Abstract
Various photosynthetic organisms have evolved to absorb light in different regions of the visible light spectrum, thus adapting to the various lighting conditions available on Earth. While most of these autotrophic organisms absorb wavelengths around the 700–800 nm region, some are capable of red-shifted absorptions above this range, but none as remarkably as Blastochloris viridis whose main absorption is observed at 1015 nm, approximately 220 nm (0.34 eV) lower in energy than their main constituent pigments, BChl-b, whose main absorption is observed at 795 nm. The structure of its light harvesting 1-reaction center was recently elucidated by cryo-EM; however, the electronic structure details behind this red-shifted absorption remain unattended. We used hybrid quantum mechanics/molecular mechanics (QM/MM) calculations to optimize one of the active centers and performed classical molecular dynamics (MD) simulations to sample conformations beyond the optimized structure. We did excited state calculations with the time-dependent density functional theory method at the CAM-B3LYP/cc-pVDZ level of theory. We reproduced the near IR absorption by sequentially modifying the number of components involved in our systems using representative structures from the calculated MD ensemble. Natural transition orbital analysis reveals the participation of the BChl-b fragments to the main transition in the native structure and the structures obtained from the QM/MM and MD simulations. H-bonding pigment–protein interactions play a role on the conformation stabilization and orientation; however, the bacteriochlorin ring conformations and the exciton delocalization are the most relevant factors to explain the red-shifting phenomenon.
Graphical Abstract

INTRODUCTION
Photosynthesis is the cardinal photophysical process that sustains life on Earth and possibly the one most related to its origin and early evolution,1 specifically when it comes to oxygenic photosynthesis.2,3 Overall, photosynthesis is a thermodynamically unfavorable process ( > 0) in which the capture of light by a light harvesting complex (LHC) promotes the enzymatic synthesis of complex products from simple molecules for the sustenance of the photosynthetic organism;1,4 however, even when most of the enzymatic and redox processes involved in the synthesis part have been mapped out, the mechanism of energy transfer upon photon capture remains an open avenue for research. Unveiling the electronic mechanisms of how it works could lead to a better understanding of the evolution of autotroph organisms,5,6 as well as to the potential improvement of sustainable solar energy capture and storage devices.7,8
Among modern photosynthetic organisms, bacteria have structurally simple LHCs compared to the structure of chloroplasts from higher organisms such as plants; this low level of complexity poses an advantage for the computational study of electronic structure effects related to the energy absorption and transfer mechanisms involved in their life cycles. LHCs are composed of pigments embedded in proteins in specific arrangements; these photosynthetic pigments, such as chlorophylls (Chl), bacteriochlorophylls (BChl), and carotenoids, have their main absorptions in the UV–visible range of the electromagnetic spectra, with bacteriochlorophylls being the most abundant of them.9 These tetrapyrrolic, Mg2+ coordinating pigments show their main absorption bands on the red region of visible light ranging from 760 to 800 nm in solution;9 however, in their solvated natural configurations, they show a slightly red-shifted absorption of about 50 to 100 nm.10,11 Some organisms, such as certain species of cyanobacteria, have even evolved to adapt to low intensity light conditions rearranging their LHC structures to absorb infrared light by a well-established phenomenon known as far red light photoacclimation (FarLiP) in which paralagous subunits of PS-I and PS-II are expressed for the purpose of harvesting longer wavelengths in environments with low levels of white light, but even in these conditions, the maximum absorption is observed around 810 nm.12,13
In the field of agrotechnology, far-red absorbing photosynthetic pigments from cyanobacteria, such as BChl-d, have been successfully included into the LHC of plants (LHC-II), without altering their overall architecture, as a crop-boosting strategy; however, the far-red absorption only shifts the maximum absorption from 672 to 699 nm (∼27 nm).14 A few naturally occurring photosystems show a noticeable red-shift with respect to the isolated pigments in solution15,16 but none as extreme as the observed on the light harvesting 1-reaction center (LH1-RC) of Blastochloris viridis,17 whose main absorption is located at 1015 nm, more than 200 nm upward from that of bacteriochlorophyll-b (BChl-b), the main pigment present in this LH complex, which shows an absorption maximum at 795 nm in solution (MeOH).9 This remarkably low energy absorption for a photosynthetic organism remains the most dramatic example to date; the reasons behind the electronic mechanism with which B. viridis is able to absorb near-IR radiation with pigments that absorb in the visible region of the spectra have not been addressed so far.
The LH1-RC of B. viridis is composed of 17 protein subunits surrounding the so called photosynthetic pigments special pair. Each of these subunits—except for subunit—17 is made up of three α-helix structures surrounding two BChl-b and one dihydroneurosporene (DHN) molecule for a total of 34 of these photosynthetic pigments inside the LHC and 17 DHN molecules interacting between the protein structures and the main BChl-b pigments. BChl-b pigments and the DHN secondary pigments are kept into place through various interactions with the surrounding proteins, from which H-bonds between tryptophan residues and the keto groups of the pigments are the most prominent; additionally, a histidine residue coordinates the Mg2+ ion located at the center of each BChl-b molecule placing it slightly above the plane as observed in various other systems.18,19 The structural elucidation by Qian et al.17 via cryo-electron microscopy allows for the computational studies of the electronic structure influence into the red shifted absorption of interest and other photophysical phenomena; however, the 2.9 Å resolution with which it was obtained makes the structure of the pigments challenging for electronic structure calculations. Nevertheless, MD simulations can help improve the atomistic details.20
When the structure of 6ET5 was first described, five structural factors were suggested as the potential causes for this remarkably low energy absorption: (1) A short Mg2+–Mg2+ distance of 8.5 Å; (2) unusually strong protein–pigment interactions; (3) the slightly more extended π-conjugated system of BChl-b with respect to that of BChl-a, which is more abundant in other organisms; (4) strong pigment–pigment coupling; and finally, (5) the structural rigidity of the LH1-RC structure, as discussed in previous works.21–25 However, no account for the electronic reasons behind this rare low energy absorption were addressed in this or any other report. Other LHCs showing short Mg2+–Mg2+ distances are chlorosomes, none of which show a red-shifted absorption as remarkable as this, even when these distances are even shorter (∼6.4 Å).10,26 The aim of this work is to explore the electronic structure effects and the electronic transitions calculated with time-dependent density functional theory (TD-DFT) methods to assess the origin of this low energy absorption in terms of the pigment–pigment and pigment–protein interactions, exciton delocalization effects, exciton coupling effects, and dynamic effects on the molecular structures of the asymmetric section of the LH1 ring from B. viridis.
COMPUTATIONAL DETAILS
The LH1—RC structure in the 6ET5 PDB file shows a circular translational symmetry whose repeating units were described above. Atomic coordinates for the pigments embedded in one of these repeating units were taken therefrom, amounting to four BChl-b and two DHN molecules; additionally, the surrounding amino acid residues were also included to get a more detailed picture of the electronic interactions involving the pigments taking 3.0 Å as a distance criterion to choose the closest amino acid residues (see Figure 1). These residues were selected first by invoking a distance criterion. The pigment–protein interactions, such as H-bonds between pigments and their embedding surroundings, have been previously shown to be important for correctly describing the electronic spectra of photosynthetic systems, but specifically, the fifth coordination of Mg2+ ions by a histidine residue has an important impact in the correct calculation of their UV–vis spectra, and thus those specific residues were kept to maintain the correct geometry of Mg2+ ions.19,27,28
Figure 1.
