Abstract
Proteolytic cleavage of the VP4 outer capsid spike protein into VP8* and VP5* proteins is required for rotavirus infectivity and for rotavirus-induced membrane permeability. In this study we addressed the function of the VP5* cleavage fragment in permeabilizing membranes. Expressed VP5* and truncated VP5* proteins were purified by nickel affinity chromatography and assayed for their ability to permeabilize large unilamellar vesicles (LUVs) preloaded with carboxyfluorescein (CF). VP5* and VP5* truncations, but not VP4 or VP8*, permeabilized LUVs as measured by fluorescence dequenching of released CF. Similar to virus-induced CF release, VP5*-induced CF release was concentration and temperature dependent, with a pH optimum of 7.35 at 37°C, but independent of the presence of divalent cations or cholesterol. VP5*-induced permeability was completely inhibited by VP5*-specific neutralizing monoclonal antibodies (2G4, M2, or M7) which recognize conformational epitopes on VP5* but was not inhibited by VP8*-specific neutralizing antibodies. In addition, N-terminal and C-terminal VP5* truncations including residues 265 to 474 are capable of permeabilizing LUVs. These findings demonstrate that VP5* permeabilizes membranes in the absence of other rotavirus proteins and that membrane-permeabilizing VP5* truncations contain the putative fusion region within predicted virion surface domains. The ability of recombinant expressed VP5* to permeabilize membranes should permit us to functionally define requirements for VP5*-membrane interactions. These findings indicate that VP5* is a specific membrane-permeabilizing capsid protein which is likely to play a role in the cellular entry of rotaviruses.
Rotaviruses are nonenveloped icosahedral viruses with 11 double-stranded RNA gene segments inside a 70-nm triple-layered particle (TLP) (14, 45). Four capsid proteins, VP2, VP6, VP7, and VP4, are present in infectious TLPs (14, 45). Calcium chelation removes VP4 and VP7 outer capsid proteins and converts TLPs into transcriptionally active but noninfectious double-layered particles (DLPs) (7, 51). Rotavirus RNAs are transcribed within DLPs by a self-contained double-stranded RNA-dependent RNA polymerase complex (7) and are extruded from pores in the DLP (31, 46). VP7 is the major structural protein on the surface of TLPs, while dimeric VP4 spikes project from the rotavirus surface (14, 45, 47, 53, 59). VP7 is the viral glycoprotein, while VP4 lacks signal sequences and posttranslational modifications (14). VP4 and VP7 are targets of neutralizing antibodies to rotaviruses, and immune responses to these proteins protect animals from disease (14, 26, 37, 42, 43, 55).
Protease treatment of rotaviruses is required for viral infectivity. On the virion, trypsin cleaves VP4 (86 kDa) into the VP8* (28 kDa) and VP5* (60 kDa) proteins, activating the virus for infection (13, 15). The VP4 and VP8* proteins of some rotaviruses are capable of hemagglutination through viral attachment to sialic acid-containing cell surface components (17, 24, 35). Cellular integrins are also reported to mediate interactions with rotavirus outer capsid proteins (9). Proteolytically activated rotaviruses enter cells rapidly by a mechanism consistent with direct membrane penetration (25, 27, 56, 57). Cellular entry of rotaviruses occurs with a half-life of 5 min at 37°C (25, 56), and rotaviruses which are not proteolytically activated are endocytosed and degraded but do not infect cells (25, 27, 56, 57). Consistent with a direct entry mechanism, viral entry is not inhibited by lysosomotropic agents, which basify endosomes, or macrolide antibiotics, bafilomycin A, and concanamycin A, which block endosome acidification by selectively inhibiting the vacuolar H+-ATPase (11, 19, 25, 27).
Rotaviruses have also been shown to permeabilize cells, permitting 51Cr release, cell-cell fusion, ethidium bromide entry, and coentry of the cellular toxin alpha sarcin (11, 16, 25, 33, 49). TLP but not DLP rotaviruses have also been shown to permeabilize vesicles preloaded with carboxyfluorescein (CF) (40, 50). Several reports demonstrate that membrane permeability requires trypsinization of TLPs, is temperature dependent, occurs at neutral pH, and is inhibited by monoclonal antibodies (MAbs) to outer capsid proteins VP8*, VP5*, and VP7 (11, 16, 20, 21, 25, 27, 33, 40, 50). However, EGTA treatment of pretrypsinized TLPs also reportedly permits virus to permeabilize membranes or fuse cells (16, 49). EGTA solubilizes the VP4 and VP7 outer capsid proteins of the virion (7, 51), suggesting that solubilized outer capsid proteins are capable of permeabilizing membranes in the absence of particles or virion uncoating (16, 49).
In this report we demonstrate that the expressed VP5* protein from rhesus rotavirus (RRV) is capable of permeabilizing membranes in the absence of other rotavirus proteins or virion associations. We demonstrate that purified VP5* and truncated VP5* proteins containing predicted virion surface domains of VP5*, but not VP8* or VP4, permeabilize liposomes, causing the release of CF from large unilamellar vesicles (LUVs). VP5*-induced CF release is dose dependent and occurs optimally at 37°C at pH 7.35. VP5*-mediated membrane permeabilization is blocked by MAbs which recognize a conformationally determined epitope on the VP5* protein and neutralize RRV infectivity. Further, VP5* truncations which permeabilize LUVs contain the putative fusion domain of VP5* (36) but do not require the new N terminus of VP5* or downstream alpha-helical heptad repeats (residues 494 to 557) for membrane permeabilization. These findings functionally demonstrate the ability of the rotavirus VP5* protein to permeabilize membranes and suggest a required role for the VP5* cleavage product in cellular permeability during rotavirus entry.
MATERIALS AND METHODS
Reagents.
