Abstract
Human-microorganism interactions play a key role in human health. However, the underlying molecular mechanisms remain poorly understood. Small-molecules that offer a functional readout of microbe-microbe-human relationship are of great interest for deeper understanding of the inter-kingdom crosstalk at the molecular level. Recent studies have demonstrated that small-molecules from gut microbiota act as ligands for specific human G protein-coupled receptors (GPCRs) and modulate a range of human physiological functions, offering a mechanistic insight into the microbe-human interaction. To this end, we focused on analysis of bacterial metabolites that are currently recognized to bind to GPCRs and are found to activate the known downstream signaling pathways. We further mapped the distribution of these molecules across the public mass spectrometry-based metabolomics data, to identify the presence of these molecules across body sites and their association with health status. By combining this with RNA-Seq expression and spatial localization of GPCRs from a public human protein atlas database, we inferred the most predominant GPCR-mediated microbial metabolite-human cell interactions regulating gut-immune-brain axis. Furthermore, by evaluating the intestinal absorption properties and blood-brain barrier permeability of the small-molecules we elucidated their molecular interactions with specific human cell receptors, particularly expressed on human intestinal epithelial cells, immune cells and the nervous system that are shown to hold much promise for clinical translational potential. Furthermore, we provide an overview of an open-source resource for simultaneous interrogation of bioactive molecules across the druggable human GPCRome, a useful framework for integration of microbiome and metabolite cataloging with mechanistic studies for an improved understanding of gut microbiota-immune-brain molecular interactions and their potential therapeutic use.
Keywords: Human-microbiota interactions, Metabolites, G protein-coupled receptors, Gut microbiota, Inflammation, Immune system
Highlights
-
•
Microbial metabolite and GPCR interactions are crucial in gut-immune-brain axis..
-
•
Microbial small-molecules affect human biology and diseases (metabolic, neuropsychiatric, inflammatory) via GPCRs.
-
•
Non-ribosomal peptides, and microbial-human co-metabolism products play key roles in the gut-brain axis via GPCRs.
-
•
GPCRs are selectively expressed on immune, enteric, and brain cells, offering insights into gut-immune-brain interactions.
-
•
Effects of microbial molecules on human receptors and physiology remains largely unexplored.
1. Introduction
In recent years, there have been substantial advances in microbiome research with the advent of fast and cost-effective phylogenetic marker-based microbiome profiling and shotgun metagenomics sequencing approaches that can characterize microbial communities at the gene-level. However, knowledge of the precise molecular mechanisms by which gut microbiota influence human biology is still in its infancy. The human microbiota (estimated to encompass >40,000 operational taxonomic units) (The Human Microbiome Project Consortium, 2012) consists of a diverse collection of mutualistic bacteria that live on and in our bodies, which produce myriad small-molecule (defined as molecular mass <1.5 kDa) metabolites that affect nearly all aspects of human physiology, including the human immune system, metabolic functioning and nutrition (Donia and Fischbach, 2015; Garg et al., 2017; Han et al., 2021; Husted et al., 2017; Lai et al., 2021; Tan et al., 2017). As such, the gut microbiota is also called a “metabolic organ”. The human microbiota encodes a diverse collection of thus far uncharacterized metabolic pathways (Aleti et al., 2019; Donia and Fischbach, 2015; Koppel et al., 2018). Their small-molecule products are promising targets for understanding causal mechanisms as well as therapeutics for multiple reasons: First, microbial small-molecules are typically produced at concentrations comparable to those of drugs that can significantly influence physiological functions (Donia and Fischbach, 2015; Fischbach, 2018). Second, a major fraction of the microbiota-associated molecules has been shown to trigger significant biological responses locally often by accumulating in the gut, as well as affecting systemic functions by passive transportation (i.e., passive diffusion through paracellular and transcellular routes) and active transportation (i.e. transporter-mediated or receptor-mediated) from the gut into the bloodstream (Fischbach, 2018). Additionally, their concentrations vary from individual to individual by more than tenfold, which signifies microbiome-mediated inter-individual differences (Funabashi et al., 2020). Furthermore, recent large-scale culture omics studies have significantly expanded our understanding of microbial metabolite-sensing G protein-coupled receptors (GPCRs) (Chen et al., 2019; Colosimo et al., 2019). These studies have demonstrated that small-molecules produced by gut microbiota can mimic human-derived molecules and act as ligands for key human receptors, particularly GPCRs. For instance, by regulating specific human GPCRs, these microbial metabolites can modulate a range of physiological functions in different human cell types and tissues, providing a functional readout of a microbe-microbe-human relationship and offering deeper understanding of the inter-kingdom crosstalk at the molecular level (Chen et al., 2019; Cohen et al., 2017; Colosimo et al., 2019). It should be noted that some microbial molecules not only regulate GPCRs but also modulate other key protein class receptors, such as pregnane X receptor (PXR; also referred as NR1I2), farnesoid X receptor (FXR; also referred as NR1H4), human peptide transporter 1 (PEPT1; also known as SLC15A1), mammalian/mechanistic target of rapamycin complex 1 (mTORC1), and aryl hydrocarbon receptors (AhR), suggesting their relevance in the gut-immune-brain pathway (Fiorucci et al., 2018; Kanegawa et al., 2010; Koh et al., 2018; Krautkramer et al., 2020; Mencarelli et al., 2009; Mizushige et al., 2020; Moriyasu et al., 2016; Roager and Licht, 2018). Taken together, the most predominant small-molecules impact a remarkable array of host traits mediated by microbiome, thereby offering mechanistic insight into microbe-host interactions.
GPCRs represent the largest class of membrane receptors comprising seven-transmembrane helix proteins that sense a range of structurally diverse extracellular ligands. Upon sensing ligands, GPCRs bind to intracellular effectors including G proteins and arrestins, which in turn relay a variety of downstream intracellular cascades that are intimately linked with a broad range of physiological functions (Insel et al., 2019). Moreover, GPCRs have been implicated in many diseases, such as metabolic, neuropsychiatric and inflammatory diseases and many others. Consequently, GPCRs are considered to be highly attractive targets for therapeutics. This is evident from the fact that over one-third of currently available medications (approximately 527 prescribed drugs) have been developed to target GPCRs, highlighting their significance as druggable targets in pharmaceutical research (Hauser et al., 2018; Sriram and Insel, 2018). Humans contain approximately 826 GPCRs, of which 350 are non-olfactory members and among these 165 have validated drug targets; however, the remaining one-third of them are orphan receptors with no known ligands (Hauser et al., 2018). These orphan receptors represent a promising set of future targets for currently unmet medical needs and for deeper understanding of physiological functions (Laschet et al., 2018). Despite challenges in screening the orphan receptors, many orphan receptors characterized over the last decade have been associated with previously known bioactive molecules (Laschet et al., 2018). In this regard, it is reasonable to speculate that bacterial metabolites may bind orphan GPCRs and to some extent may contribute to ‘deorphanization’ of the previously uncharacterized GPCRs, for which no ligands and functions have thus far been identified.
Deciphering the full potential of the microbe-human interactions by metagenomic and metatranscriptomic approaches and by colonizing gnotobiotic animal models with human-associated bacteria is a daunting challenge due to (1) many of the uncultivable bacteria which represent most of the microbial diversity are not yet sequenced and functionally characterized and (2) enormous diversity and high inter-individual variability of the human microbiota. In this regard, metabolomics, the study of small-molecules, offers a functional readout of the microbiome and can facilitate mechanistic interrogation of small-molecules. In recent years untargeted mass spectrometry-based metabolomics has allowed researchers to measure global changes in metabolites to verify the identity of unknown molecules for isolation and structural elucidation. Most notably, advances in the molecular networking tools have expanded our ability to classify and group multiple features from the metabolomics data (e.g. precursor ion count, retention time, fragmentation pattern) into a network (Schmid et al., 2021), thus advancing the discovery of novel molecules. In addition, key advances in the computational tools to query a single MS/MS spectrum from a small-molecule of interest against all publicly available MS/MS datasets, analogous to NCBI Basic Local Alignment Search Tool (BLAST), enabled the translational potential of molecular information (Wang et al., 2020). Furthermore, evaluation of intestinal absorption properties and blood-brain barrier (BBB) permeability for small-molecules is key to understanding the interactions between the small-molecules and specific human cells as molecules cannot interact with the target cell receptors without first crossing the intestinal and BBB (van de Waterbeemd and Gifford, 2003).
Thus, in this work we, first, aim to provide the overview of the current knowledge on microbiota-associated molecules and discuss the findings from investigations of human receptor (predominantly GPCRs) and bacterial metabolite interactions known to date. Next, through multi-omics analysis, we further map the presence and distribution of specific molecules across the public mass spectrometry-based metabolomics data from several human biofluids, at the repository-scale via Global Natural Product Social Molecular Networking (GNPS) (Wang et al., 2016a). By combining this with RNA-Seq expression and spatial localization of GPCRs from the human protein atlas database, we inferred the most abundant GPCR-mediated microbial metabolite-human cell interactions regulating gut-immune-brain axis with clinical translational potential.
1.1. Current knowledge of the human microbiota-associated molecules
So far, approximately 172 potential microbial metabolites have been documented, but only a small number of microbial compounds have been studied in terms of their interaction with human receptors (Wishart et al., 2022). For this work, however, we focused on nine classes of microbial molecules with unique structures that are currently recognized to bind to human cell receptors, predominantly GPCRs. These molecules have been found to activate the known downstream signaling pathways. We further categorized these into three broad groups based on the source of the molecules (1) microbiota encoded molecules, e.g. N-acyl amides and dipeptides, (2) human molecules transformed by microbes, e.g. secondary bile acids, and (3) dietary components transformed by microbes e.g. short chain fatty acids, metabolites of aromatic amino acids (tryptophan, tyrosine and phenylalanine), histidine, and glutamate. In the following sections we discuss the three categories of microbiota-associated molecules in more detail.
1.2. Microbiota-encoded molecules
1.2.1. N-acyl amides
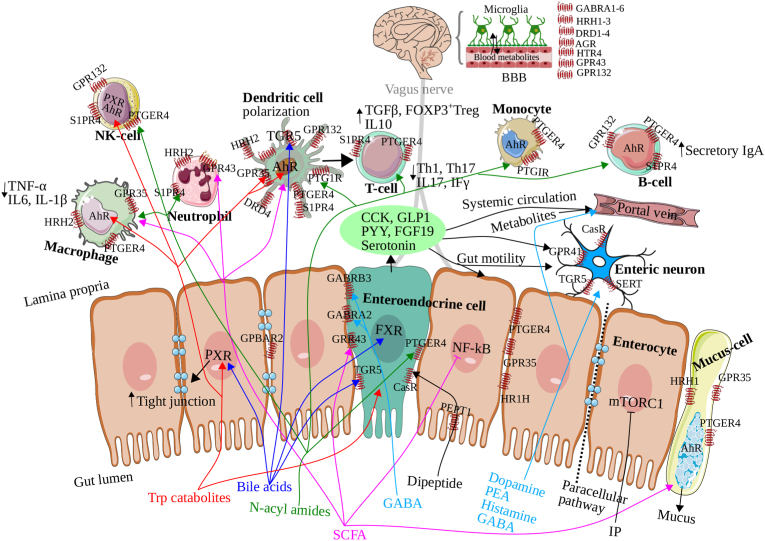
By searching the Human Microbiome Project (HMP) shotgun metagenomic data for N-acyl synthase genes, Cohen et al. identified 143 microbial genes encoding N-acyl synthases that were highly prevalent in the gut bacteria (Cohen et al., 2017). Further analysis of these genes identified six phylogenetically distinct families of N-acyl synthases that produce N-acyl amides with structural differences in the amine group and fatty acid chain as shown in Fig. 1. N-acyl amides are shown to contain either N-acyl, N-oxyacyl or N-hydroxyacyl fatty acid chains with palmitoyl and oleoyl analogues. N-acyl synthases are reported to exhibit selectivity for amine substrates glycine, serinol, alanine, lysine, ornithine and glutamine (Cohen et al., 2015, 2017) (Fig. 1). An overview of potential metabolites’ interactions with human receptors expressed on human gut, immune and brain cells, and their mechanisms of action on human functions, is displayed in Fig. 2. Notably, N-acyl amides exhibit high structural similarity with the bacterial quorum-sensing N-acyl-homoserine lactone molecules and endogenous human GPCR ligands (e.g. endogenous cannabinoid receptor agonist N-arachidonoylethanolamine) (Devane et al., 1992; Parsek et al., 1999) and are shown to mimic human ligands and activate several receptors belonging to the lipid-like GPCR family. For instance, N-acyl serinol has been shown to be an agonist of GPR119 that regulates glucose homeostasis. Additionally, GPR119 agonists also affect appetite through Glucagon-like peptide 1 (GLP-1) secretion from enteroendocrine and insulin from pancreatic β-cells (Husted et al., 2017). GLP-1 release also modulates enteric neuronal activity via GLP-1 receptors localized on enteric neurons, which is perceived by the vagus nerve, and in turn relayed to the upstream CNS (Müller et al., 2019). N-acyl alanine and N-3-hydroxypalmitoyl (also known as commendamide) are known to positively regulate GPR132. N-acyl lysine/ornithine is an agonist of S1PR4. While PTGIR is shown to be antagonized by N-acyl glutamine, PTGER4 is antagonized by N-acyl serinol, N-acyl alanine, N-acyl lysine/ornithine and N-acyl glutamine (Cohen et al., 2017).
Fig. 1.
A list of known microbial metabolites that have been shown to interact with human receptors, predominantly GPCRs. The three broad categories of microbial metabolites include: microbiota encoded molecules such as N-acyl amides and dipeptides; human molecules transformed by microbes such as secondary bile acids; dietary components transformed by microbes such as short chain fatty acids, metabolites of aromatic amino acids (e.g. tryptophan, tyrosine and phenylalanine), histidine, and glutamate. For visual simplicity, each class of molecule is shaded with distinct colors. Up or down arrows to the right of the human receptors indicate microbial metabolites with agonist and antagonist activities, respectively.
Fig. 2.
