Abstract

[FeFe] hydrogenases, metalloenzymes catalyzing proton/dihydrogen interconversion, have attracted intense attention due to their remarkable catalytic properties and (bio-)technological potential for a future hydrogen economy. In order to unravel the factors enabling their efficient catalysis, both their unique organometallic cofactors and protein structural features, i.e., “outer-coordination sphere” effects have been intensively studied. These structurally diverse enzymes are divided into distinct phylogenetic groups, denoted as Group A–D. Prototypical Group A hydrogenases display high turnover rates (104–105 s–1). Conversely, the sole characterized Group D representative, Thermoanaerobacter mathranii HydS (TamHydS), shows relatively low catalytic activity (specific activity 10–1 μmol H2 mg–1 min–1) and has been proposed to serve a H2-sensory function. The various groups of [FeFe] hydrogenase share the same catalytic cofactor, the H-cluster, and the structural factors causing the diverging reactivities of Group A and D remain to be elucidated. In the case of the highly active Group A enzymes, a well-defined proton transfer pathway (PTP) has been identified, which shuttles H+ between the enzyme surface and the active site. In Group D hydrogenases, this conserved pathway is absent. Here, we report on the identification of highly conserved amino acid residues in Group D hydrogenases that constitute a possible alternative PTP. We varied two proposed key amino acid residues of this pathway (E252 and E289, TamHydS numbering) via site-directed mutagenesis and analyzed the resulting variants via biochemical and spectroscopic methods. All variants displayed significantly decreased H2-evolution and -oxidation activities. Additionally, the variants showed two redox states that were not characterized previously. These findings provide initial evidence that these amino acid residues are central to the putative PTP of Group D [FeFe] hydrogenase. Since the identified residues are highly conserved in Group D exclusively, our results support the notion that the PTP is not universal for different phylogenetic groups in [FeFe] hydrogenases.
Keywords: hydrogenases, hydrogen, substrate transport, redox catalysis, bioinorganic chemistry, enzyme catalysis
Introduction
[FeFe] hydrogenases are metalloenzymes that catalyze the reversible interconversion of electrons and protons to dihydrogen (H2), with turnover frequencies up to 105 s–1.1 Due to their remarkable catalytic properties, these enzymes have been extensively studied and they inspire the design of new catalysts for H2 production and activation.1a,2 The reaction occurs at an iron–sulfur cofactor denoted the H-cluster, which consists of a [4Fe–4S] cluster connected via a bridging cysteine thiol to a di-iron ([2Fe]H) subsite. The Fe ions of the di-iron site, denoted as proximal (Fep) and distal (Fed) based on their location relative to the [4Fe–4S] cluster, are coordinated by CO and CN– ligands, as well as a bridging azadithiolate ligand (−SCH2NCH2S–, ADT). The catalytic efficiency of the H-cluster is partly attributable to the asymmetric ligation of the [2Fe]H subsite, which modulates the electronic structure across the metals and the ligands.3 Still, the exact nature of the [2Fe]H subsite ligand-set is not crucial for reversible catalysis,4 and through the combined efforts of bio- and synthetic chemistry, it has become evident that the activity of the H-cluster is largely governed by factors beyond the primary coordination sphere.5 A striking example of the latter is provided by the amine group of the ADT ligand, which is critical for the shuttling of protons to the apical open coordination site at Fed (indicated by * in Figure 1), the site of H2 catalysis. The ADT-amine also serves as a base during H2 oxidation and can be regarded as forming a frustrated Lewis pair (FLP) with the Fed ion enabling heterolytic H2 cleavage. The role of the protein environment has also become evident through studies of semisynthetic [FeFe] hydrogenases. Indeed, incorporation of otherwise inactive synthetic di-iron complexes into the hydrogenase active-site pocket has been shown to dramatically alter their reactivity.4,6 Analogously, bioinspired design of synthetic catalysts, incorporating noncoordinating proton relays or embedding catalysts into larger polymer frameworks, has been shown to significantly enhance their catalytic properties.7 Elucidating and optimizing these second and outer coordination sphere effects is arguably the key to understanding hydrogenases and the development of the next generation of synthetic systems for hydrogen catalysis.8
Figure 1.

Overview of the proposed proton transfer pathway (PTP) in Group D [FeFe] hydrogenase TamHydS (green) based on the YASARA-generated homology model9 versus the PTP of Group A representative, CpI (gray; pdb: 4XDC).10 The iron in the H-cluster is colored in orange and the ADT bridgehead in blue. The apical open coordination site at the distal iron (Fed) is indicated by an *. Residues belonging to the PTP of CpI are presented as sticks in gray with water molecules (red spheres) in between C299 and E279; while that of TamHydS are presented in green. In this study, the amino acids E252 and E289 are exchanged conservatively (E252D, E289D) and nonconservatively (E252V, E289A). The distance between ADT and E252 as well as between the side chains that are part of the proposed PTP of TamHydS is numbered (dark green). The distances between Fed to E252 and ADT to E289 are indicated in gray dashed lines and labels. The yellow dashed lines and numbers indicate the distances between neighboring positions of the PTP of CpI.11 The TamHydS model does not include intraprotein waters which can likely compensate for the long distances between E289, S249, and D265.
[FeFe] hydrogenases are a diverse enzyme family, reflecting their broad range of functions related to hydrogen metabolism. Various classification schemes exist, but four phylogenetically distinct groups have been identified to date and denoted Group A–D.1b,12 The “prototypical” Group A [FeFe] hydrogenases are by far the most studied. Representative examples, including e.g., Chlamydomonas reinhardtii HydA1 (CrHydA1),13Desulfovibrio desulfuricans HydAB (DdHydAB),14 and Clostridium pasteurianum and C. acetobutylicum hydrogenase I (CpI and CaI),15 are the most active [FeFe] hydrogenases identified to date. While many Group A and B [FeFe] hydrogenases are involved in H2 production, the less studied Group C enzymes are considered sensory due to the presence of a signal-transducing Per-Arnt-Sim (PAS) domain.16 Finally, the Group D [FeFe] hydrogenases are most closely related to Group C but lack the PAS domain.12a,16a Nevertheless, with the recent characterization of Thermoanaerobacter mathranii (TamHydS) as a representative, Group D enzymes are also classified as putatively sensory.9
Data on Group C and D [FeFe] hydrogenases remain limited, but the representative examples characterized to date display properties clearly distinct from previously studied “prototypical” Group A enzymes.9,16b The primary model systems for these nonprototypical [FeFe] hydrogenases include the Group C enzyme from Thermotoga maritima (TmHydS)16b,17 and the aforementioned Group D enzyme TamHydS. In particular, the reactivity of TamHydS diverges from that of other [FeFe] hydrogenases with its low H2 evolution rate and a lower Km for H2 (Km 90–100 μM) than Group A and Group C enzymes (Km > 400 μM).9,16b,18 Moreover, TamHydS displays an unusual property of being a bidirectional catalyst, but with significant overpotential requirements for both H2 oxidation and proton reduction.9,19
As all [FeFe] hydrogenases share the same H-cluster, the varied reactivity of the enzymes must be due to changes in the surrounding protein environment.12a,20 In Group A enzymes, most modifications of the conserved amino acids surrounding the active site result in loss of function and in many cases also an inability to form the H-cluster.21 However, the phylogenetic groups vary in terms of their active-site pockets, which is expected to modulate the hydrogen bonding network and electrostatics around the H-cluster. The [FeFe] hydrogenases of different phylogenetic groups also differ in their electron and substrate (H+) transport pathways.9,16b [FeFe] hydrogenases commonly feature additional FeS clusters (“F-clusters”) responsible for electron transfer during catalysis. These accessory cofactors have been shown to tune the reactivity of the H-cluster.22 However, they do not seem to be the key determinant of group-specific reactivity, as the FeS cluster content and location vary both across and within phylogenetic groups.12a In addition to rapid electron transfer, the activity of [FeFe] hydrogenase is dependent on efficient proton transfer via PTPs. Alterations of the proton transfer kinetics have been shown to change the catalytic properties in Group A enzymes11,23 and could potentially explain the activity variations between different groups.
Examples of enzymes where phylogenetic groups feature different PTPs have been previously reported. The O2-processing heme–copper oxidase (HCuO) superfamily represents one of the most well-examined examples. The variations in PTPs observed across HCuO groups have been shown to correlate with differences in biochemical properties such as O2-affinity and proton pumping stoichiometry.24 These observations support the notion that the distinct reactivity of various phylogenetic groups of hydrogenases can be partly attributable to variations of their PTPs. Still, the nature and influence of different PTPs remain an open question.
The PTP(s) of [NiFe] hydrogenases remain to be firmly established.25 In the case of [FeFe] hydrogenase, the PTP of Group A enzymes has been studied extensively in recent years and shown to be composed of conserved amino acid residues and water molecules forming a hydrogen bonding network that connects the enzyme surface to the amine of the ADT ligand of the H-cluster (Figure 1).11,23b,26 The first amino acid of this PTP, C299 (CpI numbering), is located within the H-bonding distance to the amine of the ADT ligand. Thus, in addition to proton transfer, C299CpI arguably influences the positioning of the amine base for efficient heterolytic H2 cleavage. Despite its seemingly critical importance for function, it is not conserved in Group C and Group D [FeFe] hydrogenases.9,16b,17 Instead, C299CpI is exchanged mostly into a hydrophobic alanine (e.g., Group C A131TmHydS, Group D A137TamHydS). Restoring the proton transfer pathway (PTP) of Group A by exchanging A131TmHydS into a cysteine was attempted, but the variant lowered the capacity of Group C TmHydS to oxidize and evolve H2.17 However, beyond the aforementioned cysteine, the other residues crucial for proton transfer in Group A are also mostly exchanged into nonprotonatable hydrophobic side chains in TmHydS. Similarly, in Group D TamHydS, except for a conserved glutamate residue (E282CpI, E122TamHydS) and a conservative exchange of an arginine into a lysine (R286CpI, K126TamHydS), the other residues for proton transfer in Group A are missing (Figure 1). Since the buried H-cluster requires protons to be delivered to and from the bulk solvent to be active, this raises the question of alternative pathways, especially in sensory (HydS) [FeFe] hydrogenases.
