Key Points
Question
What is the prevalence, penetrance, and expressivity of UBA1 variants in a health care system cohort?
Findings
In this retrospective observational study of 163 096 participants, 11 individuals (1 in 13 591) harbored a UBA1 disease-causing variant and all participants had hematologic features along with a broad spectrum of autoimmune, pulmonary, and dermatologic clinical manifestations.
Meaning
This study provides an estimate of the prevalence and a description of the clinical manifestations of VEXAS (vacuoles, E1-ubiquitin-activating enzyme, X-linked, autoinflammatory, somatic) syndrome, although additional studies are needed in unselected and genetically diverse populations to better define general population prevalence and phenotypic spectrum.
Abstract
Importance
VEXAS (vacuoles, E1-ubiquitin-activating enzyme, X-linked, autoinflammatory, somatic) syndrome is a disease with rheumatologic and hematologic features caused by somatic variants in UBA1. Pathogenic variants are associated with a broad spectrum of clinical manifestations. Knowledge of prevalence, penetrance, and clinical characteristics of this disease have been limited by ascertainment biases based on known phenotypes.
Objective
To determine the prevalence of pathogenic variants in UBA1 and associated clinical manifestations in an unselected population using a genomic ascertainment approach.
Design, Setting, and Participants
This retrospective observational study evaluated UBA1 variants in exome data from 163 096 participants within the Geisinger MyCode Community Health Initiative. Clinical phenotypes were determined from Geisinger electronic health record data from January 1, 1996, to January 1, 2022.
Exposures
Exome sequencing was performed.
Main Outcomes and Measures
Outcome measures included prevalence of somatic UBA1 variation; presence of rheumatologic, hematologic, pulmonary, dermatologic, and other findings in individuals with somatic UBA1 variation on review of the electronic health record; review of laboratory data; bone marrow biopsy pathology analysis; and in vitro enzymatic assays.
Results
In 163 096 participants (mean age, 52.8 years; 94% White; 61% women), 11 individuals harbored likely somatic variants at known pathogenic UBA1 positions, with 11 of 11 (100%) having clinical manifestations consistent with VEXAS syndrome (9 male, 2 female). A total of 5 of 11 individuals (45%) did not meet criteria for rheumatologic and/or hematologic diagnoses previously associated with VEXAS syndrome; however, all individuals had anemia (hemoglobin: mean, 7.8 g/dL; median, 7.5 g/dL), which was mostly macrocytic (10/11 [91%]) with concomitant thrombocytopenia (10/11 [91%]). Among the 11 patients identified, there was a pathogenic variant in 1 male participant prior to onset of VEXAS-related signs or symptoms and 2 female participants had disease with heterozygous variants. A previously unreported UBA1 variant (c.1861A>T; p.Ser621Cys) was found in a symptomatic patient, with in vitro data supporting a catalytic defect and pathogenicity. Together, disease-causing UBA1 variants were found in 1 in 13 591 unrelated individuals (95% CI, 1:7775-1:23 758), 1 in 4269 men older than 50 years (95% CI, 1:2319-1:7859), and 1 in 26 238 women older than 50 years (95% CI, 1:7196-1:147 669).
Conclusions and Relevance
This study provides an estimate of the prevalence and a description of the clinical manifestations of UBA1 variants associated with VEXAS syndrome within a single regional health system in the US. Additional studies are needed in unselected and genetically diverse populations to better define general population prevalence and phenotypic spectrum.
This retrospective observational study evaluates UBA1 variants in exome data from individuals in the Geisinger MyCode Community Health Initiative.