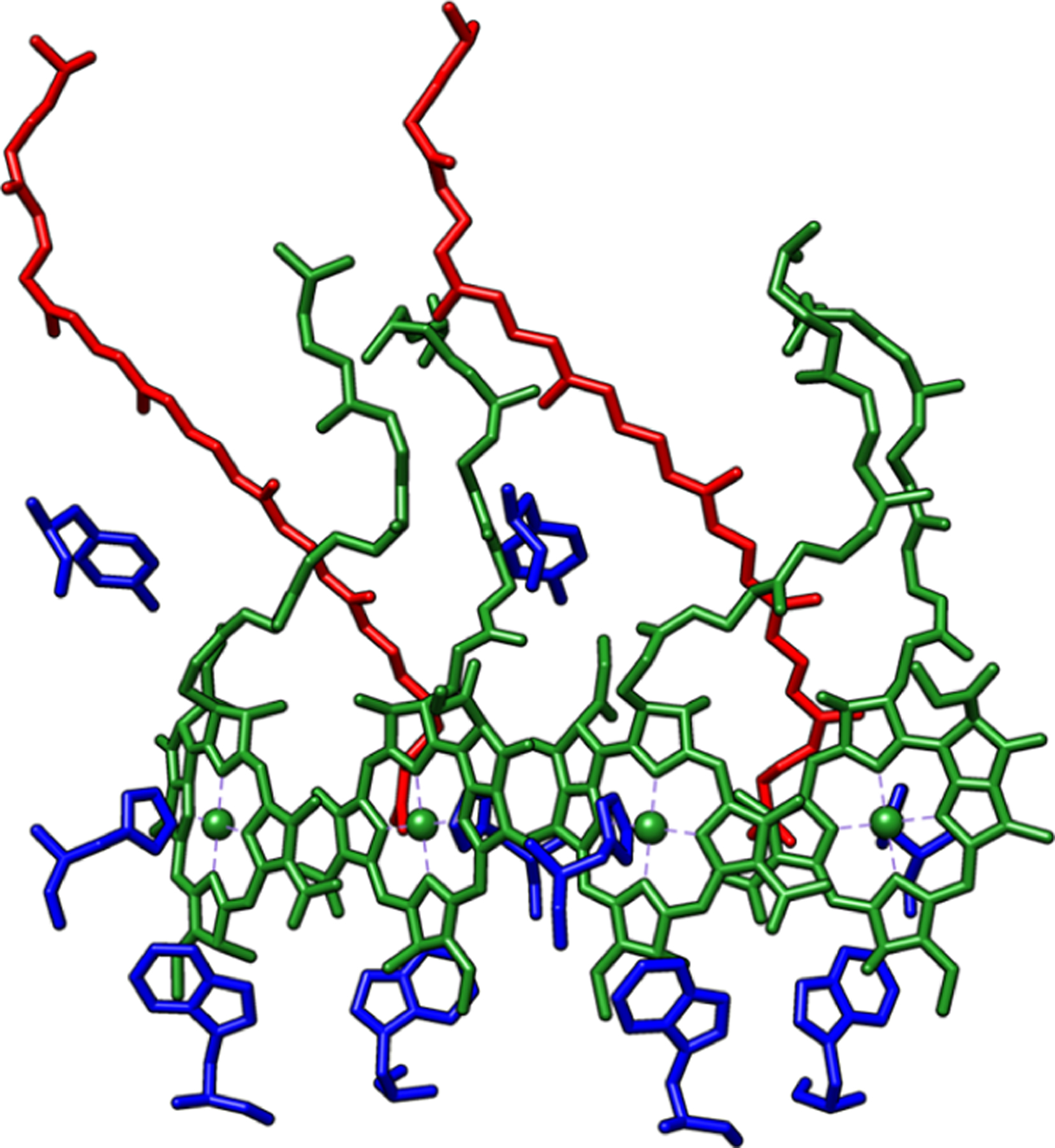
Pigments present in the asymmetric section of 6ET5 LH1-RC. BChl-b (green), DHN (red), amino acids (blue).
To reproduce the near-IR absorption, the previous arrangement shown in Figure 1 was dissected into various pigment systems of increasing complexity. In total, seven systems were built and divided into three kinds: (A) single BChl-b pigment systems; (B) two pigments (BChl-b–DHN) systems; (C) and three pigments systems comprised of two neighboring BChl-b and the surrounding DHN molecules. The first two systems from group A consist of one symmetry-independent BChl-b molecule along with their interacting amino acids (see Figure 2); these are labeled as B1 and B3 systems. The next two models from group B, consisting of one BChl-b and one DHN molecule with surrounding amino acids, were labeled B2 and B4 (see Figure 3). Finally, three models for group C, each including three pigments, were built with two BChl-b and one DHN molecules and were labeled B5, B6, and B7 (see Figure 4). The surrounding amino acids of all models were all valence completed by adding an OH fragment on the acid group, and a hydrogen atom was added to complete the primary amine on the amino group. On these seven systems, a partial geometry optimization of hydrogen atoms and sidechain amino acids (i.e., excluding those atoms from the peptidic chain) at the PM6 semiempirical level of theory was carried out and later refined by means of a second optimization with density functional theory (DFT) calculations at the PBE0/6–31G(d) level of theory. Electronic transitions and absorption spectra were calculated with TD-DFT at the CAM-B3LYP/cc-pVDZ level of theory, which allows a good description of the electron transitions and photophysical behavior of photosynthetic pigments.27,29,30 Fifteen singlet excited states were requested for the B1 to B4 systems (one and two pigments systems) and 50 singlet excited states for the B5 to B7 three-pigments systems, using in all cases the SMD31 implicit continuum dielectric model with methanol as a solvent. A control calculation was constructed from the BChl-b pigment in the B1 model with an explicit methanol molecule coordinated to the Mg2+ central ion fully optimized as a reference for comparing its behavior in solution with the same implicit continuum solvation model, SMD. For the semiempirical and DFT optimizations and TD-DFT UV–vis calculations, the Gaussian 09 rev. E.01 package was used.32 A Gaussian broadening was used for plotting the UV–vis spectra, using a bandwidth of 0.2, which was obtained by comparing the experimental bandwidth using the equation reported by Gouterman33 and taking the reported coefficient extinction values from Namsaraev,34 being a close value to accurately reproduce the absorption spectra with our calculated systems. The surrounding aminoacids from the α and β chains were included in the calculations to expand the intermolecular interactions beyond the pigment–pigment ones: histidine residues coordinating the Mg ions and triptophan and tyrosine residues interacting by H-bonds to the BChl-b pigments, according to Kimura et al.35 the tryptophan residues play an important role on the BChl-b structure stabilization and conformation in the LH1 ring of the 6ET5 complex. An electron–hole Löwdin population analysis was performed on the excited state to analyze the exciton diffusion effect into the red-shift phenomenon, using the TheoDORE 2.0.2 version program package.36
Figure 2.
