Abstract
Herpes simplex virus type 1 virions were examined by electron cryomicroscopy, allowing the three-dimensional structure of the infectious particle to be visualized for the first time. The capsid shell is identical to that of B-capsids purified from the host cell nucleus, with the exception of the penton channel, which is closed. The double-stranded DNA genome is organized as regularly spaced (∼26 Å) concentric layers inside the capsid. This pattern suggests a spool model for DNA packaging, similar to that for some bacteriophages. The bulk of the tegument is not icosahedrally ordered. However, a small portion appears as filamentous structures around the pentons, interacting extensively with the capsid. Their locations and interactions suggest possible roles for the tegument proteins in regulating DNA transport through the penton channel and binding to cellular transport proteins during viral infection.
Herpes simplex virus type 1 (HSV-1) infects about 80% of the human population and is the causative agent of many diseases, ranging from recurrent cold sores to blindness and life-threatening complications in immunosuppressed individuals (38). As one of the largest and most complex viruses, the infectious HSV-1 virion has a highly characteristic structure consisting of four compartments: envelope, tegument, capsid, and core (32). The core consists of the double-stranded DNA (dsDNA) genome of 152,000 bp, which is packaged into the preformed icosahedral capsid within the nucleus of the infected cell. The DNA has been reported to adopt a liquid crystalline organization within the capsid (3), but its precise arrangement is not known. The capsid is surrounded by a proteinaceous layer of variable thickness, called the tegument, and the entire structure is bounded by the viral envelope, a spherical lipid bilayer containing 12 or more different glycoproteins (32).
The capsid is a structurally well-defined icosahedron, an important function of which is to contain and protect the viral genome. Outside the cell, it never exists as a free entity but is always enclosed by the tegument and envelope. Tegument proteins are typically defined as being those structural proteins that are not components of purified capsids or of the envelope. Several of them have been shown to be involved in very early events during infection (2, 6, 11, 19, 24, 39), and their presence in the virion ensures their availability at this time. However, the precise roles of many tegument proteins have not yet been determined and there are several poorly understood aspects of the virus life cycle in which they are likely to be involved. Among these are packaging and release of the viral genome, transport of the capsid through the nuclear envelope and across the cytoplasm, and formation of the virion envelope (32). An insight into the nature of the tegument came from the identification of a second type of virus particle produced by infected cells, namely the L particle (33). L particles are composed of tegument and envelope but lack capsids and core and are consequently noninfectious. Their existence demonstrated that the tegument has inherent structural integrity and that its assembly could take place independently of capsids. However, tegument formation can occur in the absence of several of its major component proteins (39) and at least one of the major tegument proteins can increase severalfold in abundance (20). These observations suggest that the tegument does not have a unique geometrical organization with every protein occupying a specified position as in the capsid but rather that its constituent proteins interact in variable and possibly semirandom ways. However, due to their intimate association, it seems self-evident that the capsid and tegument will form specific interactions, although the nature of these has never been established.
The tegument is the least well characterized, structurally, of the virion compartments. It has proved intractable to analysis, and most of our information regarding its organization has been derived by the classical electron microscopic visualization techniques of thin sectioning, negative staining, and freeze etching (28, 37). Consequently, our understanding of tegument organization has lagged far behind that of the capsid, the structure of which has been determined to increasingly high resolution by electron cryomicroscopy and computer reconstruction approaches (40). In this report, we apply electron cryomicroscopy and computer reconstruction to study the three-dimensional (3D) structure of the intact, infectious HSV-1 virion. Comparisons between reconstructions of virions and purified capsids have allowed us to reveal, for the first time, details of the capsid-tegument interaction and viral DNA organization in the intact HSV-1 virion.
MATERIALS AND METHODS
Virion preparation.
To prepare virions, 80-oz roller bottles of BHK cells were infected with 0.001 PFU/cell of HSV-1 strain 17 and incubated at 31°C for 4 days. The virus was harvested from the supernatant medium and purified on 5 to 15% Ficoll gradients as described previously (33).
Electron cryomicroscopy and computer reconstruction.
A droplet of purified virions was applied to holey carbon grids and quickly frozen to liquid nitrogen temperature by using standard procedures (29). Images were taken with a dosage of 6 electrons/Å2 at ×30,000 in a JEOL 4000 electron cryomicroscope operated at 400 kV with a specimen temperature of −162°C.
Micrographs were digitized on a Zeiss SCAI microdensitometer (Carl Zeiss, Inc., Englewood, Colo.), using a 7-μm-step size. Blocks of adjacent pixels (3 by 3) were averaged to give a pixel size of 7 Å on the specimen. All data processing was carried out on an SGI Onyx2 parallel supercomputer with 24 R10000 processors (Silicon Graphics, Inc.) using parallel programs for performing refinement (40) and 3D reconstruction (17). Particle images were prescreened based on the evaluation of the defocus and image quality, using the ICE program package (41). Determination of the center and orientation parameters and their subsequent projection-based refinement were carried out by using procedures described previously (40, 42), which are based on Fourier common lines (7, 12). The parameters for 146 virion particles from 18 micrographs were determined.