Egg yolk phosphatidylcholine (PC) (l-α-lecithin; molecular weight, 760), palmitoyl-oleoyl phosphatidylcholine (POPC), phosphatidyl ethanolamine (PE), and phosphatidyl serine (PS) in chloroform were obtained from Avanti Polar Lipids, Inc. (Alabaster, Ala.). The fluorescent dye CF (molecular weight, 376) was obtained from Molecular Probes (Eugene, Oreg.), and cholesterol was from Sigma (St. Louis, Mo.). Isopropyl-β-d-thiogalactopyranoside (IPTG) was obtained from Lab Scientific Inc. (Livingston, N.J.). Neutralizing MAbs (N-MAbs) M2, M7, 2G4, and 7A12 have been previously described (36, 38, 55). N-MAbs M2, M7, and 2G4 require VP4 residues 248 to 474, within VP5*, for recognition, while N-MAb 7A12 recognizes polypeptides containing residues 55 to 222 of VP8* (38). N-MAbs have previously been demonstrated to select neutralization escape mutants with mutations in their VP4 proteins at position 188 (MAb 7A12), 388 (MAb M2), or 393 (MAbs M7 and 2G4) (36).
Cells and virus.
RRV was cultivated in MA104 cells as previously described (54). MA104 cells were grown in Dulbecco modified Eagle medium (Gibco/BRL) in the presence of 10% fetal calf serum. Cells were infected at 0.1 focus-forming unit (FFU)/cell in serum-free medium, and medium was supplemented with trypsin (0.2 μg/ml). Infected cells were disrupted by freezing and thawing three times followed by Genetron extraction and purification on CsCl gradients as previously described (54). TLPs at 1.36 g of CsCl per ml were collected, trypsin (2 μg/ml) activated at 37°C for 15 min, and used in these studies. Virus was quantitated by the Bio-Rad protein assay, and titers of infectious virus were determined by a focus assay as previously described (55).
Plasmids.
Specific primers for the 5′ and 3′ ends of the coding sequence of RRV were synthesized, with each 5′-end primer having a BamHI site and each 3′ primer having either an SstI or an XhoI site. Products containing VP4 (amino acids [aa] 1 to 776), VP8* (aa 1 to 231), VP5* (aa 248 to 776), VP5*(248 to 475), and VP5*(265 to 475) were generated by PCR and ligated into pET-6HIS plasmids by standard methods. The pET-6HIS vector is a modified version of pET30a (Novagen) containing a replacement of the NdeI-to-BamHI fragment with a methionine and six histidines in frame 1 with the BamHI site. Ligation products were transformed into Escherichia coli XL-1 Blue (Stratagene) and selected on kanamycin plates. Constructs were verified by restriction enzyme digestion, sequencing as previously described (36), and protein expression (see below).
Protein expression and purification.
pET-6HIS plasmids encoding VP4, VP8*, VP5*, VP5*(248–474), or VP5*(265–474) were transformed into BL21(DE3) cells (Novagen). Single colonies were picked and grown overnight at 37°C in L broth with 50 μg of kanamycin per ml. Overnight cultures were diluted 1:20 in L broth-kanamycin, incubated at 37°C until the optical density at 600 nm was 0.6, and induced with IPTG for 3 h. Cells were harvested by centrifugation at 3,000 × g for 15 min and sonicated in 10 mM Tris (pH 8.0)–0.1 M NaH2PO4–2 M urea. Supernatants were discarded, and pellets were solubilized in 10 mM Tris (pH 8.0)–0.1 M NaH2PO4–8 M urea (buffer B). Lysates were pelleted in a microcentrifuge for 10 min, and nickel-nitrilotriacetic acid (NTA)-agarose (Qiagen) was added to supernatants and left for 30 minutes at room temperature. NTA resin was pelleted and washed five times with buffer B containing 50 mM imidazole and one time with buffer B (pH 6.3). Proteins were eluted in buffer B containing 100 mM EDTA. Eluted proteins were dialyzed sequentially in HEPES-buffered saline (HBS) (50 mM HEPES [pH 8.0], 150 mM NaCl) containing 8, 6, 4, 2, 1, or 0.5 M urea (>4 h each) and finally against HBS.
VP5* radiolabeling and recognition.
A 50-ml culture of BL21(DE3) cells containing VP5* pET-6HIS was grown to an optical density at 600 nm of 0.6 to 0.8 at 37°C. The cells were harvested by centrifugation at 3,000 × g for 15 min, washed with phosphate-buffered saline, and radiolabeled (100 μCi of Tran35S-label per ml) in methionine- and cysteine-free Grace’s medium (GIBCO) following induction with 1 mM IPTG at 37°C. Radiolabeled expressed proteins were purified as described above on NTA resin (Qiagen). Radiolabeled VP5* was immunoprecipitated as previously described (38) with VP5*-specific (2G4) or VP8*-specific (7A12) N-MAbs (55) in radioimmunoprecipitation assay buffer containing 0.1% sodium dodecyl sulfate (SDS). Proteins were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide), and protein bands were visualized by fluorography. VP8* and VP4 proteins were recognized by MAb 7A12, and VP4 was recognized by 2G4, by enzyme-linked immunosorbent assay as previously described (55).
Preparation of liposomes.
Liposomes of PC containing CF were prepared by an extrusion method (3, 40). Briefly, 2 mg of PC was dissolved in chloroform, dried under nitrogen, and further dried under vacuum. Lipids were resuspended in 20 μl of CF (70 mM in 10 mM Tris, pH 7.35) by vortexing. Following five cycles of freezing and thawing, LUVs were prepared by extrusion through a 0.1-μm-pore-size membrane (Mini-extruder; Avanti Polar Lipids). Extravesicular fluorophore was eliminated by size exclusion chromatography on Sephadex G-50 (Pharmacia) in 10 mM Tris-HCl (pH 7.35)–140 mM NaCl (TN buffer) (3, 40). Fractions (150 μl) were collected, and 3 μl was assayed in 1.6 ml of TN buffer for fluorescence dequenching following addition of 0.125% Triton X-100. Fluorescence dequenching was monitored in a Perkin-Elmer LS-5B luminescence spectrometer at 520 nm (490 nm for excitation) (22a). Fractions with a fluorescence change ratio of >15 were pooled and used in CF release assays.
CF release assays.
The ability of RRV and purified RRV proteins to permeabilize LUVs and cause CF release was assayed by measuring the fluorescence dequenching of the released fluor, essentially as previously described (40, 50). CF-containing LUVs (5 to 10 μl) were equilibrated in 1.6 ml of TN buffer in fluorimeter cuvettes at 37°C for 3 min with constant stirring prior to addition of rotavirus (107 to 108 FFU) or rotavirus proteins (0.1 to 25 μg). CF release was monitored over time under various conditions as described in the figure legends. All experiments were repeated several times with different preparations of liposomes, proteins, and virus. At the conclusion of each experiment, LUVs were lysed with 0.1% Triton X-100 to determine the maximum dequenching within each sample (40). Samples with no added proteins or virus were incubated and similarly treated to control for spontaneous CF release from liposomes.
