Abstract
We have previously identified in the human genome a family of 200 endogenous retrovirus-like elements, the HERV-L elements, disclosing similarities with the foamy retroviruses and which might be the evolutionary intermediate between classical intracellular retrotransposons and infectious retroviruses. Southern blot analysis of a large series of mammalian genomic DNAs shows that HERV-L-related elements—so-called ERV-L—are present among all placental mammals, suggesting that ERV-L elements were already present at least 70 million years ago. Most species exhibit a low copy number of ERV-L elements (from 10 to 30), while simians (not prosimians) and mice (not rats) have been subjected to bursts resulting in increases in the number of copies up to 200. The burst of copy number in primates can be dated to shortly after the prosimian and simian branchpoint, 45 to 65 million years ago, whereas murine species have been subjected to two much more recent bursts (less than 10 million years ago), occurring after the Mus/Rattus split. We have amplified and sequenced 360-bp ERV-L internal fragments of the highly conserved pol gene from a series of 22 mammalian species. These sequences exhibit high percentages of identity (57 to 99%) with the murine fully coding MuERV-L element. Phylogenetic analyses allowed the establishment of a plausible evolutionary scheme for ERV-L elements, which accounts for the high level of sequence conservation and the widespread dispersion among mammals.
Eucaryotic genomes, from humans to yeast, contain several families of reiterated sequences displaying homology to retroviruses. These elements, named long terminal repeat (LTR) retrotransposons, are provirus-like structures bordered by two LTRs that contain two (and in some cases three) of the canonical retroviral genes, i.e., the gag and pol (and possibly env) genes. In humans, these elements have been named HERVs (for human endogenous retroviruses), and several families of such elements have been characterized (reviewed in references 18, 29, and 32). These include the HERV-K family recently demonstrated to encode the virus-like particles observed by electron microscopy in human germ line tumors and a superantigen possibly involved in autoimmune type I diabetes (9, 17). The absence of a clearly identifiable env gene in some of these elements (for instance in the yeast Ty1 and the Drosophila copia elements), as well as phylogenetic analysis of the highly conserved reverse transcriptase (RT)-containing pol genes, leads to the suggestion that some of these elements might actually be the progenitors of the modern-day retroviruses, whereas other elements would be the trace of old infections (25, 27, 33).
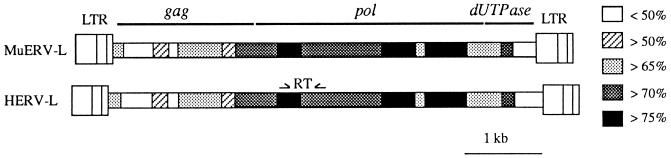
An interesting outcome of a systematic search of expressed retrovirus-like sequences in the human placenta was the discovery of a new family of moderately reiterated elements, named HERV-L, present in the human genome in 200 copies and disclosing similarities with the foamy retroviruses within their pol genes (10). No env domain could be identified within HERV-L, therefore suggesting that this element corresponds to an ancestral sequence, i.e., a retrotransposon. Interestingly, HERV-L-like sequences, so-called ERV-Ls, were also found in other mammalian species, although in general at a much lower copy number, suggesting that ERV-Ls were present before the mammalian radiation. An exception was found in mice, in which a large copy number of related sequences was also detected by a zoo blot analysis. Consistent with the recent amplification of some elements within the mouse branch, a complete proviral sequence containing fully coding gag and pol genes but no env gene has previously been isolated (2). This MuERV-L element was >70% identical to the HERV-L sequence (Fig. 1), and its gag coding sequence was found to be related to the recently cloned Fv1 resistance gene that codes for resistance to infection by leukemogenic retroviruses in some mouse strains (4).
FIG. 1.
Schematic representation and comparison of HERV-L (6,591 bp) and MuERV-L (6,471 bp) cloned elements. The positions of the oligonucleotides (see Materials and Methods) used to amplify a 360-bp fragment of the highly conserved reverse transcriptase (RT) gene are indicated.
To more precisely date the primate and murine ERV-L element bursts and the possible relationships between these elements and those in other mammals, we have now analyzed the copy number of ERV-L elements within primate and murine species and the nucleotide sequence of a central domain of the pol gene in several mammalian species. This extensive analysis allows the establishment of a plausible evolutionary scheme for these retrovirus-like endogenous elements, which would account for their high-level sequence conservation and widespread dispersion among mammals.
MATERIALS AND METHODS
DNA origin.
All DNAs were extracted from solid tissues, with the exception of the Mus dunni DNA which was extracted from fibroblasts, by standard procedures (24). Tissues from mouse laboratory strains were obtained from the animal care facility at the Institut Gustave Roussy. Feral rodent and simian tissues were provided by François Catzeflis (Institut des Sciences de l’Evolution, Montpellier, France) or given as DNA by François Bonhomme and Annie Orth (Conservatoire Génétique de Souris Sauvages [Wild Mouse Repository], UPR 9060, Université de Montpellier 2, Montpellier, France) or Jean-Louis Guénet (Institut Pasteur, Unité de Génétique des Mammifères, Paris, France). Human DNA was extracted from peripheral blood leukocytes from healthy donors. Chimpanzee DNA was extracted from peripheral blood leukocytes from healthy animals provided by Françoise Barré-Sinoussi (Institut Pasteur). Rhesus macaque (Macaca cynomolgus) DNA was prepared from tissue donated by Guy Germain (INRA, Jouy en Josas, France). DNAs from domestic animals were gifts from the Institut National de la Recherche Agronomique and Labogena (both in Jouy en Josas, France). Kangaroo (Macropus giganteus giganteus) DNA was a gift from Alexis Lecu (Zoological Institute, Vincennes, Paris, France).
