Abstract
The bovine papillomavirus E5 gene encodes a 44-amino-acid, homodimeric transmembrane protein that is the smallest known transforming protein. The E5 protein transforms cultured fibroblasts by forming a stable complex with the endogenous platelet-derived growth factor (PDGF) β receptor through transmembrane and juxtamembrane interactions, leading to sustained receptor activation. Aspartic acid 33 in the extracellular juxtamembrane region of the E5 protein is important for cell transformation and interaction with the PDGF β receptor. A. N. Meyer et al. (Proc. Natl. Acad. Sci USA 91:4634–4638, 1994) speculated that this residue interacted with lysine 499 on the receptor. We constructed E5 mutants containing all possible substitutions at position 33, as well as several double mutants containing substitutions at aspartic acid 33 and at glutamic acid 36, and we examined the ability of these mutants to transform C127 mouse fibroblasts and to bind to and induce activation of the PDGF β receptor. There was an excellent correlation between the transformation activities of the various mutants and their ability to bind to and activate the PDGF β receptor. Analysis of the mutants demonstrated that a juxtamembrane negative charge on the E5 protein was required for cell transformation and for productive interaction with the PDGF β receptor and indicated that aspartic acid 33 was more important for these activities than was glutamic acid 36. These results are consistent with the existence of an essential juxtamembrane salt bridge between lysine 499 on the PDGF β receptor and an acidic residue in the C terminus of the E5 protein and lend support to our proposed model for the complex between the E5 dimer and the PDGF β receptor.
The 44-amino-acid E5 protein of bovine papillomavirus type 1 (BPV) is the smallest known transforming protein (20). This homodimeric transmembrane protein, which is localized largely to membranes of the endoplasmic reticulum and Golgi apparatus, transforms fibroblasts by forming a stable complex with the endogenous platelet-derived growth factor (PDGF) β receptor and inducing ligand-independent receptor oligomerization and activation (1, 3, 6, 13, 17, 18, 23). The study of this unusual mechanism of receptor tyrosine kinase activation promises to lead to a greater understanding of both viral transformation and receptor biochemistry.
Experiments with chimeric and mutant receptors showed that removal of the ligand-binding domain of the PDGF β receptor does not disrupt the interaction with the E5 protein and mapped the site of interaction between the E5 protein and the PDGF β receptor to the transmembrane/juxtamembrane regions of the two proteins (2, 3, 5, 19, 22). In contrast, PDGF induces receptor activation by binding to the extracellular domain of the receptor. Therefore, the interactions by which the E5 protein induces PDGF receptor activation must be strikingly different from those utilized by PDGF. On the basis of molecular modeling and infrared spectroscopy, we developed a model for the E5 dimer that consists of two long transmembrane helices that pack together in a left-handed coiled coil (23). We have proposed that the E5 dimer interacts directly with the transmembrane/juxtamembrane domains of two PDGF β receptor molecules, with both E5 monomers contributing to a binding site on each face of the E5 dimer. Thus, the E5 dimer is thought to serve as a transmembrane scaffold for dimerization of the PDGF β receptor, allowing the receptor to undergo trans phosphorylation and activation in the absence of PDGF (13, 23). The E5 protein can also form a stable complex with the hydrophobic 16-kDa subunit of the vacuolar H+-ATPase, but there is no compelling evidence that this interaction plays a role in PDGF receptor activation or fibroblast transformation (5, 21).
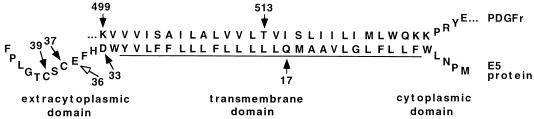
The E5 protein consists of a hydrophobic N-terminal segment of 30 amino acids that spans membranes and a hydrophilic 14-amino-acid segment at the C terminus (20, 23). Four absolutely conserved residues in the E5 protein are important for binding and activation of the PDGF β receptor and for cell transformation: the transmembrane glutamine 17, the juxtamembrane aspartic acid 33, and the C-terminal cysteines 37 and 39, which are involved in homodimerization of the E5 protein (Fig. 1) (8, 10, 14, 15). In addition, the overall hydrophobicity of the central region of the E5 protein, but not the specific amino acid sequence, is critical for cell transformation (12, 14).
FIG. 1.
Alignment of the transmembrane sequences of the E5 protein and the PDGF β receptor. The E5 protein and transmembrane region of the PDGF β receptor (PDGFr) are shown in their antiparallel orientation, and the putative transmembrane region of the E5 protein is underlined. Residues known to be critical for complex formation between the two proteins are indicated by closed arrows. Glutamic acid 36, which can substitute for a missing aspartic acid 33 in some mutants, is indicated by an open arrow.
Mutational analysis also demonstrated that a positive charge in the extracellular juxtamembrane region of the PDGF β receptor and the transmembrane threonine 513 are required for interaction with the E5 protein and for E5-induced receptor activation but not for activation by PDGF (19). Since the E5 protein is thought to be inserted in the membrane in the orientation opposite that of the PDGF β receptor, aspartate 33 of the E5 protein and lysine 499 of the receptor both lie on the extracytoplasmic face of the membrane, with glutamine 17 of the E5 protein and threonine 513 of the receptor buried in the membrane at approximately the same distance from the membrane surface (Fig. 1). These data have led to the proposal that the E5 protein and the PDGF β receptor interact directly with one another and that two pairs of interacting residues, aspartate 33-lysine 499 and glutamine 17-threonine 513, stabilize the E5 protein-PDGF β receptor complex (10, 14, 19, 23).
We began to test this proposal by analyzing a panel of E5 mutants containing every possible substitution at position 17 (10). There was an excellent correlation between the ability of these mutants to bind the receptor, induce receptor activation, and transform cells. All active E5 mutants contained a residue at position 17 that was capable of hydrogen bonding, consistent with the proposed hydrogen bond between glutamine 17 and threonine 513 of the receptor. Our previous mutational analysis of aspartic acid 33 showed that the mutation D33V abolished interaction with the PDGF β receptor and transformation of C127 fibroblasts and that the mutation D33N significantly reduced transformation (8, 15). Meyer et al. demonstrated that alanine substitutions at the negatively charged aspartic acid 33 and glutamic acid 36 inhibited transformation of NIH 3T3 cells (14). On the basis of that result, they speculated that the C-terminal aspartic acid 33 or glutamic acid 36 of the E5 protein interacted with the juxtamembrane lysine on the receptor. However, the ability of these mutants to bind to or induce activation of the PDGF β receptor was not determined, nor was the ability of amino acids other than alanine to functionally substitute for aspartic acid 33.
