:1–10. doi: 10.1006/viro.1993.1097. [DOI] [PubMed] [Google Scholar]
- 35.Teterina N L, Bienz K, Egger D, Gorbalenya A E, Ehrenfeld E. Induction of intracellular membrane rearrangements by HAV proteins 2C and 2BC. Virology. 1997;237:66–77. doi: 10.1006/viro.1997.8775. [DOI] [PubMed] [Google Scholar]
- 36.Valle R P, Falgout B. Mutagenesis of the NS3 protease of dengue virus type 2. J Virol. 1998;72:624–632. doi: 10.1128/jvi.72.1.624-632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warrener P, Collett M S. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J Virol. 1995;69:1720–1726. doi: 10.1128/jvi.69.3.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warrener P, Tamura J K, Collett M S. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J Virol. 1993;67:989–996. doi: 10.1128/jvi.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wengler G, Czaya G, Farber P M, Hegemann J H. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J Gen Virol. 1991;72:851–858. doi: 10.1099/0022-1317-72-4-851. [DOI] [PubMed] [Google Scholar]
- 41.Wengler G, Wengler G. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology. 1991;184:707–715. doi: 10.1016/0042-6822(91)90440-m. [DOI] [PubMed] [Google Scholar]
- 42.Westaway E G. Flavivirus replication strategy. Adv Virus Res. 1987;33:45–90. doi: 10.1016/s0065-3527(08)60316-4. [DOI] [PubMed] [Google Scholar]
- 43.Yao N, Hesson T, Cable M, Hong Z, Kwong A D, Le H V, Weber P C. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Mohan P M, Padmanabhan R. Processing and localization of dengue virus type 2 polyprotein precursor NS3-NS4A-NS4B-NS5. J Virol. 1992;66:7549–7554. doi: 10.1128/jvi.66.12.7549-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, Padmanabhan R. Role of protein conformation in the processing of dengue virus type 2 nonstructural polyprotein precursor. Gene. 1993;129:197–205. doi: 10.1016/0378-1119(93)90269-9. [DOI] [PubMed] [Google Scholar]
- 46.Zhao L J, Narayan O. A gene expression vector useful for protein purification and studies of protein-protein interaction. Gene. 1993;137:345–346. doi: 10.1016/0378-1119(93)90032-x. . respectively. [DOI] [PubMed] [Google Scholar]
- 47.Zhao L J, Zhang Q X, Padmanabhan R. Polymerase chain reaction-based point mutagenesis protocol. Methods Enzymol. 1993;217:218–227. doi: 10.1016/0076-6879(93)17064-c. [DOI] [PubMed] [Google Scholar]