Abstract
Interferon gamma (IFN-γ), which has been cloned in several mammalian species and recently in birds, plays a critical role in modulating immune system function. IFN-γ and tumor necrosis factor alpha (TNF-α) have been shown to be crucial in the pathogenesis of viral hepatitis and in the transient disappearance of hepatitis B virus (HBV) from the liver after adoptive transfer of HBV-specific cytotoxic T lymphocytes into HBV-transgenic mice. Similar studies in the natural animal hosts of related hepadnaviruses have been limited because the corresponding probes and recombinant cytokines were not available. For this reason, we initiated studies to clone and characterize cytokines from the duck, the natural host of the duck hepatitis B virus (DHBV). We describe here the cDNA cloning and initial characterization of the IFN-γ homologue of ducks (DuIFN-γ). The DuIFN-γ cDNA codes for a predicted mature protein of 145 amino acids with a molecular mass of 16.6 kDa. The precursor protein has 67% identity with the previously cloned chicken IFN-γ and 21 to 34% identity with mammalian IFN-γ. Recombinant DuIFN-γ induces the transcription of several IFN-inducible genes including IFN regulatory factor 1 and guanylate-binding protein, and it exhibits antiviral activity that protects duck cells from vesicular stomatitis virus-mediated lysis. Importantly, treatment of primary duck hepatocytes with recombinant DuIFN-γ inhibits DHBV replication in a dose-dependent fashion. Time course analysis revealed that IFN-γ treatment does not affect initial covalently closed circular DNA (cccDNA) conversion but inhibits the synthesis of progeny cccDNA by amplification.
Hepadnaviruses are a family of small, enveloped DNA viruses that include the human hepatitis B virus (HBV), the woodchuck hepatitis virus (WHV), and the duck hepatitis B virus (DHBV). Although hepadnaviruses are noncytopathic, they cause various degrees of liver inflammation in their corresponding hosts, depending on the strength and kinetics of the cellular immune response, which in turn determines the outcome of the infection (13). Previous studies in HBV-transgenic mice have shown that interferon gamma (IFN-γ) and tumor necrosis factor alpha (TNF-α), induced in the liver by adoptively transferred HBV-specific cytotoxic T lymphocytes (CTLs) or during infection with other viruses, modulate viral replication and gene expression and eventually lead to the elimination of viral gene products from the livers of transgenic mice (4, 11, 12, 14). Whether these cytokines act in concert or independently in vivo is uncertain. To investigate the role of such cytokines in viral clearance during natural infection and to define the antiviral mechanism(s) activated by each cytokine, the DHBV-infected duck could be a useful model, especially once the avian homologues of mammalian cytokines are defined.
Accordingly, Schultz et al. (26) recently cloned a type I IFN of a duck type (DuIFN) which was shown to have 51 to 53% amino acid sequence identity to serologically distinct chicken I IFNs. Recombinant type I DuIFN was shown to be a biologically active homologue of IFN alpha (IFN-α): it induced the transcription of the IFN-inducible Mx gene; it protected duck cells from the cytopathic effects of influenza virus, vesicular stomatitis virus (VSV), and Newcastle disease virus; and it inhibited DHBV replication in primary duck embryo hepatocytes (26). In continuation of those studies, we have recently shown that DuIFN-α suppresses the steady-state content of DHBV transcripts in primary duck hepatocytes and reduces their content of viral capsids containing pregenomic RNA (27a).
Several investigators have previously reported the cloning of chicken IFN-γ (8, 18, 33) and IFN-γ genes from other galliformes (19). In the present study, we took advantage of the chicken IFN-γ cDNA to clone the cDNA for IFN-γ of the duck. Comparative analysis of the IFN-γ proteins from avian and mammalian species revealed that duck IFN-γ has 67% amino acid sequence identity with IFN-γ of birds and 21 to 34% sequence identity with IFN-γ of mammals. Recombinant DuIFN-γ secreted by COS cells was able to induce several IFN-regulated genes in duck hepatocytes. It exhibited antiviral activity in duck and chicken cells against VSV. Furthermore, we demonstrated that recombinant DuIFN-γ rendered primary duck hepatocytes refractory to productive DHBV infection, indicating that in DHBV-infected duck hepatocytes IFN-γ alone can inhibit virus replication.
MATERIALS AND METHODS
Cell culture.
Duck embryo cells were prepared as described previously (26), and they were maintained in Dulbecco’s modified minimal essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS). COS cells were propagated in DMEM containing 5% FBS. D2 cells, chicken hepatoma cells that constitutively replicate DHBV, were maintained in DMEM/F12 medium supplemented with 10% FBS (6). Chicken CEC-32 cells were propagated in DMEM supplemented with 2% chicken serum and 8% FBS (17). Duck spleen cell cultures were established from spleens of white pekin ducks. Briefly, spleens were diced in Hank’s balanced salt solution (Gibco) and then pressed through a stainless steel sieve before they were filtered through a 70-μm-mesh-size cell strainer. The cell suspension was subjected to Ficoll density centrifugation, and the cells at interphase were washed three times with Hank’s balanced salt solution before they were adjusted to 107 cells per ml in RPMI-1640 (Gibco, Grand Island, N.Y.) supplemented with 5% normal duck serum, 2 mM l-glutamine, 20 mM HEPES, and 0.1 mM β-mercaptoethanol. Cells were cultured for the indicated times in the presence of various concentrations of phytohemagglutinin (PHA) (Difco, Detroit, Mich.).
cDNA library construction.
