Abstract
An H5N1 avian influenza A virus was transmitted to humans in Hong Kong in 1997. Although the virus causes systemic infection and is highly lethal in chickens because of the susceptibility of the hemagglutinin to furin and PC6 proteases, it is not known whether it also causes systemic infection in humans. The clinical outcomes of infection in Hong Kong residents ranged widely, from mild respiratory disease to multiple organ failure leading to death. Therefore, to understand the pathogenesis of influenza due to these H5N1 isolates, we investigated their virulence in mice. The results identified two distinct groups of viruses: group 1, for which the dose lethal for 50% of mice (MLD50) was between 0.3 and 11 PFU, and group 2, for which the MLD50 was more than 103 PFU. One day after intranasal inoculation of mice with 100 PFU of group 1 viruses, the virus titer in lungs was 107 PFU/g or 3 log units higher than that for group 2 viruses. Both types of viruses had replicated to high titers (>106 PFU/g) in the lungs by day 3 and maintained these titers through day 6. More importantly, only the group 1 viruses caused systemic infection, replicating in nonrespiratory organs, including the brain. Immunohistochemical analysis demonstrated the replication of a group 1 virus in brain neurons and glial cells and in cardiac myofibers. Phylogenetic analysis of all viral genes showed that both groups of Hong Kong H5N1 viruses had formed a lineage distinct from those of other viruses and that genetic reassortment between H5N1 and H1 or H3 human viruses had not occurred. Since mice and humans harbor both the furin and the PC6 proteases, we suggest that the virulence mechanism responsible for the lethality of influenza viruses in birds also operates in mammalian hosts. The failure of some H5N1 viruses to produce systemic infection in our model indicates that multiple, still-to-be-identified, factors contribute to the severity of H5N1 infection in mammals. In addition, the ability of these viruses to produce systemic infection in mice and the clear differences in pathogenicity among the isolates studied here indicate that this system provides a useful model for studying the pathogenesis of avian influenza virus infection in mammals.
An H5N1 avian influenza A virus was transmitted from birds to humans in 1997 in Hong Kong, infecting 18 humans, 6 of whom died (3, 4). This outbreak was unique in that the virus that was transmitted to humans is lethal in chickens (20, 22). Although the virulence of avian influenza viruses is polygenic, the susceptibility of the hemagglutinin (HA) to host proteases is the major determinant for this property. That is, influenza virus HA must be cleaved into HA1 and HA2 subunits for the virus to be infectious, as this event generates the amino terminus of HA2, which mediates the fusion of the viral envelope with the endosomal membrane (13, 15). Lethal and nonlethal avian viruses differ in this mode of activation: the HA of the former is cleaved by the ubiquitous proteases furin and PC6 (9, 19), whereas the HA of the latter is not susceptible to these proteases but rather is cleaved by proteases localized in the respiratory or intestinal organs or both (12). We have shown that pathogenic avian viruses replicate in the capillary endothelial cells of a variety of organs, leading to the hemorrhagic manifestations found in infected chickens (14). Similarly, the H5N1 Hong Kong virus replicated in the capillary endothelial cells of chickens (20, 22). Despite the high mortality rate (33%) associated with H5N1 virus infection, it is still unclear whether the virus has the potential to cause systemic infection in humans.
The clinical outcomes of H5N1 virus infection in apparently healthy humans ranged from mild respiratory symptoms to death (26). Epidemiologic studies indicate that there has been no human-to-human transmission of the H5N1 virus, suggesting that the human cases in Hong Kong originated from independent transmissions of the virus from birds. An H5N1 virus that is genetically related to that isolated from the patients was isolated from chickens in April 1997 in Hong Kong (5). In fact, H5N1 viruses cocirculating among birds in Hong Kong in December 1997 were genetically heterogeneous (4). Therefore, biologic heterogeneity among infecting strains of the H5N1 virus could have accounted for the different clinical outcomes seen in patients.
In this study, we examined the extent of biologic and genetic heterogeneities among the human H5N1 isolates in an attempt to explain the differences in clinical manifestations seen in infected patients. To this end, we investigated the virulence and pathobiological features of human H5N1 isolates in mice and chickens and established the phylogenetic relationships among these viruses.
MATERIALS AND METHODS
Viruses and cells.