Southern and slot blots.
DNA (5 μg) of each species was digested with EcoRI, fractionated on a 0.8% agarose gel, and transferred to Hybond N+ membranes (Amersham) in 10× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Membranes were subsequently hybridized with a 360-bp PCR product from an HERV-L pol gene that had been radioactively labeled with [32P]dCTP by a random priming reaction (10). One-twentieth of each digest was also loaded on Hybond N+ membranes with a slot blot apparatus (Hoefer). These blots were first probed with a radioactively labeled 2-kb PvuII cloned fragment of the FIM3 human region. This probe detects unique sequences with the same signal intensity in genomic DNA from all mammals (13a, 20) and thus acts as an internal standard to quantify DNA and allow the comparison of genomes, regardless of their sizes. Subsequently the membranes were hybridized with a 355- to 360-bp pol fragment amplified from either the human cloned element (HERV-L), the mouse cloned element (MuERV-L), or an asian rat sequence (Niviventer fulvescens clone 4, rat Nf4). Murine and rat DNAs were also hybridized with an MuERV-L LTR probe (HincII-KpnI 480-bp fragment). Copy numbers of pol and LTRs were determined by comparison with hybridizations to serial dilutions of the fragments used as probes and quantitation with a Storm 840 phosphorimager (Molecular Dynamics). Southern and slot blot hybridizations were performed at 65°C under standard conditions (7), and washes were performed in 0.5× SSC–0.1% sodium dodecyl sulfate at 65°C.
PCR and sequencing.
PCR was performed on 1 μg of genomic DNA under standard conditions with 1 U of Taq (Amersham) for 40 cycles (65°C for 1 min 30 s, 72°C for 1 min 40 s, and 93°C for 1 min 15 s), preceded by 3 cycles at a lower annealing temperature (55°C). For rabbit DNA, annealing temperatures were 60 and 50°C, respectively. The oligonucleotides used, derived from the HERV-L cloned element, were Foam51 (5′ ACTCTCGAGAAGAYAGATGGATCTTGG 3′) and Foam3′ (5′ CAGGATCCAAYCAGCMTAMTGTC 3′), where Y stands for T or C and M stands for C or G. PCR products (about 360 bp) were cloned into the pGEM-T vector (Promega) and sequenced with an Applied Biosystems model 373A automated sequencer according to the manufacturer’s instructions (Applied Biosystems-Perkin Elmer).
Phylogenetic analysis.
Multiple alignment was performed with the CLUSTAL W program (28) and refined with the editor program ED of the MUST package (21). Gaps introduced for optimal alignment were considered as missing data for the phylogenetic analysis. Phylogenetic trees were based on the analysis of nucleotide sequences with maximum likelihood (ML), maximum parsimony (MP) and distance-based methods with the programs NUCML (1), version 2.3, PAUP (26), version 3.1, and neighbor joining (NJ) (23) in the MUST package (21), version 1.0, respectively. The distances were computed with the substitution model of Kimura (16). MP trees were obtained by 10 random-addition heuristic search replicates. Bootstrap values (12) were calculated by analysis of 1,000 replicates for MP and NJ analysis. For ML analysis, only a restricted data set (34 sequences) was analyzed because of computing time limitations. Sequence alignments can be obtained from the corresponding author.
Nucleotide sequence accession numbers.
Accession no. for all sequences referred to in the text are in the EMBL data base: X89211, HERV-L; Y12713, MuERV-L; AJ233590, Mus balb 3; AJ233591, M. balb 5; AJ233592, M. balb 19; AJ233593, M. balb 20; AJ233594, M. balb 23; AJ233595, M. dunni 1; AJ233596, M. famulus 1; AJ233597, M. famulus 2; AJ233598, M. famulus 9; AJ233599, rat Rt1; AJ233600, rat Rt2; AJ233601, rat Rt3; AJ233602, rat Rt4; AJ233603, rat Rt5; AJ233604, rat Rt7; AJ233605, rat Nf1; AJ233606, rat Nf2; AJ233607, rat Nf3; AJ233608, rat Nf4; AJ233609, rat Nf5; AJ233610, rat Nf6; AJ233611, gerbil Gn2; AJ233612, gerbil Gn3; AJ233613, gerbil Gn7; AJ233614, gerbil Tg1; AJ233615, gerbil Tg5; AJ233616, gerbil Tg6; AJ233617, gerbil Tg9; AJ233618, vole Cg4; AJ233619, vole Cg7; AJ233620, vole Cg10; AJ233621, vole Cg14; AJ233622, vole Mn1; AJ233623, vole Mn3; AJ233624, vole Mn5; AJ233625, rabbit1; AJ233626, rabbit2; AJ233627, rabbit4; AJ233628, human1; AJ233629, human2; AJ233630, human4; AJ233631, human5; AJ233632, human6; AJ233633, New World monkey (NWM) As2; AJ233634, NWM As3; AJ233635, NWM As5; AJ233636, NWM As7; AJ233637, NWM As9; AJ233638, NWM Sm1; AJ233639, NWM Sm2; AJ233640, NWM Sm3; AJ233641, NWM Sm4; AJ233642, NWM Sm5; AJ233643, NWM Sm6; AJ233644, lemur Cm4; AJ233645, lemur Cm8; AJ233646, lemur Mm1; AJ233647, lemur Mm2; AJ233648, lemur Mm3; AJ233649, lemur Mm7; AJ233650, horse1; AJ233651, horse11; AJ233652, horse14; AJ233653, horse 21; AJ233654, horse24; AJ233655, horse26; AJ233656, horse27; AJ233657, donkey1; AJ233658, donkey2; AJ233659, donkey4; AJ233660, donkey6; AJ233661, pig1; AJ233662, cow1; AJ233663, cow2; AJ233664, cat1; AJ233665, dog1; AJ233666, dog2; AJ233667, dog3; AJ233668, dog5; AJ233669, dog6; AJ233670, M. saxicola 1; AJ233671, M. saxicola 3; AJ233672, M. famulus 7; AJ233673, human3; AJ233674, NWM As1.