Overview of the identified bacterial metabolites interaction with human receptors and their mechanisms of action on gut-immune-brain pathways. Our results are based on RNA-Seq expression, localization patterns of G protein-coupled receptors (GPCRs) from gut, immune and brain cells, and intestinal and blood-brain barrier (BBB) permeability. Our findings suggest that microbiota-produced N-acyl amides have the potential to cross the intestinal barrier and BBB and regulate various processes, including glucose homeostasis and immune cell differentiation via G protein-coupled receptor 132 (GPR132), sphingosine-1-phosphate receptor 4 (S1PR4), prostaglandin I2 receptor (PTGIR), prostaglandin E receptor 4 (PTGER4) expressed in intestinal, immune, and brain cells. Furthermore, these N-acyl amides stimulate enteroendocrine cells (EEC) via PTGER4, which induces the secretion of glucagon-like peptide 1 (GLP-1) (Cohen et al., 2015, 2017). We also hypothesize that microbiota-encoded dipeptides in the gut can be detected by the peptide transporter 1 (PEPT1), which is highly expressed on the microvilli of the enterocytes, and a calcium sensor receptor (CasR) localized the endocrine cells can stimulate the production of cholecystokinin (CCK) and GLP-1 (Sala-Rabanal et al., 2008; Xu et al., 2009). These hormones aid in stabilizing the intestinal barrier, regulating mucosal inflammatory responses, and preventing lipopolysaccharide-induced gut barrier dysfunction (Saia et al., 2020; Tang et al., 2015). Moreover, microbiota-transformed host molecules such as secondary bile acids activate pregnane X receptor (PXR), farnesoid X receptor (FXR), and takeda G protein-receptor-5 (TGR5, also known as G protein-coupled bile acid receptor 1 and membrane-type bile acid receptor), which play a critical role in immune tolerance (Dorrestein et al., 2021; Fiorucci et al., 2018; Mencarelli et al., 2009; Quinn et al., 2020). Interestingly, bile acids have shown both intestinal absorption and BBB permeability, suggesting the activation of these receptors in the brain by bile acids. Enterocytes have been shown to express these receptors, and upon activation, they can release fibroblast growth factor 19 (FGF19) and GLP-1, which are also capable of signaling the central nervous system (CNS). Short chain fatty acids (SCFAs) are another class of microbiota-transformed dietary molecules that interact with GPR41, GPR43, and GPR109A, which are expressed in enterocytes and immune cells and play a significant role in the gut-immune-brain pathway (Dalile et al., 2019; Rooks and Garrett, 2016). In addition, tryptophan catabolites can diffuse through the intestinal epithelium and modulate innate and adaptive immune responses via PXR, aryl hydrocarbon receptor (AhR), GPR35, and 5-hydroxytryptamine receptors (5-HT3R and 5-HT4R), expressed in the brain, enterocytes, mucus-secreting cells, dendritic cells and other immune cells (Agus et al., 2018; Cervenka et al., 2017). Dopamine regulates dopamine receptors (DRD1-4) (Rekdal et al., 2019; van Kessel et al., 2019), while phenethylamine (PEA) can readily cross the BBB and regulate mood via DRD2-4 (Chen et al., 2019). Histamine activates four histamine receptor subtypes (HRH1–4) (Barcik et al., 2017), which are highly expressed in the brain compared to epithelial and immune cells. We also report higher expression of gamma-aminobutyric acid (GABA) receptors in the brain and, to some extent, in endocrine and enterocytes. Unlike dopamine, systemic GABA can cross the BBB and directly regulate GABA receptors in the brain. GPR, G-protein coupled receptor; S1PR4, sphingosine-1-phosphate receptor 4; PTGIR, prostaglandin I2 receptor (PTGIR); PTGER4, prostaglandin E receptor 4; EEC, enteroendocrine cells; GLP-1, glucagon-like peptide 1; PEPT1, peptide transporter 1; CasR, calcium sensor receptor; CCK, cholecystokinin; PXR, pregnane X receptor; FXR, farnesoid X receptor; TGR5, Takeda G protein-receptor-5; FGF19, fibroblast growth factor 19; AhR, aryl hydrocarbon receptor; 5-HT3R and 5-HT4R, 5-hydroxytryptamine receptors; DRD1-4, dopamine receptors, HRH1–4, histamine receptors; GABA, gamma-aminobutyric acid.
Mounting evidence suggests that the aforementioned GPCRs regulate a broad range of biological processes, ranging from immune cell differentiation (S1PR4, PTGIR, PTGER4) and immune cell trafficking (S1PR4, G2A) to tissue repair (PTGIR). Notably, several disease models have shown implications of lipid-like GPCRs in the pathophysiology of obesity and diabetes (GPR119), colitis (S1PR4, PTGER4, PTGIR), autoimmunity (GPR132) and atherosclerosis (GPR132, PTGIR) (Flock et al., 2011; Kabashima et al., 2002; Manieri et al., 2015; Schulze et al., 2011).
1.2.2. Dipeptides
Recent paired genome- and metabolome-mining efforts have identified biosynthetic gene clusters encoding non-ribosomal peptide synthetases involved in the biosynthesis of structurally diverse dipeptides from the human microbiome (Aleti et al., 2022; Cao et al., 2019; Park and Crawford, 2016; Wyatt et al., 2012). However, many of these are uncharacterized. Dipeptides have been reported to play key roles in bacterial cell signaling and human neuroactive metabolism (Hatanaka et al., 2020; Kanegawa et al., 2010; Mizushige et al., 2020), and depletion of these have been previously associated with inflammatory bowel disease (Franzosa et al., 2019). In a recent multi-omics study, we identified for the first time to our knowledge several structurally distinct dipeptides in obesity-depression interrelationships, potentially derived from human, diet and microbes, of which Phe-Val (phenylalanine-valine) and Tyr-Val (tyrosine-valine) are of potential microbial origin (Aleti et al., 2022). Several animal studies have shown that aromatic amino acid containing dipeptides modulate serotonin, dopamine and gamma aminobutyric acid (GABA) metabolism in the brain (Kanegawa et al., 2010; Mizushige et al., 2020; Moriyasu et al., 2016). However, dipeptide interactions with human intestinal epithelial cells and the underlying mechanisms by which dipeptides could affect the gut-immune-brain axis remains to be established.
1.3. Human molecules transformed by microbes
1.3.1. Secondary bile acids
Microbial transformation of human bile acids has been extensively reviewed recently (Guzior and Quinn, 2021). Primary bile acids are synthesized from cholesterol within the liver (Hofmann, 1999). However, microbes in the gut are known to transform these into more chemically diverse secondary bile acids with expanded biological functions (Ridlon et al., 2006). Growing evidence suggests that bile plays a critical role in maintenance of the fine-tuned equilibrium between gut microbes, human immune response and metabolism (Brestoff and Artis, 2013; Fiorucci et al., 2018). The most abundant primary bile acids include cholic acid (CA), deoxycholic acid (DCA) and chenodeoxycholic acid (CDCA), which are conjugated with glycine or taurine before their secretion into the intestine (Guzior and Quinn, 2021). Gut microbiota transform the primary bile acids by several mechanisms such as deconjugation of the glycine and taurine through secretion of bile salt hydrolases, followed by dehydroxylation, dehydrogenation, and epimerization of the cholesterol core (Gérard, 2014; Guzior and Quinn, 2021; Quinn et al., 2020; Ridlon et al., 2014). Alterations in the chemistry and levels of secondary bile acids in the gut have been implicated in several diseases such as cirrhosis, inflammatory bowel disease and cancer (Duboc et al., 2013; Guzior and Quinn, 2021; Lleo et al., 2009). Recent studies discovered additional modification mechanisms wherein gut microbes have been shown to conjugate amino acids to primary bile acids resulting in conjugated bile acids (Dorrestein et al., 2021; Quinn et al., 2020). The most predominant amino acid conjugations were with threonine (Thr), glutamic acid (Glu), histidine (His), Phe, tryptophan (Trp), Tyr, lysine and isoleucine/leucine, although frequencies of these conjugations varied depending on the type of bile acid (Guzior and Quinn, 2021). Among these, Thr-CA, Glu-DCA, and Glu-CDCA are strong agonists of the pregnane X receptor (PXR) (Dorrestein et al., 2021), whereas Phe-CA and Tyr-CA are strong agonists of the farnesoid X receptor (FXR) (Fig. 1) (Quinn et al., 2020). Both PXR and FXR are identified as suppressors of macrophages (Fiorucci et al., 2018), dendritic cells and NK-cells (Mencarelli et al., 2009), which are crucial components of immune tolerance (Mencarelli et al., 2009).
1.4. Dietary components transformed by microbes
1.4.1. Short-chain fatty acids
Short chain fatty acids (SCFAs) such as acetate, propionate and butyrate are derived from fermentation of dietary fibers by anaerobes within the GI tract, and extensively studied for their role in gut-immune-brain communication (Dalile et al., 2019; Krautkramer et al., 2020; Rooks and Garrett, 2016). SCFAs are an important energy source for intestinal epithelial cells and therefore are absorbed by intestinal cells via both passive transportation and active transportation (e.g. monocarboxylate and sodium-coupled monocarboxylate transporters) (Sivaprakasam et al., 2017), which leads to inhibition of histone deacetylase in intestinal cells. SCFAs predominantly regulate three GPCRs—GPR41, GPR43 and GPR109A—localized on the intestinal epithelial and immune cells (Fig. 3) (Dalile et al., 2019; Rooks and Garrett, 2016). In a healthy gut, SCFAs are known to enhance epithelial barrier function and immune tolerance associated with gut homeostasis via multiple mechanisms (Dalile et al., 2019; Rooks and Garrett, 2016). For instance, SCFAs are reported to suppress the expression of nuclear factor-κB (NF-κB), stimulate mucus secreting cells for increased mucus production, and promote immunoglobulin secretion by B cells. SCFAs also regulate dendritic cells following T-cell polarization into Tregs, T helper (Th1), and Th17 cells, via cytokines such as transforming growth factor-β (TGF-β) and interleukin-10 (IL-10). SCFAs promote IL-18 production by epithelial cells, suppress proinflammatory cytokine (e.g. Tumor Necrosis Factor α (TNF-α), IL-6 and IL-1β) production by macrophages (Fig. 2). Further, SCFAs translocate to the brain and reduce neuroinflammation (Dalile et al., 2019). These underscore immunoregulatory roles of SCFAs and their therapeutic potential. In addition, SCFAs stimulate secretion of the gut hormones peptide YY (PYY), serotonin, CCK, GLP-1 from enteroendocrine cells (Dalile et al., 2019). In dysbiosis, SCFAs increase the production of proinflammatory cytokines and chemokines, such as (TNFα), IL6, CXCL1 (C-X-C Motif Chemokine Ligand 1), and CXCL10, as well as promote the production of T helper (Th) 1 and Th17 lymphocytes (Dalile et al., 2019; Rooks and Garrett, 2016) (Fig. 2).
Fig. 3.
RNA-Seq expression profiles of human receptors, mainly in intestinal cells, blood cells and brain tissues from human protein atlas. The red color intensity scale signifies a higher expression of the human receptor.
Therefore, SCFAs are implicated in a range of cardiometabolic diseases (obesity, atherosclerosis, non-alcoholic fatty liver disease, metabolic syndrome and type 2 diabetes) and neuropsychiatric disorders to a great degree in a recent literature (Parkinson's disease, Alzheimer disease, autism spectrum disorder, anxiety and depression) and cancer, to some extent (Krautkramer et al., 2020).
1.4.2. Tryptophan catabolites
The effects of gut microbial Trp metabolism on human physiology have been extensively reviewed elsewhere (Krautkramer et al., 2020; Roager and Licht, 2018). Briefly, metabolism of dietary proteins in the gut generates Trp, which is metabolized by at least three pathways—kynurenine, serotonin and protein synthesis pathways, which may be directly or indirectly regulated by gut microbiota. Trp catabolites (See Fig. 1), as signaling molecules and toxins, regulate a broad range of human functions (Krautkramer et al., 2020; Roager and Licht, 2018). Tryptamine stimulates serotonin production by enteroendocrine cells. Both tryptamine and serotonin (5-hydroxytryptamine) stimulate gut motility via serotonin type 3 receptor (5-HT3R) and serotonin type 4 receptor (5-HT4R) found on enterocytes (Bhattarai et al., 2018; Mawe and Hoffman, 2013). Moreover, serotonin signals enteric neurons via the serotonin transporter (SERT) (Mawe and Hoffman, 2013; Yano et al., 2015). Trp catabolites indole and indole derivatives e.g. indole propionic acid (IPA) and indoleacrylic acid (IA) have been shown to regulate mucosal homeostasis by reduced intestinal permeability via PXR (Fig. 2) (Krautkramer et al., 2020; Roager and Licht, 2018). Indole stimulates the release of GLP-1 by enteroendocrine cells, whereas its derivatives IPA and IA can regulate aryl hydrocarbon receptor (AhR) on immune cells and modulate innate and adaptive immune responses (Krautkramer et al., 2020; Roager and Licht, 2018). Indoxyl-sulfate (IS) generated in the liver from indole is reported to exhibit neurotoxicity (Roager and Licht, 2018). Kynurenine metabolites mediate mucosal homeostasis and human–microbiota immune tolerance via GPR35 (Agus et al., 2018; Cervenka et al., 2017). Another transporter GPR142 has been reported to be selectively expressed on the enteroendocrine cells, involved in sensing of the essential aromatic amino acids (e.g. Trp and Phe) and regulation of gut hormones (Lin et al., 2016) (See Fig. 2).
1.4.3. Tyrosine and phenylalanine metabolites
Gut microbiota have been shown to transform Phe and Tyr with their roles in human health and disease (Krautkramer et al., 2020). Gut microbes decarboxylate Phe or Tyr or levodopa (L-DOPA) to dopamine, a neurotransmitter that regulates movement and mood via dopamine receptors (DRD1-4), and which is implicated in Parkinson's disease (Rekdal et al., 2019; van Kessel et al., 2019).