Introduction
Molecular, specifically genetic, diagnoses are uncommon in rheumatologic diseases, limiting prognosis and treatment in many cases. The most common genetic causes of inflammation include MEFV (OMIM 608107) and familial Mediterranean fever,1 TNFRSF1A (OMIM 191190) and tumor necrosis factor receptor–associated periodic syndrome,2 and ADA2 (OMIM 607575) and deficiency of adenosine deaminase 2.3,4 These disorders are rare, typically early-onset, inherited, and clinically distinct from common rheumatologic conditions. Recently, VEXAS (vacuoles, E1-ubiquitin-activating enzyme, X-linked, autoinflammatory, somatic) syndrome, arising from somatic variants in UBA1 (OMIM 314370) in the bone marrow, was recognized as a novel disease entity.5 Unlike previously recognized genetic causes of inflammation, UBA1 variants are acquired and occur later in life. UBA1 is X-linked and thus phenotypic consequences occur predominantly in men or in women with monosomy of the X chromosome.6 Distinct germline UBA1 variants underlie X-linked spinal muscular atrophy and occur in different regions of the protein from VEXAS syndrome.7 Patients with VEXAS syndrome have rheumatologic, hematologic, dermatologic, and pulmonary manifestations attributed to inflammation and carry a variety of clinical diagnoses, including polyarteritis nodosa, relapsing polychondritis, giant cell arteritis, Sweet syndrome, and myelodysplastic syndrome (MDS). Prognosis in VEXAS syndrome is determined more by the molecular than the clinical diagnosis.8,9 Although the first 3 patients with VEXAS syndrome were identified through a genotype-driven strategy, subsequent ascertainment has been phenotype-driven.9
Accurate diagnoses in rheumatologic diseases can be challenging due to nonspecific overlapping clinical laboratory results and disease features, need for invasive time-sensitive biopsies, and varying natural history. There are currently no guidelines for what individuals to test for UBA1 pathogenic variants and diagnostic criteria for VEXAS syndrome do not exist. The primary objective of this study was to identify the prevalence and phenotypic spectrum of UBA1 variants in an unselected clinical cohort of participants in the MyCode Community Health Initiative at Geisinger.10
Methods
Setting and Study Participants
Participants in MyCode or their guardian or legal representative provide written informed consent and agree to provide blood, serum, and DNA samples for broad research use, including genomic analysis.10 The study population is a health system–based cohort consisting of 163 096 individuals who sought health care at a Geisinger facility and consented to participate in the MyCode Community Initiative (Table 1). Geisinger is an integrated health system in central and northeastern Pennsylvania. Enrollment in MyCode is open to all Geisinger patients regardless of their medical history. MyCode participants agree that their samples and data can be linked to Geisinger electronic health records (EHRs), and additional informed consent for this study beyond the initial written consent was deemed not to be required. UBA1 genomic ascertainment investigation was reviewed by the Geisinger Institutional Review Board and determined not to be human subject research as defined in 45CFR46.102(e) in written consent (study #2021-0818). For this study, relevant data obtained during an inpatient or outpatient encounter with a Geisinger clinician were extracted; phenotype data included clinical diagnoses, procedures, medications, and clinical laboratory results obtained from January 1, 1996, to January 1, 2022. Genetic analysis was carried out as part of the DiscovEHR collaboration between Geisinger and the Regeneron Genetics Center by microarray genotyping and exome sequencing. The cohort characteristics have been described extensively.11 Ancestry of the study participants was included to highlight potential limitations of the data. Geisinger patients provide self-reported ancestry from a drop-down list that includes “other” as an option. This information is recorded in their EHR and is the main source of race and ethnicity data. The self-reported ancestry is highly consistent with ancestry determined by genetic principal component analysis and is also consistent with regional census data. There were no inclusion or exclusion criteria based on race or ethnicity. All genetic data were generated using DNA samples from peripheral blood. The age distribution of adult MyCode participants approximates that of the Geisinger health system outpatient population,10 but is enriched for individuals older than 50 years compared with the regional population. The rank order of disease frequency in MyCode participants is nearly identical to the health system Geisinger overall adult patient population.
Table 1. Demographics of Geisinger MyCode Community Health Initiative.
| Characteristic | No. (%) |
|---|---|
| Total | 163 096 |
| Women | 99 462 (60.9) |
| Men | 63 634 (39.1) |
| Age, mean (SD), y | 59.0 (19.0) |
| Median | 61.0 |
| BMI, mean (SD)a | 31.0 (8.0) |
| Median | 29.8 |
| Ancestryb | |
| Black or African American | 3682 (2.3) |
| White | 153 073 (96.7) |
| Other | 1533 (1.0) |
| Ethnicity | |
| Hispanic | 4286 (2.7) |
| Not Hispanic or Latino | 152 524 (97.3) |
Body mass index (BMI) data were available for 97% of participants.
Patients provided self-reported ancestry from a drop-down list.
