Abstract
Pit2 is the human receptor for amphotropic murine leukemia virus (A-MuLV); the related human protein Pit1 does not support A-MuLV entry. Interestingly, chimeric proteins in which either the N-terminal or the C-terminal part of Pit2 was replaced by the Pit1 sequence all retained A-MuLV receptor function. A possible interpretation of these observations is that Pit1 harbors sequences which can specify A-MuLV receptor function when presented in a protein context other than Pit1, e.g., in Pit1-Pit2 hybrids. We reasoned that such Pit1 sequences might be identified if presented in the Neurospora crassa protein Pho-4. This protein is distantly related to Pit1 and Pit2, predicted to have a similar membrane topology with five extracellular loops, and does not support A-MuLV entry. We show here that introduction of the Pit1-specific loop 2 sequence conferred A-MuLV receptor function upon Pho-4. Therefore, we conclude that (i) a functional A-MuLV receptor can be constructed by combining sequences from two proteins each lacking A-MuLV receptor function and that (ii) a Pit1 sequence can specify A-MuLV receptor function when presented in another protein context than that provided by Pit1 itself. Previous results indicated a role of loop 4 residues in A-MuLV entry, and the presence of a Pit2-specific loop 4 sequence was found here to confer A-MuLV receptor function upon Pho-4. Moreover, the introduction of a Pit1-specific loop 4 sequence, but not of a Pit2-specific loop 4 sequence, abolished the A-MuLV receptor function of a Pho-4 chimera harboring the Pit1-specific loop 2 sequence. Together, these data suggest that residues in both loop 2 and loop 4 play a role in A-MuLV receptor function. A-MuLV is, however, not dependent on the specific Pit2 loop 2 and Pit2 loop 4 sequences for entry; rather, the role played by loops 2 and 4 in A-MuLV entry can be fulfilled by several different combinations of loop 2 and loop 4 sequences. We predict that the residues in loops 2 and 4, identified in this study as specifying A-MuLV receptor function, are to be found among those not conserved among Pho-4, Pit1, and Pit2.
Amphotropic murine leukemia virus (A-MuLV) and gibbon ape leukemia virus (GALV), both type C mammalian retroviruses, utilize related cell surface proteins for entry into cells. The human receptor for A-MuLV is Pit2 (formerly GLVR2) (31) and that for GALV is Pit1 (formerly GLVR1) (20). cDNAs encoding homologs of these proteins have been cloned from other species besides humans, e.g., Pit1 cDNAs have been cloned from mice (MusPit1, formerly Glvr-1) (8) and hamsters (HaPit1, formerly EGR) (35), and Pit2 cDNAs have been cloned from rats (RatPit2, formerly Ram-1) (18) and hamsters (HaPit2, formerly EAR) (35). Pit1 is also a receptor for feline leukemia virus subgroup B (FeLV-B) (29). In general, Pit2 homologs are not receptors for GALV or FeLV-B (6, 19, 22, 24, 27); however, one exception is HaPit2, which supports entry by GALV and the 90Z FeLV-B isolate (2, 35). For A-MuLV, only Pit2 homologs are efficient receptors, while the A-MuLV related isolate 10A1 can utilize all tested Pit1 and Pit2 homologs for entry (13, 19, 22, 35).
Pit1 and Pit2 have 62% overall amino acid identity (31) and are distantly related (both about 25% amino acid identity) to Pho-4, a sodium-dependent phosphate transporter from the filamentous fungus Neurospora crassa (8, 14, 32). The cellular function of Pit1 was suggested to be phosphate transport, based on its homology to Pho-4 (8), and both Pit1 and Pit2 were subsequently shown to be sodium-dependent phosphate transporters (10, 21, 33). On the basis of hydropathy plots, Pho-4, Pit1, and Pit2 are predicted to have 10 membrane-spanning domains, 5 extracellular loops, and 4 intracellular domains, of which the third is large and hydrophilic (8, 31). The cytoplasmic localization of the large hydrophilic domain is supported by recent biochemical data obtained on Pit2 (4).
Residues in the fourth extracellular loop of Pit1 have been shown to be critical for receptor function for GALV and FeLV-B. Chimeras between MusPit1, which does not support FeLV-B and GALV entry, and Pit1, as well as mutational analyses of these proteins, revealed that a stretch of nine amino acids, termed region A, in the C-terminal part of the fourth extracellular loop (Pit1 positions 550 through 558) was critical for receptor function for GALV and FeLV-B (9, 28). Studies on chimeras between Pit1 and Pit2 or RatPit2 confirmed the critical role of Pit1 region A in FeLV-B and GALV entry (19, 22, 27). Moreover, Eiden and colleagues showed that replacement of a lysine in the first position of Pit2 region A (position 522) by a glutamic acid (the corresponding Pit1 residue) allowed Pit2 to support GALV entry (6). Furthermore, we recently showed that the presence of 12 Pit1-specific amino acids, comprising region A, conferred GALV receptor function upon Pho-4 (25). These results demonstrate the importance of region A for infection by GALV and FeLV-B. In addition, we have recently obtained results indicating that region A also plays a role in 10A1 receptor function (13). However, as previously observed for FeLV-B (22), recent results indicate that entry by GALV and 10A1, besides region A, also is dependent on other receptor regions (3, 13).
