Summary
To analyze the differences between different-sized Acipenser dabryanus, we randomly selected 600 3-month-old A. dabryanus juveniles. Four months later, the blood and white muscle of these fish were analyzed. The results showed no significant difference in the length-weight relationship (LWR) b value between the large and small A. dabryanus. The levels of serum growth hormone (gh) and insulin-like growth factor 1 (igf1) in the large A. dabryanus were significantly lower than those in the small, whereas the activity levels of Total superoxide dismutase (T-sod) and catalase (cat) were opposite to the results of gh and igf1. A total of 212 and 245 metabolites showed significant changes in the positive and negative polarity mode, respectively. Among 3,308 proteins identified, 69 proteins showed upregulated expression, and 185 proteins showed downregulated expression. These results indicated that the growth advantage of A. dabryanus was closely related to glycolysis, protein synthesis, and antioxidant function.
Subject areas: Ichthyology, Proteomics, Metabolomics
Graphical abstract

Highlights
-
•
We explored the growth of A. dabryanus of different sizes for the first time
-
•
We compared the length-weight relationship of different sized A. dabryanus
-
•
Proteomic and metabolomic analysis revealed mechanism underlying growth difference
Ichthyology; Proteomics; Metabolomics
Introduction
Acipenser dabryanus belongs to Acipenseriformes and is a first-class protected animal in China. Our institute has been responsible for the rescue and artificial reproduction of A. dabryanus for a long time. In breeding A. dabryanus, we always find individual differences within each group; the weight of the largest fish can even exceed that of the smallest fish by more than 30 times during a certain period of time. We are very curious about this phenomenon. Fish in any group often show weight differences, and the common solution is to feed them in groups. That is, identify fish of different sizes and feed them separately to alleviate the growth of all fish inhibited by the large and potentially fast-growing fish.1 However, in the actual breeding process, even if measures are taken, extreme differences in weight will still occur. So what exactly are the changes happening in the bodies of these A. dabryanus with weight differences? The underlying molecular mechanism needs further investigation.
The length-weight relationship (LWR) is widely used to represent the growth patterns and dynamics of populations.2 It is an important parameter of fish population assessment and is an important parameter to understand the nutritional level of a species.3 LWR of fish undergoes different changes throughout the entire growth cycle, such as nutritional status, growth stage, sex, season, habitat, etc.3,4 However, we wondered whether the LWR between large and small A. dabryanus in the same environment differs.
Growth hormone (gh) and insulin-like growth factor 1 (igf1) play very important roles in fish growth.5 Gh is involved in regulating the energy balance,6 gonadal development,7 osmoregulation,8 immunity,9 and other processes in fish. The function of gh can be realized through the direct action on target cells or via its effector molecule igf1. The skeletal muscle of a fish accounts for 30%–60% of its body weight, and the growth of fish is mainly attributable to muscle growth. Moreover, gh-igf1 axis is indispensable in the regulation of skeletal muscle growth.10 Therefore, it was necessary to explore the serum gh and igf1 levels of large and small A. dabryanus.
Muscle not only accounts for most of the fish body weight but is also metabolically active tissue that greatly determines the whole-animal metabolic phenotype of most vertebrates.11 In other words, the white muscle of fish dominates the metabolic rate of the whole animal. Glycolysis is an important mechanism for glucose decomposition and energy supply in the body. In the skeletal muscle and myocardium, glycolysis is most robust, and it is weakest in the liver.12 Therefore, the white muscle of A. dabryanus was an important research object in our experiments.
Metabolomics and transcriptomics have become hot research fields in recent years. We can infer the relevant signaling pathways under specific conditions through the changes in metabolites and related protein levels. Omics-related research has been widely studied in fish, such as yellow drum (Nibea albiflora),13 discus fish (Symphysodon haraldi),14 coho salmon (Oncorhynchus kisutch),15 Takifugu fasciatus,16 and threespine sticklebacks (Gasterosteus aculeatus).17 To date, few studies on metabolome and proteome differences between different-sized fish have been reported, and therefore, we performed metabolomic and proteomic analyses to preliminarily explore and analyze the internal mechanism underlying the differences in the size of A. dabryanus. With this information, further understanding of A. dabryanus growth and development characteristics will be promoted, and the results provide strong data support and technical guidance for species protection, proliferation, and release.
Results
LWR
Through our breeding process, we have found that the individual growth of fish in the same group of A. dabryanus shows highly significant differences, even under the same breeding conditions. According to our measurement data (Table 1), the body length range of the small individuals was 16.0–22.5 cm, and the body weight range was 16.22–53.36 g (seven months old). The body length of the large individuals was 28.5–39.0 cm, and the weight range was 142.65–401.95 g (Figures 1A and 1B). The results of body weight and body length showed extremely significant differences between group A and B. According to the LWR (W = aLb), we generated growth curves for the large and small fish (Figure 1). WA = 0.004L3.024 (a: 0.002–0.008; b: 2.769–3.279; R2 = 0.902), and WB = 0.005L3.098 (a: 0.002–0.010; b: 2.875–3.320; R2 = 0.927).
Table 1.
The weight and body length of two groups
| Group | Weight (g) |
Body Length (cm) |
||||
|---|---|---|---|---|---|---|
| means ± SE | minimum | maximum | means ± SE | minimum | maximum | |
| A | 33.90 ± 1.00∗∗ | 16.22 | 53.36 | 20.0 ± 0.2∗∗ | 16.0 | 22.5 |
| B | 240.38 ± 7.22 | 142.65 | 401.95 | 33.2 ± 0.3 | 28.5 | 39.0 |
Note: Data are represented as mean ± SEM, the statistical method is independent sample t test, and “∗∗” represented highly significantly difference (p < 0.01).
Figure 1.
Length-weight relationships of Acipenser dabryanus
(A) A group, small individual.
(B) B group, large individual.
Serum hormone, enzymatic activity, and igf1 expression level
In the gh-igf1 axis, the serum levels of gh and igf1 in the small-fish group (group A) were significantly higher than those in the large-fish group (group B) (Figure 2A). The serum levels of cortisol were not significantly different between the small and large groups (Figure 2B). The activity levels of Total superoxide dismutase (T-sod) and catalase (cat) in group B were significantly higher than those in group A (Figure 2C). According to quantitative analysis (Table 2), the expression of igf1 in liver was shown in Figure 2D. The igf1 level in group A was highly significantly lower than that in group B.
