Summary
Chimeric mouse models have recently been developed to study human microglia in vivo. However, widespread engraftment of donor microglia within the adult brain has been challenging. Here, we present a protocol to introduce the G795A point mutation using CRISPR-Cas9 into the CSF1R locus of human pluripotent stem cells. We also describe an optimized microglial differentiation technique for transplantation into newborn or adult recipients. We then detail pharmacological paradigms to achieve widespread and near-complete engraftment of human microglia.
For complete details on the use and execution of this protocol, please refer to Chadarevian et al. (2023).1
Subject areas: Cell Culture, Cell Differentiation, CRISPR, Immunology, Model Organisms, Neuroscience, Single Cell, Stem Cells
Graphical abstract

Highlights
-
•
Protocol for ribonucleoprotein-mediated CRISPR/Cas9 editing of iPSCs
-
•
Generation and differentiation of CSF1R-G795A iPSC line into HPCs and iPSC-microglia
-
•
Intracranial transplantation of G795A-HPCs or G795A-iMG into xenotolerant mice
-
•
Complete CSF1Ri-mediated microglia replacement
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Chimeric mouse models have recently been developed to study human microglia in vivo. However, widespread engraftment of donor microglia within the adult brain has been challenging. Here, we present a protocol to introduce the G795A point mutation using CRISPR-Cas9 into the CSF1R locus of human pluripotent stem cells. We also describe an optimized microglial differentiation technique for transplantation into newborn or adult recipients. We then detail pharmacological paradigms to achieve widespread and near-complete engraftment of human microglia.
Before you begin
This protocol describes the generation, differentiation, and transplantation of CRISPR-edited CSF1R G795A microglia derived from human induced pluripotent stem cell (iPSC) lines. These methods can be further adapted to combine G795-variant microglia using the gRNA provided with manipulation of other genes of interest to study the function of human microglial genes in vivo or utilize microglia to deliver genetic payloads to the brain. To achieve engraftment of human microglia within the mouse brain, it is imperative to use a mouse model that is genetically immunodeficient or pharmacologically immunosuppressed to prevent xenotransplant rejection. In addition, recipient mice must express human CSF1 to enable human microglia survival (i.e., Jax #017708).
To increase the chance of biologically meaningful and reproducible results, care must be taken throughout culture, differentiation, and transplantation to ensure consistency and to avoid bacterial, fungal, or mycoplasma contamination. Consistent culture conditions (i.e., Class II Type A2 biosafety cabinet) and reagents are necessary for successful and efficient generation of clonal iPSC lines. Therefore, we recommend purchasing all reagents and plasticware before beginning and aliquoting for long-term storage per manufacturer recommendations.
Institutional permissions
Human iPSC lines were generated by the UCI-ADRC stem cell core. Subject fibroblasts or peripheral blood mononuclear cells (PBMCs) were collected under approved Institutional Review Boards and Human Stem Cell Research Oversight committee protocols. Informed consent was received from all participants. All animal procedures were conducted in accordance with the guidelines set forth by the National Institutes of Health and the University of California, Irvine Institutional Animal Care and Use Committee. Readers should acquire all necessary permissions from the relevant institutions when working with human derived cell lines and animal models.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-IBA | Abcam | Cat #ab5076 |
| Rabbit anti-Ku80 | Abcam | Cat #80592 |
| Chemicals, peptides, and recombinant proteins | ||
| mTESR-E8 | STEMCELL Technologies | Cat #05990 |
| DMEM/F-12, HEPES, no phenol red | Thermo Fisher Scientific | Cat #11039021 |
| 1-Thioglycerol (Monothioglycerol) | Sigma-Aldrich | Cat #M1753 |
| B27 Supplement | Gibco | Cat #17504044 |
| N2 Supplement | Gibco | Cat #17502048 |
| Non-essential amino acids (NEAA) | Thermo Fisher Scientific | Cat #11140050 |
| GlutaMax Supplement | Thermo Fisher Scientific | Cat #35050061 |
| Insulin-Transferrin-Selenium (ITS-G) (100×) | Thermo Fisher Scientific | Cat #41400045 |
| Human Insulin (25 mg) | Sigma-Aldrich | Cat #I2643 |
| Recombinant Human IL-34 | PeproTech | Cat #200-34 |
| Recombinant Human M-CSF | PeproTech | Cat #300-25 |
| Recombinant Human TGF-β1 | PeproTech | Cat #100-21 |
| Recombinant Human Fractalkine (CX3CL1) | PeproTech | Cat #300-31 |
| Recombinant Human OX-2/MOX1/CD200 (C-6His) | Novoprotein | Cat #C311 |
| ReLeSR | STEMCELL Technologies | Cat #05872 |
| CloneR | STEMCELL Technologies | Cat #05889 |
| Matrigel, Basement Membrane Matrix Growth Factor Reduced, Phenol Red-Free | Corning | Cat #356231 |
| Thiazovivin | STEMCELL Technologies | Cat #72252 |
| Bambanker | Wako | Cat #NC9582225 |
| DPBS, no Ca2+, no MG2+ | Thermo Fisher Scientific | Cat #14190144 |
| Alt-R® S.p. HiFi Cas9 Nuclease V3 | IDTDNA | Cat #1081061 |
| Trans-activating CRISPR RNA (tracrRNA) | IDTDNA | Cat #1072534 |
| Nuclease free duplex buffer | IDTDNA | Cat #11010301 |
| Taq PCR Master Mix | Thermo Fisher Scientific | Cat #K0172 |
| Sodium azide | Sigma-Aldrich | Cat #S2002 |
| Phosphate buffer saline (PBS) | Sigma-Aldrich | Cat #P44017 |
| Triton X-100 | Thermo Fisher Scientific | Cat #9002-93-1 |
| Donkey serum | Thermo Fisher Scientific | Cat #NC9624464 |