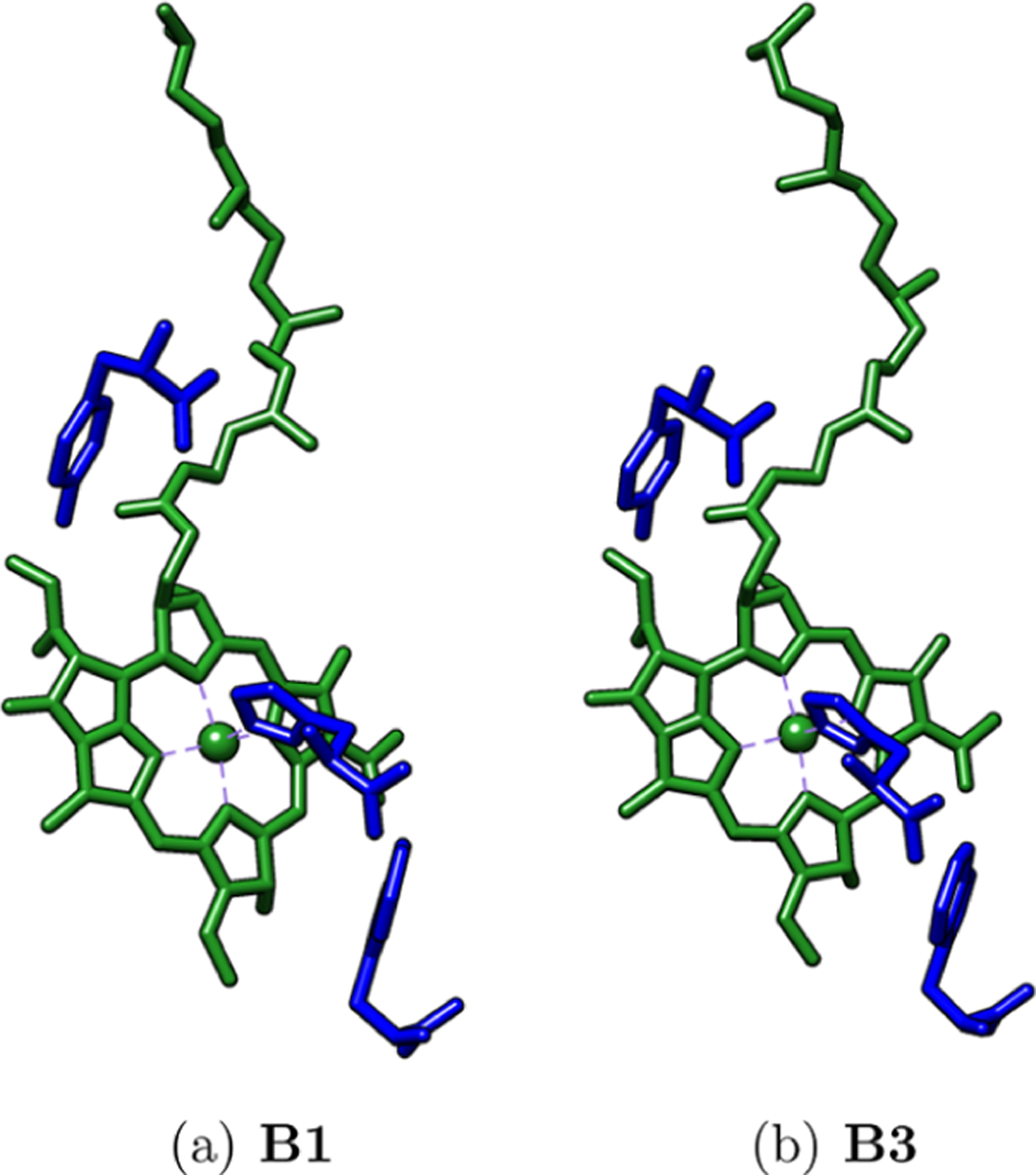
Studied A group systems. BChl-b (green), amino acids (blue).
Figure 3.

Studied B group systems. BChl-b (green), DHN (red), amino acids (blue).
Figure 4.
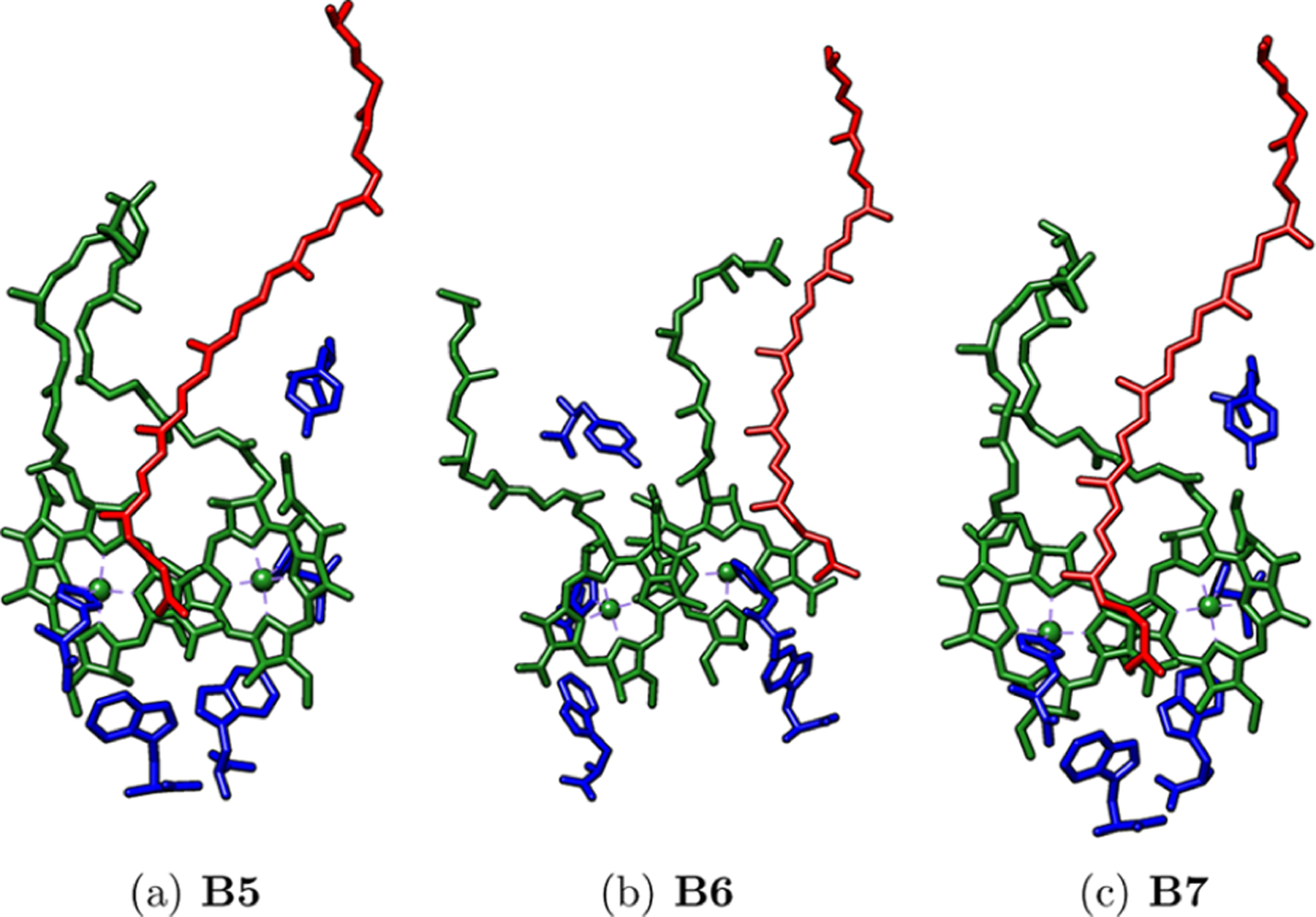
Studied C group systems. BChl-b (green), DHN (red), amino acids (blue).
Additional classical molecular dynamics (MD) and quantum mechanics/molecular mechanics (QM/MM) optimizations were performed for the B7 system. The first step was to obtain parameters for the BChl-b and DHN molecules for the amber99sb force field using the VALENCE program in TinkerTools to estimate vdW and bonded parameters and Mulliken charges to model the electrostatic contribution.37 Mulliken and ESP charges show similar values on average after QM/MM optimization (see Table S1). In all subsequent calculations, Mulliken charges were used based on a closer agreement for the pigments after optimization to the crystal structure. In our model, we have included four peptides surrounding two molecules of BChl-b and one of DHN. We used PROPKA to determine the protonation states of ionizable residues and Packmol to build the simulation unit cell.38,39 We placed the protein–pigment complex in the center of a box of 10 × 10 × 10 nm and added 33,000 TIP3P water molecules. In addition, we added five chloride ions to neutralize the charges. We performed an NPT MD simulation at 298 K and 1 atm using the Tinker 8 code to allow the water molecules to relax to equilibrate the system.37 Protein and pigments were held fixed throughout the simulation. We used the Bussi–Parinello thermostat and the Monte-Carlo barostat to control temperature and pressure, respectively.40 The simulations were carried out with the RESPA algorithm and a time step of 1 fs.41,42
The equilibrated structure was optimized using LICHEM by means of ab initio QM/MM.43,44 The QM region is composed of the atoms of the pigments, side chains of the histidine residues coordinating Mg2+ ions, and tryptophan residues interacting by H-bonds with the BChls-b from the α chains surrounding all pigments (see Figure 5a). The rest of the protein atoms as well as the water molecules and chloride atoms were treated in the MM region. To complete the residues in the QM region, we have used the pseudobond approach. Our QM/MM system in total has 434 QM atoms, 4 pseudoatoms, 12 boundary atoms, and 102,206 MM atoms. The QM region was calculated at the PBE0/6–31G(d) basis set implemented in Gaussian16,45 and the MM region was represented with the ff99SB force field using Tinker8 with the parameters for the pigments corresponding to the procedure described above. An iterative QM/MM optimization, as implemented in LICHEM, was performed for all atoms within a sphere with a radius of 30 Å centered on the center of mass of the protein–pigment complex (see Figure 5b). Atoms outside this sphere remained fixed during the optimization. The convergence criteria of the QM region were as follows: RMS deviation, 0.002 Å; RMS force, 0.005 Hartree Bohr−2; and Max force, 0.01 Hartree Bohr−2. Meanwhile, the convergence criteria of the MM region were as follows: RMS deviation, 0.1 Å; RMS force, 0.1 kcal mol−1 Å−2.
