Abstract
Human coronaviruses (HuCV) are recognized respiratory pathogens. Data accumulated by different laboratories suggest their neurotropic potential. For example, primary cultures of human astrocytes and microglia were shown to be susceptible to an infection by the OC43 strain of HuCV (A. Bonavia, N. Arbour, V. W. Yong, and P. J. Talbot, J. Virol. 71:800–806, 1997). We speculate that the neurotropism of HuCV will lead to persistence within the central nervous system, as was observed for murine coronaviruses. As a first step in the verification of our hypothesis, we have characterized the susceptibility of various human neural cell lines to infection by HuCV-OC43. Viral antigen, infectious virus progeny, and viral RNA were monitored during both acute and persistent infections. The astrocytoma cell lines U-87 MG, U-373 MG, and GL-15, as well as neuroblastoma SK-N-SH, neuroglioma H4, oligodendrocytic MO3.13, and the CHME-5 immortalized fetal microglial cell lines, were all susceptible to an acute infection by HuCV-OC43. Viral antigen and RNA and release of infectious virions were observed during persistent HuCV-OC43 infections (∼130 days of culture) of U-87 MG, U-373 MG, MO3.13, and H4 cell lines. Nucleotide sequences of RNA encoding the putatively hypervariable viral S1 gene fragment obtained after 130 days of culture were compared to that of initial virus input. Point mutations leading to amino acid changes were observed in all persistently infected cell lines. Moreover, an in-frame deletion was also observed in persistently infected H4 cells. Some point mutations were observed in some molecular clones but not all, suggesting evolution of the viral population and the emergence of viral quasispecies during persistent infection of H4, U-87 MG, and MO3.13 cell lines. These results are consistent with the potential persistence of HuCV-OC43 in cells of the human nervous system, accompanied by the production of infectious virions and molecular variation of viral genomic RNA.
Human coronaviruses (HuCV) are enveloped positive-stranded RNA viruses represented by two known viral serogroups, OC43 and 229E. These viruses were first isolated as pathogens of the respiratory tract (32, 49) and have been associated with up to one-third of common colds (58). Since their discovery, other pathologies have occasionally been associated with HuCV, such as pneumonia, meningitis, radiculitis (65), and diarrhea (64). Accumulating evidence from animal models and from studies of humans suggests a neurotropic potential for these viruses.
The murine counterpart of HuCV, mouse hepatitis virus (MHV) has been studied as an animal model of a virus-induced demyelinating disease of the central nervous system (CNS) (13, 83). The outcome of an MHV infection in the CNS is dependent upon the route and dose of inoculation and on host factors such as age, species, strain, and immune system status as well as on the genetic constitution of the virus (72, 84). After intranasal inoculation, neurotropic strains of MHV enter the brain via the olfactory nerve and then spread within the mouse CNS (7, 42, 61). Other routes have been shown to be used by MHV to gain access to the CNS, such as the hematogenous and lymphatic systems (9). Moreover, a neurotropic MHV has been shown to enter the CNS of nonhuman primates after a peripheral inoculation (14). Once in the CNS, MHV replicates in neurons, astrocytes, and oligodendrocytes (33). It was shown that the CNS of nonhuman primates is also susceptible to a coronavirus infection: after an intracerebral inoculation, a murine virus-like isolate caused a subacute panencephalitis and demyelination in these animals (56). Similar results could be envisaged for HuCV, especially the OC43 strain, which is classified within the same antigenic group as MHV and is closely linked at the molecular level (41).
Observations in humans are also consistent with HuCV neurotropism. Intrathecal anti-HuCV-OC43 antibody synthesis was detected in humans, particularly in multiple sclerosis (67) and in Parkinson’s disease patients (25), suggesting a CNS infection. Importantly, HuCV-OC43 RNA has repeatedly been detected in human brains (6, 55). Moreover, we have shown that this HuCV strain has the capacity to infect human astrocytes and microglia in primary cultures (12). In general, in vitro observations of coronavirus infections correlate with the ones occurring in vivo. For example, MHV-3 has an in vitro affinity for neurons, ependymal cells, and meningeal cells but not astrocytes and oligodendrocytes, corroborating the pattern of pathogenicity observed in vivo, where this virus induces an initial ependymitis, meningitis, and encephalitis (82). Similarly, three MHV-JHM variants that induce very different pathological patterns in vivo, demonstrated distinct corroborating in vitro growth properties on primary cultures of rat glial cells (48). Therefore, it is reasonable to assume that the characterization of HuCV replication in vitro will provide valuable data regarding its neurotropic properties, given that in vivo experiments could obviously not be performed in humans.
Of relevance to our study, MHV RNA was detected in mouse brain a long time after the initial infection (26, 33). In fact, the brain was the last organ from which MHV was cleared after an oronasal inoculation (8). Thus, this virus is able to persist in the CNS of its host (43, 70), especially in astrocytes, oligodendrocytes (63, 76), and neurons (71). It has been suggested that viral persistence observed in the CNS may play a role in chronic pathologies observed following a coronavirus infection (43). Of importance, persistent MHV infections of various neural cell lines have been observed (46, 74).
Since HuCV was detected in human brains and was able to infect human neural cells, both in primary and immortalized cultures, we speculate that HuCV could also persist in the human CNS. Collins and Sorensen (20) reported a persistent infection by HuCV-OC43 in a human glioblastoma cell line. However, more extensive studies are needed in order to elucidate the potential persistence of HuCV-OC43 in the human CNS.
Coronaviruses bear four structural proteins: spike (S), membrane (M), small membrane (E, formerly sM), and nucleocapsid (N). Some strains, such as HuCV-OC43, incorporate an additional structural protein: hemagglutinin-esterase (HE) (41). More sequence variations are usually observed within the S protein than in any other structural protein (41). The pathogenesis of MHV is dependent in part on its S glycoprotein, which has multiple important biological properties, such as receptor ligation (22, 86), mediation of fusion (19), neutralizing antibody domains (21, 81, 85), and cytotoxic T lymphocyte epitope domains (10, 15). Moreover, different laboratories have shown that important molecular determinants of neurovirulence can be localized in some regions of the MHV S glycoprotein (13, 27). Gallagher and colleagues have shown that in vitro passaging of MHV-4 in culture generates heterogeneity in the structure of the S glycoprotein (29). Moreover, multiple point and deletion mutations arose, mainly concentrated in the S and the N genes of MHV during an in vivo persistent CNS infection (1, 66). On the other hand, potential variations of the HuCV-OC43 S glycoprotein in persistent infections of the CNS have not been examined to date.
The susceptibility of various neural cell lines to coronaviruses, particularly to MHV (28), has been demonstrated. Limited results (5, 20) are available concerning the susceptibility to HuCV-OC43 infection of human continuous cell lines representative of cells of the nervous system, which would enable these to serve as invaluable in vitro models of study. We have previously demonstrated the infection of primary cultures of human brain cells with HuCV-OC43 (12). However, the limited life span of these cell cultures does not allow a study of the effects of a long-term coronavirus infection.
In the present study, cell lines representative of different neural cell types (oligodendrocytes, astrocytes, microglia, and neurons) of the human nervous system were used to evaluate the potential of HuCV-OC43 to cause an acute and/or a persistent infection. Similar experiments were also carried out with HuCV-229E; these results are presented separately (4). Viral antigens, infectious virus particles, and viral RNA (N and S1 genes) were monitored during an acute and a persistent infection by HuCV-OC43 We report that all the cell lines tested were susceptible to an acute HuCV-OC43 infection. Moreover, all but three of them sustained a persistent infection. Sequencing of the S1 gene fragment after 130 days of culture showed, in all persistently infected cell lines, mainly point mutations leading to amino acid changes. Our results are consistent with the hypothesis that HuCV-OC43, as was extensively shown for MHV, could persist in various CNS cell types, leading to production of infectious viral particles and molecular variation of viral genomic RNA.
MATERIALS AND METHODS
Viruses and cell lines.
HuCV-OC43 was originally obtained from the American Type Culture Collection (ATCC, Manassas, Va). plaque purified twice, and grown on HRT-18 cells as described previously (54). The fourth passage of HuCV-OC43 from laboratory stocks kept at −90°C, with a titer of 5.15 × 106 50% tissue culture infective doses (TCID50)/ml, was used for all experiments.
The GL-15 cell line, established from a human glioblastoma multiforme, was generated by V. Bocchini (University Medical School, Perugia, Italy) (11). The CHME-5 cell line was obtained from human fetal microglia by transfection with the large T antigen of simian virus 40 (34). The MO3.13 cell line was obtained from the fusion of a rhabdosarcoma cell line (Te671) with an adult human oligodendrocyte (51). The H4, SK-N-SH, U-373 MG, and U-87 MG cell lines were obtained from the ATCC. All of these neural cell lines were grown in Dulbecco’s modified Eagle’s medium, supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum and gentamicin (50 μg/ml; GIBCO Laboratories, Grand Island, N.Y.).
Antibodies.
Virus-specific monoclonal antibodies (MAbs) were produced in our laboratory by standard hybridoma technology. MAbs 1-10C.3, directed against the surface S glycoprotein of HuCV-OC43, and 5-11H.6, an isotypic control, were used to detect viral antigens in immunoperoxidase and immunofluorescence assays.
Acute infection.