| Isoflurane | Patterson Veterinary | Cat #14043070406 |
| Puralube Vet ointment | Dechra | Cat #17033-211-38 |
| Lidocaine 2% | Medline | Cat #17478-711-31 |
| Acetaminophen (Mapap) | Major | Cat #0904-7014-16 |
| Vetbond | 3 M | Cat #084-1469SB |
| PLX3397 | MedChemExpress | Cat #HY-16749 |
| Fluoromount-G | SouthernBiotech | Cat #0100-01 |
| Critical commercial assays | ||
| StemDiff Hematopoietic Kit (includes Basal Media, Supplement A, and Supplement B) | STEMCELL Technologies | Cat #05310 |
| Human Stem Cell Nucleofector™ Kit 2 | Lonza | Cat #VPH-5022 |
| Extracta DNA prep for PCR | Quantabio | Cat #95091 |
| MycoAlert PLUS Mycoplasma Detection Kit | Lonza | Cat #LT07-710 |
| Experimental models: Cell lines | ||
| Human: ADRC76 iPS cells | UCI-ADRC Stem Cell Core | https://stemcells.mind.uci.edu/ |
| Human: G795A-ADRC76 iPS cells | Laboratory of Mathew Blurton-Jones | https://stemcells.mind.uci.edu/ |
| Experimental models: Organisms/strains | ||
| human M-CSF knockin mouse (Rag2−/−, il2rγ−/−, CSF1h/h) | The Jackson Laboratory | JAX #017708 |
| Oligonucleotides | ||
| Primer: CSF1R-G795A_F; 5′-GAAG GCCCAAGACTAACCCT-3′ |
Chadarevian et al.1 | N/A |
| Primer: CSF1R-G795A_R; 5′-GAGG ATGCCATAGGACCAGAC-3′ |
Chadarevian et al.1 | N/A |
| ssODN: CSF1R-G795A; 5′-CCACCGGGAC GTGGCAGCGCGTAACGTGCTGTTGACCA ATGGTCATGTGGCCAAGATTGCCGACTTC GGGCTGGCTAGGGACATCATGAATGACTC CAACTACATTGTCA-3′ |
Chadarevian et al.1 | N/A |
| crRNA: CSF1R-G795; 5′-ATGGTCATGTGGCCAAGATT-3′ |
Chadarevian et al.1 | N/A |
| Primer: chr9+18565856_F; 5′-AGTAGGGCACACCCAAAGTT-3′ |
This paper | N/A |
| Primer: chr9+18565856_R; 5′-GTCTAGACAGTCTGCGTGGAG-3′ |
This paper | N/A |
| Primer: chr16-81062196_F; 5′-AGGTGGGAAAGCAGATGTCG-3′ |
This paper | N/A |
| Primer: chr16-81062196_R; 5′-GCCAAAATTGGGGCAAGGAC-3′ |
This paper | N/A |
| Software and algorithms | ||
| SnapGene 4.3.11 | Dotmatics | N/A |
| Other | ||
| 10 μL Gastight syringe, Model 1701 RN | Hamilton | Part #7653-01 |
| 30-gauge, small hub RN needle, 12 mm, Pt:4, 45° tip | Hamilton | Part #7803-07 |
| 0.2 mL PCR 8-tube strips | USA Scientific | Cat #1402-3900 |
| Nuclease-free water | Ambion | Cat #AM9937 |
| 6-well plates | Corning | Cat #3516 |
| 48-well plates | Corning | Cat #3548 |
| 12-well plates | Corning | Cat #3512 |
| 96-well plates | Corning | Cat #3596 |
| 15 mL centrifuge tube | Corning | Cat #430791 |
| 50 mL centrifuge tube | Corning | Cat #430829 |
| Cryotube vial | Thermo Fisher Scientific | Cat #374081 |
| Parafilm | Sigma-Aldrich | Cat #HS234526B |
| Sliding microtome | Leica | Cat #SM2010 R |
| 100 mL Reservoir | Thermo Fisher Scientific | Cat #07-200-130 |
| Lonza Amaxa Nucleofector II | Thermo Fisher Scientific | Cat #13458999 |
Materials and equipment
iMG-Basal Media
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM/F-12, HEPES, no phenol red (1×) | 1× | 500 mL |
| 1-Thioglycerol (Monothioglycerol) (11.5 M) | 400 mM | 17.5 μL |
| B27 Supplement (50×) | 2× | 10 mL |
| N2 Supplement#(100×) | 0.5× | 2.5 mL |
| Non-essential Amino Acids (NEAA) (100×) | 1× | 5 mL |
| GlutaMAX Supplement (100×) | 1× | 5 mL |
| Insulin-Transferrin-Selenium (ITS-G) (10×) | 2× | 10 mL |
| Human Insulin∗(5 mg/mL) | 5 μg/mL | 500 μL |
#N2 supplement should be thawed on ice and aliquoted into 2.5 mL to avoid repeated freeze/thaw.
∗Human insulin stock made by reconstituting 25 mg powder in 5 mL 0.01 N HCL.
Note: Once assembled, iMG-Basal Media is stable for up to three weeks at 4°C.
CRITICAL: 1-Thioglycerol (monothioglycerol) is harmful if swallowed and toxic by skin absorption. Contact can cause skin, eye, and respiratory irritation. Hydrochloric acid (HCL) is corrosive to the eyes, skin, and mucous membranes. Acute inhalation exposure may cause eye, nose, and respiratory tract irritation and inflammation and pulmonary edema in humans.
iMG-Complete Media
| Reagent | Final concentration | Amount |
|---|---|---|
| iMG Basal Media | 1× | 100 mL |
| Recombinant Human IL-34 | 100 ng/mL | 100 μL |
| Recombinant Human M-CSF | 25 ng/mL | 25 μL |
| Recombinant Human TGF-β1 | 50 ng/mL | 50 μL |
iMG-Maturation Media (optional)
| Reagent | Final concentration | Amount |
|---|---|---|
| iMG Basal Media | 1× | 100 mL |
| Recombinant Human IL-34 | 100 ng/mL | 100 μL |
| Recombinant Human M-CSF | 25 ng/mL | 25 μL |
| Recombinant Human TGF-β1 | 50 ng/mL | 50 μL |
| Recombinant Human Fractalkine (CX3CL1) | 100 ng/mL | 100 μL |
| Recombinant Human CD200 (C-6His) | 100 ng/mL | 100 μL |
Note: Cytokine stocks are made by reconstituting cytokines in 0.1% BSA:DPBS at 100 ng/μL and storing at −20°C for long-term storage. Cytokines are temperature sensitive and should be added fresh to iMG-Basal Media prior to use. Once combined, iMG-Complete Media and iMG-Maturation Media are stable for no more than one week at 4°C.
Step-by-step method details
Reagent preparation
Timing: 2 d
-
1.BD Matrigel Matrix Growth Factor Reduced, Phenol Red-Free.
-
a.Thaw vials of Matrigel in a bucket of ice and leave at 4°C overnight to thaw.
-
b.Check lot # per manufacturer to determine concentration: https://www.corning.com/worldwide/en/products/life-sciences/resource-library.html
-
c.Aliquot 1 mg into prechilled (−80°C) conical vials.
-
d.Store aliquots at −80°C for long-term storage.
-
e.To Matrigel-coat plates:
-
i.Remove aliquot of Matrigel from −80°C and allow it to thaw at 4°C for 1 h.
-
ii.Resuspend 1 mg Matrigel in 12 mL of 4°C cold DMEM and plate 1 mg per plate: 2 mLs per well for a 6-well plate, 1 mL per well for a 12-well plate, add 500 μL per well for a 24-well plate, 250 μL per well for a 48-well plate, and 125 μL per well for a 96-well plate.
