Abstract
Objective
The objective of this study was to investigate the genetic control of polyphenol accumulation in red raspberry (Rubus idaeus L).
Methods
The levels of total anthocyanins and 37 individual polyphenol metabolites were measured over three years in a raspberry biparental mapping population. Quantitative trait loci (QTLs) for these traits were mapped onto a high-density SNP linkage map.
Results
At least one QTL was detected for each trait, with good consistency among the years. On four linkage groups (LG), there were major QTLs affecting several metabolites. On LG1, a QTL had large effects on anthocyanins and flavonols containing a rutinoside or rhamnose group. On LG4, a QTL had large effects on several flavonols and on LG5 and LG6 QTLs had large effects on ellagic acid derivatives. Smaller QTLs were found on LG2 and LG3.
Conclusion
The identification of robust QTLs for key polyphenols in raspberry provides great potential for marker-assisted breeding for improved levels of potentially health beneficial components.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11306-023-02033-7.
Keywords: Raspberry, Polyphenols, Anthocyanins, Ellagitannins, Genetic control, Heritability, QTLs
Introduction
World raspberry production has risen steadily from 520 000 tonnes in 2010 to 896 000 tonnes in 2020 [FAO figures] with 75% of production in Europe. The value of raspberry production quadrupled in the UK from 1995 to 2015 supported by a drive for increased class I cultivation for the fresh market [DEFRA figures] although class II fruits also supply the increasing popularity of raspberry juice products (Rajauria & Tiwari, 2018) as well as traditional uses in purees, jams, and preserves.
Raspberries are valued for their distinctive flavour and colour and raspberry fruit breeding has widened its focus from traits associated with agronomic performance and disease resistance towards those associated with fruit sensory quality (Jennings, 2018; Jennings et al., 2016) and potential health benefits (e.g., Mazzoni et al., 2016). The potential health benefits of raspberries have been associated with their polyphenol content and composition (e.g., Hancock et al., 2018). The distinctive coloration of red raspberries is due to their accumulation of anthocyanins during ripening, but these are accompanied by the presence of ellagitannins, ellagic acid derivatives, and flavonols (e.g., Carvalho et al., 2013; McDougall et al., 2005, 2014; Mullen et al., 2003). The anthocyanins and ellagitannins are the major polyphenol components and have received the most attention for potential health benefits (e.g. Rao & Snyder, 2010). The ellagitannins, in particular, contribute to astringency, which is an important factor in the characteristic sensory properties of this fruit (e.g. He et al., 2015).
The levels of anthocyanins, ellagitannins and other phenolics vary between different Rubus species and indeed different raspberry varieties (e.g. Bradish et al., 2012; Deighton et al., 2000). Although phenolic content and composition can vary with agronomic practice, location of growth, abiotic and biotic stresses, there is a strong genetic element in the control of accumulation (Anttonen & Karjalainen, 2005; Connor et al. 2005; Remberg et al., 2010; Dobson et al., 2012; Bradish et al., 2012).
Previous studies have used the genetic linkage map developed from a cross between red raspberry cultivars, Glen Moy and Latham (Graham et al., 2004), followed by Quantitative Trait Loci (QTL) mapping and have identified multiple markers for a broad range of important raspberry agronomical characteristics, which can lead to improved cultivars [e.g. McCallum et al., 2018; Scolari et al., 2021]. Also using this approach, studies at the James Hutton Institute established QTLs for total anthocyanin content and for the levels of selected major individual raspberry anthocyanins (Kassim et al., 2009; McCallum et al., 2010; Paterson et al., 2013). In other studies, consistent QTLs were also established for total phenol content and total anthocyanin content in year-on-year studies (Dobson et al, 2012). As total anthocyanin content is a sub-set of total phenol content, QTLs for total phenol which did not align with those for total anthocyanin content could be related to variation in the other polyphenol components.
The genetic linkage map used in these studies based on the Glen Moy-Latham cross has subsequently been enhanced using GbS (Hackett et al., 2018), providing a higher-density GbS map that allows QTLs to be identified more precisely. The establishment of a draft genome sequence for Glen Moy aided the development of the GbS map and also allows direct assignment of SNP marker information to the Glen Moy genome scaffolds. These resources provide the ability to perform more precise QTL analysis and genes associated with QTLs can also be examined against gene expression data across fruit development to correlate expression against the timing of polyphenol accumulation. This study examines the variation in individual polyphenol components in the progeny set of this cross identified by liquid chromatography mass spectrometry (LC–MSn) across three years of cultivation. We combine this data with marker data to identify robust and consistent QTLs associated with specific polyphenol classes. The data presented offers opportunities for the development of markers to accelerate the development of raspberry varieties with specific increases in key polyphenolic antioxidants.
Materials and methods
Mapping population and map
The mapping population was the previously reported cross between the North American raspberry Latham and the Scottish raspberry Glen Moy, with 188 offspring (Graham et al. 2004). This cross has been studied in detail over many years at the James Hutton Institute, for traits such as fruit ripening and fruit characteristics including anthocyanin content (e.g. Graham et al., 2009; Kassim et al., 2009; McCallum et al., 2010). These traits have been mapped on a map of medium density, with up to 439 markers for the study of crumbly fruit by Graham et al. (2015). More recently the linkage map was extended by 2348 SNPs from genotyping-by-sequencing to give a high-density linkage map and the fruit ripening data was reanalysed using this map (Hackett et al., 2018). The high-density map has been used for the QTL interval mapping analysis below.
Metabolite analysis using liquid chromatography mass spectrometry (LC-MSn)
Fruit from the parents and offspring of the mapping population were analysed for metabolite profiles over 3 years, 2010, 2011 and 2015. In 2010, two field replicates of the population were analysed and in 2011 and 2015, three field replicates were used for each year. The analysis was carried out using LC–MSn, with one batch for each field replicate in 2010 and 2011. In 2015, extra quality control samples were included, and the samples were analysed in five batches. The weather conditions in the analysis years, 2010, 2011 and 2015 were substantially different in maximum temperature, distribution of rainfall and sun hours across the growing season (results not shown).
As before (Dobson et al., 2012), fruit was picked when ripe, placed in labelled bags stored on ice, transferred to the laboratory within one hour and frozen at − 20 °C until extraction. Whilst still frozen, a representative subsample of fruit from each progeny (5–8 berries equivalent to 7–10 g) was weighed. The selected berries were cut in half, then extracted using a glass tissue homogenizer with a PTFE pestle with an equal volume to weight of ice-cold 100% acetonitrile (ACN) containing 0.1% formic acid (FA). This method does not crush the seeds, so it is possibly not representative of some components (e.g., ellagitannins) that are known to be enriched in the seeds (e.g., Arnold et al., 2022). The homogenate was transferred in a centrifuge tube and centrifuged at 2 750 g for 10 min at 5 °C (Eppendorf 5810 R) and the supernatant aliquoted into 1 mL amounts. The extracts were analysed for total anthocyanin content as described by McDougall et al. (2005) using a standard curve of cyanidin-3-O-glucose (Extrasynthese, Genay, France). The aliquots were dried in a Speed-Vac then stored at − 20 °C.
The dried raspberry extracts were resuspended in 475 μL of 10% ACN containing 0.1% FA with vortex mixing then 25 μL of internal standard solution (0.5 mg/mL morin in methanol; Sigma Chem Co. Ltd) was added. After centrifugation at 10 000 × g for 10 min at 5 °C, the supernatants were removed and placed in 0.45 μM PTFE filter vials (Thomson Instrument Company, Bioprocess Engineering Services Ltd, Kent, UK) prior to analysis. Samples were analysed using a LC system consisting of an Accela 600 quaternary pump and Acela PDA detector coupled to an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific Ltd.). Samples (10 μL) were injected onto a 2 × 150 mm (4 μm) Synergy Hydro-RP 80 fitted with a C18 4 × 2 mm Security Guard cartridge (Phenomenex Ltd, Macclesfield, UK.). Auto-sampler and column temperatures were maintained at 6 and 30 °C, respectively. The samples were analysed at a flow rate of 200 μL/min using a binary mobile phase of (A) 0.1% aqueous formic acid and (B) 0.1% formic acid in 50% acetonitrile/water with the following gradient: 0−5 min, 5% B; 5−22 min, 5−50% B; 22−32 min, 50−100% and 32–34 min 100% B. Mass detection was carried out using an LTQ Orbitrap mass spectrometer in positive ESI mode. Two scan events were employed; full-scan analysis was followed by data-dependent MS/MS of the three most intense ions using collision energies of 45 eV source voltage (set at 3.4 kV) in wide-band activation mode. The instrument was optimized by tuning against morin at a resolution of 100 000 in a range of 80−2000 mass units. For optimal electrospray ionization, the source conditions were set at a source temperature of 280 °C, sheath gas at 60 arbitrary units, and an auxiliary gas at 5 arbitrary units. Prior to analysis, the mass accuracy of the instrument was assured by calibration following the manufacturer’s protocols. All predicted formula data presented were accurate at < 2 ppm.
The samples were analysed in a randomised order and the quality of the MS response was checked by monitoring blanks containing the internal standard and quality control samples of samples from the parental lines interspersed through the sequence of samples. After peak checking, raw peak areas for the major components were obtained using the resident Xcalibur software. After export of MS peak data to Microsoft Excel, the peak areas were ratioed against the internal standard (morin).
Metabolites were identified by their MS properties, the formulae derived from the exact mass data and fragmentation MS2 data and compared against literature on raspberry components as discussed. Only cyanidin-3-O-glucoside, pelargonidin-3-O-glucoside, cyanidin-3-O-sophoroside, quercetin-3-O-glucoside and ellagic acid were confirmed against standard compounds (Extrasynthese Ltd).
Statistical analysis
Exploratory analysis
Each metabolite was analysed using a linear mixed model, fitted using REML (GenStat 20, VSN International, 2019). The metabolites were transformed using a log10 transformation before analysis to stabilise the variance. To estimate genotype means for QTL mapping, data from each year was first analysed separately and genotype was treated as a fixed effect. For 2015, a random effect of batch in addition to that of field replicate was tested but was not significant. A mean over the years was also estimated from a mixed model with genotype, year and their interaction as fixed effects and field replicate within year as a random effect. The generalised heritabilities of the genotypes and the genotype × year interactions were estimated using the GenStat VHERITABILITY procedure. For this analysis, genotype, year, and field replicate within year were treated as random effects. Pearson correlations between the metabolites were calculated for each year separately, based on the log-transformed genotype means.
QTL analysis
For each year separately, and for the mean across years, QTL interval mapping was carried out for each metabolite using the high-density SNP linkage map and the mapping approach from Hackett et al. (2018), based on a hidden Markov model (HMM) to estimate genotype probabilities. This method was shown in that paper to give better peak resolution than software such as MapQTL and GenStat for this population, where there is much more information about markers from the Latham parent than from Glen Moy. The analysis combines information across genetic markers along the chromosome to estimate the probabilities of each possible QTL genotype for each offspring at each position. In a cross such as this with outbreeding parents, the parental genotypes at a QTL are usually represented as ab × cd, with offspring genotypes ac, ad, bc, and bd. Genetic predictors for the Latham additive effect (P1), the Glen Moy additive effect (P2) and the dominance effect (D) can then be derived from the genotype probabilities pr(ac) etc. as:
A permutation study was carried out on this population for highly multivariate imaging data (Williams et al., 2021) and estimated the genome-wide 95% LOD threshold as 3.86; the same threshold has been used for all traits here. A thinned SNP map was used to carry out a QTL × year analysis in GenStat, with a stepwise selection of multiple QTLs, as the high-density map was too computationally intensive for this analysis. This used the significance threshold of Li and Ji (2005), with a genome-wide significance of 0.05.