In this study, we examined the PTP of the putatively sensory Group D [FeFe] hydrogenase TamHydS. Key amino acids with the highest potential to be part of the PTP were identified via homology modeling and alignment. Furthermore, our phylogenetic analysis of multiple Group D sequences showed that the identified amino acids are highly conserved in Group D [FeFe] hydrogenases (Figure 2). Site-directed mutagenesis of TamHydS was used to target two proposed key amino acids (TamHydS numbering): (a) E252, the residue with the highest potential to H bond with ADT according to distance and (b) E289, the most probable proton transferring neighbor to E252 (Figure 1). These two residues were individually substituted nonconservatively into a valine (E252V), alanine (E289A), and conservatively into an aspartate (E252D and E289D). Due to their proximity to the H-cluster, the selected amino acids also provide a probe for exploring how the outer-coordination sphere can alter the electronic distribution and geometry of the [2Fe]H subsite. The catalytic rates of the variants were determined through biochemical assays and were shown to decrease relative to the wild-type enzyme in all variants. In parallel, attenuated total reflectance Fourier-transform infrared (ATR-FTIR) and electron paramagnetic resonance (EPR) spectroscopy were employed to probe the H-cluster. Our results demonstrate that variation of residues E252 and E289 causes accumulation of two distinct H-cluster states that were previously not characterized and that these residues play a crucial role for H2 catalysis in TamHydS. The identified amino acids are likely to constitute the PTP in Group D and underscore that the proton transfer pathway is not universal in [FeFe] hydrogenases.
Figure 2.

Normalized consensus logos of Groups A–D [FeFe] hydrogenase generated in Jalview using ClustalΩ sequence alignment29 of sequences retrieved from Greening et al.30 The numbering is based on the sequence of TamHydS. The larger the font of the amino acid, the more conserved it is in the specified sequence position.
Materials and Methods
General
All chemicals were purchased from Sigma-Aldrich or VWR. Protein expression was analyzed by SDS-PAGE. All anaerobic work was performed in an MBRAUN glovebox ([O2] < 10 ppm). The synthetic cofactor [Fe2(μ-ADT)(CO)4(CN)2](Et4N)2 ([2Fe]ADT) was synthesized following literature protocols with minor modifications and verified by FTIR spectroscopy.27 UV–vis spectra were obtained using an AvaSpec-ULS2048-USB2-UA-50: Avantes Fiber Optic UV/VIS/NIR.
In Silico Work and Sequence Alignment Analysis
The YASARA-generated homology model9 of Group D TamHydS was aligned with the crystal structure of Group A representative, CpI (pdb: 4XDC),10 using the MUSTANG28 multiple structural alignment algorithm in YASARA (Figure S1). Amino acid residues that could be part of the PTP were screened by checking all residues with H-bonding capacity around 5 Å, starting from the ADT of the H-cluster to the surface of the protein. From S249, the cut-off was extended up to 5.3 Å since there were no protonatable, polar, or electrically charged side chain within 5 Å. Amino acid sequence comparison of Group A–D in Figure 2 was based on a ClustalΩ sequence alignment29 of sequences retrieved from Greening et al.30 Normalized consensus logos of [FeFe] hydrogenase Groups A–D were generated in Jalview using the sequence alignment. The interatomic distances between the distal iron (Fed) and the nitrogen of the ADT bridgehead were estimated through PyMOL,31 analyzing the following crystal structures, with pdb codes: 4XDC, 6N59, 6N6P, 6NAC, 3C8Y for CpI and 6GLY, and 6GLZ for C299A and C299D, respectively. Alphafold modeling of TamHydS (RMSD = 3.692 Å vs CpI) revealed an overall similar structure albeit with differences in estimated distances between the proposed PTP residues (1.8 ± 0.7 Å, Figure S2).
Site-Directed Mutagenesis
The gene encoding for TamHydS was synthesized with a C-terminal Strep-TagII sequence and cloned in pET-11a(+) by Genscript using restriction sites NdeI and BamHI following codon optimization for expression in Escherichia coli. Parental plasmid harboring the wild-type gene was amplified by PCR with Phusion High-Fidelity to generate site-directed mutagenesis variants. Mutagenic primers were synthesized and purified by Eurofins Genomics (Ebersberg, Germany) for each of the mutants, E252V: forward 5′-GGCGTTGCGTTCGGTACCTTTAC-3′, reverse 5′-GAACGCAACGCCACCGCTG-3′; E252D: forward 5′-GGCGATGCGTTCGGTACCTTTAC-3′, reverse 5′-GAACGCATCGCCACCGCTG-3′; E289A: forward 5′-GATTTCTTTGCGGGCCTGGCGTG-3′, reverse 5′-GGCCCGCAAAGAAATCCAGGTCG-3′; E289D: forward 5′-GATTTCTTTGATGGCCTGGCGTG-3′, reverse 5′-GGCCATCAAAGAAATCCAGGTCG-3′; The PCR product was then digested with DpnI restriction enzyme. Gene integrity was verified via sequencing by Eurofins Genomics.
Generation of [2Fe]ADT-Activated TamHydS Variants
Protein purification, reconstitution of [4Fe–4S] clusters, and activation with [2Fe]ADT to generate the holo-forms of the variants were performed as previously described with minor modifications.9 In short, the expression constructs with verified sequences were transformed in chemically competent E. coli BL21(DE3) or BL21(DE3)ΔiscR32 cells (Table S1) to express the apo-forms of TamHydS lacking the di-iron subsite of the H-cluster (Figure S3). The proteins were purified via StrepTrap chromatography (StrepTrap HP (GE Healthcare) affinity column), and prior to the elution step, the column was washed with a sub-denaturing concentration of 1 M urea in 100 mM Tris, 150 mM NaCl pH 8.0.33 Comparing preparations of wild-type TamHydS (TamHydS WT) showed that the urea wash step did not change the specific activities, Fe/protein content, and spectroscopic properties as compared to preparations isolated in strict absence of urea. The yields following purification by StrepTrap chromatography varied from approximately 1–7 mg L–1 of cell culture depending on the variant. Subsequent operations after cell growth and induction were carried out under anaerobic conditions in the glovebox to prevent hydrogenase inactivation by atmospheric oxygen. The [4Fe–4S] clusters were reconstituted semienzymatically with cysteine desulfurase (E. coli IscS) (Figure S4). To generate the holo-forms of the variants for in vitro H2 evolution/oxidation assays, ATR-FTIR, and EPR analyses, the enzymes were mixed with [2Fe]ADT. After incubating the reaction mixture for 2 h, the mixture was desalted with 10 mM Tris–HCl, 2 mM sodium dithionite pH 8.0. The activated variants were then transferred into airtight serum vials before they were flash-frozen in liquid N2 and stored at −80 °C until further use.
H2-Evolution Assays
The fully reconstituted holo-enzyme was diluted to 1 μM in 180 μL phosphate buffer (100 mM, pH 6.8) in an 800 μL crimp top vial and sealed with a natural rubber/clear PTFE septum preassembled in an aluminum cap (8 mm). The reaction was initiated with the rapid addition of 10 mM methyl viologen(MV) as the electron mediator, and 100 mM sodium dithionite as the reducing agent/sacrificial electron donor.34 The mixture was incubated at 25 °C and 120 rpm. After 15 min, a 100 μL aliquot from the headspace (total volume of 620 μL) was injected into a PerkinElmer Clarus 500 gas chromatograph (GC) equipped with a thermal conductivity detector and a stainless-steel column packed with Molecular Sieve (60/80 mesh). The operational temperatures of the injection port, the oven, and the detector were 100, 80, and 100 °C, respectively. Argon was used as the carrier gas at a flow rate of 35 mL min–1. To determine the amount of H2 associated with the peak area at around 0.4 min in the chromatogram, a calibration curve made up of standard points from 0 to 60% H2 in the 620 μL headspace was generated. One unit (U) of activity catalyzes 1 μmol of H2 evolved per min under the indicated assay conditions, whereas specific activity is U per mg of the enzyme.
H2-Oxidation Assays
Benzyl viologen (BV) was used as the redox mediator. The absorbance of the mixture containing <30 μg of holo-enzyme and 1 mM BV in H2-saturated 100 mM phosphate buffer pH 6.8 was measured at 550 nm in 1 mL plastic cuvettes. A standard curve was generated to determine the molar extinction coefficient of reduced BV (ε550red = 9.12 mM–1 cm–1). The BV specific activity (U/mg) was measured by the initial rate of change of absorbance at 550 nm where one unit (U) of activity catalyzes 2 μmol of BV reduced per min (1 μmol of H2 oxidized per min) under the indicated assay conditions.
Attenuated Total Reflectance Fourier-Transform Infrared (ATR-FTIR) Spectroscopy
A solution of 1 μL enzyme (0.2–1 mM TamHydS variants) in 10 mM Tris buffer (pH 8.0) was deposited on the ATR crystal in the anaerobic atmosphere of a Braun Glove box. The ATR unit (BioRadII from Harrick) was sealed with a custom build PEEK cell that allowed for gas exchange and illumination (similar to Stripp35 and Senger et al.36) mounted in an FTIR spectrometer (Vertex V70v, Bruker). The sample was dried under 100% nitrogen gas and rehydrated with a humidified aerosol (100 mM Tris–HCl, pH 8.0) as described before.37 Spectra were recorded with 2 cm–1 resolution, a scanner velocity of 80 Hz, and averaged of varying number of scans (mostly 1000 Scans). All measurements were performed at ambient conditions (room temperature and pressure, hydrated enzyme films). Gases (N2, H2, CO) were applied at a flow rate of 0.5–1.5 L/min. D2O exchange was performed as reported previously.35 Selected samples were mixed 1:1 with a 20 mM sodium dithionite (NaDT) solution, resulting in at least 10-fold excess of NaDT (Figure S5). The data were analyzed and plotted to our protocols described previously.38
EPR Spectroscopy
X-band EPR measurements were performed on a Bruker ELEXYS E500 spectrometer equipped with a SuperX EPR049 microwave bridge and a cylindrical TE011 ER 4122SHQE cavity in connection with an Oxford Instruments continuous flow cryostat. Measuring temperatures were achieved using liquid helium flow through an ITC 503 temperature controller (Oxford Instruments). The Xepr software package (Bruker) was used for data acquisition and processing. EasySpin software version easyspin-6.0.0-dev.51 was used for spectral simulation and fitting.39 EPR samples of E252V and E289D were prepared in 100 mM Tris–HCl, pH 8.0 under a neat argon atmosphere and either directly flash-frozen (“as prepared”) or flushed with H2 or CO gas inside the EPR tube for an hour prior to freezing (“H2- or CO- flushed”). EPR samples incubated with D2 were prepared in the absence (4 μL of 1 mM enzyme in 10 mM Tris–HCl, pH 8.0 + 76 μL 100 mM Tris–HCl, pH 8.0) or presence of 95% D2O (4 μL of 1 mM enzyme in 10 mM Tris–HCl, pH 8.0 + 76 μL D2O; Figure S6). For samples that were reduced with NaDT, 1 μL of 100 mM stock solution of NaDT was added into 79 μL of 50 μM enzyme, resulting in at least 20-fold molar excess of NaDT (Figure S7).