-
iii.Wrap Matrigel coated plate with Parafilm and keep at 20°C–22°C for at least 1 h before use. Note: keep plates at 4°C for long term storage.
-
i.
-
a.
-
2.Thiazovivin (TZ).
-
a.Dissolve 1 mg Thiazovivin (TZ) in 642 mL DMSO to make 5 nM stock.
-
b.Aliquot into 20 μL aliquots and store at −20°C for long term storage.
-
c.To prepare TZ-E8 media, add 0.35 μL TZ to 10 mL of TeSR-E8 Media (0.17 μM). Note: TZ-E8 media is stable for up to two weeks at 4°C.
-
a.
CRITICAL: Thiazovivin (TZ) can cause acute toxicity if orally ingested. Wash face, hands, and any exposed skin thoroughly after exposure.
-
3.CRISPR RNA (crRNA).
-
a.Dissolve 10 nmol crRNA in 50 μL Duplex Buffer (200 μM) on ice.
-
b.Aliquot into 3 μL aliquots and store at −20°C for long term storage.
-
a.
-
4.Trans-activating CRISPR RNA (tracrRNA).
-
a.Resuspend 20 nmol tracrRNA in 100 μL Duplex Buffer (200 μM) on ice.
-
b.Aliquot into 3 μL aliquots and store at −20°C for long term storage.
-
a.
-
5.High Fidelity (HiFi) Cas9.
-
a.Thaw HiFi Cas9 Nuclease V3 on ice.
-
b.Aliquot into 6.8 μL aliquots and store at −20°C for long-term storage.
-
a.
-
6.Single-stranded Oligodeoxynucleotide template.
-
a.Resuspend 20 nmol ssODN template in 100 μL Nuclease-Free Water (200 μM).
-
b.Aliquot into 2 μL aliquots and store at −20°C for long-term storage.
-
a.
Human iPSC culture
Timing: 2 weeks
-
7.
Remove iPSC cell culture TeSR-E8 media from 4°C to warm at 20°C–22°C.
-
8.
Prepare 0.17 μM TZ-E8 Media.
-
9.Thaw and plate frozen iPSC line.
-
a.Warm frozen vial in 37°C water bath.
-
b.Transfer frozen cell suspension to 15 mL conical vial.
-
c.Add 5 mL TZ-E8 media.
-
d.Spin at 200 × g for 2 min.
-
e.Aspirate DMEM from Matrigel coated 6-well plate.
-
f.Aspirate TZ-E8 media from conical vial leaving ∼700 μL of media.
-
g.Use P1000 pipette to slowly remove excess media.
-
h.Tap conical vial with finger to break pellet into clumps.
-
i.Add total volume desired to split cell population to new wells. Note: It is generally recommended to plate human iPSCs at a seeding density between 2 × 105 – 1 × 106 viable cells per one well of a 6-well plate.
-
j.Plate cells in 6-well plate (2 mL per well) and place in 37°C, 5% CO2 incubator.
-
a.
-
10.
After 24 h, change the media to TeSR-E8 media without TZ.
-
11.
Change media every day with fresh media being sure not to disturb attached iPSCs.
-
12.Passage using ReLeSR when iPSCs reach 70% confluency in a 6-well plate.
-
a.Aspirate media from well.
-
b.Wash cells with 1 mL DPBS.
-
c.Aspirate DPBS.
-
d.Add 1 mL ReLeSR per well for 1–2 min.
-
e.Aspirate ReLeSR and allow well to sit dry for 3 min.
-
f.Add 1 mL TZ-E8 Media gently to the wall of each well.
-
g.Replace lid and tap side of plate with palm of your hand. You will see colonies begin to rise and fragment.
-
h.Transfer resuspended cells to 15 mL conical vial.
-
i.Centrifuge at 200 × g for 2 min.
-
j.Aspirate TZ-E8 media from conical vial, leaving ∼700 μL of media.
-
k.Use P1000 pipette to slowly remove excess media.
-
l.Tap conical vial with finger to break pellet into clumps.
-
m.Split iPSC suspension to 1:18 on Matrigel-coated 6-well plates for continued culture by resuspending in TZ-E8 media
-
n.After 24 h, change the media to TeSR-E8 media without TZ.
-
a.
-
13.Cryobank iPSCs in Liquid nitrogen for long-term storage.
-
a.When wells are 70% confluent, aspirate media from well.
-
b.Wash cells with 1 mL/well DPBS.
-
c.Aspirate DPBS.
-
d.Add 1 mL ReLeSR per well for 1–2 min.
-
e.Aspirate ReLeSR and allow well to sit dry for 3 min.
-
f.Add 750 μL Bambanker serum free cell freezing medium.
-
g.Replace lid and tap side of plate with palm of your hand. You will see colonies begin to rise and fragment.
-
h.Transfer resuspended cells to cryotube vial (one well per vial).
-
i.Store at −80°C for 24 h.
-
j.Transfer vials to liquid nitrogen storage the following day for long-term storage.
-
a.
Alternatives: Versene (Thermo Scientific, Cat #15040066) may also be used for human iPSC culture. Other freezing media, such as CryoStor CS10 (StemCell Technologies, Cat #07931), mFreSR (StemCell Technologies, Cat #05854), or EZ-LiFT (Sigma, Cat #SCM139), were not tested in this protocol.
Note: We strongly recommend culturing human iPSCs for 2 weeks post-thaw to allow cells to stabilize prior to cryopreservation or proceeding with CRISPR-editing.
CRISPR-edit iPSCs with CSF1R-G795 targeting RNP and template
Timing: 2 d
-
14.
Prepare CloneR-E8 media by adding 1 mL 10× CloneR supplement to 9 mL of TZ-E8 media (Figure 1).
-
15.
Aspirate TeSR-E8 media from iPSCs and plate CloneR-E8 media for at least 1 h before lifting cells for electroporation.
-
16.Generate gRNA:
-
a.Remove aliquot of crRNA and tracrRNA and place on ice to thaw.
-
b.Combine 3 μL crRNA and 3 μL tracrRNA aliquots into a PCR tube.
-
c.Heat at 95°C for 5 min to anneal.
-
d.Remove and allow to cool at 20°C–22°C for 5 min.
-
a.
-
17.Generate RNP complex:
-
a.Remove an aliquot of HiFi Cas9 and place on ice.
-
b.Combine 5.1 μL gRNA (see step 3) with 6.8 μL HiFi Cas9. Note: The recommended 1:1.2 M ratio of protein Cas9:gRNA may require further optimization if using ready-made gRNA or alternative Cas9 variants.
-
c.Gently tap to combine volume.
-
d.Allow RNP complex formation to occur at 20°C–22°C for 15 min.
-
a.
