Abstract
Retinoblastoma is a common primary intraocular malignant tumor that affects infants and young children. Radiation therapy for hereditary retinoblastoma increases the risk of secondary malignancy. The present report discusses the case of a retinoblastoma survivor who developed secondary leiomyosarcoma 42 years after receiving radiation therapy. The retinoblastoma of the patient was unilateral, and the patient had no family history of the disease. RNA and DNA panel sequencing of the leiomyosarcoma tissue was performed to elucidate the molecular mechanism of this secondary malignancy. The RNA panel sequencing detected a germline reciprocal translocation of RB1 and DMXL1, leading to a diagnosis of possible hereditary retinoblastoma. Furthermore, it detected a somatic fusion gene (RAD51-KNL1). The DNA panel sequencing identified various germline or somatic variants, including a somatic splice acceptor site mutation of TP53. We hypothesized that the molecular mechanism of the secondary malignancy of this patient was the combination of a germline reciprocal translocation of RB1 and DMXL1 and the accumulation of various somatic mutations containing the splice acceptor site mutation of TP53, which ultimately led to the development of a secondary leiomyosarcoma. Further prospective investigations are necessary to fully understand the role of reciprocal translocation of RB1 and DMXL1 or other mutations in the tumorigenesis of second malignancies in patients with hereditary retinoblastoma.
Keywords: leiomyosarcoma, retinoblastoma, RB1, DMXL1, TP53
Introduction
Retinoblastoma is a common primary intraocular malignant tumor in infants and young children. It is considered to be the most common primary malignant intraocular tumor. According to the World Health Organization, the reported incidence is 1 case per 16,000-18,000 live births, with approximately 7,000-8,000 new cases worldwide each year. It is caused by germline and somatic mutations in the RB1 gene or, rarely, somatic amplification of MYCN. Hereditary retinoblastomas are typically bilateral with multiple tumor foci in one or two eyes (1).
Survivors of hereditary retinoblastoma generally have a good prognosis but radiation therapy for hereditary retinoblastoma increases the risk of secondary malignancy (2-4). However, the precise molecular mechanism remains unknown. Here, we report on a survivor of possible hereditary retinoblastoma, diagnosed with secondary leiomyosarcoma 42 years after radiation therapy. To elucidate the molecular mechanism of secondary leiomyosarcoma, we performed an RNA panel sequencing and DNA panel sequencing on her sarcoma tissue. We could not obtain this patient's retinoblastoma tissue itself. However, we also performed reverse transcription-polymerase chain reaction (RT-PCR) analysis for the fusion genes detected by RNA sequencing to distinguish whether the fusion genes were somatic or germline mutations on both sarcoma and normal tissues, specifically lymph node and skin, respectively. We also examined short variants found in the DNA panel test by Sanger sequencing to distinguish between somatic and germline mutations.
Case report
In April 2014, a 44-year-old woman with a residual nasal cavity tumor was referred to our hospital. She had suffered from left nasal bleeding in September 2013 and consulted a nearby otolaryngologist. A tumor in her left nasal cavity was discovered (Fig. 1), and she was referred for treatment. After the embolization of the tumor, endoscopic resection of the tumor was performed in March 2014, but the tumor was located in the lateral wall of the nasal cavity. She was then transferred to our hospital for future treatment.
Figure 1.
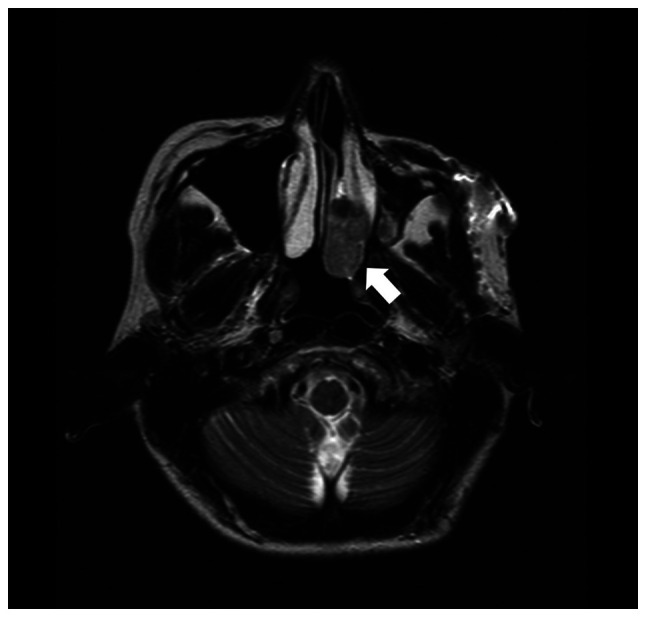
T2-weighted axial image of magnetic resonance imaging before the operation. A mild high intensity mass was recognized in the left nasal cavity (white arrow).