Total RNA was isolated from Ficoll-purified duck spleen cells 16 h postinduction with 5 μg of PHA per ml. cDNA generated from 1 μg of total RNA by using the SMART PCR cDNA Library Construction Kit (Clontech, Palo Alto, Calif.) was cloned into EcoRI-digested lambda ZAPII (Stratagene, La Jolla, Calif.). The resulting lambda phage library contained a total of 1.2 × 106 individual phages.
Isolation of cDNA clones.
Approximately 5 × 105 phages of the duck spleen cDNA library were subsequently screened with a 32P-labeled fragment of chicken IFN-γ cDNA (33). Hybridization was carried out in 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 6.8), 0.5% sodium dodecyl sulfate, 1× Denhardt’s solution, and 200 μg of denatured herring sperm DNA per ml at 56°C for 16 h. Several dozen positive clones were identified and converted to phagemids by using a helper phage according to the manufacturer’s instructions (Stratagene).
DNA sequence analysis.
The plasmids were sequenced (25) on an Applied Biosystems Model 373A Sequencer by using the Big Dye Terminator Cycle Sequencing Kit with AmpliTaq (Applied Biosystems).
Genomic Southern blot analysis.
Samples of duck liver DNA (20 μg) were digested with BglII, HindIII, and PvuII. Fragments were size fractionated by electrophoresis through a 1% agarose gel and transferred to nylon membranes in 0.4 N NaOH. A radiolabeled DNA fragment comprising the open reading frame (ORF) of DuIFN-γ was used as a hybridization probe. Restricted plasmid DNA comprising the cDNA of DuIFN-γ was subjected to threefold dilutions in buffer containing carrier DNA, and samples corresponding to 74 pg, 222 pg, 667 pg, and 2.0 ng of plasmid DNA were loaded into individual wells. Assuming that the complexity of the haploid duck genome is comparable to that of the chicken genome, which is 1.2 × 109 (3), these amounts of plasmid should yield hybridization signals that equal approximately 1, 3, 9, and 27 gene equivalents, respectively, in 20 μg of genomic duck DNA.
Computer analysis of predicted amino acid sequences.
The IFN-γ protein sequences of the various species except that of the dog IFN-γ sequence (7) were obtained from GenBank. Alignments were carried out by using CLUSTALW (version 1.74) with additional manual adjustment (32). Phylogenetic analysis also utilized CLUSTALW and the PHYLIP package (version 3.57) (9). A distance matrix was calculated from the aligned sequences by using the program PROTDIST incorporating the DAYHOFF-PAM model of amino acid replacement. A dendrogram based on these distances was generated by using the program FITCH. The tree shown was drawn by using NJPLOT (23). The program PROTPARS was used to derive maximum-parsimony phylogenetic trees.
RNA analysis.
Total RNA was prepared with the RNeasy RNA preparation kit (Qiagen Inc., Santa Clarita, Calif.). RNA was size fractionated by electrophoresis through a 1.2% formaldehyde agarose gel and blotted onto a nylon membrane. The membranes were sequentially hybridized with the indicated radiolabeled cDNA probes.
Production of recombinant DuIFN and chicken IFN in COS7 cells.
For expression of recombinant DuIFN-γ in COS7 cells, BglII/SspI fragments of clone 6.1.7 and 3.1.2, respectively, that contained the entire ORF of DuIFN-γ was cloned into the BamHI/EcoRV-restricted eukaryotic expression vector pcDNA3 (Invitrogen, Carlsbad, Calif.). Production of DuIFN-α and chicken IFN was described elsewhere (26, 27, 33). Transfection was performed by the calcium phosphate precipitation method (5′ Prime-3′Prime, Inc., Boulder, Colo.). At 72 h posttransfection, the culture supernatants were harvested and cleared of cell debris by centrifugation.
IFN titrations.
Duck embryo cells and chicken CEC-32 cells were seeded into 96-well microtiter plates and incubated in the presence of twofold serial dilutions of various IFN preparations. After 15 h of culture the cells were challenged with VSV (strain Indiana) at multiplicities of infection of 1 for duck cells and 0.01 for chicken cells. IFN titers were expressed as reciprocals of the dilutions that resulted in 50% protection against virus-induced cell lysis determined 24 h following infection. Supernatants of transfected COS cells expressing DuIFN-α served as a laboratory standard for titrations on duck cells. Chicken IFN (international standard 76/18) served as a standard for titrations on chicken cells.