-
18.Prepare 12-well plate:
-
a.Aspirate DMEM from 12-well Matrigel coated plate.
-
b.Add 1 mL CloneR-E8 media to one well per CRISPR reaction.
-
c.Place plate in 37°C, 5% CO2 incubator to pre-warm media for iPSCs.
-
a.
-
19.Collect iPSCs for electroporation:
-
a.Pre-warm 1 mL Accutase in 37°C water bath.
-
b.Aspirate CloneR-media from well.
-
c.Wash cells gently with 1 mL DPBS
-
d.Aspirate DPBS and immediately add 1 mL Accutase per 6-well plate’s well.
-
e.Transfer plate to incubator and allow it to incubate at 37°C for 2 min
-
f.Return plate to cell culture hood and aspirate accutase slowly using P1000 pipette.
-
g.Add 1 mL CloneR media.
-
h.Gently wash cells with P1000 pipette to lift adhered cells.
-
i.Transfer cells to 15 mL conical vial and count. Note: We have found consistent and efficient transfection of iPSCs with >90% viability.
-
j.Transfer 500,000 iPSCs to clean 15 mL conical vial and discard remaining cells.
-
k.Centrifuge 500,000 iPSCs at 100 × g for 2 min.
-
l.Remove supernatant slowly using P200.
-
m.Tap conical vial gently to break apart the cell pellet.
-
a.
-
20.Electroporate iPSCs with RNP complex and ssODN template.
-
a.Remove ssODN template and allow it to thaw on ice.
-
b.Combine RNP complex with ssODN template using 100 μL nucleofection buffer from Human Stem Cell Nucleofector Kit 2.
-
c.Transfer solution to cell suspension slowly.
-
d.Tap conical vial with finger gently to mix suspension. Do not triturate single-cell suspension with pipette.
-
e.Transfer suspension to electroporation cassette immediately to prevent viability loss.
-
f.Place in Amaxa nucleofector 2b and nucleofect at B-016 setting.
-
a.
Alternatives: The Lonza 4D-Nucleofector system can also be used for transfection of iPSCs in 100 μL cuvettes or 20 μL 16-well strips (Cat# AAF-1003X). Alternatively, this system can accommodate large-scale transfection of up to 1 × 109 cells (Cat#AAF-1002L) and transfection in 96-well format (Cat# AAF-1003S).
Note: The ssODN used in this protocol is specifically designed to knock-in CSF1R-G795A while simultaneously eliminating the gRNA’s PAM site. This increases efficiency by avoiding re-cutting of correctly edited alleles. For more information on this strategy, see Okamoto et al.2
Note: All components optimized for electroporation delivery using reagents listed with the Amaxa nucleofector 2b system. Any alterations to individual components or delivery system will require optimization for efficient CRISPR-Cas9 editing of iPSCs.
-
21.Plate iPSCs for overnight recovery.
-
a.Remove the 12-well plate from the incubator.
-
b.Remove cassette from Amaxa nucleofector.
-
c.Using the provided tear dropper, collect 500 μL of pre-warm CloneR-E8 media and add to electroporated cells in cassette.
-
d.Collect suspension from cassette and plate cells drop wise into a single well of the 12-well plate.
-
e.Incubate for 24 h in 37°C, 5% CO2 incubator to allow iPSCs to recover (Figure 1D).
-
a.
-
22.Single-cell plate CRISPR-edited iPSCs.
-
a.Prepare six 96-well plates for single cell-plating (D0).
-
i.Aspirate DMEM from all wells of previously prepared Matrigel-coated plates (see Regent Preparation, Step 1).
-
ii.Add 50 μL CloneR media to each well.
-
iii.Place media-filled plates in the incubator to prewarm for single-cell plating.
-
i.
-
b.Prewarm 1 mL accutase at 37°C in a water bath.
-
c.Remove the 12-well plate from incubator.
-
d.Aspirate CloneR media.
-
e.Wash cells with 1 mL DPBS.
-
f.Aspirate DPBS.
-
g.Add 500 μL warm Accutase to well.
-
h.Incubate in incubator for 1.5 min at 37°C.
-
i.Remove Accutase gently using P1000 pipette.
-
j.Add 500 μL CloneR-E8 media.
-
k.Gently flush well to lift adhered cells.
-
l.Transfer suspended cells to 15 mL conical vial and count.
-
m.Fill 100 mL reservoir with 15 mL CloneR-E8 media.
-
n.Add 100 single-celled iPSCs per plate (600 total) to CloneR-E8 media in the reservoir.
-
o.Using stereological pipette, add 15 mL additional CloneR-E8 media to the reservoir by gently moving along the length of the reservoir to mix suspension.
-
p.Remove pre-warmed 96-well plates from the incubator.
-
q.Using a multichannel pipette, plate 50 μL per well of cell suspension from reservoir.
-
r.Transfer all plates to 37°C, 5% CO2 incubator.
-
a.
Figure 1.
CRISPR-edit iPSCs with CSF1R-G795 targeting RNP and template
(A) Prepare gRNA by annealing crRNA and tracrRNA at 95°C for 5 min. Next, combine with HiFi Cas9 at RT for 15 min to generate RNP Complex.
(B) Isolate iPSCs for transfection by culturing adherent cells with prewarmed Accutase at 37°C for 3 min before resuspending cells in ClonR-E8 media to count and centrifuge 500,000 iPSCs for electroporation.
(C) Combine RNP complex, ssODN template, and iPSCs into cuvette. After electroporation, resuspend cells in ClonR-E8 media and plate in 12-well plate for recovery overnight.
(D) Representative image of CRISPR-edited iPSCs after overnight recovery in ClonR-E8 media. Scale bar, 400 μm.
Single-cell expansion and screening of CRISPR-edited iPSCs
Timing: 3–4 weeks
-
23.
After 36 h (D3), change media to TeSR-E8 media.
-
24.
Change media daily with fresh TeSR-E8 media to promote healthy growth of attached iPSCs.
-
25.
Five days after initial media change (D8), begin screening 96-well plates to identify and mark non-clonal wells (i.e., wells with either two-or-more colonies or no evidence of expansion).
-
26.
Continue screening wells over 5–7 (D13-D15) day period as clones may begin to emerge from the corner of the 96-well plate, marking wells with single clone iPSCs exhibiting typical morphology.
-
27.When iPSC clones have grown to 30%–50% confluency, passage selected wells to 48-well plates and take sample of cell suspension for screening.
-
a.One day prior, coat 48-well plates with 1 mg/plate Matrigel in DMEM.
-
b.When passaging, aspirate DMEM and add 500 μL of TZ-E8 media to each well of the 48-well plate.
-
c.Aspirate TeSR-E8 Media from all 96-well plates.
-
d.Add 100 μL DPBS to each well.
