Abstract
Two chimpanzees (Ch1535 and Ch1536) became infected with hepatitis C virus (HCV) following intrahepatic inoculation with RNA transcribed from a full-length cDNA clone of the virus. Both animals were persistently infected and have been followed for 60 weeks. They showed similar responses to infection, with transient liver enzyme elevations and liver inflammatory responses, which peaked at weeks 17 (Ch1535) and 12 (Ch1536) postinoculation (p.i.). Antibody responses to structural and nonstructural proteins were first detected at weeks 13 (Ch1535) and 10 (Ch1536) p.i. Serum RNA titers increased steadily during the first 10 to 13 weeks but decreased sharply in both animals following antibody and inflammatory responses. Despite direct evidence of humoral immune responses to multiple viral antigens, including hypervariable region 1 (HVR1), both animals remained chronically infected. Detailed sequence analysis of serum HCV RNA revealed no change in the majority HVR1 sequence in Ch1535 and a single-amino-acid mutation in Ch1536, with very little clonal variation in either animal. Full-length genome analysis at week 60 revealed several amino acid substitutions localized to antigens E1, E2, p7, NS3, and NS5. Of these, 55.6 and 40% were present as the majority sequence in serum RNA isolated at week 26 p.i. (Ch1535) and week 22 p.i. (Ch1536), respectively, and could represent immune escape mutations. Mutations accumulated at a rate of 1.57 × 10−3 and 1.48 × 10−3 nucleotide substitutions/site/year for Ch1535 and Ch1536, respectively. Taken together, these data indicate that establishment of a persistent HCV infection in these chimpanzees is not due to changes in HVR1; however, the possibility remains that mutations arising in other parts of the genome contributed to this persistence.
Hepatitis C virus (HCV), first identified in 1989 (12), is the major causative agent of parenterally transmitted non-A, non-B hepatitis. Infection occurs primarily through blood or blood-derived products but also through nonparenteral or inapparent parenteral exposure. It frequently leads to chronic hepatitis and cirrhosis and is associated with the development of hepatocellular carcinoma (50).
The viral particle consists of a nucleocapsid containing a positive-sense, single-stranded RNA genome of approximately 9,500 nucleotides (nt) (13) surrounded by an envelope derived from host membranes into which are inserted the virus-encoded glycoproteins (E1 and E2). The genome contains highly conserved untranslated regions (UTR) at both the 5′ and 3′ termini (24, 34, 58, 59) which flank a large translational open reading frame encoding a polyprotein of approximately 3,000 amino acids (13, 30, 57). This polyprotein is processed by both cellular and viral proteases to produce the structural (core, E1, and E2) and nonstructural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins of the virus (23, 26).
The mechanisms leading to viral persistence, which is associated with the more severe forms of liver disease, are as yet undefined. Any single HCV isolate exists as a quasispecies with sequence variability throughout the RNA genome (8, 56), providing a reservoir of variants any one of which can become dominant during the course of infection. This genetic variation could lead to evasion of the host immune response through the selection of neutralizing antibody or cytotoxic T-lymphocyte (CTL) escape mutants and thereby to the establishment of persistent infection. Both types of escape mutants have been reported in HCV infections. One region showing a high degree of variability, termed hypervariable region 1 (HVR1), is located in the N terminus of the E2 protein, between amino acids 384 and 410 of the polyprotein (26, 31, 41, 64), and evidence suggests that this region evolves more rapidly in vivo than the rest of the viral genome (40, 46). Observations that antibody recognizing this region changes in its specificity during the course of a chronic infection (32, 52, 60, 66) and that HVR1 appears to contain a neutralizing epitope (19, 53, 54, 70) suggest that HVR1 is subject to immune pressure and is important in the maintenance of persistent infections. In addition to antibodies, both circulating and liver-infiltrating CTLs directed to multiple gene products have been detected in chronically infected patients and chimpanzees (6, 16, 21, 27, 35–39, 49). These CTLs recognize epitopes that lie mainly within relatively conserved regions of the genome. However, escape variants have been identified and in some cases are thought to be CTL antagonists (10, 28, 63).
The quasispecies nature of infectious HCV inocula derived primarily from patient sera has made it difficult to characterize the effect of the host immune response on the molecular evolution of the virus and its persistence. The question of whether new dominant viral species that arise during infection are due to advantageous mutations that occur naturally during virus replication or are already present at a low level in the original material has been impossible to address without pure virus preparations consisting of a single clonal population.
We previously showed that HCV could be transmitted to chimpanzees and cause disease through intrahepatic inoculation of RNA transcribed from a full-length cDNA clone (33). The animals developed viremia, elevated liver transaminase levels, and serological responses to HCV antigens. The patterns of these responses were indistinguishable from those arising in animals following infection by intravenous inoculation of known infectious serum. The two chimpanzees originally transfected with the HCV RNA have both developed persistent infections and have now been followed for 14 months (60 weeks). The disease progression, antibody response, and viral evolution have been documented at the molecular and histological levels. In this study we have followed these two chimpanzees by serial liver biopsies, serum alanine amino transferase (ALT) levels; HCV-specific RNA levels; antibody to several HCV antigens, including HVR1; and extensive sequence analysis of the genome, with particular emphasis on the sequence of HVR1. In this study, we provide the first analysis of HCV evolution in vivo following inoculation with a single viral sequence.
MATERIALS AND METHODS
Chimpanzees.
Chimpanzees Ch1535 and Ch1536 were inoculated with full-length uncapped RNA transcripts by direct intrahepatic injection (33). The cDNA clone had been assembled by using early-acute-phase plasma, designated H77, obtained in 1977 from patient H (46). Both Ch1535 and Ch1536 were seronegative for all known hepatitis viruses, were negative for HCV RNA, and had normal liver enzyme levels (33) prior to inoculation. Serum was collected from these animals at weekly intervals until week 22 postinoculation (p.i.), after which samples were collected monthly. Liver biopsy specimens were collected for RNA extraction and histological analysis at weekly intervals until week 17 and then four times over the next 8 months. The biopsy specimens were formalin fixed and embedded in paraffin before being sectioned and were evaluated semiquantitatively for inflammatory features on a scale from 0 to 4 (representing none, minimal, mild, moderate, and marked). Peripheral blood mononuclear cells (PBMCs) were collected at weeks 46 and 51 for RNA extraction.
Preparation of PBMCs.
Ten milliliters of whole blood was mixed with an equal volume of phosphate-buffered saline (PBS) and layered onto 15 ml of Ficoll-Paque (Pharmacia, Uppsala, Sweden) in a 50-ml conical tube. Samples were centrifuged in a Beckman GPKR centrifuge at 2,500 rpm for 30 min. The white buffy coat was removed, and the PBMCs were washed three times in PBS. The final pellet was resuspended in RPMI 1640 containing 7% dimethyl sulfoxide (Sigma, St. Louis, Mo.) and stored at −80°C until use.
RNA extractions.
Total RNA was prepared from 100 μl of serum, approximately 1 mm3 of liver biopsy specimen, or 106 PBMCs by using TRIzol (Life Technologies, Gaithersburg, Md.) according to the manufacturer’s instructions. The RNA pellets were resuspended in 10 μl of RNasin-dithiothreitol (DTT) water (0.2 U of RNasin/μl, 10 mM DTT) (Promega, Madison, Wis.) and stored at −80°C until use. Negative controls, in the form of serum from Ch1535 and Ch1536 prior to inoculation or from uninoculated chimpanzees, were included in all extractions.
RT-PCR.
