Abstract
The ongoing degradation of natural systems and other environmental changes has put our society at a crossroad with respect to our future relationship with our planet. While the concept of One Health describes how human health is inextricably linked with environmental health, many of these complex interdependencies are still not well‐understood. Here, we describe how the advent of real‐time genomic analyses can benefit One Health and how it can enable timely, in‐depth ecosystem health assessments. We introduce nanopore sequencing as the only disruptive technology that currently allows for real‐time genomic analyses and that is already being used worldwide to improve the accessibility and versatility of genomic sequencing. We showcase real‐time genomic studies on zoonotic disease, food security, environmental microbiome, emerging pathogens, and their antimicrobial resistances, and on environmental health itself – from genomic resource creation for wildlife conservation to the monitoring of biodiversity, invasive species, and wildlife trafficking. We stress why equitable access to real‐time genomics in the context of One Health will be paramount and discuss related practical, legal, and ethical limitations.
Keywords: global health, nanopore sequencing, nature conservation, One Health, real‐time genomics
Subject Categories: Chromatin, Transcription & Genomics; Evolution & Ecology
The One Health concept describes the inextricable link between human and environmental health. This review discusses how real‐time genomics can improve our understanding of the interdependencies within ‘One Health’ and enable timely assessments and interventions in many contexts.

Introduction
The COVID‐19 pandemic has catapulted the concept of One Health into the center of public attention, showcasing how human health is inextricably linked with the health of our planet (de León et al, 2021; van Oosterhout, 2021; One Health High‐Level Expert Panel et al, 2022). Although we do not fully understand the connection between the emergence of zoonotic diseases, wild habitat destruction, and biodiversity loss (Keesing & Ostfeld, 2021), projected environmental changes will undoubtedly place an additional burden on planetary and human health. Society is now at a crossroad with respect to our future relationship with planetary health, and our window to act is rapidly closing. This urgency is reflected by the ongoing political, public, and scientific discussions led by the United Nations (UN) Convention on Biological Diversity (CBD), the International Union for Conservation of Nature (IUCN Red List, 2012), the Rockefeller Foundation–Lancet Commission on Planetary Health (Whitmee et al, 2015), the Earth Biogenome Project (EBP; Lewin et al, 2018), and governing bodies such as the UN Conferences of the Parties (COP) for biodiversity protection (COP15, 2022) and climate changes (COP26, 2021).
Molecular biology can offer practical solutions to environmental challenges, yet it is often discounted by many frontline strategies (Rodríguez‐Martínez et al, 2022). Here, we describe how the advent of real‐time genomic analyses can benefit One Health, showing how it enables timely and in‐depth ecosystem health assessments. We discuss how real‐time genomics is becoming instrumental in guiding efficient intervention strategies, presenting examples and highlighting potential future trajectories and limiting factors (Fig 1).
Figure 1. The One Health concept.
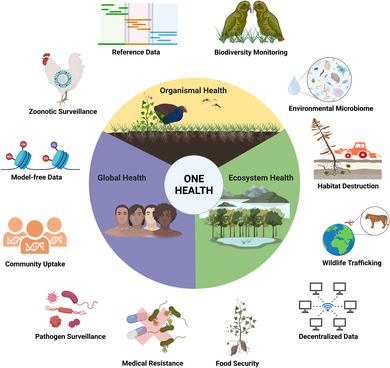
The One Health concept affirms that global human, organismal, and ecosystem health are inextricably linked (inner circle). The application of real‐time genomic approaches can help us understand and support One Health at the intersection of these different health concepts (outer circle).
Real‐time genomics
The need for real‐time genomics has been made clear by recent outbreaks of emerging infectious diseases. Given the exponential growth rates, high transmission potential, and frequent instances of drug resistance of the causative pathogens, it is important to achieve diagnosis turnaround times of hours rather than days or weeks, which precludes the option of transporting samples to large international centers (Gardy & Loman, 2018). In the past two decades, the development of increasingly smaller and cheaper bench‐top sequencing instruments for the first time allowed the use of next‐generation sequencing technologies in local laboratories and clinical settings (Quick et al, 2014).
A new milestone was reached nearly a decade ago when Oxford Nanopore Technologies released its highly portable and cost‐efficient real‐time genomic sequencing device, the MinION (Quick et al, 2014; Ip et al, 2015; Fig 2A). Nanopores are tiny purpose‐mutated protein pores that enable the sequencing of nucleotides by measuring the disruption of their internal ionic current while DNA and RNA strands pass through them as “squiggle” signal. As specific combinations of nucleotides result in characteristic disruptions of the ionic current, this squiggle signal can be base called rapidly into genomic data using dedicated algorithms such as efficient neural networks (Wick et al, 2019; Fig 2B). As such, squiggle signal is model‐free and can incorporate any chemical characteristics of the DNA and RNA strands down to atomic resolution, such as epigenomic modifications. In combination with powerful and parallelizable computers such as graphics processing units (GPUs), this basecalling can happen rapidly, at the speed of sequencing itself. Genomic data can thus both be generated and analyzed in real time and at the point‐of‐care, for example, in the clinical or fieldwork setting (Quick et al, 2016). As nanopore sequencing remains the only disruptive technology to date that allows for portable real‐time genomic analyses, it has been leveraged worldwide to break down barriers and improve the accessibility and versatility of genomic sequencing.
Figure 2. Real‐time nanopore sequencing technology.
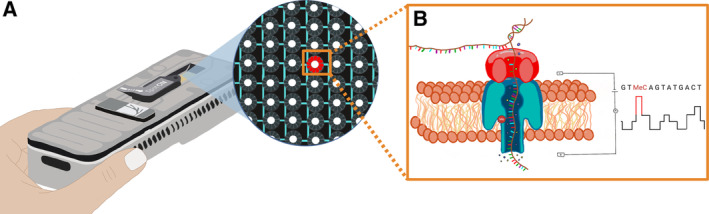
(A) The portable nanopore sequencing device MinION (version Mk1b); the disposable flow cell consists of a fluid‐impermeable polymer membrane with sequencing wells that contain nanopores and that are connected to electrical current sensing circuits to take snapshots of the electrical state of the well at a fixed sampling rate (currently 4 or 5 kHz). (B) An electrical potential across the membrane ensures an ionic flow through the nanopores. When a single‐nucleotide strand passes through the nanopore at a controlled translocation speed, this results in a characteristic disruption of the ionic current that can be basecalled into genomic information such as nucleotide sequence composition and epigenetic modifications. As nucleotides would naturally move through the nanopores too fast for the electrical circuits to detect differences in the ionic current due to individual bases, a helicase is added to the nucleotide strands as an adapter protein. This helicase docks to the nanopores, unwinds the double‐stranded DNA if applicable, and ratchets a single‐nucleotide strand through the nanopore at a controlled translocation speed. While the standard speed has been set to 400 b/s, temperature control can marginally impact translocation speed and, with that, sequencing accuracy. At a sampling rate of 4 kHz, each base is therefore assessed by approximately 10 electrical snapshots.
Advantages of real‐time genomics
Nanopore‐based real‐time genomics offers unique advantages. First, real‐time sequencing and basecalling allow for selective sequencing (aka “adaptive sampling”) by rejecting or accepting nucleotide sequences after minimal sequencing effort when the sequence matches a target of interest (Kovaka et al, 2021; Payne et al, 2021). Such computationally informed enrichment enables researchers to cost‐effectively sequence the genomic data from a specific organism (Urban et al, 2023), taxonomic group (Bao et al, 2021), or genomic region (Payne et al, 2021).
Real‐time sequencing of native DNA strands further enables the retrieval of long sequencing reads including “ultra‐long” reads of several hundred kilobases (kb; Jain et al, 2018) and “whales” of several Megabases (Mb) in length (Payne et al, 2019). Such long reads enable previously impossible genomic assemblies (e.g., of centromeres and other long tandem repeat arrays) and can improve the quality and contiguity of existing assemblies. They furthermore allow for the phasing of variants and the creation of near‐complete metagenome‐assembled genomes (MAGs) from mixed organismal communities (Jain et al, 2018; Nurk et al, 2022; Sereika et al, 2022).
The native sequencing extends to RNA sequencing, where nanopore sequencing can yield reliable abundance estimates of full‐length transcripts, without introducing biases due to reverse transcription or amplification (Garalde et al, 2018). Direct RNA sequencing has, for example, led to the fast discovery of previously undetected viral quasi‐species (Viehweger et al, 2019). Finally, the model‐free nature of nanopore squiggle data means that raw data can a posteriori be used to retrieve more accurate data and more information than just nucleotide sequence composition, just by training new basecalling algorithms (preprint: Stoiber et al, 2017; Wan et al, 2022).