Results and discussion
LC–MSn analysis of polyphenol metabolites
Thirty-seven metabolites were detected in all 3 years (Table 1). These comprised 12 anthocyanins, ten of which could be putatively identified from previous work on red raspberries (Bradish et al., 2012; McDougall et al., 2005; Mullen et al., 2003) i.e. cyanidin sophoroside, cyanidin glucoside, cyanidin sophoroside rhamnose /cyanidin glucosyl rutinoside, cyanidin sambubioside, cyanidin rutinoside/pelargonidin sophoroside, cyanidin sambubioside rhamnoside/cyanidin xylosyl rutinoside, pelargonidin glucoside, pelargonidin glucosyl rutinoside, pelargonidin sambubioside and pelargonidin rutinoside). Three of these anthocyanin components could not be distinguished by their molecular formulae or their MS2 properties so are labelled with both possible structures (e.g. CyRut/PelSoph). Two other anthocyanin-like components (CyC28H30O17 and CyC34H40O21) were identified as possible cyanidin derivatives due to their fragmentation to m/z 287 [which is consistent with cyanidin (Cy)] and were named with their putative molecular formula. Four major ellagitannins were noted (Lambertianin C, Sanguiin H2, Sanguiin H6 and Sanguiin H10) and five ellagic acid derivatives (ellagic acid, ellagic acid acetyl pentose, ellagic acid pentose 1, ellagic acid pentose 2, and methyl ellagic acid pentose) (Gasperotti et al., 2010; McDougall et al., 2014). As the homogenisation method did not crush the seeds, the metabolic profile probably more reflects the flesh of the raspberries and may not capture the entirety of ellagitannin constituents, which are known to be enriched in the seeds (Arnold et al., 2022). However, the extractable components should be consistent across the progeny. Sixteen flavonol derivatives were noted (i.e. kaempferol glucuronide, galactoside and glucoside but also quercetin dihexosides, glucuronide, galactoside, glucoside, hexosyl rhamnosides, glucosyl rutinosides, pentose and malonyl glucoside). The putative identification of these components was also aided by SOPs delivered through the EUBERRY project (Dobson et al., 2010).
Table 1.
Components identified in raspberry progeny
| Peak | RT | PDA max | m/z [M + H]+ | MS2 | Predicted formula [M + H]+ | Putative identity | Short name |
|---|---|---|---|---|---|---|---|
| A1 | 4.23 | 515 | 611.1603 | 287 | C27H31O16 | Cyanidin3-O-sophorosidea | CySoph |
| A2 | 4.52 | 515 | 449.1075 | 287 | C21H21O11 | Cyanidin-3-O-glucosidea | CyGlu |
| A3 | 4.62 | 515 | 757.2184 | 287, 611 | C33H41O20 | Cyanidin sophoroside rhamnoside/ Cyanidin glucosyl rutinoside |
CySophRham/ CyGluRut |
| A4 | 4.69 | 515 | 581.1503 | 287 | C26H29O15 | Cyanidin sambubioside | CySam |
| A5 | 4.94 | 505 | 639.1566 | 287 | C28H31O17 | Cyanidin derivative | CyC28H30O17 |
| A6 | 4.97 | 510 | 595.1660 | 271, 287, 449 | C27H31O15 | Cyanidin rutinoside/ Pelargonidin sophoroside | CyRut/PelSoph |
| A7 | 5.05 | 510 | 727.2079 | 287, 433, 581 | C32H39O19 | Cyanidin sambubioside rhamnoside/ Cyanidin xyloside rutinoside |
CyXylRut/ CySamRham |
| A8 | 5.22 | 510 | 433.1127 | 271 | C21H21O10 | Pelargonidin-3-O-glucosidea | PelGlu |
| A9 | 5.26 | 510 | 785.2134 | 287, 639 | C34H41O21 | Cyanidin derivative | CyC34H40O21 |
| A10 | 5.44 | 510 | 741.2240 | 271, 417, 595 | C33H41O19 | Pelargonidin sophoroside rhamnoside/ Pelargonidin glucosyl rutinoside | PelGluRut/ PelSophRham |
| A11 | 5.76 | 500 | 565.1540 | ND | C26H29O14 | Pelargonidin sambubioside | PelSam |
| A12 | 5.73 | 510 | 579.1711 | 271, 433 | C27H31O14 | Pelargonidin rutinoside | PelRut |
| Peak | RT | PDA max | m/z [M − H]− | MS2 | Predicted formula [M − H]− | Putative identity | Short name |
|---|---|---|---|---|---|---|---|
| 13 | 5.98 | 270 | 783.0574* | 1265, 935, 933, 897, 633, 301 | C68H47O44 | Sanguiin H10 | Sanguiin H10 |
| 14 | 6.12 | 355 | 771.1839 | 301, 591 | C33H39O21 | Quercetin glucosyl rutinoside | QGluRut1 |
| 15 | 6.34 | 350 | 771.1887 | 301, 591 | C33H39O21 | Quercetin glucosyl rutinoside | QGluRut2 |
| 16 | 6.44 | 350 | 625.1330 | 301, 445, 505 | C27H29O17 | Quercetin dihexoside | QDihex1 |
| 17 | 6.60 | 350 | 625.1331 | 301 | C27H29O17 | Quercetin dihexoside | QDihex2 |
| 18 | 7.14 | 355 | 433.0353 | 301 | C19H13O12 | Ellagic acid pentoside | EllagicPent1 |
| 19 | 7.43 | 355 | 433.0347 | 301 | C19H13O12 | Ellagic acid pentoside | EllagicPent2 |
| 20 | 7.22 | 260 | 1401.5901* | 633, 933,1099, 1250, 1567, 1867 | C123H79O78 | Lambertianin C | Lambertianin |
| 21 | 7.31 | 365 | 300.9947 | 185, 229, 257, 284 | C14H5O8 | Ellagic acida | Ellagic |
| 22 | 7.47 | 260 | 551.0307* | 529 | C48H31O31 | Sanguiin H2 | Sanguiin H2 |
| 23 | 7.55 | 260 |
1869.3168 (934.059*) |
301, 633, 1235, 1567 | C82H53O52 | Sanguiin H6 | Sanguiin H6 |
| 24 | 7.77 | 350 | 595.1219 | 301, 505 | C26H27O16 | Quercetin hexosylpentoside | QHexPent |
| 25 | 7.81 | 355 | 609.1374 | 301, 463 | C27H29O16 | Quercetin hexosylrhamnoside | QHexRham1 |
| 26 | 7.98 | 355 | 463.0823 | 301 | C21H19O12 | Quercetin galactoside | QGal |
| 27 | 8.15 | 355 | 477.0608 | 301 | C21H17O13 | Quercetin glucuronide | QGlcU |
| 28 | 8.17 | 350 | 609.1381 | 301, 463 | C27H29O16 | Quercetin hexosyl rhamnoside | QHexRham2 |
| 29 | 8.28 | 355 | 463.0824 | 301 | C21H19O12 | Quercetin-3-O-glucosidea | QGlu |
| 30 | 8.81 | 355 | 433.0713 | 301 | C20H17O11 | Quercetin pentoside | QPent |
| 31 | 8.84 | 355 | 447.0866 | 285 | C21H19O11 | Kaempferol galactoside | KGal |
| 32 | 8.94 | 350 | 549.0812 | 505, 301 | C24H21O15 | Quercetin malonyl glucoside | QMalGlu |
| 33 | 8.96 | 350 | 447.0511 | 301, 315 | C20H16O12 | Methylellagic acid pentoside | MeEllPent |
| 34 | 9.09 | 365 | 475.0455 | 301 | C21H15O13 | Ellagic acid acetyl pentoside | EllAcetylPent |
| 35 | 9.17 | 345 | 461.0656 | 285 | C21H17O12 | Kaempferol glucuronide | KGlcU |
| 36 | 9.18 | 345 | 447.0862 | 285 | C21H19O11 | Kaempferol glucoside | KGlu |
| 37 | 9.41 | 360 | 463.0812 | 301 | C21H19O12 | Quercetin hexoside | QHex |
*Doubly charged ion, m/z [M-H]2−
acomponent confirmed against standard
MS2 fragments in bold are the dominant products
Photodiode array (PDA) profiles (280 nm; Fig. S1A) showed the major components in the parents. However, profiles at 520 nm (Fig. S1B) highlighted the anthocyanins with Latham generally showing higher levels of some of the later eluting anthocyanins (A5–A12).
Exploratory analysis of polyphenol metabolite data
Summary statistics for the metabolite data and for the total anthocyanins are presented in Table 2. No samples from the Glen Moy parent were analysed in 2011. The mixed model analysis showed few statistically significant (p < 0.05) differences between the levels in the parents. For example, CySam and QPent were significantly higher in Glen Moy, whilst CyRut/PelSoph, PelGlu, PelRut, PelGluRut, KGlcU, QGlcU, QGluRut1, QGluRut2 and QDihex1 had significantly higher levels in Latham.
Table 2.