Reverse transcription (RT) of purified RNA was carried out with the First-Strand cDNA synthesis kit (Pharmacia) according to the manufacturer’s instructions and 10 pg of random hexamers in a 15- or 33-μl reaction volume. Nested PCRs, consisting of 40 cycles each, were carried out on the RT products with the Expand High Fidelity PCR system (Boehringer Mannheim, Indianapolis, Ind.). The 50-μl reaction mixtures contained 5 μl of RT reaction mixture; 300 μM (each) dATP, dCTP, dGTP, and dTTP; and 200 μM (each) forward and reverse primers.
Amplification of HVR1 was carried out with the following forward and reverse primers: outer sense primer E1/1314, AACTGGTCCCCTACGGCAGCGTTGGTGGTA (nt 1314 to 1343); outer antisense primer E2/1754, AGGACAGCCTGAAGAGTTGAATTTGTGGCG (nt 1754 to 1724); inner sense primer E1/1345, CTCAGCTGCTCCGGATCCCACAAGCCATCA (nt 1345 to 1374); and inner antisense primer E2/1680, ATTTGTGCTGATAGAAGAGCCCTGCTAACCAACCGGTGTT (nt 1680 to 1641). The HCV genome was amplified by nested PCR as overlapping 400- to 1,500-bp fragments. The primer sequences were based on the cDNA consensus clone (designated p90) used for the synthesis of the original RNA transcripts inoculated into Ch1535 and Ch1536 (33).
Cloning and sequence analysis of PCR products.
Following PCR, the samples were analyzed by ethidium bromide-stained agarose gel electrophoresis. For cloning, 2 μl of the 50-μl PCR mixture were ligated into the TA cloning vector (Invitrogen Corporation, Carlsbad, Calif.) according to the manufacturer’s instructions. For sequencing, PCR products generated from serum RNA were purified on Qiaquick purification columns (Qiagen Inc., Santa Clarita, Calif.). Recombinant plasmids from bacterial transformants were prepared with the Qiagen Qiawell 8 plasmid kit and sequenced. Sequence analysis was carried out on PCR products and clones with the PRISM dye terminator sequencing kit and the PRISM DNA sequencer (Perkin-Elmer [PE]-Applied Biosystems, Foster City, Calif.) according to the manufacturer’s instructions.
RNA quantification with fluorogenic probe.
RNA levels in serum samples were quantified with the PRISM 7700 sequence detection system (PE-Applied Biosystems) (5, 22, 25). Duplicate RNA samples were amplified with the TaqMan EZ RT-PCR kit and a TaqMan fluorogenic probe labeled with FAM (6-carboxy-fluorescein) and TAMRA (6-carboxytetramethylrhodamine) (PE-Applied Biosystems) at the 5′ and 3′ ends, respectively. Reactions were performed with 50-μl mixtures containing the following: 5 μl of template RNA; 10 μl of 5× TaqMan EZ buffer; 2.5 mM Mn(OAc)2; 300 μM (each) dATP, dCTP, dGTP, and dTTP; 300 nM (each) primers TaqMan/243 and TaqMan/390; 100 nM fluorogenic probe (TaqMan/336); and 5 U of recombinant Tth (rTth) DNA polymerase. The primer and probe sequences were as follows: sense primer TaqMan/243, AAGACTGCTAGCCGAGTAGTGTT (melting temperature [Tm] = 60°C) (nt 243 to 265); antisense primer TaqMan/390, GGTTGGTGTTACGTTTGGTTT (Tm = 59°C) (nt 390 to 370); and probe TaqMan/336, TGCACCATGAGCACGAATCCTAAA (Tm = 69°C) (nt 336 to 359). Primer and probe sequences and Tm values were determined with Primer Express (PE-Applied Biosystems).
RNA standards, run in duplicate in every reaction, were prepared from RNA transcripts produced with the RiboMAX T7 large-scale RNA production system (Promega) and the p90 full-length plasmid linearized with BsmI. The 100-μl reaction mixture, containing 20 μl of 5× T7 buffer; 7.5 mM (each) rATP, rCTP, rGTP, and rUTP; 2 μg of DNA; and 10 μl of enzyme mixture was incubated at 37°C for 2 h and then treated with 10 U of RNase-free DNase (Promega) for 30 min at 37°C. Following TRIzol extraction, the washed RNA was resuspended in RNasin-DTT water. The concentration was calculated by optical density readings at 260 nm (OD260), and the RNA was stored in aliquots of 100 ng in liquid nitrogen until use. Standards of 105, 104, 103, 102, 101, and 100 RNA copies/μl were prepared by serial dilution of the stock RNA in RNasin-DTT water containing 10 μg of tRNA/ml and stored at −80°C. The absence of DNA at these dilutions was confirmed by performing control reactions omitting the RT step.
Enzyme-linked immunosorbent assay (ELISA) for E1E2-specific antibodies.
Recombinant HCV E1, E2, and p7 (amino acids 170 to 809) was produced by cloning this region of the p90 consensus clone into a Sindbis virus expression system (7). Proteins were purified from infected BHK cells by lectin-affinity chromatography as previously described (51). Chimpanzee and human sera, diluted 1:10 and 1:50, respectively, were tested for HCV-specific antibodies in 96-well plates (Maxisorp immunoplates; Nunc Inc., Naperville, Ill.) with 1 μg of purified protein/well, as previously described (51). Mean OD values were expressed as P/N ratios calculated by dividing the OD405 for test sera by that obtained for preimmune serum. The cutoff value was taken as a P/N value of 2.
Peptide ELISA for the detection of HVR1-specific antibodies.
Biotinylated peptides covering amino acids 384 to 410 of the HCV polyprotein were synthesized on a Millipore 950 PepSynthesizer.
The ELISA was based upon that described by van Doorn et al. (61) with some modifications. Specifically, sera were diluted 1:10 in blocking buffer (PBS containing 5% nonfat milk, 0.05% Tween 20, and 4% goat serum). Following incubation with horseradish peroxidase-labeled goat anti-human immunoglobulin (KPL, Gaithersburg, Md.) diluted 1:1,000 in blocking buffer, samples were developed with 100 μl of 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) peroxidase substrate (KPL, Gaithersburg, Md.) at room temperature. The reactions were stopped by the addition of 1% sodium dodecyl sulfate, and the OD was read at 405 nm. Mean OD values from duplicate samples were expressed as P/N ratios, calculated by dividing the OD405 for test sera by that obtained for preimmune serum from the chimpanzee. The cutoff value was taken to be a P/N value of 2.
Blocking HVR1 antibodies.
Serum was incubated with 1 μg of either HVR1-p90 or H8, a negative peptide, at 37°C for 60 min. The serum was then used in a HVR1 or E1E2 antibody ELISA as described above.
RESULTS
Two chimpanzees, Ch1535 and Ch1536, were inoculated as previously described to test the infectivity of RNA transcribed in vitro from cloned HCV cDNA (33) generated from a patient serum sample (designated H77) (2, 20). For the purposes of this study the infectious cDNA clone will be referred to as p90.
Monitoring of chimpanzees.
