Abstract
B cells have previously been identified as the major hematopoietic cell type harboring latent gammaherpesvirus 68 (γHV68) (N. P. Sunil-Chandra, S. Efstathiou, and A. A. Nash, J. Gen. Virol. 73:3275–3279, 1992). However, we have shown that γHV68 efficiently establishes latency in B-cell-deficient mice (K. E. Weck, M. L. Barkon, L. I. Yoo, S. H. Speck, and H. W. Virgin, J. Virol. 70:6775–6780, 1996), demonstrating that B cells are not required for γHV68 latency. To understand this dichotomy, we determined whether hematopoietic cell types, in addition to B cells, carry latent γHV68. We observed a high frequency of cells that reactivate latent γHV68 in peritoneal exudate cells (PECs) derived from both B-cell-deficient and normal C57BL/6 mice. PECs were composed primarily of macrophages in B-cell-deficient mice and of macrophages plus B cells in normal C57BL/6 mice. To determine which cells in PECs from C57BL/6 mice carry latent γHV68, we developed a limiting-dilution PCR assay to quantitate the frequency of cells carrying the γHV68 genome in fluorescence-activated cell sorter-purified cell populations. We also quantitated the contribution of individual cell populations to the total frequency of cells carrying latent γHV68. At early times after infection, the frequency of PECs that reactivated γHV68 correlated very closely with the frequency of PECs carrying the γHV68 genome, validating measurement of the frequency of viral-genome-positive cells as a measure of latency in this cell population. F4/80-positive macrophage-enriched, lymphocyte-depleted PECs harbored most of the γHV68 genome and efficiently reactivated γHV68, while CD19-positive, B-cell-enriched PECs harbored about a 10-fold lower frequency of γHV68 genome-positive cells. CD4-positive, T-cell-enriched PECs contained only a very low frequency of γHV68 genome-positive cells, consistent with previous analyses indicating that T cells are not a reservoir for γHV68 latency (N. P. Sunil-Chandra, S. Efstathiou, and A. A. Nash, J. Gen. Virol. 73:3275–3279, 1992). Since macrophages are bone marrow derived, we determined whether elicitation of a large inflammatory response in the peritoneum would recruit additional latent cells into the peritoneum. Thioglycolate inoculation increased the total number of PECs by about 20-fold but did not affect the frequency of cells that reactivate γHV68, consistent with a bone marrow reservoir for latent γHV68. These experiments demonstrate γHV68 latency in two different hematopoietic cell types, F4/80-positive macrophages and CD19-positive B cells, and argue for a bone marrow reservoir for latent γHV68.
Murine gammaherpesvirus 68 (γHV68) was isolated from a bank vole and infects outbred and inbred mice. The genomic sequence of γHV68 is available and confirms its close relationship with other gammaherpesviruses (33). γHV68 can acutely infect multiple organs of mice, including the spleen, liver, lungs, kidneys, adrenals, heart, and thymus (20, 28). Infection has been associated with splenomegaly, pneumonitis, and a fatal arteritis in mice lacking responsiveness to gamma interferon (28, 30, 35, 36). An association between γHV68 infection and the development of lymphomas has also been reported (27). γHV68 establishes a latent infection in the spleen (28, 29, 35), and B cells have been implicated as the predominant latently infected hematopoietic cell type in vivo (29). Because of its genomic structure and association with lymphomas and evidence that it establishes a latent infection in B lymphocytes, γHV68 has been suggested as a murine model for Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (17, 23, 29, 33).
To examine the role of B cells in γHV68 infection and latency, we previously analyzed γHV68 infection in B-cell-deficient mice. B-cell-deficient mice lack mature B cells by virtue of a homozygous mutation in the transmembrane exon of the μ heavy-chain gene (11). γHV68 can efficiently establish a latent infection in B-cell-deficient mice (35), thus demonstrating that B lymphocytes are not required for establishment of latency by γHV68. More recently, we have shown that peritoneal exudate cells (PECs) harbor a higher frequency of cells that reactivate γHV68 than the spleen, in both B-cell-deficient and normal C57BL/6 mice (36a). PECs carry latent γHV68 after either intraperitoneal (i.p.) or intranasal inoculation with γHV68, demonstrating that establishment of latency at this site is independent of the route of inoculation. The finding that the PEC population in B-cell-deficient mice is composed largely of macrophages raised the issue of whether macrophages are a reservoir for latent γHV68. Here we provide evidence, obtained by employing latently infected PECs isolated from C57BL/6 mice, that macrophages are the major cellular reservoir of latent γHV68 in the peritoneum. The identification of macrophages as a site of γHV68 latency likely explains the previously observed efficient establishment of latency in B-cell-deficient mice (35) and demonstrates that γHV68 has a broader cellular tropism for establishment of latency in hematopoietic cells than does EBV. In addition, macrophages may also account for the previously identified γHV68 reactivation from a plastic-adherent cell population isolated from the spleens of latently infected mice (29).
MATERIALS AND METHODS
Mice, infections, and organ harvests.
Normal and B-cell-deficient (MuMT; C57BL/6J-Igh-6tm1Cgn) (11) C57BL/6 mice were purchased from Jackson Laboratory (Bar Harbor, Maine). The mice were then bred and maintained at Washington University, St. Louis, Mo., in accordance with all university and federal guidelines. Mice were infected with 106 PFU of γHV68 in Dulbecco modified Eagle medium plus 10% fetal calf serum in a 1-ml volume by i.p. inoculation. γHV68 strain WUMS (ATCC VR1465) was used for all infections. The viral stock was passaged once on NIH 3T12 cells for amplification. At various times postinfection, mice were sacrificed by cervical dislocation after metofane anesthesia, and PECs were harvested by peritoneal lavage with 10 to 15 ml of medium (8). Thioglycolate elicitation of PECs was accomplished by injection of 3 ml of 3% thioglycolate (Becton-Dickinson, Cockeysville, Md.) into the peritoneum 4 days prior to harvesting of PECs. Thioglycolate injection has been shown to result in accumulation predominantly of neutrophils 1 to 2 days postinjection, followed by accumulation predominantly of inflammatory macrophages 2 to 4 days postinjection (18).
Purification of specific cell populations by FACS and differential analysis of cell types.
For fluorescence-activated cell sorter (FACS) sorting, PECs were stained as previously described (8, 18). Cells were blocked in FACS blocking buffer (HBSS [1× HBSS is 0.15 M NaCl plus 0.015 M sodium citrate] with 5% bovine serum albumin, 10% normal rabbit serum, 10% normal goat serum, and 0.5-mg/ml mouse immunoglobulin G) for 30 to 60 min at 4°C prior to staining to block Fc receptors. All antibodies were diluted in FACS blocking buffer. All primary antibodies used for staining were rat anti-mouse monoclonal antibodies which were purified as previously described (34). F4/80 staining of macrophages was performed with a 50% (vol/vol) dilution of F4/80 supernatant (ATCC HB198) (1). All other antibodies were diluted to 10 μg/ml. CD4-positive T cells were stained with antibody GK1.5 (ATCC TIB207) (6). CD8+ T cells were stained with antibody 53-6.72 (ATCC TIB105) (13). CD19 staining of B cells was performed by using phycoerythrin-conjugated anti-CD19 (Pharmingen, San Diego, Calif.). All other antibody staining was detected by using phycoerythrin-conjugated goat anti-rat immunoglobulin G (heavy and light chains) (Caltag, Burlingame, Calif.). Cells were sorted on a FACS Vantage (Becton-Dickinson, San Jose, Calif.) or analyzed by using a FACScan (Becton-Dickinson). Data were analyzed by using Cell Quest Software (Becton-Dickinson). Postsorting FACS analysis of sorted populations demonstrated >95% purity. Morphological analysis of sorted populations by differential counting was performed on Wright’s-stained cytospin preparations as previously described (8).