The 3D reconstructions were generated by the Fourier-Bessel synthesis method (7). Prior to the merging of particle images for 3D reconstruction, the Fourier transforms of individual images were scaled by K
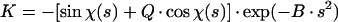 |
1 |
where s is the Fourier spatial frequency, Q is the fraction of the amplitude contrast relative to phase contrast, B is the amplitude decay factor, and
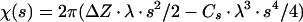 |
2 |
where ΔZ is the defocus value, λ is the wavelength of the electron beam, and Cs is the spherical aberration coefficient of the objective lens. To prevent the amplification of noise, the Fourier terms were excluded for those spatial frequency regions where the value of | sin χ(s) + Q · cos χ(s) | is less than 0.15. Because 18 micrographs of different defocuses (ranging from 1.6 to 2.7 μm) were used, no data up to 20 Å were missed in the final reconstruction. An amplitude contrast Q of 8% and a B factor of 200 Å2 were used.
The effective resolution was assessed by calculating the phase difference between two independent reconstructions from arbitrarily split data sets (44). The extent of icosahedral symmetry was evaluated by calculating the disagreement factor between two independent reconstructions (43). The structural components of interest were computationally extracted and visualized by using the Explorer software package (NAG Inc., Downer’s Grove, Ill.) with custom-designed modules (8). All surface representations of the reconstructions were displayed at one standard deviation (1ς) above the average density unless otherwise specified.
RESULTS
Electron cryomicroscopy and reconstruction of HSV-1 virions.
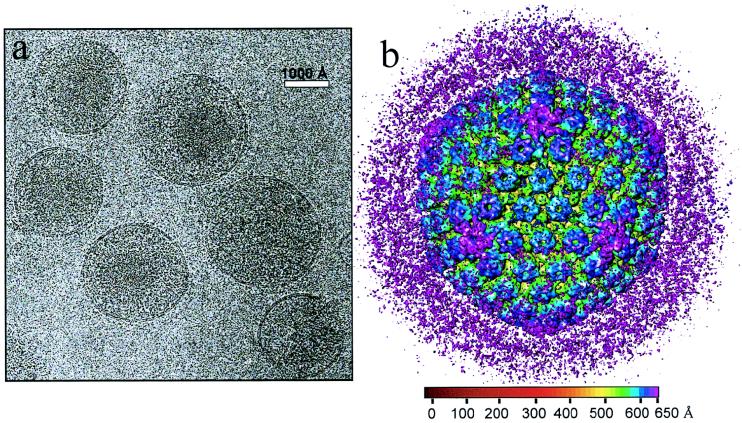
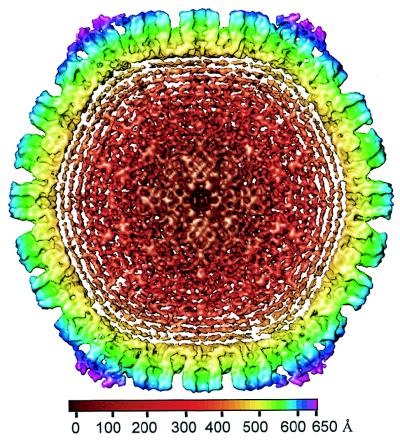
We imaged HSV-1 virions embedded in vitreous ice by using a 400-kV electron cryomicroscope. Most of the particles appeared intact, having a sharply defined envelope with an average diameter of 2,000 Å (Fig. 1a), but in some the envelopes were broken. We selected the intact virion images for further data processing and computationally boxed out the regions around the apparent center of the capsid to a radius of 1,050 Å. This choice of particle radius ensured that the tegument and viral envelope were included in the data analysis. After six iterative cycles of projection-based refinement, a 3D reconstruction to an effective resolution of 20 Å was obtained by merging 146 particles selected from 18 electron micrographs. In order to visualize the extent of icosahedrally ordered material, we displayed the 3D reconstruction out to the radius of the virion at a relatively low mass density threshold of 0.7ς (standard deviation) (Fig. 1b). In the radial coloring scheme used here, the hexons appear in blue and extend outward from the triplexes (green) and the capsid floor (yellow). Densities lying outside the nucleocapsid, which has a radius of 625 Å (29), are shown in purple (Fig. 1b). Most of the density lying outside the surface of the capsid appears as unconnected masses, and comparisons of different reconstructions revealed that their locations were not consistent, demonstrating a lack of icosahedral symmetry in the glycoprotein-containing envelope and the bulk of the tegument layer.
FIG. 1.
Electron cryomicrograph (a) and reconstruction (b) of HSV-1 virions. (a) The electron micrographs of ice-embedded HSV-1 virions were recorded at 400 kV in a JEOL 4000 electron cryomicroscope at ×30,000 magnification, using a dose of 6 electrons/Å2. The underfocus value of this image was determined to be 2.7 μm. Scale bar, 1,000 Å. (b) Shaded surface representation of the 3D map of the HSV-1 virion viewed along a threefold symmetry axis. The map is displayed at 0.7ς and colored radially using the color scheme shown at the bottom. In this scheme, all mass outside a radius of 650 Å is colored purple.
Radial distribution of DNA and proteins in the HSV-1 virion.
