Abstract
Drug resistance and disease progression are common in multiple myeloma (MM) patients, underscoring the need for new therapeutic combinations. A high-throughput drug screen in 47 MM cell lines and in silico Huber robust regression analysis of drug responses revealed 43 potentially synergistic combinations. We hypothesized that effective combinations would reduce MYC expression and enhance p16 activity. Six combinations cooperatively reduced MYC protein, frequently over-expressed in MM and also cooperatively increased p16 expression, frequently downregulated in MM. Synergistic reductions in viability were observed with top combinations in proteasome inhibitor-resistant and sensitive MM cell lines, while sparing fibroblasts. Three combinations significantly prolonged survival in a transplantable Ras-driven allograft model of advanced MM closely recapitulating high-risk/refractory myeloma in humans and reduced viability of ex vivo treated patient cells. Common genetic pathways similarly downregulated by these combinations promoted cell cycle transition, whereas pathways most upregulated were involved in TGFβ/SMAD signaling. These preclinical data identify potentially useful drug combinations for evaluation in drug-resistant MM and reveal potential mechanisms of combined drug sensitivity.
Keywords: Myeloma, MYC, p16, dinaciclib, entinostat
1. Introduction
Multiple myeloma (MM), a neoplastic clonal proliferation of plasma cells, is one of the most common hematologic malignancies1,2, with an estimated 35,730 new cases and 15,590 deaths in the United States for 2023.3 Development of therapeutics such as proteasome inhibitors (PI), immunomodulatory drugs, monoclonal antibodies, chimeric antigen receptor (CAR) T-cell therapy, bispecific T-cell engagers (BiTEs), and autologous stem cell transplantation, has resulted in prolonged survival of MM patients.1,2,4-6 However, no therapy thus far is curative, and relapsed and/or refractory multiple myeloma (RRMM) eventually develops.1,2 Thus, combining targeted agents has become important in treating RRMM and oncology in general.7 Combining drugs circumvents or slows tumor resistance by utilizing synergy, wherein the effect of two or more drugs is greater than the sum of individual drug effects. Furthermore, identifying mechanisms of molecular synergy provides a biologic rationale for proposed combinations and is essential for predicting clinical responses.8
Previous research demonstrated efficacy and drug synergy for the combination of mTOR inhibitors (mTORi) and histone deacetylase inhibitors (HDACi) in human MM cell lines, a diverse set of cancer cell types (NCI60), and MM cells isolated from patients.7,8 A proposed mechanism of action of synergy in the combination – degradation of the transcription factor MYC – was elucidated via a biologically integrated, network-based approach using MM cell lines and patient datasets.6 MYC activation is found in both MM and smoldering myeloma (SMM) patients and is associated with disease progression.10-15 Additionally, NRAS and KRAS are frequently mutated in both SMM and MM, associated with disease progression, and increase the stabilization of MYC via phosphorylation of serine 62.8,10,16 Furthermore, retroviral constructs overexpressing MYC alone or RAS alone were not very effective in inducing mouse plasma cell tumors compared with constructs that overexpressed both MYC and RAS which produced tumors in less than 30 days.17,18 Targeting MYC directly, unfortunately, has remained a key challenge in oncology due to large protein-protein interaction interfaces, lack of deep protein pockets, and nuclear localization.19,20 Finding drug combinations that indirectly target MYC, while acting upon their own respective direct targets, serves as a useful alternative strategy and provides additional opportunities for synergy.15,21 Additionally, loss of function of cell cycle transition checkpoint control gene p16 can result in uncontrolled cell proliferation, invasion, and metastasis in cancers, and inactivation contributes to disease progression.22-24 The ability to cooperatively increase p16 activity, therefore, was proposed as useful additional screening measure for top drug combinations.
We employed an agnostic discovery approach to identify drug combinations targeting MM by utilizing a high-throughput drug screen of a compound library of ~1900 small molecules25 in 47 MM cell lines and in silico regression analysis to identify drug combinations predicted to reduce MM viability. Considering the importance of MYC in MM progression, we identified candidate tool combinations that cooperatively reduce MYC protein expression and increase p16 expression, while synergistically reducing the viability of both proteasome inhibitor (PI) resistant and PI sensitive MM cell lines in vitro. We then evaluated the efficacy of our top drug combinations in prolonging viability in a transplantable Ras-driven allograft model of advanced MM that closely recapitulates high-risk/refractory myeloma in humans.26,27 The ability of the combinations to reduce the viability of patient cells ex vivo was also evaluated. Finally, gene set overrepresentation analysis of MM cells treated with the combinations revealed commonly affected pathways. Through expansion of the in vitro findings, extension to unique in vivo models, and investigation into the mechanisms of synergistic drug responses, this study provides strong preclinical rationale for further evaluation of novel drug combinations in treating MM.
2. Materials and methods
A high-throughput drug screen25 was employed to identify single agents from a pool of 1900 compounds that were effective, at a concentration of 2 μM or less, in reducing the viability of at least 25 of the 47 multiple myeloma (MM) cell lines tested. Drugs that were both effective and had different mechanisms of action were paired for Huber robust regression analysis. The 43 selected combinations were highly correlated in their responses (r2 value ≥ 0.5). These combinations were tested (Western blot) for their ability to cooperatively reduce MYC protein expression relative to single agent and/or control and increase protein expression of CDKN2A (p16). Of the six combinations that reduced MYC and increased p16 protein expression, three (CDKi/HDACi, TOP2Ai/AURKAi, TOP2Ai/HSP90i) were selected for further efficacy evaluation via testing for their ability to reduce MYC and increase p16 protein expression in cell lines inherently resistant and sensitive to the drugs used in the original high-throughput screen (based on mean IC50 for all 1920 drugs), as well as cell lines with acquired resistance to commonly used therapies against MM (proteasome inhibitor (oprozomib), corticosteroid (dexamethasone), and an anthracycline (doxorubicin)). Additionally, combinations were evaluated for their ability to cooperatively reduce viability of L363 MM cells in monoculture or co-cultured with human bone marrow HS-5 cells. Further, efficacy of the combinations was evaluated in vivo in sublethally irradiated C57BL/6J mice injected intravenously with murine Vk*MYC; NrasLSL Q61R/+; IgG1-Cre (VQ) cells harvested from bone marrow of donor mice, and ex vivo in CD138+ and CD138− MM cells obtained fresh from bone marrow biopsies of human SMM patients. Finally, functional enrichment analysis was used to find common genetic pathways similarly affected in L363 MM cells treated with the drug combinations. Detailed experimental methods, as well as functional enrichment analysis methods, are available in the supplemental methods section.
3. Results
3.1. Drug screen reveals combinations that cooperatively target MYC and CDKN2A (p16) and reduce viability of MM cells
Drugs from our high-throughput screen were selected for activity based on the following criteria: single agent dose-response curve class of −1.1 (complete response), IC50 < 2.0 μM, and activity in at least 25 of the 47 MM cell lines tested (Fig.1, Table S1). In addition, drugs paired for regression analysis were selected as having different mechanisms of action, i.e., not targeting the same pathway. Huber robust regression analysis (Fig.S1A) of each drug versus every other drug was utilized to generate a Pearson correlation coefficient (r2) for each drug pair. Forty-three combinations were noted with r2 values of at least 0.5 (Table 1).
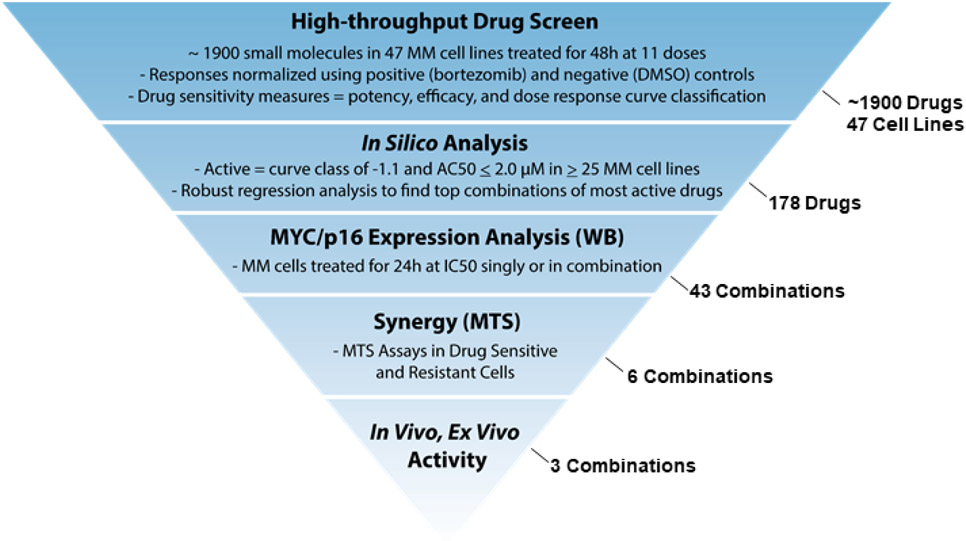
Figure 1.