Results are expressed as percentages of total fluorescence dequenching resulting from Triton X-100 addition. The percent fluorophore dequenching was calculated according to the formula percent release = (Ft − F0)/(FT − F0)] × 100, where F0 is the background fluorescence, Ft is the fluorescence at time t, and FT is the total fluorescence of the sample (40).
The ability of VP5 to permeabilize liposomes was determined in 32 separate experiments, using freshly prepared CF-loaded liposomes and at least 21 different preparations of VP5. In order to verify that CF was specifically released by VP5, each lot of VP5 was tested for the ability of MAb 2G4 to block its activity. No lots of VP5 caused CF release that was not blocked by prior addition of MAb 2G4. Results from replicate samples assayed during the course of these experiments never varied by more than 5%.
RESULTS
Protein expression and purification.
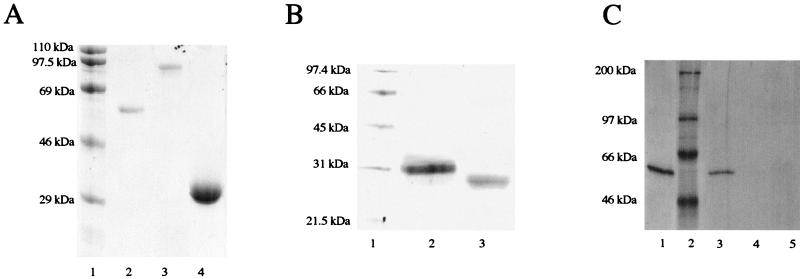
VP5*, VP8*, and VP4 were expressed in pET-6HIS plasmids fused to an upstream methionine and six histidine residues. Following IPTG induction, VP5* protein was expressed at high levels in BL21(DE3) bacteria (Fig. 1A). The N-terminal six-histidine tag conferred protein binding to nickel, and induced proteins were purified on a nickel-NTA-agarose resin (Fig. 1A). Purified VP5* was eluted, dialyzed into HBS, and tested for its recognition by VP5*-specific N-MAbs 2G4, M2, and M7 (36, 55). All three N-MAbs immunoprecipitated VP5* (results for 2G4 are shown Fig. 1C), while the VP8*-specific N-MAb, 7A12, failed to recognize VP5*. VP5* truncations containing residues 248 to 474 or 265 to 474 were similarly expressed and purified from pET-6HIS plasmids (Fig. 1B). Both truncated proteins are recognized by MAbs 2G4, M2, and M7 (not shown). Since MAbs 2G4, M2, and M7 recognize a conformational epitope of VP5* present on RRV (38, 55), this indicates that purified expressed VP5* proteins are present in a conformation similar to that of the virion outer capsid.
FIG. 1.
Expression and purification of proteins VP5*, VP4, and VP8* (A) and VP5*(248–474) and VP5*(265–474) (B) were expressed in BL21(DE3) from pET-6HIS plasmids. Bacteria were induced with IPTG (1 mM) and grown for 3 h prior to pelleting and sonication. Proteins were purified on Ni-NTA-agarose (Qiagen), and crude or purified samples were analyzed by SDS-PAGE (10% polyacrylamide for panel A and 12% polyacrylamide for panel B) and stained with Coomassie blue. (A) Lanes: 1, molecular mass standards; 2, VP5; 3, VP4; 4, VP8. (B) Lanes: 1, molecular mass standards; 2, VP5*(248–474); 3, VP5*(265–474). (C) VP5* was radiolabeled following IPTG induction of pET-6HIS VP5* plasmids in BL21(DE3) with 100 μCi of Tran35S-label per ml in methionine- and cysteine-free Grace’s medium. VP5* was purified on Ni-NTA-agarose as described above. VP5* was analyzed directly by SDS-PAGE (10% polyacrylamide) (lane 1) or immunoprecipitated with MAb 264 (lane 3), MAb 7A12 (lane 4), or normal mouse serum (lane 5). Lane 2, molecular mass standards. Proteins were visualized by fluorography.
VP5* permeabilize LUVs.
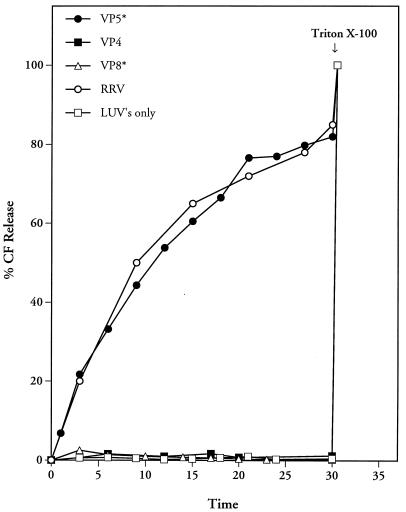
Trypsinized rotavirus TLPs as well as outer capsid proteins released from the virion by calcium chelation are capable of permeabilizing membranes (40, 49, 50). In these experiments the ability of the purified expressed VP5* protein to permeabilize membranes was tested by using LUVs preloaded with CF. Fluorescence dequenching and an increase in fluorescence at 520 nm result from permeabilization of CF-loaded LUVs (40). Similar to the case for trypsinized TLPs, addition of purified VP5* protein to LUVs resulted in 80 to 90% CF release in 30 min at 37°C (Fig. 2). In contrast, LUVs alone or the addition of VP8* or VP4 proteins did not cause CF release from LUVs (Fig. 2). Triton X-100 addition at the conclusion of experiments completely released CF from LUVs, demonstrating that the assay mixtures contained equivalent amounts of CF-loaded LUVs.
FIG. 2.
VP5* permeabilizes LUVs. Purified LUVs containing CF (70 mM) were incubated in 1.6 ml of TN buffer at 37°C with or without prior addition of 100 μl of trypsin-activated RRV (0.5 mg/ml; 109 FFU/ml) or nickel affinity-purified VP4 (5 μg; 37.5 nM), VP8* (10 μg; 240 nM), or VP5* (4 μg; 44 nM). VP4 and VP8* failed to induce CF release over the entire range of protein concentrations tested (VP8*, 5 to 25 μg [120 to 600 nM]); VP4, 1 to 10 μg [7.5 to 75 nM]). Fluorescence dequenching of samples was monitored at various times at 520 nm in an LS-5B luminescence spectrometer following excitation at 490 nm. After 30 min, Triton X-100 (0.125%) was added in order to assess the total CF content of each sample, which was quantitated as described in Materials and Methods. This experiment was repeated three times with similar results.
