Abstract
Sendai virus (SeV) is highly pathogenic for mice. In contrast, mice (including SCID mice) infected with simian virus 5 (SV5) showed no overt signs of disease. Evidence is presented that a major factor which prevented SV5 from productively infecting mice was its inability to circumvent the interferon (IFN) response in mice. Thus, in murine cells that produce and respond to IFN, SV5 protein synthesis was rapidly switched off. In marked contrast, once SeV protein synthesis began, it continued, even if the culture medium was supplemented with alpha/beta IFN (IFN-α/β). However, in human cells, IFN-α/β did not inhibit the replication of either SV5 or SeV once virus protein synthesis was established. To begin to address the molecular basis for these observations, the effects of SeV and SV5 infections on the activation of an IFN-α/β-responsive promoter and on that of the IFN-β promoter were examined in transient transfection experiments. The results demonstrated that (i) SeV, but not SV5, inhibited an IFN-α/β-responsive promoter in murine cells; (ii) both SV5 and SeV inhibited the activation of an IFN-α/β-responsive promoter in human cells; and (iii) in both human and murine cells, SeV was a strong inducer of the IFN-β promoter, whereas SV5 was a poor inducer. The ability of SeV and SV5 to inhibit the activation of IFN-responsive genes in human cells was confirmed by RNase protection experiments. The importance of these results in terms of paramyxovirus pathogenesis is discussed.
Paramyxoviruses show marked differences in host range. For example, simian virus 5 (SV5, or canine parainfluenza virus) causes kennel cough in dogs (18) and also naturally infects monkeys and humans (8, 10) but causes only self-limiting infections in mice (27, 39). In contrast, while there is no evidence that wild rodents are infected with sendai virus (SeV), and its natural host remains unknown, SeV causes serious outbreaks of disease in colonies of laboratory mice and rats. It can also infect a number of other rodents, including hamsters, guinea pigs, and rabbits. Furthermore, it appears that SeV was prevalent among pigs in Japan in the 1950s but has subsequently disappeared from the pig population (12).
The ability of viruses to infect and cause disease in a given species of animals is undoubtedly a consequence of very complicated interactions between the virus and host, both at the molecular level and at the level of the whole organism. Thus, many factors may contribute to differences in pathogenesis and host range between two related viruses, including virus cell tropism, the cytopathic effects of virus infection, and the sensitivity of virus replication to adaptive and innate immune responses. Many of the studies on paramyxovirus pathogenesis have concentrated on the importance of the virus glycoproteins in determining cell tropism (24). Thus, for example, the pathogenicity of SeV is clearly affected by the properties of the fusion protein (35, 36). Like other paramyxoviruses, the precursor form of the fusion protein, F0, has to be cleaved into two disulphide-linked subunits to attain biological activity (9, 29). However, in SeV the cleavage domain is not recognized by proteases in the trans-Golgi compartments of most tissue culture cells. The restricted pneumotropism of SeV in mice can also be partially explained by the observation that while infectious virus is produced in mouse lungs, cells from nonpermissive tissues do not cleave the fusion protein (36). A consequence of the inability of tissue culture cells to cleave F0 relevant to the studies reported here is that in the various cell lines used there was little infectious virus produced and no obvious cell-to-cell spread of SeV. However, although undoubtedly important for virus pathogenesis, given the rapid mutation rate of RNA viruses, it seems likely that if entry into cells was the only major constraint on host range, paramyxoviruses would cross species barriers much more readily than they do, especially if the host cell receptor is a common determinant such as sialic acid.
Much less is known about how immune responses, including cellular antiviral responses induced by the interferons (IFNs), influence paramyxovirus pathogenesis and host range. Indeed, given that many viruses have evolved specific mechanisms for countering the IFN response (23, 30, 31), the efficiency with which viruses counter host cell restrictions imposed on viral infection by cellular antiviral defense mechanisms might be expected to be an important contributory factor to virus pathogenesis. Since paramyxoviruses, like all viruses, have evolved their replication strategies in vivo, we speculated that they, too, may have evolved some mechanism for countering, or minimizing, IFN-induced antiviral responses.
There are two types of IFN. Alpha/beta IFN (IFN-α/β) is produced as a direct response to viral infection and consists of two major subclasses, the products of the IFN-α multigene family, which are synthesized predominantly by leukocytes, and the product of the single IFN-β gene, which is synthesized by most cell types, but especially by fibroblasts. IFN-γ consists of the product of the IFN-γ gene and is synthesized by activated T lymphocytes. The effects of IFNs are initiated by the binding to their cellular receptors. Although there are distinct receptors for IFN-α/β and IFN-γ, there is partial overlap in their signal transduction pathways, and a number of genes are induced by both types (34). The induced genes play major roles in the antiviral defense mechanism. For example, both IFN-α/β and IFN-γ induce the synthesis of the enzyme PKR. Prior to viral infection, this enzyme is inactive, but as a consequence of double-stranded RNA production during infection, PKR becomes activated and can switch off translation, thus limiting the reproductive capacity of the virus (7). IFNs also induce 2′5′-oligoadenylate synthetase (23), which, together with RNase L, results in accelerated RNA degradation and thus also an inhibition of protein synthesis. Other cellular antiviral products induced by IFN include the Mx proteins, but their mechanisms of action are poorly understood. The general importance of the IFNs in controlling virus infection can be deduced from the fact that transgenic mice lacking IFN-α/β receptors, IFN-γ receptors, or both are unable to cope with a variety of different virus infections (21, 38).
The means by which IFNs induce transcription has been elucidated in detail. IFNs-α/β induce the assembly of a heterotrimeric transcription factor (ISGF3) containing a DNA-binding subunit, p48, and the tyrosine-phosphorylated signal transducers and activators of transcription, STAT1 and STAT2 (3, 11). ISGF3 binds to the IFN-stimulated response element (ISRE) in target genes and activates transcription (33). IFN-γ induces the formation of homodimeric STAT1, which binds to the gamma-activated sequence in the regulatory regions of target genes and activates transcription. Although many viruses encode gene products which interfere with the biological activity of the IFNs or the cellular antiviral mechanisms induced by IFNs (23, 30, 31), it is less clear whether viruses have developed strategies to block transcriptional responses to IFNs and thus prevent the synthesis of the antiviral products.