Summary statistics for metabolite levels and total anthocyanin data in the parents and offspring across the 3 years of harvest
| Short name | Year | Latham | Moy | Mean | Min | Max | SD | H_G | H_GY | BM |
|---|---|---|---|---|---|---|---|---|---|---|
| TA | 10 | 10.481 | 6.922 | 8.765 | 1.924 | 25.761 | 3.820 | 0.593 | 0.096 | N |
| 11 | 16.084 | NA | 9.703 | 0.000 | 45.447 | 5.284 | N | |||
| 15 | 8.812 | 10.093 | 8.320 | 1.584 | 26.799 | 3.777 | N | |||
| CySoph | 10 | 7.631 | 7.195 | 8.366 | 1.826 | 26.167 | 4.424 | 0.715 | 0.181 | N |
| 11 | 7.301 | NA | 6.047 | 0.815 | 20.078 | 2.989 | N | |||
| 15 | 5.257 | 6.135 | 6.151 | 1.076 | 15.954 | 2.508 | N | |||
| CyGlu | 10 | 2.132 | 1.950 | 2.139 | 0.503 | 9.710 | 1.190 | 0.771 | 0.089 | N |
| 11 | 1.421 | NA | 1.406 | 0.258 | 5.012 | 0.826 | N | |||
| 15 | 1.393 | 1.894 | 1.643 | 0.257 | 4.603 | 0.856 | N | |||
| CySophRham/ CyGluRut | 10 | 6.590 | 2.520 | 3.364 | 0.000 | 16.180 | 2.999 | 0.887 | 0.186 | Y |
| 11 | 5.014 | NA | 1.925 | 0.000 | 11.318 | 1.870 | Y | |||
| 15 | 2.637 | 2.579 | 1.526 | 0.000 | 7.150 | 1.626 | Y | |||
| CySam | 10 | 0.044 | 0.252 | 0.160 | 0.022 | 0.759 | 0.116 | 0.799 | 0.119 | N |
| 11 | 0.036 | NA | 0.108 | 0.000 | 0.561 | 0.090 | N | |||
| 15 | 0.025 | 0.159 | 0.109 | 0.007 | 0.378 | 0.079 | N | |||
| CyC28H30O17 | 10 | 0.146 | 0.197 | 0.322 | 0.000 | 1.465 | 0.239 | 0.634 | 0.066 | N |
| 11 | 0.086 | NA | 0.130 | 0.007 | 0.574 | 0.097 | N | |||
| 15 | 0.190 | 0.287 | 0.320 | 0.047 | 1.486 | 0.196 | N | |||
| CyRut/ PelSoph | 10 | 3.075 | 1.117 | 1.868 | 0.114 | 6.489 | 1.174 | 0.830 | 0.053 | N |
| 11 | 2.796 | NA | 1.165 | 0.056 | 4.504 | 0.806 | N | |||
| 15 | 1.840 | 1.305 | 1.025 | 0.069 | 5.221 | 0.748 | N | |||
| CyXylRut/ CySamRham | 10 | 0.063 | 0.112 | 0.095 | 0.000 | 0.576 | 0.101 | 0.879 | 0.155 | Y |
| 11 | 0.042 | NA | 0.047 | 0.000 | 0.417 | 0.060 | Y | |||
| 15 | 0.023 | 0.078 | 0.042 | 0.000 | 0.301 | 0.054 | Y | |||
| PelGlu | 10 | 0.308 | 0.063 | 0.189 | 0.014 | 1.406 | 0.195 | 0.795 | 0.205 | N |
| 11 | 0.264 | NA | 0.123 | 0.000 | 1.512 | 0.179 | N | |||
| 15 | 0.224 | 0.148 | 0.144 | 0.010 | 0.975 | 0.130 | N | |||
| CyC34H40O21 | 10 | 0.129 | 0.065 | 0.131 | 0.000 | 0.646 | 0.136 | 0.885 | 0.109 | Y |
| 11 | 0.051 | NA | 0.038 | 0.000 | 0.343 | 0.049 | Y | |||
| 15 | 0.099 | 0.107 | 0.081 | 0.000 | 0.978 | 0.106 | Y | |||
| PelGluRut | 10 | 1.807 | 0.149 | 0.350 | 0.000 | 3.078 | 0.397 | 0.891 | 0.135 | Y |
| 11 | 1.965 | NA | 0.239 | 0.000 | 1.979 | 0.310 | Y | |||
| 15 | 1.066 | 0.218 | 0.166 | 0.000 | 1.582 | 0.224 | Y | |||
| PelSam | 10 | 0.004 | 0.002 | 0.010 | 0.000 | 0.139 | 0.016 | 0.718 | 0.227 | Y |
| 11 | 0.000 | NA | 0.004 | 0.000 | 0.113 | 0.012 | Y | |||
| 15 | 0.005 | 0.005 | 0.007 | 0.000 | 0.053 | 0.008 | N | |||
| PelRut | 10 | 0.221 | 0.030 | 0.078 | 0.000 | 0.514 | 0.091 | 0.880 | 0.099 | Y |
| 11 | 0.155 | NA | 0.035 | 0.000 | 0.587 | 0.060 | Y | |||
| 15 | 0.123 | 0.082 | 0.038 | 0.000 | 0.554 | 0.063 | Y | |||
| SanguiinH10 | 10 | 0.294 | 0.722 | 0.397 | 0.154 | 1.000 | 0.114 | 0.572 | 0.046 | N |
| 11 | 0.104 | NA | 0.131 | 0.037 | 0.352 | 0.055 | N | |||
| 15 | 0.225 | 0.309 | 0.301 | 0.073 | 0.854 | 0.112 | N | |||
| QGluRut1 | 10 | 0.074 | 0.023 | 0.016 | 0.000 | 0.149 | 0.021 | 0.847 | 0.134 | Y |
| 11 | 0.065 | NA | 0.012 | 0.000 | 0.073 | 0.016 | Y | |||
| 15 | 0.081 | 0.045 | 0.023 | 0.000 | 0.171 | 0.031 | Y | |||
| QGluRut2 | 10 | 0.158 | 0.070 | 0.064 | 0.000 | 0.491 | 0.070 | 0.894 | 0.164 | Y |
| 11 | 0.136 | NA | 0.033 | 0.000 | 0.201 | 0.036 | Y | |||
| 15 | 0.119 | 0.073 | 0.048 | 0.000 | 0.301 | 0.057 | Y | |||
| QDihex1 | 10 | 0.097 | 0.015 | 0.022 | 0.000 | 0.156 | 0.026 | 0.861 | 0.139 | Y |
| 11 | 0.069 | NA | 0.019 | 0.000 | 0.211 | 0.023 | Y | |||
| 15 | 0.140 | 0.035 | 0.043 | 0.000 | 0.194 | 0.042 | Y | |||
| QDihex2 | 10 | 0.046 | 0.024 | 0.050 | 0.001 | 0.417 | 0.068 | 0.837 | 0.171 | N |
| 11 | 0.032 | NA | 0.030 | 0.000 | 0.377 | 0.046 | N | |||
| 15 | 0.047 | 0.024 | 0.055 | 0.003 | 0.380 | 0.062 | N | |||
| EllagicPent1 | 10 | 0.162 | 0.215 | 0.171 | 0.047 | 0.360 | 0.058 | 0.798 | 0.016 | N |
| 11 | 0.063 | NA | 0.069 | 0.018 | 0.157 | 0.026 | N | |||
| 15 | 0.194 | 0.242 | 0.243 | 0.075 | 0.568 | 0.095 | N | |||
| EllagicPent2 | 10 | 0.073 | 0.031 | 0.048 | 0.000 | 0.160 | 0.040 | 0.843 | 0.107 | Y |
| 11 | 0.026 | NA | 0.022 | 0.002 | 0.075 | 0.016 | Y | |||
| 15 | 0.120 | 0.044 | 0.095 | 0.009 | 0.413 | 0.079 | Y | |||
| Lambertianin | 10 | 2.699 | 3.578 | 2.457 | 0.847 | 4.286 | 0.629 | 0.648 | 0.071 | N |
| 11 | 0.578 | NA | 0.579 | 0.151 | 1.315 | 0.222 | N | |||
| 15 | 0.917 | 1.223 | 1.086 | 0.419 | 2.111 | 0.321 | N | |||
| Ellagic | 10 | 0.057 | 0.040 | 0.044 | 0.009 | 0.155 | 0.021 | 0.446 | 0.000 | N |
| 11 | 0.013 | NA | 0.011 | 0.001 | 0.101 | 0.008 | N | |||
| 15 | 0.391 | 0.284 | 0.308 | 0.039 | 1.505 | 0.219 | N | |||
| SanguiinH2 | 10 | 0.036 | 0.034 | 0.029 | 0.012 | 0.058 | 0.008 | 0.544 | 0.130 | N |
| 11 | 0.017 | NA | 0.015 | 0.003 | 0.033 | 0.006 | N | |||
| 15 | 0.018 | 0.014 | 0.015 | 0.007 | 0.048 | 0.005 | N | |||
| SanguiinH6 | 10 | 5.496 | 5.965 | 5.020 | 2.147 | 7.419 | 0.914 | 0.630 | 0.085 | N |
| 11 | 1.297 | NA | 1.323 | 0.604 | 2.889 | 0.391 | N | |||
| 15 | 2.918 | 2.992 | 2.977 | 1.695 | 4.682 | 0.598 | N | |||
| QHexPent | 10 | 0.008 | 0.075 | 0.037 | 0.000 | 0.193 | 0.041 | 0.864 | 0.250 | Y |
| 11 | 0.005 | NA | 0.028 | 0.000 | 0.134 | 0.031 | Y | |||
| 15 | 0.014 | 0.078 | 0.064 | 0.001 | 0.245 | 0.060 | N | |||
| QHexRham1 | 10 | 0.074 | 0.035 | 0.021 | 0.000 | 0.147 | 0.031 | 0.802 | 0.278 | Y |
| 11 | 0.027 | NA | 0.012 | 0.000 | 0.077 | 0.017 | Y | |||
| 15 | 0.041 | 0.044 | 0.023 | 0.000 | 0.156 | 0.032 | N | |||
| QGal | 10 | 0.232 | 0.086 | 0.072 | 0.001 | 0.376 | 0.069 | 0.838 | 0.182 | Y |
| 11 | 0.106 | NA | 0.053 | 0.000 | 0.263 | 0.052 | Y | |||
| 15 | 0.142 | 0.109 | 0.104 | 0.000 | 0.450 | 0.096 | N | |||
| QGlcU | 10 | 1.356 | 0.431 | 0.457 | 0.038 | 1.186 | 0.244 | 0.417 | 0.128 | N |
| 11 | 0.506 | NA | 0.280 | 0.015 | 1.042 | 0.151 | N | |||
| 15 | 0.633 | 0.345 | 0.516 | 0.096 | 1.356 | 0.204 | N | |||
| QHexRham2 | 10 | 0.067 | 0.036 | 0.034 | 0.000 | 0.145 | 0.031 | 0.894 | 0.148 | Y |
| 11 | 0.031 | NA | 0.015 | 0.000 | 0.105 | 0.016 | Y | |||
| 15 | 0.038 | 0.039 | 0.023 | 0.000 | 0.178 | 0.027 | Y | |||
| QGlu | 10 | 0.124 | 0.097 | 0.062 | 0.011 | 0.255 | 0.035 | 0.590 | 0.101 | N |
| 11 | 0.058 | NA | 0.045 | 0.002 | 0.164 | 0.025 | N | |||
| 15 | 0.057 | 0.060 | 0.077 | 0.015 | 0.221 | 0.035 | N | |||
| QPent | 10 | 0.009 | 0.134 | 0.064 | 0.000 | 0.315 | 0.076 | 0.857 | 0.219 | Y |
| 11 | 0.002 | NA | 0.046 | 0.000 | 0.261 | 0.051 | Y | |||
| 15 | 0.013 | 0.095 | 0.107 | 0.001 | 0.431 | 0.101 | Y | |||
| KGal | 10 | 0.029 | 0.007 | 0.006 | 0.000 | 0.050 | 0.008 | 0.846 | 0.185 | Y |
| 11 | 0.010 | NA | 0.005 | 0.000 | 0.050 | 0.006 | Y | |||
| 15 | 0.014 | 0.010 | 0.009 | 0.000 | 0.062 | 0.010 | Y | |||
| QMalGlu | 10 | 0.038 | 0.033 | 0.018 | 0.001 | 0.087 | 0.011 | 0.616 | 0.051 | N |
| 11 | 0.018 | NA | 0.014 | 0.000 | 0.046 | 0.009 | N | |||
| 15 | 0.013 | 0.018 | 0.018 | 0.001 | 0.061 | 0.012 | N | |||
| MeEllPent | 10 | 0.055 | 0.074 | 0.062 | 0.015 | 0.169 | 0.029 | 0.817 | 0.000 | N |
| 11 | 0.018 | NA | 0.022 | 0.004 | 0.069 | 0.011 | N | |||
| 15 | 0.079 | 0.082 | 0.086 | 0.018 | 0.251 | 0.041 | N | |||
| EllAcetylPent | 10 | 0.087 | 0.123 | 0.072 | 0.003 | 0.235 | 0.048 | 0.815 | 0.214 | N |