Primary duck hepatocytes and DHBV.
Ducklings were purchased from Metzer Farms (Redlands, Calif.), and primary hepatocytes were prepared from 1- to 2-week-old ducklings by perfusion of the liver with collagenase as described previously (24). Cells were suspended in Leibowitz-15 medium (L-15; Gibco) supplemented with 5% FBS, 0.5 g of glucose per liter, 15 mM HEPES, 10−5 M dexamethasone (Sigma), 1 mg of insulin (Sigma) per liter, 5 × 104 U of penicillin per liter, 50 mg of streptomycin per liter, and 10 ml of Fungizone (Gibco) per liter. They were seeded into 60-mm-diameter dishes such that they reached confluence the following day, when the medium was replaced with L-15 medium supplemented with 1% dimethyl sulfoxide instead of FBS. DHBV used to infect 2-day-old cultures was concentrated from supernatants of D2 cells by precipitation with 10% polyethylene glycol 8000 (31).
Infection and IFN treatment of primary hepatocytes.
Hepatocyte cultures were infected 2 days after plating with an amount of DHBV derived from 10 ml of D2 cell culture medium. Recombinant DuIFN-γ produced in COS cells was added to the cultures at various concentrations starting 1 day prior to infection, on the day of infection, or 1 or 2 days thereafter. Medium and IFN were renewed every 24 h throughout the experiment. Cultures were monitored for cytotoxicity related to DuIFN-γ treatment by daily visual inspection. There was no evidence of toxicity at any concentration used in this experiment.
Isolation and analysis of viral DNA from infected hepatocytes.
Hepatocytes were analyzed for viral cccDNA and replicative intermediates according to the method described by Summers et al. (30). A hybridization standard run on each gel was used to compare the intensity of signals on different blots. Covalently closed circular DNA (cccDNA), relaxed circular DNA (rcDNA), and single-stranded DNA (ssDNA) levels of the different samples can be compared directly. Note that the signal intensities of the cccDNA samples are fivefold lower than those of the replicative intermediate DNAs.
Nucleotide sequence accession number.
The DuIFN-γ sequence determined in this study has been assigned GenBank accession no. AF087134.
RESULTS
Identification of a cDNA encoding DuIFN-γ.
A cDNA library was constructed from total RNA isolated from PHA-stimulated duck spleen cells. Screening of approximately 5 × 105 phages of the amplified library with a radiolabeled chicken IFN-γ cDNA probe yielded 27 positive clones. They were rescued to Bluescript phagemids and further analyzed by Southern blot hybridization by using radiolabeled chicken IFN-γ cDNA as a probe. Sequencing of two of these clones, designated 6.1.7 and 3.1.2, respectively, revealed that they contained a 1.4-kb cDNA insert with an ORF starting at position 119. The deduced proteins consist of 164 amino acids, the N-terminal 19 residues of which most likely constitute a signal peptide. The putative mature protein is composed of 145 amino acids with a predicted molecular mass of 16.6 kDa, which, like most of the known IFN-γ proteins, displays three N-linked glycosylation sites. The two clones revealed an almost identical sequence throughout the coding region, except for a T to C transition at position 496 and an A to G transition at position 595 in 3.1.2. The latter transition changes the arginine codon to a glycine codon in clone 3.1.2. To determine the prevalence of these amino acid residues among other clones encoding DuIFN-γ, nine more clones were sequenced. They all possessed an A at position 496, and thus their sequences resembled the sequence of 6.1.7. The nucleotide sequence of 6.1.7 and its deduced amino acid sequence are shown in Fig. 1.
FIG. 1.
Nucleotide sequence and the deduced amino acid sequence of the full-length cDNA encoding DuIFN-γ. Numbers at the left of the sequence indicate the positions of nucleotides; numbers on the right indicate amino acid positions. The predicted mature protein starts with the cysteine residue at position 20 and is marked by an arrowhead. The three potential N-linked glycosylation sites are underlined. In the 3′ noncoding region the polyadenylation signal sequence (AATAAA) is highlighted by a broken line. Positions of restriction sites used for determination of the structural organization of the IFN-γ gene are indicated.
In contrast to type I IFN genes, the IFN-γ genes of mammals and chicken contain introns. To determine the structural organization and the number of IFN-γ-related genes in duck DNA a radioactively labeled duck IFN-γ probe was hybridized to Southern blots of genomic DNA digested with various restriction endonucleases. BglII and HindIII cleave in the 5′ and 3′ noncoding region of the DuIFN-γ cDNA (Fig. 1) but are not located in the DNA fragment which was used as a probe. A unique, intronless gene would thus have appeared as a single hybridizing DNA fragment of 1.3 kb; instead, the hybridizing fragment was about 3.5 kb, indicating that the DuIFN-γ gene contains one or more introns. By comparing the intensity of this band to appropriate plasmid standards, we concluded that the duck genome most likely contains a single IFN-γ gene (data not shown).