Limiting-dilution ex vivo reactivation assay to detect latent virus.
MEF (mouse embryonic fibroblast) cells were obtained from BALB/c mice and maintained as previously described (35). Limiting-dilution analysis to detect reactivation from latency was performed as previously described (35). Briefly, serial twofold dilutions of test cells harvested from mice were plated onto indicator MEF cells in 96-well tissue culture plates. The wells were scored microscopically for a viral cytopathic effect (CPE) after 3 weeks. A maximum of 100,000 cells was plated per well, as greater numbers of cells were toxic to the MEF monolayer. To detect the presence of preformed infectious virus in the test cell populations, the cells were killed prior to plating by mechanical disruption in 1/3× Dulbecco modified Eagle medium in the presence of 0.5-mm silica beads in a Mini-Beadbeater-8 (Biospec Products, Bartlesville, Okla.).
Detection of γHV68 DNA by nested PCR.
Nested PCR to detect the ORF50 gene of γHV68 was shown to have a sensitivity of one copy of γHV68 DNA. The sequences of the outer PCR primers employed were 5′-AACTGGAACTCTTCTGTGGC-3′ and 5′-GGCCGCAGACA TTTAATGAC-3′, which amplify a 586-bp product. The sequences of the inner PCR primers employed were 5′-CCCCAATGGTTCATAAGTGG-3′ and 5′-ATCAGCACGCCATCAACATC-3′, which amplify a 382-bp product. Primers were synthesized by GIBCO BRL. Each PCR mixture contained 50 mM KCl, 10 mM Tris-HCl (pH 8.5), 0.1% Triton X-100, 1.5 mM MgCl2, 0.2 mM nucleotides, each primer at 1 ng/μl, and 1 U of Taq polymerase (Promega). PCRs were performed on a Perkin-Elmer 9600 GENEAMP thermocycler. The initial round of PCR was performed with a 20-μl (total volume) reaction mixture with 45 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, followed by extension at 72°C for 5 min. The conditions for the second round of PCR were identical, except that the reaction was amplified for 25 cycles. For the second round, 1 μl of the first-round PCR product was amplified in a 10-μl (total volume) reaction mixture. Second-round PCR products were visualized by electrophoresis on a 2% agarose gel stained with ethidium bromide. Plasmid pBamHIN containing ORF50 of γHV68, kindly provided by Stacey Efstathiou (7), was used to determine the sensitivity of the nested PCR for detection of γHV68 DNA. pBamHIN was quantitated spectrophotometrically and diluted in mouse liver DNA or tRNA (0.5 mg/ml) in Tris-EDTA. One microgram of total nucleic acid from serial 10-fold dilutions of pBamHIN in mouse liver DNA or tRNA was analyzed by nested PCR in a series of control PCRs.
Estimation of the frequency of latently infected cells harboring the γHV68 genome.
To determine the frequency of cells carrying the γHV68 genome in PECs from latently infected mice, nested PCR (single-copy sensitivity) was performed on serial dilutions of cells by previously published methods (18). In some cases, an adaptation of the published method was used, as follows. To keep the total cell number constant for each PCR, test cells were diluted in an isotonic medium (150 mM KCl, 10 mM Tris-HCl [pH 7.5], 1.5 mM MgCl2) in a background of uninfected MEF cells. Serial fourfold dilutions of cells were made, ranging from 10,000 test cells to 1 test cell per PCR, with a total of 10,000 cells (MEF plus test cells) per PCR. Twelve to 24 PCRs were analyzed per cell concentration. Five-microliter cell dilutions were added to PCR tubes containing 5 μl of lysis buffer (10 mM Tris-HCl [pH 8.5], 1.5 mM MgCl2, 1% Nonidet P-40, 1% Tween 20, 0.2-mg/ml proteinase K) and lysed overnight at 56°C. Proteinase K was inactivated at 95°C for 15 min, and 10 μl of adjusted PCR cocktail (25 mM KCl, 10 mM Tris-HCl [pH 9.0], 0.5% Triton X-100, 1.5 mM MgCl2, 2× deoxynucleoside triphosphates, primers, and Taq) was added directly to each cell lysate so that the final PCR conditions were as described above. A nested PCR was performed as described above. The dilutional nested-PCR method was shown to have a sensitivity of one copy of γHV68 DNA in a background of 10,000 cells by spiking dilutions of plasmid pBamHIN into 10,000 uninfected MEF cells prior to cell lysis. These controls for one-copy sensitivity, as well as negative controls of MEF cells alone or water alone, were included for each set of PCRs performed. In addition, one-copy sensitivity for γHV68 DNA was observed when 106 copies of pBamHIN plasmid DNA were spiked into naive spleen cells and subsequently diluted, demonstrating that target DNA was not destroyed during the lysis procedure.
RESULTS AND DISCUSSION
Analysis of cell types present in the resident PEC population before and after γHV68 infection of C57BL/6 mice.
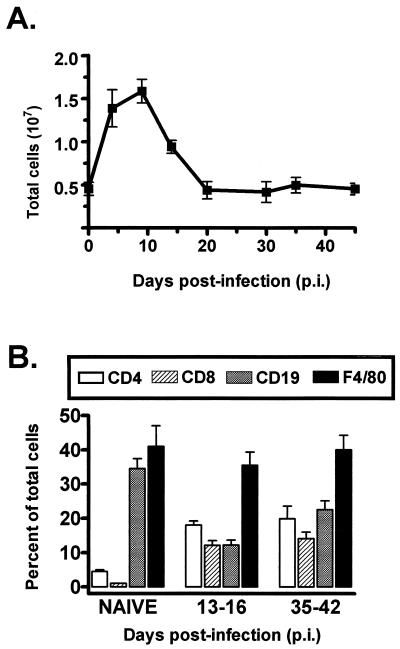
Since we have shown that PECs harbor a high frequency of latently γHV68 infected cells in C57BL/6 mice (36a), we quantitated the cell types and total numbers of cells present in PEC samples at different times postinfection (Fig. 1). In uninfected mice, we recovered an average of 4 × 106 cells from the peritoneum (Fig. 1A), of which 35 to 40% were identified as B cells (by FACS analysis staining with antibodies directed against CD19; see Materials and Methods) and 40 to 50% were identified as macrophages (F4/80-positive staining population; see Materials and Methods) (Fig. 1B). A low number of T cells were present in PECs from uninfected mice (Fig. 1B). Upon γHV68 infection, the number of PECs increased substantially, peaking at 1.6 × 107 at day 10 postinfection, and then dropped to 4 × 106 by day 20 postinfection (Fig. 1A). The influx of cells into the peritoneum after γHV68 infection included a significant increase in the number of T cells present (both CD4+ and CD8+). However, macrophages remained the predominant cell type present in PEC samples at all times postinfection (Fig. 1B).
FIG. 1.
Cell counts and FACS analysis of PECs isolated from C57BL/6 mice before and after infection with γHV68. (A) Total PECs harvested by peritoneal lavage from uninfected C57BL/6 mice or from C57BL/6 mice at various times post-i.p. infection with 106 PFU of γHV68 were counted. The value for each time point is the average of cell counts from eight or nine separate experiments, except those for days 20 and 30 postinfection, which are averages of two experiments. Data were pooled from groups of mice infected for periods of time up to 2 days apart. Error bars represent the standard error of the mean. (B) FACS analysis of PECs isolated from naive or γHV68-infected C57BL/6 mice was performed by using antibodies specific for CD4 T cells, CD8 T cells, B cells (CD19), or macrophages (F4/80). Shown are the percentages of total PECs for each cell type on the indicated days postinfection. The data are averages of four separate experiments. The error bars represent the standard error of the mean.