For detection of viral antigens, approximately 1 × 106 to 1.5 × 106 cells (obtained by trypsinization of cell monolayers grown on plastic 75-cm2 flasks, followed by pelleting) were mixed with an equal volume of HuCV-OC43 virus stock, diluted to provide a multiplicity of infection (MOI) of 0.1 or 1.0, in the presence of L-1-tosylamide-2-phenylethyl-chloromethyl ketone-treated trypsin (10 U/ml; Sigma-Aldrich Canada Ltd.). Twenty-five microliters of this suspension was deposited into each well of a 12-well glass slide (Flow, ICN Biomedical Canada Ltd., Mississauga, Ontario, Canada), and infection progressed for up to 4 days at 33°C prior to fixation in acetone at −20°C for 30 min. Slides were kept at −70°C until an immunofluorescence assay could be performed.
For quantitation of infectious virus production, cell lines at 60 to 80% confluence in 25-cm2 flasks were infected with virus stock diluted to provide an MOI of 0.1, in the presence of L-1-tosylamide-2-phenylethyl-chloromethyl ketone-treated trypsin (10 U/ml; Sigma-Aldrich Canada Ltd.). Infection was carried out at 33°C for up to 4 days, and supernatants and cell monolayers taken at different time points were harvested and kept at −70°C until infectious virus titers could be quantitated.
For studies of viral RNA, cell monolayers at 60 to 80% confluence in 150-cm2 tissue culture-treated petri dishes were infected at an MOI of 0.01, in the presence of L-1-tosylamide-2-phenylethyl-chloromethyl ketone-treated trypsin (10 U/ml; Sigma-Aldrich Canada Ltd.) for 1 day at 33°C. Then, cell monolayers were washed twice with phosphate-buffered saline (PBS), pH 7.4, and kept at −90°C until RNA could be extracted.
Persistent infection.
Cell lines at 60 to 80% confluence in 25-cm2 culture flasks were infected with 0.5 ml of viral suspension, providing an MOI of 0.5, in the presence of L-1-tosylamide-2-phenylethyl-chloromethyl ketone-treated trypsin (10 U/ml; Sigma-Aldrich Canada Ltd.) and incubated for 2 h at 33°C with periodical agitation. Cell monolayers were then washed with PBS and grown in regular cell culture medium at 37°C. Cells were passaged every 4 to 8 days. Samples of supernatants and cells were kept at −70°C for viral titration by an immunoperoxidase assay at each passage, and cells at each fifth passage were kept for RNA extraction. Twenty-five microliters of cell samples from each passage at a concentration of 0.5 × 106 to 1.5 × 106 cells/ml was deposited into each well of a 12-well glass slide (Flow, ICN Biomedical Canada Ltd.), incubated for 24 to 48 h at 37°C, and then fixed in acetone at −20°C for 30 min and kept at −70°C until immunofluorescence could be performed.
Detection of viral antigens by immunofluorescence.
The immunofluorescence assay was performed as described previously (68). Briefly, primary antibody (MAbs 1-10C.3 for HuCV-OC43 and 5-11H.6 as an isotypic control) was added to thawed slides and incubated for 1 h at 37°C. This was followed by three washes in PBS, the addition of the fluorescein-conjugated mouse-specific goat secondary antibody (Cappel, Durham, N.C.), and a 30-min incubation at 37°C. After three washes in PBS, slides were mounted with glycerol-PBS (9:1). Slides were stored at 4°C until observation with a Leitz fluorescence microscope (Dialux 20 model) could be performed.
Immunoperoxidase assay for quantitation of infectious virus titers.
The immunoperoxidase assay for quantitation of infectious virus titers was performed as described previously (68). Briefly, susceptible HRT-18 cells were inoculated with serial logarithmic dilutions of samples in a 96-well Linbro plate (Flow, ICN Biomedical Canada Ltd.). After 4 to 5 days of incubation at 33°C in 5% (vol/vol) CO2, cells were washed with PBS and fixed with 0.3% (vol/vol) hydrogen peroxide (Sigma-Aldrich Canada Ltd.) in methanol for 30 min. After washing with PBS, they were incubated for 2 h at 37°C with an appropriate antiviral MAb (1-10C.3). Cells were then washed five times with PBS, and horseradish peroxidase-conjugated goat anti-mouse immunoglobulins (Cappel) were added and incubated for 2 h at 37°C. Bound antibodies were detected by incubation with 0.025% (wt/vol) 3,3′-diamino-benzidine-tetrahydrochloride (Bio-Rad, Richmond, Calif.) and 0.01% (vol/vol) hydrogen peroxide (Sigma-Aldrich Canada Ltd.) in PBS. The chromogenic reaction was stopped with deionized water. Infectious virus titers were calculated by the Karber method. Negative controls consisted of noninfected cells.
Preparation of RNA, RT, and PCR.
To extract total cellular RNA from acutely or persistently infected or control cells, cell monolayers were lysed with GIT buffer (4 M guanidine isothiocyanate, 2.5 mM sodium acetate, 12 mM β-mercaptoethanol). Lysates were passed through a 26-gauge needle at least six times and then layered onto a cesium chloride cushion (5.7 M cesium chloride, 2.5 mM sodium acetate) for a 12- to 20-h centrifugation at 150,000 × g. Supernatant was removed, and the RNA pellet was resuspended in sterile distilled and deionized H2O. The pair of primers used for amplification of HuCV-OC43 as well as the one for the control housekeeping gene (encoding glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) are described in Table 1. Forty picomoles of the inverse-complementary primer or 100 pmol of oligo(dT) (Roche Diagnostics, Laval, Québec, Canada) was incubated with 2 to 5 μg of total cellular RNA at 65°C for 5 min to denature RNA, followed by a 30-min slow cooldown to 37°C for annealing. Reverse transcription (RT) with Expand Moloney murine leukemia virus reverse transcriptase (50 U; Roche Diagnostics) was performed at 42°C for 90 min in the presence of 60 U of RNA Guard (Pharmacia, Baie d’Urfé, Québec, Canada), 0.4 mM (each) deoxynucleoside triphosphate (Na salt; Roche Diagnostics), 1× reverse transcriptase buffer (50 mM Tris-HCl, 40 mM KCl, 5 mM MgCl2, 0.5% [vol/vol] Tween 20 [pH 8.3]), and 10 mM dithiothreitol (Roche Diagnostics). For PCR, 1 of 10 or 1 of 5 of the synthesized cDNAs was incubated in the presence of 20 pmol (HuCV-OC43 S1 and N) or 50 pmol (GAPDH) of the sense and antisense primers, 1.5 mM (HuCV-OC43 S1 or GAPDH) or 2.5 mM (HuCV-OC43 N) MgCl2 (Roche Diagnostics), 1× PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl; Roche Diagnostics) and 0.4 mM (each) deoxynucleoside triphosphate (Li salt; Roche Diagnostics) at 94°C for 5 min and at 60°C (HuCV-OC43 N), 50°C (GAPDH), or 37°C (HuCV-OC43 S1) for another 5 min. After the addition of Expand high-fidelity PCR system DNA polymerase (Taq and Pwo DNA polymerases) (2.5 U; Roche Diagnostics), 30 cycles of 2 min at 72°C, 1 min at 94°C, and 2 min at 60°C (HuCV-OC43 N), 50°C (GAPDH), or 37°C (HuCV-OC43 S1) were performed, with a final elongation step of 10 min at 72°C. The DNA amplicons were separated by electrophoresis in a 1.5% (wt/vol) agarose gel with ethidium bromide (1 μg/ml).
TABLE 1.
RT-PCR and sequencing primers
| RNA amplified or sequenced | Primer sequence | Primer designation | Nucleotides |
|---|---|---|---|
| RT-PCR HuCV-OC43 N gene | 5′-CCCAAGCAAACTGCTACCTCTCAG-3′ | O1 sense | 215–238a |
| 5′-GTAGACTCCGTCAATATCGGTGCC-3′ | O3 antisense | 497–520a | |
| RT-PCR HuCV-OC43 S1 gene portion | 5′-TCTGCCAAGAGTCTTGCTCG-3′ | OC43-S1PH sense | 54–74b |
| 5′-AGCATAGCAGCTGTTGACGG-3′ | OC43 S1PI antisense | 2081–2100b | |
| RT-PCR GAPDH gene | 5′-GTGAAGGTCGGAGTCAACG-3′ | GAPDH-H sense | 10–68c |
| 5′-CACCTGGTGCTCAGTGTAGC-3′ | GAPDH-I antisense | 824–843c | |
| Sequencing S1 gene portion | 5′-ATAATATGTGCGAGTACCCAC-3′ | S-OC43-A sense | 528–539b |
| 5′-GATAAGTCGGTGCCCTCTC-3′ | S-OC43-B sense | 1058–1076b | |
| 5′-CTTGTGATAATTTGTGCACTCC-3′ | S-OC43-C sense | 1593–1614b | |
| 5′-CGAACAGTGCTCACCTATGC-3′ | S-OC43-E antisense | 1671–1690b | |
| 5′-GACTGCAAATAGCCCAAATTACC-3′ | S-OC43-F antisense | 1265–1287b | |
| 5′-TACCATCTTGATTGAAAGCGAG-3′ | S-OC43-G antisense | 857–878b | |
| 5′-GTGTTGAATTGATTGTACGTGG-3′ | S-OC43-H antisense | 455–477b |
Cloning and nucleotide sequencing.