Figure 5.

Structures used in QM/MM calculations. (a) Close up of the pigments and residues included in the QM region (represented as lines). Pink and yellow spheres represent pseudobond and boundary atoms, respectively. (b) A 30 Å sphere centered on the center of mass used to separate active/inactive atoms during QM/MM optimization.
To see the effect that geometry has on excitations, we also did an MD simulation to allow the relaxation of the protein and pigments. Our simulation started with an additional 2 ns equilibration step. The simulation conditions were performed with the same protocol as described above, with all restraints removed except for an added harmonic potential to maintain the distance of the histidines with Mg ions in the BChl-b molecules as in the experimental structure. MD simulations were carried out for 45 ns under standard conditions, with snapshots recorded every 25 ps, giving a total of 1800 structures. From these structures, we selected 10 representative structures of the main changes in RMSD and total potential energy based on a k-means clustering analysis over three observed regions of the resulting ensemble (see Figure 8).
Figure 8.

Structure sampling of the MD structures based on k-means clustering analysis.
RESULTS AND DISCUSSION
For the reference control system, the excited states for an isolated BChl-b optimized in methanol with an explicit MeOH molecule coordinated to Mg2+ (BChl-b—MeOH) were calculated; it is observed that the main Qy absorption appears at 756 nm, in which the experimental spectra for BChl-b in methanol appears at 795 nm ( = 38 nm, = 0.08 eV),9 far from the near-IR region. An RMSD value of 4.604 Å was obtained between B1 BChl-b and the corresponding native structure from 6ET5 as the reference, which means that significant structural changes not only on the phytyl chain but also on bond distances and angles of the tetrapyrrolic section are observed for the optimized structure (see molecular coordinates and structure comparison on SI Figure S8 and Tables S2 and S3). Also, a difference of 165 nm (0.3 eV) is shown between the Qy absorptions of the BChl-b–MeOH and the one in B1, as a consequence of these geometry changes when the BChl-b is fully optimized.
As a reference for analyzing the accuracy of our results, the maximum absorption from the experimental UV–vis spectrum of 6ET5 was used (see Table 1). Only for group C, the three pigments systems, B5 and B7, was the Qy band with a wavelength larger than 1000 nm observed (see Table 1 and Figure 6). The photosynthetic pigment arrangement looks similar between B5 and B7 systems in the sense that the DHN molecule lies between the two BChl-b pigments, whereas in B6, it is located on one side. Since B6 does not show a Qy band above 1000 nm (943 nm), we can presume that the pigment arrangement from B5 and B7 are mostly responsible for the low energy absorption under study.
Table 1.
Red-Shifted Absorptions of Studied Systemsa
| system | (nm) | f | (nm) | (eV) | (eV) |
|---|---|---|---|---|---|
| B1 | 922 | 0.41 | 93 | 1.34 | 0.12 |
| B2 | 937 | 0.48 | 78 | 1.32 | 0.10 |
| B3 | 919 | 0.40 | 96 | 1.35 | 0.13 |
| B4 | 936 | 0.50 | 79 | 1.32 | 0.10 |
| B5 | 1045 | 0.83 | 30 | 1.19 | 0.03 |
| B6 | 943 | 0.92 | 72 | 1.31 | 0.09 |
| B7 | 1026 | 0.82 | 11 | 1.21 | 0.01 |
| experimental | 1015 | 1.22 |
was calculated with experimental absorption as the reference, and was calculated converting into and taking the experimental absorption as reference.
Figure 6.

UV–vis spectra of B1 to B7 models and the experimental spectrum. Arbitrary units were set by taking the experimental 1015 nm absorption band as the maximum amplitude reference for the other absorptions. (a) Full UV–vis spectra of B1 to B7 models vs experimental spectrum. (b) Zoom in on the ∼1000 nm region of the UV–vis spectra.
It is noteworthy that in their native conformations and with the selected intermolecular interactions, most systems show a low energy Qy band in the range between 900 and 1050 nm. Systems in group A (single pigment systems), namely, B1 and B3, have similar displacements with respect to the experimental Qy band and 93 (0.12 eV) and 96 nm (0.13 eV) higher in energy with respect to the sought after 1015 nm band, respectively. Similarly, group B (two pigments systems), namely, B2 and B4, shows a similar behavior with a slightly closer agreement to the experimentally observed band, 78 and 79 nm (0.10 eV in both cases), respectively. Comparing the results for these two groups, we can observe that the presence of the carotenoid DHN molecule shifts the Qy band slightly to longer wavelengths; nevertheless, care must be observed when jumping to conclusions about their involvement: a Löwdin population analysis of the group B shows how the involvement of the secondary pigment, DHN, is negligible next to that of the main BChl-b pigment (see Table 2), so despite being slightly closer to the experimental absorption compared to group A, the presence of DHN seems to have little to do with this fact.
Table 2.
Electron–Hole Populations by Molecular Fragment for the Qy Absorption Bands on B2 and B4 Systemsa
| system | molecule fragment | hole | electron |
|---|---|---|---|
| B2 | BChl-b | 1.020 | 1.019 |
| DHN | 0.004 | 0.004 | |
| B4 | BChl-b | 1.018 | 1.017 |
| DHN | 0.007 | 0.007 |
Amino acid populations are negligible, (<10−4). These data were obtained with TheoDORE 2.0.2.
The closest agreement is observed for two systems within group C (three pigments systems), B5 and B7 with and 11 nm (0.03 and 0.01 eV), rendering B7 as the one with the best agreement and the better chance to explain the origin of the red-shifted absorption of B. viridis.
From the previous control calculations, which lacked the amino acid residues, we decided to assess the influence of the pigment–residue interactions on the main absorption; these neighboring tryptophan and tyrosine residues were removed from the B7 system. The resulting absorption of interest shows only a small blue-shift from the B7 system, which includes the amino acids (see Table 3), while at the same time shows an increase in the oscillator strength, pointing toward the idea that the π – π stacking between BChl-b pigments, and not the protein–pigment interactions, is the main cause behind the red-shifting of the Qy band of 6ET5; although, the effects due to the surrounding proteins are not negligible.
Table 3.
B7 Red-Shifted Absorption with and without Tyrosine and Tryptophan Amino Acids
| system | λ (nm) | E (eV) | f |
|---|---|---|---|
| with amino acids | 1026 | 1.21 | 0.83 |
| without amino acids | 1016 | 1.22 | 0.87 |
It has to be noted that for the B2, B4, B5, and B7 systems, a spurious band at 770 nm, absent in the experimental spectra, is observed with a large absorption; the same occurs at 540 nm for B6. By analyzing the NTOs for these bands, it is shown that in all of these states, the DHN molecules play an important role in a charge transfer state (see Figure 7), the same holds true for the transition from the 540 nm band of B6. This phenomenon is already reported on plants and bacteria.46–48 However, it is also known that the energy transfer from the BChl Soret band to carotenoids has an extremely low efficiency;49 in fact, when the DHN molecule is deleted, the 770 nm band is lost (e.g., see the case of system B7 in Figure S12 of the SI).
Figure 7.

Corresponding NTOs for ∼770 nm for the B7 system.