-
e.Aspirate DPBS and add 100 μL of ReLeSR.
-
f.After 1 min, aspirate ReLeSR and allow wells to sit dry.
-
g.After 2 min dry, add 100 μL of TZ-E8 media to each well.
-
h.Using clean pipette tip per well, flush well with TZ-media and transfer 50 μL of suspension to 48-well plate and remaining 50 μL of suspension to 8-strip PCR tube for genetic screening.
-
i.Repeat for each clonal well of interest.
-
j.Place 48-well plate in the 37°C, 5% CO2 incubator and change the media daily.
-
a.
-
28.Identify correctly edited clones.
-
a.Isolate DNA from individual clone samples using Extracta DNA prep for PCR.
-
i.Centrifuge 8-strip tubes using tabletop centrifuge.
-
ii.Aspirate remaining media from each tube using fresh P200 pipette tip.
-
iii.Add 25 μL of Extraction Buffer.
-
iv.Incubate at 95°C for 15 min using a thermocycler.
-
v.Add 25 μL of Stabilization Buffer to each sample.
-
i.
-
b.PCR amplify targeted CSF1R sequence of each clone using the reaction below.
Reagent Amount Isolated iPSC clone DNA 2 μL CSF1R-G795A_F (100 μM) 1 μL CSF1R-G795A_R (100 μM) 1 μL 2× Taq PCR Master Mix 25 μL ddH2O up to 50 μL Steps Temperature Time Cycle Initial Denaturing 95°C 3 min 1 Denaturing 95°C 0:30 s 35 cycles Annealing 60°C 0:30 s Extension 72°C 0:35 s Final Extension 72°C 10 min 1 Hold 4°C N/A N/A -
i.Send PCR product for Sanger Sequencing with G795A_F primer.
-
ii.Interpret chromatograms (i.e., SnapGene Viewer) to identify iPSC colonies with Homozygous G795A knock-in.
-
i.
-
c.When 48-well plates are 70% confluent, passage correctly edited clones using ReLeSR to 2× wells of 12-well Matrigel-coated plate.
-
d.When 12-well plates are 70% confluent, freeze one well in a single cryovial and passage second well to two 6-well Matrigel-coated plates for expansion and banking per clone.
-
e.When 6-well plates are 70% confluent, cryobank all wells.Note: It is important to analyze correctly edited clones for off-target effects caused by unintended modifications within the genome or single cell-induced stress. Therefore, array comparative genomic hybridization analysis3,4 and sanger sequencing of the following predicted off-target sites are recommended before proceeding with differentiation: chr9:+18565856 and chr16:-81062196.Note: We strongly recommend testing correctly edited clones for mycoplasma contamination (MycoAlert Plus; Lonza, Cat #LT07-710) before proceeding with G795A-iPSC differentiation.
-
a.
G795A-iPSC differentiation into hematopoietic progenitor cells (HPCs) and iPSC-Microglia (iMG)
Timing: 4–6 weeks
-
29.Plating iPSCs for Differentiation:
-
a.One day prior, coat a 6-well plate with 1 mg Matrigel per plate in DMEM.
-
b.Aspirate TeSR-E8 media.
-
c.Wash cells with 1 mL DPBS
-
d.Aspirate DPBS and add 1 mL ReLeSR.
-
e.After 1–2 min, remove ReLeSR and allow the plate to rest dry for an additional 3 min.
-
f.Add 1 mL TZ-E8 media.
-
g.Close lid and tap side of plate to lift aggregates of iPSCs.
-
h.Transfer suspended cells to 15 mL conical vial and triturate gently twice.
-
i.Count aggregates by plating 10 μL of suspension onto empty 96-well plate’s well and examine under microscope. You will need 80 colonies of ∼100 cells each per 35 mm well, or 480 colonies total per 6-well plate.
-
j.Collect total number of colonies and transfer them to clean 15 mL conical vial.
-
k.Resuspend in 2 mL/well TZ-E8 media by adding with a serological pipette directly to conical vial.
-
l.Collect and plate 2 mL/well containing 80 colonies per well Matrigel coated 6-well plate.
-
a.
Note: Plating density is very important for robust differentiation of HPCs. Therefore, it is recommended to plate multiple wells at varying densities (ie. 50, 80, or 120 colonies of ∼100 cells each per 35 mm well) to then choose appropriate wells for differentiation.
-
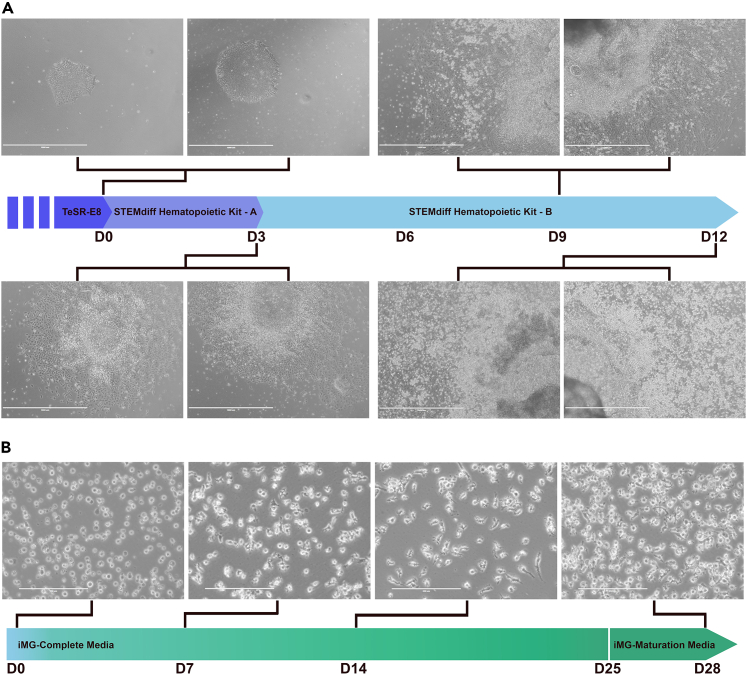
30.Differentiation of G795A-iPSCs into HPCs using STEMdiff Hematopoietic Kit5 (Figure 2A).
-
a.After 24 h (D0), change media to Medium A (Basal medium plus Supplement A at 1:200 dilution, 2 mL/well of a 6-well).
-
b.48 h after original media change (D2), replace 50% medium A using P1000 pipette to remove 1 mL/well and add 1 mL fresh Medium A per well.
-
c.24 h after 50% medium change (D3), gently remove all media from each well using P1000 pipette and add 2 mL/well Medium B (Basal medium plus supplement B at 1:200).
-
d.Without disturbing adhered clumps, add 1 mL/well Medium B every 48 h (D5, D7, and D9).
-
e.HPCs will begin to appear and lift from adhered colonies (D10) at which point medium and cells can be collected using a serological pipette and transferred to a 50 mL conical vial.