| 11 | 0.027 | NA | 0.021 | 0.000 | 0.092 | 0.017 | N | |||
| 15 | 0.080 | 0.116 | 0.078 | 0.002 | 0.284 | 0.051 | N | |||
| KGlcU | 10 | 0.210 | 0.038 | 0.044 | 0.001 | 0.204 | 0.034 | 0.367 | 0.077 | N |
| 11 | 0.068 | NA | 0.028 | 0.000 | 0.169 | 0.021 | N | |||
| 15 | 0.073 | 0.032 | 0.047 | 0.003 | 0.177 | 0.030 | N | |||
| KGlu | 10 | 0.043 | 0.027 | 0.008 | 0.000 | 0.080 | 0.013 | 0.311 | 0.472 | Y |
| 11 | 0.014 | NA | 0.010 | 0.000 | 0.045 | 0.008 | N | |||
| 15 | 0.016 | 0.016 | 0.020 | 0.001 | 0.071 | 0.012 | N | |||
| QHex | 10 | 0.025 | 0.010 | 0.012 | 0.001 | 0.038 | 0.008 | 0.763 | 0.092 | N |
| 11 | 0.010 | NA | 0.005 | 0.000 | 0.022 | 0.004 | N | |||
| 15 | 0.013 | 0.007 | 0.011 | 0.002 | 0.045 | 0.007 | N |
10, 11 and 15 represent samples from years 2010, 2011 and 2015
TA = total anthocyanin content by the colorimetric method, values in mg cyanidin glucoside equivalents/100 g fresh weight. Metabolites are denoted by their abbreviated names, see Table 1. NA = value not available due to no Moy samples that year. Mean, Min, Max, SD = mean, minimum, maximum and standard deviation of the offspring
H_G = generalised heritability of the genotype (log10 scale); H_GY = generalised heritability of the genotype × year effect (log10 scale). BM = Y if the metabolite distribution was clearly bimodal, N otherwise. Metabolites in bold were significantly higher in that parent (p < 0.05) in that year
Histograms of the distributions of the log-transformed metabolites showed several metabolites had bimodal distributions, as recorded in Table 2. This was usually consistent across all three years, although for some there was no clear bimodality in 2015. As discussed below, these distributions are associated with major QTLs detected on linkage groups (LG) 1, 4 and 6. The generalised heritability for the genotypes varied from 0.31 to 0.89 (Table 2). The highest heritabilities were found mainly for the metabolites with bimodal distributions: the anthocyanins PelRut, PelGluRut, CySophRham/CyGluRut, CyXylRut/CySamRham, CyC34H40O21, the flavonols KGal, QGal, QPent, QHexPent, QDihex1, QDihex2, QGluRut1, QGluRut2, QHexRham1, QHexRham2 and the ellagic acid derivative EllagicPent2. The lowest generalised heritabilities were found for KGlu, KGlcU, QGlcU and Ellagic, with values of 0.31–0.45. The generalised heritability of the genotype x year effect was much lower, varying from 0.00 to 0.28 apart from for KGlu, estimated as 0.47.
Because of the bimodal distributions for several metabolites, a simple correlation coefficient does not represent the relationship among the metabolites well in some cases. Supplementary Figures S2A, S2B and S2C show scatter plot matrices for the anthocyanins, flavonols and ellagitannins/ellagic acid derivatives respectively, based on the log-transformed genotype means for 2010 and Figure S2D shows relationships between the flavonols and anthocyanins. The figures for the other years (not shown) are similar. As an example, consider the correlation between CySoph and CyXylRut/CySamRham in Fig. S2A. The Pearson correlation coefficient was − 0.50 but the scatter plot showed that some genotypes had very low levels of CyXylRut/CySamRham, and the other genotypes formed two clusters, within each of which the correlation with CySoph was positive. Another interesting pair were CySoph and CySam. The individual distributions for these metabolites were unimodal but the scatter plot showed two clear clusters. Likewise, MeEllPent and EllAcetylPent in Fig. S2C showed clear clustering and a positive correlation within the clusters, but unimodal individual distributions and a negative correlation overall. These plots will be discussed in more detail in the QTL results below.
QTL mapping
The QTL analysis generally showed a good consistency over years, but there were also some significant QTL × year interactions. These were however generally associated with different sizes of QTL effects, rather than a different direction of the effect. QTLs were detected in each year on raspberry LG1-LG6, but not on LG7 (Table 3). This is in accordance with previous studies on this population (e.g. Hackett et al., 2018), which have seldom found QTLs on LG7. Figure 1 shows linkage maps with one- and two-LOD support intervals for the QTLs. The term ‘major’ QTL is used to refer to a QTL with LOD greater than 10, ‘strong’ QTL for a QTL with LOD between 5 and 10 and ‘minor’ QTL for a QTL with LOD above the permutation threshold of 3.8 but below 5. The results presented below are from the over-year averages unless otherwise stated. Linkage groups LG1, LG4 and LG6, with QTLs large enough to give bimodal distributions for some metabolites, are discussed first, followed by the groups with smaller QTLs.
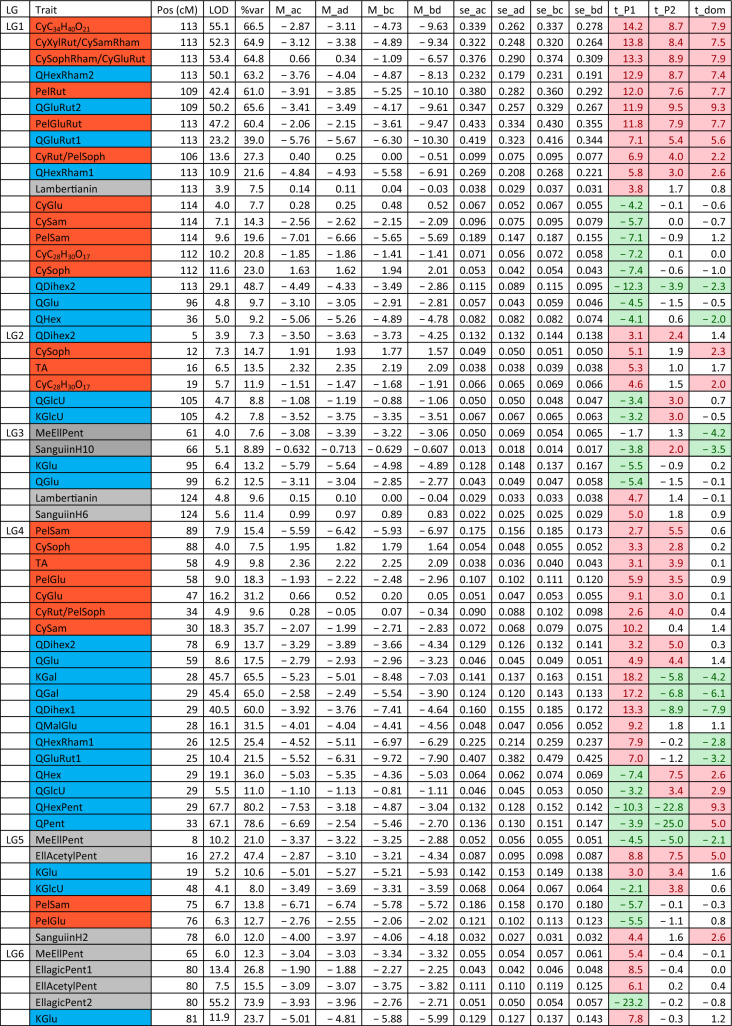
Table 3.
QTLs detected for each metabolite. This is based on the average values for each genotype over the three years
Pos is the position of the maximum LOD, %var is the %variance explained by that QTL. M_ac, M_ad, M_bc and M_bd are the estimated means for offspring genotypes ac, ad, bc and bd respectively and se_ac etc.are the corresponding standard errors. t_P1 is a t-test for the significance of the Latham additive effect, t_P2 is a t-test for the Glen Moy additive effect and t_dom for the dominance effect. The significant effects are shaded red if they are positive or green if they are negative. Anthocyanin components are shaded in red, flavonols in blue, with ellagitannins and ellagic acid derivatives in grey
Fig. 1.


Linkage maps showing QTL locations. The boxes show the one-LOD support interval, and the whiskers show the two-LOD interval. Major QTLs are shown as solid boxes, strong QTLs as diagonally filled boxes and minor QTLs as unfilled boxes. Red = anthocyanins, blue = flavonols, black = other traits. A–F show linkage groups (LG) 1–6
LG1
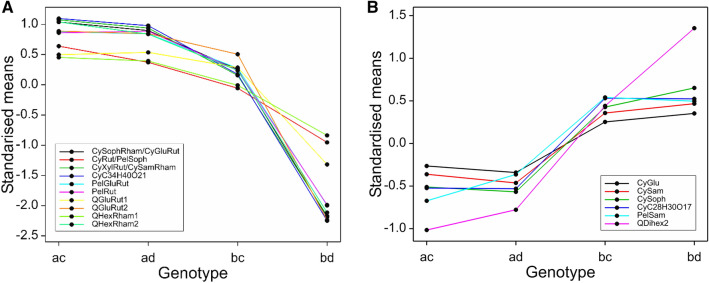
There was a very significant region at the bottom of LG1 for all three years, with many metabolites mapping to positions from 106 to 113 cM, with LODs up to 55.1 and the % variance explained (R2) by this QTL up to 67%. This is close to the RibHLH marker at 113 cM, which was identified by McCallum et al. (2010) as close to a QTL for a smaller set of anthocyanins than measured in this study. One group of metabolites had major QTLs (explaining between 22 and 67% of the trait variance) and showed a pattern where the levels were close to zero for one genotype (bd), higher for bc and highest for ac and ad (Fig. 2A). This pattern was shown by the anthocyanins, CyC34H40O21, CySophRham/CyGluRut, CyXylRut/CySamRham, PelGluRut, PelRut and CyRut/PelSoph and the flavonols, QHexRham2, QGluRut2, QGluRut1 and QHexRham1. All these flavonoid components contain attached rhamnose groups and even the unidentified cyanidin derivative CyC34H40O21 gave a neutral loss of 146 under MS fragmentation which is consistent with the presence of a rhamnose group (Table 1). The presence of the major QTLs is responsible for the bimodal distribution shown by all these metabolites apart from CyRut/ PelSoph. The genotype bd, with the lowest values, represents a clear separate cluster in the scatter plot matrices (see supplementary Figs S2A, S2B, & S2D). The metabolites are all positively correlated, both for the overall correlation and for the correlation within the genotypes ac, ad and bc, which have higher values of each metabolite than bd (Fig. 2A).
Fig. 2.