Prediction workflow used to find top drug combinations inhibiting MM cell growth. IC50 = half maximal inhibitory concentration, WB = western blot, MTS = tetrazolium-based cell proliferation assay. Numbers indicated on the right denote the numbers of drugs, combinations, or cell lines investigated in each phase of the prediction workflow.
Table 1.
Top Drug Combinations Inhibiting MM Cells Based on Huber Robust Regression
| r2 | Compound 1 Target Gene |
Compound 1 Name | Compound 2 Name | Compound 2 Target Gene |
Cmpd 1 Count |
Cmpd 2 Count |
|---|---|---|---|---|---|---|
| 0.836 | ITK | NCGC00344999-01 = ITK(1) | VER-82576 | HSP90AB1 | 39 | 33 |
| 0.822 | HSP90AB1 | Ganetespib | BS-194 | CDK1 | 35 | 46 |
| 0.804 | HSP90AB1 | SNX-5422 | NCGC00344999-01 = ITK(1) | ITK | 35 | 39 |
| 0.765 | HSP90AB1 | SNX-5422 | NCGC00188382-01 = ITK(2) | ITK | 35 | 45 |
| 0.732 | HDAC1 | Romidepsin | SR-3306 | MAPK8 | 45 | 46 |
| 0.714 | TUBB | XRP-44X | BMS-3 | LIMK1 | 28 | 37 |
| 0.702 | HSP90AB1 | SNX-5422 | NCGC00344990-01 = ITK(3) | ITK | 35 | 38 |
| 0.678 | TUBB | XRP-44X | ON-01910 | PLK1 | 28 | 37 |
| 0.671 | IKBKB | IMD-0354 | Niclosamide | STAT3 | 45 | 41 |
| 0.658 | HSP90AB1 | Ganetespib | Dacinostat | HDAC1 | 35 | 44 |
| 0.655 | AURKA | Alisertib | Doxorubicin | TOP2A | 29 | 46 |
| 0.645 | PIM3 | GDC-0349 | PP242 | MTOR | 34 | 34 |
| 0.644 | TLR7 | CPG-52364 | Sepantronium bromide | BIRC5 | 44 | 36 |
| 0.643 | ITK | NCGC00344990-01 = ITK(3) | VER-82576 | HSP90AB1 | 38 | 33 |
| 0.634 | HSP90AB1 | AT-13387AU | NCGC00344990-01 = ITK(3) | ITK | 39 | 38 |
| 0.634 | CDK1 | BS-194 | Dacinostat | HDAC1 | 46 | 44 |
| 0.626 | MAPK8 | TCS JNK 5a | Indibulin | TUBB | 33 | 28 |
| 0.616 | CDK1 | 7-Hydroxystaurosporine | PIK-75 | PIK3CA | 37 | 33 |
| 0.614 | ITK | NCGC00188382-01 = ITK(2) | Methylrosaniline chloride | TNFRSF1A | 45 | 36 |
| 0.611 | PIK3CA | PIK-75 | Flavopiridol | CDK1 | 33 | 44 |
| 0.606 | TUBB | XRP-44X | IVX-214 | PLK1 | 28 | 30 |
| 0.602 | TUBB | 4-Demethylepipodophyllotoxin | Picropodophyllin | IGF1R | 40 | 38 |
| 0.602 | MET | Tivantinib | 4-Demethylepipodophyllotoxin | TUBB | 28 | 40 |
| 0.597 | TUBB | XRP-44X | Picropodophyllin | IGF1R | 28 | 38 |
| 0.594 | MAPK8 | SR-3306 | Pracinostat | HDAC1 | 46 | 34 |
| 0.583 | PLK1 | IVX-214 | Noscapine | TUBB | 30 | 36 |
| 0.571 | CDK1 | BS-194 | VER-82576 | HSP90AB1 | 46 | 33 |
| 0.563 | TUBB | Lexibulin hydrochloride | ON-01910 | PLK1 | 27 | 37 |
| 0.557 | HSP90AB1 | SNX-5422 | Methylrosaniline chloride | TNFRSF1A | 35 | 36 |
| 0.556 | TUBB | XRP-44X | AST-1306 | ERBB2 | 28 | 28 |
| 0.540 | HSP90AB1 | Geldanamycin | Idarubicin hydrochloride | TOP2A | 34 | 41 |
| 0.540 | HSP90AB1 | CNF-2024 | Idarubicin hydrochloride | TOP2A | 35 | 41 |
| 0.536 | TUBB | XRP-44X | Tivantinib | MET | 28 | 28 |
| 0.535 | MCL1 | VU0482089-2 | AV-412 | EGFR | 29 | 33 |
| 0.534 | HSP90AB1 | CNF-2024 | Doxorubicin | TOP2A | 35 | 45 |
| 0.531 | MCL1 | VU0482089-2 | 1-alpha-Hydroxyergocalciferol | VDR | 29 | 28 |
| 0.524 | PIM3 | GDC-0349 | GNE-493 | PIK3CA | 34 | 40 |
| 0.524 | IGF1R | Picropodophyllin | Noscapine | TUBB | 38 | 36 |
| 0.521 | CDK1 | R-547 | PIK-75 | PIK3CA | 29 | 33 |
| 0.516 | HDAC1 | Abexinostat | 3-Methyladenine | PIK3CA | 43 | 47 |
| 0.515 | MAPK8 | TCS JNK 5a | Noscapine | TUBB | 33 | 36 |
| 0.508 | SRC | KX-01 | Indibulin | TUBB | 38 | 28 |
| 0.504 | TUBB | Lexibulin hydrochloride | Merck-22-6 | AKT1 | 27 | 25 |
The top 43 combinations of tool compounds were subsequently evaluated for cooperative reduction of MYC protein expression in L363 MM cells via western blot (WB) assay when treated at their IC50 doses for 24 hours. Ten of the 43 combinations cooperatively reduced expression of MYC (Table 2), and six of the ten combinations also increased p16 expression (Table 2, Fig. S1B-G). Representative MYC and p16 western blots are shown for one of the four combinations which reduced MYC protein but did not increase p16 expression compared to single agent in Fig. S1H.
Table 2.
Combinations Reducing MYC (n = 10) and Increasing p16 (n = 6) Protein Expression in L363 MM Cells
| Reduced MYC Protein Expression vs. Single Agent/Control in L363 MM Cells | |||||
|---|---|---|---|---|---|
| Drug 1 | Primary Target 1 | Drug 2 | Primary Target 2 | r2 (Robust Regr.) |
†p16 vs. Ctrl |
| BS-194 | CDK1 | Dacinostat | HDAC1 | 0.634 | ✓ |
| Alisertib | AURKA | Doxorubicin | TOP2A | 0.655 | ✓ |
| Geldanamycin | HSP90 | Idarubicin HCl | TOP2A | 0.540 | ✓ |
| Noscapine | TUBB | IVX-214 | PLK1 | 0.583 | ✓ |
| XRP-44X | TUBB | IVX-214 | PLK1 | 0.606 | ✓ |
| BS-194 | CDK1 | VER-82576 | HSP90 | 0.571 | ✓ |
| AT13387AU | HSP90 | ITK(3) | ITK | 0.634 | x |
| SNX-5422 | HSP90 | ITK(3) | ITK | 0.702 | x |
| VER-82576 | HSP90 | ITK(1) | ITK | 0.836 | x |
| VU-482089 | MCL1 | AV-412 | EGFR | 0.535 | x |
Compounds of the same drug classes were selected for further testing of the six combinations including drugs already approved to treat MM or other hematologic malignancies and/or drugs currently in clinical trials to treat MM, as well as novel combinations not yet investigated in treating MM. Out of the six combinations that simultaneously reduced MYC and increased p16 protein expression, three combinations were found to be synergistic in their ability to inhibit myeloma growth (Fig.2A-F, Fig.S2E-G). Four additional drug combinations were either additive (TUBBi + PLK1i; CDK1i + HSP90i) or antagonistic (TUBBi + PLK1i; MCL1i + EGFRi) in their effect on myeloma cells in vitro, Fig.S2A-D). Notably, the MCL1i + EGFRi combination that decreased MYC, but not p16, expression (Fig.S1H) was the most antagonistic (Fig.S2D). The three synergistic combinations were the cyclin dependent kinase inhibitor (CDKi) dinaciclib + the histone deacetylase inhibitor (HDACi) entinostat and a topoisomerase II inhibitor (TOP2Ai) doxorubicin combined with either the aurora kinase A inhibitor (AURKAi) alisertib or heat shock protein 90 inhibitor (HSP90i) SNX-2112. Increased phosphorylation of MYC at serine 62 contributes to MYC stabilization, while increased phosphorylation of MYC at threonine 58 is known to destabilize MYC.8,16 Treatment with these three combinations did not increase MYC stability in L363 cells, nor did the combinations destabilize MYC, as evidenced by WB of phosphorylated MYC at serine 62 and threonine 58, respectively (Fig.S1I).