RESULTS
ERV-L elements: conservation within placental mammals with amplification in simians and mice.
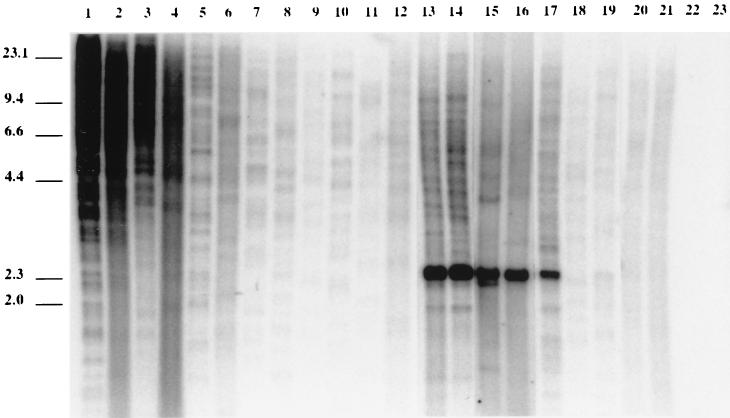
As illustrated in Fig. 2, a Southern blot analysis of DNA from a series of animal species with an HERV-L probe in the pol gene shows a dual pattern: all mammals tested display a limited number of hybridizing bands, and a limited number of species (e.g., simians [lanes 1 to 4] and mice [lanes 13 to 17]) disclose a much higher hybridization intensity. A similar pattern is observed upon rehybridization of this zoo blot with a probe corresponding to the murine homolog of HERV-L (MuERV-L 360-bp pol probe; data not shown), therefore suggesting that the variations in intensity and number of bands reflect an intrinsic difference in the number of copies. More distantly related vertebrates such as birds (chicken) and fish (salmon), as well as a nonplacental mammal (kangaroo), were negative in this Southern blot analysis as well as in a PCR analysis with oligonucleotides derived from the HERV-L pol sequence (Fig. 2; data not shown). The occurrence of hybridizing bands in six different mammalian orders tested (primates, Rodentia, Carnivora, Lagomorpha, Perissodactyla, and Artiodactyla) therefore strongly suggests that ERV-L sequences are derived from an ancestral genomic element, which was most probably present at a low copy number before the radiation of mammals. A striking feature of the natural distribution of the ERV-L sequences within (placental) mammals is the occurrence of amplifications from a limited number of copies (10 to 30) to at least 100 to 200 copies, both within the primate and the murine branches. A detailed analysis of these events has therefore been performed to date both of them and to determine, by sequence comparison, how these sequences have emerged.
FIG. 2.
Southern blot analysis of HERV-L pol-related sequences in various vertebrates. EcoRI-digested DNAs were hybridized with a 360-bp HERV-L pol fragment. Species of samples are as follows: lane 1, Homo sapiens (hominoid); lane 2, Pan troglodytes (hominoid); lane 3, Macaca cynomolgus (Old World monkey); lane 4, Alouatta seniculus (New World monkey); lane 5, Microcebus murinus (prosimian); lane 6, Cheirogaleus medius (prosimian); lane 7, Felis cattus (cat); lane 8, Canis familiaris (dog); lane 9, Oryctolagus cuniculus (rabbit); lane 10, Bos taurus (cow); lane 11, Sus scrofa domesticus (pig); lane 12, Equus caballus (horse); lane 13, C57BL/6 mouse (laboratory strain); lane 14, Mus musculus domesticus (Northern African and Near Eastern house mouse); lane 15, M. (Pyromys) saxicola (southeast Asian mouse); lane 16, M. (Coelomys) famulus (southeast Asian mouse); lane 17, Mus (Nannomys) minutoïde (African pygmy mouse); lane 18, N. fulvescens (Asian rat); lane 19, Rattus tanezumi (Asian rat); lane 20, Microcus nivalis (meadow vole); lane 21, Clethrionomys glareolus (bank vole); lane 22, Oncorhynchus sp. (salmon); and lane 23, Gallus domesticus (chicken). Numbers to the left are molecular size markers (in kilobase).
Characterization of the simian and murine bursts.