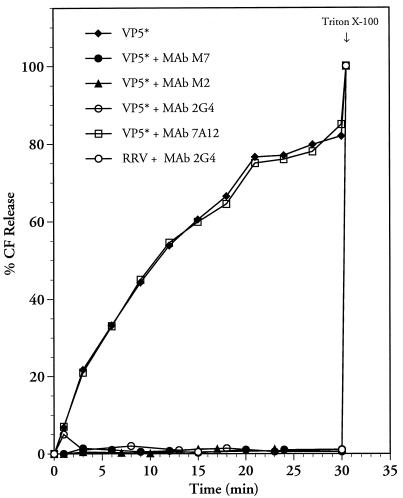
Neutralizing MAbs to VP5* completely inhibit CF release.
VP5*-specific MAbs were used to demonstrate that LUVs were specifically permeabilized by the VP5* protein. VP5*-specific antibodies M2, 2G4, and M7 neutralize RRV infectivity through the recognition of a conformationally determined epitope on the virion surface (38, 55). VP5*-specific N-MAbs (2G4, M2, and M7) or a VP8*-specific N-MAb (7A12) (1 μl of ascites fluid) was incubated with VP5* for 30 min at room temperature prior to the addition of VP5* to LUVs. RRV was incubated with MAb 2G4 for 15 min prior to addition to CF-containing liposomes as previously described (50). Addition of the VP8*-specific MAb, 7A12, to VP5* had no effect on VP5*-induced CF release (Fig. 3). In contrast, addition of VP5*-specific N-MAbs completely blocked the ability of VP5* to permeabilize LUVs. The ability of 2G4 to inhibit rotavirus-mediated CF release in this experiment corresponds to the results of Ruiz et al. (50). These findings demonstrate the specificity of VP5*-induced membrane permeabilization in the CF release assay and further indicate that VP5*-specific N-MAbs block the ability of VP5* to permeabilize membranes.
FIG. 3.
VP5*-specific N-MAbs block VP5*-induced membrane permeability. VP5*-specific (2G4, M2, and M7) or VP8*-specific (7A12) MAbs were incubated with purified VP5* (4 μg; 44 nM) or 100 μl of trypsin-activated RRV (0.5 mg/ml; 109 FFU/ml) at room temperature for 30 min prior to addition to CF-loaded LUVs in 1.6 ml of TN buffer at 37°C. At various time points, fluorescence dequenching of samples was monitored in a fluorimeter at an excitation wavelength of 490 nm and an emission wavelength of 520 nm. After 30 min, Triton X-100 (0.125%) was added in order to assess the total CF content of each sample, which was quantitated as described in Materials and Methods. This experiment was repeated two times with similar results. The specific function of each lot of VP5 was tested with MAb 2G4 in a CF release assay (see Materials and Methods).
Determinants of VP5*-induced membrane permeability.
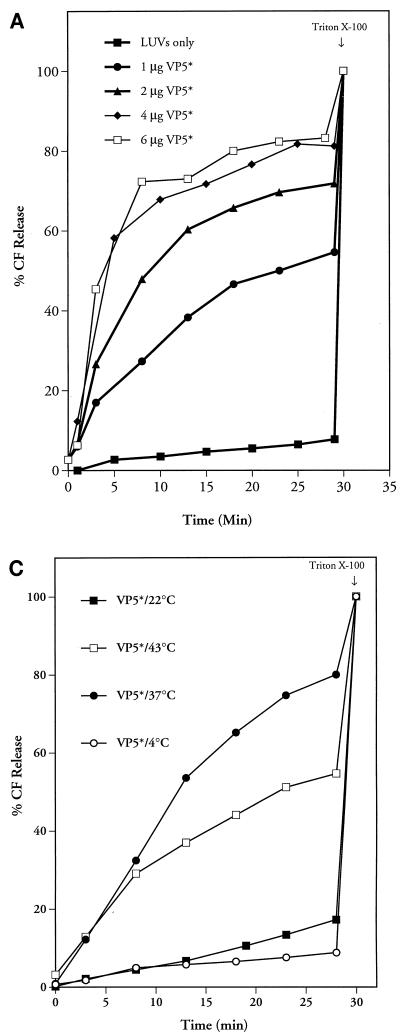
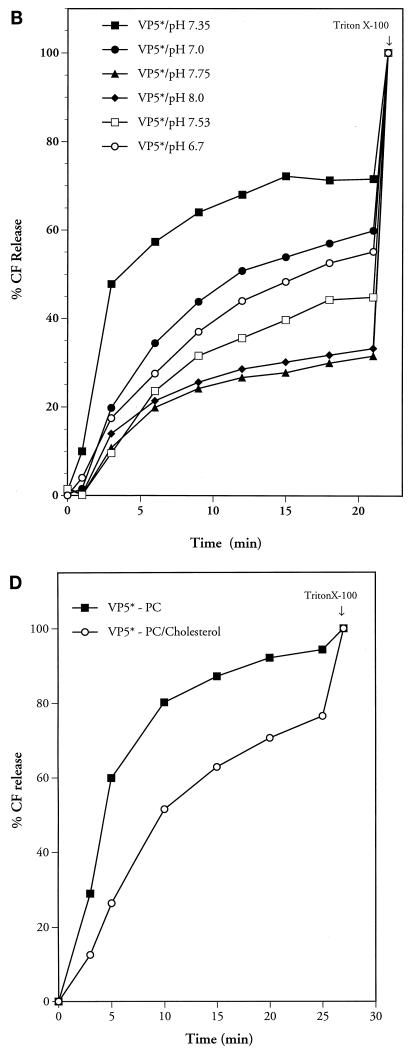
The requirements for VP5*-induced membrane permeability were tested under a number of conditions. Addition of increasing amounts of VP5* to membranes demonstrated that VP5*-induced CF release is concentration dependent (Fig. 4A). Equilibration of liposomes in buffers of various pHs or temperatures further demonstrated that VP5*-induced membrane permeability is optimal at neutral pH (pH 7.35) at 37°C (Fig. 4B and C).