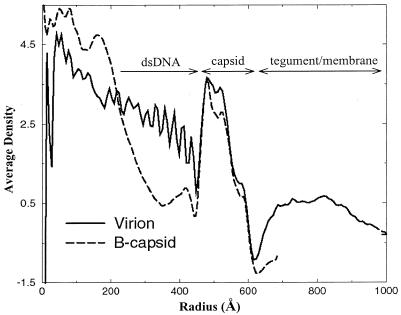
Figure 2 shows the average density distribution in the virion reconstruction as a function of particle radius. For comparison, we plotted the radial density distribution for the HSV-1 B-capsid structure computed to the same 20-Å resolution (42, 43). The B-capsids, which were purified from the nuclei of infected cells, comprise the outer icosahedral capsid shell and a proteinaceous core or scaffold but lack the viral DNA, tegument, and envelope (26). The density distribution profile of the virion resembles that of the B-capsid in having three peaks of density in the region of the capsid shell. However, the relative heights of the peaks are altered, with the outer two being elevated in the virion profile, indicating the presence of additional density in the outer regions of the virion capsid. Inside the capsid shell of the virion, the density attributable to the viral DNA appears as a series of peaks (up to 10) spaced ∼26 Å apart. Unlike the internal scaffold of the B-capsid, the DNA density fills the internal capsid space out to the inner floor of the capsid shell.
FIG. 2.
Averaged density distribution of the virion and B-capsid reconstructions as a function of particle radius. For the virion, the radial dimensions of the tegument and membrane, capsid shell, and dsDNA are indicated. The B-capsid profile terminates at 700 Å, which is at the limit of the boxed-out area.
The tegument can be seen in the virion as a region of relatively low density extending out to a radius of approximately 1,000 Å. The decline in tegument density beyond about 880-Å radius reflects the variation in size of individual virions and the displacement of the capsid from the center of the virion (Fig. 1a). In the region immediately surrounding the capsid (between radii of 600 to 635 Å), the density distribution reaches a minimum, indicating that there is little continuity between the capsid and tegument components.
Visualization of tegument-capsid interactions.
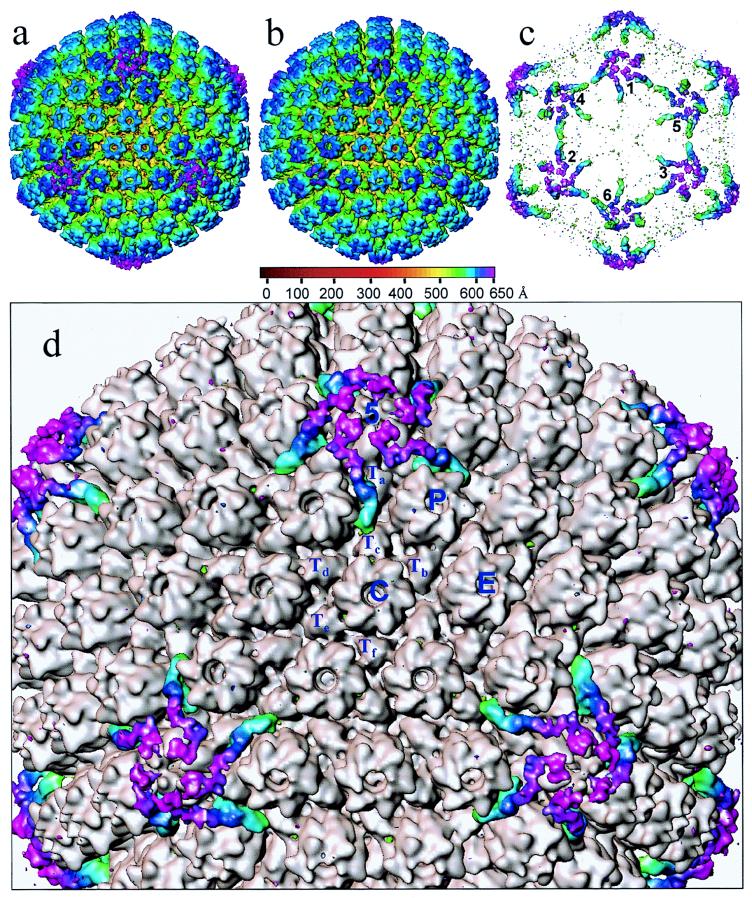
Figure 3a shows a surface view of the virion reconstruction contoured at 1ς. At this density threshold, which is higher than that used for Fig. 1, the nonicosahedrally related material beyond a radius of 650 Å has disappeared. This map reveals the characteristic T=16 icosahedral lattice previously seen in biochemically purified HSV-1 capsids (35, 42, 44). In order to highlight variations between virions and purified capsids, a difference map (Fig. 3c) was computed between the virion map (Fig. 3a) and a purified B-capsid map (Fig. 3b) reconstructed to the same resolution. This map demonstrated that their overall morphologies are similar except in localized regions around the pentons and their neighboring triplexes and in the space within the capsid shell (not shown for clarity). The capsids have identical internal and external diameters, and the organization of the subunits is unchanged. In particular, the hexons appear indistinguishable, indicating that they are not altered by the presence of the tegument.
FIG. 3.
Visualization of icosahedrally ordered tegument. (a) HSV-1 virion reconstruction displayed at 1.0ς. At this density threshold, the nonicosahedrally related densities in the tegument/membrane layers are not visible. (b) HSV-1 B-capsid reconstruction at the same resolution as the virion reconstruction (20 Å) displayed also at 1.0ς. (c) Difference map between the virion and B-capsid reconstructions. In this display, the densities inside the capsid shell are excluded for clarity. Consequently, differences occurring at all of the pentonal positions including those on the hemisphere facing toward (labeled 1, 2, and 3) and away from (labeled 4, 5, and 6) the observer can be seen. (d) Superposition of the difference map on the B-capsid reconstruction (gray). The tegument proteins are highlighted in color. Labeled are the unique structural components in one asymmetric unit (the unique building block of the entire icosahedron) of the T=16 capsid, which comprises 1 penton (5) subunit, 2½ hexons (P, C, and E), and 5 triplexes (Ta, Tb, Tc, Td, Te, and Tf).
triplexes (Ta, Tb, Tc, Td, Te, and Tf).