In this study, we determined the functional range of amino acids at position 33 which allowed a productive interaction between the E5 protein and the PDGF β receptor, leading to cell transformation. We constructed and analyzed a panel of mutants containing all possible amino acids at position 33 of the E5 protein as well as several double mutants with mutations at both positions 33 and 36. These experiments revealed an absolute requirement for a juxtamembrane negative charge on the E5 protein for interaction with the PDGF β receptor and cell transformation.
MATERIALS AND METHODS
Construction of mutant E5 genes.
Fourteen of the mutations at position 33 (alanine, phenylalanine, glycine, histidine, lysine, leucine, methionine, asparagine, proline, glutamine, arginine, serine, threonine, and valine) were constructed in the vector pBPV-H11 by using codon-cassette mutagenesis, a method we previously described in detail (9). Standard PCR-based subcloning procedures were used to subclone the mutant E5 genes into the retroviral vector pRVY-BPV-E5 (3). The remaining five position 33 mutants (cysteine, glutamic acid, isoleucine, tryptophan, and tyrosine), the glutamic acid 36-to-alanine mutant, and four of the double mutants (proline 33/alanine 36, asparagine 33/alanine 36, lysine 33/alanine 36, and glutamic acid 33/alanine 36) were made by using a QuikChange kit (Stratagene) directly in the vector pRVY-BPV-E5. pRVY-BPV-E5-D33P/E36A was used as a template to construct the proline 33/aspartic acid 36 double mutant. The DNA sequence of the entire E5 coding region was confirmed for each mutant. Retroviral DNA containing each mutant was introduced into packaging cell lines, and stable cell lines producing high-titer retrovirus stocks were obtained after selection for hygromycin resistance as described previously (3, 10). Four position 33 mutations (glutamic acid, proline, arginine, and valine) were also introduced into the vector pPava2 by using the QuikChange method, and the E5 genes were sequenced. Recombinant BPV/simian virus 40 (SV40) virus stocks were generated from the resulting plasmids as described previously (15). Details of the mutagenesis and subcloning procedures are available on request.
Cell lines and tissue culture.
C127 and COS7 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and antibiotics (DME-10). To assay the mutants for focus-forming activity, 60-mm-diameter dishes of C127 cells were infected with retroviruses (approximately 104 CFU) encoding the mutant or wild-type E5 genes. Cells were passaged and incubated in the absence of drugs to select for focus formation or in medium containing 350 U of hygromycin B per ml. Foci and drug-resistant colonies were counted 2 to 3 weeks after infection. To calculate focus-forming efficiencies, the number of foci that formed was normalized for the number of hygromycin-resistant colonies that arose in parallel in the same infection. Cell lines were established from pools of >100 hygromycin-resistant colonies and grown in medium containing hygromycin.
For the morphological reversion assay, C127 cells were maintained in the presence or absence of the PDGF receptor kinase inhibitor AG1295 (50 mM in DME-10; Calbiochem) for 3 to 5 days, after which the cells were photographed or processed for phosphotyrosine blotting as detailed below.
Metabolic labeling.
C127 cells were labeled with [14C]leucine as previously described (10). For harvest, cells were rinsed twice with cold phosphate-buffered saline (140 mM NaCl, 27 mM KCl, 1.5 mM KH2PO4, 8.1 mM Na2HPO4) supplemented with 1 mM phenylmethylsulfonyl fluoride, and 10 mM iodoacetamide (to prevent postextraction dimer formation). Cells were then lysed immediately in 1 ml of cold radioimmunoprecipitation assay (RIPA) buffer (20 mM morpholinepropanesulfonic acid [MOPS; pH 7.0], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) containing 1 mM phenylmethylsulfonyl fluoride, 10 mM iodoacetamide, 20 μg of aprotinin, and 20 μg of leupeptin and then incubated for 20 min on ice. Lysates were cleared by centrifugation at 14,000 × g for 30 min at 4°C and stored at −70°C.
Protein extracts and immunoprecipitation.
C127 extracts were prepared in RIPA buffer or CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} buffer as described previously (10). The PDGF β receptor was immunoprecipitated as previously described (10) by adding 1 μl of antibody α-PR-C3a or -B3a (which recognize the C-terminal 13 amino acids of the receptor) per 100 μg of protein extract. Immunoprecipitation of the E5 protein and associated PDGF β receptor was performed as described previously (10) by adding 1 μl of E5 antiserum (which recognizes the 16 C-terminal amino acids of the E5 protein) per 100 μg of RIPA (for E5 immunoblotting or to precipitate labeled E5 protein) or CHAPS (for co-immunoprecipitation assays) protein extract.
Electrophoresis and immunoblotting.
Samples were boiled in 2× Laemmli sample buffer with β-mercaptoethanol, except for the metabolically labeled samples used for detection of dimer, which were boiled in sample buffer without any reducing agents. Samples were then electrophoresed in 7.5 or 15% polyacrylamide gels containing SDS, to detect PDGF β receptor or E5 protein, respectively. Phosphotyrosine, PDGF β receptor, and E5 immunoblotting were performed as described previously (10). Levels of receptor tyrosine phosphorylation were quantitated with a PhosphorImager (Molecular Dynamics). Proteins were detected with 125I-protein A (ICN). To detect radiolabeled E5 protein, the gel was dried and exposed to a PhosphorImager.
Immunofluorescence.
COS7 cells grown to 50% confluence on glass coverslips were infected at a multiplicity of ∼1 with BPV/SV40 recombinant viruses containing the wild-type or various mutant E5 genes. Three days after infection, the cells were immunostained with anti-E5 antibody exactly as described previously (10), and fluorescent photomicrographs were obtained.
