Abstract
Cancer metastasis is the primary cause of cancer-related deaths. The seeding of primary tumours at a secondary site is a highly inefficient process requiring substantial alterations in the genetic architecture of cancer cells. These alterations include significant changes in global gene expression patterns. MicroRNAs are small, non-protein coding RNAs which play a central role in regulating gene expression. Here, we focus on microRNA determinants of cancer metastasis and examine microRNA dysregulation in metastatic cancer cells. We dissect the metastatic process in a step-wise manner and summarise the involvement of microRNAs at each step. We also discuss the advantages and limitations of different microRNA-based strategies that have been used to target metastasis in pre-clinical models. Finally, we highlight current clinical trials that use microRNA-based therapies to target advanced or metastatic tumours.
Keywords: Cancer therapy, cancer, metastasis, microRNA
Introduction
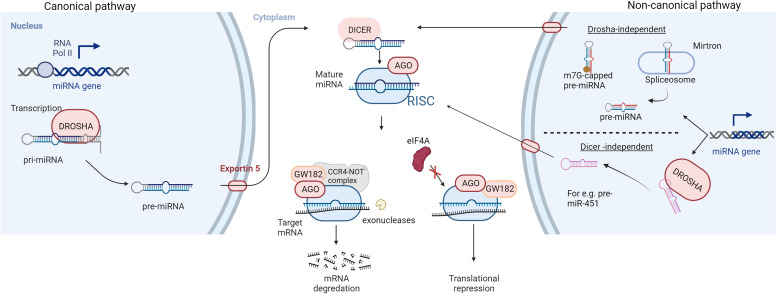
In 1993, a revolutionary discovery of the first microRNA (miRNA) while studying nematode Caenorhabditis elegans was made by Ambros et al. (Ref. 1). This finding revealed an essential part of the non-coding genome that plays a key role in post-transcriptional gene regulation (Ref. 1). Fast track 29 years, now over 2600 mature human miRNA sequences have been identified (Ref. 2). MicroRNAs are small, endogenous, single-stranded, non-protein coding RNA molecules of 19 to 24 nucleotides. MiRNAs account for approximately 3% of the human genome and are evolutionary conserved across mammals (Refs 3, 4). The canonical miRNA biogenesis pathway (Fig. 1) starts with RNA polymerase II-mediated transcription of the primary miRNA gene (pri-miRNA). The pri-miRNA is characterised by a hairpin structure with a 5′ cap and polyadenylation site at the 3′ (Ref. 5). Drosha, an RNase III protein, then cleaves the pri-miRNA, releasing a precursor loop (pre-miRNA) of approximately 70–100 nucleotides (Ref. 6). Upon release, the pre-miRNA is exported to the cytoplasm via Exportin 5 (Ref. 7). In the cytoplasm, pre-miRNA is cleaved by an RNase III called Dicer, to produce a double-stranded mature miRNA of approximately 22 nucleotides in length (Ref. 8). The non-canonical pathways utilise different combination of proteins during the biogenesis steps. These pathways can be further classified as Drosha- and Dicer- independent pathways. For instance, miRNAs produced from introns of messenger RNAs or mirtrons can bypass Drosha-mediated processing (Ref. 9). 7-methylguanosine (m7G)-capped pre-miRNA have been identified which are nascent RNAs directly exported to the cytoplasm through exportin 1 without the need for Drosha (e.g. pre-miR-320) (Ref. 10). Another example is the biogenesis of miR-451 which is independent of Dicer processing but requires Drosha and Argonuate 2 (AGO2) protein (Ref. 11).
Fig. 1.
Biogenesis and mechanism of action of miRNAs: RNA polymerase II-mediated transcription forms the primary miRNA (pri-miRNA) which is cleaved by an RNase III enzyme (DROSHA) to produce a precursor miRNA (pre-miRNA) in the canonical pathway of miRNA biogenesis. The pre-miRNA is exported to the cytoplasm via exportin 5, for further processing by RNase III DICER to form a mature miRNA duplex. Non-canonical pathways are independent of Drosha or Dicer processing. The miRNA duplex is then unwound whereby the guide strand along with Argonaute (AGO) proteins form a miRNA-induced silencing complex (RISC). The RISC complex binds to target sequences of mRNA leading to translation repression or degradation. AGO recruits GW182 which forms a complex with CCR4-NOT making the target mRNA susceptible to cleavage by exonucleases while hindrance to the binding of eukaryotic initiation factor-4A (eIF4A) to the target mRNA leads to translational inhibition.
The double-stranded mature miRNA is unwound, and the opposite strand is degraded. The remaining strand, the mature or the guide strand, is the final single-stranded miRNA molecule which forms a complex with the AGO proteins called the RNA-induced silencing complex (RISC) (Ref. 3). There are four mammalian AGO proteins (AGO1–4) but only AGO2 is able to cleave the target mRNA complementary to the miRNA (Ref. 12). The RISC complex binds to target messenger RNAs (mRNAs) which possess sequences complementary with the miRNA (Ref. 13). The binding of the RISC complex to mRNAs is mediated by a 6–8 nucleotide long region within the miRNA called the seed sequence or miRNA binding site (Ref. 14). The resulting miRNA-mRNA duplex leads to an inhibition or in some cases enhancement, of translation (Ref. 14). The miRNA target sites are usually located at the 3′-untranslated region (3′-UTR) of mRNA and the binding of the RISC complex leads to gene silencing by translation repression and mRNA decay (Ref. 15). Mechanistically, the AGO protein recruits the GW182 which interacts with the polyadenylate-binding protein PABPC to induce mRNA deadenylation (Ref. 16). This promotes decapping of the mRNA and makes it susceptible to degradation by 5′-3′ exoribonucleases (Ref. 17). For translational repression, GW182 recruits carbon catabolite repressor protein 4 complexes which in turn recruit RNA helicases like DDX6 (Refs 18, 19). MicroRNA-mediated inhibition of mRNA translation initiation results from the interference with the eukaryotic initiation factors eIF4A-I and eIF4A-II (Ref. 15). Although exact molecular details remain to be uncovered, existing evidences suggest that the RISC complex dissociates initiation factors from target mRNAs inhibiting the assembly of the translation initiation complex (Refs 20, 21).
Although a perfect complementarity with mRNAs is optimal for the function of miRNAs, some miRNAs can regulate mRNAs with partial complementarity (Ref. 22). Consequently, miRNA-mediated regulation of gene expression affects almost every fundamental cellular process, such as development, differentiation, proliferation, metabolism and apoptosis (Ref. 23). Unsurprisingly, the dysregulation of miRNA profile also significantly correlates with the onset and progression of cancer (Ref. 24). The impact of individual miRNAs on cancer often differs between cancer types. A miRNA can be tumour suppressive, oncogenic or a regulator of metastasis (Ref. 25). This review will focus on the role of miRNA in cancer metastasis. We will investigate miRNA dysregulation in different stages of the metastatic process and examine molecular drivers affected. We will summarise pre-clinical studies that have successfully employed miRNA-based therapies for metastatic cancer and highlight their limitations. Finally, we will examine miRNA-based therapies for the treatment of advanced and metastatic cancer that are currently in clinical trials.
Metastasis
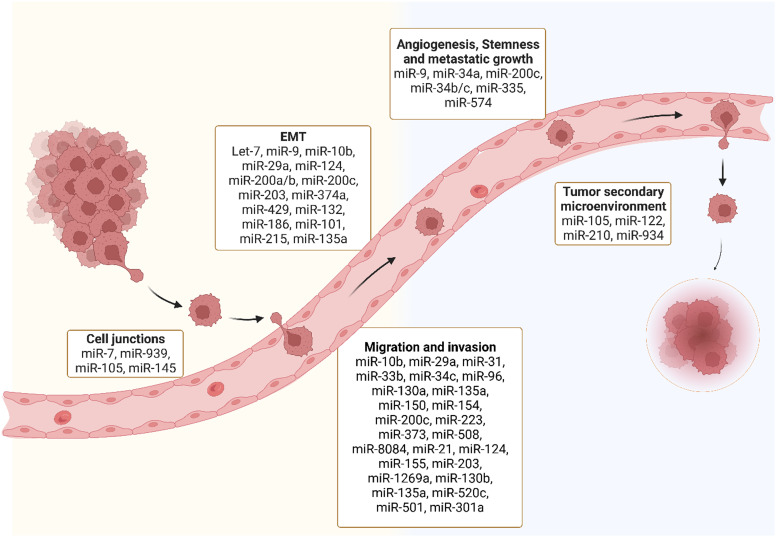
The development of secondary tumours in distant organs, i.e. metastasis, is a hallmark of cancer (Ref. 26). The spread of cancer cells to secondary sites is the main cause of cancer-related morbidity and mortality (Ref. 27), yet we are only beginning to unravel the molecular mechanisms that drive metastasis (Ref. 28). Several phases are involved in the development of secondary tumours (Ref. 29). First, cancer cells loose adhesion factors and detach from the primary tumour allowing penetration and intravasation into the circulatory and lymphatic systems (Ref. 29). The cells in the vasculature called circulating tumour cells (CTCs), exploit mechanisms like cell cycle arrest, to evade and survive immune surveillance (Ref. 30). There is significant research aimed at studying and characterising CTCs which is beyond the scope of this review. Secondly, this process is followed by the extravasation and infiltration of the cells into distant capillary beds (Ref. 31). Finally, invasion and proliferation of the tumour in distant organs occurs (Ref. 31). Metastasis and the establishment of secondary tumours is a very inefficient process as the majority of tumour cells in circulation are eliminated (Ref. 32). The establishment of a microenvironment for cancer cells to seed, known as a premetastatic niche, is essential for the development of secondary cancer (Ref. 33) (Fig. 2). The most common locations of metastasis in the body are liver, bone, lung, nervous system, pleura and peritoneum (Ref. 34). In this review, we will focus on the role of miRNAs in metastasis. We will discuss different stages in the metastatic process and summarise miRNAs that have been reported to be involved in each of these steps.
Fig. 2.
MiRNAs and different stages of metastasis: Several miRNAs are dysregulated throughout different stages of the metastatic process including disruption of tight junctions, epithelial to mesenchymal transition (EMT), migration and invasion, angiogenesis, stemness and metastatic growth and tumour secondary microenvironment.
MicroRNA dysregulation in cancer metastasis
MiRNAs are regulators of virtually every cellular process, including the ones that lead to the development of metastasis (Ref. 35). MiRNA can target the mRNA of tumour suppressors or oncogenes implicated at different phases of the developing metastatic tumour (Table 1) (Ref. 36). Genetic mutations in cellular pathways resulting from the dysregulation of miRNA have been widely identified in the metastatic pathways (Ref. 37). Here we discuss miRNA dysregulation and how it impacts key stages of metastasis.
Table 1.