The patient had a history of retinoblastoma of the left eye at 2 years old. She received radiotherapy at the age of two, and her left eyeball was removed at the age of 24. Her family had no family history of retinoblastoma. Other information, such as total radiation dose and chemotherapy, could not be obtained.
Our hospital pathologists re-examined the previously resected nasal tumor and diagnosed leiomyosarcoma. An endoscopic local examination did not identify the residual tumor, and imaging examinations revealed no metastatic lesions. Her laboratory tests yielded almost normal values except for lactate dehydrogenase was 263 U/l (normal range: 119-229 U/l).
In July 2014, partial removal of the left maxilla, including the pharyngeal opening of the auditory tube and palatine tonsils, was performed, where the location of the residual tumor was suspected. Left submandibular lymph nodes were also dissected. Pathological examination of the resected tissue revealed residual lesions of leiomyosarcoma in the inferior nasal concha. There was no metastases in the dissected lymph nodes (Sample 1).
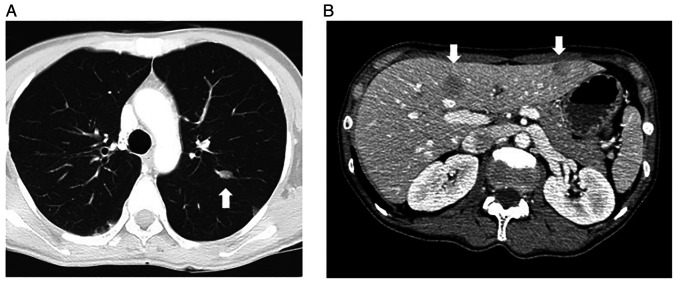
After the operation, she received four courses of adjuvant chemotherapy consisting of ifosfamide (2 g/m2 per day, infusions performed on days 1-5) and doxorubicin (30 mg/m2 per day, infusions performed on days 1-2) every 28-day cycle. However, several subcutaneous metastatic nodules occurred in May 2016. In September 2016, a total of five subcutaneous lesions were resected, two from the anterior chest, and one from the right shoulder, left upper arm, and left lumbar region (Sample 2). All of the lesions were metastatic leiomyosarcomas. In November 2016, multiple metastases were detected in her lung (Fig. 2A) and liver (Fig. 2B) via computed tomography. The first-line systemic chemotherapy, consisting of gemcitabine (900 mg/m2/day; infusions performed on day 1 and 8) and docetaxel (70 mg/m2; infusions performed on day 8) every 28-day cycle, was started, but it was discontinued after two cycles due to the suspected pneumonitis caused by these anticancer drugs. The second-line chemotherapy consisting of eribulin mesylate (1.4 mg/m2/day; infusions performed on day 1 and 8 of every 21-day cycle) was started, but it was discontinued during the second cycle due to progressive disease. The third-line therapy consisting of pazopanib, 600 mg/day orally, was started in April 2017 but was discontinued due to high fever, thrombocytopenia, and punctate erythema throughout the body. At that time, a skin biopsy was performed on her left lower leg (Sample 3), but no metastases of leiomyosarcoma were found. The fourth line of chemotherapy, trabectedin (1.2 mg/m2), was started, but after 2 cycles, there was evidence of progressive disease. A reduced dose of pazopanib, 200-400 mg/day, was re-introduced, and it was continued for approximately 2 years. However, her leiomyosarcoma gradually increased, and she died in January 2020.
Figure 2.
CT scan of the (A) lung and (B) liver at the time of metastasis. The metastatic lesions are indicated by white arrows.
Methods for pathological studies
Immunohistochemical staining was completed using a Ventana BenchMark GX system (Ventana Medical Systems Inc.). These evaluations used the following primary antibodies: pan Keratin (AE1/AE3/PCK26 [prediluted], Ventana Medical Systems Inc.), smooth muscle actin (SMA, 1A4 [prediluted] Ventana Medical Systems Inc.), desmin (DE-R-11 [prediluted] Ventana Medical Systems Inc.), MyoD1 (EP212 [prediluted] Ventana Medical Systems Inc.), p53 (DO7 [prediluted], Ventana Medical Systems Inc.), and retinoblastoma transcriptional corepressor 1 (Rb1, G3-245, 1:200; BD Pharmingen).