RESULTS
To investigate the role of position 33 in E5 transformation, we constructed all possible substitutions at this position in the E5 protein and examined the biological and biochemical effects of the mutants in C127 murine fibroblasts. To compare the transformation efficiency of the mutant E5 proteins with that of the wild-type protein, focus formation assays were performed by infecting cells with retroviruses expressing the mutant or wild-type E5 gene. After passage, infected cells were either selected for a cotransduced hygromycin resistance gene or incubated at confluence in the absence of drugs to select for foci. To correct for differences in titers of the viral stocks, the number of foci obtained with each stock was normalized to the number of drug-resistant colonies that grew. In addition, hygromycin-resistant colonies were pooled to obtain stable cell lines for biochemical analysis.
Transformation of C127 fibroblasts by the position 33 mutants.
Five of the nineteen position 33 mutants transformed C127 cells in focus formation assays. The only mutant that transformed cells better than the wild-type E5 protein was the glutamic acid mutant, which induced twice as many foci as the wild type. Interestingly, this is the only mutant that, like the wild-type E5 protein, contains a negative charge at position 33. The proline mutant induced approximately 90% as many foci as the wild-type protein, and the asparagine, lysine, glutamine, and threonine mutants transformed approximately 10 to 40% as well as wild-type E5. The remaining mutants were essentially defective for transformation. The transformation data are summarized in Table 1.
TABLE 1.
Biological activities of position 33 mutants
| Construct | Focus formationa | P-Tyrb | Complex formationc |
|---|---|---|---|
| RVY | 0 ± 0 | 8 ± 2 | NA |
| WT | 100 | 100 | ++ |
| D33A | 1 ± 1 | 8 ± 3 | − |
| D33C | 0 ± 0 | 15 ± 11 | − |
| D33F | 1 ± 1 | 4 ± 1 | − |
| D33G | 0 ± 0 | 8 ± 0 | +/− |
| D33H | 4 ± 5 | 12 ± 6 | − |
| D33I | 0 ± 0 | 8 ± 3 | +/− |
| D33L | 0 ± 0 | 16 ± 5 | +/− |
| D33M | 0 ± 0 | 9 ± 3 | − |
| D33R | 3 ± 4 | 12 ± 3 | +/− |
| D33S | 2 ± 2 | 19 ± 11 | +/− |
| D33V | 0 ± 0 | 9 ± 6 | +/− |
| D33W | 0 ± 0 | 18 ± 7 | +/− |
| D33Y | 0 ± 0 | 9 ± 6 | +/− |
| D33T | 13 ± 11 | 21 ± 3 | + |
| D33K | 16 ± 10 | 48 ± 16 | +/− |
| D33N | 30 ± 8 | 52 ± 12 | + |
| D33Q | 43 ± 11 | 67 ± 12 | + |
| D33P | 87 ± 7 | 87 ± 10 | ++ |
| D33D | 92 ± 13 | ||
| D33E | 203 ± 44 | 127 ± 37 | ++ |
Average focus formation (corrected for virus titer), expressed as percentage of the wild-type value ± standard deviation of the mean. D33D is a reconstructed wild-type E5 gene. In a typical experiment, the wild-type E5 retrovirus induced 16 transformed foci/1,000 hygromycin-resistant colonies.
Average tyrosine phosphorylation of mature PDGF β receptor as determined by PhosphorImager analysis, expressed as percentage of the wild-type value ± standard deviation of the mean.
Complex formation between the E5 protein and the PDGF β receptor, scored as follows: −, no binding above control; +/−, trace binding; +, readily detectable binding; ++, levels of binding similar to that observed with wild-type E5 protein; NA, not applicable.
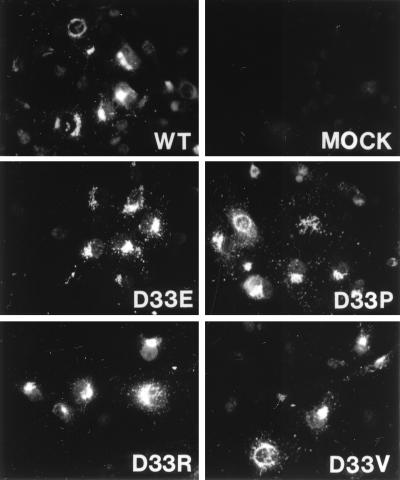
C127 cells stably expressing the wild-type E5 protein acquire a characteristic transformed appearance: the cells become elongated, refractile, and grow very densely in a criss-cross pattern. Cells stably expressing the glutamic acid and the proline mutants appeared similar to cells expressing the wild-type E5 protein (Fig. 2). Cells expressing the other, poorly transforming mutants displayed an intermediate phenotype, and cells expressing the transformation-defective mutants were indistinguishable from nontransformed parental C127 cells (data not shown).
FIG. 2.
Photomicrographs of C127 cells in the presence or absence of a PDGF receptor-specific kinase inhibitor. Transformed C127 cells stably expressing the wild-type (WT) E5 protein the proline (D33P) or glutamic acid (D33E) E5 mutant, or the polyomavirus middle T (mT) oncoprotein were seeded at subconfluence in 24-well plates and incubated in medium containing (+) or lacking (−) AG1295.
We showed previously that treatment of E5-transformed C127 cells with the PDGF receptor-specific kinase inhibitor AG1295 led to decreased tyrosine phosphorylation of the PDGF β receptor and reversal of the transformed morphology (10). When plated in the presence of AG1295, cells expressing the wild-type E5 protein or the glutamic acid or proline mutant underwent a reversion in morphology and appeared very similar to parental C127 cells (Fig. 2). In contrast, C127 fibroblasts transformed by the polyomavirus middle T antigen, an unrelated viral oncoprotein, or p185Neu*, a different activated receptor tyrosine kinase, did not undergo morphologic reversion upon treatment with the kinase inhibitor (Fig. 2 and reference 10). Treatment with the kinase inhibitor also caused a marked decrease in tyrosine phosphorylation of the PDGF β receptor in cells transformed by the wild-type or mutant E5 protein (data not shown). These results indicated that a functional PDGF receptor was required for the transformation-competent position 33 E5 mutants to transform C127 cells and argued against the possibility that these mutants caused transformation via a target other than the PDGF receptor.