Microbial-human co-metabolism of Tyr or Phe has been also shown to generate neurotoxic metabolites, 4-ethylphenyl sulfate (4EPS) and p-cresyl sulfate, which have been associated with neurodevelopmental disorders such as autism spectrum disorder and autoimmune disorders such as multiple sclerosis (Hsiao et al., 2013; Ntranos et al., 2022; Pascucci et al., 2020). However, the underlying molecular mechanisms of these neurotoxic metabolites in the gut-brain axis remain unclear. Culture-based approaches have confirmed the production of phenethylamine (PEA) from Phe, L-DOPA and Tyr. Recent microbiome-focused metabolomics approaches have confirmed gut microbial contribution in the production of additional Phe derivatives such as N-(cinnamoyl)glycine and hippuric acid, although their role in human health remains to be investigated (Han et al., 2021).
1.4.4. Histidine derivatives
The gut microbiota decarboxylate dietary-derived histidine to histamine, which plays a key role in mucosal immune response, via four histamine receptor subtypes (HRH1–4) (Barcik et al., 2017). Recently, another histidine derivative, imidazole propionate, was reported to impair insulin signaling via the mTORC1 signaling pathway and was associated with type-2 diabetes mellitus (Koh et al., 2018).
1.4.5. Gamma-aminobutyric acid
Gamma-aminobutyric acid (GABA) is produced by gut bacteria by decarboxylation of glutamate. GABA has been shown to reduce stress-induced corticosterone and plays an important role in anxiety and depression via GABA receptors (GABAR1-6) (Yunes et al., 2016). GABA is also reported to regulate intestinal barrier stabilization via CCK secretion by enterocytes, increase expression of tight junction proteins and increase mucus production by stimulating mucus secreting cells (Fig. 2) (Braun et al., 2015). GABA has been shown to suppress proinflammatory cytokine (e.g. IL-1β) production by macrophages and regulate dendritic cells via anti-inflammatory cytokines TGFβ (Bajic et al., 2019).
2. Methods
2.1. Analysis and integration of multi-omics data in this study
First, we selected small-molecules that were previously reported to bind with GPCRs. Second, we extracted spectral data of these molecules of interest from the reference spectral libraries available in GNPS public metabolomics data (Jarmusch et al., 2020; Wang et al., 2016b). Further, we utilized Mass Search Tool (MASST) and Reanalysis of Data User Interface (ReDU) (Jarmusch et al., 2020) to search these spectral data against public tandem mass spectrometry (MS/MS) data repositories. This provided a unique way to explore the presence of specific molecules across body sites and their association with health status. Next, by combining this with RNA-Seq expression and spatial localization of GPCRs from the publicly available human protein atlas database, we inferred potential molecular interactions regulating gut-immune-brain axis. In particular, we focused on gut microbial small-molecule interactions with human intestinal epithelial cells (e.g. enterocytes, endocrine and mucus-secreting cells), immune cells (e.g. macrophages, monocytes, T-cells, B-cells, NK-cells, neutrophils and dendritic cells) and the nervous system, to provide mechanistic insight into gut-immune-brain interactions. In this study, we examined a total of 34 receptors using transcriptomic evidence from the human protein atlas database (Thul and Lindskog, 2018). Of these, 18 receptors also had protein level evidence based on antibody detection, which includes GPR132, S1PR4, PTGER4, CasR, GPBAR1, HTR4, NR112, AhR, DRD1, DRD2, DRD5, HRH1, HRH2, GABRA1, GABRA2, GABRA3, GABRA5, and GABRB2 (Thul and Lindskog, 2018). Conversely, the expression of other 16 receptors, including GPR119, PTGIR, SLC15A1, GPR142, NR1H4, FFAR3, FFAR2, GPR35, DRD3, DRD4, HRH3, HRH4, GABRA4, GABRA6, GABRB1, and GABRB3, was supported by transcriptomic evidence (Thul and Lindskog, 2018). Furthermore, by computational analysis of intestinal absorption properties and blood-brain barrier (BBB) permeability for small-molecules, we evaluated potential interactions between the microbial molecules and specific human cell receptors. Finally, we identified specific GPCR-mediated microbial metabolite-human cell interactions that are shown to hold much promise for clinical translational potential, as direct targeting of these may achieve more efficacious and rapid processes. In the following section, we describe the key findings of the interactions between GPCR and microbial metabolites in the gut-immune-brain axis, which were identified in this study by integrating RNA-Seq data from human cell receptors and microbial metabolites from metabolomics.
2.2. Querying microbial molecule MS/MS spectra in GNPS public data
First, spectral data from molecules previously known to interact with GPCRs were extracted from the reference spectral libraries available in Global Natural Product Social Molecular Networking (GNPS) (Wang et al., 2016a). Next, using the metabolomics spectral search tool MASST (Wang et al., 2020), spectral data were searched for MS/MS matches against public metabolomics data from GNPS/MassIVE (mass spectrometry interactive virtual environment) data repositories to evaluate the occurrence of the specific molecule across 52,875 samples (as of March 2023). The resulting annotations obtained were at level 2 according to the 2007 metabolomics standards initiative (Sumner et al., 2007). Finally, by using the Reanalysis of Data User Interface (ReDU) infrastructure (Jarmusch et al., 2019), sample metadata was extracted for the GNPS/MASST analysis to link the presence of specific molecules with disease state, phenotype, biospecimen, and other metadata. The frequency of each molecule was calculated by counting its occurrence in a total of 52,875 samples. We then computed the corresponding percentage by dividing the number of occurrences by the total number of samples. A Heatmap was then generated based on the corresponding percentages of molecule occurrences with a column Z-score scaling method using the tidyverse package in R software (version 3.6.3) within RStudio (version February 1, 5019).
2.3. RNA-seq expression and spatial localization analysis of human receptors from human protein atlas
Protein-coding RNA-Seq data for a subset of human receptors of interest expressed in different tissue samples such as human intestinal epithelial cells (enterocytes, endocrine and mucus-secreting cells), immune cells (macrophages, monocytes, T-cells, B-cells, NK-cells, neutrophils, and dendritic cells) and different regions of brain tissue was extracted from the human protein atlas database version 21 (Thul and Lindskog, 2018). Next, we conducted a comparative analysis of the consensus gene expression levels of each human receptor across different tissues using transcriptomic data from two public databases, namely the Human Protein Atlas (HPA) and the Genotype-Tissue Expression Project (GTEx) (Lonsdale et al., 2013; Thul and Lindskog, 2018). The consensus normalized expression was determined as the maximum Transcripts Per Kilobase Million (nTPM) value for each gene in both datasets (Thul and Lindskog, 2018). Finally, a heatmap was generated on nTPM values with a row Z-score scaling method, representing gene expression data with the tidyverse package in R software (version 3.6.3) in RStudio (version February 1, 5019). In particular, we used the global scaling method (row Z-score scaling method) to calculate the mean and standard deviation across all expression values for all genes and transform the expression values by the global mean and standard deviation. This method allows for the comparison of expression levels between genes, as all genes are transformed in the same way. However, it is important to note that outliers in the sample, such as genes with very high expression, can skew the estimates of mean and standard deviation, which can then dominate the heatmap. Therefore, interpretation of the expression values in the heatmap were done with caution.
2.4. Evaluation of human blood-brain barrier and intestinal absorption properties of microbial molecules
To assess the human blood-brain barrier (BBB) and intestinal absorption properties of microbial molecules, SMILES (Simplified Molecular-Input Line-Entry System) for specific molecules were either downloaded from the PubChem database or generated based on the structure and queried via admetSAR tool against the ADMET (absorption, distribution, metabolism, excretion, and toxicity) structure and activity relationship database. This contained the well-curated ADMET profiles for more than 96,000 unique compounds with known absorption properties (Yang et al., 2019). In addition, prediction accuracy of the intestinal absorption and BBB permeability properties enabled prediction of the ADMET profiles even for novel microbial molecules.
3. Results and discussion
The identified human GPCR-microbial metabolite interactions for the gut-immune-brain pathway are described in the following.
3.1. GPCR-N-acyl amides interactions
As aforementioned, N-acyl amides have been shown to mimic human ligands and activate multitude of receptors belonging to the lipid-like GPCRs—GPR119, GPR132, S1PR4, PTGIR, and PTGER4 (Cohen et al., 2015, 2017). An overview of GPCR-N-acyl amides interactions with these receptors expressed on human gut, immune and brain cells, and their mechanisms of action on human functions, is displayed in Fig. 2. In this study, we evaluated the selective expression of these GPCRs from human intestinal epithelial cells (enterocytes, endocrine and mucus-secreting cells), immune cells (macrophages, monocytes, T-cells, B-cells, NK-cells, neutrophils and dendritic cells) and different brain regions based on the RNA-Seq data from the human protein atlas to identify the most predominantly expressed GPCRs (See Fig. 3). We found that GPR119 is poorly expressed, although GPR119 was previously reported as a key target for obesity and diabetes (Overton et al., 2008) (Fig. 3). GPR132 is highly expressed on immune cells such as dendritic and NK-cells, and to some extent on B-cells, macrophages and brain regions such as midbrain and corpus callosum. S1PR4 is highly expressed on most of the immune cells, however, with reduced expression on monocytes. While PTGER4 is expressed on immune and intestinal cells, PTGIR is expressed selectively on monocytes and dendritic cells (Fig. 3). In support of the potential biologic role of N-acyl amides, our in silico findings show that N-acyl amides have potential to cross the intestine barrier and BBB (predicted in Table 1), suggesting that N-acyl amides can diffuse through the intestinal barrier and seep into the bloodstream where they can regulate intestinal and blood cells or directly evoke GPCRs in the brain by diffusing through the BBB.
Table 1.
Evaluation of human blood-brain barrier (BBB) and intestinal absorption properties (or ADMET chemical profiles) of microbial small-molecules. As indicated, most of the microbiota-associated molecules have the potential to cross the intestinal epithelial layer as well as BBB, except kynurenic acid, dopamine and phenylethanolamine.
| Microbial metabolite ligands | Intestinal Absorption |
Blood-brain barrier permeability |
|||
|---|---|---|---|---|---|
| Absorption | Probability | Absorption | Probability | ||
| Microbiota-encoded | N-acyl serinol | Yes | 0.98 | Yes | 0.76 |
| N-acyl alanine | Yes | 0.9 | Yes | 0.93 | |
| N-acyl lysine/ornithine | Yes | 0.94 | Yes | 0.9 | |
| N-oxyacyl glutamine | Yes | 0.65 | Yes | 0.86 | |
| N-acyl glycine/commendamide | Yes | 0.74 | Yes | 0.84 | |
| Phe-Val | Yes | 0.91 | Yes | 0.59 | |
| Tyr-Val | Yes | 0.81 | Yes | 0.82 | |
| Microbial transformation of host molecules | Glycocholic acid | Yes | 0.95 | Yes | 0.84 |
| Phenylalanocholic acid | – | – | – | – | |
| Tyrosocholic acid | Yes | 0.93 | Yes | 0.71 | |
| Leucocholic acid | – | – | – | – | |
| Microbial transformation of dietary precursors | Acetate | Yes | 0.95 | Yes | 0.97 |
| Butyrate | Yes | 0.96 | Yes | 0.98 | |
| Propionate | Yes | 0.96 | Yes | 0.97 | |
| Tryptophan | Yes | 0.95 | Yes | 0.98 | |
| Tryptamine | Yes | 1 | Yes | 0.98 | |
| Serotonin | Yes | 1 | Yes | 0.98 | |
| Kynurenic acid | Yes | 0.98 | No | 0.55 | |
| Indole acetic acid | Yes | 1 | Yes | 0.99 | |
| Indole propionic acid | Yes | 1 | Yes | 0.98 | |
| Indolelactic acid | Yes | 1 | Yes | 0.87 | |
| Indoxyl sulfate | Yes | 1 | Yes | 0.87 | |
| Indoleacrylic acid | Yes | 1 | Yes | 0.97 | |
| P-cresol | Yes | 1 | Yes | 0.89 | |
| 4-ethylphenyl sulfate | Yes | 0.98 | Yes | 0.94 | |
| Dopamine | Yes | 0.96 | No | 0.84 | |
| Histamine | Yes | 0.98 | Yes | 0.86 | |
| Phenylalanine | Yes | 0.97 | Yes | 0.59 | |
| N-Cinnamoylglycine | Yes | 0.97 | Yes | 0.93 | |
| Hippuric acid | Yes | 0.94 | Yes | 0.93 | |
| Phenylethanolamine | Yes | 0.98 | No | 0.56 | |
| Gamma-aminobutyric acid | Yes | 0.93 | Yes | 0.91 | |
By querying the spectral data from these molecules of interest against 52,875 samples from GNPS public metabolomics data (Wang et al., 2016a), we evaluated the presence of specific molecules across human tissue and body fluid types associated with disease conditions (Fig. 4A and B). We found that commendamide is enriched in feces and colon samples associated with sleep deprivation and circadian rhythm disorders in comparison to healthy or no disease reported samples, warranting further investigation. Due to the absence of reference spectra for N-acyl amides other than commendamide in the public metabolomics data repository, we refrained from searching for other N-acyl amides. Therefore, it is imperative to synthesize these molecules and obtain reference MS/MS spectra to facilitate their identification in future studies.
Fig. 4.
Representation of microbial molecule MS/MS spectra in GNPS public data. A) Occurrence of spectral matches for each molecule found across tissue and body fluid types. B) Proportion of spectral matches for each molecule detected across disease conditions. The red color intensity scale signifies a higher expression of the human receptor.
3.2. GPCR-dipeptides interactions
In a recent multi-omics study we identified aromatic amino acid containing dipeptides Phe-Val and Tyr-Val potentially derived from microbiota (Aleti et al., 2022). However, their interactions with human intestinal epithelial cells and their effect on gut-immune-brain axis is not known. Given that selective expression of specific receptors on the intestinal epithelial and endocrine cells is responsible for the uptake and active transportation of the dietary peptides, we speculate that it is likely to be the case of microbial peptides as well as the possibility of passive transportation through the intestinal barrier and BBB (See Fig. 2).