-
f.Centrifuge at 300 × g for 5 min.
-
g.You may replate Medium B supernatant to your wells to collect additional cells after 48 h (D12) or discard supernatant if not.
-
h.HPCs may be collected for transplantation, cryobanked in Bambanker for long-term storage, or immediately transferred to a Matrigel-coated 6-well plate for iMG differentiation.
-
a.
-
31.Differentiation of G795A-HPCs into G795A-iMG (Figure 2B).
-
a.One day prior, coat a 6-well plate with 1 mg Matrigel per plate in DMEM.
-
b.If proceeding immediately from HPC differentiation, count suspended cells and plate 1 × 106 HPCs per Matrigel coated 6-well plate in 2 mL/well iMG-Complete Media (D0).
-
c.If frozen, thaw HPCs into Matrigel coated 6-well plate (D0).
-
i.Warm frozen vial in 37°C water bath.
-
ii.Transfer vial to TC hood and remove suspended cells to 15 mL conical vial.
-
iii.Add 1 mL iMG-Basal Media drop wise to the wall of 15 mL conical vial.
-
iv.Add an additional 5 mL iMG-Basal Media to the wall of 15 mL conical vial.
-
v.Centrifuge at 300 × g for 5 min.
-
vi.Aspirate supernatant leaving <500 μL of iMG-Basal Media.
-
vii.Tap conical vial with finger to break apart cell pellet.
-
viii.Resuspend HPCs in 1 mL iMG-Complete Media and count.
-
ix.Resuspend total suspension to plate 1 × 106 HPCs per 6-well plate in 2 mL/well iMG-Complete Media.
-
i.
-
d.Add 1 mL/well fresh iMG-Complete Media every 48 h for 10 days (D2, D4, D6, D8, D10).
-
e.On day 12, carefully collect 6 mL per well using serological pipette and transfer suspension to 50 mL conical vial to centrifuge at 300 × g for 5 min.
-
f.Aspirate supernatant, resuspend any cell pellet in fresh 6 mL iMG-Complete Media, and plate 1 mL/well (D12).
-
g.Continue to add 1 mL/well fresh iMG-Complete Media every 48 h for an additional 12 days (D14, D16, D18, D20, D22, D24).
-
a.
-
32.Maturation of G795A-iMG (optional).
-
a.If further maturation of iMGs is desired, on day 25, carefully collect 7 mL per well using serological pipette and transfer suspension to 50 mL conical vial to centrifuge at 300 × g for 5 min.
-
b.Aspirate supernatant, resuspend any cell pellet in fresh 12 mL iMG-Maturation Media, and plate 2 mL/well (D25).
-
c.Add 1 mL/well fresh iMG-Maturation Media after 48 h (D27).
-
d.On day 28, iMG are ready for transplantation or cryostorage (D28).
-
a.
Note: Microglia maturation has been shown to increase transcripts related to the microglia sensome without altering levels of core microglial genes in vitro as described in Abud et al.6 However, maturation is not necessary for successful post-transplantation engraftment and subsequent microglia replacement in vivo.
Figure 2.
G795A-iPSC differentiation into Hematopoietic progenitor cells (HPCs) and iPSC-Microglia (iMG)
(A) Schematic shows the differentiation of G795A-iPSCs through the mesoderm lineage (D0-D3) and hematopoiesis (D3-D12) using the STEMdiff Hematopoietic Kit. Primitive HPCs are visible by D9 (±2 days) and ready for collection on D10 and/or D12. Scale bar, 1000 μm.
(B) G795A-HPCs are transferred directly - or thawed if frozen - into iMG-Complete Media for microglia differentiation. To further mature in vitro, G795A-iMG can be cultured in iMG-Maturation Media for an additional 3 days (D25-D28). Scale bar, 200 μm.
Transplantation of G795A-HPCs or G795A-iMGs
Timing: 30 min/mouse
-
33.
One day prior, coat a 6-well plate with 1 mg Matrigel per plate in DMEM.
-
34.If frozen, thaw vial of G795A-HPCs or G795A-iMGs into Matrigel coated 6-well plate.
-
a.Warm frozen vial in 37°C water bath.
-
b.Transfer vial to TC hood and remove suspended cells to 15 mL conical vial.
-
c.Add 1 mL iMG-Basal Media drop wise to the wall of 15 mL conical vial.
-
d.Add an additional 5 mL iMG-Basal Media to the wall of 15 mL conical vial.
-
e.Centrifuge at 300 × g for 5 min.
-
f.Aspirate supernatant leaving <500 μL of iMG-Basal Media.
-
g.Tap conical vial with finger to break apart cell pellet.
-
h.Resuspend HPCs or iMG in 1 mL iMG-Complete Media and count.
-
i.Resuspend total suspension to plate 4 × 107 HPCs or iMG per 6-well plate in 2 mL/well iMG-Complete Media.
-
a.
-
35.
Allow cells to recover for 24 h in incubator. Note: G795A-HPCs may be transplanted immediately after thaw if viability >90%.
-
36.Prepare cells for transplantation.
-
a.After 24 h, collect cells using a serological pipette and add them to a 50 mL conical vial.
-
b.Add 1 mL DPBS to each well.
-
c.Using P1000 pipette, flush each well and add the remaining cells to suspension.
-
d.Centrifuge at 300 × g for 5 min.
-
e.Aspirate supernatant.
-
f.Resuspend cells in 1 mL DPBS and transfer to a sterile 1.5 mL Eppendorf tube.
-
g.Count cells and centrifuge Eppendorf tube at 300 × g for 5 min.
-
h.Resuspend cell pellet at 6.25 × 104 cells/μL in DPBS and transfer tube to ice bucket for transplantation.
-
a.
-
37.Neonatal intracerebroventricular transplantation.
-
a.Place P0 to P2 xenotransplantable pups (i.e., Jax #017708) in a clean cage over a heating pad with a nestle from the home cage.
-
b.When ready, transfer pups onto the ice for 2–3 min to induce hypothermic anesthesia.
-
c.Free-hand transplant 5 × 105 cells/pup at 8 sites using a 30-gauge needle affixed to a 10 μL Hamilton syringe as described in Hasselmann et al.7
-
i.Perform bilateral injections at 2/5th of the distance from the lambda suture to each eye, injecting into the lateral ventricles at 3 mm and into the overlying superior cortex at 1 mm, and into the posterior cortex in line with the forebrain injection sites, and perpendicular to lambda at a 45° angle.
-
ii.Each site receives 1 μL of cell suspension followed by 1 min diffusion time at rest before moving to the next injection site.
 CRITICAL: Pup injections should occur smoothly with constant flow at each injection site. Rapid injection of cell suspension will result in spillover leading to poor delivery and, therefore, poor engraftment. It is crucial to practice and develop reproducible delivery of cells for experimentally consistent levels of engraftment.