Receptor regions involved in A-MuLV entry have also been investigated by using chimeras between Pit1 and Pit2 homologs from different species (6, 19, 22, 27). Pit1-Pit2 chimeras harboring the region A sequence from Pit2 and RatPit2 were found to support A-MuLV entry (19, 22). Moreover, insertion of a threonine residue in MusPit1 region A (MusPit1 position 554) resulted in a functional A-MuLV receptor (13). Thus, region A in loop 4 also plays a role in A-MuLV receptor function (13). However, interestingly, chimeras in which the N-terminal two-thirds (comprising loops 1, 2, and 3 and the large intracellular domain) or the C-terminal third (comprising loops 4 and 5) of Pit2 or RatPit2 were exchanged for the corresponding Pit1 sequences all retained A-MuLV receptor function (6, 19, 22). Thus, identical region A sequences are present both in Pit1 with no A-MuLV receptor function and in a chimeric A-MuLV receptor. Moreover, comparable chimeras in which the N- or C-terminal parts were derived from HaPit2 only afforded low levels of A-MuLV infection compared to the Pit1-Pit2 and Pit1-RatPit2 hybrids, although HaPit2 is as efficient an A-MuLV receptor as is Pit2 (6). As previously noted by us (13, 22) and others (12), these results suggest that A-MuLV receptor function is defined by a combination of both N- and C-terminal receptor determinants, with region A being a C-terminal determinant of A-MuLV receptor function (13, 19, 22). These observations thus also suggest that Pit1 harbors sequences which may specify A-MuLV receptor function when presented in a protein context different from that of Pit1, e.g., in the Pit1-Pit2 hybrids. This would imply that Pit1 also harbors sequences which are incompatible with A-MuLV receptor function, in that Pit1 itself is not an efficient A-MuLV receptor. In order to investigate whether Pit1 harbors sequences which can specify A-MuLV receptor function, we reasoned that a new protein context for Pit1 sequences should be provided by a protein which is structurally related to Pit1 but is not itself an A-MuLV receptor, requirements fulfilled by Pho-4 (8, 25). We therefore tested a series of chimeras with the backbone provided by Pho-4 and each harboring an individual Pit1 loop sequence (Fig. 1) for their ability to support A-MuLV entry. Indeed, the presence of a Pit1-specific sequence in loop 2, but not in loops 1, 3, 4, or 5, conferred A-MuLV receptor function upon Pho-4. As mentioned above, the presence of Pit2 region A in loop 4 of Pit1 was sufficient to make it a functional A-MuLV receptor. We therefore tested the influence of loop 4 sequences on A-MuLV receptor function in a Pho-4 backbone. A Pit2 loop 4 sequence was found to confer A-MuLV receptor function upon Pho-4. Moreover, the presence of a Pit1-specific loop 4 sequence, but not a Pit2-specific loop 4 sequence, together with the Pit1 loop 2 sequence in Pho-4, abolished A-MuLV receptor function. These results show that the ability of residues in Pit1 loop 2 to specify A-MuLV receptor function is dependent on the residues present in loop 4. We therefore suggest that both loop 2 and loop 4 are involved in A-MuLV entry and that specific combinations of loop 2 and loop 4 sequences specify A-MuLV receptor function.
FIG. 1.
Sequence comparison between Pit1, Pit2, and Pho-4 putative extracellular loops. Numbers at the left of the sequences correspond to the positions of the first and last amino acids shown. Underlined sequences are those exchanged between Pho-4 and Pit1 and/or Pit2 in the chimeras shown in Fig. 2 and 3; however, in the chimeras harboring the Pit1 loop 4 sequence, only Pho-4 residues V450 through G463 were exchanged for Pit1 residues L545 through S556. Dots indicate amino acid identity to Pit1. Gaps introduced for alignment are indicated by dashes. Region A is a 9-amino-acid stretch first identified in Pit1 (D550 through V558) as critical for GALV and FeLV-B entry (9, 28). Amino acids A559 in Pit1, A531 in Pit2, and D466 in Pho-4 are not part of the loop sequence proposed in Johann et al. (8).
MATERIALS AND METHODS
Expression plasmids.
The plasmid pOJ72 (25) harbors the open reading frame of Pho-4, originally derived from the pBJ005 plasmid (14) obtained from R. L. Metzenberg, in the eucaryotic expression vector pcDNA1ARtkpA (31). With pOJ72 as a template, Pho-4-Pit2 chimeras were constructed by replacing the Pho-4 sequences indicated in Fig. 1 with the indicated Pit2 sequences via site-directed mutagenesis according to the method of Kunkel et al. (11). Restriction enzyme fragments harboring the mutations were cloned back into pOJ72; the integrity of these fragments was verified by nucleotide sequence analyses. The designations used for the Pho-4-derived chimeras are to be read as follows, e.g., the chimera Pho/P1(2)/P2(4) harbors the Pit1 loop 2 and Pit2 loop 4 sequences indicated in Fig. 1 in the Pho-4 backbone.
To construct pA16, encoding Pho/P2(2), amino acids 117 through 149 in loop 2 of Pho-4 were replaced by amino acids 117 through 143 in loop 2 of Pit2. A BalI-HpaI fragment harboring the mutated sequence was then used to replace the corresponding pOJ72 fragment, yielding the pA16 plasmid. pA17, encoding Pho/P2(4), was constructed by replacing amino acids 450 through 466 in loop 4 of Pho-4 by amino acids 517 through 531 in loop 4 of Pit2. An AflIII-AflIII fragment harboring the mutated sequence was then cloned back into an AflIII-digested pOJ72 vector, resulting in the intermediate plasmid p49A. Finally, a KpnI-EcoNI fragment of p49A harboring the mutated sequence was cloned into a KpnI-EcoNI-digested pOJ72 vector, resulting in pA17. To construct pA18, encoding Pho/P2(2+4), an AflIII-AflIII fragment of pA17 was replaced by the corresponding pA16 fragment harboring the Pit2 loop 2 sequence. pA19, encoding Pho/P2(2)/P1(4), was constructed by cloning an AflIII-AflIII fragment of pA16 harboring the Pit2 loop 2 sequence into AflIII-digested pOJ79 (25) vector, encoding Pho/P1(4) and thus harboring the Pit1 loop 4 sequence. To construct pA20, which encodes Pho/P1(2)/P2(4), an AflIII-AflIII fragment of pOJ86 (23), encoding Pho/P1(2) and thus harboring the Pit1 loop 2 sequence, was cloned into AflIII-digested p49A. Finally, a KpnI-EcoNI fragment of the resulting plasmid harboring the loop 2 and 4 mutations was cloned into a KpnI-EcoNI-digested pOJ72 vector, resulting in pA20.