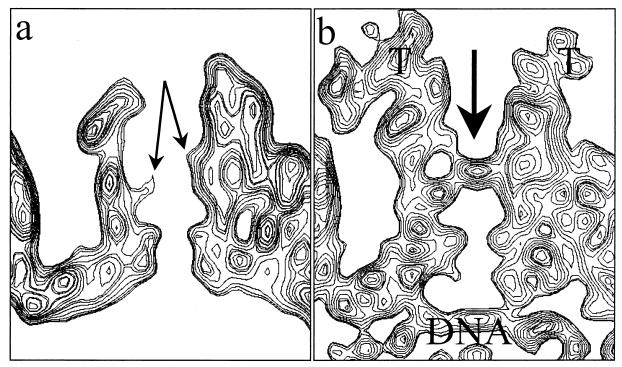
By contrast, there are marked differences between the two maps in the region of the pentons, which are highlighted in color in the superposition of the difference map on the B-capsid map (Fig. 3d). The most obvious difference is the presence of additional material extending from the surface of the pentons. The extra density appears as a continuous, convoluted ribbon, approximately 200 Å long and 40 Å thick. This density extends from the interface between the upper domains of two adjacent VP5 subunits in the penton and connects to the peripentonal triplex (Ta) and its nearest neighbor, triplex Tc, at different sites on their upper surfaces (Fig. 4). It also makes contact with the middle domains of two subunits in the P hexon (Fig. 4, upper left). The approximate mass of this extra material is 170 to 200 kDa. The presence of such external densities in the virion reconstruction, which were not seen previously in reconstructions of A-, B-, or C-capsids, suggests that they represent tegument proteins.
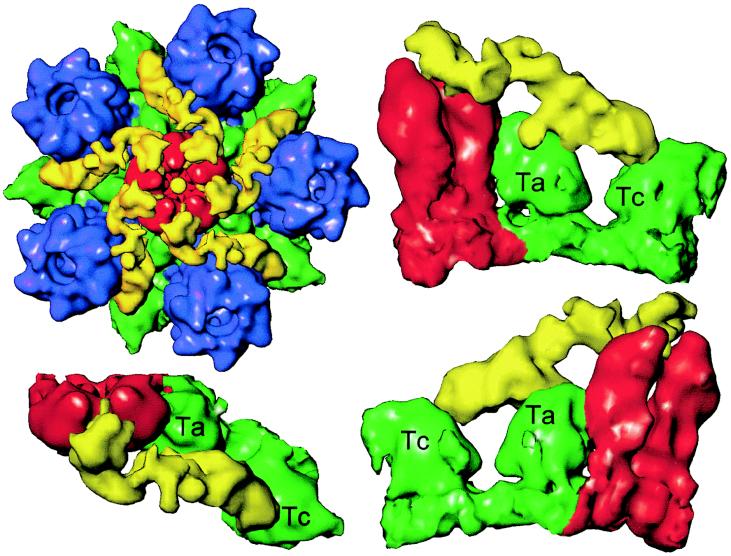
FIG. 4.
Capsid-tegument interactions. (Upper left) Close-up view of a computationally isolated portion of the superposition map (Fig. 3), including the penton (red), the five P hexons (blue), and the Ta and Tc triplexes (green). The additional density (yellow) that is not present in the B-capsid is attributed to tegument proteins, which clearly make contact with the penton, hexon, and triplexes. (Lower left panel and right column) Enlarged top and two side views of the tegument density interacting with its adjacent penton subunits and triplexes Ta and Tc. In these views, the P hexon, which also interacts with the tegument density, is removed for clarity.
Configuration of the penton channel.
A less obvious difference between the virion and purified B-capsid structures is the closure of the penton channel. As is evident in the comparisons of the sectional views of pentons from the B-capsid and the virion (Fig. 5), the closure is in a region that is constricted in the B-capsid by a protrusion from the middle domain of VP5 (Fig. 5a) (44). In addition, there are strong densities beneath the penton channel in the virion (Fig. 5b) that can be attributed to the viral DNA. No such densities are seen at equivalent positions in the hexon channel.
FIG. 5.
Closure of the penton channel in the virion. (a) Sectional view of the B-capsid penton. In the middle of the channel, densities from the VP5 subunits (arrows) protrude inward, resulting in a constriction. (b) Sectional view of the virion capsid penton. The arrow indicates the closure in the axial channel at the region that is constricted in the B-capsid channel. Also indicated are the densities attributed to tegument (T) and viral DNA.
Organization of viral dsDNA.
