Abstract
Background
Tissue necrosis releases cell-free deoxyribonucleic acid (cfDNA), leading to rapid increases in plasma concentration with clearance independent of kidney function.
Aim
To explore the diagnostic role of cfDNA in acute myocardial infarction (AMI).
Methods
This systematic review and meta-analysis included studies of cfDNA in patients with AMI and a comparator group without AMI. The quality assessment of diagnostic accuracy studies-2 (QUADAS-2) tool was used, with AMI determined from the criteria of the original study. Standardised mean differences (SMD) were obtained using a random-effects inverse variance model. Heterogeneity was reported as I2. Pooled sensitivity and specificity were computed using a bivariate model. The area under the curve (AUC) was estimated from a hierarchical summary receiver operating characteristics curve.
Results
Seventeen studies were identified involving 1804 patients (n = 819 in the AMI group, n = 985 in the comparator group). Circulating cfDNA concentrations were greater in the AMI group (SMD 3.47 (95%CI: 2.54–4.41, p < 0.001)). The studies were of variable methodological quality with substantial heterogeneity (I2 = 98%, p < 0.001), possibly due to the differences in cfDNA quantification methodologies (Chi2 25.16, p < 0.001, I2 = 92%). Diagnostic accuracy was determined using six studies (n = 804), which yielded a sensitivity of 87% (95%CI: 72%-95%) and specificity of 96% (95%CI: 92%-98%). The AUC was 0.96 (95%CI: 0.93–0.98). Two studies reported a relationship between peak cfDNA and peak troponin. No studies reported data for patients with pre-existing kidney impairment.
Conclusion
Plasma cfDNA appears to be a reliable biomarker of myocardial injury. Inferences from existing results are limited owing to methodology heterogeneity.
Keywords: Acute myocardial infarction, Ischaemic heart disease, Cell-free deoxyribonucleic acid (cfDNA), Circulating deoxyribonucleic acid, Cardiac biomarker, Cardiac-specific cfDNA
1. Introduction
Cell-free DNA (cfDNA) was first described by Mandel and Metais in 1948 [1]. Its origin is probably linked to three phenomena: cellular apoptosis, necrosis and, to a lesser extent, extracellular vesicle active secretion [2]. Healthy individuals have low plasma concentrations of cfDNA because of rapid elimination mainly by macrophages in the liver and spleen [3].
Cardiac troponin is established as the ‘gold standard’ for the diagnosis of acute myocardial infarction (AMI). However, this biomarker has three main limitations. Firstly, the major limitation of standard cardiac troponin assays (cTn) (cardiac troponin I (cTnI) and cardiac troponin T (cTnT)) is a delay to increased plasma concentration of 4–10 h after the onset of chest pain [4]. This has led to the development of high-sensitivity cardiac troponin (hs-cTn) assays (high-sensitivity cardiac troponin T (hs-cTnI) and high-sensitivity cardiac troponin I (hs-cTnI)), with plasma concentrations detected within three hours of symptoms onset [5]. Secondly, cardiac troponin concentration are challenging to interpret in patients with chronic kidney disease (CKD), a common comorbidity in patients with AMI [6]. Lastly, there is an extended window during which troponin concentrations are increased, making it challenging to determine ongoing ischaemia in patients with an AMI.
AMI is caused by prolonged ischaemia time, rapidly depleting cellular adenosine triphosphate (ATP), triggering an ischemic cascade and eventual induction of cell death through apoptosis or necrosis [7]. The disruption of the membrane of necrotic cardiomyocytes causes DNA fragments to leak out and circulate in the peripheral circulation. Circulating cfDNA is detected within two hours from the onset of chest pain [8], and is not increased in patients with kidney dysfunction [9], [10].
This systematic review aimed to clarify the potential role of cfDNA as an early biomarker in diagnosing patients with AMI. The primary hypothesis was that cfDNA concentration is greater in those patients presenting with AMI compared to a non-AMI comparator group.
2. Methods
This review was conducted according to the Preferred Reporting Items for Systematic reviews and Meta-Analysis (PRISMA)–Diagnostic Test Accuracy (DTA) statements [11], [12] (Table S1 and S2). Details of the protocol were registered prospectively with PROSPERO (CRD42022333190).
2.1. Data source and study search
A comprehensive search strategy was conducted using three databases (MEDLINE, Embase and the Cochrane Library) from inception to 17 May 2022. The medical subject heading terms and keywords included a broad range of descriptors for “acute myocardial infarction” and “cfDNA”. Additional articles were identified by manually reviewing the references of the included articles. No language restrictions were applied. Further details are available in Table S3.
2.2. Eligibility criteria - study selection
The inclusion criteria were: (1) studies that reported cfDNA concentrations at admission in patients with AMI and (2) a comparator group. Exclusion criteria were: (1) reviews, letters, case series and case reports: and (2) studies that did not contain a comparator group. Two reviewers (ET and JZ) independently screened titles and abstracts and consecutively assessed the full texts of potentially relevant articles. Any reviewer disagreement was resolved by consensus.
2.3. Data extraction
The following study data were extracted in duplicate by two authors (ET and JZ): first author, publication year, country and region, hospital, funding source, study methodology, age, gender, sample size, time from admission to when blood samples were collected, processing time, DNA extraction methods, cfDNA quantification methods, admission cfDNA concentration, cfDNA diagnostic test accuracy performance, the correlation between cfDNA level and cardiac biomarkers, and data relating cfDNA to complications post AMI were collected. When necessary, the authors of the included studies were contacted to provide additional details.
2.4. Risk of bias (quality) assessment
Methodological quality was independently assessed by ET and JZ using the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool [13]. The QUADAS-2 tool estimates the risk of bias across the following domains: (1) Patient section; (2) Index test; (3) Reference standard, and (4) Flow and timing and applicability concerns across the following domains: (1) Patient selection; (2) Index tests and (3) Reference standard. Questionnaire details are available in Table S4.
Pre-analytical parameters can significantly affect the results of cfDNA quantification. DNA sampling and pre-processing factors were thoroughly assessed in all the included studies [14]. Five checkpoints were considered (1) the type of medium for analysis (plasma); (2) the type of sample tube; (3) if the sample was analysed within four hours following venepuncture; (4) the number and speed of centrifugation, and (5) Storage conditions before nucleic acid extracts (-20 °C or −80 °C freeze). Five points were deemed high-quality cfDNA processing, four to three were deemed acceptable quality, and two to zero were considered poor quality [15] Table S5.
2.5. Statistical analyses
For studies that used more than one cfDNA quantification method, data from the method that provided the highest area under the Receiver Operative Characteristic (ROC) curve value was used. If the original data were reported as medians and interquartile range or medians and 95% confidence interval (CI) or means and standard error, they were transformed into means and standard deviations. Differences in cfDNA concentrations between the AMI and the comparator groups were quantified as Hedges’ g standardised mean differences (SMD), with 95%CI combined using a random-effects inverse variance model, and findings graphically represented using forest plots [16]. Heterogeneity was tested using the Chi2 and I2 statistic, with heterogeneity defined as Chi2 p < 0.10 or I2 > 50%. Sources of heterogeneity were explored using subgroup analysis stratified according to (1) the type of quantification method used (optical technique, quantitative polymerase chain reaction (qPCR), DNA methylation analysis), (2) whether AMI was defined according to the European Society of Cardiology (ESC) or American Heart Association (AHA) or American College of Cardiology (ACC) consensus guidelines (yes, no) and (3) whether there was funding support (yes, no).
To determine diagnostic accuracy, an AMI was determined from the criteria of the original study. Estimated pooled sensitivities, specificities, and AUCs were obtained using bivariate modelling [17] and graphically depicted using the summary ROC curve. Sensitivity analysis was performed for studies that measured cardiomyocyte-specific cfDNA and studies that did not define AMI according to the ESC or AHA or ACC consensus guidelines. Publication bias was assessed through visual inspection of funnel plots and Egger’s regression test. Statistical analyses were conducted with Review Manager version 5.4 and R software using packages ‘mada’ and ‘dmetatools’.