Results
In Silico Analysis
To identify the amino acids that could constitute the PTP of Group D TamHydS, the YASARA-generated homology model9 of TamHydS was aligned with the crystallographic structure of Group A representative CpI (pdb: 4XDC),10 with an RMSD of 1.541 Å (Figure S1). Amino acid residues potentially involved in proton transfer were identified by screening all residues with H-bonding capacity around a 5 Å distance, starting from the ADT of the H-cluster and progressing to the surface of the enzyme. The E252 residue (TamHydS numbering unless otherwise stated) is the sole candidate for proton transfer to the H-cluster with a distance of 3.4 Å to the ADT moiety (Figure 1). As the closest protonatable residue to E252, we identified E289 (3.5 Å). From E289, the next residue identified was S249 (4.1 Å to E289). From S249, the cut-off was extended up to 5.3 Å since there were no protonatable side chains within 5 Å, resulting in D265 as the last proton transfer relay closest to the enzyme surface (5.2 Å to S249). Starting from the H-cluster, the proposed PTP of TamHydS thus consists of E252 → E289 → S249 → D265. The distances between E289, S249, and D265 are slightly longer than the expected H-bonding distance. However, the model does not include intraprotein waters which can likely compensate for and modulate the elongated distances between E289, S249, and D265, as seen in structures of the PTP in Group A [FeFe] hydrogenases.11,40
To support the PTP proposed from the structural analysis, sequences from Group D were aligned to generate a consensus sequence, which defines the most common amino acid residues at positions 252, 289, 249, and 265 of TamHydS. The alignment of multiple Group D member sequences shows that the amino acids most proximal to the H-cluster, E252 and E289, are highly conserved (Figure 2). Similarly, the final residue of the proposed pathway (D265) is also well-conserved, while the intermediate residue (S249) is moderately conserved, showing a strong preference for residues with hydroxy groups (i.e. serine and threonine). These amino acids are not conserved in Group A, B, or C, with the exception of the glutamate at position 289. Thus, our sequence alignment analysis strongly indicates that the potential PTP identified here is exclusive for Group D [FeFe] hydrogenases.
Preparation of Variants and Their Catalytic Properties
TamHydS amino acids E252 and E289 were exchanged conservatively to aspartic acid (E252D, E289D) and nonconservatively to valine or alanine (E252V, E289A) through site-directed mutagenesis to assess their role in H2-processing activity rates. Valine was selected for position 252 as this is found in the corresponding position of prototypical Group A hydrogenases (Figure 2), while alanine was selected for position 289 as a classical “loss-of-function” variation. The variants were expressed heterologously in E. coli and anaerobically purified to produce the apo-variants that lack the di-iron subsite (Figure S3 for gels). After reconstituting the four [4Fe–4S] clusters (Figure S4 for UV–vis spectra of the reconstituted enzyme, Table S1 for iron quantification before and after reconstitution), the apo-variants were mixed with the synthetic [2Fe]H subsite precursor [Fe2(μ-ADT)(CO)4(CN)2](Et4N)2 ([2Fe]ADT) to generate the corresponding holo-variants. Although no reference is available of TamHydS isolated from its native host, this is expected to yield biologically relevant samples considering the well-conserved nature of the H-cluster maturation machinery (HydEFG).9
The effects of TamHydS
variants E252V, E252D, E289A, and E289D on the rates for reduction
of H+ (H2 evolution) and oxidation of H2 were studied in solution assays, each performed in pH 6.8
and at 25 °C ( = −403 mV vs SHE). Methyl viologen
(MV, E° = −446 mV vs SHE) and benzyl
viologen (BV, E° = −359 mV vs SHE) were
employed as the electron donor and acceptor, respectively. All the
variants negatively impacted the H+/H2-interconversion
activities compared to the wild-type TamHydS enzyme
(TamHydS WT) (Figure 3). H2-oxidation was the most severely affected,
with E252V/D and E289A/D displaying <0.01 and ≤3% residual
activities, respectively. In terms of H2-evolution, the
nonconservative (E252V) and conservative (E252D) substitutions are
the least functional variants with specific activities that range
from 10 to 20% with respect to TamHydS WT. On the
other hand, the conservative exchange of E289 to glutamate had the
least effect (E289D retaining 70% residual activity). This is followed
by the nonconservative exchange to an alanine (E289A) at 30% residual
activity. In general, the substitution of E252 had the greater impact
in both catalytic directions, most likely since it is closer to the
ADT ligand than E289. A similar decrease in activity has been reported
for variants of Group A hydrogenases where amino acids in their PTP
were varied.11,41
= −403 mV vs SHE). Methyl viologen
(MV, E° = −446 mV vs SHE) and benzyl
viologen (BV, E° = −359 mV vs SHE) were
employed as the electron donor and acceptor, respectively. All the
variants negatively impacted the H+/H2-interconversion
activities compared to the wild-type TamHydS enzyme
(TamHydS WT) (Figure 3). H2-oxidation was the most severely affected,
with E252V/D and E289A/D displaying <0.01 and ≤3% residual
activities, respectively. In terms of H2-evolution, the
nonconservative (E252V) and conservative (E252D) substitutions are
the least functional variants with specific activities that range
from 10 to 20% with respect to TamHydS WT. On the
other hand, the conservative exchange of E289 to glutamate had the
least effect (E289D retaining 70% residual activity). This is followed
by the nonconservative exchange to an alanine (E289A) at 30% residual
activity. In general, the substitution of E252 had the greater impact
in both catalytic directions, most likely since it is closer to the
ADT ligand than E289. A similar decrease in activity has been reported
for variants of Group A hydrogenases where amino acids in their PTP
were varied.11,41
Figure 3.

Comparison of the specific H2-oxidation and -evolution activities of TamHydS variants quantified using benzyl viologen (BV, E° = −359 mV vs SHE) and methyl viologen (MV, E° = −446 mV vs SHE) as external redox partners versus the wild-type TamHydS enzyme (WT) (note the break of the Y-axis). All reactions were performed in 100 mM potassium phosphate buffer pH 6.8. H2 oxidation (yellow bars) was measured by reducing 1 mM BV at 550 nm in H2-saturated buffer. The amount of H2 produced from 10 mM MV + 100 mM sodium dithionite was measured by gas chromatography (green bars).34 Specific activity (U/mg) is one unit (U) of activity that catalyze 1 μmol of H2 oxidized/evolved per min under the indicated assay conditions over the amount of enzyme in the reaction mixture (mg). The absolute values of the specific activities are tabulated in Table S2. Error bars indicate standard error, with n = 6 (three technical repeats each for two biological samples).
In addition to the change in overall activity, it is also noteworthy that the variants display a distinct change in apparent catalytic bias. Under the employed solution assay conditions, the TamHydS WT displays significantly higher rates for H2 oxidation than H+ reduction. However, this bias is suppressed in all studied variants and even switched to an apparent bias toward H+ reduction in three cases (variants E252V/D and E289D).
Spectroscopic Properties
FTIR Spectroscopy
The effect of each amino acid variation on the H-cluster was examined using ATR-FTIR spectroscopy. Enzyme solutions were dried on the ATR crystal surface and rehydrated under a humidified N2 aerosol as reported previously.37TamHydS WT accumulates the oxidized state (Hox) under a nitrogen atmosphere (Figure 4 top black spectrum) and the reduced state (Hred) under a reducing hydrogen atmosphere (Figure 4 top magenta spectrum).9 No distinct features attributable to either of these two states were observed in any of the variants studied here. Instead, the variants showed two FTIR signatures indicative of redox states previously not identified in prototypical [FeFe] hydrogenase (Figure 4 bottom spectra and Figure S8). The states are denoted as State 1 with bands at 2120, 2098, 2016, 1999, and 1852 cm–1 (blue bars Figure 4), and State 2 shifted to lower energies (red-shifted) with bands at 2082, 2071, 1959, 1948, and 1771 cm–1 (red bars Figure 4). We note additional small peaks (indicated by *) from minor H-cluster species that we were unable to assign completely. Under an N2 atmosphere, the nonconservative exchanges of E252 and E289 appeared to stabilize specific states. E252V was found to predominantly yield State 2, while E289A was found to be predominantly in State 1 (Figure 4 bottom black spectra). Meanwhile, both the conservative exchanges of E252D and E289D showed a mixture of State 1 and State 2, with a slight preference for the latter state.
Figure 4.

FTIR spectra of wild-type TamHydS and variants exposed to N2 and H2. For TamHydS wild-type (WT) and each variant, the FTIR spectra under N2 (black spectra) and H2 atmosphere (magenta and red spectra, respectively) are shown. TamHydS WT accumulates the oxidized state (Hox) under a nitrogen atmosphere (top black spectra) and the reduced state (Hred) under a reducing hydrogen atmosphere (top magenta spectrum) (data shown taken from ref (9).). In the TamHydS variants, two IR signatures indicative of different redox states are observed. State 1 with bands at 2120, 2098, 2016, 1999, and 1852 cm–1 (indicated with blue vertical bars) and State 2 shifted to lower energies (red-shifted) with bands at 2082, 2071, 1959, 1948, and 1771 cm–1 (indicated with red vertical bars). Upon H2 exposure (red spectra), State 2 is populated in varying fractions. The E252 variants show only diminutive changes (see also Figure S9). The redox state distribution in the E289 variants clearly shifts to State 2 upon H2 exposure. Peaks of unassigned H-cluster species are labeled with an *.