H5N1 influenza A viruses isolated from patients during the Hong Kong outbreak in 1997 were obtained from the Centers for Disease Control and Prevention (CDC) through the courtesy of Nancy Cox and are listed in Table 1. In the text, these viruses are designated with “HK” plus the field number; for example, HK156 represents A/Hong Kong/156/97 (H5N1). They were isolated and propagated in Madin-Darby canine kidney (MDCK) cells. A/Udorn/307/72 (H3N2) (Udorn) was obtained from Robert G. Webster, St. Jude Children’s Research Hospital, and had been isolated and grown in embryonated eggs. MDCK cells were cultured in minimal essential medium with 5% newborn calf serum. All of the experiments with live H5N1 viruses isolated in Hong Kong were done in a biosafety level 3 containment laboratory approved for such use by the CDC and the U.S. Department of Agriculture.
TABLE 1.
Clinical and laboratory features of the human H5N1 viruses used in this study
| Virus | Casea | Age (yr)/ genderab | Date of onset in 1997a | Clinical outcomea | Plaque size in MDCK cellsc | Passage historyad | MLD50 (PFU) | CLD50 (PFU) | PB2 amino acid at position 627 |
|---|---|---|---|---|---|---|---|---|---|
| Group 1 (pathogenic in mice) | |||||||||
| A/Hong Kong/156/97 | 1 | 3/M | May 9 | Died | Pinpoint (∼20%), medium (∼80%) | C2/C3 | 0.6 | 2.3 × 102 | Glue |
| A/Hong Kong/483/97 | 3 | 13/F | November 20 | Died | Large | C1/C3 | 0.3 | 1.7 × 102 | Lys |
| A/Hong Kong/485/97 | 7 | 24/F | December 4 | Critical/ discharged | Medium | C2/C2 | 1.1 | NTf | Lys |
| A/Hong Kong/491/97 | 9 | 4/M | December 10 | Discharged | Large | X/E1C1 | 11 | NT | Glu |
| A/Hong Kong/514/97 | 13 | 25/F | December 17 | Died | Large | X/C2 | 1.0 | NT | Lys/Glug |
| A/Hong Kong/516/97 | 12 | 60/F | December 16 | Died | Large | X/C3 | 5.9 | NT | Glu |
| Group 2 (less pathogenic in mice) | |||||||||
| A/Hong Kong/481/97 | 2 | 2/M | November 6 | Discharged | Pinpoint | X/C3 | 6.0 × 103 | NT | Glu |
| A/Hong Kong/482/97 | 4 | 54/M | November 24 | Died | Pinpoint | C1/C3 | 9.3 × 103 | NT | Glu |
| A/Hong Kong/486/97 | 5h | 5/F | December 7 | Discharged | Pinpoint | C2/C2 | >6.0 × 104 | 2.3 × 102 | Glu |
| A/Hong Kong/488/97 | 8h | 2/M | December 12 | Discharged | Pinpoint | C1/C2 | 4.2 × 105 | NT | Glu |
Information obtained from the CDC.
M, male; F, female.
Pinpoint, diameter of <0.5 mm; medium, diameter of 0.5 to 2.0 mm; large, diameter of >2.0 mm.
Passage history in Hong Kong/in the United States. C, MDCK cells; E, eggs; X, unknown, but most likely less than two passages in MDCK cells. The number indicates the number of passages.
All 21 PCR clones had Glu.
NT, not tested.
Two PCR clones had Lys and one had Glu.
From a familial cluster.
Experimental infections.
To determine the dose lethal for 50% of mice (MLD50), we infected 6-week-old female BALB/c mice, anesthetized with methoxyflurane, intranasally with 50 μl of serial 10-fold dilutions of virus and observed them for 2 weeks. Virus titers in organs were determined by use of MDCK cells 1, 3, 6, and 10 days after intranasal inoculation of the mice with 100 PFU. To determine the dose lethal for 50% of chickens (CLD50), we infected 2-week-old specific-pathogen-free chickens (SPAFAS) intranasally and orally with 100 μl of serial 10-fold dilutions of virus and observed them for 2 weeks. Virus titers in the organs of chickens infected with 105 PFU were determined as described above.
Immunohistochemistry.
Formalin-fixed and paraffin-embedded sections were deparaffinized, hydrated, and treated with 0.1% Triton X-100 in phosphate-buffered saline–bovine serum albumin. Endogenous peroxidases were quenched with 0.3% peroxide in methanol. Sections were incubated with a monoclonal antibody (NP5/1) specific for the nucleoprotein of influenza virus at a dilution of 1:400 for 1 h at room temperature. Bound antibody was detected by the peroxidase-labeled streptavidin-biotin staining method (Vectastain ABC kit; Vector Laboratories) with diaminobenzidine as the substrate. Sections were counterstained with hematoxylin.