Expression and localization of the position 33 mutants.
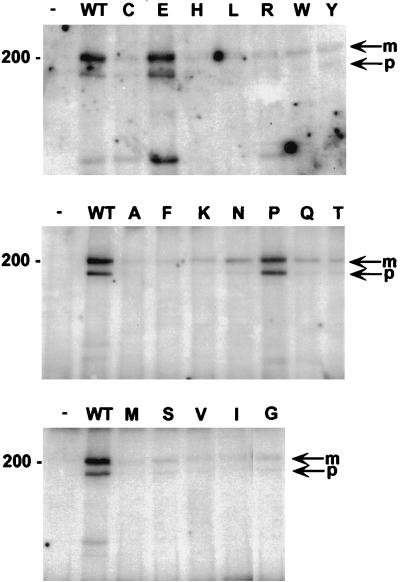
Stable cell lines generated with each of the mutant viruses were used to examine expression of the E5 protein, PDGF β receptor tyrosine phosphorylation, and complex formation between the mutant E5 protein and the PDGF β receptor. The E5 protein was detected either by immunoblotting (for the lysine and asparagine mutants) or by immunoprecipitation from cell lines after metabolic labeling with [14C]leucine. All of the mutant E5 proteins accumulated in C127 cells, and there was no correlation between the level of E5 protein and the transformation phenotype of the mutants (Fig. 3). For example, the transformation-defective mutants D33A, D33I, and D33L accumulated to higher levels than the wild-type E5 protein. Thus, the differences noted above in transforming activities were not due to differences in the level of expression of the mutant E5 proteins. For cell lines expressing several representative mutants, we also examined the relative amounts of monomeric and dimeric E5 protein by carrying out gel electrophoresis under nonreducing conditions. The ratio of dimeric to monomeric E5 protein did not vary among the position 33 mutants examined, with the wild-type and all tested mutant E5 proteins being almost exclusively dimeric (data not shown).
FIG. 3.
Expression of the mutant E5 proteins in C127-derived cell lines. RIPA extracts of stable C127 cell lines labeled with [14C]leucine were immunoprecipitated with anti-E5 antibody. After electrophoresis under reducing conditions, labeled proteins were detected with a PhosphorImager. Extracts from cell lines established by infection with the empty retrovirus vector (−) or with retrovirus expressing the wild-type (WT) E5 gene are also shown. The size of the marker (in kilodaltons) is shown on the left. Expression of the asparagine and lysine mutants was determined by immunoblotting and is not shown.
We used immunofluorescence to determine the subcellular localization of the wild-type E5 protein and representative transformation-competent (glutamic acid and proline) and transformation-defective (arginine and valine) mutants. We used COS monkey cells for these experiments because we are not able to detect the E5 protein in transformed C127 cells by immunofluorescence. Recombinant BPV/SV40 virus stocks expressing the various E5 genes were generated and used to infect COS cells. Three days after infection, the cells were fixed, permeabilized, and stained with anti-E5 antiserum. The E5 protein was detected by indirect immunofluorescence (Fig. 4). There was low background staining in mock-infected cells and bright staining in a discrete perinuclear location for cells expressing the wild-type E5 protein (Fig. 4, WT). This staining pattern is thought to represent Golgi localization (1). All four mutants showed staining indistinguishable from wild-type staining, indicating that the position 33 mutations did not alter the intracellular localization of the E5 proteins and that improper localization of the mutant E5 proteins did not affect their ability to transform cells.
FIG. 4.
Localization of mutant E5 proteins in COS7 cells. COS7 cells were mock infected or infected with BPV/SV40 recombinant viruses expressing the wild-type or indicated mutant E5 proteins. After 3 days, the cells were fixed, permeabilized, stained with anti-E5 antibody, and visualized by immunofluorescence.
Binding and activation of PDGF β receptor by the position 33 mutants.
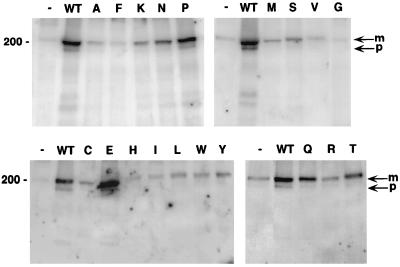
To assess the ability of the mutant E5 proteins to form a complex with the PDGF β receptor, coimmunoprecipitation experiments were carried out. Extracts of C127 cells stably expressing the E5 mutants were prepared in CHAPS buffer and immunoprecipitated with the E5 antiserum, and PDGF β receptor in the immunoprecipitate was detected by immunoblotting with receptor-specific antiserum (Fig. 5). No PDGF β receptor was immunoprecipitated with the E5 antiserum from cells infected with the empty vector. Both the slowly migrating mature form of the PDGF β receptor and a more rapidly migrating precursor form of the receptor containing incompletely processed carbohydrates were coimmunoprecipitated from cells expressing the wild-type E5 protein. These two forms of the PDGF β receptor were also coimmunoprecipitated from extracts prepared from cells expressing several mutants. Under these conditions, the two mutants with the highest transforming activity, the glutamic acid and proline mutants, bound the receptor as well as did the wild-type E5 protein. The four mutants with moderate transformation defects—lysine, asparagine, glutamine, and threonine—bound the receptor much less well than the wild-type E5 protein, whereas the nontransforming mutants bound little or no PDGF β receptor. Thus, the amount of PDGF β receptor coimmunoprecipitated with the mutant E5 proteins correlated well with transforming activity.
FIG. 5.
Complex formation between position 33 mutant E5 proteins and the PDGF β receptor in C127 cells. CHAPS extracts of C127 cells stably expressing various E5 proteins were immunoprecipitated with anti-E5 antibody, and precipitated proteins were resolved by electrophoresis and transferred to membranes. Membranes were probed with anti-PDGF receptor antibody to detect receptors associated with the E5 protein. The letter above each lane indicates the amino acid at position 33 of each mutant E5 protein. Extracts from cells expressing the wild-type (WT) E5 protein or no E5 protein (−) are also shown. Bands corresponding to the mature (m) and precursor (p) forms of the PDGF β receptor are indicated by arrows at the right; sizes of markers (in kilodaltons) are shown on the left. The figure is a composite of several independent immunoprecipitations, but positive and negative controls processed in parallel with each set of immunoprecipitations were included.