In this report, we present evidence that both SV5 and SeV have the ability to circumvent IFN-induced antiviral responses by blocking IFN signalling in cells from their natural host. We also show that SV5 cannot properly overcome these antiviral responses in murine cells, thus offering an explanation as to why SV5 establishes only a self-limiting infection in mice. However, since we also demonstrate that SeV can block IFN signalling in human cells, clearly other factors influence virus host range.
MATERIALS AND METHODS
Cells and viruses.
Murine BF cells (cloned from a primary cell culture of a BALB/c mouse embryo), human MRC 5 (human fetal lung fibroblasts), 2fTGH (19) and MG-63 (ATCC CRL 1650), 2D9 (human glioblastoma cell line) and HFF (human foreskin fibroblasts) cells, and monkey Vero cells were grown as monolayers in 25- or 75-cm2 tissue culture flasks or on 9-cm-diameter plastic petri dishes (Nunc) in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (growth medium). We are grateful to Tony Meager, National Institute for Biological Standards and Control, Potters Bar, United Kingdom, for providing the MRC 5, 2D9, and HFF cells. All cell lines were negative for mycoplasmas as screened by 4′,6-diamidino-2-phenylindole (DAPI) staining. Mouse cells were treated with recombinant human αA/D IFN (rHuIFN-αA/D) (28) kindly supplied by Hoffmann-La Roche Inc. (Nutley, N.J.) at 100 or 1,000 IU/ml (see text) in medium containing 2% bovine serum (maintenance medium). Human cells were treated with either rHuIFN-αA/D or Wellferon (a mixture of IFN-α subtypes produced by lymphoblastoid cells [lot 72, a kind gift of Glaxo-Wellcome]) added to cells at 1,000 IU/ml in maintenance medium.
The strain of SV5, designated W32, was grown and titrated under appropriate conditions in Vero cells with maintenance medium. Sendai virus, strain H was grown in eggs and titrated in Vero cells in the presence of trypsin.
Antibodies.
A detailed description of the monoclonal antibodies (MAbs) to SV5 and their nomenclature has been given elsewhere (26). The MAbs to SeV were a kind gift from Allen Portner (St. Jude Children’s Research Hospital, Memphis Tenn.).
Detection of the P protein in SV5-infected SCID mice.
SCID mice (1), while anesthetized, were infected by inhalation of 5 × 106 PFU of the W3 strain of SV5 in 75 ml of culture medium. At various times after infection, the mice were killed, and the lungs were removed and frozen at −70°C until required. The relative amount of the P protein in the lungs was estimated by Western blot analysis as has been described in detail elsewhere (39).
Preparation of radiolabelled antigen extracts, immunoprecipitation, and SDS-polyacrylamide gel electrophoresis.
BF cell monolayers (with or without IFN; see text) in 25-cm2 tissue culture flasks were infected with 5 PFU of SV5 or SeV per cell. After an adsorption period of 2 h at 37°C, the inoculum was removed and replaced with maintenance medium. At various times postinfection (p.i.) the cells were radioactively labelled for 2 h with l-[35S]methionine (500 Ci/mmol; Amersham International, Ltd.) in tissue culture medium containing one-tenth the normal concentration of methionine (i.e., 1.5 mg/liter). At the end of the labelling interval, the cells were washed in ice-cold phosphate-buffered saline (PBS) and lysed into immune precipitation buffer (10 mM Tris-HCl [pH 7.8], 5 mM EDTA, 0.5% Nonidet P-40, 0.65 M NaCl, and 0.1% sodium dodecyl sulfate (SDS); 4 × 106 to 6 × 106 cells per ml of buffer) by sonication with an ultrasonic probe. Soluble antigen extracts were obtained after the particulate material was pelleted from the total cell antigen extracts by centrifugation at 400,000 × g for 30 min. Immune complexes were formed by incubating (for 2 h at 4°C) 1-ml samples of the soluble antigen extracts with an excess of either anti-SV5 MAbs to the HN, F, P, M, and NP proteins (1 ml of concentrated tissue culture fluid) or individual anti-SeV MAbs to the HN, F, and P proteins (1 ml of ascitic fluid). The immune complexes were isolated (13) on an excess of a fixed suspension of the Cowan A strain of Staphylococcus aureus (20 μl of a 10% [wt/vol] suspension per μl of concentrated tissue culture fluid or ascitic fluid for 30 min at 4°C). The proteins in the immune complexes were dissociated by heating (100°C for 5 min) in gel electrophoresis sample buffer (0.05 M Tris-HCl [pH 7.0], 0.2% SDS, 5% 2-mercaptoethanol, and 5% glycerol) and analyzed by electrophoresis through SDS–15% polyacrylamide gel cross-linked with N,N′-methylene-bisacrylamide. After electrophoresis, gels were fixed stained and dried; labelled polypeptides were visualized by autoradiography, and the amount of radioactivity in each polypeptide was estimated by phosphoimage analysis.
Immunofluorescence.
Cells to be stained for immunofluorescence were grown on multispot microscope slides (C.A. Hendley Ltd., Essex, United Kingdom). The cells were treated and stained with specific MAbs as has been described in detail elsewhere (25). Briefly, monolayers were fixed with 5% formaldehyde, 2% sucrose in PBS for 10 min at 20°C, permeabilized with 0.5% Nonidet P-40, 10% sucrose in PBS for 5 min at 20°C, and washed three times in PBS containing 1% calf serum. Cells were stained by indirect immunofluorescence with the appropriate MAbs with rhodamine-conjugated rabbit anti-mouse immunoglobulin. In addition, cells were stained with the DNA-binding fluorochrome DAPI. Following staining for immunofluorescence, the monolayers of cells were examined with a Nikon Microphot-FXA immunofluorescence microscope.
Plasmid DNAs.
Descriptions of the plasmids used have been given elsewhere. Briefly, the control HSV TK plasmid contains a −105 to −15 fragment (tkΔ −105) of the herpes simplex virus (HSV) thymidine kinase (tk) promoter fused to −17 of the firefly luciferase (4) cassette [the full name of the plasmid is ptkΔ(−105)lucter] (14). The IFN-β promoter containing plasmid contains IFN-β sequences from −125 to +72 fused to the firefly luciferase gene [full name, pIFΔ(−125)lucter] (14). The IFN-α/β-responsive plasmid [termed p(9-27)4tkΔ(−39)lucter] (15) contained four tandem repeat sequences of the ISRE from the IFN-inducible gene, 9-27, fused to the firefly luciferase gene. pJATlacZ, a plasmid used as a transfection standard, contains a β-galactosidase gene under the control of the rat β-actin promoter (16).