Monitoring environmental health
Species extinction
The present rate of environmental change is the fastest the Earth has experienced since the last mass extinction approximately 65 million years ago (Mya). Climate change, habitat destruction, pollution, invasive species, overexploitation, and other human‐mediated threats have already resulted in a significantly elevated extinction rate of species which has been recognized as the planet's 6th mass extinction (Barnosky et al, 2011; IUCN Red List, 2012). According to the Red List, the population size of 34,432 species (47.6%) is declining, compared to only 1,271 species (1.8%) with increasing population numbers and 36,636 (50.6%) that are stable (IUCN Red List, 2012). The reduction in the effective population size increases genetic drift and the rate of inbreeding, resulting in a loss of genetic diversity. Inbreeding further leads to an increase in the genetic load that becomes expressed (i.e., the realized load; Bertorelle et al, 2022), resulting in inbreeding depression. The effects of such genomic erosion can be felt many generations after immediate threats have abated. Even when successful conservation manages to recover the population numbers after a bottleneck, the species may still be at high risk of extinction (Jackson et al, 2022). As current policy‐making heavily depends on present and past estimates of species extinction risks (COP15, 2022), real‐time genomic approaches can help rapidly assess the true impact of environmental change on ecosystem composition and functioning by comprehensively describing the genomic erosion of species that could spiral them into an extinction vortex and thereby remedying its potential impacts (preprint: van Oosterhout et al, 2022; Theissinger et al, 2023).
Cataloging reference data
Whole‐genome data of threatened species are urgently needed to assess such genomic erosion (preprint: van Oosterhout et al, 2022; Theissinger et al, 2023). Importantly, existing conservation and extinction assessments are taxonomically biased to well‐studied taxa such as vertebrates (Cowie et al, 2022). Ambitious projects such as the EBP intend to catalog all eukaryotic biodiversity and provide a reference for future genomics‐based biodiversity studies (Lewin et al, 2018; Ebenezer et al, 2022; Formenti et al, 2022). While the real‐time component can speed up such efforts, the long and ultra‐long reads produced by nanopore sequencing can greatly facilitate the generation of reference genomic data across the tree of life by providing an anchor for high‐quality, haplotype‐resolved reference genomes. This is particularly relevant to polyploid taxa which represent a significant proportion of eukaryotic life and are particularly difficult to assemble. Moreover, single reference genomes represent a very limited portion of all genomic variation within a species. To better identify structural variation relevant for conservation (Qin et al, 2021), it will be necessary to expand from simplified haploid reference genome toward characterizing pan‐genomes (Bayer et al, 2020).
The portability of nanopore sequencers hereby enables generating such reference data in situ, meaning that, for the first time in the genomic era, the data can be produced close to the species' origin, putting such research in line with CITES (CITES, 1983) and CARE (Global Indigenous Data Alliance, 2022) initiatives as well as data sovereignty principles, as, for example, specified by the Nagoya Protocol on Access and Benefit Sharing (CBD, 2010). The recently launched ORG.one project is an initiative that promotes the uptake of reference genomic data production for endangered animal species. It does this by facilitating the usage of nanopore sequencing for creating high‐quality data with additional support for downstream computational processing and assembly of such data through tailored algorithms and provision of adequate computing power (Oxford Nanopore Technologies, 2022b).
In situ biodiversity monitoring
The portable character of real‐time nanopore sequencing allows for fast in situ biodiversity assessments, with the potential to directly impact wildlife conservation decisions in the field (Blanco et al, 2020; Pomerantz et al, 2022). Nanopore sequencing has already been leveraged for species identification in the field, to generate genomic data for endangered and cryptic species, perform rapid census reports, monitor hybridization zones, and detect the presence of invasive species (Menegon et al, 2017; Pomerantz et al, 2018; Maestri et al, 2019; Blanco et al, 2020; Egeter et al, 2022; Urban et al, 2023). These applications have proven especially valuable when it comes to direct conservation management adjustments in remote environments, where sample storage and transport would be prohibitive for any genomic study (Krehenwinkel et al, 2019; Watsa et al, 2020), and in countries where access to laboratory facilities and conventional genomic sequencing approaches remains challenging (Hetu et al, 2019). This holds the promise of having local conservationists and communities of indigenous people monitor and manage the biodiversity of the ecosystems they live in, and of supporting the democratization of molecular analyses through local research and teaching (Blanco et al, 2020; Watsa et al, 2020) as well as data and benefit sharing (Mc Cartney et al, 2022) (Box 1).
Box 1. The In Situ Lab Initiative.
The In Situ Lab Initiative (ISL) was established in 2020 as a complementary model to present‐day global One Health programs for emerging disease detection in humans that are implemented in a centralized, top–down fashion. The ISL aims to empower local stakeholders such as universities, zoos, conservation non‐governmental organizations, and governments to update their wildlife or environmental surveillance efforts with modern, low‐cost, and portable molecular toolkits to engage in One Health projects in ways that are meaningful to their constituents – which is not necessarily in accordance with the goals of the international community. The ISL facilitates efforts to establish decentralized wildlife surveillance labs worldwide by fostering a network of labs that curate, standardize, and share protocols. Participants of the ISL agree to maintain certain standards of biosafety, data management, protocol sharing, and project management. Over time, as the diffuse partner network expands, and projects in one location overlap with those in another, collaboration results in convergence on best practices. A versatile and competent network of locally run laboratories can redirect resources and respond effectively to gather information on emergent diseases or ecological threats. With functioning labs in Peru, Ecuador (Fig 3C), and soon Indonesia, Vietnam, and Rwanda, the ISL aligns with the Nagoya Protocol by avoiding the exportation of genetic samples and focusing on benefit sharing within communities by centering their participation in community‐driven laboratory‐based investigations. Central to this effort is nanopore sequencing, which allows for all genetic research to be carried out at the site of sample origin, creating positive conservation incentives for genetic resources.
Long nanopore sequencing reads further assist such biodiversity assessments by allowing targeting full‐length marker genes in metabarcoding studies (Krehenwinkel et al, 2019) or complete mitochondrial genomes (Malukiewicz et al, 2021), which can increase the taxonomic resolution of genetic studies. Thanks to the advent of nanopore selective sequencing based on digital sequence information, it has become feasible to enrich environmental genomic material extracted from water, soil, or fecal samples for gene region‐ or species‐specific targets, allowing for non‐invasive genomic biodiversity monitoring (Wanner et al, 2021). Given that long reads contain several genetic variants, they can not only distinguish between species but also reliably perform individual identification based on haplotypes of several kb in length. This has been shown to work for soil environmental DNA monitoring of the critically endangered kākāpō (Strigops habroptilus) in Aotear New Zealand (Fig 3A; Urban et al, 2023), with the potential promise of extending non‐invasive biodiversity monitoring to within‐species assessments of genetic diversity and genomic erosion.
Figure 3. Applications of real‐time genomics for One Health at the point‐of‐care.

(A) In situ applications on remote islands in Aotearoa New Zealand through the employment of portable laboratory, sequencing, and computational equipment (photo credit: Kākāpō Recovery Team, Department of Conservation, New Zealand). (B) Food security application and community involvement on subsistence farms in Tanzania to rapidly diagnose African cassava mosaic viruses in cassava plants (photo credit: Laura Boykin Okalebo; Jo‐Ann L Stanton). (C) Biodiversity monitoring applications for the conservation of critically endangered wildlife in Ecuador (top) and Aotearoa New Zealand (bottom) (photo credits: The In Situ Laboratory Initiative; Kākāpō Recovery Team).
Combating wildlife trafficking
International wildlife trafficking is one of the largest organized transnational crimes, involving the smuggling, poaching, capture, or collection of protected species (Smart et al, 2021). Besides representing one of the major threats to biodiversity, it also entails a biosecurity risk because the unregulated trade of animals can mediate disease transmission (Rush et al, 2021). While local and international laws and regulations prosecute wildlife trafficking (CITES, 1983), actual prosecution of wildlife crime is very often impaired due to improper identification of the species or population, and as a result the country or area of origin after confiscation (Gouda et al, 2020).