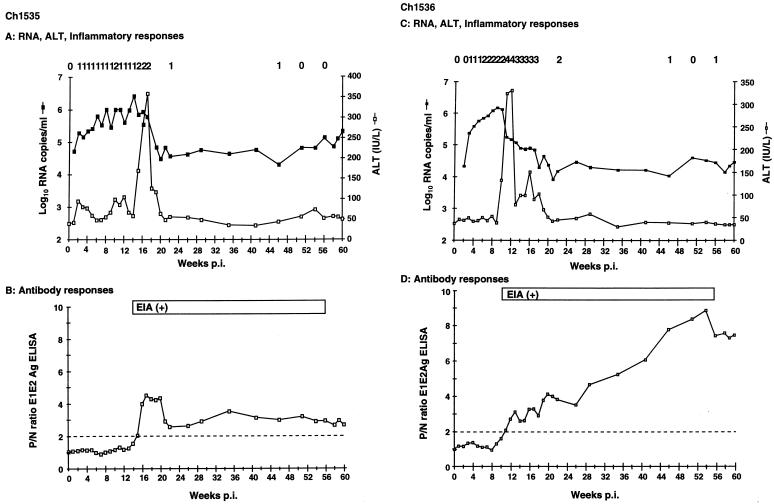
Analysis of ALT levels, liver histology, and serum RNA copy number, using the PRISM 7700 sequence detection system, showed that both animals developed similar and classical responses to HCV infection (Fig. 1A and C). Serum RNA was detectable within 1 (Ch1535) or 2 (Ch1536) weeks p.i., with titers beginning at between 104 and 105 copies/ml and reaching peaks of greater than 106 copies/ml at 14 (Ch1535) and 9 (Ch1536) weeks p.i. In both animals the increase in ALT levels correlated with a decrease in serum RNA. ALT levels returned to normal by week 21; liver enzymes were monitored throughout the study, but no further elevations were observed.
FIG. 1.
Clinical, virologic, and serologic responses in Ch1535 (A and B) and Ch1536 (C and D) following inoculation with RNA transcripts representing the HCV infectious clone. (A and C) Serum RNA, ALT, and inflammatory responses (numbers across the top of the graph), graded from 0 to 4 (representing none, minimal, mild, moderate, and marked). (B and D) Antibody responses to recombinant E1E2 antigen (open squares) and positivity in commercial EIAs [EIA (+)]. Anti-E1E2 antibody responses are represented as P/N ratios, with the cutoff of 2 (twice the value of the preserum sample) indicated by the dotted line.
Inflammatory changes observed in the liver biopsies generally followed the pattern of liver enzymes in terms of peak severity and endurance (Fig. 1A and C). Early biopsies (at 2 to 3 weeks p.i.) showed focal apoptosis of liver cells and mitotic activity without evidence of inflammation. Mild piecemeal necrosis first appeared in Ch1535 at week 10 and in Ch1536 at week 6, with maximum inflammatory responses between weeks 15 and 17 (Ch1535) and 11 and 12 (Ch1536). Inflammation in Ch1535 never advanced beyond the mild level initially seen at week 10, while that in Ch1536 was characterized by numerous foci of lobular inflammation, frequent apoptotic hepatocytes, and mild piecemeal necrosis in many portal areas (data not shown). Similar to the ALT levels, inflammation in both animals had decreased by week 22 and was scored as minimal or absent in subsequent biopsies.
Seroconversion, determined by commercial enzyme immunoassay (EIA) and E1E2 ELISA, took place at 13 to 15 (Ch1535) and 10 to 11 (Ch1536) weeks p.i. (Fig. 1B and D). Initially anti-E1E2 antibody levels were comparable in both animals. Whereas the level in Ch1535 began to decrease at week 21, remaining at a P/N of 3, that in Ch1536 steadily increased to a P/N of more than 8 at week 60 (Fig. 1B and D). In spite of this elevation in serum antibody, Ch1536 remained positive for HCV RNA, although at a level approximately 10-fold lower than that of Ch1535.
Despite a sharp decrease in serum HCV RNA levels following the immune responses, neither Ch1535 nor Ch1536 completely cleared the infection but became persistently infected, with RNA titers remaining at approximately 105 (Ch1535) and 104 (Ch1536) copies/ml until the end of the study at 60 weeks p.i. This was in the presence of detectable humoral immune responses to several HCV antigens determined by an anti-E1E2 antibody ELISA and two commercial assays, HCV EIA 2.0 (Abbott Laboratories, Abbott Park, Ill.) and HCV 3.0 ELISA (Ortho Diagnostic Systems Inc., Raritan, N.J.) (Fig. 1B and D). The Abbott and Ortho assays contain recombinant proteins corresponding to regions of the HCV core, NS3, and NS4 and to core, NS3, and NS5, respectively.
Antibody to HVR1.
A peptide ELISA was developed to assess the antibody response to HCV HVR1. We wished to establish how antibody to this region correlated with antibody responses to other viral antigens and how it might correlate with the evolution of HVR1 or development of escape mutants.
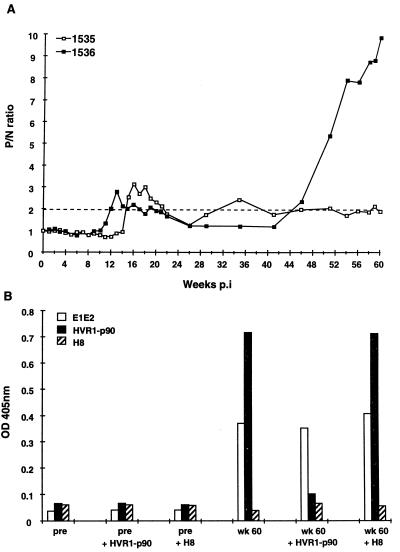
Figure 2A shows the antibody responses to the p90 HVR1 sequence measured for Ch1535 and Ch1536. The timing of the responses to this epitope in both animals paralleled those to full-length E1E2 and nonstructural proteins (Fig. 1B and D). The first detectable antibody appeared 1 to 2 weeks after the peak in serum RNA, with the response in Ch1536 preceding that in Ch1535 by 3 weeks. HVR1 antibody levels decreased during subsequent weeks in both animals, at some points dropping below the cutoff P/N value of 2. Ch1536 began to show an increase in HVR1-specific antibody at week 46; anti-HVR1 antibody in 1535 remained low or undetectable.
FIG. 2.
(A) HVR-1-specific antibody responses in Ch1535 and Ch1536 as determined by peptide ELISA, with a 20-mer peptide representing the predicted amino acid sequence encoded by the HCV infectious clone p90 (HVR1-p90). The values are expressed as P/N, with the cutoff of 2 indicated by the dotted line. The data are representative of three separate experiments. (B) ELISA blocking assay. HVR1-specific antibodies were blocked in serum from Ch1536 obtained at week 60 p.i. HVR1-p90 represents the sequence expressed by the infectious clone. H8 represents a negative peptide that does not cross-react with antibody to HVR1-p90 and is not recognized by serum from Ch1536. The data are representative of three separate experiments. pre, preimmune serum.
To determine whether the increased anti-E1E2 antibody titer in Ch1536 after week 46 was directed against epitopes other than that represented by the HVR1-p90 peptide, a blocking experiment was performed with Ch1536 serum from week 60 and preimmune serum. Figure 2B shows that without preincubation with peptide week 60 serum from Ch1536 reacted strongly with both E1E2 antigen (Ag) and peptide HVR1-p90 but showed no cross-reaction with a negative peptide, H8. Preincubation with peptide HVR1-p90 reduced the reactivity to HVR1-p90 to almost background level, whereas the reactivity to the E1E2 Ag was only minimally affected. Preincubation with peptide H8 had no effect on the reactivity of the serum with either peptide HVR1-p90 or E1E2 Ag. Therefore, even though the serum had been depleted of antibodies specific to HVR1-p90, there remained a significant titer of antibodies to other E1E2 epitopes, which contributed to the elevated E1E2 antibody levels observed in Ch1536.
Sequence analysis of HCV HVR1.
Evidence suggests that HVR1 is a target for neutralizing antibodies which drive selection for new HVR1 sequences that represent immune escape variants (32, 52, 53, 60, 66). Based on sequence analysis of this region during chronic infections in humans and chimpanzees, it has been hypothesized that the emergence of such HVR1 variants may be required for the establishment of persistence (32, 60, 66). We analyzed variation in HVR1 in both animals by sequencing a 233-bp RT-PCR product encompassing the HVR1 sequence plus 60 to 80 5′ and 3′ flanking nucleotides. In addition to examining the HVR1 sequences present in HCV RNA from serum, we also analyzed RNA obtained from liver biopsy specimens and PBMCs.