3. Result
3.1. Study selection
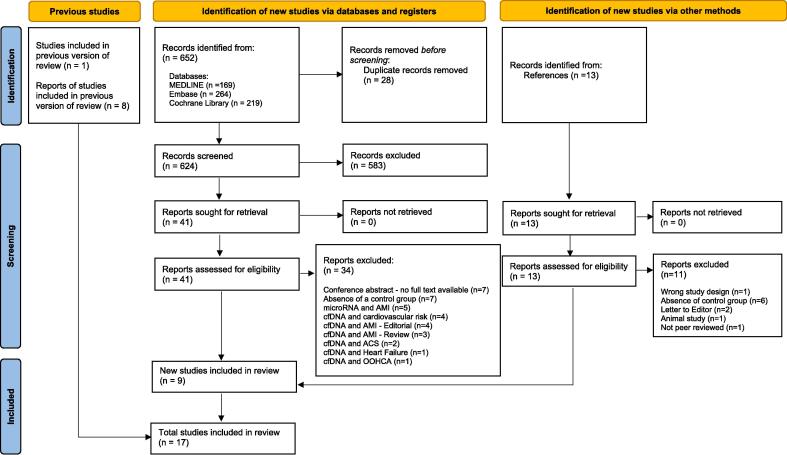
A total of 652 potential studies were identified after duplicates were removed. Of those, 583 were excluded during the screening phase, with 62 studies fully appraised. Seventeen studies [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34] were included in this review (Fig. 1).
Fig. 1.
PRISMA 2020 flow diagram for updated systematic reviews.
3.2. Study characteristics
Seventeen studies [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34] comprising 1804 participants (AMI: n = 819, comparator: n = 985) from five countries were included from 2003 to 2022. Ten studies were conducted in China [21], [22], [24], [25], [26], [27], [28], [31], [32], [33], three in Greece [18], [19], [23], two in Israel [29], [34], one in Egypt [20] and one in Russia [30].
Different definitions of AMI were used. Seven studies [20], [22], [23], [26], [27], [31], [32] defined AMI based on the ESC and/or ACC and/or AHA guidelines. Two studies [18], [28] used the universal definition of myocardial infarction. One study [19] included patients thrombolysed within 6 h of chest pain. One study [34] included coronary angiography evidence of coronary obstruction or thrombus. Two studies [24], [25] established their own AMI criteria based on clinical symptoms, 12-lead ECG and positive cardiac biomarkers (cardiac troponin, Creatinine Kinase-MB (CK-MB) and myoglobin). One study [33] included AMI defined by the Chinese Medical Association. Three studies [21], [29], [30] did not document the parameters used for the definition of AMI. The main characteristics of the included studies are outlined in Table 1 and Table S6.
Table 1.
Study characteristics of the included studies.
| Author | Year | Country | Study type | Number of patients | Age, years mean (SD) |
Male, n (%) | Time of blood taken | Reference tests for the diagnosis of AMI |
|---|---|---|---|---|---|---|---|---|
| Agiannitopoulos | 2020 | Greece | Case-control | 80 AMI 50 controls |
62.1 (11.0) 59.3 (9.8) |
NR | NR | STEMI defined based on the Third Universal definition of Myocardial Infarction |
| Antonatos | 2006 | Greece | Case-control | 13 AMI 30 controls |
NR NR |
7 (54%) NR |
On admission and for 5 consecutive days | AMI patients who underwent thrombolysis within six hours of chest pain |
| Arafat | 2017 | Egypt | Case-control | 50 AMI 30 controls |
57.7 (9.6) 54.6 (10.1) |
40 (80%) 22 (73%) |
On admission and Day 3 | AMI defined based on the joint ESC/ACC criteria |
| Chang | 2003 | China | Case-control | 55 AMI 274 controls |
NR NR |
NR NR |
NR | Did not define |
| Cui | 2013 | China | Case-control | 49 AMI 60 controls |
NR 56.7 (11.1) |
NR 34 (56.7%) |
Within 6 h after the onset of chest pain | STEMI defined based on the ACC/AHA 2007 guidelines |
| Destouni | 2009 | Greece | Case-control | 47 AMI 100 controls |
61.7 (11.8) 65.4 (6.5) |
34 (72%) 44 (44%) |
On admission, Day 1, Day 2, Day 3, Day 4 and Day 5 | AMI defined based on the joint ESC/ACC 2000 consensus guidelines |
| Jing | 2011 | China | Case-control | 22 AMI 60 controls |
NR 18–56 |
NR 45 (75%) |
Within 6 h of admission after onset of chest pain. 5 random patients had blood samples collected on admission, and Day 1, Day 2 and Day 3 after PCI. |
AMI defined according to the clinical symptoms, 12-lead ECG and positive cardiac biomarkers (cTnI, CK-MB, MYO) |
| Lou | 2015 | China | Case-control | 120 AMI 60 controls |
40–80 40–80 |
60 (50%) 30 (50%) |
Within 6 h of admission after onset of chest pain | AMI defined according to clinical symptoms, 12-lead ECG and positive cardiac biomarkers (cTnI, CK-MB, MYO, LDH) |
| Qin | 2016 | China | Cohort study | 38 AMI 32 controls |
54.8 (14.3) 59.3 (12.7) |
31 (82%) 25 (78%) |
On admission, 12 h post PCI, 24 h post PCI, 48 h post PCI | AMI defined based on the joint ESC and ACC/AHA guidelines |
| Rainer | 2006 | China | Case-control | 10 AMI 21 controls |
68 (13) 64 (8) |
NR 8 (38%) |
On admission and 6 h later, then followed up for 2 years | STEMI - AMI defined based on the joint ESC/ACC 2000 consensus definition |
| Ren | 2022 | China | Observational | 20 AMI (cohort 1) 20 AMI (cohort 2) 116 AMI (cohort 3) 25 controls |
NR | NR | Cohort 1: after PCI Cohort 2: On admission, Day 1 post PCI, Day 2 post PCI Cohort 3: within 24 h of symptoms onset |
AMI defined based on Fourth Universal definition of Myocardial infarction |
| Shimony | 2010 | Israel | Observational | 16 AMI 47 controls |
50.3 (12.5) 26.3 (4.7) |
14 (93%) 25 (53%) |
On admission, and three additional time points, 5–8 h apart | STEMI – Did not define |
| Veiko | 2007 | Russia | Observational | 7 AMI 18 controls |
78 (7) 38 (16) |
4 (57%) 11 (61%) |
Within 3 h after the development of AMI | Did not define |
| Wang | 2015 | China | Case-control | 25 AMI 25 controls |
59.3 (13.4) 64.5 (12) |
21 (84%) 18 (72%) |
Within 8 h of admission and Day 2 post PCI | AMI defined based on the joint ESC/ and ACC/AHA guidelines |
| Xie | 2018 | China | Case-control | 100 AMI 30 controls |
59 (10) 58 (5) |
71 (71%) 15 (50%) |
Onset of AMI, 5 consecutive days and monthly intervals for 5 months | AMI defined based on the joint ESC/ACC 2000 consensus guidelines |
| Xu | 2012 | China | Case-control | 40 AMI 40 controls |
44–66 40–60 |
28 (70%) 22 (55%) |
Within 48 h after onset of chest pain and approximately 8 weeks post treatment | AMI defined based on the Guidelines for the Diagnosis and Treatment of Acute Myocardial Infarction, revised by the Cardiovascular Branch of the Chinese Medical Association |
| Zemmour | 2018 | Israel | Observational | 31 AMI 83 controls |
NR 35.3 |
NR 37 (45%) |
Within 13 h of onset of chest pain and 6 h intervals up to 60hr from hospital admission | AMI diagnosed with STE > 1 mm in two or more contiguous leads. Final diagnosis was determined after coronary angiography with evidence of coronary obstruction or thrombus. |
ACC: American College of Cardiology, AHA: American Heart Association, ESC: European Society of Cardiology, NR: Not Reported.