The following pcDNA1ARtkpA expression plasmids have been described elsewhere: pOJ93, pOJ86, pOJ88, and pOJ95, which encode Pho/P1(1), Pho/P1(2), Pho/P1(3), and Pho/P1(5), respectively, (23); pOJ87 and pOJ79, which encode Pho/P1(2+4) and Pho/P1(4), respectively (25); and pOJ74 and pOJ75, which encode Pit2 and Pit1, respectively (22).
Cell culture.
Mus dunni tail fibroblasts (MDTF) (ATCC CRL-2017) were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 100 IU/ml of penicillin, and 100 μg of streptomycin per ml (DMEM-FBS-PS). Chinese hamster ovary, CHO K1 (ATCC CCL-61), and dog osteosarcoma, D17 (ATCC CCL-183), cells were grown in α-modified minimal essential medium supplemented with 10% FBS and antibiotics as above (αMEM-FBS-PS). GALV (SEATO) and A-MuLV (4070A) pseudotypes of the β-galactosidase encoding vector G1BgSvN (15) were obtained from the producer cell lines PG13GBN (15, 16) and PA317GBN (15, 17), respectively. Both producer cell lines were maintained in DMEM supplemented with 10% newborn calf serum and antibiotics (DMEM-NCS-PS). GALV and A-MuLV pseudotypes were harvested in DMEM-NCS-PS and αMEM-FBS-PS, respectively, filtered (0.45-μm pore size), and stored at −80°C until use. 10A1 virus pseudotypes of the β-galactosidase-encoding vector LNPOZ (1) were obtained as previously described (13).
Transient-transfection and infection assay.
MDTF and CHO K1 cells were seeded at 4 × 104 cells/60-mm-diameter dish. The next day, the cells were transfected by the calcium phosphate-DNA precipitation method (7). Each precipitate contained 10 μg of a CsCl gradient-purified expression plasmid and 5 μg of CsCl gradient-purified pUC19 plasmid as carrier in 1 ml. From each precipitate, aliquots of 200 μl corresponding to 2 μg of expression plasmid were added to 60-mm-diameter dishes with CHO K1 or MDTF cells. Three independent precipitates were made per construct to be tested. At 48 h after transfection, the cells were challenged with GALV (MDTF), A-MuLV (CHO K1), or 10A1 (CHO K1) pseudotypes in producer cell supernatant adjusted to 8 μg of Polybrene per ml. Challenging of transfected cells with 10A1 pseudotypes was performed in the presence of medium conditioned with CHO K1 cells to specifically block the endogenous 10A1 receptors (19). After 4 h, fresh media were added, and the cells were incubated for another 44 h, at which time they were fixed in 0.05% glutaraldehyde and assayed for β-galactosidase activity with X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) as a substrate (30). The dishes were examined by using a light microscope, and the number of blue (stained) cells per dish was counted.
Virus titer.
All pseudotypes were titrated on D17 cells. Cells were exposed to serial dilutions of supernatant from producer cells in medium adjusted to 8 μg of Polybrene per ml for 4 h, fresh media were added, and 44 h later the cells were assayed for β-galactosidase activity as described above. The titers of 10A1, GALV, and A-MuLV pseudotypes used were 1 × 104, 1.5 × 104, and the range of 1 × 104 to 4 × 104 CFU/ml, respectively.
RESULTS
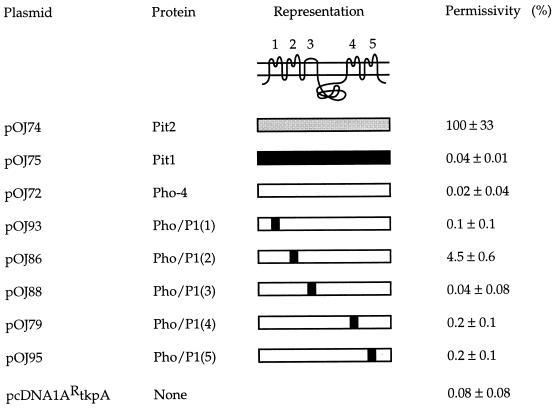
In an attempt to assess whether Pit1 harbors sequences which can specify A-MuLV receptor function when presented in a protein context different from Pit1 itself, Pho-4 chimeras each harboring a different Pit1 loop sequence, i.e., Pho/P1(1), Pho/P1(2), Pho/P1(3), Pho/P1(4), and Pho/P1(5), were tested for their ability to support A-MuLV infection. As shown in Fig. 1, the loop sequences exchanged in the chimeras were those least conserved between Pho-4 and Pit1. A-MuLV receptor function of the chimeras was investigated by transfecting expression plasmids encoding chimeric or wild-type proteins into nonpermissive cells and assaying the susceptibility of these cells to infection with A-MuLV pseudotypes of a β-galactosidase-encoding vector. The results obtained are shown in Fig. 2.
FIG. 2.