A list of microRNAs involved at different stages of cancer metastasis
| miRNA | miRNA expression | Pro/anti | Target genes/ effect | Primary tumour | Ref. |
|---|---|---|---|---|---|
| Cell Junction | |||||
| miR-105 | High | Anti-cell junction | Targets ZO-1 to destroy natural barrier | Breast | (Ref. 40) |
| miR-486-5p | High | Anti-cell junction | Targets CADM1 to destroy natural barrier | NSCLC | (Ref. 41) |
| 939-5p | High | Anti-cell junction | Downregulates CDH5/ enhance cell migration | Breast | (Ref. 42) |
| miR-145 | Low | Pro-cell junction | Increased focal junction protein, paxillin increasing migration | CRC | (Ref. 43) |
| miR-7 | Low | Pro-cell junction | increases FAK expression increasing migration | Colon | (Ref. 44) |
| Epithelial to mesenchymal transition | |||||
| Let-7 | Low | Anti-EMT | Increase m-MYC, HMGA2 & KRAS expression | PDAC | (Ref. 57) |
| miR-9 | High | Pro-EMT | Reduced CDH1 downregulates E-cadherin | Breast | (Ref. 54) |
| miR-132 | Low | Anti-EMT | Increase ZEB2, downregulation of E-cadherin, upregulation of vimentin & fibronectin. | CRC | (Ref. 50) |
| miR-132 | Low | Anti-EMT | Increase ZEB2, downstream downregulation of E-cadherin & upregulation of vimentin. | NSCLC | (Ref. 53) |
| miR-186-5p | Low | Anti-EMT | Increased ZEB1 expression | CRC | (Ref. 52) |
| miR-200 family | Low | Anti-EMT | Increased ZEB1 & ZEB2 expression | CRC pancreatic lung breast gastric | (Refs 45–49) |
| miR-203 | Low | Anti-EMT | Increased Annexin-A4 expression | Gastric | (Ref. 56) |
| miR-215 | Low | Anti-EMT | Increased ZEB2 expression | NSCLC | (Ref. 51) |
| miR-374a | High | Pro-EMT | Suppress WIF1, PTEN, WNT5A, to activate Wnt/B-catenin signalling | Breast | (Ref. 55) |
| Migration and Invasion | |||||
| miR-10b | High | Pro-migration & invasion | Suppression of HOXD10 | Breast Gastric CRC | (Refs 58-60) |
| miR-21-5p | High | Pro-migration & invasion | Suppression of PDCD4 | Breast | (Ref. 63) |
| miR-29c | Low | Anti-migration & invasion | Upregulation of Integrin B1 & MMP2 | Lung | (Ref. 66) |
| miR-96 | High | Pro-migration & invasion | Inhibition of PTPN9 | Breast | (Ref. 61) |
| miR-124 | Low | Anti-migration & invasion | Increased ROCK1 expression | CRC | (Ref. 68) |
| miR-135a | High | Pro-migration & invasion | Suppression of HOXA10 | Breast | (Ref. 62) |
| miR-150-5p | Low | Anti-migration & invasion | Increased HMGA2 expression | Breast | (Ref. 64) |
| miR-203 | Low | Anti-migration & invasion | Increased EIF5A2 expression | CRC | (Ref. 67) |
| miR-373 | Low | Anti-migration & invasion | Increased BRF2 expression | NSCLC | (Ref. 65) |
| miR-520c | High | pro migration & invasion | suppression of IRF2 | gastric | (Ref. 69) |
| Angiogenesis, Stemness and Metastatic Growth | |||||
| miR-30c | High | Pro-stemness | Increased OCT4, SOX2, KLF4 and NANOG | Breast | (Ref. 70) |
| miR-129-2 | Low | Pro-metastatic growth | Increased SOX4 expression | PDAC | (Ref. 71) |
| miR-145 | Low | Anti-angiogenesis | Increased N-RAS and VEGF signalling pathways | Uveal melanoma | (Ref. 72) |
| miR-200c | Low | Anti-stemness | Increased SOX2 & KLF4 stemness gene expression | Breast | (Ref. 70) |
| miR-335 | Low | Pro-metastatic growth | Increased SOX4 expression | PDAC | (Ref. 71) |
| miR-574-5p | High | Pro-angiogenesis | Suppression of PTPRU/ enhanced tyrosine phosphorylation of β-catenin | NSCLC | (Ref. 73) |
| Tumour Microenvironment | |||||
| miR-301a-3p | High | Pro-tumour microenvironment | Mediates M2 macrophage via PTEN/PI3Kℽ | Pancreatic | (Ref. 74) |
| miR-130b-3p | High | Pro-tumour microenvironment | Mediates M2 macrophages and cancer cells communication | Gastric | (Ref. 75) |
| miR-934 | High | Pro-tumour microenvironment | Downregulation of PTEN and activation of PI3K/AKT signalling pathway | CRC | (Ref. 76) |
| miR-148b | Low | Anti-tumour microenvironment | Increased CSF1 enhancing TAM infiltration | HCC | (Ref. 77) |
| miR-501-3p | High | Pro-tumour microenvironment | TGFBR3 inhibition; activating TGF-B signalling pathway | PDAC | (Ref. 78) |
| miR-214 | High | Pro-tumour microenvironment | Induction of Tregs secreted IL-10 | Breast HCC NSCLC Pancreatic | (Ref. 79) |
MiRNAs and cell junctions
MiRNAs regulate the transcription of cellular junction proteins critical for signalling communication, growth and migration (Ref. 38). For metastasis to occur, the disruption of cellular junctions is essential (Ref. 39). Several miRNAs regulate the expression of zonula occluden-1 (ZO-1), a major component of tight junctions (Ref. 40), e.g. miR-105 regulates the metastasis of breast cancer by inhibiting ZO-1 (Ref. 40). Furthermore, overexpression of miR-105 induces the metastasis of cancer cells to distant organs including the liver (Ref. 40). The cell-cell adhesion for the adaption of the premetastatic niche is promoted by oncogenic miR-105 through the tight junction protein ZO-1 (Ref. 40).
A highly elevated expression of miR-486-5p in CD31+ vascular endothelial (VE) cells increases permeability and promotes non-small cell lung cancer (NSCLC) metastasis (Ref. 41). The transfection of human VE cells, with miR-486-5p antagomirs, targets CADM1 and destroys the tight junctions of VE cells. Similarly, the downregulation of adherens junction protein, VE cadherin (CDH5), by miRNA, enhances breast cancer cell migration (Ref. 42). MDA-MB-231-GFP breast cancer cells transfected with miR-939 mimics, show higher migration through the endothelial barrier which is mediated by a downregulation of CDH5 (Ref. 42). MiR-145 negatively correlates with the focal junction protein paxillin in colorectal cancer (CRC) (Ref. 43). Delivery of miR-145 mimics downregulates paxillin and inhibits cell proliferation, migration and invasion (Ref. 43). Similarly, miR-7 targets focal adhesion kinase (FAK) expression to suppress colon cancer proliferation, migration and invasion (Ref. 44). HCT-8 and Caco-2 colon cancer cell lines transfected with miR-7 mimics have been shown to have a negative correlation with colon cancer metastasis (Ref. 44).
MiRNAs in epithelial to mesenchymal transition (EMT)
Epithelial to mesenchymal transition (EMT) of cells is mostly controlled by zinc finger E-box binding protein transcription factors (TFs) ZEB1 and ZEB2 and leads to an upregulation of vimentin and N-cadherin and repression of E-cadherin (Ref. 45). The miR-200 family (miR-200a, miR-200b, miR-200c, miR-124, miR-429) has been extensively investigated as a regulator of ZEB1 and ZEB2 in cancer that mostly metastasises to the liver, including colorectal (Ref. 45), pancreatic (Ref. 46), lung (Ref. 47), breast (Ref. 48) and gastric cancers (Ref. 49). ZEB1 and ZEB2 are direct targets of several miRNAs in CRC and NSCLC. Zheng et al. reported a significant downregulation of miR-132 in CRC and furthermore used a luciferase activity assay to demonstrate miR-132-mediated regulation of ZEB2 (Ref. 50). MiR-215-mediated regulation of ZEB2 expression has been demonstrated in CRC while low levels of miR-186-5p in CRC inhibit EMT by targeting ZEB1 (Refs 51, 52). Additionally, it was reported that miR-132 directly targets ZEB2 in NSCLC driving EMT (Ref. 53). There are other miRNAs that promote EMT in breast cancer. MiR-9 repression of E-cadherin, increases EMT in breast cancer (Ref. 54) while oncogene miR374a targets negative regulators of Wnt/B-catenin pathway including WIF1, PTEN and WNT5A and activates the Wnt/B-catenin pathway to promote EMT in breast cancer (Ref. 55). The downregulation of miR-203 promotes EMT in gastric cancer by releasing the repression of its target gene Annexin A4 (Ref. 56). In pancreatic ductal adenocarcinoma (PDAC), EMT is driven by an increase in the expression of c-MYC, HMGA2 and KRAS mediated by a reduction in Let-7 miRNA expression (Ref. 57).
MiRNAs in cancer cell migration and invasion
MiR-10b was the first miRNA to be associated with metastasis in patients with advanced breast cancer (Ref. 58). MiR-10b promotes the migration and invasion in different cancer types including colorectal, breast and gastric cancers by regulating HOXD10 (Refs 58–60). In breast cancer, migration and invasion is enhanced by an upregulation of miR-96, miR-135a and miR-21 and the downregulation of miR-150. Similarly, in breast cancer, oncogenic miR-96 targets PTPN9 (Ref. 61) and miR-135a represses HOXA10 to drive the migration and invasion (Ref. 62). Another oncogene miR-21-5p upregulates the expression of programmed cell death protein 4 (PDCD4) to increase breast cancer invasion and migration (Ref. 63). Tumour suppressor miR-150, which targets HMGA2, is aberrantly expressed in breast cancer and promotes migration and invasion (Ref. 64). In NSCLC, a downregulation of miR-373 which regulates TFIIB-related factor 2 (BRF2), promotes cell migration and invasion (Ref. 65). Multiple investigations of cancers metastasising to the liver including pancreatic cancer and lung cancer, demonstrate that cell migration and invasion is controlled by miR-29c-mediated regulation of MMP2 (Ref. 66). Low levels of miR-124 and miR-203 promote migration and invasion in CRC by targeting ROCK1 and eukaryotic initiation factor 5A2, respectively (Refs 67, 68). Similarly, gastric cancer migration and invasion is driven by the dysregulation in miR-520c-mediated regulation of IRF2 (Ref. 69).
MiRNAs in stemness, angiogenesis and metastatic growth
Key events of metastatic growth and tumour mediated angiogenesis are regulated by miRNAs. Downregulation of miR-200c and overexpression of miR-30c targets stemness-related genes in breast cancer influencing secondary tumour growth (Ref. 70). Oncogene miR-9-mediated repression of E-Cadherin contributes to an overexpression of vascular endothelial growth factor (VEGF) leading to an increase in angiogenesis which is essential for the growth of secondary tumour (Ref. 54). The co-repression of miR-129-2 and miR-335 significantly upregulates oncogenic SOX4 driving metastatic growth in PDAC (Ref. 71). Likewise, VEGF overexpression leading to angiogenesis, tumour growth and invasion in uveal melanoma was significantly suppressed by miR-145 mimics directly targeting N-RAS and VEGF signalling pathways (Ref. 72). Upregulation of miR-574-5p promotes metastatic growth in NSCLC enhancing tyrosine phosphorylation of B-catenin via the repression of protein tyrosine phosphate receptor type U (PTPRU) (Ref. 73).