Proteins in the human gut are sensed by a proton-dependent amino-acid transporter PEPT1, which is crucially involved in active transportation of the dietary oligo peptides (2–4 amino acids), predominantly dipeptides from the small intestinal lumen (Sala-Rabanal et al., 2008; Xu et al., 2009). In agreement with our findings, RNA-Seq analysis revealed PEPT1 is highly expressed on the enterocytes and has been shown to be localized on the microvilli of the enterocytes mediating oligopeptide uptake by enterocytes (Fig. 2, Fig. 3). A calcium sensor CasR, reported to be localized on the basolateral membrane of the enterocytes and endocrine cells, albeit expressed only on endocrine cells as indicated in Fig. 3, is also involved in sensing amino acids and oligo-peptides in conjunction with PEPT1 (Diakogiannaki et al., 2013; Husted et al., 2017). While CasR stimulates the secretion of cholecystokinin (CCK) and GLP-1 by enteroendocrine cells, it regulates nutrient uptake and intestinal barrier stabilization by enterocytes’ CCK secretion. In particular, CCK regulates the mucosal inflammatory response and prevents the gut barrier dysfunction induced by lipopolysaccharide (Saia et al., 2020; Tang et al., 2015).
Dipeptides exhibited intestinal absorption properties and BBB permeability (Table 1), suggesting that dipeptides can cross the intestinal barrier by passive transportation and indirectly affect the CNS via enteric neuron modulation by GLP-1 release or directly to some extent via PEPT1 and CasR receptors in the BBB and brain. We found that Phe-Val is mainly enriched in saliva, gastrointestinal (GI) tract and brain samples associated with obesity and circadian rhythm disorders, whereas high levels of Tyr-Val are found in urine, feces, GI tract and brain of healthy individuals (Fig. 4A and B).
3.3. GPCR, PXR, and FXR-bile acids interactions
While most primary bile acid is reabsorbed within the ileum for reuse, the remaining 5% activate bile acid receptors such as PXR, FXR and TGR5 (GPBAR1) in the liver and GI tract (Sepe et al., 2016). The absorption of bile acids is in line with our in silico prediction of intestinal absorption properties for the bile acids (Table 1). Notably, we also found that bile acids exhibit BBB permeability, suggesting that bile acids not only act in the gut, but also enter the systemic circulation and may evoke PXR, FXR and TGR5 receptors in the brain directly. In the gut, activation of intestinal PXR, FXR and TGR5 receptors can result in the release of fibroblast growth factor 19 (FGF19) and GLP-1, both capable of signaling the CNS. Based on the RNA-Seq analysis of GPCRs, TGR5 is highly expressed on immune cell subsets such as dendritic cells and enteroendocrine cells of the gut. Previous studies have shown that TGR5 is also abundantly expressed on enteric neurons (RNA-Seq data not available for enteric neurons) (Fig. 2, Fig. 3). Bile acid receptors PXR, FXR and TGR5 are reported as a negative regulator of macrophages (Fiorucci et al., 2018), dendritic cells and NK-cells (Mencarelli et al., 2009), contributing to immune tolerance in the liver and intestine (Mencarelli et al., 2009). Recently, Campbell et al. identified bile acids as immunosuppressive agents altering dendritic cell-mediated T-cell stimulation, by decreasing dendritic cell costimulatory molecules expression and proinflammatory cytokine production, which in turn promotes generation of FoxP3+ T-regulatory cells (Tregs) (Campbell et al., 2020). Tyr-CA is found in GI tract samples associated with ulcerative colitis, whereas high levels of Phe-CA are detected in urine and GI tract samples of ulcerative colitis, Crohn's disease and obesity (Fig. 4A and B). Although these findings expand the diversity of microbial bile acids, their implications on human physiology and health remain largely unknown, warranting new avenues in human-microbe communication research. The absence of reference spectra for other bile acids in public metabolomics data repositories highlights the need for further synthesis and acquisition of reference spectra to support future studies.
3.4. GPCR-SCFAs interactions
Short chain fatty acids (SCFAs) are known to play a key role in gut-immune-brain communication (Dalile et al., 2019; Krautkramer et al., 2020; Rooks and Garrett, 2016). Corroborating the intestinal absorption findings (as predicted in Table 1), SCFAs are absorbed by intestinal cells via both passive transportation and active transportation, leading to suppression of histone deacetylase in intestinal cells (Sivaprakasam et al., 2017). SCFAs have been shown to interact with GPCRs—GPR41, GPR43 and GPR109A—localized on the intestinal epithelial and immune cells (Dalile et al., 2019; Rooks and Garrett, 2016). Based on RNA-Seq analysis, while GPR41 appears to be poorly expressed, GPR43 and GPR109A are highly expressed on the neutrophil, endocrine cells and midbrain (Fig. 3). Further, SCFAs translocate to the brain and reduce neuroinflammation (corroborating the BBB permeability findings of Table 1) (Dalile et al., 2019), underscoring their role in gut-immune-brain pathway and their therapeutic use. We did not detect SCFAs in the metabolomics data due to their lower mass range in mass spectra, which contained a high number of interfering peaks from solvents and additives.
3.5. GPCR, PXR, AhR-tryptophan catabolite interactions
In agreement with the existing evidence, except kynurenic acid all Trp catabolites displayed strong intestinal absorption and BBB permeability (Table 1) (Krautkramer et al., 2020; Roager and Licht, 2018). Tryptamine and serotonin are known to activate receptors 5-HT3R and 5-HT4R (Bhattarai et al., 2018; Mawe and Hoffman, 2013), and further RNA-Seq analysis in our study revealed that these receptors are expressed in the basal ganglia in the brain and in the intestinal and immune cells to some extent (See Fig. 2). Trp catabolites have been shown to reduce intestinal permeability via PXR, which is highly expressed on the enterocytes and NK-cell (Fig. 2, Fig. 3) (Krautkramer et al., 2020; Roager and Licht, 2018). Indole derivatives IPA and IA can diffuse through the intestinal epithelium and regulate aryl hydrocarbon receptor (AhR) on immune cells (macrophages and dendritic cells; see Fig. 3) and modulate innate and adaptive immune responses. Kynurenine metabolites regulate mucosal homeostasis and human–microbiota immune tolerance via GPR35 (Agus et al., 2018; Cervenka et al., 2017), which is expressed on dendritic, enterocytes and mucus-secreting cells as shown in Fig. 3. Another transporter GPR142 previosuly reported to be selectively expressed on the enteroendocrine cells, involved in sensing of the essential aromatic amino acids and regulation of gut hormones, is poorly expressed on the intestinal cells (Lin et al., 2016) (Fig. 3). Our examination indicates high levels of Trp in blood, colon, liver, and gallbladder samples associated with depression (Fig. 4A and B). However, this is in contrast to previous studies, wherein lower Trp availability or metabolism and kynurenine pathway dysregulation has been associated with depression, prompting further determination of absolute concentration (Chen et al., 2021; Hunt et al., 2020). Indoxyl sulfate is found in nearly all the tissues and body fluids examined and appears to be enriched in COVID-19 infection as well as healthy samples (Fig. 4A and B). The public metabolomics samples did not reveal any tryptophan catabolites other than Trp and indoxyl sulfate, indicating that the LC-MS method used in these studies may not be optimal for detection of other tryptophan catabolites.
3.6. GPCR-tyrosine and phenylalanine metabolite interactions
Gut microbiota convert Phe, Tyr, and L-DOPA into dopamine, a neurotransmitter that regulates dopamine receptors (DRD1-4) (Rekdal et al., 2019; van Kessel et al., 2019).. Although RNA-Seq analysis in our study revealed that dopamine receptors are highly expressed in most of the brain regions (Fig. 3), systemic dopamine cannot pass the BBB (Table 1), and thus gut microbial potential for dopamine synthesis has been extensively studied with regard to the bioavailability of L-DOPA in Parkinson's disease and for the adverse effects of dopamine (Rekdal et al., 2019; van Kessel et al., 2019). Although alteration of the neuroactive potential of the gut microbiota is linked with variation in neurotransmitter levels in the brain (Valles-Colomer et al., 2019), it remains to be seen if neurotransmitters, particularly dopamine in the intestine, can evoke the enteric dopaminergic system. Furthermore, culture-based methods have validated the biosynthesis of PEA from Phe, L-DOPA and Tyr. PEA readily crosses the BBB (Table 1) and regulates mood via DRD2-4 (Chen et al., 2019). The absence of any tyrosine and phenylalanine metabolites in the public metabolomics samples suggests that the LC-MS method used in these studies may not be optimal for detection of these metabolites.
3.7. GPCR interactions with histidine and GABA
Gut microbiota convert dietary histidine to histamine, which can activate four histamine receptor subtypes (Barcik et al., 2017). We found greater expression of histamine receptors (HRH1–4) in the brain than epithelial and immune cells (Fig. 3). We report higher expression of GABA receptors in the brain and, to some extent, in endocrine and enterocytes (Fig. 3). Unlike dopamine, systemic GABA can cross the BBB (Table 1) and directly regulate GABA receptors in the brain. Based on the public metabolomics samples, the lack of histidine and GABA molecules indicates that the LC-MS method utilized in these studies may not be the most suitable for detecting these particular metabolites.
Overall, it is important to note that there is a higher overlap in the expression profiles of these receptors in the intestine and gut cells compared to immune cells, suggesting their role in the gut-brain connection (as shown in Fig. 3).
3.8. Approaches to enable surveillance of human microorganism interactions through GPCRs
Although “omics'’ approaches provide unparalleled associations between changes in the microbiome or metabolome and health, they are limited in their ability to reveal the mechanistic details of how the microbiome or metabolome might modulate human physiology. In this case, animal models have demonstrated the ability to link a particular metabolite to organism-level phenotypes. However, it is crucial to identify the human target to gain a better understanding of the underlying mechanism of action. To this end, the luciferase reporter gene-based cell assay that facilitates simultaneous profiling of 315 human GPCR targets via a G protein-independent β-arrestin recruitment assay has been currently benchmarked (Chen et al., 2019; Kroeze et al., 2015).
As there are several types of G proteins based on their alpha-subunit and each GPCR initiates a distinct downstream signaling, screening many molecules across the entire collection of GPCRs to identify potential agonists or antagonists is a cumbersome task and demands multipronged assays. However, β-arrestins have been shown to directly interact with nearly every GPCR. In this regard, GPCR-β-arrestin interactions can be utilized as a streamlined readout for identification of agonists across the entire repertoire of GPCRs. Previously, Chen et al. (2019) used a PRESTO-Tango assay, a transcriptional outcome-based β-arrestin recruitment assay (Kroeze et al., 2015) to screen the microbial metabolites against the entire collection of non-olfactory GPCRs (Chen et al., 2019; Kroeze et al., 2015). Although PRESTO-Tango can be used to study the interactions between GPCRs and their ligands, this method has some limitations, such as not being time-resolved and having a delayed measurement, leading to low signal-to-noise ratios for some receptors. As a result, alternative methods such as BRET-based assays have been developed in recent years to dissect receptor signaling through canonical G protein coupling (Avet et al., 2022; Namkung et al., 2016). It is important to address these limitations, particularly when conducting experimental follow-up for beta-arrestin recruitment and the dissection of downstream signaling pathways. The BRET-based assays allow for the measurement of protein-protein interactions in real-time, which improves the accuracy and reliability of the results, and thus enables us to study the activation of GPCRs and their downstream signaling at the sub-cellular level (Avet et al., 2022).
Thus, the PRESTO-Tango and BRET-based assays will enable simultaneous interrogation of small-molecules across the targetable human GPCRome, a useful framework for integration of metabolite cataloging studies with mechanistic studies for an improved understanding of microbiota-human molecular interactions and provide new avenues for developing novel microbial small-molecule therapeutics. It will be possible to better understand potential mechanistic details of the molecular interactions, particularly among gut microbes, intestinal epithelial cells, immune cells and the nervous system for the discovery of therapeutic compounds.
4. Conclusions
Our findings provide novel insights into the most abundant human-microbe interactions with clinical implications and show that GPCRs are expressed widely across organ systems but selectively on specific human cells, including immune, enteric and brain cells, offering clues to gut-immune-brain interactions. The expression and activity of GPCRs undergo changes throughout development and aging, which can significantly impact health and disease across the life course. Therefore, it is crucial to investigate these changes in GPCR expression profiles to gain a better understanding of their role in the gut-immune-brain axis and their potential contribution to various physiological and pathological processes. The microbial small-molecules we have described in this study play a role in human biology and a range of diseases such as metabolic, neuropsychiatric and inflammatory conditions mediated by GPCRs. Our findings of GPCR and bacterial metabolite interactions in conjunction with their localization and abundance profiles reveal that N-acyl amides and microbial peptidic molecules, particularly derived from non-ribosomal peptide synthetases, and other well-known microbial molecules such as secondary bile acids and recent microbial-human co-metabolism of tryptophan, tyrosine and phenylalanine, are highly relevant in health versus disease. Further, these may play a key role in the gut-immune-brain axis, warranting further investigation.
In addition, to date about 172 potential microbial metabolites are reported, however, only a handful of the microbial molecules reviewed in this study have been investigated against human receptors directly (Wishart et al., 2022). In addition, only about 10% of molecular spectra from human metabolomes can be annotated, leaving the rest as non-assignable or dark matter—many molecules are still awaiting discovery, and their effect on human receptors and physiology remains unexplored. In this regard, we speculate that several uncharacterized bacterial metabolites may bind GPCRs and may contribute to ‘deorphanization’ of the so far untapped trove of GPCRs. Such metabolites and their human interactions may offer promising targets for microbiome therapeutics and therefore open novel avenues for prevention and intervention of human diseases.
Declaration of competing interest
None.
Acknowledgements
Aleti was supported by Wayne State University Endowment Fund (Hong) and the Kavli Institute for Brain and Mind (KIBM) Innovative Research Grant (Aleti). Hong was supported by NIH R01-HL90975 and R01-90975S1. Troyer was supported by a trainee fellowship from NIMH T32-MH018399.