Transient transfections.
BF cells or 2fTGH cells were transfected with 0.5 μg of DNA and 2 μl of Lipofectamine (Life Technologies Inc.) according to the manufacturer’s instructions. After 18 h, the cells were infected with SeV or SV5 and induced with 1,000 U of rHuIFN-αA/D per ml at 18 or 24 h p.i. Cells were lysed at 4 h after induction by IFN. Lysates were prepared and assayed for luciferase and β-galactosidase activity as described previously (14). The relative expression levels were calculated by dividing the luciferase values by the β-galactosidase values. The experiments presented were repeated several times with equivalent results.
RNA isolation and RNase protection.
Total cellular RNA was prepared from 9-cm-diameter plastic petri dishes of confluent cultures of mouse or human cells treated as indicated and analyzed by RNase protection as described previously (42) with probes for human IFN-β (42), mouse IFN-β (5), human IRF-1 and -6-16 (7a), or human and mouse γ-actin (5).
RESULTS
Replication of SV5 in SCID mice.
We previously reported that following intranasal infection of immunocompetent mice with SV5 there was a wave of virus RNA transcription and protein synthesis. However, infected mice showed no overt signs of disease and cleared the infection by 7 days postinfection (27). Adoptive transfer experiments demonstrated that virus clearance was primarily dependent on CD8+-T-cell responses. In mice which had been immunocompromised by X-irradiation, SV5 established a prolonged infection, but again these mice showed no overt signs of disease, and the virus was eventually cleared once the immune system in these mice began to recover (39). To further examine the replication of SV5 in immunocompromised animals, SCID mice (1) were infected with SV5, and at various times p.i. groups of mice were sacrificed, and the presence of the P protein in the lungs was detected by Western blot analysis (Fig. 1). There was an obvious increase in the amount of the P protein in lung extracts between 0 and 4 days p.i. (Fig. 1, compare lanes 2 to 4), demonstrating that the mice had been infected with SV5 and that some virus protein synthesis had taken place. After this time, the amount of P protein remained relatively constant until 13 days p.i.; thereafter, a decrease in the amount of P was detected. At no time throughout the infection did any of the SCID mice show overt signs of disease. These results emphasized the restricted nature of SV5 replication in mice and clearly demonstrated that in the absence of an adaptive immune response, other responses are capable of limiting the replication and spread of SV5 in mice.
FIG. 1.
Autoradiogram of a Western blot used to detected the P and V proteins of SV5 in extracts of BF cells infected with SV5 for 24 h (lane 1) or in lung extracts of mock-infected SCID mice (lane 2) or SCID mice that were infected with SV5 for 1, 4, 9, 13, or 21 days (lanes 3 to 7, respectively).
Comparisons between the replication of SV5 and SeV in murine cells that produce and respond to IFN-α/β.
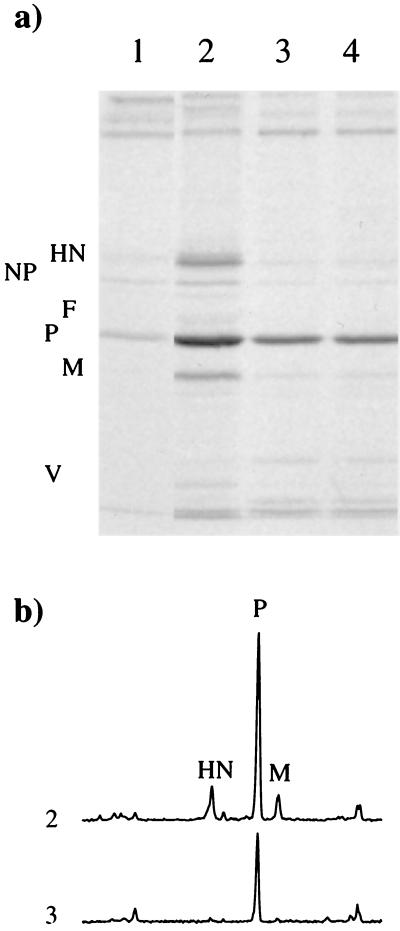
The inability of SV5 to cause overt disease in mice clearly contrasts with the potentially lethal disease that can follow SeV infection of mice. We have previously shown that SV5 can replicate efficiently in mouse cells if these cells are rendered unable to respond to IFN-α/β either by the use of anti IFN-β antibodies or by IFN-α/β receptor knockouts (40), suggesting that one possible reason for difference in pathogenicity between SV5 and SeV lies in differences in their interaction with cellular antiviral responses induced by IFNs. In agreement with our previous results (6, 40), following infection of murine BF cells with SV5 at 24 h p.i., there was a significant level of virus protein synthesis at 24 h p.i. (Fig. 2, lane 2). However, by 3 days p.i., at a time when the cells had begun to produce and respond to IFN, there was a dramatic reduction in the level of ongoing SV5 protein synthesis (Fig. 2, lane 3). Addition of exogenous IFN to the culture medium of the infected cells at 24 h p.i. did not further reduce the amount of protein synthesis observed at 3 days p.i. compared to that observed in untreated cells (Fig. 2, compare lanes 3 and 4). Phosphoimage analysis of the amount of radioactivity in the different immunoprecipitated virus proteins demonstrated that initially there was a greater reduction in the amount of the HN, F, and M proteins being made than in the P proteins (or NP as detected in total cell lysates; data not shown). Pretreatment of BF cells with IFN 24 h prior to infection with SV5 also dramatically reduced the amount of virus protein synthesis observed at 24 h p.i. compared with that observed in untreated cells (Fig. 2, compare lanes 1 and 2). Immunofluorescence data suggested that the virus protein synthesis observed in IFN-pretreated cells at 24 h p.i. was occurring primarily in a small percentage of cells which appeared to be making normal levels of virus proteins (data not shown).
FIG. 2.
Analysis of 35S-labelled polypeptides present in immune precipitates (a) formed by the reaction of a pool of MAbs specific for the HN, NP, F, M, and P or V proteins of SV5 with soluble antigen extracts made from BF cells infected with SV5 for 1 day (lanes 1 and 2) or 3 days (lanes 3 and 4). The cells were pretreated with IFN-α/β (100 IU/ml) 24 h prior to infection (lane 1) or left untreated (lanes 2 to 4). At 24 h p.i., exogenous rHuIFN-alphaA/D (100 IU/ml) was added to the culture medium of cells used to make the extract shown in lane 4. The amount of 35S label in the precipitated polypeptides was quantitated by phosphoimage analysis, and the profiles of lanes 2 and 3 are shown in panel b.