MiRNAs in tumour microenvironment
MiRNA is an important mediator of the crosstalk between the tumour microenvironment and tumour cells, playing an important role in metastasis progression. Tumour-associated macrophages (TAM) are key components of the tumour microenvironment, regulated by miRNAs, to exhibit pro-tumour activity in the microenvironment. For example, highly expressed miR-301a in pancreatic cancer cells induces M2 macrophage polarisation via the PTEN/PI13Kℽ signalling pathway to promote pancreatic cancer cell metastasis (Ref. 74). Similarly, miR-130-3p upregulated in gastric cancer mediates communication between M2 macrophages and cancer cells in the tumour microenvironment by promoting the expression of mixed lineage leukaemia 3 (MLL3) gene and grainyhead-like 2 (GRHL2) gene (Ref. 75). Tumour-derived exosomal miR-934 promotes liver metastasis of CRC by regulating the interaction between TAMs and the metastatic microenvironment (Ref. 76). Downregulated miR-148b expression negatively correlates with the upregulation of colony-stimulating factor-1 (CSF1), promoting CSF1 signalling and inducing TAM infiltration to promote hepatocellular carcinoma (HCC) metastasis (Ref. 77). Furthermore, M2 macrophage-derived exosomal miR-501-3p downregulated TGFRR3 to promote liver and lung metastasis of PDAC in nude mice by activating the TGF-β signalling pathway (Ref. 78). Upregulated miR-214 negatively correlates with PTEN in several cancers including breast, HCC, NSCLC and pancreatic cancer. The upregulated miR-214 promotes regulatory T-cells (Tregs) which secret high levels of IL-10 and enhance immune suppression for metastatic progression (Ref. 79).
Regulation of metastatic miRNAs
The inter-regulation between miRNAs and different TFs lead to a finely tuned and spatio-temporally regulated transcriptional and post-transcriptional gene regulation system which gets perturbed during metastasis (Ref. 80). Evidences suggest that the dysregulation of miRNAs in cancer can occur at the genomic level. An example is the frequently lost genomic locus of miR-146a in acute myeloid leukaemia (Ref. 81). However, aberrations at the transcription level are widely studied and thought to be more impactful. For instance, tumour suppressive TF p53 regulates the expression of the miR-16, miR-145 and miR-34 family (Ref. 82). While miR-145 is repressed by the oncogenic RAS-responsive element-binding protein 1 (RREB1) (Ref. 83). Other reported TFs that regulate miR-145 include CCAAT/enhancer-binding protein beta, beta-catenin/T cell factor 4 and forkhead TFs FOXO1 and FOXO3 (Refs 84, 85). The oncogenic c-Myc TF suppresses expression of miRNAs 29, 30 and let-7 family (Refs 86–88). ZEB1 and ZEB2, key activators of EMT repress the expression of miR-200 family of genes (Ref. 89) including miR-200c (Ref. 90). Similarly, studies have demonstrated that nuclear receptors, especially estrogen receptor (ER) and androgen receptor (AR) can directly regulate the transcriptional activity of miRNAs in cancer by binding to promoter or repressor regions. For instance, ER binds to the promoter region of the miR-221 and inhibits its expression in breast cancer (Ref. 91). Interestingly in prostate cancer, a negative feedback loop that regulates miR-135a and AR protein expression in an androgen-dependent manner was identified. Here, androgen stimulates the expression of miR-135a which inhibits AR expression. In turn, AR binds to the miR-135a locus and controls its expression (Ref. 92). Some studies have also indicated roles of epigenetic factors like DNA methylation in the regulation of metastatic miRNAs like miR-200c (Refs 93, 94). Similarly, the promoter of miR-34a is hypermethylated in ovarian cancer (Ref. 95). Other factors that modulate miRNA activity like regulation and post-translational modifications of AGO proteins (Ref. 96), miRNA transport to the cytoplasm and regulation of miRNA–mRNA interactions need to be further explored in the context of metastasis.
Several recent studies have also compared changes in miRNA expression in primary and secondary tumours to identify potential drivers of metastasis. For instance, a study involving 33 CRC patients with metastasis and 14 patients without metastasis revealed differential expression of 17 miRNAs and their 198 predicted targets. There was a strong association of the target genes with cancer progression and metastasis (Ref. 97). In another study involving metastatic breast cancer patients, the upregulation of miR-342-3p and miR-187-3p was associated with an increased progression-free survival (PFS) and overall survival (OS); while, the downregulation of miR-301a-3p was associated with a higher PFS and OS (Ref. 98). In addition to being therapeutic targets, studies in several different cancer types have identified differences in expression levels of several miRNA indicating that they may serve as diagnostic or prognostic markers (Refs 99–106).
MiRNA-based therapies
Endogenous miRNAs play a crucial role in maintaining cellular homoeostasis (Ref. 107). The genomic and transcriptomic alterations in cancer cells can perturb the global miRNA expression profile causing genome-wide transcriptional changes (Ref. 108). These changes can lead to an upregulation of oncogenes and/or a downregulation of tumour suppressors which is critical for metastasis (Ref. 108). Most miRNA-targeted cancer therapies focus on restoration or inhibition of dysregulated miRNAs (Ref. 109) but recently, miRNA-based detargeting strategies have been utilised for cell/tissue-specific targeted therapies (Ref. 110). Table 2 provides a comprehensive list of miRNA-based therapeutic strategies which will be discussed in this section in detail.
Table 2.
MiRNA-based therapies for cancer metastasis: Dysregulated miRNAs are potential therapeutic targets to treat metastatic cancer
| MiRNA-based therapies | miRNA | Cancer | Therapy effects | Model | Ref. |
|---|---|---|---|---|---|
| MiRNA replacement | |||||
| miRNA mimics | miR-149-3p | Breast | Regulation of Foxp1 expression in CD8+ T-Cells | Homograft mouse model | 82 |
| miR-140-5p | NSCLC | Higher adhesion, changes to EMT | A549 & SK-MES1 cell lines | 83 | |
| miR-195-5p miR-101-3p miR-338-5p | Lung | Combination delivery yields reduced tumour growth | Animal model | 84 | |
| miR-34a | Lung | combination radiation/mimic regulates double-strand break repair | 85 | ||
| MiRNA inhibition | |||||
| ASOs | miR-21 | NSCLC | Regulation of PTEN | H1650 cell line | 88 |
| miR-21 | Colon | Reduced VEGF expression | HCT116 cell line | 96 | |
| miR-21 miR-10b | TNBC | Multiple oncogenic miRNAs targeted for cumulative therapeutic effects | Mouse model | 97 | |
| Antagomir | miR-210 | Pancreatic | Combined delivery of miR-210 and siRNA-KRAS reduces metastasis. | Mouse model | 89 |
| MiRNA Sponges | miR-10b | Breast | Upregulation of HOXD10 inhibiting cancer growth and proliferation, migration and invasion. | MDA-MB-231 and MCF7 | 99 |
| miR-155 miR-21 miR-221 miR-22 | Breast Pancreatic | Multi-potent sponge inhibits multiple miRNA regulating Foxo3a, PTEN and RhoA | Cell lines | 100 | |
| MiRNA Mask | miR-522 | NSCLC | Protects DENN2D expression to reduce migration and invasion | Cell lines | 91 |
| LNA anti-miR | miR-21 | CRC | Inhibits cell proliferation | LS174T | 92 |
| miR-9 | Gastric | Promotes CDH1 expression to re-establish E-cadherin expression | Cell lines | 101 | |
| miR-663a miR-4787-5p | Pancreatic | Reduced TGFβ1-induced EMT | Orthotopic mouse model | 102 | |
| Small RNA Zipper | miR-221 | Breast | Inhibits miRNA to increase target genes | cell lines | 93 |
| MiRNA-mediated Detargeting | |||||
| miRNA Detargeting | miR-122 miR-7 miR-148a | Pancreatic | Successful detargeting of the liver, brain and GT tract | Cell lines and xenograft models | 104 |
| miR-148a miR-216a | Metastatic PDAC Liver | Successful targeting for reduced cancer growth and metastasis | 105 | ||
| miR-122a miR-199a | HCC | Prevented off-targeting effects by sparing normal liver cells. | Cell lines | 106–108 | |
MiRNA replacement
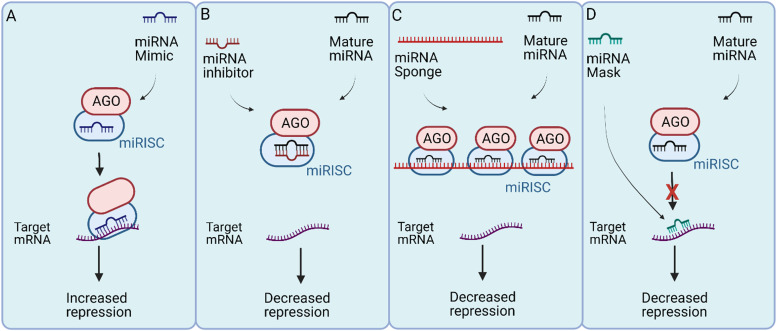
The restoration of miRNAs that are downregulated in cancer is one approach to target metastasis (Ref. 4). MiRNAs in metastatic cells can be restored using miRNA mimics (Ref. 111) which are small synthetic RNA duplexes containing an antisense strand with the same sequence as the endogenous miRNA (Ref. 112). To increase stability of the duplex and to enhance cellular uptake, the sense strand can be chemically modified. The sense strand may also contain several mismatches to minimise off-target effects (Ref. 112). Like the naturally occurring miRNA, these miRNA mimics are loaded into the RISC complex and inhibit downstream targets (Fig. 3a) (Ref. 112). MiRNA mimics have been widely studied for therapeutic purposes in both in vitro and in vivo cancer models (Ref. 113). For instance, miR-149-3p mimics suppress breast cancer growth and metastasis by regulating inhibitory receptors and Foxp1 gene expression in CD8+ T cells in a homograft mouse model (Ref. 113). Treatment with miR-149-3p mimics reduced the apoptosis of CD8+ T cells which mediate the immune surveillance of cancer cells (Ref. 113). The promotion of CD8+ T cells resulted in the death of 4T1 mouse breast cancer cells (Ref. 114). Similarly, a reduction of migration and invasion in A549 and SK-MES1 squamous carcinoma NSCLC cell lines was observed after the transfection of miR-140-5p mimics (Ref. 114). The authors also reported a higher adhesion to an artificial extracellular matrix (ECM), indicating a change in EMT (Ref. 114). A combined delivery of three miRNA mimics, miR-195-5p, miR-101-3p and miR-338-5p, is more effective in reducing tumour growth and the number of metastatic nodules in animal models of lung cancer (Ref. 115). Similarly, delivering miR-34a mimic sensitises primary and metastatic derived lung cancer cell lines to radiotherapy, in vitro and in vivo (Ref. 116). Several other miRNA replacement therapies are currently being tested in both clinical and pre-clinical settings.