FIG. 4.
Conditions of VP5*-induced membrane permeability. VP5* was added to CF-loaded LUVs under various conditions. (A) Increasing amounts of purified VP5* (1 μg [11 nM], 2 μg [22 nM], 4 μg [44 nM], or 6 μg [66 nM]) were added to LUVs. This experiment was repeated two times with similar results. (B) VP5* (4 μg; 44 nM) was incubated with LUVs in TN buffer at various pHs, and results were corrected for spontaneous CF release at identical pHs. This experiment was repeated six times with similar results. (C) VP5* (4 μg; 44 nM) and LUVs were incubated as described above at 4, 22, 37, or 43°C. This experiment was repeated five times with similar results. (D) Egg yolk PC or PC-cholesterol (10:1) LUVs were preloaded with CF and purified by Sephadex G-50 chromatography. This experiment was repeated three times with similar results. The ability of the VP5* (4 μg; 44 nM) to permeabilize PC liposomes with or without the inclusion of cholesterol was monitored over time. Fluorescence dequenching of samples was monitored in a fluorimeter at an excitation wavelength of 490 nm and an emission wavelength of 520 nm. After 22 min (B), 27 min (D), or 30 min (A and C), the total CF content of each sample was assessed by addition of Triton X-100 (0.125%) and quantitated as described in Materials and Methods.
Rotaviruses are sensitive to calcium concentration and are converted from infectious TLPs to noninfectious but transcriptionally active DLPs by reduced intracellular [Ca2+] (7, 51). However, the addition of 5 mM CaCl2, MgCl2, MnCl2, or ZnCl2 to CF-containing LUVs had no effect on VP5*-induced CF release (not shown). VP5* also permeabilized LUVs with various lipid compositions, including POPC, PS, or PE at a 1:1 ratio with PC (not shown). Additionally there was no detectable difference in VP5*-induced CF release from PC or POPC CF-loaded LUVs. Cholesterol has also been suggested to be a requirement for rotavirus-mediated membrane permeability (16, 21). Figure 4D demonstrates the ability of the VP5* protein to permeabilize PC liposomes with or without the inclusion of cholesterol (PC/cholesterol ratio, 10:1).
VP5* truncations permeabilize LUVs.
Single amino acid changes within the VP5* protein at residues 388 and 393 are selected by N-MAbs M2, M7, and 2G4 within the putative fusion domain of VP5* (36). Further mapping of these antibodies by deletional analysis defined that the VP5*(248–474) fragment (residues 248 to 474) was required for recognition by the VP5*-specific N-MAbs (38). Additionally, cleavage of VP4 following residue 247 has recently been shown to be required for virus-like particle (VLP)-mediated membrane permeabilization (20). In order to determine whether the N terminus of VP5* is required for VP5* function and whether VP5*(248–474) MAb recognition domains are sufficient for VP5*-induced membrane permeabilization, we investigated the ability of truncated VP5* proteins to permeabilize LUVs. Similar to VP5*, both VP5*(248–474) and VP5*(265–474) were capable of permeabilizing LUVs (Fig. 5). Further, the ability of the VP5* truncation proteins to permeabilize membranes was completely inhibited by prior incubation with VP5*-specific N-MAb 2G4 (Fig. 5). This demonstrates that VP5* truncations containing predicted extravirion domains of VP5* are functional in membrane permeabilization and that a VP5* truncation containing residues 265 to 474 is recognized by N-MAb 2G4. Additionally, N-terminal residues of VP5*, resulting from trypsin cleavage of VP4 after residue 247 on the virion, are not required for purified VP5* to permeabilize membranes.
FIG. 5.
VP5* truncations permeabilize LUVs. Purified LUVs containing CF were incubated in 1.6 ml of TN buffer at 37°C for 30 min with purified VP5* (6 μg; 67 nM) or truncation VP5*(248–474) (3 μg; 72 nM) or VP5*(265–474) (3 μg; 78 nM). As for Fig. 3, aliquots were also incubated with VP5*-specific N-MAb 2G4 at room temperature for 30 min prior to incubation with LUVs. Fluorescence dequenching of samples was monitored in a fluorimeter at an excitation wavelength of 490 nm and an emission wavelength of 520 nm. After 22 min, the total CF content of each sample was assessed by addition of Triton X-100 (0.125%) and quantitated as described in Materials and Methods. This experiment was repeated three times with similar results.
DISCUSSION
Although fusion proteins which mediate the entry of enveloped viruses have been extensively investigated, the ability of nonenveloped viruses to cross cellular membranes and enter cells is poorly understood. In this study we have demonstrated that the purified rotavirus VP5* protein is capable of permeabilizing membranes in the absence of other rotavirus proteins. Like virus-induced membrane permeability, VP5*-induced membrane permeability is optimal at 37°C at neutral pH (40). VP5* truncations containing predicted extravirion domains are similarly active in permeabilizing membranes. These findings implicate the outer capsid protein VP5* in permeabilizing membranes during rotavirus entry and further suggest that trypsin cleavage of VP4 may be required to effect conformational changes in VP5* which permit VP5*-membrane interactions.
VP5*-mediated membrane permeabilization is blocked by N-MAbs which recognize a conformationally determined epitope of VP5* on the virion (38, 55). This demonstrates that apparently native VP5* is functional in permeabilizing membranes and further suggests that VP5*-specific N-MAbs may neutralize rotavirus by preventing virus-membrane interactions and cellular entry. Neutralizing antibodies to VP7 or VP8* have also been demonstrated to inhibit TLP-induced membrane permeability (11, 16, 33, 50). However, the function of VP5* in permeabilizing membranes suggests that antibodies to VP8* and VP7 may block membrane permeability indirectly, perhaps by altering conformational changes in VP5*, by sterically inhibiting VP5*-membrane interactions, or by preventing cell attachment or virion uncoating. However, our studies do not exclude the possibility that VP8* or VP7 has membrane-permeabilizing functions, and a role for VP7 in permeabilizing membranes has been suggested (4). In these studies there was no evidence that VP8* or uncleaved VP4 was capable of permeabilizing membranes.