The in situ application of real‐time nanopore sequencing to wildlife trafficking opens new avenues for direct application to confiscated samples at borders and airports. Traditional genetic methods such as the sequencing of targeted marker genes or of mitochondrial DNA have become part of the daily toolkit of wildlife forensics (Wasser et al, 2015; Smart et al, 2021). Portable sequencing technologies that produce long reads can further increase the taxonomic resolution and accuracy when determining the place of origin of the confiscated sample. This has previously been demonstrated through ex situ next‐generation sequencing of confiscated live chimpanzees (Fontsere et al, 2022). Here, long‐read sequencing can potentially be applied for reintroducing such confiscated individuals to their populations of origin, as well as for detecting poaching hotspots so that authorities can enforce laws in order to protect wildlife. The same technology has been used to help identify CITES‐listed shark species on a food market in India through genome skimming (Johri et al, 2019). Such in situ monitoring at locations of high human and trade traffic can be extended to monitor the spread of invasive species and infectious disease, which might be associated with any wildlife material.
Application of model‐free sequencing
The model‐free nature of nanopore sequencing means that any modification of the genomic or transcriptomic material can be detected – as long as a basecalling algorithm can be trained to detect such a modification beyond the variation in the sequence composition (preprint: Stoiber et al, 2017; Wan et al, 2022). This has been leveraged to call epigenetic modifications such as DNA methylation and histone modifications (Simpson et al, 2017; preprint: Stoiber et al, 2017; Yue et al, 2022), which provide another rich level of information for assessing biodiversity and its function. Epigenetic variation represents an important part of biodiversity and can provide information about the ecological or environmental components of species and populations (Moore et al, 2013; Lacal & Ventura, 2018). Such modifications can influence an offspring's attributes and fitness conditions, potentially impacting several generations (Bošković & Rando, 2018) and further contributing to a potential extinction vortex. Epigenetic changes can further be used to identify species‐ and population‐specific adaptations to changing environments such as climate change (Lighten et al, 2016), or exposure to toxic substances (Fernández et al, 2014). Nanopore sequencing has already been used to study the epigenomic landscape of medaka fish (Leger et al, 2022) and to evaluate functionally important phenotypes in bacteria (Beaulaurier et al, 2019).
Model‐free sequencing of native genomic material has the potential to de novo discover any modification of the nucleotide strand down to the atomic level (preprint: Stoiber et al, 2017). This has, for example, been leveraged to distinguish bacterial from human DNA on the squiggle signal level, allowing for efficient enrichment of microbial genomic information through nanopore selective sequencing (Bao et al, 2021). In such cases, the analysis of the basecalling algorithms through explainable machine learning (ML) approaches can teach us more about hidden molecular differences between taxonomic groups. We envision that the usage of squiggle data for functional assessments of genomic data will open up new possibilities for epigenetic and comparative genomic research.
Environmental, organismal, and human health
The bidirectional relationship between environmental and human health
In contrast to several organisms that face an elevated extinction risk, many pathogenic organisms such as fungi, bacteria, and viruses are thriving as a result of the rapidly changing environmental conditions. The extremely high biomass and population densities of our livestock and crops (Bar‐On et al, 2018) in combination with their relatively low levels of genetic diversity (Zhang et al, 2018) make them susceptible to emerging infectious diseases (van Oosterhout, 2021). Habitat destruction and wildlife trade further increase the risk of pathogen spillover events from wildlife to livestock (Rush et al, 2021). In turn, close contacts between humans and our livestock promote the evolution of zoonotic disease. Hybridization between previously isolated pathogens allows for genetic introgression, potentially resulting in hybrid speciation in pathogens of humans (Tichkule et al, 2022), animals (Borlase et al, 2021), and plants (preprint: Mathers et al, 2022; Rogério et al, 2022). All these developments are shifting the dynamic co‐evolutionary equilibria in favor of pathogens (van Oosterhout, 2021), putting severe pressure on our society and environment, both now and in the future.
Given the global scale and complex nature of the interdependencies between environmental, organismal, and human health, real‐time genomics has the potential to provide fast diagnostic tools at the point‐of‐care anywhere in the world. Nanopore sequencing has been adopted by a wide variety of stakeholders, including environmental scientists, conservationists, genetic engineers, and health practitioners, to save valuable time and implement appropriate control measures – in the clinical, veterinarian, agricultural, environmental, biodiversity monitoring, or wildlife health setting (Quick et al, 2016, 2017; Vanmechelen et al, 2017; Theuns et al, 2018; Kafetzopoulou et al, 2019; Freed et al, 2020; Rambo‐Martin et al, 2020; Street et al, 2020; Charalampous et al, 2021; Vandenbogaert et al, 2022).
Zoonotic disease
Zoonoses are infectious diseases caused by host switching of pathogenic viruses, bacteria, fungi, or protists (Jones et al, 2008). The spillover of a zoonotic virus from bats to humans via a still unknown animal species has been suggested as the potential source of the recent COVID‐19 pandemic caused by the SARS‐CoV‐2 virus (Yoo & Yoo, 2020; Temmam et al, 2022), showcasing the potential impact of zoonoses on a global scale. In this context, real‐time genomics can tackle zoonoses in remote areas (Gardy & Loman, 2018), while long sequencing reads can help identify novel genomic variants and distinguish genuine recombinants from chimeras, i.e., sequencing or assembly artifacts that can be generated when analyzing mixed infections.
Shortly after the first release of nanopore sequencing technology, an in situ real‐time genomic surveillance program was established to track the Ebola virus epidemic in West Africa (Quick et al, 2016). It was further used to track the Zika virus epidemic in Brazil (Faria et al, 2016; Quick et al, 2017), the COVID‐19 pandemic (Fauver et al, 2020; Freed et al, 2020; Meredith et al, 2020), and several other pathogens, including viruses causing Lassa and yellow fever, avian influenza, and rabies (Kafetzopoulou et al, 2019; Brunker et al, 2020; Hill et al, 2020; Rambo‐Martin et al, 2020; Crossley et al, 2021). The rapid operability has also made nanopore technology the first go‐to tool in the multi‐country monkeypox outbreak in 2022, resulting in the first draft genome of the virus shortly after the beginning of the outbreak (Isidore et al, 2022). Real‐time genomics can help inform medical responses, vaccine development, and public health management by providing a better understanding of transmission routes and frequency. These whole‐genome assessments can detect various pathogenic species across diverse taxonomic groups through metagenomic approaches, which can simultaneously identify and characterize diverse microorganisms (e.g., viral, bacterial, or fungal) with precision and even detect yet‐to‐emerge pathogens (Ko et al, 2022).
In situ real‐time genomics can generate genomic data of pathogens in a decentralized manner, improving the surveillance in previously neglected geographic regions and low‐income countries. Existing large‐scale viral monitoring projects such as Prezode (Peyre et al, 2021), the Global Virome Project (Carroll et al, 2018), or Virion (Carlson et al, 2022) have already made use of publicly available big data to detect novel potentially pathogenic viruses or viral variants (Carroll et al, 2018; Albery et al, 2021). Nanopore sequencing with its potential for automatization and decentralized deployment provides a unique opportunity to further augment and federate worldwide data and use it to enable predictions of novel threats to human health through ML applications (Carlson, 2020). We envision that such decentralized data production in combination with efficient ML can help enable instant, globalized communication about public health risks.
Food security
The challenge of securing food supply for human society is growing in both size and inequity and is directly linked to many other One Health‐related problems. Real‐time genomics can improve global food security and contribute to economic stability, for example, by reducing crop loss through diagnosing plant diseases and pests accurately and early (Boykin et al, 2018). The creation of global genomic reference data can further inform and accelerate global food production, ultimately democratizing the benefits of genomic research. For example, nanopore sequencing has revolutionized genome sequencing of important plant species such as crops, enabling the accurate assembly of their often large and highly repetitive genomes (Ibe, 2022). Nanopore sequencing has also shown a link between antimicrobial resistance and bacterial virulence in livestock and their human farmers (Viñes et al, 2021), highlighting the role of livestock as a reservoir of pathotypes with zoonotic potential and as a potential source of food insecurity.
The potential impact of diagnostic sequencing in real time was convincingly demonstrated in its application to the cassava plant in Tanzania in 2017 and 2018, which feeds 800 million people worldwide (Fig 3B; Boykin et al, 2019). During a visit to a subsistence farm run by a women's chama (Swahili for collective), portable DNA extraction and nanopore sequencing were used to identify particular strains of African cassava mosaic virus in cassava plants. Rapid diagnosis allowed replacing the crop with two cassava varieties tolerant to the identified viral strains in time for the 2018 harvest. This ensured food security as the virus‐tolerant cassava varieties produced around 35 tons per hectare for sale, whereas previous crops from the chama had not yielded enough harvest for the market. When considering the average market value of cassava in 2018, the associated production costs, and the household incomes in Tanzania (National Bureau of Statistics, 2019), this real‐time genomics‐informed intervention provided the chama with surplus income equivalent to approximately 3.7 times the average monthly income in Tanzania for 2018 (also see “Extant Challenges”).