Permissivity for A-MuLV infection in CHO K1 cells transiently expressing Pit1, Pit2, Pho-4, or chimeras. The representation at the top shows the putative topology of Pit1, Pit2, and Pho-4; the five extracellular loops are numbered 1 through 5. Pit1 is represented by a black bar, Pit2 by a gray bar, and Pho-4 by a white bar. Black squares in the individual chimeras indicate the loops in which the Pho-4 sequence is replaced by the corresponding Pit1 sequence as indicated in Fig. 1. pcDNA1ARtkpA is the empty vector. The permissivity data are the averages of three independent transfections ± the standard deviations. The number of blue cells per 60-mm-diameter dish transfected with pOJ74 (Pit2) was assigned a value of 100% (ca. 9,000 blue cells), and all other averages and standard deviations were normalized to this value. The experiment was repeated with similar results.
A chimeric A-MuLV receptor derived from two proteins each lacking A-MuLV receptor function.
As previously observed, Pho-4 did not support A-MuLV infection (Fig. 2) (25). Pho-4 chimeras harboring Pit1 sequences in loops 1, 3, 4, or 5 [chimeras Pho/P1(1), Pho/P1(3), Pho/P1(4), and Pho/P1(5), respectively] also were not efficient A-MuLV receptors. The lack of A-MuLV receptor function observed with these constructs was not due to lack of chimeric proteins on the cell surface, in that the Pho-4-Pit1 chimeras shown in Fig. 2 all support infection by 10A1 (23); furthermore, chimera Pho/P1(4) supports GALV entry (25). Interestingly, the Pho-4 chimera harboring the Pit1 loop 2 sequence, Pho/P1(2), supported A-MuLV infection at about 4.5% of the Pit2 level, which is more than 200 times above the background level (the level obtained on cells transfected with pOJ72 which encodes Pho-4) (Fig. 2). Therefore, a Pit1 loop 2 sequence can confer A-MuLV receptor function upon Pho-4, although neither Pho-4 nor Pit1 themselves are efficient A-MuLV receptors. Furthermore, this result indicates that residues in loop 2 play a role in A-MuLV receptor function.
To further investigate the role of loop 2 in A-MuLV entry, we tested whether the corresponding Pit2-specific sequence conferred A-MuLV receptor function upon Pho-4. Interestingly, the chimera Pho/P2(2) featuring the Pit2 loop 2 sequence did not efficiently support A-MuLV entry (Fig. 1 and 3). When tested in parallel for 10A1 and A-MuLV receptor function in CHO K1 cells in an independent experiment, Pho/P2(2) supported entry by 10A1 at a level comparable to that of Pit1 (data not shown); thus, the inability of Pho/P2(2) to allow A-MuLV infection is not due to the lack of chimeric protein on the cell surface.
FIG. 3.
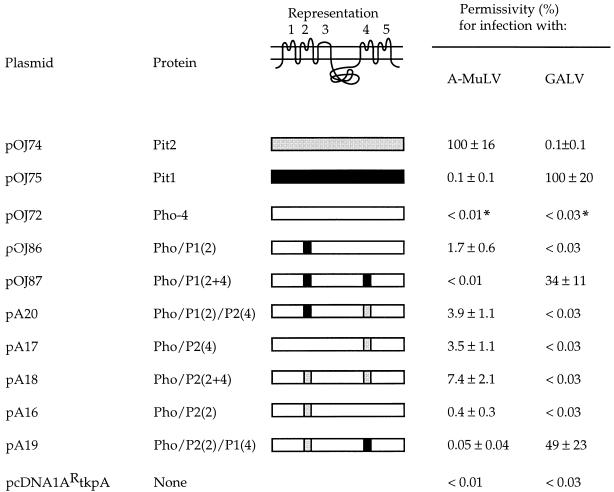
Permissivity for A-MuLV and GALV infection in CHO K1 or MDTF cells transiently expressing Pit1, Pit2, Pho-4, or chimeras. CHO K1 or MDTF cells were transfected with the DNA of the indicated plasmids and were challenged with A-MuLV or GALV pseudotypes, respectively, as described in Materials and Methods. The amino acids exchanged in the chimeras are as indicated in Fig. 1. The representation at the top shows the putative topology of Pit1, Pit2, and Pho-4; the five putative extracellular loops are numbered 1 through 5. The permissivity data are the averages of three independent transfections ± the standard deviations. The number of blue cells per 60-mm-diameter dish transfected with the wild-type receptor for a given virus was assigned a value of 100% (ca. 2,700 and 1,000 blue cells per dish for A-MuLV and GALV, respectively), and all other averages and standard deviations were normalized to this value. The experiment was repeated with similar results. ∗, Values based on one blue cell in three 60-mm-diameter dishes.
Pit1 loop 4, but not Pit2 loop 4, interferes with A-MuLV receptor function in a Pho-4 backbone.
The presence of a Pit1-specific loop 2 sequence conferred A-MuLV receptor function upon Pho-4, although Pit1 itself does not efficiently support A-MuLV entry (Fig. 2). This result suggests the existence of a Pit1 sequence(s) which is incompatible with A-MuLV receptor function. We and others previously found that a C-terminal A-MuLV receptor determinant is positioned in loop 4, in that replacement of Pit1 loop 4 residues 550 through 558 (region A) for the corresponding Pit2 residues resulted in an efficient A-MuLV receptor with a Pit1 backbone (19, 22). We therefore wanted to investigate the possible influence of loop 4 sequences on the receptor function of chimeras with a Pho-4 backbone.
A chimera harboring a Pit2-specific loop 4 sequence, Pho/P2(4), was found to support A-MuLV entry at a level of 3.5% of the Pit2 level (Fig. 1 and 3). Thus, the presence of the Pit2 loop 4 sequence conferred A-MuLV receptor function upon Pho-4, in agreement with the previous results obtained with Pit1-Pit2 chimeras and a MusPit1 mutant, showing that residues in loop 4 can specify A-MuLV receptor function (13, 19, 22). Moreover, a chimera harboring Pit2 loop 2 sequence together with the Pit2 loop 4 sequence, Pho/P2(2+4), supported A-MuLV entry at a level of 7.4% of that obtained by Pit2 (Fig. 3). In addition, the presence of the Pit2 loop 4 sequence together with Pit1 loop 2 sequence in Pho-4, chimera Pho/P1(2)/P2(4), was compatible with A-MuLV receptor function, in that the chimera supported A-MuLV entry at a level of 3.9% of the Pit2 level (Fig. 3). Thus, all of the tested chimeras harboring the Pit2-specific loop 4 sequence were A-MuLV receptors.