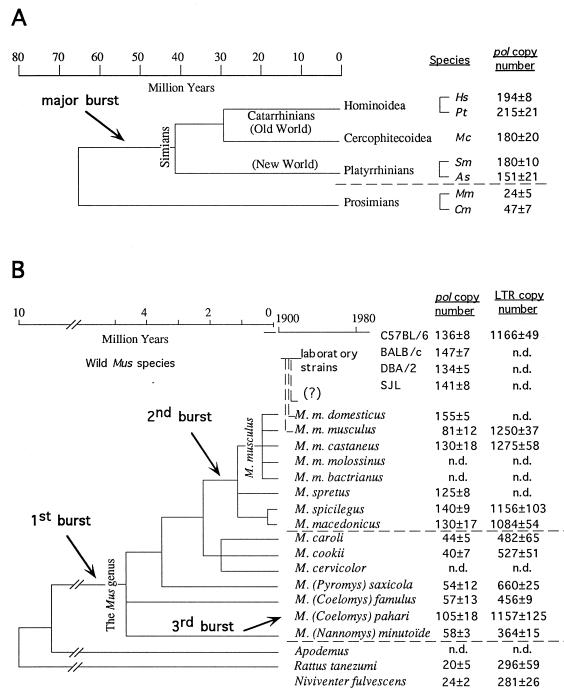
Slot blot analysis of the genomes of simian and prosimian species for the copy number of ERV-L sequences, with an HERV-L pol probe and as an internal standard a probe for the FIM3 gene (which discloses identical signals in all mammals; see Materials and Methods), showed that amplification of the ERV-L sequences within the primate branch is a very ancient event (Fig. 3A). Lack of amplification is observed only in the lemurian species, which possess 25 to 50 copies/genome, and a major burst (>100 copies) most probably took place before the separation of the Old and New World monkey branches, between 45 and 65 million years (MYrs) ago. Additional and more limited bursts could have occurred in some branches, resulting in approximately 200 copies in hominoids (Fig. 3A).
FIG. 3.
(A) Copy number determination of ERV-L elements (pol probe) in primates. The arrow indicates the major primate burst (>100 copies). Cm, Cheirogaleus medius; Mm, Microcebus murinus; As, Alovatta seniculus; Sm, Saguinus midas; Mc, Macaca cynomolgus; Pt, Pan troglodytes; Hs, Homo sapiens. (B) Copy number determination of ERV-L elements (pol probe) and ERV-L LTRs (LTR probe) for feral and laboratory mice as well as for two Asian rats. Arrows indicate the proposed dating of the two major bursts and the burst observed in M. pahari (3). Phylogenetic links were established by Sage et al. (22) and Boursot et al. (5). All data are expressed as means ± standard errors of the means (n = 3). n.d., not determined.
Amplification of the ERV-L sequences in the mouse branch has evidently occurred recently, as no amplification can be observed within rats (Fig. 2), the rodents most closely related to mice and which diverged from them only 10 to 12 MYrs ago (6). Moreover, the Southern blot of the murine DNAs shows a very intense 2.3-kb signal, which is consistent with a rather homogenous family of sequences and a recent amplification event (conversely, the absence of a single band in the primates cannot be taken as a sign of heterogeneity and might simply result from the absence of the appropriate restriction sites within the proviral sequence). Analysis of the number of ERV-L copies in several mice and rat species (Fig. 3B) was performed by slot blot analysis, as for the primate species, with an MuERV-L and a rat (Niviventer fulvescens clone 4, rat Nf4) ERV-L probe from the pol gene (360-bp probe; see Materials and Methods). The murine and rat probes yielded similar results (within 10%), and data from the mouse probe are given in Fig. 3B. Mice from the subgenus Mus (sensu stricto) all exhibit a high copy number (approximately 140 copies) with the exception of Mus musculus musculus (81 ± 12 copies) and the Asian M. caroli and M. cookii species (44 ± 5 and 40 ± 7 copies, respectively). A moderate copy number is also found in the three other subgenera of the Mus genus (sensu lato), i.e., Pyromys, Coelomys, and Nannomys, with the exception of M. (Coelomys) pahari, which possess 105 ± 18 ERV-L copies. For the Rattus genus, the number of ERV-L copies is about 25, a low copy number like that observed for most other mammals. Comparison of these data with the consensus phylogeny of the Murinae (Fig. 3B) leads to the simple proposal that two successive bursts must have occurred, resulting in, respectively, 25 and 100 newly generated copies, which can be dated at about 4 to 10 MYrs for the first one and less than 2 MYrs for the second (referred to as “mice1” and “mice2” bursts, respectively, hereafter). The independent additional burst detected in the M. (Coelomys) pahari strain (see above) as well as in M. dunni mice—which are Asian mice of the subgenus Mus (117 ± 9 copies [data not shown], as measured in this unique case from long-term cultured fibroblasts and not from animals)—would also attest to a potentially active inherited ERV-L element present in most—if not all—mouse branches. No specific additional increase in ERV-L copy number could be detected among laboratory mice.
Rehybridization of the murine slot blots with an LTR probe from MuERV-L (Fig. 3B) gave similar results for the dating of the different bursts (including the third independent burst observed in M. [Coelomys] pahari), but with LTR copy numbers approximately 10 times higher than those observed with the pol probe. This higher copy number most probably corresponds to the existence of “solo LTRs,” which are commonly observed with most retrotransposons and are generated through homologous recombination between LTRs upon transposon excision. It is also noteworthy that for the M. musculus musculus mice, the LTR copy number observed is similar to that obtained for the other mice of the M. musculus group, while the pol copy number was clearly lower (i.e., 81 ± 12 [Fig. 3B], data obtained from the inbred M. musculus musculus PWK strain and confirmed with two other inbred M. musculus musculus strains, i.e., MPB and MBS; data not shown), possibly indicating that the rate of homologous recombination is not the same in all strains. In any case, these results strongly suggest that the total number of ERV-L copies that have been integrated into the mouse and rat genomes throughout evolution should be much higher than the number detected at the present time. A search of GenBank also revealed solo HERV-L LTRs in the human genome (data not shown).