Methods for genetic studies. Extraction of RNA and DNA from formalin-fixed, paraffin-embedded (FFPE) tissues
RNAs were extracted from slices of FFPE tissue using Maxwell RSC Instrument and Maxwell RCS RNA FFPE Kit (Promega) and their concentrations were measured with the QuantiFluor RNA System (Promega). The DV200 score for quality assessment of each RNA was determined using Bioanalyzer 2100 and RNA 6000 Nano/Pico Kit. DNA was extracted from slices of FFPE tissue with QIAamp DNA FFPE Advanced UNG Kit (Qiagen) and its concentration was measured using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Inc.). The quality of DNA was confirmed using 1% agarose gel electrophoresis.
RNA panel analysis. The extracted tumor RNA was converted to a next-generation sequencing (NGS) library with the TruSight RNA Pan-Cancer Panel (RS-303-1002, Illumina) following their reference guide (Document # 1000000001632 v01). The quality and quantity of the library were measured using Agilent DNA1000 Kit (Agilent) and the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Inc.). Libraries from 12 samples were equimolarly pooled and diluted to a concentration of 1.5 pM. 1.3 ml of the diluted library was sequenced on a NextSeq 550 instrument using the NextSeq 500/550 Mid Output Kit v2.5 (20024905, Illumina). The data were analyzed using RNA-Seq Alignment (version 2.0.2) of the BaseSpace Sequence Hub (https://basespace.illumina.com/, Illumina).
RT-PCR analysis. RNAs were converted to cDNA with a random hexamer and SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific, Inc.) as described in the manufacturer's instructions. The cDNAs were used as templates for PCR reactions with Platinum SuperFi DNA Polymerase and SuperFi GC Enhancer (Thermo Fisher Scientific, Inc.). The primer sequences are as follows: RB1exon8_F, 5'-GATACAAGAATTATTGAAGTTCTCTGTAAAGAAC-3'; DMXL1exon9_R, 5'-CAAGTGATCTCCTCCGACCTCT-3'; DMXL1exon9_F, 5'-GAACACTCCACTGCATGCC-3'; RB1exon9_R, 5'-AGTCCATTAGATGTTACAAGTCCA-3'; RAD51exon5_F, 5'-ACCCAGATCTGTCATACGCT-3'; KNL1utr5_R, 5'-TGCCGTCTTCTAACAGGTCT-3'. The expected amplicon sizes are 97 bp (RB1exon8-DMXL1exon9), 139 bp (DMXL1exon9-RB1exon9), 125 bp (RB1exon8-RB1exon9), 121 bp (RAD51exon5-KNL1utr5). The cycling profile was as follows: 30 s at 98˚C for initial denaturation, followed by 40 or 45 cycles of 10 s at 98˚C for denaturation, 10 s at 60˚C for annealing, and 30 s at 72˚C for extension. The PCR products were separated on a 2% agarose gel electrophoresis and stained with GelGreen (Biotium). The low molecular weight DNA ladder (cat. no. N3233S; New England BioLabs, Inc.) was used.
DNA panel analysis. The extracted tumor DNA was sheared using the Covaris M220 according to TruSight Tumor 170 DNA Shearing Quick Guide (Part Number: 010515 Rev B, Covaris) with slight modification (Repeat/Iterations: 16) and checked the distribution of the fragments using Bioanalyzer 2100 and High Sensitivity DNA Kit (Agilent Technologies). The sheared DNA was converted to an NGS library with the TruSight Oncology 500 Kit (Illumina) following the reference guide (Document # 1000000067621 v04, Illumina). Sequencing was performed on a NextSeq 550 instrument, and the data were analyzed using the Local Run Manager TruSight Oncology 500 Analysis Module on the sequencer. After filtering out candidate germline variants using public databases, 1000 genomes (https://www.internationalgenome.org/) (5) and gnomAD exome/genom (https://gnomad.broadinstitute.org/) (6) according to the Workflow Guide of the above analysis module (Document # 10000000099600 v00, Illumina), we annotated the pathogenicity of the detected variants using knowledge bases, OncoKB (v3.8, https://www.oncokb.org/) (7), Cancer Genome Interpreter (https://www.cancergenomeinterpreter.org/home) (8), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) (9), COSMIC (https://cancer.sanger.ac.uk/cosmic) (10). We used ANNOVAR (https://annovar.openbioinformatics.org/en/latest/) (11) to annotate the ClinVar data (version 20210501).
Sanger sequencing. To determine whether the short variants detected by DNA panel sequencing were somatic or germline mutations, we performed PCR followed by Sanger sequencing on the normal lymph node tissue (Sample 1). Each mutant gene was amplified from DNA using appropriate primer sets (Table SI).