Our B7 system model, built with the native cryo-EM structure, reproduces the red-shifted band observed experimentally for the LH1-RC of B. viridis. However, by the very nature of the cryo-EM technique, the pigments are fitted on the protein structure by taking a molecular sampling from other LH1-RC structures and using force field densities into protein cavities; in the particular case of 6ET5, the resolution is 2.9 Å, which means that the atomic coordinates for the pigments may not be entirely accurate for a detailed quantum description of their photophysics.17,50 Beyond the structural reliability of these fitted structures, for a complete ligand–pigment description of a biological system, the structural parameters must be refined for the study of these quantum mechanics effect at an atomistic level. Taking the B7 system as our starting point, we include the surrounding proteins to make a QM/MM description of the protein effect on the red-shift phenomenon (see Figure 5). The calculated UV–vis spectra based on the QM/MM optimized structure does not show the previously obtained spurious 770 nm band explaining the DHN electronic transitions, appearing now at 489 nm (2.53 eV) and resulting on an energy difference of 0.29 eV from the experimental DHN band in methanol (439 nm, 2.82 eV)51 and on an energy difference of 0.42 eV from the maximum absorption attributed to the DHN residues on the 6ET5 structure (420 nm, 2.95 eV).17 Nevertheless, the maximum absorption peak is observed at 828 nm (1.49 eV), not consistent with the expected electronic structure effects on the red-shifting absorption. The RMSD value of the pigment with respect to the initial coordinates is 0.393 Å, indicating that a slight geometrical change on the pigments affects its photophysical properties, and we realize that this minimum cannot reproduce the red-shifting effect. Based on these results, we decided to investigate the spectroscopic effects of structures at room temperature based on a thermalized ensemble from an MD simulation.
After a 45 ns MD simulation, 10 structures based on a k-means clustering were randomly selected and their first 10 excited states were calculated (see Figure 8). The red-shifted absorption appears in all these systems (see Table 4) with oscillator strength values related to a strong absorption.
Table 4.
Excitation Energies, Oscillator Strengths, Wavelengths, Exciton Energy Couplings, and RMSD of each BChl-b Fragment of the 10 MD Structures and the Optimized B7 System with QM/MM Methoda
| RMSD |
RMSD |
|||||
|---|---|---|---|---|---|---|
| representative structure | excitation energy (eV) | f | λ (nm) | exciton coupling energy (eV) | BChl-b 1 (Å) | BChl-b 2 (Å) |
| 1 | 1.20 | 0.43 | 1031 | −0.0098 | 0.878 | 0.269 |
| 2 | 1.36 | 1.29 | 913 | 0.0011 | 1.011 | 0.318 |
| 3 | 1.20 | 0.32 | 1034 | −0.0053 | 0.713 | 0.315 |
| 4 | 1.24 | 0.80 | 996 | 0.0310 | 0.547 | 0.295 |
| 5 | 1.27 | 0.57 | 977 | −0.0085 | 0.423 | 0.248 |
| 6 | 1.20 | 0.88 | 1033 | −0.0275 | 0.573 | 0.302 |
| 7 | 1.21 | 0.87 | 1028 | 0.0294 | 0.435 | 0.204 |
| 8 | 1.30 | 0.39 | 956 | 0.0108 | 0.736 | 0.355 |
| 9 | 1.18 | 0.89 | 1046 | 0.0356 | 0.367 | 0.209 |
| 10 | 1.25 | 0.70 | 992 | 0.0358 | 0.745 | 0.232 |
| B7 QM/MM | 1.49 | 1.05 | 839 | 0.0255 | 0.000 | 0.000 |
An optimized B7 structure was used for RMSD calculations as the reference.
Exciton population analyses were performed on each one of the sampled structures from the MD simulations, and the results are collected in Table 5. Therein, we observe that electron–hole populations vary for each system, reflecting the different processes of exciton oscillation and diffusion between the two pigments. These results suggest that there is a direct relationship between the oscillator strength and the exciton diffusion, e.g., system 2 exhibits a significant hole population localized over the DHN fragment, which relates to an oscillator strength value larger than unity. This is a typical signature of the main energy absorption of carotenoid systems when their photophysical properties are studied with quantum mechanical methods.52–54 It is noteworthy that the more delocalized the exciton between the BChl-b fragments is, the larger their corresponding oscillator strength value, as it is noted for systems 4, 6, 7, 9, and 10.
Table 5.
Electron–Hole Populations of the 10 Calculated Systems from the B7 Structurea
| representative structure | molecule fragment | hole | electron |
|---|---|---|---|
| 1 | BChl-b 1 | 0.03 | 0.09 |
| BChl-b 2 | 0.993 | 0.93 | |
| DHN | 0.05 | 0.005 | |
| 2 | BChl-b 1 | 0.6 | 0.79 |
| BChl-b 2 | 0.005 | 0.03 | |
| DHN | 0.4 | 0.19 | |
| 3 | BChl-b 1 | 0.008 | 0.01 |
| BChl-b 2 | 1.01 | 1.00 | |
| DHN | 0.008 | 0.006 | |
| 4 | BChl-b 1 | 0.54 | 0.52 |
| BChl-b 2 | 0.48 | 0.49 | |
| DHN | 0.0009 | 0.002 | |
| 5 | BChl-b 1 | 1.01 | 1.01 |
| BChl-b 2 | 0.01 | 0.01 | |
| DHN | 0.006 | 0.007 | |
| 6 | BChl-b 1 | 0.58 | 0.58 |
| BChl-b 2 | 0.43 | 0.44 | |
| DHN | 0.006 | 0.005 | |
| 7 | BChl-b 1 | 0.55 | 0.55 |
| BChl-b 2 | 0.47 | 0.47 | |
| DHN | 0.002 | 0.002 | |
| 8 | BChl-b 1 | 0.02 | 0.02 |
| BChl-b 2 | 0.99 | 1.01 | |
| DHN | 0.001 | 0.001 | |
| 9 | BChl-b 1 | 0.75 | 0.75 |
| BChl-b 2 | 0.27 | 0.27 | |
| DHN | 0.003 | 0.003 | |
| 10 | BChl-b 1 | 0.17 | 0.17 |
| BChl-b 2 | 0.85 | 0.83 | |
| DHN | 0.001 | 0.002 |
These data were obtained with TheoDORE 2.0.2.
These sampled structures accurately reproduce the main absorption in the near IR region. Taking system 7 as a representative example whose calculated UV–vis spectrum can be observed to match the experimental one (see Figure 9a), a spurious band at 835 nm (1.48 eV) is observed yet once again as in the case of B7, but as in the previous case, removing the DHN molecule yields a matching spectra free from that carotene signature band (see Figure 9b).
Figure 9.
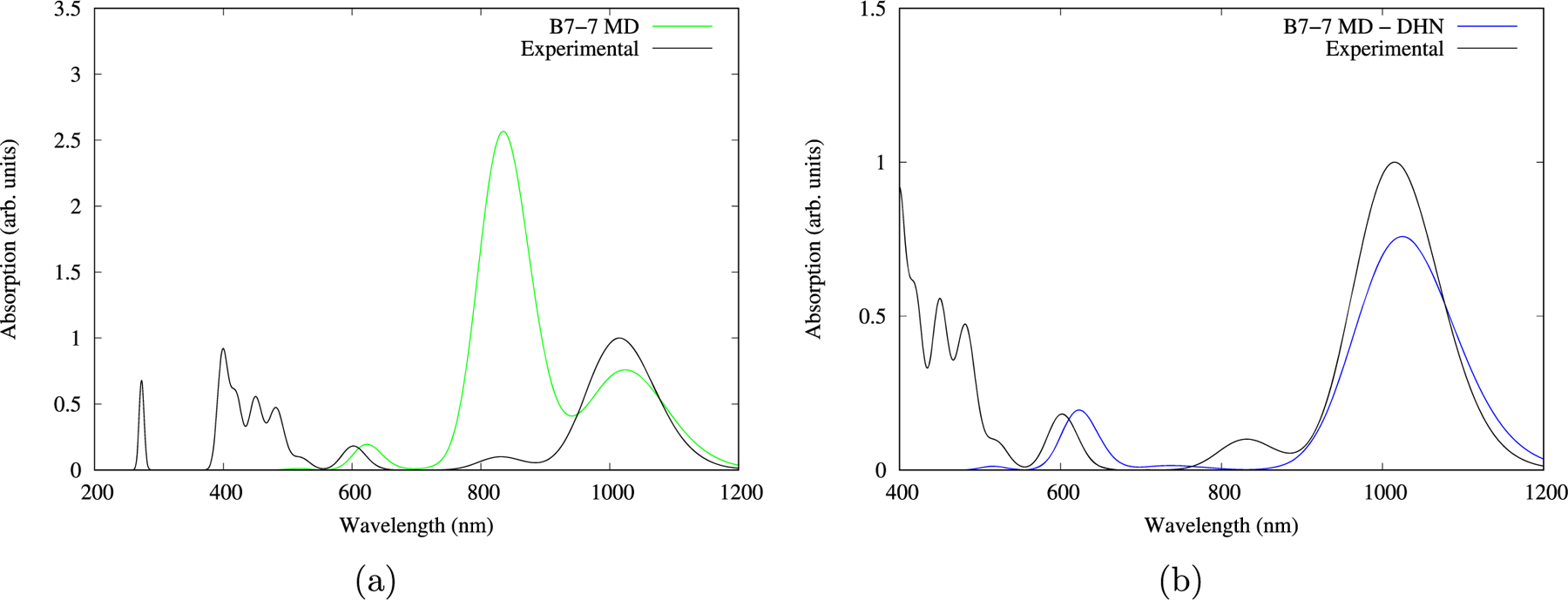
Calculated spectra of representative structure 7 vs the experimental spectrum. (a) Calculated spectrum (on green) including the DHN molecule fragment. (b) Calculated spectrum (blue) excluding the DHN molecule fragment.