Figure 2.
Serum gh, igf1, cortisol level and the activities of serum T-sod and cat of Acipenser dabryanus
(A) A group, small individual.
(B) B group, large individual; a: the concentration of gh and igf1; b: the concentration of cortisol; c: the activities of T-sod and cat. Data are represented as mean ± SEM, the statistical method is independent sample t test, and “∗∗” represented highly significantly difference (p < 0.01)).
Table 2.
Primer sequence, optimal annealing temperatures (OAT) of genes selected for analysis by real-time PCR
| Name | Sequence(5′–3′) | OAT (°C) | efficiency |
|---|---|---|---|
| igf1-QF | GCCATCCAAATCCTCACGCTC | 59 | 104.5% |
| igf1-QR | CTGTTGCCTGTGTTCCCGC | ||
| β-actin-QF | GACCGAGGCACCCCTGAAC | 54.9 | 102.4% |
| β-actin-QR | GATGGGCACTGTGTGTGTGAC |
Metabolomic analysis
Overview of the metabolite data
A total of 966 metabolites were identified, including 470 in the positive polarity mode (pos) (Table S2) and 496 in the negative polarity mode (neg) (Table S3). The metabolome is easily disrupted by external factors and changes rapidly. Quality control (QC) is therefore a necessary step to obtain stable and accurate metabolomic analysis results, and the Pearson correlation between the samples is shown in Figure S1. The R2 values of the pos and neg QC samples were between 0.99 and 0.992 and 0.989–0.991, respectively. Principal-component analysis (PCA) of total samples showed that the QC samples clustered together and that the group A and group B samples clustered together, with the distribution identified at a nearly 95% confidence level.
The PCA method was used to observe the overall distribution trend between the two groups of samples. The PCA and PCA-discriminate analysis (DA) score plots of groups A and B in pos and neg and a sorting-verification diagram are shown in Figures 3A–3D. After dimensionality reduction, the principal components accounted for 60.49% (pos) and 58.22% (neg) of the total variables.
Figure 3.
Identification and analysis of differentially expressed metabolites (DEMs)
(A and B) PCA score plots of A and B group following positive (A) and negative (B).
(C and D) PLS-DAscore plots of A and B group following positive (C) and negative (D).
(E and F) Volcano map of A and B group following positive (E) and negative (F). (Gray in the volcano plot indicates metabolites with no significant difference, red indicates upregulated and blue indicates downregulated; n = 6.).
Differential metabolites
The identified metabolites with differential levels of abundance are shown in Tables S4 and S5. In the pos, a total of 212 metabolites showed significant changes, of which 126 metabolite levels were increased and of which 86 metabolite levels were decreased. In neg group, a total of 245 metabolites were significantly changed, of which the levels of 95 metabolites were increased and those of 150 metabolites were decreased.
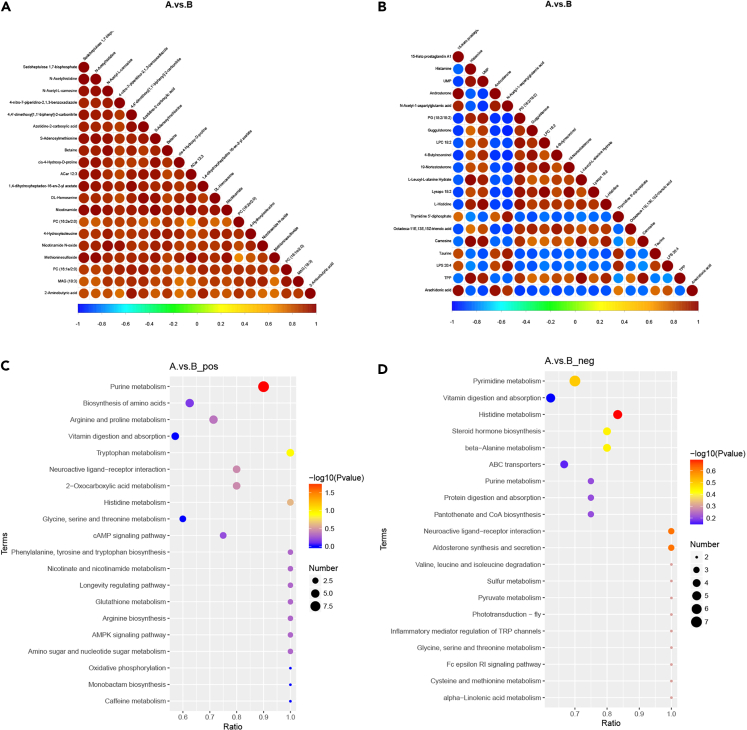
We generated volcano maps to show a more intuitive view of the overall distribution of the differentially abundant metabolites (Figures 3E and 3F). The cluster heatmap of the differentially abundant metabolites is shown in Figures S2A and S2B. The correlations among the 20 most differentially abundant metabolites are shown in Figures 4A and 4B arranged in descending p value order.
Figure 4.
Visualization of DEMs
(A and B): correlation analysis of DEMs (when the linear relationship between the two metabolites is enhanced, the positive correlation tends to 1, and the negative correlation tends to −1, significance level <0.05 is the threshold of significant correlation, (A: A group, small individual; B: B group, large individual; a: positive mode, b: negative mode).
(C and D) KEGG pathway enrichment analysis (C: positive mode, D: negative mode; only show the results of top20; n = 6.).
KEGG enrichment analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the differentially abundant metabolites is shown in Table S5. According to the enrichment results, the bubble diagram of the 20 most enriched KEGG pathways is shown in Figures 4C and 4D. There are significant differences in 13 annotated signal pathways of pos metabolites and 15 annotated signal pathways of neg metabolites. The signaling pathways with the most differential metabolites were purine metabolism (pos) and pyrimidine metabolism (neg).
Proteomic analysis
Overview of the proteomic data
A total of 16,307 peptides were identified, and 3,308 proteins were identified among them (Tables S6 and S7). The PCA is shown in Figure S3. The samples in group A clustered together and those in group B clustered together with 95% confidence.