Data availability
RNA-Seq expression profiles from all human receptors used in this study, and mass spectral profiles for each molecule found across tissue and body fluid types, code/scripts, and data underlying heatmap figures are publicly available from the GitHub repository (https://github.com/gajenderaleti/GPCR-MicrobialMetaboliteInteractions). In addition, MASST analysis for each molecule are available at Global Natural Product Social Molecular Networking (GNPS) with the following task IDs: GABA: 87b3aac5b86b4124be0cb4aaa6679d11; Hippuric acid: df76ea23ecd54b988f1116321dd4ae46; Phenylathylamine: 6a4ed755c33f4df3bb71dbe629bdeac5; Phe: 4e52e1644a224971ab4f4749744e6ed8; Histamine: e7f992ade5234b63906e1b8a4d3a12c0; Dopamine: 27df5f82bf074195bdccf75f82398fb2; p-cresol sulfate: 8c99e2bae67f4dc09156f84bfe417970; Indoleacrylic acid: ef7e85bf5d324613887e7d026cc56212; Indoxylsulfate: e65dfe111f8943169f39b639a8bc53c8; Indolepropionic acid: d38f7ce18898460bb64f476d90f5c62a; Indolelactic acid: e083444bfa104fed9394943011b7e563; Indoleacetic acid: e8bfa90bfb184205a862444d150653e3; Tryptophan: f18c2a7c189a4d1b9d698c6bff526a5d; Tyrptamine: dc36f9351f4a442ba185d10b0d481739; kynurenic acid: 02a8416a52f8463e9cb7aeb4088d9863; Serotonin: af3282ea885243a9b631f60c5d6b0841 Tyrosine cholic acid: 0e486c6ecd9f42f08dd466e67d6997c8; Phenylalanine cholic acid: 3188dfca7a814e30b46f534f8ffdcb53; Glutamic acid chenodeoxy cholic acid: 6beb245285ad4926aa8d19c514878319; Commendamide: 2214cf1de9a54e419fc6a527cd87bd48; Tyr-Val: 3956099739214030bc76dd95dd1bf894; Phe-Val: 1f1269480de44f378f5b469bc3b28b0aRNA-Seq expression profiles from all human receptors used in this study, and mass spectral profiles for each molecule found across tissue and body fluid types, code/scripts, and data underlying heatmap figures are publicly available from the GitHub repository (https://github.com/gajenderaleti/GPCR-MicrobialMetaboliteInteractions). In addition, MASST analysis for each molecule are available at Global Natural Product Social Molecular Networking (GNPS) with the following task IDs: GABA: 87b3aac5b86b4124be0cb4aaa6679d11 Hippuric acid: df76ea23ecd54b988f1116321dd4ae46 Phenylathylamine: 6a4ed755c33f4df3bb71dbe629bdeac5 Phe: 4e52e1644a224971ab4f4749744e6ed8 Histamine: e7f992ade5234b63906e1b8a4d3a12c0 Dopamine: 27df5f82bf074195bdccf75f82398fb2 p-cresol sulfate: 8c99e2bae67f4dc09156f84bfe417970 Indoleacrylic acid: ef7e85bf5d324613887e7d026cc56212 Indoxylsulfate: e65dfe111f8943169f39b639a8bc53c8 Indolepropionic acid: d38f7ce18898460bb64f476d90f5c62a Indolelactic acid: e083444bfa104fed9394943011b7e563 Indoleacetic acid: e8bfa90bfb184205a862444d150653e3 Tryptophan: f18c2a7c189a4d1b9d698c6bff526a5d Tyrptamine: dc36f9351f4a442ba185d10b0d481739 kynurenic acid: 02a8416a52f8463e9cb7aeb4088d9863 Serotonin: af3282ea885243a9b631f60c5d6b0841 Tyrosine cholic acid: 0e486c6ecd9f42f08dd466e67d6997c8 Phenylalanine cholic acid: 3188dfca7a814e30b46f534f8ffdcb53 Glutamic acid chenodeoxy cholic acid: 6beb245285ad4926aa8d19c514878319 Commendamide: 2214cf1de9a54e419fc6a527cd87bd48 Tyr-Val: 3956099739214030bc76dd95dd1bf894 Phe-Val: 1f1269480de44f378f5b469bc3b28b0aRNA-Seq expression profiles from all human receptors used in this study, and mass spectral profiles for each molecule found across tissue and body fluid types, code/scripts, and data underlying heatmap figures are publicly available from the GitHub repository (https://github.com/gajenderaleti/GPCR-MicrobialMetaboliteInteractions). In addition, MASST analysis for each molecule are available at Global Natural Product Social Molecular Networking (GNPS) with the following task IDs: GABA: 87b3aac5b86b4124be0cb4aaa6679d11 Hippuric acid: df76ea23ecd54b988f1116321dd4ae46 Phenylathylamine: 6a4ed755c33f4df3bb71dbe629bdeac5 Phe: 4e52e1644a224971ab4f4749744e6ed8 Histamine: e7f992ade5234b63906e1b8a4d3a12c0 Dopamine: 27df5f82bf074195bdccf75f82398fb2 p-cresol sulfate: 8c99e2bae67f4dc09156f84bfe417970 Indoleacrylic acid: ef7e85bf5d324613887e7d026cc56212 Indoxylsulfate: e65dfe111f8943169f39b639a8bc53c8 Indolepropionic acid: d38f7ce18898460bb64f476d90f5c62a Indolelactic acid: e083444bfa104fed9394943011b7e563 Indoleacetic acid: e8bfa90bfb184205a862444d150653e3 Tryptophan: f18c2a7c189a4d1b9d698c6bff526a5d Tyrptamine: dc36f9351f4a442ba185d10b0d481739 kynurenic acid: 02a8416a52f8463e9cb7aeb4088d9863 Serotonin: af3282ea885243a9b631f60c5d6b0841 Tyrosine cholic acid: 0e486c6ecd9f42f08dd466e67d6997c8 Phenylalanine cholic acid: 3188dfca7a814e30b46f534f8ffdcb53 Glutamic acid chenodeoxy cholic acid: 6beb245285ad4926aa8d19c514878319 Commendamide: 2214cf1de9a54e419fc6a527cd87bd48 Tyr-Val: 3956099739214030bc76dd95dd1bf894 Phe-Val: 1f1269480de44f378f5b469bc3b28b0aRNA-Seq expression profiles from all human receptors used in this study, and mass spectral profiles for each molecule found across tissue and body fluid types, code/scripts, and data underlying heatmap figures are publicly available from the GitHub repository (https://github.com/gajenderaleti/GPCR-MicrobialMetaboliteInteractions). In addition, MASST analysis for each molecule are available at Global Natural Product Social Molecular Networking (GNPS) with the following task IDs: GABA: 87b3aac5b86b4124be0cb4aaa6679d11 Hippuric acid: df76ea23ecd54b988f1116321dd4ae46 Phenylathylamine: 6a4ed755c33f4df3bb71dbe629bdeac5 Phe: 4e52e1644a224971ab4f4749744e6ed8 Histamine: e7f992ade5234b63906e1b8a4d3a12c0 Dopamine: 27df5f82bf074195bdccf75f82398fb2 p-cresol sulfate: 8c99e2bae67f4dc09156f84bfe417970 Indoleacrylic acid: ef7e85bf5d324613887e7d026cc56212 Indoxylsulfate: e65dfe111f8943169f39b639a8bc53c8 Indolepropionic acid: d38f7ce18898460bb64f476d90f5c62a Indolelactic acid: e083444bfa104fed9394943011b7e563 Indoleacetic acid: e8bfa90bfb184205a862444d150653e3 Tryptophan: f18c2a7c189a4d1b9d698c6bff526a5d Tyrptamine: dc36f9351f4a442ba185d10b0d481739 kynurenic acid: 02a8416a52f8463e9cb7aeb4088d9863 Serotonin: af3282ea885243a9b631f60c5d6b0841 Tyrosine cholic acid: 0e486c6ecd9f42f08dd466e67d6997c8 Phenylalanine cholic acid: 3188dfca7a814e30b46f534f8ffdcb53 Glutamic acid chenodeoxy cholic acid: 6beb245285ad4926aa8d19c514878319 Commendamide: 2214cf1de9a54e419fc6a527cd87bd48 Tyr-Val: 3956099739214030bc76dd95dd1bf894 Phe-Val: 1f1269480de44f378f5b469bc3b28b0aRNA-Seq expression profiles from all human receptors used in this study, and mass spectral profiles for each molecule found across tissue and body fluid types, code/scripts, and data underlying heatmap figures are publicly available from the GitHub repository (https://github.com/gajenderaleti/GPCR-MicrobialMetaboliteInteractions). In addition, MASST analysis for each molecule are available at Global Natural Product Social Molecular Networking (GNPS) with the following task IDs: GABA: 87b3aac5b86b4124be0cb4aaa6679d11 Hippuric acid: df76ea23ecd54b988f1116321dd4ae46 Phenylathylamine: 6a4ed755c33f4df3bb71dbe629bdeac5 Phe: 4e52e1644a224971ab4f4749744e6ed8 Histamine: e7f992ade5234b63906e1b8a4d3a12c0 Dopamine: 27df5f82bf074195bdccf75f82398fb2 p-cresol sulfate: 8c99e2bae67f4dc09156f84bfe417970 Indoleacrylic acid: ef7e85bf5d324613887e7d026cc56212 Indoxylsulfate: e65dfe111f8943169f39b639a8bc53c8 Indolepropionic acid: d38f7ce18898460bb64f476d90f5c62a Indolelactic acid: e083444bfa104fed9394943011b7e563 Indoleacetic acid: e8bfa90bfb184205a862444d150653e3 Tryptophan: f18c2a7c189a4d1b9d698c6bff526a5d Tyrptamine: dc36f9351f4a442ba185d10b0d481739 kynurenic acid: 02a8416a52f8463e9cb7aeb4088d9863 Serotonin: af3282ea885243a9b631f60c5d6b0841 Tyrosine cholic acid: 0e486c6ecd9f42f08dd466e67d6997c8 Phenylalanine cholic acid: 3188dfca7a814e30b46f534f8ffdcb53 Glutamic acid chenodeoxy cholic acid: 6beb245285ad4926aa8d19c514878319 Commendamide: 2214cf1de9a54e419fc6a527cd87bd48 Tyr-Val: The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). https://doi.org/10.1038/nature11234
References
- Agus A., Planchais J., Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Aleti G., Baker J.L., Tang X., Alvarez R., Dinis M., Tran N.C., Melnik A.V., Zhong C., Ernst M., Dorrestein P.C., Edlund A. Identification of the bacterial biosynthetic gene clusters of the oral microbiome illuminates the unexplored social language of bacteria during health and disease. mBio. 2019;10:1–19. doi: 10.1128/MBIO.00321-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleti G., Kohn J.N., Troyer E.A., Weldon K., Huang S., Tripathi A., Dorrestein P.C., Swafford A.D., Knight R., Hong S. Salivary bacterial signatures in depression-obesity comorbidity are associated with neurotransmitters and neuroactive dipeptides. BMC Microbiol. 2022;22 doi: 10.1186/S12866-022-02483-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avet C., Mancini A., Breton B., Gouill C.L., Hauser A.S., Normand C., Kobayashi H., Gross F., Hogue M., Lukasheva V., St-Onge S., Carrier M., Heroux M., Morissette S., Fauman E., Fortin J.P., Schann S., Leroy X., Gloriam D.E., Bouvier M. Effector membrane translocation biosensors reveal G protein and Parrestin coupling profiles of 100 therapeutically relevant GPCRs. Elife. 2022;11 doi: 10.7554/ELIFE.74101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic S.S., Djokic J., Dinic M., Veljovic K., Golic N., Mihajlovic S., Tolinacki M. GABA-producing natural dairy isolate from artisanal zlatar cheese attenuates gut inflammation and strengthens gut epithelial barrier in vitro. Front. Microbiol. 2019;10 doi: 10.3389/FMICB.2019.00527/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcik W., Wawrzyniak M., Akdis C.A., O'Mahony L. Immune regulation by histamine and histamine-secreting bacteria. Curr. Opin. Immunol. 2017;48:108–113. doi: 10.1016/J.COI.2017.08.011. [DOI] [PubMed] [Google Scholar]
- Bhattarai Y., Williams B.B., Battaglioli E.J., Whitaker W.R., Till L., Grover M., Linden D.R., Akiba Y., Kandimalla K.K., Zachos N.C., Kaunitz J.D., Sonnenburg J.L., Fischbach M.A., Farrugia G., Kashyap P.C. Gut microbiota produced tryptamine activates an epithelial G-protein coupled receptor to increase colonic secretion. Cell Host Microbe. 2018;23:775. doi: 10.1016/J.CHOM.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun H.S., Sponder G., Pieper R., Aschenbach J.R., Deiner C. GABA selectively increases mucin-1 expression in isolated pig jejunum. Genes & Nutrition. 2015;10 doi: 10.1007/S12263-015-0497-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff J.R., Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 2013;14:676. doi: 10.1038/NI.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C., McKenney P.T., Konstantinovsky D., Isaeva O.I., Schizas M., Verter J., Mai C., Jin W.B., Guo C.J., Violante S., Ramos R.J., Cross J.R., Kadaveru K., Hambor J., Rudensky A.Y. Bacterial metabolism of bile acids promotes peripheral Treg cell generation. Nature. 2020;581:475. doi: 10.1038/S41586-020-2193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Shcherbin E., Mohimani H. A metabolome- and metagenome-wide association network reveals microbial natural products and microbial biotransformation products from the human microbiota. mSystems. 2019;4 doi: 10.1128/msystems.00387-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervenka I., Agudelo L.Z., Ruas J.L. Kynurenines: tryptophan's metabolites in exercise, inflammation, and mental health. Science. 2017;357 doi: 10.1126/SCIENCE.AAF9794/ASSET/42BAD7E6-DCAB-4B50-9BCB-6550F5B016B6/ASSETS/GRAPHIC/357_AAF9794_FA.JPEG. [DOI] [PubMed] [Google Scholar]