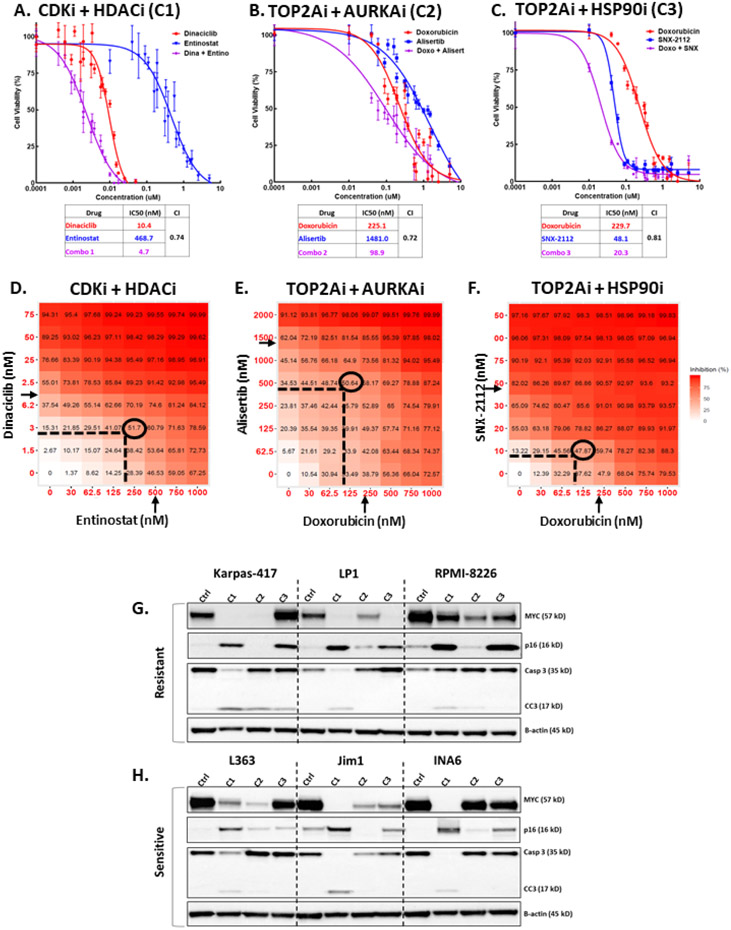
Figure 2.
High-throughput drug screen reveals combinations that synergistically reduce viability of MM cells and cooperatively target MYC and p16. A-C) Dose-response curves for top 3 drug combinations in L363 MM cells. Cell viability was assessed with MTS assay 48h after treatment with escalated dose concentrations of either drug individually or in combination at a 1:1 molar ratio. Each data point represents mean of 4 wells and error bars indicate replicate standard deviation. Each table lists IC50 (in nM) values for individual drugs and combinations. Chou-Talalay computation of combination indices (CI) for treated cells are shown for 50% affected fraction 48 hours post-exposure. Synergy is interpreted as CI<1.0. D-F) Graphical depiction of dose-matrix analyses for the top drug combinations in L363 MM cells. Percent inhibition of cell growth is shown for each different combination of doses and colorized in red. Cells were treated for 48 hours with different concentrations of each drug (indicated by X- and Y-axes) singly or in combination. Arrows indicate individual drug IC50 (half-maximal inhibitory concentration) as determined by MTS dose-response assay, ovals surround optimal dose for combinations, as determined by synergy scoring. G-H) Representative WB analysis of MM cell lines resistant or sensitive to all 1920 drugs used in the screen (see Table 3). Three resistant and three sensitive cell lines were treated for 24 hours with the top 3 drug combinations at the IC50 concentration (the concentration at half-maximal activity; equal to IC50) for each line (C1 = dinaciclib (CDKi) + entinostat (HDACi), C2 = Doxorubicin (TOP2Ai) + Alisertib (AURKAi), C3 = Doxorubicin + SNX-2112 (HSP90i). Lysates of treated cells were probed for MYC, p16, total and cleaved caspase 3 (Casp3 and CC3, respectively), and β-actin.
3.2. Evaluation of synergy of top drug combinations
Dose-response curves were generated for the top 3 drug combinations in L363 MM cells, treated for 48 hours with each drug singly or in combination at a 1:1 molar ratio (Fig.2A-C). All three combinations synergistically reduced the viability of L363 MM cells, as determined by Chou-Talalay combination indices (CI) less than 1.28 An 8x8 dose matrix combination response screen of each of the top three combinations at 7 different concentrations, and all iterations thereof, was performed in L363 MM cells to ascertain the activity and synergy across a spectrum of doses. Heatmaps (Fig. 2D-F) indicate the percent inhibition of treated cells vs. vehicle control after 48 hours. For each of the top combinations, the summary synergy scores are indicative of synergistic interaction between the drugs (average HSA score >10) and synergy was also achieved at a lower concentration than the half maximal inhibitory concentration (IC50) of each individual agent, indicating lower concentrations of each drug may be used in combination to generate a pharmacologically achievable reduction in MM viability. Surface plots of the excess inhibition of highest single agent (HSA) are shown in Fig.S2E-G, with the CDKi/HDACi combination achieving the highest average HSA score of 15.
3.3. The top drug combinations reduce MYC expression and increase p16 expression in MM cell lines, regardless of inherent resistance or sensitivity
The 47 cell lines were ranked for their overall sensitivity/resistance based on the mean IC50 for all 1920 compounds in the initial high-throughput screen (Table 3). The ability of the top drug combinations (CDKi/HDACi, TOP2Ai/AURKAi, and TOP2Ai/HSP90i) to reduce MYC protein while increasing p16 was evaluated in cell lines inherently more resistant (Karpas417, RPMI8226, LP1), as well as cell lines inherently more sensitive (INA6, JIM1, L363), to the compounds from the high-throughput drug screen (Fig.2G-H). Of the three top combinations, CDKi/HDACi was the most effective overall at reducing MYC protein, increasing p16, and inducing apoptosis (as indicated by cleavage of caspase 3) in all three inherently more sensitive cell lines and in two (Karpas-417, LP1) of the three resistant cell lines.
Table 3.
Multiple Myeloma Cell Lines (n = 47) Ranked by Mean IC50
| Cell Line | Mean IC50 for Drugs in High-Throughput Screen |
|
|---|---|---|
| KMS21BM_JCRB | 10.123 |
|
| Karpas417_ECACC | 4.309 | |
| MMM1_PLB | 3.449 | |
| RPMI8226_ATCC | 2.866 | |
| OCIMY1_PLB | 2.675 | |
| JJN3_DSMZ | 2.583 | |
| KMS26_JCRB | 2.566 | |
| OPM2_DSMZ | 2.526 | |
| EJM_DSMZ | 2.363 | |
| SKMM1_PLB | 2.357 | |
| U266_ATCC | 2.218 | |
| LP1_DSMZ | 1.932 | |
| MOLP8_DSMZ | 1.766 | |
| KMS28PE_JCRB | 1.690 | |
| MM1R_ATCC | 1.683 | |
| NCIH929_DSMZ | 1.659 | |
| MM1S_ATCC | 1.481 | |
| KMS34_JCRB | 1.365 | |
| FR4_PLB | 1.252 | |
| Karpas620_DSMZ | 1.140 | |
| H1112_PLB | 1.094 | |
| ARP1_JJKsccF8 | 1.074 | |
| OCIMY7_PLB | 1.024 | |
| PE2_PLB | 1.018 | |
| KMS12BM_JCRB | 0.949 | |
| PCM6_Riken | 0.877 | |
| KMS12PE_JCRB | 0.835 | |
| KHM11_PLB | 0.825 | |
| ARD_JJKsccE7 | 0.758 | |
| JIM3_ECACC | 0.736 | |
| KMM1_JCRB | 0.687 | |
| KMS28BM_JCRB | 0.680 | |
| AMO1_DSMZ | 0.639 | |
| XG6_PLB | 0.605 | |
| OCIMY5_PLB | 0.598 | |
| OPM1_PLB | 0.573 | |
| KMS20_JCRB | 0.547 | |
| Delta47_JCRB | 0.537 | |
| KMS27_JCRB | 0.522 | |
| UTMC2_PLB | 0.503 | |
| L363_DSMZ | 0.482 | |
| Karpas25_ECACC | 0.464 | |
| JIM1_ECACC | 0.447 | |
| INA6_PLB | 0.391 | |
| XG1_PLB | 0.332 | |
| VP6_DJ | 0.318 | |
| KMS11_JCRBsus | 0.294 |
3.4. Evaluation of microenvironment effects of drug combinations and in vitro tolerability
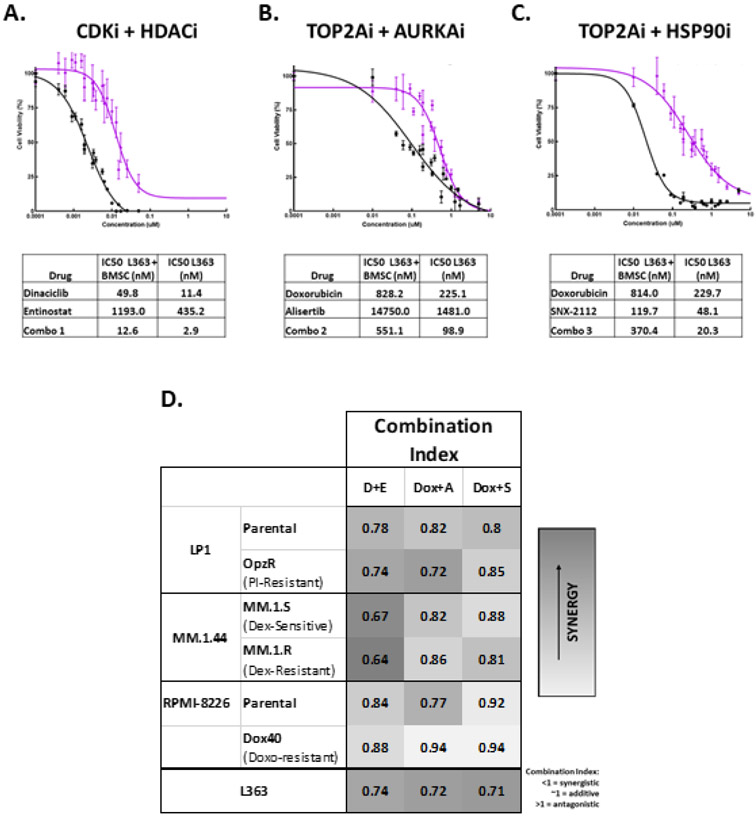
To test if the drug combinations retain activity in a co-culture system simulating the protective interaction of MM cells within the tumor microenvironment, L363 MM cells were treated with increasing doses of each drug treated singly or in combination at a 1:1 molar ratio for 48 hours with a feeder layer of immortalized human HS-5 bone marrow stromal cells (BMSC) – (Fig.3A-C). For the CDKi/HDACi and TOP2Ai/AURKAi combinations, although co-culture with BMSCs provided some protection of tumor cells when treated with single agents, the protection effect was partially overcome by combination treatment (Fig.3A, B). The TOP2Ai/HSP90i combination did not reduce MM viability more than single agent HSP90i (SNX-2112) treatment alone (Fig.3C). In dose response cell viability assays of H1634 non-neoplastic human fibroblasts and L363 MM cells treated at escalating doses of each of the combinations for 48 hours, the IC50 in H1634 cells is either much higher than in L363 MM cells or undetermined as an IC50 couldn’t be established (Fig.S2H-J). For example, the IC50 for the CDKi/HDACi combination of dinaciclib and entinostat is 4.9 nM in L363 cells, while the IC50 for the same combination is 335.0 nM in H1634 fibroblasts. The marked increase in combination IC50 in non-neoplastic cells is suggestive of a favorable therapeutic window for combination treatment.