Protein difference analysis
The protein difference analysis results are shown in Table S8. Among the 3,308 proteins, the expression of 69 proteins was upregulated with a fold change (FC) ≥ 2.0 and p value ≤ 0.05, and the expression of 185 proteins was downregulated with an FC ≤ 0.50 and p value ≤ 0.05. A differential protein volcano map is shown in Figures 5A, and a heatmap is shown in Figure S4.
Figure 5.
Identification and analysis of differentially expressed proteins (DEPs)
(A) Volcano map of DEPs (A: A group, small individual. B: B group, large individual).
(B) The enriched GO terms of DEPs (The enrichment results of three categories are shown in the figure, and each category can display up to 20 types. The percentage of ordinate represents x/n in the table. p < 0.05).
(C) KEGG pathway enrichment analysis of DEPs (only show the results of top20; n = 3.).
Gene Ontology (GO) enrichment analysis (GO and KEGG functional analysis of differentially expressed proteins [DEPs])
A GO function significant enrichment analysis revealed the GO function terms significantly enriched with differential proteins compared with all the identified protein backgrounds, indicating the biological functions that are significantly related to different proteins. As shown in Figure 5B, the GO annotation data are classified into three categories: biological process (BP), cellular component (CC), and molecular function (MF). According to the number and significance of the DEPs, among the three GO categories, the three most enriched terms were carbohydrate metabolic process, cellular component biogenesis, and cellular component assembly in BP; intracellular membrane-bounded organelle, intracellular organelle part, and nucleus in CC; calcium ion binding, DNA binding, and extracellular matrix structural constituent in MF. Significant differences were found in 20 of the 119 annotated signaling pathways (Figure 5C). The three signaling pathways with the greatest DEPs enrichment were human papillomavirus infection, PI3K-Akt signaling pathway, and ECM-receptor interaction.
Combined analysis of proteomic and metabolomic data
Combined analysis of DEPs and differentially expressed metabolites (DEMs) KEGG pathway of proteome and metabolome was shown in Figures S5 and S6. The three signaling pathways with the greatest DEPs enrichment were PI3K-Akt signaling pathway, alcoholism, and biosynthesis of amino acids. The greatest DEM enrichment was purine metabolism.
Discussion
During the process of fish culture, individual differences in fry and differences in feeding ability cause the growth of the fish in the same pond to be significantly different after a period of time. This outcome is also very common in our breeding program of A. dabryanus. However, the reasons for these differences are complex. We gained more information through this experiment, including not only the apparent weight differences but also the internal regulatory changes and even the changes in related molecules in key signaling pathways.
LWR is an important parameter to describe the growth of fish in a population, and it can be affected by species, age, gonad maturity, sex, temperature, season, habitat, health status, and other factors.18 The b values of groups A and B were not significantly different from 3, indicating that the fish in these two groups nearly reached an isokinetic growth rate. However, the weight of the fish measured in the same period was significantly different.
To explore the reasons for the difference in body weight, we measured gh and igf1 levels in serum and performed a metabolome and proteome analysis of the muscles of the fish in the two size-based groups. The gh-igf1 axis plays an essential role in regulating the growth of fish.19 However, we observed that the levels of serum gh and igf1 in the small-individual-fish group were significantly higher than those in the large-individual-fish group. We speculate that the reason for this outcome may be related to the intake, utilization, and transformation of food because the intake and utilization efficiency of exogenous nutrients are key factors of physical changes. Although we employed a strategy for feeding the fish to satiety, the appetite of each fish and the digestion and absorption capacity for nutrients are also factors. Peng et al.20 showed that neuropeptide Y (npy) stimulates gh release in a dose-dependent manner in female goldfish. Himick et al.21 found in a study of goldfish (Carassius auratis) that intraperitoneal or intraventricular injection of cholecystokinin-octapeptide (CCK-8s) increased the serum gh level. In the process of breeding and production, feeding regulation is often one of the important factors affecting food intake, and appetite is affected by brain and gut peptides; their functions in the central and peripheral environment may involve different mechanisms. As appetite regulators, npy and cholecystokinin (cck) were also found to be involved in regulating the gh level,20,21,22 which may be a very important clue for future exploration and research. Igf-1 is the main effector of gh, and it is an endocrine mediator of growth and development.23 We know that igf1 is widely distributed in many tissues, among which the liver is one of the important tissues for igf1 synthesis. After igf1 synthesis in the liver, it can be transported to target cells through the blood.24 However, the mRNA level of igf1 in the liver is opposite to the concentration of igf1 in the serum. To our knowledge, gh-igf1 exhibits negative feedback regulation, and excessive circulating IGF1 can directly inhibit GH secretion.24 Therefore, we inferred that high levels of igf1 mRNA in group B may lead to the synthesis and release of a large amount of igf1, ultimately inhibiting the synthesis and release of gh by the pituitary gland. This can also explain why the serum gh levels in group B are lower than those in group A. However, the reason for the changes in igf1 level needs further investigation.
Through observations of the metabolome, betaine has been determined to be a precursor of S-adenosylmethionine (SAM).25 SAM, the principal biological methyl donor,26 is an essential functional group for protein and fat metabolism and DNA and RNA methylation.25 Although betaine is metabolized mainly in the liver and kidney, it has been reported to accumulate in fish muscles.25,27 In addition, betaine is involved in antioxidant and cell osmotic pressure regulation.25,28 Zhang et al. (2016) believed that the antioxidant mechanism of betaine is realized through SAM; however, betaine can form an electronegative protective film around a cell, preventing free radicals from contacting the cell membrane and inhibiting reactive oxygen species (ROS) generation.28 Our results showed that betaine and SAM levels were significantly increased in group B. This finding suggested that the metabolism of the proteome and fat in the large individuals was greater than that in the small individuals, and we speculated that the muscle antioxidant capacity of the large fish was also relatively high. The results of our serum T-sod and cat activity measurements seemed to confirm this assumption. In contrast, we found that the content of proline (Pro) in the muscle of the group B fish was significantly increased compared with that in the A group fish (Figure 6). Pro is involved in protecting carp red blood cells from oxidative damage,29 and it can be produced through ornithine metabolism in mammals.30 Pro stimulates the synthesis and deposition of collagen via the transforming growth factor β (TGF-β)/Smad pathway in the swim bladder of Chu’s croaker (Nibea coibor)31,32 and has been shown to function in regulating and repairing wounds.33
Figure 6.