Results from pathological studies
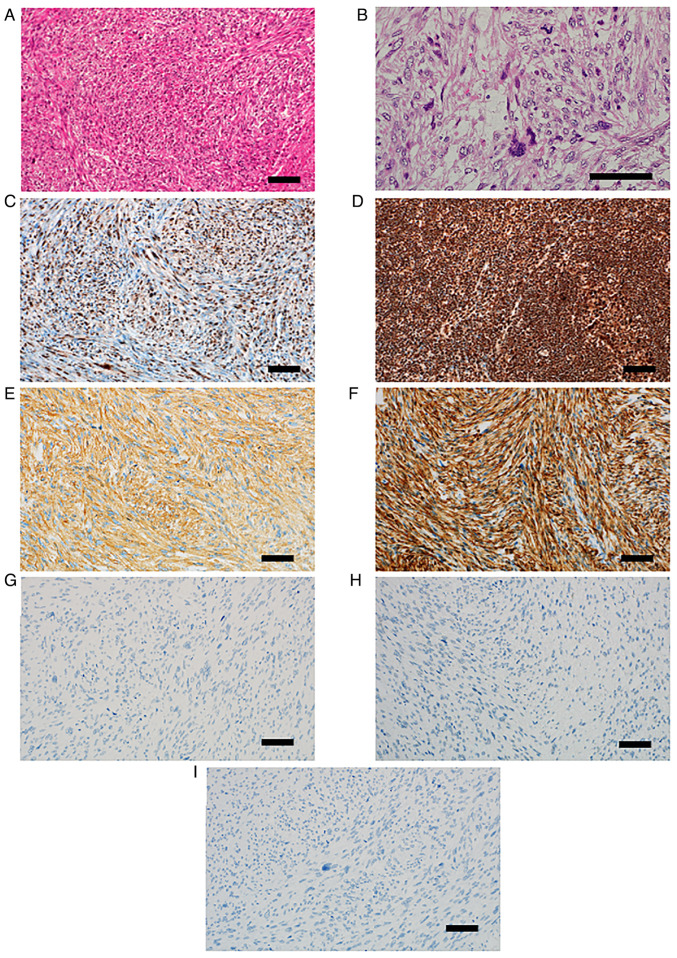
Pathological examination of the resected tissue revealed residual lesions of leiomyosarcoma in the inferior nasal concha, in which atypical spindle-shaped cells intermingle in bundles proliferated (Fig. 3A). These tumor cells have oval to spindle-shaped nuclei which contain small nucleoli, and eosinophilic cytoplasm. The cellularity of the tumor cells was moderate. Tumor cells often show strong nuclear atypia and mitotic figures were observed with a frequency of 30/10 high-power field (HPF) (Fig. 3B). The immunohistochemical staining identified that approximately 50% of tumor cells lacked nuclear retinoblastoma transcriptional corepressor 1 (Rb1) protein expression (Fig. 3C). In contrast, in normal lymph node tissue (Sample 1), Rb1 protein expression was observed in more than 90% of the component cell nuclei (Fig. 3D). Tumor cells were diffusely positive for smooth muscle actin (Fig. 3E) and desmin (Fig. 3F). Pan Keratin (Fig. 3G) and MyoD1 (Fig. 3H) were completely negative. p53 also showed complete loss of expression (Fig. 3I).
Figure 3.
Histopathological findings. (A) Spindle-shaped tumor cells, which had oval to spindle-shaped atypical nuclei and eosinophilic cytoplasm, intermingled in bundles. (B) Tumor cells often showed strong nuclear atypia with occasional mitotic figures. (C) Approximately 50% of tumor cells lacked nuclear Rb1 protein expression. (D) By contrast, in normal lymph node tissue, Rb1 protein expression was observed in >90% of the component cell nuclei. (E) Tumor cells were diffusely positive for SMA. (F) Desmin was also diffusely positive. (G) Pan keratin (AE1/AE3) was negative. (H) MyoD1 was also negative. (I) p53 was negative. Scale bar, 100 µm. Rb1, retinoblastoma transcriptional corepressor 1; SMA, smooth muscle actin; MyoD1, myogenic differentiation 1.