The environmental microbiome
Real‐time metagenomics surveillance approaches can also be used to describe the natural environmental microbiome, for example, from non‐invasive samples such as air, water, or soil, and help us better understand the functional interactions between humans, the environmental microbiome, and ecological change (Gowers et al, 2019; Haan & Drown, 2021; Edwards et al, 2022). Increased anthropogenic pressures and rapid climate change can also leave their footprint on these microbial communities. For example, antimicrobials such as antibiotics, antifungals, and disinfectants have been overused in the clinical and agricultural setting, leading to resistances and environmental pollution. When coupled with extreme weather patterns and higher temperatures, it can lead to the spread of superbugs, i.e., microorganisms that are resistant to most medications (UNEP, 2023).
Real‐time environmental metagenomics has the potential to unmask such complex relationships between human and environmental health in the context of One Health. Known and novel pathogens and their transmission dynamics have been identified from freshwater or wastewater sources to describe potential human health consequences (Izquierdo‐Lara et al, 2021; Urban et al, 2021), and harmful algal blooms that can directly influence ecosystem services have been detected early (Hatfield et al, 2020; preprint: Koeppel et al, 2022). Beyond taxonomic assignments of microorganisms, the long reads of nanopore sequencing have been used to detect medical resistance‐ and virulence‐associated genes and gene clusters, allowing for direct functional predictions and transmission surveillance – for example for drug resistance profiling of tuberculosis‐causing Mycobacterium (Chan et al, 2020), malaria‐causing Plasmodium falciparum (Runtuwene et al, 2018), Leishmaniasis‐causing Leishmania infantum (Martí‐Carreras et al, 2022), and a wide range of other taxa (Ashton et al, 2015; Břinda et al, 2020; Bokma et al, 2021).
A vision for equitable and inclusive One Health
Accessibility
Real‐time genomics has the potential to increase equity and inclusion with respect to access to One Health research through its disruptive and distributed nature. Nanopore sequencing has been developed with the aim of being accessible to “anyone, anywhere”, which has been achieved through reduced upfront investment costs and by pairing it with portable DNA and RNA extraction and data analysis approaches (Palatnick et al, 2020; Oxford Nanopore Technologies, 2022b). This has had important implications for shifting the current hierarchical genomic framework with high‐volume large sequencing centers to highly distributed low‐volume bespoke sequencing. This solves logistically difficult storage and transport of samples, reduces the risk of invasive pathogens being transported along with the samples, and circumvents issues related to permits and travel restrictions.
Community
The in situ application of real‐time genomics for One Health enables its uptake by communities themselves, reducing the harmful practices of neocolonialism and helicopter science (Adame, 2021; Haelewaters et al, 2021), with the potential to democratize and diversify scientific practice (Nagaraj et al, 2020). If used in the right way, in situ applications can support indigenous rights – such as demonstrated by the concept of “ahi kā” or “keeping the home fires burning” by Māori communities in Aotearoa New Zealand. This means that genomic material and data can remain in the hands of the involved communities, who can subsequently be in control of their own diagnostics and maintain self‐determination.
Māori, like many other indigenous communities globally, are a recognized force in the front lines of biosecurity surveillance and conservation management (Lambert & Mark‐Shadbolt, 2021). In Te Ao Māori (the world of the Mori), the entire Earth is known as Papatūānuku, the Earth mother, and all life depends upon Papatūānuku for their wellbeing. People have the option of caring for her to maintain their own health or abandoning her to concentrate on their own short‐term needs. By always keeping in mind the needs of Papatūānuku and the requirements of her immediate whānau, Māori have for a long time been advocates of the importance of One Health, while these interdependencies between environmental and human health have been gradually de‐emphasized by many other cultures.
However, Māori continue to have limited access to the latest technologies in disease diagnostics (Palmer et al, 2020), often due to financial and technical limitations. Efforts to protect their culturally significant species, the environments they exist in, and customary harvesting practices require more accessible tools and training opportunities so that indigenous communities can contribute to, and benefit from, a better, more inclusive biosecurity and conservation system. Emerging pests and pathogens are of great concern both economically as well as presenting a conservation threat to already endangered species. One example is the plant pathogen Phytophthora agathidicida, which is the accepted causal agent of kauri (Agathis australis) forest diebacks (Weir et al, 2015), but which is not well‐understood with respect to transmission and possible prevention. As many of the infected and vulnerable kauri forests are managed by Māori, portable real‐time genomics can provide an accessible and accurate diagnostic tool for the effective identification of emerging pests and pathogens of economic and cultural importance.
Extant challenges
While real‐time portable genomics through nanopore sequencing has led to increased accessibility, decreased initial financial investment, and offers many opportunities for improving our global understanding of One Health, nanopore sequencing still faces substantial financial, technological, and ethical limitations.
Nanopore sequencing does not require immense upfront investment costs when the freely available portable sequencing devices are being used, but the regular consumable costs can be substantial, especially for low‐ and middle‐income countries. For portable applications, the lowest‐capacity sequencing flow cell is called a Flongle, which costs about US$ 7 each, has an output of hundreds of Mb, and does not have any specific storage requirements. The MinION flow cell costs about US$ 900, has a typical yield of about 5–15 Gb, and has to be stored at fridge temperature. For high‐throughput sequencing devices, the P2 Solo (US$ 10,455 + 1,000 USD per year) is the most affordable, with additional costs of US$ 1,400 per PromethION flow cell resulting in a yield of 50–150 Gb. While optimization of these costs through multiplexing, re‐usage of flow cells, and optimization of DNA extraction and sequencing protocols can result in reasonable costs per base (Blanco et al, 2020), pilot studies usually have to be conducted to understand the sequencing throughput and the necessary sequencing depth for each new sample type. This creates uncertainty for the user with respect to financial considerations and data storage. Simultaneously, many countries do not yet have access to reliable suppliers of nanopore sequencing material, resulting in often substantially increased material and shipping costs, or a complete lack of accessibility.
While these financial limitations will have to be resolved in the future, the advantage of real‐time in situ application has to be taken into account for any economic considerations. For example, if we consider sequencing costs in the African cassava mosaic virus study described earlier (see “Food security”), we estimated processing costs of US$ 42 per sample. These costs were about a tenth the cost of Illumina sequencing in Tanzania (quoted at US$ 386 in 2018), and the additional turn‐around time of Illumina sequencing at an offshore site would have prevented the replanting of different cassava varieties and therefore any immediate economic and societal benefits for the chama. In other words, where the costs can be met, the benefit of in situ sequencing is substantial.
Many practical challenges prevent the widespread and large‐scale uptake of real‐time genomic technology. The lack of stable electrical supply can negatively affect the storage of temperature‐sensitive reagents, which poses a big limitation on long‐term field studies in remote areas (Pomerantz et al, 2018; Blanco et al, 2020). A lack of internet connection and large amounts of data that accumulate over long periods of time can be limiting factors for the subsequent bioinformatic analysis of sequencing data (Blanco et al, 2020). Another important technological limitation of nanopore sequencing has been the inflated sequencing read error rate of up to 8% (Urban et al, 2021), which mainly stemmed from difficulties of the nanopores to accurately distinguish homopolymers (Delahaye & Nicolas, 2021) with potential implications for faulty assemblies, false‐positive variant calling, and frameshift errors. Thanks to the latest Kit 14 nanopore sequencing chemistry (introduced in 2022) together with duplex sequencing (basecalling the forward and reverse‐complementary DNA strands in tandem), this sequencing read error rate has decreased to 0.6% (Oxford Nanopore Technologies, 2022a). This means that it is now possible to generate highly accurate and complete assemblies without the need for short‐read polishing (Sereika et al, 2022). This high sequencing accuracy is very promising for any future application of nanopore sequencing in the space of One Health.
Community uptake and empowerment to routinely use real‐time genomic‐based diagnostics remain difficult. Access to real‐time genomic technology remains hampered by a lack of automatization, both on the laboratory and on the computational level. Therefore, the actual application of nanopore sequencing still requires advanced molecular biology and analytical skills. Even the application of standard bioinformatic pipelines implemented in Oxford Nanopore Technologies' EPI2ME (EPI2ME Labs, 2023) still requires knowledge about the underlying analysis pipeline and databases for comprehensive interpretation. If this, however, leads to the application of a “lab‐in‐a‐suitcase” without appropriate community engagement and involvement, it could threaten the idea of self‐determined independent applications by local communities and researchers and further perpetuate helicopter research (Haelewaters et al, 2021). The difficulty of analyzing and interpreting nanopore data has, for example, been highlighted after in‐field sequencing in Africa (Boykin et al, 2019). Subsequent courses organized by the African BecA‐ILRI hub as a 3‐month hybrid training program for African scientists allowed them to learn the basic applications of nanopore sequencing and to simultaneously apply this real‐time genomic technology to study the genetic potential of crops and livestock in the context of food security (BecA‐ILRI hub, 2023). Such training needs to be standardized and made available worldwide (at community‐affordable costs) to enable truly global and equitable access to the advances of real‐time genomic research for One Health.