Antioxidant enzymes and some metabolites may affect the antioxidant function of the fish (A vs. B)
Glycolysis plays an important role in the anabolism and catabolism of organisms. We found significant changes in the enzymes of the glycolysis pathway in the muscle proteome (Figure 7). Among the 10 detected enzymes, 7 in group B were significantly increased, and the levels of the three other proteins showed no significant difference. This outcome showed that glycolysis was active in the muscles of the large individuals, and it also proved the management viewpoint of Sun et al.34 that glycolysis may play an important role in the superior growth of fish. The dual role of glycolysis, which degrades glucose to produce ATP, provides raw materials for synthetic reactions, such as the formation of long-chain fatty acids. However, the level of fructose-1,6-bisphosphatase was significantly increased in group B, which is one of the key enzymes involved in the glycolysis.35 The activation of the glycolysis pathway also led to the activation of multiple biological processes, including gluconeogenesis.36 Our results also indicated that the glucose metabolism of large individuals is more active than that of small individuals. In fact, it is easy to understand that skeletal muscle is one of the important glycogen reserve tissues of the body and that large individuals contain significantly more muscle than small individuals. Surprisingly, the 3 enzymes in glycolysis (pyruvate kinase, 6-phosphofructokinase, and hexokinase), which are key enzymes in irreversible reactions during glycolysis, had no significant change between group A and B. We speculated that as key steps in the regulation process, it may remain constant under certain conditions.
Figure 7.
Differentially expressed proteins in glycolysis pathway between A and B group
(Red means upregulation, green means no change; A vs. B).
Mammalian target of rapamycin (mTOR) is a serine/threonine phosphatidylinositol kinase-related protein kinase that is relatively conserved in structure and function and is a key regulator of cell growth, immunity, protein synthesis, metabolism, and other processes.37,38,39 The PI3K/AKT-mTOR signaling pathway is thought to regulate the growth of skeletal muscle.10 The results of our proteome analysis showed that the mTOR level in group B was significantly higher than that in group A. On the other hand, we observed changes in the protein transfer pathway of the endoplasmic reticulum (ER). The export of protein from the ER to the Golgi apparatus is mediated by the coat protein II (COPII) complex. COPII consists of three parts, a guanosine triphosphate (GTP)-binding protein, Sar1, and coat protein complexes Sec23/24 and Sec13/31.40 Sec23 is the GTPase-activating protein (GAP) for Sar1, and Sec24 can combine with cargo protein or a cargo protein receptor to envelope cargo protein into vesicles.41 Activated Sar1-GTP recruits the Sec23/Sec24 dimer, which functions in cargo binding. Sar1-Sec23/24-cargoes form prebudding complexes.41 Our results showed that the levels of Sar1 and Sec24 in group B were significantly higher than those in group A. This may suggest that the large fish undergo greater protein synthesis and metastasis in the ER than the small fish.
The increase in mTOR level may be closely related to glycolysis. Using Jurkat cells, Ramanathan et al.37 identified that mTOR regulated glycolysis and respiratory metabolism by forming complexes with mitochondrial outer membrane proteins. Although our proteome data showed significant changes in the levels of many enzymes in the glycolytic pathway, unfortunately, the metabolome data did not show the same results, which may have been related to the broad-spectrum metabolome sequencing method we chose. We did not target the metabolites in the glycometabolism pathway. The data we obtained sent us in a totally new direction. The molecules that activate the mTOR pathway are complex, and perhaps through more in-depth research in the future, we can more clearly explain why there was no change in the upstream (pi3k/akt) and downstream signaling molecules (4ebp/s6k) in the mTOR signaling pathway.
However, it is puzzling that our results showed that the serum levels of gh and igf1 in group B were significantly lower than those in group A. We proposed that small fish secrete more gh and igf1 than large fish because they do need these hormones to regulate growth with their body type, but it is possible that a lack of nutrient substrates needed for growth leads to the difference in the size of fish at the same growth stage. The lack of nutrients is closely related to the intake, digestion, and absorption of food. In other words, the feed conversion efficiency may be different in fish of different sizes. In combination with the result showing that the b value in the relationship between body length and weight of small fish was close to 3, this result indicates that the metabolic process in the small individual fish may have been blocked, with the body weight not increasing in the process of average growth. Whether the nutrient synthesis and decomposition rates of two different sizes of fish are equal, it is a possible explanation that we need to determine in future research.
In general, the aims we studied with same group of A. dabryanus of different sizes helped us further understand the changes in their growth and metabolism. We hope that through an in-depth exploration into these findings in the future, we can put forward some guiding suggestions for production practice that will also benefit fish programs in addition to that of A. dabryanus. In fact, according to the results, we have provided some hints, including but not limited to, appetite regulation, gluconeogenesis, protein synthesis, and antioxidant pathways, as directions with which to carry out more in-depth research in the future.
Limitations of the study
Due to insufficient sample size for small individuals, we are unable to conduct further targeted analysis of metabolites in the glucose metabolism pathway. In addition, if the sample size is sufficient, transcriptome sequencing is also necessary.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Healthy 3-month-old A. dabryanus | The Fishery Institute of the Sichuan Academy of Agricultural Sciences | 202012 |
| Critical commercial assays | ||
| TMT® Mass Tagging Kits and Reagents | Thermo | N/A |
| Q Exactive™ HF-X | Thermo Fisher | N/A |
| Vanquish UHPLC | Thermo Fisher | N/A |
| RNAiso Plus | Takara | 9109 |
| PrimeScriptTM Rt reagent Kit with GDNA Eraser | Takara | RR047A |
| TB Green Premix Ex TaqTM II(Tli RNasaH Plus) | Takara | RR820A |
| Deposited data | ||
| Raw and analyzed data: metabolome data | This paper | NGDC:OMIX004496(https://ngdc.cncb.ac.cn/) |
| Raw and analyzed data: proteome data | This paper | NGDC:OMIX002794(https://ngdc.cncb.ac.cn/) |
| Oligonucleotides | ||
| Primers for igf1/β-actin, see Table 2 | This paper | N/A |
| Software and algorithms | ||
| Spss 25 | This paper | https://www.ibm.com/cn-zh/spss |
| GraphPad Prism 5 | This paper | https://www.graphpad.com/ |
| Proteome Discoverer 2.2 | PD2.2, Thermo | |
| STRING DB | This paper | http://STRING.embl.de/ |
| mzCloud | This paper | (https://www.mzcloud.org/) |
Resource availability
Lead contact
Further information and request for resources and reagents should be directed to and will be fulfilled by Quan Gong (admiral671@163.com)
Materials availability
Information of materials, and their possible availability, can be requested from the lead contact.