Sequencing analysis.
Viral RNA was isolated from the virus-containing culture fluid of MDCK cells as previously described (1). The cDNA was synthesized by use of reverse transcriptase and an oligonucleotide complementary to the conserved 3′ end of the viral RNA as described by Katz et al. (10). Genes were amplified by PCR with this cDNA, gene-specific oligonucleotide primers, and LA Taq polymerase (Panvera). PCR products were cloned into a plasmid and sequenced with an Autosequencer (Applied Biosystems Inc.) in accordance with the protocol recommended by the manufacturer. The sequences of the oligonucleotide primers will be supplied upon request. At least three independent cDNA clones were sequenced for each gene. When one of the cDNA clones contained a different nucleotide at a given position, it was considered to be an error introduced by the polymerase during PCR, unless otherwise stated. Phylogenetic analysis of the sequence data was performed with Clustal W software, version 1.6 (23), which relies on the neighbor-joining method (17) to generate phylogenetic trees.
RESULTS
Replication in tissue cultures and eggs.
The virus titers of stocks measured as PFU in MDCK cells and 50% egg infectious doses were similar to each other. There was an appreciable difference in plaque size among the H5N1 viruses in MDCK cells. Five viruses—HK483, HK485, HK491, HK514, and HK516—produced medium to large plaques, whereas four produced pinpoint plaques (Table 1). HK156 displayed a mixed population with respect to plaque size: ∼20% pinpoint and ∼80% medium plaques. These results suggested biologic differences among these viruses.
Human H5N1 isolates differ in virulence for mice.
One explanation for the differences in clinical manifestations among the patients in Hong Kong may be that the H5N1 viruses differ in their ability to replicate in mammalian hosts. Thus, we compared the virulence of 10 Hong Kong viruses in mice. HK156, HK483, HK485, HK491, HK514, and HK516 (group 1) were highly pathogenic, with an MLD50 of less than 11 PFU, whereas the remaining four viruses (group 2) were clearly less pathogenic, as indicated by an MLD50 of greater than 103 PFU. There was, in addition, a correlation between virulence and plaque size (Table 1), with the less pathogenic viruses tending to produce pinpoint plaques, while the pathogenic viruses produced medium to large plaques. Only the HK156 virus, which was highly lethal for mice, produced a mixed population of plaques. Regardless of their pathogenicity, all of the viruses induced disease symptoms within 24 h, including rapid breathing, ruffled fur, and hunched posture. In contrast, other mouse-adapted human viruses, such as A/WSN/33 (H1N1) and A/PR/8/34 (H1N1), usually do not induce these effects until 3 days after infection (11a), even though they are lethal for mice.
To further examine the pathogenicity of the Hong Kong isolates, we compared the kinetics of virus growth in mouse organs by infecting the animals with 100 PFU of a representative virus from each group, HK483 and HK486. We also tested the index Hong Kong H5N1 isolate, HK156, and a non-mouse-adapted human virus, Udorn. Both the HK156 isolate and the HK483 isolate replicated to extremely high titers (>107 PFU/g) in the lungs on day 1 and continued to replicate at this level until day 3, with titers remaining high even on day 6 (Table 2). In nasal turbinates, both viruses reached maximal titers on day 3 and remained at this level until day 6. The HK483 virus spread to other organs, including the liver, kidney, pancreas, and intestine, but the virus titers in these organs were at least 2 log units lower than those in respiratory organs. This virus was recovered at high titers from the brain on day 6. HK156 also spread to other organs, but to a lesser extent than did HK483. In contrast, HK486 replication in the lungs was at least 103-fold lower than that of HK483 on day 1, although it reached more than 106 PFU/g on days 3 and 6. In nasal turbinates, HK486 replicated only marginally, and it did not spread to other organs. The human virus Udorn replicated only in the upper and lower respiratory tracts. On day 10, we were unable to recover any virus from mice. Because both HK156 and HK483 killed all of the mice by day 8, we were unable to test virus titers in mice on day 10. Thus, the group 1 H5N1 strains were pathogenic for mice, causing systemic infection, whereas the less pathogenic strain (group 2) replicated only in respiratory organs. The data also suggest that group 1 viruses replicate more rapidly in the primary replication site in mice (i.e., lungs) than do group 2 viruses.
TABLE 2.