Conclusion
We envision that the use of real‐time genomic technologies and their application at the point‐of‐care can improve our understanding of complex interdependencies within the One Health concept, directly informing management decisions in situ in clinical, veterinarian, agricultural, conservation, and environmental applications. A paradigm shift is required to really ensure an equitable and global uptake of real‐time genomics. To support decentralized capacities, this will have to involve tackling global disadvantages and rendering the technology flexible with respect to commercialization, and compatible with different cultural models and infrastructure. To increase the accessibility of the technology, further development and automation are required, which would enable its application at airports, in hospitals, at remote wildlife camps, in local communities, or in daily conservation and biosecurity surveillance. The global and decentralized genomic data created this way hold the promise of widening our horizon to implications of the One Health concept that have so far been hidden from us, and would enable us to further adapt our attitude towards the interdependencies between human and environmental health.
Author contributions
Lara Urban: Conceptualization; supervision; visualization; writing – original draft; project administration; writing – review and editing. Albert Perlas: Conceptualization; writing – original draft. Olga Francino: Conceptualization; writing – original draft. Joan Martí‐Carreras: Conceptualization; writing – original draft. Brenda A Muga: Conceptualization; writing – original draft. Jenniffer W Mwangi: Conceptualization; writing – original draft. Laura Boykin Okalebo: Conceptualization; writing – original draft. Jo‐Ann L Stanton: Conceptualization; writing – original draft. Amanda Black: Conceptualization; writing – original draft. Nick Waipara: Conceptualization; writing – original draft. Claudia Fontsere: Conceptualization; writing – original draft. David Eccles: Conceptualization; visualization; writing – original draft. Harika Urel: Visualization; writing – original draft. Tim Reska: Visualization. Hernán E Morales: Conceptualization; writing – original draft. Marc Palmada‐Flores: Conceptualization; writing – original draft. Tomas Marques‐Bonet: Conceptualization; writing – original draft. Mrinalini Watsa: Writing – original draft. Zane Libke: Writing – original draft. Gideon Erkenswick: Writing – original draft. Cock van Oosterhout: Conceptualization; supervision; writing – original draft.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Mol Syst Biol. (2023) 19: e11686
References
- Adame F (2021) Meaningful collaborations can end ‘helicopter research’. Nature 10.1038/d41586-021-01795-1 [DOI] [PubMed] [Google Scholar]
- Albery GF, Becker DJ, Brierley L, Brook CE, Christofferson RC, Cohen LE, Dallas TA, Eskew EA, Fagre A, Farrell MJ et al (2021) The science of the host–virus network. Nat Microbiol 6: 1483–1492 [DOI] [PubMed] [Google Scholar]
- Ashton PM, Nair S, Dallman T, Rubino S, Rabsch W, Mwaigwisya S, Wain J, O'Grady J (2015) MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol 33: 296–300 [DOI] [PubMed] [Google Scholar]
- Bao Y, Wadden J, Erb‐Downward JR, Ranjan P, Zhou W, McDonald TL, Mills RE, Boyle AP, Dickson RP, Blaauw D et al (2021) SquiggleNet: real‐time, direct classification of nanopore signals. Genome Biol 22: 298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnosky AD, Matzke N, Tomiya S, Wogan GOU, Swartz B, Quental TB, Marshall C, McGuire JL, Lindsey EL, Maguire KC et al (2011) Has the earth's sixth mass extinction already arrived? Nature 471: 51–57 [DOI] [PubMed] [Google Scholar]
- Bar‐On YM, Phillips R, Milo R (2018) The biomass distribution on earth. Proc Natl Acad Sci U S A 115: 6506–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer PE, Golicz AA, Scheben A, Batley J, Edwards D (2020) Plant pan‐genomes are the new reference. Nat Plants 6: 914–920 [DOI] [PubMed] [Google Scholar]
- Beaulaurier J, Schadt EE, Fang G (2019) Deciphering bacterial epigenomes using modern sequencing technologies. Nat Rev Genet 20: 157–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BecA‐ILRI hub (2023) BecA‐ILRI hub . https://www.ilri.org/research/programs/beca-ilri-hub
- Bertorelle G, Raffini F, Bosse M, Bortoluzzi C, Iannucci A, Trucchi E, Morales HE, van Oosterhout C (2022) Genetic load: genomic estimates and applications in non‐model animals. Nat Rev Genet 23: 492–503 [DOI] [PubMed] [Google Scholar]
- Blanco MB, Greene LK, Rasambainarivo F, Toomey E, Williams RC, Andrianandrasana L, Larsen PA, Yoder AD (2020) Next‐generation technologies applied to age‐old challenges in Madagascar. Conserv Genet 21: 785–793 [Google Scholar]
- Bokma J, Vereecke N, Nauwynck H, Haesebrouck F, Theuns S, Pardon B, Boyen F (2021) Genome‐wide association study reveals genetic markers for antimicrobial resistance in mycoplasma bovis. Microbiol Spectr 9: e00262‐21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlase A, Rudge JW, Léger E, Diouf ND, Fall CB, Diop SD, Catalano S, Sène M, Webster JP (2021) Spillover, hybridization, and persistence in schistosome transmission dynamics at the human–animal interface. Proc Natl Acad Sci U S A 118: e2110711118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bošković A, Rando OJ (2018) Transgenerational epigenetic inheritance. Annu Rev Genet 52: 21–41 [DOI] [PubMed] [Google Scholar]
- Boykin L, Ghalab A, Marchi BRD, Savill A, Wainaina JM, Kinene T, Lamb S, Rodrigues M, Kehoe M, Ndunguru J et al (2018) Real time portable genome sequencing for global food security. F1000Res 7: 1101 [Google Scholar]
- Boykin L, Sseruwagi P, Alicai T, Ateka E, Mohammed IU, Stanton J‐AL, Kayuki C, Mark D, Fute T, Erasto J et al (2019) Tree lab: portable genomics for early detection of plant viruses and pests in sub‐Saharan Africa. Gene 10: 632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Břinda K, Callendrello A, Ma KC, MacFadden DR, Charalampous T, Lee RS, Cowley L, Wadsworth CB, Grad YH, Kucherov G et al (2020) Rapid inference of antibiotic resistance and susceptibility by genomic neighbour typing. Nat Microbiol 5: 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunker K, Jaswant G, Thumbi SM, Lushasi K, Lugelo A, Czupryna AM, Ade F, Wambura G, Chuchu V, Steenson R et al (2020) Rapid in‐country sequencing of whole virus genomes to inform rabies elimination programmes. Wellcome Open Res 5: 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CJ (2020) From PREDICT to prevention, one pandemic later. Lancet Microbe 1: e6–e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CJ, Gibb RJ, Albery GF, Brierley L, Connor RP, Dallas TA, Eskew EA, Fagre AC, Farrell MJ, Frank HK et al (2022) The global Virome in one network (VIRION): an atlas of vertebrate‐virus associations. MBio 13: e02985‐21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D, Daszak P, Wolfe ND, Gao GF, Morel CM, Morzaria S, Pablos‐Méndez A, Tomori O, Mazet JAK (2018) The global Virome project. Science 359: 872–874 [DOI] [PubMed] [Google Scholar]
- CBD (2010) The Nagoya Protocol on Access and Benefit‐sharing . Convention on Biological Diversity Website; Secretariat of the Convention on Biological Diversity. https://www.cbd.int/abs/
- Chan WS, Au CH, Chung Y, Leung HCM, Ho DN, Wong EYL, Lam TW, Chan TL, Ma ESK, Tang BSF (2020) Rapid and economical drug resistance profiling with Nanopore MinION for clinical specimens with low bacillary burden of mycobacterium tuberculosis. BMC Res Notes 13: 444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charalampous T, Alcolea‐Medina A, Snell LB, Williams TGS, Batra R, Alder C, Telatin A, Camporota L, Meadows CIS, Wyncoll D et al (2021) Evaluating the potential for respiratory metagenomics to improve treatment of secondary infection and detection of nosocomial transmission on expanded COVID‐19 intensive care units. Genome Med 13: 182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CITES (1983) Convention on International Trade in Endangered Species of Wild Fauna and Flora | CITES . https://cites.org/eng/disc/text.php [DOI] [PubMed]
- COP15 (2022) Nations Adopt Four Goals, 23 Targets for 2030 In Landmark UN Biodiversity Agreement . https://www.cbd.int/article/cop15-cbd-press-release-final-19dec2022