Results from genetic studies. RNA panel sequencing and RT-PCR
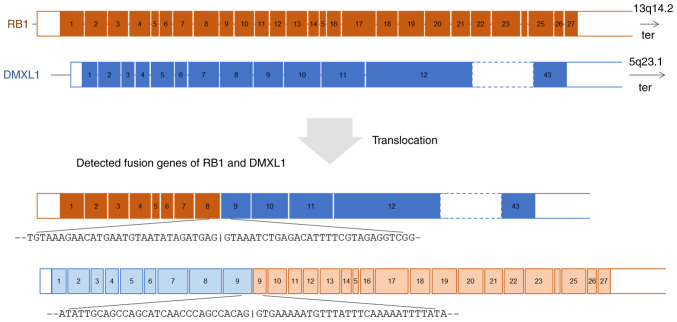
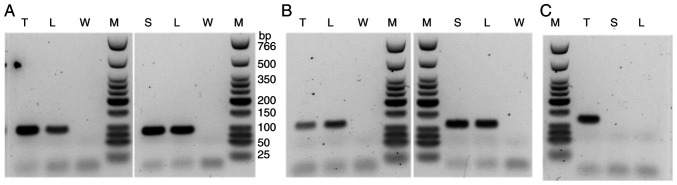
RNA panel sequencing of the metastatic leiomyosarcoma (Sample 2) identified a novel reciprocal translocation between 13q14.2 and 5q23.1 and a frameshift fusion gene RAD51 (exon5; NM_002875.5)-KNL1 (exon2; NM_144508.5). This reciprocal translocation resulted in two fusion genes: RB1(exon 8; NM_000321.3)-DMXL1(exon 9; NM_001290321.3) and DMXL1(exon 9)-RB1(exon 9) (Fig. 4). The exon 9 of DMXL1 was included in both fusions. The fusion of RB1(exon 8)-DMXL1(exon 9) was validated by RT-PCR analysis revealing a rearrangement between these loci. Similar rearrangement was identified in Sample 1 (normal lymph node) and Sample 3 (skin) (Fig. 5A). Wild-type RB1 genes were proven by RT-PCR of RB1 exon 8-RB1 exon 9 in the leiomyosarcoma, the lymph node, and the skin (Fig. 5B). On the other hand, the RAD51-KNL1 fusion gene was identified only in the sarcoma tissue and not in normal samples (Fig. 5C).
Figure 4.
RNA panel sequencing of the metastatic leiomyosarcoma (sample 2). A novel reciprocal translocation between RB1 and DMXL1 was identified. ter, the direction to the telomere.
Figure 5.
After reverse transcription of sample RNAs, synthesized cDNAs were used as templates for PCR of (A) RB1 exon 8-DMXL1 exon 9 (amplicon size, 97 bp), (B) RB1 exon 8-RB1 exon 9 (amplicon size, 125 bp) and (C) RAD51 exon 5-KNL1 5' untranslated region (amplicon size, 121 bp). The marker lane was run on the same gel for the data shown in (B) (left panel) and (C). L, sample 1 (lymph node); T, sample 2 (tumor); S, sample 3 (skin); W, water; M, low molecular weight DNA ladder.
DNA panel sequencing and Sanger sequencing. DNA panel sequencing identified eight short variants: CDK12(Arg271Lys), CDK6(Val62Gly), HGF(Asp330Val), KDM5A(Thr1343Ile), LRP1B(Cys138Gly), MSH2(Met688Ile), SNCAIP(Glu87Lys), and TP53 (splice acceptor of intron 8). Only the variants in HGF, KDM5A, LRP1B, and MSH2 were detected in the normal lymph node (Table I). The panel analysis identified fourteen copy number changed genes. Nine of these genes, AR, FGF23, FGF4, FGF6, FGF7, FGFR1, KRAS, NRG1, and RAF1, were amplified. The other five genes, ATM, FGF5, FGF8, MDM4, and PTEN, were lost (Table II).
Table I.
Short variants detected by DNA panel sequencing.
| Gene | Variant | VAF | OncoKB | CGI | COSMIC | COSMIC_id | ClinVar |
|---|---|---|---|---|---|---|---|
| CDK12 | Arg271Lys | 0.418 | Passenger | ||||
| CDK6 | Val62Gly | 0.408 | Driver | ||||
| HGF | Asp330Val | 0.416 | Driver | ||||
| KDM5A | Thr1343Ile | 0.333 | Passenger | ||||
| LRP1B | Cys138Gly | 0.613 | Passenger | ||||
| MSH2 | Met688Ile | 0.611 | Driver | Pathogenic | COSV 51886508 | US | |
| SNCAIP | Glu87Lys | 0.035 | Passenger | Pathogenic | COSV 54401827 | ||
| TP53 | Splice acceptor of intron 8 | 0.849 | Driver |
Blank cells mean that the records do not exist in the database. VAF, variant allele fraction; OncoKB, Memorial Sloan Kettering's Precision Oncology Knowledge Base; CGI, Cancer Genome Interpreter; COSMIC, Catalogue of Somatic Mutations in Cancer; US, uncertain significance.
Table II.