A similar set of experiments, whose results are shown in Fig. 3, were carried out following infection of BF cells with SeV. It is clear that while pretreatment of the cells with IFN reduced the amount of SeV protein synthesis (compare untreated and treated cells at 24 h p.i.; Fig. 3, lanes 1 and 2) the levels of SeV protein synthesis in untreated cells were similar at 1 and 3 days p.i. (Fig. 3, compare lanes 2 and 3). This result contrasts with the result seen with SV5 and suggests that SeV-infected cells are insensitive to the downregulation of gene expression induced by any IFN-α/β that is produced. Comparative measurements of the relative amounts of IFN secreted into the culture medium of SV5- and SeV-infected BF cells demonstrated that the continued replication of SeV in BF cells could not be explained by the fact that SeV-infected cells produced less IFN than SV5-infected cells. SeV-infected cultures induced slightly more IFN than SV5-infected cultures (e.g., at 48 h p.i., SV5-infected cultures had produced 25 to 50 IU of IFN, and SeV cultures had produced 100 to 200 IU of IFN per 105 cells, respectively). Furthermore, the addition of exogenous IFN to the culture medium of SeV-infected BF cells at 24 h p.i. did not reduce the levels of virus protein synthesis observed at 3 days p.i. (Fig. 3, lane 4). Phosphoimage analysis of the amount of radioactivity in the P, F, and HN proteins also showed that there was no significant change in the relative amounts of these proteins being synthesized at 1 and 3 days p.i. (data not shown).
FIG. 3.
Analysis of 35S-labelled polypeptides present in immune precipitates formed by the reaction of MAbs specific for the P (a), HN (b), and F (c) proteins of SeV with soluble antigen extracts made from BF cells infected with SeV for 1 day (lanes 1 and 2) or 3 days (lanes 3 and 4). The cells were pretreated with IFN 24 h (100 IU/ml) prior to infection (lane 1) or left untreated (lanes 2 to 4). At 24 h p.i., exogenous IFN (100 IU/ml) was added to the culture medium of cells used to make the extract shown in lane 4.
Immunofluorescence analysis of BF cells infected with SeV.
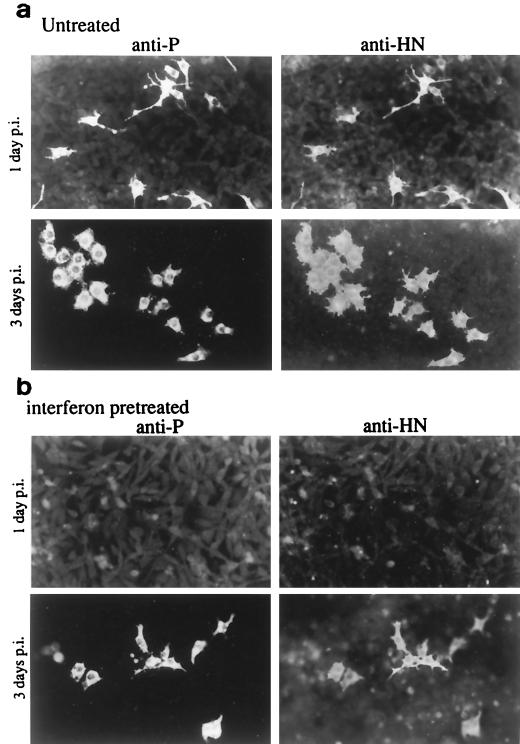
Previous immunofluorescence analysis of BF cells infected with SV5 revealed that while all the virus proteins could be detected at 1 day p.i., with time, and following the production of IFN, reduction in the percentage of cells positive for HN was much more rapid than that of cells positive for P. Thus, at 4 days p.i., while >90% of the cells remained positive for P, only 0.1 to 1.0% of the cells were positive for HN. Indeed, the cellular antiviral responses induced by IFN were so effective against SV5 that by 14 days p.i., the majority of the cells had cleared the infection (40). To compare the effects of IFN on individual cells infected with SeV, monolayers of BF cells were infected at a low multiplicity of infection (MOI), and the presence of the HN and P proteins was detected by immunofluorescence. In contrast to the situation with SV5, at 3 days p.i., every SeV-infected cell that was positive for P was also positive for HN (Fig. 4a). Furthermore, not only did SeV protein synthesis continue in the presence of IFN (Fig. 3), but the BF cells also survived the infection. Indeed, it can be seen in Fig. 4a that at 3 days p.i., all the infected cells appear to have divided (every infected cell is one of a pair of infected cells). Pretreatment of BF cells with IFN did, however, significantly reduce the number of cells that expressed SeV proteins. Nevertheless, unlike the situation with SV5, even in IFN-pretreated monolayers, at 3 days p.i., significant numbers of cells that were positive for both the HN and the P proteins were detected (Fig. 4b).
FIG. 4.
Photographs showing the localization of the P and HN proteins in monolayers of BF cells untreated (a) or treated with IFN (b) 24 h prior to infection with SeV. Monolayers were fixed at 1 and 3 days p.i. prior to staining with the appropriate MAbs.
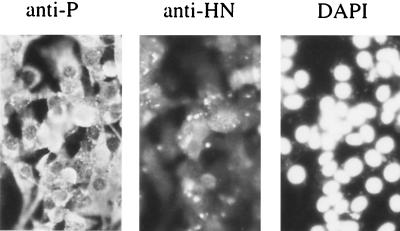
To confirm that BF cells did not die following infection with SeV, monolayers of BF cells were infected with SeV at 5 to 10 PFU/cell. Immunofluorescence analysis at 24 h p.i. confirmed that all the cells were infected with virus. The monolayer remained intact 4 days p.i.; the infected cells could be continuously passaged. Again in contrast to the situation with SV5, upon passage, the majority of cells remained positive for both the P and the HN proteins (Fig. 5).
FIG. 5.
Photographs showing the localization of the P and HN proteins of SeV in BF cells infected at a high MOI with SeV and passaged twice over a 2-week period. The cells were also stained with DAPI. As can be seen, all the cells remained infected with SeV.
Interaction of SV5 and SeV with human cells that produce and respond to IFN.