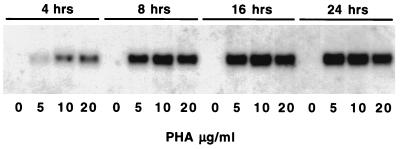
In order to confirm that this gene is expressed during mitogenic stimulation of lymphocytes, we analyzed total RNA isolated from PHA-stimulated and unstimulated spleen cells by Northern blotting. The steady-state levels of DuIFN-γ mRNA increased in PHA-stimulated spleen cells in a time- and dose-dependent fashion, being detectable as early as 4 h after induction, attaining peak levels at around 16 h, and persisting for at least 24 h (Fig. 2).
FIG. 2.
Kinetics of DuIFN-γ mRNA expression in duck spleen cells stimulated with PHA. Northern blot analysis was carried out on total RNA extracted from Ficoll-purified spleen cells that were cultured for 4, 8, 16, and 24 h in the presence of 0, 5, 10, or 20 μg of PHA per ml. RNA samples (5 μg) were fractionated on a 1.2% formaldehyde agarose gel and blotted onto a nylon membrane, and the membrane was hybridized with the DuIFN-γ cDNA probe.
Sequence homologies between avian and mammalian IFN-γ.
Comparison of the nucleotide and amino acid sequences of DuIFN-γ and chicken IFN-γ, which was used for screening the duck cDNA library, revealed 80% identity at the nucleotide level and 67% identity at the amino acid level. Overall, as expected, the amino acid sequence of the characterized duck IFN-γ showed a higher degree of identity to avian (67%) than to mammalian (21 to 34%) sequences (Fig. 3 and 4). We carried out phylogenetic analysis in order to determine the levels of evolutionary relatedness among the IFN-γ proteins of the different species by using the sequences shown in Fig. 3 and several other IFN-γ sequences obtained from GenBank. The dendrogram depicted the IFN-γ sequences as forming four separate clusters, comprising the IFN-γ sequences of birds, rodents, primates, and ungulates (Fig. 4). Maximum-parsimony analysis produced one parsimonious phylogenetic tree, which showed the same branching arrangements as the tree based on genetic distances. We were unable to place the root on this evolutionary tree because of the lack of a sequence from reptiles that might be ancestral.
FIG. 3.
Alignment of duck IFN-γ with human IFN-γ and other avian IFN-γ sequences. Sequences were obtained from GenBank and aligned by using the CLUSTALW (1.74) multiple sequence alignment program. The sequences shown are duck IFN-γ, chicken IFN-γ (X99774), turkey IFN-γ (AJ000715B), pheasant IFN-γ (AJ001289), quail IFN-γ (AJ001678), guinea fowl IFN-γ (AJ001263), and human IFN-γ (X13274). Asterisks indicate identity, colons indicate conservative substitutions, and periods indicate semiconservative substitutions. Gaps introduced to optimize the alignment are shown as dashes.
FIG. 4.
Phylogenetic tree of IFN-γ polypeptides. The dendrogram is based on distance analysis of predicted IFN-γ polypeptide sequences aligned with CLUSTALW. Sequences were obtained from GenBank. The sequences for which data are shown are duck IFN-γ, chicken IFN-γ (accession no. X99774), turkey IFN-γ (AJ000715B), pheasant IFN-γ (AJ001289), quail IFN-γ (AJ001678), guinea fowl IFN-γ (AJ001263), gerbil IFN-γ (L37782), mouse IFN-γ (K00083), rat IFN-γ (AF010466), woodchuck IFN-γ (Y14138), rabbit IFN-γ (AB010386), marmoset IFN-γ (X64659), rhesus macaque IFN-γ (L26024), mangabey IFN-γ (L26025), human IFN-γ (X13274), sheep IFN-γ (X52640), goat IFN-γ (U34232), bovine IFN-γ (Z54144), red deer IFN-γ (X63079), swine IFN-γ (X53085), cat IFN-γ (D30619), and horse IFN-γ (A11777). Dog IFN-γ is described in reference 7.
Biological activities of recombinant DuIFN-γ produced by transfected COS cells.
To determine which of the obtained clones coded for an active IFN-γ, we cloned fragments of the two clones encoding the entire ORFs into a eukaryotic expression vector. Supernatants of COS cells transfected with either of these constructs protected duck fibroblasts from lysis by VSV. However, several independent experiments revealed that supernatants from COS cells transfected with clone 3.1.2 contained a fourfold-higher antiviral titer than supernatants from COS cells transfected with clone 6.1.7 (the titers were 2,048 and 512 U per ml, respectively). Furthermore, supernatants resulting from transfection with clone 3.1.2 were also able to protect chicken cells from virus-mediated lysis, although the antiviral titer was 16-fold less than the titer on duck cells (Table 1). Supernatants from COS cells transfected with an irrelevant plasmid failed to protect duck and chicken cells. These results indicate that both clones have the potential to code for antivirally active proteins. Taken together, the results indicate that recombinant duck IFN-γ seems to be a less potent antiviral agent in this assay than duck IFN-α. Similar results were obtained with chicken IFNs that were produced in the same way and were assayed on chicken cells.