PCR products of the HuCV-OC43 S1 gene were ligated into pGEM-T or pGEM-T Easy vector (Promega, Madison, Wis.) according to the manufacturer’s instructions. The ligation mixture was then transformed into competent XL-1 blue bacteria. Plasmidic DNA was prepared with a plasmid mini kit (Qiagen, Mississauga, Ontario, Canada) and sequenced with an automated sequencer (ABI 310 genetic analyzer; Perkin-Elmer) in both directions by using a universal primer, SP6, and internal primers (Table 1). Nucleotide sequences and predicted amino sequences were analyzed with Geneworks software for Macintosh (version 2.5.1; Oxford Molecular Ltd., Oxford, United Kingdom).
RESULTS
Acute infection of neural cell lines.
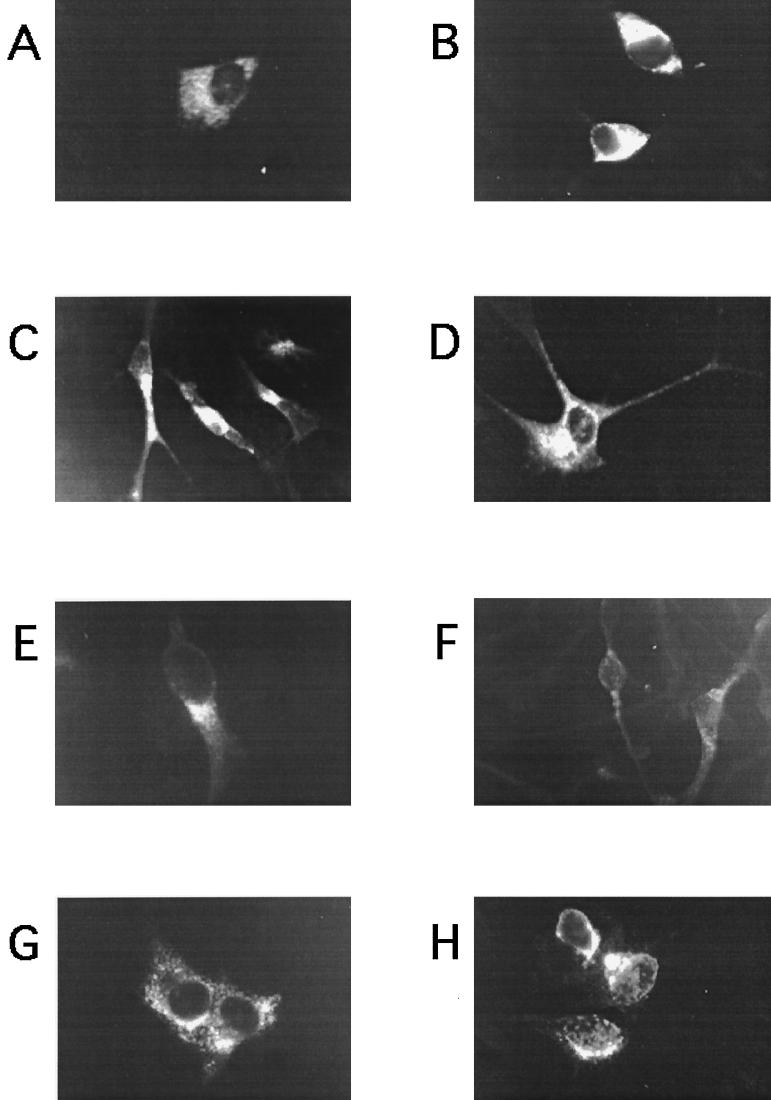
We have previously reported that HuCV-OC43 can infect primary cultures of human brain cells (12). However, the limited life span of these cell cultures did not allow us to study the effects of a long-standing coronavirus infection. Only limited results are available concerning the susceptibility to HuCV infection of immortalized human cell lines representative of cells of the nervous system (5, 20). Therefore, the susceptibility to an acute HuCV-OC43 infection of various available cell lines was evaluated, and persistent infections were established and characterized in the virus-susceptible cell lines. This HuCV-OC43 virus was initially adapted in our laboratory to replicate in the HRT-18 cell line, a human adenocarcinoma of colon and rectum. Therefore, these cells were used as a positive control for infection. Expression of the HuCV S antigen in acutely infected cells was verified by an indirect immunofluorescence assay using a specific MAb. For all cell lines tested, no signal was detected on noninfected cells or with an isotypic control MAb on infected cells (data not shown). Representative results obtained at an MOI of 1.0 for at least three independent experiments are presented in Fig. 1. The appropriateness of the indirect immunofluorescence test for the detection of HuCV-OC43 infection is indicated by its specificity but also, as is evident in Fig. 1H, by a characteristic cytoplasmic signal observed in infected HRT-18 cells (50). Both neuronal cell lines (SK-N-SH cells and H4 cells [Fig. 1A and B, respectively) were susceptible to HuCV-OC43 infection, to a comparable level. HuCV-OC43 antigens were also detected in all three astrocytic cell lines (U-373 MG cells, U-87 MG cells, and GL-15 cells [Fig. 1C to E, respectively]). However, a lower percentage of infected cells was observed for the GL-15 cell line, compared to the other astrocytic cell lines. The oligodendrocytic MO3.13 (Fig. 1F) and microglial CHME-5 (Fig. 1G) cell lines were also positive after HuCV-OC43 infection. Overall, all neural cell lines tested were susceptible to HuCV-OC43 infection, as detected by an indirect immunofluorescence assay.
FIG. 1.
Detection of human coronavirus antigens by indirect immunofluorescence on cells acutely infected by HuCV-OC43, using virus-specific MAb (1.10C.3). (A) H4 cells; (B) SK-N-SH cells; (C) U-373 MG cells; (D) U-87 MG cells; (E) GL-15 cells; (F) MO3.13 cells; (G) CHME-5 cells; (H) HRT-18 cells.
Infectious HuCV-OC43 virions were also produced by all susceptible cell lines. An assay to determine the kinetics of virion release was performed between 2 and 96 h of infection. The titers measured (data not shown) were not due to input viruses, since a strong increase in virus titers was observed from the 6 to 12 h time points. Most of the time, extracellular virus production paralleled intracellular production but was at a higher level. For most cell lines (H4, SK-N-SH, and MO3.13), maximal virus production was observed after 20 to 40 h and then a plateau effect with a slow decrease was seen for the remainder of the infection (data not shown). For the U-373 MG cell line, viral titers continued to increase until the end of the experiment (96 h). Maximal viral titers observed for all neural cell lines varied between 3.0 and 4.0 log TCID50/ml and were lower than those obtained with the HRT-18 cell line (7 log TCID50/ml). Overall, neural cell lines were able to produce infectious virions but to a lower level compared to the reference cell line.
Detection of coronavirus antigens in neural cell lines during persistent infection.
MHV can establish a persistent infection in different cell lines (17, 46, 59, 74) with various effects and is also involved in in vivo persistent infection in animals (1, 70). Since primary cultures of human brain cells cannot tolerate long-term culture, immortalized neural cell lines were used to verify the potential of a sustained persistent HuCV-OC43 infection in the CNS. Cell lines susceptible to an acute infection were infected and then cultured for several passages. Cells were collected for the detection of coronavirus antigens by indirect immunofluorescence at each cell passage. Representative results obtained for at least two independent experiments are presented in Fig. 2. Coronavirus antigens were detected in at least 1 to 5% of persistently infected HRT-18 cells (Fig. 2E). The neuronal H4 cells (Fig. 2A) and the astrocytic U-87 MG cells (Fig. 2D) showed the presence of HuCV-OC43 antigens at all cell passages tested. However, the percentage of positive cells varied from 1 to 10% for persistently infected H4 cells and 5 to 50% for U-87 MG cells. In the case of the persistently infected astrocytic U-373 MG cells, some cell passages (15 to 20) were negative, although the signal was detected at passage 21, at which >50% of cells were positive for viral antigens. Subsequently, the percentage of positive cells dropped to 5%. The MO3.13 oligodendrocytic cell line sustained a persistent infection in >50% of the cells in the monolayer, as detected by indirect immunofluorescence. Moreover, starting at the fifth passage, cytopathic effects were repeatedly observed: growth was slowed, and cells appeared bigger than normal, had fused, and contained oversized vacuoles until the end of the culture period, i.e., for more than 140 days (noninfected and persistently infected MO3.13 cells [Fig. 3A and B, respectively). The observed oversized vacuoles in persistently infected MO3.13 cells contained coronavirus antigens that were detected by immunofluorescence (Fig. 2B). On the other hand, the CHME-5, GL-15, and SK-N-SH cell lines did not show any HuCV-OC43 antigens after the second passage, even though they were all acutely infected by the same virus. Overall, HuCV-OC43 easily established a persistent infection in various neural cell lines representative of all four cellular phenotypes.
FIG. 2.
Detection of HuCV antigens by indirect immunofluorescence on cells persistently infected by HuCV-OC43, using virus-specific MAb (1.10C.3). (A) H4 cells, passage 40; (B) MO3.13 cells, passage 21; (C) U-373 MG cells, passage 22; (D) U-87 MG cells, passage 24; (E) HRT-18 cells, passage 40; (F) isotypic control MAb (5-11H.6) on virus-infected U-373 MG cells.