Figure 3.
Evaluation of synergy in top drug combinations. A-C) Dose-response curves, along with tables listing IC50 in nM, for L363 MM cells in monoculture and L363 MM cells cocultured with HS-5 bone marrow derived stromal cells (BMSC) treated for 48 hours with increasing concentrations of the top 3 drug combinations (A = CDKi/HDACi, B = TOP2Ai/AURKAi, C = TOP2Ai/HSP90i). Black curves represent percent viability following combination treatment, relative to DMSO-treated control, in L363 cells cultured alone, purple curves show combination dose responses for L363 cocultured with human HS-5 BMSC. D) Heat-map depicting Chou-Talalay combination index scores for the following parental MM cell lines and their drug-resistant counterparts: LP1-Parental and LP1-OpzR (resistance induced via prolonged incubation with the proteasome inhibitor (PI) oprozomib (Opz)), Dexamethasone (Dex)-sensitive MM.1.S and Dex-resistant MM.1.R cells, RPMI-8226-Parental and RPMI-8226-Dox40 (treatment-induced resistance to the topoisomerase inhibitor doxorubicin), all compared to CI scores of L363. D+E = dinaciclib (CDKi) + entinostat (HDACi), Dox + A = doxorubicin (TOP2Ai) + alisertib (AURKAi), Dox + S = doxorubicin + SNX-2112 (HSHP90i). Darker gray = more synergy.
3.5. Drug combinations are synergistic in MM cells with induced resistance to common therapies
The ability of the top combinations to synergistically reduce the viability of MM cells with acquired resistance was assessed in three drug resistant cell line model pairs (Fig.3D). In these models, LP1, MM.1.44 (commonly referred to as MM.1), or RPMI-8226 MM cells developed resistance to common treatments for MM via prolonged incubation with either the PI oprozomib, the corticosteroid dexamethasone, or the TOP2Ai doxorubicin, respectively, for up to 24 weeks.29,30 Resistance to respective treatments was confirmed with cell viability dose response curves for sensitive vs. resistant MM cell lines treated with increased doses of each agent (LP1-Parental and LP1-OpzR PI resistant with oprozomib; MM.1.S and MM.1R corticosteroid resistant with dexamethasone; RPMI-8226-Parental and RPMI-8226-Dox40 TOP2Ai resistant with doxorubicin) – (Fig.S2K-M). IC50 concentrations for each combination in each parental and resistant cell line are shown in Table S2. As shown in the heat-map depicting CI scores for the parental MM cell lines and their drug-resistant counterparts, all three combinations were synergistic in all of the cell lines (CI<1) regardless of sensitivity or induced resistance to indicated agents (Fig.3D). The CDKi/HDACi combination of dinaciclib and entinostat had greatest synergy in LP1-Parental and PI-resistant cells, as well as in dexamethasone-sensitive MM.1.S and dexamethasone-resistant MM.1.R cells. The TOP2Ai-resistant RPMI-8226-Dox40 cell line was still particularly resistant to combination treatments compared to the RPMI-8226-Parental MM cells.
3.6. Comparison of top drug combinations in a novel allograft mouse model (VQ) of MM
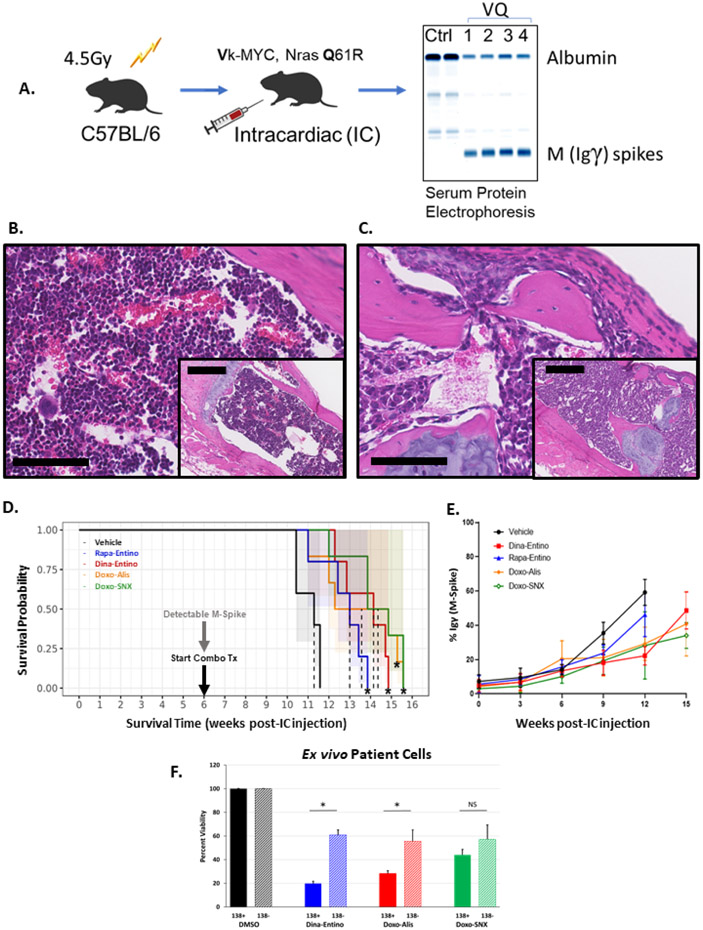
Assessments of the in vivo activity of the drug combinations was made in a 16-week efficacy study in C57BL/6J mice sublethally irradiated then injected IC with murine Vk*MYC; Nras LSL Q61R/+; IgG1-Cre (VQ) tumor cells harvested from bone marrow of donor mice (Fig.4A). 26,27 After 6-8 weeks, serum M-spikes, indicating an increase in gamma immunoglobulin on serum protein electrophoresis, were evident in VQ mice (Fig.4A; Fig.S3A). VQ cells completely obliterated normal hematopoietic cells within sternal bone marrow (Fig.4B-C) and even induced bony lysis of the cortex (Fig.S3B, bony lesion), similar to what is observed in the bone and marrow of humans affected by MM. Treatment with the top drug combinations, and mTORi/HDACi (rapamycin and entinostat), commenced when M-spikes were first detected in mice (Fig.4D). Survival of mice injected with VQ cells and treated with the combinations was significantly prolonged compared to control mice and was also slightly longer compared to the combination of rapamycin (mTORi) and entinostat (HDACi).8,9 Further, the mean M-spike percentage in treated mice remained lower than that of control mice until the final weeks of treatment (Fig.4E). Additionally, CDK, TOP2A, and HSP90 inhibitors, along with the HDAC inhibitor quisinostat, effectively reduced VQ cell line (4935) viability at 100 nM and 1000 nM, while AURKA and other HDAC inhibitors minimally or moderately decreased viability at one or both concentrations (Table S3). Although all combinations reduced Ki67 positive nuclei in a subset of splenic metastases (Fig.S3D-H), the reduction was only significant in the CDKi/HDACi group. In agreement with the pSMAD3 data seen in L363 cells, the only combination leading to increases in pSMAD3 signaling in VQ splenic metastases was that of doxorubicin and alisertib (Fig.S4B-F). None of the evaluated drug combinations significantly reduced the body weights in treated mice compared to those treated with vehicle (Fig.S3C), and there was no microscopic evidence of combination-related toxicity in evaluated tissues.