4. Risk of bias within studies
4.1. QUADAS-2 methodological quality
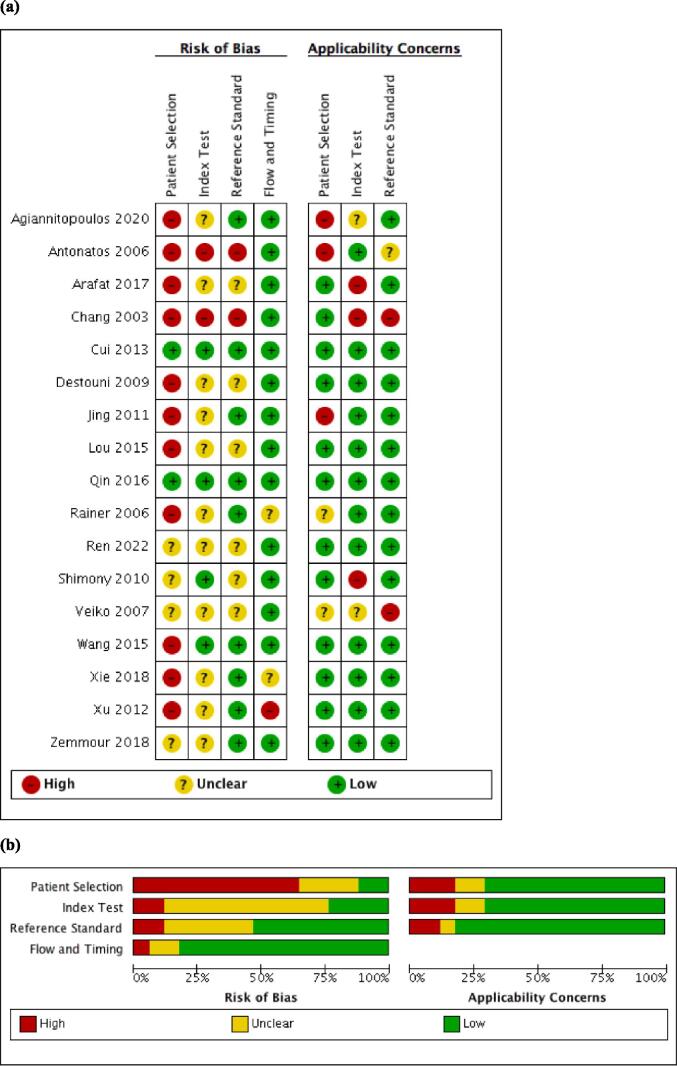
Methodological quality varied. Most of the studies [18], [19], [20], [22], [23], [24], [25], [26], [27], [28], [29], [31], [32] excluded patients with comorbidities that could foreseeably raise cfDNA, which may have introduced selection bias. However, the patients included matched the review questions (i.e. no major concerns for the applicability domain). The interval between the index test and reference standard was sufficiently short to avoid changes in disease status in>80% of the studies, therefore having a low risk of bias for the flow and timing domain (Fig. 2 and Table S4).
Fig. 2.
Methodological quality of the included studies (a) individual assessment (b) Summary of methodological quality of included studies.
4.2. cfDNA pre-analytical considerations for DNA processing
DNA processing quality was considered high quality in two studies [31], [34], acceptable quality in ten studies [19], [22], [23], [24], [25], [26], [27], [28], [32], [33] and poor quality in five studies [18], [20], [21], [29], [30] (Table S6).
5. Results of individual studies
5.1. Techniques for the detection of cfDNA
Eleven studies [18], [19], [21], [23], [26], [27], [28], [31], [32], [33], [34] applied cfDNA quantitative methods that required DNA to be extracted before cfDNA quantification. Various DNA extraction methods contribute to a broad range of cfDNA concentrations [35]. cfDNA can be detected by an array of quantitative techniques: (1) Optical technique - (i) Fluorometric quantification - Qubit™ ssDNA and dsDNA assay kit [18], PicoGreen® dsDNA assay [21], [32], SYBR®Gold nucleic acid gel stain [29], Hoechst 33,528 nucleic acid stain [30] (ii) Spectrophotometric quantification - NanoDrop® 1000 spectrometer for nucleic acid quantification [18] (iii) Luminometric quantification [22], [24]; (2) quantitative polymerase chain reaction (qPCR) method (i) probe-based [18], [19], [23], [27], [31] (ii) dye-based [20], [25], [26] (iii) not otherwise specified [34] and (3) DNA methylation analysis [28], [34]. Only two studies [28], [34] inferred the origins of cfDNA and quantified cardiomyocyte-specific cfDNA concentrations (Table 2).
Table 2.
Studies reporting cfDNA concentration in AMI group versus comparator group.
| Author | Year | Number | cfDNA |
p-value | ||
|---|---|---|---|---|---|---|
| DNA extraction method | cfDNA quantification method | cfDNA concentration (unit) on admission | ||||
| Agiannitopoulos | 2020 | 80 AMI 50 controls |
QIAamp Circulating Nucleic Acid Kit (Qiagen) | Fluorometric quantification using Qubit® 3.0 fluorometer of Qubit™ ssDNA assay | AMI: 48.5 ng/μl (3.3)∼ Controls: 8.2 ng/μl (0.3)∼ |
p < 0.05 |
| Fluorometric quantification using Qubit® 3.0 fluorometer of Qubit™ dsDNA assay | AMI: 6.7 ng/μl (0.3)∼ Controls: 1.2 ng/μl (0.1)∼ |
p < 0.05 | ||||
| NanoDrop® 1000 spectrophotometer for nucleic acid quantification, (the user selects an option to specify between RNA, DNA and ssDNA | AMI: 22.0 ng/μl (1.2)∼ Controls: 7.8 ng/μl (0.2)∼ |
p < 0.05 | ||||
| probe-based qPCR using Rotor-Gene 6000 system with the TaqMan Copy Number Reference Assay TERT (This assay targets Telomerase reverse transcriptase (TERT) gene) | AMI: 42.8 ng/μl (2.9)∼ Controls: 1.4 ng/μl (0.5)∼ |
p < 0.05 | ||||
| Antonatos | 2006 | 13 AMI 30 controls |
QIAamp Blood Kit (Qiagen) | probe-based? qPCR using LightCycler™ - reagent not reported | AMI: 6873 log GE/ml (3 5 7)‘ Controls: 4112 log GE/ml (2 3 4)‘ |
p < 0.05 |
| Arafat | 2018 | 50 AMI 30 controls |
QIAamp Min Elute Virus Spin kit (Qiagen) | SYBR-green mix dye-based qPCR with Low ROx (Primer: L1PA2 (90) using 96-well plates in a 7500 qPCR system | AMI: 179.3 ng/ml (151.1) Controls: 6.8 ng/ml (2.6) |
P = 0.001 |
| SYBR-green mix dye-based qPCR with Low ROx (Primer: L1PA2 (2 2 2) using 96-well plates in a 7500 qPCR system | AMI: 114.1 ng/ml (83.3)‘ Controls: 3.2 ng/ml (1.8) |
P = 0.001 | ||||
| Chang | 2003 | 55 AMI 274 controls |
QIAamp 96 Spin Blood DNA extraction Kit | Fluorometric quantification of PicoGreen® dsDNA assay (PicogGreen® reagent is a florescent probe that binds to dsDNA and create a nucleic acid stain by forming a highly luminescent complex, used for quantitating dsDNA) | AMI: 510 ng/ml (3 9 8)∼ Controls: 36 ng/ml (23.8)∼ |
NR |
| Cui | 2013 | 49 AMI 60 controls |
Not required | LMAX® microplate luminometer for the quantification of branched DNA-based Alu assay Duration for results to be obtained: 2-3 h |
AMI: 5745 ng/ml (4013–8643)* Controls: 118 ng/ml (81–221)* |
p < 0.05 |
| Destouni | 2009 | 47 AMI 100 controls |
QIAamp DNA Blood MiniKit (Qiagen) | probe-based qPCR using LightCycler™ with a primer for the amplification of a 189 bp fragment of the beta-globin gene | AMI: 196 GE/ml (1.4–496212.1)* Controls: 31 GE/ml (6.5–266.1)* |
p < 0.05 |
| Jing | 2011 | 22 AMI 60 controls |
Not required | LMAX® microplate luminometer for the quantification of branched DNA-based Alu assay | AMI: 4439 ng/ml (2100.4–9435.5)* Controls: 117 ng/ml (80.8–218.6)* |
p < 0.05 |
| Lou | 2015 | 120 AMI 60 controls |
NR | SYBR-green mix dye-based qPCR with Alu-based qPCR assay (Primer size 201 bp) using an ABI 7500 sequence detector [Alu1] Running time required: 30 min |
AMI: 4.2 log copies/ml (0.2)∼ Controls: 1.8 log copies/ml (0.2)∼ |