Establishment of latency in the peritoneum of B-cell-deficient mice.
To determine if B cells account (or are required) for generation of latent infection of PECs, we analyzed γHV68 latency in PECs isolated from B-cell-deficient mice. The presence of latent γHV68 was quantitated by using a limiting-dilution reactivation assay (see Materials and Methods). To detect the presence of preformed infectious virus, cells are subjected to mechanical disruption by using a protocol which kills >99% of the cells present but has a less-than-twofold effect on the titer of preformed infectious virus (35). Since reactivation from latency requires the presence of live cells, only preformed infectious virus can be detected after mechanical disruption of the cells. Reactivated γHV68 is scored 2 to 3 weeks after plating of the latently infected cells by the presence of a viral CPE on the MEF monolayer. Consistent with our previous analyses (36a), we observed a high frequency of PECs that reactivated γHV68 (Fig. 2A).
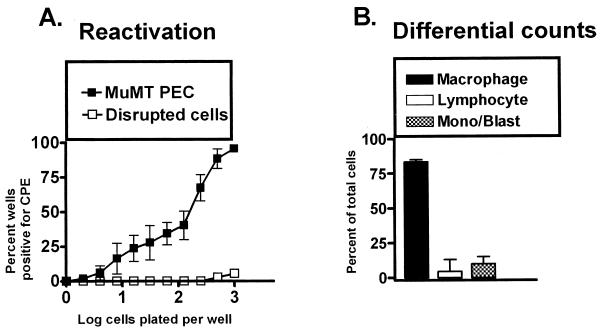
FIG. 2.
PECs from B-cell-deficient mice (MuMT[11]) harbor latent γHV68. (A) Limiting-dilution analysis to quantitate the frequency of cells that reactivate γHV68 was performed by using PECs from B-cell-deficient mice 5 to 10 weeks postinfection with γHV68. Shown are percentages of wells that scored positive for viral CPE 3 weeks after plating as a function of the number of cells plated per well. Twenty-four wells were plated per cell dilution in each experiment. Shown as open symbols are the results obtained when cells were killed by mechanical disruption prior to plating, which indicates that no preformed infectious virus was present in the samples analyzed. The data are averages of four separate experiments. Cells from 6 to 10 mice were pooled and assayed per experiment. The error bars represent the standard error of the mean. (B) Pre- and postsorting differential analysis of Wright’s-stained PECs from B-cell-deficient mice 5 to 10 weeks postinfection with γHV68. PECs were categorized by morphological criteria as macrophages, lymphocytes, or monocytes and/or lymphoblasts. Based on morphological criteria, monocytes could not always be distinguished from lymphoblasts. The data shown are averages of nine separate experiments. Cells from 6 to 10 mice were pooled and assayed per experiment. The error bars represent the standard error of the mean.
The detection of a low level of preformed infectious virus in B-cell-deficient mouse PECs in the experiment shown in Fig. 2A (open symbols) most likely represents a failure to adequately disrupt all γHV68 latently infected cells, since we have consistently failed to detect preformed infectious virus under these conditions in other experiments (35, 36a). Indeed, as shown in Fig. 3, no preformed infectious virus was detected with latently infected PECs harvested from normal C57BL/6 mice, even when 105 cell equivalents were plated. It should be noted that the subsequent analyses of latently infected cell populations reported here were carried out with cells isolated from C57BL/6 mice. Based on our previously determined sensitivity of the limiting dilution assay (0.2 PFU, in which 0.2 PFU was detected in 62.5% of the wells) (35, 36a), the data shown in Fig. 2 indicate that there is <0.05 PFU of virus detected in the mechanically disrupted sample when 103 cell equivalents was plated. The estimated frequency of latently infected PECs in the B-cell-deficient mice is ca. 1 in 100 cells, and thus we estimate that there is <1 PFU per 200 latently infected cells (for the data shown in Fig. 3, we estimate that there is <1 PFU per 20,000 latently infected cells). Thus, preformed infectious virus can account for, at most, 2.5% of the CPE in wells receiving latent cells in Fig. 2 (0.025% of the CPE in wells receiving latent cells in Fig. 3). This verifies that we were measuring latently infected cells in this assay.
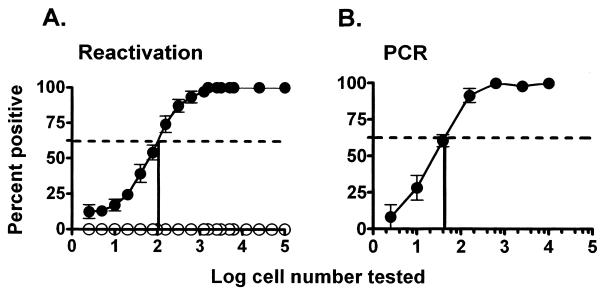
FIG. 3.
Relationship between the frequency of cells reactivating γHV68 and the frequency of cells carrying the γHV68 genome in C57BL/6 PECs 9 to 10 days postinfection. Shown is the percentage of wells in which γHV68 reactivation was detected (A) or the percentage of PCRs which were positive for the presence of the viral genome (B) as a function of the number of cells analyzed. For each cell number, 24 wells in the reactivation analysis (A) or 12 to 24 PCRs (B) were analyzed in each experiment. The data presented are averages of seven separate experiments, and each experiment involved a pool of three mice. The dotted line indicates 62.5%, which was used to calculate the frequency of reactivating or genome-positive cells by Poisson distribution. The error bars represent the standard error of the mean. (A) Frequency of cells that reactivated γHV68 assessed by using the limiting-dilution reactivation assay as described in Materials and Methods. The results of the reactivation assay using disrupted cells, representing the presence of preformed infectious virus, are shown as open symbols. (B) Frequency of cells carrying the γHV68 genome determined by limiting-dilution PCR analysis. Each point represents 84 to 168 separate PCRs.
We evaluated the types of cells in different populations by using differential counting. Note that all differential counts were read without knowledge of the identity of the sample. It is not always possible to distinguish lymphoblasts from immature monocytes on morphologic grounds. Cells in this category are therefore listed as monocytes or lymphoblasts. Differential analysis of the cells present in the PECs isolated from B-cell-deficient mice revealed that >75% were macrophages, while <5% were lymphocytes and ca. 5% were either monocytes or lymphoblasts (Fig. 2B). Thus, γHV68 can efficiently establish a latent infection in the peritoneum of B-cell-deficient mice, demonstrating that a cell other than a B cell harbors latent γHV68 in this site. Because PECs from B-cell-deficient mice are composed predominantly of macrophages, we focused on determining whether latent γHV68 was present in a macrophage-enriched population.
Correlation between the frequency of cells reactivating γHV68 and the frequency of cells carrying the γHV68 genome in PECs isolated from C57BL/6 mice 9 to 10 days postinfection.