Figure 4.
Evaluating the top drug combinations in a novel allograft mouse model of MM and human myeloma cells ex vivo. A) Illustration of VQ inoculation scheme. 6–8-week-old C57BL/6J mice were sublethally irradiated then injected intracardiac with 5x10^6 Vk*MYC; NrasLSL Q61R/+; IgG1-Cre (VQ) cells harvested from bone marrow of donor mice. After 6-8 weeks, serum M-spikes, as evidenced by the γ immunoglobulin band on serum protein electrophoresis, are evident. Once M-spikes were detected in mice, treatment with the top drug combinations commenced. B-C) Photomicrographs (bar =100 μm; inset bar = 250 μm; H&E stain) of C57BL/6J mouse sternum 12 weeks post-IC injection with either saline (B, normal bone marrow) or VQ cells (C, marrow replaced by neoplastic plasma cells). D) Survival plots of C57BL/6J mice injected with 5x10^6 VQ cell IC, treated with the top drug combinations vs a previously investigated combination of rapamycin (mTORi) and entinostat (HDACi), all compared to DMSO-treated control mice. CDKi (dinaciclib, 20 mg/kg, IP, 3x/week + HDACi (entinostat, 20 mg/kg, PO 5x/week; TOP2Ai, (doxorubicin,4 mg/kg IV 1x/week) + AURKAi (alisertib, 30 mg/kg PO 5x/week); or HSP90i (SNX-2112, 20 mg/kg, PO, 3x/week), n= 5. * indicates significantly prolonged survival vs. DMSO-treated control mice (p < 0.01, Log-Rank test). E) Graphical representation of mean M-spike percentage for each treatment group of mice administered one of the top drug combinations, combined mTORi-HDACi, or DMSO control. Each data point represents mean M-spike percentage of all mice for a given time point, error bars indicate standard deviation amongst group mice. F) Viability of human CD138 positive (MM) and CD138 negative cells extracted from bone marrow of smoldering multiple myeloma patients (n=3). Cells were selected for CD138 status using magnetic-activated cell sorting (MACS). CD138 positive and negative cells were treated with the top 3 combinations of dinaciclib (10 nM) and entinostat (500 nM), doxorubicin (225 nM) and alisertib (2 μM), or doxorubicin (225 nM) and SNX-2112 (50 nM) for 48 hours. Solid bars indicate average viability for each combination in CD138 positive cells, hash-marked bars indicate the average viability for CD138 negative cells. Error bars = standard deviation. * =p <0.001; NS = no significance p > 0.05 by unpaired two-tailed Student’s t test.
3.7. Drug combinations are effective at selectively reducing the viability of human CD138+ MM cells ex vivo
The capability of the combinations to selectively reduce the viability of MM cells was evaluated in bone marrow biopsy samples obtained from patients with confirmed smoldering multiple myeloma (SMM). Cells expressing the CD138 surface maker (plasma cells) or cells negative for CD138 (non-plasma cells) from human SMM biopsies were treated with the combinations of dinaciclib (10 nM) and entinostat (500 nM), doxorubicin (225 nM) and alisertib (2 uM), or doxorubicin (225 nM) and SNX-2112 (50 nM) for 48 hours; viability was compared to that of control CD138 positive or CD138 negative cells treated with DMSO) (n = 3) (Fig.4F). Overall, all three combinations effectively reduced the viability of CD138 positive MM cells compared to DMSO alone. However, the CDKi/HDACi and TOP2Ai/AURKAi combinations were more selective in reducing MM viability compared to the TOP2Ai/HSP90i combination.
3.8. Genetic pathways commonly affected by the top combinations
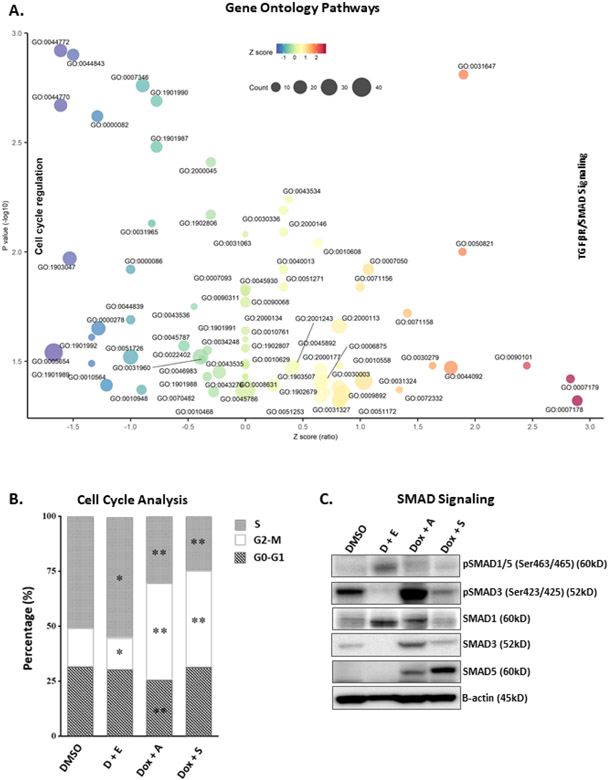
To determine if common genetic pathways are similarly affected by the top drug combinations, L363 MM cells were treated for 48 hours with single agent IC50 concentrations of the top combinations (CDKi/HDACi, TOP2Ai/AURKAi, and TOP2Ai/HSP90i). Two separate CDKi/HDACi combinations (dinaciclib/entinostat and dinaciclib/mocetinostat) were included for the most promising combination; 48-hour dose response results for dinaciclib and mocetinostat in L363 was similar to dinaciclib/entinostat (Fig.S4). RNAs from each treatment were analyzed via the Nanostring nCounter® digital gene expression codeset system, and counts were normalized to mRNA of housekeeping genes. Of the 143 genes whose expressions were significantly changed by the top drug combinations versus control (Fig.S5A, Table S4), a total of 125 genes were significantly upregulated or downregulated simultaneously by all combination treatments (Fig.S5B). Next, 78 of the concordantly upregulated or downregulated genes were found to induce a two-fold change in gene expression across all the combination treatments (38 up-regulated and 40 down-regulated genes). DAVID pathway analysis determined the overrepresented GO functions in the concordant response signature of the 78 genes (Fig.5A), and enriched GO terms were selected with nominal p-values<0.05. For each over-represented GO term a z-score was computed based on the number of up- and down-regulated genes according to the formula (up minus down)/sqrt(total) proposed by Walter et al. and visualized with bubble-plots.31 The pathways most downregulated by all combinations (GO:0005654, GO:0044770, GO:0044772) were those implicated in promoting cell cycle transition (Fig.5A, Table S5), indicating a common beneficial effect in preventing MM cell growth. Some of the genes most downregulated by the drug combinations whose lower expression is linked to better prognosis include CCND2, HISTH1H3H, EIF4EBP1, NRAS, TCF3 E2F1, and CDK4 (Fig.S6). Flow cytometric analysis was utilized to investigate the effects of the top combinations on cell cycle transition (Fig.5B). TOP2Ai/AURKAi, and TOP2Ai/HSP90i combinations induced G2-M arrest, whereas the CDKi/HDACi combination increased the percentage of cells in S-phase and decreased the percentage of cells in G2-M phase (Fig.5B).
Figure 5.
Pathways most commonly affected by the top 3 combinations. A) Bubble plot visualization of top pathways commonly affected by the top 3 combinations (CDKi/HDACi, TOP2Ai/AURKAi, and TOP2Ai/HSP90i) using single agent IC50 concentrations of each drug. Purified RNA from each sample was analyzed using the NanoString nCounter® system. Raw counts were normalized to mRNA of five housekeeping genes (ZNF384, MRPS5, CNOT4, NUBP1, and SF3A3). Statistical significance was based on a false discovery rate of 5%. The concordant response signature includes genes significantly changed in the same direction by each combination treatment (Total=125, 49 up-regulated and 76 down-regulated genes), and that reached 2-fold change in the average fold-change profile calculated across all the combination treatments (Total=78, 38 up-regulated and 40 down-regulated genes). Fisher’s exact test was used to determine overrepresented Gene Ontology (GO) functions in the concordant response signature of 78 genes. Enriched GO terms were selected with nominal p-values less than 0.05. For each overrepresented GO term a z-score was computed based on the number of up-regulated and down-regulated genes according to the formula (up-down)/sqrt(total) proposed by Walter et al.31 The pathways most downregulated by all combinations include those involved in cell cycle regulation. All top combinations also commonly increased TGFβ and SMAD3 signaling pathways. B) Cell cycle distribution of PI/RNase-stained L363 cells treated for 24 hours with DMSO or IC50 concentrations of the top three combinations. D + E = dinaciclib + entinostat; Dox + A = doxorubicin + alisertib; Dox + S = doxorubicin + SNX-2112. * = p<0.05, ** = p<0.001 (unpaired Student’s t test). C) Representative WB analysis of SMAD signaling effector molecules in L363 MM cells treated for 48 hours with the top 3 drug combinations at the IC50 concentrations. Lysates of treated cells were probed for pSMAD1/5 (Ser463/465), pSMAD3 (Ser423/425), SMAD1, SMAD3, SMAD5, and β-actin.