Exome Sequencing and Variant Detection
Exome sequencing was performed on genomic DNA extracted from peripheral blood using previously reported protocols (additional information appears in the eMethods in Supplement 1) and variant calling was performed in collaboration with Regeneron Genetics Center as described previously.12,13 Individuals with variants at previously reported pathogenic and likely pathogenic positions in UBA1, including those listed both for somatic and germline diseases, were obtained from ClinVar and the literature (https://www.ncbi.nlm.nih.gov/clinvar/) and evaluated for pathogenicity using the guidelines for interpretation of sequence variants from the American College of Medical Genetics and Genomics and the Association of Molecular Pathology.14 A subset of individuals was sequenced separately twice as part of continued quality improvement studies; all data were consistent between sequencing studies, with data included in this article from higher-depth sequencing. Prevalence estimates included patients with variants at known pathogenic or likely pathogenic positions and an individual with a highly suspicious variant of uncertain significance. Variants of uncertain significance were defined as (1) missense or consensus splice site variants detected in men with age of blood sample collection older than 50 years and (2) variants not found in any annotated sequenced relative within DiscovEHR and less than or equal to 4 DiscovEHR participants with complete sample collection data. Variants of uncertain significance were filtered for number of reads supporting the alternate allele of greater than or equal to 3, read depth (total number of sequencing reads over a position) greater than or equal to 20, variant allele fraction (VAF; ratio of alternative allele to total reads) greater than or equal to 0.2, genotype quality (confidence of variant on sequencing data) greater than or qual to 50, and allele frequency in the Genome Aggregation Database less than 0.00001.15 Variants with VAF of 0.8 or more in all men and VAF of 0.4 to 0.6 in all women were excluded. Highly suspicious variants of uncertain significance were defined by meeting the above sequence and variant criteria and were found in a participant with overlapping features of VEXAS syndrome and in vitro data supporting a catalytic defect. Overlapping features for VEXAS syndrome included concomitant clinical features in 2 different organ systems known to be associated with VEXAS syndrome (rheumatologic, hematologic, dermatology, pulmonary) along with supportive laboratory test result abnormalities (anemia and/or thrombocytopenia). To assess whether UBA1 variants of uncertain significance were catalytically defective (compared with wild-type variants) and thus altered UBA1 activity, complementation (rescue) assays were performed using an established method in Chinese hamster ovary cells. In this cell line, endogenous UBA1 can be destabilized at restrictive temperatures and ubiquitylation and ubiquitin-conjugating enzyme (E2) charging are impaired and can be rescued by UBA1 wild-type, but not catalytically impaired, versions.16
Outcomes
For individuals with a genetic variant of interest, all available medical record data were reviewed. Clinical profiles of these individuals were outcomes of this study. This included structured data extracts and manual reviews of all Geisinger medical records within the Geisinger EHR system. The available data spanned 4 to 25 years (median, 12 years) and were reviewed by Geisinger clinical data analysts and practicing internal medicine physicians (eTable 1 in Supplement 1). Structured EHR review included only prespecified data elements, including all assigned International Classification of Diseases, Tenth Revision (ICD-10) visit diagnoses, prescribed medications, and any procedures performed. Consultations with rheumatology, dermatology, pulmonology, and hematology were also recorded. All data from the following laboratory tests were collected and analyzed: complete blood cell count, erythrocyte sedimentation rate, and C-reactive protein. All structured data related to VEXAS syndrome were included in the article in aggregate after manual review of complete raw data (Tables 1 and 2; eTable 1 in Supplement 1). Manual chart review was defined as a chart review without constraints performed by a Geisinger internal medicine physician using a standard case report form as a basis, and clinical data were detailed in deidentified patient-level descriptive summaries (eTable 2 in Supplement 1). Structured chart review served as the basis for Table 1 with manual chart review for additional information. Centralized review of peripheral blood and bone marrow aspirate smears, histologic sections, and immunohistochemical and cytogenetic studies performed at the time of diagnosis were performed by a Geisinger pathologist (eTable 3 in Supplement 1). Given the lack of VEXAS syndrome ICD-10 code or diagnostic criteria, a UBA1 carrier was considered to have VEXAS syndrome if there were concomitant clinical features in 2 different organ systems known to be associated with VEXAS syndrome (rheumatologic, hematologic, dermatology, pulmonary) along with supportive laboratory test result abnormalities (anemia and/or thrombocytopenia). In addition to the clinical outcomes described above, prevalence of VEXAS syndrome was an outcome of this study.
Table 2. Demographics of UBA1 Pathogenic or Likely Pathogenic Variant Carriers.
| Participant/sex | Age at symptom onset, y | Age at death, y | Duration in care, ya | Age at sample collection, y | Identified variants | Variant allele fractionb |
|---|---|---|---|---|---|---|
| 1/Male | 50-59 | 60-69 | 10 | 60-69 | c.121A>G; p.Met41Val | 0.13 |
| 2/Male | 50-59 | Living | 4 | 50-59 | c.167C>T; p.Ser56Phe | 0.79 |
| 3/Male | 50-59 | 60-69 | 19 | 60-69 | c.121A>C; p.Met41Leu | 0.30 |
| 4/Male | 60-69 | 80-89 | 18 | 80-89 | c.122T>C; p.Met41Thr | 0.38 |
| 5/Male | 60-69 | Living | 4 | 60-69 | c.121A>C; p.Met41Leu | 0.74 |
| 6/Female | 60-69 | 70-79 | 15 | 70-79 | c.121A>G; p.Met41Val | 0.19 |
| 7/Male | 70-79 | 80-89 | 27 | 80-89 | c.121A>C; p.Met41Leu | 0.52 |
| 8/Male | 70-79 | 70-79 | 25 | 70-79 | c.121A>G; p.Met41Val | 0.57 |
| 9/Male | 70-79 | 80-89 | 6 | 70-79 | c.121A>C; p.Met41Leu | 0.09 |
| 10/Female | 70-79 | 80-89 | 4 | 70-79 | c.118-2A>G | 0.21 |
| 11/Male | 80-89 | 80-89 | 12 | 80-89 | c.167C>T; p.Ser56Phe | 0.04 |
| 12/Malec | 60-69 | Living | 2 | 70-79 | c.1861A>T; p.Ser621Cys | 0.20 |
Time during which visits were covered at Geisinger system for any clinical indication.