We also tested the receptor function of chimeras harboring a Pit1-specific loop 4 sequence. As mentioned above, chimeras harboring either a Pit1 loop 4 sequence, Pho/P1(4), or a Pit2 loop 2 sequence, Pho/P2(2), were not efficient A-MuLV receptors, and a chimera harboring Pit1 loop 4 and Pit2 loop 2 sequences together, Pho/P2(2)/P1(4), did also not support A-MuLV entry (Fig. 3). Pho/P2(2)/P1(4) supported entry by GALV (Fig. 3), thus demonstrating the presence of chimeric protein on the cell surface. Whether the presence of a Pit1-specific loop 4 sequence would influence the A-MuLV receptor function introduced by the presence of the Pit1-specific loop 2 sequence in Pho-4 was investigated by analyzing the receptor function of the chimera Pho/P1(2+4), which harbors the Pit1 loop 4 sequence together with Pit1 loop 2. Cells transfected with a construct encoding this chimera did not support A-MuLV entry (Fig. 3). The chimera supports 10A1 entry (23); moreover, it was shown to be an efficient GALV receptor in a parallel experiment (Fig. 3), and the lack of A-MuLV receptor function of chimera Pho/P1(2+4) is therefore not due to the lack of cell surface expression of the protein. Thus, the introduced Pit1 loop 4 sequence manifests a negative effect on the A-MuLV receptor function introduced by the Pit1 loop 2 sequence, and it may thus indeed represent a Pit1 sequence that is incompatible with A-MuLV receptor function when present in Pit1.
Specific combinations of loop 2 and loop 4 sequences determine A-MuLV receptor function.
The results obtained from the receptor function analyses of Pho-4 chimeras harboring loop sequences from Pit1 and/or Pit2 are summarized in Table 1. All Pho-4 chimeras harboring Pit2 sequence in loop 4 were functional A-MuLV receptors, while the presence of Pit1 sequence in loop 4 was incompatible with A-MuLV receptor function. The Pho-4 loop 4 sequence was compatible with A-MuLV receptor function only when the divergent part of loop 2 was derived from Pit1.
TABLE 1.
Ability of combinations of loop sequences in a Pho-4 backbone to support A-MuLV entrya
| Loop 2 sequence origin | Loop 4 sequence origin
|
||
|---|---|---|---|
| Pho-4 | Pit1 | Pit2 | |
| Pho-4 | Nob | No | Yes |
| Pit1 | Yes | No | Yes |
| Pit2 | No | No | Yes |
The loop sequences present in the chimeras are indicated in Fig. 1. The loop 2 and loop 4 sequences were derived from receptors Pit1 and Pit2 and N. crassa protein Pho-4 as indicated in the table.
No, this combination of loop sequences does not confer A-MuLV receptor function upon Pho-4.
DISCUSSION
Pit2 is the human receptor for A-MuLV, while the related protein Pit1 supports entry by GALV and FeLV-B but not by A-MuLV. We (22) and others (6, 19) have observed that when the N-terminal two-thirds or the C-terminal third of Pit2 or RatPit2 were exchanged for the corresponding Pit1 sequences, all chimeric proteins retained A-MuLV receptor function. A possible interpretation of these results is that Pit1 harbors sequences which can specify A-MuLV receptor function when presented in a protein context other than Pit1 itself, e.g., in Pit1-Pit2 hybrids. We have shown here that the introduction of the Pit1-specific loop 2 sequence conferred A-MuLV receptor function upon the N. crassa protein, Pho-4 (Fig. 2), although neither Pit1 nor Pho-4 are efficient A-MuLV receptors. Therefore, first, a Pit1 sequence can specify A-MuLV receptor function when presented in a protein context other than that provided by Pit1 itself. Second, a functional retroviral receptor can be constructed by combining sequences from two proteins, each lacking receptor function for the retrovirus in question. Finally, as previously shown for GALV (25), the fungal phosphate transporter Pho-4 can serve as a receptor backbone for the mammalian retrovirus A-MuLV.
The observation that the introduction of a Pit1-specific loop 2 sequence confers A-MuLV receptor function upon Pho-4 indicates a role of loop 2 in A-MuLV entry. Recent studies by Leverett and coworkers showed that A-MuLV receptor function is conferred upon Pit1 by introduction of C-terminal Pit2-specific loop 2 residues (Pit2 positions 121 through 141) (chimera Pit1-4 [12]). Moreover, we find that exchange of the C-terminal part of Pit1 loop 2 for the corresponding Pit2 sequence (Pit2 positions 130 through 141) results in a functional A-MuLV receptor (5). These results demonstrate that residues in the second extracellular loop are important for A-MuLV receptor function and suggest that loop 2 harbors an N-terminal determinant of A-MuLV receptor function.