- Chen H., Nwe P.K., Yang Y., Rosen C.E., Bielecka A.A., Kuchroo M., Cline G.W., Kruse A.C., Ring A.M., Crawford J.M., Palm N.W. A forward chemical genetic screen reveals gut microbiota metabolites that modulate host physiology. Cell. 2019;177:1217. doi: 10.1016/J.CELL.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.M., Bao C.H., Wu Y., Liang S.H., Wang D., Wu L.Y., Huang Y., Liu H.R., Wu H.G. Tryptophan-kynurenine metabolism: a link between the gut and brain for depression in inflammatory bowel disease. J. Neuroinflammation. 2021;18(1 18):1–13. doi: 10.1186/S12974-021-02175-2. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen L.J., Esterhazy D., Kim S.H., Lemetre C., Aguilar R.R., Gordon E.A., Pickard A.J., Cross J.R., Emiliano A.B., Han S.M., Chu J., Vila-Farres X., Kaplitt J., Rogoz A., Calle P.Y., Hunter C., Bitok J.K., Brady S.F. Commensal bacteria produce GPCR ligands that mimic human signaling molecules. Nature. 2017;549:48. doi: 10.1038/NATURE23874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen L.J., Kang H.S., Chu J., Huang Y.H., Gordon E.A., Reddy B.V.B., Ternei M.A., Craig J.W., Brady S.F. Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E4825–E4834. doi: 10.1073/PNAS.1508737112/-/DCSUPPLEMENTAL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo D.A., Kohn J.A., Luo P.M., Piscotta F.J., Han S.M., Pickard A.J., Rao A., Cross J.R., Cohen L.J., Brady S.F. Mapping interactions of microbial metabolites with human G-protein-coupled receptors. Cell Host Microbe. 2019;26:273. doi: 10.1016/J.CHOM.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalile B., Van Oudenhove L., Vervliet B., Verbeke K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019;16(8 16):461–478. doi: 10.1038/s41575-019-0157-3. 2019. [DOI] [PubMed] [Google Scholar]
- Devane W.A., Hanuš L., Breuer A., Pertwee R.G., Stevenson L.A., Griffin G., Gibson D., Mandelbaum A., Etinger A., Mechoulam R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–1949. doi: 10.1126/SCIENCE.1470919. [DOI] [PubMed] [Google Scholar]
- Diakogiannaki E., Pais R., Tolhurst G., Parker H.E., Horscroft J., Rauscher B., Zietek T., Daniel H., Gribble F.M., Reimann F. Oligopeptides stimulate glucagon-like peptide-1 secretion in mice through proton-coupled uptake and the calcium-sensing receptor. Diabetologia. 2013;56:2688. doi: 10.1007/S00125-013-3037-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia M.S., Fischbach M.A. Small molecules from the human microbiota. Science (New York, N.Y.) 2015;349 doi: 10.1126/SCIENCE.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrestein P., Gentry E., Collins S., Panitchpakdi M., Belda-Ferre P., Stewart A., Wang M., Jarmusch A., Avila-Pacheco J. A synthesis-based reverse metabolomics approach for the discovery of chemical structures from humans and animals. 2021. [DOI]
- Duboc H., Rajca S., Rainteau D., Benarous D., Maubert M.A., Quervain E., Thomas G., Barbu V., Humbert L., Despras G., Bridonneau C., Dumetz F., Grill J.P., Masliah J., Beaugerie L., Cosnes J., Chazouillères O., Poupon R., Wolf C., Mallet J.M., Langella P., Trugnan G., Sokol H., Seksik P. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–539. doi: 10.1136/GUTJNL-2012-302578. [DOI] [PubMed] [Google Scholar]
- Fiorucci S., Biagioli M., Zampella A., Distrutti E. Bile acids activated receptors regulate innate immunity. Front. Immunol. 2018;9:1. doi: 10.3389/FIMMU.2018.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach M.A. Microbiome: focus on causation and mechanism. Cell. 2018;174:785. doi: 10.1016/J.CELL.2018.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flock G., Holland D., Seino Y., Drucker D.J. GPR119 regulates murine glucose homeostasis through incretin receptor-dependent and independent mechanisms. Endocrinology. 2011;152:374. doi: 10.1210/EN.2010-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa E.A., Sirota-Madi A., Avila-Pacheco J., Fornelos N., Haiser H.J., Reinker S., Vatanen T., Hall A.B., Mallick H., McIver L.J., Sauk J.S., Wilson R.G., Stevens B.W., Scott J.M., Pierce K., Deik A.A., Bullock K., Imhann F., Porter J.A., Zhernakova A., Fu J., Weersma R.K., Wijmenga C., Clish C.B., Vlamakis H., Huttenhower C., Xavier R.J. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nature Microbiology. 2019;4:293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi M., Grove T.L., Wang M., Varma Y., McFadden M.E., Brown L.C., Guo C., Higginbottom S., Almo S.C., Fischbach M.A. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature. 2020;582:566. doi: 10.1038/S41586-020-2396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N., Luzzatto-Knaan T., Melnik A.V., Caraballo-Rodríguez A.M., Floros D.J., Petras D., Gregor R., Dorrestein P.C., Phelan V.V. Natural products as mediators of disease. Nat. Prod. Rep. 2017;34:194. doi: 10.1039/C6NP00063K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens. 2014;3:14. doi: 10.3390/PATHOGENS3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzior D.V., Quinn R.A. Review: microbial transformations of human bile acids. Microbiome. 2021;9 doi: 10.1186/S40168-021-01101-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Van Treuren W., Fischer C.R., Merrill B.D., DeFelice B.C., Sanchez J.M., Higginbottom S.K., Guthrie L., Fall L.A., Dodd D., Fischbach M.A., Sonnenburg J.L. A metabolomics pipeline for the mechanistic interrogation of the gut microbiome. Nature. 2021;595(7867 595):415–420. doi: 10.1038/s41586-021-03707-9. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka M., Morita H., Aoyagi Y., Sasaki K., Sasaki D., Kondo A., Nakamura T. Effective bifidogenic growth factors cyclo-Val-Leu and cyclo-Val-Ile produced by Bacillus subtilis C-3102 in the human colonic microbiota model. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-64374-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser A.S., Chavali S., Masuho I., Jahn L.J., Martemyanov K.A., Gloriam D.E., Babu M.M. Pharmacogenomics of GPCR drug targets. Cell. 2018;172:41. doi: 10.1016/J.CELL.2017.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A.F. The continuing importance of bile acids in liver and intestinal disease. Arch. Intern. Med. 1999;159:2647–2658. doi: 10.1001/ARCHINTE.159.22.2647. [DOI] [PubMed] [Google Scholar]
- Hsiao E.Y., McBride S.W., Hsien S., Sharon G., Hyde E.R., McCue T., Codelli J.A., Chow J., Reisman S.E., Petrosino J.F., Patterson P.H., Mazmanian S.K. The microbiota modulates gut physiology and behavioral abnormalities associated with autism. Cell. 2013;155:1451. doi: 10.1016/J.CELL.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt C., Macedo e Cordeiro T., Suchting R., de Dios C., Cuellar Leal V.A., Soares J.C., Dantzer R., Teixeira A.L., Selvaraj S. Effect of immune activation on the kynurenine pathway and depression symptoms – a systematic review and meta-analysis. Neurosci. Biobehav. Rev. 2020;118:514. doi: 10.1016/J.NEUBIOREV.2020.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husted A.S., Trauelsen M., Rudenko O., Hjorth S.A., Schwartz T.W. GPCR-mediated signaling of metabolites. Cell Metabol. 2017;25:777–796. doi: 10.1016/J.CMET.2017.03.008. [DOI] [PubMed] [Google Scholar]
- Insel P.A., Sriram K., Gorr M.W., Wiley S.Z., Michkov A., Salmerón C., Chinn A.M. GPCRomics: an approach to discover GPCR drug targets. Trends Pharmacol. Sci. 2019;40:378. doi: 10.1016/J.TIPS.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmusch A.K., Wang M., Aceves C.M., Advani R.S., Aguire S., Aksenov A.A., Aleti G., Aron A.T., Bauermeister A., Bolleddu S., Bouslimani A., Rodriguez A.M.C., Chaar R., Coras R., Elijah E.O., Ernst M., Gauglitz J.M., Gentry E.C., Husband M., Jarmusch S.A., Jones K.L., Kamenik Z., Gouellec A.L., Lu A., McCall L.-I., McPhail K.L., Meehan M.J., Melnik A.V., Menezes R.C., Giraldo Y.A.M., Nguyen N.H., Nothias L.F., Nothias-Esposito M., Panitchpakdi M., Petras D., Quinn R., Sikora N., Hooft J.J.J. van der, Vargas F., Vrbanac A., Weldon K., Knight R., Bandeira N., Dorrestein P.C. Repository-scale Co- and Re-analysis of tandem mass spectrometry data. bioRxiv. 2019 doi: 10.1101/750471. [DOI] [Google Scholar]
- Jarmusch A.K., Wang M., Aceves C.M., Advani R.S., Aguirre S., Aksenov A.A., Aleti G., Aron A.T., Bauermeister A., Bolleddu S., Bouslimani A., Caraballo Rodriguez A.M., Chaar R., Coras R., Elijah E.O., Ernst M., Gauglitz J.M., Gentry E.C., Husband M., Jarmusch S.A., Jones K.L., Kamenik Z., Le Gouellec A., Lu A., McCall L.I., McPhail K.L., Meehan M.J., Melnik A.V., Menezes R.C., Montoya Giraldo Y.A., Nguyen N.H., Nothias L.F., Nothias-Esposito M., Panitchpakdi M., Petras D., Quinn R.A., Sikora N., van der Hooft J.J.J., Vargas F., Vrbanac A., Weldon K.C., Knight R., Bandeira N., Dorrestein P.C. ReDU: a framework to find and Re-analyze public mass spectrometry data. Nat. Methods. 2020;17:901. doi: 10.1038/S41592-020-0916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashima K., Saji T., Murata T., Nagamachi M., Matsuoka T., Segi E., Tsuboi K., Sugimoto Y., Kobayashi T., Miyachi Y., Ichikawa A., Narumiya S. The prostaglandin receptor EP4 suppresses colitis, mucosal damage and CD4 cell activation in the gut. J. Clin. Investig. 2002;109:883. doi: 10.1172/JCI14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanegawa N., Suzuki C., Ohinata K. Dipeptide Tyr-Leu (YL) exhibits anxiolytic-like activity after oral administration via activating serotonin 5-HT1A, dopamine D1 and GABAA receptors in mice. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2010;584:599–604. doi: 10.1016/j.febslet.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Koh A., Molinaro A., Ståhlman M., Khan M.T., Schmidt C., Mannerås-Holm L., Wu H., Carreras A., Jeong H., Olofsson L.E., Bergh P.O., Gerdes V., Hartstra A., de Brauw M., Perkins R., Nieuwdorp M., Bergström G., Bäckhed F. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell. 2018;175:947–961. doi: 10.1016/J.CELL.2018.09.055/ATTACHMENT/638B8BF7-511C-4CF3-80D5-20C8375AA8D6/MMC3.DOCX. e17. [DOI] [PubMed] [Google Scholar]
- Koppel N., Rekdal V.M., Balskus E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science (New York, N.Y.) 2018;356:1246–1257. doi: 10.1126/SCIENCE.AAG2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautkramer K.A., Fan J., Bäckhed F. Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 2020;19(2 19):77–94. doi: 10.1038/s41579-020-0438-4. 2020. [DOI] [PubMed] [Google Scholar]
- Kroeze W.K., Sassano M.F., Huang X.P., Lansu K., McCorvy J.D., Giguère P.M., Sciaky N., Roth B.L. PRESTO-TANGO: an open-source resource for interrogation of the druggable human GPCR-ome. Nat. Struct. Mol. Biol. 2015;22:362. doi: 10.1038/NSMB.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Liu C.W., Yang Y., Hsiao Y.C., Ru H., Lu K. High-coverage metabolomics uncovers microbiota-driven biochemical landscape of interorgan transport and gut-brain communication in mice. Nat. Commun. 2021;12 doi: 10.1038/S41467-021-26209-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laschet C., Dupuis N., Hanson J. The G protein-coupled receptors deorphanization landscape. Biochem. Pharmacol. 2018;153:62–74. doi: 10.1016/J.BCP.2018.02.016. [DOI] [PubMed] [Google Scholar]