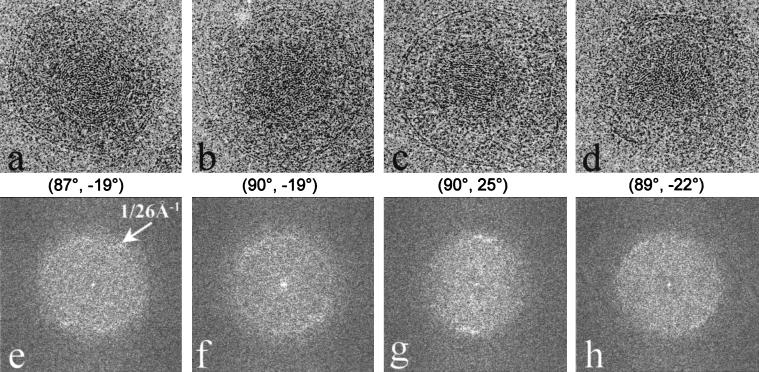
The radial density plot shown in Fig. 2 revealed that the density inside the capsid shell of the virion, which is contributed by the viral DNA, appears as a regular pattern of peaks spaced ∼26 Å apart. A cross-sectional view of the virion reconstruction along a twofold axis (Fig. 6) exhibits high-density features organized as multiple shells inside the inner surface of the capsid floor. At least six concentric shells can be easily distinguished before the pattern becomes indistinct toward the center of the capsid (Fig. 6). The same 26-Å spacing can be seen in raw images of the particles (Fig. 7a to d) and in their computed diffraction patterns (Fig. 7e to h). The angular distribution patterns of the diffraction intensities at this spacing appear to vary, not only among particles in different orientations but also among particles in icosahedrally equivalent orientations (Fig. 7e to h). Since these orientations were determined by assuming icosahedral symmetry, variations in the DNA density distributions among particles of similar orientations suggest a lack of icosahedral symmetry in the DNA organization. To confirm this, we calculated the degree of correlation between the DNA density distributions in two different reconstructions (data not shown). Our results show that the DNA densities indeed correlate poorly between reconstructions, and thus they are not icosahedrally organized. Therefore, in our reconstruction, the concentrically arranged layers of DNA have been smeared out into uniform shells of density by icosahedral averaging.
FIG. 6.
Central cross-section (100 Å thick) through the virion reconstruction as viewed along a twofold axis. The concentric shells of density inside the capsid are attributable to the viral DNA. The spacing between the layers is 26 Å. The map is colored radially, using the color scheme shown at the bottom.
FIG. 7.
Organization of the dsDNA inside the HSV-1 virion. (a through d) Representative projection views of raw virion images, revealing characteristic patterns of DNA, including circular ring (a), dotted (b), striation (c), and dotted-striation (d) patterns. The orientations [(θ, φ), listed below each image] were determined by assuming icosahedral symmetry. Although the orientations are similar for panels a and b and for panels c and d, their DNA projections are strikingly different. (e through f) Computed diffraction patterns of panels a through d, respectively, showing distinctive patterns due to differences in the DNA genome projections. All the diffraction patterns show 1/26 Å−1 spacing (arrows in panel e) with different distributions.
DISCUSSION
HSV-1 is among the largest spherical viruses and has a very complex structure and composition (32). Due to its large size, lack of uniformity, and sensitivity to structural damage, it is difficult to study the molecular structure of the intact virion by X-ray crystallography. Electron cryomicroscopy has proved an effective method for studying the icosahedral shells of biochemically purified HSV-1 capsids to a relatively high resolution (40). However, these studies have not yielded information on proteins external to the capsid shell or on the genome inside the capsid shell. We have now carried out the first attempt to analyze the structure of the entire HSV-1 virion, using a 400-kV electron cryomicroscope that produces micrographs of sufficiently high quality (Fig. 1a) to allow 3D reconstruction from particles at different orientations (Fig. 1b).
Our analysis of intact virions demonstrated that the bulk of the tegument is not icosahedrally ordered, thereby providing the first quantitative verification that the tegument has a largely asymmetric or unstructured organization. However, a small amount of tegument material with icosahedral symmetry is resolved where it makes contact with the outer capsid surface (Fig. 3a and c). This is the first direct structural evidence for the existence of ordered tegument proteins and their well-defined interactions with capsid proteins. Such localized symmetry in an otherwise nonicosahedral structure bears a close resemblance to the situation recently described for the internal B-capsid scaffold (43).
The pattern of interaction between the capsid and tegument (Fig. 4) is very interesting, being confined to the vicinity of the pentonal vertices and apparently involving contacts between the tegument protein or proteins and the penton and some P (peripentonal) hexon subunits (composed of VP5) and their adjacent triplexes (VP19C and VP23). Although we cannot exclude the possibility that contact between tegument and capsid may occur at other locations, the virtual identity of the virion capsid and B-capsid structures away from the pentonal vertices, even when examined at a lower density threshold (Fig. 1b), implies that such contacts must either be very tenuous or unrelated to icosahedral symmetry. The restriction of tegument contacts to the pentons would be consistent with the observation of tightly attached tegument material at the vertices of capsids in negative stain and freeze-etching images of detergent-treated equine herpesvirus virions (37). It is interesting to consider why tegument binding might be limited to the capsomeres and triplexes around the pentonal vertices, when apparently similar contact sites are present on other capsomeres and triplexes throughout the capsid. For example, the tegument proteins occupy the space between the upper domains of two adjacent penton VP5 subunits (Fig. 4) but not those between adjacent hexon VP5 subunits. In the hexon, this position is already occupied by VP26, making it unavailable for tegument binding (42). Therefore, it is probably the availability of these sites on the penton VP5 subunits and their unique spatial relationship with the other contact sites on the middle domain of hexon VP5 and on Ta and Tc that determines their favorable interactions with the tegument proteins. Binding of the tegument proteins to these positions does not direct the formation of pentons, since stable capsids can be assembled in their absence. Thus, the locations of these tegument proteins are presumably related to other properties of the capsid, including, as discussed below, DNA packaging and cytoplasmic transport.