Sequence analysis of ERV-L elements.
To get further insight into the history of ERV-L elements, which were identified according to their abilities to hybridize to the MuERV-L or HERV-L probe, PCR amplification of a 360-bp domain centered in the pol gene, i.e., within the most conserved domain of retroid elements (11, 33), was performed under moderately stringent amplification conditions (see Materials and Methods). Fragments could be amplified for all placental mammals tested (6 orders and 22 species), ranging from 317 to 366 bp. Up to 12 different clones of amplified pol fragments were sequenced for each species.
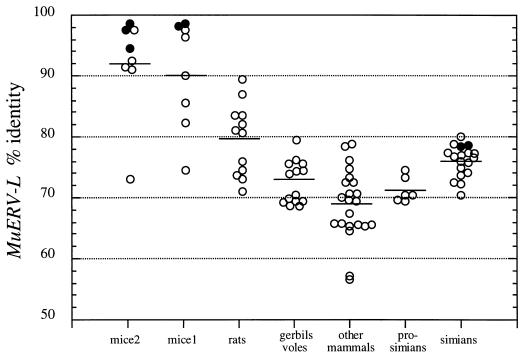
Among the 202 sequences obtained only two sequences were unrelated to pol genes. In several cases, within given species, several sequences were very similar or identical to each other (most probably corresponding to the same genomic copy), and only in those cases where similarity was less than 98% within a given species were clones considered independent and retained for the sequence analysis below. A total of 87 sequences, representing one to six sequences per species, was then retained for sequence comparison (Fig. 4). An important outcome of this comparison is that all sequences display a high percentage of identity to MuERV-L, even for distantly related species, ranging from 57% (one cow and one dog sequence) to 99% (average, 76% ± 8%), whereas all of them were only about 40% similar to sequences from other families of retroviral elements. These results are compatible with the presence of a rather homogenous family of ERV-L-related elements within placental mammals, as already suggested from the hybridization data. Among all sequences, coding sequences were found only in humans (two sequences) and mice (five sequences among four species including mice from the mice1 and mice2 groups; see above), i.e., within species where bursts have occurred. Mouse sequences make clusters with the highest percentages of identity (>90%), but a few sequences, in both the mice1 and mice2 groups, have reduced identity (but still in the range of ERV-L sequences from species without bursts) and could possibly correspond to sequences not resulting from the bursts. Sequences from other species exhibit more dispersed values (with the exception of the simian sequences), with lower averages of identity, the highest ones being observed for rats, as expected for a species closely related to mice. To further characterize these ERV-L sequences, a phylogenetic analysis was therefore performed.
FIG. 4.
Percentages of identity between 87 ERV-L mammal sequences and MuERV-L. mice2 and mice1 refer to mice subjected to 2 bursts or 1 burst, respectively. Other mammals includes cows, pigs, cats, dogs, donkeys, horses, and rabbits. Black circles indicate entirely coding sequences. The average value for each group is indicated by a horizontal line.
Phylogenetic analysis of ERV-L sequences.
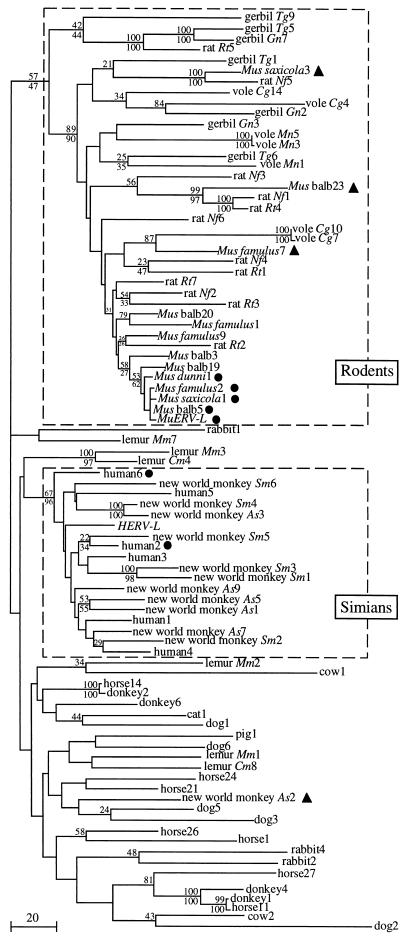
Phylogenetic trees with the above-mentioned 87 ERV-L sequences were constructed by using either the MP or NJ method, with support for individual branches investigated by bootstrap analysis (see Materials and Methods). One of the most parsimonious trees, obtained by the MP method with 1,000 bootstrap replicates, is shown in Fig. 5. Bootstrap values for the nodes are indicated when larger than 20%, together with the values obtained with the NJ method. We checked that the overall organization of the tree is conserved with the ML method, which is less sensitive to the variation of evolution rates (14) but which could be used only on a reduced sample (34 representative sequences selected) because of computing time limitations. The major feature recovered by all methods is that sequences from species subjected to bursts (i.e., simians and mice) each belong to well-defined and separated groups. As illustrated in Fig. 5, all mouse ERV-L sequences belong to a distinct rodent monophyletic group, including species without bursts (rats, voles, and gerbils). Within this rodent group, most mouse sequences are clustered and display rather short branches, indicating a low mutation rate. As expected, the shortest mouse branches are those for coding sequences (M. saxicola 1, M. balb 5, M. famulus 2, M. dunni 1, and MuERV-L; black circles), corresponding to those with the highest percentage of identity to MuERV-L in Fig. 4, while the other noncoding sequences display longer branches. This group of clustered sequences most probably corresponds to sequences with bursts that have emerged from a common ancestral progenitor. Other mouse sequences (M. famulus 7, M. balb 23, and M. saxicola 3; black rectangles) corresponding to those exhibiting the lowest percentages of identity to MuERV-L in Fig. 4 are distributed throughout the rodent group and exhibit longer branches. These sequences might derive from ERV-L elements already present before the mouse bursts and not associated with it. As for the rodent sequences, the phylogenetic analyses consistently associate the simian sequences within a monophyletic group, whereas the prosimian sequences, which have not been subjected to the simian burst, are excluded from this group. One interesting exception among simian sequences is the New World monkey As2 sequence (black triangles), which falls outside the simian group and might correspond, as proposed for some mouse sequences, to an ancestral sequence not subjected to the simian burst. The other simian sequences, clustered in the simian group, display relatively long branches, as expected for sequences derived from an old burst (>45 MYrs, according to the analysis in the previous section). Among them, only “human2” and “human6,” which display relatively shorter branches, are coding ones, consistent with a similar observation for the mouse sequences.