Exposure to a H2 (1 atm) atmosphere led to a clear shift in the relative redox state population in favor of State 2 in the E289 variants. The E252 variants displayed only a minor (E252V) or no (E252D) shift of the redox state equilibrium toward State 2 (Figure 4 bottom red spectra). The stronger response to H2 exposure of the E289 variants correlates with their observed H2 oxidation capability, compared to the undetectable H2 oxidation activities of the E252 variants (Figure 3). Notably, while TamHydS WT has a low CO affinity,9 none of the variants bind external CO to form the inhibited “Hox-CO” state (data not shown). Instead, the exposure to CO led to a slow conversion of the enzyme population from State 2 to State 1 (representative spectra for E252V are shown in Figure S9C,D). Reduction using sodium dithionite caused a shift toward State 2 (Figure S5).
The observed red-shift of cofactor ligand bands upon H2 exposure (approximately Δ30 cm–1 for CN and Δ50–80 cm–1 for CO ligands) indicates a reduction at the di-iron site when compared to similar shifts observed for Group A [FeFe] hydrogenases.42 For TamHydS WT, the CN band intensities for Hox, Hred (Figure 4 top spectra) and Hox-CO states are more or less even.9 In contrast, for both State 1 and State 2, the area of the lower wavenumber CN band (2098 and 2071 cm–1 for State 1 and State 2, respectively) exceeds the area of the higher wavenumber CN bands (2120 and 2082 cm–1) greatly, suggestive of a difference in the hydrogen bonding strength of the CN ligands and/or an alteration in cofactor geometry or ligand binding.43 Moreover, for State 2 the relative band area of the two terminal CO ligands (1959 and 1948 cm–1) are inverted (larger band area for the higher wavenumber band) when compared to all redox states known for TamHydS WT and State 1 (2016 and 1999 cm–1) hinting in a similar direction (Figure 4). Another peculiarity of the band pattern of State 2 is the large distance between the terminal CO ligand bands (1959, 1948 cm–1) and the band assigned to μCO (1771 cm–1). Although State 2 accumulates under reducing conditions, H/D exchange revealed no specific shifts of cofactor ligand bands for State 1 or State 2 indicative of e.g., (terminal) hydride binding (Figure S5).41,44
EPR Spectroscopy
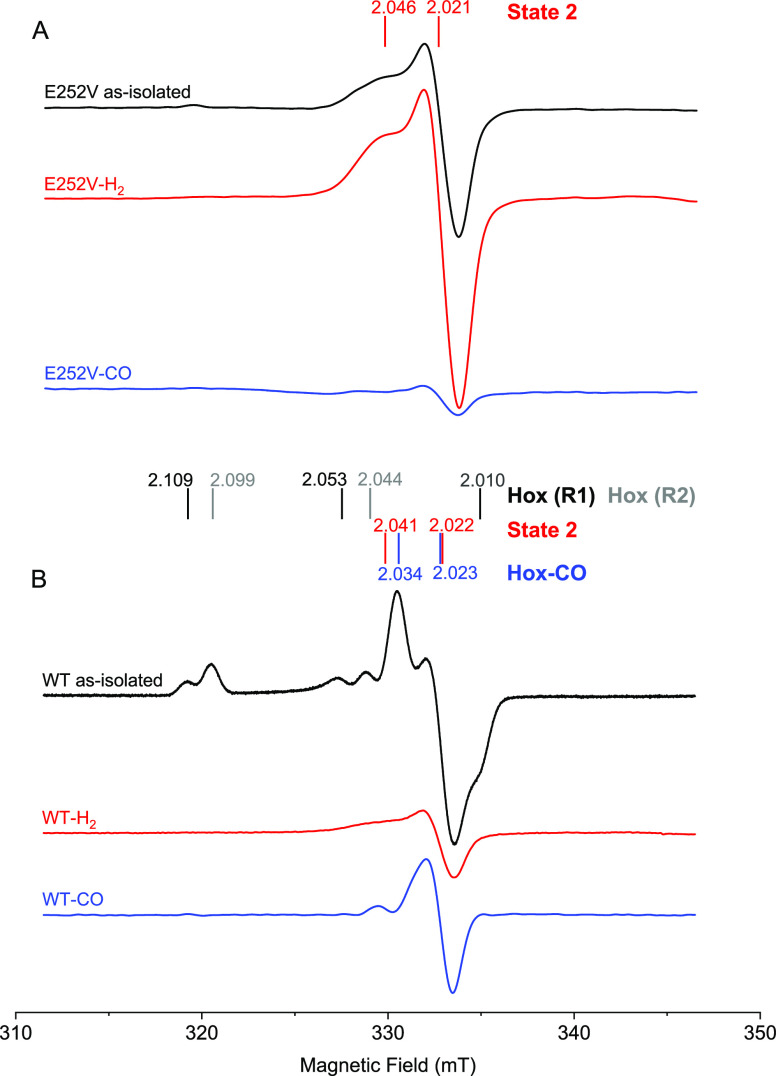
The electronic configuration of State 1 and State 2 was further probed using EPR spectroscopy. The E252V variant was selected as a representative example as it displayed a high fraction of State 2. X-band EPR spectra recorded at 21 K of this “as-isolated” E252V sample were dominated by a narrow axial signal, (Figure 5A, black spectrum). This is in contrast to reference EPR spectra recorded of analogously prepared samples of TamHydS WT (Figure 5B), which shows a mixture of oxidized resting states associated with two Hox components and a CO-inhibited state Hox-CO in the g ∼ 2 region (Table S3).9
Figure 5.
EPR spectra of TamHydS variant E252V and wild-type (WT). (A) EPR measurements of E252V prepared under a neat argon atmosphere (“as-isolated,” black spectrum) revealed a dominant pseudo-axial signal, assigned to State 2 (g∥ = 2.046 and g⊥ = 2.021), which increases after H2-flushing (red spectrum) and disappears after flushing with CO (blue spectrum). (B) As-isolated spectrum of TamHydS WT (black) reveals rhombic features of the H-cluster in the Hox states (two components denoted as R1 and R2 with the corresponding g-values in black and gray respectively) and an axial feature corresponding to Hox-CO (g-values in blue). TamHydS WT samples incubated under H2 (red spectrum) and CO (blue spectrum) shown for reference. Data adapted from Land et al.,9 revising the g-values assigned to Hox-CO (g∥ = 2.034 and g⊥ = 2.023) and assigning the previously unknown signal that appears when WT is flushed with H2 to State 2 (g∥ = 2.041 and g⊥ = 2.022). Spectra recorded with the following settings: T = 21 K; modulation frequency = 100 kHz; amplitude = 10 G; microwave frequency = 9.4 GHz; microwave power = 16 μW.
The amplitude of the axial signal in E252V was further increased after continuous flushing with H2 (Figure 5A, red spectrum). Consequently, it is assigned to State 2, in accordance with the red-shifted features observed by FTIR spectroscopy following H2 treatment. Simulation of the State 2 EPR signal showed that it can be described as a pseudo-axial EPR signal with g∥ = 2.046, g⊥ = 2.0225, 2.0196 (average 2.021) (Figure S10). In addition to the narrow axial signal, spectra of H2-treated samples of E252V collected at 10 K also displayed features attributable to reduced F-clusters ([4Fe–4S]+) (Figures S10 and S11). This shows that the H-cluster remains capable of shuttling electrons into the accessory clusters despite modifications to the putative PTP. As observed by ATR-FTIR spectroscopy, observation of State 2 is not strictly dependent on H2 treatment, as the same signal was found in E252V (and E289D) when treated with sodium dithionite (Figure S5 and S7). Finally, apart from variations in relative intensities, no distinct differences in the signal shape were observed for the State 2 EPR signal in E252V and E289D when comparing samples generated under H2 to D2, in the absence or presence of 95% D2O in the buffer (Figure S6).
Continuous flushing of E252V samples under a neat CO atmosphere resulted in a significant decrease in the amplitude of the axial EPR signal, with no new distinct features appearing in the spectrum (Figure 5A, blue spectrum). Notably, E252V did not show any signs of CO-inhibited state, Hox-CO, even when it was flushed with CO gas for an hour. This supports the notion that CO treatment induces the formation of State 1 (Figure 5A, blue spectrum) as seen by FTIR spectroscopy (Figure S9C,D) and that State 1 is EPR-silent.
In summary, the data from FTIR and low-temperature EPR measurements revealed that the variants do not accumulate any states commonly observed for the H-cluster in [FeFe] hydrogenases. However, we note that an axial signal strikingly similar to that of State 2 was previously reported for TamHydS WT (Figure 5 and Table S3).9
Discussion
Based on studies of Group A enzymes, [FeFe] hydrogenases are generally considered to be fast and reversible catalysts, highly sensitive to CO inhibition.1a,2a,12a The recent characterization of Group C and D [FeFe] hydrogenases has shown that this is not always the case.9,16b,19 Elucidating key reactivity differences between [FeFe] hydrogenases and the structural factors promoting this is arguably central to our understanding of biological H2-metabolism. In parallel, it is expected to provide guiding principles in the preparation of molecular-based catalysts for redox catalysis.
Through homology modeling and sequence alignment, we have now identified a putative PTP unique for Group D [FeFe] hydrogenases. Employing TamHydS as a Group D representative, variants were generated through site-directed mutagenesis that target E252 and E289, the two putative proton relay sites most proximal to the H-cluster. The variants were characterized biochemically and spectroscopically, and all variants were found to have a significant impact on H2 catalysis and the H-cluster redox state configuration. In all cases, the overall rates for H2 oxidation and production decreased, with variation of the proposed proton relay partner of the ADT-amine (E252), having the most prominent effect. In addition, the variants also resulted in a shift in apparent catalytic bias of the enzyme toward H2 evolution. With regard to the latter point, arguably the most striking observation was made for E289D which retained around 70% of the H2 production capacity of TamHydS WT, while significantly decreasing H2 oxidation rates (≤3% relative to TamHydS WT).