Candidate tegument proteins.
The tegument is a complex structure which contains at least 18 different viral proteins (32). The functions of most of these and their structural relationships within the tegument are still poorly defined; however, a number of them have been shown to be nonessential for virus replication and therefore seem unlikely to be candidates to form the major connection between tegument and capsid. Earlier morphological and biochemical studies provide some indications regarding which tegument protein is being resolved in our reconstruction of the intact virion.
Biochemically, the essential tegument protein VP1-3 has been shown to bind very tightly to the capsid. Thus, detergent treatment of virions removes the envelope and solubilizes some tegument proteins but leaves others (notably VP1-3) in an insoluble, capsid/tegument fraction (31, 36), while more vigorous treatment results in the loss of virtually all envelope and tegument proteins except for VP1-3 (14). Since it has been shown (37) that tegument attached at the pentons was also resistant to removal by detergent, it seems highly likely that the detergent-insoluble VP1-3 is located at or near these positions. Thus, VP1-3 represents a good candidate for the icosahedrally ordered tegument in our reconstruction. The virion has been reported to contain approximately 120 to 200 molecules of VP1-3 (16), considerably more than the 60 copies that would be expected if it was bound in a 1:1 ratio to the pentonal VP5 subunits. However, VP1-3 is present in both virions and in L particles (33), which lack capsids, indicating that it does not have an exclusive association with capsids. With a predicted molecular size of about 336 kDa, VP1-3 is larger than the estimated mass of each tegument density seen here (170 to 200 kDa). Therefore, if these densities do represent VP1-3, this suggests that part of the protein may not be resolved in our reconstruction.
VP1-3 is an interesting but poorly characterized protein. It is by far the largest HSV protein. The gene encoding it (UL36) (21, 22) has recognizable, although poorly conserved, counterparts in all other mammalian and avian herpesviruses examined to date. It has an essential function, and a temperature-sensitive mutant (tsB7) with a mutation in the UL36 gene (2, 18) is defective for the release of DNA from infecting capsids. Thus, at nonpermissive temperatures, tsB7 virions fuse with the cell, liberating tegument and capsids into the cytoplasm. The capsids are transported to the nuclear pore but fail to release the viral genome into the nucleus. Since the penton has been suggested to be the route by which the virus DNA leaves the capsid (23), an interaction between VP1-3 and the penton would place it in an appropriate position to influence the passage of the viral genome. When the HSV virion enters the infected cell by fusing with the plasma membrane, the viral envelope is removed, the capsid plus tegument enters the cytoplasm, and many of the tegument proteins disassociate from the capsid. The phenotype of tsB7 suggests that VP1-3 remains associated with the capsid at least until it reaches the nuclear pore.
HSV capsids are transported across the cytosol on the microtubule network, and the transport is mediated through an interaction with the microtubule motor protein, dynein, which attaches to the vertices of the capsid. It has been suggested that VP1-3 might be involved in this interaction (30), and the prominent locations of the ordered masses surrounding the penton (Fig. 4) support the possibility that the interaction of dynein with the vertex could occur via the tegument proteins rather than with VP5 directly.
Status of the penton channel.
In all of the biochemically purified capsid types that have been examined, there is an open channel through the penton (Fig. 5) (1, 34, 44) and studies on purified A-capsids have shown that this is the largest channel to penetrate the capsid shell (44). This observation is consistent with it being the port through which the viral DNA is packaged, as suggested by Newcomb and Brown (23) following studies of the effects of denaturants on DNA-containing C-capsids. However, in order to retain the DNA within the capsid after packaging, the closure of such large holes would be necessary and our reconstruction reveals that the penton channel is indeed blocked in the virion.
The amount of additional mass in the penton channel is very small (<10 kDa), suggesting the closure is most likely due to movement of the middle domain of VP5 rather than the presence of another protein. However, at the current resolution, we cannot unambiguously distinguish between these possibilities. If this central mass does represent a domain of VP5, the protein would have to undergo significant conformational changes in order to adopt the different structures seen in the B-capsid and the mature virion. Binding of the tegument proteins to the penton might trigger such changes after packaging of DNA has taken place. Following infection of a new cell, release of the DNA would require the penton channels to open again. This could be achieved through a reversal of the above process, in which separation of the tegument protein from the penton VP5 results in the channel reverting to its original state.
Arrangement of dsDNA inside the virion capsid.
As in dsDNA bacteriophage, the HSV-1 genome is packaged into a preformed icosahedral capsid. Several models have been proposed to describe how DNA might be organized within a capsid, among them the spool model, hairpin model, ball-of-string model, and toroid model. Early studies by electron microscopy and low-angle X-ray scattering both indicated that in bacteriophage, the DNA is wound into a spool-like structure (9, 10, 25). A recent study, which used computer-generated projections of a mathematical model of spooled DNA to replicate features seen in electron microscopic images of bacteriophage T7 capsids, provided strong evidence in support of this arrangement (5).