- Lin H.V., Efanov A.M., Fang X., Beavers L.S., Wang X., Wang J., Gonzalez Valcarcel I.C., Ma T. GPR142 controls tryptophan-induced insulin and incretin hormone secretion to improve glucose metabolism. PLoS One. 2016;11 doi: 10.1371/JOURNAL.PONE.0157298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lleo A., Selmi C., Invernizzi P., Podda M., Coppel R.L., Mackay I.R., Gores G.J., Ansari A.A., Van de Water J., Gershwin E. Apotopes and the biliary specificity of primary biliary cirrhosis. Hepatology (Baltimore, Md. 2009;49:871. doi: 10.1002/HEP.22736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., Foster B., Moser M., Karasik E., Gillard B., Ramsey K., Sullivan S., Bridge J., Magazine H., Syron J., Fleming J., Siminoff L., Traino H., Mosavel M., Barker L., Jewell S., Rohrer D., Maxim D., Filkins D., Harbach P., Cortadillo E., Berghuis B., Turner L., Hudson E., Feenstra K., Sobin L., Robb J., Branton P., Korzeniewski G., Shive C., Tabor D., Qi L., Groch K., Nampally S., Buia S., Zimmerman A., Smith A., Burges R., Robinson K., Valentino K., Bradbury D., Cosentino M., Diaz-Mayoral N., Kennedy M., Engel T., Williams P., Erickson K., Ardlie K., Winckler W., Getz G., DeLuca D., MacArthur Daniel, Kellis M., Thomson A., Young T., Gelfand E., Donovan M., Meng Y., Grant G., Mash D., Marcus Y., Basile M., Liu J., Zhu J., Tu Z., Cox N.J., Nicolae D.L., Gamazon E.R., Im H.K., Konkashbaev A., Pritchard J., Stevens M., Flutre T., Wen X., Dermitzakis E.T., Lappalainen T., Guigo R., Monlong J., Sammeth M., Koller D., Battle A., Mostafavi S., McCarthy M., Rivas M., Maller J., Rusyn I., Nobel A., Wright F., Shabalin A., Feolo M., Sharopova N., Sturcke A., Paschal J., Anderson J.M., Wilder E.L., Derr L.K., Green E.D., Struewing J.P., Temple G., Volpi S., Boyer J.T., Thomson E.J., Guyer M.S., Ng C., Abdallah A., Colantuoni D., Insel T.R., Koester S.E., Little A Roger, Bender P.K., Lehner T., Yao Y., Compton C.C., Vaught J.B., Sawyer S., Lockhart N.C., Demchok J., Moore H.F. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580. doi: 10.1038/NG.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manieri N.A., Mack M.R., Himmelrich M.D., Worthley D.L., Hanson E.M., Eckmann L., Wang T.C., Stappenbeck T.S. Mucosally transplanted mesenchymal stem cells stimulate intestinal healing by promoting angiogenesis. J. Clin. Investig. 2015;125:3606. doi: 10.1172/JCI81423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawe G.M., Hoffman J.M. Serotonin signaling in the gastrointestinal tract:: functions, dysfunctions, and therapeutic targets. Nat. Rev. Gastroenterol. Hepatol. 2013;10:473. doi: 10.1038/NRGASTRO.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencarelli A., Renga B., Migliorati M., Cipriani S., Distrutti E., Santucci L., Fiorucci S. The bile acid sensor farnesoid X receptor is a modulator of liver immunity in a rodent model of acute hepatitis. J. Immunol. 2009;183:6657–6666. doi: 10.4049/JIMMUNOL.0901347. [DOI] [PubMed] [Google Scholar]
- Mizushige T., Uchida T., Ohinata K. Dipeptide tyrosyl-leucine exhibits antidepressant-like activity in mice. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-59039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyasu K., Ichinose T., Nakahata A., Tanaka M., Matsui T., Furuya S. The dipeptides ile-tyr and ser-tyr exert distinct effects on catecholamine metabolism in the mouse brainstem. International Journal of Peptides. 2016;2016 doi: 10.1155/2016/6020786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller T.D., Finan B., Bloom S.R., D'Alessio D., Drucker D.J., Flatt P.R., Fritsche A., Gribble F., Grill H.J., Habener J.F., Holst J.J., Langhans W., Meier J.J., Nauck M.A., Perez-Tilve D., Pocai A., Reimann F., Sandoval D.A., Schwartz T.W., Seeley R.J., Stemmer K., Tang-Christensen M., Woods S.C., DiMarchi R.D., Tschöp M.H. Glucagon-like peptide 1 (GLP-1) Mol. Metabol. 2019;30:72. doi: 10.1016/J.MOLMET.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namkung Y., Le Gouill C., Lukashova V., Kobayashi H., Hogue M., Khoury E., Song M., Bouvier M., Laporte S.A. Monitoring G protein-coupled receptor and β-arrestin trafficking in live cells using enhanced bystander BRET. Nat. Commun. 2016;7 doi: 10.1038/NCOMMS12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntranos A., Park H.J., Wentling M., Tolstikov V., Amatruda M., Inbar B., Kim-Schulze S., Frazier C., Button J., Kiebish M.A., Lublin F., Edwards K., Casaccia P. Bacterial neurotoxic metabolites in multiple sclerosis cerebrospinal fluid and plasma. Brain. 2022;145:569–583. doi: 10.1093/BRAIN/AWAB320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton H.A., Fyfe M.C.T., Reynet C. GPR119, a novel G protein-coupled receptor target for the treatment of type 2 diabetes and obesity. Br. J. Pharmacol. 2008;153:S76. doi: 10.1038/SJ.BJP.0707529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.B., Crawford J.M. Pyrazinone protease inhibitor metabolites from Photorhabdus luminescens. J. Antibiot. 2016;69:616–621. doi: 10.1038/ja.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek M.R., Val D.L., Hanzelka B.L., Cronan J.E., Greenberg E.P. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4360. doi: 10.1073/PNAS.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascucci T., Colamartino M., Fiori E., Sacco R., Coviello A., Ventura R., Puglisi-Allegra S., Turriziani L., Persico A.M. P-Cresol alters brain dopamine metabolism and exacerbates autism-like behaviors in the BTBR mouse. Brain Sci. 2020;10 doi: 10.3390/BRAINSCI10040233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn R.A., Melnik A.V., Vrbanac A., Fu T., Patras K.A., Christy M.P., Bodai Z., Belda-Ferre P., Tripathi A., Chung L.K., Downes M., Welch R.D., Quinn M., Humphrey G., Panitchpakdi M., Weldon K.C., Aksenov A., da Silva R., Avila-Pacheco J., Clish C., Bae S., Mallick H., Franzosa E.A., Lloyd-Price J., Bussell R., Thron T., Nelson A.T., Wang M., Leszczynski E., Vargas F., Gauglitz J.M., Meehan M.J., Gentry E., Arthur T.D., Komor A.C., Poulsen O., Boland B.S., Chang J.T., Sandborn W.J., Lim M., Garg N., Lumeng J.C., Xavier R.J., Kazmierczak B.I., Jain R., Egan M., Rhee K.E., Ferguson D., Raffatellu M., Vlamakis H., Haddad G.G., Siegel D., Huttenhower C., Mazmanian S.K., Evans R.M., Nizet V., Knight R., Dorrestein P.C. Global chemical impact of the microbiome includes novel bile acid conjugations. Nature. 2020;579:123. doi: 10.1038/S41586-020-2047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekdal V.M., Bess E.N., Bisanz J.E., Turnbaugh P.J., Balskus E.P. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science (New York, N.Y.) 2019;364:1055. doi: 10.1126/SCIENCE.AAU6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon J.M., Kang D.J., Hylemon P.B. Bile salt biotransformations by human intestinal bacteria. JLR (J. Lipid Res.) 2006;47:241–259. doi: 10.1194/JLR.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- Ridlon J.M., Kang D.J., Hylemon P.B., Bajaj J.S. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 2014;30:332. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roager H.M., Licht T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks M.G., Garrett W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016;16:341. doi: 10.1038/NRI.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saia R.S., Ribeiro A.B., Giusti H. Cholecystokinin modulates the mucosal inflammatory response and prevents the lipopolysaccharide-induced intestinal epithelial barrier dysfunction. Shock. 2020;53:242–251. doi: 10.1097/SHK.0000000000001355. [DOI] [PubMed] [Google Scholar]
- Sala-Rabanal M., Loo D.D.F., Hirayama B.A., Wright E.M. Molecular mechanism of dipeptide and drug transport by the human renal H+/oligopeptide cotransporter hPEPT2. Am. J. Physiol. Ren. Physiol. 2008;294:1422–1432. doi: 10.1152/AJPRENAL.00030.2008/SUPPL_FILE/SALA-RABANAL_SUPPL_REVISED.PDF. [DOI] [PubMed] [Google Scholar]
- Schmid R., Petras D., Nothias L.F., Wang M., Aron A.T., Jagels A., Tsugawa H., Rainer J., Garcia-Aloy M., Dührkop K., Korf A., Pluskal T., Kameník Z., Jarmusch A.K., Caraballo-Rodríguez A.M., Weldon K.C., Nothias-Esposito M., Aksenov A.A., Bauermeister A., Albarracin Orio A., Grundmann C.O., Vargas F., Koester I., Gauglitz J.M., Gentry E.C., Hövelmann Y., Kalinina S.A., Pendergraft M.A., Panitchpakdi M., Tehan R., Le Gouellec A., Aleti G., Mannochio Russo H., Arndt B., Hübner F., Hayen H., Zhi H., Raffatellu M., Prather K.A., Aluwihare L.I., Böcker S., McPhail K.L., Humpf H.U., Karst U., Dorrestein P.C. Ion identity molecular networking for mass spectrometry-based metabolomics in the GNPS environment. Nat. Commun. 2021;12 doi: 10.1038/S41467-021-23953-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze T., Golfier S., Tabeling C., Räbel K., Gräler M.H., Witzenrath M., Lipp M. Sphingosine-1-phospate receptor 4 (S1P4) deficiency profoundly affects dendritic cell function and TH17-cell differentiation in a murine model. Faseb. J. 2011;25:4024–4036. doi: 10.1096/FJ.10-179028. [DOI] [PubMed] [Google Scholar]
- Sepe V., Renga B., Festa C., Finamore C., Masullo D., Carino A., Cipriani S., Distrutti E., Fiorucci S., Zampella A. Investigation on bile acid receptor regulators. Discovery of cholanoic acid derivatives with dual G-protein coupled bile acid receptor 1 (GPBAR1) antagonistic and farnesoid X receptor (FXR) modulatory activity. Steroids. 2016;105:59–67. doi: 10.1016/J.STEROIDS.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Sivaprakasam S., Bhutia Y.D., Yang S., Ganapathy V. Short-chain fatty acid transporters: role in colonic homeostasis. Compr. Physiol. 2017;8:299. doi: 10.1002/CPHY.C170014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram K., Insel P.A. G protein-coupled receptors as targets for approved drugs: how many targets and how many drugs? Mol. Pharmacol. 2018;93:251–258. doi: 10.1124/MOL.117.111062/-/DC1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner L.W., Amberg A., Barrett D., Beale M.H., Beger R., Daykin C.A., Fan T.W.M., Fiehn O., Goodacre R., Griffin J.L., Hankemeier T., Hardy N., Harnly J., Higashi R., Kopka J., Lane A.N., Lindon J.C., Marriott P., Nicholls A.W., Reily M.D., Thaden J.J., Viant M.R. Proposed minimum reporting standards for chemical analysis: chemical analysis working group (CAWG) metabolomics standards initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J.K., McKenzie C., Mariño E., Macia L., Mackay C.R. Metabolite-sensing G protein–coupled receptors—facilitators of diet-related immune regulation. 2017. 371–402. [DOI] [PubMed]
- Tang C., Ahmed K., Gille A., Lu S., Gröne H.J., Tunaru S., Offermanns S. Loss of FFA2 and FFA3 increases insulin secretion and improves glucose tolerance in type 2 diabetes. Nat. Med. 2015;21(2 21):173–177. doi: 10.1038/nm.3779. 2015. [DOI] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul P.J., Lindskog C. The human protein atlas: a spatial map of the human proteome. Protein Sci. : A Publication of the Protein Society. 2018;27:233. doi: 10.1002/PRO.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valles-Colomer M., Falony G., Darzi Y., Tigchelaar E.F., Wang J., Tito R.Y., Schiweck C., Kurilshikov A., Joossens M., Wijmenga C., Claes S., Van Oudenhove L., Zhernakova A., Vieira-Silva S., Raes J. The neuroactive potential of the human gut microbiota in quality of life and depression. Nature Microbiology. 2019;4:623–632. doi: 10.1038/s41564-018-0337-x. [DOI] [PubMed] [Google Scholar]
- van de Waterbeemd H., Gifford E. ADMET in silico modelling: towards prediction paradise? Nat. Rev. Drug Discov. 2003;2(3 2):192–204. doi: 10.1038/nrd1032. 2003. [DOI] [PubMed] [Google Scholar]
- van Kessel S.P., Frye A.K., El-Gendy A.O., Castejon M., Keshavarzian A., van Dijk G., El Aidy S. Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of Parkinson's disease. Nat. Commun. 2019;10 doi: 10.1038/S41467-019-08294-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Carver J.J., Phelan V.V., Sanchez L.M., Garg N., Peng Y., Nguyen D.D., Watrous J., Kapono C.A., Luzzatto-Knaan T., Porto C., Bouslimani A., Melnik A.V., Meehan M.J., Liu W.T., Crüsemann M., Boudreau P.D., Esquenazi E., Sandoval-Calderón M., Kersten R.D., Pace L.A., Quinn R.A., Duncan K.R., Hsu C.C., Floros D.J., Gavilan R.G., Kleigrewe K., Northen T., Dutton R.J., Parrot D., Carlson E.E., Aigle B., Michelsen C.F., Jelsbak L., Sohlenkamp C., Pevzner P., Edlund A., McLean J., Piel J., Murphy B.T., Gerwick L., Liaw C.C., Yang Y.L., Humpf H.U., Maansson M., Keyzers R.A., Sims A.C., Johnson A.R., Sidebottom A.M., Sedio B.E., Klitgaard A., Larson C.B., Boya C.A.P., Torres-Mendoza D., Gonzalez D.J., Silva D.B., Marques L.M., Demarque D.P., Pociute E., O'Neill E.C., Briand E., Helfrich E.J.N., Granatosky E.A., Glukhov E., Ryffel F., Houson H., Mohimani H., Kharbush J.J., Zeng Y., Vorholt J.A., Kurita K.L., Charusanti P., McPhail K.L., Nielsen K.F., Vuong L., Elfeki M., Traxler M.F., Engene N., Koyama N., Vining O.B., Baric R., Silva R.R., Mascuch S.J., Tomasi S., Jenkins S., Macherla V., Hoffman T., Agarwal V., Williams P.G., Dai J., Neupane R., Gurr J., Rodríguez A.M.C., Lamsa A., Zhang C., Dorrestein K., Duggan B.M., Almaliti J., Allard P.M., Phapale P., Nothias L.F., Alexandrov T., Litaudon M., Wolfender J.L., Kyle J.E., Metz T.O., Peryea T., Nguyen D.T., VanLeer D., Shinn P., Jadhav A., Müller R., Waters K.M., Shi W., Liu X., Zhang L., Knight R., Jensen P.R., Palsson B., Pogliano K., Linington R.G., Gutiérrez M., Lopes N.P., Gerwick W.H., Moore B.S., Dorrestein P.C., Bandeira N. Sharing and community curation of mass spectrometry data with GNPS. Nat. Biotechnol. 