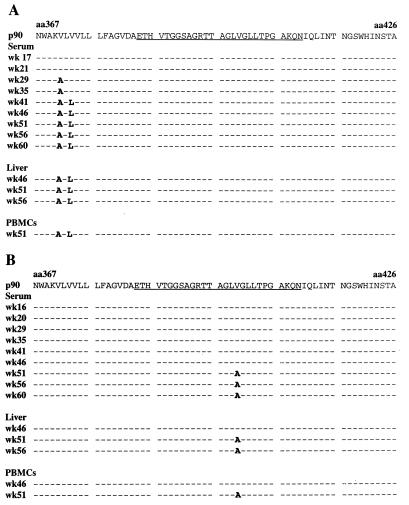
The predicted HVR1 amino acid sequences obtained from serum-, liver-, and PBMC-derived RNAs from Ch1535 and Ch1536 are shown in Fig. 3. This region of the genome was amplified and sequenced for all serum samples from the animals; however, for clarity, the data shown are from approximately monthly intervals and represent the sequence obtained from the PCR product. Surprisingly, there was very little change in the overall HVR1 sequence in either chimpanzee. Ch1535 had no substitutions in HVR1 either at the nucleotide or the amino acid level, although two substitutions were detected upstream of HVR1 in the E1 signal sequence at weeks 29 (V371A) and 41 (V373L) (Fig. 3A) which were maintained throughout the remainder of the study. The wild-type sequence was detected in Ch1536 until week 51 (Fig. 3B), at which point a single-nucleotide mutation leading to a V400A substitution was observed. Clonal analysis showed that this represented 63.6% (7 of 11) of the sequences; the remaining 36.4% (4 of 11) of the clones encoded the wild-type sequence. This mutation persisted as the majority sequence, although the wild-type HVR1 was still present in 30% of the clones analyzed at week 60. Sequence analysis of HVR1 PCR products obtained from liver biopsy specimens and PBMCs correlated with the sequences observed in the serum (Fig. 3A and B); no product could be obtained for RNA isolated from the PBMCs of Ch1535 at week 46.
FIG. 3.
Predicted amino acid sequence of HVR1 in Ch1535 (A) and Ch1536 (B) serum, liver, and PBMCs at selected time points following inoculation with the infectious clone. The sequence p90 corresponds to the predicted amino acid sequence encoded by the RNA transcripts used for inoculations. HVR1 is underlined. The sequence displayed is the majority sequence as determined by analysis of purified PCR product. Dashes represent identical residues. aa, amino acid.
The above data represents the majority sequence obtained from PCR products. To determine the degree of heterogeneity in this region, products amplified from serum RNA obtained throughout the study were cloned and sequenced. Conditions were used such that a minimum of 60 cDNA molecules were amplified to ensure an adequate representation of sequences. In all samples throughout the study there was little, if any, variability. Point mutations at the nucleotide level, some of which resulted in amino acid substitutions, were observed in individual clones in both animals at various sample dates. However, none of these were detected at more than one time point and they were often located outside of the HVR1 coding region. At week 60 p.i., 4 of 39 clones (10.3%) from Ch1535 contained five point mutations leading to amino acid sequences different from the majority sequence shown in Fig. 3A; three of these (E384K, A392T, and L403P) were located in HVR1. All clones contained the new sequence at amino acids 371 and 373 which had emerged at weeks 29 and 41, respectively (Fig. 3A). In Ch1536 the clonal variation was greater: 15 of 30 clones (50%) contained amino acid substitutions different from the majority detected in serum RNA at week 60 p.i. (Fig. 3B). However, 10 of these contained the wild-type V400 residue. Two clones contained a H386R substitution, and the remaining mutations (T385A and A407V) were located in individual clones. In the analysis of HVR1 no mutations common to both animals were detected at either the amino acid or the nucleotide level.
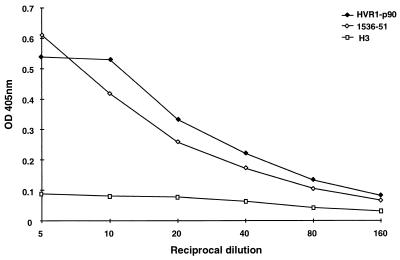
The V400A mutation observed in Ch1536 at week 51 followed a sharp increase in HVR1-specific antibody (Fig. 2). To determine whether this substitution created an antibody escape mutant, the peptide representing amino acids 384 to 410 of this isolate (designated 1536-51) was synthesized and tested against sera from Ch1535 and Ch1536. Although serum samples from Ch1536 taken throughout the study showed identical reactivities to peptides 1536-51 and HVR1-p90 (data not shown), the V400A mutation could have arisen early in infection, without detection in sequence analysis, and generated specific antibody to this peptide. However, the reactivity of serum from Ch1535 at week 17, in which the 1536-51 sequence has never been detected, was also assessed and found to be comparable for the two peptides (Fig. 4). These data suggest that this is not an antibody escape mutant.
FIG. 4.
Reactivity of serum from Ch1535 at week 17 p.i. following serial dilution with peptides p90, 1536-51, and H3 (negative or nonreactive peptide sequence).
HCV genome sequence.
The major HCV genome sequence present in both animals at week 60 was determined from overlapping 400- to 1,500-bp fragments generated by RT-PCR. The fragments were sequenced in the forward and reverse directions to provide data from nt 42 to 9418, terminating just prior to the poly(U) tract in the 3′ UTR. To safeguard against errors incorporated by the polymerase used in PCR amplification giving false mutations, this analysis was carried out twice on each animal with separate preparations of RNA.
For both animals the calculated rates at which mutations accumulated were similar, 1.57 × 10−3 (Ch1535) and 1.48 × 10−3 (Ch1536) nucleotide substitutions per site per year, which compare with a value of ∼1.92 × 10−3 previously reported for analysis of two patient samples taken 20 years apart (46). Of the substitutions observed in Ch1535 and Ch1536, 52.9 (9 of 16) and 60% (9 of 15), respectively, resulted in nonsynonymous mutations, none of which were located in the core or NS4 region of either animal. Table 1 shows a summary of the number of amino acid substitutions compared to nucleotide substitutions for each viral gene product and demonstrates that not all regions accumulated mutations to the same degree in both animals. Single, though different, amino acid mutations were detected in the p7 regions of the virus isolated from both animals despite its length of only 189 nt (63 amino acids).
TABLE 1.
Number of predicted amino acid substitutions compared to nucleotide substitutions at week 60 p.i.
| HCV region | No. of changes/total for each region (%)
|
|||
|---|---|---|---|---|
| Ch1535
|
Ch1536
|
|||
| Nucleotide | Amino Acid | Nucleotide | Amino acid | |
| Core | 1/573 (0.18) | 0/191 | 0/573 | 0/191 |
| E1 | 2/576 (0.35) | 2/192 (1.04) | 1/576 (0.17) | 1/192 (0.52) |
| E2 | 3/1,089 (0.28) | 2/363 (0.55) | 3/1,089 (0.28) | 2/363 (0.55) |
| p7 | 2/189 (1.06) | 1/63 (1.59) | 1/189 (0.53) | 1/63 (1.59) |
| NS2 | 2/651 (0.31) | 1/217 (0.46) | 1/651 (0.15) | 0/217 |
| NS3 | 3/1,893 (0.16) | 2/631 (0.32) | 2/1,893 (0.11) | 1/631 (0.16) |
| NS4A | 0/164 | 0/54 | 0/164 | 0/54 |
| NS4B | 1/784 (0.13) | 0/261 | 0/784 | 0/261 |
| NS5A | 1/1,343 (0.08) | 0/448 | 3/1,343 (0.22) | 2/448 (0.45) |
| NS5B | 2/1,773 (0.11) | 1/591 (0.17) | 5/1,773 (0.85) | 3/591 (0.51) |
| Totala | 17/9,376 (0.18) | 9/3,011 (0.3) | 16/9,363 (0.17) | 10/3,011 (0.33) |
Totals do not include silent mutations marking 5′ and 3′ variations.