To determine the frequency of cells carrying the γHV68 genome in purified cell populations, we developed a sensitive nested-PCR assay to detect the presence of the γHV68 genome in serial dilutions of cells (see Materials and Methods). This assay reproducibly detected 10 copies of a target plasmid, diluted in a background of DNA prepared from 104 uninfected cells, in 100% of the assays (55 PCRs). In addition, this assay was able to detect a single copy of the target plasmid (in a background of DNA prepared from 104 uninfected cells) in 54% of the assays (37 of 68 PCRs) and 0.1 copy of the target plasmid in 3.7% of the assays (2 of 54 PCRs). The frequency of detection of a single copy of the target plasmid compares well with the 63% predicted by Poisson distribution and indicates that this assay can reproducibly detect a single copy of the γHV68 genome in a background of cellular DNA prepared from 104 uninfected cells. This PCR assay was used to directly compare the frequency of γHV68 genome-positive PECs to the frequency of PECs reactivating γHV68 by using cells harvested from latently infected C57BL/6 mice 9 to 10 days postinfection (Fig. 3). As shown in Fig. 3A, mechanical disruption of C57BL/6 PECs resulted in complete loss of detectable virus, while plating of live cells readily revealed the presence of reactivated latent γHV68. Notably, the frequency of cells that reactivated γHV68 in this population was ca. 1 in 100 cells. The estimated frequency of γHV68 genome-positive cells in PEC samples was ca. 1 in 50 cells (Fig. 3B). Thus, the frequency of genome-positive PECs correlates very closely with the frequency of cells that reactivate γHV68 in vitro (compare Fig. 3A and B). Notably, we have observed a more complex relationship between the frequency of cells carrying the γHV68 genome and the frequency of cells reactivating γHV68 at late times postinfection, particularly with latently infected splenocytes (36a). However, as shown here for C57BL/6 mice, at early times postinfection, the frequency of cells reactivating γHV68 and the frequency of cells carrying the γHV68 genome are nearly identical. This assured us that quantitation of the frequency of cells carrying the γHV68 genome was a good indicator of latently infected cells, as opposed to dead-end genomes (i.e., cells harboring the viral genome but unable to reactivate the virus). Thus, we focused our initial identification of the cell types harboring the viral genome in PEC samples harvested 9 to 15 days postinfection.
F4/80-positive cells harbor the γHV68 genome.
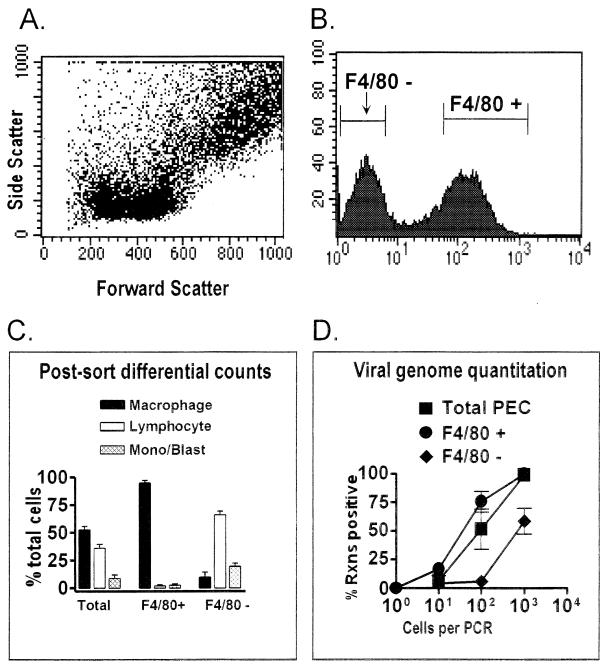
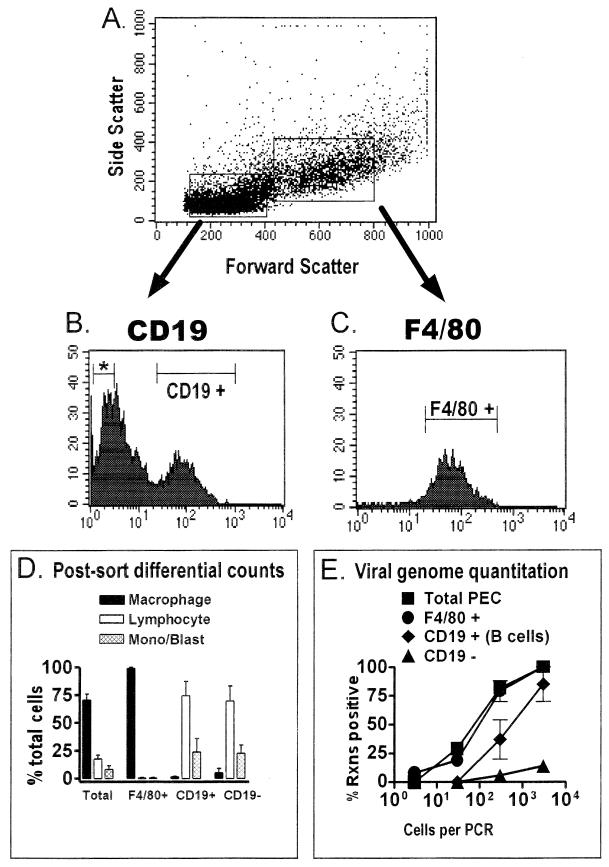
F4/80 is a cell surface marker found on macrophages, as well as some eosinophils (1, 15). Eosinophils are a minor cell population in the peritoneum and are not enriched in PECs sorted for F4/80 expression (see below and reference 18). F4/80-positive and F4/80-negative cell populations were isolated by FACS (Fig. 4A and B), and these populations were subsequently analyzed by differential analysis (Fig. 4C). Post-FACS differential analysis of these populations indicated that >95% of the F4/80-positive cells were macrophages and <5% were lymphocytes or monocytes-lymphoblasts. Thus, this fractionation resulted in a ca. twofold enrichment of macrophages and, importantly, a >10-fold depletion of lymphocytes. The F4/80-negative cell population contained 5 to 10% macrophages, ca. 70% lymphocytes, and ca. 20% cells that were monocytes and/or lymphoblasts (Fig. 4C). Total PECs and F4/80-positive PECs (enriched for macrophages and depleted of lymphocytes) had similar frequencies of γHV68 genome-positive cells (Fig. 4D). The high frequency of γHV68-positive cells in the F4/80-positive, lymphocyte-depleted, fraction (ca. 1 in 50 cells) argues that F4/80-positive macrophages harbor latent γHV68. Note that there is a less-than-twofold increase in the proportion of macrophages in F4/80-positive compared to total PECs (Fig. 4C), explaining the minimal change in the frequency of genome-positive cells observed between total and F4/80-positive PECs. Consistent with macrophages as a site of γHV68 latency, the F4/80-negative cell population contained an at least 10-fold lower frequency of γHV68-positive cells than either F4/80-positive or total PECs (Fig. 4D), despite the fact that this cell population was enriched for lymphocytes. Since there were >30-fold more lymphocytes in the F4/80-negative population than the F4/80-positive population, it is clear that lymphocytes (e.g., B cells) cannot account for the majority of γHV68 genome-bearing cells. From this analysis, it is unclear whether the residual viral genome present in the F4/80-negative fraction represented contaminating latently infected macrophages in this cell population or represented another latently infected cell type (e.g., B lymphocytes).
FIG. 4.