Data from Table 5 are consistent with the NTO description of the main transitions. Using system 7 as a representative example, the main transitions are explained by two simultaneous electronic excitations (see Figure 10), with each transition localized on each BChl-b pigment. The coexistence of both electronic transitions is equivalent to the exciton delocalization, such as the ones described by the electronic transitions in sampled system 9 (see Figure 12); also, the sampled system 6 NTOs correspond to a charge transfer process between both BChl-b pigments on one of the two electronic transitions (see Figure 11), with this process being involved on the red-shifting effects, like the one present on the photosystem-II oxygenic photosynthetic structure.13 The rest of the NTOs associated to the electronic transitions on the sampled structures are reported in the SI (Figures S2–S11).
Figure 10.

NTOs for the 1.21 eV absorption for representative structure 7.
Figure 12.
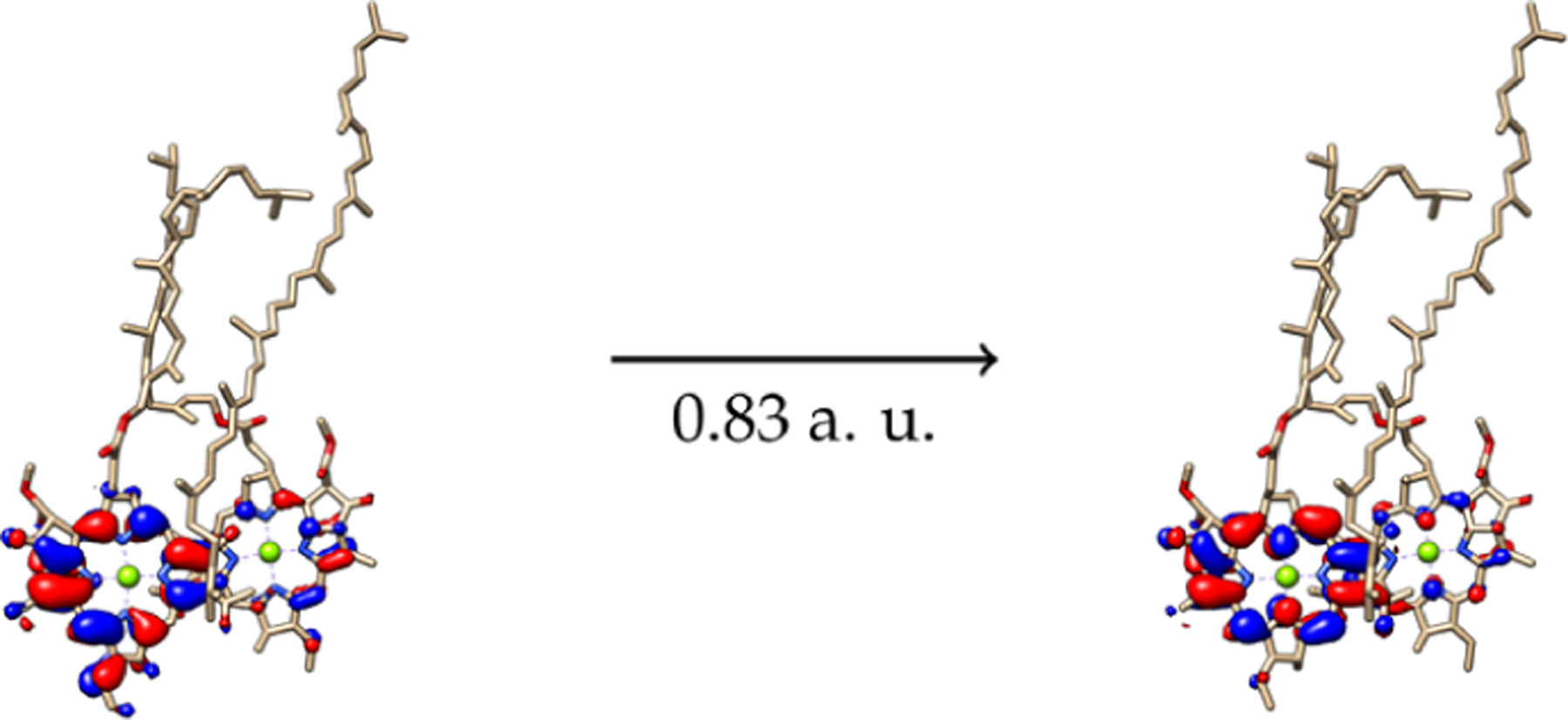
NTOs for the 1.25 eV absorption for representative structure 9.
Figure 11.

Charge transfer NTOs for the 1.20 eV absorption for representative structure 6.
The obtained data suggests that slight structural changes in the bacteriochlorin rings directly impact the red-shifting effect: we report RMSD values of each BChl-b fragment of the representative MD structures using the B7 optimized structure with the QM/MM method as the reference on Table 4 together with the exciton energy couplings. In general, it can be observed that small fluctuations on the bacteriochlorin rings drives to a lower absorption energy compared to the optimized B7 energy minimum. Also, the absorption energy seems to increase for those structures with an RMSD value greater than 1 Å as it is the case for structure 2, the one with the most important structural deviations from the original cryo-EM determination, in comparison to the other structures from the MD sampling and the higher excitation energy of all structures. The calculated exciton coupling energy between the BChl-b pigments on each structure from the MD sampling show no correlation between the red-shifting effect and the exciton coupling energies between these pigments. Therefore, we can conclude that a structural effect on the BChl-b rings is important to explain the ∼1000 nm absorption band on Blastochloris viridis’ LH1-RC structure.
CONCLUSIONS
We reproduced the red-shifted absorption of the Blastochloris viridis LH1-RC and explored the structural effects behind them using its cryo-EM structure (PDB ID: 6ET5) by taking the structure system B7 from 7 systems divided on A, B, and C groups, and by performing TD-DFT, QM/MM calculations, and MD simulations, we explored the structural quantum and dynamic effects behind this red-shifting absorption phenomenon. We discussed and concluded that the dynamic effect is determinant to understand this low energy absorption since the RMSD values of the bacteriochlorin rings obtained from the MD simulation realize that the minimal conformation changes due to dynamic effects are determinant on the explanation of the red-shifted absorption of the LH1-RC Blastochloris viridis structure. The exciton delocalization and oscillation between BChl-b photosynthetic pigments are also a consequence of the dynamic effects as the NTO analysis shows on the 10 representative structures from the MD simulation. The protein effect on pigment stabilization are important for the π – π-stacking pigment–pigment interaction; however, the bacteriochlorin conformation changes are enough to red-shift the Qy absorption and probably explain it to other LH1-RC complexes like the one on Halorhodospira halochloris, also containing a BChl-b pigment and whose structure has not been determined. Our results underscore the role of subtle structural differences in electronic transitions in these systems and the importance of sampling multiple structures.