The ratio of alternative to total reads. Variant annotation to cDNA used transcript ID NM_003334.4 and to protein used NP_003325.2 ID for UBA1.
Denotes a participant with a highly suspicious variant of uncertain significance deemed to be pathogenic.
Statistical Analysis
Laboratory values were obtained and analyzed as continuous variables with means and SDs. Prevalence estimates and 95% CIs were calculated using Poisson test in the “statsmodels” package for Python, version 0.13.2.
Results
UBA1 Pathogenic and Likely Pathogenic Variant Identification
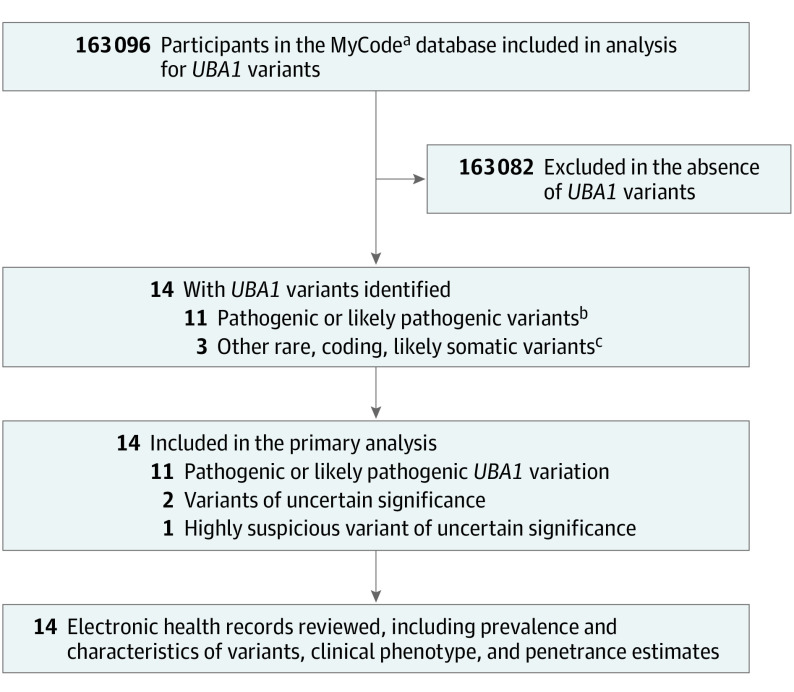
The study population of 163 096 individuals was interrogated for UBA1 variants at positions reported as pathogenic or likely pathogenic either from ClinVar or publications (Figure and Table 1). A total of 11 individuals were identified with pathogenic or likely pathogenic variants in UBA1 (NM_003334.4) that were likely somatic based on VAFs ranging from 0.04 to 0.79 (Table 2). Eight individuals (73%) had variants at codon 41 (c.121 A>C; p.Met41Leu, c.121A>G; p.Met41Val, c.122T>C; p.Met41Thr), 2 (18%) had a missense variant at codon 56 (c.167C>T; p.Ser56Phe), and 1 (9%) had a canonical splice site variant at a position reported pathogenic (c.118-2A>G).17 Two of the 11 variant carriers (18%) were women without evidence of aneuploidy on single nucleotide polymorphism array or exome sequencing. Clinical UBA1 variant testing was not performed for any individual. On structured data extraction and physician EHR review, 6 of these 11 individuals (55%) had assigned clinical diagnoses associated with VEXAS syndrome (Table 3). Although 55% of UBA1 variant carriers in this cohort had a VEXAS-associated clinical diagnosis, all individuals had nondiagnostic clinical manifestations consistent with VEXAS syndrome identified both by structured data extraction and manual EHR review (eTable 1 and 2 and eMethods in Supplement 1).5 All variant carriers were older than 50 years at onset of symptoms, which is consistent with previous reports (Table 2). No pathogenic variants for X-linked spinal muscular atrophy were identified within the cohort.
Figure. Outline of the Study Design.
aDiscovEHR project within the MyCode Community Health Initiative, an ongoing collaboration between Geisinger and the Regeneron Genetics Center.
bPathogenic or likely pathogenic variants in UBA1 based on ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) or American College of Medical Genetics and Genomics/Association for Molecular Pathology criteria.
cMissense or consensus splice site variants of uncertain significance detected in men older than 50 years at blood sample collection, likely somatic based on variant allele fraction.