F4/80-positive peritoneal macrophages from latently infected C57BL/6 mice harbor the γHV68 genome. PECs collected from C57BL/6 mice 9 to 15 days postinfection with γHV68 were stained with F4/80. The F4/80-negative and F4/80-positive cell populations were separated by FACS sorting, and the frequency of cells carrying the γHV68 genome was quantitated by PCR. The results shown are averages of four separate experiments. In three experiments, cells were sorted as shown. In one experiment, cells were pregated into lymphocyte-enriched or macrophage-enriched populations prior to F4/80 sorting, as shown in Fig. 5 and 6. Data from the four experiments were comparable. (A) Dot plot showing forward and side scatter characteristics of peritoneal cells from a representative experiment. (B) Results of F4/80 staining and gates used for FACS sorting of F4/80-negative and F4/80-positive cell populations from a representative experiment. Cell counts are shown on the y axis, and mean fluorescence intensity is shown on the x axis. Gates for sorting were drawn tightly to prevent contamination of sorted populations. For the four experiments performed, 39 to 42% of the PECs were sorted as F4/80 negative and 39 to 44% of the PECs were sorted as F4/80 positive. (C) Pre- and postsorting differential analysis of Wright’s-stained cells from total PECs and F4/80-positive and F4/80-negative PECs. Presorting and postsorting populations were categorized by morphological criteria as macrophages, lymphocytes, or monocytes-lymphoblasts (Mono/Blast). Based on morphological criteria, monocytes could not always be distinguished from lymphoblasts. (D) Limiting-dilution quantitation of the frequency of γHV68 genome-positive cells by using total PECs and F4/80-positive and F4/80-negative PEC populations. Tenfold dilutions of each cell population were tested for the presence of the γHV68 genome by nested PCR as described in Materials and Methods. The data in panels C and D are averages of four experiments. The error bars represent the standard error of the mean. Rxns, reactions.
B cells, as well as macrophages, harbor latent γHV68.
To further address the question of whether B lymphocytes might also be a reservoir of latent γHV68 in the peritoneum, both B cells (CD19 positive) and macrophages (F4/80 positive) were recovered from latently infected PECs. In this analysis, we took advantage of the distinct differences in the forward versus side scatter of macrophages and lymphocytes. Thus, PECs were presorted by using gates that enriched for either lymphocytes or macrophages, and then these populations were sorted for either F4/80-positive cells or for CD19-positive and CD19-negative cells (Fig. 5). This two-stage approach was used to diminish contamination of the sorted lymphocyte populations with macrophages. Postsorting differential analysis of the F4/80-positive population demonstrated that this population was highly (>98%) enriched for macrophages, with few contaminating lymphocytes (Fig. 5D). Conversely, the CD19-positive population was composed almost entirely (∼75%) of lymphocytes and a population of cells that were either monocytes or lymphoblasts (∼25%), with only a very small percentage of contaminating macrophages (Fig. 5D). The CD19-negative fraction sorted from the lymphocyte gate contained a slightly higher frequency of contaminating macrophages (∼5%) but was still significantly enriched for lymphocytes (∼70%) and monocytes-lymphoblasts (∼25%) (Fig. 5D). As previously observed (Fig. 4D), analysis of the macrophage-enriched population (F4/80 positive) revealed a frequency of cells harboring the γHV68 genome that was similar to the frequency observed in unsorted PECs (Fig. 5D). This was expected, since in these experiments, the total cell population was ∼70% macrophages, and thus there was a less-than-twofold enrichment for macrophages. However, there was a significant (eightfold) depletion of lymphocytes from F4/80-positive cells, demonstrating that depleting lymphocytes did not significantly reduce the frequency of γHV68 genome-positive cells. This finding, obtained by using a more rigorous two-stage FACS sorting protocol, supports the conclusion drawn from data in Fig. 4 that macrophages harbor latent γHV68.
FIG. 5.
The frequency of macrophages harboring the viral genome is higher than the frequency of B cells harboring the viral genome in the peritoneum of latently infected C57BL/6 mice. PECs isolated from C57BL/6 mice 13 to 15 days postinfection with γHV68 were stained with F4/80 (specific for macrophages) or with a CD19-specific antibody (specific for B cells), and relevant cell populations were isolated by FACS sorting. The results shown represent two separate experiments. (A) Cells were pregated into lymphocyte-enriched or macrophage-enriched populations based on forward scatter and side scatter characteristics, as shown for a representative experiment. (B) PECs from the lymphocyte-enriched population were sorted into CD19-negative (denoted by an asterisk) and CD19-positive fractions. Shown are the results of CD19 staining and the gates used for FACS sorting of CD19-positive and CD19-negative cell populations from a representative experiment. Gates for sorting were drawn tightly to prevent contamination of sorted populations. By these criteria, 40 to 50% of cells from the lymphocyte-enriched gate were sorted as CD19 negative and 16 to 24% of the cells from the lymphocyte-enriched gate were sorted as CD19 positive. (C) F4/80-positive PECs were sorted from the macrophage-enriched population. Shown are the results of F4/80 staining and the gate used for FACS sorting of F4/80-positive cells from a representative experiment. For the two experiments performed, 91 to 94% of the PECs from the macrophage-enriched gate were sorted as F4/80 positive. (D) Pre- and postsorting differential analysis of Wright’s-stained cells. Cells were categorized by morphological criteria as macrophages, lymphocytes, or monocytes-lymphoblasts (Mono/Blast). Based on morphological criteria, monocytes could not always be distinguished from lymphoblasts. (E) Limiting-dilution nested PCR analysis to quantitate the frequency of γHV68 genome-positive cells in the total PECs and the F4/80-positive, CD19-positive, and CD19-negative populations. Tenfold dilutions of each cell population were tested for the presence of the γHV68 genome by nested PCR. The data in panels D and E are averages of two separate experiments. The error bars represent the standard error of the mean. Rxns, reactions.
Comparison of the CD19-positive and CD19-negative populations sorted from the lymphocyte gate revealed that CD19-positive cells also harbored the γHV68 genome (as observed in the F4/80-negative population shown previously [Fig. 4]). However, the frequency of cells carrying the γHV68 genome among CD19-positive cells was ca. 10-fold lower than the frequency observed in the macrophage-enriched F4/80-positive population. The CD19-negative fraction harbored a very low frequency of γHV68 genome-positive cells, which may reflect <1% contamination of this population with latently infected macrophages and/or B cells. Thus, this analysis indicates that macrophages are the major reservoir, and B cells are a minor reservoir, for latent γHV68 in the peritoneum of C57BL/6 mice.
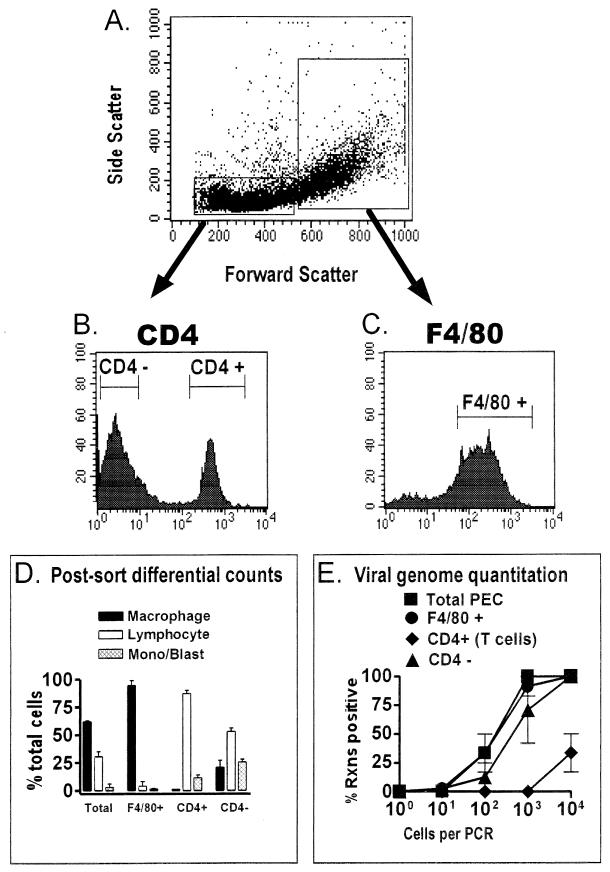
CD4 T cells do not harbor latent γHV68.