Experimental mode and study participant details
Animals: the F2 generation of A. dabryanus.
Age: 3-month-old to 7-month-old.
Institutional permission: Animal Care Advisory Committee of the Sichuan Academy of Agricultural Sciences (20201229001A).
Sex: indeterminacy(Undifferentiated gonads)
Method details
Experimental fish and sample collection
The F2 generation of A. dabryanus reproduced by The Fishery Institute of the Sichuan Academy of Agricultural Sciences was used. All experimental protocols were approved by the Animal Care Advisory Committee of the Sichuan Academy of Agricultural Sciences (20201229001A).
A total of 600 (3-month-old) A. dabryanus form the same parents were randomly distributed in equal number to 10 tanks (3∗1.5∗1 m). The mean weight of the fish was 24.65 ± 0.71 g, and mean body length was 19.4 ± 0.2 cm. The fish were fed at 8:00 a.m. and 18:00 p.m. the same diet to apparent satiation throughout the growth cycle. Dissolved oxygen content was 6.24 ± 0.53 mg L−1, pH was 6.90 ± 0.04, NH4+-N content was 0.066 ± 0.003 mg L−1, and NO2-N content was 0.001 ± 0.000 mg L−1. After 4 months, The water temperature was 27.5 ± 5°C, We selected the fish with the smallest and largest body weight from the 10 tanks, with a proportion of about 10% of the total antity. Based on the actual range of results, the approximate standard is that group A (control qugroup) is 33.90 ± 1.00 g and group B is 240.38 ± 7.22 g. We select fish in ascending or descending order of weight, and finally select 63 fish for each group. The fish were anesthetized with M222, and then, their body lengths and weights were measured. We selected 6 fish that were close to the average weight of each group separately. Blood was collected from the fishtail vein of the fish in two groups (group A: small fish; group B: large fish). The samples were centrifuged at 3000 rpm for 10 min, and serum was collected and stored at −20°C for the determination of gh, igf1 and cortisol levels using ELISA kits (Mei mian, China), and T-sod and cat activity levels were measured (Nanjing Jiancheng, China). The muscle from 6 fish in each size-based group (small and large) was sampled rapidly and then stored at −80°C.
RNA extraction and quantitative RT-PCR
The procedures of RNA isolation, reverse transcription, and quantitative real-time PCR were similar to those previously described by Wu et al.42 Specific primers for igf1 (in liver) was listed in Table 2. The real-time PCR mixture consisted of 2 μL of the first strand cDNA sample, 1 μL each of forward and reverse primers from 20 μM stocks, 6 μL of RNase-free ddH2O, and 10 μL of 2x TB Green Premix Ex TaqII (TakaRa, Japan). The cycling conditions were 98°C for 2 min, followed by 40 cycles of 98°C for 5 s, annealing at 59°C for 10 s, and 72°C for 15 s. The target gene mRNA concentration was normalized to the mRNA concentration of the reference gene β-actin; for detailed methods, refer to Wu et al.42 The expression results were calculated using the 2−ΔΔCT method after verification that the primers amplified with an efficiency of approximately 100%.
Metabolome material preparation and analysis methods
Sample preparation
Six white muscle tissues were individually ground with liquid nitrogen, and then, the samples were resuspended in methanol. After vortex oscillation, the sample was allowed to stand for 5 min in an ice bath and then centrifuged. The supernatant was diluted with liquid chromatography–mass spectrometry (LC‒MS) grade water until the methanol content was 53%. After centrifugation, the supernatant was injected into an LC‒MS/MS system for analysis43
Ultrahigh performance liquid chromatography-tandem mass spectrometry (UHPLC‒MS/MS) analysis
UHPLC‒MS/MS analysis was performed according to method of Zhou et al.44 The eluents and chromatographic gradient elution procedure are shown in Table S1. The parameters of the Q ExactiveTMHF-X mass spectrometer were set as follows: Scanning range: m/z100-1500; Spray Voltage: 3.2 kV; Sheath gas flow rate: 40arb; Aux Gasflow rate: 10 arb; Capillary Temp: 320°C. Polarity: positive: negative; MS/MS secondary scanning: data-dependent scans.
Metabolite identification and annotation
Offline data (.Raw) file was imported into Compound Discoverer library search software to determine the retention time, mass load ratio and other parameters. Relevant parameters were chosen in reference to those of Zhang et al.45 Then, the identification and quantitative results of the data were obtained. The identified metabolites were annotated for function and classification. The main databases for this analysis were the Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.genome.jp/kegg/pathway.html), Human Metabolome Database (HMDB) (https://hmdb.ca/metabolites), LIPID MAPS, etc. The KEGG pathways were analyzed according to method of Liu et al.46
Screening and analysis of differential metabolites
Principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) were performed to determine the overall distribution trend between the two groups of samples, which enabled the prediction for a sample category. Relevant parameter settings were obtained from Zhang et al.45 The screening of differential metabolites mainly refers to three parameters: VIP, FC and p-value. VIP refers to the variable importance in the projection of the first principal component of PLS-DA model, and VIP value represents the contribution of metabolites to the grouping; FC refers to the fold change, which is the ratio of the mean value of all biological repeated quantitative values of each metabolite in the comparison group; p-value is calculated by t-test, indicating the significance level of the difference. Set the threshold value as VIP >1.0, FC > 1.5 or FC < 0.667 and p-value < 0.05. Cluster analysis was used to judge the metabolic patterns of metabolites under different experimental conditions.