In projection, the herpesvirus genomes show characteristic patterns (Fig. 7), similar to those expected for the spool model proposed for bacteriophage T7 (5). Furthermore, the concentric spherical shells and the 26-Å spacing observed inside the HSV-1 virion (Fig. 6 and 7e to f) are strikingly similar to those seen in packaged phage DNA (5, 9). Although the outermost layer of the HSV DNA density is in contact with the floor domain of the penton subunit (Fig. 5b), we have not yet demonstrated any icosahedral tendency in the viral genome. A lack of icosahedral symmetry is also a feature of the dsDNA-containing phages. The close parallels between the HSV-1 and T7 DNA data strongly suggest that the spool model may also describe the organization of the HSV-1 genome. Interestingly, the spooled organization of the viral DNA suggested by our results is similar in several aspects to the toroid model originally proposed by Furlong et al. (13) which may account for certain striking images of apparently toroidal cores in thin sections of capsids. However, unlike bacteriophage T7 (27), HSV does not appear to possess a central protein plug, or spindle, inside the capsid around which the genome is arranged.
According to the spool model, DNA passes into the capsid through a unique entry port and then wraps around the inner surface of the capsid shell. It accumulates one layer at a time, with the layers becoming less well ordered as their distance from the shell increases (15). The orientation of the spool along a definite axis accounts for the changing appearance of the DNA when viewed from different, although possibly icosahedrally equivalent, directions (Fig. 7). During unpacking, the process is reversed and the DNA uncoils from the inside of the capsid outward. The spacing between adjacent close-packed dsDNA duplexes in bacteriophage is about 25 Å (9). This agrees very well with the pattern we see in HSV and suggests that the genome is packed as extended, predominantly naked DNA. As shown in the radially averaged density distribution in Fig. 2, up to 10 concentric DNA layers can be distinguished inside the capsid, occupying ∼90% of the internal volume. Based on the 3.4-Å pitch (rise per base pair) of B-form DNA (4) and our observed distance of 26 Å between adjacent DNA duplexes, we estimate that a close-packed HSV-1 genome of 152 kb would occupy a total volume of 3.0 × 108 Å3. This is about 75% of the total volume available inside the HSV-1 capsid. Therefore, it seems probable that the DNA occupying the inner portions of the capsid is packed at a lower density.
ACKNOWLEDGMENTS
We thank Matthew Dougherty for help in graphics display. D.H.C. is a visiting graduate student of K. H. Kuo from the Department of Materials Physics, University of Science & Technology, Beijing, and the Beijing Laboratory of Electron Microscopy, Institute of Physics, Chinese Academy of Sciences, China.
This work was supported by grants from the NIH (AI38469 and RR02250) and the NSF (BIR-9413229) and by the Human Frontier Science Program (RG-537/96).
REFERENCES
- 1.Baker T S, Newcomb W W, Booy F P, Brown J C, Steven A C. Three-dimensional structures of maturable and abortive capsids of equine herpesvirus 1 from cryoelectron microscopy. J Virol. 1990;64:563–573. doi: 10.1128/jvi.64.2.563-573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in the release of viral DNA and in other stages of the viral reproductive cycle. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Booy F P, Newcomb W W, Trus B L, Brown J C, Baker T S, Steven A C. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantor C R, Schimmel P R. Structures of nucleic acids. In: Cantor C R, Schimmel P R, editors. Biophysical chemistry. Part 1. The conformation of biological macromolecules. Vol. 1. San Francisco, Calif: W. H. Freeman and Company; 1980. pp. 155–190. [Google Scholar]
- 5.Cerritelli M E, Cheng N, Rosenberg A H, McPherson C E, Booy F P, Steven A C. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–280. doi: 10.1016/s0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 6.Coulter L, Moss H, Lang J, McGeoch D. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J Gen Virol. 1993;74:387–395. doi: 10.1099/0022-1317-74-3-387. [DOI] [PubMed] [Google Scholar]
- 7.Crowther R A. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Phil Trans R Soc London Ser B. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- 8.Dougherty M T, Chiu W. Using animation to enhance 3D visualization: a strategy for a production environment. In: Bailey G W, Alexander K B, Jerome W G, Bond M G, McCarthy J J, editors. Microscopy and microanalysis 1998. Atlanta, Ga: Springer; 1998. pp. 452–453. [Google Scholar]
- 9.Earnshaw W, Harrison S C. DNA arrangement in isometric phage heads. Nature. 1977;268:598–602. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- 10.Earnshaw W, King J, Harrison S C, Eiserling F A. The structural organization of DNA packed within the heads of T4 wild type, isometric, and giant bacteriophages. Cell. 1978;14:559–568. doi: 10.1016/0092-8674(78)90242-8. [DOI] [PubMed] [Google Scholar]
- 11.Fenwick M L, Everett R D. Inactivation of the shutoff gene (UL41) of herpes simplex virus types 1 and 2. J Gen Virol. 1990;71:2961–2967. doi: 10.1099/0022-1317-71-12-2961. [DOI] [PubMed] [Google Scholar]
- 12.Fuller S D, Butcher S J, Cheng R H, Baker T S. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 13.Furlong D, Swift H, Roizman B. Arrangement of herpesvirus deoxyribonucleic acid in the core. J Virol. 1972;10:1071–1074. doi: 10.1128/jvi.10.5.1071-1074.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson W, Roizman B. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J Virol. 1972;10:1044–1052. doi: 10.1128/jvi.10.5.1044-1052.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison S C. Packaging of DNA into bacteriophage heads: a model. J Mol Biol. 1983;171:577–580. doi: 10.1016/0022-2836(83)90045-1. [DOI] [PubMed] [Google Scholar]