Copy number changes.
| Gene | Amp/Loss | OncoKB | CGI |
|---|---|---|---|
| AR | Amp | LO | Known |
| ATM | Loss | LO | Known |
| FGF23 | Amp | Predicted passenger | |
| FGF4 | Amp | Inconclusive | Known |
| FGF5 | Loss | Predicted passenger | |
| FGF6 | Amp | Predicted passenger | |
| FGF7 | Amp | Predicted passenger | |
| FGF8 | Loss | Predicted passenger | |
| FGFR1 | Amp | O | Known |
| KRAS | Amp | LO | Predicted driver |
| MDM4 | Loss | Predicted passenger | |
| NRG1 | Amp | Predicted passenger | |
| PTEN | Loss | O | Known |
| RAF1 | Amp | O | Known |
Amp, amplification; OncoKB, Memorial Sloan Kettering's Precision Oncology Knowledge Base; LO, likely oncogenic (OncoKB); O, oncogenic (OncoKB); CGI, Cancer Genome Interpreter.
Discussion
Retinoblastoma is a common primary intraocular malignant tumor in infants and young children. Knudson's ‘two-hit hypothesis’ accurately explains the development mechanism of retinoblastoma (12,13). Briefly, in hereditary retinoblastoma, individuals carry a germline heterozygous alteration in RB1. Somatic inactivation of the second RB1 allele results in the development of retinoblastoma.
To diagnose hereditary retinoblastoma in clinical practice, various sequencing methods are used to identify a heterozygous germline pathogenic variant in RB1. Pathogenic variants in RB1 show various types of mutations, including single nucleotide variations, small indels, and large deletions/duplications (14). Schieffer et al (15) performed RNA sequencing on the tumor tissue of an infant with sporadic and intracranial retinoblastoma and reported multiple fusion genes, including RB1-SIAH3. However, the reports of fusion genes, including RB1, are rare.
We did not have this patient's retinoblastoma tissue, so the secondary malignancy (leiomyosarcoma) and normal tissues (lymph node and skin) were used for genetic testing. RNA sequencing is a useful tool for detecting fusion genes. Using this method on the leiomyosarcoma tissue, reciprocal translocations RB1(exon 8)-DMXL1(exon 9) and DMXL1(exon 9)-RB1(exon 9), and fusion gene of RAD51 (exon5)-KNL1 (exon2), were identified. To the best of our knowledge, these are novel fusion genes. RT-PCR confirmed the presence of RB1(exon 8)-DMXL1(exon9) in the leiomyosarcoma, lymph node, and skin samples, identifying this fusion gene as a germline mutation. DMXL1(exon 9)-RB1(exon 9) was confirmed in leiomyosarcoma and lymph node but not in the skin. However, this fusion gene is also probably a germline mutation. Because RNA quality was poorest in the skin (DV200 score: 35 for tumor, 36 for lymph node, 15 for skin), that RNA was highly fragmented, which may explain the false-negative results. Since wild-type RB1 genes were proven by RT-PCR of RB1 exon 8-RB1 exon 9 in the leiomyosarcoma, the lymph node, and the skin, the reciprocal translocation between RB1 and DMXL1 is regarded as heterozygous alteration, and, thus, we considered that her retinoblastoma was likely hereditary. On the other hand, RAD51-KNL1 was identified in leiomyosarcoma but not lymph node or skin. Therefore, this fusion gene is regarded as a somatic mutation.
The protein encoded by RB1 is a tumor suppressor that is a regulator of the G1/S transition of the cell cycle (https://www.uniprot.org/uniprot/P06400) (16). On the other hand, the protein encoded by DMXL1 is a member of the WD repeat superfamily of proteins with regulatory functions. DMXL1 is expressed in various tissue types, including several types of eye tissue, and it has been associated with ocular phenotypes (https://genome-asia.ucsc.edu/cgi-bin/hgGene?hgg_gene=ENST00000311085.8&hgg_chrom=chr5&hgg_start=119071489&hgg_end=119249127&hgg_type=knownGene&db=hg38) (17). The role of chimeric RB1-DMXL1 genes in the formation of retinoblastoma or secondary leiomyosarcoma is difficult to understand, but this fusion gene predicts the possibility of dysfunction of the RB1 gene. Dommering et al (18) investigated RB1 mutations in 44 hereditary retinoblastoma patients with a second primary malignancy and found an increased risk of second primary malignancy among carriers of one of the 11 recurrent CGA>TGA nonsense RB1 mutations. In their samples, one case had a deletion of exon 9-27, which was associated with an epithelial second primary malignancy. In our case, we may have had a similar mechanism of RB1 gene dysfunction. The protein coded by RAD51 plays an important role in homologous strand exchange, a key step in DNA repair through homologous recombination (https://www.uniprot.org/uniprot/Q06609) (16). The protein coded by KNL1 is essential for spindle-assembly checkpoint signaling and correct chromosome alignment (https://www.uniprot.org/uniprot/Q8NG31) (16). RAD51-KNL1 fusion causes a frameshift and may result in the loss of both proteins.