TABLE 1.
Antiviral activity of DuIFN and chicken IFN
| Species | Type of IFN | IFN titer (U/ml)a
|
|
|---|---|---|---|
| Duck fibroblasts | Chicken fibroblasts | ||
| Duck | IFN-γ 6.1.7 | 512 | ND |
| IFN-γ 3.1.2 | 2,048 | 128 | |
| IFN-α | 25,000 | 2,048 | |
| Chicken | IFN-γ | NDb | 128 |
| IFN-α | ND | 50,000 | |
Recombinant duck and chicken IFNs were assayed for their ability to inhibit the cytopathic effect of VSV in duck and chicken cells.
ND, not detectable.
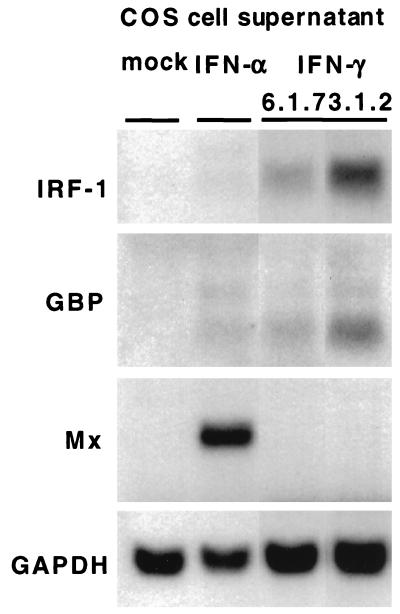
A typical feature of mammalian IFN-γ is that it induces a set of genes (e.g., the genes encoding IFN regulatory factor 1 [IRF-1] and guanylate binding protein [GBP]) that is weakly or not at all induced by type I IFN. Other genes (e.g., the Mx gene) are preferentially induced by IFN type I (2, 29). To determine whether these genes were controlled similarly in duck cells, we monitored whether their transcription was induced in response to recombinant DuIFN-γ or DuIFN-α. Primary duck hepatocytes were incubated in the presence of 1/50 dilutions of COS cell supernatants containing the IFNs for 16 h before RNA was isolated and subjected to Northern blotting. The genes encoding IRF-1 and GBP are preferentially induced by IFN-γ, while Mx gene transcription is induced by IFN-α. None of these transcripts could be detected in hepatocytes that were incubated with supernatants from COS cells transfected with an irrelevant plasmid (Fig. 5). These results indicate that recombinant duck IFNs induce the same set of genes as their mammalian equivalents and emphasize that, despite the low sequence conservation of the IFN proteins, the various components of the avian IFN system are functionally well conserved.
FIG. 5.
Induction of IFN-regulated genes by recombinant DuIFNs in primary duck hepatocytes. Cell cultures were incubated for 16 h with 1/50 dilutions of 72-h culture supernatants from COS cells transfected with the expression constructs 6.1.7 and 3.1.2, respectively, both encoding DuIFN-γ, and with similar supernatants from COS cells transfected with an expression construct encoding DuIFN-α or from mock-transfected COS cells. Samples of total RNA (20 μg) were subjected to Northern blot analysis. The blot was sequentially hybridized with radiolabeled chicken IRF-1 (16), duck Mx (1), chicken GBP (28), and chicken glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (22) cDNA probes.
Recombinant DuIFN-γ inhibits the replication of DHBV in primary duck hepatocytes.
In order to evaluate the antiviral potential of recombinant DuIFN-γ toward a productive DHBV infection in vitro, we exposed primary duck hepatocytes to various concentrations of recombinant DuIFN-γ starting 1 day before infection, on the day of infection, or on day 1 or day 2 thereafter. Treated cells were harvested 4 days after infection and analyzed for the presence of cccDNA and replicative intermediates. Untreated cells were harvested on 4 consecutive days following infection to monitor the accumulation of intracellular viral DNA. Southern blot analysis of viral DNA revealed that exposure to DuIFN-γ-inhibited the accumulation of viral intermediates and cccDNA in a dose- and time-dependent fashion (Fig. 6). Daily visual inspection of the cell cultures revealed no evidence of DuIFN-γ-related cytotoxicity. Furthermore, determination of the optical density at 260 nm of the cccDNA preparations (which are principally composed of rRNA) from IFN-treated and untreated cells revealed similar nucleic acid contents (not shown), suggesting that all samples contained similar numbers of cells. We conclude from these observations that the reduction of the amounts of various viral DNAs in IFN-treated cells is not due to a cytotoxic effect.
FIG. 6.