2016;34:828. doi: 10.1038/NBT.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Carver J.J., Phelan V.V., Sanchez L.M., Garg N., Peng Y., Nguyen D.D., Watrous J., Kapono C.A., Luzzatto-Knaan T., Porto C., Bouslimani A., Melnik A.V., Meehan M.J., Liu W.T., Crüsemann M., Boudreau P.D., Esquenazi E., Sandoval-Calderón M., Kersten R.D., Pace L.A., Quinn R.A., Duncan K.R., Hsu C.C., Floros D.J., Gavilan R.G., Kleigrewe K., Northen T., Dutton R.J., Parrot D., Carlson E.E., Aigle B., Michelsen C.F., Jelsbak L., Sohlenkamp C., Pevzner P., Edlund A., McLean J., Piel J., Murphy B.T., Gerwick L., Liaw C.C., Yang Y.L., Humpf H.U., Maansson M., Keyzers R.A., Sims A.C., Johnson A.R., Sidebottom A.M., Sedio B.E., Klitgaard A., Larson C.B., Boya C.A.P., Torres-Mendoza D., Gonzalez D.J., Silva D.B., Marques L.M., Demarque D.P., Pociute E., O'Neill E.C., Briand E., Helfrich E.J.N., Granatosky E.A., Glukhov E., Ryffel F., Houson H., Mohimani H., Kharbush J.J., Zeng Y., Vorholt J.A., Kurita K.L., Charusanti P., McPhail K.L., Nielsen K.F., Vuong L., Elfeki M., Traxler M.F., Engene N., Koyama N., Vining O.B., Baric R., Silva R.R., Mascuch S.J., Tomasi S., Jenkins S., Macherla V., Hoffman T., Agarwal V., Williams P.G., Dai J., Neupane R., Gurr J., Rodríguez A.M.C., Lamsa A., Zhang C., Dorrestein K., Duggan B.M., Almaliti J., Allard P.M., Phapale P., Nothias L.F., Alexandrov T., Litaudon M., Wolfender J.L., Kyle J.E., Metz T.O., Peryea T., Nguyen D.T., VanLeer D., Shinn P., Jadhav A., Müller R., Waters K.M., Shi W., Liu X., Zhang L., Knight R., Jensen P.R., Palsson B., Pogliano K., Linington R.G., Gutiérrez M., Lopes N.P., Gerwick W.H., Moore B.S., Dorrestein P.C., Bandeira N. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Jarmusch A.K., Vargas F., Aksenov A.A., Gauglitz J.M., Weldon K., Petras D., da Silva R., Quinn R., Melnik A.V., van der Hooft J.J.J., Caraballo-Rodríguez A.M., Nothias L.F., Aceves C.M., Panitchpakdi M., Brown E., Di Ottavio F., Sikora N., Elijah E.O., Labarta-Bajo L., Gentry E.C., Shalapour S., Kyle K.E., Puckett S.P., Watrous J.D., Carpenter C.S., Bouslimani A., Ernst M., Swafford A.D., Zúñiga E.I., Balunas M.J., Klassen J.L., Loomba R., Knight R., Bandeira N., Dorrestein P.C. Mass spectrometry searches using MASST. Nat. Biotechnol. 2020;38:23. doi: 10.1038/S41587-019-0375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Guo A.C., Oler E., Wang F., Anjum A., Peters H., Dizon R., Sayeeda Z., Tian S., Lee B.L., Berjanskii M., Mah R., Yamamoto M., Jovel J., Torres-Calzada C., Hiebert-Giesbrecht M., Lui V.W., Varshavi Dorna, Varshavi Dorsa, Allen D., Arndt D., Khetarpal N., Sivakumaran A., Harford K., Sanford S., Yee K., Cao X., Budinski Z., Liigand J., Zhang L., Zheng J., Mandal R., Karu N., Dambrova M., Schiöth H.B., Greiner R., Gautam V. Hmdb 5.0: the human metabolome database for 2022. Nucleic Acids Res. 2022;50:D622–D631. doi: 10.1093/NAR/GKAB1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt M.A., Mok M.C.Y., Junop M., Magarvey N.A. Heterologous expression and structural characterisation of a pyrazinone natural product assembly line. Chembiochem. 2012;13:2408–2415. doi: 10.1002/cbic.201200340. [DOI] [PubMed] [Google Scholar]
- Xu L., Haworth I.S., Kulkarni A.A., Bolger M.B., Davies D.L. Mutagenesis and cysteine scanning of transmembrane domain 10 of the human dipeptide transporter. Pharmaceut. Res. 2009;26:2358. doi: 10.1007/S11095-009-9952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Lou C., Sun L., Li J., Cai Y., Wang Z., Li W., Liu G., Tang Y. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019;35:1067–1069. doi: 10.1093/BIOINFORMATICS/BTY707. [DOI] [PubMed] [Google Scholar]
- Yano J.M., Yu K., Donaldson G.P., Shastri G.G., Ann P., Ma L., Nagler C.R., Ismagilov R.F., Mazmanian S.K., Hsiao E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunes R.A., Poluektova E.U., Dyachkova M.S., Klimina K.M., Kovtun A.S., Averina O.V., Orlova V.S., Danilenko V.N. GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe. 2016;42:197–204. doi: 10.1016/J.ANAEROBE.2016.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA-Seq expression profiles from all human receptors used in this study, and mass spectral profiles for each molecule found across tissue and body fluid types, code/scripts, and data underlying heatmap figures are publicly available from the GitHub repository (https://github.com/gajenderaleti/GPCR-MicrobialMetaboliteInteractions). In addition, MASST analysis for each molecule are available at Global Natural Product Social Molecular Networking (GNPS) with the following task IDs: GABA: 87b3aac5b86b4124be0cb4aaa6679d11; Hippuric acid: df76ea23ecd54b988f1116321dd4ae46; Phenylathylamine: 6a4ed755c33f4df3bb71dbe629bdeac5; Phe: 4e52e1644a224971ab4f4749744e6ed8; Histamine: e7f992ade5234b63906e1b8a4d3a12c0; Dopamine: 27df5f82bf074195bdccf75f82398fb2; p-cresol sulfate: 8c99e2bae67f4dc09156f84bfe417970; Indoleacrylic acid: ef7e85bf5d324613887e7d026cc56212; Indoxylsulfate: e65dfe111f8943169f39b639a8bc53c8; Indolepropionic acid: d38f7ce18898460bb64f476d90f5c62a; Indolelactic acid: e083444bfa104fed9394943011b7e563; Indoleacetic acid: e8bfa90bfb184205a862444d150653e3; Tryptophan: f18c2a7c189a4d1b9d698c6bff526a5d; Tyrptamine: dc36f9351f4a442ba185d10b0d481739; kynurenic acid: 02a8416a52f8463e9cb7aeb4088d9863; Serotonin: af3282ea885243a9b631f60c5d6b0841 Tyrosine cholic acid: 0e486c6ecd9f42f08dd466e67d6997c8; Phenylalanine cholic acid: 3188dfca7a814e30b46f534f8ffdcb53; Glutamic acid chenodeoxy cholic acid: 6beb245285ad4926aa8d19c514878319; Commendamide: 2214cf1de9a54e419fc6a527cd87bd48; Tyr-Val: 3956099739214030bc76dd95dd1bf894; Phe-Val: 1f1269480de44f378f5b469bc3b28b0aRNA-Seq expression profiles from all human receptors used in this study, and mass spectral profiles for each molecule found across tissue and body fluid types, code/scripts, and data underlying heatmap figures are publicly available from the GitHub repository (https://github.com/gajenderaleti/GPCR-MicrobialMetaboliteInteractions). In addition, MASST analysis for each molecule are available at Global Natural Product Social Molecular Networking (GNPS) with the following task IDs: GABA: 87b3aac5b86b4124be0cb4aaa6679d11 Hippuric acid: df76ea23ecd54b988f1116321dd4ae46 Phenylathylamine: 6a4ed755c33f4df3bb71dbe629bdeac5 Phe: 4e52e1644a224971ab4f4749744e6ed8 Histamine: e7f992ade5234b63906e1b8a4d3a12c0 Dopamine: 27df5f82bf074195bdccf75f82398fb2 p-cresol sulfate: 8c99e2bae67f4dc09156f84bfe417970 Indoleacrylic acid: ef7e85bf5d324613887e7d026cc56212 Indoxylsulfate: e65dfe111f8943169f39b639a8bc53c8 Indolepropionic acid: d38f7ce18898460bb64f476d90f5c62a Indolelactic acid: e083444bfa104fed9394943011b7e563 Indoleacetic acid: e8bfa90bfb184205a862444d150653e3 Tryptophan: f18c2a7c189a4d1b9d698c6bff526a5d Tyrptamine: dc36f9351f4a442ba185d10b0d481739 kynurenic acid: 02a8416a52f8463e9cb7aeb4088d9863 Serotonin: af3282ea885243a9b631f60c5d6b0841 Tyrosine cholic acid: 0e486c6ecd9f42f08dd466e67d6997c8 Phenylalanine cholic acid: 3188dfca7a814e30b46f534f8ffdcb53 Glutamic acid chenodeoxy cholic acid: 6beb245285ad4926aa8d19c514878319 Commendamide: 2214cf1de9a54e419fc6a527cd87bd48 Tyr-Val: 3956099739214030bc76dd95dd1bf894 Phe-Val: 1f1269480de44f378f5b469bc3b28b0aRNA-Seq expression profiles from all human receptors used in this study, and mass spectral profiles for each molecule found across tissue and body fluid types, code/scripts, and data underlying heatmap figures are publicly available from the GitHub repository (https://github.com/gajenderaleti/GPCR-MicrobialMetaboliteInteractions). In addition, MASST analysis for each molecule are available at Global Natural Product Social Molecular Networking (GNPS) with the following task IDs: GABA: 87b3aac5b86b4124be0cb4aaa6679d11 Hippuric acid: df76ea23ecd54b988f1116321dd4ae46 Phenylathylamine: 6a4ed755c33f4df3bb71dbe629bdeac5 Phe: 4e52e1644a224971ab4f4749744e6ed8 Histamine: e7f992ade5234b63906e1b8a4d3a12c0 Dopamine: 27df5f82bf074195bdccf75f82398fb2 p-cresol sulfate: 8c99e2bae67f4dc09156f84bfe417970 Indoleacrylic acid: ef7e85bf5d324613887e7d026cc56212 Indoxylsulfate: e65dfe111f8943169f39b639a8bc53c8 Indolepropionic acid: d38f7ce18898460bb64f476d90f5c62a Indolelactic acid: e083444bfa104fed9394943011b7e563 Indoleacetic acid: e8bfa90bfb184205a862444d150653e3 Tryptophan: f18c2a7c189a4d1b9d698c6bff526a5d Tyrptamine: dc36f9351f4a442ba185d10b0d481739 kynurenic acid: 02a8416a52f8463e9cb7aeb4088d9863 Serotonin: af3282ea885243a9b631f60c5d6b0841 Tyrosine cholic acid: 0e486c6ecd9f42f08dd466e67d6997c8 Phenylalanine cholic acid: 3188dfca7a814e30b46f534f8ffdcb53 Glutamic acid chenodeoxy cholic acid: 6beb245285ad4926aa8d19c514878319 Commendamide: 2214cf1de9a54e419fc6a527cd87bd48 Tyr-Val: 3956099739214030bc76dd95dd1bf894 Phe-Val: 1f1269480de44f378f5b469bc3b28b0aRNA-Seq expression profiles from all human receptors used in this study, and mass spectral profiles for each molecule found across tissue and body fluid types, code/scripts, and data underlying heatmap figures are publicly available from the GitHub repository (https://github.com/gajenderaleti/GPCR-MicrobialMetaboliteInteractions). In addition, MASST analysis for each molecule are available at Global Natural Product Social Molecular Networking (GNPS) with the following task IDs: GABA: 87b3aac5b86b4124be0cb4aaa6679d11 Hippuric acid: df76ea23ecd54b988f1116321dd4ae46 Phenylathylamine: 6a4ed755c33f4df3bb71dbe629bdeac5 Phe: 4e52e1644a224971ab4f4749744e6ed8 Histamine: e7f992ade5234b63906e1b8a4d3a12c0 Dopamine: 27df5f82bf074195bdccf75f82398fb2 p-cresol sulfate: 8c99e2bae67f4dc09156f84bfe417970 Indoleacrylic acid: ef7e85bf5d324613887e7d026cc56212 Indoxylsulfate: e65dfe111f8943169f39b639a8bc53c8 Indolepropionic acid: d38f7ce18898460bb64f476d90f5c62a Indolelactic acid: e083444bfa104fed9394943011b7e563 Indoleacetic acid: e8bfa90bfb184205a862444d150653e3 Tryptophan: f18c2a7c189a4d1b9d698c6bff526a5d Tyrptamine: dc36f9351f4a442ba185d10b0d481739 kynurenic acid: 02a8416a52f8463e9cb7aeb4088d9863 Serotonin: af3282ea885243a9b631f60c5d6b0841 Tyrosine cholic acid: 0e486c6ecd9f42f08dd466e67d6997c8 Phenylalanine cholic acid: 3188dfca7a814e30b46f534f8ffdcb53 Glutamic acid chenodeoxy cholic acid: 6beb245285ad4926aa8d19c514878319 Commendamide: 2214cf1de9a54e419fc6a527cd87bd48 Tyr-Val: 3956099739214030bc76dd95dd1bf894 Phe-Val: 1f1269480de44f378f5b469bc3b28b0aRNA-Seq expression profiles from all human receptors used in this study, and mass spectral profiles for each molecule found across tissue and body fluid types, code/scripts, and data underlying heatmap figures are publicly available from the GitHub repository (https://github.com/gajenderaleti/GPCR-MicrobialMetaboliteInteractions). In addition, MASST analysis for each molecule are available at Global Natural Product Social Molecular Networking (GNPS) with the following task IDs: GABA: 87b3aac5b86b4124be0cb4aaa6679d11 Hippuric acid: df76ea23ecd54b988f1116321dd4ae46 Phenylathylamine: 6a4ed755c33f4df3bb71dbe629bdeac5 Phe: 4e52e1644a224971ab4f4749744e6ed8 Histamine: e7f992ade5234b63906e1b8a4d3a12c0 Dopamine: 27df5f82bf074195bdccf75f82398fb2 p-cresol sulfate: 8c99e2bae67f4dc09156f84bfe417970 Indoleacrylic acid: ef7e85bf5d324613887e7d026cc56212 Indoxylsulfate: e65dfe111f8943169f39b639a8bc53c8 Indolepropionic acid: d38f7ce18898460bb64f476d90f5c62a Indolelactic acid: e083444bfa104fed9394943011b7e563 Indoleacetic acid: e8bfa90bfb184205a862444d150653e3 Tryptophan: f18c2a7c189a4d1b9d698c6bff526a5d Tyrptamine: dc36f9351f4a442ba185d10b0d481739 kynurenic acid: 02a8416a52f8463e9cb7aeb4088d9863 Serotonin: af3282ea885243a9b631f60c5d6b0841 Tyrosine cholic acid: 0e486c6ecd9f42f08dd466e67d6997c8 Phenylalanine cholic acid: 3188dfca7a814e30b46f534f8ffdcb53 Glutamic acid chenodeoxy cholic acid: 6beb245285ad4926aa8d19c514878319 Commendamide: 2214cf1de9a54e419fc6a527cd87bd48 Tyr-Val: The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). https://doi.org/10.1038/nature11234