- COP26 (2021) The Glasgow Climate Pact . UN Climate Change Conference (COP26) at the SEC – Glasgow 2021. https://ukcop26.org/the-glasgow-climate-pact/
- Cowie RH, Bouchet P, Fontaine B (2022) The Sixth Mass Extinction: fact, fiction or speculation? Biol Rev 97: 640–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley BM, Rejmanek D, Baroch J, Stanton JB, Young KT, Killian ML, Torchetti MK, Hietala SK (2021) Nanopore sequencing as a rapid tool for identification and pathotyping of avian influenza a viruses. J Vet Diagn Invest 33: 253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahaye C, Nicolas J (2021) Sequencing DNA with nanopores: troubles and biases. PLoS One 16: e0257521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebenezer TE, Muigai AWT, Nouala S, Badaoui B, Blaxter M, Buddie AG, Jarvis ED, Korlach J, Kuja JO, Lewin HA et al (2022) Africa: sequence 100,000 species to safeguard biodiversity. Nature 603: 388–392 [DOI] [PubMed] [Google Scholar]
- Edwards A, Soares A, Debbonaire A, Edwards Rassner SM (2022) Before you go: a packing list for portable DNA sequencing of microbiomes and metagenomes. Microbiology 168: 001220 [DOI] [PubMed] [Google Scholar]
- Egeter B, Veríssimo J, Lopes‐Lima M, Chaves C, Pinto J, Riccardi N, Beja P, Fonseca NA (2022) Speeding up the detection of invasive bivalve species using environmental DNA: a Nanopore and Illumina sequencing comparison. Mol Ecol Resour 22: 2232–2247 [DOI] [PubMed] [Google Scholar]
- EPI2ME Labs (2023) EPI2ME Labs . https://labs.epi2me.io/
- Faria NR, Sabino EC, Nunes MRT, Alcantara LCJ, Loman NJ, Pybus OG (2016) Mobile real‐time surveillance of Zika virus in Brazil. Genome Med 8: 97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauver JR, Petrone ME, Hodcroft EB, Shioda K, Ehrlich HY, Watts AG, Vogels CBF, Brito AF, Alpert T, Muyombwe A et al (2020) Coast‐to‐coast spread of SARS‐CoV‐2 during the early epidemic in the United States. Cell 181: 990–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández AF, Toraño EG, Urdinguio RG, Lana AG, Fernández IA, Fraga MF (2014) The epigenetic basis of adaptation and responses to environmental change: perspective on human reproduction. Adv Exp Med Biol 753: 97–117 [DOI] [PubMed] [Google Scholar]
- Fontsere C, Kuhlwilm M, Morcillo‐Suarez C, Alvarez‐Estape M, Lester JD, Gratton P, Schmidt JM, Dieguez P, Aebischer T, Álvarez‐Varona P et al (2022) Population dynamics and genetic connectivity in recent chimpanzee history. Cell Genom 2: 100133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formenti G, Theissinger K, Fernandes C, Bista I, Bombarely A, Bleidorn C, Ciofi C, Crottini A, Godoy JA, Höglund J et al (2022) The era of reference genomes in conservation genomics. Trends Ecol Evol 37: 197–202 [DOI] [PubMed] [Google Scholar]
- Freed NE, Vlková M, Faisal MB, Silander OK (2020) Rapid and inexpensive whole‐genome sequencing of SARS‐CoV‐2 using 1200 bp tiled amplicons and Oxford Nanopore rapid barcoding. Biol Methods Protoc 5: bpaa014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A et al (2018) Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods 15: 201–206 [DOI] [PubMed] [Google Scholar]
- Gardy JL, Loman NJ (2018) Towards a genomics‐informed, real‐time, global pathogen surveillance system. Nat Rev Genet 19: 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Indigenous Data Alliance (2022) CARE Principles . Global Indigenous Data Alliance Website https://www.gida-global.org/care
- Gouda S, Kerry RG, Das A, Chauhan NS (2020) Wildlife forensics: a boon for species identification and conservation implications. Forensic Sci Int 317: 110530 [DOI] [PubMed] [Google Scholar]
- Gowers G‐OF, Vince O, Charles J‐H, Klarenberg I, Ellis T, Edwards A (2019) Entirely off‐grid and solar‐powered DNA sequencing of microbial communities during an ice cap traverse expedition. Gene 10: 902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan TJ, Drown DM (2021) Unearthing antibiotic resistance associated with disturbance‐induced permafrost thaw in interior Alaska. Microorganisms 9: 116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haelewaters D, Hofmann TA, Romero‐Olivares AL (2021) Ten simple rules for global north researchers to stop perpetuating helicopter research in the global south. PLoS Comput Biol 17: e1009277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield RG, Batista FM, Bean TP, Fonseca VG, Santos A, Turner AD, Lewis A, Dean KJ, Martinez‐Urtaza J (2020) The application of Nanopore sequencing technology to the study of dinoflagellates: aproof of concept study for rapid sequence‐based discrimination of potentially harmful algae. Front Microbiol 11: 844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetu M, Koutouki K, Joly Y (2019) Genomics for all: international Open Science genomics projects and capacity building in the developing world. Front Genet 10: 95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SC, de Souza R, Thézé J, Claro I, Aguiar RS, Abade L, Santos FCP, Cunha MS, Nogueira JS, Salles FCS et al (2020) Genomic surveillance of yellow fever virus epizootic in São Paulo, Brazil, 2016–2018. PLoS Pathog 16: e1008699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibe CN (2022) Democratizing plant genomics to accelerate global food production. Nat Genet 54: 911–913 [DOI] [PubMed] [Google Scholar]
- Ip CLC, Loose M, Tyson JR, de Cesare M, Brown BL, Jain M, Leggett RM, Eccles DA, Zalunin V, Urban JM et al (2015) MinION Analysis and Reference Consortium: Phase 1 data release and analysis. F1000Res 4: 1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidore J, Borges V, Pinto M, Ferreira R, Sobral D, Nunes A, Dourado Santos J, José Borrego M, Núncio S, Pelerito A et al (2022, May 19) First draft genome sequence of Monkeypox virus associated with the suspected multi‐country outbreak, May 2022 (confirmed case in Portugal) . Virological Genome Report. https://virological.org/t/first-draft-genome-sequence-of-monkeypox-virus-associated-with-the-suspected-multi-country-outbreak-may-2022-confirmed-case-in-portugal/799
- IUCN Red List (2012) IUCN red list categories and criteria, version 3.1, second edition . IUCN. https://portals.iucn.org/library/node/10315
- Izquierdo‐Lara R, Elsinga G, Heijnen L, Munnink BBO, Schapendonk CME, Nieuwenhuijse D, Kon M, Lu L, Aarestrup FM, Lycett S et al (2021) Monitoring SARS‐CoV‐2 circulation and diversity through community wastewater sequencing, The Netherlands and Belgium. Emerg Infect Dis 27: 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson HA, Percival‐Alwyn L, Ryan C, Albeshr MF, Venturi L, Morales HE, Mathers TC, Cocker J, Speak SA, Accinelli GG et al (2022) Genomic erosion in a demographically recovered bird species during conservation rescue. Conserv Biol 36: e13918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Koren S, Miga KH, Quick J, Rand AC, Sasani TA, Tyson JR, Beggs AD, Dilthey AT, Fiddes IT et al (2018) Nanopore sequencing and assembly of a human genome with ultra‐long reads. Nat Biotechnol 36: Article 4–Article 345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri S, Solanki J, Cantu VA, Fellows SR, Edwards RA, Moreno I, Vyas A, Dinsdale EA (2019) ‘Genome skimming’ with the MinION hand‐held sequencer identifies CITES‐listed shark species in India's exports market. Sci Rep 9: 4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P (2008) Global trends in emerging infectious diseases. Nature 451: 990–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafetzopoulou LE, Pullan ST, Lemey P, Suchard MA, Ehichioya DU, Pahlmann M, Thielebein A, Hinzmann J, Oestereich L, Wozniak DM et al (2019) Metagenomic sequencing at the epicenter of the Nigeria 2018 Lassa fever outbreak. Science 363: 74–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keesing F, Ostfeld RS (2021) Impacts of biodiversity and biodiversity loss on zoonotic diseases. Proc Natl Acad Sci U S A 118: e2023540118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko KKK, Chng KR, Nagarajan N (2022) Metagenomics‐enabled microbial surveillance. Nat Microbiol 7: 486–496 [DOI] [PubMed] [Google Scholar]
- Koeppel AF, Goodrum W, Steffen M, Wurch L, Turner SD (2022) Environmental DNA sequencing data from algal blooms in Lake Erie using Oxford Nanopore MinION. bioRxiv 10.1101/2022.03.12.483776 [PREPRINT] [DOI] [PMC free article] [PubMed]