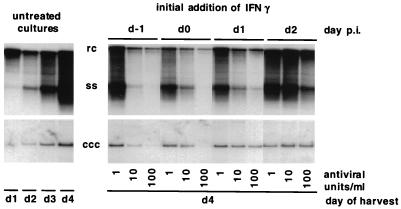
Effect of IFN-γ on the accumulation of cccDNA and replicative intermediates in primary duck hepatocytes infected with DHBV. Primary duck hepatocytes were cultured in the presence of specified concentrations of recombinant DuIFN-γ. IFN was first added either 1 day prior to infection (d−1), on the day of infection (d0), or 1 or 2 days thereafter (d1 and d2, respectively). cccDNA and replicative intermediates were isolated from IFN-treated cells 4 days postinfection (p.i.). Untreated cultures were harvested on the day indicated. Sample loaded is equivalent to one-fifth of a 60-mm-diameter plate. Replicative intermediates are shown on the upper panel, and cccDNA is shown on the lower panel. Gel migration positions of relaxed circular (rc), single-stranded (ss), and covalently closed circular (ccc) forms of viral DNA are indicated.
Viral DNA levels were maximally reduced when IFN was added to the culture medium either prior to infection or at the time of infection. In cultures treated with 100 U per ml of IFN-γ, cccDNA levels remained 10- to 20-fold lower than in the untreated culture. However, on longer exposures of the autoradiographs, cccDNA can be detected at day 4 after infection when IFN is added 1 day before infection (data not shown). The signal intensity is comparable to the cccDNA signal intensity we obtained from untreated cells harvested 1 day after infection (Fig. 6; compare lane 1 with lane 7). This suggests that initial cccDNA conversion from input virus can occur in the presence of DuIFN-γ, while the synthesis of progeny cccDNA by amplification is inhibited (Fig. 6; compare lanes 1 to 4 with lane 7). The accumulation of ssDNA and rcDNA was virtually abolished in these cultures, and this probably accounts for the lack of cccDNA amplification. When the DuIFN-γ was added to the hepatocytes after infection the accumulation of viral DNA, especially ssDNA, was retarded, suggesting that DuIFN-γ inhibited an early step in the virus replication cycle.
DISCUSSION
Recently, there has been considerable interest in the ability of cytokines to modulate viral infection, a role that has been highlighted by observations in HBV transgenic mice, in which adoptively transferred hepatitis B surface antigen-specific CTLs inhibit HBV replication by a noncytolytic process that is mediated by IFN-γ and TNF-α secreted by the CTLs after antigen recognition (11, 14). In subsequent studies it was demonstrated that HBV replication is also abolished after interleukin-12 treatment of transgenic mice due to its ability to induce IFN-γ in the liver (5). Additionally, these cytokines also inhibit hepatic HBV replication during lymphocytic choriomeningitis virus- (12), adenovirus-, and murine cytomegalovirus-induced hepatitis (4), although in these infections IFN type I also mediates the antiviral effect.
These findings prompted us to clone, sequence, and produce recombinant duck IFN-γ to permit analysis of the involvement of IFN-γ in the establishment and clearance of a natural hepadnavirus infection. In this report we describe the cloning and characterization of a duck IFN-γ cDNA. Two of the identified clones (6.1.7 and 3.1.2) appeared identical throughout the coding region except for an A to G transition that conferred an amino acid change from Arg to Gly in clone 3.1.2. Whether this nucleotide change represents an allelic difference or a reverse transcriptase or Taq polymerase error in the construction of the cDNA library remains unknown. Because additionally sequenced clones were identical to clone 6.1.7 with respect to this amino acid residue, we assume that clone 6.1.7 encoded the putative DuIFN-γ. In addition, a basic residue in this position is conserved in other avian IFN-γ proteins. Both cDNAs gave rise to biologically active proteins, indicating that this amino acid residue is not crucial for biological activity. This is consistent with the observation that after deletion of up to 11 amino acids from the carboxy terminus of human IFN-γ the protein still retains its biological activity (10).
IFNs generally have been considered to be host species specific, yet it is known that several IFN-α proteins show various degrees of cross-species activity. DuIFN-γ 3.1.2 shows a significant degree of antiviral activity on chicken cells, whereas cross-species activity could not be detected with the DuIFN-γ clone, 6.1.7, that differs from the clone 3.1.2 in just one amino acid residue. We therefore speculate that this amino acid residue might mediate species specificity.