p < 0.05 |
| SYBR-green mix dye-based qPCR with Alu-based qPCR assay (Primer size 170 bp) using an ABI 7500 sequence detector [Alu2] | AMI: 5.4 log copies/ml (0.1)∼ Controls: 2.9 log copies/ml (0.4)∼ |
p < 0.05 | ||||
| SYBR-green mix dye-based qPCR with Alu-based qPCR assay (Primer size 147 bp) using an ABI 7500 sequence detector [Alu3] | AMI: 5.1 log copies/ml (0.3)∼ Controls: 1.7 log copies/ml (0.3)∼ |
p < 0.05 | ||||
| SYBR-green mix dye-based qPCR with Alu-based qPCR assay (Primer size 113 bp) using an ABI 7500 sequence detector [Alu4] | AMI: 7.0 log copies/ml (0.2)∼ Controls: 2.9 log copies/ml (0.1)∼ |
p < 0.05 | ||||
| SYBR-green mix dye-based qPCR with Alu-based qPCR assay (Primer size 76 bp) using an ABI 7500 sequence detector [Alu5] | AMI: 7.21 log copies/ml (0.17)∼ Controls: 3.79 log copies/ml (0.14)∼ |
p < 0.05 | ||||
| Alu = log (Alu1 + Alu2 + Alu3 + Alu4 + Alu 5) | AMI: 7.90 log copies/ml (0.17) Controls: 4.67 log copies/ml (0.24)∼ |
p < 0.05 | ||||
| Qin | 2016 | 38 AMI 32 controls |
DNeasy Blood and Tissue Kit (Qiagen) | mtDNA was measured with a SYBR® green dye-based qPCR assay using an ABI PRISM 7300 sequence detection system. The primer sequence was the human NADH dehydrogenase 1 gene | AMI: 478 copies/μl (1 0 6)∼ Controls: 157 copies/μl (97)∼ |
p < 0.05 |
| Rainer | 2006 | 10 AMI 21 controls |
QIAamp DNA Blood Kit (Qiagen) | probe-based qPCR analysis was performed using a PE Applied Biosystems 7700 sequence detector with a TaqMan probe for the beta-globin gene. | AMI: 660 kGE/l (440–1120)^ Controls: 350 kGE/l (250–610)^ |
p < 0.05 |
| Ren | 2022 | 20 AMI 25 controls |
QIAamp Circulating Nucleic Acid Kit (Qiagen) | cfDNA methylation analysis (genome wide method): cfDNA concentration quantification method is not specified. | AMI: 6.5 ng/ml Controls: 6.3 ng/ml |
p = 0.21 |
| 116 AMI 25 controls |
QIAamp Circulating Nucleic Acid Kit (Qiagen) | cfDNA Methylation analysis (targeted method): cfDNA concentration quantification method is not specified. cfDNA underwent bisulfite conversion with EZ-96 Methylation Direct™ MagPrep. Bisulfite-converted cfDNA was interrogated using the TaqMan assay targeted at CORO6 locus as a heart-specific hypermethylation marker. CORO6 ddPCR assay was used to provide quantification of cardiomyocyte-specific cfDNA. | AMI: 1.0 copies/ml (0.8–2.0)* Controls: 0 copies / ml (0–0.9)* |
P = 0.001 | ||
| Shimony | 2010 | 16 AMI 47 controls |
Not required | Fluorometric quantification of SYBR®Gold Nucleic Acid Gel Stain (SYBR® Gold is a dye that exhibits florescence enhancement upon binding to nucleic acids (dsDNA or ssDNA or RNA)). | AMI: 735 ng/ml (3 5 0)∼ Controls: 471 ng/ml (2 0 3)∼ |
p < 0.05 |
| Veiko | 2008 | 7 AMI 18 controls |
NR | Fluorometric quantification of Hoechst 33,528 nucleic acid stain (Hoechst 33,528 emits blue fluorescence when bound to dsDNA) | AMI: 862 ng/ml (6 0 8)∼ Controls: 161 ng/ml (70)∼ |
NR |
| Wang | 2015 | 25 AMI 25 controls |
QIAamp DNA blood Mini kit (Qiagen) | probe-based qPCR using LightCycle®96 sequence detection system with primer for the beta-globin gene | AMI: 0.5 ng/μl (0.1)‘ Controls: 0.2 ng/μl (0.0)‘ |
p < 0.05 |
| mtDNA was measured with probe-based qPCR using LightCycle®96 sequence detection system with primer for the human NADH dehydrogenase 1 gene | AMI: 3.8 ng/μl (0.4)‘ Controls: 1.9 ng/μl (0.3)‘ |
p < 0.05 | ||||
| Xie | 2018 | 100 AMI 30 controls |
QIAamp Circulating Nucleic Acid Kit (Qiagen) | Fluorometric quantification of PicoGreen® dsDNA assay (PicoGreen® dsDNA quantitation reagent is a florescent probe that binds to dsDNA and create a nucleic acid stain by forming a highly luminescent complex, used for quantitating dsDNA) | AMI: 5071 ng/mL (3709 – 6363)*€ Controls: 974 ng/ml (3 2 8)∼ |
p < 0.05+ |
| Xu | 2012 | 40 AMI 40 controls |
Magnetic bead method | qPCR not otherwise specified | AMI: 253.6 ng/ml (45.7)∼ Controls: 21.5 ng/ml (10.7)∼ |
p < 0.05- |
| Zemmour | 2018 | 31 AMI 83 controls |
QIAsymphony Circulating DNA Kit | DNA Methylation analysis (targeted method): cfDNA measured using Qubit® dsDNA HS Assay Kit, followed by bisulfite conversion with EZ DNA Methylation Gold™ and PCR amplification. Bisulfite-treated cfDNA is interrogated using two methylation sensitive TaqMan™ probes to cover informative CpG sites in the FAMA1010A locus using prespecified primers. ddPCR™ Supermix for Probes was used and analysed with QuantaSoft analysis software to quantify cardiomyocyte specific cfDNA (unmethylated FAM101A sequencing-based assay) | AMI: 90.0 copies/ml (17.5–324.7)*€ Controls: 16.0 copies/ml (–22.4 – 51.2)*€ |
p < 0.05+ |
ddPCR: digital droplet Polymerase Chain Reaction, qPCR: Real-Time Polymerase Chain Reaction or Quantitative Polymerase Chain Reaction or quantitative real-time polymerase chain reaction (gives faster more detailed real time results used to quantify nucleic acid, NR: Not Reported, AMI - Acute Myocardial Infarction, STEMI – ST Elevated Myocardial Infarction, TnI Troponin I, cTnT cardiac troponin T, cTnI cardiac troponin I, CI: confidence Interval, KGE/l: kilogenome equivalents per liter, GE/ml: genomic equivalent per milliliter, ng/ml: nanograms per milliliter, ng/μL: nanograms per microliter, SE: standard error.
*median (interquartile range), ‘mean (standard error), ∼mean (standard deviation),^median (95% CI), +Mann-Whitney, - T-test.