A Genotype means from the over-years analysis (on log10 transformed data) for metabolites at the QTLs at the bottom of LG1. The genotype bd results in lower levels of these metabolites than the other genotypes. Each genotype mean is standardised by subtracting the overall mean and then dividing by √(QTL residual mean square) for comparability. B Genotype means from the over-years analysis (on log10 transformed data) for metabolites at the QTL at the bottom of LG1. The ac and ad genotypes have lower levels of the components, an effect mainly due the Latham parent. Each genotype mean is standardised by subtracting the overall mean and then dividing by √(QTL residual mean square) for comparability
A further set of metabolites mapped to the same location (109–114 cM) on LG1, but with smaller QTLs, and gave a different pattern of segregation and more variation over the years (Fig. 2B). All these components are flavonoid glucosides with glucose groups directly attached to the flavonoid aglycone (e.g. CyGlu) or with further glycosyl groups attached to the flavonoid glucosyl core (e.g. cyanidin 3-sambubioside; CySam or cyanidin sophoroside; CySoph). A major QTL for the flavonol QDihex2 was also detected in this region for all three years. Major QTLs were detected in 2010, with smaller QTLs that are consistent in position and sign in 2011 and 2015, for the anthocyanins CySoph, CyC28H30O17, and PelSam. A major QTL for CySam and strong QTLs for CyGlu and PelGlu were also detected in this region in 2010 only, together with a minor QTL nearby, at 96 cM, for QGlu in 2010 only. For all these metabolites, the difference in the genotype means is due to the allele received from Latham, with lower values for the ‘a’ allele (in contrast to the flavonoids shown in Fig. 2A), where the ‘a’ allele had high levels.
All the anthocyanins that map here showed positive correlations (Fig S2A), particularly PelGlu and PelSam, with a correlation of 0.90 in 2010, and CyGlu and CySam, with a correlation of 0.88. The correlations were also high in 2011 and 2015. These correlations may be expected as the glucosides could be metabolic precursors of the sambubioside derivatives, which have xylose groups added at position 2 of the glucose moiety which is attached to the anthocyanidin (Bradish et al., 2012). In addition, there was a strong correlation between PelGluRut and PelRut (0.95) which also could reflect a -precursor relationship (Fig. S2A). The strong correlation (0.93) between the potential cyanidin derivative CYC34H44O21 and CyXylRut could also suggest a precursor-intermediate relationship. However, the strong correlation (0.95) between PelRut and CyXylRut is not so readily explained.
For some pairs of metabolites, the overall correlations do not give a satisfactory representation of the true relationships between these anthocyanins and those with the alternative pattern above. For example, the overall correlation between CyXylRut/CySamRham and CySoph was − 0.50 in 2010 (Fig. S2A). However, apart from the cluster of values from genotype bd, which has very low levels of CyXylRut/CySamRham (see red arrow in Fig. S2A), the relationship within the other three clusters is positive. Indeed, CySam and CyXylRut/CySamRham (overall r = − 0.18 in 2010) and CyGlu and CyXylRut/CySamRham (overall r = − 0.10 in 2010) showed the same pattern with low levels of CyXylRut/CySamRham due to the genotype bd.
Supplementary Figure S2D shows the relationships between anthocyanins and flavonols. There were many near-zero correlations, but some components showed high positive correlations. For example, CySophRham/CyGluRut had strong correlations with QGluRut2 (r = 0.97), QHexRham2 (0.97) and QGluRut1 (0.72). A similar pattern of correlation was seen between PelGluRut with QGluRut2 (0.90), QHexRham (0.89) and QGluRut1 (0.61) and between PelRut with QGluRut2 (0.91), QHexRham (0.93) and QGluRut1 (0.71). In all these cases, the lower correlation with QGluRut1 was influenced by low levels associated with one genotype. The biochemical feature that links these components is the addition of similar glycosyl groups to the flavonol or anthocyanidin core, so it is intriguing to suggest that these steps are under common genetic control. In addition, QDihex2 also had a correlation of 0.82 with CySoph, again suggesting a commonality of added glycosyl groups.
There were some candidate genes underlying this QTL region (LG1, 106–114 cM) that could be involved in the control of anthocyanin and flavonol biosynthesis. Firstly, there was a homologue (> 80% homology with genes from Rosa chinensis) of a leucine-rich repeat protein kinase that has been associated with the accumulation of polyphenols in Tannat grapes (Da Silva et al., 2013). There was a homologue of a MYB39 transcription factor (> 80% homology to genes from R. chinensis and Fragaria vesca) that has been associated with microRNA control of phenylpropanoid biosynthesis in Arabidopsis (AT3G61250; Sharma et al., 2016) and another MYB3-like transcription factor (homology to AT1G22640.1; ≥ 85% homology with R. chinensis and F. vesca genes) that was upregulated in polyphenol-rich blueberry skins vs flesh (Plunkett et al., 2018). There was a homologue of a C2H2-type zinc finger protein (> 75% homology to R. chinensis and F. vesca genes), which was found to be upregulated in ripe versus unripe grapes (Ahn et al., 2019). Also, there was a homologue to a structural biosynthetic gene that could be directly involved in the formation of the anthocyanidins, i.e. an anthocyanidin reductase (AT1G61720.1; ANR; > 80% homology with Prunus genes), which was upregulated in the polyphenol-rich skin of blueberries versus blueberry flesh (Plunkett et al., 2018) and implicated in polyphenol accumulation in grapes (Bogs et al., 2005).
Also, there was a homologue to an Arabidopsis gene (~ 80% homology to R. chinensis and F. vesca genes) that encoded a UDP-glycosyl transferase (UDPGT) that has been implicated in polyphenol biosynthesis in ripe grape berries (Ahn et al., 2019). This could be responsible for the attachment of glycosyl groups directly to the flavonoid aglycone (e.g. to the cyanidin group in CyGlu) and/or in adding extra glycosyl groups to the flavonoid glucoside (as discussed for components in Fig. 2B).
Despite the QTLs for individual anthocyanins, no QTL was detected for Total Anthocyanin content (TA) on LG1. This appears to be because the increase in the levels of some of the individual anthocyanins is balanced by the decrease in the levels of others. This was also the case in a previous study (McCallum et al., 2010) which examined levels of eight major raspberry anthocyanins. A strong QTL for QHex was also detected at 36 cM on LG1. This was an effect mainly of the Latham parent.
LG4
There was a very significant area on LG4 from 25 to 33 cM for all three years, especially for the flavonol metabolites, with QTLs with LODS up to 67.7 and R2 up to 80%. This is close to the region detected by Kassim et al. (2009) and McCallum et al. (2010). Markers ERubLR_SQ8.1_H09HMGR, JHIRi49055_GIG and ERubLR_11_2C02_ADH are in this region, also marker RiCAD is nearby. The flavonol metabolites mapping to this region showed three distinct genotype patterns. The most significant metabolites were QHexPent and QPent, which had major QTLs in this region in all three years (with LODs of 67.1 to 67.7 and R2 from 78.6 to 80.2%) with associated bimodal distributions. For these metabolites genotype ac had the lowest level, then bc, with ad and bd highest (Fig. 3A). They are also very highly correlated (r = 0.90 in 2010; Fig. S2B) which could indicate a precursor- product relationship. QHex also had a major QTL here but the genotype pattern was different, with ad having the lowest level, ac and bd having intermediate levels and bc having the highest level. QGlcU also had a strong QTL in this region with the same genotype pattern as QHex.
Fig. 3.
A Genotype means from the over-years analysis (on log10 transformed data) for the QTL at 25–33 cM on LG4 (and the metabolites where the QTL effects are different from Fig. B). Each genotype mean is standardised by subtracting the overall mean and then dividing by √(QTL residual mean square) for comparability. B Genotype means from the over-years analysis (on log10 transformed data) for the QTL at 25–33 cM on LG4. Each genotype mean is standardised by subtracting the overall mean and then dividing by √(QTL residual mean square) for comparability. The levels of the metabolites were lowest in genotype bc
The relationship between the pentose containing flavonols and the genotypes could be explained if this QTL overlaid genes involved in sugar addition to the flavonols. However, the metabolic relationship between QHex and QGlcU is less obvious.
Other flavonols KGal, QGal and QDihex1 also had major QTLs in this region, with LODs of 40.5 to 45.7 and R2 from 60.0 to 65.5%. For these, the lowest levels were for genotype bc, higher for bd and the highest for ac and ad (Fig. 3B). This region was associated with the observed bimodal distribution for these metabolites. KGal, QGal and QDihex1 were positively correlated, with correlations of at least 0.88 in 2010 (Fig S2B), and similarly high for the other years. QHexRham1 and QGluRut1 also had major QTLs here (LODs of 10.4–12.5) with a similar genotype pattern. QMalGlu also mapped with a major QTL here, with a similar pattern but less distinction between bc and bd. It is intriguing that these flavonol components share attached hexosyl groups (i.e. either glucosyl or galactosyl) and perhaps this region controls the addition of such sugar groups.
Scatterplots of these flavonols show a strong structure with up to four groups corresponding closely to the genotypes at about 30 cM on LG4 (Fig. 4). This is generally backed up by the scatter plots for flavonols shown in Fig. S2B. The ac genotype (black) and ad genotype (red), which inherit the “a” allele from Latham, both had high levels of KGal, QGal, QDihex1, QMalGlu and QHex but the ac genotype had much higher levels than the ad genotype for QPent and QHexPent. The ac genotype and bc genotype (green) inheriting allele c from Glen Moy both had high QPent and QHexPent but the bc genotype (green) had lower levels of KGal, QGal, QDihex1 and QMalGlu. There were strong correlations (~ 0.9) between the levels of QGal and QDihex1 across the progeny in 2010, which could reflect precursor relationships (Fig. 4 & Fig. S2). This fits with the addition of a second hexose group to QGal. A similar precursor-product relationship may exist between the kaempferol and quercetin hexosides 1 (probably Gal), which differ by one hydroxyl group on the A-ring. Indeed, a similar strong correlation (r = 0.92) was noted between KGlcU and QGlcU (Fig. S2c). This could suggest metabolic interconversion of kaempferol to quercetin glycosides.
Fig. 4.
Scatterplot matrix of flavonols with major QTLs on LG4, labelled by the genotypes at 30 cM (based on SNPs s340_p9738_F13 from Latham and n1_9428_p6492_F60 from Glen Moy). The colour coding reflects the alleles inherited from the parents, ac, ad, bc & bd
Strong QTLs were also detected for QGlu and QDihex, but these were well separated from the above, with peak LODs at 59 cM and 78 cM respectively. Both had highest levels for genotype ac, intermediate for ad and bc and lowest for bd.
Some QTLs for anthocyanins were also detected on LG4, with good consistency over years, but the QTL positions for each anthocyanin varied more. A major QTL for CySam mapped to 30 cM with LOD 18.3, and R2 of 35.7%. This was mainly an effect of the Latham parent. Another major QTL for CyGlu was mapped to 47 cM, with LOD 16.2, and R2 of 31.2%, and a strong QTL for PelGlu and a minor QTL for Total Anthocyanin mapped to 58 cM. All these showed a similar pattern of genotype means, with ac having the highest level, ad and bc having intermediate levels and bd having the lowest level. PelSam had a strong QTL at 89 cM and CySoph had a minor QTL at 88 cM, with similar patterns to the previous three anthocyanins.