Previous results indicate that sequences in loop 4 constitute a C-terminal determinant of A-MuLV receptor function, e.g., Pit2 and RatPit2 region A in the C-terminal part of loop 4 was found to confer A-MuLV receptor function upon Pit1 (19, 22), and a single amino acid insertion in MusPit1 region A resulted in an A-MuLV receptor (13). We have here tested whether loop 4 sequences also play a role in A-MuLV receptor function in chimeras with a Pho-4 backbone. We found that the presence of a Pit2 loop 4 sequence conferred A-MuLV receptor function upon Pho-4, chimera Pho/P2(4) (Fig. 3), thus confirming the involvement of loop 4 sequences in receptor function for A-MuLV. The importance of loop 4 sequences in A-MuLV entry was further demonstrated by the result obtained with the chimera Pho/P1(2+4). This chimera supports infection by GALV and 10A1 and is thus present on the cell surface; however, it does not support A-MuLV infection in spite of the Pit1 loop 2 sequence present. Thus, loop 4 sequences can not only specify but also interfere with A-MuLV receptor function. Interestingly, the presence of the Pit2 loop 4 sequence or the Pho-4 loop 4 sequence did not interfere with the A-MuLV receptor function introduced by the Pit1 loop 2 sequence, chimeras Pho/P1(2)P2(4) and Pho/P1(2), respectively, as both of these chimeras support A-MuLV entry. The ability of the Pit1 loop 2 sequence to confer receptor function on Pho-4 is thus dependent on the sequence present in loop 4. Moreover, comparison of the A-MuLV receptor function of all the tested chimeras with a Pho-4 backbone revealed that whether a given loop 4 sequence will support A-MuLV entry may also be dependent on the sequence present in loop 2 (Table 1). Thus, although the presence of the Pit2 loop 4 sequence was compatible with A-MuLV infection and the presence of the Pit1 loop 4 sequence was incompatible with A-MuLV entry in all chimeras, the Pho-4 loop 4 sequence was compatible with A-MuLV receptor function only when the loop 2 sequence was derived from Pit1.
To summarize, we suggest that, first, A-MuLV receptor function is defined by sequences in both loop 2 and loop 4. Second, specific combinations of loop 2 and 4 sequences specify A-MuLV receptor function, i.e., whether a given loop 2 sequence will specify A-MuLV receptor function is dependent on the sequence present in loop 4 and vice versa; thus, sequences in loops 2 and 4 interdependently determine A-MuLV receptor function. Third, the observed interdependency of sequences in loops 2 and 4 in determining A-MuLV receptor function suggests that residues in loops 2 and 4 are spatially proximate.
Presently, the specific roles of loops 2 and 4 in A-MuLV entry are not known. It is possible that residues from loops 2 and 4 together create a nonlinear viral recognition site. However, it is also possible that the loop 2 and 4 sequences compatible with A-MuLV entry allow the SU protein to interact with another part(s) of the receptor, which then is directly critical for binding of the SU protein. Such a sequence(s) may be conserved between all wild-type and chimeric A-MuLV receptors, including Pho-4-derived chimeras. Interestingly, there is complete amino acid identity between all Pit1 and Pit2 homologs and Pho-4 in the six most N-terminal residues of loop 1 (Fig. 1). Moreover, the 10, 9, and 7 most N-terminal residues of loops 2, 4, and 5 in these proteins show 5, 7, and 5 conserved residues, respectively (Fig. 1) (8, 18, 20, 26, 31, 34, 35). However, other loop residues are also conserved between all known wild-type and chimeric A-MuLV receptors, and further investigations need to be performed in order to assess the possible role of conserved loop residues in A-MuLV entry. Besides, since the conserved N-terminal loop sequences are present in all Pit1 and Pit2 homologs and Pho-4, it is also possible that these might play a common role in receptor function for all viruses which can utilize wild-type and/or chimeric Pit proteins as receptors, that is, A-MuLV, 10A1, GALV, and FeLV-B (2, 3, 6, 8, 9, 12, 13, 19, 20, 22, 23, 25–29, 31, 34, 35).
An interesting observation comes from a comparison of data presented here with recent data published by Leverett and coworkers (12). As mentioned above, the presence of Pit2 loop 2 residues 121 through 141 can confer A-MuLV receptor function upon Pit1 (12); however, the presence of the same Pit2 loop 2 sequence cannot confer this function upon the chimera Pho/P1(4)P2(2) (Fig. 1 and 3). Thus, in the Pit1 backbone, the Pit2 loop 2 and the Pit1 loop 4 sequences are compatible with A-MuLV receptor function, but in the Pho-4 backbone, they are incompatible. This observation is not trivial, in that other combinations of loop 2 and 4 sequences in Pho-4 backbones support A-MuLV infection. This observation therefore suggests that the ability of specific combinations of loop 2 and 4 sequences to support A-MuLV entry also is dependent on the receptor backbone, which here means the sequences outside the divergent parts of loops 2 and 4. The receptor backbone is the major determinant of the structural organization of the receptor proteins and may thus influence the relative positioning of residues in loop 2 and loop 4 with respect to each other. Therefore, the backbone of the receptor might be expected to influence the A-MuLV receptor function no matter whether loop 2 and 4 residues are directly involved in binding of the viral SU protein or create a binding site in another part of the receptor as discussed above.
A role of protein backbone in modulating receptor function is also supported by data previously published by Miller and Miller (19). They reported on a chimera (RGG) harboring the N-terminal part of RatPit2 (encompassing loops 1 and 2) and supporting low levels of infection by A-MuLV. However, when the middle part of RatPit2 (encompassing loop 3 and the large intracellular domain) was introduced into Pit1 in addition to the N terminus of RatPit2, the resulting chimera (RRG) supported A-MuLV infection at the wild-type level, although the middle part of RatPit2 alone did not confer A-MuLV receptor function upon Pit1 (chimera GRG). Recently, Leverett and coworkers (12) obtained similar results with chimeras between Pit1 and Pit2. Thus, since the Pit2 sequence encompassing loop 3 and the large intracellular domain alone cannot confer A-MuLV receptor function upon Pit1, we suggest that these sequences influence receptor function by modulating the structural organization of the proteins. In line with this hypothesis, Tailor and Kabat (27) have pointed out the possibility that the large intracellular domain plays a role in receptor function due to its conceivable influence on overall receptor folding.