KEGG enrichment analysis
The main biological functions of differential metabolites can be determined by KEGG pathway enrichment analysis. Where N is the number of metabolites involved in KEGG metabolic pathway among all metabolites, n is the number of differential metabolism in N, y is the number of metabolites annotated to a KEGG pathway, and X is the number of differential metabolites enriched to a KEGG pathway. If the ratio condition X/N > Y/N is met, the pathway is a KEGG enrichment pathway. With the help of hypergeometric test method, the p-value value of pathway enrichment is obtained. Taking p-value ≤0.05 as the threshold, the KEGG pathway meeting this condition is defined as the KEGG pathway significantly enriched in different metabolites.
Proteomic material preparation and analysis methods
Sample preparation and tandem mass tag (TMT) labeling
Each sample was ground into powder at a low temperature, and then, protease was added for treatment. The sample was centrifuged at 4°C and 12,000 × g for 15 min and the supernatant was added with 10 mM DTT and react at 56°C for 1 h, then sufficient IAM and react were added at room temperature without light for 1 h. We added 4 times the volume of - 20°C precooled acetone, precipitated at - 20°C for at least 2 h, centrifuged at 4°C and 12,000 × g for 15 min, and collected the precipitate. After that, we added 1 mL −20°C precooled acetone to resuspend and clean the precipitation, centrifuged at 4°C and 12,000 × g for 15 min, collected the precipitation, air dry, and added an appropriate amount of protein solution (8 M Urea 100 mM TEAB, pH = 8.5) dissolved protein precipitation.
TMT labeling, each protein sample was taken and the volume was made up to 100 μL with DB dissolution buffer (8 M Urea, 100 mM TEAB, pH 8.5). Trypsin and 100 mM TEAB buffer were added, sample was mixed and digested at 37°C for 4h. And then, trypsin and CaCl2 were added, sample was digested overnight. Formic acid was mixed with digested sample, adjusted pH under 3, and centrifuged at 12000 g for 5 min at room temperature. The supernatant was slowly loaded to the C18 desalting column, washed with washing buffer (0.1% formic acid, 3% acetonitrile) 3 times, then eluted by some elution buffer (0.1% formic acid, 70% acetonitrile). The eluents of each sample were collected and lyophilized. 100 μL of 0.1 M TEAB buffer was added to reconstitute, and 41 μL of acetonitrile-dissolved TMT labeling reagent was added, sample was mixed with shaking for 2 h at room temperature. Then, the reaction was stopped by adding 8% ammonia. All labeling samples were mixed with equal volume, desalted and lyophilized.
Proteome data processing
Using the protein database, we accessed the database search software Proteome Discoverer 2.2 (PD 2.2, Thermo) to search the spectrum of each run. Bioinformatics analyses were performed according to the method of Zhou et al.47 Interproscan software was used to annotate the GO and IPR functions (including Pfam, prints, ProDom, smart, PROSITE and Panther databases), COG and KEGG were used to analyze the functional protein families and pathways of the identified proteins. Volcanic map analysis, cluster heatmap analysis and pathway enrichment analysis of GO, IPR and KEGG were carried out for DPE, and string DB software was used to predict possible protein-protein interactions(http://STRING.embl.de/).
Quantification and statistical analysis
All data are reported as the means±standard errors (SEs). The relationship between body length and body weight was fitted using a power exponent function (W = aLb). Under the condition that all data conformed to a normal distribution and homogeneity of variance, an independent sample T test method was performed; in contrast, for nonparametric analysis, the Mann‒Whitney U test was performed. p < 0.05 indicates a significant difference, p < 0.01 indicates an extremely significant difference, and p > 0.05 indicates no significant difference. SPSS 25 was used for the statistical analysis of the data.
Acknowledgments
This work was supported by the Sichuan Science and Technology Program (2021YFYZ0015), the Sichuan Science and Technology Program (2023ZHCG0095), the National Modern Agricultural Industrial Technology System of the Sichuan Freshwater Fish Innovation Team, and Natural Science Foundation of Sichuan Province (2022NSFSC0124), the Key Laboratory of Sichuan Province for Fishes Conservation and Utilization in the Upper Reaches of the Yangtze River, Neijiang Normal University(NJTCCJSYSYS08).
Author contributions
Xiaoyun Wu: Writing – review & editing, Writing – original draft, Conceptualization. Jiansheng Lai: Conceptualization, Resources, Formal analysis. Yeyu Chen: Methodology, Formal analysis. Ya Liu: Validation. Mingjiang Song: Methodology. Feiyang Li: Validation. Pengcheng Li: Resources. Qingzhi Li: Conceptualization, Funding acquisition. Quan Gong: Resources, Writing – review & editing, Conceptualization.
Declaration of interests
The authors declare no competing interests.
Published: July 18, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107413.
Supplemental information
Data and code availability
-
•
Raw data derived from A. dabryanus samples have been deposited at National Genomics Data Center (NGDC), and accession numbers are listed in the key resources table, and are publicly available as of the date of publication.
-
•
This study did not generate original code.
-
•
Any additional information required to reanalyze the data reported in this work is available from the lead contact upon reasonable request.