FIG. 3.
Cytopathic effects of a persistent HuCV-OC43 infection on MO3.13 cells. (A) Noninfected cells; (B) HuCV-OC43-infected MO3.13 cells, passage 5.
Detection of infectious virions during persistent infection.
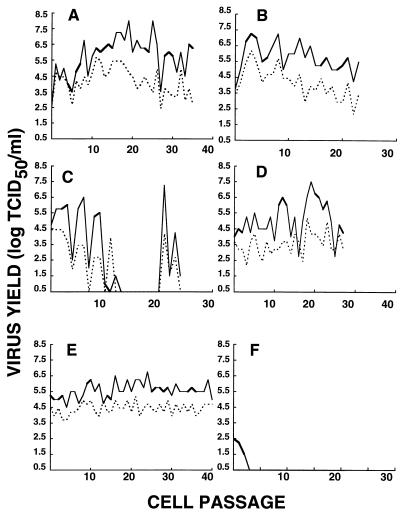
Pearson and Mims showed that a persistent HuCV-OC43 infection does not necessarily correlate with production of infectious virus (60). Therefore, we verified the presence of infectious viral particles in the supernatant and in the intracellular compartment of all persistently infected cell lines (Fig. 4). No infectious virions were detected after the third passage of infected SK-N-SH cells. Thus, no persistent infection could be established for this cell line (Fig. 4F). Similar results were obtained with GL-15 and CHME-5 cells (data not shown). All persistently infected cell lines produced infectious virions at least until the 25th passage (∼130 days). In most cases, viral titers obtained from the extracellular compartment were at a higher level than those for intracellular infectious viruses. During persistent infection, continuous production of infectious virions of at least 3.5 and even up to 8.0 log TCID50/ml was detected in the supernatants of persistently infected H4 cells (Fig. 4A). In the case of persistently infected U-373 MG cells (Fig. 4C), no virus was detected between cell passages 13 and 20, despite the presence of viruses at the beginning and at the end of the culture. This correlated with the absence of HuCV-OC43 antigen at the same cell passages, as monitored by immunofluorescence. The negative results obtained by both methods may relate to the sensitivity of these tests. Negligible, undetectable amounts of virus may have been present at these specific cell passages, since infectious virions as well as viral antigens were detected later on. Alternatively, only viral RNA may have subsisted at these cell passages. At the end of the culture period, approximately 5% of the U-373 MG cells were infected, as shown by immunofluorescence. Persistently infected U-87 MG cells (Fig. 4D) produced infectious virions in various amounts (from 2.5 to 7.5 TCID50/ml). In the case of persistently infected MO3.13 cells (Fig. 4B), relatively stable amounts of virions were detected, both intracellularly and extracellularly. Even though the persistent HuCV-OC43 infection produced cytopathic effects on MO3.13 cells, infectious virions were released from these cells. All HuCV-OC43 persistently infected cell lines were positive for expression of coronavirus antigen and release of infectious virus (a summary of results is presented in Table 2).
FIG. 4.
Yield of infectious virions from persistent HuCV-OC43 infections of various neural cell lines. (A) H4 cells; (B) MO3.13 cells; (C) U-373 MG cells; (D) U-87 MG cells; (E) HRT-18 cells; (F) SK-N-SH cells. Solid lines, supernatant (extracellular virus); dotted lines, cell lysate (intracellular virus).
TABLE 2.
Susceptibility of neural cell lines to acute and persistent infection by HuCV-OC43
| Cell line
|
Infectiona
|
||
|---|---|---|---|
| Line designation | Phenotype | Acute | Persistent |
| U-87 MG | Astrocytoma | 2+ | 2+ |
| U-373 MG | Astrocytoma (grade III) | 2+ | 2+ |
| GL-15 | Astrocytic (glioblastoma multiforme, positive for glial fibrillary acidic protein marker) | 1+ | − |
| MO3.13 | Oligodendrocytic (adult oligodendrocyte fused with a rhabdosarcoma) | 2+ | 3+ |
| CHME-5 | Microglial (fetal microglia transfected with large T antigen of simian virus 40) | 1+ | − |
| H4 | Neuroglial (ganglioglioma) | 2+ | 2+ |
| SK-N-SH | Neuroblastoma | 2+ | − |
Results obtained from an indirect immunofluorescence assay and an infectious virus titer assay. −, noninfectable or no persistence after fifth passage; 1+, <10% of cells infected; 2+, between 10 and 50% of cells infected; 3+, >50% of cells infected.
Detection of viral RNA during persistent infection.
We have also verified the presence of coronavirus RNA by RT-PCR as another indicator of viral replication. RT-PCR is more sensitive than the indirect immunofluorescence assay (68). Moreover, it could detect RNA while no antigen or virion was produced. The N protein was chosen since it is usually present at high levels during a coronavirus infection. Moreover, the 3′-coterminal nested set structure of subgenomic coronavirus mRNAs results in the presence of this gene on all viral RNAs (41). RNA from acutely infected, noninfected, and persistently infected cells at different cell passages was extracted, reverse transcribed, and amplified with specific N primers (Table 1). GAPDH RNA was detected in each case (data not shown), indicating that RNA was suitable for amplification even from noninfected cells in which no viral RNA could be detected. Viral RNA was detected in comparable amounts at every cell passage tested for persistently infected H4, MO3.13, U-87 MG, and HRT-18 cells (Fig. 5). However, in the case of the persistently infected U-373 MG cells, we observed less viral RNA at passages 15 and 20, even though the same amount of total RNA was used in all cases, corroborating negative results for infectious virus and viral antigen at cell passages 13 to 20.
FIG. 5.
Detection of the N protein gene by RT-PCR during persistent HuCV-OC43 infection of various cell lines. One tenth of the PCR amplicon obtained was loaded onto a 1.5% (wt/vol) agarose gel for electrophoretic separation. (A) H4 cells; (B) MO3.13 cells; (C) U-373 MG cells; (D) U-87 MG cells; (E) HRT-18 cells. Lanes: N, noninfected cells; A, acutely infected cells; -, negative control for reverse transcriptase and for PCR; numbers, cell passage at which RNA was extracted.
Sequencing of viral S1 gene and a region of the viral N gene from persistently infected cells.
Different laboratories have shown the presence of important determinants of neurovirulence within the S glycoprotein of MHV, especially its N-terminal half, designated S1. We looked for variations within the S1 portion of the S gene during persistent infection with HuCV-OC43, given that this region was shown in MHV to be hypervariable (33). First, an RT-PCR for this gene was performed (primers are listed in Table 1), and then 2,045-bp amplicons were cloned and sequenced in both directions. Sequencing results from acute and persistent infections at passage 42 for HRT-18, passage 36 for H4, passage 28 for U-87 MG, passage 25 for U-373 MG, and passage 24 for MO3.13 were compared to the sequence we previously reported (53). Results were confirmed by reading both nucleic acid strands to reduce errors introduced by automatic sequence analysis and by examining three clones for each cell line and infection. For all acute and persistent infections, the HuCV-OC43 S1 gene presented four point mutations compared to our published sequence (53): they were at positions 894, 895, 949, and 2025, leading to two amino acid changes (Table 3). On the other hand, Künkel and Herrler have also published a sequence of the HuCV-OC43 S glycoprotein (40) that showed these same point mutations. Moreover, they have also published sequences from two different isolates (39) that are identical to our sequence at these positions. We conclude that the input virus we used for acute and persistent infections was identical in all cases but bore sequence differences compared to the S1 gene sequence already published by our group.
TABLE 3.
Characterization of sequence changes in S1 during persistent HCV-OC43 infection
| Cell line(s) | Infection | Position of mutationa | Codon change | Amino acid change |
|---|---|---|---|---|
| HRT-18, H4, U-87 MG, U-373 MG, and MO3.13 | Acute and persistent (all clones) | 894–895b | GAA→GTT | E→V |
| 949b | GCG→GCA | No change | ||
| 2025b | ATA→ACA | I→T | ||
| HRT-18 | Persistent (passage 42) | 279c | AGT→ACT | S→T |
| 1496 | AAT→CAT | N→H | ||
| 1740cf | CGA→CAA | R→Q | ||
| H4g | Persistent (passage 36) | 102e | GAT→GCT | D→A |
| 134c | CCT→TCT | P→S | ||
| 579b | CAT→CGT | H→R | ||
| 752 | TAT→CAT | Y→H | ||
| 1445cf | CTT→TTT | L→F | ||
| U-87 MG | Persistent (passage 28) | 102c | GAT→GTT | D→V |
| 110f | AAT→TAT | N→Y | ||
| 279c | AGT→ACT | S→T | ||
| 579c | CAT→CGT | H→R | ||
| 773d | CAC→TAC | H→Y | ||
| U-373 MG | Persistent (passage 25) | 102c | GAT→GTT | D→V |
| 279c | AGT→ACT | S→T | ||
| 579c | CAT→CGT | H→R | ||
| 752 | TAT→CAT | Y→H | ||
| MO3.13 | Persistent (passage 24) | 474e | ACA→ATA | T→I |
| 579c | CAT→CGT | H→R | ||
| 752f | TAT→CAT | Y→H | ||
| 1488 | GCT→GTT | A→V | ||
| 1496 | AAT→CAT | N→H | ||
| 1811 | TTT→ATT | F→I | ||
| 2079 | TTT→TAT | F→Y |
Reference 53.