We identified eight short variants and fourteen copy number changed genes using DNA panel sequencing. To predict the involvement of these mutations in carcinogenesis of secondary leiomyosarcoma, we used cancer knowledge databases: OncoKB (7), Cancer Genome Interpreter (8), ClinVar (9), and COSMIC (10) (Table I). Based on these results, we speculated that short variants of CDK6, HGF, MSH2, SNCAIP, and TP53 might be involved in malignant transformation. In the copy number change, amplification of AR, FGFR1, KRAS, RAF1, and loss of ATM and PTEN were suspected.
To determine whether the eight short variants detected by DNA panel sequencing were somatic or germline mutations, Sanger sequencing was performed on the normal lymph node (Sample 1). Four variants, TP53 (splice acceptor of intron 8), CDK12(Arg271Lys), CDK6(Val62Gly), and SNCAIP(Glu87Lys), were somatic mutations. The other four variants, HGF(Asp330Val), KDM5A(Thr1343Ile), LRP1B(Cys138Gly), and MSH2(Met688Ile), were germline mutations detected by DNA panel sequencing. The variant allele fraction of TP53 was 0.85, which is a very high value. This mutation was not detected in the normal lymph node by Sanger sequencing. Therefore, we concluded that it is a somatic mutation and speculated that there was a loss of heterozygosity. This mutation is in the splice acceptor site and is expected to cause an exon skipping or intron retention of mRNA, resulting in dysfunction of TP53 protein.
Schaefer et al (19) reported a high frequency of co-inactivation of TP53 and RB1 in local recurrent or distant metastatic leiomyosarcomas using immunohistochemistry. Using whole-exome and transcriptome sequencing, Chudasama et al (20) reported that leiomyosarcoma tumors are characterized by substantial mutational heterogeneity, near-universal inactivation of TP53, as well as RB1, widespread DNA copy number alterations including chromothripsis, and frequent whole-genome duplication. As a molecular mechanism of our patient's secondary leiomyosarcoma tumorigenesis, we speculate that the patient had a reciprocal translocation of the RB1 and DMXL1 as a germline mutation and various somatic mutations containing the splice acceptor site mutation of TP53 accumulated, resulting in the secondary leiomyosarcoma. Radiation therapy is thought to trigger somatic mutations. However, her sarcoma developed 42 years after the radiotherapy, and the involvement of age-related exposure to environmental carcinogens cannot be ruled out.
This study has limitations. First, this study is based on only one case report. Therefore, further prospective investigations are needed to elucidate the role of reciprocal translocation of the RB1 and DMXL1 or other mutations in tumorigenesis of second malignancy in patients with hereditary retinoblastoma. Second, after adjuvant chemotherapy, the leiomyosarcoma tissue (Sample 2) was analyzed for mutation analysis. Therefore, it cannot be ruled out that anticancer drugs may have affected the somatic mutations in leiomyosarcoma. Third, a detailed family history could not be confirmed due to the small number of her relatives, no family history of retinoblastoma could be found, and no genetic test was performed on her relatives. Therefore, it is possible that the reciprocal translocation of the RB1 and DMXL1 is a de novo mutation rather than hereditary. Fourth, we did not perform the fluorescence in situ hybridization (FISH) of RB1-DMXL1 fusion gene. Therefore, this fusion gene could not be visualized in the tissues.
In summary, we report the case of a woman with a history of possible hereditary retinoblastoma diagnosed with secondary leiomyosarcoma 42 years after radiotherapy. She had several germline mutations, including a reciprocal translocation of RB1 and DMXL1. It is speculated that the addition of some somatic mutations, such as the splice acceptor site mutation of TP53, might have precipitated the development of the second malignant leiomyosarcoma. Further prospective investigations are needed to elucidate the role of reciprocal translocation of the RB1 and DMXL1 or other mutations in the tumorigenesis of second malignancy in patients with hereditary retinoblastoma.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available due to containing information that could compromise the privacy of the research participant and her relatives but are available from the corresponding author on reasonable request.
Authors' contributions
TY conceived the present study. Data acquisition, analysis and interpretation were performed by HN and YK. TW, HiT, SN, MW, SK, HaT, RS, ST and YH collected clinical data. TY and HN confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The tissues of the patient were collected after written informed consent was obtained from the patient according to the protocol approved by the Osaka International Cancer Institute (Osaka, Japan).