Table 3. Clinical Findings in UBA1 Pathogenic or Likely Pathogenic Variant Carriers.
| Participant | Clinical diagnoses | Clinical characteristicsa | Laboratory findingsb | Medications (ever) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Attributed to VEXAS | Rheumatologic | Hematologic | Autoimmune involvement | Dermatologic involvement | Pulmonary involvement | Hematologic involvement | Hemoglobin, g/dL (nadir) | Platelet count, ×109/L (nadir) | MCV, fL (peak) | ESR, mm/h (peak) | Bone marrow biopsy results | Specialist visits | ||
| 1 | None | None | None | Yes | Yesc | Yesc | Yes | 10.2 | 82 | 115.5 | 104 | NA | NA | Methylprednisone |
| 2 | Yes | None | Myelofibrosis,c MDS | No | Yes | Yes | Yes | 5.5 | 8 | 100.9 | NA | Cytoplasmic vacuoles (myeloid and erythroid) and megaloblastic changes | Hematologist, pulmonologist | Ruxolitinib |
| 3 | Yes | None | MDS | Yes | No | Yes | Yes | 6 | 28 | 119 | NA | Cytoplasmic vacuoles (myeloid) and megaloblastic changes | Hematologist, pulmonologist | Prednisone |
| 4 | Yes | Giant cell arteritisc | None | Yes | Yes | Yes | Yes | 7.8 | 118 | 126.4 | 106 | NA | Rheumatologist, pulmonologist | Prednisone |
| 5 | Yes | Dermatomyositis | MDS | Yesc | Yes | No | Yes | 7.5 | 55 | 116 | 17 | Cytoplasmic vacuoles (myeloid) and megaloblastic changes | Hematologist | Methylprednisone, prednisone, mycophenolate mofetil |
| 6 | Yes | Psoriasis,c polymyalgia rheumaticac | Chronic myeloid leukemia,c MGUSc | Yes | Yes | Yes | Yes | 7.5 | 18 | 126 | NA | Rheumatologist, hematologist, pulmonologist | Colhicine, prednisone, methylprednisolone | |
| 7 | None | None | None | Yes | Yes | Yes | Yes | 10 | 125 | 115.8 | NA | NA | Rheumatologist, hematologist, pulmonologist | |
| 8 | Yes | Sarcoidosis | MDSc | Yes | Yes | Yes | Yes | 7.3 | 102 | 109.3 | 107 | Cytoplasmic vacuoles (myeloid) | Rheumatologist, hematologist, pulmonologist | Methylprednisone, prednisone |
| 9 | None | None | None | Yesc | No | Yes | Yes | 7.4 | 91 | 93.1 | 37 | Cytoplasmic vacuoles (myeloid) and megaloblastic changes | Hematologist, pulmonologist | Methylprednisone, prednisone |
| 10 | None | None | None | Yesc | Yes | Yes | Yes | 9 | 178 | 97.8 | NA | NA | Rheumatologist, pulmonologist | Prednisone |
| 11 | None | None | None | No | No | Yes | Yes | 7.9 | 83 | 97.3 | NA | NA | Pulmonologist | |
| 12d | None | Granulomatosis with polyangiitisc | None | Yes | Yes | No | Yes | 13.9 | 86 | 95.9 | NA | NA | Rheumatologist, hematologist | |
| Total, % | 55 | 36 | 45 | 82 | 73 | 91 | 100 | Mean (SD), 8.3 (2.2) | Mean (SD), 81.2 (48.7) | Mean (SD), 109.4 (12.0) | ||||
Abbreviations: MDS, myelodysplastic syndrome; MGUS, monoclonal gammopathy of undetermined significance.
Based on International Classification of Diseases, Tenth Revision (ICD-10) codes and clinical review. Hematologic involvement was defined as anemia and/or thrombocytopenia. Dermatologic and pulmonary diagnoses listed when possible, with details for ICD coding and clinical diagnoses in eTables 1-2 in Supplement 1.
References ranges: hemoglobin, 12-17 g/dL; platelet count, 150-400 ×109/L; mean corpuscular volume (MCV), 80-95 fL; erythrocyte sedimentation rate (ESR), 0-30 mm/h. Mean (SD) patient values, excluding patient with highly suspicious variant: hemoglobin, 7.8 (1.5) g/dL; MCV, 110.6 (11.7) fL; platelet count, 80.7 (51.0) ×109/L.
Clinical findings absent on ICD-10 codes but present on chart review.
Participant with a highly suspicious variant of uncertain significance; not included in total percentages.