To examine the ability of the various mutants to induce activation of the PDGF β receptor, the receptor was immunoprecipitated from extracts of the stable cell lines by using a PDGF receptor-specific antiserum, and tyrosine phosphorylation of the receptor was determined by immunoblotting with a monoclonal antibody that recognizes phosphotyrosine. Representative results for the entire set of mutants are shown in Fig. 6; the results from multiple independently derived cell lines expressing each mutant were quantitated, and the averages are shown in Table 1. The wild-type E5 protein induced tyrosine phosphorylation of both mature and immature forms of the receptor. Overall, the levels of receptor tyrosine phosphorylation induced by the various E5 mutants correlated well with the transforming activities of the mutants. The glutamic acid mutant and the proline mutant induced high levels of receptor phosphorylation, similar to the levels seen with the wild-type E5 protein. The glutamine, lysine, and asparagine mutants, which displayed intermediate transforming activity, also induced receptor phosphorylation, although to lower levels than did the wild-type protein. Cells expressing the threonine mutant consistently exhibited lower levels of receptor phosphorylation than did cells expressing the other transformation-competent mutants, although the phosphorylation levels were above the background phosphorylation seen in cells expressing vector alone. The defective mutants induced little or no receptor phosphorylation above background levels.
FIG. 6.
Tyrosine phosphorylation of the PDGF β receptor by position 33 mutant E5 proteins in C127 cells. RIPA extracts (500 μg of protein) of C127 cells expressing no E5 protein (−), the wild-type (WT) E5 protein, or the position 33 mutants (indicated by the letter above each lane) were precipitated with anti-PDGF receptor antibody. Proteins were resolved by electrophoresis, transferred to membranes, and probed with antiphosphotyrosine antibodies to detect tyrosine-phosphorylated receptor. Bands corresponding to the mature (m) and precursor (p) forms of the PDGF β receptor are indicated by arrows at the right; sizes of markers (in kilodaltons) are shown on the left. The figure is a composite of several independent immunoprecipitations, but positive and negative controls processed in parallel with each set of immunoprecipitations were included.
In general, the correlation between C127 cell transformation, complex formation, and receptor activation was strong (Table 1), and in no case did transformation occur in the absence of receptor phosphorylation and binding. These results are further evidence that position 33 of the E5 protein plays an important role in mediating the interaction with the PDGF receptor.
Analysis of the double mutants at positions 33 and 36.
Several E5 mutants without a negative charge at position 33 transformed cells and interacted productively with the PDGF β receptor. The proline mutant in particular displayed considerable activity. At face value, this result is not consistent with the simple model that an electrostatic interaction between a negatively charged residue at position 33 in the E5 protein and the positively charged lysine 499 in the receptor is critical for complex formation. We hypothesized that in these mutants the negatively charged glutamic acid 36 was able to compensate for the absence of a negative charge at position 33. To test this possibility, we constructed and analyzed a series of double mutants with substitutions at both positions 33 and 36. Four double mutants were made in which a glutamic acid-to-alanine substitution at position 36 was combined with a mutation of aspartic acid 33 to glutamic acid, proline, asparagine, or lysine. As a control, glutamic acid 36 in the wild-type protein was mutated to alanine. We also made a fifth double mutant, proline 33/aspartic acid 36, to test whether, in the context of the proline 33 mutation, an aspartic acid at position 36 could substitute for the glutamic acid. The double mutant E5 proteins were stably expressed in C127 cells (Fig. 3).
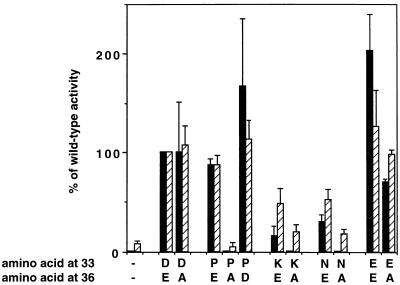
The alanine mutation at position 36 eliminated the transforming activity of E5 mutants containing proline, asparagine, or lysine at position 33 but permitted substantial transforming activity if the amino acid at position 33 had a negative charge (i.e., if it was the wild-type aspartic acid or glutamic acid) (Fig. 7). Thus, the double mutants without a juxtamembrane negative charge were transformation defective, whereas a negative charge at position 36 was not required for efficient transformation if there was one at position 33. In addition, restoration of a negative charge to the transformation-defective D33P/E36A mutant to generate D33P/E36D resurrected robust transforming activity. The biochemical analysis of cells expressing these various mutants is shown in Fig. 8 and summarized in Fig. 7. The transformation-competent mutants (E36A, D33E/E36A, and D33P/E36D) induced high levels of receptor tyrosine phosphorylation (lanes 3, 13, and 15), but the three defective double mutants (D33P/E36A, D33N/E36A, and D33K/E36A) induced substantially less receptor phosphorylation than the corresponding position 33 single mutants (compare, for example, lane 6 to lane 7 and lane 8 to lane 9 in Fig. 8A). We also compared the abilities of three representative mutants, E36A, D33P, and D33P/E36A, to form complexes with the receptor. Whereas the E36A mutant and the transformation-competent D33P single mutant bound the receptor about as well as the wild-type when measured by coimmunoprecipitation analysis, the transformation-defective D33P/E36A mutant bound substantially less receptor (Fig. 8B). Our results indicate that E5 proteins with a negative charge at position 33 displayed high transforming activity regardless of the presence of a negative charge at position 36. In addition, several mutants without a negative charge at position 33 transformed cells only if they retained a negative charge at position 36. Therefore, at least one negative juxtamembrane charge, at either position 33 or position 36, was necessary for the E5 protein to interact with and induce activation of the PDGF β receptor and to transform cells.
FIG. 7.