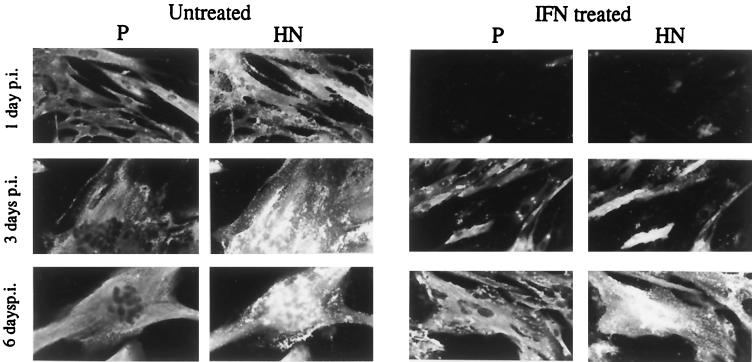
In a series of experiments analogous to those described above, the interaction of SV5 and SeV with human cells that produce and respond to IFN was examined. In contrast to the situation in murine BF cells, once established in human cells, SV5 or SeV protein synthesis continued, even in the presence of IFN. However, as with BF cells, if the cells were pretreated with IFN 24 h prior to infection, there was a marked reduction in the number of cells that expressed detectable levels of virus proteins at 24 h p.i. These results are illustrated for SV5 in Fig. 6. In this experiment, the culture medium of MRC-5 cells was supplemented with IFN 24 h prior to infection with SV5 or left untreated. The cells were then infected with SV5 at an MOI of 5. At 1, 3, and 6 days p.i., the monolayers were fixed and stained with MAbs specific for the HN and P proteins. As illustrated, pretreatment of the cells with IFN significantly reduced the percentage of cells that expressed SV5 proteins at 24 h p.i. However, in contrast to the situation which occurred in BF cells, SV5 eventually managed to overcome the antiviral response induced by IFN and by 3 days p.i., the majority of cells that were pretreated with IFN were positive for both the HN and P proteins. However, even at 6 days p.i., the level of virus-induced cell-cell fusion observed in the IFN-pretreated monolayers was not as extensive as that observed in untreated monolayers at 3 days p.i. Furthermore, the IFN-treated monolayers survived the infection better than the untreated monolayers, in which the majority of cells fused and eventually detached from the culture dish. These observations held true for two independent human cell lines that respond to IFN, namely, HFF and 2D9 cells.
FIG. 6.
Photograph showing the localization of the P and HN proteins of SV5 in human MRC-5 cells pretreated with IFN (100 IU/ml) 24 h prior to infection or left untreated. Cells were fixed at 1, 3, and 6 days p.i. prior to staining with the appropriate MAbs.
Effects of SeV and SV5 infection on IFN-α/β signalling in murine and human cells.
To begin to elucidate the molecular basis for the differential sensitivity of SV5 and SeV to IFN in human and murine cells, a series of experiments were undertaken which examined the effect of SeV and SV5 infections on the activation of a synthetic promoter containing multimers of the well-defined ISRE from the 9-27 gene. This promoter is linked to a luciferase reporter gene, and the resultant IFN-α/β-responsive plasmid was transiently transfected into mouse or human cell lines. As a control for any general effect of virus infection on promoter activity, cells were also transfected with a control plasmid that expressed the luciferase gene under the control of the HSV TK promoter. To control for transfection efficiencies, cells were cotransfected with a control plasmid that expressed β-galactosidase under the control of the rat β-actin promoter. The luciferase results were then corrected for relative β-galactosidase activity.
(i) Promoter activity in murine cells.
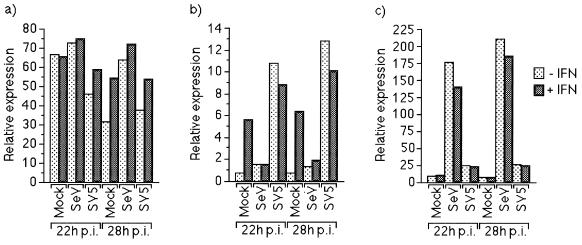
We first examined the responsiveness of the reporter gene in mouse BF cells. At 18 h posttransfection, the cells were infected with either SeV or SV5, and at 18 and 24 h p.i., IFN-α/β was added to the culture medium. Four hours later (i.e., at 22 or 28 h p.i.), the cells were harvested, and the level of luciferase activity measured. Figure 7b shows that the IFN-α/β-responsive promoter was strongly activated both in SV5-infected cells and in mock-infected cells that were treated with IFN. In marked contrast, little or no activation of the IFN-α/β-responsive promoter was observed in SeV-infected cells, irrespective of IFN treatment. The strong activation of the IFN-α/β-responsive promoter in SV5-infected cells in the absence of exogenous IFN was shown to be a result of SV5-infected cells secreting IFN-β, since activation could be blocked by adding IFN-β antibodies into the culture medium (data not shown). The lack of activation of the IFN-α/β-responsive promoter in BF cells infected with SeV was not a general consequence of virus infection on the activation of cellular genes, as neither infection with SeV or SV5 had any striking effect on the level of luciferase activity when the gene was under the control of the HSV TK promoter (Fig. 7a).
FIG. 7.
SeV, but not SV5, can block activation of the Type I IFN-responsive promoter in BF cells. BF cells were transfected with 0.3 and 0.1 μg of control plasmids, pUC13 and pJATlacZ, respectively, and 0.1 μg of one of the HSV TK promoter containing-plasmid (a), the IFN-α/β-responsive plasmid (b), or the IFN-β promoter-containing plasmid (c). At 16 h posttransfection, the cells were infected with SeV or SV5. Eighteen or twenty-four hours postinfection, the culture medium was supplemented with IFN or left untreated as indicated. Four hours later, luciferase and β-galactosidase activities in cellular lysates were measured. Luciferase activity, expressed in relative light units, was normalized to β-galactosidase activity.
The induction of IFN-β by SV5-infected BF cells was confirmed by analyzing (i) the activity of the IFN-β promoter in transient transfections (Fig. 7c) and (ii) BF cell RNA for the presence of specific IFN-β transcripts with RNase protection (Fig. 8). Strikingly, when SeV-infected BF cells were examined, SeV was a much stronger inducer of the IFN-β promoter than SV5 (Fig. 7c) and also produced more IFN-β transcripts than SV5-infected cells (Fig. 8). The failure of SeV-infected cells to respond to the substantial levels of IFN-α/β they produce provides a striking demonstration of the effectiveness of the SeV-induced block.
FIG. 8.