It has previously been suggested by us (13, 22) and others (12), based on results obtained on Pit1-Pit2, Pit1-RatPit2, and Pit1-HaPit2 chimeras and on a MusPit1 mutant, that A-MuLV receptor function is defined by a combination of sequences in the N- and C-terminal parts of the receptors. We have here elaborated on this model and suggest that specific combinations of loop 2 and loop 4 sequences determine A-MuLV receptor function. Moreover, as discussed above, comparison of the data presented here on Pho-4-derived chimeras with the result obtained with a Pit1-Pit2 chimera harboring a Pit2 loop 2 sequence (12) suggests that the receptor backbone, which here means receptor sequences outside the divergent regions of loops 2 and 4, modulates A-MuLV receptor function determined by sequences in these two loops.
Recently, Tailor and Kabat (27) studied the ability of viruses carrying chimeric A-MuLV-FeLV-B SU proteins to utilize Pit1-RatPit2 chimeras as receptors. For some of these chimeras, they found that the presence of FeLV-B and A-MuLV VRAs and the C-terminal parts (comprising loops 4 and 5) from Pit1 and Pit2, respectively, correlated with high infection levels. A similar correlation was found between the presence of FeLV-B and A-MuLV VRBs and the N-terminal parts (comprising loops 1 and 2) from Pit1 and Pit2, respectively. Based on these observations the authors suggested that A-MuLV VRA interacts with Pit2 loops 4 and 5 and A-MuLV VRB interacts with Pit2 loop 2 and that both these interactions are required for A-MuLV entry. The majority of the chimeric receptors were not tested for A-MuLV receptor function in this study. However, several observations on A-MuLV receptor function of the same Pit1-RatPit2 chimeras by Miller and Miller (19) and of similar chimeras between Pit1 and Pit2 by us (22) and others (6) appear to be in disagreement with the model proposed by Tailor and Kabat (27). For example, a chimera harboring RatPit2 loops 1, 2, and 3 and Pit1 loops 4 and 5 support wild-type level of infection by A-MuLV (chimera RRG in Miller and Miller [19]). Moreover, a chimera harboring Pit1 loops 1, 2, and 3 and RatPit2 loops 4 and 5 is also an efficient A-MuLV receptor (chimera GGR in Miller and Miller [19]). In addition, introduction of a Pit2 loop 2 sequence only (5, 12) or of a Pit2 loop 4 sequence only (19, 22) in Pit1 both resulted in functional A-MuLV receptors. Thus, A-MuLV can use these chimeras for entry, although neither of them harbors both loop 2 and loops 4 and 5 from Pit2. These observations can, however, be explained by the elaborated model on A-MuLV receptor function presented here. Thus, although we also suggest that loop 2 and 4 sequences both are involved in A-MuLV receptor function, we furthermore suggest, based on the data presented here, that A-MuLV is not dependent on the specific Pit2 loop 2 and Pit2 loop 4 sequences for entry. Rather, loops 2 and 4 interdependently determine A-MuLV receptor function, and the role played by loops 2 and 4 in A-MuLV entry can be fulfilled by several different combinations of loop 2 and loop 4 sequences. We also predict that the residues in loop 2 and loop 4, identified in this study as specifying A-MuLV receptor function, are to be found among those not conserved among Pho-4, Pit1, and Pit2.
As mentioned above, chimeric proteins in which either the N-terminal or the C-terminal parts of Pit2 or RatPit2 were replaced by the corresponding Pit1 sequences retained A-MuLV receptor function (6, 19, 22). However, comparable chimeras between Pit1 and HaPit2 supported A-MuLV infection less efficiently, although HaPit2 is as efficient an A-MuLV receptor as is Pit2 (6). Interestingly, in the extracellular loops, HaPit2 only differs from both Pit2 and RatPit2 in loops 2, 4, and 5 (18, 35). It remains to be investigated whether the differences in loops 2 and 4 can account for the lower efficiency by which Pit1-HaPit2 chimeras support A-MuLV entry compared to Pit1-Pit2 and Pit1-RatPit2 chimeras. However, it is possible that study of the loop 2 and 4 sequence differences present among naturally occurring efficient A-MuLV receptors might provide further insight into which combinations of loop 2 and 4 residues will allow for A-MuLV receptor function and thus also elucidate the role of loops 2 and 4 sequences in A-MuLV entry.
ACKNOWLEDGMENTS
We thank Maribeth V. Eiden for the PG13GBN and PA317GBN cell lines. We furthermore thank Ann Hørning and Bente Andersen for excellent technical assistance.
This work was supported by the Karen Elise Jensen Foundation and the Danish Biotechnology Programme.
REFERENCES
- 1.Adam M A, Ramesh N, Miller A D, Osborne W R. Internal initiation of translation in retroviral vectors carrying picornavirus 5′ nontranslated regions. J Virol. 1991;65:4985–4990. doi: 10.1128/jvi.65.9.4985-4990.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boomer S, Eiden M, Burns C C, Overbaugh J. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J Virol. 1997;71:8116–8123. doi: 10.1128/jvi.71.11.8116-8123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaudry G J, Eiden M V. Mutational analysis of the proposed gibbon ape leukemia virus binding site in Pit1 suggests that other regions are important for infection. J Virol. 1997;71:8078–8081. doi: 10.1128/jvi.71.10.8078-8081.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chien M L, Foster J L, Douglas J L, Garcia J V. The amphotropic murine leukemia virus receptor gene encodes a 71-kilodalton protein that is induced by phosphate depletion. J Virol. 1997;71:4564–4570. doi: 10.1128/jvi.71.6.4564-4570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dreyer, K., and L. Pedersen. Unpublished results.