C127 cell transformation and PDGF β receptor tyrosine phosphorylation induced by position 33 single mutants and position 33/position 36 double mutants. Retroviral stocks expressing wild-type or mutant E5 proteins were used to infect C127 cells, and focus formation was measured and expressed as a percentage of wild-type activity (black bars). Extracts of stable cell lines were immunoprecipitated with anti-PDGF receptor antibody, blotted with antiphosphotyrosine antibodies, and quantitated by PhosphorImager analysis to measure receptor phosphorylation, which is expressed as a percentage of receptor tyrosine phosphorylation induced by wild-type E5 (shaded bars). The first set of lanes shows cells infected with empty vector, and the second set of lanes (D33/E36) shows cells expressing the wild-type E5 protein. No focus-forming activity was detectable for the empty vector (−/−), D33P/E36A, D33K/E36A, and D33N/E36A. The results of multiple experiments have been combined, and the error bars represent the standard deviation of the mean.
FIG. 8.
Biochemical analysis of the position 33/position 36 double mutants. (A) RIPA extracts (500 μg of protein) of C127 cells expressing no E5 protein (−), the wild-type (WT) E5 protein, or single or double E5 mutants (indicated above each lane) were precipitated with anti-PDGF receptor antibody. Proteins were resolved by electrophoresis, transferred to membranes, and probed with antiphosphotyrosine antibodies to detect tyrosine-phosphorylated receptor. Bands corresponding to the mature (m) and precursor (p) forms of the PDGF β receptor are indicated by arrows at right. Sizes of markers (in kilodaltons) are shown on the left. The right and left panels are the results of two independent immunoprecipitations, but the positive and negative controls were processed in parallel with both sets of immunoprecipitations. (B) CHAPS extracts (1,000 μg of protein) of C127 cells stably expressing various E5 proteins were immunoprecipitated with anti-E5 antibody, and precipitated proteins were resolved by electrophoresis. Membranes were probed with anti-PDGF receptor antibody to detect receptors associated with the E5 protein. Bands corresponding to the mature (m) and precursor (p) forms of the PDGF β receptor are indicated by arrows at right, and size of markers (in kilodaltons) is shown on the left.
DISCUSSION
To clarify the role of aspartic acid 33 in E5 transformation and to test the hypothesis that there is an essential interaction between this residue and lysine 499 in the PDGF β receptor, we constructed and analyzed the effects of all possible substitutions at this position of the E5 protein. The glutamic acid mutant transformed cells approximately twice as well as the wild type, the proline mutant transformed approximately as well as the wild type, and four hydrophilic substitutions resulted in substantially lower but detectable transforming activity. The glutamic acid and proline mutants efficiently bound the PDGF β receptor and induced high levels of receptor tyrosine phosphorylation, while the four mutants with moderate transformation defects bound the receptor much less well than the wild-type E5 protein and induced lower levels of receptor phosphorylation. All other position 33 mutants failed to transform C127 cells and were significantly impaired in the ability to bind and activate the PDGF β receptor. In addition, a kinase inhibitor specific for the PDGF receptor reduced receptor tyrosine phosphorylation and led to reversion of the transformed phenotype in cells expressing the proline and glutamic acid mutants. These results highlighted the importance of the residue at position 33 of the E5 protein in cell transformation and binding and activation of the PDGF β receptor and provided further evidence that this receptor is the main target of the E5 protein in murine fibroblasts.
All of the mutant E5 proteins accumulated in cells, and representative mutants localized normally and formed dimers at levels similar to those for the wild-type protein. Therefore, altered stability, dimerization, or localization did not appear to be responsible for the phenotypes of the various mutants. It also seems unlikely that altered orientation of the E5 protein in the membrane was responsible for the behavior of the mutants. The difference in the charge of the N-terminal versus C-terminal juxtamembrane segment of single-span transmembrane proteins has been proposed to be the primary determinant of orientation (7). However, transforming activity of the E5 mutants did not correlate in any simple way with juxtamembrane charge, since transformation was severely impaired by most neutral amino acids at position 33, even though other neutral amino acids (and lysine, a basic one) at this position allowed transformation. Furthermore, replacing the negative charge at position 33 with alanine inhibited transformation, whereas replacing the negative charge at position 36 did not inhibit. Other models propose that the sequence of N-terminal segment or the length of the hydrophobic domain are crucial for specifying orientation of type II membrane proteins (4, 16), but the mutations studied here did not affect either of these segments.
We previously showed that a positive charge in the extracytoplasmic juxtamembrane domain of the PDGF β receptor is required for a productive interaction between the E5 protein and the receptor (19). Here we showed that a negative charge in the corresponding region of the E5 protein was also required for this interaction. Evidently, either aspartic acid or glutamic acid can function at position 33, since a negative charge at position 36 was not required when either of these acidic amino acids occupied position 33. Several amino acids without a negative charge, most notably proline, could also substitute for the wild type aspartic acid 33, but a negative charge at position 36 was required for the transforming activity of these position 33 mutants. Thus, a negative charge in the juxtamembrane region of the E5 protein and a positive charge in this region of the PDGF receptor are necessary for the productive interaction between these two proteins and for cell transformation. The simplest explanation for these results is that the E5 protein and the PDGF β receptor contact one another directly and that this complex is stabilized by a salt bridge between oppositely charged residues in the juxtamembrane region.
We have not been able to reconstitute this interaction by swapping the positive and negative charges on the E5 protein and the PDGF receptor. There are numerous possible explanations for the failure of these mutants to complement each other. For example, changing the sequence context of the juxtamembrane charges may alter their translational position relative to the negatively charged membrane surface, thereby preventing the interaction.
These experiments confirm and extend our earlier findings that mutations at position 33 impair C127 cell transformation and productive interaction with the PDGF β receptor (8, 15). Meyer et al. reported that the mutants D33A and E36A were both able to transform NIH 3T3 cells (14); in our experiments the mutant D33A was defective for C127 cell transformation. It is possible that differences between the cell types and transformation assays used account for the difference observed in the activity of the D33A mutant. In addition, the D33A mutant used by Meyer et al. contained a second mutation, substituting a glutamic acid for glutamine at position 17 (14). We showed previously that the Q17E mutation leads to an approximate doubling of transforming activity in C127 cells (10), and it is possible that in their experiments the Q17E mutation compensated for the loss of the negative charge at position 33. In any event, Meyer et al. (14) speculated that either aspartic acid 33 or glutamic acid 36 of the E5 protein interacted with the juxtamembrane lysine on the PDGF β receptor and concluded that the aspartic acid was probably more important than the glutamic acid, conclusions consistent with the biochemical analysis reported here. The importance of aspartic acid 33 compared to glutamic acid 36 is also suggested by the absolute conservation of aspartic acid 33 in the E5 proteins of all of the fibropapillomaviruses, in contrast to the absence of a negative charge at position 36 in the other E5 proteins, including the deer papillomavirus E5 protein, which also activates the PDGF β receptor and transforms C127 cells (11).