- 16.Heine J W, Honess R W, Cassai E, Roizman B. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol. 1974;14:640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson O, Govindan V, Park Y, Zhou Z H. The 4th International Conference on High Performance Computing. Los Alamitos, Calif: IEEE Computer Society Press; 1997. Custom virtual memory policy for an image reconstruction application; pp. 517–521. [Google Scholar]
- 18.Knipe D M, Batterson W, Nosal C, Roizman B, Buchan A. Molecular genetics of herpes simplex virus. VI. Characterization of a temperature-sensitive mutant defective in the expression of all early viral gene products. J Virol. 1981;38:539–547. doi: 10.1128/jvi.38.2.539-547.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemaster S, Roizman B. Herpes simplex virus phosphoproteins. II. Characterization of the virion protein kinase and of the polypeptides phosphorylated in the virion. Virology. 1980;35:798–811. doi: 10.1128/jvi.35.3.798-811.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leslie J, Rixon F J, McLauchlan J. Overexpression of the herpes simplex virus type 1 tegument protein VP22 increases its incorporation into virus particles. Virology. 1996;220:60–68. doi: 10.1006/viro.1996.0286. [DOI] [PubMed] [Google Scholar]
- 21.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 22.McNabb D S, Courtney R J. Analysis of the UL36 open reading frame encoding the large tegument protein (ICP1/2) of herpes simplex virus type 1. J Virol. 1992;66:7581–7584. doi: 10.1128/jvi.66.12.7581-7584.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newcomb W W, Brown J C. Induced extrusion of DNA from the capsid of herpes simplex virus type 1. J Virol. 1994;68:433–440. doi: 10.1128/jvi.68.1.433-440.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Post L E, Mackem S, Roizman B. Regulation of α genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with α gene promoters. Cell. 1981;24:555–565. doi: 10.1016/0092-8674(81)90346-9. [DOI] [PubMed] [Google Scholar]
- 25.Richards K, Williams R, Calendar R. Mode of DNA packing within bacteriophage heads. J Mol Biol. 1973;78:255–259. doi: 10.1016/0022-2836(73)90114-9. [DOI] [PubMed] [Google Scholar]
- 26.Rixon F J. Structure and assembly of herpesviruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 27.Roeder G S, Sadowski P D. Bacteriophage T7 morphogenesis: phage-related particles in cells infected with wild-type and mutant T7 phage. Virology. 1977;76:263–285. doi: 10.1016/0042-6822(77)90302-6. [DOI] [PubMed] [Google Scholar]
- 28.Roizman B, Furlong D. The replication of herpesviruses. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. New York, N.Y: Plenum Press; 1974. pp. 229–403. [Google Scholar]
- 29.Schrag J D, Prasad B V V, Rixon F J, Chiu W. Three-dimensional structure of the HSV-1 nucleocapsid. Cell. 1989;56:651–660. doi: 10.1016/0092-8674(89)90587-4. [DOI] [PubMed] [Google Scholar]
- 30.Sodeik B, Ebersold M W, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spear P G, Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972;9:143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steven A C, Spear P G. Herpesvirus capsid assembly and envelopment. In: Chiu W, Burnett R M, Garcea R, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1997. pp. 312–351. [Google Scholar]
- 33.Szilágyi J F, Cunningham C. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J Gen Virol. 1991;72:661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 34.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 35.Trus B L, Homa F L, Booy F P, Newcomb W W, Thomsen D R, Cheng N, Brown J C, Steven A C. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vernon S K, Lawrence W C, Long C A, Cohen G H, Rubin B A. Herpesvirus vaccine development: studies of virus morphological components. In: Voller A, Friedman H, editors. New trends and developments in vaccines. Lancaster, Pa: MTP Press; 1978. pp. 179–210. [Google Scholar]
- 37.Vernon S K, Lawrence W C, Long C A, Rubin B A, Sheffield J B. Morphological components of herpesvirus. IV. Ultrastructural features of the envelope and tegument. J Ultrastruct Res. 1982;81:163–171. doi: 10.1016/s0022-5320(82)90072-7. [DOI] [PubMed] [Google Scholar]
- 38.Whitley R J. Herpes simplex viruses. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2297–2342. [Google Scholar]
- 39.Zhang Y, McKnight K L C. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J Virol. 1993;67:1482–1492. doi: 10.1128/jvi.67.3.1482-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Z H, Chiu W, Haskell K, Spears H J, Jakana J, Rixon F J, Scott L R. Refinement of herpesvirus B-capsid structure on parallel supercomputers. Biophys J. 1998;74:576–588. doi: 10.1016/S0006-3495(98)77816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Z H, Hardt S, Wang B, Sherman M B, Jakana J, Chiu W. CTF determination of images of ice-embedded single particles using a graphics interface. J Struct Biol. 1996;116:216–222. doi: 10.1006/jsbi.1996.0033. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Z H, He J, Jakana J, Tatman J D, Rixon F J, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Z H, Macnab S J, Jakana J, Scott L R, Chiu W, Rixon F J. Identification of the sites of interaction between the scaffold and outer shell in herpes simplex virus-1 capsids by difference electron imaging. Proc Natl Acad Sci USA. 1998;95:2778–2783. doi: 10.1073/pnas.95.6.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Z H, Prasad B V V, Jakana J, Rixon F, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:458–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]