The specific nucleotide and amino acid substitutions detected in the chimpanzees are shown in Table 2. No identical mutations were found in both animals except for silent markers included in the original inocula to distinguish 5′ and 3′ UTR variants. The 5′ UTR adaptations consisted of additional bases at the extreme 5′ termini; these clones were identified by an A-to-T substitution at nt 518. The majority sequence at this site was a T in both animals, indicating that these adaptations were not lethal to viral replication. The low RNA titers in both chimpanzees prevented any direct analysis of the 5′ termini to determine whether one or all of the four potential sequences were viable.
TABLE 2.
Sequence changes in HCV genome at week 60 p.i.
| HCV region | Ch1535
|
Ch1536
|
||||||
|---|---|---|---|---|---|---|---|---|
| Nucleotide position | Nucleotide change | Amino acid position | Amino acid change | Nucleotide position | Nucleotide change | Amino acid position | Amino acid change | |
| 5′ NCRa | ||||||||
| Core | 362 | T-C | 518b | A-T | ||||
| 518b | A-T | |||||||
| E1 | 1453 | T-C | 371 | V-A | 1134 | C-A | 265 | L-I |
| 1458 | G-C | 373 | V-L | |||||
| E2 | 1613 | C-T | 1540 | T-C | 400 | V-A | ||
| 1773 | A-G | 478 | S-G | 1728 | A-G | 463 | T-A | |
| 2499 | G-A | 720 | V-I | 2468 | C-T | |||
| p7 | 2603 | C-T | 2718 | A-G | 793 | M-V | ||
| 2638 | T-C | 766 | V-A | |||||
| NS2 | 2916 | G-A | 859 | V-M | 3047 | T-C | ||
| 3173 | T-C | |||||||
| NS3 | 3632 | C-G | 3883 | G-A | 1181 | R-K | ||
| 4938 | G-C | 1533 | A-P | 5042 | T-C | |||
| 5244 | G-A | 1635 | V-I | |||||
| NS4A | ||||||||
| NS4B | 6104 | C-T | ||||||
| NS5A | 6947 | G-A | 7382 | T-C | ||||
| 7386 | A-G | 2349 | T-A | |||||
| 7575 | G-A | 2412 | A-T | |||||
| NS5B | 7796 | G-T | 2485 | Q-H | 7698 | C-A | 2453 | H-N |
| 8624 | G-A | 7707 | C-A | 2456 | L-M | |||
| 8004 | G-A | 2555 | D-N | |||||
| 8054c | G-A | |||||||
| 9135 | C-T | |||||||
| 9376 | G-A | TGA | TAA | |||||
NCR, noncoding region.
Silent marker mutation for RNA transcripts from cDNA clones containing additional 5′ bases.
Silent marker mutation for RNA transcripts from cDNA clones with 75-base (A8054) versus 133-base (G8054) poly(U/UC) tracts in 3′ UTR (33).
The two types of 3′ UTRs tested in the chimpanzee inoculations contained a 75- or 133-base poly(U/UC) tract, distinguished by an A or G residue, respectively, at position 8054. At week 60, the predominant RNA from Ch1535 contained the marker for the 133-base clone while Ch1536 RNA contained that for the 75-base clone (Table 2).
To assess whether any of the amino acid mutations listed in Table 2 arose earlier rather than later in infection, the corresponding regions from Ch1535 and Ch1536 serum RNA, isolated at 26 and 22 weeks p.i., respectively, were amplified by RT-PCR and sequenced. These sample dates were chosen because they occurred at the point where serum RNA titers in both animals had stopped decreasing and antibody, ALT, and inflammatory responses had appeared to subside (Fig. 1). Of the mutations observed in the animals at week 60, 55.6 and 40% were already present as the majority sequence in Ch1535 and Ch1536, respectively, at these earlier time points. In the case of Ch1535, these amino acid substitutions were located in E1 (V371A), E2 (S478G), p7 (V766A), and NS3 (A1533P and V1635I), while those in Ch1536 were located in E1 (L265I), E2 (T463A), NS3 (R1181K), and NS5B (H2453N). These regions could represent targets for antibody or CTL responses.
DISCUSSION
In this report, we show that two chimpanzees inoculated with an infectious clone of HCV have remained persistently infected over a period of more than 1 year. The disease progression was similar to that observed in animals inoculated with infectious serum (17–19, 47, 55, 62) and was characterized by detectable viremia from weeks 1 and 2 p.i., relatively rapid viral replication leading to transient increases in ALT levels, liver inflammation between weeks 9 and 20, and antibody responses to both structural and nonstructural gene products from week 10 until the end of the study. The onset of these responses was followed by a significant decrease in viremia, but the virus was not cleared (Fig. 1A to F), leading to a persistent infection in both animals. This may be a feature of the infectious clone or mode of infection or a characteristic of the chimpanzees used. Future experiments with other infectious clones in different animals will provide the answers to these questions.
Persistent HCV infection occurs in more than 85% of patients (3) and 30% of chimpanzees (4), even though they often have both humoral and cellular immune responses to several viral Ags. The mechanism by which HCV eludes the host response is not well understood. Observations that HCV exists as a heterogeneous population in natural infections and reports of escape mutants at the antibody and CTL levels (10, 28, 53, 63, 66) provide evidence that immune escape may contribute to persistence either by repeated selection of preexisting viruses or by the incorporation of advantageous mutations to generate new variants. The animals in this study were inoculated with RNA transcribed from a cDNA clone representing a single viral genomic sequence. The only potential source of mutations would have been during in vitro transcription, with T7 RNA polymerase, or in vivo during HCV replication. For these variants to become dominant, both of these mechanisms would need to generate replication-competent viruses with some growth advantage over the wild type either through host adaptation or immune escape.
The mechanisms involved in clearance of virus are also poorly understood. A CD4 T-cell response to a conserved epitope contained within the NS3 region has been detected in a majority of patients with varied haplotypes who have resolved acute infections (14, 15). The epitope was shown to have high binding affinity for several common HLA-DR alleles and may play an important role in viral clearance. It has also been suggested that low or late anti-envelope antibody responses may correlate with chronicity while earlier responses, within 3 to 6 months of infection, are associated with resolution of acute infections (1, 68, 69). This could also be true of cellular responses. Ch1535 and Ch1536 developed detectable antibodies to several viral antigens within 9 to 13 weeks p.i. (Fig. 1B and D and 2) however, quantitative analyses of antibodies to the E1E2 region and HVR1 indicated that the responses were low during this time and therefore may not have been adequate to clear the virus, even though serum RNA titers were reduced significantly (Fig. 1A and C). Similarly, although the elevated ALT levels and inflammatory responses are indicative of a cellular response, this may also have been too weak to completely clear the virus.
There is evidence to suggest that the HCV HVR1 contains an antibody neutralization epitope. When subjected to immune pressure, new HVR1 sequences can be selected that represent immune escape variants (32, 52, 53, 60, 66). Observations of humans and chimpanzees have shown that the dominant sequence for this region changes during the course of an infection, leading to the suggestion that the emergence of HVR1 variants may be required for the establishment of persistence (32, 60, 66). Our data clearly show that mutations in HVR1 are not required for persistence of HCV in infected chimpanzees and that, in this instance, HVR1 does not accumulate mutations at an unusually high rate relative to the rest of the viral genome. In one animal, Ch1535, after 60 weeks of infection in the presence of specific antibody responses, the dominant HVR1 sequence was still identical to that in the original inoculum (Fig. 3A). Ch1536 incorporated a single-amino-acid mutation in HVR1 (Fig. 3B) which did not appear to represent an escape mutant. Virus persisted in this animal without diminution of viremia despite a marked increase in anti-E1E2, non-HVR1-reactive antibody. Analysis of multiple clones at week 60 did not demonstrate any significant degree of variability in either animal and certainly nothing similar to that associated with natural isolates (44, 67).