Supplementary Material
ACKNOWLEDGMENTS
We thank DGTIC–UNAM for granting access to the Miztli supercomputer, Citlalit Martínez for keeping our local computing facilities running properly, and to DGAPA–UNAM for funding under project PAPIIT–IN208219. G.M.S. thanks CONACYT for doctoral scholarship under contract 771371. J.S.L. acknowledges the DGAPA–UNAM postdoctoral funding. This work was partially funded by R01GM108583 to G.A.C. G.A.C. and J.N. acknowledge computational time provided by the University of Texas at Dallas CyberInfrastructure Facilities and the University of North Texas CASCaM CRUNTCh3 high-performance cluster partially supported by NSF grant nos. CHE-1531468 and OAC-2117247. Additional computing time from XSEDE Project TG-CHE160044 is gratefully acknowledged.
Footnotes
The authors declare no competing financial interest.
Contributor Information
Gustavo Mondragón-Solórzano, Centro Conjunto de Investigación en Química Sustentable UAEM, UNAM, Toluca de Lerdo 50200, México; Instituto de Química. Universidad Nacional Autónoma de México, CDMX 04510, México.
Jacinto Sandoval-Lira, Centro Conjunto de Investigación en Química Sustentable UAEM, UNAM, Toluca de Lerdo 50200, México; Instituto de Química. Universidad Nacional Autónoma de México, CDMX 04510, México; Departamento de Ingeniería Ambiental, Instituto Tecnológico Superior de San Martín Texmelucan, C.P. 74120 San Martín Texmelucan, Puebla, México.
Jorge Nochebuena, Department of Physics, University of Texas at Dallas, Richardson, Texas 75801, United States.
G. Andrés Cisneros, Department of Physics and Department of Chemistry and Biochemistry, University of Texas at Dallas, Richardson, Texas 75801, United States
Joaquín Barroso-Flores, Centro Conjunto de Investigación en Química Sustentable UAEM, UNAM, Toluca de Lerdo 50200, México; Instituto de Química. Universidad Nacional Autónoma de México, CDMX 04510, México.
REFERENCES
- (1).Green NJ; Xu J; Sutherland JD Illuminating Life’s Origins: UV Photochemistry in Abiotic Synthesis of Biomolecules. J. Am. Chem. Soc 2021, 143, 7219–7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Cardona T; Rutherford AW Evolution of Photochemical Reaction Centres: More Twists? Trends Plant Sci 2019, 24, 1008–1021. [DOI] [PubMed] [Google Scholar]
- (3).Cardona T; Sánchez-Baracaldo P; Rutherford AW; Larkum AW Early Archean origin of Photosystem II. Geobiology 2019, 17, 127–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Osterloh FE Photocatalysis versus Photosynthesis: A Sensitivity Analysis of Devices for Solar Energy Conversion and Chemical Transformations. ACS Energy Lett 2017, 2, 445–453. [Google Scholar]
- (5).Olson JM Photosynthesis in the Archean era. Photosynth. Res 2006, 88, 109–117. [DOI] [PubMed] [Google Scholar]
- (6).Valleau S; Studer RA; Häse F; Kreisbeck C; Saer RG; Blankenship RE; Shakhnovich EI; Aspuru-Guzik A Absence of Selection for Quantum Coherence in the Fenna–Matthews–Olson Complex: A Combined Evolutionary and Excitonic Study. ACS Cent. Sci 2017, 3, 1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Dau H; Fujita E; Sun L Artificial Photosynthesis: Beyond Mimicking Nature. ChemSusChem 2017, 10, 4228–4235. [DOI] [PubMed] [Google Scholar]
- (8).Barroso-Flores J Evolution of the Fenna–Matthews–Olson Complex and Its Quantum Coherence Features. Which Led the Way? ACS Cent. Sci 2017, 3, 1061–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Grimm B; Porra RJ; Rüdiger; Scheer H Advances in Photosynthesis and Respi-ration; Advances in Photosynthesis and Respiration; Springer Netherlands: Dordrecht, 2006; Vol. 25. [Google Scholar]
- (10).Pšencík J; Butcher SJ; Tuma R In The Structural Basis of Biological Energy Generation; Hohmann-Marriott MF, Ed.; Springer Netherlands: Dordrecht, 2014; pp. 77–109. [Google Scholar]
- (11).Vulto SIE; Neerken S; Louwe RJW; de Baat MA; Amesz J; Aartsma TJ Excited-State Structure and Dynamics in FMO Antenna Complexes from Photosynthetic Green Sulfur Bacteria. J. Phys. Chem. B 1998, 102, 10630–10635. [Google Scholar]
- (12).Ho MY; Niedzwiedzki DM; MacGregor-Chatwin C; Gerstenecker G; Hunter CN; Blankenship RE; Bryant DA Extensive remodeling of the photosynthetic apparatus alters energy transfer among photosynthetic complexes when cyanobacteria acclimate to far-red light. Biochim. Biophys. Acta, Bioenerg 2020, 2020, 148064. [DOI] [PubMed] [Google Scholar]
- (13).Sirohiwal A; Pantazis DA The Electronic Origin of Far-Red-Light-Driven Oxygenic Photosynthesis. Angew. Chem., Int. Ed 2022, 61, No. e202200356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Elias E; Liguori N; Saga Y; Schäfers J; Croce R Harvesting Far-Red Light with Plant Antenna Complexes Incorporating Chlorophyll d. Biomacromolecules 2021, 22, 3313–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tani K; Kanno R; Makino Y; Hall M; Takenouchi M; Imanishi M; Yu L-J; Overmann J; Madigan MT; Kimura Y; Mizoguchi A; Humbel BM; Wang-Otomo Z-Y Cryo-EM structure of a Ca2+−bound photosynthetic LH1-RC complex containing multiple αβ-polypeptides. Nat. Commun 2020, 11, 4955–4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ma F; Yu LJ; Wang-Otomo ZY; Van Grondelle R The origin of the unusual Qy red shift in LH1-RC complexes from purple bacteria Thermochromatium tepidum as revealed by Stark absorption spectroscopy. Biochim. Biophys. Acta, Bioenerg 2015, 1847, 1479–1486. [DOI] [PubMed] [Google Scholar]
- (17).Qian P; Siebert CA; Wang P; Canniffle DP; Hunter CN Cryo-EM structure of the Blastochloris viridis LH1–RC complex at 2.9 Å. Nature 2018, 556, 203–208. [DOI] [PubMed] [Google Scholar]
- (18).Cogdell RJ; Howard TD; Isaacs NW; McLuskey K; Gardiner AT Structural factors which control the position of the Qy absorption band of bacteriochlorophyll a in purple bacterial antenna complexes. Photosynth. Res 2002, 74, 135–141. [DOI] [PubMed] [Google Scholar]
- (19).Edwards WD; Zerner MC Electronic structure of model chlorophyll systems. Int. J. Quantum Chem 1983, 23, 1407–1432. [Google Scholar]
- (20).Igaev M; Kutzner C; Bock LV; Vaiana AC; Grubmüller, H. Automated cryo-EM structure refinement using correlation-driven molecular dynamics. eLife 2019, 8, No. e43542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Llansola-Portoles MJ; Li F; Xu P; Streckaite S; Ilioaia C; Yang C; Gall A; Pascal AA; Croce R; Robert B Tuning antenna function through hydrogen bonds to chlorophyll a. Biochim. Biophys. Acta, Bioenerg 2020, 2020, 148078. [DOI] [PubMed] [Google Scholar]
- (22).Polívka T; Frank HA Molecular factors controlling photosynthetic light harvesting by carotenoids. Acc. Chem. Res 2010, 43, 1125–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Scholes GD; Rumbles G Materials for Sustainable Energy; Co-Published with Macmillan Publishers Ltd, UK, 2010; pp. 12–25. [Google Scholar]