As a negative control for F4/80 and CD19 cell sorting, we also sorted for CD4-positive T cells. This sorting provides a control for the remote possibility that all positive sortings (e.g., F4/80-positive and CD19-positive sortings) artifactually enrich for γHV68 genome-positive cells. T cells have never been implicated as a reservoir for latent γHV68. Indeed, an analysis of cell populations isolated from a latently infected spleen found little evidence of γHV68 latency in the T-cell-enriched fraction (29). CD4-positive and CD4-negative cell populations were recovered from the lymphocyte gate (Fig. 6A and B) as described above, and F4/80-positive cells were recovered from the macrophage gate (Fig. 6A and C). The postsorting differential analysis demonstrated that the F4/80-positive population was >95% macrophages, while the CD4-positive population was >85% lymphocytes (the remainder was monocytes and/or lymphoblasts) (Fig. 6D). The CD4-negative population was heavily contaminated with macrophages (∼20%) and was only enriched about twofold for lymphocytes over the unfractionated population (Fig. 6D). Analysis of the frequency of γHV68 genome positive cells demonstrated again that the F4/80-positive fraction was nearly indistinguishable from the unfractionated PECs (Fig. 6E). As expected, the CD4-positive cell population was significantly depleted of cells harboring the viral genome (>100-fold lower frequency of γHV68-positive cells) (Fig. 6E). Consistent with the notion that B cells and macrophages harbor the viral genome, the CD4-negative population retained a relatively high frequency of γHV68 genome-positive cells (Fig. 6E).
FIG. 6.
CD4-positive T cells from latently infected C57BL/6 mice do not harbor the γHV68 genome. PECs collected from C57BL/6 mice 13 to 15 days postinfection with γHV68 were stained with F4/80 or with a CD4-specific antibody for FACS sorting. The results shown represent two separate experiments. (A) Cells were pregated into lymphocyte-enriched or macrophage-enriched populations based on forward scatter and side scatter characteristics, as shown for a representative experiment. (B) PECs from the lymphocyte-enriched population were sorted into CD4-negative and CD4-positive fractions as described in Materials and Methods. Shown are the results of CD4 staining and the gates used for FACS sorting of CD4-positive and CD4-negative cell populations from a representative experiment. Gates for sorting were drawn tightly to prevent contamination of sorted populations. By these criteria, 62 to 67% of the cells from the lymphocyte-enriched gate were sorted as CD4 negative and 28% of the cells from the lymphocyte-enriched gate were sorted as CD4 positive. (C) F4/80-positive PECs were sorted from the macrophage-enriched population. Shown are the results of F4/80 staining and the gate used for FACS sorting of F4/80-positive cells from a representative experiment. For the two experiments performed, 75 to 85% of the PECs were sorted as F4/80 positive. (D) Pre- and postsorting differential analysis of Wright’s-stained cells. Cells were categorized by morphological criteria as macrophages, lymphocytes, or monocytes-lymphoblasts (Mono/Blast). Based on morphological criteria, monocytes could not always be distinguished from lymphoblasts. (E) Limiting-dilution PCR analysis to quantitate the frequency of γHV68 genome-positive cells in total PECs and F4/80-positive, CD4-positive, and CD4-negative PECs. Tenfold dilutions of each cell population were tested for the presence of the γHV68 genome by nested PCR. The data in panels D and E are averages of two separate experiments. The error bars represent the standard error of the mean. Rxns, reactions.
γHV68 genome-positive macrophages reactivate γHV68 in explant cultures.
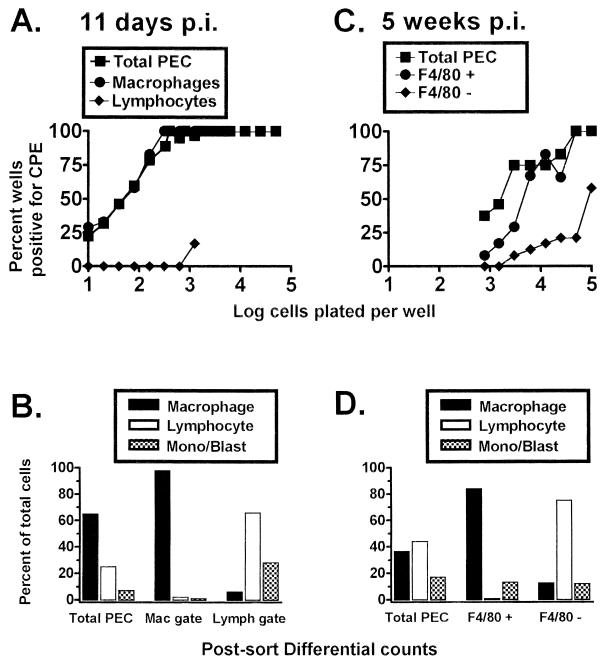
While the above-described analyses provide strong evidence that macrophages harbor the γHV68 genome, they do not directly address the issue of whether these cells are latently infected (i.e., whether they can reactivate γHV68). However, the close correlation between the frequency of cells harboring the γHV68 genome and the frequency of cells reactivating γHV68 (Fig. 3), coupled with the observation that nearly all of the γHV68 genome-positive cells are F4/80 positive (Fig. 4, 5, and 6), makes it very likely that macrophages are latently infected by γHV68. To formally address the question of whether the viral genome-positive, macrophage-enriched (lymphocyte-depleted) population can reactivate γHV68, PECs isolated either 11 days or 5 weeks postinfection were fractionated by FACS sorting (Fig. 7). Consistent with the viral genome analysis, using PECs isolated 11 days postinfection, cells recovered from the macrophage gate (which were judged by postsorting differential analysis to be >98% macrophages and significantly depleted of lymphocytes; Fig. 7B) reactivated γHV68 with a frequency indistinguishable from that of the unsorted cell population (Fig. 7A). However, the frequency of cells reactivating γHV68 was low among cells recovered from the lymphocyte gate (Fig. 7A). Although the frequency of cells reactivating γHV68 was about a log unit lower, a very similar profile was obtained by using PECs isolated 5 weeks postinfection (Fig. 7C). F4/80-positive cells, which were enriched for macrophages and significantly depleted of lymphocytes (Fig. 7D), displayed a reactivation frequency only slightly lower than that of unfractionated PECs (Fig. 7C). However, the F4/80-negative cell population, which was enriched for lymphocytes and partially depleted of macrophages (Fig. 7D), exhibited a significant reduction in the frequency of cells reactivating γHV68 (Fig. 7C). Based on this analysis, γHV68-positive macrophages are capable of reactivating the virus and can thus be defined as a true reservoir of γHV68 latency.
FIG. 7.
Peritoneal macrophages from C57BL/6 mice harbor latent γHV68, as detected by an ex vivo reactivation assay. Limiting-dilution analysis was used to quantitate the frequency of cells that reactivate γHV68 by using FACS-sorted PEC populations isolated from γHV68-infected C57BL/6 mice. (A) Limiting-dilution reactivation analysis to determine the frequency of cells that reactivate γHV68 by using PECs from C57BL/6 mice 11 days postinfection (p.i.). PECs were FACS sorted into macrophage- or lymphocyte-enriched populations based on forward and side scatter characteristics (as for Fig. 5A and 6A). (B) Pre- and postsorting Wright’s differential staining analysis of total and fractionated PECs isolated from C57BL/5 mice 11 days postinfection. Cells were categorized by morphological criteria as macrophages (Mac), lymphocytes (Lymph), or monocytes-lymphoblasts (Mono/Blast). (C) Limiting-dilution reactivation analysis to determine the frequency of cells that reactivate γHV68 by using PECs collected from C57BL/6 mice 5 weeks postinfection. Cells were stained with F4/80, and the F4/80-negative and F4/80-positive cell populations were separated by FACS sorting as described in Materials and Methods. (D) Pre- and postsorting Wright’s differential staining analysis of total and fractionated PECs isolate from C57BL/6 mice 5 weeks postinfection. For the limiting-dilution reactivation analyses shown in panels A and C, the percentage of wells that scored positive for viral CPE 3 weeks after plating is plotted as a function of the number of cells plated per well. Twenty-four wells were plated per cell dilution. Each graph represents a single experiment. Cells from 4 to 10 mice were pooled and assayed per experiment.