Patient consent for publication
Written informed consent was obtained from the patient's brother to publish her data and associated images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Eagle RC Jr, Chévez-Barrios P, Li B, Al Hussaini M, Wilson M. Retinoblastoma. In: WHO classification of tumours of the eye. Grossniklaus HE, Eberhart CG and Kivelä TT (eds). 4th edition. International Agency for Research on Cancer, Lyon, pp111-117, 2018. [Google Scholar]
- 2.Kleinerman RA, Schonfeld SJ, Sigel BS, Wong-Siegel JR, Gilbert ES, Abramson DH, Seddon JM, Tucker MA, Morton LM. Bone and soft-tissue sarcoma risk in long-term survivors of hereditary retinoblastoma treated with radiation. J Clin Oncol. 2019;37:3436–3445. doi: 10.1200/JCO.19.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Temming P, Arendt M, Viehmann A, Eisele L, Le Guin CH, Schündeln MM, Biewald E, Astrahantseff K, Wieland R, Bornfeld N, et al. Incidence of second cancers after radiotherapy and systemic chemotherapy in heritable retinoblastoma survivors: A report from the German reference center. Pediatr Blood Cancer. 2017;64:71–80. doi: 10.1002/pbc.26193. [DOI] [PubMed] [Google Scholar]
- 4.Wong FL, Boice JD Jr, Abramson DH, Tarone RE, Kleinerman RA, Stovall M, Goldman MB, Seddon JM, Tarbell N, Fraumeni JF Jr, Li FP. Cancer incidence after retinoblastoma. Radiation dose and sarcoma risk. JAMA. 1997;278:1262–1267. doi: 10.1001/jama.278.15.1262. [DOI] [PubMed] [Google Scholar]
- 5.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. 1000 Genomes Project Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, et al. OncoKB: A precision oncology knowledge base. JCO Precis Oncol. 2017;2017(PO.17.00011) doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamborero D, Rubio-Perez C, Deu-Pons J, Schroeder MP, Vivancos A, Rovira A, Tusquets I, Albanell J, Rodon J, Tabernero J, et al. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 2018;10(25) doi: 10.1186/s13073-018-0531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, et al. COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(e164) doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knudson AG Jr. Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer. 2001;1:157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- 14.Singh J, Mishra A, Pandian AJ, Mallipatna AC, Khetan V, Sripriya S, Kapoor S, Agarwal S, Sankaran S, Katragadda S, et al. Next-generation sequencing-based method shows increased mutation detection sensitivity in an Indian retinoblastoma cohort. Mol Vis. 2016;22:1036–1047. [PMC free article] [PubMed] [Google Scholar]
- 15.Schieffer KM, Feldman AZ, Kautto EA, McGrath S, Miller AR, Hernandez-Gonzalez ME, Lahaye S, Miller KE, Koboldt DC, Brennan P, et al. Molecular classification of a complex structural rearrangement of the RB1 locus in an infant with sporadic, isolated, intracranial, sellar region retinoblastoma. Acta Neuropathol Commun. 2021;9(61) doi: 10.1186/s40478-021-01164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navarro Gonzalez J, Zweig AS, Speir ML, Schmelter D, Rosenbloom KR, Raney BJ, Powell CC, Nassar LR, Maulding ND, Lee CM, et al. The UCSC genome browser database: 2021 update. Nucleic Acids Res. 2021;49:D1046–D1057. doi: 10.1093/nar/gkaa1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dommering CJ, Marees T, van der Hout AH, Imhof SM, Meijers-Heijboer H, Ringens PJ, van Leeuwen FE, Moll AC. RB1 mutations and second primary malignancies after hereditary retinoblastoma. Fam Cancer. 2012;11:225–233. doi: 10.1007/s10689-011-9505-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer IM, Lundberg MZ, Demicco EG, Przybyl J, Matusiak M, Chibon F, Ingram DR, Hornick JL, Wang WL, Bauer S, et al. Relationships between highly recurrent tumor suppressor alterations in 489 leiomyosarcomas. Cancer. 2021;127:2666–2673. doi: 10.1002/cncr.33542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chudasama P, Mughal SS, Sanders MA, Hübschmann D, Chung I, Deeg KI, Wong SH, Rabe S, Hlevnjak M, Zapatka M, et al. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat Commun. 2018;9(144) doi: 10.1038/s41467-017-02602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due to containing information that could compromise the privacy of the research participant and her relatives but are available from the corresponding author on reasonable request.