Kinetics of virus replication in micea
| Day | Virus | Virus titer (mean log PFU ± SD/g) in:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Lungs | Nasal turbinates | Spleen | Liver | Kidneys | Brain | Pancreas | Colon | ||
| 1 | HK156 | 7.2 ± 0.4 | 3.6 ± 0.3 | 2.8 | 2.8 | — | — | — | — |
| HK483 | 7.0 ± 0.5 | 2.5 | 2.2 ± 0.7 | — | 1.9 ± 0.3 | — | — | — | |
| HK486 | 3.6 ± 0.3 | — | — | — | — | — | — | — | |
| Udorn | 4.6 ± 0.8 | 1.5 | — | — | — | — | — | — | |
| 3 | HK156 | 7.8 ± 0.2 | 6.0 ± 0.6 | 2.2 ± 0.5 | — | — | — | — | — |
| HK483 | 7.3 ± 0.3 | 5.6 ± 0.7 | 5.3 ± 0.4 | 2.6 ± 0.1 | 2.1 ± 0.8 | — | 2.5 | 2.5 | |
| HK486 | 6.1 ± 0.5 | 1.5, 1.5 | — | — | — | — | — | — | |
| Udorn | 6.3 ± 0.2 | 6.7, 6.3 | — | — | — | — | — | — | |
| 6 | HK156 | 7.8b | 7.4b | — | — | — | — | — | — |
| HK483 | 6.5 ± 0.3 | 6.0 ± 0.1 | 2.0 ± 0.4 | — | 3.9 | 4.9 ± 0.0 | — | 2.5 | |
| HK486 | 6.1 ± 0.8 | 2.2 | — | — | — | — | — | — | |
| Udorn | 4.9 ± 0.7 | 4.4, 4.9 | — | — | — | — | — | — | |
BALB/c mice, anesthetized with methoxyflurane, were infected intranasally with 50 μl of virus (100 PFU). Three mice from each virus-infected group were sacrificed on days 1, 3, 6, and 10 postinfection for virus titration. None of the mice infected with HK156 or HK483 survived more than 8 days after infection. Virus was not recovered from any of the mice infected with HK486 or Udorn on day 10. When virus was not recovered from all three mice, individual titers are reported. —, no virus isolation.
Only one mouse survived until day 6.
Immunohistochemical analysis of virus replication in the brains and hearts of mice.
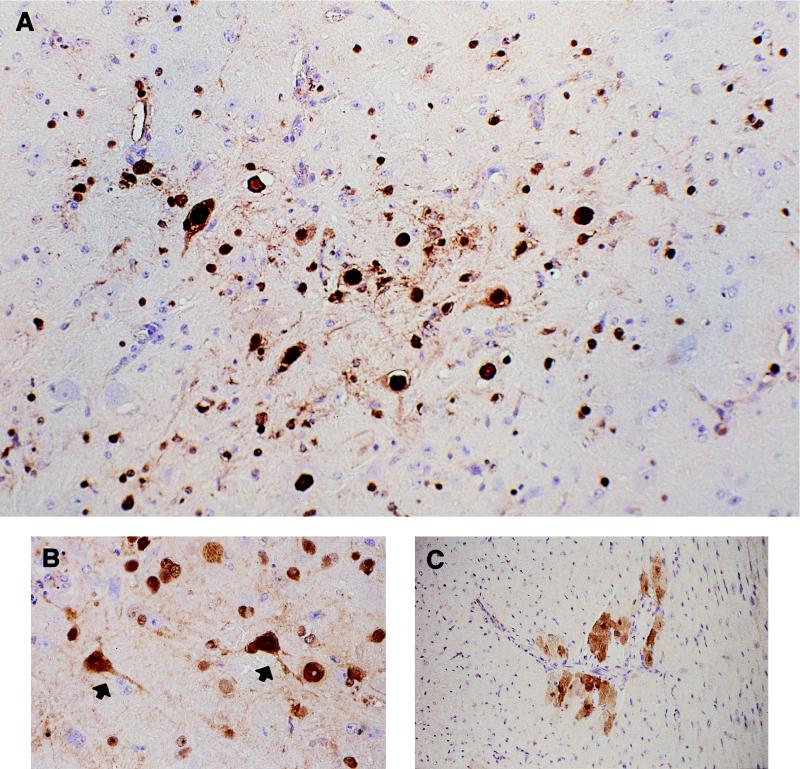
To test whether HK483 indeed replicated in the mouse brain or whether its recovery from this organ was simply due to viremia, we performed immunohistochemical analysis of organs from HK483-infected mice by using a monoclonal antibody specific for the nucleoprotein of influenza A virus. On day 5 (Fig. 1A and B) and day 6 (data not shown), tissue specimens from the brain stem at the metencephalon showed multiple foci that stained intensely with this antibody in both the nucleus and the cytoplasm of neurons as well as glial cells. Because lesions were also apparent by histopathologic examination in the hearts of mice infected with HK483 on day 6 (data not shown), we also elected to examine hearts by immunohistochemical methods. Necrotic cardiac myofibers were positive for viral antigen in both the nucleus and the cytoplasm (Fig. 1C). These results prove that the H5N1 virus causes systemic infection in mice.