References
- 1.Barki A., Harpaz S., Hulata G., Karplus I. Effects of larger fish and size grading on growth and size variation in fingerling silver perch. Aquacult. Int. 2000;8:391–401. [Google Scholar]
- 2.Tran H.D., Hai H.N., Ha L.M. REG STUD MAR SCI; 2021. Length-weight Relationship and Condition Factor of the Mudskipper (Periophthalmus modestus) in the Red River Delta. [Google Scholar]
- 3.Sileesh M., Kurup B.M., Korath A. Length at maturity and relationship between weight and total length of five deep-sea fishes from the, Andaman and Nicobar Islands of India, North-eastern Indian Ocean. J. Mar. Biol. Assoc. U. K. 2020;100:639–644. [Google Scholar]
- 4.Keyombe J.L., Obegi B., Waithaka E. Length-weight relationship and condition factor of Clarias gariepinus in Lake Naivasha, Kenya. AkiNik Publications. 2015;2:382–385. [Google Scholar]
- 5.Zhong H., Yi Z., Liu S., Min T., Yu L., Zhen L., Zhang C., Wei D., Jie H., Song C. Elevated expressions of GH/IGF axis genes in triploid crucian carp. Gen. Comp. Endocrinol. 2012;178 doi: 10.1016/j.ygcen.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Rousseau K., Dufour S. Comparative Aspects of GH and Metabolic Regulation in Lower Vertebrates. Neuroendocrinology. 2007;86:165–174. doi: 10.1159/000101029. [DOI] [PubMed] [Google Scholar]
- 7.Singh A.K., Lal B. Seasonal and circadian time-dependent dual action of GH on somatic growth and ovarian development in the Asian catfish, Clarias batrachus (Linn.): role of temperature. Gen. Comp. Endocrinol. 2008;159:98–106. doi: 10.1016/j.ygcen.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Sakamoto T., Mccormick S.D. Prolactin and growth hormone in fish osmoregulation. Gen. Comp. Endocrinol. 2006;147:24–30. doi: 10.1016/j.ygcen.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Yada T. Immune Function and Growth Hormone in Fish. Fish. Sci. 2002;68:761–764. [Google Scholar]
- 10.Fuentes E.N., Valdés J.A., Molina A., Björnsson B.T. Regulation of skeletal muscle growth in fish by the growth hormone - Insulin-like growth factor system. Gen. Comp. Endocrinol. 2013;192:136–148. doi: 10.1016/j.ygcen.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Davies R., Moyes C.D. Allometric scaling in centrarchid fish: origins of intra-and inter-specific variation in oxidative and glycolytic enzyme levels in muscle. J. Exp. Biol. 2007;210:3798–3804. doi: 10.1242/jeb.003897. [DOI] [PubMed] [Google Scholar]
- 12.Wang G.Y., Liu B., Xie J., Ge X.P. Research progress of several carbohydrate metabolic key enzymes in fish. J Shanghai Ocean Uni. 2008;3:377–383. [Google Scholar]
- 13.Maha I.F., Xie X., Zhou S., Yu Y., Liu X., Zahid A., Lei Y., Ma R., Yin F., Qian D. Skin metabolome reveals immune responses in yellow drum Nibea albiflora to Cryptocaryon irritans infection. Fish Shellfish Immunol. 2019;94:661–674. doi: 10.1016/j.fsi.2019.09.027. [DOI] [PubMed] [Google Scholar]
- 14.Wen B., Zhou J.Q., Gao J.Z., Chen H.R., Shen Y.Q., Chen Z.Z. Sex-dependent changes in the skin mucus metabolome of discus fish (Symphysodon haraldi) during biparental care. J. Proteonomics. 2020;221 doi: 10.1016/j.jprot.2020.103784. [DOI] [PubMed] [Google Scholar]
- 15.Causey D.R., Kim J.H., Stead D.A., Martin S.A.M., Devlin R.H., Macqueen D.J. Proteomic comparison of selective breeding and growth hormone transgenesis in fish: Unique pathways to enhanced growth. J. Proteonomics. 2019;192:114–124. doi: 10.1016/j.jprot.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen X., Zhang X., Hu Y., Xu J., Wang T., Yin S. iTRAQ-based quantitative proteomic analysis of Takifugu fasciatus liver in response to low-temperature stress. J. Proteonomics. 2019;201:27–36. doi: 10.1016/j.jprot.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Li J., Levitan B., Gomez-Jimenez S., Kültz D. Development of a gill assay library for ecological proteomics of threespine sticklebacks (Gasterosteus aculeatus) Mol. Cell. Proteomics. 2018;17:2146–2163. doi: 10.1074/mcp.RA118.000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tesch F.W. Blackwell; 1971. Age and Growth in Fish Production in Freshwaters; pp. 98–130. [Google Scholar]
- 19.Zhang Y., Shi M., Mao X., Kou Y., Liu J. Physiological role of the growth axis in embryonic development and early growth of Cynoglossus semilaevis. Biotechnol. Biofuels. 2019;12:287. [Google Scholar]
- 20.Peng C., Humphries S., Peter R.E., Rivier J.E., Blomqvist A.G., Larhammar D. Actions of goldfish neuropeptide Y on the secretion of growth hormone and gonadotropin-II in female goldfish. Gen. Comp. Endocrinol. 1993;90:306–317. doi: 10.1006/gcen.1993.1086. [DOI] [PubMed] [Google Scholar]
- 21.Himick B.A., Peter R.E. CCK/gastrin-like immunoreactivity in brain and gut, and CCK suppression of feeding in goldfish. Am. J. Physiol. 1994;267:841–851. doi: 10.1152/ajpregu.1994.267.3.R841. [DOI] [PubMed] [Google Scholar]
- 22.Volkoff H., Canosa L.F., Unniappan S., Cerdá-Reverter J.M., Bernier N.J., Kelly S.P., Peter R.E. Neuropeptides and the control of food intake in fish. Gen. Comp. Endocrinol. 2005;142:3–19. doi: 10.1016/j.ygcen.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Wood A.W., Duan C., Bern H.A. Insulin-like growth factor signaling in fish. Int. Rev. Cytol. 2005;243:215–285. doi: 10.1016/S0074-7696(05)43004-1. [DOI] [PubMed] [Google Scholar]
- 24.Reinecke M. Influences of the environment on the endocrine and paracrine fish growth hormone-insulin-like growth factor-I system. J. Fish. Biol. 2010;76:1233–1254. doi: 10.1111/j.1095-8649.2010.02605.x. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann L., Brauers G., Gehrmann T., Häussinger D., Mayatepek E., Schliess F., Schwahn B.C. Osmotic regulation of hepatic betaine metabolism. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;304:835–846. doi: 10.1152/ajpgi.00332.2012. [DOI] [PubMed] [Google Scholar]