FIG. 5.
Phylogenetic tree of 87 ERV-L elements. BALB/c (laboratory strain), Mus balb; Rattus tanezumi, rat Rt; Niviventer fulvescens, rat Nf; Clethrionomys glareolus, vole Cg; Microtus nivalis, vole Mn; Tatera gambiana, gerbil Tg; Gerbillus nigeriae, gerbil Gn. Clone numbers are indicated. HERV-L and MuERV-L correspond to the cloned human and BALB/c mouse elements, respectively (2, 10). This tree comprises 87 taxa and is 3,663 steps long, with a consistency index of 0.323 and a retention index of 0.484. Bootstrap proportions are indicated when greater than 20% with the values for the MP and NJ methods above and below the nodes, respectively. Bar, 20 substitutions. Rodents and primate monophyletic groups are boxed. Black circles indicate coding sequences, and triangles indicate candidate burst-unrelated sequences among mouse and simian sequences.
Finally, for the other mammalian sequences, the phylogenetic analyses display a rather poorly defined organization. For instance, the five dog sequences are dispersed among mammalian sequences, and the two cow sequences are neither close to one another nor to the single type of sequence that was identified in pigs, as one would expect for elements derived from the same Artiodactyla branch. In addition, several branches are not very well supported statistically (bootstrap values, <50%) and are rather long. However, a few sequences still exhibit very high percentages of identity (they are joined by short branches): for instance, the donkey2 and horse14 (99% identity) or the donkey4, donkey1, and horse11 (96 and 99% identity) sequences. Such sequences, found in closely related species, might be inherited from a common ancestor and would correspond to so-called orthologous sequences. Orthologous sequences can also be observed among the primate and rodent sequences, with for instance lemur Mm3 and lemur Cm4 (90% identity) or rat Nf1 and rat Rt4 (95%), and within the high-copy-number simian group, the simian New World monkeys As3 and Sm4 (97%).
DISCUSSION
ERV-L sequences are widespread and ancient.
By Southern and slot blot analyses of DNAs from a series of mammals, as well as by partial sequencing of a pol domain that is highly conserved among retroid elements, we have shown that ERV-L retrovirus-like sequences are present at a low copy number among all mammals tested, including six different orders (primates, Rodentia, Carnivora, Lagomorpha, Perissodactyla, and Artiodactyla), with high sequence conservation. The occurrence of ERV-L sequences within all mammals tested strongly suggests that these sequences predate the placental-mammalian radiation (70 MYrs ago), most probably deriving from one or a few common progenitor(s). A recent search for retroviral elements within a large series of vertebrates further provides evidence for possible progenitors of spumaviruses and ERV-L-like elements in nonmammalian branches, i.e., in birds and amphibians (15). Actually, such elements still display 46 to 51% identity to MuERV-L (they were not sorted out under the rather stringent PCR and Southern blot hybridization conditions used in our investigation of salmon and chicken DNAs). Altogether, these data strongly suggest that ERV-L elements could be very ancient occupants in living species, as also shown, although not strongly documented for mammals, for the human HERV-I elements (15, 19).
ERV-L elements remained active through mammalian evolution.
We have shown that major amplifications in copy number of ERV-Ls have occurred in the course of evolution of mammals, namely, in the simian and mouse branches, thus demonstrating that some ERV-L sequences must have remained active for a long period of mammalian evolution. The strong conservation among sequences from the simian and mouse bursts further strongly suggests that the functional sequences involved in the generation of these bursts are very closely related. In addition, the mouse sequences are more closely related to the rat sequences than to the simian sequences with bursts. This strongly suggests that the mouse bursts originated from a rodent-inherited sequence, rather than from a horizontally transferred active simian element. Although the horizontal transmission of retroviral elements has been described, for instance, in the case of an endogenous type C retrovirus transferred from Mus cervicolor to primates (3) or the endogenous baboon BaEV retroviral element transferred between primate species (30), bursts of transposition, associated with the transcriptional activation of genomic elements, are common features of transposable elements, resulting in large increases in genomic copy number (31). The transposition of resident ERV-L copies is also consistent with the nature of these elements: indeed, the two cloned and entirely sequenced ERV-L elements, HERV-L and MuERV-L, both lack an envelope gene, and it is therefore likely that elements of the ERV-L family are not infectious (lack of an env gene is also consistent with ERV-L elements being ancient, primitive retrotransposons; see the introduction). Although horizontal transfers have been reported even for class II (non-RT) transposons, which are unambiguously not infectious, such as the P and Mariner elements in Drosophila (8), recent phylogenetic analyses of endogenous retrovirus-like elements in vertebrates (15) suggest that interspecies transfers should not be so frequent in the course of evolution.