- 6.Eiden M V, Farrell K B, Wilson C A. Substitution of a single amino acid residue is sufficient to allow the human amphotropic murine leukemia virus receptor to also function as a gibbon ape leukemia virus receptor. J Virol. 1996;70:1080–1085. doi: 10.1128/jvi.70.2.1080-1085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorman C. High efficiency gene transfer into mammalian cells. In: Glover D M, editor. DNA cloning. 2. A practical approach. Oxford, United Kingdom: IRL Press; 1985. pp. 143–190. [Google Scholar]
- 8.Johann S V, Gibbons J J, O’Hara B. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J Virol. 1992;66:1635–1640. doi: 10.1128/jvi.66.3.1635-1640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johann S V, van Zeijl M, Cekleniak J, O’Hara B. Definition of a domain of GLVR1 which is necessary for infection by gibbon ape leukemia virus and which is highly polymorphic between species. J Virol. 1993;67:6733–6736. doi: 10.1128/jvi.67.11.6733-6736.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kavanaugh M P, Miller D G, Zhang W, Law W, Kozak S L, Kabat D, Miller A D. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 12.Leverett B D, Farrell K B, Eiden M V, Wilson C A. Entry of amphotropic murine leukemia virus is influenced by residues in the putative second extracellular domain of its receptor, Pit2. J Virol. 1998;72:4956–4961. doi: 10.1128/jvi.72.6.4956-4961.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lundorf M D, Pedersen F S, O’Hara B, Pedersen L. Single amino acid insertion in loop 4 confers amphotropic murine leukemia virus receptor function upon murine Pit1. J Virol. 1998;72:4524–4527. doi: 10.1128/jvi.72.5.4524-4527.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mann B J, Bowman B J, Grotelueschen J, Metzenberg R L. Nucleotide sequence of pho-4+, encoding a phosphate-repressible phosphate permease of Neurospora crassa. Gene. 1989;83:281–289. doi: 10.1016/0378-1119(89)90114-5. [DOI] [PubMed] [Google Scholar]
- 15.McLachlin J R, Mittereder N, Daucher M B, Kadan M, Eglitis M A. Factors affecting retroviral vector function and structural integrity. Virology. 1993;195:1–5. doi: 10.1006/viro.1993.1340. [DOI] [PubMed] [Google Scholar]
- 16.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 18.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller D G, Miller A D. A family of retroviruses that utilize related phosphate transporters for cell entry. J Virol. 1994;68:8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Hara B, Johann S V, Klinger H P, Blair D G, Rubinson H, Dunn K J, Sass P, Vitek S M, Robins T. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 21.Olah Z, Lehel C, Anderson W B, Eiden M V, Wilson C A. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J Biol Chem. 1994;269:25426–25431. [PubMed] [Google Scholar]
- 22.Pedersen L, Johann S V, van Zeijl M, Pedersen F S, O’Hara B. Chimeras of receptors for gibbon ape leukemia virus/feline leukemia virus B and amphotropic murine leukemia virus reveal different modes of receptor recognition by retrovirus. J Virol. 1995;69:2401–2405. doi: 10.1128/jvi.69.4.2401-2405.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedersen, L., M. D. Lundorf, M. van Zeijl, F. S. Pedersen, and B. O’Hara. Unpublished data.
- 24.Pedersen, L., and B. O’Hara. Unpublished results.
- 25.Pedersen L, van Zeijl M, Johann S V, O’Hara B. Fungal phosphate transporter serves as a receptor backbone for gibbon ape leukemia virus. J Virol. 1997;71:7619–7622. doi: 10.1128/jvi.71.10.7619-7622.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneiderman R D, Farrell K B, Wilson C A, Eiden M V. The Japanese feral mouse Pit1 and Pit2 homologs lack an acidic residue at position 550 but still function as gibbon ape leukemia virus receptors: implications for virus binding motif. J Virol. 1996;70:6982–6986. doi: 10.1128/jvi.70.10.6982-6986.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tailor C S, Kabat D. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J Virol. 1997;71:9383–9391. doi: 10.1128/jvi.71.12.9383-9391.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tailor C S, Takeuchi Y, O’Hara B, Johann S V, Weiss R A, Collins M K. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infections. J Virol. 1993;67:6737–6741. doi: 10.1128/jvi.67.11.6737-6741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi Y, Vile R G, Simpson G, O’Hara B, Collins M K, Weiss R A. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol. 1992;66:1219–1222. doi: 10.1128/jvi.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner D L, Cepko C L. A common progenitor for neurons and glia persists in rat retina late in development. Nature (London) 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- 31.van Zeijl M, Johann S V, Closs E, Cunningham J, Eddy R, Shows T B, O’Hara B. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Versaw W K, Metzenberg R L. Repressible cation-phosphate symporters in Neurospora crassa. Proc Natl Acad Sci USA. 1995;92:3884–3887. doi: 10.1073/pnas.92.9.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson C A, Eiden M V, Anderson W B, Lehel C, Olah Z. The dual-function hamster receptor for amphotropic murine leukemia virus (MuLV), 10A1 MuLV, and gibbon ape leukemia virus is a phosphate symporter. J Virol. 1995;69:534–537. doi: 10.1128/jvi.69.1.534-537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson C A, Farrell K B, Eiden M V. Comparison of cDNAs encoding the gibbon ape leukemia virus receptor from susceptible and non-susceptible murine cells. J Gen Virol. 1994;75:1901–1908. doi: 10.1099/0022-1317-75-8-1901. [DOI] [PubMed] [Google Scholar]
- 35.Wilson C A, Farrell K B, Eiden M V. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J Virol. 1994;68:7697–7703. doi: 10.1128/jvi.68.12.7697-7703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]