Reference 40.
Reference 39; isolates CU and VA.
Reference 39; isolate VA.
Reference 39; isolates CU and VA (a point mutation at the same position but for a different amino acid).
Point mutation observed in two out of three sequenced clones.
One H4 cell line demonstrated an in-frame deletion of 15 nucleotides (471 to 485), leading to a 5-amino-acid deletion.
No consensus concerning mutations from a nucleotide to a specific one or for specific amino acid changes was observed. Point mutations at positions 102, 279, 579, 752, and 1496 were shared by viral RNA extracted from several infected cell lines. Other point mutations were specific to only one cell line, such as the ones at positions 110 and 773 for the U-87 MG cells and positions 474, 1488, 1811, and 2079 for the MO3.13 cells. It is intriguing that the cell line in which cytopathic effects were detected (MO3.13; Fig. 3) presented the highest number of point mutations and also more distinct mutations compared to the other cell lines. A point mutation at position 579 was only found on viral RNA extracted from neural cell lines and was not observed in persistently infected HRT-18 cells. No mutation appeared essential for persistent infection since none was present on every infected cell line, with the exception of the one at position 579 for neural cell lines. One cell line (H4) demonstrated an in-frame deletion of 15 nucleotides, leading to a 5-amino-acid deletion. Some regions of the S1 gene were more prone to mutations than others: no mutations or deletions were observed between positions 773 and 1445, whereas mutations were observed within other regions of the amplified gene. For some point mutations (Table 3), exactly the same ones or mutations at the same position but not accompanied by the same amino acid change, were observed by Künkel and Herrler with their CU isolate, obtained from HuCV-OC43 passaged in MDCK cells, and their VA isolate from HuCV-OC43 passaged on Vero cells (39). These two clones were reported to have a less efficient ability to recognize 9-O-acetylated sialic acid, compared to the ATCC strain (39). However, specific amino acids involved were not identified (39). We conclude that these positions are predisposed to changes during cell passage on different cell lines.
The frequency of isolated mutations, found only in one out of three clones of the sequenced S1 gene, was between 2.1 × 10−4 and 8.6 × 10−4 for acute infections and 0 to 10.9 × 10−4 for persistent infections (data not shown). These isolated mutations were probably not representative of viral genomes but due to RT-PCR, since almost the same frequencies were observed in acute and persistent infections. Some point mutations were detected in two out of three sequenced clones from persistently infected cells, such as those at positions 1445 for H4 cells, 110 for U-87 MG cells, and 752 for MO3.13 cells (Table 3). These variations among the sequenced clones suggest that part of the viral genome population has mutated at these positions during persistent infection.
The amplified HuCV-OC43 N fragment (positions 215 to 520) was also sequenced. One point mutation, at position 401, was observed in all clones sequenced either from acutely or persistently infected cells (GGT to CGT [GGT→CGT], leading to an amino acid change from a glycine to an arginine), compared to the published sequenced (35). Thus, it must have been present in the input virus used and was not induced by our experiments.
DISCUSSION
Limited data (20) have been available concerning the susceptibility of human cell lines representative of nervous system cells to infection by HuCV-OC43. We have shown in the present study that cell lines representative of astrocytes (U-87 MG, U-373 MG, and GL-15), oligodendrocytes (MO3.13), neurons (H4 and SK-N-SH), and microglia (CHME-5) were all susceptible to an acute HuCV-OC43 infection, as illustrated by the detection of viral antigen (Fig. 1) and the release of infectious virions (a summary of results is given in Table 2). Some astrocytic cell lines (U-87 MG and U-373 MG), as well as an oligodendrocytic cell line (MO3.13) and one neuronal cell line (H4), also sustained a persistent HuCV-OC43 infection.
These results can be compared with data from infections performed on primary cultures of human neural cells. We previously showed that fetal astrocytes as well as adult astrocytes were infectable by HuCV-OC43, with concomitant release of detectable infectious virions only from fetal astrocytes. Therefore, available data is consistent with the possibility that astrocytes are susceptible to HuCV-OC43 infection, given infection of such cells in primary cultures at either the fetal or adult stage (12) and the susceptibility of astrocytoma cells (20). Interestingly, the murine MHV-JHM strain was shown to infect astrocytes in vivo (77). Moreover, during the chronic pathology induced in infected mice, they were shown to represent the predominant cells expressing inflammatory cytokines such as interleukins 1β and 6, as well as nitric oxide (76), that are probably involved in tissue damage, such as demyelination. Moreover, in situ hybridization combined with immunohistochemistry performed on brains from nonhuman primates infected intracerebrally showed that astrocytes are the target cells in white matter during an acute MHV-JHM infection (57). In addition, in asymptomatic MHV-JHM-infected mice, astrocytes apparently constitute a reservoir for persisting viruses (63). Thus, our current results combined with published work are consistent with a potential role for astrocytes in acute CNS infection with HuCV-OC43. These cells would also presumably represent a potential HuCV-OC43 reservoir in the human CNS, as was shown for MHV in mice.
The CHME-5 cell line was only susceptible to an acute HuCV-OC43 infection. We previously reported that primary cultures of human microglia were susceptible to HuCV-OC43 infection (12). Adult human microglia are among the numerous cell types (others are oligodendrocytes, astrocytes, ependymal cells, perivascular macrophages, and neurons) shown to be infected after intracranial MHV inoculation in mice (33). Microglia were susceptible to HuCV-OC43 infection, but our experiments with an immortalized cell line do not favor this cell type as a reservoir for virus during persistence in the CNS. Collins (18) has recently reported the susceptibility of human macrophages to an acute HuCV-OC43 infection. Since microglia and macrophages are closely related, her observations support our results.
Although we have previously shown that primary cultures of oligodendrocytes were not positive for viral antigen when infected with HuCV-OC43 (12), we now show that the MO3.13 oligodendrocytic cell line sustained acute and persistent infections by both HuCV-229E (4) and HuCV-OC43 (present study). Thus, more-extensive studies in primary cultures and on brain sections will be needed to confirm the potential susceptibility of myelin-synthesizing oligodendrocytes to HuCV infection. These studies are in progress.
For neuronal infection, we tested two different immortalized cell lines: SK-N-SH and H4. Both cell lines were susceptible to an acute infection as shown by detection of viral antigen and release of infectious virions. However, SK-N-SH cells did not sustain a persistent infection, while H4 cells did. Primary cultures of fetal neurons were previously shown to be negative for HuCV-OC43 (12). However, these cultures were in limited supply and multiple experiments could not be performed. Neurons are the site of coronavirus replication in mice (33) as well as the site of viral persistence in rats (71). Moreover, a HuCV-OC43 infection of primary cultures of mouse brain was shown to lead to production of infectious virus by neurons (60). Again, more work will be needed to better characterize potential neuronal susceptibility to HuCV infection. Moreover, with the H4 cell line being from a neuroglioma (ganglioglioma) that consists of a mixture of cells of neuronal and glial origins, we cannot strongly conclude that neurons can be persistently infected by HuCV-OC43. Since, to our knowledge, no human neuronal cell lines from the CNS are available (neuroblastomas are from the peripheral nervous system), we have performed our experiments with the H4 cell line. More experiments should be performed when a CNS neuronal cell line becomes available, to validate the possibility that neurons could serve as a reservoir for HuCV-OV43.
Cell lines presenting lower percentages of infected cells during an acute infection (GL-15 and CHME-5) did not sustain a persistent infection (Table 2). These results suggest that a minimal percentage of acutely infected cells was necessary for establishment of a persistent HuCV-OC43 infection. Given the fact that astrocytes and neurons do not constitute completely homogeneous populations in the nervous system, the difference in susceptibility to acute and persistent infection by HuCV-OC43 could be explained by the different original neuronal or astrocytic cell that was immortalized, giving rise to the cell lines we have used. Additional experiments defining the exact phenotype of the cell lines would be necessary to fully explain the difference in susceptibility to HuCV-OC43 infections, such as number of viral receptors, secretion of viral inhibitors such as interferon, or induction of apoptosis in infected cells. The differences observed between immortalized and primary cells for neuronal and oligodendrocytic cell types could be due to different degrees of differentiation of the neural cells we have used as immortalized cell lines compared to primary cultures. Also, it is possible that the sensitivities of our assays were insufficient to detect a low level of infection by this virus. Nonetheless, we cannot rule out the possibility that the immortalized cell lines have acquired new properties, such as susceptibility to HuCV infections.