The pathways most upregulated by all combinations (GO:0007178 and GO:0007179) were involved in TGFβ/SMAD signaling. WB analysis of SMAD signaling effector molecules in L363 cells showed an increase in pSMAD1/5 signaling with CDKi/HDACi treatment, driven primarily by SMAD1; whereas pSMAD3 signaling was increased with the TOP2Ai/AURKAi treatment and total SMAD5 was increased with TOP2Ai/HSP90i (Fig.5C).
3.9. Co-targeting the TGFβ pathway along with the CDKi/HDACi drug combination
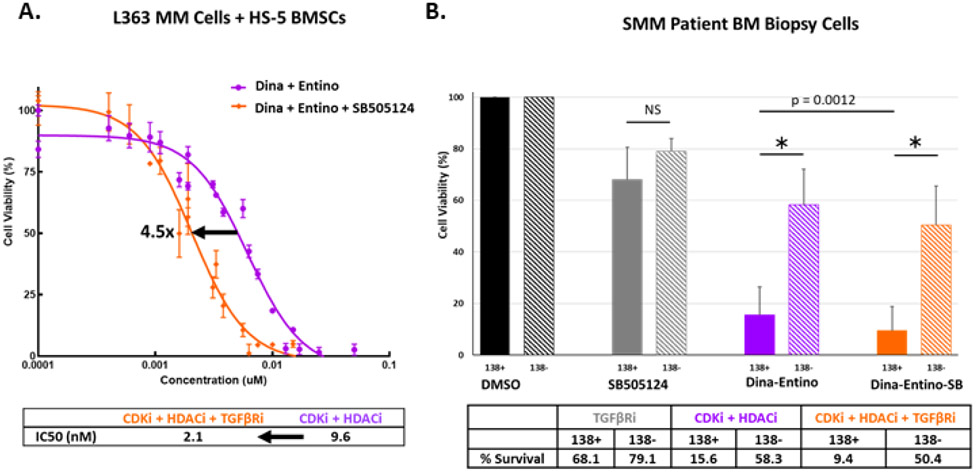
Since, upregulating the transforming growth factor β (TGFβ) pathway was a common, potentially deleterious, effect of the three combinations (Fig.5), we tested the addition of a drug targeting the TGFβ pathway to determine if it would be effective in further reducing the viability of L363 MM cells co-cultured with HS-5 BMSCs (Fig.6A). Indeed, the addition of the TGFβ-receptor inhibitor SB505124 was effective at cooperatively reducing L363 MM cell viability in co-culture with HS5 cells. The IC50 value for triple therapy (CDKi /HDACi + TGFβi) was 4.5 times lower than that of the CDKi/HDACi combination alone. Additionally, in 48-hour viability assays of human SMM patient bone marrow biopsy cells, the addition of SB505124 to the CDKi/HDACi combination effectively reduced the viability of CD138 positive cells by almost 40%, while relatively sparing CD138 negative cells (n=3) (Fig.6B).
Figure 6.
Co-targeting the TGFβ pathway with combination therapy. A) Dose-response curves for L363 MM cells, co-cultured with HS-5 BMSCs for 48 hours with escalated doses of either combined CDKi/HDACi of dinaciclib and entinostat (purple curve) or combined CDKi/HDACi + TGFβRi (orange curve; dinaciclib + entinostat + SB505124) at a 1:1 or 1:1:1 molar ratio (in nM), respectively. Arrow = IC50 shift. B) Viability of human CD138 positive (MM) and CD138 negative cells extracted from bone marrow of SMM patients (n=3). Cells were selected for CD138 status using MACS. CD138 positive and negative cells were treated with the SB505124 (TGFβRi – 5 uM), dinaciclib (CDKi – 10 nM) and entinostat (HDACi – 500 nM), or CDKi + HDACi + TGFβRi for 48 hours. Solid bars indicate the average viability of CD138 positive cells and hash-marked bars represent the average viability for CD138 negative cells. Error bars = standard deviation. * = p <0.001; NS = no significance p > 0.05 by unpaired two-tailed Student’s t test. p = 0.0012 indicates p-value for significance in comparison of CD138 positive cells treated with CDKi + HDACi versus CDKi + HDACi + TGFβRi.
4. Discussion
To find cooperative drug combinations for MM patients, we used a multilayered drug combination prediction workflow based on a high-throughput drug screen. Single agents effective in most of the tested MM cell lines were identified, followed by in silico Huber robust regression analysis of all active drugs against each other to select combinations of drugs that reduced MYC protein expression while increasing p16 expression. This strategy overcomes the limitations of other regression models, such as ordinary least squares, in that it is not overly affected by extreme outliers.32 Additionally, Huber robust regression analysis enabled the prediction of potential combinations, regardless of drug target, to treat a wide variety of MM subtypes without the cumbersome and time-consuming step of individually testing each of the approximately 5 million potential drug combinations/cell lines in vitro.32-34 This process identified 43 potentially cooperative combinations based on individual drug activity across most of the MM cell lines tested. Many of the target classes associated with the 43 drug combinations (Table 1) are proposed targets for chemotherapeutics used in MM and/or other cancers33, providing additional confidence in our combination discovery approach.
The efficacy of the top synergistic drug combinations (CDKi/HDACi, TOP2Ai/AURKAi, and TOP2Ai/HSP90i) was investigated in cell lines with inherent sensitivity or resistance to all drugs in the high-throughput screen. These combinations were generally non-toxic in non-neoplastic human fibroblasts suggesting favorable safety margins for their use in patients. Together, these data provide a relatively agnostic approach to identify drug combinations for further preclinical development for myeloma therapy, especially in cases resistant to first-line treatments.
The combination of a CDK inhibitor with an HDAC inhibitor was effective overall in inhibiting myeloma cells, and in simultaneously reducing MYC protein expression while increasing p16 expression and inducing apoptosis in a variety of MM cell lines with inherent sensitivity or resistance to most drugs in the high-throughput screen. This was the most synergistic combination overall in MM cell lines with induced resistance to common first-line chemotherapeutics.
Dinaciclib (Merck & Co.) inhibits CDK1,2,5,9.34-36 Entinostat (Syndax), a class I HDAC inhibitor, inhibits HDAC1 and 3.37-40 Both drugs are in clinical trials for cancer. Dinaciclib has a favorable safety profile in combination with the proteasome inhibitor bortezomib and the corticosteroid dexamethasone for treating patients with relapsed MM in a phase I clinical trial (NCT01711528). Additionally, dinaciclib is being investigated in phase I-III clinical trials in combination with other agents aimed at treating hematologic malignancies (NCT0348520, NCT01650727, NCT01580228), solid tumors (NCT01434316), metastatic triple negative breast cancer (NCT01624441), and pancreatic cancer (NCT01783171). Entinostat is in a phase III clinical trial in combination with hormone therapy in treating patients with recurrent hormone receptor-positive breast cancer (NCT02115282) and has been investigated as a combined agent with immune checkpoint inhibitors (PD-1/PD-L1 antagonists in particular) in clinical trials treating patients with various solid tumors (NCT02437136).39-42 Further understanding of how this combination affects the proposed targets and determining a cooperative response signature for the combination will provide insight into on/off-target effects, mechanism(s) of action, and biomarkers of a combined response, and may facilitate circumvention of certain side-effects of treating MM by lowering doses of the single agents.
Therapeutic efficacy of the top combinations was assessed in a transplantable mouse model (Vk*MYC; NrasLSL Q61R/+; IgG1-Cre) of highly malignant MM that more closely recapitulates human high-risk/refractory MM in an immunocompetent animal.26,27 This model developed M-spikes, similar to those seen in human patients, at approximately 6 weeks and served as a means of tracking progression of disease and response to treatment.43,44 Additionally, the histologic appearance of myeloma within the bone marrow of mice in this model closely resembles that of myeloma in human patients, with neoplastic plasma cells disrupting marrow architecture and lysing bone.45 In the present study, all 3 combinations extended survival and were superior to treatment with the mTORi/HDACi combination.8,9
The combinations were assessed in freshly isolated myeloma cells from human patients to ensure that their efficacy was not limited to cultured cell lines and engineered mouse cells. When human SMM patient bone marrow biopsy samples were separated into plasma cells (CD138+) and nonplasma cells (CD138−) the top combinations effectively reduced the viability of human CD138+ myeloma cells. The CDKi/HDACi and TOP2Ai/AURKAi combinations were more selective than the TOP2A/HSP90i combination when comparing CD138+ MM cell viability to that of non-MM CD138-negative cells. The difference in combination-induced toxicity between neoplastic and non-neoplastic bone marrow cells may suggest that these treatments would impart less off-target bone marrow depletion. Treating ex vivo patient samples and co-cultures of myeloma cells and HS-5 stromal cells with a TGFβR1 inhibitor (SB505124) suggests that combining this agent with dinaciclib and entinostat could help to prevent potentially deleterious effects of TGFβ receptor signaling in the tumor microenvironment.46 Future directions include investigating the mechanisms of drug synergy for the top combination of CDKi/HDACi, as well as investigating efficacy of additional TGFβR or SMAD inhibition in vivo.