It is possible that the anti-HVR1 antibody responses in Ch1535 and Ch1536 were insufficient for the selection of HVR1 variants or escape mutants during the course of this infection. However, anti-HVR1 antibodies became elevated in one animal, Ch1536, at week 51 p.i. and remained high until the end of the study, but this did not result in significant sequence heterogeneity nor in the selection of HVR1 escape mutants. These data suggest that HVR1 escape mutants could be, in natural infections, a selection of preexisting variants which become dominant when the major species is suppressed by the antibody response in the host. The possibility that this principle would also apply in humans is supported by a recent study examining the evolution of HVR1 in a cohort of patients infected from a common source (42). The study suggested that multiple sequences, or lineages, could coexist for long periods during chronic infection and that variant sequences within infected individuals had diverged many years before in previous hosts, not since the infection took place. In addition, the amino acid substitutions in HVR1 were constrained in that there appeared to be variable and nonvariable sites. At the variable positions substitutions were confined to residues with similar characteristics.
The mutations detected in the viral genome RNA isolated at week 60 from both animals (Tables 1 and 2) were not evenly distributed among the different gene products. Substitutions were clustered within the envelope, NS2, NS3, and NS5 regions, though the clustering differed in the two animals. The function of the p7 protein is unidentified and it has yet to be established whether it forms part of the viral envelope; however, despite the short length of this region, serum RNA from both chimpanzees at week 60 carried mutations that resulted in amino acid substitutions.
Only further vaccine or neutralization experiments with single-sequence viruses will determine directly which, if any, of the observed substitutions in Ch1535 and Ch1536 represent escape mutations and therefore may have contributed to persistent infection in these animals. The identification of antibody epitopes in the E1E2 region of HCV has been difficult due to the conformational nature of these proteins. Linear B-cell epitopes other than those in HVR1 have been identified in E1 and E2 (11, 43, 48), although none of the mutations observed in Ch1535 and Ch1536 were located within these epitopes. Numerous reports have been published describing major histocompatibility complex class I CTL-recognizing Ags of HCV (6, 9, 16, 21, 29, 35–37, 39, 45, 49); taken together, these reports show that CTLs to all viral proteins have been detected. There is some evidence for the existence of CTL escape variants in experimental chimpanzee infections (65) and natural human infections (10), and an association has been observed between the presence of peptide variants and CTL responses, suggesting that some form of selection may take place, although early in infection (10).
If the mutations observed in Ch1535 and Ch1536 arose early in infection, shortly after the immune responses occurred, there is a strong possibility that they represent escape mutants, whereas if they arose late they are probably more a consequence of persistence than the cause. Analysis of serum RNA from earlier time points revealed that none of the mutations detected at week 60 were present before the peaks in ALT. However, several were present in both animals as the majority sequence following the stabilization of ALT and viremia. These mutations were located in regions that could be targets for antibody or CTL responses. Although none of the mutations, nor any of those detected at week 60 in either chimpanzee, were located in previously identified immune epitopes, they could reflect adaptation to the animal-specific HCV immune responses that enabled the virus to evade the host response well enough to remain in circulation. The Patr alleles of these animals are known to be different (49a), and further characterization is under way, together with observation of B and T cell responses, to determine whether any of the mutations observed could represent immune escape variants.
The overall mutation rate for HCV was calculated as 1.57 × 10−3 and 1.48 × 10−3 nucleotide substitutions per site per year for Ch1535 and Ch1536, respectively. This is similar to that previously reported for samples taken 20 years apart from a chronically infected patient (∼1.9 × 10−3 base substitutions per genome site per year), although that study found a significantly high rate of change in HVR1 (28.2% variation at the nucleotide level [46]), probably due to preexisting variants. It should be noted that these analyses are measuring the rates at which mutations accumulate over time and are influenced not only by mutation frequency during RNA replication but also by the relative fitness of these mutations in vivo.
With the development of an infectious clone for HCV we can begin to analyze the effects of the host immune response on viral evolution and the role this plays in persistence. The use of early-acute-phase serum from these transfected chimpanzees also provides an inoculum that, although it cannot be guaranteed to consist of a single viral sequence, will have a sequence variability as low as can be achieved with current approaches and will therefore be invaluable in addressing further the questions surrounding the mechanisms that allow persistence and reinfection and the feasibility of an effective vaccine.
ACKNOWLEDGMENTS
We thank Phil Snoy, Kamela Evans-Davis, and Ray Olsen for excellent veterinary work, Michael Klutch for valuable assistance with automated sequencing, and Barbara Rehermann for helpful discussion and analysis of chimpanzee Patr alleles.
The work was supported in part by PHS grants CA57973 and AI40034.
REFERENCES
- 1.Allander T, Beyene A, Jacobson S H, Grillner L, Persson M A. Patients infected with the same hepatitis C virus strain display different kinetics of the isolate-specific antibody response. J Infect Dis. 1997;175:26–31. doi: 10.1093/infdis/175.1.26. [DOI] [PubMed] [Google Scholar]
- 2.Alter H J, Purcell R H, Holland P V, Popper H. Transmissible agent in non-A, non-B hepatitis. Lancet. 1978;i:459–463. doi: 10.1016/s0140-6736(78)90131-9. [DOI] [PubMed] [Google Scholar]
- 3.Alter M J, Margolis H S, Krawczynski K, Judson F N, Mares A, Alexander W J, Hu P Y, Miller J K, Gerber M A, Sampliner R E, Meeks E L, Beach M J. The natural history of community-acquired hepatitis C in the United States. N Engl J Med. 1992;327:1899–1905. doi: 10.1056/NEJM199212313272702. [DOI] [PubMed] [Google Scholar]
- 4.Bassett S E, Brasky K M, Lanford R E. Analysis of hepatitis C virus-inoculated chimpanzees reveals unexpected clinical profiles. J Virol. 1998;72:2589–2599. doi: 10.1128/jvi.72.4.2589-2599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassler H A, Flood S J A, Livak K J, Marmaro J, Knorr R, Batt C A. Use of fluorogenic probe in a PCR-based assay for the detection of Listeria monocytogenes. Appl Environ Microbiol. 1995;61:3724–3728. doi: 10.1128/aem.61.10.3724-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battegay M, Fikes J, Di B A, Wentworth P A, Sette A, Celis E, Ching W M, Grakoui A, Rice C M, Kurokohchi K, Berzofsky J A, Hoofnagle J H, Feinstone S M, Akatsuka T. Patients with chronic hepatitis C have circulating cytotoxic T cells which recognize hepatitis C virus-encoded peptides binding to HLA-A2.1 molecules. J Virol. 1995;69:2462–2470. doi: 10.1128/jvi.69.4.2462-2470.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bredenbeek P J, Frolov I, Rice C M, Schlesinger S. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukh J, Miller R H, Purcell R H. Genetic heterogeneity of the hepatitis C virus. Princess Takamatsu Symp. 1995;25:75–91. [PubMed] [Google Scholar]