- Kovaka S, Fan Y, Ni B, Timp W, Schatz MC (2021) Targeted nanopore sequencing by real‐time mapping of raw electrical signal with UNCALLED. Nat Biotechnol 39: 431–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krehenwinkel H, Pomerantz A, Henderson JB, Kennedy SR, Lim JY, Swamy V, Shoobridge JD, Graham N, Patel NH, Gillespie RG et al (2019) Nanopore sequencing of long ribosomal DNA amplicons enables portable and simple biodiversity assessments with high phylogenetic resolution across broad taxonomic scale. GigaScience 8: giz006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacal I, Ventura R (2018) Epigenetic inheritance: concepts, mechanisms and perspectives. Front Mol Neurosci 11: 292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Mark‐Shadbolt M (2021) Indigenous knowledges of forest and biodiversity management: How the watchfulness of Māori complements and contributes to disaster risk reduction . https://www.preventionweb.net/publication/indigenous-knowledges-forest-and-biodiversity-management-how-watchfulness-maori
- Leger A, Brettell I, Monahan J, Barton C, Wolf N, Kusminski N, Herder C, Aadepu N, Becker C, Gierten J et al (2022) Genomic variations and epigenomic landscape of the Medaka inbred Kiyosu‐Karlsruhe (MIKK) panel. Genome Biol 23: 58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de León EA, Shriwise A, Tomson G, Morton S, Lemos DS, Menne B, Dooris M (2021) Beyond building back better: imagining a future for human and planetary health. Lancet Planet Health 5: e827–e839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin HA, Robinson GE, Kress WJ, Baker WJ, Coddington J, Crandall KA, Durbin R, Edwards SV, Forest F, Gilbert MTP et al (2018) Earth BioGenome project: sequencing life for the future of life. Proc Natl Acad Sci U S A 115: 4325–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighten J, Incarnato D, Ward BJ, van Oosterhout C, Bradbury I, Hanson M, Bentzen P (2016) Adaptive phenotypic response to climate enabled by epigenetics in a K‐strategy species, the fish Leucoraja ocellata (Rajidae). R Soc Open Sci 3: 160299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestri S, Cosentino E, Paterno M, Freitag H, Garces JM, Marcolungo L, Alfano M, Njunjić I, Schilthuizen M, Slik F et al (2019) A rapid and accurate MinION‐based workflow for tracking species biodiversity in the field. Gene 10: 468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malukiewicz J, Cartwright RA, Dergam JA, Igayara CS, Nicola PA, Pereira LMC, Ruiz‐Miranda CR, Stone AC, Silva DL, Silva FFRD et al (2021) Genomic skimming and nanopore sequencing uncover cryptic hybridization in one of world's most threatened primates. Sci Rep 11: 17279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí‐Carreras J, Carrasco M, Gómez‐Ponce M, Noguera‐Julián M, Fisa R, Riera C, Alcover MM, Roura X, Ferrer L, Francino O (2022) Identification of leishmania infantum epidemiology, drug resistance and pathogenicity biomarkers with nanopore sequencing. Microorganisms 10: 2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers TC, Wouters RHM, Mugford ST, Biello R, Oosterhout CV, Hogenhout SA (2022) Hybridisation has shaped a recent radiation of grass‐feeding aphids. bioRxiv 10.1101/2022.09.27.509720 [PREPRINT] [DOI] [PMC free article] [PubMed]
- Mc Cartney AM, Anderson J, Liggins L, Hudson ML, Anderson MZ, TeAika B, Geary J, Cook‐Deegan R, Patel HR, Phillippy AM (2022) Balancing openness with Indigenous data sovereignty: an opportunity to leave no one behind in the journey to sequence all of life. Proc Natl Acad Sci U S A 119: e2115860119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegon M, Cantaloni C, Rodriguez‐Prieto A, Centomo C, Abdelfattah A, Rossato M, Bernardi M, Xumerle L, Loader S, Delledonne M (2017) On site DNA barcoding by nanopore sequencing. PLoS One 12: e0184741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith LW, Hamilton WL, Warne B, Houldcroft CJ, Hosmillo M, Jahun AS, Curran MD, Parmar S, Caller LG, Caddy SL et al (2020) Rapid implementation of SARS‐CoV‐2 sequencing to investigate cases of health‐care associated COVID‐19: a prospective genomic surveillance study. Lancet Infect Dis 20: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LD, Le T, Fan G (2013) DNA methylation and its basic function. Neuropsychopharmacology 38: 23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj A, Shears E, de Vaan M (2020) Improving data access democratizes and diversifies science. Proc Natl Acad Sci 117: 23490–23498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Bureau of Statistics (2019) The 2017‐18 household budget survey: key indicators report . https://www.nbs.go.tz/index.php/en/census-surveys/poverty-indicators-statistics/household-budget-survey-hbs/413-the-2017-18-household-budget-survey-key-indicators-report
- Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A et al (2022) The complete sequence of a human genome. Science 376: 44–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- One Health High‐Level Expert Panel , Adisasmito WB, Almuhairi S, Behravesh CB, Bilivogui P, Bukachi SA, Casas N, Becerra NC, Charron DF, Chaudhary A et al (2022) One health: a new definition for a sustainable and healthy future. PLoS Pathog 18: e1010537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosterhout C (2021) Mitigating the threat of emerging infectious diseases; a coevolutionary perspective. Virulence 12: 1288–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosterhout C, Speak SA, Birley T, Bortoluzzi C, Percival‐Alwyn L, Urban L, Groombridge JJ, Segelbacher G, Morales HE (2022) Genomic erosion in the assessment of species extinction risk and recovery potential. bioRxiv 10.1101/2022.09.13.507768 [PREPRINT] [DOI]
- Oxford Nanopore Technologies (2022a) Oxford Nanopore tech update: new duplex method for Q30 nanopore single molecule reads, PromethION 2, and more . Oxford Nanopore Technologies. https://nanoporetech.com/about-us/news/oxford-nanopore-tech-update-new-duplex-method-q30-nanopore-single-molecule-reads-0
- Oxford Nanopore Technologies (2022b) ORG.one . Oxford Nanopore Technologies Website. https://nanoporetech.com/oo
- Palatnick A, Zhou B, Ghedin E, Schatz MC (2020) iGenomics: comprehensive DNA sequence analysis on your smartphone. GigaScience 9: giaa138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Mercier OR, King‐Hunt A, Palmer S, Mercier OR, King‐Hunt A (2020) Towards rangatiratanga in pest management? Māori perspectives and frameworks on novel biotechnologies in conservation. Pac Conserv Biol 27: 391–401 [Google Scholar]
- Payne A, Holmes N, Rakyan V, Loose M (2019) BulkVis: a graphical viewer for Oxford nanopore bulk FAST5 files. Bioinformatics 35: 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne A, Holmes N, Clarke T, Munro R, Debebe BJ, Loose M (2021) Readfish enables targeted nanopore sequencing of gigabase‐sized genomes. Nat Biotechnol 39: 442–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyre M, Vourch G, Lefrançois T, Martin‐Prevel Y, Soussana J‐F, Roche B (2021) PREZODE: preventing zoonotic disease emergence. Lancet 397: 792–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz A, Peñafiel N, Arteaga A, Bustamante L, Pichardo F, Coloma LA, Barrio‐Amorós CL, Salazar‐Valenzuela D, Prost S (2018) Real‐time DNA barcoding in a rainforest using nanopore sequencing: opportunities for rapid biodiversity assessments and local capacity building. GigaScience 7: giy033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz A, Sahlin K, Vasiljevic N, Seah A, Lim M, Humble E, Kennedy S, Krehenwinkel H, Winter S, Ogden R et al (2022) Rapid in situ identification of biological specimens via DNA amplicon sequencing using miniaturized laboratory equipment. Nat Protoc 17: 1415–1443 [DOI] [PubMed] [Google Scholar]
- Qin P, Lu H, Du H, Wang H, Chen W, Chen Z, He Q, Ou S, Zhang H, Li X et al (2021) Pan‐genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 184: 3542–3558 [DOI] [PubMed] [Google Scholar]