FIG. 1.
Immunohistochemical analysis of mice infected with a mouse-pathogenic Hong Kong H5N1 virus. Mice were infected intranasally with 100 PFU of the mouse-pathogenic HK483 virus. Mice were sacrificed, and brains (day 5; A and B) and hearts (day 6; C) were processed for identification of influenza virus replication with a monoclonal antibody specific for nucleoprotein. (A and B) Nonsuppurative encephalitis showing intense nuclear and slightly less intense cytoplasmic staining (brown) of influenza virus nucleoprotein in neurons (arrows) and glial cells of the brain stem at the metencephalon. Magnifications, ×100 (A) and ×400 (B). (C) Staining (brown) of the nucleoprotein in the nucleus and the cytoplasm of necrotic cardiac myofibers. Magnification, ×200.
Comparison of virulence for chickens.
Because the index human isolate of the Hong Kong H5N1 virus, HK156, retained its virulence in chickens (22), we compared the virulence of representative viruses from group 1 and group 2 in chickens. As shown by CLD50 values, the three viruses tested—HK156, HK483, and HK486—were equally pathogenic for chickens (Table 1). Interestingly, the two group 1 viruses (HK156 and HK483) were less pathogenic for chickens than they were for mice (MLD50, <1 PFU; CLD50, ∼200 PFU).
To determine the extent of virus growth, we infected chickens with 105 PFU of HK483 and HK486 and examined the virus titers in organs 1 day after infection. None of the infected chickens were alive by day 3. A relatively high virus dose was selected for these experiments because the dose used in mice (100 PFU) is lower than the CLD50. Both viruses were recovered from all organs tested (Table 3), indicating that they replicate systemically within 1 day. These results confirm that the H5N1 viruses from human patients retain their virulence in chickens (22), although the dose required to kill chickens is higher than that for mice.
TABLE 3.
Systemic replication of human H5N1 viruses in chickensa
| Virus | Virus titer (mean log PFU ± SD/g) in:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Lungs | Nasal turbinates | Spleen | Liver | Kidneys | Brain | Pancreas | Colon | |
| HK483 | 6.5 ± 0.6 | 6.8 ± 0.4 | 5.2 ± 0.3 | 5.1 ± 0.3 | 4.9 ± 0.5 | 3.6 ± 0.7 | 4.8 ± 0.2 | 5.1 ± 0.5 |
| HK486 | 5.3 ± 0.9 | 4.1 ± 1.3 | 5.1 ± 0.7 | 5.3 ± 0.7 | 4.2 ± 0.9 | 3.7 ± 0.9 | 3.8 ± 0.9 | 4.8 ± 0.3 |
Specific-pathogen-free chickens (2 weeks old) were infected intranasally and orally with 100 μl of virus (total, 105 PFU). Three chickens from each virus-infected group were sacrificed 1 day after infection for virus titration. By day 3, all of the chickens infected with either HK483 or HK486 had died.
Phylogenetic analysis.