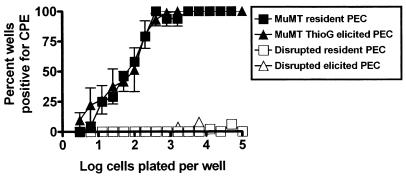
Thioglycolate-elicited PECs harbor the same frequency of γHV68-reactivating cells as resident PECs.
We have shown that γHV68 latency in the peritoneum is established regardless of whether mice are inoculated by the i.p. or the intranasal route (36a), indicating that the establishment of latency at this site is not dependent on i.p. inoculation. However, the derivation of the latently infected macrophages remains unclear. Since it is known that inflammatory peritoneal macrophages are derived from bone marrow (22, 31, 32) and we have shown that bone marrow harbors latent γHV68 (36a), we addressed the question of whether thioglycolate treatment would alter the frequency of γHV68 genome-positive cells in the peritoneum. B-cell-deficient mice were used in this analysis because (i) we have shown that the frequency of cells reactivating γHV68 is much higher in B-cell-deficient mice than in normal C57BL/6 mice at late times postinfection (36a) and (ii) we were able to analyze the dynamics of latency in the macrophage compartment independent of B cells. PECs were recovered from thioglycolate-treated and control B-cell-deficient mice 4 to 7 weeks postinfection. The average number of resident PECs recovered from the control B-cell-deficient mice was 1.4 × 106 ± 0.4 × 106, while the average number of elicited PECs recovered from the thioglycolate-treated, B-cell-deficient mice was 3.1 × 107 ± 0.7 × 107. Thus, there was an about 20-fold increase in the number of PECs upon thioglycolate elicitation. As shown in Fig. 8, mechanical disruption of the PECs revealed no detectable preformed γHV68 in either resident or elicited PECs. The frequencies of cells reactivating γHV68 from resident and elicited PECs were indistinguishable, despite the 20-fold increase in the total number of cells after thioglycolate elicitation (Fig. 8). This result cannot be explained by a 20-fold increase in the reactivation frequency of resident latently infected PECs after thioglycolate elicitation, since we have shown that the frequency of resident PECs that reactivate the virus correlates closely with the frequency of PECs that harbor the viral genome (Fig. 3 and data not shown). Thus, this result indicates that there is a cellular source of latent γHV68 that seeds the peritoneum after thioglycolate elicitation, presumably the bone marrow (22, 31, 32). Similar frequencies of reactivation were also seen in resident versus elicited PECs recovered from latently infected C57BL/6 mice (data not shown).
FIG. 8.
The frequencies of cells reactivating γHV68 are similar in resident and thioglycolate-elicited PECs. Ex vivo limiting-dilution reactivation analysis was used to determine the frequency of cells that reactivate latent γHV68 by using PECs from B-cell-deficient mice (MuMT[11]) ranging from days 31 to 57 postinfection with γHV68 i.p. Latently infected mice were either left untreated (resident PECs) or injected 4 days prior to harvest with 3 ml of thioglycolate i.p. (ThioG-elicited PECs) as described in Materials and Methods. The results of the reactivation analysis using disrupted cells, representing the presence of preformed infectious virus, are shown as open symbols. In resident (unelicited) PECs, there was an average of 1.4 × 106 cells per mouse. After thioglycolate elicitation, there was an average of 3.1 × 107 cells per mouse. The data shown are averages of three separate experiments. For each experiment, PECs from two to nine mice in the thioglycolate-elicited group or 9 to 17 mice in the unelicited groups were pooled. Error bars represent the standard error of the mean. Similar frequencies of γHV68 reactivation were also seen when resident and elicited PECs from C57BL/6 mice were compared (one experiment, data not shown).
Conclusions.
We have demonstrated that macrophages in the peritoneum of normal and B-cell-deficient C57BL/6 mice harbor latent γHV68. In addition, consistent with previous analyses (29), we also found that CD19-positive B cells carry the γHV68 genome during latent infection. Quantitation of the frequency of genome-positive cells revealed that B cells were a relatively minor reservoir of γHV68 latency in PECs compared to macrophages. Recent studies have also provided evidence that γHV68 establishes a latent infection in lung epithelial cells (25). The latter studies are problematic, since the identification of latently infected cells relied on in situ detection of cells containing viral tRNA-like gene transcripts, in conjunction with the failure to detect by in situ hybridization cells expressing the viral glycoprotein H (gH) and thymidine kinase gene transcripts (presented as evidence to rule out the presence of lytic infection). Since the viral tRNA-like genes are abundantly expressed during lytic infection, (2, 36b), it is difficult to rule out the possibility that in situ detection of the viral tRNA-like transcripts is more sensitive than detection of gH and thymidine kinase gene transcripts. Indeed, while these investigators detected the presence of linear viral genomes in lung tissue (diagnostic of ongoing virus replication), they were unable to detect lytically infected cells by using the gH and thymidine kinase probes. Notwithstanding this limitation, it is clear that γHV68 establishes latency in multiple cell types: peritoneal macrophages, B cells, and perhaps lung epithelial cells. The observation of multiple cellular reservoirs of latent γHV68 is similar to the observations of latent EBV in both B cells and epithelial cells (10, 21), that Kaposi’s sarcoma-associated herpesvirus can latently infect both B cells and endothelial or spindle cells (3–5, 9, 14, 16, 19, 24, 26, 37), and of murine cytomegalovirus latency in both macrophages and endothelial cells (12, 18). It remains to be determined whether macrophages are a major reservoir of γHV68 latency in other latently infected tissues (e.g., spleen and lung).
As shown in Fig. 7, the frequency of PECs reactivating γHV68 from normal mice 5 weeks postinfection was significantly lower than that observed at early times. However, we have determined that as in B-cell-deficient mice, the frequency of viral genome-positive cells remains fairly constant for >150 days postinfection in normal C57BL/6 mice, indicating that there is a decrease in the efficiency of virus reactivation upon explanation of cells from normal mice into tissue culture (36a). This decrease in reactivation efficiency observed in normal but not B-cell-deficient C57BL/6 mice may reflect the establishment of a distinct form of viral latency during the course of infection in normal C57BL/6 mice.
The identification of peritoneal macrophages as a reservoir of latent γHV68 raises the question of whether these cells harbor latent γHV68 for an extended period (e.g., >5 weeks postinfection). While we have not directly addressed the turnover of latently infected macrophages, we have determined that the frequency of cells carrying the γHV68 genome (as well as the frequency of cells that reactivate γHV68) in B-cell-deficient mouse PECs remains relatively constant for >150 days postinfection (36a). Since macrophages likely compose the entire γHV68 latency reservoir in B-cell-deficient mouse PECs, this indicates that macrophages are a long-term latency reservoir for γHV68. Notably, the finding that thioglycolate elicitation of inflammatory PECs resulted in no alteration in the frequency of latently γHV68-infected cells in the peritoneum (Fig. 8) suggests that the source of latent macrophages is an infected bone marrow precursor. Additional characterization of γHV68-infected cells in the bone marrow is required to further address this issue.