CRITICAL: Pup injections should occur smoothly with constant flow at each injection site. Rapid injection of cell suspension will result in spillover leading to poor delivery and, therefore, poor engraftment. It is crucial to practice and develop reproducible delivery of cells for experimentally consistent levels of engraftment.
-
i.
-
d.Place transplanted pups on clean kimwipe over heating pad and cover with bedding from home cage during recovery. Note: Mice should remain on heating pad until awake, active, and vocal. Color will return to bright pink tone after awakening from hypothermic induced anesthesia.
-
e.Return transplanted pups back to their home cages and wean at P21.
-
a.
-
38.Adult intracranial transplantation.
-
a.Anesthetize adult mice (2 mo old) for 12 min and keep them under continuous isoflurane sedation after transferring them to a stereotaxic frame.
-
b.Prepare mouse for surgery.
-
i.After mounting the mouse to the frame and placing the nose cone, tighten ear bars making sure to keep head level.
-
ii.Using razor, shave fur from snout to ears and sanitize skin with iodine wipes.
-
iii.Apply local anesthetic before making an incision along the scalp to reveal the skull.
-
i.
-
c.Identify coordinates for drilling.
-
i.Attach 30-gauge needle affixed to a 10-mL Hamilton syringe to stereotaxic frame.
-
ii.Identify bregma suture at 0 and move to the following coordinates: anteroposterior, 2.06 mm; and mediolateral, ± 1.75 mm.
-
iii.Mark skull with sharpie and gently use Dremel to shave small indention in the skull at coordinates.
-
iv.Remove syringe and wash with DPBS before collecting 4 μL of suspended cells for transplantation.
-
v.Reattach syringe to stereotaxic frame and re-zero at bregma.
-
i.
-
d.Transplant G795A-HPCs or G795A-iMGs.
-
i.From bregma, navigate to the following coordinates: anteroposterior, 2.06 mm; and mediolateral, + 1.75 mm.
-
ii.Lower syringe along the dorsoventral axis 1.85 mm then rise to 1.75 mm to create a small pocket to receive injection in the dorsal hippocampus.
-
iii.Inject 2 μL of cell suspension at a rate of 1 μL/30 s.
-
iv.Wait 4 min after injection to allow transplanted volume to diffuse into the targeted region.
-
v.Rise along the dorsoventral axis to 0.95 mm to inject into the parietal association cortex.
-
vi.Inject 2 μL of cell suspension at a rate of 1 μL/30 s.
-
vii.Wait 4 min after injection to allow transplanted volume to diffuse into the targeted region.
-
viii.Elevate needle along dorsoventral axis until clear of mouse and remove from stereotaxic frame.
-
ix.Clean syringe with consecutive washes of DPBS, 70% (vol/vol) ethanol, and DPBS before collecting 4 μL cell suspension for transplantation into the alternative hemisphere.
-
x.From bregma, navigate to following coordinates: anteroposterior, 2.06 mm; and mediolateral, - 1.75 mm.
-
xi.Repeat injection steps to deliver 2 μL into the dorsal hippocampus and parietal association cortex.
-
i.
-
e.Prepare mice for postoperative care.
-
i.After transplantation, gently bring tissue surrounding the incision together and apply Vetbond adhesive.
-
ii.Remove mouse from the stereotaxic and transfer to clean cage on heating pad to recover.
-
iii.Provide mice with systemic analgesia, such as 2 mg/mL Acetaminophen diluted in water for 10 d.
-
i.
-
a.
Note: Successful xenotransplantation of human iMG requires immune compromised (i.e., Rag2−/− and IL2rγ−/−) mice that also express human colony stimulating factor 1 (CSF-1).
CSF1Ri-mediated microglia replacement in G795A transplanted mice
Timing: 4–8 weeks
-
39.
For mice transplanted as pups, provide 290 mg/kg PLX3397 chow ad libitum for at least 4 weeks after weaning.
-
40.
For mice transplanted as adults, provide 290 mg/kg PLX3397 chow ad libitum for at least 8 weeks (Figure 3).
Note: Adjustment to dose, duration, and the specific CSF1R antagonist used are likely possible, but must be empirically determined.
Note: For immunofluorescence staining of engrafted human G795A-microglia (Figure 3), mice were perfused with PBS and isolated brains were drop fixed in 4% (wt/vol) PFA for 36 h then cryoprotected in a 30% (wt/vol) sucrose at 4°C. Brains were sectioned coronally into 30-μm-thick slices on a freezing microtome and stored in a solution of 0.05% NaN3 in 1× PBS as free-floating slices. For staining, tissue was blocked for 1 h in 1× PBS, 0.2% Triton X-100, and 10% donkey serum. Immediately following blocking, brain sections were labeled with goat anti-IBA1 (1:200; Abcam; ab5076) and rabbit anti-Ku80 (1:100; Abcam; ab80592) diluted in 1× PBS and 1% donkey serum and incubated overnight on a shaker at 4°C. After washing, sections were incubated with AlexaFluor-conjugated secondary antibodies (1:400) for 1 h, then washed three times with PBS before mounting with Fluoromount-G. Immunofluorescent sections were visualized and captured using an Olympus FX 1200 confocal microscope.
Figure 3.
CSF1Ri-mediated Microglia Replacement in G795A-iMG transplanted adult mice
(A) IBA1+/Ku80+ human G795A-iMG engraft minimally in the occupied adult murine brain.
(B) G795A-iMG proliferate from the injection site and replace IBA+/Ku80- murine microglia under constant CSF1Ri treatment.
(C) 60 days of 290 mg/kg PLX3397 treatment is sufficient to replace murine microglia throughout the Striatum, Medial Septum, Corpus Callosum, Cortex, Thalamus, and Brain Stem. Brain stitch scale bar, 500 μm. Representative 20× scale bar, 100 μm.
Expected outcomes
Prior to treatment with CSF1R antagonists, G795A-HPCs will engraft into the xenotransplantation-compatible murine brain more robustly than fully differentiated G795A-iMGs. Furthermore, both cell types will engraft more robustly in neonatal pups compared to adult mice. However, we have observed that once the recommended dose and duration of PLX3397 treatment has been concluded a near complete replacement of endogenous microglia with human microglia will be achieved within 4 weeks for pup transplanted mice and 8 weeks for adult transplanted mice (Figure 3), regardless of whether G795A-HPCs or G795A-iMGs are used.