In addition to decreasing catalytic rates, altering the putative PTP clearly had a direct effect on the accumulation of H-cluster states of TamHydS. The variants form almost exclusively two previously unidentified states (States 1 and 2, Figures 4 and 5). Conversely, the redox states observed for TamHydS WT and most Group A [FeFe] hydrogenases, i.e., the well-known Hox and Hred states, are absent.9,45 The exact nature of the H-cluster species denoted as State 1 and State 2 herein remains to be fully elucidated. Still, the spectroscopy data reported here do provide initial insight. In short, State 2 (S = 1/2) is an EPR-active species that forms under reducing conditions while State 1 (S = 0) is EPR-silent and is relatively more oxidized. The IR band pattern of State 1 shows some similarities with the so-called Hinact and Hhyd states previously observed in Group A [FeFe] hydrogenases (Figure S12 for the structures of the H-cluster states).2a,41,42,46 The EPR-silent nature of this state implies an oxidized [4Fe–4S]H cluster ([4Fe–4S]H2+), yielding an overall oxidation state analogous to the previously reported inhibited “over-oxidized” Hinact state (i.e., [4Fe–4S]2+-[FeIIFeII]),2a,42a,46a−46e although we stress that the ligation of the [2Fe]H subsite is not necessarily identical. The red-shift observed by FTIR spectroscopy indicates that State 2 reflects a reduced state relative to State 1. Given the axial and relatively narrow nature of the H2-induced EPR signal, the unpaired electron likely resides on the [2Fe]H subsite rather than the iron–sulfur cluster, as the latter usually displays broader rhombic signals in its reduced ([4Fe–4S]H+) state.9,12a Based on these observations, we tentatively assign State 2 a formal oxidation state analogous to that of Hox (i.e. [4Fe–4S]2+-[FeIFeII]), which can explain the strong red-shift of the μCO ligand by 81 cm–1 in particular. However, the well-known active-ready resting-state Hox generally gives rise to a rhombic EPR signal, as also reported previously in the case of TamHydS WT.9 A somewhat related observation of unusual EPR properties of Hox-related states has recently been reported for the Group A [FeFe] hydrogenase from C. beijerinkii (CbHydA1). In the latter case, two distinct EPR signals were observed for the Hox state, but only a single set of FTIR bands, an observation attributed to structural flexibility around the [4Fe–4S] component of the H-cluster.47 Assuming that State 2 does indeed correspond to a similar mixed valence I,II formal oxidation state of the [2Fe]H subsite, the shift from rhombic to axial EPR signal as well as the distinct differences observed in the FTIR spectra indicates a change in the geometry and/or electron configuration of the H-cluster as compared to the standard Hox state.9 Moreover, State 2 accumulates under H2 in at least three of the variants (E289A/D and E252V), while incubation under a CO atmosphere results in (slow) conversion to State 1. This behavior is in stark contrast to what is commonly observed for the Hox state, which generally converts to Hred and Hhyd species under H2 and Hox-CO under CO.
The lower catalytic rates as well as the altered properties of the H-cluster of the variants support a critical influence of the varied residues. The larger decrease of H2 oxidation rates is in line with an impaired proton transport affecting the faster reaction more strongly than the intrinsically slower proton reduction reaction (Figure 3). However, the effects on bias as well as stabilization of new H-cluster states are difficult to reconcile with an exclusive substrate (proton) transfer influence.11,48 It is well established that bias in hydrogenases can further be controlled by, e.g., accessory FeS clusters, which govern the intramolecular electron transfer rates.22,49 In parallel, a variation in bias has previously also been observed in Group A [FeFe] hydrogenase variants in which the terminal PTP residue (C299CpI) was exchanged into serine or aspartate,11,50 an effect attributed to changes in relative acidity of the proton donor and the [2Fe]H subsite.50 Still, also these two latter models struggle to fully rationalize the observed reactivity of the variants. Instead, we propose that the variations of the PTP studied here cause a structural change at the H-cluster, resulting in a disruption of the ADT-amine and Fed FLP interaction (Figure 6A,B). More specifically, the E252 residue is likely contributing to the stabilization of an ADT geometry where the amine is positioned at an optimal distance from Fed for heterolytic H2 cleavage. Removing this carboxylate (E252V), or even shortening the residue by one methylene group (E252D), releases strain from the cofactor facilitating formation of a conformation with decreased activity. Similarly, variations of the second target residue, E289, are likely to not only impair proton transfer but also influence the positioning of the ADT ligand either directly or via interactions with E252. Changes in the H-bonding network around the [2Fe]H subsite could also rationalize the changes in relative intensities of the CN bands between TamHydS WT and the variants, observed by FTIR spectroscopy. Hints of similar structural effects are found in crystallographic data on CpI, which show that variations of the closest PTP residue to the ADT ligand (C299ACpI and C299DCpI) slightly reduced the distance between the Fed and ADT-amine by 0.3–0.4 Å. Moreover, variation of the next proton transfer relay (E279DCpI) showed modified configurations of the other PTP side chains.11
Figure 6.

Proposed models illustrating the structural changes at the H-cluster by the PTP variants. (A) Schematic model illustrating an optimal FLP interaction between the ADT-amine and the distal Fe (Fed) of the H-cluster. The glutamate at position 252 (E252) of TamHydS WT likely fine-tunes the positioning of the ADT-amine, either through direct H-bonding or indirectly via intermediate water molecules, thus ensuring an optimal FLP distance from Fed for heterolytic H2 cleavage. (B) Absence of H-bonding provided by the carboxylate side chain upon exchange of E252 to V252 shifts the ADT-amine toward a relatively farther distance from Fed, releasing cofactor strain and consequently elongates the FLP distance. (C) Reduced steric hindrance, due to a relaxed FLP geometry as illustrated in (B), could facilitate the binding of exogenous ligands to Fed, thereby, facilitating the formation of inhibited states. The binding of an aqua (or hydroxide) ligand is proposed for State 1.
Beyond the geometry argument outlined above, additional contributions from changes in electrostatics cannot be excluded. However, as the site of variation (E252 or E289) had a greater impact on reactivities than the nature of variation (conservative or nonconservative), we consider electrostatics unlikely to be a dominant factor. Assuming an impaired FLP geometry in the variants, it follows that the change in bias can be motivated with different rate-determining steps depending on the direction of the catalytic cycle. During H2 oxidation, the heterolytic cleavage of H2 appears to be an important contributor to the overall rate. Conversely, the H–H bond formation step appears to contribute less during the reverse reaction, as the variants have a relatively smaller effect on the rates of H2 production.
As State 1 and 2 are found to accumulate in variants with impaired function, care needs to be taken when attributing them catalytic relevance. Still, in particular the E289 variants do display significant H+ reduction activities despite only displaying these two states in FTIR spectroscopy almost exclusively. Moreover, our earlier study on TamHydS has shown that upon treatment with H2, an axial EPR signal with g∥ = 2.041 and g⊥ = 2.022 dominates the spectrum (Figure 5B).9 The identity of the H2-induced species in TamHydS WT (denoted A1) reported in the study by Land et al.9 is most likely analogous to State 2. In Group A [FeFe] hydrogenases, variations in the PTP close to the H-cluster (e.g., C299CpI) facilitate the accumulation of a terminal hydride species under a H2 atmosphere.41,44 Subsequent studies have shown that the latter state, denoted Hhyd, accumulates also in wild-type Group A enzymes, strongly supporting its catalytic relevance.37,41,51 The absence of any distinct isotope effect on the FTIR and EPR spectra in the presence of D2 or D2O argues against ligation of a terminal hydride ligand in either State 1 or 2. Still, it is tempting to consider State 2 as reflecting a catalytically active H-cluster state. Conversely, the Hinact-like State 1 is most likely an inhibited species. Assuming that State 1 formation is associated with the binding of an extra ligand, this process would be facilitated by the removal of steric hindrance. Considering the disrupted FLP model, an increased ADT-amine and Fed distance could then also rationalize the apparent facile formation of this state. Again comparing to earlier studies of Group A [FeFe] hydrogenases, it has been reported that exchange of the terminal PTP cysteine residue to alanine facilitates the binding of exogenous cyanide ligands to the [2Fe]H subsite.52 This was attributed to a decreased electron density at the [2Fe]H subsite following the cysteine to alanine exchange, but we note that a steric rationale might also be applicable. Ligand binding to the [2Fe]H subsite during State 1 formation would also provide an explanation for its sluggish reactivity toward H2, as particularly noted for the E252 variants (Figure 4). As no potential inhibitor is added to the enzyme, we propose that the most likely incoming ligand would be H2O. State 1 would then represent an inhibited state, coordinating an aqua or hydroxido ligand analogous to earlier models of the so-called Hinact state (Figure 6C).42a,53 Further studies are clearly needed to fully elucidate the electronic properties and ligand geometry of these states as well as their catalytic relevance.
In conclusion, this study has provided a first insight into the PTP in Group D [FeFe] hydrogenase. This study also showed how a relatively conservative change in ≥5 Å distance from the H-cluster (E289D) can significantly alter the apparent catalytic bias of the enzyme. However, the PTP effects are likely convoluted with effects on the H-cluster itself. This study also supports the notion that the route of proton transfer is not universal across phylogenetic groups of [FeFe] hydrogenases. Indeed, aligning the amino acids that are part of the proposed PTP in Group D TamHydS with the corresponding amino acids in Group A, B and C reveal that they are unlikely to share a similar PTP. Why the two putatively sensory groups (Group C and D) of [FeFe] hydrogenases have major differences in their PTP remains to be resolved. When analyzing the sequences of Group C from Greening et al.30 and further extracting the Group D sequences from that data using their differences as presented in Calusinska et al.,16a one can see that both groups are present in around 25% of all organisms (including Thermoanaerobacter mathranii) in that dataset. This indicates that they actually have different functions and if they are both sensory, they could sense H2 at different concentrations and/or induce different cellular responses. Furthermore, in some rare instances, organisms also encode multiple isozymes of Group C or Group D, which strengthens the hypothesis that one organism can encode and benefit from several sensory [FeFe] hydrogenases.
Future work aims to closely examine the role of the next amino acids proposed to complete the PTP (S249 and D265), confirm the changes in the local structure (including E252 and E289), and identify geometrical considerations of the hydrogen bonding network through high-resolution structural studies. The identification of this possible alternative PTP paves the way to elucidate how the protein environment promotes the diverging reactivity of different groups of [FeFe] hydrogenase. By extension, this can be expected to expand the tool-kit in the design of site-isolated catalysts coupling proton transfer and redox chemistry.