5.2. cfDNA concentration in AMI versus comparator group
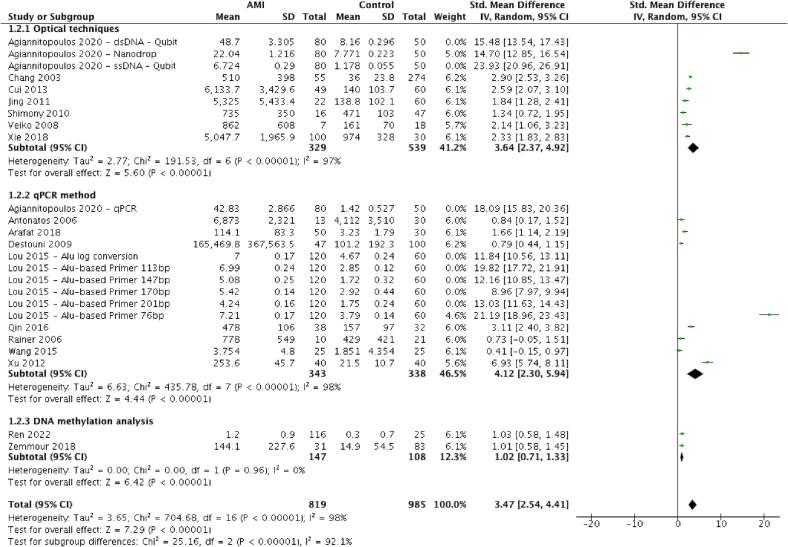
From 17 studies [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34] involving 1804 patients, cfDNA concentrations were substantially greater in AMI patients than in the comparator group (pooled SMD 3.47 (95% CI 2.54–4.41, p < 0.001)). The heterogeneity of these studies was high (I2 = 98%, p < 0.001). Subgroup analysis suggests that heterogeneity could partly be explained by the different cfDNA quantification methods used (Test for subgroup differences: Chi2 25.16, p < 0.001, I2 = 92%, Fig. 3) but not explained by whether studies defined AMI according to consensus guidelines or if there was funding support (Fig. S1 and Fig. S2). The funnel plot for assessing publication bias was asymmetrical (Egger’s test, p < 0.001, Fig. S3). On visual inspection, the inverted funnel shape poorly captured the distribution of included studies, with zero studies falling within the expected funnel region. Accordingly, publication bias could not be excluded.
Fig. 3.
Forest plot of cfDNA levels in AMI versus the comparator group with subgroup analysis stratified according to the different cfDNA quantification methods.
5.3. Diagnostic performance of cfDNA in AMI
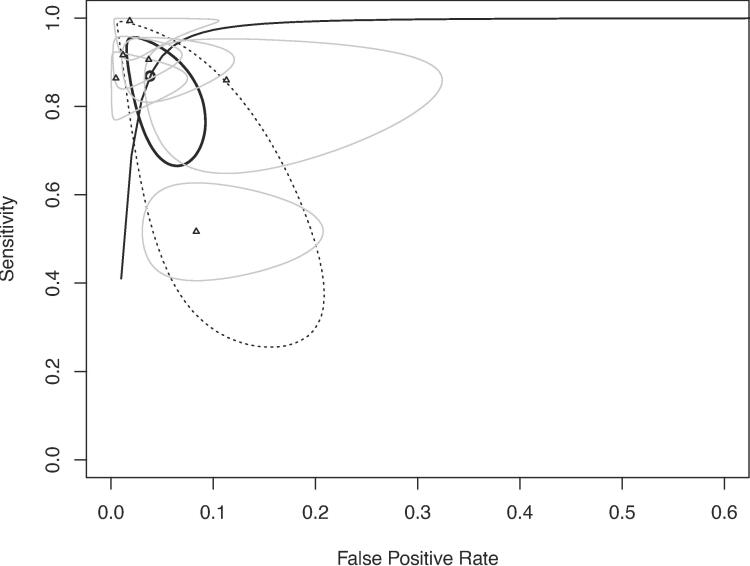
Six studies [18], [22], [25], [28], [32], [34] with a total sample size of n = 804 (AMI: n = 496, comparator: n = 308) could be used for diagnostic performance. The overall diagnostic accuracy of cfDNA, when compared to the ‘gold standard’ in each study, yielded a sensitivity of 87% (95%CI 72%-95%), specificity of 96% (95%CI 92%-98%) and AUC of 0.96 (95%CI 0.93–0.98) (Fig. 4 and Table 3). Two studies [25], [34] that did not use consensus guidelines for the diagnoses of AMI were removed as part of a sensitivity analysis. The four remaining studies [18], [22], [28], [32] yielded a sensitivity of 89% (95%CI 62%-98%), specificity of 96% (95%CI 92%-99%) and AUC of 0.98 (95%CI 0.92–––0.99) (Fig. S4). Cardiomyocyte-specific cfDNA studies [28], [34] were removed as part of a sensitivity analysis. The remaining four studies [18], [22], [25], [32] provided a pooled sensitivity, specificity and AUC of 90% (95%CI 86%-93%) and 98% (95%CI 95%-99%) and 0.97 (95%CI 0.88–0.99), respectively (Fig. S5). Three studies [18], [22], [25] provided pooled estimates of sensitivity, specificity and AUC for cardiac troponins (cTnT [18], cTnI [22], [25]) as 80% (95%CI 52%-94%), 97% (95%CI 86%-99%) and 0.96 (95%CI 0.78–0.99), respectively (Fig. S6).
Fig. 4.
Sroc of six studies with pooled estimates of auc, sensitivity and specificity for cfdna in the diagnosis of ami.
Table 3.
Studies reporting Diagnostic Test Accuracy data for cfDNA in AMI.
| Author | Year | Number | Outcome to discriminate |
cfDNA |
Troponin |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cfDNA quantification method | AUC (95%CI), p-value | Cut-off, units | Sensitivity | Specificity | AUC (95%CI) | Cut-off | Sensitivity | Specificity | ||||
| Agiannitopoulos | 2020 | 80 AMI 50 controls |
AMI | Qubit® 3.0 ssDNA assay | 0.99 (0.997–1.00), p < 0.001 | 20.45 ng/μl | 100% | 99% | 0.94 (0.87–1.00), p < 0.001* | NR | NR | NR |
| Qubit® 3.0 dsDNA assay | 0.94 (0.89–0.98), p < 0.001 | 3.43 ng/μl | 100% | 98% | ||||||||
| NanoDrop® | 0.99 (0.99–1.00), p < 0.001 | 15.4 ng/μl | 100% | 92% | ||||||||
| qPCR TaqMan Copy Number Reference Assay TERT | 0.99 (0.99–1.00), p < 0.001 | 13.70 ng/μl | 100% | 99% | ||||||||
| Cui | 2013 | 49 AMI 60 controls |
ACS | branched DNA-based Alu assay | 0.98 (0.96–1.00) | NR | 92% | 98% | 0.74 (0.68–0.80)** | NR | 57% | 98% |
| STEMI from ACS | branched DNA-based Alu assay | 0.90 (0.84–0.95) | NR | NR | NR | 0.76 (0.67–0.84)** | NR | NR | NR | |||
| Lou | 2015 | 120 AMI 60 controls |
AMI | Alu-based qPCR assay (Primer size 201 bp) | 0.89 (0.82–0.95), p < 0.001 | 3.71 log copies/ml | 77% | 96% | 0.90 (0.85–0.96), p < 0.001** | 1.35 ng/ml | 80% | 84% |
| Alu-based qPCR assay (Primer size 170 bp) | 0.76 (0.66–0.85), p < 0.001 | 1.93 log copies/ml | 100% | 49% | ||||||||
| Alu-based qPCR assay (Primer size 147 bp) | 0.86 (0.79–0.93), p < 0.001 | 0.22 log copies/ml | 100% | 60% | ||||||||
| Alu-based qPCR assay (Primer size 113 bp) | 0.94 (0.89–0.99), p < 0.001 | 3.73 log copies/ml | 87% | 96% | ||||||||
| Alu-based qPCR assay (Primer size 76 bp) | 0.97 (0.93–1.00), p < 0.001 | 6.13 log copies/ml | 87% | 100% | ||||||||
| Alu = log (Alu1 + Alu2 + Alu3 + Alu4 + Alu 5) | 0.93 (0.87–0.99), p < 0.001 | 6.40 log copies/ml | 92% | 96% | ||||||||
| Ren | 2022 | 116 AMI 25 controls |
AMI | methylated CORO6 ddPCR assay | 0.68 (0.59–0.78), p = 0.0037 | NR | NR | NR | NR | NR | NR | NR |
| Xie | 2018 | 100 AMI 30 controls |
AMI | PicoGreen® dsDNA assay | 0.96 (0.90–0.99) | 110 ng | NR | NR | NR | NR | NR | NR |
| Zemmour | 2018 | 31 AMI 83 controls |
STEMI | demethylated FAM101A sequencing-based assay | 0.94 (0.90–0.98), p < 0.001 | NR | NR | NR | NR | NR | NR | NR |
*Cardiac troponin T, **Cardiac Troponin I, NR: Not Reported.