ERV-L sequence conservation via transposition.
One rather striking feature of our results is the observation of a high level of sequence conservation among ERV-L elements. It is generally admitted that such sequence conservation is a common feature of functional genes, whereas noncoding sequences as well as pseudogenes diverge more rapidly, due to the lack of any selection pressure. The status of retrovirus-like elements, and more generally of transposable elements, is rather unique, as they can be considered neither classical genes nor pseudogenes. Transposons are most probably not necessary for their hosts, as their copy number is extremely variable among species and, for a given family of elements, can even be null. Conversely, their maintenance in a functional state over millions of years and the high level of sequence conservation among elements from distantly related species, as shown in the present investigation, are characteristic features of classical genes. High-level sequence conservation among transposable elements is in general interpreted in terms of horizontal transmission. According to this scheme the occurrence of almost identical elements among distantly related species is simply accounted for, and the interpretation is further supported by examples where a given element is present within some species and absent from others, independently of the expected phylogenetic relationships between these species. In case of the ERV-L elements, which were found in all mammalian species tested, such an interpretation is not likely, not only for species where bursts had occurred (see above) but also for species with a low copy number. Rather, a scheme in which sequence conservation is the unnecessary consequence of the transpositional activity of the transposable element itself (which requires both transcription activity and coding capacity), independent of any possible selective pressure imposed by the host, appears more plausible and would account for the data. According to this scheme, a functional sequence present in an ancestor of mammalian species would survive only if active, simply by generating a sufficiently high number of copies, so that despite the fact that such elements are submitted to genetic drift, as any pseudogene-like sequence, and also to elimination through transposon excision, there still remain functional copies of the founder element. Evidence for the transpositional activity of ERV-L elements is provided by the occurrence of bursts in both the primate and mouse branches, which resulted in large increases in ERV-L copy numbers. In fact, the transpositional activity should be even more intense than suspected from the simple assay for pol-containing elements. We found an almost 10-fold excess of LTR-hybridizing elements within both humans and mice, most likely corresponding to solo LTRs generated by the excision of proviruses via homologous LTR recombination. A similar excess was also observed in rats, i.e., for a species without a burst. The continuous replacement of excised transposable elements by newly transposed copies from functional ERV-L elements would then simply account for the high level of sequence conservation as observed for genes. Conversely, one can expect that the lack of transpositional activity, for instance, because of transcriptional silencing, would result in the elimination of functional copies of the element within a given species. In this respect, it is possible that some mammalian branches, in which we could not detect any transposition burst nor sequences with strong identity with functional ERV-L sequences, are dead ends for ERV-L elements. Interestingly, the high level of sequence conservation among ERV-L elements is reminiscent of the homogenization process observed in several multigene families, including pseudogene families, which has been termed concerted evolution (13). This phenomenon has been interpreted as resulting from the partial or total exchange of a sequence between copies by gene conversion or unequal crossing over (that is, concerted evolution sensu stricto) or alternatively from the net result of turnover mechanisms, implying some kind of equilibrium between the insertion of new copies and the removal of older ones. In the case of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) pseudogene family, it was indeed shown that pseudogenes do not evolve independently but are subjected to homogenization and that pseudogene evolution is driven in the long term by the active founder gene (13). Such dual effects, i.e., passive homogenization between pseudogenes and active homogenization driven by the founder gene—via the generation of new pseudogenes and elimination of others—would also fit for a transposable element, with the only—but fundamental—difference that the founder gene would not be, in the case of transposons, a gene submitted to selective pressure for its maintenance in the genome, but simply one—or a few—copies that were active when they entered the host organism.
In conclusion, the present investigation has provided evidence that some ERV-L elements have entered genomes before the mammalian radiation and that they have maintained at least some copies of the element in a functional state, most probably by the simple virtue of their capacity to replicate and generate multiple safety copies within the host genomes, so that these otherwise dispensable genetic elements have escaped genetic drift and elimination.
ACKNOWLEDGMENTS
We are grateful to F. Catzeflis, F. Bonhomme, A. Orth, and J. L. Guénet for providing DNA and/or tissues of primates and rodents and for helpful suggestions and to P. Dessen for assistance and discussions at the Infobiogen GIS.
This work was supported by the CNRS (ACC-SV3), by grants from the ARC and the Ligue Nationale contre le Cancer, and by a fellowship to L.B. from the Fondation pour la Recherche Médicale.