- Quick J, Quinlan AR, Loman NJ (2014) A reference bacterial genome dataset generated on the MinION™ portable single‐molecule nanopore sequencer. GigaScience 3: 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J, Loman NJ, Duraffour S, Simpson JT, Severi E, Cowley L, Bore JA, Koundouno R, Dudas G, Mikhail A et al (2016) Real‐time, portable genome sequencing for Ebola surveillance. Nature 530: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, Oliveira G, Robles‐Sikisaka R, Rogers TF, Beutler NA et al (2017) Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc 12: 1261–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo‐Martin BL, Keller MW, Wilson MM, Nolting JM, Anderson TK, Vincent AL, Bagal UR, Jang Y, Neuhaus EB, Davis CT et al (2020) Influenza avirus field surveillance at a swine‐human interface. MSphere 5: e00822‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Martínez M, Nielsen J, Dupont S, Vamathevan J, Glover BJ, Crosswell LC, Rouse B, Luisi BF, Bowler C, Gasser SM et al (2022) Molecular biology for green recovery—a call for action. PLoS Biol 20: e3001623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogério F, van Oosterhout C, Ciampi‐Guillardi M, Correr FH, Hosaka GK, Cros‐Arteil S, Rodrigues Alves Margarido G, Massola Júnior NS, Gladieux P (2022) Means, motive and opportunity for biological invasions: genetic introgression in a fungal pathogen. Mol Ecol 32: 2428–2442 [DOI] [PubMed] [Google Scholar]
- Runtuwene LR, Tuda JSB, Mongan AE, Makalowski W, Frith MC, Imwong M, Srisutham S, Nguyen Thi LA, Tuan NN, Eshita Y et al (2018) Nanopore sequencing of drug‐resistance‐associated genes in malaria parasites, Plasmodium falciparum . Sci Rep 8: 8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush ER, Dale E, Aguirre AA (2021) Illegal wildlife trade and emerging infectious diseases: pervasive impacts to species, ecosystems and human health. Animals 11: 1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sereika M, Kirkegaard RH, Karst SM, Michaelsen TY, Sørensen EA, Wollenberg RD, Albertsen M (2022) Oxford Nanopore R10.4 long‐read sequencing enables the generation of near‐finished bacterial genomes from pure cultures and metagenomes without short‐read or reference polishing. Nat Methods 19: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W (2017) Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods 14: 407–410 [DOI] [PubMed] [Google Scholar]
- Smart U, Cihlar JC, Budowle B (2021) International wildlife trafficking: a perspective on the challenges and potential forensic genetics solutions. Forensic Sci Int Genet 54: 102551 [DOI] [PubMed] [Google Scholar]
- Stoiber M, Quick J, Egan R, Lee JE, Celniker S, Neely RK, Loman N, Pennacchio LA, Brown J (2017) De novo identification of DNA modifications enabled by genome‐guided Nanopore signal processing. bioRxiv 10.1101/094672 [PREPRINT] [DOI]
- Street TL, Barker L, Sanderson ND, Kavanagh J, Hoosdally S, Cole K, Newnham R, Selvaratnam M, Andersson M, Llewelyn MJ et al (2020) Optimizing DNA extraction methods for Nanopore sequencing of Neisseria gonorrhoeae directly from urine samples. J Clin Microbiol 58: e01822‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmam S, Vongphayloth K, Baquero E, Munier S, Bonomi M, Regnault B, Douangboubpha B, Karami Y, Chrétien D, Sanamxay D et al (2022) Bat coronaviruses related to SARS‐CoV‐2 and infectious for human cells. Nature 604: 330–336 [DOI] [PubMed] [Google Scholar]
- Theissinger K, Fernandes C, Formenti G, Bista I, Berg PR, Bleidorn C, Bombarely A, Crottini A, Gallo GR, Godoy JA et al (2023) How genomics can help biodiversity conservation. Trends Genet 10.1016/j.tig.2023.01.005 [DOI] [PubMed] [Google Scholar]
- Theuns S, Vanmechelen B, Bernaert Q, Deboutte W, Vandenhole M, Beller L, Matthijnssens J, Maes P, Nauwynck HJ (2018) Nanopore sequencing as a revolutionary diagnostic tool for porcine viral enteric disease complexes identifies porcine kobuvirus as an important enteric virus. Sci Rep 8: 9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichkule S, Cacciò SM, Robinson G, Chalmers RM, Mueller I, Emery‐Corbin SJ, Eibach D, Tyler KM, van Oosterhout C, Jex AR (2022) Global population genomics of two subspecies of Cryptosporidium hominis during 500 years of evolution. Mol Biol Evol 39: msac056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNEP (2023) Bracing for superbugs: strengthening environmental action in the one health response to antimicrobial resistance . UNEP – UN Environment Programme. http://www.unep.org/resources/superbugs/environmental-action
- Urban L, Holzer A, Baronas JJ, Hall MB, Braeuninger‐Weimer P, Scherm MJ, Kunz DJ, Perera SN, Martin‐Herranz DE, Tipper ET et al (2021) Freshwater monitoring by nanopore sequencing. Elife 10: e61504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban L, Miller AK, Eason D, Vercoe D, Shaffer M, Wilkinson SP, Jeunen G‐J, Gemmell NJ, Digby A (2023) Non‐invasive real‐time genomic monitoring of the critically endangered kākāpō. Elife 12: RP84553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbogaert M, Kwasiborski A, Gonofio E, Descorps‐Declère S, Selekon B, Nkili Meyong AA, Ouilibona RS, Gessain A, Manuguerra J‐C, Caro V et al (2022) Nanopore sequencing of a monkeypox virus strain isolated from a pustular lesion in the Central African Republic. Sci Rep 12: 10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanmechelen B, Bertelsen MF, Rector A, van den Oord JJ, Laenen L, Vergote V, Maes P (2017) Identification of a novel species of papillomavirus in giraffe lesions using nanopore sequencing. Vet Microbiol 201: 26–31 [DOI] [PubMed] [Google Scholar]
- Viehweger A, Krautwurst S, Lamkiewicz K, Madhugiri R, Ziebuhr J, Hölzer M, Marz M (2019) Direct RNA nanopore sequencing of full‐length coronavirus genomes provides novel insights into structural variants and enables modification analysis. Genome Res 29: 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viñes J, Cuscó A, Napp S, Alvarez J, Saez‐Llorente JL, Rosàs‐Rodoreda M, Francino O, Migura‐Garcia L (2021) Transmission of similar Mcr‐1 carrying plasmids among different Escherichia coli lineages isolated from livestock and the farmer. Antibiotics 10: 313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan YK, Hendra C, Pratanwanich PN, Göke J (2022) Beyond sequencing: machine learning algorithms extract biology hidden in Nanopore signal data. Trends Genet 38: 246–257 [DOI] [PubMed] [Google Scholar]
- Wanner N, Larsen PA, McLain A, Faulk C (2021) The mitochondrial genome and Epigenome of the Golden lion tamarin from fecal DNA using Nanopore adaptive sequencing. BMC Genomics 22: 726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasser SK, Brown L, Mailand C, Mondol S, Clark W, Laurie C, Weir BS (2015) Genetic assignment of large seizures of elephant ivory reveals Africa's major poaching hotspots. Science 349: 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watsa M, Erkenswick GA, Pomerantz A, Prost S (2020) Portable sequencing as a teaching tool in conservation and biodiversity research. PLoS Biol 18: e3000667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Paderes EP, Anand N, Uchida JY, Pennycook SR, Bellgard SE, Beever RE (2015) A taxonomic revision of Phytophthora clade 5 including two new species, Phytophthora agathidicida and P. cocois . Phytotaxa 205: 21–38 [Google Scholar]
- Whitmee S, Haines A, Beyrer C, Boltz F, Capon AG, Dias BFS, Ezeh A, Frumkin H, Gong P, Head P et al (2015) Safeguarding human health in the Anthropocene epoch: report of the Rockefeller Foundation–lancet commission on planetary health. Lancet 386: 1973–2028 [DOI] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Holt KE (2019) Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol 20: 129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HS, Yoo D (2020) COVID‐19 and veterinarians for one health, zoonotic‐ and reverse‐zoonotic transmissions. J Vet Sci 21: e51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X, Xie Z, Li M, Wang K, Li X, Zhang X, Yan J, Yin Y (2022) Simultaneous profiling of histone modifications and DNA methylation via nanopore sequencing. Nat Commun 13: 7939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Peng W‐F, Hu X‐J, Zhao Y‐X, Lv F‐H, Yang J (2018) Global genomic diversity and conservation priorities for domestic animals are associated with the economies of their regions of origin. Sci Rep 8: 11677 [DOI] [PMC free article] [PubMed] [Google Scholar]