Fig. 3.
MiRNA-based therapies. (a) MiRNA replacement with mimics function like an overexpression of endogenous miRNA and increase the degradation or repression of target mRNAs. (b) The miRNA inhibitor approach minimises the binding of miRNA-induced silencing complex (miRISC) to target mRNAs. Different strategies used for miRNA inhibition includes antisense oligonucleotides (ASOs), antagomir antisense oligonucleotides, locked nucleic acid (LNA), antisense oligonucleotide and small RNA zippers. (c) MiRNA sponge binds to the miRISC complex reducing its binding to the target mRNA. (d) MiRNA mask prevents the miRISC from binding to the mRNA by ‘masking’ the miRNA binding site.
MiRNA inhibition
Another approach to target metastasis is to inhibit upregulated oncogenic miRNAs (Ref. 117). The inhibition of oncogenic miRNAs overexpressed during metastasis can restore silenced tumour suppressors (Ref. 117). MiRNA inhibitors are single-stranded oligonucleotides complimentary to an endogenous miRNA (Ref. 4). These inhibitors can bind to endogenous miRNAs and inhibit their incorporation into the RISC complex (Fig. 3b) (Ref. 118). Several types of miRNA inhibitors have shown therapeutic advantages both in vitro and in vivo including antisense oligonucleotides (ASOs) (Ref. 119), antagomirs (Ref. 120), miRNA sponges (Ref. 121), miRNA masks (Ref. 122), locked nucleic acid (LNA) anti-miRNAs (Ref. 123) and small miRNA Zippers (Ref. 124).
Synthetic Antisense Oligonucleotides (ASOs)
ASOs are single-stranded, chemically modified DNA molecules, 20-25 nucleotides in length, with a full complementarity to a target miRNA (Ref. 125). ASOs inhibit the binding of mature miRNA to its target mRNA by producing an ASO-miRNA duplex which can lead to the cleavage of the miRNA and the upregulation of the target mRNA (Ref. 125). ASOs have already been approved by the Food and Drug Administration for the treatment of Duchenne muscular dystrophy and spinal muscular atrophy whereby exon skipping strategies are utilised to restore the dystrophin expression (Ref. 126). For cancer therapy and metastasis inhibition, some pre-clinical studies have been reported with ASOs. For example, Ge et al. designed an ASO to target miR-21 which is overexpressed in NSCLC. MiR-21 regulates the activity of PTEN, a regulator of invasion and a metastasis promoter (Ref. 119). The ASO-based drug was successful in reducing miR-21 expression and induced apoptosis in H1650 NSCLC cell line (Ref. 119). Likewise, the transfection of synthetic ASOs targeting miR-21 significantly reduced migration and invasion of HCT116 human colon carcinoma cell line accompanied by a reduction in the expression of VEGF which is critical for colon cancer metastasis (Ref. 127). Multiple oncogenic miRNAs can also be targeted simultaneously for additive therapeutic effects (Ref. 128). ASOs targeting miR-21 and miR-10b have been successfully delivered in cell lines and tumour xenografts for triple-negative breast cancer (TNBC) (Ref. 128). The simultaneous delivery of these miRNAs induces cancer apoptosis and inhibits tumour growth and metastasis in a mouse model of TNBC (Ref. 128).
Antagomir antisense oligonucleotides
Antagomirs are artificially synthesised single stranded RNA of 23 nucleotides length complementary to a miRNA. Antagomirs can be chemically modified with a cholesterol moiety for greater stability (Ref. 120). In a mouse model of pancreatic cancer, a cholesterol-modified polymetric CXCR4 antagonist was delivered with nanoparticles via an intraperitoneal delivery to localise efficacy and limit systemic side (Ref. 120). The co-delivery of antagomirs against miR-210 and siRNA against KRAS to this model demonstrated a reduced metastatic activity (Ref. 120). Of particular importance was the complete inhibition of liver metastasis, the primary metastatic site of PDAC (Ref. 120).
MiRNA sponges antisense oligonucleotide
MiRNA sponges are short, synthetic transcripts with the same sequence as the 3′UTR of mRNAs targeted by the miRNA. Acting as a decoy, sponges inhibit the ability of miRNAs to regulate their target mRNAs (Fig. 3c) (Ref. 121). There are some reports where miRNA sponging has been successfully performed for a single miRNA (Ref. 129). Liang, Zhang, Zhou, Wu, Lin and Liu (Ref. 130) designed a miRNA sponge plasmid to target miR-10b in metastatic breast cancer cell lines, MDA-MB-231 and MCF-7, demonstrating an inhibition of miR-10b and upregulation of its target HOXD10. This resulted in an inhibition of cancer growth and proliferation as well as a reduction in migration and invasion (Ref. 130). Additionally, a multi-potent miRNA sponge that simultaneously inhibits 4 oncogenic miRNAs, miR-155, miR-21, miR-221 and miR-222 was developed (Ref. 131). This multi-potent miRNA sponge was successful in inhibiting multiple oncogenic miRNAs, thus promoting anti-tumour effects in human breast cancer and pancreatic cancer cells (Ref. 131). Results demonstrated the multi-potent miRNA sponge to be more effective in inhibiting proliferation when compared to single miRNA-targeted sponges and demonstrated a 1.3-2.3-fold change in the protein levels of Foxo3a, PTEN and RhoA which are associated with an increased metastatic potential (Ref. 131).
MiRNA-masking antisense oligonucleotide
MiRNA-Masking (miR-Mask) is an inverted approach to protect mRNAs from miRNA-mediated repression (Ref. 122). In this approach, the miR-Masking oligonucleotides shield the miRNA binding sites of the mRNA to be protected (Fig. 3d) (Ref. 122). A full complementarity is required for better specificity (Ref. 122). This approach inhibits miRNA-mediated repression of targeted mRNAs without effecting the expression and potentially important functions of a miRNA (Ref. 122). Zhang et al. studied the effects of a miR-mask designed to complement the miR-522 binding site within DENND2D for the treatment of NSCLC and observed a reduced cell migration and invasion in NSCLC cells (Ref. 122).
Locked nucleic acid (LNA) antisense oligonucleotide
Another alternative oligonucleotide designed to inhibit miRNA oncogenic function are locked nucleic acid anti-miRs (LNA-i-miR) (Ref. 123). LNAs are chemically modified by connecting the 2′ oxygen and 4′ carbon to form an extra methylene bridge locking the ribose ring (Ref. 123). This leads to a higher thermal and in vivo stability and a greater binding affinity with mRNA targets (Ref. 123). LNA against miR-21 was effective in reducing the invasiveness and inhibited the proliferation of human colorectal adenocarcinoma cells (Ref. 123). Likewise, Lima et al. devised a strategy whereby LNA was efficient even when delivered at a low dose. In their study, miR-9 was targeted with LNAs to promote the expression of CDH1 for the reestablishment of E-cadherin in human gastric cancer cells (Ref. 132). In another study, the delivery of LNA-i-miRs against miR-663a and miR-4787-5p reduced tumour burden and metastasis in an orthotopic mouse model of pancreatic cancer by decreasing TGFβ1-induced EMT (Ref. 133).
Small RNA zippers
In this approach, oligonucleotides complementary to the second and the first half of a miRNA are synthesised and delivered into the cells (Ref. 124). Small RNA zippers connect multiple copies of a miRNA end-to-end by forming a duplex of multiple miRNA copies and inhibit the function of the target miRNA (Ref. 124). Like chemically modified LNAs, small RNA zippers have increased affinity, specificity and stability (Ref. 124). A 70–90% inhibition of miR-221 and miR-17 and rescue of their target genes was observed in breast cancer cell lines using miRNA zippers (Ref. 124). Further, the oncogenic effects of miR-221 were reversed by miR-221 zippers as demonstrated by the cell migration assay. However, the in vivo applications of miRNA zippers are yet to be tested (Ref. 124).
MiRNA-mediated detargeting
Unlike miRNA replacement or inhibition therapies, this approach utilises the binding sites of miRNAs that are downregulated in cancer for detargeting the therapy from the normal cells and thereby reduce off-target effects. This approach is mostly useful for genetic therapies which utilise therapeutic gene transfer (Ref. 134). For instance, Baertsch et al. utilised three miRNAs; miR-122, miR-7, miR-148a, expressed at high levels in the liver, brain and the gastrointestinal tract, respectively, and demonstrated successful detargeting of these organs for a measles virus-mediated oncolytic virotherapy of pancreatic cancer in cell lines and murine xenograft models (Ref. 135). In another example, the binding sites of miRNAs downregulated in PDAC, miR-148a and miR-216a, were used for detargeting in locally advanced and metastatic pancreatic and liver cancer (Ref. 136). 8-miR148aT demonstrated detargeting effects by repressing all miR-148/152 family members in the pancreas and liver (Ref. 136). This study demonstrated that this method was highly efficient for targeted therapies as a significant decrease in cancer growth and metastasis was observed (Ref. 136). To prevent off-targets effects in the liver after suicide gene therapy, the binding sites of miR-122a and miR-199a, which are significantly downregulated in HCC, were used in multiple studies. Adeno-associated virus-based vectors were used to deliver miRNA122a and/or miRNA199a-regulated the suicide gene therapy system cytosine deaminase (CD)/ 5-fluorocytosine (Refs 137–139). Limited killing of normal liver cells with this system demonstrated an efficient liver detargeting using the binding sites of these miRNAs (Refs 137–139).
MiRNA-based therapies in clinical trials
MiRNA-based strategies have demonstrated therapeutic potential in a range of conditions including advanced cancers (Ref. 140). In fact, several miRNA-targeted therapeutics are at different phases of clinical development for the treatment of advanced cancers and metastases (Ref. 141). For cancer therapy, miRNA therapeutics are injected directly into the site of the tumour which can increase the specificity, efficacy and reduce off-target effects (Ref. 112). Below we summarise clinically applied miRNA-targeted therapies (Table 3). Although not exclusive to metastasis, these therapies have shown promise in treating advanced cancers including those with metastasis to secondary organs.
Table 3.