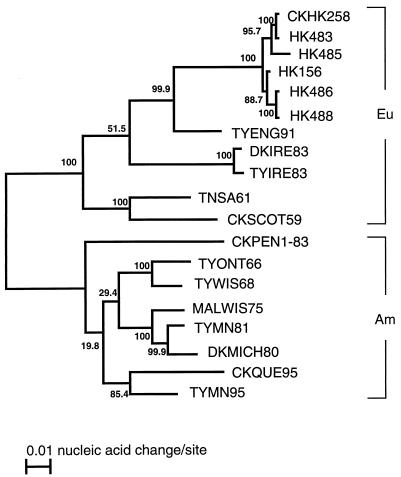
It is possible that the difference in mouse virulence among the human H5N1 isolates is attributable to genetic reassortment with other avian or human viruses. We therefore sequenced portions of the PA, PB1, PB2, NP, NA, and NS genes and the entire HA and M genes from two group 1 and two group 2 viruses. We then analyzed these sequences phylogenetically, together with the published sequences of HK156 and A/chicken/Hong Kong/258/97 (H5N1), which was isolated in April 1997 and is closely related to HK156 (5). Regardless of the genes tested, all of the Hong Kong H5N1 viruses examined formed a lineage that was distinct from those of other viruses. As an example, the phylogenetic tree of the HA genes is shown in Fig. 2. Although the H5N1 viruses were divided into two sublineages, they did not correlate with mouse virulence. Instead, each of the two sublineages contained viruses from both group 1 (highly pathogenic) and group 2 (less pathogenic). Interestingly, HK483 was most closely related to A/chicken/Hong Kong/258/97; only 4 nucleotides were different in the HA genes of these viruses. HK486 and HK488 were also highly similar; there were only 4 nucleotide differences between the two viruses among 6,321 nucleotides compared.
FIG. 2.
Phylogenetic relationships of the Hong Kong H5N1 virus HA genes. The nucleotide sequences of the entire HA genes were analyzed with the Clustal W program (23), which uses the algorithm of Myers and Miller (16). Numbers at nodes represent bootstrap values as a percentage of 1,000 resamplings of the data set. The lengths of the horizontal lines are proportional to the minimum number of nucleotide differences required to join nodes and HA sequences. Vertical lines are for spacing branches and labels; their lengths are not important. Strain abbreviations are explained elsewhere (5). Eu, Eurasian lineage; Am, American lineage.
Amino acid comparison of HA, PB2, and M1 proteins.
Because previous work had suggested that the HA and M1 proteins can affect the virulence of influenza A viruses in mice (18), we sought to compare the deduced amino acid sequences of group 1 (HK156, HK483, and HK485) and group 2 (HK486 and HK488) viruses. In the HA, one amino acid of HK156 differed from the published sequence (at the fourth position in the signal peptide—Ile in our sequence versus Thr in the sequence reported by Subbarao et al. [22]); all other amino acids were identical. None of the amino acid differences correlated with mouse virulence. Claas et al. (5) reported that A/chicken/Hong Kong/258/97 (H5N1), which is closely related to the human H5N1 isolates, contains a glycosylation site at position 156, whereas the human isolate HK156 does not. We found that HK483 and HK485 also possess this glycosylation site (N156ST for HK483 and N156SS for HK485), in contrast to HK486 and HK488 (N156SA for both viruses). Thus, the presence or absence of the glycosylation site cannot account for the observed differences in mouse virulence.
Alterations of amino acids at positions 41 and 139 in the M1 protein are thought to affect the virulence of influenza A viruses in mice (18, 24). Hence, we compared the M1 amino acid sequences of all of the viruses listed in Table 1. There were no differences at these two positions in any of the viruses.
Because the amino acid residue at position 627 of PB2 determines the replicative ability of the virus in MDCK cells (21), we also compared the amino acids at this position for all of the test viruses. Interestingly, amino acids at this position differed among the Hong Kong H5N1 viruses. Although each of the viruses that produced pinpoint plaques had Glu and three of those producing medium to large plaques had Lys, some of the viruses that formed large plaques (i.e., HK491 and HK516) also had Glu (Table 1). HK156, which produced a mixed population of plaques, had Glu at this position in all of the 21 PCR clones examined, while 2 of the HK514 PCR clones had Lys and another had Glu. Thus, the amino acid residue at position 627 of PB2 does not appear to correlate directly with plaque size or with virulence in mice.
DISCUSSION
We demonstrate here that the H5N1 viruses isolated from Hong Kong residents are biologically heterogeneous with respect to their replicative potential in mammalian hosts. Although some of the viruses were relatively benign, others were extremely pathogenic for mice (MLD50, <1 PFU). Moreover, the virulent HK483 strain caused systemic infection, replicating in the brains and hearts of mice. Influenza A viruses have also been isolated from the brains of seals (25), but it is not clear whether they replicated in that tissue or were present there due to viremia. Thus, our report presents the first direct evidence of systemic infection of a mammal with an avian influenza virus. Since mice possess the same proteases (i.e., furin and PC6) that allow systemic replication of influenza viruses in chickens, we suggest that the susceptibility of the HA to these proteases is critical for influenza viruses to cause systemic infection in both avian and mammalian hosts, but other viral factors also contribute to this ability.