5.4. Relationship between cfDNA and other cardiac biomarkers
Zemmour, et al. [34] described time to first increase of cfDNA concentrations within two hours from the onset of chest pain (similar to hs-cTn assays [36]), whereas conventional cTn assays only detects rises 4–10 h [4] after.Xie, et al. reported a significant association between cfDNA taken on admission and cTnI 6 h later (n = 100, Spearman correlation R = 0.57 (95%CI 0.41–0.69)) [32]. As expected, four studies [20], [21], [22], [24] reported no relationship between admission cfDNA and troponin concentrations. When assessing the association between peak cfDNA and peak troponin concentrations, a strong association was described in two studies [22], [29] (Shimony, et al.: Peak cfDNA and peak cTnT: r = 0.65 (95%CI 0.23–0.87), p = 0.006; Cui, et al.: peak cfDNA and peak cTnI: r = 0.72) (Table S7).
Peak cfDNA concentrations were significantly greater in patients with complicated post-AMI courses than those with an uncomplicated one [19], [23] (Table S8). Similarly, Xie, et al. reported 1.8 fold greater cfDNA concentrations in the group experiencing one or more adverse events than the stable group (p < 0.001) [32].
5.5. Relationship between initial cfDNA and complicated admission
Rainer, et al. reported that admission cfDNA concentrations were two-fold greater in patients who developed heart failure (1060 vs 500 kGE/l, p = 0.009) and patients who later re-infarcted (1000 vs 530 kGE/l, p = 0.029); and three-fold greater in patients who had a cardiac arrest in that admission (1350 vs 525 kGE/l, p = 0.04) [27]. Similarly, Cui, et al. reported a significant difference in cfDNA concentration between the groups stratified according to the Global Registry of Acute Coronary Events (GRACE) scores (GRACE score < 100: median 1792 ng/ml (IQR 1198–3649); GRACE score 100–200: median 2635 ng/ml (IQR 2345–5220); GRACE score > 200: 8162 (IQR 2688–9206); p < 0.001) [22].
5.6. cfDNA and kidney impairment
None of the extracted studies included only CKD patients or reported outcomes in the subgroup of CKD patients with AMI.
6. Discussion
The main finding is that plasma cfDNA is an emerging biomarker of myocardial injury. Three major observations supported this concept. Firstly, cfDNA concentrations were more than three-fold greater in those patients presenting with AMI compared to a non-AMI comparator group. Secondly, some studies reported strong associations between peak cfDNA and cTn concentrations [22], [29], suggesting that cfDNA increases due to cfDNA leak from necrotic cardiomyocytes during an AMI [32], [37]. Thirdly, cfDNA demonstrated excellent diagnostic accuracy for AMI, performing similarly to conventional cTn and hs-cTn assays [5].
The latter observation is from the pooled sensitivity and specificity of cfDNA, which were 87% and 96%, respectively, extending improved sensitivity compared with conventional cTn assays but similar to hs-cTn assays [5]. This observation could attribute this higher sensitivity to the diagnosis of AMI in early presenters. cfDNA concentrations are increased within two hours from the onset of chest pain in those with AMI [8], [34], whereas conventional cTn levels require 4–10 h after the onset of symptoms before concentrations increase [4].
The rapid increase in cfDNA concentrations from the onset of symptoms, similar to hs-cTn, is of major clinical relevance. Early identification of AMI facilitates earlier commencement of effective treatment, which is vital to reduce mortality [38]. Point-of-care hs-cTnI is currently being investigated to reduce time-to-invention compared to central laboratory processes [39]. While traditional DNA extraction methods were labour intensive and led to barriers to the rapid measurement of cfDNA; three included studies [24], [25], [29] described quantification methods without the need for prior DNA extraction. Point-of-care devices to measure cfDNA have been developed [40]. Prospective studies are needed to evaluate the accuracy and speed of these devices to determine AMI.
This study provides an expanded update to a 2015 mini-review [41]. In the previous review, two studies [30], [33] before 2015 met the inclusion criteria but were not included. Comprehensive risks of bias were not assessed, and the definitions of AMI, which varied between studies, were not extracted or evaluated in the 2015 review; consequently, the potential impact of these factors on the conclusion remains uncertain. The present review was performed according to the PRISMA guidelines with risks of bias assessed using the QUADAS-2 tool and an inclusion of two pioneering studies that analysed the performance of cardiomyocyte-specific cfDNA as a biomarker of AMI.
Patients with kidney impairment have high mortality following AMI [42] but are less likely to receive invasive coronary angiography than those without [6]. The reason/s for this remains unclear, but there may be a potential bias against investigating patients with kidney disease [6], [42]. Because hs-cTn concentrations are frequently chronically elevated in CKD, alternative diagnostic methodologies are vital. The clearance of cfDNA is mainly through the liver and spleen [3], and are not increased in patients with kidney dysfunction [9], [10], as such, cfDNA is a potential diagnostic biomarker of AMI in patients with CKD. Hitherto, none of the included studies focused on CKD patients.
There remain two main challenges to the widespread use of cfDNA. First, interindividual variability due to various disease conditions (such as cancers [43], autoimmune diseases [44], solid organ transplants [45], sepsis [46] and trauma [15]) that can increase plasma cfDNA concentrations. Therefore, DNA methylation analysis to determine cardiomyocyte-specific cfDNA represents a promising technique. In 2018, Zemmour, et al. reported the first cardiomyocyte-specific cfDNA marker, demethylated FAM101A, in diagnosing ST-elevation myocardial infarction. They reported excellent diagnostic accuracy [34]. Ren, et al. validated Zemmour’s study and established another cardiomyocyte-specific cfDNA marker, methylated CORO6, which can provide an even more rapid result [28].
Second, the half-life of cfDNA ranges between several minutes and two hours [3], comparable with well-established markers of necrosis such as troponin [47] and CK-MB [48]. Nonetheless, biomarkers may persist for days despite their short biological half-life. Zemmour, et al. reported that after percutaneous coronary intervention, cfDNA concentrations increase rapidly before returning to baseline after one to two days. Meanwhile, cardiac troponin reduces over the next four to ten days [49] despite its short biological half-life [47]. The rapid change in cfDNA concentration should be considered an advantage to monitor the immediate treatment response and serve as a marker for adverse events after AMI. Nonetheless, time to dominant peak of plasma concentration cfDNA remains unclear and contradictory, indicating both earlier [8], [26], [28] and later [21], [29], [41] than that of troponin and CK-MB. Further studies are required to assess the kinetic pattern and molecular pathways [50] of cfDNA in AMI.