SeV and SV5 induce IFN-β mRNA in murine BF cells. BF cells were infected with either SeV or SV5 for 24 h. Twenty micrograms of total cellular RNA from cells infected with SeV or SV5 was mapped with RNase protection probes corresponding to mouse IFN-β (Mif) or γ-actin mRNAs, and the protected fragments are indicated at the right.
(ii) Promoter activity in human cells.
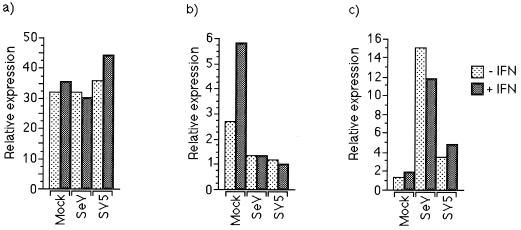
In a set of experiments similar to those described above, the ability of SeV and SV5 to interfere with the activation of the IFN-α/β-responsive promoter was examined in human cells. 2fTGH cells were transfected with the appropriate plasmids and infected 18 h posttransfection with SV5 or SeV or mock infected. At 24 h p.i., the culture medium on transfected cells was supplemented with IFN-α/β. At 28 h p.i., the cells were harvested and the relative levels of promoter activation were estimated by measuring luciferase activity. Infection with neither SeV nor SV5 had a marked effect on the activity of a control plasmid in which the luciferase gene was under the control of the HSV TK promoter (Fig. 9a). However, in striking contrast to the result seen in BF cells both SV5 and SeV inhibited the activation of the IFN-α/β-responsive promoter in 2fTGH cells (Fig. 9b). Again, both viruses induced the activity of the IFN-β promoter, with SeV being much more effective than SV5 (Fig. 9c); these results were also reflected at the level of IFN-β-specific transcripts (data not shown).
FIG. 9.
SeV and SV5 block activation of the IFN-α/β-responsive promoter in 2fTGH cells. Cells were transfected with 0.3 and 0.1 μg of control plasmids pUC13 and pJATlacZ, respectively, and 0.1 μg of one of the HSV TK promoter-containing plasmid (a), the IFN-α/β-responsive plasmid (b), or the IFN-β promoter-containing plasmid (c). At 16 h postinfection, cells were infected with either SeV or SV5, and at 18 or 24 h p.i. the culture medium was supplemented with IFN or left untreated as indicated. Four hours later, luciferase and β-galactosidase activities in cellular lysates were measured. Luciferase activity, expressed in relative light units, was normalized to β-galactosidase activity.
SV5 and SeV inhibit the induction of the IFN-responsive 6-16 gene.
Type I IFN induces the transcription of a number of cellular genes, including 6-16 and IRF-1. To determine whether the transient transfection experiments described above mirrored the induction of IFN-α/β-responsive genes in situ, the relative levels of 6-16 and IRF-1 mRNA in human cells infected with SeV and SV5 following treatment with IFN-α/β was determined by RNase protection. It is clear from the results presented in Fig. 10 that SeV and SV5 infection inhibited IFN-α/β induction of 6-16 mRNA. IFN-α/β induction of IRF-1 was significantly less marked than 6-16, a consequence of the kinetics of IFN induction in which IRF-1 mRNA levels decline significantly from their peak by 18 h. Nevertheless, infection with both SV5 and SeV appeared to reduce the level of IRF-1 mRNA to that observed in untreated, mock-infected cultures. In contrast, no obvious effect of virus infection on the relative amounts of actin mRNA was observed.
FIG. 10.
SeV and SV5 block the induction of IFN-responsive gene mRNAs in MG-63 cells. MG-63 cells were infected with either SeV or SV5, and at 24 h p.i. the culture medium was supplemented with IFN for 4 h or left untreated. Twenty micrograms of total cellular RNA from cells was mapped with RNase protection probes corresponding to human IFN-β (5′IF), 6-16, IRF-1, or γ-actin mRNAs, and the protected fragments are indicated at the right.
DISCUSSION
The results presented here clearly demonstrate that both SV5 and SeV are capable of continued virus protein synthesis in the presence of IFN in cells derived from species that they naturally infect. We also present evidence that both SV5 and SeV circumvent the IFN response by interfering with the transcriptional activation of IFN-responsive genes. While many viruses have the ability to inhibit IFN responses, they usually achieve this by blocking enzymes such as PKR (23, 30, 31). There is, however, a precedent for a virus blocking IFN signalling, namely, human herpesvirus 8 (HHV-8) encodes an IFN regulatory factor (vIRF), which inhibits responses to both IFN-α/β and IFN-γ (41). Clearly there are potentially many advantages to the ability of a virus to block IFN signalling. In addition to the ability of IFN to induce genes such as those encoding PKR, 2′,5′-oligoadenylate synthetase, and the Mx proteins, IFN upregulates class 1 MHC molecules, making cells more susceptible to cytotoxic T lymphocyte activity. Furthermore, IFNs activate monocytes/macrophages, cytotoxic T cells, and NK cells and are critical mediators of inflammatory immune responses. The ability of SV5 and SeV to inhibit IFN signalling appears to be dependent upon virus gene expression. However, since these viruses remain sensitive to pretreatment of cells with IFN, it seems surprising that they have not also evolved a mechanism to inhibit or prevent IFN production. However, the efficiency with which different isolates and strains of viruses induce IFN may vary (17), and low producers may be selected for in natural infections. Also, by specifically blocking the induction of IFN, e.g., by inhibiting NF-κB activity, virally infected cells may become sensitive to apoptosis (37).
It is not clear from these results why some IFN-pretreated cells began to synthesize virus proteins while others did not. In the case of SV5 infection of IFN-pretreated human and canine cells, the virus eventually managed to spread to cells that did not originally support virus replication. Given that these cells were initially resistant to SV5 infection, the question arises as to how the virus managed to spread from an infected cell to a neighboring cell in an antiviral state. Presumably, either the infected cell produced an amount of infectious virus so large that it overwhelmed the defense mechanisms of adjacent cells or some of the contents of the infected cell, including perhaps virus-encoded products which inhibited the IFN response, were transferred to the neighboring cell as a result of cell-cell fusion. There was no spread of SeV from cell to cell, even though SeV protein synthesis, once initiated, was clearly uninhibited by IFN. These latter results can be explained by the observation that in the tissue culture cells used, the SeV F protein was not cleaved and therefore remained nonfunctional (9, 29).