Acknowledgments
The authors would like to thank Prof. Sven T. Stripp for insightful discussions related to the preparation of this manuscript. The Swedish Energy Agency (STEM, project No. 48574-1 to G.B.), the European Union’s Horizon 2020 research and innovation program (Marie Skłodowska Curie grant No. 897555 to M.S.), and Stiftelsen Olle Engkvist Byggmästare (project No 220-0226 to G.B. and M.S.) are gratefully acknowledged for funding.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscatal.3c02314.
Alignment of CpI and TamHydS, SDS-PAGE gels, details on FeS cluster reconstitution and Fe content of TamHydS variants, tabulated specific activity data, as well as additional FTIR and EPR spectroscopy data, and schematic representations of proposed catalytic cycles of [FeFe] hydrogenase (PDF)
Author Contributions
† P.R.C. and K.W. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- a Lubitz W.; Ogata H.; Rüdiger O.; Reijerse E. Hydrogenases. Chem. Rev. 2014, 114, 4081–4148. 10.1021/cr4005814. [DOI] [PubMed] [Google Scholar]; b Benoit S. L.; Maier R. J.; Sawers R. G.; Greening C. Molecular Hydrogen Metabolism: a Widespread Trait of Pathogenic Bacteria and Protists. Microbiol. Mol. Biol. Rev. 2020, 84, e00092-19 10.1128/mmbr.00092-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Lorenzi M.; Berggren G.. [FeFe] Hydrogenases and Their Functional Models. In Comprehensive Coordination Chemistry III, Constable E. C., Parkin G., Que L. Jr, Eds.; Elsevier, 2021; pp. 731–756. [Google Scholar]; b Kleinhaus J. T.; Wittkamp F.; Yadav S.; Siegmund D.; Apfel U.-P. [FeFe]-Hydrogenases: maturation and reactivity of enzymatic systems and overview of biomimetic models. Chem. Soc. Rev. 2021, 50, 1668–1784. 10.1039/D0CS01089H. [DOI] [PubMed] [Google Scholar]
- Reijerse E. J.; Pelmenschikov V.; Birrell J. A.; Richers C. P.; Kaupp M.; Rauchfuss T. B.; Cramer S. P.; Lubitz W. Asymmetry in the Ligand Coordination Sphere of the [FeFe] Hydrogenase Active Site Is Reflected in the Magnetic Spin Interactions of the Aza-propanedithiolate Ligand. J. Phys. Chem. Lett. 2019, 10, 6794–6799. 10.1021/acs.jpclett.9b02354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi M.; Gellett J.; Zamader A.; Senger M.; Duan Z.; Rodríguez-Maciá P.; Berggren G. Investigating the role of the strong field ligands in [FeFe] hydrogenase: spectroscopic and functional characterization of a semi-synthetic mono-cyanide active site. Chem. Sci. 2022, 13, 11058–11064. 10.1039/D2SC02271K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stripp S. T.; Duffus B. R.; Fourmond V.; Léger C.; Leimkühler S.; Hirota S.; Hu Y.; Jasniewski A.; Ogata H.; Ribbe M. W. Second and Outer Coordination Sphere Effects in Nitrogenase, Hydrogenase, Formate Dehydrogenase, and CO Dehydrogenase. Chem. Rev. 2022, 122, 11900–11973. 10.1021/acs.chemrev.1c00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Berggren G.; Adamska A.; Lambertz C.; Simmons T. R.; Esselborn J.; Atta M.; Gambarelli S.; Mouesca J. M.; Reijerse E.; Lubitz W.; Happe T.; Artero V.; Fontecave M. Biomimetic assembly and activation of [FeFe]-hydrogenases. Nature 2013, 499, 66–69. 10.1038/nature12239. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Esselborn J.; Lambertz C.; Adamska-Venkatesh A.; Simmons T.; Berggren G.; Noth J.; Siebel J.; Hemschemeier A.; Artero V.; Reijerse E.; Fontecave M.; Lubitz W.; Happe T. Spontaneous activation of [FeFe]-hydrogenases by an inorganic [2Fe] active site mimic. Nat. Chem. Bio. 2013, 9, 607–609. 10.1038/nchembio.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Siebel J. F.; Adamska-Venkatesh A.; Weber K.; Rumpel S.; Reijerse E.; Lubitz W. Hybrid [FeFe]-Hydrogenases with Modified Active Sites Show Remarkable Residual Enzymatic Activity. Biochemistry 2015, 54, 1474–1483. 10.1021/bi501391d. [DOI] [PubMed] [Google Scholar]; d Kertess L.; Wittkamp F.; Sommer C.; Esselborn J.; Rüdiger O.; Reijerse E. J.; Hofmann E.; Lubitz W.; Winkler M.; Happe T.; et al. Chalcogenide substitution in the [2Fe] cluster of [FeFe]-hydrogenases conserves high enzymatic activity. Dalton Trans. 2017, 46, 16947–16958. 10.1039/C7DT03785F. [DOI] [PubMed] [Google Scholar]
- a Helm M. L.; Stewart M. P.; Bullock R. M.; DuBois M. R.; DuBois D. L. A Synthetic Nickel Electrocatalyst with a Turnover Frequency Above 100,000 s–1 for H2 Production. Science 2011, 333, 863–866. 10.1126/science.1205864. [DOI] [PubMed] [Google Scholar]; b Brezinski W. P.; Karayilan M.; Clary K. E.; Pavlopoulos N. G.; Li S.; Fu L.; Matyjaszewski K.; Evans D. H.; Glass R. S.; Lichtenberger D. L.; et al. [FeFe]-Hydrogenase Mimetic Metallopolymers with Enhanced Catalytic Activity for Hydrogen Production in Water. Angew. Chem. Int. Ed. 2018, 57, 11898–11902. 10.1002/anie.201804661. [DOI] [PubMed] [Google Scholar]; c Simmons T. R.; Berggren G.; Bacchi M.; Fontecave M.; Artero V. Mimicking hydrogenases: From biomimetics to artificial enzymes. Coord. Chem. Rev. 2014, 270-271, 127–150. 10.1016/j.ccr.2013.12.018. [DOI] [Google Scholar]; d Karayilan M.; Brezinski W. P.; Clary K. E.; Lichtenberger D. L.; Glass R. S.; Pyun J. Catalytic Metallopolymers from [2Fe-2S] Clusters: Artificial Metalloenzymes for Hydrogen Production. Angew. Chem. Int. Ed. 2019, 58, 7537–7550. 10.1002/anie.201813776. [DOI] [PubMed] [Google Scholar]
- Ginovska-Pangovska B.; Dutta A.; Reback M. L.; Linehan J. C.; Shaw W. J. Beyond the Active Site: The Impact of the Outer Coordination Sphere on Electrocatalysts for Hydrogen Production and Oxidation. Acc. Chem. Res. 2014, 47, 2621–2630. 10.1021/ar5001742. [DOI] [PubMed] [Google Scholar]
- Land H.; Sekretareva A.; Huang P.; Redman H. J.; Németh B.; Polidori N.; Mészáros L. S.; Senger M.; Stripp S. T.; Berggren G. Characterization of a putative sensory [FeFe]-hydrogenase provides new insight into the role of the active site architecture. Chem. Sci. 2020, 11, 12789–12801. 10.1039/D0SC03319G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esselborn J.; Muraki N.; Klein K.; Engelbrecht V.; Metzler-Nolte N.; Apfel U. P.; Hofmann E.; Kurisu G.; Happe T. A structural view of synthetic cofactor integration into [FeFe]-hydrogenases. Chem. Sci. 2016, 7, 959–968. 10.1039/C5SC03397G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J.; Senger M.; Esselborn J.; Engelbrecht V.; Wittkamp F.; Apfel U.-P.; Hofmann E.; Stripp S. T.; Happe T.; Winkler M. Crystallographic and spectroscopic assignment of the proton transfer pathway in [FeFe]-hydrogenases. Nat. Commun. 2018, 9, 4726. 10.1038/s41467-018-07140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Land H.; Senger M.; Berggren G.; Stripp S. T. Current State of [FeFe]-Hydrogenase Research: Biodiversity and Spectroscopic Investigations. ACS Catal. 2020, 10, 7069–7086. 10.1021/acscatal.0c01614. [DOI] [Google Scholar]; b Poudel S.; Tokmina-Lukaszewska M.; Colman D. R.; Refai M.; Schut G. J.; King P. W.; Maness P.-C.; Adams M. W. W.; Peters J. W.; Bothner B.; et al. Unification of [FeFe]-hydrogenases into three structural and functional groups. Biochim. Biophys. Acta 2016, 1860, 1910–1921. 10.1016/j.bbagen.2016.05.034. [DOI] [PubMed] [Google Scholar]
- Happe T.; Naber J. D. Isolation, characterization and N-terminal amino acid sequence of hydrogenase from the green alga Chlamydomonas reinhardtii. Eur. J. Mol. Biol. Biochem. 1993, 214, 475–481. 10.1111/j.1432-1033.1993.tb17944.x. [DOI] [PubMed] [Google Scholar]
- Glick B. R.; Martin W. G.; Martin S. M. Purification and properties of the periplasmic hydrogenase from Desulfovibrio desulfuricans. Can. J. Microbiol. 1980, 26, 1214–1223. 10.1139/m80-203. [DOI] [PubMed] [Google Scholar]
- Nakos G.; Mortenson L. Subunit structure of azoferredoxin from Clostridium pasteurianum W5. Biochemistry 1971, 10, 455–458. 10.1021/bi00779a016. [DOI] [PubMed] [Google Scholar]
- a Calusinska M.; Happe T.; Joris B.; Wilmotte A. The surprising diversity of clostridial hydrogenases: a comparative genomic perspective. Microbiology 2010, 156, 1575–1588. 10.1099/mic.0.032771-0. [DOI] [PubMed] [Google Scholar]; b Chongdar N.; Birrell J. A.; Pawlak K.; Sommer C.; Reijerse E. J.; Rüdiger O.; Lubitz W.; Ogata H. Unique Spectroscopic Properties of the H-Cluster in a Putative Sensory [FeFe] Hydrogenase. J. Am. Chem. Soc. 2018, 140, 1057–1068. 10.1021/jacs.7b11287. [DOI] [PubMed] [Google Scholar]