An interesting pattern was noted in the scatterplots for the data in 2010 (Fig. S2a, top left box) for CySoph and CySam. CySam is a disaccharide consisting of xylose and glucose attached to cyanidin whereas CySoph is cyanidin with an attached disaccharide of glucose. These showed two distinct parallel clusters related to the genotypes. The marker that corresponds best to the separation is JHIRi49055_GIG at 26 cM on LG4, which segregates in Latham only: the clusters correspond closely to the offspring receiving the a and b alleles from Latham. Similar patterns were observed in the other years (not shown).
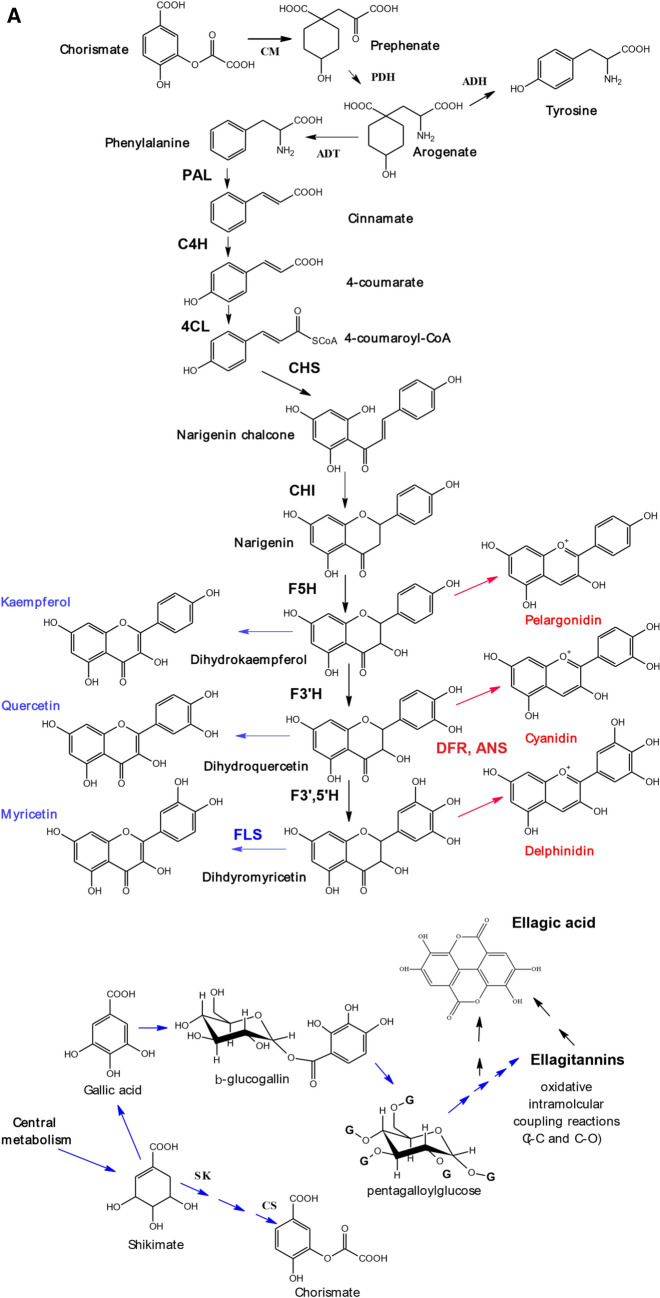
There were some genes underlying the QTL region at 25–33 cM on LG4 that have been implicated in the control of polyphenol biosynthesis. There was a homologue of a cytochrome P450 that has been implicated in anthocyanin accumulation in purple Brassicas (Zhu et al, 2021). Also, there was a gene with homology to a dihydroflavonol 4-reductase (DFR)-like protein in Arabidopsis (Yuan et al., 2007) (also 85% homology with the R. chinensis gene) which could be involved in the final steps of anthocyanidin biosynthesis (see scheme 1A).
Scheme 1.
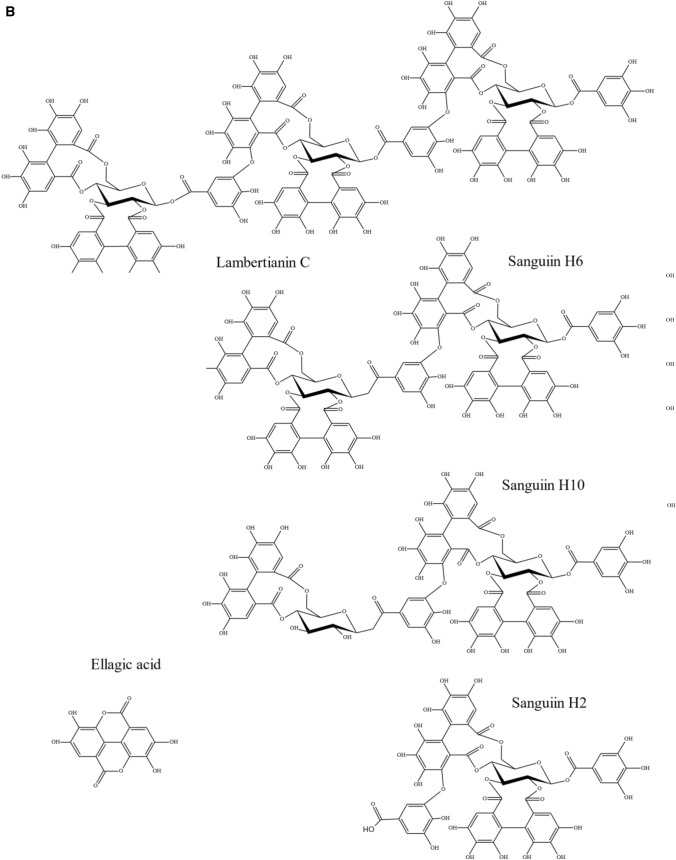
A Pathway for anthocyanin and flavonol biosynthesis. Insert of possible biosynthetic pathway to ellagitannins and ellagic acid. Enzymes are abbreviated as follows: ADH, arogenate dehydrogenase; ADT, arogenate dehydratase; ANS, anthocyanin synthase; CHI, chalcone isomerase; CHS, chalcone synthase; C4H, cinnamate-4-hydroxylase; 4CL, 4-coumarate ligase; CM, chorismate mutase; DFR, dihydroflavonol reductase; F3H, flavonone-3-hydroxylase; F5H, ferulate 5-hydroxylase; F3′H, flavonoid-3′-hydroxylase; F3′5′H, flavonoid-3′,5′-hydroxylase; FLS, flavonol synthase; PAL, phenylalanine ammonia lyase; PDH, prephenate dehydratase; SK = Shikimate kinase and CS = chorismate synthase. For clarity, only enzymes that produce the flavonoid aglycones are shown, not the glycosyltransferases and acyl transferases that decorate the aglycones. Methylated anthocyanidins (i.e. peonidin, petunidin and malvidin) and flavonols (isorhamnetin) are also not shown as they are not present in raspberry. B Structures of ellagitannins and ellagic acid found in raspberry
However, most interestingly there were homologues of genes encoding uridine diphosphate glycosyltransferases (UDPGTs) which have been implicated in the glycosylation of flavonoids, and in particular anthocyanins. All the homologies were > 80% with the genes from R. chinensis or F. vesca. Indeed, one hit was noted with UDPGT 74E2 (AT1G05680.1) which has been described as an anthocyanin 3-O-glucoside: 2″-O-xylosyl-transferase that converts cyanidin 3-O-glucoside to cyanidin 3-O-xylosyl-glucoside in Arabidopsis [AT5G54060(UF3GT) (arabidopsis.org)]. This is interesting as CySam (or cyanidin 3-O-xylosyl-glucoside) has a major QTL in this region. Also, the transfer of xylose would also be relevant to the formation of the flavonol pentoses (QPent and QHexPent), which also have major QTLs here. It would be intriguing if this UDPGT homologue could also be involved with the transferal of hexosyl units to form the other anthocyanidin and flavonol hexose components associated with this QTL region.
In 2010 at 77 cM on LG4, there was a strong QTL for the ellagitannins Lambertianin C and Sanguiin H10 and minor QTLs for Sanguiin H6, and Sanguiin H2. These had minor QTLs in a similar region in 2011 (77–89 cM) and there was a minor QTL for Lambertianin C at 84 cM in 2015 only. However, no significant QTLs were detected here for the over-years means. These metabolites are all positively correlated from 0.52 up to a maximum of 0.86 for the correlation between Lambertianin C and Sanguiin H6 in 2010 (Fig. S2c). Correlations for other years were similar (results not shown). Ellagitannins (ETs) originate from gallic acid, itself formed from the central metabolite, shikimate (Scheme 1A). The production of galloyl glucose is the first committed step in the formation of Ets (e.g. Schulenburg et al., 2016) then further galloyl groups are added to form pentagalloyl glucose. Ets are then produced by defined intramolecular oxidative coupling reactions on pentagalloyl glucose with a C–C coupling of galloyl groups producing the hexahydroxydiphenic (HHDP) acid unit characteristic of Ets (Yamada et al. 2017). Indeed, pentagalloyl glucose with two HHDP groups is also called casuarictin. The high correlation between Sanguiin H6 and Lambertianin C (R = 0.86) may reflect a precursor relationship as Sanguiin H6 can be described as a dimer of casuarictin linked through the gallic acid residue and a HHDP acid unit whereas Lambertianin C has been described as a trimer of casuarictin (Scheme 1B). The slightly less strong correlation (R = 0.76) between Sanguiin H2 and Sanguiin H6 also suggests a precursor relationship and indeed Sanguiin H6 differs from H2 by the addition of an HHDP-glucose unit (Scheme 1B). The less strong correlation (R = 0.56) between Sanguiin H6 and Sanguiin H10 may also indicate a less immediate precursor relationship as they differ by a HHDP unit.
Ellagic acid arises when HHDP groups released from Ets undergo spontaneous lactonization (Schulenburg et al., 2016). The correlation between the ET Sanguiin H2 and ellagic acid was relatively high (R = 0.72), which may indicate some metabolic connection. The correlations were less strong for the larger Ets (Sanguiin H10 (R = 0.40), Sanguiin H6 (R = 0.40) and Lambertianin C (R = 0.21) and ellagic acid, which implies a greater metabolic distance between the larger Ets and ellagic acid.
The correlations between ellagic acid and some of the ellagic acid derivatives was also interesting. There was a strong correlation between EllagicPent1 and MeEll pent (R = 0.82), which suggests a precursor relationship between these compounds. However, the correlation between EllagicPent2 and MeEll pent was negative (R = -0.29). The consensus is that the first eluting ellagic pentose is likely to be a xyloside and the latter eluting one an arabinoside (Gasperotti et al., 2010; Mullen et al., 2003). Therefore, this correlation may reflect that the pentose attached to EllagicPent1 is most probably xylose and arabinose in EllagicPent2. Indeed, there was a negative correlation between Ellagic pent 1 and 2 (R = − 0.6) with a bimodal pattern (Fig. S2C).