ACKNOWLEDGMENTS
This work was supported by grants R01 CA74730 (H.W.V.), R01 CA43143 (S.H.S.), R01 CA52004 (S.H.S.), R01 CA58524 (S.H.S.), and K08 AI01279 (K.E.W.).
We acknowledge helpful discussions with members of the Speck and Virgin labs, as well as discussions that occurred during lab meetings shared with David Leib. Finally, we acknowledge Parveen Chand and Debbie Wyman for help with FACS sorting of cell populations.
REFERENCES
- 1.Austyn J M, Gordon S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol. 1981;11:805–815. doi: 10.1002/eji.1830111013. [DOI] [PubMed] [Google Scholar]
- 2.Bowden R J, Simas J P, Davis A J, Efstathiou S. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J Gen Virol. 1997;78:1675–1687. doi: 10.1099/0022-1317-78-7-1675. [DOI] [PubMed] [Google Scholar]
- 3.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 4.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 5.Decker L L, Shankar P, Khan G, Freeman R B, Dezube B J, Lieberman J, Thorley-Lawson D A. The Kaposi’s sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J Exp Med. 1996;184:283–288. doi: 10.1084/jem.184.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dialynas D, Quan Z, Wall K, Pierres A, Quintans J, Loken M, Fitch F. Characterization of the murine T cell surface molecule, designated L3T4, identified by monoclonal antibody GK1.5. J Immunol. 1983;131:2445–2451. [PubMed] [Google Scholar]
- 7.Efstathiou S, Ho Y M, Minson A C. Cloning and molecular characterization of the murine herpesvirus 68 genome. J Gen Virol. 1990;71:1355–1364. doi: 10.1099/0022-1317-71-6-1355. [DOI] [PubMed] [Google Scholar]
- 8.Heise M T, Virgin H W., IV The T-cell-independent role of gamma interferon and tumor necrosis factor alpha in macrophage activation during murine cytomegalovirus and herpes simplex virus infection. J Virol. 1995;69:904–909. doi: 10.1128/jvi.69.2.904-909.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi’s sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2343–2396. [Google Scholar]
- 11.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 12.Koffron A J, Hummel M, Patterson B K, Yan S, Kaufman D B, Fryer J P, Stuart F P, Abecassis M I. Cellular localization of latent murine cytomegalovirus. J Virol. 1998;72:95–103. doi: 10.1128/jvi.72.1.95-103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ledbetter J, Herzenberg L. Xenogeneic monoclonal antibodies to mouse lymphoid differentiation antigens. Immunol Rev. 1979;47:63–90. doi: 10.1111/j.1600-065x.1979.tb00289.x. [DOI] [PubMed] [Google Scholar]
- 14.Li J J, Huang Y Q, Cockerell C J, Friedman-Kien A E. Localization of human herpes-like virus type 8 in vascular endothelial cells and perivascular spindle-shaped cells of Kaposi’s sarcoma lesions by in situ hybridization. Am J Pathol. 1996;148:1741–1748. [PMC free article] [PubMed] [Google Scholar]
- 15.McGarry M P, Stewart C C. Murine eosinophil granulocytes bind the murine macrophage-monocyte specific monoclonal antibody F4/80. J Leukocyte Biol. 1991;50:471–478. doi: 10.1002/jlb.50.5.471. [DOI] [PubMed] [Google Scholar]
- 16.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi’s sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nash A A, Usherwood E J, Stewart J P. Immunological features of murine gammaherpesvirus infection. Semin Virol. 1996;7:125–130. [Google Scholar]
- 18.Pollock J L, Presti R M, Paetzold S, Virgin H W. Latent murine cytomegalovirus infection in macrophages. Virology. 1997;227:168–179. doi: 10.1006/viro.1996.8303. [DOI] [PubMed] [Google Scholar]
- 19.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S-J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajcani J, Blaskovic D, Svobodova J, Ciampor F, Huckova D, Stanekova D. Pathogenesis of acute and persistent murine herpesvirus infection in mice. Acta Virol. 1985;29:51–60. [PubMed] [Google Scholar]
- 21.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 22.Scher M G, Unanue E R, Beller D I. Regulation of macrophage populations. III. The immunologic induction of exudates rich in Ia-bearing macrophages is a radiosensitive process. J Immunol. 1982;128:447–450. [PubMed] [Google Scholar]
- 23.Simas J P, Efstathiou S. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 1998;6:276–282. doi: 10.1016/s0966-842x(98)01306-7. [DOI] [PubMed] [Google Scholar]
- 24.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart J P, Usherwood E J, Ross A, Dyson H, Nash T. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J Exp Med. 1998;187:1941–1951. doi: 10.1084/jem.187.12.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sturzl M, Blasig C, Schreier A, Neipel F, Hohenadl C, Cornali E, Ascherl G, Esser S, Brockmeyer N H, Ekman M, Kaaya E E, Tschachler E, Biberfeld P. Expression of HHV-8 latency-associated T0.7 RNA in spindle cells and endothelial cells of AIDS-associated, classical and African Kaposi’s sarcoma. Int J Cancer. 1997;72:68–71. doi: 10.1002/(sici)1097-0215(19970703)72:1<68::aid-ijc10>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 27.Sunil-Chandra N P, Arno J, Fazakerley J, Nash A A. Lymphoproliferative disease in mice infected with murine gammaherpesvirus 68. Am J Pathol. 1994;145:818–826. [PMC free article] [PubMed] [Google Scholar]
- 28.Sunil-Chandra N P, Efstathiou S, Arno J, Nash A A. Virological and pathological features of mice infected with murine gammaherpesvirus 68. J Gen Virol. 1992;73:2347–2356. doi: 10.1099/0022-1317-73-9-2347. [DOI] [PubMed] [Google Scholar]
- 29.Sunil-Chandra N P, Efstathiou S, Nash A A. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J Gen Virol. 1992;73:3275–3279. doi: 10.1099/0022-1317-73-12-3275. [DOI] [PubMed] [Google Scholar]
- 30.Usherwood E J, Ross A J, Allen D J, Nash A A. Murine gammaherpesvirus-induced splenomegaly: a critical role for CD4 T cells. J Gen Virol. 1996;77:627–630. doi: 10.1099/0022-1317-77-4-627. [DOI] [PubMed] [Google Scholar]
- 31.van Furth R. Origin and kinetics of monocytes and macrophages. Semin Hematol. 1970;7:125–141. [PubMed] [Google Scholar]
- 32.van Furth R, Cohn Z A. The origin and kinetics of mononuclear phagocytes. J Exp Med. 1968;128:415–433. doi: 10.1084/jem.128.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Virgin H W, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Virgin H W, Tyler K L. Role of immune cells in protection against and control of reovirus infection in neonatal mice. J Virol. 1991;65:5157–5164. doi: 10.1128/jvi.65.10.5157-5164.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weck K E, Barkon M L, Yoo L I, Speck S H, Virgin H W., IV Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J Virol. 1996;70:6775–6780. doi: 10.1128/jvi.70.10.6775-6780.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weck K E, Dal Canto A J, Gould J D, O’Guin A K, Roth K A, Saffitz J E, Speck S H, Virgin H W. Murine gammaherpesvirus 68 causes large vessel arteritis in mice lacking interferon-gamma responsiveness: a new model for virus induced vascular disease. Nat Med. 1997;3:1346–1353. doi: 10.1038/nm1297-1346. [DOI] [PubMed] [Google Scholar]
- 36a.Weck, K. E., S. S. Kim, H. W. Virgin, and S. H. Speck. Submitted for publication.
- 36b.Weck, K. E., H. W. Virgin, and S. H. Speck. Unpublished data.
- 37.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi’s sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]