- 9.Cerny A, McHutchison J G, Pasquinelli C, Brown M E, Brothers M A, Grabscheid B, Fowler P, Houghton M, Chisari F V. Cytotoxic T lymphocyte response to hepatitis C virus-derived peptides containing the HLA A2.1 binding motif. J Clin Investig. 1995;95:521–530. doi: 10.1172/JCI117694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang K M, Rehermann B, McHutchison J G, Pasquinelli C, Southwood S, Sette A, Chisari F V. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J Clin Investig. 1997;100:2376–2385. doi: 10.1172/JCI119778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ching W M, Wychowski C, Beach M J, Wang H, Davies C L, Carl M, Bradley D W, Alter H J, Feinstone S M, Shih J W. Interaction of immune sera with synthetic peptides corresponding to the structural protein region of hepatitis C virus. Proc Natl Acad Sci USA. 1992;89:3190–3194. doi: 10.1073/pnas.89.8.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 13.Choo Q L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby A, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghton M. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diepolder H M, Gerlach J T, Zachoval R, Hoffmann R M, Jung M C, Wierenga E A, Scholtz S, Santantonio T, Houghton M, Southwood S, Sette A, Pape G R. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J Virol. 1997;71:6011–6019. doi: 10.1128/jvi.71.8.6011-6019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diepolder H M, Zachoval R, Hoffmann R M, Wierenga E A, Santantonio T, Jung M C, Eichenlaub D, Pape G R. Possible mechanism involving T-lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet. 1995;346:1006–1007. doi: 10.1016/s0140-6736(95)91691-1. [DOI] [PubMed] [Google Scholar]
- 16.Erickson A L, Houghton M, Choo Q L, Weiner A J, Ralston R, Muchmore E, Walker C M. Hepatitis C virus-specific CTL responses in the liver of chimpanzees with acute and chronic hepatitis C. J Immunol. 1993;151:4189–4199. [PubMed] [Google Scholar]
- 17.Farci P, Alter H J, Govindarajan S, Wong D C, Engle R, Lesniewski R, Mushahwar I K, Desai S M, Miller R H, Ogata N, Purcell R H. Lack of protective immunity against reinfection with hepatitis C virus. Science. 1992;258:140. doi: 10.1126/science.1279801. [DOI] [PubMed] [Google Scholar]
- 18.Farci P, Alter H J, Wong D C, Miller R H, Govindarajan S, Engle R, Shapiro M, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter H J, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci USA. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feinstone S M, Alter H J, Dienes H P, Shimizu Y, Popper H, Blackmore D, Sly D, London W T, Purcell R H. Non-A, non-B hepatitis in chimpanzees and marmosets. J Infect Dis. 1981;144:588–598. doi: 10.1093/infdis/144.6.588. [DOI] [PubMed] [Google Scholar]
- 21.Ferrari C, Valli A, Galati L, Penna A, Scaccaglia P, Giuberti T, Schianchi C, Missale G, Marin M G, Fiaccadori F. T-cell response to structural and nonstructural hepatitis C virus antigens in persistent and self-limited hepatitis C virus infections. Hepatology. 1994;19:286–295. [PubMed] [Google Scholar]
- 22.Gibson U E M, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 23.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han J H, Shyamala V, Richman K H, Brauer M J, Irvine B, Urdea M S, Tekamp-Olson P, Kuo G, Choo Q L, Houghton M. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at the 3′ end. Proc Natl Acad Sci USA. 1991;88:1711–1715. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 26.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem Biophys Res Commun. 1991;175:220–228. doi: 10.1016/s0006-291x(05)81223-9. [DOI] [PubMed] [Google Scholar]
- 27.Hiroishi K, Kita H, Kojima M, Okamoto H, Moriyama T, Kaneko T, Ishikawa T, Ohnishi S, Aikawa T, Tanaka N, Yazaki Y, Mitamura K, Imawari M. Cytotoxic T lymphocyte response and viral load in hepatitis C virus infection. Hepatology. 1997;25:705–712. doi: 10.1002/hep.510250336. [DOI] [PubMed] [Google Scholar]
- 28.Kaneko T, Moriyama T, Udaka K, Hiroishi K, Kita H, Okamoto H, Yagita H, Okumura K, Imawari M. Impaired induction of cytotoxic T lymphocytes by antagonism of a weak agonist borne by a variant hepatitis C virus epitope. Eur J Immunol. 1997;27:1782–1787. doi: 10.1002/eji.1830270728. [DOI] [PubMed] [Google Scholar]
- 29.Kaneko T, Nakamura I, Kita H, Hiroishi K, Moriyama T, Imawari M. Three new cytotoxic T cell epitopes identified within the hepatitis C virus nucleoprotein. J Gen Virol. 1996;77:1305–1309. doi: 10.1099/0022-1317-77-6-1305. [DOI] [PubMed] [Google Scholar]
- 30.Kato N, Hijikata M, Nakagawa M, Ootsuyama Y, Muraiso K, Ohkoshi S, Shimotohno K. Molecular structure of the Japanese hepatitis C viral genome. FEBS Lett. 1991;280:325–328. doi: 10.1016/0014-5793(91)80322-t. [DOI] [PubMed] [Google Scholar]
- 31.Kato N, Ootsuyama Y, Ohkoshi S, Nakazawa T, Sekiya H, Hijikata M, Shimotohno K. Characterization of hypervariable regions in the putative envelope protein of hepatitis C virus. Biochem Biophys Res Commun. 1992;189:119–127. doi: 10.1016/0006-291x(92)91533-v. [DOI] [PubMed] [Google Scholar]
- 32.Kato N, Sekiya H, Ootsuyama Y, Nakazawa T, Hijikata M, Ohkoshi S, Shimotohno K. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J Virol. 1993;67:3923–3930. doi: 10.1128/jvi.67.7.3923-3930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolykhalov A A, Agapov E V, Blight K, Mihalik K, Feinstone S M, Rice C M. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 34.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koziel M J, Dudley D, Afdhal N, Choo Q L, Houghton M, Ralston R, Walker B D. Hepatitis C virus (HCV)-specific cytotoxic T lymphocytes recognize epitopes in the core and envelope proteins of HCV. J Virol. 1993;67:7522–7532. doi: 10.1128/jvi.67.12.7522-7532.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koziel M J, Dudley D, Afdhal N, Grakoui A, Rice C M, Choo Q L, Houghton M, Walker B D. HLA class I-restricted cytotoxic T lymphocytes specific for hepatitis C virus. Identification of multiple epitopes and characterization of patterns of cytokine release. J Clin Investig. 1995;96:2311–2321. doi: 10.1172/JCI118287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koziel M J, Dudley D, Wong J T, Dienstag J, Houghton M, Ralston R, Walker B D. Intrahepatic cytotoxic T lymphocytes specific for hepatitis C virus in persons with chronic hepatitis. J Immunol. 1992;149:3339–3344. [PubMed] [Google Scholar]
- 38.Koziel M J, Wong D K, Dudley D, Houghton M, Walker B D. Hepatitis C virus-specific cytolytic T lymphocyte and T helper cell responses in seronegative persons. J Infect Dis. 1997;176:859–866. doi: 10.1086/516546. [DOI] [PubMed] [Google Scholar]
- 39.Kurokohchi K, Akatsuka T, Pendleton C D, Takamizawa A, Nishioka M, Battegay M, Feinstone S M, Berzofsky J A. Use of recombinant protein to identify a motif-negative human cytotoxic T-cell epitope presented by HLA-A2 in the hepatitis C virus NS3 region. J Virol. 1996;70:232–240. doi: 10.1128/jvi.70.1.232-240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurosaki M, Enomoto N, Marumo F, Sato C. Rapid sequence variation of the hypervariable region of hepatitis C virus during the course of chronic infection. Hepatology. 1993;18:1293–1299. [PubMed] [Google Scholar]
- 41.Major M E, Feinstone S F. Molecular virology of hepatitis C. Hepatology. 1997;25:1527–1538. doi: 10.1002/hep.510250637. [DOI] [PubMed] [Google Scholar]
- 42.McAllister J, Casino C, Davidson F, Power J, Lawlor E, Yap P L, Simmonds P, Smith D B. Long-term evolution of the hypervariable region of hepatitis C virus in a common-source-infected cohort. J Virol. 1998;72:4893–4905. doi: 10.1128/jvi.72.6.4893-4905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mink M A, Benichou S, Madaule P, Tiollais P, Prince A M, Inchauspe G. Characterization and mapping of a B-cell immunogenic domain in hepatitis C virus E2 glycoprotein using a yeast peptide library. Virology. 1994;200:246–255. doi: 10.1006/viro.1994.1182. [DOI] [PubMed] [Google Scholar]
- 44.Nakajima N, Hijikata M, Yoshikura H, Shimizu Y K. Characterization of long-term cultures of hepatitis C virus. J Virol. 1996;70:3325–3329. doi: 10.1128/jvi.70.5.3325-3329.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson D R, Marousis C G, Davis G L, Rice C M, Wong J, Houghton M, Lau J Y. The role of hepatitis C virus-specific cytotoxic T lymphocytes in chronic hepatitis C. J Immunol. 1997;158:1473–1481. [PubMed] [Google Scholar]
- 46.Ogata N, Alter H J, Miller R H, Purcell R H. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:3392–3396. doi: 10.1073/pnas.88.8.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prince A M, Brotman B, Huima T, Pascual D, Jaffery M, Inchauspé G. Immunity in hepatitis C infection. J Infect Dis. 1992;165:438–443. doi: 10.1093/infdis/165.3.438. [DOI] [PubMed] [Google Scholar]
- 48.Ray R, Khanna A, Lagging L M, Meyer K, Choo Q L, Ralston R, Houghton M, Becherer P R. Peptide immunogen mimicry of putative E1 glycoprotein-specific epitopes in hepatitis C virus. J Virol. 1994;68:4420–4426. doi: 10.1128/jvi.68.7.4420-4426.1994. . (Erratum, 68:6136.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rehermann B, Chang K M, McHutchison J G, Kokka R, Houghton M, Chisari F V. Quantitative analysis of the peripheral blood cytotoxic T lymphocyte response in patients with chronic hepatitis C virus infection. J Clin Investig. 1996;98:1432–1440. doi: 10.1172/JCI118931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a.Rehermann, B. Personal communication.