Supplementary Material
Highlights.
High-throughput screen identifies drug combinations for multiple myeloma (MM)
Top combinations cooperatively reduce MYC and increase p16 protein expression
Combinations prolong survival in a Ras-driven allograft model of advanced MM
Combinations reduce viability off ex vivo treated smoldering MM patient cells
Commonly affected genes decrease cell cycle transition and increase SMAD signaling
Acknowledgements
The authors acknowledge Robert Hawley, Ke Zhang and Doug Lowy for their helpful insights. TJP performed doctoral research as a fellow in the NIH Comparative Biomedical Scientist Training Program supported by the NCI in Partnership with Purdue University.
Financial support:
NIH Intramural Research Programs of the National Cancer Institute and the National Center for Advancing Translational Sciences, MMRF Trainee fellowship grant to JKS, a MMRF Cell Line Characterization Award to JKS and R01CA152108, Trillium Fund, UWCCC Developmental Therapeutics Program Pilot Award, and Immunotherapy Pilot Award to JZ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests: The authors declare no competing financial interests.
Conflicts of Interest
The authors report no conflicts of interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon request.
References
- 1.Goldschmidt H, Ashcroft J, Szabo Z, Garderet L. Navigating the treatment landscape in multiple myeloma: which combinations to use and when? Ann Hematol. 2018. Nov;23(1)1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar SK, Rajkumar S. The multiple myelomas - current concepts in cytogenetic classification and therapy. Nat Rev Clin Oncol. 2018. Jul;15(7):409–21. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023. Jan;73(1):17–48. [DOI] [PubMed] [Google Scholar]
- 4.Castella M, Fernández de Larrea C, Martín-Antonio B. Immunotherapy: a novel era of promising treatments for multiple myeloma. Int J Mol Sci. 2018. Nov;19(11):3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raje N, Berdeja J, Lin Y, Siegel D, Jagannath S, Madduri D, Liedtke M, Rosenblatt J, Maus MV, Turka A, Lam LP, Morgan RA, Friedman K, Massaro M, Wang J, Russotti G, Yang Z, Campbell T, Hege K, Petrocca F, Quigley MT, Munshi N, Kochenderfer JN. Anti-BCMA CAR T-Cell Therapy bb2121 in Relapsed or Refractory Multiple Myeloma. N Engl J Med. 2019. May 2;380(18):1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah N, Aiello J, Avigan DE, Berdeja JG, Borrello IM, Chari A, Cohen AD, Ganapathi K, Gray L, Green D, Krishnan A, Lin Y, Manasanch E, Munshi NC, Nooka AK, Rapoport AP, Smith EL, Vij R, Dhodapkar M. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of multiple myeloma. J Immunother Cancer. 2020. Jul;8(2):e000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dancey J, Chen H. Strategies for optimizing combinations of molecularly targeted anticancer agents. Nat Rev Drug Discov. 2006. Aug;5(8):649–59. [DOI] [PubMed] [Google Scholar]
- 8.Simmons JK, Michalowski AM, Gamache BJ, DuBois W, Patel J, Zhang K, Gary J, Zhang S, Gaikwad SM, Connors D, Watson N, Leon E, Chen JQ, Kuehl WM, Lee MP, Zingone A, Landgren O, Ordentlich P, Huang J, Mock BA. Cooperative targets of combined mTOR/HDAC inhibition promote MYC degradation. Mol Cancer Ther. 2017. Sep;16(9):2008–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simmons JK, Patel J, Michalowski AM, Zhang S, Wei BR, Sullivan P, Gamache B, Felsenstein K, Kuehl WM, Simpson RM, Zingone A, Landgren O, Mock BA. TORC1 and class I HDAC inhibitors synergize to suppress mature B cell neoplasms. Mol Oncol. 2014. Mar;8(2):261–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jovanović KK, Roche-Lestienne C, Ghobrial IM, Facon T, Quesnel B, Manier S. Targeting MYC in multiple myeloma. Leukemia. 2018. Jun;32(6):1295–306. [DOI] [PubMed] [Google Scholar]
- 11.Chesi M, Robbiani DF, Sebag M, Chng WJ, Affer M, Tiedemann R, Valdez R, Palmer SE, Haas SS, Stewart AK, Fonseca R, Kremer R, Cattoretti G, Bergsagal PL. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of postgerminal center malignancies. Cancer Cell. 2008. Feb;13:167–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chng WJ, Huang GF, Chung TH, Ng SB, Gonzalez-Paz N, Troska-Price T, Mulligan G, Chesi M, Bergsagel PL, Fonseca R. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia. 2011. Jun;25(6):1026–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misund K, Keane N, Stein CK, Asmann YW, Day G, Welsh S, Wier SV, Riggs D, Ahmann G, Chesi M, Viswanatha D, Kumar SK, Dispenzieri A, Gonzalez-Calle V, Kyle RA, O’Dwyer M, Rajikumar SV, Kortum KM, Keats J, MMRFCoMMpass Network, Fonseca R, Stewart AK, Kuehl WM, Braggio E, Bergsagel PL. MYC dysregulation in the progression of multiple myeloma. Leukemia. 2020. Jan 34(1):322–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bustoros M, Sklavenitis-Pistofidis R, Park J, Redd R, Zhitomirsky B, Dunford AJ, Salem K, Tai YT, Anand S, Mouhieddine TH, Chavda SJ, Boehner C, Elagina L, Neuse CJ, Cha J, Rahmat M, Taylor-Weiner A, Van Allen E, Kumar S, Kastritis E, Leshchiner I, Morgan EA, Laubach J, Casneuf T, Richardson P, Munshi NC, Anderson KC, Trippa L, Aguet F, Stewart C, Dimopoulos MA, Yong K, Bergsagel PL, Manier S, Getz G, Ghobrial IM. Genomic Profiling of Smoldering Multiple Myeloma Identifies Patients at a High Risk of Disease Progression. J Clin Oncol. 2020. Jul 20;38(21):2380–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y, Sun F, Cheng Y, Holmes B, Dhakal B, Gera JF, Janz S, Lichtenstein A. Critical Role for Cap-Independent c-MYC Translation in Progression of Multiple Myeloma. Mol Cancer Ther. 2022. Apr 1;21(4):502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000. Oct 1;14(19):2501–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mock B, Wax J, Clynes R, Marcu KB, Potter M. The genetics of susceptibility to RIM-induced plasmacytomagenesis. Curr Top Microbiol Immunol. 1988;141:125–7. [DOI] [PubMed] [Google Scholar]
- 18.Clynes R, Stanton LW, Wax J, Smith-Gill S, Potter M, Marcu KB. Synergy of an IgH promoter-enhancer-driven c-myc/v-Ha-ras retrovirus and pristane in the induction of murine plasmacytomas. Curr Top Microbiol Immunol. 1988;141:115–24. [DOI] [PubMed] [Google Scholar]
- 19.Whitfield JR, Beaulieu ME, Soucek L. Strategies to Inhibit Myc and Their Clinical Applicability. Front Cell Dev Biol. 2017. Feb 23;5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dang CV, Reddy EP, Shokat KM, Soucek L. Drugging the 'undruggable' cancer targets. Nat Rev Cancer. 2017. Aug;17(8):502–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011. Sep 16;146(6):904–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S, Ramsay ES, Mock BA. Cdkn2a, the cyclin-dependent kinase inhibitor encoding p16INK4a and p19ARF, is a candidate for the plasmacytoma susceptibility locus, Pctr1. Proc Natl Acad Sci U S A. 1998. Mar 3;95(5):2429–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang SL, DuBois W, Ramsay ES, Bliskovski V, Morse HC 3rd, Taddesse-Heath L, Vass WC, DePinho RA, Mock BA. Efficiency alleles of the Pctr1 modifier locus for plasmacytoma susceptibility. Mol Cell Biol. 2001. Jan;21(1):310–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Paz N, Chng WJ, McClure RF, Blood E, Oken MM, Van Ness B, James CD, Kurtin PJ, Henderson K, Ahmann GJ, Gertz M, Lacy M, Dispenzieri A, Greipp PR, Fonseca R. Tumor suppressor p16 methylation in multiple myeloma: biological and clinical implications. Blood. 2007. Feb 1;109(3):1228–32. [DOI] [PubMed] [Google Scholar]