A list of cancer therapy clinical trials utilising miRNA-based strategies
| MiR/Clinical trial identifier | Study | Condition | Drug | Phase |
|---|---|---|---|---|
| miR-34 NCT01829971 | A Multicenter Phase I Study of MRX34, MicroRNA miR-RX34 Liposomal Injection | Primary Liver Cancer, SCLC Lymphoma Melanoma, Multiple Myeloma, Renal Cell Carcinoma, NSCLC | MRX34 | Phase 1 |
| miR-16 NCT02369198 | MesomiR 1: A Phase I Study of TargomiRs as 2nd or 3rd Line Treatment for Patients with Recurrent MPM and NSCLC | Malignant Pleural Mesothelioma, Non-Small Cell Lung Cancer | TargomiRs | Phase 1 |
| miR-155 NCT03603431 | Safety, Tolerability and Pharmacokinetics of MRG-106 in Patients with Mycosis Fungoides (MF), CLL, DLBCL or ATLL | Cutaneous T-cell Lymphoma (CTCL), Mycosis Fungoides (MF), Chronic Lymphocytic Leukaemia (CLL) | Cobomarsen | Phase 1 |
| miR-155 NCT03713320 | SOLAR: Efficacy and Safety of Cobomarsen (MRG-106) vs. Active Comparator in Subjects with Mycosis Fungoides | Cutaneous T-Cell Lymphoma/Mycosis Fungoides | Cobomarsen Vorinostat | Phase 2 |
| miR-155 NCT03837457 | PRISM: Efficacy and Safety of Cobomarsen (MRG-106) in Subjects with Mycosis Fungoides Who Have Completed the SOLAR Study (PRISM) | Cutaneous T-Cell Lymphoma/Mycosis Fungoides | Cobomarsen | Phase 2 |
| miR-193a-3p NCT04675996 | First-in-Human Study of INT-1B3 in Patients with Advanced Solid Tumours | Advanced Solid Tumour | INT-1B3 | Phase 1 |
| miR-221 NCT04811898 | A Dose Escalation Study of LNA-i-Mir-221 for Cancer Treatment | Multiple Myeloma, Refractory Hepatocarcinoma Advanced Solid Tumour | LNA-i-miR-221 | Phase 1 |
Clinical miRNA replacement
Tumour suppressor miR-15/16 family is downregulated in various cancers including lung and colon cancer that metastasise to the liver (Ref. 142). Downregulation of miR15/16 can increase drivers of metastasis including tumour growth, angiogenesis, EMT and stemness (Ref. 141). In pre-clinical studies, miR-16 mimic replacement safely inhibited growth and metastasis of Malignant Pleural Mesothelioma (MPM) and NSCLC xenograft tumours (Ref. 143). TargomiRs are minicells coated with an anti-EGFR-specific antibody carrying miR-16 mimics for a cancer-targeted delivery (Ref. 143). A Phase 1 clinical trial of TargomiR initiated in 2014 demonstrated the safety of the approach in 26 patients. Of the 22 patients who were assessed for response, one showed a partial response and 15 had stable disease (Ref. 144).
Tumour suppressor miR-34a downregulates the expression of oncogenes including MET, MYC, PDGFR-α, CDK4/6 and BCL2 (Ref. 145). Both in vitro and in vivo studies have reported that miR-34a mimics can reduce tumour growth, migration and invasion and metastasis (Ref. 146). Anti-tumour activity of co-injecting let-7 and miR-34 was demonstrated in multiple NSCLC cell lines (Ref. 147). The anti-tumour activity of this combinatorial therapy was also tested in KrasLSD−G12D/+; p53flx, flx mouse model of NSCLC (Ref. 147). A second in vivo study was then initiated using lipid-based delivery agent (NOV340) to deliver miR-34a mimics (Ref. 147). A Phase I clinical trial with MRX34, a liposomal formulation of a synthetic, double-stranded miR-34a mimic, was initiated for patients with HCC and unresectable liver metastasis. (Refs 147, 148). Unfortunately, adverse immune-mediated toxicities precluded the trial advancing to phase II (Clinical Trial identifiers: NCT01829971, NCT02862145) (Ref. 148).
Tumour suppressor miR-193a-3p is downregulated in a range of cancers including HCC (Ref. 149), NSCLC (Ref. 150) and TNBC (Ref. 151). The repression of miR-193a-3p in these cancers decreases apoptosis, increases cell proliferation and migration tumour growth and metastasis (Ref. 152). Targets of miR-193a-3p play an important role in malignant cell behaviour including KRAS (Ref. 153), ERBB (Ref. 154), and S6K2 (Ref. 155) in lung cancer, PLAU in bladder cancer (Ref. 156), MCL-1 in glioma (Ref. 157), CCND1 in prostate cancer (Ref. 158), RAB27B in osteosarcoma (Ref. 159) and SRSF2 in HCC (Ref. 160). Telford et al. reported that miR-193a-3p mimics reduce cancer cell proliferation/survival by inducing cell cycle arrest, apoptosis, increased cell senescence, DNA damage and inhibit migration (Ref. 161). INT-1B3, a 193a-3p mimic replacement drug, consists of a lipid nanoparticle-based delivery system (Ref. 152). In preclinical studies with tumour bearing mice, systemic injection of INT-1B3 shows significant anti-tumour activity (Ref. 152). A phase 1/1b clinical trial to investigate the safety, preliminary efficacy, pharmacokinetics and pharmacodynamics of INT-1B3 is currently ongoing (Clinical Trial identifier: NCT04675996) (Ref. 152).
Clinical miRNA inhibition
Oncogenic miR-221-222 cluster located on the X chromosome is highly expressed in several solid tumours such as lung cancer (Ref. 162), breast cancer (Ref. 163), HCC (Ref. 164), and glioblastoma (Ref. 165) as well as haematological malignancies including myeloma (Ref. 166). In advanced cancers, upregulation of miR-221 interferes with the expression of its targets p27, p57, PUMA and PTEN promoting tumour growth (Ref. 152). Di Martino et al. reported anticancer effects of anti-miR221-targeted LNAs both in vitro and in vivo. LNA-i-miR-221 is a 13-mer antisense oligonucleotide that uses LNA technology and phosphorothioate backbone chemistry for increased affinity for miR221 targeting (Ref. 166). A phase I clinical trial of LNA-i-miR-221 will administer the drug via an intravenous injection to patients with multiple myeloma and advanced solid tumour (Clinical Trial identifier: NCT04811898).
Oncogene miR-155 regulates immune cell function and its overexpression affects multiple genes associated with the promotion of solid tumours including breast cancer, lung cancer, liver cancer as well as haematological malignancies including leukaemia (Ref. 167). Upregulation of miR-155 has been linked to JAK/STST, NK-KB and PI3K/AKT survival pathways stimulating T-cell receptors (Ref. 168). MiR-155 inhibition reduces proliferation and increases apoptosis in T-cell lymphoma cell lines (Ref. 168). In xenografts of B-cell lymphoma, miR-155 silencing with a LNA delivered systemically, reduced tumour burden and metastasis (Ref. 169). In another preclinical study, an anti-miR-155 molecule with a peptide nucleic acid backbone was used for greater sensitivity and efficient delivery to treat haematological malignancies (Ref. 170). These preclinical studies lead to the development of Cobomarsen which is a single-stranded, chemically modified miR-155-targeting molecule with chemical modifications for increased stability (Ref. 168). A Phase 1 trial of Cobomarsen in patients with cutaneous T-cell lymphoma (CTCL) [mycosis fungoides (MF) subtype] was initiated in 2016 and reported some therapeutic benefit for 95% of all enrolled patients with increased benefits reported for subjects who underwent more than one cycle (Ref. 168). Encouraging early data was followed by phase 2 clinical trial which started in 2019 but was terminated in 2020 due to commercial reasons (Clinical Trial identifiers: NCT03603431, NCT03713320, NCT03837457).
Challenges for miRNA-based cancer therapies
There are several challenges in using mi-RNA based therapies including insufficient delivery to the target tissue (cancer), stability in the biological system, immune responses and unwanted off-targeting. Arguably the primary challenge for miRNA-based cancer therapies is their efficient delivery to target tissues. Tumours can have poor blood perfusion and the complexity of ECM often hinder the delivery of miRNA-based therapies. In addition, scavenging cells like TAMs, neutrophils and monocytes can prevent the miRNA carrying vehicle from reaching cancer cells (Ref. 171). To overcome these challenges, different modes of delivery are being investigated, including both viral and non-viral vector systems (Refs 172, 173). The more common viral vectors are derived from adeno-associated viruses, adenoviruses and lentiviruses, while non-viral vectors can include exosomes, polymers and liposomes (Refs 172, 173). Viral vectors have a high efficiency and have been successfully used in several clinical trials. They also form the basis of a number of FDA approved therapies. (Ref. 174). Non-viral approaches may also be beneficial. The biocompatibility and biodegradability of polymers and liposomes are some of their advantages (Ref. 175). The conjugate vehicles have selective targeting and high stability due to the use of lipid or receptor binding molecules (Ref. 176). Naturally occurring exosomes are an advantage due to their immune compatibility (Ref. 177). Both viral and non-viral systems have inherent advantages and shortcomings, therefore the choice of the delivery system should be based on the overall design of a study.
Once delivered to the target tissue, the effect of miRNA therapeutics in non-cancer cells needs to be prevented. Similarly, effects on off-target mRNAs within cancer cells is another concern as miRNAs can target multiple transcripts simultaneously (Ref. 178). MiRNA imperfect complementarity to the targeted mRNA 3′UTR has the capacity to indiscriminately silence off-target genes (Ref. 179). Another cause of off-targeting is the possibility of artificial exogenous miRNA competing with endogenous miRNA creating a dysregulation in gene expression (Ref. 179). Off-target effects can induce the silencing of tumour suppressors or activation of oncogenes in normal cells (Ref. 180). For instance, miR-15/16 cluster regulate a large proportion of the whole transcriptome in leukaemia cells (Ref. 181). Thus, these miRNAs would not likely be used as therapeutic targets. The use of cancer targeted viral and non-viral delivery vectors can also reduce unwanted off-target effects (Refs 173, 182). Furthermore, cancer cell-specific regulatory elements like tumour specific promoters can be used in the delivery system (Ref. 183). Vigilant bioinformatic and wet-lab studies need to be performed with proposed inhibitors or mimics to identify any potential off-target effects in pre-clinical studies.
Both the miRNA and the delivery vector can elicit an immune response (Ref. 184). MiRNA duplexes can trigger toll like receptor response (Refs 171, 184) leading to an interferon response against miRNA therapeutics (Ref. 171). MiRNA therapeutics designed with certain chemical modifications can mitigate these immune responses (Ref. 171). Chemical modifications can enhance the miRNA stability in vivo. For instance, miRNAs without chemical modification of the ribose 2′-OH are prone to nuclease-mediated degradation and have a short half-life when injected systemically. Three generations of ASOs modification techniques developed (Ref. 1) First-generation modifications substitute phosphodiester backbone with phosphothiorate to increase in vivo stability; (Ref. 2) Second-generation modifications substitute the 2′-O-alkyl group of the sugar moieties with 2′-OMe, 2′-O-methoxyethyl (2′MOE) or 2′-Fluoro to enhance efficacy and bioavailability and to reduce the immune stimulation and toxicity; (Ref. 3) Third-generation modifications are the chemical alteration to the furanose ring with 2′4′-methylene producing LNAs to reduce nuclease degradation and increase membrane penetration (Ref. 126).