- 26.Barve S., Moghe A., Gobejishvili L., Barve S.J., Mcclain C.J. S-adenosylmethionine (SAM) deficiency in alcoholic/non- alcoholic steatohepatitis: potential therapeutic role of SAM. Trop. Gastroenterol. 2011 [Google Scholar]
- 27.Ito Y., Suzuki T., Shirai T., Hirano T. Presence of cyclic betaines in fish. Comp. Biochem. Physiol. Part B Comparative Biochemistry. 1994;109:115–124. [Google Scholar]
- 28.Zhang M., Zhang H., Li H., Lai F., Li X., Tang Y., Min T., Wu H. Antioxidant mechanism of betaine without free radical scavenging ability. J. Agric. Food Chem. 2016;64:7921–7930. doi: 10.1021/acs.jafc.6b03592. [DOI] [PubMed] [Google Scholar]
- 29.Li H.T., Feng L., Jiang W.D., Liu Y., Jiang J., Li S.H., Zhou X.Q. Oxidative stress parameters and anti-apoptotic response to hydroxyl radicals in fish erythrocytes: Protective effects of glutamine, alanine, citrulline and proline. Aquat. Toxicol. 2013;126:169–179. doi: 10.1016/j.aquatox.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Lodato R.F., Smith R.J., Valle D., Phang J.M., Aoki T.T. Regulation of proline biosynthesis: The inhibition of pyrroline-5-carboxylate synthase activity by ornithine. Metabolism. 1981;30:908–913. doi: 10.1016/0026-0495(81)90070-6. [DOI] [PubMed] [Google Scholar]
- 31.Rong H., Lin F., Limbu S.M., Lin Z., Bi B., Dou T., Zhao L., Wen X. Effects of dietary proline on swim bladder collagen synthesis and its possible regulation by the TGF-β/Smad pathway in spotted drum, Nibea diacanthus. Aquacult. Nutr. 2020;26:1792–1805. [Google Scholar]
- 32.Rong H., Zhang H., Ning L., Wu K., Limbu S.M., Shi Q., Qin C., Wen X. The transforming growth factor beta (TGF-β/Smads) pathway regulates collagen synthesis and deposition in swim bladder of Chu's croaker (Nibea coibor) stimulated by proline. Aquaculture. 2022 [Google Scholar]
- 33.Ponrasu T., Jamuna S., Mathew A., Madhukumar K.N., Ganeshkumar M., Iyappan K., Suguna L. Efficacy of L-proline administration on the early responses during cutaneous wound healing in rats. Amino Acids. 2013;45:179–189. doi: 10.1007/s00726-013-1486-0. [DOI] [PubMed] [Google Scholar]
- 34.Sun Y., Huang Y., Hu G., Zhang X., Ruan Z., Zhao X., Guo C., Tang Z., Li X., You X., et al. Comparative transcriptomic study of muscle provides new insights into the growth superiority of a novel grouper hybrid. PLoS One. 2016;11 doi: 10.1371/journal.pone.0168802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nordlie R.C., Foster J.D., Lange A.J. Regulation of glucose production by the liver. Annu. Rev. Nutr. 1999;19:379–406. doi: 10.1146/annurev.nutr.19.1.379. [DOI] [PubMed] [Google Scholar]
- 36.Cohen G.N., Cohen G.N. Glycolysis, gluconeogenesis and glycogen and cellulose synthesis. Microbial Biochemistry. 2014:73–83. [Google Scholar]
- 37.Ramanathan A., Schreiber S.L. Direct control of mitochondrial function by mTOR. Proc. Natl. Acad. Sci. USA. 2009;106:22229–22232. doi: 10.1073/pnas.0912074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X., Proud C.G. The mTOR Pathway in the Control of Protein Synthesis. Physiology. 2006;21:362–369. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 39.Weichhart T., Hengstschläger M., Linke M. Regulation of innate immune cell function by mTOR. Nat. Rev. Immunol. 2015;15:599–614. doi: 10.1038/nri3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuoka K., Orci L., Amherdt M., Bednarek S.Y., Hamamoto S., Schekman R., Yeung T. COPII-Coated Vesicle Formation Reconstituted with Purified Coat Proteins and Chemically Defined Liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 41.Jennifer G., D'Arcangelo, Kyle R., Stahmer, Elizabeth and A. Vesicle-mediated export from the ER: COPII coat function and regulation. Biochim. Biophys. Acta Mol. Cell Res. 2013:2464–2472. doi: 10.1016/j.bbamcr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X.Y., Chen Y.Y., Lai J.S., Liu Y., Long Z.H. Effects of starvation and refeeding on growth performace, appetite, GH-IGFs axis levels and digestive function of Acipenser dabryanus. Br. J. Nutr. 2020;126:1–39. doi: 10.1017/S0007114520004389. [DOI] [PubMed] [Google Scholar]
- 43.Want E.J., Masson P., Michopoulos F., Wilson I.D., Theodoridis G., Plumb R.S., Shockcor J., Loftus N., Holmes E., Nicholson J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013;8:17–32. doi: 10.1038/nprot.2012.135. [DOI] [PubMed] [Google Scholar]
- 44.Jie Zhou J.G.Q.C., Baosong Wang X.H.Q.Z., Wang Integrated metabolomics and transcriptomics analyses reveal anthocyaninand carotenoid biosynthesis involved in color development in willow bark. Research. 2022 [Google Scholar]
- 45.Zhang W., Tan B., Ye G., Wang J., Dong X., Yang Q., Chi S., Liu H., Zhang S., Zhang H. Identification of potential biomarkers for soybean meal-induced enteritis in juvenile pearl gentian grouper, Epinephelus lanceolatus♂× Epinephelus fuscoguttatus. Aquaculture. 2019;512 [Google Scholar]
- 46.Liu Y., Kong Z., Liu J., Zhang P., Wang Q., Huan X., Li L., Qin P. Non-targeted metabolomics of quinoa seed filling period based on liquid chromatography-mass spectrometry. Food Res. Int. 2020;137 doi: 10.1016/j.foodres.2020.109743. [DOI] [PubMed] [Google Scholar]
- 47.Zhou T., Chen G., Wang Y., Chen M., Zou G., Liang H., Liang H. Tandem mass Tag-Based quantitative proteomics analysis of gonads reveals new lnsight into sexual reversal mechanism in Chinese soft-shelled turtles. Biology. 2022;11:1081. doi: 10.3390/biology11071081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw data derived from A. dabryanus samples have been deposited at National Genomics Data Center (NGDC), and accession numbers are listed in the key resources table, and are publicly available as of the date of publication.
-
•
This study did not generate original code.
-
•
Any additional information required to reanalyze the data reported in this work is available from the lead contact upon reasonable request.