REFERENCES
- 1.Adachi J, Hasegawa M. Computer monographs. Vol. 28. Tokyo, Japan: Institute of Statistical Mathematics; 1996. MOLPHY: programs for molecular phylogenetics based on maximum likelihood, version 2.3. [Google Scholar]
- 2.Bénit L, de Parseval N, Casella J-F, Callebaut I, Cordonnier A, Heidmann T. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J Virol. 1997;71:5652–5257. doi: 10.1128/jvi.71.7.5652-5657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benveniste R E, Callahan R, Sherr C J, Chapman V, Todaro G J. Two distinct endogenous type C viruses isolated from the asian rodent Mus cervicolor: conservation of virogene sequences in related rodent species. J Virol. 1977;21:849–862. doi: 10.1128/jvi.21.3.849-862.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Best S, le Tissier P, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 5.Boursot P, Auffray J C, Britton-Davidian J, Bonhomme F. The evolution of house mice. Annu Rev Ecol Syst. 1993;24:119–152. [Google Scholar]
- 6.Catzeflis F M, Aguilar J P, Jaeger J J. Muroid rodents: phylogeny and evolution. Tree. 1992;7:122–126. doi: 10.1016/0169-5347(92)90146-3. [DOI] [PubMed] [Google Scholar]
- 7.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark J B, Kidwell M G. A phylogenetic perspective on P-transposable element evolution in Drosophila. Proc Natl Acad Sci USA. 1997;94:11428–11433. doi: 10.1073/pnas.94.21.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conrad B, Weissmahr R N, Böni J, Arcari R, Schüpbach J, Mach B. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell. 1997;90:303–313. doi: 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 10.Cordonnier A, Casella J-F, Heidmann T. Isolation of novel human endogenous retrovirus-like elements with foamy virus-related pol sequence. J Virol. 1995;69:5890–5897. doi: 10.1128/jvi.69.9.5890-5897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doolittle R F, Feng D F, Johnson M S, McClure M A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;40:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Meunier P, Etienne-Julan M, Fort P, Piechaczyk M, Bonhomme F. Concerted evolution in the GAPDH family of retrotransposed pseudogenes. Mamm Genome. 1993;4:695–703. doi: 10.1007/BF00357792. [DOI] [PubMed] [Google Scholar]
- 13a.Gisselbrecht, S. Personal communication.
- 14.Hasegawa M, Fujiwara M. Relative efficiencies of the maximum likelihood, maximum parsimony, and neighbor-joining methods for estimating protein phylogeny. Mol Phylogenet Evol. 1993;2:1–5. doi: 10.1006/mpev.1993.1001. [DOI] [PubMed] [Google Scholar]
- 15.Herniou E, Martin J, Miller K, Cook J, Wilkinson M, Tristem M. Retroviral diversity and distribution in vertebrates. J Virol. 1998;72:5955–5966. doi: 10.1128/jvi.72.7.5955-5966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 17.Löwer R, Boller K, Hasenmaier B, Korbmacher C, Mueller-Lantzsch N, Löwer J, Kurth R. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci USA. 1993;90:4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Löwer R, Löwer J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin J, Herniou E, Cook J, Waugh O’Neill R, Tristem M. Human endogenous retrovirus type I-related viruses have an apparently widespread distribution within vertebrates. J Virol. 1997;71:437–443. doi: 10.1128/jvi.71.1.437-443.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen V C, Fichelson S, Gross M S, Sola B, Bordereaux D, de Tand M F, Guilhot S, Gisselbrecht S, Frézal J, Tambourin P. The human homologues of Fim1, Fim2/cFms, and Fim3, three retroviral integration regions involved in mouse myeloblastic leukemias, are respectively located on chromosomes 6p23, 5q33, and 3q27. Hum Genet. 1989;81:257–263. doi: 10.1007/BF00279000. [DOI] [PubMed] [Google Scholar]
- 21.Philippe H. MUST, a computer package of management utilities for sequences and trees. Nucleic Acids Res. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sage J, Yuan L, Martin L, Mattei M G, Guénet J L, Liu J G, Hoög C, Rassoulzadegan M, Cuzin F. The Sycp1 loci of the mouse genome: successive retropositions of a meiotic gene during the recent evolution of the genus. Genomics. 1997;44:118–126. doi: 10.1006/geno.1997.4832. [DOI] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Shih A, Misra R, Rush M G. Detection of multiple, novel reverse transcriptase coding sequences in human nucleic acids: relation to primate retroviruses. J Virol. 1989;63:64–75. doi: 10.1128/jvi.63.1.64-75.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swofford D L, Nei M. PAUP: phylogenetic analysis using parsimony, version 3.1.1. Champaign, Ill: Illinois Natural History Survey; 1987. [Google Scholar]
- 27.Temin H M. Origin of retroviruses from cellular moveable genetic elements. Cell. 1980;21:599–600. doi: 10.1016/0092-8674(80)90420-1. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urnovitz H B, Murphy W H. Human endogenous retroviruses: nature, occurrence, and clinical implications in human disease. Clin Microbiol Rev. 1996;9:72–99. doi: 10.1128/cmr.9.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Kuyl A C, Dekker J T, Goudsmit J. Distribution of baboon endogenous virus among species of African monkeys suggests multiple ancient cross-species transmissions in shared habitats. J Virol. 1995;69:7877–7887. doi: 10.1128/jvi.69.12.7877-7887.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waugh O’Neill R J, O’Neill M J, Marshall Graves J A. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature. 1998;393:68–72. doi: 10.1038/29985. [DOI] [PubMed] [Google Scholar]
- 32.Wilkinson D A, Mager D L, Leong J A C. Endogenous human retroviruses. In: Levy J A, editor. The Retroviridae. New York, N.Y: Plenum Press; 1994. pp. 465–535. [Google Scholar]
- 33.Xiong Y, Eickbush T H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]