Although SeV prevented the activation of IFN-responsive genes in both human and murine cells, SV5 failed to inhibit the activation of IFN-responsive genes in murine cells, thereby explaining why SV5 protein synthesis was switched off in murine BF cells but not in human cells. Quantitation of the amount of different SV5 proteins made with time in BF cells after IFN treatment revealed that synthesis of HN and M was more sensitive than the synthesis of the P (or NP) protein to the effects of IFN. Since we have previously reported that the reduction in virus protein synthesis observed in BF cells correlated with a reduction in the amount of viral mRNA (6), one possible explanation for these results is that an antiviral mechanism(s) induced by IFN directly or indirectly affects virus transcription and the processivity of the virus polymerase, resulting in premature termination of transcription, thus favoring the expression of genes nearer the 3′ end of the virus genome. As BF cells were originally derived from BALB/c mice, the effect of IFN on SV5 protein synthesis could not have been mediated through the induction of the Mx proteins, as the gene encoding Mx in BALB/c mice has a large deletion, inactivating any product made (32).
We have previously shown that SV5 can replicate efficiently in BF cells if cultured in the presence of anti IFN-β antibodies and in cells derived from IFN-α/β receptor knockout mice (40). It is thus clear that the mechanistic requirements for SV5 transcription, replication, and virus production can be met in murine cells. Thus, the inability of SV5 to establish a truly productive infection even in SCID mice is probably due to the virus’s inability to overcome the IFN-induced cellular antiviral responses. In an attempt to adapt SV5 to replicate in mice, persistently infected BF cells have been passaged and virus variants have been selected (40). However, a fusogenic variant, termed W3-f, that was isolated from these cells after 30 passages remained as sensitive to IFN as the parental W3 isolate (40). Sequence analysis revealed multiple mutations in the HN and F genes of W3-f, most of which were silent, but three of which gave rise to amino acid substitutions in HN (4a). However, the fact that multiple mutations accumulated in W3-f suggests that it is extremely difficult for SV5 to adapt its mechanism for interfering with IFN signalling in human and canine cells to function correctly in murine cells. Although currently under investigation, neither the cellular target(s) nor the virus gene products involved in this process have yet been identified. However, given the evolutionary divergence between SV5 and SeV, it seems likely that the ability to block IFN signalling may be a general mechanism by which paramyxoviruses overcome IFN responses. Nevertheless, since SV5 failed to block IFN signalling in murine cells, the molecular basis for the block must be subject to species-specific effects such as protein-protein interactions.
From these results, it appears that one of the factors which limits the host range of paramyxoviruses is their ability to interact and overcome the IFN response. However, the fact that SeV blocks the IFN response in human cells and yet, as far as is known, does not naturally infect humans, emphasizes the point that other factors must also influence host range. Nevertheless, once the virus factor(s) which inhibit IFN signalling have been identified and characterized, this information may be useful for designing safe attenuated viruses and predicting whether a chimeric virus may be capable of replication in a given host. For example, if neither of the virus glycoproteins was responsible for inhibiting the activation of IFN-responsive genes, a SeV chimeric virus, in which the glycoprotein genes are replaced by those of SV5, might be expected to productively infect mice. (Whether such a virus would cause disease is another matter, as pathogenicity is subject to many considerations.) However, the converse would not be true, i.e., a chimeric SV5 virus with SeV glycoprotein genes would not replicate efficiently in mice. If this principle can be established, then attenuated viruses, which could potentially be developed as vaccines, may be genetically engineered by selectively knocking out or altering genes that encode the products which interfere with the IFN response.
ACKNOWLEDGMENTS
Lynsey Didcock is grateful to the MRC for a research studentship, Dan Young has been supported by a grant from the Wellcome Trust and BBSRC, and Steve Goodbourn is supported by a Wellcome Trust University Award.
We are indebted to Allen Portner for providing the MAbs to SeV, to Dan Kolakofsky and Dominique Garcin for the original SeV stocks used in these experiments, and to Tony Meager for providing the MRC 5, HFF, and 2D9 cells.
REFERENCES
- 1.Bosma G C, Custer R P, Bosma M J. A severe combined immunodeficiency mutation in mice. Nature. 1983;301:479–482. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 2.Choppin P W. Multiplication of a myxovirus (SV5) with minimal cytopathic effects and without interference. Virology. 1964;23:224–233. doi: 10.1016/0042-6822(64)90286-7. [DOI] [PubMed] [Google Scholar]
- 3.Darnell J E, Jr, Kerr I M, Stark G R. JAK-STAT pathways and transcriptional activation in response to interferon and other extracellular signalling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 4.de Wet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Didcock, L., and R. E. Randall. Unpublished data.
- 5.Enoch T, Zinn K, Maniatis T. Activation of the human beta-interferon gene requires an interferon-inducible factor. Mol Cell Biol. 1986;6:801–810. doi: 10.1128/mcb.6.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fearns R, Young D F, Randall R E. Evidence that the paramyxovirus SV5 can establish quiescent infections by remaining inactive in cytoplasmic inclusion bodies. J Gen Virol. 1994;75:3525–3529. doi: 10.1099/0022-1317-75-12-3525. [DOI] [PubMed] [Google Scholar]
- 7.Gale M, Jr, Katze M G. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon induced protein kinase. Pharmacol Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 7a.Goodbourn, S. Unpublished data.