EHR and Pathology Review
Seven individuals (64%) with pathogenic or likely pathogenic variants in UBA1 had arthritis and 4 (36%) were diagnosed with rheumatologic diseases, including psoriasis, polymyalgia rheumatica, dermatomyositis, and sarcoidosis (Table 3). Only 4 individuals (36%) had features on chart review consistent with reported VEXAS flares (fevers, elevated acute phase reactants, steroid dependency) (eTable 2 in Supplement 1). All 11 individuals had anemia, 10 (91%) had macrocytosis, 5 (46%) required transfusions, 10 (91%) had thrombocytopenia, 4 (36%) had MDS, 1 (9%) had monoclonal gammopathy of undetermined significance, and 1 (9%) had chronic myelogenous leukemia (eTable 2 in Supplement 1). Eight individuals (73%) had skin involvement and 10 (91%) had pulmonary disease (Table 3). A progressive increase in mean corpuscular volume was noted in some patients, with macrocytosis only developing after symptom onset (eFigure 1 in Supplement 1). Bone marrow biopsies were performed as standard care in 6 patients, with hypercellular marrow detected in all individuals. Of those 6 patients, cytoplasmic vacuoles in myeloid and/or erythroid precursors were observed in 5 (83%), while megaloblastic changes in erythroid elements were seen in 4 (67%) (Table 3 and eTable 3 in Supplement 1). Moreover, 80% of those tested showed elevated erythrocyte sedimentation rate (Table 3).
UBA1 Variant of Uncertain Significance Query
A query was performed for novel disease-causing UBA1 variants in men. Three rare, likely somatic, missense or splicing variants in men older than 50 years were identified (eTable 4 in Supplement 1). Although none of these men had documented clinical diagnoses originally attributed to VEXAS syndrome, 1 individual (P12, c.1861A>T; p.Ser621Cys, VAF:0.2) had characteristics consistent with VEXAS syndrome with a diagnosis of granulomatosis with polyangiitis (Tables 2 and 3). Functional validation of these 3 variants in UBA1-deficient CHO cell lines showed that wild-type, p.Gly213Val, and p.Val75Ile UBA1 variants rescued E2 charging (E2-Ub/E2) but p.Ser621Cys did not, demonstrating that p.Ser621Cys caused a catalytic defect (eFigure 2 in Supplement 1). Based both on the clinical phenotype and enzymatic defect p.Ser621Cys should be considered to be highly suspicious for causing VEXAS syndrome.
Prevalence of VEXAS Syndrome
Ten men and 2 women with likely pathogenic or pathogenic variants (n = 11) or highly suspicious disease-causing variants (n = 1) were identified, giving an estimated variant prevalence in individuals older than 50 years of 1:4269 (95% CI, 1:2319-1:7859) for men and 1:26 238 (95% CI, 1:7196-1:147 669) for women or 1:7931 (95% CI, 1:4537-1:13 863) for all individuals older than 50 years. The estimated prevalence was 1:13 591 (95% CI, 1:7775-1:23 758) for the entire cohort.
Discussion
The findings of this study indicated that UBA1 pathogenic variants were present in 1 in 13 591 individuals, can precede clinical onset, and are highly penetrant. Many of the classic features and diagnoses previously recognized as VEXAS syndrome in the literature (relapsing polychondritis, polyarteritis nodosa, Sweet syndrome) were not identified in the Geisinger cohort, except for MDS. This work expands the clinical spectrum of VEXAS syndrome to include individuals older than 50 years without specific clinical rheumatologic diagnoses but with a combination of rheumatologic, hematologic, dermatologic, or pulmonary symptoms along with anemia, thrombocytopenia, and elevated inflammatory markers. Further highlighting the strength of an unbiased genomic ascertainment approach to identify patients with VEXAS syndrome from within the general population, there was a different proportion of VEXAS-defining variants compared with 2 recent phenotypically defined cohort studies of VEXAS syndrome8,9 and an additional likely disease-causing variant was identified. There was a greater proportion of euploid women with pathogenic variants in UBA1 than previously reported.18,19
At an occurrence of 1 in 13 591 across all age groups, UBA1 pathogenic variants may be more common than the reported prevalence of clinical diagnoses comprising VEXAS syndrome, including most vasculitides (granulomatosis with polyangiitis: approximately 1/18 000; polyarteritis nodosa: approximately 1/33 000),20,21 with a prevalence similar to Behçet disease (approximately 1/10 000) and MDS (approximately 1/14 000).22,23 Thus, given the current understanding of VEXAS syndrome, many of these patients may not have been identified using phenotype-based ascertainment. Despite the large number of cases reported to date, UBA1 is still not routinely offered on standard workup for myeloid neoplasms or immune dysregulation diagnostic panels. Establishing the clinical spectrum of UBA1 variants using biobanks is a necessary step to better define disease criteria and testing recommendations. Identification of UBA1 pathogenic variants defines VEXAS syndrome and can alter treatment, prognosis, and screening plans.17,24,25,26,27 Therefore, broad UBA1 testing in patients with features indicative of VEXAS, including those with nonspecific differential diagnoses, such as macrocytic anemia of uncertain etiology in association with elevated inflammatory markers, may be indicated.