This systematic review and meta-analysis has some limitations inherent to the original studies. Eight included studies [19], [21], [24], [25], [29], [30], [33], [34] applied a variety of methods to determine AMI. Given the lack of a single ‘gold standard’, the overall estimates of sensitivity or specificity may be biased, and it is not possible to determine whether the estimates would consistently overestimate or underestimate the true accuracy of cfDNA. There was also considerable heterogeneity in cfDNA extraction, cfDNA quantification and cut-off values used. It is worth noting that the development of a preferred quantification method to ensure standardisation is limited by several obstacles that include, but are not limited to, the lack of universal pre-analytical standards, a growing range of pre-analytical methods, no standard reference materials and the need for further validation of quantification methodologies to demonstrate clinical relevance. While in-depth enquiry into these factors is beyond the scope of this review, at the moment, there is no optimal approach for the quantification of total cfDNA levels, and the development of such may be imperative [51]. Furthermore, most studies included in this analysis were diagnostic case controls, which introduce spectrum bias with the possibility of overestimating diagnostic performance. Publication bias also cannot be excluded with unpublished studies not included in this meta-analysis. Lastly, no studies investigated cfDNA concentration in CKD patients with AMI.
7. Conclusion
Plasma cfDNA appears to be a reliable biomarker of myocardial injury. Inferences from existing results are limited owing to methodology heterogeneity. Peak concentrations appear to correlate with poorer clinical outcomes. Further studies to assess point-of-care devices prospectively are required to fully evaluate the efficacy and accuracy of rapid on-site defections of AMI.
Funding
No funding was received for this study.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijcha.2023.101246.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Mandel P., Metais P. Nuclear Acids in Human Blood Plasma. CR Seances Soc. Biol. Fil. 1948;142:241–243. [PubMed] [Google Scholar]
- 2.Grabuschnig S., Bronkhorst A.J., Holdenrieder S., Rodriguez I.R., Schliep K.P., Schwendenwein D., et al. Putative Origins of Cell-Free DNA in Humans: A Review of Active and Passive Nucleic Acid Release Mechanisms. Int. 2020;21:8062. doi: 10.3390/ijms21218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kustanovich A., Schwartz R., Peretz T., Grinshpun A. Life and death of circulating cell-free DNA. Cancer Biol. Ther. 2019;20(8):1057–1067. doi: 10.1080/15384047.2019.1598759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garg P., Morris P., Fazlanie A.L., Vijayan S., Dancso B., Dastidar A.G., et al. Cardiac biomarkers of acute coronary syndrome: from history to high-sensitivity cardiac troponin. Intern. Emerg. Med. 2017;12(2):147–155. doi: 10.1007/s11739-017-1612-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keller T., Zeller T., Peetz D., Tzikas S., Roth A., Czyz E., et al. Sensitive Troponin I assay in Early Diagnosis of Acute Myocardial Infarction. NEJM. 2009;361(8):868–877. doi: 10.1056/NEJMoa0903515. [DOI] [PubMed] [Google Scholar]
- 6.Murry J., Balmuri A., Saurav A., Smer A., Alla V.M. Impact of Chronic Kidney Disease on Ultilization of Coronary Angiography and Percutaneous Coronary Intervention, and Their Outcomes in Patients with Non-ST Elevation Myocardial Infarction. Am. J. Cardiol. 2018;122(11):1830–1836. doi: 10.1016/j.amjcard.2018.08.024. [DOI] [PubMed] [Google Scholar]
- 7.Bronkhorst A.J., Ungerer V., Oberhofer A., Gabriel S., Polatoglou E., Randeu H., et al. New Perspectives on the Importance of Cell-Free DNA Biology. Dignostics. 2022;12:2147. doi: 10.3390/diagnostics12092147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polina I.A., Ilatovskaya D.V., DeLeon-Pennell K.Y. Cell-free DNA as a diagnostic and prognostic marker for cardiovascular disease. Clin. Chim. Acta. 2020;503:145–501. doi: 10.1016/j.cca.2020.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korabencna M., Opatrna S., Wirth J., Rulcova K., Eiselt J., Sefrna F., et al. Cell-free plasma DNA during peritoneal dialysis and hemodialysis and in patients with chronic kidney disase. Ann. N. Y. Acad. Sci. 2008;1137:296–301. doi: 10.1196/annals.1448.014. [DOI] [PubMed] [Google Scholar]
- 10.McGuire A.L., Urosevic N., Chan D.T., Dogra G., Inglis T.J.J., Chakera A. The impact of chronic kidney disease and short-term treatment with rosiglitazone on plasma cell-free DNA levels. PPAR Res. 2014;643189 doi: 10.1155/2014/643189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339(2535):1–8. [PMC free article] [PubMed] [Google Scholar]
- 12.M.D.F. McInnes, D. Moher, B.D. Thombs, T.A. McGrath, P.M. Bossuyt, Group TP-D, Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies: The PRISMA-DTA Statement, JAMA 319(4) (2018) 388–396. [DOI] [PubMed]
- 13.Schueler S., Schuetz G.M., Dewey M. The Revised QUADAS-2 tool. Ann. Intern. Med. 2012;156(4):323. doi: 10.7326/0003-4819-156-4-201202210-00018. [DOI] [PubMed] [Google Scholar]
- 14.El Messaoudi S., Rolet F., Mouliere F., Thierry A.R. Circulating cell free DNA: Preanalytical considerations. Clin. Chim. Acta. 2013;424:222–230. doi: 10.1016/j.cca.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 15.Gogenur M., Burcharth J., Gogenur I. The role of total cell-free DNA in predicting outcomes among trauma patietns in the intensive care unit: a systematic review. Crit. Care. 2017;21(14) doi: 10.1186/s13054-016-1578-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.J.J. Deeks, J.P.T. Higgins, Statistical algorithms in Review Manager 5, Statistical Methods Group of The Cochrane Collaboration, 2010, 1–11.
- 17.Reitsma J.B., Glas A.S., Rutjes A.W.S., Scholten R.J.P.M., Bossuyt P.M., Zwinderman A.H. Bivariate analysis of sensitivity and specificity produces information summary measures in daignostic reviews. J. Clin. Epidemiol. 2005;58(10):982–990. doi: 10.1016/j.jclinepi.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 18.Agiannitopoulos K., Samara P., Papadopoulou E., Tsamis K., Mertzanos G., Babalis D., et al. Study on the admission levels of circulating cell-free DNA in patients with acute myocardial infarction using different quantification methods. Scand. J. Clin. Lab. Invest. 2020;80(4):348–350. doi: 10.1080/00365513.2020.1729400. [DOI] [PubMed] [Google Scholar]
- 19.Antonatos D., Patsilinakos S., Spanodimos S., Korkonikitas P., Tsigas D. Cell-free DNA levels as a prognostic marker in acute myocardial infarction. Ann. N. Y. Acad. Sci. 2006;1075:278–281. doi: 10.1196/annals.1368.037. [DOI] [PubMed] [Google Scholar]
- 20.Arafat E.S., Elmadbouha I., Radwan E.I., Kamal A.M., Badr E.A., Ghanayem N.M. Circulating cell-free DNA as a sensitive biomarker in patients with acute myocardial infarction. Menoufia Med J. 2017;31:772–779. [Google Scholar]
- 21.Chang C.P.Y., Chia R.H., Wu T.L., Tsao K.C., Sun C.F., Wu J.T. Elevated cell-free serum DNA detected in patients with myocardial infarction. Clin. Chim. Acta. 2003;327(1–2):95–101. doi: 10.1016/s0009-8981(02)00337-6. [DOI] [PubMed] [Google Scholar]
- 22.Cui M., Fan M., Jing R., Wang H., Qin J., Sheng H., et al. Cell-free circulating DNA: A new biomarker for the acute coronary syndrome. Cardiology (Switzerland). 2013;124(2):76–84. doi: 10.1159/000345855. [DOI] [PubMed] [Google Scholar]
- 23.Destouni A., Vrettou C., Antonatos D., Chouliaras G., Traeger-Synodinos J., Patsilinakos S., et al. Cell-free DNA levels in acute myocardial infarction patients during hospitalization. Acta Cardiol. 2009;64(1):51–57. doi: 10.2143/AC.64.1.2034362. [DOI] [PubMed] [Google Scholar]
- 24.Jing R.R., Wang H.M., Cui M., Fang M.K., Qiu X.J., Wu X.H., et al. A sensitive method to quantify human cell-free circulating DNA in blood: Relevance to myocardial infarction screening. Clin. Biochem. 2011;44(13):1074–1079. doi: 10.1016/j.clinbiochem.2011.06.083. [DOI] [PubMed] [Google Scholar]
- 25.Lou X., Hou Y., Liang D., Peng L., Chen H., Ma S., et al. A novel Alu-based real-time PCR method for the quantitative detection of plasma circulating cell-free DNA: Sensitivity and specificity for the diagnosis of myocardial infarction. Int. J. Mol. Med. 2015;35(1):72–80. doi: 10.3892/ijmm.2014.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin C., Gu J., Liu R., Xu F., Qian H., He Q., et al. Release of mitochondrial DNA correlates with peak inflammatory cytokines in patients with acute myocardial infarction. Anatol. J. Cardiol. 2016;17:224–228. doi: 10.14744/AnatolJCardiol.2016.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rainer T.H., Lam N., Man C.Y., Chiu R.W.K., Woo K.S., Lo D. Plasma b-globin DNA as a prognositc marker in chest pain patients. Clin. Chim. Acta. 2006;368:110–113. doi: 10.1016/j.cca.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 28.Ren J., Jiang L., Liu X., Liao Y., Zhao X., Tang F., et al. Heart-specific DNA methylation analysis in plasma fo rthe investigatin of myocardial damage. J. Transl. Med. 2022;20(36) doi: 10.1186/s12967-022-03234-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A. Shimony, D. Zahger, H. Gilutz, H. Goldstein, R. Ilia, G. Orlov, et al., Cell free DNA as a potential marker in acute ST elevation myocardial infarction, Eur. Heart J. 1 (2009) 642.