Limitations
Although this protocol is highly adaptable, alterations to the media (i.e., mTeSR™1 cGMP Pluripotent Stem Cell; StemCell Technologies, Cat #85857), use of antibiotics in culture (i.e., Antibiotic-Antimycotic; Gibco, Cat #15240096), gRNA or template (i.e., plasmid vector), RNP-complex delivery method (i.e., lipid based delivery vehicles), transplantation dose, number of transplantation sites (i.e., unilateral injection), or CSF1R-inhibitor (i.e., PLX5622) may significantly change the amount of time needed to generate, differentiate and replace murine microglia with human microglia. Alteration to any reagent may require further optimization and change the prep time and efficiency of clonal isolation and expansion of correctly edited G795A iPSCs. Variability in proliferation rates between iPSC lines may also contribute.
Furthermore, proliferation and eventual microglia replacement were observed under treatment of 290 mg/kg PLX3397 formulated in AIN-76A standard chow after transplantation and recovery of mice. Utilizing lower concentrations of PLX3397, alternative routes of administration (i.e., IP injections), pre-treatment methods, or different CSF1R inhibitors (i.e., PLX5622, Edicotinib, or BLZ945) may require either longer or shorter durations to achieve equivalently robust microglia replacement of the murine brain.
Troubleshooting
Problem 1
Poor viability of electroporated cells after 24 h recovery (see CRISPR-edit iPSCs with CSF1R-G795 targeting RNP and template step 19).
Potential solution
The time necessary to digest adhered iPSCs may need to be optimized per cell line and plating density. Larger colonies will require additional time to separate and lift; however, extended incubation in accutase may result in poor viability of collected cells. Therefore, it is recommended to plate iPSCs at high density (i.e., 1 × 106 viable cells per one well of a 6-well plate) for enzymatic digestion.
Problem 2
iPSC clones differentiate when passage from 96-well plate to 48-well plate (see single-cell expansion and screening of CRISPR-edited iPSCs step 27).
Potential solution
If passaged too late, iPSCs can become overgrown and stressed within the 96-well plate. It is important to passage before visible signs of cell death or excessive cell-shedding. This is especially important if working with a highly proliferative iPSC line. Therefore, it is recommended to passage earlier to ensure robust growth and maintenance of iPSC clones in culture.
Problem 3
Low yield of G795-variant HPCs (see G795A-iPSC differentiation into Hematopoietic progenitor cells and iPSC-Microglia step 30).
Potential solution
We have found culturing iPSCs in a richer media (i.e., mTeSR1; STEMCELL Technologies #8585) for at least three passages prior to plating iPSCs for HPC differentiation primes the cells for more robust differentiation and yields a greater number of HPCs on Day 10.
Problem 4
Poor engraftment of G795A-iMGs (see transplantation of G795A-HPCs or G795A-iMGs step 38).
Potential solution
iMGs prepared for transplantation should be kept on ice prior to surgery. However, an extended period of suspension in DPBS may lead to cell death. Therefore, we recommend keeping iMGs on ice for no more than 4 h before transplantation.
Problem 5
Incomplete microglia replacement (see CSF1Ri-mediated Microglia Replacement in G795A transplanted mice step 39–40).
Potential solution
The treatment paradigm tested in this protocol has been reproduced using three different iPSC lines in two mouse models by two independent laboratories. However, different genetic backgrounds may contribute to altered rates of iMG proliferation in vivo. Therefore, an extended period of treatment may be required to achieve complete microglia replacement.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mathew Blurton-Jones (mblurton@uci.edu).
Materials availability
G795A-iPSC cell lines generated in this study are available upon request from the lead contact via email or https://stemcells.mind.uci.edu/.
Acknowledgments
This work was supported by NIH T32 AG073088 (J.P.C.), NIH R01 AG061895 (H.D.), NIH T32 GM008076 (S.I.L.), The Klingenstein-Simons fellowship in neuroscience (F.C.B.), NIH P30 AG066519 (M.B.-J.), NIH R43 NS125730 (M.B.-J.), and NIH R01 DA048813 (M.B.-J.). Graphical abstract and Figure 1 were created with BioRender.com.
Author contributions
J.P.C. drafted the protocol and constructed the figures. M.B.-J. and H.D. reviewed and revised the protocol. S.I.L. and F.C.B. independently validated the microglia replacement protocol. All authors read and approved the final manuscript.
Declaration of interests
M.B.-J., J.P.C., and H.D. are co-inventors on a pending patent filed by the University of California Regents (application 63/169,578) related to genetic modification of cells to confer resistance to CSF1R antagonists. M.B.-J. is a co-inventor of patent WO/2018/160496, related to the differentiation of human pluripotent stem cells into microglia. M.B.-J. is a co-founder of NovoGlia Inc. F.C.B. is a co-inventor on a pending patent filed by The Board of Trustees of The Leland Stanford Junior University (application 16/566,675) related to methods of microglia replacement.
Contributor Information
Jean Paul Chadarevian, Email: jchadare@uci.edu.
Mathew Blurton-Jones, Email: mblurton@uci.edu.
Data and code availability
This paper does not report original code.
References
- 1.Chadarevian J.P., Lombroso S.I., Peet G.C., Hasselmann J., Tu C., Marzan D.E., Capocchi J., Purnell F.S., Nemec K.M., Lahian A., et al. Engineering an inhibitor-resistant human CSF1R variant for microglia replacement. J. Exp. Med. 2023;220 doi: 10.1084/jem.20220857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okamoto S., Amaishi Y., Maki I., Enoki T., Mineno J. Highly efficient genome editing for single-base substitutions using optimized ssODNs with Cas9-RNPs. Sci. Rep. 2019;9:4811. doi: 10.1038/s41598-019-41121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucito R., Healy J., Alexander J., Reiner A., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaffer L.G., Bejjani B.A., Torchia B., Kirkpatrick S., Coppinger J., Ballif B.C. The identification of microdeletion syndromes and other chromosome abnormalities: cytogenetic methods of the past, new technologies for the future. Am. J. Med. Genet. C Semin. Med. Genet. 2007;145c:335–345. doi: 10.1002/ajmg.c.30152. [DOI] [PubMed] [Google Scholar]
- 5.McQuade A., Coburn M., Tu C.H., Hasselmann J., Davtyan H., Blurton-Jones M. Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol. Neurodegener. 2018;13:67. doi: 10.1186/s13024-018-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abud E.M., Ramirez R.N., Martinez E.S., Healy L.M., Nguyen C.H.H., Newman S.A., Yeromin A.V., Scarfone V.M., Marsh S.E., Fimbres C., et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron. 2017;94:278–293.e9. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasselmann J., Coburn M.A., England W., Figueroa Velez D.X., Kiani Shabestari S., Tu C.H., McQuade A., Kolahdouzan M., Echeverria K., Claes C., et al. Development of a Chimeric Model to Study and Manipulate Human Microglia In Vivo. Neuron. 2019;103:1016–1033.e10. doi: 10.1016/j.neuron.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This paper does not report original code.