Limitations
This study has several limitations. First, this study is based on a single-center regional cohort that may not be representative of other geographic locations and ancestries due to a population that is predominantly of European ancestry. Second, this work is limited to clinical care received within a single health system and cannot account for missing data from clinical findings, treatment, or testing provided by an external clinician not captured in the Geisinger EHR available as part of the study. Third, UBA1 somatic variant detection on exome data may be limited due to sequencing depth and coverage, use of peripheral blood rather than bone marrow samples, and the possibility of novel and currently unrecognized pathogenic variants. Fourth, prevalence may be overestimated if highly suspicious variants are not validated as causal in other studies. Although clinical diagnoses were likely underrecorded because of diagnostic and physician coding challenges and cohort-specific approaches and characteristics, both in this study and those for other diagnoses, the reported UBA1 variant prevalence is also likely underestimated due to the requirements for sequence depth to detect low-level somatic variants. Additional cohort analyses will be critical to continue to evaluate and better understand the prevalence, penetrance, and expressivity of UBA1 variants in diverse populations.
Conclusions
This study provides an estimate of the prevalence and a description of the clinical manifestations of UBA1 variants associated with VEXAS syndrome within a single regional health system in the United States. Additional studies are needed in unselected and genetically diverse populations to better define general population prevalence and phenotypic spectrum.
eMethods
eFigure 1. Macrocytosis in individuals with UBA1 pathogenic variants
eFigure 2. UBA1 mutant S621C shows decreased Ubiquitin-conjugating enzyme (E2) charging in vitro
eTable 1. Structured ICD 10 Code Electronic Health Queries for individuals with UBA1 pathogenic/likely pathogenic variants
eTable 2. Manual review of EHR for individuals with UBA1 pathogenic/likely pathogenic variants
eTable 3. Bone marrow biopsy features in individuals with UBA1 pathogenic/likely pathogenic variants
eTable 4. Variants of Uncertain Significance in UBA1 and associated phenotypes based on structured and manual review of EHR
Nonauthor collaborators
Data sharing statement
References
- 1.The International FMF Consortium . Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. Cell. 1997;90(4):797-807. doi: 10.1016/S0092-8674(00)80539-5 [DOI] [PubMed] [Google Scholar]
- 2.McDermott MF, Aksentijevich I, Galon J, et al. Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell. 1999;97(1):133-144. doi: 10.1016/S0092-8674(00)80721-7 [DOI] [PubMed] [Google Scholar]
- 3.Zhou Q, Yang D, Ombrello AK, et al. Early-onset stroke and vasculopathy associated with mutations in ADA2. N Engl J Med. 2014;370(10):911-920. doi: 10.1056/NEJMoa1307361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navon Elkan P, Pierce SB, Segel R, et al. Mutant adenosine deaminase 2 in a polyarteritis nodosa vasculopathy. N Engl J Med. 2014;370(10):921-931. doi: 10.1056/NEJMoa1307362 [DOI] [PubMed] [Google Scholar]
- 5.Beck DB, Ferrada MA, Sikora KA, et al. Somatic mutations in UBA1 and severe adult-onset autoinflammatory disease. N Engl J Med. 2020;383(27):2628-2638. doi: 10.1056/NEJMoa2026834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stubbins RJ, McGinnis E, Johal B, et al. VEXAS syndrome in a female patient with constitutional 45,X (Turner syndrome). Haematologica. 2022;107(4):1011-1013. doi: 10.3324/haematol.2021.280238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramser J, Ahearn ME, Lenski C, et al. Rare missense and synonymous variants in UBE1 are associated with X-linked infantile spinal muscular atrophy. Am J Hum Genet. 2008;82(1):188-193. doi: 10.1016/j.ajhg.2007.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrada MA, Savic S, Cardona DO, et al. Translation of cytoplasmic UBA1 contributes to VEXAS syndrome pathogenesis. Blood. 2022;140(13):1496-1506. doi: 10.1182/blood.2022016985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georgin-Lavialle S, Terrier B, Guedon AF, et al. ; French VEXAS group; GFEV, GFM, CEREMAIA, MINHEMON . Further characterization of clinical and laboratory features in VEXAS syndrome: large-scale analysis of a multicentre case series of 116 French patients. Br J Dermatol. 2022;186(3):564-574. doi: 10.1111/bjd.20805 [DOI] [PubMed] [Google Scholar]