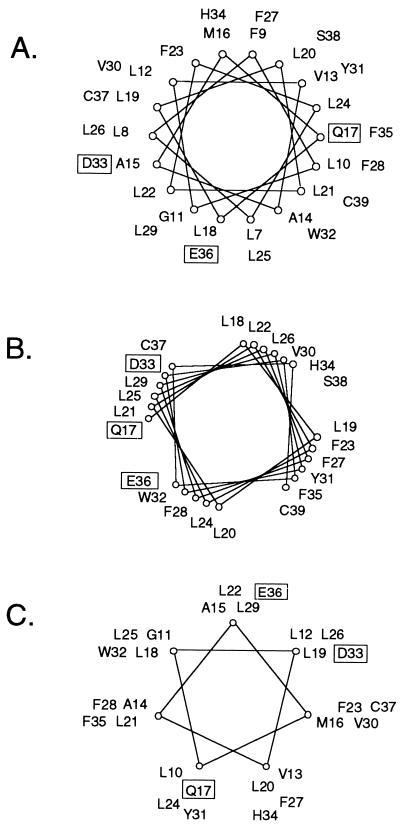
The analysis of the double mutants suggests that the lysine on the PDGF β receptor interacted with glutamic acid 36 in the transformation-competent E5 mutants without a negative charge at position 33. Our previous spectroscopic analysis indicated that the E5 protein is largely α-helical and that the α-helical segment spans the membrane and includes aspartic acid 33 and possibly glutamic acid 36 as well (23). Helical secondary structure in the juxtamembrane region of the wild-type E5 protein would place aspartic acid 33 and glutamic acid 36 on the same face of the helix (Fig. 9A), with the potential for the glutamic acid to contact the lysine on the receptor with only a modest change in the configuration of the E5 protein. Presumably, the transformation-competent mutants without a negative charge at position 33 assumed a conformation that steered the negatively charged residue at position 36 into position to interact with the lysine, whereas the defective position 33 mutants failed to do so. In contrast, if the E5 protein had β-sheet structure in this region, then aspartic acid 33 and glutamic acid 36 would not be in near alignment, and it would be more difficult to imagine how the glutamic acid 36 could substitute for the missing aspartic acid 33.
FIG. 9.
Helical wheel diagrams of the E5 protein in a canonical α-helix (A), a right-handed coil-coil (B), and a left-handed coiled-coil (C). Since paired transmembrane helices typically pack in either right- or left-handed coiled-coil arrangements, the diagrams having 3.9 (B) or 3.5 (C) residues per turn provide a better representation of which groups would line the dimer interface as the two helices coil about one another. Specific amino acids involved in E5-PDGF β receptor complex formation are boxed.
If aspartic acid 33 forms a salt bridge with the receptor, it must be oriented away from the E5 dimer interface. This is consistent with our result that the identity of the residue at position 33 did not influence E5 dimerization. In contrast, we previously demonstrated that some position 17 mutations had marked effects on dimerization, suggesting that glutamine 17 is at least partially buried in the dimer interface, where it can contribute to the stability of the E5 dimer (10, 12, 23). These considerations suggest that aspartic acid 33 and glutamine 17 are situated on opposite faces of the E5 helix, the arrangement that would result if the E5 dimer exists as a left-handed coiled-coil (Fig. 9C). In contrast, a right-handed coiled-coil would place these two residues on the same face of the helix (Fig. 9B). Our spectroscopic data and the molecular modeling also predicted that the E5 dimer would assume a left-handed coiled-coil conformation (23).
The data reported here strongly suggest the existence of a direct interaction between the juxtamembrane lysine on the PDGF β receptor and a negatively charged juxtamembrane residue on the E5 protein. We previously demonstrated that PDGF receptor binding and activation required a residue at position 17 of the E5 protein that can form hydrogen bonds, presumably with threonine 513 of the PDGF β receptor (10). Finally, transforming activity displays relatively relaxed requirements for the precise sequence of hydrophobic residues in the transmembrane domain of the E5 protein (12, 14). Taken together, these and previous studies have identified the minimal requirements of the dimeric E5 protein for interaction with the PDGF β receptor and cell transformation: a hydrophobic transmembrane domain whose sequence may vary considerably, a hydrogen-bonding residue at position 17, and a juxtamembrane extracytoplasmic negative charge. It may be possible to design heterologous peptides incorporating these features which could bind the PDGF β receptor and perhaps serve as starting points for the design of peptides which could interact with and influence the activity of a variety of receptor tyrosine kinases. In addition, detailed characterization of the interaction between the E5 protein and the PDGF β receptor may reveal general principles governing assembly of transmembrane protein complexes.
ACKNOWLEDGMENTS
We thank Venkat Reddy for technical assistance, Karl Haglund for assistance in the initial phases of this work, Lisa Petti for helpful discussions, Edward Goodwin for critical reading of the manuscript, and Jan Zulkeski for assistance in the preparation of the manuscript.
O.K. was supported in part by an MSTP grant. This work was supported by grant CA37157 from the National Cancer Institute.