Although the majority of human influenza A viruses do not replicate systemically in mice (11, 11a), A/WSN/33 (H1N1) (6), which was derived by repeated passage of A/WS/33 (H1N1) in mouse brain, replicates in this organ upon intracerebral inoculation. A/WSN/33 (H1N1) replicates in systemic organs, including the brain, even upon intranasal inoculation of mice (2). Interestingly, A/WSN/33 does not have multiple basic amino acids at the HA cleavage site, a property required for cleavage by the ubiquitous furin and PC6 proteases (9, 19). We have recently demonstrated that NA binds and sequesters plasminogen, leading to higher local concentrations of this ubiquitous protease precursor and thus to increased cleavage of the HA (8). Therefore, regardless of the mechanism (recognition of multiple basic amino acids by furin and PC6 proteases or sequestration of plasminogen by NA), HA cleavage by ubiquitous proteases seems a prerequisite for systemic influenza virus infection of animals.
Titration of viruses in mouse organs revealed that the pathogenic H5N1 viruses differed in replicative ability from the less pathogenic viruses. The former group caused systemic infection, whereas the latter remained in the respiratory tract. This outcome is inconsistent with the properties of the HK486 HA, which possesses multiple basic amino acids at the cleavage site and can be cleaved by the ubiquitous proteases (as indicated by plaque formation without trypsin [data not shown]). Thus, viral properties other than HA cleavage must contribute to the difference in virulence among these H5N1 viruses in mice. Comparison of the amino acid sequences of the HA and M1 proteins failed to disclose changes that might account for differences in replicative capacity.
The amino acid residue at position 627 in the PB2 protein is associated with plaque formation in MDCK cells (21). A single mutation from Glu to Lys at this position converts a non-plaque-forming virus to a plaque producer in these cells. All avian influenza A viruses contain Glu at position 627, whereas human influenza A viruses possess Lys (7). Interestingly, we found that the Hong Kong H5N1 viruses differed in the amino acid residue at this position, although this difference was not directly correlated with plaque size in MDCK cells or with virulence in mice. Nonetheless, because all viruses that produced pinpoint plaques in this study had Glu at position 627, as opposed to Lys in three of the six viruses that produced medium to large plaques, it is possible that amino acid substitutions in PB2 or other viral proteins allowed the remaining medium- to large-plaque producers with 627-Glu in PB2 (i.e., HK156, HK491, and HK516) to increase their replicative potential in these cells. It will be important to perform reassortment studies to identify the viral gene products that determine the difference in mouse virulence between the two groups of H5N1 viruses.
Despite their clustering in phylogenetic trees, the human H5N1 isolates were genetically divergent enough to form sublineages (4). One explanation for this genetic diversity is that the H5N1 virus had been circulating among Hong Kong poultry as early as March 1997 (when the outbreak of highly pathogenic avian H5N1 virus infections was identified on Hong Kong chicken farms), leaving adequate time for the generation of multiple sublineages. Two of the viruses (i.e., HK486 and HK488), isolated from two cousins, are genetically and biologically closely related, albeit not identical. This finding suggests that either a virus was transmitted from one cousin to the other or that both patients acquired the virus from the same avian source.
It is premature to conclude that the human H5N1 isolates differ in their virulence potential, as mouse pathogenicity may not extrapolate directly to humans. However, we find it intriguing that four of the six viruses in group 1 (highly virulent in mice) were isolated from patients who eventually died, whereas three of the four viruses in group 2 (less virulent in mice) were isolated from patients who eventually recovered (Table 1). Definitive studies for determining virulence heterogeneity will have to be conducted in primates. The disease outcomes of patients cannot be used as a criterion for evaluating virus virulence because of the heterogeneous nature of the genetic background and health conditions of patients. However, the ability of the viruses to produce systemic infection in mice and the clear differences in pathogenicity among the isolates studied indicate that our system provides a useful model for molecular analysis of avian influenza virus infection in mammals.
ACKNOWLEDGMENTS
We gratefully acknowledge Nancy Cox for the Hong Kong H5N1 viruses and Susan Watson and John Gilbert for editing the manuscript.
Support for this work was provided by National Institute of Allergy and Infectious Diseases Public Health Service research grants AI-29599 and AI-33898.