The present results illustrate, for the first time, that IFN-γ has an inhibitory effect on hepadnavirus replication in infected cells. An antiviral effect of IFN-γ has been reported in HBV transgenic mice (4, 5, 11, 14) and in DHBV- and HBV-transfected cell lines (15, 21). However, neither of these systems allows examination of the factors involved in the establishment or clearance of hepadnavirus infection. The present experiments reveal for the first time that IFN-γ alone is capable of inhibiting DHBV infection. This inhibition is most prominent when IFN treatment is started before the establishment of infection. Our findings also provide some insight into the step in DHBV infection that is affected by IFN-γ treatment. The observation that cccDNA was readily detectable even when infection followed the initiation of treatment by 24 h indicates that DuIFN-γ inhibits DHBV replication after initial conversion of the rcDNA of the infecting virus into cccDNA but before the appearance of progeny cccDNA in the nucleus. This is in agreement with the time course of initial cccDNA conversion (20) and with recent data obtained from DHBV-infected duck hepatocytes treated with IFN-α (27a). The particular step in DHBV infection that is sensitive to IFN-γ treatment cannot be identified with certainty at this time. It appears that single-stranded replicative DNA (ssDNA) intermediates are more profoundly reduced than the more mature rcDNA and the cccDNA forms, especially in the early stages of infection. This is compatible with the notion that most of the rcDNA present early in the infection probably comes from the infecting virus rather than reflecting newly synthesized viral DNA. Thus, DuIFN-γ treatment seems to inhibit the accumulation of ssDNA-containing immature viral capsids or an earlier step in the viral replication cycle. Of course, it is also possible that DuIFN-γ might also inhibit the conversion of ssDNA to rcDNA, but this hypothesis was not examined in the present experiments.
Little is known about the specific mechanism with which IFN-γ inhibits DHBV replication. Indeed, IFN-γ could either bind directly to the hepatocyte and induce cellular genes that interfere with viral replication or it could stimulate nonparenchymal cells (e.g., macrophages) that contaminate the primary duck hepatocyte cultures, to secrete antiviral products that are responsible for the effect. Obviously, further studies are needed to clarify this important issue. Nonetheless, the efficiency with which low concentrations of IFN-γ inhibit the productive infection of hepatocytes by DHBV suggests that this cytokine could modulate the DHBV life cycle in the infected animal. The availability of these reagents now permits the role of IFN-γ in the outcome of DHBV infection to be examined both in vitro and in vivo.
ACKNOWLEDGMENTS
We thank Jesse Summers, Michael Roggendorf, and Kirsten Weining for discussion and helpful comments and Jennifer Newmann for assistance with manuscript preparation.
This research was supported by grant R37 CA40489 from the National Institutes of Health. U. Schultz was partially supported by a fellowship from the Deutsche Forschungsgemeinschaft (1152/1-1).
Footnotes
Manuscript no. 11943-MEM from The Scripps Research Institute.
REFERENCES
- 1.Bazzigher L, Schwarz A, Staeheli P. No enhanced influenza virus resistance of murine and avian cells expressing cloned duck Mx protein. Virology. 1993;195:100–112. doi: 10.1006/viro.1993.1350. [DOI] [PubMed] [Google Scholar]
- 2.Briken V, Ruffner H, Schultz U, Schwarz A, Reis L F L, Strehlow I, Decker T, Staeheli P. Interferon regulatory factor 1 is required for mouse Gbp gene activation by gamma interferon. Mol Cell Biol. 1995;15:975–982. doi: 10.1128/mcb.15.2.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burt D W, Bumstead N, Bitgood J J, Ponce de Leon F A, Crittenden L B. Chicken genome mapping: a new era in avian genetics. Trends Genet. 1995;11:190–194. doi: 10.1016/s0168-9525(00)89042-3. [DOI] [PubMed] [Google Scholar]
- 4.Cavanaugh V J, Guidotti L G, Chisari F V. Inhibition of hepatitis B virus replication during adenovirus and cytomegalovirus infections in transgenic mice. J Virol. 1998;72:2630–2637. doi: 10.1128/jvi.72.4.2630-2637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavanaugh V J, Guidotti L G, Chisari F V. Interleukin-12 inhibits hepatitis B virus replication in transgenic mice. J Virol. 1997;71:3236–3243. doi: 10.1128/jvi.71.4.3236-3243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Condreay L D, Wu T T, Aldrich C E, Delaney M A, Summers J, Seeger C, Mason W S. Replication of DHBV genomes with mutations at the sites of initiation of minus- and plus-strand DNA synthesis. Virology. 1992;188:208–216. doi: 10.1016/0042-6822(92)90751-a. [DOI] [PubMed] [Google Scholar]
- 7.Devos K, Duerinck F, Van Audenhove K, Fiers W. Cloning and expression of the canine interferon-gamma gene. J Interferon Res. 1992;12:95–102. doi: 10.1089/jir.1992.12.95. [DOI] [PubMed] [Google Scholar]