REFERENCES
- 1.Burkhardt A, Willingham M, Gay C, Jeang K-T, Schlegel R. The E5 oncoprotein of bovine papillomavirus is oriented asymmetrically in Golgi and plasma membranes. Virology. 1989;170:334–339. doi: 10.1016/0042-6822(89)90391-7. [DOI] [PubMed] [Google Scholar]
- 2.Cohen B D, Goldstein D J, Rutledge L, Vass W C, Lowy D R, Schlegel R, Schiller J. Transformation-specific interaction of the bovine papillomavirus E5 oncoprotein with the platelet-derived growth factor receptor transmembrane domain and the epidermal growth factor receptor cytoplasmic domain. J Virol. 1993;67:5303–5311. doi: 10.1128/jvi.67.9.5303-5311.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drummond-Barbosa D, Vaillancourt R R, Kazlauskas A, DiMaio D. Ligand-independent activation of the platelet-derived growth factor β receptor: requirements for bovine papillomavirus E5-induced mitogenic signaling. Mol Cell Biol. 1995;15:2570–2581. doi: 10.1128/mcb.15.5.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eusebio A, Friedberg T, Spiess M. The role of the hydrophobic domain in orienting natural signal sequences within the ER membrane. Exp Cell Res. 1998;241:181–185. doi: 10.1006/excr.1998.4042. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein D J, Andresson T, Sparkowski J J, Schlegel R. The BPV-1 E5 protein, the 16 kDa membrane pore-forming protein and the PDGF receptor exist in a complex that is dependent on hydrophobic transmembrane interactions. EMBO J. 1992;11:4851–4859. doi: 10.1002/j.1460-2075.1992.tb05591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein D J, Li W, Wang L-M, Heidaran M A, Aaronson S A, Shinn R, Schlegel R, Pierce J H. The bovine papillomavirus type 1 E5 transforming protein specifically binds and activates the β-type receptor for platelet-derived growth factor but not other tyrosine kinase-containing receptors to induce cellular transformation. J Virol. 1994;68:4432–4441. doi: 10.1128/jvi.68.7.4432-4441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartmann E, Rapoport T A, Lodish H F. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horwitz B H, Burkhardt A L, Schlegel R, DiMaio D. 44-amino-acid E5 transforming protein of bovine papillomavirus requires a hydrophobic core and specific carboxyl-terminal amino acids. Mol Cell Biol. 1988;8:4071–4078. doi: 10.1128/mcb.8.10.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kegler-Ebo D M, Docktor C M, DiMaio D. Codon cassette mutagenesis: a general method to insert or replace individual codons by using universal mutagenic cassettes. Nucleic Acids Res. 1994;22:1593–1599. doi: 10.1093/nar/22.9.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein O, Polack G W, Surti T, Kegler-Ebo D, Smith S O, DiMaio D. Role of glutamine 17 of the bovine papillomavirus E5 protein in platelet-derived growth factor β receptor activation and cell transformation. J Virol. 1998;72:8921–8932. doi: 10.1128/jvi.72.11.8921-8932.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulke R, DiMaio D. Biological properties of the deer papillomavirus E5 gene in mouse C127 cells: growth transformation, induction of DNA synthesis, and activation of the platelet-derived growth factor receptor. J Virol. 1991;65:4943–4949. doi: 10.1128/jvi.65.9.4943-4949.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulke R, Horwitz B H, Zibello T, DiMaio D. The central hydrophobic domain of the bovine papillomavirus E5 transforming protein can be functionally replaced by many random sequences containing a glutamine. J Virol. 1992;66:505–511. doi: 10.1128/jvi.66.1.505-511.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai C C, Henningson C, DiMaio D. Bovine papillomavirus E5 protein induces oligomerization and trans-phosphorylation of the platelet-derived growth factor β receptor. Proc Natl Acad Sci USA. 1998;95:15241–15246. doi: 10.1073/pnas.95.26.15241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer A N, Xu Y-F, Webster M K, Smith A S, Donoghue D J. Cellular transformation by a transmembrane peptide: structural requirements for the bovine papillomavirus E5 oncoprotein. Proc Natl Acad Sci USA. 1994;91:4634–4638. doi: 10.1073/pnas.91.11.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilson L A, Gottlieb R, Polack G W, DiMaio D. Mutational analysis of the interaction between the bovine papillomavirus E5 transforming protein and the endogenous β receptor for platelet-derived growth factor in mouse C127 cells. J Virol. 1995;69:5869–5874. doi: 10.1128/jvi.69.9.5869-5874.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parks G D. Differential effects of changes in the length of a signal/anchor domain on membrane insertion, subunit assembly, and intracellular transport of a type II integral membrane protein. J Biol Chem. 1996;271:7187–7195. doi: 10.1074/jbc.271.12.7187. [DOI] [PubMed] [Google Scholar]
- 17.Petti L, Nilson L, DiMaio D. Activation of the platelet-derived growth factor receptor by the bovine papillomavirus E5 protein. EMBO J. 1991;10:845–855. doi: 10.1002/j.1460-2075.1991.tb08017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petti L, DiMaio D. Stable association between the bovine papillomavirus E5 transforming protein and activated platelet-derived growth factor receptor in transformed mouse cells. Proc Natl Acad Sci USA. 1992;89:6736–6740. doi: 10.1073/pnas.89.15.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petti L M, Reddy V, Smith S O, DiMaio D. Identification of amino acids in the transmembrane and juxtamembrane domains of the platelet-derived growth factor receptor required for productive interaction with the bovine papillomavirus E5 protein. J Virol. 1997;71:7318–7327. doi: 10.1128/jvi.71.10.7318-7327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlegel R, Wade-Glass M, Rabson M S, Yang Y-C. The E5 transforming gene of bovine papillomavirus encodes a small hydrophobic protein. Science. 1986;233:464–467. doi: 10.1126/science.3014660. [DOI] [PubMed] [Google Scholar]
- 21.Sparkowski J, Mense M, Anders J, Schlegel R. E5 oncoprotein transmembrane mutants dissociate fibroblast transforming activity from 16-kilodalton protein binding and platelet-derived growth factor receptor binding and phosphorylation. J Virol. 1996;70:2420–2430. doi: 10.1128/jvi.70.4.2420-2430.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staebler A, Pierce J H, Brazinski S, Heidaran M A, Li W, Schlegel R, Goldstein D J. Mutational analysis of the β-type platelet-derived growth factor receptor defines the site of interaction with the bovine papillomavirus type 1 E5 transforming protein. J Virol. 1995;69:6507–6517. doi: 10.1128/jvi.69.10.6507-6517.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Surti T, Klein O, Ascheim K, DiMaio D, Smith S O. Structural models of the bovine papillomavirus E5 protein. Proteins Struct Funct Genet. 1998;33:601–612. [PubMed] [Google Scholar]