Antibodies to VP5* were previously used to generate neutralization escape mutants with single amino acid changes within the VP5* protein selected by N-MAbs M2 M7 and 2G4 at residues 388 and 393 (36). Further mapping of these antibodies by deletional analysis defined the VP5*(248–474) fragment (residues 248 to 474) required for recognition by the VP5*-specific N-MAbs (38). Two truncated VP5* proteins were functionally analyzed in these experiments, the VP5*(248–474) fragment and a short N-terminal truncation of VP5*(265–474). Both VP5 truncations were capable of permeabilizing membranes, but permeabilization was completely blocked by preincubating truncations with N-MAb 2G4.
We previously reported that VP5 contains a hydrophobic putative internal “fusion” domain (VP5 HD) (residues 385 to 404) which has homology with the fusion domains of the E1 proteins of Sindbis virus and Semliki Forest virus (36). Interestingly, the identical GGA sequence required for the function of the Semliki Forest virus E1 fusion peptide (residues 90 to 92) is present in the VP5* HD and conserved in all rotaviruses (28, 29, 30, 32, 36). An additional diglycine motif flanks the VP5* HD, and five helix-breaking glycine and proline residues, which are highly conserved among rotaviruses, confer a random coiled structure to the VP5* HD. Glycine residues have recently been identified as key elements in vesicular stomatitis virus G protein-induced membrane fusion (6).
Although rotaviruses do not normally fuse cells, studies using a rotavirus cell-cell fusion assay showed a requirement for cholesterol for rotavirus-induced cell fusion (16, 20, 21). Cholesterol is not required for rotavirus permeabilization of model membranes (40, 50), and from our studies, VP5* proteins containing VP5* HD mediate membrane permeabilization which is cholesterol independent. As a result, cholesterol may be a specific requirement of the cell-cell fusion assay but not a specific requirement of VP5*-induced membrane permeabilization.
Recently, VLPs with mutagenized VP4 trypsin cleavage sites were used to demonstrate that cleavage following residue 247 was required for VLP-induced cell-cell fusion (20). The specificity suggested that the new amino terminus of VP5* was required for membrane permeabilization and that additional residues present on VP5* might block VP5* membrane functions. However, in this study each of the expressed VP5* proteins contains an additional methionine and six histidine residues added to the N terminus of VP5*, and the addition of these residues does not block VP5*-mediated membrane permeabilization. In addition, the VP5*(265–474) truncation, with the N-terminal 17 residues of VP5* deleted, is functional in membrane permeabilization. These results suggest that specific VP4 cleavage is a requirement for the function of VP5* on the virion and is not a functional requirement of the VP5* polypeptide.
Other nonenveloped viruses, like poliovirus, reovirus, and adenovirus, have been suggested to mediate interactions with membranes during viral entry (2, 5, 8, 12, 18, 22, 23, 39, 41, 48, 52, 58). However, specific proteins from these viruses have not been demonstrated to have membrane-permeabilizing activity. The ability of the rotavirus VP5* protein to permeabilize membranes suggests that additional nonenveloped viruses are likely to have specific proteins which effect membrane permeability during viral entry.
Hypotheses for how rotavirus outer capsid proteins permeabilize membranes have also been suggested (16, 33, 49, 50). Reduced intracellular calcium levels within early endosomes have been suggested to uncoat rotavirus outer capsid proteins, which then permeabilize and lyse the early endosome, effecting viral entry (49). It has also been suggested that outer capsid proteins on the virion first permeabilize cellular membranes, permitting calcium efflux from the early endosome (49). The reduced-calcium environment causes subsequent conformational changes, and outer capsid uncoating disrupts the early endosomal membrane attached to the virion (49). It is also possible that a focal permeabilization of the cellular membrane occurs and permits a localized calcium flux in the virion which effects virion uncoating and membrane disruption.
The ability of VP5* to permeabilize membranes is consistent with all of these possibilities. However, it has yet to be determined whether VP5* lyses or selectively permeabilizes membranes. The delivery of rotavirus 50-nm particles into cells is relatively complex, involving both membrane permeability and conformational changes in the virion which could contribute to particle entry and membrane disruption. In contrast, the use of purified VP5* in studies of membrane permeability should permit us to define VP5* functions in membrane interactions and discern the role of VP5* in the rotavirus entry process. Additionally, the ability to selectively mutagenize VP5* should further define the domains and residues of VP5* which are required for membrane binding and permeability.
ACKNOWLEDGMENTS
We are grateful to Erwin London and Irina Gavrilovskaya for insightful discussions of these studies. We thank Mary Ellen Fay and Yildiz Farooqui for technical assistance.
This work was supported by a Merit Award from the Veterans Administration and by NIH grants R01-AI31016 and RO3-AI42150 to E.R.M.