Concluding remarks
The inability of current therapies to effectively treat advanced or metastatic cancers stems from an incomplete understanding of the molecular mechanisms governing metastasis. Understanding molecular drivers of cancer metastasis can provide opportunities to develop novel therapeutic approaches. MiRNAs play a central role in regulating gene expression, thus, the dysregulation of miRNAs in metastasising cells warrants special attention. The dysregulation of several miRNAs is observed at every step of the metastatic process and restoring their levels is an attractive therapeutic avenue. Depending on the cancer type, dysregulated miRNAs can function as an oncogene or tumour suppressor. These dysregulations can lead to significant alterations in the expression of downstream target genes. Therapeutic approaches that utilise miRNAs aim to restore the normal levels of dysregulated miRNAs using miRNA mimics and inhibitors. There are several studies which report a successful use of miRNA mimics and inhibitors in pre-clinical in vitro and in vivo studies for targeting both primary tumours and metastasis for different cancer types. This has led to the initiation of human clinical trials using miRNA-based therapies for both solid and blood cancers. However, there are both technical and practical limitations for delivering miRNA-based therapies to patients. The in vivo stability and delivery of these miRNAs at therapeutic levels to the target tissue is a major issue for clinical applications. Similarly, therapy-induced toxicity and potential off-target effects are major concerns. There are several ongoing developments in this area to increase the stability of miRNAs mostly involving chemical modifications. Similarly, developments in the field of both viral and non-viral vector-based delivery can make the therapy cancer-specific and reduce off-target effects. Further research needs to be performed in order to identify novel miRNAs which control metastasis and potential therapeutic target; only then will the full potential of miRNA-based therapies for cancer metastasis be realised.
Abbreviations: BRF2, TFIIB-related factor 2, CADM1, Cell Adhesion Molecule 1, CDH1, Cadherin-1 or Epithelial cadherin, CDH5, Cadherin 5 or VE-Cadherin, CSF1, Colony-stimulating factor-1, EIF5A2, Eukaryotic Translation Initiation Factor 5A2, FAK, Focal adhesion kinase, HMGA2, High-mobility group AT-hook 2, HOXA10, Homeobox A10, HOXD10, Homeobox D10, IRF2, Interferon regulatory factor 2, IL-10, Interleukin 10, KLF4, Krüppel-like factor 4, KRAS, Kirsten rat sarcoma viral oncogene homologue, MMP2, Matrix metalloproteinase-2, MYC, MYC proto-oncogene, bHLH transcription factor, NANOG, Nanog Homeobox, N-RAS, Neuroblastoma RAS viral oncogene homologue, Oct-4, Octamer-binding transcription factor 4, PDCD4, Programmed Cell Death 4, PTEN, Phosphatase and tensin homologue, PTPN9, Tyrosine-protein phosphatase non-receptor type 9, PTPRU, Protein Tyrosine Phosphatase Receptor Type U, ROCK1, Rho Associated Coiled-Coil Containing Protein Kinase 1, SOX2, SRY-Box Transcription Factor 2, SOX4, SRY-Box Transcription Factor 4, TGF-β, Transforming growth factor beta, TGFBR3, Transforming growth factor beta receptor 3, VEGF, Vascular Endothelial Growth Factor, WIF1, WNT Inhibitory Factor 1, Wnt5a, Wnt Family Member 5A, ZEB1, Zinc finger E-box binding homeobox 1, ZEB2, Zinc finger E-box-binding homeobox 2, ZO-1, Zonula occludens-1. Cancers: Breast, Colon, CRC, Colorectal cancer, Gastric, HCC, Hepatocellular carcinoma, NSCLC, non-small cell lung cancer, pancreatic, PDAC, Pancreatic ductal adenocarcinoma, uveal melanoma.
Acknowledgements
The figures in this manuscript were drawn with Biorender.
Conflict of interest
The authors declare no competing interests.
References
- 1.Rawat M et al. (2019) MicroRNA in pancreatic cancer: from biology to therapeutic potential. Genes (Basel) 10, 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozomara A, Birgaoanu M and Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Research 47, D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mishra S, Yadav T and Rani V (2016) Exploring miRNA based approaches in cancer diagnostics and therapeutics. Critical Reviews in Oncology/Hematology 98, 12–23. [DOI] [PubMed] [Google Scholar]
- 4.To KKW et al. (2020) Advances in the discovery of microRNA-based anticancer therapeutics: latest tools and developments. Expert Opinion On Drug Discovery 15, 63–83. [DOI] [PubMed] [Google Scholar]
- 5.Bautista-Sánchez D et al. (2020) The promising role of miR-21 as a cancer biomarker and Its importance in RNA-based therapeutics. Molecular Therapy. Nucleic Acids 20, 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iorio MV and Croce CM (2012) MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Molecular Medicine 4, 143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iqbal MA et al. (2019) MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Molecular Aspects of Medicine 70, 3–20. [DOI] [PubMed] [Google Scholar]
- 8.Acunzo M et al. (2015) MicroRNA and cancer – a brief overview. Advances in Biological Regulation 57, 1–9. [DOI] [PubMed] [Google Scholar]
- 9.Ruby JG, Jan CH and Bartel DP (2007) Intronic microRNA precursors that bypass Drosha processing. Nature 448, 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie M et al. (2013) Mammalian 5'-capped microRNA precursors that generate a single microRNA. Cell 155, 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheloufi S et al. (2010) A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465, 584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J et al. (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science (New York, N.Y.) 305, 1437–1441. [DOI] [PubMed] [Google Scholar]
- 13.Gurbuz N and Ozpolat B (2019) MicroRNA-based targeted therapeutics in pancreatic cancer. Anticancer Research 39, 529–532. [DOI] [PubMed] [Google Scholar]
- 14.Hayes J, Peruzzi PP and Lawler S (2014) MicroRNAs in cancer: biomarkers, functions and therapy. Trends in Molecular Medicine 20, 460–469. [DOI] [PubMed] [Google Scholar]
- 15.Jonas S and Izaurralde E (2015) Towards a molecular understanding of microRNA-mediated gene silencing. Nature Reviews Genetics 16, 421–433. [DOI] [PubMed] [Google Scholar]
- 16.Braun JE et al. (2011) GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Molecular Cell 44, 120–133. [DOI] [PubMed] [Google Scholar]
- 17.Braun JE et al. (2012) A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5' exonucleolytic degradation. Nature Structural & Molecular Biology 19, 1324–1331. [DOI] [PubMed] [Google Scholar]
- 18.Fabian MR et al. (2011) miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nature Structural & Molecular Biology 18, 1211–1217. [DOI] [PubMed] [Google Scholar]
- 19.Mathys H et al. (2014) Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Molecular Cell 54, 751–765. [DOI] [PubMed] [Google Scholar]
- 20.Fukao A et al. (2014) MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target mRNAs in humans. Molecular Cell 56, 79–89. [DOI] [PubMed] [Google Scholar]
- 21.Fukaya T, Iwakawa HO and Tomari Y (2014) MicroRNAs block assembly of eIF4F translation initiation complex in Drosophila. Molecular Cell 56, 67–78. [DOI] [PubMed] [Google Scholar]
- 22.Ganju A et al. (2017) miRNA nanotherapeutics for cancer. Drug Discovery Today 22, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barger JF and Nana-Sinkam SP (2015) MicroRNA as tools and therapeutics in lung cancer. Respiratory Medicine 109, 803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue J et al. (2017) MicroRNA-targeted therapeutics for lung cancer treatment. Expert Opinion on Drug Discovery 12, 141–157. [DOI] [PubMed] [Google Scholar]
- 25.Kim J et al. (2018) MicroRNAs and metastasis: small RNAs play big roles. Cancer and Metastasis Reviews 37, 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajasegaran Y et al. (2021) Footprints of microRNAs in cancer biology. Biomedicines 9, 1494–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dillekås H, Rogers MS and Straume O (2019) Are 90% of deaths from cancer caused by metastases? Cancer Medicine 8, 5574–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fares J et al. (2020) Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduction and Targeted Therapy 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seyfried TN and Huysentruyt LC (2013) On the origin of cancer metastasis. Critical Reviews in Oncogenesis 18, 43–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeeshan R and Mutahir Z (2017) Cancer metastasis – tricks of the trade. Bosnian Journal of Basic Medical Sciences 17, 172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welch DR and Hurst DR (2019) Defining the hallmarks of metastasis. Cancer Research 79, 3011–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labelle M and Hynes RO (2012) The initial hours of metastasis: the importance of cooperative host-tumor cell interactions during hematogenous dissemination. Cancer Discovery 2, 1091–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y and Cao X (2016) Characteristics and significance of the pre-metastatic niche. Cancer Cell 30, 668–681. [DOI] [PubMed] [Google Scholar]
- 34.Riihimäki M et al. (2018) Clinical landscape of cancer metastases. Cancer Medicine 7, 5534–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solé C and Lawrie CH (2019) MicroRNAs and metastasis. Cancers (Basel) 12, 96–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng Y and Croce CM (2016) The role of MicroRNAs in human cancer. Signal Transduction and Targeted Therapy 1, 15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iorio MV and Croce CM (2012) Causes and consequences of microRNA dysregulation. Cancer Journal 18, 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhuang Y et al. (2016) MicroRNA regulation of endothelial junction proteins and clinical consequence. Mediators of Inflammation 2016, 5078627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin TA (2014) The role of tight junctions in cancer metastasis. Seminars in Cell & Developmental Biology 36, 224–231. [DOI] [PubMed] [Google Scholar]
- 40.Zhou W et al. (2014) Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 25, 501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun B, Han Y and Shi M (2021) Stromal-derived miR-486-5p promotes metastasis of non-small-cell lung cancer cells by targeting the CADM1/tight junctions axis in vascular endothelial cells. Cell Biology International 45, 849–857. [DOI] [PubMed] [Google Scholar]
- 42.Di Modica M et al. (2017) Breast cancer-secreted miR-939 downregulates VE-cadherin and destroys the barrier function of endothelial monolayers. Cancer Letters 384, 94–100. [DOI] [PubMed] [Google Scholar]
- 43.Qin J et al. (2015) MicroRNA-145 suppresses cell migration and invasion by targeting paxillin in human colorectal cancer cells. International Journal of Clinical and Experimental Pathology 8, 1328–1340. [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng CY et al. (2016) MicroRNA-7 suppresses human colon cancer invasion and proliferation by targeting the expression of focal adhesion kinase. Molecular Medicine Reports 13, 1297–1303. [DOI] [PubMed] [Google Scholar]
- 45.Vu T and Datta PK (2017) Regulation of EMT in colorectal cancer: a culprit in metastasis. Cancers (Basel) 9, 171–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gui Z et al. (2017) Oridonin inhibition and miR-200b-3p/ZEB1 axis in human pancreatic cancer. International Journal of Oncology 50, 111–120. [DOI] [PubMed] [Google Scholar]
- 47.Liu C et al. (2018) Roles of miR-200 family members in lung cancer: more than tumor suppressors. Future Oncology (London, England) 14, 2875–2886. [DOI] [PubMed] [Google Scholar]
- 48.Ji H et al. (2019) miR-124 regulates EMT based on ZEB2 target to inhibit invasion and metastasis in triple-negative breast cancer. Pathology Research and Practice 215, 697–704. [DOI] [PubMed] [Google Scholar]
- 49.Yu L et al. (2022) Complete loss of miR-200 family induces EMT associated cellular senescence in gastric cancer. Oncogene 41, 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng YB et al. (2014) miR-132 inhibits colorectal cancer invasion and metastasis via directly targeting ZEB2. World Journal of Gastroenterology 20, 6515–6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen DL et al. (2017) Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics 7, 4836–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J et al. (2018) MiR-186-5p upregulation inhibits proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cell by targeting ZEB1. Archives of Biochemistry and Biophysics 640, 53–60. [DOI] [PubMed] [Google Scholar]