- Chongdar N.; Rodríguez-Maciá P.; Reijerse E. J.; Lubitz W.; Ogata H.; Birrell J. A. Redox tuning of the H-cluster by second coordination sphere amino acids in the sensory [FeFe] hydrogenase from Thermotoga maritima. Chem. Sci. 2023, 14, 3682–3692. 10.1039/D2SC06432D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmond V.; Baffert C.; Sybirna K.; Dementin S.; Abou-Hamdan A.; Meynial-Salles I.; Soucaille P.; Bottin H.; Léger C. The mechanism of inhibition by H2 of H2-evolution by hydrogenases. Chem. Commun. 2013, 49, 6840–6842. 10.1039/C3CC43297A. [DOI] [PubMed] [Google Scholar]
- Fasano A.; Land H.; Fourmond V.; Berggren G.; Léger C. Reversible or Irreversible Catalysis of H+/H2 Conversion by FeFe Hydrogenases. J. Am. Chem. Soc. 2021, 143, 20320–20325. 10.1021/jacs.1c09554. [DOI] [PubMed] [Google Scholar]
- Mészáros L. S.; Land H.; Redman H. J.; Berggren G. Semi-synthetic hydrogenases—in vitro and in vivo applications. Curr. Opin. Green Sustain. Chem. 2021, 32, 100521 10.1016/j.cogsc.2021.100521. [DOI] [Google Scholar]
- Knörzer P.; Silakov A.; Foster C. E.; Armstrong F. A.; Lubitz W.; Happe T. Importance of the protein framework for catalytic activity of [FeFe]-hydrogenases. J. Biol. Chem. 2012, 287, 1489–1499. 10.1074/jbc.M111.305797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Caserta G.; Papini C.; Adamska-Venkatesh A.; Pecqueur L.; Sommer C.; Reijerse E.; Lubitz W.; Gauquelin C.; Meynial-Salles I.; Pramanik D.; Artero V.; Atta M.; del Barrio M.; Faivre B.; Fourmond V.; Léger C.; Fontecave M. Engineering an [FeFe]-Hydrogenase: Do Accessory Clusters Influence O2 Resistance and Catalytic Bias?. J. Am. Chem. Soc. 2018, 140, 5516–5526. 10.1021/jacs.8b01689. [DOI] [PubMed] [Google Scholar]; b Gauquelin C.; Baffert C.; Richaud P.; Kamionka E.; Etienne E.; Guieysse D.; Girbal L.; Fourmond V.; André I.; Guigliarelli B.; et al. Roles of the F-domain in [FeFe] hydrogenase. Biochim. Biophys. Acta 2018, 1859, 69–77. 10.1016/j.bbabio.2017.08.010. [DOI] [PubMed] [Google Scholar]; c Artz J. H.; Mulder D. W.; Ratzloff M. W.; Lubner C. E.; Zadvornyy O. A.; LeVan A. X.; Williams S. G.; Adams M. W. W.; Jones A. K.; King P. W.; et al. Reduction Potentials of [FeFe]-Hydrogenase Accessory Iron–Sulfur Clusters Provide Insights into the Energetics of Proton Reduction Catalysis. J. Am. Chem. Soc. 2017, 139, 9544–9550. 10.1021/jacs.7b02099. [DOI] [PubMed] [Google Scholar]
- a Morra S.; Giraudo A.; Di Nardo G.; King P. W.; Gilardi G.; Valetti F. Site Saturation Mutagenesis Demonstrates a Central Role for Cysteine 298 as Proton Donor to the Catalytic Site in CaHydA [FeFe]-Hydrogenase. PLoS One 2012, 7, e48400 10.1371/journal.pone.0048400. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Morra S.; Maurelli S.; Chiesa M.; Mulder D. W.; Ratzloff M. W.; Giamello E.; King P. W.; Gilardi G.; Valetti F. The effect of a C298D mutation in CaHydA [FeFe]-hydrogenase: Insights into the protein-metal cluster interaction by EPR and FTIR spectroscopic investigation. Biochim. Biophys. Acta 2016, 1857, 98–106. 10.1016/j.bbabio.2015.10.005. [DOI] [PubMed] [Google Scholar]
- Lee H. J.; Reimann J.; Huang Y.; Ädelroth P. Functional proton transfer pathways in the heme–copper oxidase superfamily. Biochim. Biophys. Acta 2012, 1817, 537–544. 10.1016/j.bbabio.2011.10.007. [DOI] [PubMed] [Google Scholar]
- a Fdez Galván I.; Volbeda A.; Fontecilla-Camps J. C.; Field M. J. A QM/MM study of proton transport pathways in a [NiFe] hydrogenase. Proteins 2008, 73, 195–203. 10.1002/prot.22045. [DOI] [PubMed] [Google Scholar]; From NLM; b Teixeira V. H.; Soares C. M.; Baptista A. M. Proton pathways in a [NiFe]-hydrogenase: A theoretical study. Proteins 2008, 70, 1010–1022. 10.1002/prot.21588. [DOI] [PubMed] [Google Scholar]; c Matias P. M.; Soares C. M.; Saraiva L. M.; Coelho R.; Morais J.; Le Gall J.; Carrondo M. A. [NiFe] hydrogenase from Desulfovibrio desulfuricans ATCC 27774: gene sequencing, three-dimensional structure determination and refinement at 1.8 Å and modelling studies of its interaction with the tetrahaem cytochrome c3. J. Biol. Inorg. 2001, 6, 63–81. 10.1007/s007750000167. [DOI] [PubMed] [Google Scholar]; d Tai H.; Hirota S.; Stripp S. T. Proton Transfer Mechanisms in Bimetallic Hydrogenases. Acc. Chem. Res. 2021, 54, 232–241. 10.1021/acs.accounts.0c00651. [DOI] [PubMed] [Google Scholar]
- a Senger M.; Viktor E.; Konstantin L.; Jifu D.; Wittkamp F.; Knör G.; Apfel U.; Happe T.; Winkler M.; Heberle J.; et al. How FeFe -Hydrogenase Facilitates Bidirectional Proton Transfer. J. Am. Chem. Soc. 2019, 141, 17394–17403. 10.1021/jacs.9b09225. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Cornish A. J.; Ginovska B.; Thelen A.; Da Silva J. C. S.; Soares T. A.; Raugei S.; Dupuis M.; Shaw W. J.; Hegg E. L. Single-Amino Acid Modifications Reveal Additional Controls on the Proton Pathway of [FeFe]-Hydrogenase. Biochemistry 2016, 55, 3165–3173. 10.1021/acs.biochem.5b01044. [DOI] [PubMed] [Google Scholar]; c Sode O.; Voth G. A. Electron transfer activation of a second water channel for proton transport in [FeFe]-hydrogenase. J. Chem. Phys. 2014, 141, 22D527. 10.1063/1.4902236. [DOI] [PubMed] [Google Scholar]; d Long H.; King P. W.; Chang C. H. Proton transport in Clostridium pasteurianum [FeFe] hydrogenase I: a computational study. J. Phys. Chem. B 2014, 118, 890–900. 10.1021/jp408621r. [DOI] [PubMed] [Google Scholar]; e Ginovska-Pangovska B.; Ho M.-H.; Linehan J. C.; Cheng Y.; Dupuis M.; Raugei S.; Shaw W. J. Molecular dynamics study of the proposed proton transport pathways in [FeFe]-hydrogenase. Biochim. Biophys. Acta 2014, 1837, 131–138. 10.1016/j.bbabio.2013.08.004. [DOI] [PubMed] [Google Scholar]; f Cornish A. J.; Gärtner K.; Yang H.; Peters J. W.; Hegg E. L. Mechanism of proton transfer in [FeFe]-hydrogenase from Clostridium pasteurianum. J. Biol. Chem. 2011, 286, 38341–38347. 10.1074/jbc.M111.254664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Rauchfuss T. B. Iron Carbonyl Sulfides, Formaldehyde, and Amines Condense To Give the Proposed Azadithiolate Cofactor of the Fe-Only Hydrogenases. J. Am. Chem. Soc. 2002, 124, 726–727. 10.1021/ja016964n. [DOI] [PubMed] [Google Scholar]
- Konagurthu A. S.; Whisstock J. C.; Stuckey P. J.; Lesk A. M. MUSTANG: a multiple structural alignment algorithm. Proteins 2006, 64, 559–574. 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- Sievers F.; Wilm A.; Dineen D.; Gibson T. J.; Karplus K.; Li W.; Lopez R.; McWilliam H.; Remmert M.; Söding J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening C.; Biswas A.; Carere C. R.; Jackson C. J.; Taylor M. C.; Stott M. B.; Cook G. M.; Morales S. E. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 2016, 10, 761–777. 10.1038/ismej.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger, LLC . The PyMOL Molecular Graphics System, Version 2.5.4; Schrodinger, LLC, 2022. [Google Scholar]
- Akhtar M. K.; Jones P. R. Deletion of iscR stimulates recombinant clostridial Fe-Fe hydrogenase activity and H2-accumulation in Escherichia coli BL21(DE3). Appl. Microbiol. Biotechnol. 2008, 78, 853–862. 10.1007/s00253-008-1377-6. [DOI] [PubMed] [Google Scholar]
- Belval L.; Marquette A.; Mestre P.; Piron M.-C.; Demangeat G.; Merdinoglu D.; Chich J.-F. A fast and simple method to eliminate Cpn60 from functional recombinant proteins produced by E. coli Arctic Express. Protein Expression Purif. 2015, 109, 29–34. 10.1016/j.pep.2015.01.009. [DOI] [PubMed] [Google Scholar]
- a Adams M. W.; Mortenson L. E. The physical and catalytic properties of hydrogenase II of Clostridium pasteurianum. A comparison with hydrogenase I. J. Biol. Chem. 1984, 259, 7045–7055. 10.1016/S0021-9258(17)39835-6. [DOI] [PubMed] [Google Scholar]; b Adams M. W.; Eccleston E.; Howard J. B. Iron-sulfur clusters of hydrogenase I and hydrogenase II of Clostridium pasteurianum. Proc. Natl. Acad. Sci. U. S. A. 1989, 86, 4932–4936. 10.1073/pnas.86.13.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stripp S. T. In Situ Infrared Spectroscopy for the Analysis of Gas-processing Metalloenzymes. ACS Catal. 2021, 11, 7845–7862. 10.1021/acscatal.1c00218. [DOI] [Google Scholar]