Different strains and variants of the same virus (MHV) demonstrate different neurotropic properties. MHV-4 infects mainly neurons (24) in intracerebrally inoculated mice, MHV-JHM is found mainly in oligodendrocytes of spinal cord and brain of the same animals (80), and MHV-A59 infects mostly neurons and neural processes and terminals after an intranasal inoculation of mice (42). The genetic constitution of various viral strains and variants affects the neurotropism and the pathology observed, and these differences could appear during in vivo and in vitro infections (78, 79). Thus, it was important to look at genetic variations of HuCV in the context of human neurotropism, given the variety of results obtained with different MHV strains. Indeed, multiple point and deletion mutations within the N and S genes were reported to occur during a 42-day period postinfection in mice, suggesting a rapid evolution of viral genomic RNA (1, 66). Moreover, important molecular determinants of MHV pathogenesis are located within the S protein (47, 52). We looked at the S1 gene fragment since most of the mutations observed in MHV were concentrated within this fragment of the spike protein. The selection of virus variants with distinct fixed mutations and the evolution of more-virulent phenotypes was shown to arise in cell lines persistently infected with MHV (16, 17, 30). The mutation frequencies and rates at which they appear were shown to vary at different genomic positions (17). However, no specific mutation was necessary for the establishment of in vivo persistence (66), even though a variety of mutations in the N and S genes was observed in mice, suggesting the development of quasispecies (66). On the other hand, during a persistent infection of rats by MHV, the S gene was reported to be stable, arguing in this case against the emergence of quasispecies (75). The generation of diverse genomic populations during a persistent infection could depend on host factors, explaining differences observed between mice and rats.
The error rate of retrovirus reverse transcriptase enzymes has been estimated at 10−4 on complex RNA template (69). The Expand high fidelity PCR system DNA polymerase (Taq Pwo DNA polymerases; Roche Diagnostics) we used has a calculated error rate of 8.5 × 10−6. Since we performed 30 cycles, the total error rate of our RT-PCR could be estimated at 3.6 × 10−4 [=1 × 10−4 + (8.5 × 10−6 × 30 cycles)]. Variations between molecular clones of virus genome generated by RT-PCR should be greater than that expected from the error rate of reverse transcriptase and Taq (69) to be considered as mutations representative of the viral population. However, when we added mutations found on two and three out of three molecular clones from persistently infected cells, these mutation rates were 2.5 × 10−3 to 3.4 × 10−3 for neural cells, corresponding to a 6.9- to 9.2-fold increase compared to the estimated error rate for RT-PCR. Substitutions present in more than one clone are likely to represent mutations actually present in the virus population (69). The mutations we observed in only one out of three clones (data not shown) were probably due to RT-PCR-induced errors since the rate of these variations was comparable to the error rate expected and these rates were not significantly different between acute and persistent infections. However, point mutations representative of the viral population were found within two or three clones sequenced in both directions. These point mutations would lead to amino acid changes as shown by the predicted sequence (Table 3). Mutations present in two out of three sequenced molecular clones suggest a variation within the viral population (Table 3) since not every viral genome has acquired these mutations. A larger sample of sequenced clones will be necessary for clearly evaluating the generation of viral quasispecies during a persistent HuCV-OC43 infection. Nonetheless, our results do favor the emergence of quasispecies during a persistent infection by HuCV-OC43 in neural cells, especially in the H4, U-87 MG, and MO3.13 cell lines. In these cells, at least one point mutation was observed in two out of three clones, meaning that the entire viral population did not mutate at this position (Table 3). The significance of the point mutations we observed is not yet established. We can speculate that they favor viral replication within these particular cell lines. Some mutations were prone to arise, since we observed them in many different cell lines and some were even observed after cell culture on different cell lines by another group (39).
Studies with other viruses have demonstrated that specific point mutations are critical for viral tropism and that the mutations arising are not necessarily numerous. For example, lymphocytic choriomeningitis virus (LCMV), an RNA virus, exhibits minimal genetic drift during chronic infection in its natural host, the mouse. Moreover, a single point mutation in its genome was associated with an organ-specific selection (2). Such studies are still to come for HuCV. The development of an in vitro model of persistent HuCV infection in neural cells will now allow us to explore mechanisms underlying acute and persistent CNS infections with HuCV. Kinetics of production of viral proteins and RNA, involvement of interferons, and the number of viral receptors on cells are all avenues that could now be investigated.
Numerous in vitro and in vivo experiments have proven the neurotropism of murine coronaviruses. However, a coronavirus infection does not necessarily yield severe pathological symptoms. Indeed, it was shown that MHV-JHM could persist and replicate within neurons and oligodendrocytes of rats without causing cell degeneration and death or eliciting an immune attack (70). On the other hand, an MHV-JHM mutant was shown to establish reproducible persistent infections in mice and to induce progression from acute demyelinating disease to a chronic recurring form (37). MHV-JHM also establishes a persistent infection in the retina when injected by the intravitreal route, i.e., into another immunologically privileged site in the body, where viral RNA was detected by in situ hybridization up to 60 days postinfection (38). The neurotropism and persistence of HuCV remain to be explored in more depth. However, we predict from our results that several kinds of cells in the human CNS (astrocytes, oligodendrocytes, and some neurons) could presumably be used as a potential reservoir during a persistent HuCV-OC43 infection.
Other viruses have been identified as persisting viruses in the human CNS. For example, Borna disease viruses cause CNS disease in several vertebrate species that is characterized by behavioral abnormalities. Recent studies suggest a potential role for these viruses in human mental health (31). Also, human immunodeficiency virus is known to induce dementia in AIDS patients, and JC virus, a polyomavirus, is associated with most cases of progressive multifocal leukoencephalopathy (3). Finally, persistent measles infection of the human brain is associated with subacute sclerosing panencephalitis and measles inclusion body encephalitis (45). A variety of pathological effects could be induced by persistent infections at this anatomical site, where the maintenance of intact neurons is primordial. The mechanisms by which coronavirus RNA persists for long periods in the CNS are still unclear. Similarly, Sindbis virus was shown to persist in the mouse brain for a long period, in a nonproductive form (44). An endogenous reverse transcriptase that could generate cDNA transcripts from LCMV in a host-specific manner during an acute LCMV infection was reported. This cDNA was shown to persist long after the original acute infection had waned below detectable levels (36). Indeed, numerous unanswered questions have arisen with the increasing evidence of a persistent presence of viruses in various organs.
It is striking that coronavirus RNA was detected in both rodents and humans for long periods after primary infection (1, 4, 6, 55, 66). It is unlikely that all human brain tissues that tested positive for HuCV RNA in different laboratories (6, 55) were from humans infected by HuCV just prior to death. A low level of coronavirus antigens might be expressed in order to escape from immune surveillance during a persistent infection, since most often only viral RNA and neither infectious viruses nor viral antigens are detected for such long periods. Detailed in situ detection of HuCV in human brain sections would confirm the neurotropism observed in vitro with primary and immortalized cultures and provide clues for the outcome of the presence of HuCV in human CNS. These studies are in progress.
ACKNOWLEDGMENTS
This work was supported by grant MT-9203 from the Medical Research Council of Canada to P.J.T., who also gratefully acknowledges a senior scholarship award from the Fonds de la Recherche en Santé du Québec (FRSQ). N.A. is grateful to the Institut Armand-Frappier as well as to the Multiple Sclerosis Society of Canada for studentship support.
We thank Francine Lambert for excellent technical assistance. We thank Julie Edwards for critically reviewing the manuscript.
REFERENCES
- 1.Adami C, Pooley J, Glomb J, Stecker E, Fazal F, Fleming J O, Baker S C. Evolution of mouse hepatitis virus (MHV) during chronic infection: quasispecies nature of the persisting MHV RNA. Virology. 1995;209:337–346. doi: 10.1006/viro.1995.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed R, Hahn C S, Somasundaram T, Villarete L, Matloubian M, Strauss J H. Molecular basis of organ-specific selection of viral variants during chronic infection. J Virol. 1991;65:4242–4247. doi: 10.1128/jvi.65.8.4242-4247.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aksamit A R., Jr Progressive multifocal eukoencephalopathy: a review of the pathology and pathogenesis. Microsc Res Tech. 1995;32:302–311. doi: 10.1002/jemt.1070320405. [DOI] [PubMed] [Google Scholar]
- 4.Arbour N, Ekandé S, Côté G, Lachance C, Chagnon F, Tardieu M, Cashman N R, Talbot P J. Persistent infection of human oligodendrocytic and neuroglial cell lines by human coronavirus 229E. J Virol. 1999;73:3326–3337. doi: 10.1128/jvi.73.4.3326-3337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arbour N, Talbot P J. Persistent infection of neural cell lines by human coronaviruses. Adv Exp Med Biol. 1998;440:575–581. doi: 10.1007/978-1-4615-5331-1_75. [DOI] [PubMed] [Google Scholar]
- 6.Arbour, N., and P. J. Talbot. Unpublished data.