- 53.You J et al. (2014) MiR-132 suppresses the migration and invasion of lung cancer cells via targeting the EMT regulator ZEB2. PLoS One 9, e91827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma L et al. (2010) miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nature Cell Biology 12, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai J et al. (2013) MicroRNA-374a activates Wnt/β-catenin signaling to promote breast cancer metastasis. Journal of Clinical Investigation 123, 566–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J et al. (2019) miR-203 inhibits the invasion and EMT of gastric cancer cells by directly targeting annexin A4. Oncology Research 27, 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y et al. (2017) Lin28B facilitates the progression and metastasis of pancreatic ductal adenocarcinoma. Oncotarget 8, 60414–60428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim J et al. (2016) Ablation of miR-10b suppresses oncogene-induced mammary tumorigenesis and metastasis and reactivates tumor-suppressive pathways. Cancer Research 76, 6424–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y et al. (2016) miR-10b promotes invasion by targeting HOXD10 in colorectal cancer. Oncology Letters 12, 488–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang YY et al. (2015) MicroRNA-10b promotes migration and invasion through Hoxd10 in human gastric cancer. World Journal of Surgical Oncology 13, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong Y et al. (2016) miR-96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Scientific Reports 6, 37421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y et al. (2012) miRNA-135a promotes breast cancer cell migration and invasion by targeting HOXA10. BMC Cancer 12, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J et al. (2021) Andrographolide suppresses the growth and metastasis of luminal-like breast cancer by inhibiting the NF-κB/miR-21-5p/PDCD4 signaling pathway. Frontiers in Cell and Developmental Biology 9, 643525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Z et al. (2019) Long intergenic Non-coding RNA 01121 promotes breast cancer cell proliferation, migration, and invasion via the miR-150–5p/HMGA2 axis. Cancer Management and Research 11, 10859–10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L et al. (2018) MicroRNA-373 inhibits cell proliferation and invasion via targeting BRF2 in human non-small cell lung cancer A549 cell line. Cancer Research and Treatment: Official Journal of Korean Cancer Association 50, 936–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H et al. (2013) miRNA-29c suppresses lung cancer cell adhesion to extracellular matrix and metastasis by targeting integrin β1 and matrix metalloproteinase2 (MMP2). PLoS One 8, e70192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deng B et al. (2016) MiRNA-203 suppresses cell proliferation, migration and invasion in colorectal cancer via targeting of EIF5A2. Scientific Reports 6, 28301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou L et al. (2016) MicroRNA-124 (MiR-124) inhibits cell proliferation, metastasis and invasion in colorectal cancer by downregulating rho-associated protein kinase 1(ROCK1). Cellular Physiology and Biochemistry 38, 1785–1795. [DOI] [PubMed] [Google Scholar]
- 69.Li YR et al. (2016) MicroRNA-520c enhances cell proliferation, migration, and invasion by suppressing IRF2 in gastric cancer. FEBS Open Bio 6, 1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Rahimi M et al. (2020) Down-Regulation of miR-200c and Up-regulation of miR-30c target both stemness and metastasis genes in breast cancer. Cell Journal 21, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang HY et al. (2012) SOX4 Transcriptionally regulates multiple SEMA3/plexin family members and promotes tumor growth in pancreatic cancer. PLoS One 7, e48637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang JY et al. (2020) MicroRNA-145 suppresses uveal melanoma angiogenesis and growth by targeting neuroblastoma RAS viral oncogene homolog and vascular endothelial growth factor. Chinese Medical Journal (English 133, 1922–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou R et al. (2016) MicroRNA-574-5p promotes metastasis of non-small cell lung cancer by targeting PTPRU. Scientific Reports 6, 35714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang X et al. (2020) Correction: hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kγ to promote pancreatic cancer metastasis. Cancer Research 80, 922. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Y et al. (2020) M2 macrophage-derived extracellular vesicles promote gastric cancer progression via a microRNA-130b-3p/MLL3/GRHL2 signaling cascade. Journal of Experimental & Clinical Cancer Research: CR 39, 134. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Zhao S et al. (2020) Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. Journal of Hematology & Oncology 13, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ke M et al. (2019) MicroRNA-148b-colony-stimulating factor-1 signaling-induced tumor-associated macrophage infiltration promotes hepatocellular carcinoma metastasis. Biomedicine & Pharmacotherapy 120, 109523. [DOI] [PubMed] [Google Scholar]
- 78.Yin Z et al. (2019) Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-β signaling pathway. Journal of Experimental & Clinical Cancer Research: CR 38, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yin Y et al. (2014) Tumor-secreted miR-214 induces regulatory T cells: a major link between immune evasion and tumor growth. Cell Research 24, 1164–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu F (2017) Genomic regulation of MicroRNA expression in disease development. Methods in Molecular Biology 1617, 159–167. [DOI] [PubMed] [Google Scholar]
- 81.Zhao JL and Starczynowski DT (2014) Role of microRNA-146a in normal and malignant hematopoietic stem cell function. Frontiers in Genetics 5, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He L et al. (2007) A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kent OA, Fox-Talbot K and Halushka MK (2013) RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene 32, 2576–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zeinali T et al. (2019) Regulatory mechanisms of miR-145 expression and the importance of its function in cancer metastasis. Biomedicine & Pharmacotherapy 109, 195–207. [DOI] [PubMed] [Google Scholar]
- 85.Gan B et al. (2010) FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell 18, 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chang TC et al. (2008) Widespread microRNA repression by Myc contributes to tumorigenesis. Nature Genetics 40, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Molenaar JJ et al. (2012) LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nature Genetics 44, 1199–1206. [DOI] [PubMed] [Google Scholar]
- 88.Zhang X et al. (2012) Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-cell lymphomas. Cancer Cell 22, 506–523. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89.Guan T et al. (2018) ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8. Journal of Experimental Medicine 215, 1153–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen P et al. (2017) MiR-200c is a cMyc-activated miRNA that promotes nasopharyngeal carcinoma by downregulating PTEN. Oncotarget 8, 5206–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Di Leva G et al. (2010) MicroRNA cluster 221–222 and estrogen receptor alpha interactions in breast cancer. Journal of the National Cancer Institute 102, 706–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Coarfa C et al. (2016) Comprehensive proteomic profiling identifies the androgen receptor axis and other signaling pathways as targets of microRNAs suppressed in metastatic prostate cancer. Oncogene 35, 2345–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davalos V et al. (2012) Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 31, 2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ceppi P et al. (2010) Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Molecular Cancer Research 8, 1207–1216. [DOI] [PubMed] [Google Scholar]
- 95.Schmid G et al. (2016) Expression and promotor hypermethylation of miR-34a in the various histological subtypes of ovarian cancer. BMC Cancer 16, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheng N, Li Y and Han ZG (2013) Argonaute2 promotes tumor metastasis by way of up-regulating focal adhesion kinase expression in hepatocellular carcinoma. Hepatology 57, 1906–1918. [DOI] [PubMed] [Google Scholar]
- 97.Lee J et al. (2020) Identifying metastasis-initiating miRNA-target regulations of colorectal cancer from expressional changes in primary tumors. Scientific Reports 10, 14919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martinez-Gutierrez AD et al. (2019) miRNA profile obtained by next-generation sequencing in metastatic breast cancer patients is able to predict the response to systemic treatments. International Journal of Molecular Medicine 44, 1267–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McGuire A, Brown JA and Kerin MJ (2015) Metastatic breast cancer: the potential of miRNA for diagnosis and treatment monitoring. Cancer and Metastasis Reviews 34, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zografos E et al. (2019) Prognostic role of microRNAs in breast cancer: a systematic review. Oncotarget 10, 7156–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sasaki R, Osaki M and Okada F (2019) MicroRNA-based diagnosis and treatment of metastatic human osteosarcoma. Cancers (Basel) 11, 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gajos-Michniewicz A and Czyz M (2019) Role of miRNAs in melanoma metastasis. Cancers (Basel) 11, 326–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Santos JMO et al. (2018) The role of MicroRNAs in the metastatic process of high-risk HPV-induced cancers. Cancers (Basel) 10, 493–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu SG et al. (2019) MicroRNA in lung cancer metastasis. Cancers (Basel) 11, 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhu Z et al. (2020) Identifying the key genes and microRNAs in prostate cancer bone metastasis by bioinformatics analysis. FEBS Open Bio 10, 674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Song Z et al. (2018) Elementary screening of lymph node metastatic-related genes in gastric cancer based on the co-expression network of messenger RNA, microRNA and long non-coding RNA. Brazilian Journal of Medical and Biological Research 51, e6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rupaimoole R et al. (2016) miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discovery 6, 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Raue R et al. (2021) Therapeutic targeting of MicroRNAs in the tumor microenvironment. International Journal of Molecular Sciences 22, 2210–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dhungel B, Ramlogan-Steel CA and Steel JC (2018) MicroRNA-regulated gene delivery systems for research and therapeutic purposes. Molecules 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hosseinahli N et al. (2018) Treating cancer with microRNA replacement therapy: a literature review. Journal of Cellular Physiology 233, 5574–5588. [DOI] [PubMed] [Google Scholar]
- 112.van Rooij E and Kauppinen S (2014) Development of microRNA therapeutics is coming of age. EMBO Molecular Medicine 6, 851–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang M et al. (2019) miR-149-3p reverses CD8(+) T-cell exhaustion by reducing inhibitory receptors and promoting cytokine secretion in breast cancer cells. Open Biology 9, 190061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Flamini V, Jiang WG and Cui Y (2017) Therapeutic role of MiR-140-5p for the treatment of non-small cell lung cancer. Anticancer Research 37, 4319–4327. [DOI] [PubMed] [Google Scholar]
- 115.Liu SH et al. (2021) Systematic identification of clinically relevant miRNAs for potential miRNA-based therapy in lung adenocarcinoma. Molecular Therapy. Nucleic Acids 25, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cortez MA et al. (2015) In vivo delivery of miR-34a sensitizes lung tumors to radiation through RAD51 regulation. Molecular Therapy. Nucleic Acids 4, e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nguyen DD and Chang S (2017) Development of novel therapeutic agents by inhibition of oncogenic MicroRNAs. International Journal of Molecular Sciences 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rupaimoole R and Slack FJ (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nature Reviews. Drug Discovery 16, 203–222. [DOI] [PubMed] [Google Scholar]