- 30.Veiko N., Bulycheva N., Roginko O., Veiko R., Ershova E., Kozdoba O., et al. Ribosomal repeat in cell fre DNA as a marker for cell death. Biochem. (Moscow) Suppl. Series B: Biomed. Chem. 2008;2:198–207. [PubMed] [Google Scholar]
- 31.Wang L., Xie L., Zhang Q., Cai X., Tang Y., Wang L., et al. Plasma nuclear and mitochondrial DNA levels in acute myocardial infarction patients. Coron. Artery Dis. 2015;26(4):296–300. doi: 10.1097/MCA.0000000000000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie J., Yang J., Hu P. Correlations of Circulating Cell-Free DNA with Clinical Manifestations in Acute Myocardial Infarction. Am. J. Med. Sci. 2018;356(2):121–129. doi: 10.1016/j.amjms.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Xu Y., Liu B., Zhao Z., Wang G., Zhu X., Pan S., et al. Clinical significance of peripheral blood circular DNA level measurements in patients with acute myocardial infarction. Zhongguo Yi Liao Qi Xie Za Zhi. 2012;36(6):456–458. [PubMed] [Google Scholar]
- 34.Zemmour H., Planer D., Magenheim J., Moss J., Neiman D., Gilon D., et al. Non-invasive detection of human cardiomyocyte death using methylation patterns of circulating DNA. Nat. Commun. 2018;9:1443. doi: 10.1038/s41467-018-03961-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Leest P., Boonstra P.A., ter Elst A., van Kempen L.C., Tibbesma M., Koopmans J., et al. Comparison of Circulating Cell-Free DNA Extraction Methods for Downstream Analysis in Cancer patients. Cancers (Basel). 2020;12(5):1222. doi: 10.3390/cancers12051222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubini Gimenez M., Twerenbold R., Reichlin T., Wildi K., Haaf P., Schaefer M., et al. Direct comparison of high-sensitivity-cardiac troponin I vs. T for the early diagnosis of acute myocardial infarction. Eur. Heart J. 2014;35(34):2303–2311. doi: 10.1093/eurheartj/ehu188. [DOI] [PubMed] [Google Scholar]
- 37.Vukajlovic J.T., Simic I., Milosevic-Djordjevic O. DNA and chromosomal damage in peripheral blood lymphocytes in patients with acute coronary syndrome undergoing a coronary angiography. Anatol. J. Cardiol. 2021;25:243–249. doi: 10.14744/AnatolJCardiol.2020.39479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cannon C.P., Gibson M., Lambrew C.T., Shouultz D.A., Levy D., French W.J., et al. Relationship of Symptom-Onset-to-Balloon Time and Door-to-Balloon Time with Mortality in Patients Undergoing Angioplasty for Acute Myocardial Infarction. J. Am. Med. Assoc. 2000;283(22):2941–2947. doi: 10.1001/jama.283.22.2941. [DOI] [PubMed] [Google Scholar]
- 39.Boeddinghaus J., Nestelberger T., Koechlin L., Wussler D., Lopez-Ayala P., Walter J.E., et al. Early Diagnosis of Myocardial Infarction with Point-of-Care High-Sensitivity Cardiac Troponin I. J. Am. Coll. Cardiol. 2019;75(10) doi: 10.1016/j.jacc.2019.12.065. [DOI] [PubMed] [Google Scholar]
- 40.Damodara S., Arora J., Liaw P.C., Fox-Robichaud A.E., Selvaganapathy P.R. Single-step measurement of cell-free DNA for sepsis prognosis using a thread-based microfluidic device. Microchim. Acta. 2022;189(146) doi: 10.1007/s00604-022-05245-1. [DOI] [PubMed] [Google Scholar]
- 41.Lippi G., Sanchis-Gomar F., Cervellin G. Cell-free DNA for diagnosing myocardial infarction: Not ready for prime time. Clin. Chem. Lab. Med. 2015;53(12):1895–1901. doi: 10.1515/cclm-2015-0252. [DOI] [PubMed] [Google Scholar]
- 42.Shroff G.R., Li S., Herzog C.A. Trends in mortality following acute myocardial infarction among dialysisk patients in the United States over 15 years. JAHA. 2015;4(10) doi: 10.1161/JAHA.115.002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.M.L.H.-P. Cisneros-Villanueva, M. Rios-Romero, A. Cedro-Tanda, C.A. Ruiz-Villavicencio, K. Page, et al., Cell-free DNA analysis in current cancer trials: a review. Br. J. Cancer 126 (2021) 391–400. [DOI] [PMC free article] [PubMed]
- 44.Duvvuri B., Lood C. Cell-Free DNA as a Biomarker in Autoimmune Rheumatic Diseses. Front. Immunol. 2019 doi: 10.3389/fimmu.2019.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knight S.R., Thorne A., Lo Faro M.L. Donor-specific Cell-free DNA as a biomarker in Solid Organ Transplantation. A Systematic Review. Transplantation. 2019;103(2):273–283. doi: 10.1097/TP.0000000000002482. [DOI] [PubMed] [Google Scholar]
- 46.Duplessis C., Gregory M., Frey K., Bell M., Truong L., Schully K., et al. Evaluating the discriminating capacity of cell death (apoptotic) biomarkers in sepsis. J. Intensive Care. 2018;6(72) doi: 10.1186/s40560-018-0341-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Babuin L., Jaffe A.S. Troponin: the biomarker of choice for the detection of cardiac injury. CMAJ. 2005;173(10):1191–1202. doi: 10.1503/cmaj.050141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.S.S. Smith, G.L. Walter, R.M. Walker, Chapter 18 - Clinical Pathology in Non-Clinical Toxicology Testing, Third Edition ed. Hascheck W, Rousseux CG, Wallig MA, editors, 2013.
- 49.M. Stark, C. Kerndt, S. Sharma, Troponin. Island T, editor. Treasure Island: StatPearls Publishing, 2021. [PubMed]
- 50.Tian Y., Charles E.J., Yan Z., Wu D.L., French B.A., Kron I.L., et al. The myocardial infarct-exacerbating effect of cell-free DNA is mediated by HMGB1-RAGE-TLR9 pathway. J. Thorac. Cardiovasc. Surg. 2019;157(6):2256–2269. doi: 10.1016/j.jtcvs.2018.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.A.J. Bronkhorst, V. Ungerer, S. Holdenrieder, Comparison of methods for the qquantification of cell-free DNA isolated from cell culture supernatant, Tumour Biol. 41(8) (2019) 1010428319866369. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.