- 7.Barnett E M, Perlman S. The olfactory nerve and not the trigeminal nerve is the major site of CNS entry for mouse hepatitis virus, strain JHM. Virology. 1993;194:185–191. doi: 10.1006/viro.1993.1248. [DOI] [PubMed] [Google Scholar]
- 8.Barthold S W, de Souza M S, Smith A L. Susceptibility of laboratory mice to intranasal and contact infection with coronaviruses of other species. Lab Anim Sci. 1990;40:481–485. [PubMed] [Google Scholar]
- 9.Barthold S W, Smith A L. Viremic dissemination of mouse hepatitis virus-JHM following intranasal inoculation of mice. Arch Virol. 1992;122:35–44. doi: 10.1007/BF01321116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergmann C C, Yao Q, Lin M, Stohlman S A. The JHM stain of mouse hepatitis virus induces a spike protein-specific Db-restricted cytotoxic T cell response. J Gen Virol. 1996;77:315–325. doi: 10.1099/0022-1317-77-2-315. [DOI] [PubMed] [Google Scholar]
- 11.Bocchini V, Casalone R, Collini P, Rebel G, Lo Curto F. Changes in glial fibrillary acidic protein and karyotype during culturing of two cell lines established from human glioblastoma multiforme. Cell Tissue Res. 1991;265:73–81. doi: 10.1007/BF00318141. [DOI] [PubMed] [Google Scholar]
- 12.Bonavia A, Arbour N, Yong V W, Talbot P J. Infection of primary cultures of human neural cells by human coronaviruses 229E and OC43. J Virol. 1997;71:800–806. doi: 10.1128/jvi.71.1.800-806.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchmeier M J, Dalziel R G, Koolen M J M, Lampert P W. Molecular determinants of CNS virulence of the coronavirus mouse hepatitis virus-4. In: Brinton M A, Rueckert R R, editors. Positive-strand RNA viruses. New York, N.Y: Alan R. Liss, Inc.; 1987. pp. 409–422. [Google Scholar]
- 14.Cabirac G F, Soike K F, Zhang J-Y, Hoel K, Butunoi C, Cai G-Y, Johnson S, Murray R S. Entry of coronavirus into primate CNS following peripheral infection. Microb Pathog. 1994;16:349–357. doi: 10.1006/mpat.1994.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castro R F, Perlman S. CD8+ T-cell epitopes within the surface glycoprotein of a neurotropic coronavirus and correlation with pathogenicity. J Virol. 1995;69:8127–8131. doi: 10.1128/jvi.69.12.8127-8131.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W, Baric R S. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J Virol. 1996;70:3947–3960. doi: 10.1128/jvi.70.6.3947-3960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W, Baric R S. Function of a 5′-end genomic RNA mutation that evolves during persistent mouse hepatitis virus infection in vitro. J Virol. 1995;69:7529–7540. doi: 10.1128/jvi.69.12.7529-7540.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins A R. Human macrophages are susceptible to coronavirus OC43. Adv Exp Med Biol. 1998;440:635–639. doi: 10.1007/978-1-4615-5331-1_82. [DOI] [PubMed] [Google Scholar]
- 19.Collins A R, Knobler R L, Powell H, Buchmeier M J. Monoclonal antibodies to murine hepatitis virus-4 (strain JHM) define the viral glycoprotein responsible for attachment and cell-cell fusion. Virology. 1982;119:358–371. doi: 10.1016/0042-6822(82)90095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins A R, Sorensen O. Regulation of viral persistence in human glioblastoma and rhabdosarcoma cells infected with coronavirus OC43. Microb Pathog. 1986;1:573–582. doi: 10.1016/0882-4010(86)90042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daniel C, Anderson R, Buchmeier M J, Fleming J O, Spaan W J M, Wege H, Talbot P J. Identification of an immunodominant linear neutralization domain of the S2 portion of the murine coronavirus spike glycoprotein and evidence that it forms part of a complex tridimensional structure. J Virol. 1993;67:1185–1194. doi: 10.1128/jvi.67.3.1185-1194.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dveksler G S, Pensiero M N, Cardellichio C B, Williams R K, Jiang G S, Holmes K V, Dieffenbach C W. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J Virol. 1991;65:6881–6891. doi: 10.1128/jvi.65.12.6881-6891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ercolani L, Florence B, Denaro M, Alexander M. Isolation and complete sequence of a functional human glyceraldehyde-3-phosphate dehydrogenase gene. J Biol Chem. 1988;263:15335–15341. [PubMed] [Google Scholar]
- 24.Fazakerley J K, Parker S E, Bloom F, Buchmeier M J. The V5A13.1 envelope glycoprotein deletion mutant of mouse hepatitis virus type-4 is neuroattenuated by its reduced rate of spread in the central nervous system. Virology. 1992;187:178–188. doi: 10.1016/0042-6822(92)90306-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fazzini E, Fleming J, Fahn S. Cerebrospinal fluid antibodies to coronavirus in patients with Parkinson’s disease. Mov Disord. 1992;7:153–158. doi: 10.1002/mds.870070210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleming J O, Houtman J J, Alaca H, Hinze H C, McKenzie D, Aiken J, Bleasdale T, Baker S. Persistence of viral RNA in the central nervous system of mice inoculated with MHV-4. Adv Exp Med Biol. 1994;342:327–332. doi: 10.1007/978-1-4615-2996-5_50. [DOI] [PubMed] [Google Scholar]
- 27.Fleming J O, Trousdale M D, Bradbury J, Stohlman S A, Weiner L P. Experimental demyelination induced by coronavirus JHM (MHV-4): molecular identification of a viral determinant of paralytic disease. Microb Pathog. 1987;3:9–20. doi: 10.1016/0882-4010(87)90033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flintoff W F, Van Dinter S. Several rat cell lines share a common defect in their inability to internalize murine coronaviruses efficiently. J Gen Virol. 1989;70:1713–1724. doi: 10.1099/0022-1317-70-7-1713. [DOI] [PubMed] [Google Scholar]
- 29.Gallagher T M, Parker S E, Buchmeier M J. Neutralization-resistant variants of a neurotropic coronavirus are generated by deletions within the amino-terminal half of the spike glycoprotein. J Virol. 1990;64:731–741. doi: 10.1128/jvi.64.2.731-741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gombold J L, Hingley S T, Weiss S R. Fusion-defective mutants of mouse hepatitis virus A59 contain a mutation in the spike protein cleavage signal. J Virol. 1993;67:4504–4512. doi: 10.1128/jvi.67.8.4504-4512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez-Dunia D, Sauder C, de la Torre J C. Borna disease virus and the brain. Brain Res Bull. 1997;44:647–664. doi: 10.1016/S0361-9230(97)00276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamre D, Procknow J J. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 33.Houtman J J, Fleming J O. Pathogenesis of mouse hepatitis virus-induced demyelination. J Neurovirol. 1996;2:361–376. doi: 10.3109/13550289609146902. [DOI] [PubMed] [Google Scholar]
- 34.Janabi N, Peudenier S, Héron B, Ng K H, Tardieu M. Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglia cells with the SV40 large T antigen. Neurosci Lett. 1995;195:105–108. doi: 10.1016/0304-3940(94)11792-h. [DOI] [PubMed] [Google Scholar]
- 35.Kamahora T, Soe L H, Lai M M C. Sequence analysis of nucleocapsid gene and leader RNA of human coronavirus OC43. Virus Res. 1989;12:1–9. doi: 10.1016/0168-1702(89)90048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klenerman P, Hengartner, Zinkernagel R M. A non-retroviral RNA virus persists in DNA form. Nature. 1997;390:298–301. doi: 10.1038/36876. [DOI] [PubMed] [Google Scholar]
- 37.Knobler R L, Lampert P W, Oldstone M B A. Virus persistence and recurring demyelination produced by a temperature-sensitive mutant of MHV-4. Nature. 1982;298:279–280. doi: 10.1038/298279a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komurasaki Y, Nagineni C N, Wang Y, Hooks J J. Virus RNA persists within the retina in coronavirus-induced retinopathy. Virology. 1996;222:446–450. doi: 10.1006/viro.1996.0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Künkel F, Herrler G. Structural and functional analysis of the S proteins of two human coronavirus OC43 strains adapted to grow in different cells. Arch Virol. 1996;141:1123–1131. doi: 10.1007/BF01718615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Künkel F, Herrler G. Structural and functional analysis of the surface protein of human coronavirus OC43. Virology. 1993;195:195–202. doi: 10.1006/viro.1993.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavi E, Fishman P S, Highkin P S, Weiss S R. Limbic encephalitis after inhalation of a murine coronavirus. Lab Investig. 1988;58:31–36. [PubMed] [Google Scholar]
- 43.Lavi E, Gilden D H, Highkin M K, Weiss S R. Persistence of mouse hepatitis virus A59 RNA in a slow virus demyelinating infection in mice as detected by in situ hybridization. J Virol. 1984;51:563–566. doi: 10.1128/jvi.51.2.563-566.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levine B, Griffin D E. Persistence of viral RNA in mouse brains after recovery from acute alphavirus encephalitis. J Virol. 1992;66:6429–6435. doi: 10.1128/jvi.66.11.6429-6435.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liebert U G. Measles virus infections of the central nervous system. Intervirology. 1997;40:176–184. doi: 10.1159/000150544. [DOI] [PubMed] [Google Scholar]