- 8.Digby M R, Lowenthal J W. Cloning and expression of the chicken interferon-gamma gene. J Interferon Cytokine Res. 1995;15:939–945. doi: 10.1089/jir.1995.15.939. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 10.Gray P W, Goeddel D V. Cloning and expression of murine immune interferon cDNA. Proc Natl Acad Sci USA. 1983;80:5842–5846. doi: 10.1073/pnas.80.19.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guidotti L G, Ando K, Hobbs M V, Ishikawa T, Runkel L, Schreiber R D, Chisari F V. Cytotoxic T lymphocytes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice. Proc Natl Acad Sci USA. 1994;91:3764–3768. doi: 10.1073/pnas.91.9.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guidotti L G, Borrow P, Hobbs M V, Matzke B, Gresser I, Oldstone M B, Chisari F V. Viral cross talk: intracellular inactivation of the hepatitis B virus during an unrelated viral infection of the liver. Proc Natl Acad Sci USA. 1996;93:4589–4594. doi: 10.1073/pnas.93.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guidotti L G, Chisari F V. To kill or to cure: options in host defense against viral infection. Curr Opin Immunol. 1996;8:478–483. doi: 10.1016/s0952-7915(96)80034-3. [DOI] [PubMed] [Google Scholar]
- 14.Guidotti L G, Ishikawa T, Hobbs M V, Matzke B, Schreiber R, Chisari F V. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity. 1996;4:25–36. doi: 10.1016/s1074-7613(00)80295-2. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi Y, Koike K. Interferon inhibits hepatitis B virus replication in a stable expression system of transfected viral DNA. J Virol. 1989;63:2936–2940. doi: 10.1128/jvi.63.7.2936-2940.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jungwirth C, Rebbert M, Ozato K, Degen H J, Schultz U, Dawid I B. Chicken interferon consensus sequence-binding protein (ICSBP) and interferon regulatory factor (IRF) 1 genes reveal evolutionary conservation in the IRF gene family. Proc Natl Acad Sci USA. 1995;92:3105–3109. doi: 10.1073/pnas.92.8.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaaden O R, Lange S, Stiburek B. Establishment and characterization of chicken embryo fibroblast clone LSCC-H32. In Vitro. 1982;18:827–834. doi: 10.1007/BF02796323. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser P, Wain H M, Rothwell L. Structure of the chicken interferon-gamma gene, and comparison to mammalian homologues. Gene. 1998;207:25–32. doi: 10.1016/s0378-1119(97)00600-8. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser P, Sonnemans D, Smith L E. Avian IFN-γ genes: sequence analysis suggests probable cross-species reactivity among galliformes. J Interferon Cytokine Res. 1998;18:711–719. doi: 10.1089/jir.1998.18.711. [DOI] [PubMed] [Google Scholar]
- 20.Kock J, Schlicht H J. Analysis of the earliest steps of hepadnavirus replication: genome repair after infectious entry into hepatocytes does not depend on viral polymerase activity. J Virol. 1993;67:4867–4874. doi: 10.1128/jvi.67.8.4867-4874.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavine J E, Ganem D. Inhibition of duck hepatitis B virus replication by interferon-gamma. J Med Virol. 1993;40:59–64. doi: 10.1002/jmv.1890400112. [DOI] [PubMed] [Google Scholar]
- 22.Panabieres F, Piechaczyk M, Rainer B, Dani C, Fort P, Riaad S, Marty L, Imbach J L, Jeanteur P, Blanchard J M. Complete nucleotide sequence of the messenger RNA coding for chicken muscle glyceraldehyde-3-phosphate dehydrogenase. Biochem Biophys Res Commun. 1984;118:767–773. doi: 10.1016/0006-291x(84)91461-x. [DOI] [PubMed] [Google Scholar]
- 23.Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 24.Pugh J C, Summers J W. Infection and uptake of duck hepatitis B virus by duck hepatocytes maintained in the presence of dimethyl sulfoxide. Virology. 1989;172:564–572. doi: 10.1016/0042-6822(89)90199-2. [DOI] [PubMed] [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1997;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz U, Kock J, Schlicht H J, Staeheli P. Recombinant duck interferon: a new reagent for studying the mode of interferon action against hepatitis B virus. Virology. 1995;212:641–649. doi: 10.1006/viro.1995.1522. [DOI] [PubMed] [Google Scholar]
- 27.Schultz U, Rinderle C, Sekellick M J, Marcus P I, Staeheli P. Recombinant chicken interferon from Escherichia coli and transfected COS cells is biologically active. Eur J Biochem. 1995;229:73–76. doi: 10.1111/j.1432-1033.1995.0073l.x. [DOI] [PubMed] [Google Scholar]
- 27a.Schultz, U. Submitted for publication.
- 28.Schwemmle M, Kaspers B, Irion A, Staeheli P, Schultz U. Chicken guanylate-binding protein. Conservation of GTPase activity and induction by cytokines. J Biol Chem. 1996;271:10304–10308. doi: 10.1074/jbc.271.17.10304. [DOI] [PubMed] [Google Scholar]
- 29.Staeheli P. Interferon-induced proteins and the antiviral state. Adv Virus Res. 1990;38:147–200. doi: 10.1016/s0065-3527(08)60862-3. [DOI] [PubMed] [Google Scholar]
- 30.Summers J, Smith P M, Horwich A L. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J Virol. 1990;64:2819–2824. doi: 10.1128/jvi.64.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Summers J, Smith P M, Huang M J, Yu M S. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weining K C, Schultz U, Munster U, Kaspers B, Staeheli P. Biological properties of recombinant chicken interferon-gamma. Eur J Immunol. 1996;26:2440–2447. doi: 10.1002/eji.1830261026. [DOI] [PubMed] [Google Scholar]