REFERENCES
- 1.Arias C F, Romero P, Alvarez V, Lopez S. Trypsin activation pathway of rotavirus infectivity. J Virol. 1996;70:5832–5839. doi: 10.1128/jvi.70.9.5832-5839.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumenthal R, Seth P, Willingham M C, Pastan I. pH-dependent lysis of liposomes by adenovirus. Biochemistry. 1986;25:2231–2237. doi: 10.1021/bi00356a057. [DOI] [PubMed] [Google Scholar]
- 3.Butko P, Huang F, Pusztai-Carey M, Surewicz W K. Membrane permeabilization induced by cytolytic delta-endotoxin CytA from Bacillus thuringiensis var. israelensis. Biochemistry. 1996;35:11355–11360. doi: 10.1021/bi960970s. [DOI] [PubMed] [Google Scholar]
- 4.Charpilienne A, Abad M J, Michelangeli F, Alvarado F, Vasseur M, Cohen J, Ruiz M C. Solubilized and cleaved VP7, the outer glycoprotein of rotavirus, induces permeabilization of cell membrane vesicles. J Gen Virol. 1997;78:1367–1371. doi: 10.1099/0022-1317-78-6-1367. [DOI] [PubMed] [Google Scholar]
- 5.Chow M, Newman J F, Filman D, Hogle J M, Rowlands D J, Brown F. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature. 1987;327:482–486. doi: 10.1038/327482a0. [DOI] [PubMed] [Google Scholar]
- 6.Cleverley D Z, Lenard J. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc Natl Acad Sci USA. 1998;95:3425–3430. doi: 10.1073/pnas.95.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen J, Laporte J, Charpilienne A, Scherrer R. Activation of rotavirus RNA polymerase by calcium chelation. Arch Virol. 1979;60:177–186. doi: 10.1007/BF01317489. [DOI] [PubMed] [Google Scholar]
- 8.Cotten M, Saltik M, Kursa M, Wagner E, Maass G, Birnstiel M L. Psoralen treatment of adenovirus particles eliminates virus replication and transcription while maintaining the endosomolytic activity of the virus capsid. Virology. 1994;205:254–261. doi: 10.1006/viro.1994.1641. [DOI] [PubMed] [Google Scholar]
- 9.Coulson B S, Londrigan S L, Lee D J. Rotavirus contains integrin ligand sequences and a disintegrin-like domain that are implicated in virus entry into cells. Proc Natl Acad Sci USA. 1997;94:5389–5394. doi: 10.1073/pnas.94.10.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford S E, Labbe M, Cohen J, Burroughs M H, Zhou Y J, Estes M K. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J Virol. 1994;68:5945–5922. doi: 10.1128/jvi.68.9.5945-5952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuadras M A, Arias C F, Lopez S. Rotaviruses induce an early membrane permeabilization of MA104 cells and do not require a low intracellular Ca2+ concentration to initiate their replication cycle. J Virol. 1997;71:9065–9074. doi: 10.1128/jvi.71.12.9065-9074.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curry S, Chow M, Hogle J M. The poliovirus 135S particle is infectious. J Virol. 1996;70:7125–7131. doi: 10.1128/jvi.70.10.7125-7131.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espejo R T, Lopez S, Arias C. Structural polypeptides of simian rotavirus SA11 and the effect of trypsin. J Virol. 1981;37:156–160. doi: 10.1128/jvi.37.1.156-160.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estes M K. Rotaviruses and their replication. In: Fields B N, Knipe D M, editors. Virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1329–1352. [Google Scholar]
- 15.Estes M K, Graham D Y, Mason B B. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J Virol. 1981;39:879–888. doi: 10.1128/jvi.39.3.879-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falconer M M, Gilbert J M, Roper A M, Greenberg H B, Gavora J S. Rotavirus-induced fusion from without in tissue culture cells. J Virol. 1995;69:5582–5591. doi: 10.1128/jvi.69.9.5582-5591.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiore L, Greenberg H B, Mackow E R. The VP8* fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology. 1991;181:553–563. doi: 10.1016/0042-6822(91)90888-i. [DOI] [PubMed] [Google Scholar]
- 18.Fricks C E, Hogle J M. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol. 1990;64:1934–1945. doi: 10.1128/jvi.64.5.1934-1945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuhara N, Yoshie O, Kitaoka S, Konno T, Ishida N. Evidence for endocytosis-independent infection by human rotavirus. Arch Virol. 1987;97:93–99. doi: 10.1007/BF01310737. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert J, Greenberg H. Cleavage of rhesus rotavirus VP4 after arginine 247 is essential for rotavirus-like particle-induced fusion from without. J Virol. 1998;72:5323–5327. doi: 10.1128/jvi.72.6.5323-5327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert J, Greenberg H. Virus-like particle-induced fusion form without in tissue culture cells: role of outer-layer proteins VP4 and VP7. J Virol. 1997;71:4555–4563. doi: 10.1128/jvi.71.6.4555-4563.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greber U F, Willetts M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–486. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- 22a.Hargland R P. Polar tracers. In: Spence M T Z, editor. Handbook of fluorescent probes and research chemicals. 6th ed. Portland, Ore: Molecular Probes, Inc.; 1990. pp. 331–338. [Google Scholar]
- 23.Honda M, Hu P C, Huang C H, Matsui H, Lemon S M. A replication-deficient adenovirus enhances liposome-mediated nucleic acid transfer into a stable cell line expressing T7 RNA polymerase. J Virol Methods. 1996;58:41–51. doi: 10.1016/0166-0934(95)01986-3. [DOI] [PubMed] [Google Scholar]
- 24.Kalica A R, Flores J, Greenberg H B. Identification of the rotaviral gene that codes for hemagglutination and protease-enhanced plaque formation. Virology. 1983;125:194–205. doi: 10.1016/0042-6822(83)90073-9. [DOI] [PubMed] [Google Scholar]
- 25.Kaljot K T, Shaw R D, Rubin D H, Greenberg H B. Infectious rotavirus enters cells by direct cell membrane penetration, not by endocytosis. J Virol. 1988;62:1136–1144. doi: 10.1128/jvi.62.4.1136-1144.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapikian A Z, Chanock R M. Rotaviruses. In: Fields B N, Knipe D M, editors. Virology. 3rd ed. Vol. 2. New York, N.Y: Raven Press; 1996. pp. 1353–1404. [Google Scholar]
- 27.Keljo D J, Kuhn M, Smith A. Acidification of endosomes is not important for the entry of rotavirus into the cell. J Pediatr Gastroenterol Nutr. 1988;7:257–263. doi: 10.1097/00005176-198803000-00016. [DOI] [PubMed] [Google Scholar]
- 28.Kielian M. Membrane fusion and the alphavirus life cycle. Adv Virus Res. 1995;45:113–151. doi: 10.1016/s0065-3527(08)60059-7. [DOI] [PubMed] [Google Scholar]