REFERENCES
- 1.Bean W J, Sriram G, Webster R G. Electrophoretic analysis of iodine-labeled influenza virus RNA segments. Anal Biochem. 1980;102:228–232. doi: 10.1016/0003-2697(80)90343-7. [DOI] [PubMed] [Google Scholar]
- 2.Castrucci M R, Kawaoka Y. Biologic importance of neuraminidase stalk length in influenza A virus. J Virol. 1993;67:759–764. doi: 10.1128/jvi.67.2.759-764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Isolation of avian influenza A (H5N1) viruses from humans—Hong Kong, May–December 1997. Morbid Mortal Weekly Rep. 1997;46:1204–1207. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Update: isolation of avian influenza A (H5N1) viruses from humans—Hong Kong, 1997–1998. Morbid Mortal Weekly Rep. 1998;46:1245–1247. [PubMed] [Google Scholar]
- 5.Claas E C J, Osterhaus A D M E, Van Beek R, De Jong J C, Rimmelzwaan G F, Senne D A, Krauss S, Shortridge K F, Webster R G. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 6.Francis T, Moore A E. A study of the neurotropic tendency in strains of the virus of epidemic influenza. J Exp Med. 1940;72:717–728. doi: 10.1084/jem.72.6.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorman O T, Donis R O, Kawaoka Y, Webster R G. Evolution of influenza A virus PB2 genes: implications for evolution of the ribonucleoprotein complex and origin of human influenza A virus. J Virol. 1990;64:4893–4902. doi: 10.1128/jvi.64.10.4893-4902.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goto H, Kawaoka Y. A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc Natl Acad Sci USA. 1998;95:10224–10228. doi: 10.1073/pnas.95.17.10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horimoto T, Nakayama K, Smeekens S P, Kawaoka Y. Proprotein-processing endoproteases PC6 and furin both activate hemagglutinin of virulent avian influenza viruses. J Virol. 1994;68:6074–6078. doi: 10.1128/jvi.68.9.6074-6078.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katz J M, Wang M, Webster R G. Direct sequencing of the hemagglutinin gene of influenza (H3N2) virus in original clinical samples reveals sequence identity with mammalian cell-grown virus. J Virol. 1990;64:1808–1811. doi: 10.1128/jvi.64.4.1808-1811.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawaoka Y. Equine H7N7 influenza A viruses are highly pathogenic in mice without adaptation: potential use as an animal model. J Virol. 1991;65:3891–3894. doi: 10.1128/jvi.65.7.3891-3894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Kawaoka, Y. Unpublished data.
- 12.Klenk H-D, Garten W. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 1994;2:39–43. doi: 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 13.Klenk H D, Rott R, Orlich M, Blodorn J. Activation of influenza A viruses by trypsin treatment. Virology. 1975;68:426–439. doi: 10.1016/0042-6822(75)90284-6. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi Y, Horimoto T, Kawaoka Y, Alexander D J, Itakura C. Pathological studies of chickens experimentally infected with two highly pathogenic avian influenza viruses. Avian Pathol. 1996;25:285–304. doi: 10.1080/03079459608419142. [DOI] [PubMed] [Google Scholar]
- 15.Lazarowitz S G, Choppin P W. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975;68:440–454. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- 16.Myers E W, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 17.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 18.Smeenk C A, Brown E G. The influenza virus variant A/FM/1/47-MA possesses single amino acid replacements in the hemagglutinin, controlling virulence, and in the matrix protein, controlling virulence as well as growth. J Virol. 1994;68:530–534. doi: 10.1128/jvi.68.1.530-534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stieneke-Grober A, Vey M, Angliker H, Shaw E, Thomas G, Roberts C, Klenk H D, Garten W. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992;11:2407–2414. doi: 10.1002/j.1460-2075.1992.tb05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suarez D L, Perdue M L, Cox N, Rowe T, Bender C, Huang J, Swayne D E. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J Virol. 1998;72:6678–6688. doi: 10.1128/jvi.72.8.6678-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subbarao E K, London W, Murphy B R. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 23.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward A C. Neurovirulence of influenza A virus. J Neurovirol. 1996;2:139–151. doi: 10.3109/13550289609146876. [DOI] [PubMed] [Google Scholar]
- 25.Webster R G, Hinshaw V S, Bean W J, Van Wyke K L, Geraci J R, St. Aubin D J, Peturson G. Characterization of an influenza A virus from seals. Virology. 1981;113:712–724. doi: 10.1016/0042-6822(81)90200-2. [DOI] [PubMed] [Google Scholar]
- 26.Yuen K Y, Chan P S, Peiris M, Tsang D C, Que T L, Shortridge K F, Cheung P T, To W K, Ho E F, Sung R, Cheng A B, Mena I the H5N1 Study Group. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]