LG6
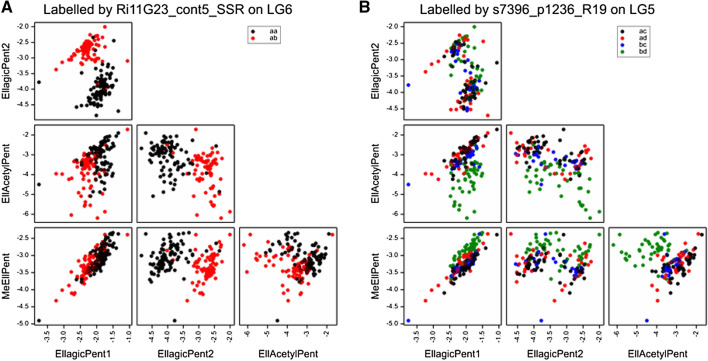
LG6 has few markers from Glen Moy and so most QTLs are due to differences between the alleles inherited from Latham. LG6 shows a very significant area for ellagic acid derivatives close to 80 cM in each year. There was a major QTL for EllagicPent2 (arabinose) for each year (giving it a bimodal distribution), with LOD 55 for the over-years average and R2 of 74%. This is in the same region as a QTL for total phenol content identified in previous work (Dobson et al., 2012). EllagicPent1 (xylose) also had a major QTL here, with LOD 13.4, and R2 of 27%, but with the opposite effect to EllagicPent2. MeEllPent and EllAcetylPent also have strong QTLs with the same direction as EllagicPent1 (Xyl) in this region, and the flavonol Kglu has a major QTL (LOD 11.9, R2 24%). Figure 5A shows a scatter plot of the over-year genotype means for the ellagic acid derivatives, labelled by marker Ri11G23_cont5_SSR at 80 cM, displaying the separation of the distribution of EllagicPent2 (Ara) between the two genotypes. The bimodal distribution for EllagicPent2 (Ara) complicates correlations; for example, the overall correlation between EllagicPent1 (Xyl) and EllagicPent2 (Ara) in 2010 was -0.60, but within each of the clusters corresponding to the genotypes of Ri11G23_cont5_SSR the correlation is positive. There was also a strong QTL for the ellagitannin Sanguiin H10 in 2010 only, with the peak LOD of 5.9 at 71 cM.
Fig. 5.
Scatterplots of the ellagic acid derivatives, labelled by markers close to the major QTLs on A LG6 and B LG5. The colour coding reflects the alleles inherited from the parents, ac, ad, bc & bd
There are some genes underlying the QTL region (75–80 cM) on LG6 that could be implicated in the control of transfer of sugars to flavonoids. Once again there are homologues (> 80% homology) with Arabidopsis genes that encode uridine diphosphate glycosyltransferases (UDPGTs), one of which has been described as specific for the transfer of glucose to quercetin to form quercetin 7-O-glucoside (AT1G05675.1–rabidopsis.org) but another is described as specific for transfer to benzoic acids, including salicylates (AT2G43820.1). However, homologues in other species have not been described with these specificities so it is possible that the raspberry gene encodes a transferase that could be involved in the transfer of pentoses (arabinose or xylose) to ellagic acid.
LG5
LG5 also has major QTLs in the over-years analysis for ellagic acid derivatives EllAcetylPent (LOD 27.2, R2 47%) and MeEllPent (LOD 10.2, R2 21%) at 8–16 cM. For EllAcetylPent, offspring with genotype ac had the highest level, those with genotype ad and bc had intermediate levels and bd had the lowest level. The pattern was reversed for MeEllPent. Figure 5B shows a scatter plot of the ellagic acid derivatives, labelled by marker s7396_p1236_R19 at 17 cM on LG5. Neither EllAcetylPent nor MeEllPent have bimodal distributions, but their scatterplot shows a separation of the bd genotype (shown in green) from the other three genotypes. Once again, these metabolites have an overall negative correlation (Fig. S2C), but a positive correlation within the QTL genotype clusters. KGlu has a strong QTL nearby, at 19 cM, with a similar pattern to EllAcetylPent.
There were also strong QTLs in the over-years analysis for PelSam and PelGlu mapping to 75–76 cM on LG5, with similar effects due to the allele from Latham.
LG2
No major QTLs for metabolites were detected on LG2. Strong QTLs (LODs 5.7–7.3) were detected in the over-years analysis in the region 12–19 cM, for the anthocyanins CySoph, CyC28H30O17 and total anthocyanins. All showed a similar pattern of effects, with ac and ad having the highest levels, bc intermediate and bd lowest. A minor QTL with a similar pattern was detected nearby, at 5 cM, for QDihex2.
A second region was found at the other end of LG2 for KGlcU and QGlcU, centred at 105 cM. This matches with the position of a QTL previously identified for total phenol content (Dobson et al., 2012). However, that study was based on fewer progeny and the significance is lower. For these flavonols, genotype bc had the highest level, genotypes ac and bd were intermediate and genotype ad was lowest.
LG3
Unlike the other linkage groups, the QTLs on LG3 were not consistent from year to year. No QTLs for were detected here in 2015, few in 2011 but more in 2010. Therefore, results for this linkage group are given from the analysis of data from each year separately in Fig. 1 and in the discussion below.
In 2010, a strong QTL was detected for total anthocyanins at 101 cM, together with a strong QTL for CyGlu at 111 cM. This matches with a QTL for total anthocyanins noted in previous studies (Dobson et al., 2012; McCallum et al., 2010). A strong QTL for QGlu and minor QTLs for QDihex2, KGlu and Lambertianin C were also detected in the region 99–111 cM. All were predominantly Latham effects, and all had effects in the same direction, apart from Lambertianin C, which showed the opposite effect. There was also a minor QTL detected for PelGlu at 76 cM, where the significant effect was from Glen Moy. Finally, there was a minor QTL for KGlcU at 8 cM (a significant effect of Latham). In 2011, a strong QTL was detected for Sanguiin H10 at 53 cM, and minor QTLs were detected for MeEllagicPent and EllagicPent1 (presumed xyloside) at 61 cM, all with effects from both parents. Interestingly, the QTL for Sanguiin H10 overlaps a QTL for total phenol content noted previously (Dobson et al., 2012), albeit from a smaller progeny set. There was also a minor QTL at KGlu at 98 cM, an effect of Latham.
Conclusions
Major and consistent QTLs were established for many of the polyphenol components of red raspberry, which have been previously implicated in health beneficial effects. This indicates that there is considerable genetic control of the levels of these components and provides confidence that markers could be deployed in breeding programmes to accelerate breeding of cultivars with defined polyphenol profiles. Many of these QTLs have underlying candidate genes that could influence accumulation of specific polyphenols. For example, the QTL region at 25–33 cM on LG4 had major QTLs for flavonoid components (QHexPent, QPent and CySam) which contained pentose sugars and was underlaid by a homologue of a UDP-glycosyltransferase implicated in the transfer of xylosyl groups to produce CySam from CyGlu in Arabidopsis [AT5G54060(UF3GT) (arabidopsis.org)]. This region could be examined for markers which could be used in accelerated breeding of new raspberry varieties with elevated levels of these specific anthocyanins and flavonols. Future work could examine the expression of some of the candidate genes indicated across the ripening stages of the parents (e.g., Scolari et al., 2021) to confirm if their expression matched the known accumulation of the specific polyphenols.
There was good correlation across years between various ellagic acid derivatives which suggested metabolic inter-connectivity. Similar inter-connectivity was noted for flavonol components, some following expected biosynthetic pathways but others less easily explainable. The correlations in levels of ellagitannins could reflect metabolic patterns and inter-relationships in the synthesis of these large polyphenol components. This is interesting because the biosynthetic pathways that lead to the formation of large ellagitannin structures are still not well defined despite pioneering work which suggested controlled oxidation of pentagalloyl glucose groups by specific laccases (see Niemetz & Gross, 2005). Closer examination of the genes that underly the strong QTLs for the ellagitannins, Sanguiin H6 and Lambertianin C, on LG3 at 124 cM may reveal candidate genes involved in the formation of these abundant and biologically active components. Once again, backing up with candidate gene expression studies could be interesting as ellagitannins are known to be accumulated early in the fruit formation (Beekwilder et al., 2005).
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This research was partly funded by Scottish Government’s Rural and Environmental Science and Analytical Services Division (RESAS) under Theme 3 Improving Food and Drink (2016–2021) and continues in current RESAS funding (2022–27).
Author contributions
All authors contributed to the study concept and design. Material preparation, data collection and some analyses were performed by CA and GD. LC–MS analysis and data analysis was carried out by GD and JWA, with oversight from GMD. QTL mapping and associated statistical analysis was carried out by CAH, with SV and CJA providing support. The first draft of the manuscript was written by GMD, CAH and JG. All authors commented on various versions of the manuscript. All authors read and approved the final manuscript.
Data availability
The metabolomic data may be made available on request to the corresponding author.
Declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This article does not contain any studies with human and/or animal participants performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahn SY, Kim SA, Yun HK. Differentially expressed genes during berry ripening in de novo RNA assembly of Vitis flexuosa fruits. Horticulture, Environment, and Biotechnology. 2019;60:531–553. doi: 10.1007/s13580-019-00148-2. [DOI] [Google Scholar]
- Anttonen MJ, Karjalainen RO. Environmental and genetic variation of phenolic compounds in red raspberry. Journal of Food Composition and Analysis. 2005;18:759–769. doi: 10.1016/j.jfca.2004.11.003. [DOI] [Google Scholar]
- Arnold H, Dobson G, Foito A, Austin C, Sungurtas J, Allwood JW, Stewart D, McDougall GJ. Assessing available phytochemicals from commercial blackcurrant and raspberry pomaces. Journal of Berry Research. 2022;12:415–431. doi: 10.3233/JBR-220017. [DOI] [Google Scholar]
- Beekwilder J, Jonker H, Meesters P, Hall RD, van der Meer IM, Ric de Vos CH. Antioxidants in raspberry: On-line analysis links antioxidant activity to a diversity of individual metabolites. Journal of Agriculture and Food Chemistry. 2005;53:3313–3320. doi: 10.1021/jf047880b. [DOI] [PubMed] [Google Scholar]
- Bogs J, Downey MO, Harvey JS, Ashton AR, Tanner GJ, Robinson SP. Proanthocyanidin synthesis and expression of genes encoding Leucoanthocyanidin reductase and Anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiology. 2005;139:652–663. doi: 10.1104/pp.105.064238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradish CM, Perkins-Veazie P, Fernandez GE, Xie G, Jia W. Comparison of flavonoid composition of red raspberries (Rubus idaeus L.) grown in the Southern United States. Journal of Agriculture and Food Chemistry. 2012;60:5779–5786. doi: 10.1021/jf203474e. [DOI] [PubMed] [Google Scholar]
- Carvalho E, Franceschi P, Feller A, Palmieri L, Wehrens R, Martens S. A targeted metabolomics approach to understand differences in flavonoid biosynthesis in red and yellow raspberries. Plant Physiology and Biochemistry. 2013;72:79–86. doi: 10.1016/j.plaphy.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Connor AM, McGhie TK, Stephens MJ, Hall HK, Alspach PA. Variation and heritability estimates of anthocyanins and their relationship to antioxidant activity in a red raspberry factorial mating design. Journal of the American Society for Horticultural Science. 2005;130:534–542. doi: 10.21273/JASHS.130.4.534. [DOI] [Google Scholar]
- Da Silva C, Zamperin G, Ferrarini A, Minio A, Dal Molin A, Venturini L, Buson G, Tononi P, Avanzato C, Zago E, Boido E, Dellacassa E, Gaggero C, Pezzotti M, Carrau F, Delledonne M. The high polyphenol content of grapevine cultivar Tannat berries is conferred primarily by genes that are not shared with the reference genome. The Plant Cell. 2013;25(4777):4788. doi: 10.1105/tpc.113.118810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighton N, Brennan R, Finn C, Davies HV. Antioxidant properties of domesticated and wild Rubus species. Journal of the Science of Food and Agriculture. 2000;80:1307–1313. doi: 10.1002/1097-0010(200007)80:9<1307. [DOI] [Google Scholar]
- Department for Environment, Food & Rural Affairs (2022), Agriculture in the United Kingdom data sets https://www.gov.uk/government/statistical-data-sets/agriculture-in-the-united-kingdom
- Dobson, G., McDougall, G. J. & Stewart, D. (2010) Analysis of polyphenolics in raspberry, blackberry, blueberry, black currant and strawberry by high pressure liquid chromatography-photodiode array-mass spectrometry (HPLC-PDA-MS/MS). https://www.euberry.univpm.it/sites/www.euberry.univpm.it/files/euberry/documenti/JHI%20SOP%20compilation%20130412.pdf
- Dobson P, Graham J, Stewart D, Brennan R, Hackett CA, McDougall GJ. Over-seasons analysis of quantitative trait loci affecting phenolic content and antioxidant capacity in raspberry. Journal of Agriculture and Food Chemistry. 2012;60:5360–5366. doi: 10.1021/jf3005178. [DOI] [PubMed] [Google Scholar]
- Gasperotti M, Masuero D, Vrhovsek U, Guella G, Mattivi F. Profiling and accurate quantification of Rubus ellagitannins and ellagic acid conjugates using direct UPLC-Q-TOF HDMS and HPLC-DAD analysis. Journal of Agriculture and Food Chemistry. 2010;58:4602–4616. doi: 10.1021/jf904543w. [DOI] [PubMed] [Google Scholar]
- Graham J, Hackett CA, Smith K, Woodhead M, Hein I, McCallum S. Mapping QTLs for developmental traits in raspberry from bud break to ripe fruit. Theoretical and Applied Genetics. 2009;118:1143–1155. doi: 10.1007/s00122-009-0969-6. [DOI] [PubMed] [Google Scholar]
- Graham J, Smith K, MacKenzie K, Jorgenson L, Hackett C, Powell W. The construction of a genetic linkage map of red raspberry (Rubus idaeus subsp. idaeus) based on AFLPs, genomic-SSR and EST-SSR markers. Theoretical and Applied Genetics. 2004;109:740–749. doi: 10.1007/s00122-004-1687-8. [DOI] [PubMed] [Google Scholar]
- Graham J, Smith K, McCallum S, Hedley PE, Cullen DW, Dolan A, Milne L, McNicol JW, Hackett CA. Towards an understanding of the control of ‘crumbly’ fruit in red raspberry. Springerplus. 2015;4:223. doi: 10.1186/s40064-015-1010-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett CA, Milne L, Smith K, Hedley P, Morris J, Simpson CG, Preedy K, Graham J. Enhancement of Glen Moy x Latham raspberry linkage map using GbS to further understand control of developmental processes leading to fruit ripening. BMC Genetics. 2018;19:59. doi: 10.1186/s12863-018-0666-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock RD, Petridis A, McDougall GJ. Raspberry fruit chemistry in relation to fruit quality and human nutrition. In: Graham J, Brennan R, editors. Raspberry. Cham: Springer; 2018. [Google Scholar]
- He M, Tian H, Luo X, Qi X, Chen X. Molecular progress in research on fruit astringency. Molecules. 2015;20:1434–1451. doi: 10.3390/molecules20011434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings SN. Advances in Rubus breeding. In: Graham J, Brennan R, editors. Raspberry. Cham: Springer; 2018. [Google Scholar]
- Jennings SN, Graham J, Ferguson L, Young V. New developments in raspberry breeding in Scotland. Acta Horticulture. 2016;1133:23–28. doi: 10.17660/ActaHortic.2016.1133.4. [DOI] [Google Scholar]
- Kassim A, Poette J, Paterson A, Zait D, McCallum S, Woodhead M, et al. Environmental and seasonal influences on red raspberry anthocyanin antioxidant contents and identification of quantitative trait loci (QTL) Molecular Nutrition & Food Research. 2009;53:625–634. doi: 10.1002/mnfr.200800174. [DOI] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multi-locus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Mazzoni L, Perez-Lopez P, Giampieri F, Alvarez-Suarez JM, Gasparini M, Forbes-Hernandez TY, et al. The genetic aspects of berries, from field to health. Journal of the Science of Food and Agriculture. 2016;96:365–371. doi: 10.1002/jsfa.7216. [DOI] [PubMed] [Google Scholar]
- McCallum S, Simpson C, Graham J. QTL Mapping and marker assisted breeding in Rubus spp. In: Graham J, Brennan R, editors. Raspberry. Cham: Springer; 2018. [Google Scholar]
- McCallum S, Woodhead M, Hackett CA, Kassim A, Paterson A, Graham J. Genetic and environmental effects influencing fruit colour and QTL analysis in raspberry. Theoretical and Applied Genetics. 2010;121:611–627. doi: 10.1007/s00122-010-1334-5. [DOI] [PubMed] [Google Scholar]
- McDougall GJ, Conner S, Pereira-Caro G, Gonzalez-Barrio R, Brown EM, Verrall S, et al. Tracking (poly)phenol components from raspberries in ileal fluid. Journal of Agriculture and Food Chemistry. 2014;62:7631–7641. doi: 10.1021/jf502259j. [DOI] [PubMed] [Google Scholar]
- McDougall GJ, Dobson P, Smith P, Blake A, Stewart D. Assessing potential bioavailability of raspberry anthocyanins using an in vitro digestion system. Journal of Agriculture and Food Chemistry. 2005;53:5896–5904. doi: 10.1021/jf050131p. [DOI] [PubMed] [Google Scholar]
- Mullen W, Lean ME, Crozier A. Rapid characterization of anthocyanins in red raspberry fruit by high-performance liquid chromatography coupled to single quadrupole mass spectrometry. Journal of Chromatography A. 2002;966:63–70. doi: 10.1016/s0021-9673(02)00699-4. [DOI] [PubMed] [Google Scholar]
- Mullen W, Yokota T, Lean ME, Crozier A. Analysis of ellagitannins and conjugates of ellagic acid and quercetin in raspberry fruits by LC-MSn. Phytochemistry. 2003;64:617–624. doi: 10.1016/S0031-9422(03)00281-4. [DOI] [PubMed] [Google Scholar]
- Niemetz R, Gross GG. Enzymology of gallotannin and ellagitannin biosynthesis. Phytochemistry. 2005;66:2001–2011. doi: 10.1016/j.phytochem.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Paterson A, Kassim A, McCallum S, Woodhead M, Smith K, Zait D, Graham J. Environmental and seasonal influences on red raspberry flavour volatiles and identification of quantitative trait loci (QTL) and candidate genes. Theoretical & Applied Genetics. 2013;126:33–48. doi: 10.1007/s00122-012-1957-9. [DOI] [PubMed] [Google Scholar]
- Plunkett BJ, Espley RV, Dare AP, Warren BAW, Grierson ERP, Cordiner S, et al. MYBA from blueberry (Vaccinium section Cyanococcus) is a subgroup 6 type R2R3MYB transcription factor that activates anthocyanin production. Frontiers in Plant Science. 2018;11:1300. doi: 10.3389/fpls.2018.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajauria G, Tiwari BK. Fruit juices, extraction, composition, quality, and analysis. London, UK: Academic Press; 2018. [Google Scholar]
- Rao AV, Snyder DM. Raspberries and human health: A review. Journal of Agriculture and Food Chemistry. 2010;58:3871–3883. doi: 10.1021/jf903484g. [DOI] [PubMed] [Google Scholar]
- Remberg SF, Sønsteby A, Aaby K, Heide OM. Influence of post flowering temperature on fruit size and chemical composition of glen ample raspberry (Rubus idaeus L.) Journal of Agriculture and Food Chemistry. 2010;58:9120–9128. doi: 10.1021/jf101736q. [DOI] [PubMed] [Google Scholar]
- Schulenburg K, Feller A, Hoffmann T, Schecker JH, Martens S, Schwab W. Formation of β-glucogallin, the precursor of ellagic acid in strawberry and raspberry. Journal of Experimental Botany. 2016;67:2299–2308. doi: 10.1093/jxb/erw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolari LM, Hancock RD, Hedley PE, Morris J, Smith K, Graham J. Combining QTL mapping and gene expression analysis to elucidate the genetic control of ‘crumbly’ fruit in red raspberry (Rubus idaeus L.) Agronomy. 2021;11:794–799. doi: 10.3390/agronomy11040794. [DOI] [Google Scholar]
- Sharma D, Tiwari M, Pandey A, Bhatia C, Sharma A, Trivedi PK. MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiology. 2016;171:944–959. doi: 10.1104/pp.15.01831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Food and Agriculture Organization (FAO) database (2022). https://www.fao.org/faostat/en/
- VSN International . Genstat for Windows. 20. Hemel Hempstead, UK: VSN International; 2019. [Google Scholar]
- Williams D, Hackett CA, Karley A, McCallum S, Smith K, Britten A. Seeing the wood for the trees, hyperspectral imaging for high throughput QTL detection in raspberry, a perennial crop species. Fruit Research. 2021 doi: 10.48130/FruRes-2021-0007. [DOI] [Google Scholar]
- Yamada H, Hirokane T, Ikeuchi K, Wakamori S. Fundamental methods in ellagitannin synthesis. Natural Product Communications. 2017 doi: 10.1177/1934578X1701200846. [DOI] [Google Scholar]
- Yuan T, Fujioka S, Takatsuto S, Matsumoto S, Gou X, He K, et al. BEN1, a gene encoding a dihydroflavonol 4-reductase (DFR)-like protein, regulates the levels of brassinosteroids in Arabidopsis thaliana. The Plant Journal. 2007;51:220–233. doi: 10.1111/j.1365-313X.2007.03129.x. [DOI] [PubMed] [Google Scholar]
- Zhu M-C, Hu R, Zhao H-Y, Tang Y-S, Shi X-T, Jiang H-Y, et al. Identification of quantitative trait loci and candidate genes controlling seed pigments of rapeseed. Journal of Integrative Agriculture. 2021;20:2862–2879. doi: 10.1016/S2095-3119(20)63377-9. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metabolomic data may be made available on request to the corresponding author.