- 29.Kielian M, Helenius A. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J Cell Biol. 1985;101:2284–2291. doi: 10.1083/jcb.101.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kielian M, Klimjack M R, Ghosh S, Duffus W A. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J Cell Biol. 1996;134:863–872. doi: 10.1083/jcb.134.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawton J A, Estes M K, Prasad B V. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 32.Levy-Mintz P, Kielian M. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J Virol. 1991;65:4292–4300. doi: 10.1128/jvi.65.8.4292-4300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liprandi F, Moros Z, Gerder M, Ludert J E, Pujol F H, Ruiz M C, Michelangeli F, Charpilienne A, Cohen J. Productive penetration of rotavirus in cultured cells induces coentry of the translation inhibitor alpha-sarcin. Virology. 1997;237:430–438. doi: 10.1006/viro.1997.8803. [DOI] [PubMed] [Google Scholar]
- 34.Lopez S, Lopez I, Romero P, Mendez E, Soberon X, Arias C F. Rotavirus YM gene 4: analysis of its deduced amino acid sequence and prediction of the secondary structure of the VP4 protein. J Virol. 1991;65:3738–3745. doi: 10.1128/jvi.65.7.3738-3745.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackow E R, Barnett J W, Chan H, Greenberg H B. The rhesus rotavirus outer capsid protein VP4 functions as a hemagglutinin and is antigenically conserved when expressed by a baculovirus recombinant. J Virol. 1989;63:1661–1668. doi: 10.1128/jvi.63.4.1661-1668.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mackow E R, Shaw R D, Matsui S M, Vo P T, Dang M N, Greenberg H B. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc Natl Acad Sci USA. 1988;85:645–649. doi: 10.1073/pnas.85.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackow E R, Vo P T, Broome R, Bass D, Greenberg H B. Immunization with baculovirus-expressed VP4 protein passively protects against simian and murine rotavirus challenge. J Virol. 1990;64:1698–1703. doi: 10.1128/jvi.64.4.1698-1703.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackow E R, Yamanaka M Y, Dang M N, Greenberg H B. DNA amplification-restricted transcription-translation: rapid analysis of rhesus rotavirus neutralization sites. Proc Natl Acad Sci USA. 1990;87:518–522. doi: 10.1073/pnas.87.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moscufo N, Yafal A G, Rogove A, Hogle J, Chow M. A mutation in VP4 defines a new step in the late stages of cell entry by poliovirus. J Virol. 1993;67:5075–5078. doi: 10.1128/jvi.67.8.5075-5078.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nandi P, Charpilienne A, Cohen J. Interaction of rotavirus particles with liposomes. J Virol. 1992;66:3363–3367. doi: 10.1128/jvi.66.6.3363-3367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nibert M L, Schiff L A, Fields B N. Mammalian reoviruses contain a myristoylated structural protein. J Virol. 1991;65:1960–1967. doi: 10.1128/jvi.65.4.1960-1967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Offit P A, Blavat G. Identification of the two rotavirus genes determining neutralization specificities. J Virol. 1986;57:376–378. doi: 10.1128/jvi.57.1.376-378.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Offit P A, Clark H F, Blavat G, Greenberg H B. Reassortant rotaviruses containing structural proteins vp3 and vp7 from different parents induce antibodies protective against each parental serotype. J Virol. 1986;60:491–496. doi: 10.1128/jvi.60.2.491-496.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patton J T, Hua J, Mansell E A. Location of intrachain disulfide bonds in the VP5** and VP8** trypsin cleavage fragments of the rhesus rotavirus spike protein VP4. J Virol. 1993;67:4848–4855. doi: 10.1128/jvi.67.8.4848-4855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prasad B V, Chiu W. Structure of rotavirus. Curr Top Microbiol Immunol. 1994;185:9–29. doi: 10.1007/978-3-642-78256-5_2. [DOI] [PubMed] [Google Scholar]
- 46.Prasad B V, Rothnagel R, Zeng C Q, Jakana J, Lawton J A, Chiu W, Estes M K. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 47.Prasad B V, Wang G J, Clerx J P, Chiu W. Three-dimensional structure of rotavirus. J Mol Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez E, Everitt E. Adenovirus uncoating and nuclear establishment are not affected by weak base amines. J Virol. 1996;70:3470–3477. doi: 10.1128/jvi.70.6.3470-3477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruiz M C, Abad M J, Charpilienne A, Cohen J, Michelangeli F. Cell lines susceptible to infection are permeabilized by cleaved and solubilized outer layer proteins of rotavirus. J Gen Virol. 1997;78:2883–2893. doi: 10.1099/0022-1317-78-11-2883. [DOI] [PubMed] [Google Scholar]
- 50.Ruiz M C, Alonso-Torre S R, Charpilienne A, Vasseur M, Michelangeli F, Cohen J, Alvarado F. Rotavirus interaction with isolated membrane vesicles. J Virol. 1994;68:4009–4016. doi: 10.1128/jvi.68.6.4009-4016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruiz M C, Charpilienne A, Liprandi F, Gajardo R, Michelangeli F, Cohen J. The concentration of Ca2+ that solubilizes outer capsid proteins from rotavirus particles is dependent on the strain. J Virol. 1996;70:4877–4883. doi: 10.1128/jvi.70.8.4877-4883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seth P. Mechanism of adenovirus-mediated endosome lysis: role of the intact adenovirus capsid structure. Biochem Biophys Res Commun. 1994;205:1318–1324. doi: 10.1006/bbrc.1994.2809. [DOI] [PubMed] [Google Scholar]
- 53.Shaw A L, Rothnagel R, Chen D, Ramig R F, Chiu W, Prasad B V. Three-dimensional visualization of the rotavirus hemagglutinin structure. Cell. 1993;74:693–701. doi: 10.1016/0092-8674(93)90516-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaw R D, Stoner-Ma D L, Estes M K, Greenberg H B. Specific enzyme-linked immunoassay for rotavirus serotypes 1 and 3. J Clin Microbiol. 1985;22:286–291. doi: 10.1128/jcm.22.2.286-291.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaw R D, Vo P T, Offit P A, Coulson B S, Greenberg H B. Antigenic mapping of the surface proteins of rhesus rotavirus. Virology. 1986;155:434–451. doi: 10.1016/0042-6822(86)90205-9. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki H, Kitaoka S, Konno T, Sato T, Ishida N. Two modes of human rotavirus entry into MA 104 cells. Arch Virol. 1985;85:25–34. doi: 10.1007/BF01317003. [DOI] [PubMed] [Google Scholar]
- 57.Suzuki H, Kitaoka S, Sato T, Konno T, Iwasaki Y, Numazaki Y, Ishida N. Further investigation on the mode of entry of human rotavirus into cells. Arch Virol. 1986;91:135–144. doi: 10.1007/BF01316734. [DOI] [PubMed] [Google Scholar]
- 58.Tosteson M T, Chow M. Characterization of the ion channels formed by poliovirus in planar lipid membranes. J Virol. 1997;71:507–511. doi: 10.1128/jvi.71.1.507-511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeager M, Berriman J A, Baker T S, Bellamy A R. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 1994;13:1011–1018. doi: 10.1002/j.1460-2075.1994.tb06349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]