- 46.Lucas A, Coulter M, Anderson R, Dales S, Flintoff W. In vivo and in vitro models of demyelinating diseases. II. Persistence and host-regulated thermosensitivity in cells of neural derivation infected with mouse hepatitis and measles viruses. Virology. 1978;88:325–337. doi: 10.1016/0042-6822(78)90289-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Makino S, Fleming J O, Keck J G, Stohlman S A, Lai M M C. RNA recombination of coronaviruses: localization of neutralizing epitopes and neuropathogenic determinants on the carboxyl terminus of peplomers. Proc Natl Acad Sci USA. 1987;84:6567–6571. doi: 10.1073/pnas.84.18.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Massa P T, Wege H, ter Meulen V. Growth pattern of various JHM coronavirus isolates in primary rat glial cell cultures correlates with differing neurotropism in vivo. Virus Res. 1988;9:133–144. doi: 10.1016/0168-1702(88)90028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McIntosh K, Becker W B, Chanock R M. Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc Natl Acad Sci USA. 1967;58:2268–2273. doi: 10.1073/pnas.58.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McIntosh K, McQuillin J, Reed S E, Gardner P S. Diagnosis of human coronavirus infection by immunofluorescence: method and application to respiratory disease in hospitalized children. J Med Virol. 1978;2:341–346. doi: 10.1002/jmv.1890020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McLaurin J, Trudel G C, Shaw I T, Antel J P, Cashman N R. A human glial hybrid cell line differentially expressing genes subserving oligodendrocyte and astrocyte phenotype. J Neurobiol. 1995;26:283–293. doi: 10.1002/neu.480260212. [DOI] [PubMed] [Google Scholar]
- 52.Morris V L, Tieszer C, Mackinnon J, Percy D. Characterization of coronavirus JHM variants isolated from Wistar Furth rats with a viral-induced demyelinating disease. Virology. 1989;169:127–136. doi: 10.1016/0042-6822(89)90048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mounir S, Talbot P J. Molecular characterization of the S protein gene of human coronavirus OC43. J Gen Virol. 1993;74:1981–1987. doi: 10.1099/0022-1317-74-9-1981. [DOI] [PubMed] [Google Scholar]
- 54.Mounir S, Talbot P J. Sequence analysis of the membrane protein gene of human coronavirus OC43 and evidence for O-glycosylation. J Gen Virol. 1992;73:2731–2736. doi: 10.1099/0022-1317-73-10-2731. [DOI] [PubMed] [Google Scholar]
- 55.Murray R S, Brown B, Brian D, Cabirac G F. Detection of coronavirus RNA and antigen in multiple sclerosis brain. Ann Neurol. 1992;31:525–533. doi: 10.1002/ana.410310511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murray R S, Cai G Y, Hoel K, Zhang J-Y, Soike K F, Cabirac G F. Coronavirus infects and causes demyelination in primate central nervous system. Virology. 1992;188:274–284. doi: 10.1016/0042-6822(92)90757-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murray R S, Cai G Y, Soike K F, Cabirac G F. Further observations on coronavirus infection of primate CNS. J Neurovirol. 1997;3:71–75. doi: 10.3109/13550289709015795. [DOI] [PubMed] [Google Scholar]
- 58.Myint S H. Human coronaviruses—a brief review. Rev Med Virol. 1994;4:35–46. [Google Scholar]
- 59.Okumura A, Machii K, Azuma S, Toyoda Y, Kyuwa S. Maintenance of pluripotency in mouse embryonic stem cells persistently infected with murine coronavirus. J Virol. 1996;70:4146–4149. doi: 10.1128/jvi.70.6.4146-4149.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pearson J, Mims C A. Differential susceptibility of cultured neural cells to the human coronavirus OC43. J Virol. 1985;53:1016–1019. doi: 10.1128/jvi.53.3.1016-1019.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perlman S, Evans G, Afifi A. Effect of olfactory bulb ablation on spread of a neurotropic coronavirus into the mouse brain. J Exp Med. 1990;172:1127–1132. doi: 10.1084/jem.172.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perlman S, Jacobsen G, Afifi A. Spread of a neurotropic murine coronavirus into the CNS via the trigeminal and olfactory nerves. Virology. 1989;170:556–560. doi: 10.1016/0042-6822(89)90446-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perlman S, Ries D. The astrocyte is a target cell in mice persistently infected with mouse hepatitis virus, strain JHM. Microb Pathog. 1987;3:309–314. doi: 10.1016/0882-4010(87)90064-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Resta S, Luby J P, Rosenfeld C R, Siegel J D. Isolation and propagation of a human enteric coronavirus. Science. 1985;229:978–981. doi: 10.1126/science.2992091. [DOI] [PubMed] [Google Scholar]
- 65.Riski H, Hovi T. Coronavirus infections of man associated with diseases other than the common cold. J Med Virol. 1980;6:259–265. doi: 10.1002/jmv.1890060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rowe C L, Baker S C, Nathan M J, Fleming J O. Evolution of mouse hepatitis virus: detection and characterization of spike deletion variants during persistent infection. J Virol. 1997;71:2959–2969. doi: 10.1128/jvi.71.4.2959-2969.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salmi A, Ziola B, Hovi T, Reunanen M. Antibodies to coronaviruses OC43 and 229E in multiple sclerosis patients. Neurology. 1982;32:292–295. doi: 10.1212/wnl.32.3.292. [DOI] [PubMed] [Google Scholar]
- 68.Sizun J, Arbour N, Talbot P J. Comparison of immunofluorescence with monoclonal antibodies and RT-PCR for the detection of human coronaviruses 229E and OC43 in cell culture. J Virol Methods. 1998;72:145–152. doi: 10.1016/S0166-0934(98)00013-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith D B, McAllister J, Casino C, Simmonds P. Virus ‘quasispecies’: making a mountain out of a molehill? J Gen Virol. 1997;78:1511–1519. doi: 10.1099/0022-1317-78-7-1511. [DOI] [PubMed] [Google Scholar]
- 70.Sorensen O, Coulter-Mackie M B, Puchalski S, Dales S. In vivo and in vitro models of demyelinating disease. IX. Progression of JHM virus infection in the central nervous system of the rat during overt and asymptomatic phases. Virology. 1984;137:347–357. doi: 10.1016/0042-6822(84)90227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sorensen O, Dales S. In vivo and in vitro models of demyelinating disease: JHM virus in the rat central nervous system localized by in situ cDNA hybridization and immunofluorescent microscopy. J Virol. 1985;56:434–438. doi: 10.1128/jvi.56.2.434-438.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sorensen O, Dugre R, Percy D, Dales S. In vivo and in vitro models of demyelinating disease: endogenous factors influencing demyelinating disease caused by mouse hepatitis virus in rats and mice. Infect Immun. 1982;37:1248–1260. doi: 10.1128/iai.37.3.1248-1260.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stewart J N, Mounir S, Talbot P J. Human coronavirus gene expression in the brains of multiple sclerosis patients. Virology. 1992;191:502–505. doi: 10.1016/0042-6822(92)90220-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stohlman S A, Weiner L P. Stability of neurotropic mouse hepatitis virus (JHM strain) during chronic infection of neuroblastoma cells. Arch Virol. 1978;57:53–61. doi: 10.1007/BF01315637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stühler A, Flory E, Wege H, Lassmann H, Wege H. No evidence for quasispecies populations during persistence of the coronavirus mouse hepatitis virus JHM: sequence conservation within the surface glycoprotein S in Lewis rats. J Gen Virol. 1997;78:747–756. doi: 10.1099/0022-1317-78-4-747. [DOI] [PubMed] [Google Scholar]
- 76.Sun N, Grzybicki D, Castro R F, Murphy S, Perlman S. Activation of astrocytes in the spinal cord of mice chronically infected with a neurotropic coronavirus. Virology. 1995;213:482–493. doi: 10.1006/viro.1995.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun N, Perlman S. Spread of a neurotropic coronavirus to spinal cord white matter via neurons and astrocytes. J Virol. 1995;69:633–641. doi: 10.1128/jvi.69.2.633-641.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taguchi F, Massa P T, ter Meulen V. Characterization of a variant virus isolated from neural cell culture after infection of mouse coronavirus JHMV. Virology. 1986;155:267–270. doi: 10.1016/0042-6822(86)90187-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taguchi F, Siddell S G, Wege H, ter Meulen V. Characterization of a variant virus selected in rat brains after infection by coronavirus mouse hepatitis virus JHM. J Virol. 1985;54:429–435. doi: 10.1128/jvi.54.2.429-435.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takahashi K, Goto N, Ishida T, Katami K, Fujiwara K. Acute demyelination in mice inoculated intraspinally with mouse hepatitis virus, JHM strain. Jpn J Exp Med. 1981;51:323–330. [PubMed] [Google Scholar]
- 81.Talbot P J, Salmi A A, Knobler R L, Buchmeier M J. Topographical mapping of epitopes on the glycoproteins of murine hepatitis virus-4 (strain JHM): correlation with biological activities. Virology. 1984;131:250–260. doi: 10.1016/0042-6822(84)90032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tardieu M, Boespflug O, Barbé T. Selective tropism of a neurotropic coronavirus for ependymal cells, neurons, and meningeal cells. J Virol. 1986;60:574–582. doi: 10.1128/jvi.60.2.574-582.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Watanabe R, Wege H, Ter Meulen V. Adoptive transfer of EAE-like lesions from rats with coronavirus-induced demyelinating encephalomyelitis. Nature. 1983;305:150–153. doi: 10.1038/305150a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wege H. Immunopathological aspects of coronavirus infections. Springer Semin Immunopathol. 1995;17:133–148. doi: 10.1007/BF00196162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wege H, Winter J, Meyermann R. The peplomer protein E2 of coronavirus JHM as a determinant of neurovirulence: definition of critical epitopes by variant analysis. J Gen Virol. 1988;69:87–98. doi: 10.1099/0022-1317-69-1-87. [DOI] [PubMed] [Google Scholar]
- 86.Yokomori K, Lai M M C. Mouse hepatitis virus utilizes two carcinoembryonic antigens as alternative receptors. J Virol. 1992;66:6194–6199. doi: 10.1128/jvi.66.10.6194-6199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]