- 25.Mathews Griner LA, Guha R, Shinn P, Young RM, Keller JM, Liu D, Goldlust IS, Yasgar A, McKnight C, Boxer MB, Duveau DY, Jiang JK, Michael S, Mierzwa T, Huang W, Walsh MJ, Mott BT, Patel P, Leister W, Maloney DJ, Leclair CA, Rai G, Jadhav A, Peyser BD, Austin CP, Martin SE, Simeonov A, Ferrer M, Staudt LM, Thomas CJ. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci U S A. 2014. Feb 11;111(6):2349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen Z, Rajagopalan A, Flietner ED, Yun G, Chesi M, Furumo Q, Burns RT, Papadas A, Ranheim EA, Pagenkopf AC, Morrow ZT, Finn R, Zhou Y, Li S, You X, Jensen J, Yu M, Cicala A, Menting J, Mitsiades CS, Callander NS, Bergsagel PL, Wang D, Asimakopoulos F, Zhang J. Expression of NrasQ61R and MYC transgene in germinal center B cells induces a highly malignant multiple myeloma in mice. Blood. 2021. Jan 7;137(1):61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flietner E, Wen Z, Rajagopalan A, Jung O, Watkins L, Weisner J, You X, Yun Z, Sun Y, Kingstad-Bakke B, Callander NS, Rapraeger A, Suresh M, Asimakopoulos F, Zhang J. Ponatinib sensitizes myeloma cells to MEK inhibition in the high-risk VQ model. Sci Rep. 2022;12(1):10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010. Jan 15;70(2):440–46. [DOI] [PubMed] [Google Scholar]
- 29.Greenstein S, Krett NL, Kurosawa Y, Ma C, Chauhan D, Hideshima T, Anderson KC, Rosen ST. Characterization of the MM.1 human multiple myeloma (MM) cell lines: a model system to elucidate the characteristics, behavior, and signaling of steroid-sensitive and -resistant MM cells. Exp Hematol. 2003. Apr;31(4):271–82. [DOI] [PubMed] [Google Scholar]
- 30.Riz I, Hawley RG. Increased expression of the tight junction protein TJP1/ZO-1 is associated with upregulation of TAZ-TEAD activity and an adult tissue stem cell signature in carfilzomib-resistant multiple myeloma cells and high-risk multiple myeloma patients. Oncoscience. 2017. Aug 1;4(7-8):79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015. Sep 1;31(17):2912–4. [DOI] [PubMed] [Google Scholar]
- 32.Sliwoski G, Kothiwale S, Meiler J, Lowe WE Jr. Computational methods in drug discovery. Pharmacol Rev. 2014. Jan;66(1):334–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinstein ZB, Bender A, Cokol M. Prediction of synergistic drug combinations. Curr Opin Syst Biol. 2017. May 11;4(1):24–28. [Google Scholar]
- 34.Gayvert KM, Aly O, Platt J, Bosenberg MW, Stern DF, Elemento O. A computational approach for identifying synergistic drug combinations. PLoS Comput Biol. 2017. Jan 13;13(1):e1005308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ovejero S, Moreaux J. Multi-omics tumor profiling technologies to develop precision medicine in multiple myeloma. Explor Target Antitumor Ther. 2021;2:65–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar SK, LaPlant B, Chng WJ, Zonder J, Callander N, Fonseca R, Fruth B, Roy V, Erlichman C, Stewart AK; Mayo Phase 2 Consortium. Dinaciclib, a novel CDK inhibitor, demonstrates encouraging single-agent activity in patients with relapsed multiple myeloma. Blood. 2015. Jan 15;125(3):443–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parry D, Guzi T, Shanahan F, Davis N, Prabhavalkar D, Wiswell D, Seghezzi W, Paruch K, Dwyer MP, Doll R, Nomeir A, Windsor W, Fischmann T, Wang Y, Oft M, Chen T, Kirschmeier P, Lees EM. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol Cancer Ther. 2010. Aug;9(8):2344–53. [DOI] [PubMed] [Google Scholar]
- 38.Kumar SK, Rajkumar V, Kyle RA, van Duin M, Sonneveld P, Mateos MV, Gay F, Anderson K. Multiple myeloma. Nat Rev Dis Primers. 2017. Jul 20;3:17046. [DOI] [PubMed] [Google Scholar]
- 39.De Souza C, Chatterji BP. HDAC inhibitors as novel anti-cancer therapeutics. Recent Pat Anticancer Drug Discov. 2015;10(2):145–62. [DOI] [PubMed] [Google Scholar]
- 40.Connolly RM, Rudek MA, Piekarz R. Entinostat: a promising treatment option for patients with advanced breast cancer. Future Oncol. 2017. Jun;13(13):1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connolly RM, Zhao F, Miller KD, Lee MJ, Pikarz RL, Smith KL, Brown-Glaberman UA, Winn JS, Faller BA, Adedayo AO, Burkard ME, Budd GT, Levine EG, Royce ME, Kaufman PA, Thomas A, Trepel JB, Wolff AC, Sparano JA. E2112: Randomized phase III trial of endocrine therapy plus entinostat or placebo in hormone receptor–positive advanced breast cancer. A trial of the ECOG-ACRIN cancer research group. J Clin Oncol. 2021. 39:28, 3171–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hellmann MD, Jänne PA, Opyrchal M, Hafez N, Raez LE, Gabrilovich DI, Wang F, Trepel JB, Lee MJ, Yuno A, Lee S, Brouwer S, Sankoh S, Wang L, Tamang D, Schmidt EV, Meyers ML, Ramalingam SS, Shum E, Ordentlich P. Entinostat plus pembrolizumab in patients with metastatic NSCLC previously treated with anti-PD-(L)1 therapy. Clin Cancer Res. 2021. Feb 15;27(4):1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajkumar SV, Kyle RA, Therneau TM, Melton LJ 3rd, Bradwell AR, Clark RJ, Larson DR, Plevak MF, Dispenzieri A, Katzmann JA. Serum free light chain ratio is an independent risk factor for progression in monoclonal gammopathy of undetermined significance. Blood. 2005. Aug 1;106(3):812–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dejoie T, Corre J, Caillon H, Hulin C, Perrot A, Caillot D, Boyle E, Chretien ML, Fontan J, Belhadj K, Brechignac S, Decaux O, Voillat L, Rodon P, Fitoussi O, Araujo C, Benboubker L, Fontan C, Tiab M, Godmer P, Luycx O, Allangba O, Pignon JM, Fuzibet JG, Legros L, Stoppa AM, Dib M, Pegourie B, Orsini-Piocelle F, Karlin L, Arnulf B, Roussel M, Garderet L, Mohty M, Meuleman N, Doyen C, Lenain P, Macro M, Leleu X, Facon T, Moreau P, Attal M, Avet-Loiseau H. Serum free light chains, not urine specimens, should be used to evaluate response in light-chain multiple myeloma. Blood. 2016. Dec 22;128(25):2941–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujino M. The histopathology of myeloma in the bone marrow. J Clin Exp Hematop. 2018;58(2):61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong M, Blobe GC. Role of transforming growth factor-beta in hematologic malignancies. Blood. 2006. Jun 15;107(12):4589–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, Van Wier S, Tiedemann R, Shi CX, Sebag M, Braggio E, Henry T, Zhu YX, Fogle H, Price-Troska T, Ahmann G, Mancini C, Brents LA, Kumar S, Greipp P, Dispenzieri A, Bryant B, Mulligan G, Bruhn L, Barrett M, Valdez R, Trent J, Stewart AK, Carpten J, Bergsagel PL. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007. Aug;12(2):131–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Jadhav A, Southal N, Huang R, Nguyen DT. A grid algorithm for high throughput fitting of dose-response curve data. Curr Chem Genomics. 2010. Oct 21;4:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002. Jun 18;3(7):RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995. 57(1), 289–300. [Google Scholar]
- 53.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 54.R Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. (2020). URL https://www.R-project.org/. [Google Scholar]
- 55.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003. Nov;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010. Nov 15;5(11):e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keats JJ, Craig DW, Liang W, Venkata Y, Kurdoglu A, Aldrich J, Auclair D, Allen K, Harrison B, Jewell S, Kidd PG, Correll M, Jagannath S, Siegel DS, Vij R, Orloff G, Zimmerman TM, Mmrf CoMMpass Network, Capone W, Carpten J, Lonial S. Interim analysis of the mmrf commpass trial, a longitudinal study in multiple myeloma relating clinical outcomes to Genomic and immunophenotypic profiles. Blood. 2013. Nov;122(21), 532–532. [Google Scholar]
- 58.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015. Jan 15;31 (2):166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turner S (n.d.). annotables: R data package for annotating/converting Gene IDs. Github. Retrieved January 21, 2022, from https://github.com/stephenturner/annotables. [Google Scholar]
- 60.Andersen P, Gill R. Cox's regression model for counting processes, a large sample study. Ann Stat. 1982. Jan 10;1100–1120. [Google Scholar]
- 61.Therneau TM, Grambsch PM (2000). Modeling Survival Data: Extending the Cox Model. Springer, New York. ISBN 0-387-98784-3. [Google Scholar]
- 62.Scheinin I, Kalimeri M, Jagerroos V, Parkkinen J, Tikkanen E, Würt Pz Kangas A. ggforestplot: Forestplots of Measures of Effects and Their Confidence Intervals. 2022. https://nightingalehealth.github.io/ggforestplot/index.html, https://github.com/nightingalehealth/ggforestplot. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon request.