- 119.Ge JH et al. (2019) An antisense oligonucleotide drug targeting miR-21 induces H1650 apoptosis and caspase activation. Technology in Cancer Research & Treatment 18, 1533033819892263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xie Y et al. (2020) Stromal modulation and treatment of metastatic pancreatic cancer with local intraperitoneal triple miRNA/siRNA nanotherapy. ACS Nano 14, 255–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tay FC et al. (2015) Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Advanced Drug Delivery Reviews 81, 117–127. [DOI] [PubMed] [Google Scholar]
- 122.Zhang T et al. (2016) Downregulation of miR-522 suppresses proliferation and metastasis of non-small cell lung cancer cells by directly targeting DENN/MADD domain containing 2D. Scientific Reports 6, 19346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nedaeinia R et al. (2016) Locked nucleic acid anti-miR-21 inhibits cell growth and invasive behaviors of a colorectal adenocarcinoma cell line: LNA-anti-miR as a novel approach. Cancer Gene Therapy 23, 246–253. [DOI] [PubMed] [Google Scholar]
- 124.Meng L et al. (2017) Small RNA zippers lock miRNA molecules and block miRNA function in mammalian cells. Nature Communications 8, 13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bajan S and Hutvagner G (2020) RNA-Based Therapeutics: from antisense oligonucleotides to miRNAs. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rinaldi C and Wood MJA (2018) Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nature Reviews. Neurology 14, 9–21. [DOI] [PubMed] [Google Scholar]
- 127.Tao YJ et al. (2015) Antisense oligonucleotides against microRNA-21 reduced the proliferation and migration of human colon carcinoma cells. Cancer Cell International 15, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Devulapally R et al. (2015) Polymer nanoparticles mediated codelivery of antimiR-10b and antimiR-21 for achieving triple negative breast cancer therapy. ACS Nano 9, 2290–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ebert MS and Sharp PA (2010) MicroRNA sponges: progress and possibilities. RNA 16, 2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liang AL et al. (2016) MiRNA-10b sponge: an anti-breast cancer study in vitro. Oncology Reports 35, 1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jung J et al. (2015) Simultaneous inhibition of multiple oncogenic miRNAs by a multi-potent microRNA sponge. Oncotarget 6, 20370–20387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lima JF et al. (2018) Targeting miR-9 in gastric cancer cells using locked nucleic acid oligonucleotides. BMC Molecular Biology 19, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mody HR et al. (2016) Inhibition of S-adenosylmethionine-dependent methyltransferase attenuates TGFβ1-induced EMT and metastasis in pancreatic cancer: putative roles of miR-663a and miR-4787-5p. Molecular Cancer Research 14, 1124–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kopp F et al. (2013) De-targeting by miR-143 decreases unwanted transgene expression in non-tumorigenic cells. Gene Therapy 20, 1104–1109. [DOI] [PubMed] [Google Scholar]
- 135.Baertsch MA et al. (2014) MicroRNA-mediated multi-tissue detargeting of oncolytic measles virus. Cancer Gene Therapy 21, 373–380. [DOI] [PubMed] [Google Scholar]
- 136.Bofill-De Ros X, Gironella M and Fillat C (2014) miR-148a- and miR-216a-regulated oncolytic adenoviruses targeting pancreatic tumors attenuate tissue damage without perturbation of miRNA activity. Molecular Therapy 22, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dhungel B et al. (2018) miRNA122a regulation of gene therapy vectors targeting hepatocellular cancer stem cells. Oncotarget 9, 23577–23588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dhungel B et al. (2018) MicroRNA199a-based post-transcriptional detargeting of gene vectors for hepatocellular carcinoma. Molecular Therapy. Nucleic Acids 13, 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dhungel B, Ramlogan-Steel CA and Steel JC (2018) Synergistic and independent action of endogenous microRNAs 122a and 199a for post-transcriptional liver detargeting of gene vectors. Scientific Reports 8, 15539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fortunato O and Iorio MV (2020) The therapeutic potential of MicroRNAs in cancer: illusion or opportunity? Pharmaceuticals (Basel) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ivkovic C, et al. T (2017) microRNAs as cancer therapeutics: a step closer to clinical application. Cancer Letters 407, 113–122. [DOI] [PubMed] [Google Scholar]
- 142.Reid G et al. (2013) Restoring expression of miR-16: a novel approach to therapy for malignant pleural mesothelioma. Annals of Oncology: Official Journal of the European Society for Medical Oncology 24, 3128–3135. [DOI] [PubMed] [Google Scholar]
- 143.Reid G et al. (2016) Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics 8, 1079–1085. [DOI] [PubMed] [Google Scholar]
- 144.van Zandwijk N et al. (2017) Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. The Lancet. Oncology 18, 1386–1396. [DOI] [PubMed] [Google Scholar]
- 145.Hong DS et al. (2020) Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. British Journal of Cancer 122, 1630–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Reda El Sayed S et al. (2021) MicroRNA therapeutics in cancer: current advances and challenges. Cancers (Basel) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kasinski AL et al. (2015) A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene 34, 3547–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Beg MS et al. (2017) Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investigational New Drugs 35, 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Grossi I et al. (2017) Biological function of MicroRNA193a-3p in health and disease. International Journal of Genomics 2017, 5913195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gao X et al. (2018) The clinical value of miR-193a-3p in non-small cell lung cancer and its potential molecular mechanism explored in silico using RNA-sequencing and microarray data. FEBS Open Bio 8, 94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yu M et al. (2019) PTP1B markedly promotes breast cancer progression and is regulated by miR-193a-3p. FEBS Journal 286, 1136–1153. [DOI] [PubMed] [Google Scholar]
- 152.van den Bosch MTJ et al. (2021) Transcriptome-wide analysis reveals insight into tumor suppressor functions of 1B3, a novel synthetic miR-193a-3p mimic. Molecular Therapy. Nucleic Acids 23, 1161–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fan Q et al. (2017) MiR-193a-3p is an important tumour suppressor in lung cancer and directly targets KRAS. Cellular Physiology and Biochemistry 44, 1311–1324. [DOI] [PubMed] [Google Scholar]
- 154.Liang H et al. (2015) miR-193a-3p functions as a tumor suppressor in lung cancer by down-regulating ERBB4. Journal of Biological Chemistry 290, 926–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yu T et al. (2015) MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene 34, 413–423. [DOI] [PubMed] [Google Scholar]
- 156.Lv L et al. (2014) The DNA methylation-regulated miR-193a-3p dictates the multi-chemoresistance of bladder cancer via repression of SRSF2/PLAU/HIC2 expression. Cell death & disease 5, e1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kwon JE et al. (2013) Ionizing radiation-inducible microRNA miR-193a-3p induces apoptosis by directly targeting Mcl-1. Apoptosis 18, 896–909. [DOI] [PubMed] [Google Scholar]
- 158.Liu Y et al. (2017) MicroRNA-193a-3p inhibits cell proliferation in prostate cancer by targeting cyclin D1. Oncology Letters 14, 5121–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pu Y et al. (2016) MiR-193a-3p and miR-193a-5p suppress the metastasis of human osteosarcoma cells by down-regulating Rab27B and SRR, respectively. Clinical & Experimental Metastasis 33, 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bader AG (2012) miR-34 - a microRNA replacement therapy is headed to the clinic. Frontiers in Genetics 3, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Telford BJ et al. (2021) Multi-modal effects of 1B3, a novel synthetic miR-193a-3p mimic, support strong potential for therapeutic intervention in oncology. Oncotarget 12, 422–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Garofalo M et al. (2021) Correction: MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene 40, 1204. [DOI] [PubMed] [Google Scholar]
- 163.Miller TE et al. (2008) MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. Journal of Biological Chemistry 283, 29897–29903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Callegari E et al. (2012) Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic model. Hepatology 56, 1025–1033. [DOI] [PubMed] [Google Scholar]
- 165.Zhang CZ et al. (2010) MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Molecular Cancer 9, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Di Martino MT et al. (2013) In vitro and in vivo anti-tumor activity of miR-221/222 inhibitors in multiple myeloma. Oncotarget 4, 242–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Higgs G and Slack F (2013) The multiple roles of microRNA-155 in oncogenesis. Journal of Clinical Bioinformatics 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Seto AG et al. (2018) Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. British Journal of Haematology 183, 428–444. [DOI] [PubMed] [Google Scholar]
- 169.Zhang Y et al. (2012) LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood 120, 1678–1686. [DOI] [PubMed] [Google Scholar]
- 170.Cheng CJ et al. (2015) MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature 518, 107–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yu HR, Huang LH and Li SC (2018) Roles of microRNA in the immature immune system of neonates. Cancer Letters 433, 99–106. [DOI] [PubMed] [Google Scholar]
- 172.Bulcha JT et al. (2021) Viral vector platforms within the gene therapy landscape. Signal Transduction and Targeted Therapy 6, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Santana-Armas ML and Tros de Ilarduya C (2021) Strategies for cancer gene-delivery improvement by non-viral vectors. International Journal of Pharmaceutics 596, 120291. [DOI] [PubMed] [Google Scholar]
- 174.Lundstrom K (2018) Viral vectors in gene therapy. Diseases (basel, Switzerland) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Karlsen TA and Brinchmann JE (2013) Liposome delivery of microRNA-145 to mesenchymal stem cells leads to immunological off-target effects mediated by RIG-I. Molecular Therapy 21, 1169–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Biscans A et al. (2019) Diverse lipid conjugates for functional extra-hepatic siRNA delivery in vivo. Nucleic Acids Research 47, 1082–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Li X et al. (2019) Challenges and opportunities in exosome research-perspectives from biology, engineering, and cancer therapy. APL Bioengineering 3, 011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bandi N and Vassella E (2011) miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Molecular Cancer 10, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kara G, Calin GA and Ozpolat B (2022) RNAi-based therapeutics and tumor targeted delivery in cancer. Advanced Drug Delivery Reviews 182, 114113. [DOI] [PubMed] [Google Scholar]
- 180.Seok H et al. (2018) Evaluation and control of miRNA-like off-target repression for RNA interference. Cellular and Molecular Life Sciences 75, 797–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pepe F et al. (2022) A large fraction of trisomy 12, 17p(-), and 11q(-) CLL cases carry unidentified microdeletions of miR-15a/16-1. Proceedings of the National Academy of Sciences of the USA 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Segal M and Slack FJ (2020) Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opinion on Drug Discovery 15, 987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Dhungel B et al. (2018) Evaluation of the Glypican 3 promoter for transcriptional targeting of hepatocellular carcinoma. Gene Therapy 25, 115–128. [DOI] [PubMed] [Google Scholar]
- 184.Ceppi M et al. (2009) MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proceedings of the National Academy of Sciences of the USA 106, 2735–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]