- 8.Goswami K K A, Lange L S, Mitchell D N, Cameron K R, Russell W C. Does simian virus 5 infect humans? J Gen Virol. 1984;65:1295–1303. doi: 10.1099/0022-1317-65-8-1295. [DOI] [PubMed] [Google Scholar]
- 9.Homma M, Ohuchi M. Trypsin action on the growth of Sendai virus in tissue culture cells. III. Structural difference of Sendai viruses grown in eggs and tissue culture cells. J Virol. 1973;12:1457–1465. doi: 10.1128/jvi.12.6.1457-1465.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsiung G D. Parainfluenza-5 virus. Infection of man and animals. Prog Med Virol. 1972;14:241–274. [PubMed] [Google Scholar]
- 11.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 12.Ishida N, Homma M. Sendai virus. Adv Virol Res. 1978;23:349–383. doi: 10.1016/s0065-3527(08)60103-7. [DOI] [PubMed] [Google Scholar]
- 13.Kessler S W. Rapid isolation of antigens from cells with a staphylococcus protein A-antibody absorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975;115:1617–1627. [PubMed] [Google Scholar]
- 14.King P, Goodbourn S. The β-interferon promoter responds to priming through multiple independent regulatory elements. J Biol Chem. 1994;269:30609–30615. [PubMed] [Google Scholar]
- 15.King P, Goodbourn S. STAT1 is inactivated by a caspase. J Biol Chem. 1998;273:8699–8704. doi: 10.1074/jbc.273.15.8699. [DOI] [PubMed] [Google Scholar]
- 16.Masson N, Ellis M, Goodbourn S, Lee K A W. Cyclic-AMP response element-binding protein and the catalytic subunit of protein kinase A are present in F9 embryonal carcinoma cells but are unable to activate the somatostatin promoter. Mol Cell Biol. 1992;12:1096–1102. doi: 10.1128/mcb.12.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattana P, Viscomi G C. Variations in the interferon-inducing capacity of Sendai virus subpopulations. J Interferon Cytokine Res. 1998;18:399–405. doi: 10.1089/jir.1998.18.399. [DOI] [PubMed] [Google Scholar]
- 18.McCandlish L A P, Thompson H, Cornwell H J C, Wright N G. A study of dogs with kennel cough. Vet Rec. 1978;102:298–301. doi: 10.1136/vr.102.14.293. [DOI] [PubMed] [Google Scholar]
- 19.McKendry R, John J, Flavell D, Muller M, Kerr I M, Stark G R. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc Natl Acad Sci USA. 1991;88:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrison T, Portner A. Paramyxoviruses. New York, N.Y: Plenum Publishing Corporation; 1991. Structure, function, and intracellular processing of the glycoproteins of Paramyxoviridae. In D. W. Kingsbury (ed.) [Google Scholar]
- 21.Müller U, Steinhoff U, Reis L, Hemmi S, Pavloviv J, Zinkernagel R, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;256:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 22.Player M R, Torrence P F. The 2-5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol Ther. 1998;78:55–113. doi: 10.1016/S0163-7258(97)00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ploegh H L. Viral strategies of immune evasion. Science. 1998;280:248–253. doi: 10.1126/science.280.5361.248. [DOI] [PubMed] [Google Scholar]
- 24.Portner A. Monoclonal antibodies as probes of the antigenic structure and functions of Sendai virus glycoproteins. In: Bishop D H L, Compans R W, editors. Nonsegmented negative strand viruses. New York, N.Y: Elsevier/North-Holland; 1984. [Google Scholar]
- 25.Randall R E, Dinwoodie N. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins. Evidence that ICP4 is associated with progeny virus DNA. J Gen Virol. 1986;67:2163–2177. doi: 10.1099/0022-1317-67-10-2163. [DOI] [PubMed] [Google Scholar]
- 26.Randall R E, Young D F, Goswami K K A, Russell W C. Isolation and characterisation of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J Gen Virol. 1987;68:2769–2780. doi: 10.1099/0022-1317-68-11-2769. [DOI] [PubMed] [Google Scholar]
- 27.Randall R E, Young D, Southern J A. Immunization with solid matrix-antibody-antigen complexes containing surface or internal virus structural proteins protects mice from infection with the paramyxovirus, simian virus 5. J Gen Virol. 1988;69:2517–2526. doi: 10.1099/0022-1317-69-10-2517. [DOI] [PubMed] [Google Scholar]
- 28.Rehberg E, Kelder B, Hoal E G, Pestka S. Specific molecular activities of recombinant and hybrid interferons. J Biol Chem. 1982;257:11497–11502. [PubMed] [Google Scholar]
- 29.Scheid A, Choppin P W. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis and infectivity by cleavage of an inactive precursor protein of Sendai virus. Virology. 1974;57:475–490. doi: 10.1016/0042-6822(74)90187-1. [DOI] [PubMed] [Google Scholar]
- 30.Seow H F. Pathogen interactions with cytokines and host defence: an overview. Vet Immunol Immunopathol. 1998;63:139–148. doi: 10.1016/s0165-2427(98)00090-7. [DOI] [PubMed] [Google Scholar]
- 31.Smith G L. Virus strategies for evasion of the host response to infection. Trends Microbiol. 1994;2:81–88. doi: 10.1016/0966-842x(94)90539-8. [DOI] [PubMed] [Google Scholar]
- 32.Staeheli P, Grob R, Meier E, Sutcliffe J G, Haller O. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol Cell Biol. 1988;8:4518–4523. doi: 10.1128/mcb.8.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stark G R, Kerr I M. Interferon-dependent signalling pathways: DNA elements, transcription factors, mutations, and effects of viral proteins. J Interferon Res. 1992;12:147–151. doi: 10.1089/jir.1992.12.147. [DOI] [PubMed] [Google Scholar]
- 34.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreider R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 35.Tashiro M, Homma M. Pneumotropism of Sendai virus in relation to protease-mediated activation in mouse lung. Arch Virol. 1983;77:127–137. doi: 10.1128/iai.39.2.879-888.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tashiro M, Yamakawa M, Tobita K, Klenk K-D, Rott R, Seto J T. Organ tropism of Sendai virus in mice: proteolytic activation of the fusion glycoprotein in mouse organs and budding site at the bronchial epithelium. J Virol. 1990;64:3627–3634. doi: 10.1128/jvi.64.8.3627-3634.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Antwerp D J, Martin S J, Verma I M, Green D R. Inhibition of TNF-induced apoptosis by NF-kappa B. Trends Cell Biol. 1998;3:107–111. doi: 10.1016/s0962-8924(97)01215-4. [DOI] [PubMed] [Google Scholar]
- 38.Van den Broek M F, Müller U, Huang S, Aguet M, Zinkernagel R M. Antiviral defense in mice lacking both alpha/beta and gamma interferon receptors. J Virol. 1995;69:4792–4796. doi: 10.1128/jvi.69.8.4792-4796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young D F, Randall R E, Hoyle J A, Souberbielle B E. Clearance of a persistent paramyxovirus infection is mediated by cellular immune responses but not by serum neutralizing antibody. J Virol. 1990;64:5403–5411. doi: 10.1128/jvi.64.11.5403-5411.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young D F, Didcock L, Randall R E. Isolation of highly fusogenic variants of simian virus 5 from persistently infected cells that produce and respond to interferon. J Virol. 1997;71:9333–9342. doi: 10.1128/jvi.71.12.9333-9342.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimring J C, Goodbourn S, Offerman M K. Human Herpesvirus 8 encodes and interferon regulatory factor (IRF) homolog that represses IRF-1 mediated transcription. J Virol. 1998;72:701–707. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zinn K, DiMaio D, Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983;34:865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]