- 10.Carey DJ, Fetterolf SN, Davis FD, et al. The Geisinger MyCode community health initiative: an electronic health record-linked biobank for precision medicine research. Genet Med. 2016;18(9):906-913. doi: 10.1038/gim.2015.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staples J, Maxwell EK, Gosalia N, et al. Profiling and leveraging relatedness in a precision medicine cohort of 92,455 exomes. Am J Hum Genet. 2018;102(5):874-889. doi: 10.1016/j.ajhg.2018.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carruth ED, Qureshi M, Alsaid A, et al. ; Regeneron Genetics Center . Loss-of-function FLNC variants are associated with arrhythmogenic cardiomyopathy phenotypes when identified through exome sequencing of a general clinical population. Circ Genom Precis Med. 2022;15(4):e003645. doi: 10.1161/CIRCGEN.121.003645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Hout CV, Tachmazidou I, Backman JD, et al. ; Geisinger-Regeneron DiscovEHR Collaboration; Regeneron Genetics Center . Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature. 2020;586(7831):749-756. doi: 10.1038/s41586-020-2853-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karczewski KJ, Francioli LC, Tiao G, et al. ; Genome Aggregation Database Consortium . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434-443. doi: 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handley-Gearhart PM, Trausch-Azar JS, Ciechanover A, Schwartz AL. Rescue of the complex temperature-sensitive phenotype of Chinese hamster ovary E36ts20 cells by expression of the human ubiquitin-activating enzyme cDNA. Biochem J. 1994;304(Pt 3):1015-1020. doi: 10.1042/bj3041015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourbon E, Heiblig M, Gerfaud Valentin M, et al. Therapeutic options in VEXAS syndrome: insights from a retrospective series. Blood. 2021;137(26):3682-3684. doi: 10.1182/blood.2020010177 [DOI] [PubMed] [Google Scholar]
- 18.Tsuchida N, Kunishita Y, Uchiyama Y, et al. Pathogenic UBA1 variants associated with VEXAS syndrome in Japanese patients with relapsing polychondritis. Ann Rheum Dis. 2021;80(8):1057-1061. doi: 10.1136/annrheumdis-2021-220089 [DOI] [PubMed] [Google Scholar]
- 19.Poulter J, Morgan A, Cargo C, Savic S; UKGCA/VEXAS Consortium . A high-throughput amplicon screen for somatic UBA1 variants in cytopenic and giant cell arteritis cohorts. J Clin Immunol. 2022;42(5):947-951. doi: 10.1007/s10875-022-01258-w [DOI] [PubMed] [Google Scholar]
- 20.Watts RA, Hatemi G, Burns JC, Mohammad AJ. Global epidemiology of vasculitis. Nat Rev Rheumatol. 2022;18(1):22-34. doi: 10.1038/s41584-021-00718-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panupattanapong S, Stwalley DL, White AJ, Olsen MA, French AR, Hartman ME. Epidemiology and outcomes of granulomatosis with polyangiitis in pediatric and working-age adult populations in the United States: analysis of a large national claims database. Arthritis Rheumatol. 2018;70(12):2067-2076. doi: 10.1002/art.40577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neukirchen J, Schoonen WM, Strupp C, et al. Incidence and prevalence of myelodysplastic syndromes: data from the Düsseldorf MDS-registry. Leuk Res. 2011;35(12):1591-1596. doi: 10.1016/j.leukres.2011.06.001 [DOI] [PubMed] [Google Scholar]
- 23.Ma X. Epidemiology of myelodysplastic syndromes. Am J Med. 2012;125(7)(suppl):S2-S5. doi: 10.1016/j.amjmed.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck DB, Grayson PC, Kastner DL. Mutant UBA1 and severe adult-onset autoinflammatory disease: reply. N Engl J Med. 2021;384(22):2164-2165. [DOI] [PubMed] [Google Scholar]
- 25.Diarra A, Duployez N, Fournier E, et al. Successful allogeneic hematopoietic stem cell transplantation in patients with VEXAS syndrome: a 2-center experience. Blood Adv. 2022;6(3):998-1003. doi: 10.1182/bloodadvances.2021004749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Hakim A, Poulter JA, Mahmoud D, et al. Allogeneic haematopoietic stem cell transplantation for VEXAS syndrome: UK experience. Br J Haematol. 2022;199(5):777-781. doi: 10.1111/bjh.18488 [DOI] [PubMed] [Google Scholar]
- 27.Heiblig M, Ferrada MA, Koster MJ, et al. Ruxolitinib is more effective than other JAK inhibitors to treat VEXAS syndrome: a retrospective multicenter study. Blood. 2022;140(8):927-931. doi: 10.1182/blood.2022016642 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods
eFigure 1. Macrocytosis in individuals with UBA1 pathogenic variants
eFigure 2. UBA1 mutant S621C shows decreased Ubiquitin-conjugating enzyme (E2) charging in vitro
eTable 1. Structured ICD 10 Code Electronic Health Queries for individuals with UBA1 pathogenic/likely pathogenic variants
eTable 2. Manual review of EHR for individuals with UBA1 pathogenic/likely pathogenic variants
eTable 3. Bone marrow biopsy features in individuals with UBA1 pathogenic/likely pathogenic variants
eTable 4. Variants of Uncertain Significance in UBA1 and associated phenotypes based on structured and manual review of EHR
Nonauthor collaborators
Data sharing statement