- Senger M.; Mebs S.; Duan J.; Wittkamp F.; Apfel U.-P.; Heberle J.; Haumann M.; Stripp S. T. Stepwise isotope editing of [FeFe]-hydrogenases exposes cofactor dynamics. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 8454–8459. 10.1073/pnas.1606178113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger M.; Kernmayr T.; Lorenzi M.; Redman H. J.; Berggren G. Hydride state accumulation in native [FeFe]-hydrogenase with the physiological reductant H2 supports its catalytic relevance. Chem. Commun. 2022, 58, 7184–7187. 10.1039/D2CC00671E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Senger M.; Mebs S.; Duan J.; Shulenina O.; Laun K.; Kertess L.; Wittkamp F.; Apfel U.-P.; Happe T.; Winkler M.; et al. Protonation/reduction dynamics at the [4Fe–4S] cluster of the hydrogen-forming cofactor in [FeFe]-hydrogenases. Phys. Chem. Chem. Phys. 2018, 20, 3128–3140. 10.1039/C7CP04757F. [DOI] [PubMed] [Google Scholar]; b Senger M.; Laun K.; Wittkamp F.; Duan J.; Haumann M.; Happe T.; Winkler M.; Apfel U. P.; Stripp S. T. Proton-Coupled Reduction of the Catalytic [4Fe-4S] Cluster in [FeFe]-Hydrogenases. Angew. Chem. Int. Ed. 2017, 56, 16503–16506. 10.1002/anie.201709910. [DOI] [PubMed] [Google Scholar]
- a Stoll S.; Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 2006, 178, 42–55. 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]; b Stoll S.Computational Modeling and Least-Squares Fittingof EPR Spectra. In Multifrequency Electron Paramagnetic Resonance; Wiley, 2014; pp. 69–138. [Google Scholar]
- Duan J.; Hemschemeier A.; Burr D. J.; Stripp S. T.; Hofmann E.; Happe T. Cyanide Binding to [FeFe]-Hydrogenase Stabilizes the Alternative Configuration of the Proton Transfer Pathway. Angew. Chem. Int. Ed. 2023, 62, e202216903 10.1002/anie.202216903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M.; Senger M.; Duan J.; Esselborn J.; Wittkamp F.; Hofmann E.; Apfel U.-P.; Stripp S. T.; Happe T. Accumulating the hydride state in the catalytic cycle of [FeFe]-hydrogenases. Nat. Commun. 2017, 8, 16115. 10.1038/ncomms16115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Roseboom W.; De Lacey A. L.; Fernandez V. M.; Hatchikian E. C.; Albracht S. P. J. The active site of the [FeFe]-hydrogenase from Desulfovibrio desulfuricans. II. Redox properties, light sensitivity and CO-ligand exchange as observed by infrared spectroscopy. J. Biol. Inorg. 2006, 11, 102–118. 10.1007/s00775-005-0040-2. [DOI] [PubMed] [Google Scholar]; b Laun K.; Baranova I.; Duan J.; Kertess L.; Wittkamp F.; Apfel U.-P.; Happe T.; Senger M.; Stripp S. T. Site-selective protonation of the one-electron reduced cofactor in [FeFe]-hydrogenase. Dalton Trans. 2021, 50, 3641–3650. 10.1039/D1DT00110H. [DOI] [PubMed] [Google Scholar]
- Redman H. J.; Huang P.; Haumann M.; Cheah M. H.; Berggren G. Lewis acid protection turns cyanide containing [FeFe]-hydrogenase mimics into proton reduction catalysts. Dalton Trans. 2022, 51, 4634–4643. 10.1039/D1DT03896F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder D. W.; Ratzloff M. W.; Bruschi M.; Greco C.; Koonce E.; Peters J. W.; King P. W. Investigations on the Role of Proton-Coupled Electron Transfer in Hydrogen Activation by [FeFe]-Hydrogenase. J. Am. Chem. Soc. 2014, 136, 15394–15402. 10.1021/ja508629m. [DOI] [PubMed] [Google Scholar]
- Land H.; Ceccaldi P.; Meszaros L. S.; Lorenzi M.; Redman H. J.; Senger M.; Stripp S. T.; Berggren G. Discovery of novel [FeFe]-hydrogenases for biocatalytic H2-production. Chem. Sci. 2019, 10, 9941–9948. 10.1039/c9sc03717a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Rodríguez-Maciá P.; Galle L. M.; Bjornsson R.; Lorent C.; Zebger I.; Yoda Y.; Cramer S. P.; DeBeer S.; Span I.; Birrell J. A. Caught in the Hinact: Crystal Structure and Spectroscopy Reveal a Sulfur Bound to the Active Site of an O2-stable State of [FeFe] Hydrogenase. Angew. Chem. Int. Ed. 2020, 59, 16786–16794. 10.1002/anie.202005208. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Corrigan P. S.; Tirsch J. L.; Silakov A. Investigation of the Unusual Ability of the [FeFe] Hydrogenase from Clostridium beijerinckii to Access an O2-Protected State. J. Am. Chem. Soc. 2020, 142, 12409–12419. 10.1021/jacs.0c04964. [DOI] [PubMed] [Google Scholar]; c Rodríguez-Maciá P.; Reijerse E. J.; van Gastel M.; DeBeer S.; Lubitz W.; Rüdiger O.; Birrell J. A. Sulfide Protects [FeFe] Hydrogenases From O2. J. Am. Chem. Soc. 2018, 140, 9346–9350. 10.1021/jacs.8b04339. [DOI] [PubMed] [Google Scholar]; d Morra S.; Arizzi M.; Valetti F.; Gilardi G. Oxygen Stability in the New [FeFe]-Hydrogenase from Clostridium beijerinckii SM10 (CbA5H). Biochemistry 2016, 55, 5897–5900. 10.1021/acs.biochem.6b00780. [DOI] [PubMed] [Google Scholar]; e Albracht S. P. J.; Roseboom W.; Hatchikian E. C. The active site of the [FeFe]-hydrogenase from Desulfovibrio desulfuricans. I. Light sensitivity and magnetic hyperfine interactions as observed by electron paramagnetic resonance. J. Biol. Inorg. 2006, 11, 88–101. 10.1007/s00775-005-0039-8. [DOI] [PubMed] [Google Scholar]; f Mebs S.; Kositzki R.; Duan J.; Kertess L.; Senger M.; Wittkamp F.; Apfel U.-P.; Happe T.; Stripp S. T.; Winkler M.; et al. Hydrogen and oxygen trapping at the H-cluster of [FeFe]-hydrogenase revealed by site-selective spectroscopy and QM/MM calculations. Biochim. Biophys. Acta 2018, 1859, 28–41. 10.1016/j.bbabio.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Corrigan P. S.; Majer S. H.; Silakov A. Evidence of Atypical Structural Flexibility of the Active Site Surrounding of an [FeFe] Hydrogenase from Clostridium beijerinkii. J. Am. Chem. Soc. 2023, 145, 11033–11044. 10.1021/jacs.2c13458. [DOI] [PubMed] [Google Scholar]
- Lampret O.; Duan J.; Hofmann E.; Winkler M.; Armstrong F. A.; Happe T. The roles of long-range proton-coupled electron transfer in the directionality and efficiency of [FeFe]-hydrogenases. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 20520–20529. 10.1073/pnas.2007090117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Abou Hamdan A.; Dementin S.; Liebgott P.-P.; Gutierrez-Sanz O.; Richaud P.; De Lacey A. L.; Rousset M.; Bertrand P.; Cournac L.; Léger C. Understanding and Tuning the Catalytic Bias of Hydrogenase. J. Am. Chem. Soc. 2012, 134, 8368–8371. 10.1021/ja301802r. [DOI] [PubMed] [Google Scholar]; b Hexter S. V.; Esterle T. F.; Armstrong F. A. A unified model for surface electrocatalysis based on observations with enzymes. Phys. Chem. Chem. Phys. 2014, 16, 11822–11833. 10.1039/C3CP55230F. [DOI] [PubMed] [Google Scholar]
- Kisgeropoulos E. C.; Bharadwaj V. S.; Mulder D. W.; King P. W. The Contribution of Proton-Donor pKa on Reactivity Profiles of [FeFe]-hydrogenases. Front. Microbiol. 2022, 13, 903951 10.3389/fmicb.2022.903951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Wittkamp F.; Senger M.; Stripp S. T.; Apfel U. P. [FeFe]-Hydrogenases: recent developments and future perspectives. Chem. Commun. 2018, 54, 5934–5942. 10.1039/C8CC01275J. [DOI] [PubMed] [Google Scholar]; b Mulder D. W.; Guo Y.; Ratzloff M. W.; King P. W. Identification of a Catalytic Iron-Hydride at the H-Cluster of [FeFe]-Hydrogenase. J. Am. Chem. Soc. 2017, 139, 83–86. 10.1021/jacs.6b11409. [DOI] [PubMed] [Google Scholar]
- a Martini M. A.; Bikbaev K.; Pang Y.; Lorent C.; Wiemann C.; Breuer N.; Zebger I.; DeBeer S.; Span I.; Bjornsson R.; et al. Binding of exogenous cyanide reveals new active-site states in [FeFe] hydrogenases. Chem. Sci. 2023, 14, 2826–2838. 10.1039/D2SC06098A. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Lorenzi M.; Ceccaldi P.; Rodríguez-Maciá P.; Redman H. J.; Zamader A.; Birrell J. A.; Mészáros L. S.; Berggren G. Stability of the H-cluster under whole-cell conditions—formation of an Htrans-like state and its reactivity towards oxygen. J. Biol. Inorg. 2022, 27, 345–355. 10.1007/s00775-022-01928-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Cao Z.; Hall M. B. Modeling the Active Sites in Metalloenzymes. 3. Density Functional Calculations on Models for [Fe]-Hydrogenase: Structures and Vibrational Frequencies of the Observed Redox Forms and the Reaction Mechanism at the Diiron Active Center. J. Am. Chem. Soc. 2001, 123, 3734–3742. 10.1021/ja000116v. [DOI] [PubMed] [Google Scholar]; b Liu Z.-P.; Hu P. A Density Functional Theory Study on the Active Center of Fe-Only Hydrogenase: Characterization and Electronic Structure of the Redox States. J. Am. Chem. Soc. 2002, 124, 5175–5182. 10.1021/ja0118690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.