- (24).Thornber JP; Cogdell RJ; Seftor REB; Webster GD Further studies on the composition and spectral properties of the photochemical reaction centers of bacte-riochlorophyll b-containing bacteria. Biochim. Biophys. Acta, Bioenerg 1980, 593, 60–75. [DOI] [PubMed] [Google Scholar]
- (25).Reimers JR; Biczysko M; Bruce D; Coker DF; Frankcombe TJ; Hashimoto H; Hauer J; Jankowiak R; Kramer T; Linnanto J; Mamedov F; Müh F; Rätsep M; Renger T; Styring S; Wan J; Wang Z; Wang-Otomo ZY; Weng YX; Yang C; Zhang JP; Freiberg A; Krausz E Challenges facing an understanding of the nature of low-energy excited states in photosynthesis. Biochim. Biophys. Acta, Bioenerg 2016, 1857, 1627–1640. [DOI] [PubMed] [Google Scholar]
- (26).Fujita T; Huh J; Saikin SK; Brookes JC; Aspuru-Guzik A Theoretical characterization of excitation energy transfer in chlorosome light-harvesting antennae from green sulfur bacteria. Photosynth. Res 2014, 120, 273–289. [DOI] [PubMed] [Google Scholar]
- (27).Mondragón-Solórzano G; Barroso-Flores J Spectroscopical UV-Vis implications of an intramolecular η2-Mg coordination in bacteriochlorophyll-a from the Fenna-Matthews-Olson complex. Int. J. Quantum Chem 2018, 118, No. e25663. [Google Scholar]
- (28).Bondanza M; Cupellini L; Lipparini F; Mennucci B The Multiple Roles of the Protein in the Photoactivation of Orange Carotenoid Protein. Chem 2020, 6, 187–203. [Google Scholar]
- (29).List NH; Curutchet C; Knecht S; Mennucci B; Kongsted J Toward Reliable Prediction of the Energy Ladder in Multi-chromophoric Systems: A Benchmark Study on the FMO Light-Harvesting Complex. J. Chem. Theory Comput 2013, 9, 4928–4938. [DOI] [PubMed] [Google Scholar]
- (30).Shrestha K; Virgil KA; Jakubikova E Electronic Absorption Spectra of Tetrapyrrole-Based Pigments via TD-DFT: A Reduced Orbital Space Study. J. Phys. Chem. A 2016, 120, 5816–5825. [DOI] [PubMed] [Google Scholar]
- (31).Marenich AV; Cramer CJ; Truhlar DG Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [DOI] [PubMed] [Google Scholar]
- (32).Frisch MJ et al. Gaussian-09 Revision E.01 Gaussian, Inc.: Gaussian Inc. Wallingford CT, 2009. [Google Scholar]
- (33).Gouterman M Study of the Effects of Substitution on the Absorption Spectra of Porphin. J. Chem. Phys. 1959, 30, 1139–1161. [Google Scholar]
- (34).Namsaraev ZB Application of extinction coefficients for quantification of chlorophylls and bacteriochlorophylls. Microbiology 2009, 78, 794–797. [Google Scholar]
- (35).Kimura Y; Yamashita T; Seto R; Imanishi M; Honda M; Nakagawa S; Saga Y; Takenaka S; Yu LJ; Madigan MT; Wang-Otomo ZY Circular dichroism and resonance Raman spectroscopies of bacteriochlorophyll b-containing LH1-RC complexes. Photosynth. Res 2021, 148, 77–86. [DOI] [PubMed] [Google Scholar]
- (36).Plasser F TheoDORE: A toolbox for a detailed and automated analysis of electronic excited state computations. J. Chem. Phys 2020, 152, No. 084108. [DOI] [PubMed] [Google Scholar]
- (37).Rackers JA; Wang Z; Lu C; Laury ML; Lagardère L; Schnieders MJ; Piquemal J-P; Ren P; Ponder JW Tinker 8: Software Tools for Molecular Design. J. Chem. Theory Comput 2018, 14, 5273–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Li H; Robertson AD; Jensen JH Very fast empirical prediction and rationalization of protein pKa values. Proteins: Struct., Funct., Bioinf 2005, 61, 704–721. [DOI] [PubMed] [Google Scholar]
- (39).Martínez L; Andrade R; Birgin EG; Martínez JM PACKMOL: A package for building initial configurations for molecular dynamics simulations. J. Comput. Chem 2009, 30, 2157–2164. [DOI] [PubMed] [Google Scholar]
- (40).Bussi G; Parrinello M Accurate sampling using Langevin dynamics. Phys. Rev. E 2007, 75, No. 056707. [DOI] [PubMed] [Google Scholar]
- (41).Tuckerman M; Berne BJ; Martyna GJ Reversible multiple time scale molecular dynamics. J. Chem. Phys 1992, 97, 1990–2001. [Google Scholar]
- (42).Tuckerman ME; Berne BJ; Martyna GJ Molecular dynamics algorithm for multiple time scales: Systems with long range forces. J. Chem. Phys 1991, 94, 6811–6815. [Google Scholar]
- (43).Kratz EG; Walker AR; Lagardère L; Lipparini F; Piquemal JP; Andrés Cisneros G LICHEM: A QM/MM program for simulations with multipolar and polarizable force fields. J. Comput. Chem 2016, 37, 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Gökcan H; Vázquez-Montelongo EA; Cisneros GA LICHEM 1.1: Recent Improvements and New Capabilities. J. Chem. Theory Comput 2019, 15, 3056–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Frisch MJ et al. Gaussian∼16 Revision A.03 2016; Gaussian Inc.: Wallingford CT. [Google Scholar]
- (46).Cupellini L; Calvani D; Jacquemin D; Mennucci B Charge transfer from the carotenoid can quench chlorophyll excitation in antenna complexes of plants. Nat. Commun 2020, 11, 662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Šlouf V; Fuciman M; Dulebo A; Kaftan D; Koblížek M; Frank HA; Polívka T Carotenoid charge transfer states and their role in energy transfer processes in LH1–RC complexes from aerobic anoxygenic phototrophs. J. Phys. Chem. B 2013, 117, 10987–10999. [DOI] [PubMed] [Google Scholar]
- (48).Šlouf V; Keşan G; Litvín R; Swainsbury DJK; Martin EC; Hunter CN; Polívka T Carotenoid to bacteriochlorophyll energy transfer in the RC–LH1–PufX complex from Rhodobacter sphaeroides containing the extended conjugation ketocarotenoid diketospirilloxanthin. Photosynth. Res 2018, 135, 33–43. [DOI] [PubMed] [Google Scholar]
- (49).Razjivin A; Götze J; Lukashev E; Kozlovsky V; Ashikhmin A; Makhneva Z; Moskalenko A; Lokstein H; Paschenko V Lack of Excitation Energy Transfer from the Bacteriochlorophyll Soret Band to Carotenoids in Photosynthetic Complexes of Purple Bacteria. J. Phys. Chem. B 2021, 125, 3538–3545. [DOI] [PubMed] [Google Scholar]
- (50).Emsley P; Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Struct. Biol 2004, 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- (51).Malhotra HC; Britton G; Goodwin TW Occurrence of 1,2-dihydro-carotenoids in Rhodopseudomonas viridis. J. Chem. Soc. D 1970, 127–128. [Google Scholar]
- (52).Andreussi O; Knecht S; Marian CM; Kongsted J; Mennucci B Carotenoids and Light-Harvesting: From DFT/MRCI to the Tamm–Dancoff Approximation. J. Chem. Theory Comput 2015, 11, 655–666. 26579601 [DOI] [PubMed] [Google Scholar]
- (53).Srivastava R Physicochemical, antioxidant properties of carotenoids and its optoelectronic and interaction studies with chlorophyll pigments. Sci. Rep 2021, 11, 18365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Fuciman M; Keşan G; LaFountain AM; Frank HA; Polívka T Tuning the Spectroscopic Properties of Aryl Carotenoids by Slight Changes in Structure. J. Phys. Chem. B 2015, 119, 1457–1467. 25558974 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