- 50.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y, Choo Q L, Houghton M, Kuo G. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saito T, Sherman G J, Kurokohchi K, Guo Z P, Donets M, Yu M Y, Berzofsky J A, Akatsuka T, Feinstone S M. Plasmid DNA-based immunization for hepatitis C virus structural proteins: immune responses in mice. Gastroenterology. 1997;112:1321–1330. doi: 10.1016/s0016-5085(97)70146-x. [DOI] [PubMed] [Google Scholar]
- 52.Sekiya H, Kato N, Ootsuyama Y, Nakazawa T, Yamauchi K, Shimotohno K. Genetic alterations of the putative envelope proteins encoding region of the hepatitis C virus in the progression to relapsed phase from acute hepatitis: humoral immune response to hypervariable region 1. Int J Cancer. 1994;57:664–670. doi: 10.1002/ijc.2910570509. [DOI] [PubMed] [Google Scholar]
- 53.Shimizu Y K, Hijikata M, Iwamoto A, Alter H J, Purcell R H, Yoshikura H. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J Virol. 1994;68:1494–1500. doi: 10.1128/jvi.68.3.1494-1500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimizu Y K, Igarashi H, Kiyohara T, Cabezon T, Farci P, Purcell R H, Yoshikura H. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology. 1996;223:409–412. doi: 10.1006/viro.1996.0497. [DOI] [PubMed] [Google Scholar]
- 55.Shindo M, Di Bisceglie A M, Biswas R, Mihalik K, Feinstone S M. Hepatitis C virus replication during acute infection in the chimpanzee. J Infect Dis. 1992;166:424–427. doi: 10.1093/infdis/166.2.424. [DOI] [PubMed] [Google Scholar]
- 56.Simmonds P, Smith D B, McOmish F, Yap P L, Kolberg J, Urdea M S, Holmes E C. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J Gen Virol. 1994;75:1053–1061. doi: 10.1099/0022-1317-75-5-1053. [DOI] [PubMed] [Google Scholar]
- 57.Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka T, Kato N, Cho M J, Shimotohno K. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem Biophys Res Commun. 1995;215:744–749. doi: 10.1006/bbrc.1995.2526. [DOI] [PubMed] [Google Scholar]
- 59.Tanaka T, Kato N, Cho M J, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taniguchi S, Okamoto H, Sakamoto M, Kojima M, Tsuda F, Tanaka T, Munekata E, Muchmore E E, Peterson D A, Mishiro S. A structurally flexible and antigenically variable N-terminal domain of the hepatitis C virus E2/NS1 protein: implication for an escape from antibody. Virology. 1993;195:297–301. doi: 10.1006/viro.1993.1378. [DOI] [PubMed] [Google Scholar]
- 61.van Doorn L J, Capriles I, Maertens G, DeLeys R, Murray K, Kos T, Schellekens H, Quint W. Sequence evolution of the hypervariable region in the putative envelope region E2/NS1 of hepatitis C virus is correlated with specific humoral immune responses. J Virol. 1995;69:773–778. doi: 10.1128/jvi.69.2.773-778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Doorn L J, Quint W, Tsiquaye K, Voermans J, Paelinck D, Kos T, Maertens G, Schellekens H, Murray K. Longitudinal analysis of hepatitis C virus infection and genetic drift of the hypervariable region. J Infect Dis. 1994;169:1226–1235. doi: 10.1093/infdis/169.6.1226. [DOI] [PubMed] [Google Scholar]
- 63.Weiner A, Erickson A L, Kansopon J, Crawford K, Muchmore E, Hughes A L, Houghton M, Walker C M. Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc Natl Acad Sci USA. 1995;92:2755–2759. doi: 10.1073/pnas.92.7.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weiner A J, Brauer M J, Rosenblatt J, Richman K H, Tung J, Crawford K, Bonino F, Saracco G, Choo Q L, Houghton M. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 65.Weiner A J, Erickson A L, Kansopon J, Crawford K, Muchmore E, Houghton M, Walker C M. Association of cytotoxic T lymphocyte (CTL) escape mutations with persistent hepatitis C virus (HCV) infection. Princess Takamatsu Symp. 1995;25:227–235. [PubMed] [Google Scholar]
- 66.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, Brunetto M, Barr P J, Miyamura T, McHutchinson J, Houghton M. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wyatt C A, Andrus L, Brotman B, Huang F, Lee D H, Prince A M. Immunity in chimpanzees chronically infected with hepatitis C virus: role of minor quasispecies in reinfection. J Virol. 1998;72:1725–1730. doi: 10.1128/jvi.72.3.1725-1730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zibert A, Kraas W, Meisel H, Jung G, Roggendorf M. Epitope mapping of antibodies directed against hypervariable region 1 in acute self-limiting and chronic infections due to hepatitis C virus. J Virol. 1997;71:4123–4127. doi: 10.1128/jvi.71.5.4123-4127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zibert A, Meisel H, Kraas W, Schulz A, Jung G, Roggendorf M. Early antibody response against hypervariable region 1 is associated with acute self-limiting infections of hepatitis C virus. Hepatology. 1997;25:1245–1249. doi: 10.1002/hep.510250530. [DOI] [PubMed] [Google Scholar]
- 70.Zibert A, Schreier E, Roggendorf M. Antibodies in human sera specific to hypervariable region 1 of hepatitis C virus can block viral attachment. Virology. 1995;208:653–661. doi: 10.1006/viro.1995.1196. [DOI] [PubMed] [Google Scholar]